Abstract
Background
The contribution of genetic factors to epilepsy has long been recognized and has been estimated to play a role in 70−80% of cases. Identification of a pathogenic variant can help families to better cope with the disorder, allow for genetic counseling to determine recurrence risk, and in some cases, can directly influence treatment options. In this study, we determined the diagnostic yield of a clinical gene panel applied to an unselected cohort of epilepsy patients.
Methods
Variant reports from 339 clinically-referred epilepsy patients screened using a 110-gene panel were retrospectively reviewed. Variants were classified using ACMG guidelines.
Results
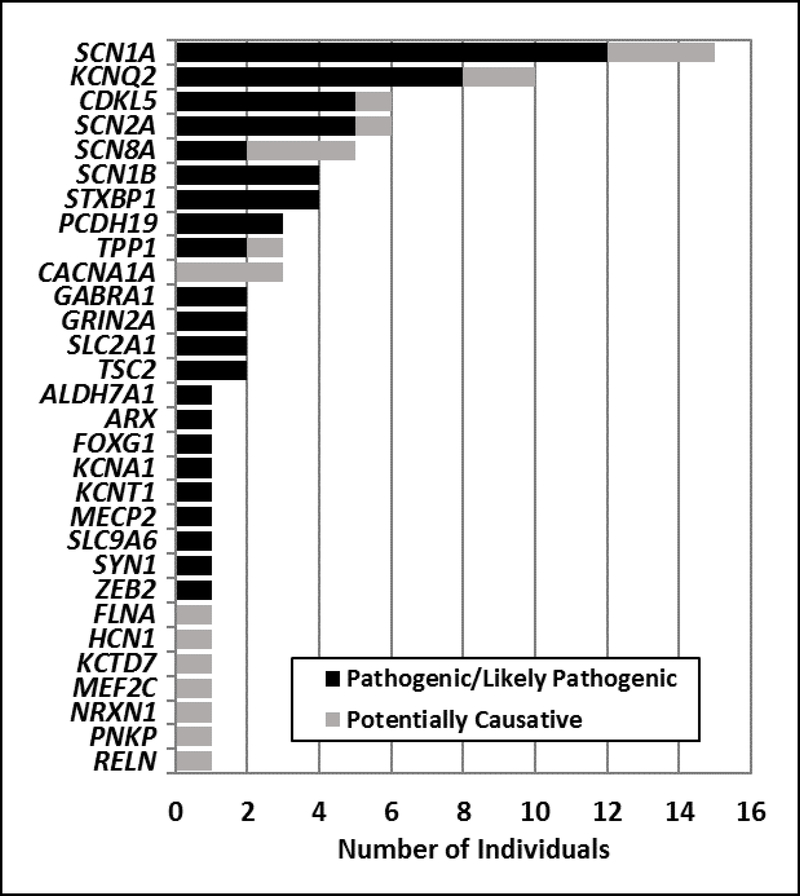
Pathogenic or likely pathogenic variants were identified in 62 individuals (18%) and potentially causative variants were identified in an additional 21 individuals (6%). Causative and potentially causative variants were most frequently identified in SCN1A (n = 15) and KCNQ2 (n = 10). Other genes in which disease-causing variants were identified in multiple individuals included CDKL5, SCN2A, SCN8A, SCN1B, STXBP1, TPP1, PCDH19, CACNA1A, GABRA1, GRIN2A, SLC2A1, and TSC2. Sixteen additional genes had variants identified in single individuals.
Conclusions
We identified 87 variants in 30 different genes that could explain disease, of which 54% were not previously reported. This study confirms the utility of targeted gene panel analysis in epilepsy and highlights several factors to improve the yield of diagnostic genetic testing, including the critical need for clinical phenotype information and parental samples, microarray analysis for whole exon deletions and duplications, and frequent update of panels to incorporate new disease genes.
Keywords: diagnostic yield, epilepsy, epileptic encephalopathy, genetic testing
INTRODUCTION
Epilepsy comprises a group of disorders that are characterized by recurrent, unprovoked seizures that collectively affect about 1% of the population. Although epilepsy can develop following insults such as head trauma, stroke, and infection, it is now believed that genetic factors may contribute to 70−80% of epilepsy cases.1
Advances in next generation sequencing have allowed for the identification of new causal genes in both familial and sporadic forms of epilepsy.2,3 To date, pathogenic variants in over one hundred different genes have been reported to cause epilepsy and seizures.4 Many of these research findings have been used to develop clinical gene panels, which are available to physicians to aid in diagnosis. These panels can range in size from dozens to hundreds of genes and may be specific to epilepsy subtype, such as early-infantile epileptic encephalopathy, or more broadly encompass many known epilepsy genes.
In this study, we investigated the diagnostic yield of one such gene panel, known as the Epilepsy and Seizure Disorders (ESD) panel, in an unselected cohort of 339 clinically-referred patients. The ESD panel uses a targeted sequencing library of 110 genes that are associated with a spectrum of epilepsies, as well as genes associated with metabolic and structural disorders that include epilepsy as a prominent symptom. Identification of a causative variant can help families to better cope with the disorder, allow for genetic counseling to determine recurrence risk, and in some cases, can directly influence treatment options.5–7 Additionally, it can end the often long and expensive diagnostic odyssey that families experience when searching for answers.
MATERIALS AND METHODS
Patients
Variant reports from 339 consecutive, clinically-referred patients screened between 2013 and 2016 were retrospectively reviewed for this study. All molecular diagnostic testing was performed at EGL Genetics, a Clinical Laboratory Improvement Amendments (CLIA) and College of American Pathologists (CAP) accredited laboratory. This study was approved by the institutional review board of Emory University.
Targeted gene panel testing
A custom-designed in-solution hybridization probe library (IDT or SureSelect, Agilent Technologies, Santa Clara, CA) was used to capture the coding exons of the 110 genes on the Epilepsy and Seizure Disorders (ESD) panel. Direct sequencing of the amplified captured regions was performed using next-generation sequencing (2×100bp, paired-end reads) on an Illumina HiSeq 2500 (Illumina, San Diego, CA) in rapid run mode. The individual DNA sequence reads were aligned to the published human genome reference (hg19 build) and variants were called using NextGENe® (SoftGenetics, State College, PA). Further analysis was performed using the EGL bioinformatics pipeline, which annotates identified variants utilizing a variety of external and internal sources. Variants are called within the coding exons and +/− 10bp of flanking intronic sequence. Relevant regions of epilepsy genes not amenable to NGS were filled in using the Sanger sequencing method. A list of the 110 genes examined as part of the ESD panel can be found in the supplemental information for this article (Supplemental materials).
Variant Evaluation
Variants were evaluated and classified by board certified clinical molecular geneticists using the ACMG guidelines for variant classification.8 Briefly, variants were classified as (1) pathogenic, (2) likely pathogenic, (3) of unknown significance, (4) likely benign, or (5) benign. For research purposes, we considered certain variants of unknown significance to be “potentially causative” if specific conditions were met, such as the variant was absent from the population, in silico analyses predicted the variant to be damaging, and/or the variant was located in a critical or evolutionarily conserved region of the protein.8 Classified variants were submitted to ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) as well as EGL Genetics’ publicly-available database EmVClass (www.egl-eurofins.com/emvclass/emvclass.php).
RESULTS
Patient demographics
Demographic information was available for approximately 80% of the 339 individuals. The age range of individuals referred for genetic testing was 2.5 months to 74 years; however, most of the referrals were children and more than half were below the age of 5 years. There were equal numbers of males and females. The amount of clinical information that accompanied each referral varied widely, with nearly 22% of the referrals lacking any clinical information about the patient. For the referrals that did include clinical information, 75% of cases reported two or more phenotypes. Aside from seizures, developmental delay was the next most common phenotype reported, followed by hypotonia, epileptic encephalopathy, intellectual disability, autism, and developmental regression. Additional phenotypes frequently encountered included ataxia, microcephaly, and spasticity. A positive family history of seizures or other neurological disorders was noted for 4% of cases.
Yield from the ESD Panel
Pathogenic or likely pathogenic variants were identified in 62 (18%) of the 339 individuals screened (Table 1). Twenty-one additional patients (6%) had potentially causative variants. Pathogenic, likely pathogenic, and potentially causative variants were identified in 30 different genes, accounting for 27% of the 110 genes on the ESD panel. Approximately 75% of the variants were in genes associated with autosomal dominant inheritance, while 17% and 8% of the variants affected X-linked and autosomal recessive genes, respectively. Pathogenic, likely pathogenic, and potentially causative variants were most frequently identified in SCN1A (n = 15) and KCNQ2 (n = 10). Other genes in which variants were identified in multiple individuals included CDKL5 (n = 6), SCN2A (n = 6), SCN8A (n = 5), SCN1B (n = 4), STXBP1 (n = 4), TPP1 (n = 3), PCDH19 (n = 3), CACNA1A (n = 3), GABRA1 (n = 2), GRIN2A (n = 2), SLC2A1 (n = 2), and TSC2 (n = 2). Sixteen additional genes had variants identified in single individuals (Figure 1, Table 1).
Table 1.
Diagnostic yield from 339 epilepsy patients screened on the Epilepsy and Seizure disorders panel.
| Pathogenic and Likely Pathogenic variants | |||||
| Case No. | Gene | Nucleotide | Protein | Classification | Phenotype (if provided) |
| 1 | ALDH7A1 | c.1093+1G>A, c.1279G>C | p.Glu427Gln | Pathogenic | Seizures, epileptic encephalopathy, macrocephaly, hypotonia, muscle weakness, and developmental delay |
| 2 | ARX | c.30C>A | p.Cys10Ter | Pathogenic | Infantile spasms, dysphagia, and poor weight gain |
| 3 | CDKL5 | c.1891_1916delATAGGGCAAGGGATGGCAGCTAGAGC | p.Ile631Glnfs*43 | Pathogenic | Unspecified Epilepsy |
| 4 | CDKL5 | c.1152C>G | p.Tyr384Ter | Pathogenic | Epileptic encephalopathy, myotonic and tonic-clonic seizures, and a course gyral pattern on brain imaging |
| 5 | CDKL5 | c.1553delC | p.Pro518Hisfs*5 | Pathogenic | Seizures and spasticity |
| 6 | CDKL5 | c.212delA | p.Asn71Thrfs*5 | Pathogenic | Infantile/epileptic spasms and hypotonia |
| 7 | CDKL5 | c.1108_1109dupAA | p.Asn370Lysfs*124 | Pathogenic | N/A |
| 8 | FOXG1 | c.648_655delTTACTACC | p.Tyr217Argfs*235 | Pathogenic | Unspecified Epilepsy |
| 9 | GABRA1 | c.335G>A | p.Arg112Gln | Likely Pathogenic | N/A |
| 10 | GABRA1 | c.640C>A | p.Arg214Ser | Likely pathogenic | Generalized convulsive epilepsy with intractable epilepsy |
| 11 | GRIN2A | c.2890delC | p.Gln964Lysfs*37 | Pathogenic | Generalized seizures and speech disturbance |
| 12 | GRIN2A | c.3813G>A | p.Trp1271Ter | Pathogenic | N/A |
| 13 | KCNA1 | c.1222G>A | p.Val408Met | Likely Pathogenic | Seizures, developmental delay, and multiple joint contractures |
| 14 | KCNQ2 | c.793G>A | p.Ala265Thr | Likely pathogenic | Neonatal seizure disorder |
| 15 | KCNQ2 | c.637C>T | p.Arg213Trp | Pathogenic | Neonatal seizure disorder |
| 16 | KCNQ2 | c.640C>T | p.Arg214Trp | Likely pathogenic | Autism, seizures, hydrocephaly, a brother with autism and epilepsy, and a father with a history of epilepsy in childhood |
| 17 | KCNQ2 | c.1118+2T>C | Pathogenic | Infantile/epileptic spasms | |
| 18 | KCNQ2 | c.701C>T | p.Thr234Ile | Likely pathogenic | Seizures and developmental delay |
| 19 | KCNQ2 | c.821C>T | p.Thr274Met | Likely pathogenic | N/A |
| 20 | KCNQ2 | c.841G>A | p.Gly281Arg | Likely pathogenic | N/A |
| 21 | KCNQ2 | c.1088A>G | p.Tyr363Cys | Likely pathogenic | Neonatal seizure disorder and epileptic encephalopathy |
| 22 | KCNT1 | c.2849G>A | p.Arg950Gln | Likely Pathogenic | N/A |
| 23 | MECP2 | c.880C>T | p.Arg294Ter | Pathogenic | Delayed milestones and intractable epilepsy |
| 24 | PCDH19 | c.814C>T | p.Gln272Ter | Pathogenic | Seizure disorder |
| 25 | PCDH19 | c.1091dupC | p.Tyr366Leufs*10 | Pathogenic | Intellectual disability, autism, and seizures |
| 26 | PCDH19 | c.1265_1266delCA | p.Thr422Asnfs*23 | Pathogenic | N/A |
| 27 | SCN1A | c.2584C>T | p.Arg862Ter | Pathogenic | Psychomotor epilepsy, intellectual disability, phenotype consistent with Dravet syndrome |
| 28 | SCN1A | c.4907G>A | p.Arg1636Gln | Likely pathogenic | Epileptic encephalopathy, myoclonic seizures, dystonia, and spasticity |
| 29 | SCN1A | c.602+1G>A | Pathogenic | Seizures and developmental delay | |
| 30 | SCN1A | c.269T>C | p.Phe90Ser | Likely pathogenic | Prolonged seizures, possible myoclonus, and a clinical suspicion for Dravet syndrome |
| 31 | SCN1A | c.1264G>A | p.Val422Met | Likely pathogenic | Infantile spasms, focal seizures, hypotonia, bilateral polydactyly, lack of coordination, and grand mal status |
| 32 | SCN1A | c.1259C>A | p.Ala420Asp | Likely pathogenic | N/A |
| 33 | SCN1A | c.302G>A | p.Arg101Gln | Pathogenic | N/A |
| 34 | SCN1A | c.5389G>C | p.Ala1797Pro | Likely Pathogenic | Tonic-clonic and focal seizures |
| 35 | SCN1A | c.5348C>T | p.Ala1783Val | Pathogenic | N/A |
| 36 | SCN1A | c.4812delG | p.Trp1604Ter | Pathogenic | N/A |
| 37 | SCN1A | c.5563C>T | p.Pro1855Ser | Likely Pathogenic | N/A |
| 38 | SCN1A | c.1076A>G | p.Asn359Ser | Likely Pathogenic | N/A |
| 39 | SCN1B | c.347delC | p.Ser116Trpfs*31 | Pathogenic | Hypotonia and seizures |
| 40 | SCN1B | c.653delG | p.Ser218Thrfs*21 | Likely Pathogenic | Developmental delay, seizures with an abnormal EEG, and high myopia |
| 41 | SCN1B | c.363C>G | pCys121Trp | Pathogenic | N/A |
| 42 | SCN1B | c.363C>G | pCys121Trp | Pathogenic | N/A |
| 43 | SCN2A | c.5387_5390dup AGAT | p.Met1797Ilefs*5 | Pathogenic | Seizures and intellectual disability |
| 44 | SCN2A | c.5645G>A | p.Arg1882Gln | Likely pathogenic | Lack of normal physiological development, autism spectrum disorder, and intractable epilepsy |
| 45 | SCN2A | c.2558G>A | p.Arg853Gln | Pathogenic | Failure to thrive, developmental delay, speech delay, autism, seizures, dystonia, microcephaly, and gastroesophageal reflux disease |
| 46 | SCN2A | c.1178C>A | p.Thr393Lys | Likely Pathogenic | Seizures and developmental regression |
| 47 | SCN2A | c.2713A>G | p.Lys905Glu | Likely Pathogenic | Epileptic encephalopathy |
| 48 | SCN8A | c.3985A>G | p.Asn1329Asp | Likely pathogenic | Intractable epilepsy |
| 49 | SCN8A | c.2287A>G | p.Ile763Val | Likely Pathogenic | Seizures and developmental delay |
| 50 | SLC2A1 | c.997C>T | p.Arg333Trp | Pathogenic | Febrile seizures, ataxia, hypotonia, hypermobility, and a family history of seizures. |
| 51 | SLC2A1 | c.1006C>G | p.Leu336Val | Likely Pathogenic | N/A |
| 52 | SLC9A6 | c.508–1G>A | Pathogenic | N/A | |
| 53 | STXBP1 | c.1029+1G>T | Pathogenic | Seizures, developmental delay, and cognitive impairment. | |
| 54 | STXBP1 | c.364C>T | p.Arg122Ter | Pathogenic | Infantile/epileptic spasms and tonic seizures |
| 55 | STXBP1 | c.548T>C | p.Leu183Pro | Likely Pathogenic | N/A |
| 56 | STXBP1 | c.704G>A | p.Arg235Gln | Likely Pathogenic | Generalized, absence, and tonic-clonic seizures, infantile spasms, hypotonia, developmental delay, and cerebral palsy |
| 57 | SYN1 | c.377G>A | p.Trp126Ter | Pathogenic | N/A |
| 58 | TPP1* | c.509–1G>C | Pathogenic | N/A | |
| 59 | TPP1 | c.509–1G>C, c.1016G>A | p.Arg339Gln | Pathogenic | Seizures and speech delay |
| 60 | TSC2 | c.4415delG | p.Gly1472Alafs*4 | Pathogenic | Seizure disorder and intellectual disability |
| 61 | TSC2 | c.3598C>T | p.Arg1200Trp | Pathogenic | Infantile spasms and global developmental delay |
| 62 | ZEB2 | c.1876G>T | p.Gly626Ter | Pathogenic | Developmental delay and psychomotor epilepsy |
| Potentially Causative Variants | |||||
| Case No. | Gene | Nucleotide | Protein | Classification | Phenotype (if provided) |
| 63 | CACNA1A | c.5017C>T | p.Arg1673Cys | Unknown | Generalized convulsive epilepsy and hemiplegia |
| 64 | CACNA1A | c.4177G>A | p.Val1393Met | Unknown | Global developmental delay, seizures, and tremor |
| 65 | CACNA1A | c.4177G>A | p.Val1393Met | Unknown | N/A |
| 66 | CDKL5 | c.541G>A | p.Glu181Lys | Unknown | N/A |
| 67 | HCN1 | c.990G>C | p.Trp330Cys | Unknown | Epileptic encephalopathy, abnormal EEG, and other clinical features suggestive of Ohtahara syndrome |
| 68 | FLNA | c.4237G>A | p.Glu1413Lys | Unknown | Hypotonia, epilepsy, and abnormal MRI |
| 69 | KCNQ2 | c.1627G>A | p.Val543Met | Inherited | Developmental delay, seizures, a ventricular septal defect, unilateral cryptorchidism, and macrocephaly |
| 70 | KCNQ2 | c.1627G>A | p.Val543Met | Unknown | N/A |
| 71 | KCTD7 | c.190A>G, c.793G>A | p.Thr64Ala, p.Gly265Arg | Inherited from both parents | Status epilepticus, generalized, tonic-clonic, and myoclonic seizures |
| 72 | MEF2C | c.121T>C | p.Cys41Arg | Unknown | Generalized and myoclonic seizures, intellectual disability, hypotonia, spasticity, and muscle weakness |
| 73 | NRXN1 | c.3619C>T | p.Arg1207Ter | Unknown | Autism, epilepsy, and developmental delay |
| 74 | PNKP* | c.1324G>A | p.Gly442Ser | Unknown | N/A |
| 75 | RELN | c.1817C>T, c.2201T>A | p.Thr606Ile, p.Val734Asp | Inherited from both parents | Cerebral palsy, abnormal EEG, developmental delay, and lack of coordination |
| 76 | SCN1A | c.638C>G | p.Ser213Trp | Unknown | Febrile and afebrile seizures and developmental delay |
| 77 | SCN1A | c.1703G>A | p.Arg568Gln | Unknown | Seizures |
| 78 | SCN1A | c.2923G>C | p.Val975Leu | Unknown | Seizures and developmental delay |
| 79 | SCN2A | c.4156T>G | p.Cys1386Gly | Unknown | Seizures, speech and developmental regression |
| 80 | SCN8A | c.491C>T | p.Thr164Met | Inherited from affected mother | Focal seizures, developmental delay, failure to thrive, and a maternal family history of seizures |
| 81 | SCN8A | c.605T>A | p.Ile202Asn | Unknown | Seizures, developmental delay, dysmorphic features, pica, paroxysmal behavior with nonspecific encephalopathy on EEG, and a brother who also has seizures |
| 82 | SCN8A | c.1241A>T | p.Tyr414Phe | Unknown | N/A |
| 83 | TPP1*‡ | c.523C>T | p.Arg175Cys | Unknown | Infantile spasms, learning disability, developmental delay, seizures, and hypotonia |
N/A, phenotype was not available
Indicates a homozygous variant
This individual also had a novel homozygous missense variant in SLC25A22
Figure 1.
Genes with pathogenic/likely pathogenic (black) and potentially causative (grey) variants detected in 339 clinically-referred epilepsy patients.
DISCUSSION
In this study, we determined the diagnostic yield of a clinical gene panel in an unselected cohort of 339 physician-referred epilepsy patients. This is one of the largest unselected epilepsy cohorts to be examined in this manner. We detected pathogenic or likely pathogenic variants in 18% of screened individuals, consistent with two recent studies that used gene panels to screen selected cohorts of individuals with severe, early-onset seizures, developmental delay, and intellectual disability.9,10
We also observed potentially causative variants in an additional 6% of screened cases (21 individuals). These variants occurred in many of the same genes as the pathogenic and likely pathogenic variants (Figure 1), but were generally novel missense changes, which are more difficult to interpret clinically and will require additional functional evidence to be classified as pathogenic/likely pathogenic. Classification of these variants was often made more challenging due to a lack accompanying clinical information and parental samples to assist with the determination of variant inheritance.
The majority of the disease-causing variants identified in our study were detected in a small number of genes. SCN1A had the largest number of variants, followed by KCNQ2, together accounting for nearly 30% of the cases with a pathogenic, likely pathogenic, or potentially causative variant. Furthermore, multiple individuals had variants in CDKL5, SCN2A, SCN8A, STXBP1, and PCDH19, genes which are associated with epileptic encephalopathies, and in which pathogenic variants are consistently identified using gene panels and whole exome sequencing in different epilepsy studies.2,3,9 We also observed two or more variants each in CACNA1A, GABRA1, GRIN2A, SCN1B, SLC2A1, TPP1, and TSC2. Pathogenic variants in these genes can cause severe seizures and epileptic encephalopathy, but each gene has also been associated with other clinical phenotypes, for instance movement disorders, migraine, and milder forms of epilepsy. Although we were limited in the amount of clinical information provided with each case, the identified genes would suggest that the individuals with pathogenic or potentially causative variants are most likely affected with severe forms of childhood epilepsy. This highlights the benefit of genetic testing in children with severe epilepsy.
Sixteen additional genes had variants that were seen in only one patient. This low frequency may be explained for some genes, such as ZEB2 (Mowat-Wilson syndrome) and MECP2 (Rett syndrome), which typically present with very distinctive phenotypes that may prompt gene-specific testing rather than broader screening on an epilepsy gene panel. In other instances, affected individuals may be less likely to be referred for an epilepsy-specific gene panel. For example, individuals with pathogenic variants in FLNA and RELN, which are associated with periventricular nodular heterotopia and lissencephaly, respectively, may be referred for testing on a brain malformations panel rather than an epilepsy panel. On the other hand, the individuals with variants in these genes that were referred for screening on an epilepsy-specific panel may represent cases with atypical presentation. For example, we identified the compound heterozygous RELN variants, Thr606Ile and Val734Asp, in an individual with epilepsy, cerebral palsy, and developmental delay, but without any significant magnetic resonance imaging (MRI) findings (Table 1). These individual cases emphasize the utility of referring individuals with unexplained epilepsy for a gene panel rather than testing single genes.
The epilepsy panel used in this study contained 110 genes curated for association with epilepsy and seizures (Supplemental materials). Because of their known role in epilepsy, the majority of these genes are present on other commercially-available epilepsy panels. Therefore, it is likely that a similar diagnostic yield would be obtained if the cohort examined in this study was tested using another epilepsy panel, especially since most of the pathogenic variants were identified in a small number of genes (Figure 1).
Whole exome sequencing (WES), which examines all known coding regions in the genome, has the potential to result in a higher diagnostic yield compared to gene panels; however, clinical laboratories are currently only able to assign pathogenicity to variants in known epilepsy genes. Furthermore, WES is more expensive and time-consuming, and is most efficient when both parents are also screened (trio format).
Based on this study, we identified several factors that could improve the diagnostic yield of gene panel testing for epilepsy. First, although parental samples are not routinely included when performing gene panel analysis, follow up testing on parental samples is critical for the reclassification of variants of unknown significance identified in the proband. It would be beneficial to collect parent samples at the same time as the proband sample when sending for testing in order to avoid delays in follow up of variants that require further analysis. Greater availability of parental samples might have increased the yield in our cohort by allowing for the identification of de novo variants and phasing of recessive variants. Second, the diagnostic yield would also likely have been higher if more samples were accompanied by clinical information since such knowledge can facilitate variant interpretation by allowing for comparisons with previously reported cases. This information is also helpful for identifying individuals with atypical clinical presentations that may expand the phenotypic spectrum of a given gene. The expansion of clinical spectrum has already been demonstrated for many epilepsy genes, including SCN2A and CACNA1A.11,12 Third, although recent publications have demonstrated the ability of next-generation sequencing (NGS) to detect intragenic copy number variation, these algorithms are still in the development stage and need confirmatory testing.13 Therefore, exon level copy number analysis for single and multiple exon deletions and duplications is still beneficial for identifying variants that may be missed by NGS. For instance, there were three individuals in our cohort that had concurrent deletion/duplication testing that detected copy number variants that could explain their epilepsy. Finally, it is important to frequently update gene panels to incorporate newly identified epilepsy genes. Many laboratories now perform sequencing on large sets of known disease genes (referred to as a Mendeliome) but only analyze a subset of those genes according to the test ordered. In negative cases, with appropriate consent, the expanded gene set could be evaluated to identify variants in genes not previously associated with epilepsy. Even as whole exome and genome sequencing approaches become more commonly used for genetic testing, it will remain important to have up-to-date epilepsy gene lists to aid in variant prioritization and phenotype definition.
Supplementary Material
Acknowledgments
FUNDING
This work was supported by funding from Children’s Healthcare of Atlanta to AE, the Emory University Research Council to AE and JJA, and a National Institutes of Health training grant appointment (5T32GM008490-23) to KMB.
Footnotes
CONFLICTS OF INTERESTS
CdS and JJA are employed by and receive a salary from EGL Genetics. The Epilepsy and Seizure Disorders panel is among EGL Genetics’ commercially available tests. MH is a former employee of EGL Genetics. KMB and AE have no conflicts of interest.
SUPPLEMENTAL MATERIAL
S1. Genes of the Epilepsy and Seizure Disorders panel
REFERENCES
- 1.Myers CT, Mefford HC. Advancing epilepsy genetics in the genomic era. Genome medicine. 2015;7(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carvill GL, Heavin SB, Yendle SC, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nature genetics. 2013;45(7):825–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epi KC, Epilepsy Phenome/Genome P, Allen AS, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501(7466):217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Lin ZJ, Liu L, et al. Epilepsy-associated genes. Seizure. 2017;44:11–20. [DOI] [PubMed] [Google Scholar]
- 5.Berkovic SF. Genetics of Epilepsy in Clinical Practice. Epilepsy currents / American Epilepsy Society. 2015;15(4):192–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pierson TM, Yuan H, Marsh ED, et al. GRIN2A mutation and early-onset epileptic encephalopathy: personalized therapy with memantine. Annals of clinical and translational neurology. 2014;1(3):190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boerma RS, Braun KP, van de Broek MP, et al. Remarkable Phenytoin Sensitivity in 4 Children with SCN8A-related Epilepsy: A Molecular Neuropharmacological Approach. Neurotherapeutics : the journal of the American Society for Experimental NeuroTherapeutics. 2016;13(1):192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine : official journal of the American College of Medical Genetics. 2015;17(5):405–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trump N, McTague A, Brittain H, et al. Improving diagnosis and broadening the phenotypes in early-onset seizure and severe developmental delay disorders through gene panel analysis. Journal of medical genetics. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Kong W, Gao Y, et al. Gene Mutation Analysis in 253 Chinese Children with Unexplained Epilepsy and Intellectual/Developmental Disabilities. PloS one. 2015;10(11):e0141782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura K, Kato M, Osaka H, et al. Clinical spectrum of SCN2A mutations expanding to Ohtahara syndrome. Neurology. 2013;81(11):992–998. [DOI] [PubMed] [Google Scholar]
- 12.Epi KC. De Novo Mutations in SLC1A2 and CACNA1A Are Important Causes of Epileptic Encephalopathies. American journal of human genetics. 2016;99(2):287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Epilepsy Phenome/Genome Project Epi KC. Copy number variant analysis from exome data in 349 patients with epileptic encephalopathy. Annals of neurology. 2015;78(2):323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.