Abstract
The systemic delivery of tamoxifen (Tam) to activate inducible CreERT2-loxP transgenic mouse systems is now widely used in neuroscience studies. This critical technological advancement allows temporal control of DNA-cre recombination, avoidance of embryonically lethal phenotypes, and minimization of residual cell labeling encountered in constitutively active drivers. Despite its advantages, the use of Tam has the potential to cause long-lasting, uncharacterized side effects on the transcriptome and epigenome in the CNS, given its mixed estrogen receptor (ER) agonist/antagonist actions. With the welcome focus on including both sexes in biomedical studies and efforts to understand sex differences, Tam administration could also cause sexually divergent responses that would confound studies. To examine these issues, epigenetic and transcriptomic profiles were compared in C57BL/6 J female and male hippocampus, cortex, and retina 1 month after a 5-day Tam treatment typical for cre induction, or vehicle control (sunflower seed oil). Cytosine methylation and hydroxymethylation levels, in both CG and non-CG contexts, were unchanged as determined by oxidative bisulfite sequencing. Long-lasting Tam transcriptomic effects were also not evident/minimal. Furthermore, there is no evidence of sexually divergent responses with Tam administration and Tam did not alter sex differences evident in controls. Combined with recently reported data that Tam alone does not cause long-lasting changes in behavior and neurogenesis, our findings provide confidence that Tam can be used as a cre-recombinase inducer without introducing significant confounds in transcriptomic and epigenomic neuroscience studies, particularly those focused on genomic and transcriptomic aspects of the aging brain.
Electronic supplementary material
The online version of this article (10.1007/s11357-019-00090-2) contains supplementary material, which is available to authorized users.
Keywords: Transgenic, Tamoxifen, Sex differences, Epigenome, Methylation, Transcriptome, Hippocampus, Cortex, Retina
Introduction
Epigenetic mechanisms, such as chromatin structure and DNA modifications, affect DNA accessibility and act as regulators of genomic organization and gene expression. Methylcytosine (mC) and hydroxymethylcytosine (hmC) can be dynamic or long-lasting DNA modifications that can also be passed on to daughter cells (Holliday 2006; Law and Jacobsen 2010; Allis and Jenuwein 2016; Masser et al. 2018). Recently, mC and hmC modifications have become a focus of neuroscience and aging research studies, along with other fields. Of particular interest in aging studies, specific genomic sites of differential methylation patterns in human and animal models have been identified (Horvath et al. 2012; Lister et al. 2013; Rakyan et al. 2010; Maegawa et al. 2010; Masser et al. 2017). Also, as biomedical research has begun to address the under-representation of female animal models in studies [(Clayton and Collins 2014)], developmental sex divergences in CNS DNA methylation have been identified (Nugent et al. 2015). Moreover, sex differences in hippocampal CG and non-CG (CH) methylation are evident in early adulthood and persist into advanced age in mouse and man (Masser et al. 2017). In addition to differences in the epigenome, sex-specific transcriptome profiles are evident. For instance, sexually divergent expression of the MHCI pathway across the CNS with aging (Mangold et al. 2017a), and significant sex differences in gene expression in the retina (Du et al. 2017) throughout the life span have been reported. Two understudied neuroscience questions are (1) whether sex-dependent differences in the epigenome and transcriptome are cell type-specific and (2) how sex affects DNA modifications and transcriptomic profiles with brain aging. The development of cell type-specific reporter Cre-lox mouse models that label specific cell types and allow paired analysis of the methylome and transcriptome upon induction of cre-recombinase with Tam could make the study of these questions feasible. Yet, if and how the introduction of Tam as an inducer of recombination affects the epigenetic and transcriptional processes in the brain has not been addressed. These control studies are needed for the field to have confidence that Tam induction models are not introducing a confounding variable.
Tam is a selective estrogen receptor modulator (SERM) that exerts agonist or antagonist activity on estrogen receptors (ERs) in a tissue-specific manner. This response depends on the complexity of ER signaling, its distribution, ligand-binding specificity, and interactions with co-activators or co-repressors (Pfaffl et al. 2001; Tee et al. 2004; Kuiper et al. 1997; Bramlett and Burris 2002; Webb et al. 2003). Since the early 1970s, Tam has been widely used as a peripheral ER antagonist for the treatment of breast cancer while identified as an ER agonist associated with increased endometrial cancer [(Maximov et al. 2013; van Leeuwen et al. 1994; Fornander et al. 1989)]. Within the brain, Tam may have ER-mixed antagonist-agonist properties (Newhouse and Dumas 2015; Haynes and Dowsett 1999), possibly due to expression of classical ERs by glial cells (Garcia-Ovejero et al. 2005), enhanced under several pathological conditions (Khalaj et al. 2013; Sakuma et al. 2009; Takahashi et al. 2004; Lu et al. 2003; Blurton-Jones and Tuszynski 2001). The expression of ERα has been localized in the cellular processes and soma of microglial cells in the hippocampus of adult mice and rat cerebellum, consistent with non-classical and understudied mechanisms of action of ERs (Marin et al. 2006; Beyer et al. 2003; Suuronen et al. 2005).
During the last two decades, Tam has become an important reagent applied in the analysis of gene functions, as well as cell-specific tagging in inducible conditional mouse mutants (Indra et al. 1999; Srinivasan et al. 2016; Jahn et al. 2018; Fonseca et al. 2017; Feil et al. 1997). Temporally controlled recombination of floxed target genes achieved by the CreERT2/loxP system circumvents embryonic lethal or aberrant phenotypes associated with constitutively activated cre recombinase during development (Jahn et al. 2018; Patel et al. 2017). CreERT2 is a recombinant fusion protein of the cre DNA recombinase and a mutant ligand-binding domain of the human ER. Similarly, to the endogenous ER, CreERT2 is trapped in the cytosol by heat shock proteins. While this protein complex is impervious to natural ligands, it is highly sensitive to synthetic estrogen antagonists, such as the Tam active metabolite, 4-OH-Tam, which in term activates the CreERT2 for recombination of floxed alleles in the cell nucleus. The cross of mice expressing cell-specific TAM-inducible cre recombinases with floxed allele mice such as INTACT (isolation of nuclei tagged in specific cell types) [(Mo et al. 2015)], TRAP (translating ribosome affinity purification) (Heiman et al. 2008), RiboTag (Sanz et al. 2009), and most recently developed NuTRAP (nuclear tagging and translating ribosome affinity purification) (Roh et al. 2017), are promising approaches for interrogating the sex- and age-dependent differences in transcriptomes and epigenomes of specific cell types in the adult CNS. In these models, upon Tam treatment, the tagging of cell-specific nuclei and/or ribosomes and downstream isolation of nucleic acids is only possible in floxed mice that co-express a specific cre transgene but not in the cre-negative counter-parts. In this context, the execution of parallel cre-negative controls is no longer a viable control as there will be no tagged ribosomes or nuclei for isolation.
A potential disadvantage of the use of Tam as inducer of CreERT2 in neuroscience research is the possibility of long-term effects on the normal brain function, proteins, or nucleic acids. In regard to this concern, Tam metabolism in mouse brains was reported to be age-, strain-, and dose-dependent (Patel et al. 2017). Another study suggested neither the route of application nor sex of treated mice influenced the response to Tam, and based on an end-point analysis 15 days post-treatment, concluded that short Tam treatments used to activate CreERT2 models have no significant persistent impact on adult neurogenesis or behavioral testing (Rotheneichner et al. 2017).
The purpose of the present study is to examine the long-lasting effects of Tam on the existing sex differences in the transcriptome as well as in genomic methylation and hydroxymethylation patterns in the brain. To do so, and in an attempt to separate the effect of Tam from the effects of Cre- or floxed-transgene expression, we performed paired analysis of cytosine methylation and hydroxymethylation levels, in both CG and non-CG contexts and RNA-seq transcriptomic profiles in hippocampus, cortex, and retina tissues from female and male C57BL/6 J mice treated with Tam or with its vehicle (Veh, sunflower seed oil).
No significant, long-lasting effects of Tam treatment in the transcriptome or epigenome were found in any of the tissues analyzed. Combined with recently reported data that Tam alone does not cause long-lasting changes in behavior and neurogenesis, our findings provide confidence that Tam can be used as a cre-recombinase inducer without introducing significant confounds in cell-specific transcriptomic and epigenomic neuroscience studies, including those exploring mechanisms to be targeted in aging (Unnikrishnan et al. 2017; Ashpole et al. 2017; Deepa et al. 2017; Podlutsky et al. 2017), where timely controlled tamoxifen induction could be of benefit in elucidating cell-specific contributions to the development or delay of any of the hallmarks of aging.
Methods
Animals
Male (n = 8) and female (n = 7) C57BL/6 J mice were purchased from the Jackson Laboratory (stock number: 000664; Bar Harbor, ME), housed in the animal facility at the University of Oklahoma Health Sciences Center and maintained under specific pathogen-free (SPF) conditions in a HEPA barrier environment. At 3 months of age, mice were divided into 4 experimental groups: female + Veh (n = 3), female + Tam (n = 4), male + Veh (n = 4), and male + Tam (n = 4) and subjected to Tam or Veh treatment. One month following Tam treatment, mice were euthanized, and hippocampi, cortices, and retina were harvested for further analyses. All procedures with mice were approved by the Institutional Animal Care and Use Committee at the University of Oklahoma Health Sciences Center.
Tamoxifen treatment
At 3 months of age, mice received a single daily intraperitoneal (ip) injection of 100 μl Tam solubilized in 100% sunflower seed oil by sonication (100 mg/kg body weight, 20 mg/ml stock solution, Sigma, St. Louis, MO) for five consecutive days (Srinivasan et al. 2016). Control mice received 100 μl of Veh, 100% sunflower seed oil, by ip delivery.
Genomic DNA and total RNA isolations
Genomic DNA (gDNA) and total RNA (RNA) were isolated from hippocampal, cortical, and retinal frozen tissue using AllPrep DNA/RNA Mini kits (Qiagen, Germantown, MD) as described previously (Imperio et al. 2016; Mangold et al. 2017b). gDNA was quantified by Qubit fluorescent assays (dsDNA BR assay kit, Thermofisher Scientific, Grand Island, NY) and RNA was quantified with a Nanodrop 2000c spectrophotometer (Thermofisher Scientific). gDNA and RNA quality (> 8 DNA and RNA integrity numbers) were assessed by genomic DNA D1000 and RNA ScreenTape assays, with a 2200 Tapestation analyzer (Agilent Technologies, Santa Clara, CA).
Library construction and oxidative bisulfite sequencing
For each hippocampal, cortical, and retinal sample, 1 μg of gDNA was brought up to 50 μl volume with 1× low-EDTA TE buffer and sheared with a Covaris E220 sonicator (Covaris, Inc., Woburn, MA) to an average base pair size of 200 using the following settings: intensity of 5, duty cycle of 10%, 200 cycles per burst, 2 cycles of 60 s, at 7 °C. The size of sheared products was confirmed by capillary electrophoresis (DNA D1000, Agilent). gDNA fragments were cleaned by an Agencourt bead-based purification protocol, after which gDNA was quantified (Qubit, Thermofisher Scientific). Two aliquots of 200 ng gDNA fragments were prepared in a 12 μl volume to which 1 μl of spike-in control DNA (0.08 ng/ul) with known levels of specific mC, hmC, and fC at individual sites was added. End repair, ligation of methylated adaptors (#L2V11DR-BC 1–96 adaptor plate, NuGEN, Tecan Genomics, Inc., Redwood City, CA) and final repair were performed according to manufacturer’s instructions (Ovation Ultralow Methyl-Seq Library System (NuGEN). Of the two DNA aliquots per sample, one was oxidized and then bisulfite-converted and the other only bisulfite-converted with the True Methy oxBS module (NuGEN) with desulfonation and purification. Twenty-two microliters of libraries was eluted from the magnetic beads. qPCR was used to determine the number (N) of PCR cycles required for library amplification. Bisulfite-converted samples were amplified for 7 cycles while oxidative bisulfite-converted samples were amplified for 11 cycles [95 °C—2 min, N (95 °C—15 s, 60 °C–1 min, 72 °C–30 s)]. Amplified libraries were purified with Agencourt beads and eluted in low-EDTA TE buffer. TapeStation HD1000 was used to validate and quantify libraries. Amplified libraries were normalized to a concentration of 4 nM and pooled, denatured, and diluted to 12 pM for sequencing on MiSeq (Illumina, San Diego, CA) according to manufacturer’s guidelines with the exception of a custom sequencing primer (MetSeq Primer) that was spiked in with the Illumina Read 1 primer to a final concentration of 0.5 μM.
OxBS-seq data analysis
Prior to alignment paired-end reads were adaptor-trimmed and filtered using FASTQ Toolkit in Basespace (Illumina, San Diego, CA). End-trimming removed 3 bp and 4 bp from the 5′ and 3′ end of each paired-end read, respectively. Only reads with a Q score ≥ 25 and matching length criteria were used for mapping. Alignment of trimmed bisulfite-converted sequences was carried out using MethylSeq in Basespace (Illumina) (Krueger and Andrews 2011) against the mouse reference genome (GRCm38/mm10). Methylation call percentages for each CpG and non-CpG (CH) site within the genome were calculated by dividing the methylated counts over the total counts for that site in the oxidative bisulfite-converted libraries (OXBS). Genome-wide CpG and CH methylation levels were calculated separately. Hydroxymethylation levels in CpG (hmCG) and CH (hmCH) contexts were calculated by subtracting call levels from the oxidative bisulfite-converted (OXBS) libraries from the bisulfite-converted (BS) libraries. BAM files generated by MethylSeq (Basespace, Illumina) were run through MethylKit in R (Akalin et al. 2012) to generate context-specific (CpG/CH) coverage text files. Methylation percentages for the X and Y chromosomes were calculated separately as previously described [(Hadad et al. 2016)]. Briefly, coverage files were filtered to X and Y chromosomes to calculate sex-chromosome-specific methylation and hydroxymethylation levels in the same manner as genome-wide levels. Bisulfite conversion efficiency for C, mC, and hmC was estimated using CEGX spike-in control sequences (Supplemental Fig. 1). Untrimmed fastq files were run through CEGX QC v0.2 which output a fastqc_data.txt file containing the conversion mean for C, mC, and hmC.
Library construction and RNA-seq
Stranded RNA sequencing (RNA-Seq) (TruSeq v2, Illumina) was performed on 500 ng of total RNA from each sample and brain region from males and females, VEH- and TAM-treated mice (n = 3–4/group). Library construction was performed in a stranded manner to retain the directionality of the transcripts (Sultan et al. 2012). Each sequencing library was prepared from RNA from a single animal to provide biological variance. Illumina Truseq Stranded HT library generation was performed according to manufacturer’s instructions. Briefly, polyA containing mRNA was purified using oligo-dT attached magnetic beads. mRNA was then chemically fragmented and cDNA synthesized. For strand-specificity, the incorporation of dUTP instead of dTTP in the second strand cDNA synthesis does not allow amplification past this dUTP with the polymerase. Following cDNA synthesis, each product underwent end repair process, the addition of a single ‘A’ base, and finally ligation of adapters. The cDNA products were further purified and enriched using PCR to make the final library for sequencing. Library sizing was performed by TapeStation (Agilent Technologies) and libraries were quantified by qPCR (Kappa Biosystems, Inc., Wilmington, MA). The cDNA library was then sequenced using an Illumina Hiseq 2500 at the PennState College of Medicine Genome Sciences Facility in a 2 × 100 bp fashion for the hippocampus. RNA-seq libraries from cortex and retina tissues were sequenced using an Illumina NextSeq (2 × 75bp) at the Oklahoma Medical Research Foundation Genomics Facility.
RNA-Seq data analysis
Following sequencing, reads were trimmed, aligned, differential expression statistics and correlation analyses were performed in Strand NGS software package (Agilent). Reads were aligned against the Mm10 build of the mouse genome (2014.11.26). Alignment and filtering criteria included: adapter trimming, fixed 2 bp trim from 5′ and 6 bp from 3′ ends, a maximum number of one novel splice allowed per read, a minimum of 90% identity with the reference sequence, a maximum of 5% gap, trimming of 3′ end with Q < 30. The alignment was performed directionally with read 1 aligned in reverse and read 2 in forward orientation. Reads were filtered based on the mapping status and only those reads that aligned normally (in the appropriate direction) were retained. Normalization was performed with the DESeq algorithm [(Anders and Huber 2010)]. Transcripts with an average read count value > 20 in at least 100% of the samples in at least one group were considered expressed at a level sufficient for quantitation per tissue and those transcripts below this level were considered not detected/not expressed and excluded, as these low levels of reads are close to background and are highly variable. For statistical analysis of differential expression, a two-way ANOVA with the factors of sex and treatment and a Benjamini-Hochberg multiple testing correction followed by Student-Newman Keuls post hoc test was used. For those transcripts meeting this statistical criterion, a fold change >|1.25| cutoff was used to eliminate those genes which were statistically significant but unlikely to be biologically significant and orthogonally confirmable due to their very small magnitude of change. Visualizations of hierarchical clustering and principal components analysis were performed in Strand Next Generation Analysis Software (NGS) (Version 3.1, Bangalore, India). The entirety of the sequencing data is available for download in FASTQ format from NCBI Sequence Read Archive (GSE135752 and GSE135208).
Quantitative PCR
Confirmation of gene expression levels was performed with quantitative PCR (qPCR) as previously described [(Du et al. 2017; Masser et al. 2014)]. cDNA was synthesized from purified RNA with the ABI High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems Inc., Foster City, CA) from the following RNA amounts: 1 μg from hippocampus, 500 ng from retina, and 200 ng from cortex. qPCR was performed with gene-specific primer-probe fluorogenic exonuclease assays (TaqMan, Life Technologies, Waltham, MA, Supplemental Table 1) and the QuantStudio™ 12 K Flex Real-Time PCR System (Applied Biosystems). Relative gene expression (RQ) was calculated with Expression Suite v 1.0.3 software using the 2−ΔΔCt analysis method with GAPDH as an endogenous control. Statistical analysis of the qPCR data was performed using SigmaPlot 12.5 (SyStat Software, San Jose, CA). Two-way ANOVA analyses were performed with the factors of sex and treatment. Post-hoc All Pairwise comparisons with factors of sex and treatment and interactions of both factors were performed with all pairwise multiple comparison procedures (Holm-Sidak method) with overall significance level = 0.05.
Protein extraction and western blotting
Hippocampus, cortex, and retina samples were harvested and homogenized in RIPA buffer (Thermo Scientific) supplemented with 1× Halt™ protease inhibitor cocktail (Thermo Scientific) by sonication (Sonic dismembrator Model 100, Fisher Scientific). The supernatants from tissue homogenates were assayed for protein content using a BCA protein method (Thermo Scientific). Equal amounts of protein per sample (20 μg) were resolved on SDS 4–20% gradient polyacrylamide gels (Thermo Scientific) prior to transferring onto nitrocellulose membranes. Proteins were detected using the following primary antibodies: anti-BANP antibody (Abcam, Cambridge, MA, # ab105404, 1:2000), anti-FOS (Sigma-Aldrich, # SAB4500995, 1:1000), and anti-β actin (Abcam, #ab6276, 1:10,000). For secondary chemiluminiscent detection, blots were incubated with corresponding HRP-conjugated secondary antibodies (GE Healthcare Bio-Sciences, Pittsburgh, PA). Imaging of the blots and analysis of band intensities were performed by conventional image analysis using a Kodak in vivo imaging system F Pro and Carestream MI SE 4.4 SE version software (Carestream Health, Inc., Rochester, NY) [(Chucair-Elliott et al. 2017)]. Two-way ANOVA analyses were performed with the factors of sex and treatment. Post-hoc all pairwise comparisons with factors of sex and treatment and interactions of both factors were performed with all pairwise multiple comparison procedures (Holm-Sidak method) with overall significance level = 0.05.
Results
For the study of the potential interactions of Tam systemic treatment with sex in the mouse brain at the epigenetic and transcriptomic levels, the terminology follows recommendations stated by McCarthy et al. (McCarthy et al. 2012). We define a sex difference as a difference between males and females that persists throughout life (such as gene expression level of a particular gene), a sexual dimorphism as a binomial difference between males and females (such as Y chromosome encoded gene expression is only in males). Sex divergences are emergent, such as expression of a gene at a young age that is the same in males and females but becomes different at old age [(McCarthy et al. 2012)].
Previously observed X-linked sex differences and Y-linked dimorphisms are unaffected by tamoxifen
To test whether Tam had long-lasting effects on the existing sex differences/dimorphisms in brain gene expression, expression of genes located on the X- and Y- chromosomes [(Du et al. 2017; Armoskus et al. 2014; Snell and Turner 2018) was measured by qPCR in hippocampus, cortex, and retina samples from female and male C57BL/6 J mice 30 days after a 5-day course of Tam or Veh treatment.
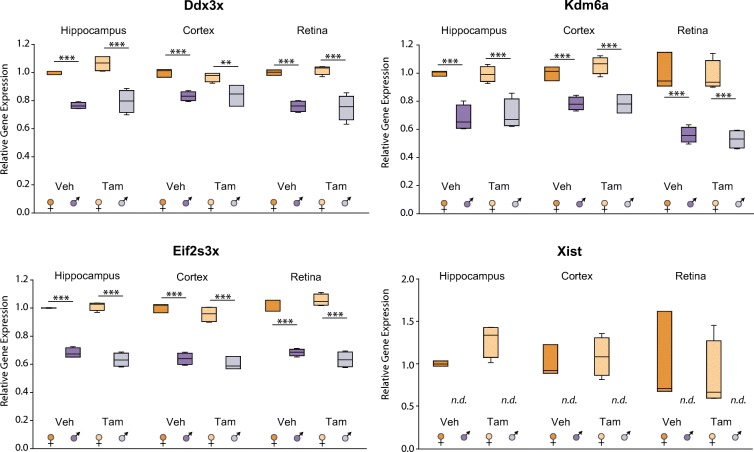
The X-linked target genes selected were Ddx3x, with diverse roles in RNA splicing and export, translation initiation, cell cycle regulation, and apoptosis (Cordin et al. 2006; Su et al. 2015; Sun et al. 2008); Kdm6a (Utx), a histone lysine demethylase that catalyzes removal of H3K27me2 and me3, and possibly also functions as a transcriptional activator (Agger et al. 2007; Lee et al. 2007; Xu et al. 2008, 2006); Eif2s3x, which regulates the rate of protein translation (Xu et al. 2006); and Xist, necessary and sufficient to initiate X-inactivation (Gayen et al. 2016). As shown in Fig. 1, 1 month after cessation of treatment, the sex differences and divergences of all X-linked transcripts tested between males and females across hippocampus, cortex, and retina tissues was unchanged. Differential expression by sex was independent of treatment. Males treated with Veh had significantly lower expression of Ddx3x, Kdm6a, Eeif2s3x, and no detectable expression of Xist compared to Veh-treated females. Tam treatment did not significantly affect such differences.
Fig. 1.
Previously observed X-linked sex differences are unaffected by Tam. At 3 months of age, male and female C57BL/6 J mice (n = 3–4/group) were subjected to daily, ip delivery of Tam or Veh (control) for 5 consecutive days. One month after cessation of treatment, hippocampus, cortex, and retina were harvested and RNA processed for qPCR analysis of X chromosome-linked candidate genes. TaqMan probes were used to target Ddx3x, Kdm6a, Eif2s3x, Xist genes, and GAPDH as housekeeping gene. Box plot expresses average normalized RQ for each group ± SEM. **p < 0.01, *** p < 0.001, by two-way ANOVA with factors of sex and treatment followed by all pairwise multiple comparison procedures (Holm-Sidak method)
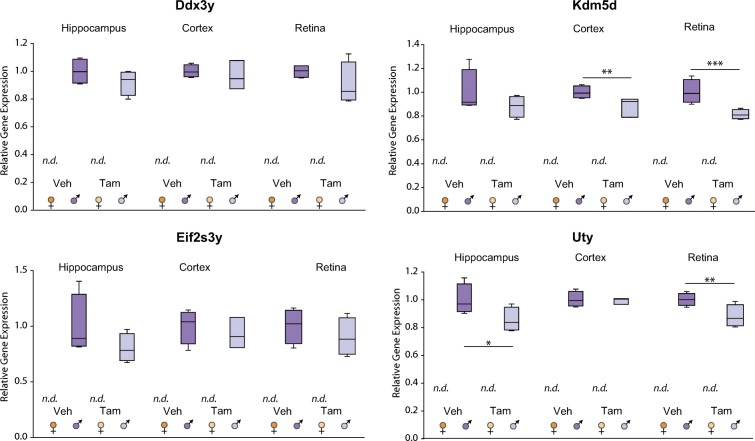
For the analysis of Y-linked genes, the following target genes were selected: Ddx3y, a Y chromosome homolog of Ddx3x, and regulator of spermatogenesis and sperm development in mice (Mazeyrat et al. 1998); Kdm5d, required for demethylation of H3K4me3 and H3K4me1 [(Blair et al. 2011; Mizukami et al. 2019)]; Eif2s3y, a Y chromosomal homolog of the X-linked Eif2s3x [(Abdelhaleem 2005)]; with role in spermatogenesis (Kopsida et al. 2009; Mazeyrat et al. 2001); and Uty, a Utx paralogue on the Y chomosome [(Xu et al. 2008)]. As previously shown in the mouse CNS (Du et al. 2017; Armoskus et al. 2014; Xu et al. 2008), such Y-linked genes are exclusively expressed in males. The expression of Kdm5d was slightly (< 20%) and significantly lower in cortex and retina of Tam-treated male mice compared to controls. Similarly, Tam treatment resulted in slightly decreased Uty expression in male retina and hippocampus. Despite small changes in the transcription of two of the Y-linked genes with treatment, Tam did not generally affect the sexually dimorphic gene expression tested in the CNS (Fig. 2).
Fig. 2.
Previously observed Y-linked sex dimorphisms are unaffected by Tam. At 3 months of age, male and female C57BL/6 J mice (n = 3–4/group) were subjected to daily, ip delivery of Tam or Veh (control) for 5 consecutive days. One month after cessation of treatment, hippocampus, cortex, and retina were harvested and RNA processed for qPCR analysis of Y chromosome-linked candidate genes. TaqMan assay probes were used to target Ddx3y, Kdm5d, Eif2s3y, subunit 3, and Uty transcripts. Box plots express average normalized RQ for each group ± SEM. *p < 0.05, **p < 0.01, ** p < 0.001, comparing male and female groups under the same treatment by two-way ANOVA followed by all pairwise multiple comparison procedures (Holm-Sidak method). n.d. expression not detected
Tamoxifen does not interact with sex differences in the hippocampus, cortex, and retina
To examine the long-term effects of Tam treatment on the female and male transcriptomes across different regions of the CNS, a discovery approach was taken. Hippocampi, cortices, and retinae collected 30 days after cessation of ip treatment with Tam or Veh and were processed for RNA-Seq analysis.
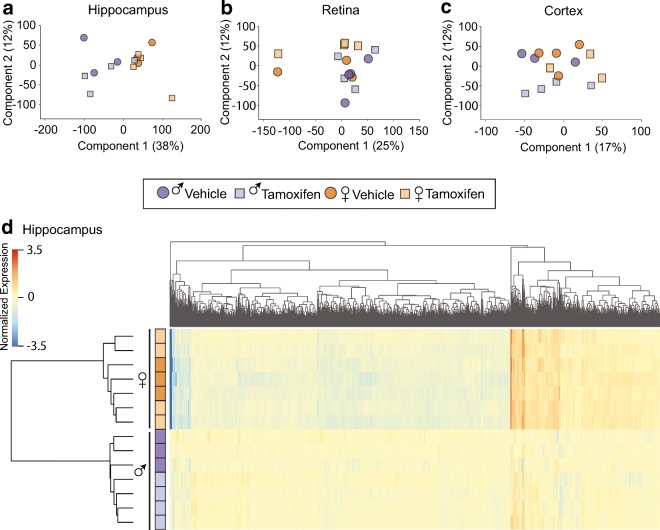
An average of 19 million reads per sample passed filtering and alignment criteria for cortex and retina samples. Hippocampal samples were sequenced to a great depth averaging 44 million reads per sample. There was no difference in the number of reads mapped per group within each tissue and equivalent number of reads mapped to the positive and negative strands. Using a read count cutoff of > 20 for all samples in at least one group, 13,163 transcripts with mouse RefSeq annotations were called as expressed in the retina, 13,237 cortex, and 14,046 hippocampus. Principal component analysis (PCA) on samples for each tissue revealed no clustering by treatment and some segregation by sex, as expected (Fig. 3a–c). Differentially expressed transcripts between groups were identified through a two-way ANOVA, with factors of treatment and sex, followed by the Benjamini-Hochberg multicomparison test (p < 0.05). These transcripts were further filtered to only those that had a > |1.25| fold difference between the groups that had a significant post-hoc test result. For hippocampus, 2073 genes by sex and no significant differences with treatment or for an interaction effect were found. Similarly, hierarchical clustering on the hippocampus samples demonstrated that the individual samples were separated by sex with samples from Tam- and Veh-treated mice interspersed by sex (Fig. 3d).
Fig. 3.
Tam does not interact with sex differences in hippocampus, retina, or cortex. At 3 months of age, male and female C57BL/6 J mice (n = 3–4/group) were subjected to daily, ip delivery of Tam or Veh (control) for 5 consecutive days. Following 1 month after cessation of treatment, their CNS tissues were harvested and processed for RNA-Seq analysis. a Principal component analysis (PCA) of transcriptome profile of hippocampus shows the consistent sex differences we have described previously but Veh- and Tam-treated samples cluster together. PCA of retina (a) and cortex (c) did not show clustering by treatment, with female and male samples interspersed with treatment. d Heatmap presentation of genes with significant differences in expression analysis (two-way ANOVA, factors of treatment and sex, p < 0.05, Benjamini-Hochberg multiple testing correction) revealed 2073 genes differentially expressed by sex and no significant differences with treatment or for an interaction effect in the hippocampus. By hierarchical clustering samples were separated by sex with samples from Tam- and Veh-treated mice interspersed by sex
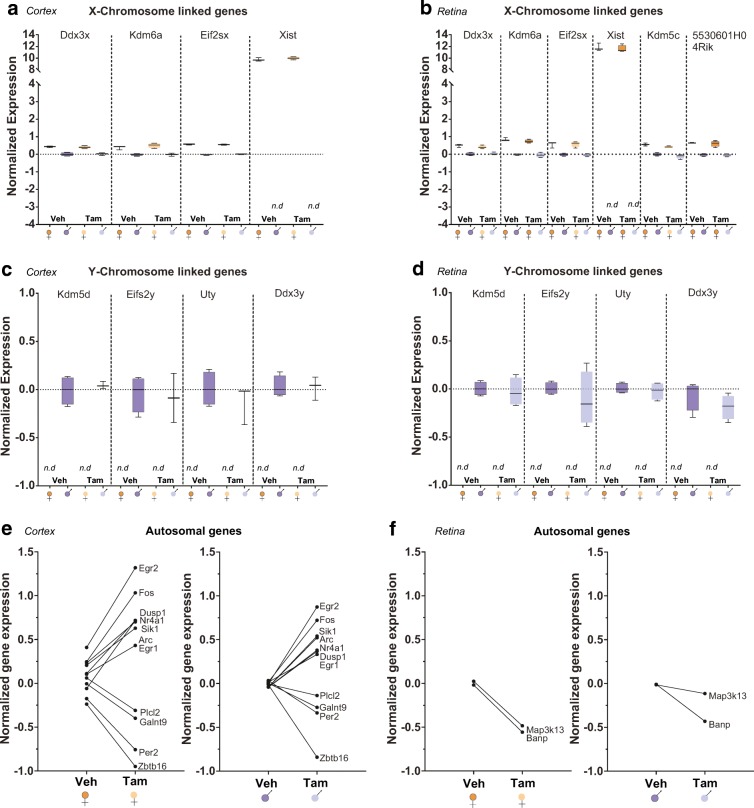
As previously published for cortex (Mangold et al. 2017b) and retina (Du et al. 2017), the sex differences in the transcriptome were maintained across tissues and treatments in our study. There were no significant interactions between Tam treatment and sex in either of these CNS tissues (Fig. 4). Cortex differentially expressed 8 genes for sex (Fig. 4a, c) and 11 genes for treatment (Fig. 4e). The number of genes differentially expressed in the retina was 10 for sex (Fig. 4b, d) and 2 for treatment (Fig. 4f). Collectively, the RNA-seq analysis identified no significant interaction effect of Tam treatment with sex differences and only minimal effects of Tam administration.
Fig. 4.
Tam treatment does not introduce significant interactions effect with sex in retina or cortex. At 3 months of age, male and female C57BL/6 J mice (n = 3–4/group) were subjected to daily, ip delivery of Tam or Veh (control) for 5 consecutive days. One month after cessation of Tam treatment RNA-Seq was performed. Both in cortex and retina Tam treatment did not alter known sex differences in expression of X (a. b) or Y chromosome encoded genes (c, d). A few differences in gene expression were observed with TAM treatment in the cortex (e) and retina (f), but these were generally equivalent in males and females. RNA-Seq data presented as normalized gene expression as described in detail in the “Methods” section.
Tamoxifen has minimal effect on the expression of selected autosomal transcripts across hippocampus, cortex, and retina tissues
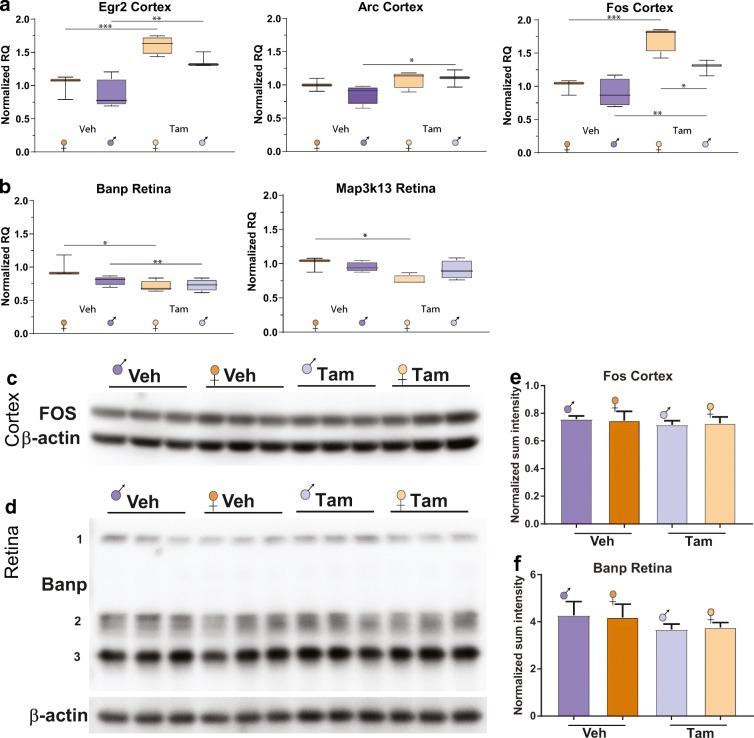
To further assess potential interactions between Tam and sex shown by our RNA-seq results, differentially expressed autosomal genes with Tam administration in the cortex and retina were examined by qPCR. For cortex, the target genes were Egr2, associated with the onset of myelination of the peripheral nervous system and hindbrain development and for T and B cell development and differentiation [(Taefehshokr et al. 2017)]; Fos, which helps the formation of the transcription factor AP-1 to regulate a wide array of genes in response to many stimuli [(Coulon et al. 2010)]; and Arc, implicated in neuronal activation, synaptic plasticity, and a variety of memory tasks (Bramham et al. 2008; Gao et al. 2018) (Fig. 5a). For retina, the target genes were Banp, which functions as anti-tumorigenic and modulator of the cell cycle through interaction with and stabilization of p53 [(Malonia et al. 2011)]; and Map3k13, reported to activate both the NFkB and JNK pathways (Keshet and Seger 2010) (Fig. 5b). The qPCR data analysis showed significantly higher expression of Egr2, Arc, and Fos genes in the cortex of Tam-treated males and/or females compared to their same-sex Veh-treated counterparts, with only sex and treatment interactions for Fos expression (Fig. 5a). In the retina, Tam had a slight effect decreasing the transcript levels of Banp compared to same-sex control groups. Map3k13 transcription was slightly lower in Tam-treated females compared to their respective VEH-control (Fig. 5b). For hippocampus, no autosomal genes were found to be differentially expressed with treatment by RNA-seq, thus no qPCR data is shown.
Fig. 5.
Tam has minimal effect on the expression of selected autosomal transcripts across cortex and retina tissues. To confirm RNA-Seq findings qPCR was performed for selected gene targets. In the cortex, significant changes in Egr2, Arc, and Fos expression were evident (a). Only minimal interactions of sex with TAM treatment were evident, less than 25% differences. In the retina, very small differences with Tam treatment were observed for Banp and Map3k13. To further examine Tam effects, where specific antibodies were available, Fos and Banp protein expression in cortex (c) and retina (d) respectively were examined. No significant differences with Tam were observed. Box plots express average normalized RQ for each group (n = 3–4) ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, comparing male and female groups under the same treatment by two-way ANOVA followed by all pairwise multiple comparison procedures (Holm-Sidak method). e–f The ratio of signal intensity of Fos and Banp bands relative to β-actin bands in cortex (e) and retina (f) by WB densitometry analysis is expressed as average normalized sum intensity for each group (n = 3) ± SEM. Statistical analysis was performed by two-way ANOVA followed by all pairwise multiple comparison procedures (Holm-Sidak method)
In order to assess whether the minimal effect of Tam on the gene expression of selected autosomal transcripts has repercussions at the protein level, WB was performed on cortex and retina protein extracts from all experimental groups. For the protein analysis, and based on significance of changes by qPCR, Fos was selected for cortex, and Banp for retina. In cortex, a single Fos detected protein band showed at ~ 55 kDa, consistent with predicted size (Fig. 5c). Of note, for Banp (with an expected band size of ~ 56 kDa on WB of specific cell lysates and tissues, and with variable reported sizes from 50 to 71 KDa), in retina detected bands exhibited three different sizes that were summed within each sample for densitometric quantification (band 1:~ 170 kDa, dimer bands 2: ~ 64 kDa, and single/dimer bands 3: ~ 40 kDa) (Fig. 5d). The detected band size for β actin was ~ 42 kDa, as expected.
The densitometry analysis revealed no significant differences between groups in the protein content of the protein tested in the cortex (Fig. 5e), or retina (Fig. 5f). In conclusion, the minimal differences introduced by Tam at the transcript level were not recapitulated at the protein level across the cortex and retina tissues in our study.
TAM does not alter genome-wide methylation and hydroxymethylation
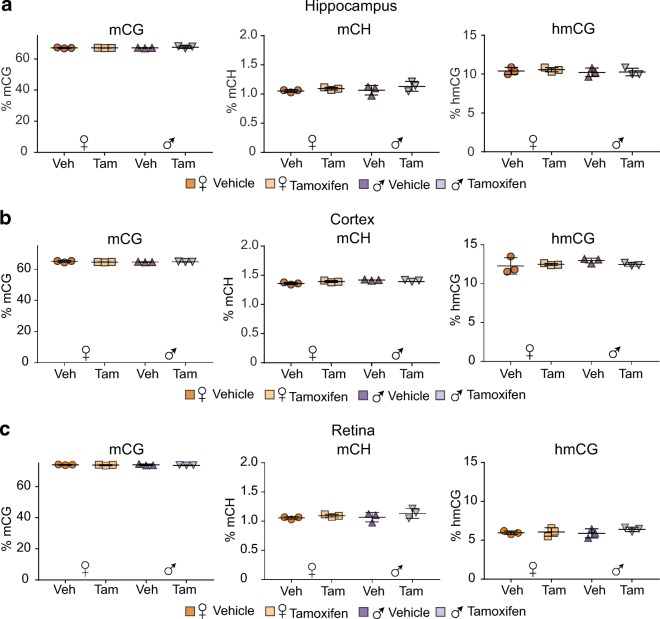
To our knowledge, there is no published control data on whether Tam administration affects DNA modifications. To assess levels of methylation (mC) and hydroxymethylation (hmC) in both CG and CH contexts whole-genome oxidative bisulfite sequencing was performed across sexes, treatments, and brain regions. Across all sexes, treatment groups, and brain regions equivalent methylation (~65%) were observed in the CG context (Fig. 6). Non-CpG methylation (mCH) was not altered by Tam treatment and was equivalent in males and females with higher levels in the cortex than hippocampus or retina. For hydroxymethylation in the CG context (hmCG), no effect of treatment of sex was evident with the highest hmCG levels being observed in the cortex followed by the hippocampus and then retina. No non-CpG hydroxymethylation was observed, in agreement with prior studies [(Hadad et al. 2016; Wen et al. 2014)].
Fig. 6.
Tam does not alter genome methylation and hydroxymethylation. At 3 months of age, male and female C57BL/6 J mice (n = 3–4/group) were subjected to daily, ip delivery of Tam or Veh (control) for 5 consecutive days. One month after cessation of treatment mC and hmC were analyzed in the hippocampus (a), cortex (b), and retina (c) by context (CpG and nonCpG (CH)) DNA isolated from tissues was subjected to oxidative bisulfite conversion, library construction, and whole-genome oxidative bisulfite sequencing. No differences with treatment or sex in mCG, hmCG, or mCH were observed across all neural tissues assayed. hmCH was undetectable (not shown). Statistical analysis was performed by two-way ANOVA, with factors of treatment and sex, p < 0.05, Benjamini-Hochberg multiple testing correction
TAM does not alter sex-chromosome methylation and hydroxymethylation
We have previously reported no age or sex differences in mCG-specific methylation levels in autosomes, but higher X chromosome methylation in females compared to males in hippocampus [(Hadad et al. 2016)]. To elucidate whether Tam could disrupt such sex differences, the methylation and hydroxymethylation patterns, specifically within X- and Y-chromosomes, were analyzed. Whole-genome oxidative bisulfite sequencing (WG-oxBS) data revealed male hippocampus had slightly and yet significantly lower mCG, while higher levels of mCH and hmCG in the X-chromosome compared to the female hippocampus (Fig. 7a). Such differential pattern of mCG, mCH, and hmCG in males and females was mirrored in cortex samples (Fig. 7B). Of interest, male retina samples had higher X-chromosome mCG, mCH, and hmCG levels than female retina (Fig. 7c). There was no effect of Tam on X-chromosome mCG, mCH, or hmCG, across the three CNS tissues. Analysis of male hippocampus, cortex, and retina showed Y-chromosome mCG, hmCG, and mCH were also unaffected by Tam-treatment (Fig. 8a–8c). In summary, the epigenetic data generated in this study supports no interactions between Tam and the existing sex differences in the brain at the level of DNA modifications.
Fig. 7.
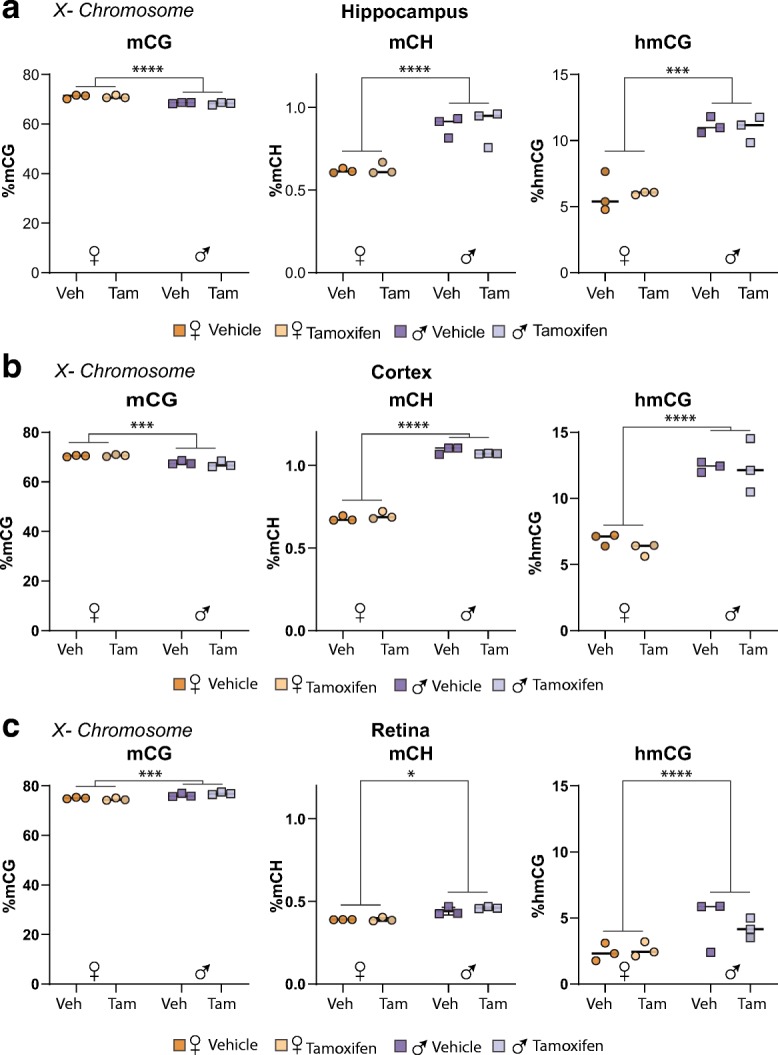
Tam does not alter X-chromosome methylation and hydroxymethylation. At 3 months of age, male and female C57BL/6 J mice (n = 3–4/group) were subjected to daily, ip delivery of Tam or Veh (control) for 5 consecutive days. Following 1 month after cessation of treatment, their hippocampi (a), cortices (b), and retinae (c) were harvested and processed for oxidative bisulfite conversion, library construction, and whole-genome oxidative bisulfite sequencing. Sex differences were observed in X-chromosome mCG, mCH, and hmCG across all neural tissues assayed. No differences with in X-chromosome mCG, hmCG, or mCH were observed in any neural tissues assayed. Statistical analysis was performed by two-way ANOVA, with factors of treatment and sex, p < 0.05, Benjamini-Hochberg multiple testing correction
Fig. 8.
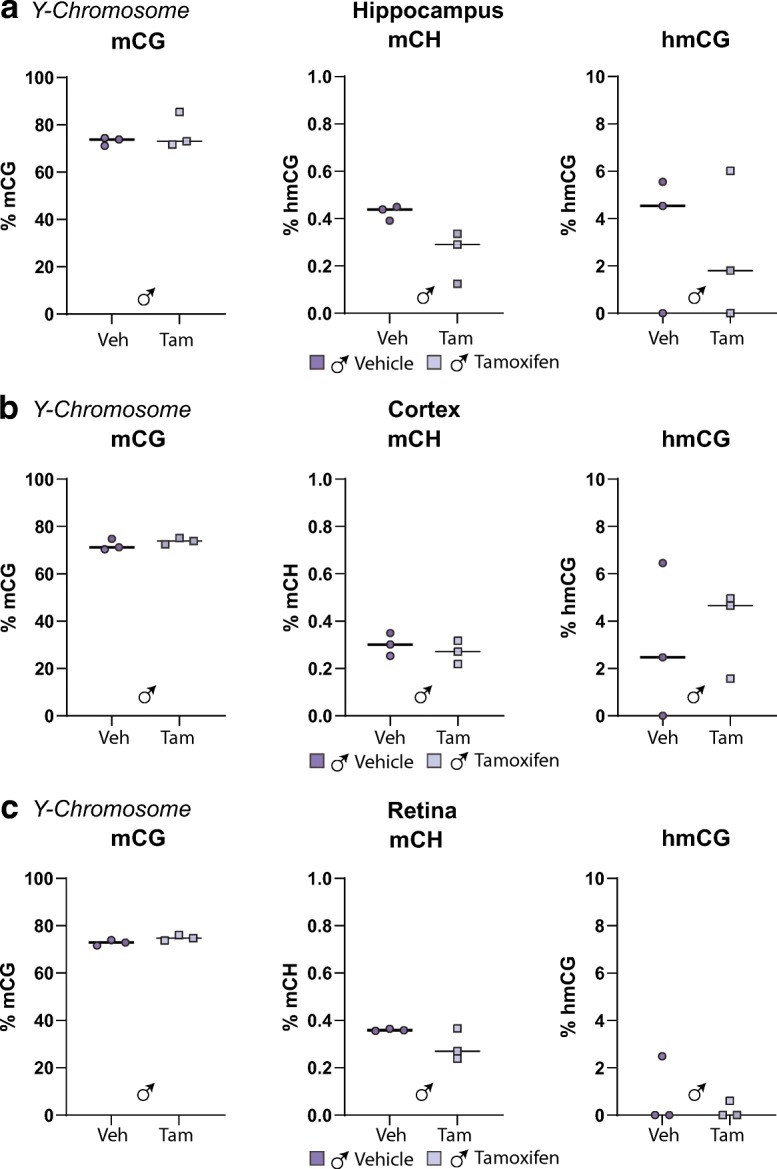
Tam does not alter Y-chromosome methylation and hydroxymethylation. At 3 months of age, male and female C57BL/6 J mice (n = 3–4/group) were subjected to daily, ip delivery of Tam or Veh (control) for 5 consecutive days. Following 1 month after cessation of treatment, their hippocampi (a), cortices (b), and retinae (c) were harvested and processed for oxidative bisulfite conversion, library construction, and whole-genome oxidative bisulfite sequencing. No differences with in Y-chromosome mCG, hmCG, or mCH were observed in any neural tissues assayed. Statistical analysis was performed by two-way ANOVA, with factors of treatment and sex, p < 0.05, Benjamini-Hochberg multiple testing correction
Discussion
Recently, significant contributions have been made in geroscience research that suggest potential pathways as targets for intervention to increase the lifespan and counteract frailty [(Ashpole et al. 2017; Deepa et al. 2017; Podlutsky et al. 2017)]. In this context, and taking into account the need for sex differences/dimorphisms to be accounted in animal research [(Masser et al. 2017; Clayton and Collins 2014; Ashpole et al. 2017)], well-controlled in vivo systems are a priority to avoid misinterpretation of data, specifically at the molecular level, as discussed below.
In light of the increasing interest in sex effects on the epigenome and transcriptome both in normal aging and disease settings, transgenic inducible cre mouse models that allow for manipulation of specific floxed genes, or tagging of cell-specific nuclei/and or polysomes represent valuable research tools. For instance, the cross of mice expressing cell-specific Tam-inducible cre recombinases with mice with floxed alleles, such as INTACT [(Mo et al. 2015)], TRAP [(Heiman et al. 2008)], RiboTag [(Sanz et al. 2009)], and NuTRAP [(Roh et al. 2017)], is a promising strategy for interrogating the sex- and age-dependent differences in the transcriptomes and/or epigenomes of specific cell types in the adult CNS. Despite the advantages that such technical advancements have to offer to the field of neuroscience, the potential effects of Tam on the epigenetic and transcriptional processes in the brain are a concern. This concern stems from the fact that Tam could exert agonist/antagonist actions on endogenous estrogen receptors [(Pfaffl et al. 2001; Tee et al. 2004; Kuiper et al. 1997; Bramlett and Burris 2002; Webb et al. 2003; Riggs and Hartmann 2003; Wardell et al. 2014)] in addition to control of Cre-lox-mediated recombination. Such SERM-associated activities might have implications at the molecular level and consequently on cognitive function. The aim of the present study was to examine the long-lasting effects of Tam on the existing sex differences in the transcriptome as well as in genomic methylation and hydroxymethylation patterns in the brain.
To address this question, we used C57BL/6 J male and female mice, subjected to a routinely used dose and regime of Tam i.p. injections to induce cre recombinases in a variety of tissues, including CNS [(Srinivasan et al. 2016; Jahn et al. 2018; Park et al. 2018)]. In the absence of cre expression, we were able to interrogate side effects of TAM on cytosine methylation and hydroxymethylation levels, in both CG and non-CG contexts and RNA-seq transcriptomic profiles of three commonly studied CNS tissues: hippocampus, cortex, and retina. Collectively, our findings support the Tam does not affect existing sex differences, dimorphisms, and (presumably) divergences in the brain, epigenome- and trancriptome-wide. To our knowledge, this is the first report addressing potential side effects of this drug on the DNA modifications or at the entire transcriptome level. Here, we show that (1) Tam does not affect previously observed X-linked sex differences or Y-linked dimorphisms (Figs. 1 and 2), nor does it interact with sex differences in autosomal gene expression (Figs. 3, 4, and 5) in hippocampus, cortex, and retina; and (2) Tam does not alter genome-wide methylation and hydroxymethylation (Fig. 6) or previously described sex-chromosome methylation and hydroxymethylation (Figs. 7 and 8) across tested CNS tissues. Importantly, Tam administration does not demonstrate lasting sex-specific effects either.
The toxicity of Tam as a confounding factor in experiments has been a matter of concern across research animal models, tissues, and with timing of induction [(Patel et al. 2017; Rotheneichner et al. 2017; Karlsson 2006; Morales-Otal et al. 2005)]. In testis—which responds to estrogen signaling and produces endocrine factors—adverse effects of Tam have been reported [(Patel et al. 2017; Karlsson 2006; Morales-Otal et al. 2005)]. A study by Patel et al. described long-term consequences of a single Tam injection on the testis of male mice at early pubertal ages (~p21), with persistent adverse changes to both spermatogenic and endocrine functions of the testis that could be detected weeks later [(Patel et al. 2017)]. From those reported findings (Patel et al. 2017), it seems reasonable to infer that Tam induction for cre recombination at a borderline developmental time window with respect to full sexual maturation might have long-lasting effects that persist or even worsen with age and might affect almost every tissue in the body. In contrast, the experiments in this study, started at 3 months of age, and were analyzed at ~ 4 months of age, which led to no apparent dysfunctional phenotype, and most importantly to our purpose, no adverse effects of Tam on the sex-dependent differences in methylation and transcriptome signatures in the brain. Consistent with our experimental design and results, Rotheneichner et al. (Rotheneichner et al. 2017) showed neither the route of application, nor sex of treated mice influenced the response to Tam. Furthermore, Tam does not impact adult neurogenesis or behavioral performance [(Rotheneichner et al. 2017)].
In summary, no long-lasting effects of Tam treatment in the transcriptome or epigenome were found in any of the CNS tissues analyzed in our study. In agreement with recently reported data that Tam alone does not cause long-lasting changes in behavior and neurogenesis, our findings support that Tam can be safely used as a cre-recombinase inducer without introducing significant confounds in transcriptomic and epigenomic studies in the adult CNS.
Electronic supplementary material
(DOCX 516 kb).
Acknowledgements
The authors acknowledge the Laboratory for Molecular Biology and Cytometry Research at OUHSC for the use of the Core Facility which provided Illumina MiSeq Next Generation Sequencing services and the Oklahoma Medical Research Foundation Clinical Genomics Center which provided NextSeq services Computing for this project was performed at the OU Supercomputing Center for Education and Research (OSCER) at the University of Oklahoma (OU).
Support
This work was supported by grants from the National Institutes of Health (NIH) P30AG050911, R56AG059430, R01AG58430, P20GM125528, R01AG0256161, Veterans Affairs I01BX003906, Oklahoma Center for Adult Stem Cell Research (OCASCR) grant through the Oklahoma Tobacco Settlement Endowment Trust, and Presbyterian Health Foundation.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ana J. Chucair-Elliott and Sarah R. Ocanas contributed equally to this work.
References
- Abdelhaleem M. RNA helicases: regulators of differentiation. Clin Biochem. 2005;38:499–503. doi: 10.1016/j.clinbiochem.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Akalin A, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armoskus C, Moreira D, Bollinger K, Jimenez O, Taniguchi S, Tsai HW. Identification of sexually dimorphic genes in the neonatal mouse cortex and hippocampus. Brain Res. 2014;1562:23–38. doi: 10.1016/j.brainres.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashpole NM, Logan S, Yabluchanskiy A, Mitschelen MC, Yan H, Farley JA, Hodges EL, Ungvari Z, Csiszar A, Chen S, Georgescu C, Hubbard GB, Ikeno Y, Sonntag WE. IGF-1 has sexually dimorphic, pleiotropic, and time-dependent effects on healthspan, pathology, and lifespan. Geroscience. 2017;39:129–145. doi: 10.1007/s11357-017-9971-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer C, Pawlak J, Karolczak M. Membrane receptors for oestrogen in the brain. J Neurochem. 2003;87:545–550. doi: 10.1046/j.1471-4159.2003.02042.x. [DOI] [PubMed] [Google Scholar]
- Blair LP, Cao J, Zou MR, Sayegh J, Yan Q. Epigenetic regulation by lysine demethylase 5 (KDM5) enzymes in Cancer. Cancers (Basel) 2011;3:1383–1404. doi: 10.3390/cancers3011383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blurton-Jones M, Tuszynski MH. Reactive astrocytes express estrogen receptors in the injured primate brain. J Comp Neurol. 2001;433:115–123. doi: 10.1002/cne.1129. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Worley PF, Moore MJ, Guzowski JF. The immediate early gene arc/arg3.1: regulation, mechanisms, and function. J Neurosci. 2008;28:11760–11767. doi: 10.1523/JNEUROSCI.3864-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramlett KS, Burris TP. Effects of selective estrogen receptor modulators (SERMs) on coactivator nuclear receptor (NR) box binding to estrogen receptors. Mol Genet Metab. 2002;76:225–233. doi: 10.1016/s1096-7192(02)00043-4. [DOI] [PubMed] [Google Scholar]
- Chucair-Elliott AJ, Gurung HR, Carr MM, Carr DJJ. Colony stimulating Factor-1 receptor expressing cells infiltrating the cornea control corneal nerve degeneration in response to HSV-1 infection. Invest Ophthalmol Vis Sci. 2017;58:4670–4682. doi: 10.1167/iovs.17-22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton JA, Collins FS. Policy: NIH to balance sex in cell and animal studies. Nature. 2014;509:282–283. doi: 10.1038/509282a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Coulon V, Chebli K, Cavelier P, Blanchard JM. A novel mouse c-fos intronic promoter that responds to CREB and AP-1 is developmentally regulated in vivo. PLoS One. 2010;5:e11235. doi: 10.1371/journal.pone.0011235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deepa SS, Bhaskaran S, Espinoza S, Brooks SV, McArdle A, Jackson MJ, van Remmen H, Richardson A. A new mouse model of frailty: the cu/Zn superoxide dismutase knockout mouse. Geroscience. 2017;39:187–198. doi: 10.1007/s11357-017-9975-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, et al. Retinal gene expression responses to aging are sexually divergent. Mol Vis. 2017;23:707–717. [PMC free article] [PubMed] [Google Scholar]
- Feil R, Wagner J, Metzger D, Chambon P. Regulation of cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Fonseca MI, Chu SH, Hernandez MX, Fang MJ, Modarresi L, Selvan P, MacGregor GR, Tenner AJ. Cell-specific deletion of C1qa identifies microglia as the dominant source of C1q in mouse brain. J Neuroinflammation. 2017;14:48. doi: 10.1186/s12974-017-0814-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornander T, Rutqvist LE, Cedermark B, Glas U, Mattsson A, Silfverswärd C, Skoog L, Somell A, Theve T, Wilking N. Adjuvant tamoxifen in early breast cancer: occurrence of new primary cancers. Lancet. 1989;1:117–120. doi: 10.1016/s0140-6736(89)91141-0. [DOI] [PubMed] [Google Scholar]
- Gao X, Castro-Gomez S, Grendel J, Graf S, Süsens U, Binkle L, Mensching D, Isbrandt D, Kuhl D, Ohana O. Arc/Arg3.1 mediates a critical period for spatial learning and hippocampal networks. Proc Natl Acad Sci U S A. 2018;115:12531–12536. doi: 10.1073/pnas.1810125115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ovejero D, Azcoitia I, Doncarlos LL, Melcangi RC, Garcia-Segura LM. Glia-neuron crosstalk in the neuroprotective mechanisms of sex steroid hormones. Brain Res Brain Res Rev. 2005;48:273–286. doi: 10.1016/j.brainresrev.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Gayen S, Maclary E, Hinten M, Kalantry S. Sex-specific silencing of X-linked genes by Xist RNA. Proc Natl Acad Sci U S A. 2016;113:E309–E318. doi: 10.1073/pnas.1515971113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadad N, Masser DR, Logan S, Wronowski B, Mangold CA, Clark N, Otalora L, Unnikrishnan A, Ford MM, Giles CB, Wren JD, Richardson A, Sonntag WE, Stanford DR, Freeman W. Absence of genomic hypomethylation or regulation of cytosine-modifying enzymes with aging in male and female mice. Epigenetics Chromatin. 2016;9:30. doi: 10.1186/s13072-016-0080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes B, Dowsett M. Clinical pharmacology of selective estrogen receptor modulators. Drugs Aging. 1999;14:323–336. doi: 10.2165/00002512-199914050-00001. [DOI] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suárez-Fariñas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. Epigenetics: a historical overview. Epigenetics. 2006;1:76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
- Horvath S, Zhang Y, Langfelder P, Kahn RS, Boks MPM, van Eijk K, van den Berg LH, Ophoff RA. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012;13:R97. doi: 10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperio CG, McFalls AJ, Colechio EM, Masser DR, Vrana KE, Grigson PS, Freeman WM. Assessment of individual differences in the rat nucleus accumbens transcriptome following taste-heroin extended access. Brain Res Bull. 2016;123:71–80. doi: 10.1016/j.brainresbull.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn HM, Kasakow CV, Helfer A, Michely J, Verkhratsky A, Maurer HH, Scheller A, Kirchhoff F. Refined protocols of tamoxifen injection for inducible DNA recombination in mouse astroglia. Sci Rep. 2018;8:5913. doi: 10.1038/s41598-018-24085-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson S. Histopathology and histomorphometry of the urogenital tract in 15-month old male and female rats treated neonatally with SERMs and estrogens. Exp Toxicol Pathol. 2006;58:1–12. doi: 10.1016/j.etp.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Keshet Y, Seger R. The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods Mol Biol. 2010;661:3–38. doi: 10.1007/978-1-60761-795-2_1. [DOI] [PubMed] [Google Scholar]
- Khalaj AJ, Yoon J, Nakai J, Winchester Z, Moore SM, Yoo T, Martinez-Torres L, Kumar S, Itoh N, Tiwari-Woodruff SK. Estrogen receptor (ER) beta expression in oligodendrocytes is required for attenuation of clinical disease by an ERbeta ligand. Proc Natl Acad Sci U S A. 2013;110:19125–19130. doi: 10.1073/pnas.1311763110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopsida E, Stergiakouli E, Lynn PM, Wilkinson LS, Davies W. The role of the Y chromosome in brain function. Open Neuroendocrinol J. 2009;2:20–30. doi: 10.2174/1876528900902010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper GG, et al. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Croce LD, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, Yu M, Tonti-Filippini J, Heyn H, Hu S, Wu JC, Rao A, Esteller M, He C, Haghighi FG, Sejnowski TJ, Behrens MM, Ecker JR. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YP, Zeng M, Hu XY, Xu H, Swaab DF, Ravid R, Zhou JN. Estrogen receptor alpha-immunoreactive astrocytes are increased in the hippocampus in Alzheimer's disease. Exp Neurol. 2003;183:482–488. doi: 10.1016/s0014-4886(03)00205-x. [DOI] [PubMed] [Google Scholar]
- Maegawa S, Hinkal G, Kim HS, Shen L, Zhang L, Zhang J, Zhang N, Liang S, Donehower LA, Issa JPJ. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malonia SK, et al. Gene regulation by SMAR1: role in cellular homeostasis and cancer. Biochim Biophys Acta. 2011;1815:1–12. doi: 10.1016/j.bbcan.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Mangold CA, Masser DR, Stanford DR, Bixler GV, Pisupati A, Giles CB, Wren JD, Ford MM, Sonntag WE, Freeman WM. CNS-wide sexually dimorphic induction of the major histocompatibility complex 1 pathway with aging. J Gerontol A Biol Sci Med Sci. 2017;72:16–29. doi: 10.1093/gerona/glv232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold CA, Wronowski B, du M, Masser DR, Hadad N, Bixler GV, Brucklacher RM, Ford MM, Sonntag WE, Freeman WM. Sexually divergent induction of microglial-associated neuroinflammation with hippocampal aging. J Neuroinflammation. 2017;14:141. doi: 10.1186/s12974-017-0920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin R, Ramirez CM, Gonzalez M, Alonso R, Diaz M. Alternative estrogen receptors homologous to classical receptor alpha in murine neural tissues. Neurosci Lett. 2006;395:7–11. doi: 10.1016/j.neulet.2005.10.047. [DOI] [PubMed] [Google Scholar]
- Masser DR, Bixler GV, Brucklacher RM, Yan H, Giles CB, Wren JD, Sonntag WE, Freeman WM. Hippocampal subregions exhibit both distinct and shared transcriptomic responses to aging and nonneurodegenerative cognitive decline. J Gerontol A Biol Sci Med Sci. 2014;69:1311–1324. doi: 10.1093/gerona/glu091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masser DR, Hadad N, Porter HL, Mangold CA, Unnikrishnan A, Ford MM, Giles CB, Georgescu C, Dozmorov MG, Wren JD, Richardson A, Stanford DR, Freeman WM. Sexually divergent DNA methylation patterns with hippocampal aging. Aging Cell. 2017;16:1342–1352. doi: 10.1111/acel.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masser DR, Hadad N, Porter H, Stout MB, Unnikrishnan A, Stanford DR, Freeman WM. Analysis of DNA modifications in aging research. Geroscience. 2018;40:11–29. doi: 10.1007/s11357-018-0005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximov PY, Lee TM, Jordan VC. The discovery and development of selective estrogen receptor modulators (SERMs) for clinical practice. Curr Clin Pharmacol. 2013;8:135–155. doi: 10.2174/1574884711308020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazeyrat S, Saut N, Sargent CA, Grimmond S, Longepied G, Ehrmann IE, Ellis PS, Greenfield A, Affara NA, Mitchell MJ. The mouse Y chromosome interval necessary for spermatogonial proliferation is gene dense with syntenic homology to the human AZFa region. Hum Mol Genet. 1998;7:1713–1724. doi: 10.1093/hmg/7.11.1713. [DOI] [PubMed] [Google Scholar]
- Mazeyrat S, Saut N, Grigoriev V, Mahadevaiah SK, Ojarikre OA, Rattigan Á, Bishop C, Eicher EM, Mitchell MJ, Burgoyne PS. A Y-encoded subunit of the translation initiation factor Eif2 is essential for mouse spermatogenesis. Nat Genet. 2001;29:49–53. doi: 10.1038/ng717. [DOI] [PubMed] [Google Scholar]
- McCarthy MM, Arnold AP, Ball GF, Blaustein JD, De Vries GJ. Sex differences in the brain: the not so inconvenient truth. J Neurosci. 2012;32:2241–2247. doi: 10.1523/JNEUROSCI.5372-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami H, Kim JD, Tabara S, Lu W, Kwon C, Nakashima M, Fukamizu A. KDM5D-mediated H3K4 demethylation is required for sexually dimorphic gene expression in mouse embryonic fibroblasts. J Biochem. 2019;165:335–342. doi: 10.1093/jb/mvy106. [DOI] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, Eddy SR, Ecker JR, Nathans J. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron. 2015;86:1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Otal A, Retana-Marquez S, Ferreira-Nuno A, Velazquez-Moctezuma J. Testosterone levels and histological features of reproductive glands in adult male rats treated neonatally with tamoxifen. Neuro Endocrinol Lett. 2005;26:729–732. [PubMed] [Google Scholar]
- Newhouse P, Dumas J. Estrogen-cholinergic interactions: implications for cognitive aging. Horm Behav. 2015;74:173–185. doi: 10.1016/j.yhbeh.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent BM, Wright CL, Shetty AC, Hodes GE, Lenz KM, Mahurkar A, Russo SJ, Devine SE, McCarthy MM. Brain feminization requires active repression of masculinization via DNA methylation. Nat Neurosci. 2015;18:690–697. doi: 10.1038/nn.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YM, Chun H, Shin JI, Lee CJ. Astrocyte specificity and coverage of hGFAP-CreERT2 [Tg(GFAP-Cre/ERT2)13Kdmc] mouse line in various brain regions. Exp Neurobiol. 2018;27:508–525. doi: 10.5607/en.2018.27.6.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SH, O’Hara L, Atanassova N, Smith SE, Curley MK, Rebourcet D, Darbey AL, Gannon AL, Sharpe RM, Smith LB. Low-dose tamoxifen treatment in juvenile males has long-term adverse effects on the reproductive system: implications for inducible transgenics. Sci Rep. 2017;7:8991. doi: 10.1038/s41598-017-09016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Lange IG, Daxenberger A, Meyer HH. Tissue-specific expression pattern of estrogen receptors (ER): quantification of ER alpha and ER beta mRNA with real-time RT-PCR. APMIS. 2001;109:345–355. doi: 10.1034/j.1600-0463.2001.090503.x. [DOI] [PubMed] [Google Scholar]
- Podlutsky A, Valcarcel-Ares MN, Yancey K, Podlutskaya V, Nagykaldi E, Gautam T, Miller RA, Sonntag WE, Csiszar A, Ungvari Z. The GH/IGF-1 axis in a critical period early in life determines cellular DNA repair capacity by altering transcriptional regulation of DNA repair-related genes: implications for the developmental origins of cancer. Geroscience. 2017;39:147–160. doi: 10.1007/s11357-017-9966-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, Whittaker P, McCann OT, Finer S, Valdes AM, Leslie RD, Deloukas P, Spector TD. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20:434–439. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs BL, Hartmann LC. Selective estrogen-receptor modulators -- mechanisms of action and application to clinical practice. N Engl J Med. 2003;348:618–629. doi: 10.1056/NEJMra022219. [DOI] [PubMed] [Google Scholar]
- Roh HC, Tsai LTY, Lyubetskaya A, Tenen D, Kumari M, Rosen ED. Simultaneous transcriptional and Epigenomic profiling from specific cell types within heterogeneous tissues in vivo. Cell Rep. 2017;18:1048–1061. doi: 10.1016/j.celrep.2016.12.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotheneichner P, et al. Tamoxifen activation of Cre-recombinase has no persisting effects on adult neurogenesis or learning and anxiety. Front Neurosci. 2017;11:27. doi: 10.3389/fnins.2017.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma S, Tokuhara D, Hattori H, Matsuoka O, Yamano T. Expression of estrogen receptor alpha and beta in reactive astrocytes at the male rat hippocampus after status epilepticus. Neuropathology. 2009;29:55–62. doi: 10.1111/j.1440-1789.2008.00946.x. [DOI] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell DM, Turner JMA. Sex chromosome effects on male-female differences in mammals. Curr Biol. 2018;28:R1313–R1324. doi: 10.1016/j.cub.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Lu TY, Chai H, Xu J, Huang BS, Golshani P, Coppola G, Khakh BS. New transgenic mouse lines for selectively targeting astrocytes and studying calcium signals in astrocyte processes in situ and in vivo. Neuron. 2016;92:1181–1195. doi: 10.1016/j.neuron.2016.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CY, Lin TC, Lin YF, Chen MH, Lee CH, Wang HY, Lee YC, Liu YP, Chen CL, Hsiao M. DDX3 as a strongest prognosis marker and its downregulation promotes metastasis in colorectal cancer. Oncotarget. 2015;6:18602–18612. doi: 10.18632/oncotarget.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M, Dökel S, Amstislavskiy V, Wuttig D, Sültmann H, Lehrach H, Yaspo ML. A simple strand-specific RNA-Seq library preparation protocol combining the Illumina TruSeq RNA and the dUTP methods. Biochem Biophys Res Commun. 2012;422:643–646. doi: 10.1016/j.bbrc.2012.05.043. [DOI] [PubMed] [Google Scholar]
- Sun M, Song L, Li Y, Zhou T, Jope RS. Identification of an antiapoptotic protein complex at death receptors. Cell Death Differ. 2008;15:1887–1900. doi: 10.1038/cdd.2008.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suuronen T, Nuutinen T, Huuskonen J, Ojala J, Thornell A, Salminen A. Anti-inflammatory effect of selective estrogen receptor modulators (SERMs) in microglial cells. Inflamm Res. 2005;54:194–203. doi: 10.1007/s00011-005-1343-z. [DOI] [PubMed] [Google Scholar]
- Taefehshokr S, Key YA, Khakpour M, Dadebighlu P, Oveisi A. Early growth response 2 and Egr3 are unique regulators in immune system. Cent Eur J Immunol. 2017;42:205–209. doi: 10.5114/ceji.2017.69363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Tonchev AB, Koike K, Murakami K, Yamada K, Yamashima T, Inoue M. Expression of estrogen receptor-beta in the postischemic monkey hippocampus. Neurosci Lett. 2004;369:9–13. doi: 10.1016/j.neulet.2004.07.042. [DOI] [PubMed] [Google Scholar]
- Tee MK, et al. Estradiol and selective estrogen receptor modulators differentially regulate target genes with estrogen receptors alpha and beta. Mol Biol Cell. 2004;15:1262–1272. doi: 10.1091/mbc.E03-06-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unnikrishnan A, Jackson J, Matyi SA, Hadad N, Wronowski B, Georgescu C, Garrett KP, Wren JD, Freeman WM, Richardson A. Role of DNA methylation in the dietary restriction mediated cellular memory. Geroscience. 2017;39:331–345. doi: 10.1007/s11357-017-9976-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen FE, van den Belt-Dusebout AW, van Leeuwen FE, Benraadt J, Diepenhorst FW, van Tinteren H, Coebergh JWW, Kiemeney LALM, Gimbrère CHF, Otter R, Schouten LJ, Damhuis RAM, Benraadt J, Bontenbal M. Risk of endometrial cancer after tamoxifen treatment of breast cancer. Lancet. 1994;343:448–452. doi: 10.1016/s0140-6736(94)92692-1. [DOI] [PubMed] [Google Scholar]
- Wardell SE, Nelson ER, McDonnell DP. From empirical to mechanism-based discovery of clinically useful selective estrogen receptor modulators (SERMs) Steroids. 2014;90:30–38. doi: 10.1016/j.steroids.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb P, Nguyen P, Kushner PJ. Differential SERM effects on corepressor binding dictate ERalpha activity in vivo. J Biol Chem. 2003;278:6912–6920. doi: 10.1074/jbc.M208501200. [DOI] [PubMed] [Google Scholar]
- Wen L, Li X, Yan L, Tan Y, Li R, Zhao Y, Wang Y, Xie J, Zhang Y, Song C, Yu M, Liu X, Zhu P, Li X, Hou Y, Guo H, Wu X, He C, Li R, Tang F, Qiao J. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol. 2014;15:R49. doi: 10.1186/gb-2014-15-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Watkins R, Arnold AP. Sexually dimorphic expression of the X-linked gene Eif2s3x mRNA but not protein in mouse brain. Gene Expr Patterns. 2006;6:146–155. doi: 10.1016/j.modgep.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Xu J, Deng X, Watkins R, Disteche CM. Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J Neurosci. 2008;28:4521–4527. doi: 10.1523/JNEUROSCI.5382-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 516 kb).