Abstract
Osteosarcoma, a malignant primary bone tumor most commonly diagnosed in children and adolescents, has a poorly understood genetic etiology. Genome-wide association studies (GWAS) and candidate-gene analyses have identified putative risk variants in subjects of European ancestry. However, despite higher incidence among African-American and Hispanic children, little is known regarding common heritable variation that contributes to osteosarcoma incidence and clinical presentation across racial/ethnic groups. In a multi-ethnic sample of non-Hispanic white, Hispanic, African-American and Asian/Pacific Islander children (537 cases, 2165 controls), we performed association analyses assessing previously-reported loci for osteosarcoma risk and metastasis, including meta-analysis across racial/ethnic groups. We also assessed a previously described association between genetic predisposition to longer leukocyte telomere length (LTL) and osteosarcoma risk in this independent multi-ethnic dataset. In our sample, we were unable to replicate previously-reported loci for osteosarcoma risk or metastasis detected in GWAS of European-ancestry individuals in either ethnicity-stratified analyses or meta-analysis across ethnic groups. Our analyses did confirm that genetic predisposition to longer LTL is a risk factor for osteosarcoma (ORmeta: 1.22; 95% CI: 1.09–1.36; P=3.8×10−4), and the strongest effect was seen in Hispanic subjects (OR: 1.32; 95% CI: 1.12–1.54, P= 6.2×10−4). Our findings shed light on the replicability of osteosarcoma risk loci across ethnicities and motivate further characterization of these genetic factors in diverse clinical cohorts.
Keywords: Osteosarcoma, genome-wide association study, polygenic risk score, Mendelian randomization
Introduction
Osteosarcoma, a malignant primary bone tumor most commonly diagnosed in children and young adults, has a poorly understood genetic etiology1. A majority of osteosarcoma tumors occur at the metaphyses of long bones and less commonly can present at other sites such as craniofacial, chest and pelvic regions2. Although some osteosarcoma cases are attributed to inherited disorders (e.g. Li-Fraumeni syndrome, Rothmund-Thompson syndrome, hereditary retinoblastoma), most cases appear to be sporadic3. A single genome-wide association study (GWAS) of osteosarcoma risk, conducted primarily in children and adolescents, identified two putative risk alleles in subjects of European ancestry4. A separate case-only GWAS of osteosarcoma clinical characteristics among patients of European, African, and Brazilian ancestry identified a single nucleotide polymorphism (SNP) associated with osteosarcoma metastasis5. Common germline variants involved in DNA repair6, growth regulation7,8, antigen processing and presentation9, and telomere maintenance10,11 pathways have also been implicated in osteosarcoma risk.
A recent study of common genetic variants known to contribute to inter-individual variation in leukocyte telomere length (LTL) showed that genetic predisposition to longer LTL conferred an increased risk of osteosarcoma in patients of European ancestry10. Interestingly, this was the first time that germline genetic determinants of telomere length had been associated with osteosarcomagenesis or broadly with development of any adolescent-onset cancer. This observation was concurrent with studies of osteosarcoma tumor genomes where recurrent somatic mutations in ATRX were shown to promote cellular immortalization through telomere lengthening12.
Telomeres are tandem repeat sequences (TTAGGGn) that reside at the ends of chromosomes and are stabilized and regenerated by protein interactions. Telomeres help maintain genome stability during cell replication; however, each successive round of mitotic division shortens telomeres, leading to cell senescence or apoptosis13,14. Although previous epidemiologic studies have reported short LTL in association with age-related diseases including cardiovascular disease and overall mortality15–17, the direction and magnitude of association between LTL and cancer risk have been contradictory across observational studies18. Mendelian randomization has been utilized as a causal inference tool to study cancer risk factors, including to address the issues of 1) environmental confounders; 2) treatment confounders; and 3) reverse causality, e.g. LTL attrition as disease sequela. Although environmental factors may not have had time to exert confounding effects among a pediatric population, the latter issues are potential sources of bias. Specifically, telomere shortening can occur after diagnosis, potentially due to treatment,19,20 or as an early step in malignant cell transformation.21 More recent causal inference studies using Mendelian randomization approaches suggest that long LTL is a risk factor for several types of cancers22–26. Mechanistically, shorter LTL may cause premature cell death and dysfunction while longer telomere length may permit additional replicative divisions and increase risk of neoplasms. Longer LTL has been described among African-American and Hispanic individuals compared to non-Hispanic whites.27,28
There is a slightly higher incidence of osteosarcoma among African-American and Hispanic children compared to non-Hispanic white children (15% and 11% higher, respectively).2,29 However, the etiologic factors underlying this ethnic disparity (e.g. genetic variants, environmental exposures, gene-environment interactions) are unknown. Previous studies of the genetic etiology of osteosarcoma have primarily been conducted among individuals of European ancestry. It is necessary to assess how heritable variation contributes to osteosarcoma across ethnic groups since genetic associations for traits identified in one ancestry group may not replicate across other ancestry groups30. To address this knowledge gap, we assessed whether osteosarcoma susceptibility variants identified in previous studies of European-ancestry populations were associated with osteosarcoma risk in a multi-ethnic set of pediatric osteosarcoma patients and controls.
Materials and Methods
Study participants
The study was approved by the Institutional Review Boards at the University of California, Berkeley and the California Health and Human Services Agency. Newborn blood samples are obtained from all neonates born within the state of California for the purpose of disease screening by the California Department of Public Health Genetic Diseases Screening Branch. Bloodspots remaining after screening have been archived at −20°C since 1982 and are available for approved research. We linked statewide birth records (1982–2009) to cancer diagnosis data from the California Cancer Registry (CCR, for years 1988–2011). Cases are defined as patients diagnosed with osteosarcoma (ICD-O-3 codes 9180–9183, 9185–9187, and 9192–9195) before age 20, per CCR record. Controls were matched on birth year, sex, and maternal self-reported race/ethnicity, with on average four controls selected per case. We identified 537 osteosarcoma cases born in California from 1982–2009 for which a newborn bloodspot was successfully retrieved for DNA extraction and genotyping. Detailed characteristics of osteosarcoma cases have been reported previously and appear in Supplementary Table 131.
DNA extraction and genotyping
DNA extraction from bloodspots was performed using the QIAamp DNA Investigator Kit (Qiagen) as previously described31. DNA was genotyped on the Affymetrix Axiom Latino Array, with average genotype concordance >99% between duplicate samples on the same plates (n=34). Quality control procedures were performed as previously described31. In brief, iterative call-rate filtering removed SNPs with call-rates <97% and samples with call-rates <97%; SNPs with significant departure from Hardy-Weinberg equilibrium P<1.0×10−5 among non-Hispanic white controls were excluded; samples with mismatched reported versus genotyped sex were excluded; one individual of any sample pair that had an identity-by-descent proportion >0.18 was excluded. Ancestry-informative principal components were calculated separately for individuals of each self-identified racial/ethnic group, i.e. Hispanic, non-Hispanic white, African-American, and Asian/Pacific Islander using HapMap reference samples based on ASW, CEU, CHB, CHD, GIH, JPT, LWK MEX, MKK and YRI populations. Using genome-wide SNP data from HapMap Phase 3 samples, individuals showing evidence of mismatched ancestry (>3 SDs from mean MXL, CEU, ASW, or EAS values on the first three principal components) were excluded. Haplotype phasing and imputation was performed with SHAPEIT v2.79028 and Minimac3 software using phased genotype data from the 2016 release of the Haplotype Reference Consortium32. Poorly-imputed SNPs with imputation quality (info) scores less than 0.60 or posterior probabilities less than 0.90 were excluded.
Statistical analyses
Although we were underpowered to conduct a genome-wide analysis, we assessed whether osteosarcoma susceptibility variants identified in previous studies of European-ancestry populations replicate in a multi-ethnic sample. We performed single SNP association analyses assessing the two previously reported osteosarcoma risk SNPs from GWAS.4 Association analyses were performed separately for each racial/ethnic group using logistic regression, assuming an allelic additive model for 0, 1, or 2 copies of the risk allele (matching the methodology of the original GWAS report), and adjusting for sex and the first five ancestry-informative principal components for each group. Results were meta-analyzed across the four racial/ethnic groups. Resulting beta estimates represent the per-allele increase in osteosarcoma risk. We additionally identified and analyzed 18 previously-reported SNPs from candidate-gene approaches reporting significant associations.6,7,33–35
To investigate the combined genetic effect of eight telomere-length associated SNPs on osteosarcoma risk in a multi-ethnic study, we constructed polygenic scores for longer LTL for each individual by summing the number of alleles associated with longer LTL, with each allele weighted by its effect size from the ENGAGE Consortium Telomere Group36 (16 alleles from 8 unlinked SNPs). We limited our study to eight SNPs on different chromosomes for three main reasons: 1) we wished to directly match the methodology of the original polygenic score report for this replication work; 2) we wanted to avoid “double-counting” SNPs that are correlated due to linkage disequilibrium when performing the polygenic risk score association analysis; and 3) we wanted to avoid using SNP weights from effect estimates of different studies which may not scale appropriately, particularly those using different methods of LTL measurement, i.e. quantitative PCR and Southern blot of the terminal restriction fragment. Although the proportion of variation explained by the eight SNPs is small, (approximately 1–2%)36 the genotypically-estimated relative LTL across individuals ranged from 25 to 1024 base pairs. This 999 base-pair range corresponds to approximately 30 years of age-related telomere attrition (based on an average LTL attrition rate of 20–40 base pairs/year).37,38 Associations between the polygenic risk scores for longer LTL and osteosarcoma risk were calculated among each racial/ethnic group, adjusting for sex and five principal components. Resulting beta estimates represent the difference in osteosarcoma risk per one standard deviation increase in the polygenic score. Given the greater risk of osteosarcoma in males, we assessed the presence of effect modification by performing interaction analysis between sex and LTL polygenic score. We also performed single SNP association analyses assessing the eight SNPs associated with telomere length using the aforementioned method for testing single-SNP associations with osteosarcoma risk.
In a case-only single-SNP association test, we assessed a SNP previously reported to be associated with osteosarcoma metastasis in a GWAS report5. Association analyses were performed separately for each racial/ethnic group using logistic regression with tumor metastasis as the outcome, and adjusted for sex and the first five principal components for each group. Results were meta-analyzed across the four racial/ethnic groups.
Case-only associations between LTL score and clinical presentation of osteosarcoma were explored using logistic regression for tumor site and metastasis. Coding for clinical variables were obtained through the CCR Data Dictionary Web site and the National Cancer Institute Surveillance, Epidemiology, and End Results Program Coding and Staging Manual, 2015, and have previously been described in detail39. Linear regression was used to assess tumor stage, grade and extension categories (coded as ordinal variables), and age at diagnosis and tumor size (coded as linear variables). For these analyses, one estimate was obtained for all subjects with racial/ethnicity included as a covariate rather than performing stratified analyses due to limited sample size. Also included in the model were sex and the first five principal components, recalculated with HapMap reference samples based on MXL, CEU, ASW, and EAS populations combined.
Mendelian randomization analyses
A two-sample Mendelian randomization analysis was performed with summary statistics of the eight LTL associated SNPs from the ENGAGE Consortium Telomere Group,36 and with summary statistics for osteosarcoma of the same eight SNPs from the genotyped California case-control data. The Mendelian-Randomization R package was used to implement the inverse-variance weighted, MR-Egger, and weighted median methods to assess the causal relationship between LTL and osteosarcoma risk.40
Results
Of 564 osteosarcoma cases identified by the California Cancer Registry and meeting eligibility criteria, 537 were successfully linked to an archived newborn blood spot (95.2%). Of 27 cases that were not successfully linked to biospecimens, 9 were born in 1982, the first year that specimens were stored. After quality control procedures were performed, 537 osteosarcoma cases (227 Hispanic, 207 non-Hispanic white, 59 African American, and 44 Asian/Pacific Islander) and 2165 multi-ethnic controls remained for genetic analyses. Nineteen non-Hispanic white osteosarcoma patients overlapped with those previously included in both the NCI-led osteosarcoma GWAS,4 the GWAS of osteosarcoma metastasis,5 and/or the prior study of LTL genetics and osteosarcoma risk10 based on identity-by-descent measures. These were removed from replication analyses, resulting in an independent case-control dataset of 518 osteosarcoma patients and 2165 controls.
In the assessment of two SNPs previously reported to be associated with osteosarcoma risk in prior GWAS (rs7591996 and rs1906953),4 we were unable to replicate any signal in either a meta-analysis of our multi-ethnic replication set or in analyses stratified by race/ethnicity. No consistent direction of effect was observed across race/ethnicity (Table 1). In a 500kb region of chromosome 2, centered on rs7591996, no strong evidence of association was observed for any SNP (Pmin=0.0025 across 5161 SNPs). Similarly null results were observed in a 500kb region of chromosome 6 centered on rs1906953 (Pmin = 0.0043 across 5420 SNPs) (Supplementary Figures 1A and 1B). In a case-only analysis of a SNP previously associated with osteosarcoma metastasis (rs7034162),5 we observed a non-significant association in the expected direction in the meta-analysis (P=0.33) and a suggestive positive association among Hispanic patients (OR: 1.79; 95% CI: 0.90–3.55; P=0.09) (Table 1). Similar risk allele frequencies in osteosarcoma controls of each racial/ethnic group were observed as those reported in 1000 Genomes (Supplementary Table 2). One variant from prior candidate-SNP analyses was associated at P<0.05 and in the same direction as previous reports in our multi-ethnic meta-analysis of California subjects (rs6599400 upstream of FGFR3, previously evaluated in Mirabello, et al., 20116 and Naumov, et al., 201233) (Supplementary Table 8). In ethnicity-stratified analyses, two SNPs in IGF2R were associated with osteosarcoma risk in non-Hispanic whites only (rs998074 and rs998074, P=5.0×10−3) (Supplementary Table 8).
Table 1:
Replication analyses of variants associated with osteosarcoma risk and metastasis in prior GWAS among an independent multi-ethnic case-control sample
| Study | SNP | Effect in previous study |
Effect in replication study |
||
|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | ||
| Osteosarcoma risk4 | rs7591996 | 1.39 (1.23–1.54) | 1.0×10−8 | Meta-analysis: 1.05 (0.91, 1.21) Hispanic: 1.10 (0.90, 1.36) Non-Hispanic white: 0.98 (0.78, 1.23) African American: 1.03 (0.58, 1.84) Asian/Pacific Islander: 1.18 (0.68, 2.06) |
0.47 0.34 0.86 0.91 0.56 |
| Osteosarcoma risk4 | rs1906953 | 1.57 (1.35–1.83) | 8.0×10−9 | Meta-analysis: 0.99 (0.84, 1.18) Hispanic: 0.84 (0.66, 1.07) Non-Hispanic white: 1.07 (0.77, 1.49) African American: 1.42 (0.92, 2.20) Asian/Pacific Islander: 1.04 (0.62, 1.74) |
0.93 0.16 0.68 0.11 0.88 |
| Osteosarcoma metastasis5 | rs7034162 | 2.43 (1.83–3.24) | 1.2×10−9 | Meta-analysis: 1.32 (0.76, 2.28) Hispanic: 1.79 (0.90, 3.55) Non-Hispanic white: 0.36 (0.04, 1.63) African American: 1.32 (0.29, 6.01) Asian/Pacific Islander: 0.74 (0.16, 3.50) |
0.33 0.09 0.25 0.72 0.71 |
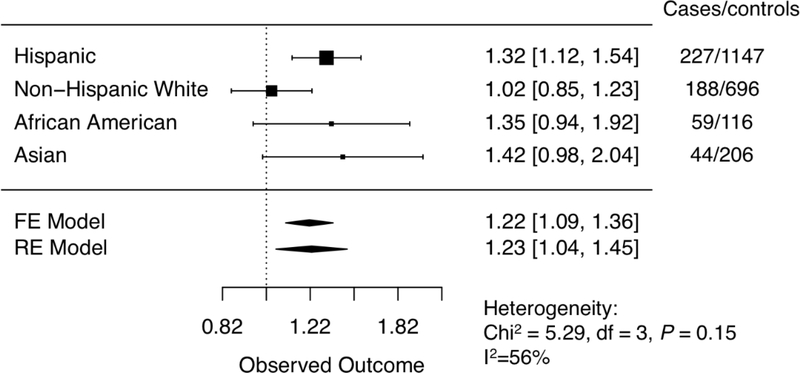
In both fixed-effects and random-effects meta-analyses including all racial/ethnic groups, the LTL score was significantly positively associated with osteosarcoma risk (ORFixed: 1.22; 95% CI: 1.09–1.36; P=3.8×10−4; ORRandom: 1.23; 95% CI: 1.04–1.45; P=0.01, respectively) (Figure 1). A formal test for interaction in a regression model assessing the P value of the cross-product term of the polygenic risk score and self-reported race/ethnicity (with non-Hispanic white subjects as the reference category) showed evidence of modification by race/ethnicity, with the risk score conferring additional risk among Hispanic individuals (ORinteraction=1.32, 95% CIinteraction= 1.04–1.68, Pinteraction = 0.024). Among control subjects, African Americans had the longest genetically-predicted LTL based on LTL polygenic scores (Supplementary Figure 2). In stratified analyses, the LTL score was significantly associated with osteosarcoma risk in Hispanic individuals, with each standard deviation increase in the score associated with a 1.32-fold increased risk of osteosarcoma (OR=1.32, 95% CI: 1.12–1.54, P= 6.2×10−4). Non-significant positive associations with comparable magnitudes of effect were also observed among African-Americans (OR=1.35, 95% CI: 0.94–1.92, P=0.10) and Asian/Pacific Islanders (OR=1.42, 95% CI: 0.98–2.04, P=0.06), yet no evidence of association was observed among non-Hispanic white subjects (OR=1.02, 95% CI: 0.85–1.23, P=0.80) (Figure 1). Tests for interaction in regression models assessing the cross-product term of the LTL polygenic risk score and subject sex showed no evidence of effect modification in stratified or meta-analyses across race/ethnic groups (Pinteraction>0.10)
Figure 1:
Forest plot of associations between osteosarcoma risk and polygenic scores for leukocyte telomere length.
Mendelian randomization analyses also showed significantly increased risk per standard deviation of LTL across subjects (ORFixed: 5.61; 95% CI: 2.21–14.24; P=2.8×10−4; ORRandom: 5.67; 95% CI: 1.52–21.21; P=9.9×10−3, respectively) (Supplementary Figure 3). In stratified analyses, each standard deviation increase in LTL in Hispanic individuals was causally associated with a 9.30-fold increased risk of osteosarcoma (OR=9.30, 95% CI: 2.61–33.1, P= 1.0×10−3). Sensitivity analyses using median-based estimation showed similar estimates as the inverse-variance weighted method (OR=14.6, 95% CI: 2.97–72.2, P= 1.0×10−3), and MR-Egger regression did not show evidence of directional pleiotropy (MR-Egger intercept P = 0.286). Note that Mendelian randomization estimates are larger in magnitude than polygenic risk score association estimates due to the different scale of the exposure being assessed (i.e. standardized polygenic score versus standardized LTL).
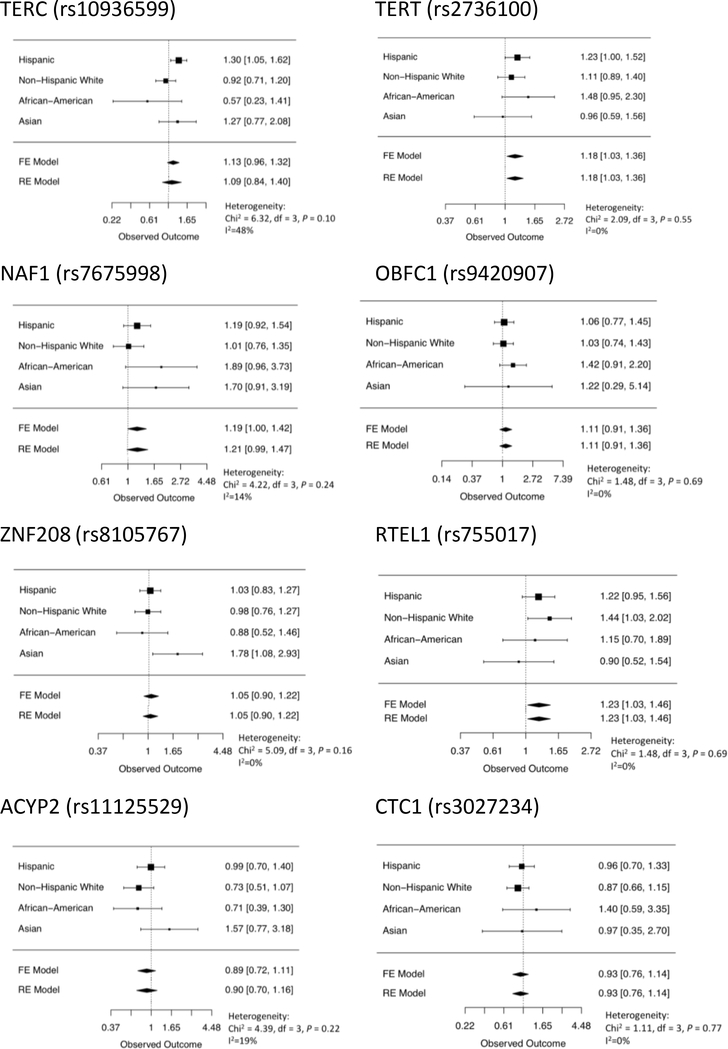
In single-SNP association analyses of the eight individual alleles contributing to the polygenic LTL score, the allele associated with longer LTL was associated with increased risk of osteosarcoma for six of the eight LTL SNPs (Figure 2). Furthermore, alleles in TERT, NAF1, and RTEL1 were nominally associated in meta-analyses across racial/ethnic groups (P<0.05) (Figure 2). Among the eight SNPs used in the polygenic risk score, similar risk allele frequencies in osteosarcoma controls of each racial/ethnic group were observed as those reported in 1000 Genomes (Supplementary Table 3).
Figure 2:
Forest plots of associations between osteosarcoma risk and SNPs previously associated with inter-individual variation in leukocyte telomere length.
We also explored whether the LTL score was associated with clinical characteristics in osteosarcoma patients. For these exploratory case-only analyses, the 19 subjects that overlapped with prior publications were returned to the dataset because no previous assessment of the relationship between LTL score and osteosarcoma clinical characteristics has been conducted to-date. Patients whose tumors were located in craniofacial bones, short bones of the lower limb, and “other sites” (e.g. ribs, pelvis, sternum) had non-significantly higher LTL scores than patients with tumors in the long bones of the upper or lower limbs, and we were unable to detect statistically significant associations with age at diagnosis, metastasis, and other tumor characteristics (Supplementary Figure 4, Supplementary Table 4). There was no evidence that ethnic differences in tumor site were confounding the observed associations (Supplementary Table 5).
Discussion
Although advances in genomic technologies have spurred progress in understanding the genetic epidemiology of many diseases, replication in independent datasets and validation in multi-ethnic patient populations are indispensable.41 We were unable to replicate two previously reported osteosarcoma risk loci first detected in a GWAS of European ancestry individuals among our multi-ethnic case-control set and observed little evidence of association in either meta-analysis or in analyses stratified by race/ethnicity. We had sufficient power to detect a nominal association (P<0.05) in the pooled/meta-analysis, as well as in race/ethnicity-stratified analyses among Hispanics and non-Hispanic white subjects, but underpowered to detect associations within other specific racial/ethnic groups (Supplementary Table 6). Osteosarcoma patients in the previous GWAS was not restricted by age at diagnosis, although the patients were diagnosed primarily at childhood and adolescence (aged <21). However, the rs1906953 SNP has been successfully replicated in two larger adult Chinese cohorts, so it is possible that our race-stratified analysis may be underpowered or sensitive to age-specific differences.42,43
A SNP previously associated with risk of metastasis in osteosarcoma patients (rs7034162) also failed to reach statistical significance in a meta-analysis of our multi-ethnic sample. However, the observed effect was in the same direction as previous reports and was suggestively associated when analysis was limited to Hispanic patients (OR: 1.79; P=0.09). Lack of power is a potential explanation for the non-significant replication, as only 64 metastatic cases were assessed, and further stratified by race/ethnicity (Supplementary Table 1), although we have approximately 80% power to detect an association at nominal significance (P<0.05) (Supplementary Table 6). Additionally, the 11.9% metastasis rate among cases in our study was lower than previous worldwide estimates of 18% among adults and 14% among children aged 18 or under,44 suggesting a potentially different distribution of cases in our study. Of note, osteosarcoma patients needed to be born in and have remained in California in order to be successfully linked across cancer and birth registries. We explored the possibility that study design limitations potentially resulted in linked cases being younger than unlinked cases and the osteosarcoma population in general, although the evidence does not support this (Supplementary Table 7). Finally, it is possible that we were better able to replicate the association with metastasis given that the SNP was previously identified in a meta-analysis of multiple ethnicities that included European, African and Brazilian patients. Our analyses failed to replicate the majority of associations from previous candidate SNP studies, suggesting that traditional candidate-gene approaches remain limited as a method of identifying novel genetic associations due to difficulty in identifying relevant genes,45 and perhaps more importantly, difficulty in identifying the functional/regulatory variants with disease relevance within those candidate genes and pathways.46
Our analyses replicate a previous report suggesting that genetic predisposition to longer LTL is a risk factor for osteosarcoma and extend this association beyond European-ancestry subjects. Furthermore, our results suggest potential effect modification by race/ethnicity, with a stronger association with LTL polygenic risk score among non-white individuals. This observation may potentially contribute to the slightly higher incidence of osteosarcoma among African-American and Hispanic children. Single-SNP analyses show contributions by variants in TERT, NAF1, and RTEL1, which were nominally associated with osteosarcoma risk in our multi-ethnic dataset. Overall, these findings are consistent with the previously described association between the polygenic score for longer LTL and osteosarcoma risk in a study of 660 non-Hispanic white patients.10 However, the single-SNP associations reaching nominal significance varied across studies, suggesting that the overall association between longer predicted LTL and osteosarcoma risk is likely due to aggregated weak effects from numerous individual SNPs, rather than any one specific SNP driving the association. Indeed, this is a noted advantage of polygenic scores over single-SNP analyses.
Although our meta-analysis across four racial/ethnic groups identified a significant positive association between longer LTL and osteosarcoma risk, as previously observed in an independent study of non-Hispanic white individuals10, no association was observed in our subset of non-Hispanic white subjects (although confidence intervals overlap with the previously described positive association, OR = 1.10; 95%CI = 1.01–1.19; P = 0.017). Possible explanations include inadequate power (this study has approximately one-third the number of non-Hispanic white cases), or differences in age of cases (the prior study was not restricted to a specific age range whereas our subjects were all <20 at diagnosis).
With this study, the association between longer predicted LTL and increased osteosarcoma risk has now been demonstrated in two independent datasets and in multiple racial/ethnic groups. This finding is in contrast to a previous smaller study that described an inverse relationship between LTL and osteosarcoma risk in which LTL was directly assessed using real-time quantitative PCR11. In this other study, LTL was measured from blood samples collected from osteosarcoma patients at the time of limb salvage surgery, which may have followed chemoradiotherapy (treatment data were not provided). Therefore, this previously reported association may be susceptible to reverse causality (e.g. iatrogenic telomere attrition or attrition as disease sequela)19,20. Because heritable genotypes are established at birth and do not vary in response to environmental factors (e.g. lifestyle factors, clinical interventions, etc.), case-control comparisons that leverage genotypes as genetic instruments are robust against reverse causality and many potential confounders. Thus, the use of a polygenic LTL score, as well as the use of the Mendelian randomization framework, is a particular advantage of our study design.
A limitation of our analysis is that our genetic estimate of telomere length is derived from a GWAS of LTL, and it is therefore unclear whether these SNPs are truly predictive of telomere length in the cancer-prone tissue of interest, i.e. bone. However, prior literature report correlations between telomere length measured in blood, lung and skeletal muscle (r=0.35–0.84),47–49 consistent with our assumption of inter-tissue correlation. Another limitation is that the polygenic risk score for LTL and the instrumental variables for Mendelian randomization analyses are based on summary statistics of SNPs associated with inter-individual variation in LTL among European-ancestry subjects.36 Our observation of an association between longer predicted LTL and increased osteosarcoma risk among the non-white subjects is particularly intriguing given the caveats associated with transferring genetic association estimates across populations with different ancestral backgrounds. In general, risk variants detected in one ethnic group may not be replicated in another ethnic group, even if the variant is a true association, due to: 1) different genetic architectures (i.e. the risk allele may be tagging a causal variant in one population, but not in others); 2) allelic heterogeneity (i.e. a phenotype is influenced by different ethnic-specific causal variants at the same locus); 3) differences in risk allele frequencies between populations, and 4) different effect sizes due to variable epistasis or gene-environment interactions across populations30.
Caution must be taken when generalizing genetic estimates or risk scores across racial/ethnic groups due to the aforementioned issues50,51. These are also important considerations with respect to the Mendelian randomization interpretations in this study, which requires that the instrumental variables comprised of the weighted LTL SNPs derived from subjects of European ancestry also predict LTL in non-European subjects. Although other studies have reported associations between polygenic risk scores built from European data and phenotypes diagnosed in non-European subjects52–54, evidence generally suggests that there is a range of performance of polygenic risk scores across racial/ethnic groups depending on the phenotype of interest. Furthermore, it was suggested by prior studies that genetic risk scores performed better when weights for effect estimates were obtained from multi-ethnic meta-analyses of GWAS data50 and that genetic risk scores developed in European ancestry populations are more accurate when applied to Hispanic subjects than subjects of other ethnicities due to the high level of European admixture in U.S. Hispanics51. It is also plausible that the telomere-associated SNPs identified in GWAS of European ancestry subjects also tag (to varying degrees) the underlying causal SNPs in other ancestral groups given the consistent positive associations observed across the various racial/ethnic groups we investigated. It remains important to consider the performance of genetic risk scores across racial/ethnic groups as there are both incidence and outcomes disparities in sarcoma patients across racial/ethnic groups.55 Understanding the underlying reasons for these disparities is essential to future screening and treatment practices.
GWAS of telomere length in non-European subjects have had varied success in reproducing associations from European-ancestry datasets. Namely, a study with Bangladeshi individuals showed directionally consistent associations with the eight previously reported signals first identified in Europeans, but only one SNP (rs8105767 in ZNF208) was nominally significant (P=0.003)56. A GWAS in Han Chinese showed directionally consistent associations with four out of five comparisons with previously reported signals in Europeans (TERT, TERC, CTC1 and ZNF676), with variants in TERT and TERC both nominally significant (1.93×10−5 and 5.57×10−3, respectively). Despite weaker associations with LTL in non-European individuals, aggregate effects across multiple LTL-associated SNPs may have allowed us to observe a significant association with osteosarcoma risk in our sample. Although many LTL effect alleles are similar in frequency between ancestry groups, as observed in our data and in 1000 Genomes (Supplementary Table 2), there remain substantial frequency differences for some alleles in our analysis. The allele frequency differences in LTL-associated SNPs between ancestral groups has previously been described with respect to anti-cancer polygenetic adaptation58. However, the different estimates between ancestry groups is not likely explained by differences in allele frequency, as discussed in a systematic review of genetic variation across ethnic groups30. Future directions for assessing cancer risk associations between ancestry groups would benefit from the identification and use of more tightly linked tagging SNPs, as well as validation of susceptibility loci for traits of interest within each ethnic group. Another valuable approach that complements variant-level association testing is to apply a gene-based association framework. A gene-based analysis of osteosarcoma risk recently reported 217 genes that achieved genome-wide significance, implicating numerous new regions of interest.59 Although our analyses involved single-locus association testing and therefore is not an appropriate framework for replicating gene-based analyses, our observation that a telomere maintenance pathway may contribute to osteosarcoma polygenicity complements findings from Yang, et al.59
In summary, we observed an association between genetic predisposition to longer LTL and increased pediatric osteosarcoma risk in a multi-ethnic case-control study. Results using a polygenic LTL score comprised of known LTL variants, a Mendelian randomization analysis, and single SNP analyses highlighted the aggregate role of genetic contributions to telomere length in conferring osteosarcoma risk. These results are consistent with the hypothesis that long telomeres – including alternative-lengthening of telomeres caused by germline ATRX deficiency – are a risk factor for osteosarcoma. A strength of our study is the diversity of populations included in the analysis. Despite the higher incidence of osteosarcoma among African-Americans and Hispanic children, few studies to-date have investigated potential genetic factors underlying osteosarcoma in populations other than non-Hispanic whites. Our findings shed light on the replicability of such genetic associations across multiple ethnicities and motivate the further characterization of these genetic factors in populations that are broadly generalizable to diverse clinical cohorts.
Supplementary Material
Key Resources Table.
| Resource | Source | Identifier |
|---|---|---|
| software | ||
| Plink | https://www.cog-genomics.org/plink/1.9/ | N/A |
Highlights.
Genetic predisposition to longer telomere length is a risk factor for childhood osteosarcoma in a multiethnic case-control population
GWAS loci for osteosarcoma risk and metastasis discovered in European-ancestry populations were not replicated in this multi-ethnic dataset
Polygenic modifiers of osteosarcoma risk should be studied in racially and ethnically diverse patient samples
ACKNOWLEDGEMENTS
The biospecimens and/or data used in this study were obtained from the California Biobank Program (SIS request number 550). The California Department of Public Health is not responsible for the results or conclusions drawn by the authors of this publication. The collection of cancer incidence data used in this study was supported by the California Department of Public Health pursuant to California Health and Safety Code Section 103885; Centers for Disease Control and Prevention’s (CDC) National Program of Cancer Registries, under cooperative agreement 5NU58DP006344; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201800032I awarded to the University of California, San Francisco, contract HHSN261201800015I awarded to the University of Southern California, and contract HHSN261201800009I awarded to the Public Health Institute, Cancer Registry of Greater California. The ideas and opinions expressed herein are those of the author(s) and do not necessarily reflect the opinions of the State of California, Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors.
FUNDING
This work was supported by the National Institutes of Health T32 CA151022-06 (CZ) and R01 CA155461 (JLW), The Children’s Health and Discovery Initiative of Translating Duke Health (JHH, KMW), and by two ‘A’ Awards from The Alex’s Lemonade Stand Foundation (AJdS, KMW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Savage SA, Mirabello L. Using epidemiology and genomics to understand osteosarcoma etiology. Sarcoma. 2011;2011:548151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009. April 1;115(7):1531–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat Rev Endocrinol. 2017;13(8):480–491. [DOI] [PubMed] [Google Scholar]
- 4.Savage SA, Mirabello L, Wang Z, et al. Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat Genet. 2013. July;45(7):799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mirabello L, Koster R, Moriarity BS, et al. A Genome-Wide Scan Identifies Variants in NFIB Associated with Metastasis in Patients with Osteosarcoma. Cancer Discov. 2015. September;5(9):920–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirabello L, Yu K, Berndt SI, et al. A comprehensive candidate gene approach identifies genetic variation associated with osteosarcoma. BMC Cancer. 2011. May 29;11:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savage SA, Woodson K, Walk E, et al. Analysis of genes critical for growth regulation identifies Insulin-like Growth Factor 2 Receptor variations with possible functional significance as risk factors for osteosarcoma. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2007. August;16(8):1667–1674. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C, Morimoto LM, de Smith AJ, et al. Genetic determinants of childhood and adult height associated with osteosarcoma risk. Cancer [Internet]. 0(0). Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/cncr.31645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C, Wiemels JL, Hansen HM, et al. Two HLA class II gene variants are independently associated with pediatric osteosarcoma risk. Cancer Epidemiol Biomark Amp Prev [Internet]. 2018. January 1; Available from: http://cebp.aacrjournals.org/content/early/2018/07/21/1055-9965.EPI-18-0306.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh KM, Whitehead TP, de Smith AJ, et al. Common genetic variants associated with telomere length confer risk for neuroblastoma and other childhood cancers. Carcinogenesis. 2016. June;37(6):576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mirabello L, Richards EG, Duong LM, et al. Telomere length and variation in telomere biology genes in individuals with osteosarcoma. Int J Mol Epidemiol Genet. 2011. January 1;2(1):19–29. [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Bahrami A, Pappo A, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014. April 10;7(1):104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shawi M, Autexier C. Telomerase, senescence and ageing. Mech Ageing Dev. 2008. January;129(1–2):3–10. [DOI] [PubMed] [Google Scholar]
- 14.Blackburn EH, Epel ES, Lin J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science. 2015. December 4;350(6265):1193–1198. [DOI] [PubMed] [Google Scholar]
- 15.Cawthon RM, Smith KR, O’Brien E, Sivatchenko A, Kerber RA. Association between telomere length in blood and mortality in people aged 60 years or older. The Lancet. 2003. February 1;361(9355):393–395. [DOI] [PubMed] [Google Scholar]
- 16.Haycock PC, Heydon EE, Kaptoge S, Butterworth AS, Thompson A, Willeit P. Leucocyte telomere length and risk of cardiovascular disease: systematic review and meta-analysis. BMJ [Internet]. 2014. July 8;349 Available from: http://www.bmj.com/content/349/bmj.g4227.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dean SG, Zhang C, Gao J, et al. The association between telomere length and mortality in Bangladesh. Aging. 2017;9(6):1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wentzensen IM, Mirabello L, Pfeiffer RM, Savage SA. The Association of Telomere Length and Cancer: a Meta-analysis. Cancer Epidemiol Biomarkers Prev. 2011. June 1;20(6):1238–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J-J, Nam C-E, Cho S-H, Park K-S, Chung I-J, Kim H-J. Telomere length shortening in non-Hodgkin’s lymphoma patients undergoing chemotherapy. Ann Hematol. 2003. August 1;82(8):492–495. [DOI] [PubMed] [Google Scholar]
- 20.Schroder CP, Wisman GBA, de Jong S, et al. Telomere length in breast cancer patients before and after chemotherapy with or without stem cell transplantation. Br J Cancer. 2001. May 18;84(10):1348–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meeker AK, Hicks JL, Iacobuzio-Donahue CA, et al. Telomere Length Abnormalities Occur Early in the Initiation of Epithelial Carcinogenesis. Clin Cancer Res. 2004. May 15;10(10):3317–3326. [DOI] [PubMed] [Google Scholar]
- 22.Haycock PC, Burgess S, Nounu A, et al. Association Between Telomere Length and Risk of Cancer and Non-Neoplastic Diseases: A Mendelian Randomization Study. JAMA Oncol. 2017. February 23; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iles MM, Bishop DT, Taylor JC, et al. The effect on melanoma risk of genes previously associated with telomere length. J Natl Cancer Inst. 2014. October;106(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh KM, Codd V, Rice T, et al. Longer genotypically-estimated leukocyte telomere length is associated with increased adult glioma risk. Oncotarget. 2015. December 15;6(40):42468–42477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ojha J, Codd V, Nelson CP, et al. Genetic variation associated with longer telomere length increases risk of chronic lymphocytic leukemia. Cancer Epidemiol Biomarkers Prev [Internet]. 2016. May 13; Available from: http://cebp.aacrjournals.org/content/early/2016/05/13/1055-9965.EPI-15-1329.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierce BL, Kraft P, Zhang C. Mendelian randomization studies of cancer risk: a literature review. Curr Epidemiol Rep. 2018. June;5(2):184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown L, Needham B, Ailshire J. Telomere Length Among Older U.S. Adults: Differences by Race/Ethnicity, Gender, and Age. J Aging Health. 2017;29(8):1350–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch SM, Peek MK, Mitra N, et al. Race, Ethnicity, Psychosocial Factors, and Telomere Length in a Multicenter Setting. PloS One. 2016;11(1):e0146723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ottaviani G, Jaffe N. The Epidemiology of Osteosarcoma In: Jaffe N, Bruland OS, Bielack S, editors. Pediatr Adolesc Osteosarcoma [Internet]. Boston, MA: Springer US; 2010. p. 3–13. Available from: 10.1007/978-1-4419-0284-9_1 [DOI] [Google Scholar]
- 30.Jing L, Su L, Ring BZ. Ethnic background and genetic variation in the evaluation of cancer risk: a systematic review. PloS One. 2014;9(6):e97522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiemels JL, Walsh KM, de Smith AJ, et al. GWAS in childhood acute lymphoblastic leukemia reveals novel genetic associations at chromosomes 17q12 and 8q24.21. Nat Commun. 2018. 18;9(1):286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarthy S, Das S, Kretzschmar W, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016. October;48(10):1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naumov VA, Generozov EV, Solovyov YN, Aliev MD, Kushlinsky NE. Association of FGFR3 and MDM2 gene nucleotide polymorphisms with bone tumors. Bull Exp Biol Med. 2012. October;153(6):869–873. [DOI] [PubMed] [Google Scholar]
- 34.Musselman JR, Bergemann TL, Ross JA, et al. Case-parent analysis of variation in pubertal hormone genes and pediatric osteosarcoma: a Children’s Oncology Group (COG) study. Int J Mol Epidemiol Genet. 2012;3(4):286–293. [PMC free article] [PubMed] [Google Scholar]
- 35.Savage SA, Burdett L, Troisi R, et al. Germ-line genetic variation of TP53 in osteosarcoma. Pediatr Blood Cancer. 2007. July;49(1):28–33. [DOI] [PubMed] [Google Scholar]
- 36.Codd V, Nelson CP, Albrecht E, et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet. 2013. April;45(4):422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fitzpatrick AL, Kronmal RA, Gardner JP, et al. Leukocyte Telomere Length and Cardiovascular Disease in the Cardiovascular Health Study. Am J Epidemiol. 2007. January 1;165(1):14–21. [DOI] [PubMed] [Google Scholar]
- 38.Codd V, Mangino M, van der Harst P, et al. Common variants near TERC are associated with mean telomere length. Nat Genet. 2010. March;42(3):197–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Endicott AA, Morimoto LM, Kline CN, Wiemels JL, Metayer C, Walsh KM. Perinatal factors associated with clinical presentation of osteosarcoma in children and adolescents. Pediatr Blood Cancer. 2017. June 1;64(6):e26349–n/a. [DOI] [PubMed] [Google Scholar]
- 40.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017. 01;46(6):1734–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sirugo G, Williams SM, Tishkoff SA. The Missing Diversity in Human Genetic Studies. Cell. 2019. March 21;177(1):26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang C, Chen H, Shao L, Dong Y. GRM4 gene polymorphism is associated with susceptibility and prognosis of osteosarcoma in a Chinese Han population. Med Oncol Northwood Lond Engl. 2014. July;31(7):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K, Zhao J, He M, Fowdur M, Jiang T, Luo S. Association of GRM4 gene polymorphisms with susceptibility and clinicopathological characteristics of osteosarcoma in Guangxi Chinese population. Tumour Biol J Int Soc Oncodevelopmental Biol Med. 2016. January;37(1):1105–1112. [DOI] [PubMed] [Google Scholar]
- 44.Marko TA, Diessner BJ, Spector LG. Prevalence of Metastasis at Diagnosis of Osteosarcoma: An International Comparison. Pediatr Blood Cancer. 2016. June;63(6):1006–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morgan TM, Krumholz HM, Lifton RP, Spertus JA. Nonvalidation of reported genetic risk factors for acute coronary syndrome in a large-scale replication study. JAMA. 2007. April 11;297(14):1551–1561. [DOI] [PubMed] [Google Scholar]
- 46.Walsh KM, Anderson E, Hansen HM, et al. Analysis of 60 reported glioma risk SNPs replicates published GWAS findings but fails to replicate associations from published candidate-gene studies. Genet Epidemiol. 2013. February;37(2):222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saferali A, Lee J, Sin DD, Rouhani FN, Brantly ML, Sandford AJ. Longer Telomere Length in COPD Patients with α1-Antitrypsin Deficiency Independent of Lung Function. PLoS ONE. 2014. April 24;9(4):e95600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alder JK, Chen JJ-L, Lancaster L, et al. Short telomeres are a risk factor for idiopathic pulmonary fibrosis. Proc Natl Acad Sci. 2008. September 2;105(35):13051–13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daniali L, Benetos A, Susser E, et al. Telomeres shorten at equivalent rates in somatic tissues of adults. Nat Commun. 2013. March 19;4:1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grinde KE, Qi Q, Thornton TA, et al. Generalizing Genetic Risk Scores from Europeans to Hispanics/Latinos. bioRxiv [Internet]. 2018. January 1; Available from: http://biorxiv.org/content/early/2018/01/04/242404.abstract [Google Scholar]
- 51.Martin AR, Gignoux CR, Walters RK, et al. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am J Hum Genet. 2017. April 6;100(4):635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klimentidis YC, Wineinger NE, Vazquez AI, de Los Campos G. Multiple metabolic genetic risk scores and type 2 diabetes risk in three racial/ethnic groups. J Clin Endocrinol Metab. 2014. September;99(9):E1814–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beecham AH, Wang L, Vasudeva N, et al. Utility of blood pressure genetic risk score in admixed Hispanic samples. J Hum Hypertens. 2016. December;30(12):772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perry DJ, Wasserfall CH, Oram RA, et al. Application of a Genetic Risk Score to Racially Diverse Type 1 Diabetes Populations Demonstrates the Need for Diversity in RiskModeling. Sci Rep. 2018. March 14;8(1):4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lazarides AL, Visgauss JD, Nussbaum DP, et al. Race is an independent predictor of survival in patients with soft tissue sarcoma of the extremities. BMC Cancer. 2018. 27;18(1):488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delgado DA, Zhang C, Chen LS, et al. Genome-wide association study of telomere length among South Asians identifies a second RTEL1 association signal. J Med Genet. 2018. January 1;55(1):64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y, Cao L, Li Z, et al. A genome-wide association study identifies a locus on TERT for mean telomere length in Han Chinese. PloS One. 2014;9(1):e85043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hansen MEB, Hunt SC, Stone RC, et al. Shorter Telomere Length in Europeans than in Africans due to Polygenetic Adaptation. Hum Mol Genet [Internet]. 2016. March 2; Available from: http://hmg.oxfordjournals.org/content/early/2016/03/01/hmg.ddw070.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Y, Basu S, Mirabello L, Spector L, Zhang L. A Bayesian Gene-Based Genome-Wide Association Study Analysis of Osteosarcoma Trio Data Using a Hierarchically Structured Prior. Cancer Inform. 2018;17:1176935118775103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.