Abstract
The basement membrane (BM) provides a physical barrier to invasion in epithelial tumors, and alterations in the molecular makeup and structural integrity of the BM have been implicated in cancer progression. Invadopodia are the invasive protrusions that enable cancer cells to breach the nanoporous basement membrane, through matrix degradation and generation of force. However, the impact of covalent cross-linking on invadopodia extension into the BM remains unclear. Here, we examine the impact of covalent cross-linking of extracellular matrix on invasive protrusions using biomaterials that present ligands relevant to the basement membrane and provide a nanoporous, confining microenvironment. We find that increased covalent cross-linking of reconstituted basement membrane (rBM) matrix diminishes matrix mechanical plasticity, or the ability of the matrix to permanently retain deformation due to force. Covalently cross-linked rBM matrices, and rBM-alginate interpenetrating networks (IPNs) with covalent cross-links and low plasticity, restrict cell spreading and protrusivity. The reduced spreading and reduced protrusivity in response to low mechanical plasticity occurred independent of proteases. Mechanistically, our computational model reveals that the reduction in mechanical plasticity due to covalent cross-linking is sufficient to mechanically prevent cell protrusions from extending, independent of the impact of covalent cross-linking or matrix mechanical plasticity on cell signaling pathways. These findings highlight the biophysical role of covalent cross-linking in regulating basement membrane plasticity, as well as cancer cell invasion of this confining tissue layer.
Keywords: Basement Membrane, Cross-linking, Invadopodia, Invasion, Breast Cancer, Biomaterials
Graphical Abstract
1. Introduction
Cancer progression is accompanied by dramatic changes to the extracellular matrix (ECM) of the tumor microenvironment, including variations in cross-linking, enzymatic modification, and proteolytic degradation of the ECM [1–3]. While the amount of covalent cross-linking mediated by lysyl oxidase (LOX) and tissue transglutaminase (tTg) family proteins is known to change at multiple stages of cancer, the evidence is mixed as to whether these changes ultimately promote or suppress cancer [2,4]. Several studies find that covalent cross-linking suppresses invasion, migration, and metastasis of cancer cells [4–7]. However, other studies have shown that covalent cross-linking promotes malignant cell phenotypes [8–11], invasion, and metastasis [10,12], and have implicated its role in increasing stiffness and intracellular tension [13–15].
There are a number of considerations regarding the complicated influence of covalent cross-linking on cancer progression. Aside from impacting matrix stiffness, covalent cross-linking can impact polymer network architecture and fibrillarity [16] as well as mechanical plasticity [17–19]. Matrix mechanical plasticity, which is distinct from matrix stiffness, refers to the ability of a material to permanently retain deformation through microstructural rearrangements in response to mechanical force [17] and is known to regulate cancer cell invasion [20]. Furthermore, covalent cross-linking not only serves to alter biophysical properties of the ECM [9]; LOX and tTg are also potent biochemical signaling molecules [11,12]. Moreover, many prior studies have examined the role of covalent cross-linking in collagen-1 matrices. While elevated covalent cross-linking of collagen-1 in the tumor stroma is a major driver of enhanced tumor stiffness and malignancy [9,15], cancer cells must first breach basement membranes, which surround carcinoma cells in epithelial tumors, before they reach the stromal tissue [21]. Basement membranes are thin, yet confining, nanoporous meshes that owe their biomechanical integrity predominantly to covalently cross-linked collagen IV networks and collagen IV-nidogen-laminin bridges [22,23]. Although this tens to hundreds of nanometers-thick tissue layer remains challenging to study, proteomic evidence suggests that, like the stromal tissue, basement membranes undergo changes in molecular makeup, proteolytic activity, and cross-linking during cancer progression [24,25]. These changes may compromise the structural integrity of basement membranes. Therefore, an investigation into the role of covalent cross-linking on cells in basement membrane-like matrices is highly relevant to elucidating the biophysics of initial invasion out of primary tumors.
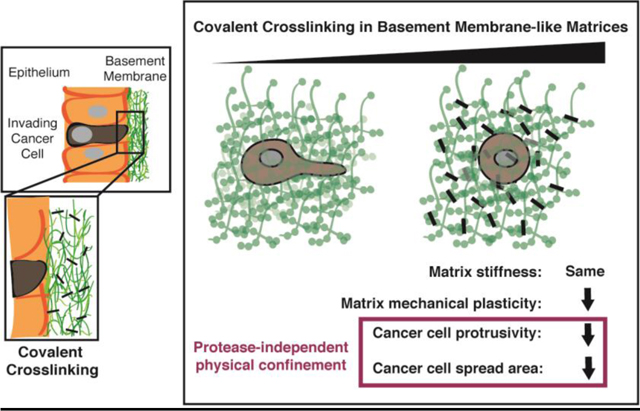
Proteases and, more recently, matrix mechanical plasticity have both been implicated in facilitating basement membrane invasion. Invadopodia are the actin-rich cellular protrusions that enable cancer cells to break through confining basement membranes. The established view has been that invadopodia facilitate this invasion primarily by using proteases to biochemically degrade BM and thereby open up a path for invasion [21,25–29]. However, protease inhibitors have not been successful in preventing invasion in clinical trials [30], and it has been suspected that invadopodia can breach basement membranes without requiring proteases by exploiting local structural defects or through physical disassembly of the matrix [22]. Recently, we showed that invadopodia can initiate protease-independent, force-dependent 3D migration through confining, basement membrane-mimicking matrix, so long as the matrix exhibits sufficient mechanical plasticity [20] (Fig. 1a). This migration mode can occur when highly plastic extracellular matrix (ECM) permanently retains deformations caused by the repeated pushing and pulling of invadopodia, leading to the formation of permanent channels through which cells can then migrate [20] (Fig 1a). By contrast, low mechanical plasticity ECM can recover from deformation, and in these matrices, cells are unable to migrate using this force-dependent migration mode [20].
Fig. 1. Protease-dependent and -independent cancer cell invasion of covalently cross-linked basement membranes.
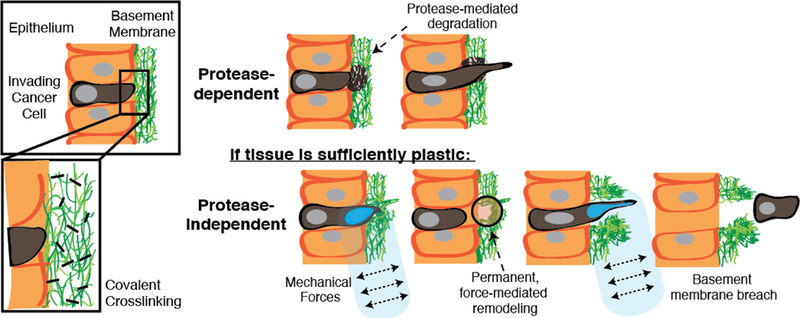
Schematic depicting basement membrane invasion, including (top) protease-dependent modes and (bottom) a protease-independent mode, recently shown, that is mediated by extracellular matrix mechanical plasticity combined with cell-generated forces [20]. The role of covalent cross-linking in mediating the second mode of invasion remains unclear.
The impact of ECM cross-linking on invadopodia has been a topic of recent study. Crosslinking of 2D gelatin substrates regulates the oscillatory speed of invadopodia formation and retraction, as well as the dynamic mode switching that allows cells to transition between protrusion extension and migration states [31,32]. Outcomes were mixed, however, from studies using 3D collagen substrates, showing that ECM cross-linking had either no effect on invadopodia dynamics [31] or a biphasic relationship with invadopodia dynamics [32]. Thus, it is not yet understood how covalent cross-linking of basement membrane impacts the extension of invasive protrusions in 3D, particularly with relevance to basement membrane invasion.
Here, we investigate the biophysical effect of covalent cross-linking on protease-independent breast cancer cell extension of invasive protrusions. Covalent cross-linking is varied in reconstituted Basement Membrane (rBM) hydrogels cross-linked with tissue transglutaminase (tTg), as well as in rBM-alginate hydrogels that exhibit physiologically relevant stiffness and biochemically inert covalent cross-linking. Both material systems are used for 3D culture of highly invasive, triple negative breast cancer cells. We find that in these materials systems, covalent cross-linking reduces matrix mechanical plasticity, restricts cell spreading, increases cell circularity, and inhibits the extension of invadopodia. Our computational model confirms that covalent cross-linking of extracellular matrices lowers the mechanical plasticity of the matrix and physically restricts the extension of invasive protrusions.
2. Results
2.1. Covalent cross-linking reduces mechanical plasticity in reconstituted Basement Membrane matrices
We first investigated the impact of matrix covalent cross-linking on cancer cell invasiveness in 3D culture using reconstituted Basement Membrane (rBM) matrices. rBM matrices are derived from the Engelbreth-Holm-Swarm (EHS) mouse tumor and contain biological signaling proteins found in physiological basement membrane matrix, including laminin, collagen IV, and nidogen [33]. Like physiological basement membrane matrix, commercially available rBM can be covalently cross-linked with tissue transglutaminase (tTg) for in vitro studies (Fig. 2a). Mechanical testing of acellular matrices showed that pure rBM matrices, and those cross-linked with different doses of tTg, exhibited similar Young’s Moduli (~ 100 Pa), a property related to stiffness (Fig. 2b, Supplementary Figure 1a–c). The loss tangent of the rBM matrix, an indicator of matrix viscosity or ability to flow, increased with the maximum covalent cross-linking tested, 500 μg/mL (Supplementary Figure 1d). In order to quantify the mechanical plasticity of the different rBM matrices, or the degree to which each hydrogel retained an applied deformation after a defined recovery period, we performed creep and recovery tests on pure rBM and tTg-cross-linked rBM [17] (Fig. 2c,d). We observed that the maximum concentration of tTg tested decreased the mechanical plasticity of rBM by about an order of magnitude (~ 80% vs. ~ 10%), and that the mechanical plasticity of rBM hydrogels exponentially decayed with increasing tTg concentration (Fig. 2e,f). These findings show that transglutaminase-mediated covalent cross-linking can significantly decrease the mechanical plasticity of rBM matrices without significantly impacting their stiffness.
Fig. 2. Covalent cross-linking of reconstituted Basement Membrane (rBM) matrices decreases their mechanical plasticity.
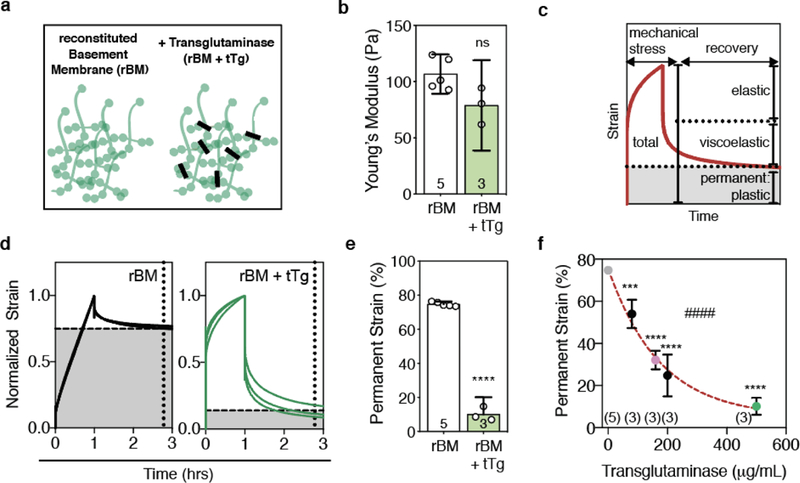
a, Schematic of rBM matrix without or with covalent cross-linking by tissue Transglutaminase (tTg). b, Young’s moduli of 8 mg/mL rBM alone or with 500 μg/mL tTg, quantified at 1% strain and 0.15 Hz. c, Cartoon depicting the elastic, viscoelastic, and plastic (permanent) portions of a material’s strain response during a creep and recovery test, during which time a mechanical stress is applied and then released. d, Normalized strain throughout creep and recovery tests on rBM and rBM with 500 μg/mL tTg. e, Permanent strain remaining in rBM or rBM with 500 μg/mL tTg, as indicated by the intersections of the dashed lines in d. Difference is significantly different (**** P < 0.0001, t-test). f, Permanent strain remaining after creep and recovery tests for rBM matrices cross-linked with the amount of tTg as indicated. Statistically significant differences are indicated (***P < 0.001, ****P < 0.0001, ANOVA; #### P < 0.0001, Spearman’s Rank Correlation). Red line indicates exponential decay fit line (R2 = 0.95). In b, e, and f, bars or markers indicate means and error bars indicate 95% confidence intervals.
2.2. Covalent cross-linking restricts cell spread area in reconstituted basement membrane matrices
We next examined the role of covalent cross-linking on highly invasive breast cancer cells (MDA-MB-231) in reconstituted basement membrane (rBM) matrices. Cells were encapsulated in rBM alone or in rBM covalently cross-linked with transglutaminase (rBM+tTg) and stimulated with EGF to activate invasive signaling pathways (Fig. 3a). Differences in cellular morphology were quantified after 24 hours. Cells in pure rBM matrices were spread with elongated protrusions, while cells in rBM+tTg exhibited significantly increased circularity and significantly reduced spread area (Fig. 3b–d, Supplementary Figure 2). Notably, the addition of broad spectrum matrix metalloprotease inhibitor GM6001 at a dose previously shown to inhibit invadopodia-mediated matrix degradation [20], resulted in cell circularity and spread areas that were similar to those in the vehicle alone conditions (Fig. 3b–d, Supplementary Figure 2). Furthermore, the addition of a strong protease inhibitor cocktail previously published [20], which contains not only a broad spectrum matrix metalloprotease inhibitor (20 μM Marimastat) but also inhibitors for aspartyl, serine, and cystine proteases (20 μM Pepstatin A, 20 μM E-64, 0.7 μM Aprotinin, 2 μM Leupeptin) also did not impact cell circularity and spread areas (Fig. 3c,d). These findings indicate that protease-mediated degradation was not responsible for the morphological differences observed, and show that covalently cross-linked, low-plasticity matrix physically confines cancer cells and restricts protrusive activity.
Fig. 3. Covalent cross-linking mediated by tissue transglutaminase physically restricts cancer cell spreading in reconstituted basement membrane matrices.
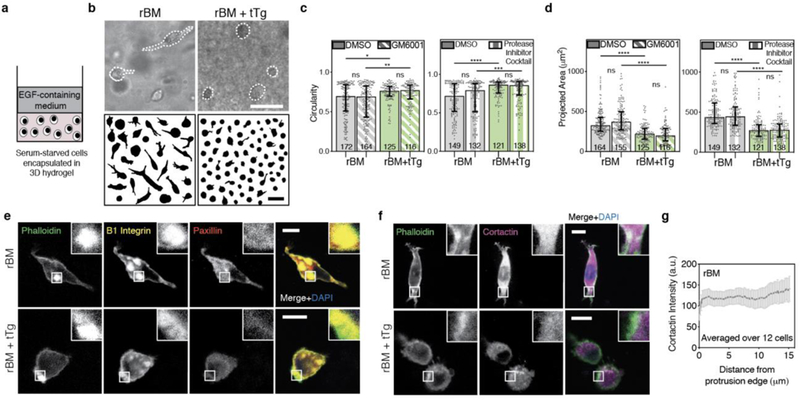
a, Schematic depicting the 3D encapsulation of MDA-MB-231 cancer cells in 8 mg/mL reconstituted basement membrane (rBM) hydrogels, without or with 500 μg/mL tissue Transglutaminase (tTg). b, After ~ 24 hours in 3D culture, cells were imaged using bright field microscopy and cell outlines were traced. Example MDA-MB-231 cells and cell outlines shown. Scale bars are 50 μm. c, MDA-MB-231 cell circularity and d, 2D-projected spread area in the different rBM matrices, in the presence of vehicle alone control, broad-spectrum matrix metalloprotease inhibitor (10 μM GM6001) or a protease inhibitor cocktail (20 μM Marimastat, 20 μM Pepstatin A, 20 μM E-64, 0.7 μM Aprotinin, and 2 μM Leupeptin). In c and d, bars indicate medians and error bars indicate interquartile ranges. Differences in morphological characteristics indicated are statistically significant (* P < 0.05, ** P < 0.01, **** P < 0.0001, Kruskal-Wallis Test). Data shown are from one representative biological replicate experiment, and additional biological replicates are shown in Supplementary Figure 2. e,f, Localization of indicated proteins on stained cryosections of MDA-MB-231 cells encapsulated for ~ 24 hours, imaged using confocal immunofluorescence. Main panel scale bar is 10 μm, and inset is 3x zoom. g, Cortactin intensity, measured from the tip of cellular protrusions toward the nucleus, averaged over protrusions from 12 cells in rBM alone matrices. Line plots mean intensity and error bars indicate 95% confidence interval.
To explore these phenotypic differences in cell spreading and circularity further, we examined the structure of adhesions and localization of the actin polymerization-related protein cortactin. Large, β1-integrin rich, adhesive plaques that were reinforced by actin and that excluded paxillin could be observed in both rBM and rBM + tTg conditions (Fig. 3e). However, highly spread cells in rBM displayed cortactin uniformly throughout the cell body, with some actin-cortactin colocalization at the neck of protrusions (Fig. 3f,g). By contrast, the confined, circular cells in rBM+tTg displayed some actin and cortactin puncta, but they did not persistently colocalize.
2.3. Design of matrices that exhibit bio-orthogonal covalent cross-linking and physiologically relevant stiffness
Next, we built on these studies in reconstituted basement membrane (rBM) matrices using engineered matrices that provided control over stiffness and employed bio-orthogonal crosslinking. Our goal was twofold. First, we aimed to design rBM-containing hydrogels that mimicked the elevated stiffness of breast tumor tissue, with a Young’s Modulus ~ 2 kPa [34]. Secondly, we aimed to investigate the role of covalent cross-linking in a bio-orthogonal manner, without varying molecules that could biochemically activate invasion-related signaling. For these studies, we utilized interpenetrating networks (IPNs) of reconstituted basement membrane (rBM) and alginate. Alginate, a polymer derived from seaweed which cannot be degraded by mammalian enzymes, is a biologically inert, mechanically tunable component of the IPNs [14,35,36]. Alginate can be cross-linked ionically through the addition of calcium ions [37]. Additionally, by modifying some alginate polymers with tetrazine and other polymers with norbornene functional groups, a combination of the two can be covalently cross-linked, in a biorthogonal and cytocompatible manner, using the click reaction between the functional groups [38]. Here, we combined this click chemistry-mediated covalent cross-linking with our recently developed hydrogels of tunable mechanical plasticity [20]. The rBM and alginate were mixed and cross-linked to form IPN hydrogels with a final concentration of 4.4 mg/mL rBM and 10 mg/mL alginate, with the IPN containing a combination of norbornene-and tetrazine-conjugated alginate (Fig. 4a, Supplementary Table 1). Like the high plasticity (HP) IPNs previously shown [20], the new low plasticity, covalently cross-linked “LP-CC” IPNs exhibited a Young’s modulus of ~ 2 kPa, which is comparable to the stiffness of malignant breast tumor tissue [34] (Fig. 4b,c). Notably, the covalently cross-linked IPN demonstrated a lower viscosity than the HP IPN, as indicated by its diminished loss modulus across a range of frequencies tested, and significantly lower loss tangent (Fig. 4b,d). We quantified the mechanical plasticity of the covalently cross-linked IPN compared to the HP IPN using creep and recovery tests. We found that the covalently cross-linked IPNs retained significantly less permanent deformation (< 10%) than the HP IPNs (~ 30%) (Fig. 4e,f). These data show that low plasticity IPN hydrogels can be formed by incorporating covalent cross-linking into our rBM and alginate IPN system. Using these materials, the impact of covalent cross-linking can be investigated in hydrogels mimicking tumor stiffness and without using bioactive cross-linking molecules.
Fig. 4. rBM-alginate IPNs with bio-orthogonal covalent cross-linking exhibit low plasticity and physiologically relevant stiffness.
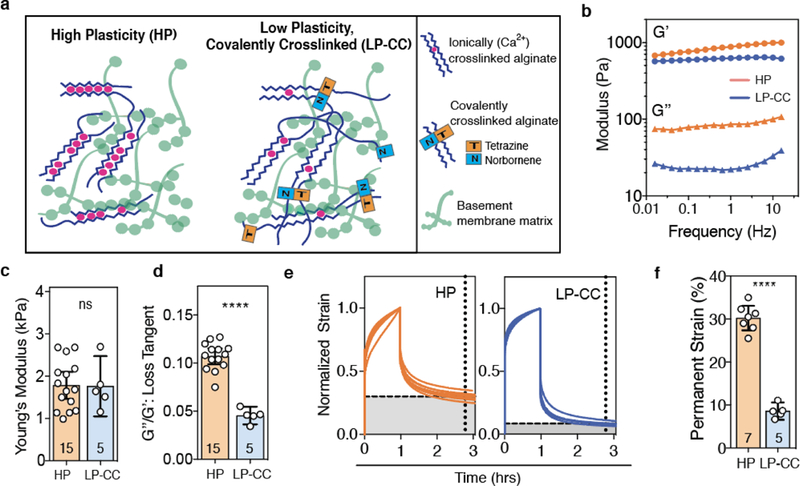
a, Approach to forming IPNs of high and low mechanical plasticity using alginate (blue) and reconstituted basement membrane (rBM) matrix (green), without or with sparse covalent cross-links in addition to ionic cross-linking (red). High plasticity (HP) IPNs were formed with low molecular weight alginate, and low plasticity, covalently cross-linked (LP-CC) IPNs were formed using high molecular weight alginate. b, Storage (G’) and loss (G”) moduli by frequency for HP and LP-CC IPNs. c, Young’s modulus and d, loss tangent of these formulations, measured at 1% strain and 0.15 Hz. e, Normalized strain throughout creep and recovery tests on HP and LP-CC IPNs. f, Permanent strain remaining in the IPNs after a recovery time, indicated by the intersecting dashed lines in e. In c, d, and f, bars indicate means and error bars indicate 95% confidence intervals. Differences indicated are significantly different (**** P < 0.0001, Student’s t-test). HP IPN mechanical testing data shown in c-f reprinted with permission from ref. [20].
2.4. Interpenetrating network (IPN) hydrogels that exhibit bio-orthogonal covalent cross-linking and low mechanical plasticity restrict cellular protrusivity
We next used these interpenetrating network (IPN) hydrogels as substrates for 3D culture to determine how covalent cross-linking impacted cancer cell phenotype in matrices exhibiting stiffness comparable to tumors. MDA-MB-231 cells were encapsulated in the high plasticity (HP) and low plasticity, covalently cross-linked (LP-CC) IPNs and stimulated with EGF to activate invasive signaling pathways. After 24 hours in culture, clear differences in cellular protrusivity emerged: while cells in HP IPNs extended protrusions, those in LP-CC IPNs adopted highly circular morphologies (Fig. 5a,b, Supplementary Fig. 3a). While spread area was somewhat reduced in LP-CC compared to HP IPNs, the reduction was not statistically significant (Fig. 5c, Supplementary Figure 3b). By contrast, cell circularity was significantly greater in LP-CC than in the HP IPNs, in both the vehicle alone and protease inhibitor conditions (Fig. 5b, Supplementary Fig. 3a). We note that cells in HP and LP IPNs occupied spread areas that were both smaller than those in pure rBM, yet cells in the HP IPN exhibited the lowest median circularity, and those in the LP-CC IPN exhibited the highest median circularity, of cells in any biomaterial tested here (Supplementary Figure 4a,b). Analysis of prior data on invasion and migration in a similar IPN materials system [20] shows that lower circularity trends with extension of oscillatory, invasive protrusions and migration, while no clear trend is observed between spread area and these outcomes (Supplementary Figure 5). Overall, these findings reinforce the notion that stiff, low plasticity matrix with covalent cross-linking physically confines cells and restricts the extension of invasive protrusions.
Figure 5. Low plasticity IPN matrix with covalent cross-links physically restricts cancer cell protrusivity, independent of proteases.
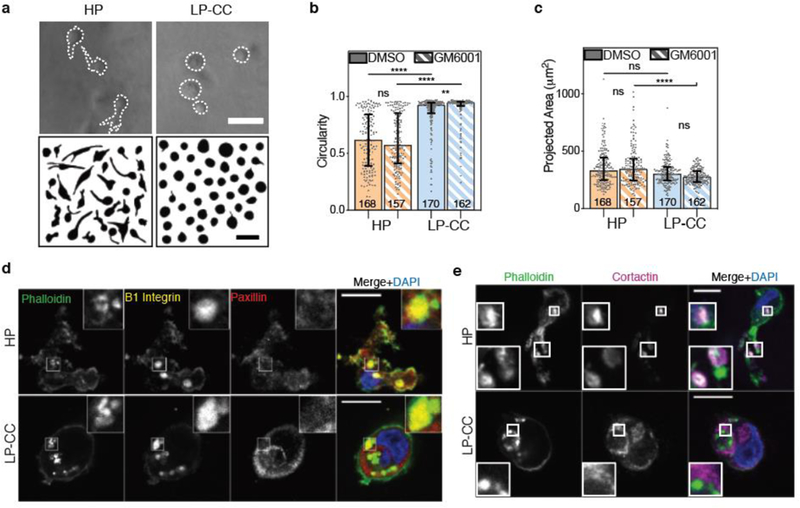
a, After ~ 24 hours in 3D culture, cells were imaged using bright field microscopy and cell outlines were traced. Example MDA-MB-231 cells and cell outlines shown. Scale bars are 50 μm. b, MDA-MB-231 cell circularity and c, 2D-projected spread area in the presence of broad-spectrum protease inhibitor (10 μM GM6001) or vehicle alone control. In b and c, bars indicate medians and error bars indicate interquartile ranges. Differences in morphological characteristics indicated are statistically significant (**** P < 0.0001, ** P < 0.01, Kruskal-Wallis Test). Data shown is from one representative biological replicate experiment, and an additional biological replicate is shown in Supplementary Figure 3. d and e, Localization of indicated proteins on stained cryosections of MDA-MB-231 cells encapsulated for ~ 24 hours, imaged using confocal immunofluorescence. Main panel scale bar is 10 μm and inset is 3x zoom.
We next examined the localization of adhesion and actin polymerization-related proteins in these LP-CC IPNs compared to the HP IPNs. Notably, MDA-MB-231 cells in both HP and LP-CC IPNs assembled β1-integrin rich plaques that were reinforced by actin and excluded paxillin (Fig. 5d). However, only cells in HP IPNs displayed robust cortactin puncta colocalized with actin puncta (Fig. 5e). By contrast, cells in LP-CC IPNs exhibited spatial decoupling of actin and cortactin. These data suggest that low plasticity matrices with covalent cross-links interfere with the localization of actin polymerization-related proteins.
2.5. A computational model reveals that reduced matrix mechanical plasticity is sufficient to block invasive protrusions
Finally, we used a computational model to determine whether cell behaviors in covalently crosslinked matrices were mediated by an active cellular response, such as via biochemical signaling pathways involving mechanotransduction of matrix plasticity [39], or whether these behaviors could be explained as a consequence of mechanical phenomena alone. For these modeling studies, a cell membrane was simplified into a series of interconnected elements, and the extracellular matrix consisted of randomly oriented polymers that were cross-linked together (Fig. 6a, Supplementary Table 2). In this model, cross-linkers unbind from fibers following Bell’s law (Eq. 1), in which an unbinding rate exponentially increases from its base rate (or zero-force unbinding rate constant, k0,u) as a force applied to the cross-linkers increases. Dependence of the unbinding rate on forces is stronger if force sensitivity (λu) is higher. We emulated effects of enhanced covalent cross-linking using two different approaches. First, k0,u was lowered because increased covalent cross-linking is expected to reduce the average rate of spontaneous cross-linker unbinding even in the absence of stress. Secondly, we reduced λu because covalent cross-linking should be less sensitive to applied forces, given that these permanent cross-links can bear and support substantial forces, unlike weak, protein-protein interactions that behave as slip bonds, unbinding at higher rates under force [40]. To understand how these two parameters, k0,u and λu, impact matrix mechanical plasticity and initial matrix stiffness, a series of creep and recovery tests were simulated on the polymer matrix in the absence of a cell (Fig. 6b). A constant shear stress of 100 Pa was applied for the first hour to induce creep, and then the stress was removed to allow the matrix to recover. Strain in the matrix was quantified during the creep and recovery periods. As expected, smaller k0,u and λu resulted in a matrix with lower mechanical plasticity (Fig. 6c–f, Supplementary Figure 6a–b). On the other hand, higher values of k0,u and λu resulted in a matrix with higher mechanical plasticity. Notably, the initial elastic moduli, related to the stiffness of the polymer matrix on short time scales, were similar for the different networks (Fig. 6d,f). This is expected, given that it takes time for cross-linkers to unbind, even in high plasticity matrices. The magnitude of permanent strain in these matrices was also dependent on the magnitude of the stress applied to the matrices (Supplementary Fig 6c,d).
Figure 6. A computational model of the extracellular matrix shows that changes in kinetics of cross-link unbinding modulate matrix mechanical plasticity.
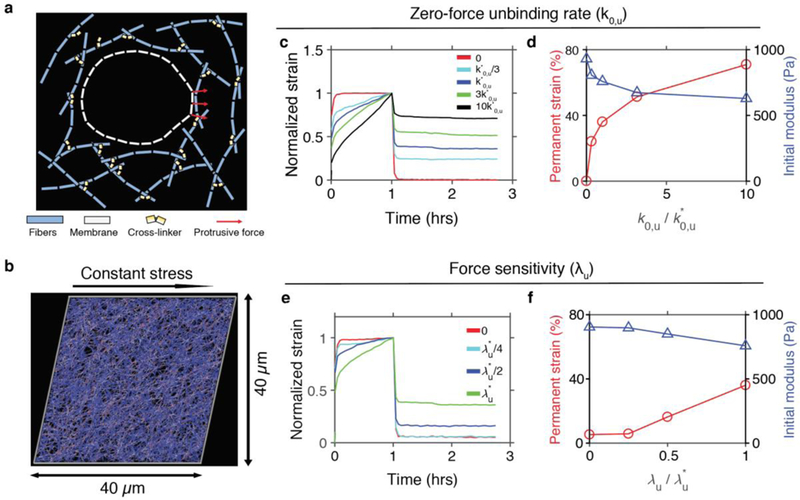
a, Schematic diagram showing elements in the model. A cell membrane is simplified into serially connected elements. A matrix is comprised of fibers (cyan) and cross-linkers (yellow). Cross-linkers can unbind from fibers in a force-dependent manner, following Bell’s law, ku = k0,u exp(Fλu/kBT). There is a volume-exclusion effect between the membrane elements and the matrix, so they do not cross over each other. A fraction of membrane nodes undergo protrusive forces in radially outward directions, resulting in local deformation of the matrix. b-f, Creep test of the matrix without a cell. b, Snapshot of a matrix undergoing a creep response. c and e, Normalized strain of matrices in response to constant stress of 100 Pa at t = 0–1 hour with various values of c, the zero-force unbinding rate constant (k0,u), and e, the force sensitivity (λu). k*0,u = 3 ×10−6 s−1 and λ*u = 100 pm are reference values. d and f, Permanent strain and initial modulus depending on variations in d, k0,u and f, λu.
Next, to determine the impact of enhanced matrix covalent cross-linking on cellular extension of invasive protrusions, we used a computational model with a cell membrane surrounded by a matrix under two matrix conditions. A matrix with “high plasticity” was modeled using high k0,u, whereas a matrix with “low plasticity” was modeled using low k0,u (Fig. 7a,b). In these studies, force sensitivity, λu, of cross-linker unbinding in both matrices was identical. In order to simulate a 3D invadopodium, which can extend to lengths on the order of tens of microns and exhibit lifetimes on the order of hours [26,41,42], a constant force of 1 nanonewton was applied to a portion of the cell membrane in radially outward directions for the first hour of computational simulations. For direct comparison of results between simulations, it was assumed that there is only one invadopodium toward the right. Note that the model is able to induce more than one invasive protrusion to simulate various cell shapes observed in experiments (Supplementary Fig. 7a). Repulsive forces acting between the membrane and the matrix enable the membrane with protrusion forces to push and deform the surrounding matrix. The portion of the membrane experiencing the forces can spontaneously change dynamically in size and shape in response to the evolving matrix deformation. Strikingly, enforcing these physical conditions alone, cells exhibited long and thin protrusions in the high plasticity matrix, whereas cells were more rounded with thicker, blunter protrusions in the low plasticity matrix (Fig. 7a,b, Supplementary Figure 7a). These morphologies qualitatively match the experimental observations. Cells in high plasticity matrices achieved substantial matrix deformation, as forces exerted on individual network cross-links were sufficient to break cross-linking points and reduce connectivity of the matrix (Fig. 7a, Supplementary Fig. 7b). Matrix deformation in the high plasticity matrix appeared to remain after the applied stress was removed, whereas little to no matrix deformation was achieved or retained in the low plasticity matrix (Fig. 7a,b). Matrix strain quantified over the duration of the simulations supported these observations; higher plasticity matrices experience more creep, greater maximum strain (~ 40%), and greater permanent strain (~ 30%) than those in lower plasticity matrices, which exhibited ~10% maximum strain and ~ 0% permanent strain (Fig. 7c,d). A change in λu led to less dramatic effects on the maximum and permanent strains (Fig. 7e,f). However, there is a tendency that higher λu results in larger maximum and permanent strains in matrices.
Figure 7. Changes in kinetics of cross-link unbinding modulate cell shapes and degree of permanent matrix deformation.
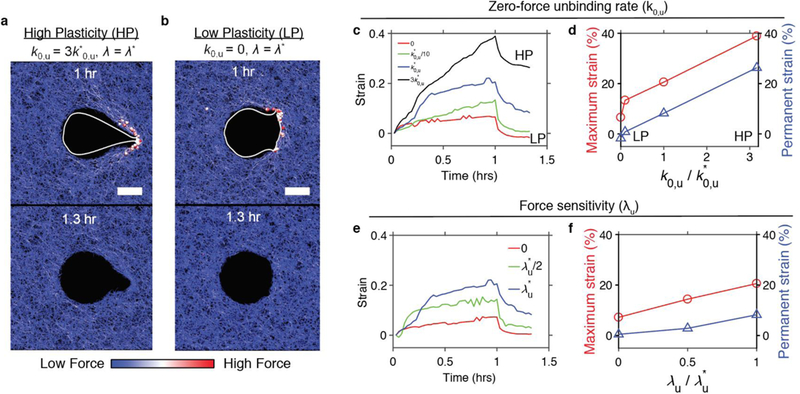
a and b, Snapshots of cell membranes making invasive protrusions into a matrix with a, high and b, low plasticity. Scale bars indicate 10 μm. Spheres indicate cross-linkers bearing forces higher than 60 pN. Color scaling represents level of forces acting on fibers and cross-linkers. c and e, Strain in a matrix during cell protrusions with a total force of 1 nN at t = 0–1 hour, and during an unloaded recovery phase of t = 1–1.3 hours, with different values of c, the zero-force unbinding rate constant (k0,u) and e, the force sensitivity (λu). d and f, Maximum and permanent strains depending on d, k0,u and f, λu. k*0,u = 3 ×10−6 s−1 and λ*u = 100 pm are reference values.
3. Discussion
Using two different material systems and a computational model, we show that covalent crosslinking reduces mechanical plasticity in basement membrane-like matrices, and diminished matrix mechanical plasticity in turn physically restricts extension of invasive protrusions. The high plasticity of pure reconstituted Basement Membrane (rBM), which has been noted previously [17], likely arises out of the weak binding interactions between matrix proteins [43]. While rBM is rich in native, bioactive signaling molecules, this loosely connected, solubilized network does not capture the structure or stiffness of basement membrane in vivo [43–46]. The monotonic dose-response relationship we obtained between transglutaminase (tTg) concentration and rBM mechanical plasticity supports the explanation that tTg-mediated covalent cross-linking lowers mechanical plasticity by reducing molecular rearrangements available to retain permanent deformation through unbinding and rebinding [17]. This dose-response relationship also revealed that a surprisingly high degree of covalent cross-linking (500 μg/mL) of tTg is required to use rBM as a model for an elastic extracellular matrix. We observed that concentrations of tTg that appeared to saturate rBM cross-linking did not significantly alter the stiffness of the rBM hydrogels compared to the pure rBM control. Indeed, all rBM formulations remained an order of magnitude less stiff than tumor tissue [34]. While the precise molecular interactions within rBM underlying this change in plasticity but not stiffness are unclear, our computational model supported these results, showing that extensive covalent cross-linking diminishes matrix plasticity but only moderately affects stiffness. This may be because stiffness relates to the instantaneous resistance of a material, whereas the effects of altered cross-linking kinetics impact material response to, and recovery from, deformation over time. Therefore, as a complement to studies in rBM and rBM+tTg, we designed a biomaterials system that would allow us to mimic the stiffness of tumor tissue and tune covalent cross-linking without altering biochemical signaling. To achieve the latter, our biomaterial formulations kept ligand density constant and incorporated biorthogonal cross-linkers.
In both kinds of basement membrane-like matrices studied here, we find that covalent cross-linking and low matrix mechanical plasticity inhibit cell protrusivity. However, the decrease in cell circularity is most pronounced in the IPN materials system. Our recent findings indicate that in confining IPNs, cells can extend long, thin invadopodia if the matrix exhibits sufficient plasticity [20]. While these protrusions may not add substantially to projected area, they add substantially to cell perimeter, and therefore cell circularity is highly penalized for long, thin protrusions. By contrast, broad changes in spread area that are seen in the rBM system can take place in otherwise highly round cells, meaning this spreading need not severely penalize circularity. Because physiological basement membranes are tens to hundreds of nanometers thick, a cellular protrusion on the order of ten microns is sufficient to puncture this thin, yet confining, tissue layer. While no migration studies were included here, data from prior experiments supports the association between cell circularity and the likelihood that a cell will extend invadopodia cyclically and subsequently migrate (Supplementary Figure 5) [47].
In all matrices investigated, large adhesion plaques resembling invadopodia precursors, with β1 integrin-rich plaques that excluded paxillin, were present [26,48]. However, across both systems, cells in covalently cross-linked matrices exhibited less actin-cortactin colocalization, suggesting reduced actin polymerization, diminished cell shape change, and decreased invasion in rBM+tTg and LP-CC matrices. Interestingly, differences in cortactin localization between the matrices that were not covalently cross-linked, the rBM and HP IPNs, support the idea that mechanisms of cell spreading may be different within the pure rBM and rBM-alginate IPN materials systems. In rBM matrices, cortactin is expressed uniformly throughout the cell body, indicating cells may spread using broad protrusions reminiscent of lobopodia, and that covalent cross-linking inhibits this mechanism [49]. By contrast, in HP IPNs, cortactin is expressed in colocalized actin-cortactin puncta, an indicator that invadopodia are the structures enabling cell protrusivity in these matrices [26,48]. Evidence showing that cells can use larger, actin-mediated structures to spread in rBM matrices, other than the invadopodia that are generally required to invade through basement membranes in vivo, supports the differences in morphological changes observed across two different materials systems, and reinforces the idea that rBM matrices do not necessarily impose the kind of structural or physical constraints that cells face when invading physiological basement membranes [21,33]. Furthermore, differences in localization of actin-related proteins indicate that cellular mechanotransduction may enable cells to sense and respond to differences in matrix mechanical plasticity. In response to varied degrees of matrix plasticity, it is also conceivable that cells could secrete and sense their own nascent matrix differently, potentially contributing to differential activation biochemical signaling pathways [50]. However, our computational model shows that active signaling feedback is not required to recapitulate cell morphologies observed, and that the use of mechanical constraints alone is sufficient to induce reductions in cell spreading and protrusivity that arise in covalently cross-linked matrices.
Our studies point toward protease-independent invasion mechanisms, mediated by generation of forces and matrix plasticity, rather than protease-dependent mechanisms as playing a key role in breaching basement membranes. We find that in both materials systems, protease inhibition by a broad spectrum protease inhibitor or a protease inhibitor cocktail failed to significantly impact cell morphology in high plasticity matrices, and failed to persistently alter the impact of covalent cross-linking on cell spreading. Prior studies, which validated that this dose of protease inhibitor (10 μM GM6001) significantly decreases matrix degradation by invasive protrusions, simultaneously showed that this treatment had no effect on the formation of the protrusions themselves [20]. However, the actin-mediated protrusions observed here could be physically inhibited by covalent cross-linking and decreased matrix mechanical plasticity. Together, these results indicate that in these basement membrane ligand-containing material systems, cancer cells are using actin-mediated protrusions to generate force in order to spread, rather than using them primarily to degrade the extracellular matrix. Indeed, there is strong support for the in vivo importance of protease-independent, force-dependent basement membrane invasion in a C. elegans developmental model of invasion. In this model, even in the absence of proteases, it is Arp 2/3, localized ATP production, and F-actin polymerization-driven force that can facilitate robust basement membrane breaching [51,52]. Such data point toward the therapeutic potential of physically reinforcing the structural integrity of basement membrane in order to physically inhibit invasion.
4. Conclusion
Here, we investigated the effect of covalent cross-linking on the extension of invasive protrusions in 3D, basement membrane ligand-containing extracellular matrices (ECM). In rBM matrices, tissue transglutaminase-mediated covalent cross-linking lowers matrix mechanical plasticity and inhibits cell spreading and protrusivity. In rBM-alginate IPNs that mimic the elevated stiffness of tumor tissue, incorporate bio-orthogonal covalent cross-links, and exhibit low mechanical plasticity, cells were also physically constrained and protein localization required for the extension of invadopodia was disrupted. Notably, protease inhibition did not alter cell spreading in matrices of high or low mechanical plasticity. Our computational model of extracellular matrix provided mechanistic insight into these findings, revealing that lower rates of cross-linker unbinding and lower cross-link force sensitivity, both aspects of covalent cross-linking, decrease matrix mechanical plasticity. Computer simulations also showed that cellular morphologies in high and low plasticity matrix could be recapitulated in large part by considering mechanical phenomena, without requiring incorporation of mechanotransduction or biochemical signaling. Altogether, these studies show that in combination with biochemical strategies inhibiting protease-mediated degradation, physical inhibition of cell spreading and protrusivity, perhaps through mechanical reinforcement of basement membrane matrix, may help to prevent cancer cell invasion.
Experimental Procedures and Computational Methods
Alginate preparation.
Sodium alginate rich in guluronic acid blocks and with a high molecular weight (FMC Biopolymer, Protanal LF 20/40, High-MW, 280 kDa) was prepared as described previously [53]. High-MW was irradiated 8 Mrad (3 or 8×106 rad) by a cobalt source to produce low-MW (35 kDa) alginates [35]. Alginate was dialyzed against deionized water for 3–4 days (molecular weight cutoff of 3500 Da), treated with activated charcoal, sterile filtered, lyophilized, and then reconstituted in serum-free DMEM (Life Technologies). Click alginate, consisting of alginate conjugated to tetrazine or norbornene, was prepared as described previously [38]. Briefly, high molecular weight alginate (FMC Biopolymer, Protanal LF 20/40, High-MW, 265 kDa) was dissolved in stirred buffer containing 0.1 M MES, 0.3 M NaCl, pH 6.5 at 0.5% w/v. Then, N-hydroxysuccinimide (NHS) and 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride were (EDC) added in 5x molar excess of the carboxylic acid groups of alginate. Next, 1 mM was added of either tetrazine (3-(p-benzyla-mino)-1,2,4,5-tetrazine, synthesized according to established protocols [54] or norbornene (1-bicyclo[2.2.1]hept-5-en-2-ylmethanamine, Norbornene; Matrix Scientific) per gram of alginate. The coupling reaction was stirred overnight and then quenched, dialyzed, charcoal-filtered, sterile-filtered, and lyophilized.
Formation of reconstituted Basement Membrane (rBM) matrices.
For rBM matrices, high concentration growth factor reduced Matrigel (Corning) was diluted to 8 mg/ml using transglutaminase from guinea pig liver (Sigma-Aldrich; 2.2 UN/mg) or vehicle-alone control. In brief, transglutaminase powder was dissolved in 50 mM Tris Buffer (pH 7.4) to make a stock solution of 1 mg/ml. Prior to mixing with Matrigel, 1 mg/ml transglutaminase solution was treated for 10 min at room temperature with a small quantity of 500 mM dithiothreitol (DTT) solution such that the final concentration of DTT was 2 mM. Then, 50 mM calcium chloride (CaCl2) was added to achieve a final concentration of 5 mM CaCl2 to activate the transglutaminase. An appropriate volume of this mix was added to Matrigel on ice and mixed thoroughly while avoiding air bubbles to achieve the desired concentration of transglutaminase. The final concentration of Matrigel was fixed at 8 mg/ml for all concentrations of transglutaminase by adding the appropriate volume of DMEM (Life Technologies).
Mechanical characterization of rBM matrices and IPNs.
Rheology measurements were made with an AR2000EX stress-controlled rheometer (TA Instruments). 8 mg/mL reconstituted Basement Membrane (rBM) matrices were formed as described previously, either with the addition of transglutaminase or vehicle alone. IPNs were made for mechanical testing as described previously [14]. rBM or IPN hydrogels were deposited directly onto the bottom plate of the rheometer immediately after mixing with cross-linker. A 25 mm flat plate was then immediately brought down, forming a 25 mm disk of gel. Mineral oil (Sigma) was applied to the edges of the gel disk to prevent dry-out. The mechanical properties were then measured over time until the storage modulus reached an equilibrium value. The storage and loss moduli at 0.15 Hz and 1% strain, a frequency and amplitude which were both within linear regimes, were recorded periodically for at least 2 hours. Elastic moduli (i.e. Young’s moduli) were calculated assuming a Poisson’s ratio of 0.5 and using the equation , where G* is the complex modulus found using the storage and loss moduli measured and using the relationship G* = (G’2 + G“;2)1/2. For plasticity experiments, this time sweep was followed by a creep-recovery test. This involved first applying a constant shear stress (10 Pa for rBM studies, or 100 Pa for IPN studies) for 1 hour while strain was recorded as a function of time. Then, the sample was unloaded (0 Pa) and strain was recorded as a function of time as the sample “recovered” from the absence of load for about 2 hours. This recovery time period was sufficient to minimize transient effects due to stress unloading, and was on the same time scale as the periodic structures and migration events observed in this study.
Cell culture.
Human breast adenocarcinoma cells MDA-MB-231 (ATCC) were cultured in high glucose Dulbecco’s Modified Eagles Medium (Hyclone) with 10% Fetal Bovine Serum (Hyclone) and 1% Penicillin/Streptomycin (Pen/Strep, Life Technologies). Cells were authenticated by the ATCC and tested to be mycoplasma negative.
3D Cell Encapsulation in rBM and IPNs.
For analysis of invasive morphology, cells were starved overnight in serum-free medium and encapsulated in rBM matrices or IPNs as follows. The 8 mg/mL rBM matrices were formed as described previously, except that the diluent of the rBM, DTT, CaCl2 mixture was a cell suspension in serum-free DMEM. For IPN studies, cells were encapsulated similarly to what has been described previously [14,36]. In brief, cells were resuspended in serum-free medium. After rBM was mixed with alginate, cells were added into this polymers mixture and deposited into a cooled syringe. The solution was then vigorously mixed with a solution containing calcium sulfate (CaSO4) and deposited into wells of a chambered coverglass (LabTek). For both rBM and IPN studies, the final concentration of cells was 0.5×106 cells/mL of IPN. The cell-laden hydrogels gelled in an incubator at 37°C and 5% CO2 for 35–45 minutes, and then stimulated with medium containing 10% FBS, 50 ng/mL EGF, and either DMSO as a vehicle-alone control or 10 μM GM6001 (Millipore) [46,55] to inhibit proteases. For studies utilizing a protease inhibitor cocktail previously published [20], either protease inhibitor cocktail (20 μM Marimastat, 20 μM Pepstatin A, 20 μM E-64, 0.7 μM Aprotinin, and 2 μM Leupeptin) or DMSO as vehicle alone control was added to the media. After one day, bright field microscopy was used to capture cell morphologies.
Immunohistochemistry
Preparation of gels.
For immunohistochemical staining, media was first removed from the gels. The gels were washed once with serum free DMEM, and then fixed with 4% paraformaldehyde in serum-free DMEM at room temperature for 45 – 60 minutes. The gels were then washed 3 times in PBS containing calcium (cPBS), and then incubated in 30% sucrose in cPBS at 4°C. The gels were then placed in a mix, which contained 50% of a 30% sucrose in cPBS solution, and the other 50% was OCT (Tissue-Tek), for at least 1 day. The media was then removed, the gels were embedded in OCT, and the gels were frozen. The frozen gels were sectioned and stained following standard immunohistochemistry protocols.
Staining sections.
The following antibodies and reagents were used for immunohistochemistry: Anti-Paxillin antibody (1:300; Abcam, Y113), Anti-β1 Integrin (1:300; Abcam, P5D2), and Anti-Cortactin (1:300; Abcam, ab33333). Negative controls, where the secondary antibody was added but the primary antibody was not, were conducted to ensure specificity of all stains. Matching secondary antibodies were purchased from Life Technologies. Alexa Fluor 488 phalloidin (Life Technologies, dilutions of 1:50 for β1 Integrin / Paxillin co-stain and 1:60 for Cortactin co-stain) was used to label the actin cytoskeleton, and DAPI was used to label the nucleus. ProLong Gold antifade reagent (Life Technologies) was used to minimize photobleaching. Images were acquired using a Leica HC PL APO 63X/1.4 NA oil immersion objective.
Image Analysis.
To quantify morphology of cancer cells, ImageJ was used to manually segment images and to calculate cell circularity, 4π * area /(perimeter2), whereby 1 indicates a perfect circle, for regions of interest. To evaluate cortactin expression in rBM matrices, cortactin intensity was measured from the tip of a protrusion toward the nucleus. For cells with multiple protrusions, cortactin intensity was measured for each protrusion. Mean cortactin intensity was then plotted by averaging intensity data of 12 cells, where at least 3 measurements (without overlapping pixels) were made per protrusion.
Computational Model
Creep Test.
To mimic a creep test in experiments, we simulated a cross-linked fiber matrix without a cell using an agent-based computational model based on Brownian dynamics (Fig. 1a). More details of the model are described in the supplementary material, and parameter values are listed in Table S2. In the model, fibers and cross-linkers are coarse-grained by cylindrical elements. Motions of the cylindrical elements are regulated by the Langevin equation. Bending and extensional forces maintain equilibrium angles and distances formed by the cylindrical elements, respectively. Formation of fibers is initiated by appearance of one cylindrical element whose length is 1.4 μm. The element is elongated by addition of identical cylindrical elements. To keep fibers short, the maximum number of cylindrical elements that can be added to the first element is limited to two, resulting in the average fiber length of ~4 μm.
Cross-linkers bind to binding sites located every 140 nm on fibers and also unbind from fibers in a force-dependent manner following Bell’s law [56]
| (1) |
where k0,u is the zero-force unbinding rate, λu represents sensitivity to applied force, and kBT is thermal energy. is a vector for a spring force acting on a cross-linker element, and r0,x1 is an equilibrium length of the cross-linker element. only when the spring force is tensile force, the unbinding rate, ku, increases beyond its base rate, k0,u. The references values of and are and respectively.
We first preassemble a matrix to perform the creep test. A cross-linked polymeric network is assembled via dynamic events of fibers and cross-linkers within a three-dimensional rectangular (3D) domain (40×40×1 μm) with a periodic boundary condition in x and y directions. Then, the preassembled matrix is loaded at the beginning of simulations for creep tests. All chains crossing the boundaries of the computational domain in the y direction are severed and clamped to the boundaries. The top boundary is displaced in the +x direction while the bottom boundary is fixed. Actual stress on the top boundary is calculated by summing all forces exerted on chains clamped to the boundary and then dividing the sum by the area of the top boundary. Position of the top boundary is continuously adjusted via feedbacks to match the actual stress with goal stress level. It is assumed that the goal stress is non-zero for the first hour to induce a creep behavior, and then becomes zero after that to evaluate recovery and plastic deformation. Initial modulus and permanent strain of the matrix are calculated in the same manner as experiments.
Cell invasion.
To investigate cell invasion, we simulated a cell membrane with protrusion surrounded by a matrix within a computational domain (100×60×1 μm) (Fig. 1A). The cell membrane is drastically simplified into serially connected rectangular solid elements whose height is the same as the domain width in the z direction (1 μm). Thus, the interconnected rectangular solid elements form a wall dividing a space in the domain into intracellular and extracellular spaces. In each simulation, a circular membrane with a radius of 10 μm is initially located at the center of the domain. Then, a matrix is assembled around the membrane as explained above. Under this condition, the number and initial length of membrane elements are 158 and 400 nm, respectively. Motions of the membrane elements are also regulated by the Langevin equation. Bending forces are exerted on membrane elements, and extensional forces prevent membrane elements from being too long (> 800 nm) or too short (< 4 nm). In most simulations, we applied 25 pN in radially outward directions to 40 membrane nodes initially located on the right side of the membrane for the first hour. Because membrane elements become much shorter, those membrane nodes with small protrusive forces can converge toward a specific location to exert stronger protrusive forces together. Repulsive forces acting between membrane elements and the matrix enable the membrane to push and deform the surrounding matrix. After one hour, protrusive forces are not applied anymore, and then the recovery and plastic deformation of the matrix are evaluated.
The matrix strain is quantified as follows. First, the closest nodes of fibers to the instantaneous center of a membrane are identified in each angular direction with resolution of 3°. These 120 fiber nodes delineate the boundary shape of a matrix around the cell membrane. Then, a distance between the membrane center and the farthest fiber node among 120, d, is used for calculating the matrix strain, ε.
| (2) |
where r0, m is the initial radius of the membrane.
Statistics.
Statistical analyses were performed using GraphPad Prism. P values provided in figure legends have been corrected for multiple comparisons, where relevant.
Supplementary Material
Acknowledgements
We acknowledge Sungmin Nam for technical assistance with tissue tranglutaminase. This work was supported by National Science Foundation Graduate Research Fellowship to K.W., a Stanford University Vice Provost for Graduate Education Diversifying Academia and Recruiting Excellence (DARE) Fellowship to K.W., a Postdoctoral Fellowship for Academic Diversity from the University of Pennsylvania, anNIH Grant [R37 CA214136] for O.C., and [1R01GM126256] FOR T.K AND P.C. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by a National Science Foundation grant [ACI-1548562], and the computations were conducted on the Comet supercomputer, which is supported by an NSF award [ACI-1341698], at the San Diego Supercomputing Center (SDSC) [57,58].
Abbreviations
- ECM
extracellular matrix
- LOX
lysyl oxidase
- tTg
Tissue Transglutaminase
- IPN Hydrogel
Interpenetrating Network Hydrogel
- HP
high plasticity
- LP-CC
low plasticity with covalent cross-links
- BM
basement membrane
- rBM
reconstituted Basement Membrane
Footnotes
Conflict of Interest
The authors declare that no conflict of interest exists.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lu P, Weaver VM, Werb Z, The extracellular matrix: A dynamic niche in cancer progression, J. Cell Biol. 196 (2012) 395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yuzhalin AE, Lim SY, Kutikhin AG, Gordon-Weeks AN, Dynamic matrisome: ECM remodeling factors licensing cancer progression and metastasis, Biochim. Biophys. Acta - Rev. Cancer. 1870 (2018) 207–228. doi: 10.1016/j.bbcan.2018.09.002. [DOI] [PubMed] [Google Scholar]
- [3].Karamanos NK, Theocharis AD, Neill T, Iozzo RV, Matrix modeling and remodeling: A biological interplay regulating tissue homeostasis and diseases, Matrix Biol. (2018). doi: 10.1016/j.matbio.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kotsakis P, Griffin M, Tissue transglutaminase in tumour progression: Friend or foe?, Amino Acids. 33 (2007) 373–384. doi: 10.1007/s00726-007-0516-1. [DOI] [PubMed] [Google Scholar]
- [5].Zhao Y, Min C, Vora SR, Trackman PC, Sonenshein GE, Kirsch KH, The lysyl oxidase pro-peptide attenuates fibronectin-mediated activation of focal adhesion kinase and p130 in breast cancer cells, J. Biol. Chem. 284 (2009) 1385–1393. doi: 10.1074/jbc.M802612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cellura D, Pickard K, Quaratino S, Parker H, Strefford JC, Thomas GJ, Mitter R, Mirnezami AH, Peake NJ, miR-19-Mediated Inhibition of Transglutaminase-2 Leads to Enhanced Invasion and Metastasis in Colorectal Cancer, Mol. Cancer Res. 13 (2015) 1095–1105. doi: 10.1158/1541-7786.MCR-14-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mangala LS, Arun B, Sahin AA, Mehta K, Tissue transglutaminase-induced alterations in extracellular matrix inhibit tumor invasion, Mol. Cancer. 4 (2005) 1–8. doi: 10.1186/1476-4598-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ranga A, Lutolf MP, Hilborn J, Ossipov DA, Hyaluronic Acid Hydrogels Formed in Situ by Transglutaminase-Catalyzed Reaction, Biomacromolecules. 17 (2016) 1553–1560. doi: 10.1021/acs.biomac.5b01587. [DOI] [PubMed] [Google Scholar]
- [9].Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM, Matrix crosslinking forces tumor progression by enhancing integrin signaling, Cell. 139 (2009) 891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shao M, Cao L, Shen C, Satpathy M, Chelladurai B, Bigsby RM, Nakshatri H, Matei D, Epithelial-to-mesenchymal transition and ovarian tumor progression induced by tissue transglutaminase, Cancer Res. 69 (2009) 9192–9201. doi: 10.1158/0008-5472.CAN-09-1257. [DOI] [PubMed] [Google Scholar]
- [11].Park MK, You HJ, Lee HJ, Kang JH, Oh SH, Kim SY, Lee CH, Transglutaminase-2 induces N-cadherin expression in TGF-β1-induced epithelial mesenchymal transition via c-Jun-N-terminal kinase activation by protein phosphatase 2A down-regulation, Eur. J. Cancer. 49 (2013) 1692–1705. doi: 10.1016/j.ejca.2012.11.036. [DOI] [PubMed] [Google Scholar]
- [12].Payne SL, Fogelgren B, Hess AR, Seftor EA, Wiley EL, Fong SFT, Csiszar K, Hendrix MJC, Kirschmann DA, Lysyl oxidase regulates breast cancer cell migration and adhesion through a hydrogen peroxide-mediated mechanism, Cancer Res. 65 (2005) 11429–11436. doi: 10.1158/0008-5472.CAN-05-1274. [DOI] [PubMed] [Google Scholar]
- [13].Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM, Tensional homeostasis and the malignant phenotype., Cancer Cell. 8 (2005) 241–54. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- [14].Chaudhuri O, Koshy ST, Branco da Cunha C, Shin J-W, Verbeke CS, Allison KH, Mooney DJ, Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium., Nat. Mater. 13 (2014) 1–9. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- [15].Chang TT, Thakar D, Weaver VM, Force-dependent breaching of the basement membrane, Matrix Biol. 57–58 (2017) 178–189. doi: 10.1016/j.matbio.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Egeblad M, Rasch MG, Weaver VM, Dynamic interplay between the collagen scaffold and tumor evolution., Curr. Opin. Cell Biol. 22 (2010) 697–706. doi: 10.1016/j.ceb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nam S, Lee J, Brownfield DG, Chaudhuri O, Viscoplasticity Enables Mechanical Remodeling of Matrix by Cells, Biophys. J. 111 (2016) 2296–2308. doi: 10.1016/j.bpj.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chaudhuri O, Viscoelastic hydrogels for 3D cell culture, Biomater. Sci. 5 (2017) 1480–1490. doi: 10.1039/c7bm00261k. [DOI] [PubMed] [Google Scholar]
- [19].Ban E, Franklin JM, Nam S, Smith LR, Wang H, Wells RG, Chaudhuri O, Liphardt JT, Shenoy VB, Mechanisms of Plastic Deformation in Collagen Networks Induced by Cellular Forces, Biophys. J. 114 (2018) 450–461. doi: 10.1016/j.bpj.2017.11.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker E-B, Friedl P, Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis., J. Cell Biol. 160 (2003) 267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rowe RG, Weiss SJ, Breaching the basement membrane: who, when and how?, Trends Cell Biol. 18 (2008) 560–574. doi: 10.1016/j.tcb.2008.08.007. [DOI] [PubMed] [Google Scholar]
- [22].Yurchenco PD, Basement membranes: cell scaffoldings and signaling platforms., Cold Spring Harb. Perspect. Biol. 3 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cosgrove D, Liu S, Collagen IV diseases: A focus on the glomerular basement membrane in Alport syndrome, Matrix Biol. 57–58 (2017) 45–54. doi: 10.1016/j.matbio.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Randles MJ, Humphries MJ, Lennon R, Proteomic definitions of basement membrane composition in health and disease, Matrix Biol. 57–58 (2017) 12–28. doi: 10.1016/j.matbio.2016.08.006. [DOI] [PubMed] [Google Scholar]
- [25].Pozzi A, Yurchenco PD, Iozzo RV, The nature and biology of basement membranes, Matrix Biol. 57–58 (2017) 1–11. doi: 10.1016/j.matbio.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Murphy DA, Courtneidge SA, The “ins” and “outs” of podosomes and invadopodia: characteristics, formation and function, Nat. Rev. Mol. Cell Biol. 12 (2011) 413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Eddy RJ, Weidmann MD, Sharma VP, Condeelis JS, Tumor Cell Invadopodia: Invasive Protrusions that Orchestrate Metastasis, Trends Cell Biol. 27 (2017) 595–607. doi: 10.1016/j.tcb.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Leong HS, Robertson AE, Stoletov K, Leith SJ, Chin CA, Chien AE, Hague MN, Ablack A, Carmine-Simmen K, McPherson VA, Postenka CO, Turley EA, Courtneidge SA, Chambers AF, Lewis JD, Invadopodia Are Required for Cancer Cell Extravasation and Are a Therapeutic Target for Metastasis, Cell Rep. 8 (2014) 1558–1570. doi: 10.1016/j.celrep.2014.07.050. [DOI] [PubMed] [Google Scholar]
- [29].Mrkonjic S, Destaing O, Albiges-Rizo C, Mechanotransduction pulls the strings of matrix degradation at invadosome, Matrix Biol. 57–58 (2017) 190–203. doi: 10.1016/j.matbio.2016.06.007. [DOI] [PubMed] [Google Scholar]
- [30].Coussens LM, Matrix Metalloproteinase Inhibitors and Cancer:Trials and Tribulations, Science (80-. ). 295 (2002) 2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- [31].Enderling H, Alexander NR, Clark ES, Branch KM, Estrada L, Crooke C, Jourquin J, Lobdell N, Zaman MH, Guelcher SA, Anderson ARA, Weaver AM, Dependence of invadopodia function on collagen fiber spacing and cross-linking: Computational modeling and experimental evidence, Biophys. J. 95 (2008) 2203–2218. doi: 10.1529/biophysj.108.133199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pourfarhangi KE, Bergman A, Gligorijevic B, ECM Cross-Linking Regulates Invadopodia Dynamics, Biophys. J. 114 (2018) 1455–1466. doi: 10.1016/j.bpj.2018.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yurchenco PD, Basement membranes: Cell scaffoldings and signaling platforms, Cold Spring Harb. Perspect. Biol. 3 (2011) 1–27. doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Acerbi I, Cassereau L, Dean I, Shi Q, Au A, Park C, Chen YY, Liphardt J, Hwang ES, Weaver VM, Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration., Integr. Biol. (Camb). 7 (2015) 1120–1134. doi: 10.1039/c5ib00040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chaudhuri O, Gu L, Darnell M, Klumpers D, Bencherif SA, Weaver JC, Huebsch N, Mooney DJ, Substrate stress relaxation regulates cell spreading, Nat. Commun. 6 (2015) 6365. doi: 10.1038/ncomms7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wisdom K, Chaudhuri O, 3D cell culture in interpenetrating networks of alginate and rBM matrix, 3D Cell Cult. Methods Protoc. Methods Mol. Biol. 1612 (2017) 29–37. doi: 10.1007/978-1-4939-7021-6. [DOI] [PubMed] [Google Scholar]
- [37].Lee KY, Mooney DJ, Alginate: properties and biomedical applications., Prog. Polym. Sci. 37 (2012) 106–126. doi: 10.1016/j.progpolymsci.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Desai RM, Koshy ST, Hilderbrand SA, Mooney DJ, Joshi NS, Versatile click alginate hydrogels crosslinked via tetrazine-norbornene chemistry, Biomaterials. 50 (2015) 30–37. doi: 10.1016/j.biomaterials.2015.01.048. [DOI] [PubMed] [Google Scholar]
- [39].Liu AP, Parekh SH, Integrative Biology New advances in probing cell – extracellular matrix interactions, Integr. Biol. 9 (2017) 383–405. doi: 10.1039/C6IB00251J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nam S, Hu KH, Butte MJ, Chaudhuri O, Strain-enhanced stress relaxation impacts nonlinear elasticity in collagen gels, Proc. Natl. Acad. Sci. 113 (2016) 201523906. doi: 10.1073/pnas.1523906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tolde O, Rösel D, Veselý P, Folk P, Brábek J, The structure of invadopodia in a complex 3D environment, Eur. J. Cell Biol. 89 (2010) 674–680. doi: 10.1016/j.ejcb.2010.04.003. [DOI] [PubMed] [Google Scholar]
- [42].Génot E, Gligorijevic B, Invadosomes in their natural habitat, Eur. J. Cell Biol. 93 (2014) 367–379. doi: 10.1016/j.ejcb.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kleinman HK, Martin GR, Matrigel: basement membrane matrix with biological activity., Semin. Cancer Biol. 15 (2005) 378–86. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- [44].Hotary K, Li X, Allen E, Stevens SL, Weiss SJ, A cancer cell metalloprotease triad regulates the basement membrane transmigration program, Genes Dev. 20 (2006) 2673–2686. doi: 10.1101/gad.1451806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rowe RG, Weiss SJ, Navigating ECM barriers at the invasive front: the cancer cell-stroma interface., Annu. Rev. Cell Dev. Biol. 25 (2009) 567–595. doi: 10.1146/annurev.cellbio.24.110707.175315. [DOI] [PubMed] [Google Scholar]
- [46].Sabeh F, Shimizu-Hirota R, Weiss SJ, Protease-dependent versus-independent cancer cell invasion programs: three-dimensional amoeboid movement revisited., J. Cell Biol. 185 (2009) 11–19. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wisdom KM, Adebowale K, Chang J, Lee JY, Nam S, Desai R, Rossen NS, Rafat M, West RB, Hodgson L, Chaudhuri O, Matrix mechanical plasticity regulates cancer cell migration through confining microenvironments, Nat. Commun. 9 (2018) 4144. doi: 10.1038/s41467-018-06641-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Moshfegh Y, Bravo-Cordero JJ, Miskolci V, Condeelis J, Hodgson L, A Trio-Rac1-Pak1 signalling axis drives invadopodia disassembly., Nat. Cell Biol. 16 (2014) 571–585. doi: 10.1038/ncb2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Petrie RJ, Gavara N, Chadwick RS, Yamada KM, Nonpolarized signaling reveals two distinct modes of 3D cell migration., J. Cell Biol. 197 (2012) 439–55. doi: 10.1083/jcb.201201124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Loebel C, Rodell CB, Chen MH, Burdick JA, Shear-thinning and self-healing hydrogels as injectable therapeutics and for 3D-printing, Nat. Protoc. 12 (2017) 1521–1541. doi: 10.1038/nprot.2017.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kelley LC, Hastie EL, Cáceres R, Chi Q, Schindler AJ, Jiang Y, Matus DQ, Plastino J, Sherwood DR, Adaptive F-actin polymerization and localized ATP production drive basement membrane invasion in the absence of MMPs, Dev. Cell. (2018) 1–16. doi: 10.1016/J.DEVCEL.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cáceres R, Bojanala N, Kelley LC, Dreier J, Manzi J, Di Federico F, Chi Q, Risler T, Testa I, Sherwood DR, Plastino J, Forces drive basement membrane invasion in Caenorhabditis elegans, Proc. Natl. Acad. Sci. (2018) 201808760. doi: 10.1073/pnas.1808760115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Rowley J, Madlambayan G, Mooney D, Alginate hydrogels as synthetic extracellular matrix materials., Biomaterials. 20 (1999) 45–53. http://www.ncbi.nlm.nih.gov/pubmed/9916770. [DOI] [PubMed] [Google Scholar]
- [54].Karver MR, Weissleder R, Hilderbrand SA, Synthesis and evaluation of a series of 1,2,4,5-tetrazines for bioorthogonal conjugation, Bioconjug. Chem. 22 (2011) 2263–2270. doi: 10.1021/bc200295y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Schoumacher M, Goldman RD, Louvard D, Vignjevic DM, Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia., J. Cell Biol. 189 (2010) 541–556. doi: 10.1083/jcb.200909113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


