Abstract
Background/Aim: Colon cancer is prone to distant metastases to other sites and the risk of recurrence is relatively high. Therefore, the identification of liver metastasis-related factors is important for the diagnosis or treatment of colon cancer. The aim of this study was to identify the metastasis-related factors that are differentially expressed in synchronous solitary liver metastasis compared to primary colon cancer. Materials and Methods: Tissues of primary colon cancer and associated with liver metastases of five patients were used for mass spectrometry. Identified proteins were validated by western blotting. The in silico analysis was performed using the STRING database and GeneMANIA. Results: We identified 58 differentially expressed proteins (DEPs), including 51 under-expressed and 7 over-expressed proteins among a total of 164 identified proteins. Major hubs of protein-protein networks were ACTC1, PRDX6, TPI1, and ALDH1A1. DEPs were located in the extracellular region and cytoplasm and were involved in the regulation of enzymatic activity. The metabolic process was significantly enriched in biological processes and an involvement in the KEGG pathway. Conclusion: These DEPs can potentially be used as biomarkers for the diagnosis of liver metastasis and they may provide a new strategy for developing anti-metastatic liver drugs in colon cancer patients.
Keywords: Colon cancer, differentially expressed proteins, gene ontology, liver metastatic cancer, mass spectrometry analysis, protein-protein interactions
Colon and rectum are one of the most common organs of cancer development, and malignant tumors of these sites are associated with high mortality rates in both men and women in Korea (1). Tumor evolution is a critical factor in chemo-responsiveness and the clinical outcome of stage IV colon cancer. More than 50% of colorectal cancer patients either have liver metastases at the time of diagnosis or subsequent metastasis develops soon after (2-4). Currently, hepatic resection is accepted as the most effective treatment for patients with liver-restricted colorectal metastases. However, experimental demonstrations of therapeutic heterogeneity associated with proteomic signatures are limited.
Progression of metastatic cancer has a decreased level of E-cadherin, or ZO-1 and increased level of N-cadherin, vimentin, Snail, and Slug (5,6). Despite the fact that these characteristics are known in the research field, most of these factors are hard to apply clinically in human biopsy samples, mainly due to difficulties in accuracy of the results and technological limitations. Thus, investigation of accessible colon cancer biomarkers or drug targets to predict liver metastasis is necessary to propose a guideline for suitable treatment or diagnosis. The expression levels of nine proteins have been detected in the SW620 and SW480 metastatic colorectal cancer cell lines using two-dimensional (2D) differential gel electrophoresis and mass spectrometry (7), while enrichment analysis of the extracellular matrix from patient samples was determined using proteomics (8). These previous studies have focused on differentially expressed proteins (DEPs), wherein the expression of factors in primary colon cancer is different from that in normal colon. However, identification of DEPs by comparative analysis between primary colon cancer and synchronous solitary liver metastasis is currently poorly studied.
With the aim of finding liver metastasis-related factors, our study has concentrated on the identification of DEPs in liver metastasis using mass spectrometry analysis. The aspect of differential expression was compared between primary colon and liver metastatic cancer. Our study indicates that identification of DEPs provides a set of potential diagnostic biomarkers and possible targets for anti-liver metastatic drugs.
Materials and Methods
Materials. All chemicals (iodoacetamide, 4-Sulfophenyl isothiocyanate, acetonitrile, a-cyano-4-hydroxycinnamicacid (CHCA), sodium bicarbonate, urea, bis-acrylamide, trifluoroacetic acid, thiourea, ammonium bicarbonate, Bradford solution, acrylamide, 3-[(3-cholamidopropy) dimethyammonio]-1-propanesulfonate (CHAPS), SDS, DTT, benzamidine, and α-cyano-4-hydroxycinnamic acid) were in grade of electrophoresis or analytical and were purchased from Sigma (St. Louis, MO, USA). Pharmalyte (pH 3.5-10) was purchased from Amersham Biosciences (Piscataway, NJ, USA). Modified porcine trypsin of sequencing grade was purchased from Promega (Madison, WI, USA). Antibodies against carbonic anhydrase 1 (CA1, sc-393490), Serpin A1 (sc-59438), N-acetylneuraminate synthase (NANS, sc-374133), transferrin (sc-365871), and GAPDH (sc-47724) were all purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA)
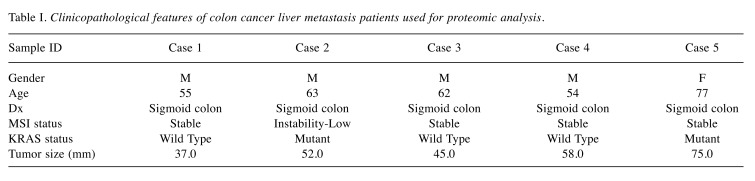
Biopsy samples. The study proposal was finalized and approved by the institutional review board of Gil Hospital, Gachon University (IBR No. GIRBA2216), prior to the investigation. Written informed consent was obtained from all participants. The samples were retrieved from the Gil Hospital, Gachon University (Incheon, Republic of Korea) between June 2015 and June 2017. The eligibility criteria for hepatic metastasis samples are as follows: i) synchronous colon cancer and single hepatic metastasis confirmed by spiral abdomino-pelvic computed tomography and liver MRI, ii) no evidence of tomography or other metastasis following positron emission tomography iii) liver metastasis as the first manifestation of M1 disease without contagious disease due to preoperative imaging, iv) no history of liver-specific therapy, v) histologically proven colorectal carcinoma, and vi) 18 years of age of older. Five of the 25 patients selected for hepatectomy specimens satisfied these criteria and progressed for analysis. Their characteristics are shown in Table I.
Table I. Clinicopathological features of colon cancer liver metastasis patients used for proteomic analysis.
Protein sample preparation for proteomics. Tissue samples were lysed in sample lysis solution (2% pharmalyte, 2 M thiourea, 4%(w/v) CHAPS, 1 mM benzamidine, 1% dithiothreitol, and 7 M urea) using a motor-driven PowerGen125 homogenizer (Fisher Scientific, Pittsburgh, PA, USA). Proteins were extracted by vortexing for 10 minutes at room temperature. Samples were centrifuged at 15,000 × g for 1 hour at 15˚C. Subsequently, the insoluble fractions were discarded and the soluble material was analyzed using 2D gel electrophoresis. Detection of protein concentration was analyzed using the Bradford method (9).
Two-dimensional PAGE and image analysis. 200 μg from each sample was loaded in IPG dry strips (24 cm, 4-10 NL IPG, Genomine, Republic of Korea) which were equilibrated for 12-16 hours with strip solution (1% pharmalyte, 1% DTT, 7 M urea and, 2 M thiourea with 2% CHAPS). Following manufacturer’s instruction, isoelectric focusing was operated at 20˚C using a Multiphor II electrophoresis system and power supply (EPS 3500 XL, Amersham Biosciences). For isoelectric focusing, the voltage increased linearly from 150 to 3,500 V for 3 hours after entering the sample, then it was constantly applied at 3,500 V, with focusing complete 96 kVh later. Prior to the 2D, strips were incubated in the equilibration buffer (6 M urea, 30% glycerol, 50 mM Tris-Cl, pH 6.8, and 2% SDS) for 10 minutes, first with 1% DTT and second with 2.5% iodoacetamide. The equilibrated strips were embedded onto SDS-PAGE gels (10-16%, 20×24 cm). SDS-PAGE gels were run with Hoefer DALT 2D systems (Amersham Biosciences), following manufacturer instructions. 2D gels were performed at 20˚C for 1,700 Vh and were then stained using Coomassie brilliant Blue G250 (CBB) (10), omitting the fixation and sensitization steps with glutaraldehyde.
Quantitative analysis of digitized images was subjected using PDQuest (version 7.0, BioRad, Hercules, CA, USA) according to the manufacturer’s protocol. The quantity of each spot was normalized by the total effective spot intensity. For significant expression, protein spots were selected variations that deviated more than twice the expression level compared to normal samples.
Mass spectrometry and analysis of heat map. Protein spots were cut out and were digested using trypsin. Trypsinized protein spots were mixed with α-cyano-4-hydroxycinnamic acid in 50% acetonitrile/ 0.1% TFA for protein identification by peptide mass fingerprinting and were analyzed by Microflex LRF 20 MALDI-TOF analysis (Bruker Daltonics, Billerica, MA, USA) (11).
The spectra were taken at 300 shots per spectrum over the m/z range 600-3,000 and corrected with a two-point internal calibration by trypsin auto-digestion peaks (m/z 842.5099, 2211.1046). A list of peaks was created using Flex Analysis 3.0 (Bruker Daltonics). The thresholds used for peak-picking were as follows: the minimum resolution of monoisotopic mass was 500 and the S/N was 5. MASCOT, a search program developed by Matrixscience (http://www.matrixscience.com/), was used to identify proteins using peptide mass fingerprinting. The following parameters were used in the database search: i) trypsin digestion, ii) a maximum of one lost cleavage, iii) a complete modification of 2-iodoacetamide (Cys), iv) oxidation (Met) as partial modification, v) monoisotopic masses, and vi) a mass tolerance of ±0.1 Da. PMF acceptance criteria were used in the probability score calculation. The heat map of 58 DEPs was generated by average linkage clustering using online Heatmapper (http://www.heatmapper.ca/) (12,13).
Western blotting. Sample extracts were lysed in RIPA lysis buffer consisting of 50 mM Tris (pH 8.0), 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, and 1X protease inhibitor mitxture (GenDEPOT, Barker, TX, USA). Tissue samples were incubated in lysis buffer for 30 minutes at 4˚C. We centrifuged lysates at 12,000 × g for 15 minutes at 4˚C. To measure protein concentrations in the collected supernatants the method of Bradford protein assay was used (Bio-Rad). Equal concentrations of protein were loaded in 12% SDS-PAGE gel and, were then transferred onto nitrocellulose membranes (Pall Gelman Laboratory, MI, USA). Prior to probing with primary the antibody at 4˚C overnight, the membranes were blocked with 8% skimmed milk or 5% bovine serum albumin. Following washing, the membranes were incubated with a peroxidase-conjugated 2nd antibody. The membranes were developed using the Amersham ECL western blotting detection reagent.
Analysis of protein-protein interaction (PPI) and gene ontology by STRING. The 58 identified DEPs were analyzed and visualized to predict PPI using the STRING software [v10.5, http://string-db.org, (14)], which employs search tools for multiple proteins. Network data from the STRING database displayed a combination of data, including text mining, neighborhood, co-expression, co-occurrence, gene fusion, and experiments. The score of minimum required interaction was medium confidence (0.400). The pathways of 58 DEPs were analyzed using GeneMANIA (15,16).
Gene Ontology (GO) by STRING was used to classify DEPs according to functional enrichments. The network of 58 DEPs was analyzed with regard to: i) cellular component, ii) molecular function, iii) biological process, and iv) KEGG pathways.
Results
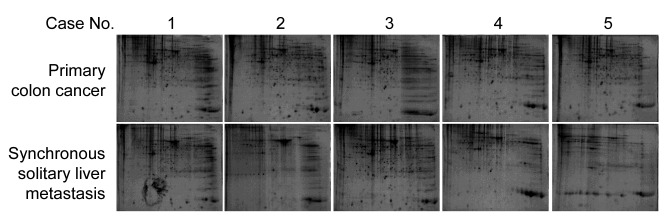
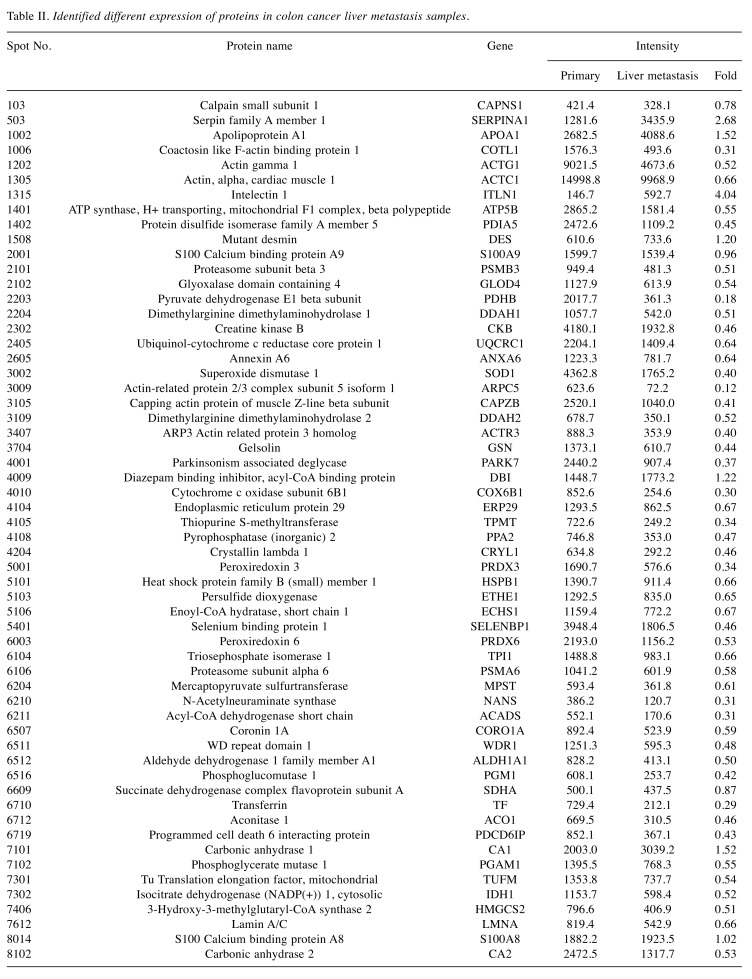
Identification of differentially expressed proteins between primary colon cancer and liver metastasis cancer. To identify distinct biomarkers for liver metastatic colon cancer, tissue lysates of primary colon cancer and liver metastasis cancer were fractionated on a 2D-PAGE gel (Figure 1). Analysis of the extracted peptides was performed using mass spectrometry. In total, 164 individual spots showed DEPs between primary colon cancer and synchronous solitary liver metastasis. Seven DEPs had higher synchronous solitary liver metastasis compared to primary colon cancer (Table II). Fifty-one DEPs had lower expression for liver metastasis compared to primary colon cancer. Figure 2 shows the heatmap of protein expression differences between primary colon cancer and liver metastatic colon cancer.
Figure 1. 2D-PAGE gel of primary colon cancer and synchronous solitary liver metastatic cancer samples from 5 patients. The protein expression patterns of the primary colon cancer and synchronous solitary liver metastasis tissues were represented by Coommassie blue staining after 2DPAGE.
Table II. Identified different expression of proteins in colon cancer liver metastasis samples.
Figure 2. Heat map analysis of 58 differentially expressed proteins (DEPs) from mass spectrometry. The relative levels of the 58 proteins found to exhibit differential expression between primary colon cancer and the isolated liver metastases were expressed as expression-based heat maps. The Y-axis is a list of 58 DEPs.

Western blotting analysis of liver metastasis-related factors. To validate the results of mass spectrometry analysis, we performed western blotting for detection of liver metastasis-related proteins (Figure 3). We randomly selected two factors of increased and decreased proteins. Serpin A1 and CA1 are as representative of up-regulated markers in liver metastasis. Serpin A1 is a serine protease inhibitor related to tumor migration. (17). Expression of Serpin A1 by mass spectrometry analysis showed a 2.68-fold increase in metastatic liver cancer over the level in primary colon cancer (Table II). Western blot analysis of Serpin A1 showed also 1.7- or 3.6-fold increase in metastatic liver cancer samples (M of cases 4 and 5) compared to that of primary colon cancer samples (P of cases 4 and 5). CA1 is a zinc metalloenzyme that catalyzes of the reversible hydration of carbon dioxide (18). Mass spectrometry analysis of CA1 showed a 1.52-fold increase in metastatic liver cancer samples (Table II). Immunoblotting of CA1 showed 1.1-fold and 7.5-fold increases in metastatic liver cancer cases 4 and 5, respectively, compared to their levels in primary colon cancer in these cases.
Figure 3. Validation of liver metastasis-related factors. Western blot analysis showed that CA1 and Serpin A1 were up-regulated and that NANS and transferrin were down-regulated in liver metastases (M) compared to primary colon cancer (P).
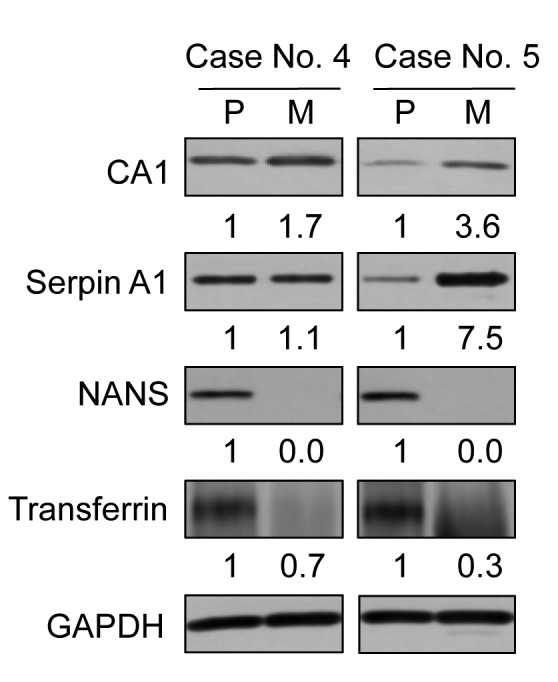
The selected decreased factors in liver metastasis were NANS and transferrin. NANS works as an enzyme in the biosynthetic pathways of sialic acids (19). In the results of mass spectrometry analysis, NANS was 0.31-fold lower in liver metastatic cancer compared to primary colon cancer (Table II). The protein levels of NANS by western blotting demonstrated almost absence of expression (0.1-fold or 0) in liver metastatic cancer. Transferrin has the function of transporting iron and manganese (20), and mass spectrometry analysis showed that it was 0.29-fold lower in liver metastatic cancer compared to primary colon cancer (Table II). Western blot analysis showed that the level of transferrin also decreased by 0.7-fold and 0.3-fold in metastatic liver cancer cases 4 and 5, respectively, compared to cases of primary colon cancer in these cases.
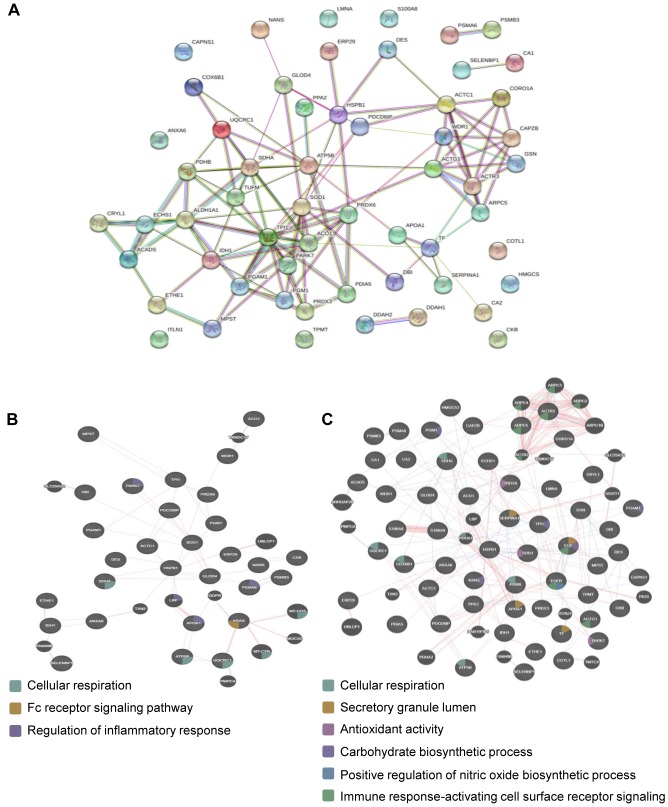
Protein-protein interaction analysis. To explore PPI networks among the identified proteins, the 58 identified DEPs were analyzed using the STRING software (Figure 4A). Eight of the 58 DEPs did not connect to any type of network (STRING interaction score=0.4). Forty-two of the DEPs were connected to networks by complex relationships. ACTC1, WDR1, HSPB1, SDHA, PRDX6, TPI1, and ALDH1A1 showed network hubs highly associated with other factors in PPI. In addition, single networks were formed between PSMA6 and PSMB3, SELENBP1 and CA1, S100A9 and S100A8, and DDAH2 and DDAH1.
Figure 4. Analysis of protein–protein interaction (PPI) by online bioinformatics. (A) PPI of DEPs analyzed against the STRING Database for association networks. Known interactions are edges of pink (experimentally determined) and deep sky blue (database obtained). Predicted interactions are edges of green (gene neighborhood), blue (gene co-occurrence), and red (gene fusions). Edges of yellowish green are text-mining. Edges of black color mean co-expression. Edges of light purple mean protein homology. (B-C) Physical interaction and involvement of DEPs in the KRAS pathway (B) or EGF and EGFR (C), as analyzed by GeneMANIA.
Progression of colon cancer related with KRAS and epidermal growth factor receptor (EGFR) (21-23). The GeneMANIA database was used to evaluate the relationship of KRAS or EGFR signaling among 58 DEPs. The network analysis of DEPs showed that KRAS associated with the Fc receptor signaling pathway and physically interacted with UQCRC1 and GLOD4 (Figure 4B). The network between DEPs and epidermal growth factor (EGF) showed that EGF physically related with ACO1, COX6B1, GSN, HSPB1, and LMNA (Figure 4C). The network analysis of DEPs with EGFR showed that EGFR physically interacted with NANS, TPI1, S100A9, GCN, ARPC5, HSPB1, and PDCD6IP (Figure 4C). Common functions of TPI1, NANS, and EGF were involved in the carbohydrate biosynthetic process.
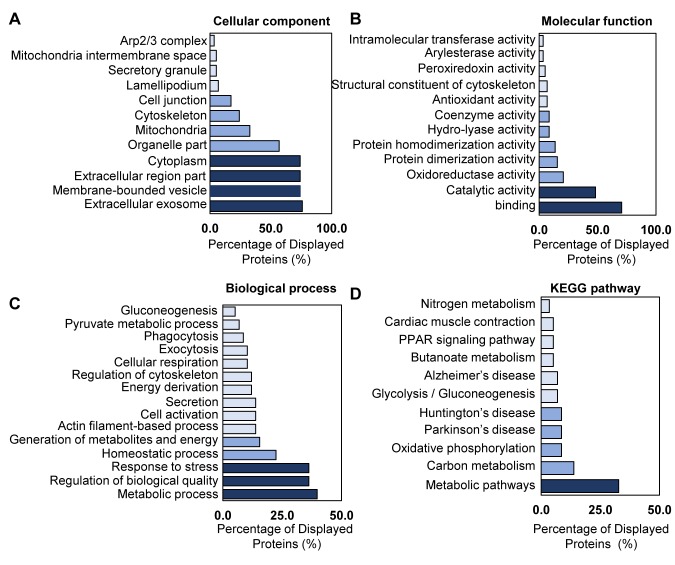
Gene ontology analysis of identified proteins in liver metastasis. To investigate gene ontology of the 58 DEPs, we used bioinformatics to profile the cellular component, biological process, molecular function, and involvement in the KEGG pathway. The GO analysis of 58 DEPs is shown in Figure 5. Most of the identified proteins were located in the cytoplasm, extracellular region including membrane-bound vesicles, and exosome (Figure 5A). Molecular function was primarily associated with binding or multiple-enzyme activity (Figure 5B). The biological process of DEPs belonged to: i) metabolic processes, ii) response to stress, and iii) homeostatic process (Figure 5C). According to the analysis of the biological process, the identified DEPs that were increased are involved in various cellular processes, including processes without actin filaments, phagocytosis, pyruvate metabolic processes, and gluconeogenesis (Figure 5C). This result matches the analysis of the cellular position and suggests that liver metastasis from colon cancer induces down-regulation of glucose-related metabolism; however, other cellular metabolisms were up-regulated. Most of the 58 DEPs had a role in the KEGG metabolic pathway (Figure 5D).
Figure 5. Analysis of GO annotation of 58 identified DEPs. A total of 58 proteins analyzed with regard to their cellular component (A) and molecular function (B). 58 DEPs were involved in a biological process (C) and in the KEGG pathway (D). These data were analyzed by the gene ontology from the STRING database.
Discussion
Patients with metastatic colorectal cancer with liver or lung metastases have a 5-year overall survival rate of 30-50% (24). Conventional chemotherapy for metastatic colorectal cancer with leucovorin and fluorouracil may extend the progression-free survival and overall survival, regardless of the use of oxaliplatin or irinotecan (25); however, the long-term results are not satisfactory. Therapeutic chemotherapy treatment for colon cancer with liver metastases is not currently possible. Stage IV colon cancer is a heterogeneous disease in terms of genomic and transcriptomic alterations with respect to patient survival and chemotherapy responses. Currently, the Colorectal Cancer Subtyping Consortium suggests four consensus molecular subtypes (CMS) that combines six independent classification systems based on gene expression analysis: i) CMS1 (MSI immune subtype; strong immune activation, microsatellite unstable, and hypermutated), ii) CMS2 (canonical subtype; MYC and WNT signal pathway activation), iii) CMS3 (metabolic subtype; metabolic dysregulation), and iv) CMS4 (mesenchymal subtype; TGF-β activation, stromal infiltration and angiogenesis) (26).
Despite this classification, CRCSC do not include an evaluation of the concordance and shifts of CMS calls in primary and matched metastatic samples, as well as the predictive value of the CRC subtypes. It is absolutely critical to understand that their marked biological differences are likely to develop new targeted therapies in CRC.
In this study, we investigated tumor metastatic contributors from primary colon cancer to liver metastasis using a proteomics approach. Samples of patients were chosen from primary colon cancer tissue to liver metastasis tissue. The 164 identified proteins included factor expression levels that were higher, lower, and unchanged. Proteins considerably increased the expression are: i) SERPINA1, ii) APOA1, iii) ITLN1, iv) DES, v) DBI, vi) SDHA, and vii) CA1.
Protein-protein interaction data show that central proteins in the networks are: i) ACTC1, ii) WDR1, iii) HSPB1, iv) SDHA, v) PRDX6, vi) TPI1, and vii) ALDH1A1 (Figure 4). Unexpectedly, one of the hub proteins, SDHA, only belongs to increased DEPs. Others are involved in decreased DEPs. This result suggests that progression of metastasis induces imbalance in protein networks changing protein expression levels. These DEPs in liver metastasis tissue may be inducers of metastasis from the colon to liver or residue products of the liver-micro environment following the establishment of metastasis at a distant site. Alternative expression of proteins in primary colon cancer induces the possibility of quickly transitioning to a new progression status that may be involved in metastasis, drug resistance, cancer stem cell, supporting cell, and adapting cell.
The metastatic feature of cancer cells is their increasing energy metabolism and decreasing immune cell-related migration (27-29). Serpin A1 and ITLN1 are involved with the regulation of the innate immune system. Parts of decreased DEPs are related to energy metabolism. These data suggest that metastatic cancers have individual different conditions with regard to energy metabolism and immune system during tumor progression.
According to the analysis of cellular components, increased DEPs (SERPINA1, APOA1, ITLN1, DES, DBI, SDHA, and CA1) and decreased DEPs were located in various regions. The main location of increased DEPs is the extracellular region and exosome or in membrane-bounded vesicles. This characteristic means it such DEPs can be used as biomarkers for the detection of liver metastatic colon cancer using serum or colon samples. Especially, the level of Serpin A1 in serum is exceptionally high in patients with prostate, colorectal, lung, breast cancers, and insulinomas (30-35). However, the areas with decreased DEPs, include: i) the actin cytoskeleton, ii) lamellipodia, iii) the Arp2/3 protein complex, iv) the proteasome complex, v) the mitochondrial matrix, and vi) the mitochondrial intermembrane space. The function of actin cytoskeleton, lamellipodia, and the Arp2/3 protein complex helps maintain cell morphology and regulates vesicle trafficking using actin filament dynamics (36,37) while the proteasome complex contributes to the degradation of proteins (38). Cancer stem-like properties have low proteasomal activity in colorectal cancer, lung cancer, and prostate cancer (39-41). The mitochondrial matrix and mitochondrial intermembrane space play a major part of ATP synthesis in cells (42,43). Mitochondrial ATP synthase is down-regulated in colon cancer treated with 5-fluorouracil resistance (44). This result suggests that liver metastasis from colon cancer reduces actin dynamics, protein degradation, and ATP synthesis.
In conclusion, our study identified DEPs between synchronous solitary liver metastasis and primary colon cancer using mass spectrometry analysis from five patient samples. Analysis of DEPs determined up-regulation of 7 proteins and down-regulation of 51 proteins. Protein-protein interactions among DEPs shows that: i) ACTC1, ii) WDR1, iii) HSPB1, iv) SDHA, v) PRDX6, vi) TPI1, and vii) ALDH1A1 are connected with other proteins. Further studies are required to investigate the functional mechanism of these identified genes in liver metastasis progression of colon cancer. This study provides a set of useful biomarkers for diagnosis of liver metastasis, which may prove to be potential new targets for anti-metastatic liver cancer treatment in colon cancer.
Conflicts of Interest
The Authors declare that they have no conflicts of interest.
Authors’ Contributions
Conceptualization, EKK, WSL and HHJ; Investigation, EKK and MJS; Resources, YJJ, WSL and HHJ; Data Curation, EKK and MJS; Writing, EKK, WSL and HHJ; Funding Acquisition, WSL and HHJ.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (NRF-2017R1D1A1B03035453) to WSL.
References
- 1.Jung KW, Park S, Kong HJ, Won YJ, Lee JY, Seo HG, Lee JS. Cancer statistics in Korea: Incidence, mortality, survival, and prevalence in 2009. Cancer Res Treat. 2012;44(1):11–24. doi: 10.4143/crt.2012.44.1.11. PMID: 22500156. DOI: 10.4143/crt.2012.44.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee WS, Kim MJ, Yun SH, Chun HK, Lee WY, Kim SJ, Choi SH, Heo JS, Joh JW, Kim YI. Risk factor stratification after simultaneous liver and colorectal resection for synchronous colorectal metastasis. Langenbecks Arch Surg. 2008;393(1):13–19. doi: 10.1007/s00423-007-0231-0. PMID: 17909846. DOI: 10.1007/s00423-007-0231-0. [DOI] [PubMed] [Google Scholar]
- 3.Curley SA. Outcomes after surgical treatment of colorectal cancer liver metastases. Semin Oncol. 2005;32(6 Suppl 9):S109–111. doi: 10.1053/j.seminoncol.2005.06.011. PMID: 16399446. DOI: 10.1053/j.seminoncol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Choti MA, Sitzmann JV, Tiburi MF, Sumetchotimetha W, Rangsin R, Schulick RD, Lillemoe KD, Yeo CJ, Cameron JL. Trends in long-term survival following liver resection for hepatic colorectal metastases. Ann Surg. 2002;235(6):759–766. doi: 10.1097/00000658-200206000-00002. PMID: 12035031. DOI: 10.1097/00000658-200206000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. PMID: 24556840. DOI: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. PMID: 19487818. DOI: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katayama M, Nakano H, Ishiuchi A, Wu W, Oshima R, Sakurai J, Nishikawa H, Yamaguchi S, Otsubo T. Protein pattern difference in the colon cancer cell lines examined by two-dimensional differential in-gel electrophoresis and mass spectrometry. Surg Today. 2006;36(12):1085–1093. doi: 10.1007/s00595-006-3301-y. PMID: 17123137. DOI: 10.1007/s00595-006-3301-y. [DOI] [PubMed] [Google Scholar]
- 8.Naba A, Clauser KR, Whittaker CA, Carr SA, Tanabe KK, Hynes RO. Extracellular matrix signatures of human primary metastatic colon cancers and their metastases to liver. BMC Cancer. 2014;14:518. doi: 10.1186/1471-2407-14-518. PMID: 25037231. DOI: 10.1186/1471-2407-14-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brody EB, Ottey F, Lagranade J. Early sex education in relationship to later coital and reproductive behavior: Evidence from jamaican women. Am J Psychiatry. 1976;133(8):969–972. doi: 10.1176/ajp.133.8.969. PMID: 942015. DOI: 10.1176/ajp.133.8.969. [DOI] [PubMed] [Google Scholar]
- 10.Oakley BR, Kirsch DR, Morris NR. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980;105(2):361–363. doi: 10.1016/0003-2697(80)90470-4. PMID: 6161559. DOI: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez J, Gharahdaghi F, Mische SM. Routine identification of proteins from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (sds-page) gels or polyvinyl difluoride membranes using matrix assisted laser desorption/ionization-time of flight-mass spectrometry (maldi-tof-ms) Electrophoresis. 1998;19(6):1036–1045. doi: 10.1002/elps.1150190619. PMID: 9638950. DOI: 10.1002/elps.1150190619. [DOI] [PubMed] [Google Scholar]
- 12.Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016;44(W1):W147–153. doi: 10.1093/nar/gkw419. PMID: 27190236. DOI: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verhaak RG, Sanders MA, Bijl MA, Delwel R, Horsman S, Moorhouse MJ, van der Spek PJ, Lowenberg B, Valk PJ. Heatmapper: Powerful combined visualization of gene expression profile correlations, genotypes, phenotypes and sample characteristics. BMC Bioinformatics. 2006;7:337. doi: 10.1186/1471-2105-7-337. PMID: 16836741. DOI: 10.1186/1471-2105-7-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. String v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. PMID: 25352553. DOI: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montojo J, Zuberi K, Rodriguez H, Bader GD, Morris Q. Genemania: Fast gene network construction and function prediction for cytoscape. F1000Res. 2014;3:153. doi: 10.12688/f1000research.4572.1. PMID: 25254104. DOI: 10.12688/f1000research.4572.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The genemania prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–220. doi: 10.1093/nar/gkq537. PMID: 20576703. DOI: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Serres F, Blanco I. Role of alpha-1 antitrypsin in human health and disease. J Intern Med. 2014;276(4):311–335. doi: 10.1111/joim.12239. PMID: 24661570. DOI: 10.1111/joim.12239. [DOI] [PubMed] [Google Scholar]
- 18.Boone CD, Pinard M, McKenna R, Silverman D. Catalytic mechanism of alpha-class carbonic anhydrases: Co2 hydration and proton transfer. Subcell Biochem. 2014;75:31–52. doi: 10.1007/978-94-007-7359-2_3. PMID: 4146373. DOI: 10.1007/978-94-007-7359-2_3. [DOI] [PubMed] [Google Scholar]
- 19.Rangarajan ES, Ruane KM, Proteau A, Schrag JD, Valladares R, Gonzalez CF, Gilbert M, Yakunin AF, Cygler M. Structural and enzymatic characterization of nans (yjhs), a 9-o-acetyl n-acetylneuraminic acid esterase from Escherichia coli o157:H7. Protein Sci. 2011;20(7):1208–1219. doi: 10.1002/pro.649. PMID: 21557376. DOI: 10.1002/pro.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunter TE, Gerstner B, Gunter KK, Malecki J, Gelein R, Valentine WM, Aschner M, Yule DI. Manganese transport via the transferrin mechanism. Neurotoxicology. 2013;34:118–127. doi: 10.1016/j.neuro.2012.10.018. PMID: 23146871. DOI: 10.1016/j.neuro.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pabla B, Bissonnette M, Konda VJ. Colon cancer and the epidermal growth factor receptor: Current treatment paradigms, the importance of diet, and the role of chemoprevention. World J Clin Oncol. 2015;6(5):133–141. doi: 10.5306/wjco.v6.i5.133. PMID: 26468449. DOI: 10.5306/wjco.v6.i5.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markman B, Javier Ramos F, Capdevila J, Tabernero J. Egfr and kras in colorectal cancer. Adv Clin Chem. 2010;51:71–119. doi: 10.1016/s0065-2423(10)51004-7. PMID: 20857619. [DOI] [PubMed] [Google Scholar]
- 23.Heinemann V, Stintzing S, Kirchner T, Boeck S, Jung A. Clinical relevance of egfr- and kras-status in colorectal cancer patients treated with monoclonal antibodies directed against the egfr. Cancer Treat Rev. 2009;35(3):262–271. doi: 10.1016/j.ctrv.2008.11.005. PMID: 19117687. DOI: 10.1016/j.ctrv.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Steele G Jr., Bleday R, Mayer RJ, Lindblad A, Petrelli N, Weaver D. A prospective evaluation of hepatic resection for colorectal carcinoma metastases to the liver: Gastrointestinal tumor study group protocol 6584. J Clin Oncol. 1991;9(7):1105–1112. doi: 10.1200/JCO.1991.9.7.1105. PMID: 2045852. DOI: 10.1200/JCO.1991.9.7.1105. [DOI] [PubMed] [Google Scholar]
- 25.Saltz LB, Cox JV, Blanke C, Rosen LS, Fehrenbacher L, Moore MJ, Maroun JA, Ackland SP, Locker PK, Pirotta N, Elfring GL, Miller LL. Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. Irinotecan study group. N Engl J Med. 2000;343(13):905–914. doi: 10.1056/NEJM200009283431302. PMID: 11006366. DOI: 10.1056/NEJM200009283431302. [DOI] [PubMed] [Google Scholar]
- 26.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, Bot BM, Morris JS, Simon IM, Gerster S, Fessler E, De Sousa EMF, Missiaglia E, Ramay H, Barras D, Homicsko K, Maru D, Manyam GC, Broom B, Boige V, Perez-Villamil B, Laderas T, Salazar R, Gray JW, Hanahan D, Tabernero J, Bernards R, Friend SH, Laurent-Puig P, Medema JP, Sadanandam A, Wessels L, Delorenzi M, Kopetz S, Vermeulen L, Tejpar S. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. PMID: 26457759. DOI: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin X, Zhang Y, Guo S, Jin H, Wang W, Yang P. Large scale systematic proteomic quantification from non-metastatic to metastatic colorectal cancer. Sci Rep. 2015;5:12120. doi: 10.1038/srep12120. PMID: 26175278. DOI: 10.1038/srep12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parks SK, Chiche J, Pouyssegur J. Disrupting proton dynamics and energy metabolism for cancer therapy. Nat Rev Cancer. 2013;13(9):611–623. doi: 10.1038/nrc3579. PMID: 23969692. DOI: 10.1038/nrc3579. [DOI] [PubMed] [Google Scholar]
- 29.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. PMID: 24664755. DOI: 10.1007/978-1-4614-3209-8_82. [DOI] [PubMed] [Google Scholar]
- 30.Abbritti RV, Polito F, Cucinotta M, Lo Giudice C, Caffo M, Tomasello C, Germano A, Aguennouz M. Meningiomas and proteomics: Focus on new potential biomarkers and molecular pathways. Cancer Genomics Proteomics. 2016;13(5):369–379. PMID: 27566655. [PMC free article] [PubMed] [Google Scholar]
- 31.Da Costa GG, Gomig TH, Kaviski R, Santos Sousa K, Kukolj C, De Lima RS, De Andrade Urban C, Cavalli IJ, Ribeiro EM. Comparative proteomics of tumor and paired normal breast tissue highlights potential biomarkers in breast cancer. Cancer Genomics Proteomics. 2015;12(5):251–261. PMID: 26417028. [PubMed] [Google Scholar]
- 32.Perez-Holanda S, Blanco I, Menendez M, Rodrigo L. Serum concentration of alpha-1 antitrypsin is significantly higher in colorectal cancer patients than in healthy controls. BMC Cancer. 2014;14:355. doi: 10.1186/1471-2407-14-355. PMID: 24886427. DOI: 10.1186/1471-2407-14-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Akawi ZJ, Abu-Awad AM, Sharara AM, Khader Y. The importance of alpha-1 antitrypsin (alpha1-at) and neopterin serum levels in the evaluation of non-small cell lung and prostate cancer patients. Neuro Endocrinol Lett. 2010;31(1):113–116. PMID: 20150872. [PubMed] [Google Scholar]
- 34.El-Akawi ZJ, Al-Hindawi FK, Bashir NA. Alpha-1 antitrypsin (alpha1-at) plasma levels in lung, prostate and breast cancer patients. Neuro Endocrinol Lett. 2008;29(4):482–484. PMID: 18766166. [PubMed] [Google Scholar]
- 35.de Sa SV, Correa-Giannella ML, Machado MC, Krogh K, de Almeida MQ, Albergaria Pereira MA, Coelho Siqueira SA, Patzina RA, Ibuki FS, Sogayar MC, Machado MC, Giannella-Neto D. Serpin peptidase inhibitor clade a member 1 as a potential marker for malignancy in insulinomas. Clin Cancer Res. 2007;13(18 Pt 1):5322–5330. doi: 10.1158/1078-0432.CCR-06-1477. PMID: 17855650. DOI: 10.1158/1078-0432.CCR-06-1477. [DOI] [PubMed] [Google Scholar]
- 36.Zhou K, Sumigray KD, Lechler T. The arp2/3 complex has essential roles in vesicle trafficking and transcytosis in the mammalian small intestine. Mol Biol Cell. 2015;26(11):1995–2004. doi: 10.1091/mbc.E14-10-1481. PMID: 25833710. DOI: 10.1091/mbc.E14-10-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henson JH, Yeterian M, Weeks RM, Medrano AE, Brown BL, Geist HL, Pais MD, Oldenbourg R, Shuster CB. Arp2/3 complex inhibition radically alters lamellipodial actin architecture, suspended cell shape, and the cell spreading process. Mol Biol Cell. 2015;26(5):887–900. doi: 10.1091/mbc.E14-07-1244. PMID: 25568343. DOI: 10.1091/mbc.E14-07-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A. Structure and function of the 26s proteasome. Annu Rev Biochem. 2018;87:697–724. doi: 10.1146/annurev-biochem-062917-011931. PMID: 29652515. DOI: 10.1146/annurev-biochem-062917-011931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munakata K, Uemura M, Tanaka S, Kawai K, Kitahara T, Miyo M, Kano Y, Nishikawa S, Fukusumi T, Takahashi Y, Hata T, Nishimura J, Takemasa I, Mizushima T, Ikenaga M, Kato T, Murata K, Carethers JM, Yamamoto H, Doki Y, Mori M. Cancer stem-like properties in colorectal cancer cells with low proteasome activity. Clin Cancer Res. 2016;22(21):5277–5286. doi: 10.1158/1078-0432.CCR-15-1945. PMID: 27166395. DOI: 10.1158/1078-0432.CCR-15-1945. [DOI] [PubMed] [Google Scholar]
- 40.Della Donna L, Lagadec C, Pajonk F. Radioresistance of prostate cancer cells with low proteasome activity. Prostate. 2012;72(8):868–874. doi: 10.1002/pros.21489. PMID: 21932424. DOI: 10.1002/pros.21489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan J, Zhang Q, Wang Y, You M. 26s proteasome activity is down-regulated in lung cancer stem-like cells propagated in vitro. PLoS One. 2010;5(10):e13298. doi: 10.1371/journal.pone.0013298. PMID: 20949018. DOI: 10.1371/journal.pone.0013298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stuart RA, Gruhler A, van der Klei I, Guiard B, Koll H, Neupert W. The requirement of matrix atp for the import of precursor proteins into the mitochondrial matrix and intermembrane space. Eur J Biochem. 1994;220(1):9–18. doi: 10.1111/j.1432-1033.1994.tb18593.x. PMID: 8119302. DOI: 10.1111/j.1432-1033.1994.tb18593.x. [DOI] [PubMed] [Google Scholar]
- 43.Glick BS, Wachter C, Reid GA, Schatz G. Import of cytochrome b2 to the mitochondrial intermembrane space: The tightly folded heme-binding domain makes import dependent upon matrix atp. Protein Sci. 1993;2(11):1901–1917. doi: 10.1002/pro.5560021112. PMID: 8268801. DOI: 10.1002/pro.5560021112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin YK, Yoo BC, Chang HJ, Jeon E, Hong SH, Jung MS, Lim SJ, Park JG. Down-regulation of mitochondrial f1f0-atp synthase in human colon cancer cells with induced 5-fluorouracil resistance. Cancer Res. 2005;65(8):3162–3170. doi: 10.1158/0008-5472.CAN-04-3300. PMID: 15833846. DOI: 10.1158/0008-5472.CAN-04-3300. [DOI] [PubMed] [Google Scholar]