Abstract
Patients with pancreatic ductal adenocarcinoma have a dismall prognosis because at the time of diagnosis, in the vast majority of patients the tumor has already disseminated to distant organs and the therapeutic benefit of approved agents such as gemcitabine is limited. Therefore, the identification and preclinical and clinical validation of therapeutic agents covering new targets is of paramount importance. In this review we have summarized microRNAs and corresponding targets which affect growth and metastasis of pancreatic tumors in preclinical mouse in vivo models. We identified four up-regulated and 16 down-regulated miRs in PDAC in comparison to corresponding normal tissues. Three sub-categories of miRs have emerged: miRs affecting tumor growth and miRs with an impact on both, tumor growth and metastasis or metastasis only. Finally, we discuss technical and therapeutic aspects of miR-related therapeutic agents for the treatment of pancreatic ductal adenocarcinoma.
Keywords: Antagomirs, KRAS oncogene, miR sponges, orthotopic xenograft models, therapeutic miR-related agents, transforming growth factor β signaling, tumor suppressor genes, review
Pancreatic ductal adenocarcinoma (PDAC) comprises 85% of pancreatic tumors, is one of the most lethal solid human tumors with a yearly prevalence of 50,000 patients in the USA and a slighthly lower death rate (1). The incidence of PDAC is increasing and in 2020, PDAC is expected to cover the second most common cancer-related death rate after lung carcinoma (2). Only 10-20% of PDACs can be resected and it is an exception to find a resected carcinoma that is still limited to the primary tumor (3). The medium survival time after diagnosis is 3-10 months in patients with unresectable tumors and 10-20 months after resection (4). Gemcitabine together with 5-Fluoro-Uracil (5-FU) and susequently FOLFIRINOX (5-FU, irinotecan, oxaliplatin and leucovorin) as well as a nanobody-based formulation of paclitaxel (nab paclitaxel), give rise to slightly improved survival rates in comparison to gemcitabine, depending on the clinical scenario (5).
Activating mutations of Kirsten rat sarcoma viral homolog (K-RAS) and inactivating mutations of tumor suppressor genes (TS) such as cyclin-dependent kinase inhibitor 2A (CDKN2A), TP53 and SMAD family number 4 (SMAD4) have been identified as the most prominent drivers of pathogenesis and metastasis of PDAC (6,7). Mutated K-RAS leads to activation of oncogenic and pro-metastatic mitogen-activated protein kinase (MAPK), phosphoinosite-3-kinase (PI3K) and ral guanine nucleotide dissociation stimulator (RALGDS) pathways (6,7). Inactivating mutations in CDKN2A prevent inhibition of cell-cycle progression from the G1 phase to the S phase of the cell cycle and proteosomal degradation of p53 inhibitor mouse double minute 2 homolog (MDM2) (6,7). TP53 mutations interfere with G1/S and G2/M arrest and with inhibition of apoptosis (6,7). Inactivating mutations in SMAD4 impair the protein’s tumor suppressor anti-metastatic function (6,7). Further treatment-obstructive features of PDAC are: intratumoral heterogeneity, pronounced desmoplastic tumor micro-environment (TME), intrinsic resistance to chemotherapy, hypovascularitity and resistance to immuno-therapy (5,8,9). Another issue complicating treatment of PDAC is its dependance on a super-enhancer based regenerative program with cellular myelocytomatosus (c-MYC) as a switch engaging and maintaining the program (10). Although, the pathogenesis of PDAC starting with pancreatic intraepithelial neoplasia can be mimicked faithfully in transgenic mice, the impact of these models on the discovery of new drugs against PDAC remains to be validated (11,12). Therefore, the identification of new targets and modalities for the treatment of PDAC is of paramount importance (13,14). In this review we focus on the role of microRNAs (miRs) on the pathogenesis and metastasis of PDACs. We restricted the selection to PDAC-related miRs with documented efficacy in preclinical in vivo models.
Metastasis of PDAC
From a preclinical point of view, PDAC-derived exosomes have been shown to be involved in metastasis of PDAC to the liver. These exosomes contain macrophage inhibitory factor (MIF), which can induce liver metastasis by mediating pre-metastatic niche formation in the liver (15). Uptake of PDAC-derived exosomes by Kupffer cells causes secretion of TGFβ1 and enhancement of fibronectin secretion by hepatic stellate cells, which are responsible for the generation of a fibrotic TME and recruitment of bone-marrow-derived macrophages (15). Blockade of MIF prevents liver pre-metastatic niche formation and metastasis (15). MIF primes the liver for metastasis and is markedly higher expressed in exosomes from PDAC patients who later develop liver metastasis (15).
From a clinical point of view, the liver was found to be the most common site of metastasis, followed by the peritoneum and the lungs. Colonization of adrenal glands and bones comprise approximately 10% of metastatic disease derived from pancreatic cancer (16-18).
microRNAs: General Issues and Involvement in Cancer
microRNAs (miRs) are non-coding RNAs in the range of 22 to 25 nucleotides (nts) which regulate gene expression at the post-transcriptional level by inducing degradation or blocking translation of corresponding mRNAs (19,20). miRs are transcribed by RNA Pol II as about 700 nts comprising pri-miRNAs precursors in the nucleus and then cleaved by RNAse of type III DROSHA, from both ends to pre-miRs of 70-100 nts in length (21,22). The pre-miRs are subsequently transferred to the cytoplasm by carrier-protein exportin-5 (21,22). Finally, RNAse type III member DICER, which acts as an endoribonuclease, cleaves the stem-loop of the pre-miRs to yield shorter, double-stranded RNAs comprising 22 to 25 nts with two unpaired 3’ nts at each end (21,22). One of the strands is finally bound to the RNA-induced silencing complex (RISC) (21,22). The RISC-miR complex matches the target mRNA in its 3’-untranslated region (3’-UTR) and induces its degradation or inhibition of its translation (21,22). Approximately 1,000 miR-genes have been identified in humans (23). The transcripts are capped at their 5’-ends, polyadenylated at their 3’-ends and sometimes they are spliced. The corresponding genes can be localized in exons, introns or intergenic regions (23). It has emerged as an important principle that one type of miR can target several distinct mRNAs and a single mRNA can be cleaved by several different types of miRs, thus enabling interference with several cancer-related cellular networks with the potential of rewiring the transformed and metastatic state (24).
The involvement of miRs in the pathogenesis of cancer has been demonstrated by the involvement of miR-15a and miR-16-1 in the pathogenesis of B-cell chronic lymphatic leukemia (B-CLL) (25-27). It was observed that tumor suppressors on chromosome 13q14, a locus containing miR-15a and miR-16-1, are frequently deleted in patients with B-CLL. In a proof-of-concept experiment, deletion of miR-15a and miR-16-1 in mice recapitulated the human disease phenotype due to circumventing cleavage of the anti-apoptotic protein B-cell lymphoma 2 (BCL2) (25-27).
We have recently reviewed the role of miRs in metastasis of breast-, ovarian- and prostate cancer (28-30). In this review, we outline the role of miRs in the pathogenesis and metastasis of PDAC. We focus on miRs with documented in vivo efficacy in pathogenesis- or metastasis-related preclinical models of PDAC.
Growth-promoting miRs Up-regulated in PDAC
miRs discussed in this chapter enhance tumor growth. Since they have not been tested in metastasis-related in vivo models, some of them might also function as mediators of metastasis.
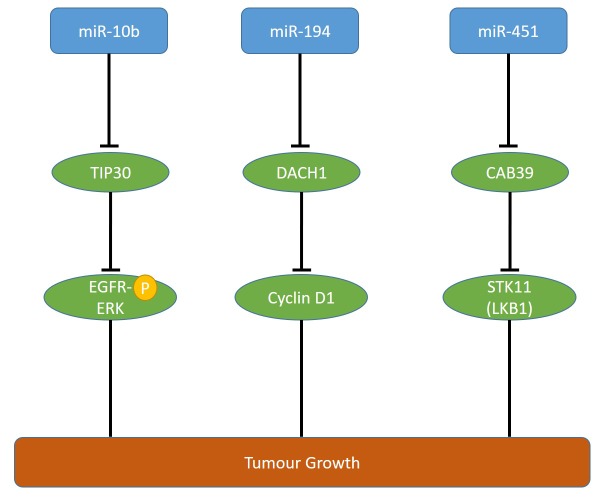
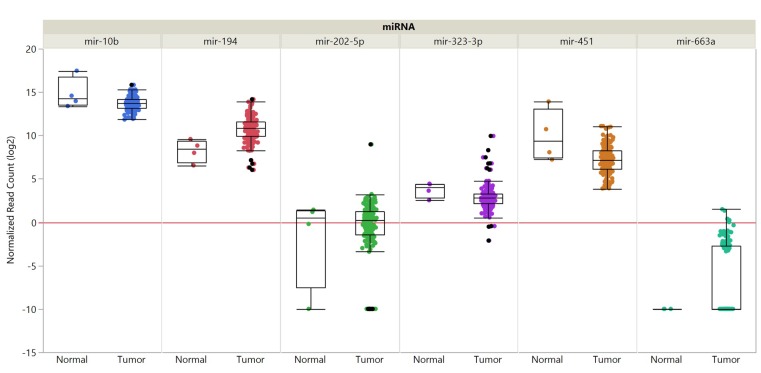
miR-10b steady-state levels are increased in PDAC patients in comparison to normal pancreatic tissues and its up-regulation correlates with a poor prognosis (31,32). miR-10 enhances epidermal growth factor (EGF)-dependent invasion and proliferation of PANC-1 and COLO-357 pancreatic carcinoma cells and targets TAT-interacting protein 30 (TIP30) directly (31). TIP30 regulates endocytosis of EGFR (31,33). miR-10b enhances the ability of EGF to activate MAPK and loss of TIP30 up-regulates epidermal growth factor receptor (EGFR) (Figure 1) (31,33). However, the mechanistic interactions underlying these phenomena have to be resolved in more detail. Pancreatic carcinoma derived T3M4 cells overexpressing miR-10b exhibit enhanced proliferation and tumor growth in an orthotopic xenograft model (31). Data derived from The Cancer Genome Atlas (TCGA) did not reveal tendency of increased steady-state levels of miR-101 in PDAC samples in comparison to normal pancreatic tissues (Figure 2).
Figure 1. Up-regulated miRs affecting growth of pancreatic ductal adenocarcinoma targeting tumor suppressors in preclinical in vivo models. miRs- 10b, -194 and -451 affect tumor growth via the outlined targets. CAB39: Calcium biding protein 39; DACH1: dachshund homolog 1; EGFR-P: phosphorylated epidermal growth factor receptor; ERK: extracellular signal regulated kinase 1; LKB1: liver kinase B1; STK11: serine-threonine kinase 11.
Figure 2. Expression of selected microRNAs in pancreatic adenocarcinoma compared to matching normal tissues. Expression data are shown for miRs-10b, -194, -202-5p, -323-3p, -367, -451 and -663a. Data from 179 pancreatic cancer samples and 4 matching normal samples derived from the PAAD cohort of The Cancer Genome Atlas (TCGA) are shown. miRNA expression was quantified by RNA sequencing and is shown as log2 of normalized read counts. The red lines indicate low versus higher expression. Expression data are shown as box plots. The line in the middle of the box represents the median values, the rectangles show the upper and lower 25% quartiles and 50% of all data points are included in the greater rectangle. All other data points, except for outliers are located within the upper and lower whiskers.
miR-194 is up-regulated in PDAC, mediates migration, proliferation and colony formation of PANC-1 cells and suppresses apoptosis of these cells (34). miR-194 transfected PANC-1 cells exhibit reduced growth in an orthotopic pancreatic mouse model in comparison to the untransfected cell line (34). miR-194 targets dachshund 1 (DACH1), a cell fate determination factor, which functions as a TS in several types of cancer (34-36) (Figure 1). Steady-state RNA levels for miR-194 derived from TCGA did reveal a tendency of increased expression of miR-194 in PDAC samples in comparison to normal pancreatic tissues (Figure 2).
Overexpression of miR-451 is related to poor prognosis in patients with PDAC (37). miR-451 promotes in vivo growth of subcutaneously implanted PANC-1 and AsPC-1 pancreatic carcinoma cells (37). Calcium binding protein 39 (CAB39) was identified as a direct target of miR-451 (37). CAB39 stimulates tumor-suppressive serine threonine kinase 11 (STK11), also referred to as liver kinase B1 (LKB1) (38,39) (Figure 1). miR-451 steady-state levels in PDAC samples in comparison to normal pancreatic tissues did not reveal a tendency of increased levels in PDAC samples (Figure 2).
Growth-inhibiting miRs Down-regulated in PDAC
miRs discussed in this chapter inhibit tumor growth. Since they have not been tested in metastasis-related in vivo models, some of them might have an impact on metastasis.
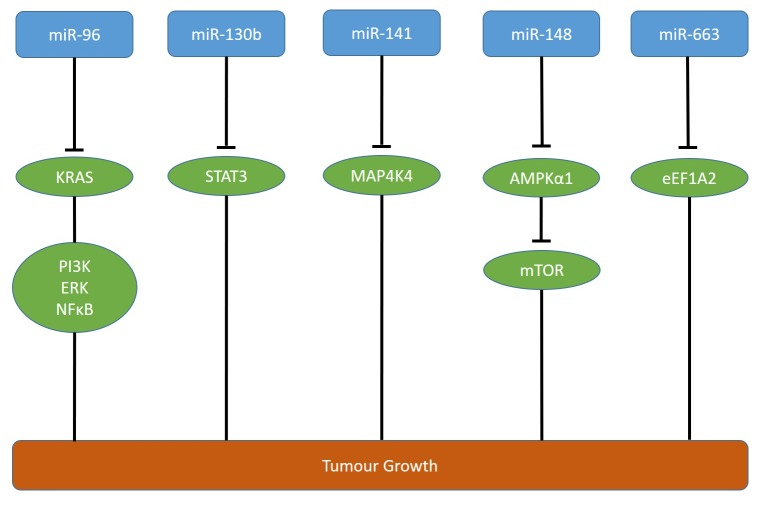
miR-96 is down-regulated in PDAC in comparison to corresponding normal pancreatic tissues (40). miR-96 inhibits invasion, migration and proliferation of MIA PaCa-2 and PANC-1 pancreatic carcinoma cells in vitro and growth of both cell lines subcutaneously injected into the flanks of nude mice (40). KRAS has been identified as a direct target of miR-96 (40) (Figure 3). KRAS is one of the three members of the RAS oncogene family which encode small GTPases involved in signaling transduction mediating cell growth, differentiation and survival (41). Oncogenic KRAS promotes pancreatic tumorigenesis through multiple pathways including PI3K/AKT, extracellular signal-regulated kinase (ERK) and nuclear factor ĸB (NFĸB) (41,42).
Figure 3. Down-regulated miRs affecting growth of pancreatic ductal adenocarcinoma-related xenografts. miRs -96, -10b, -141, -148 and -663 are down-regulated and interfere with the targets as outlined. AMPKα1: AMP-activated protein kinase; eEFA2: eukaryotic initiation factor A2; ERK: extracellular signal regulated kinase; KRAS: Kirsten rat sarcoma; mTOR: mammalian target of rapamycin; MAP4K4: mitogen-activated protein kinase kinase kinase kinase 4; NFĸB: nuclear factor ĸB; PI3K: phosphoinosite 3-kinase; STAT 3: signal transducer and activator of transcription 3.
miR-130b is down-regulated in PDAC in comparison to normal pancreatic tissues and is associated with worse prognosis (43). miR-130b suppresses proliferation and invasiveness of PANC-1 and AsPC-1 pancreatic carcinoma cells in vitro as well as growth of PANC-1 cells transfected with miR-130b in nude mice in comparison to untransfected cells (43). Signal transducer and activator of transcription (STAT3) has been identified as a direct target of miR-130b (43). STAT3 is a transcription factor which is frequently overexpressed in human cancers including PDAC, and inhibits apoptosis and promotes angiogenesis, invasion and metastasis (44,45) (Figure 3).
Decreased expression of miR-141 is asscociated with poor prognosis in patients with PDAC (46). In PANC-1 and MIA PaCa-2 transfectants, miR-141 inhibits cell proliferation, colony formation, invasion, G1-phase arrest and apoptosis (46). PANC-1 cells transfected with miR-141 exhibit reduced tumor growth as xenografts in nude mice in comparison to untransfected PANC-1 cells (46). Mitogen-activated protein kinase kinase kinase kinase 4 (MAP4K4) has been identified as a direct target of miR-141 (46) (Figure 3). MAP4K4, a serine-threonine kinase, is involved in migration, proliferation and regulation of focal adhesion dynamics (47,48). Expression levels of MAP4K4 are associated with worse prognosis in patients with stage II PDAC (49).
miR-148 is down-regulated in PDAC and correlates with lymphatic invasion, distant metastases and worse prognosis (50). In PANC-1 and BxPC-3 cells, miR-148 inhibits invasion and proliferation and induces apoptosis and cell-cycle arrest (50). miR-148 inhibits growth of PANC-1 cells subcutaneously injected into nude mice (50). 5’-AMP activated protein kinase catalytic subunit α1 (AMPKα1) was identified as a direct target of miR-148 (50) (Figure 3). AMPKα1 interferes with growth of human cancer cells by inhibition of mechanistic target of rapamycin (mTOR) signaling (51,52). AMPKα1 acts as a cellular engergy sensor conserved in all eukaryotic cells and regulates the activity of key metabolic enzymes through phosphorylation (53).
miR-663a is down-regulated in PDAC in comparison to normal pancreatic tissues and its down-regulation is associated with decreased survival (54). In PANC-1 and AsPC-1 cells, miR-663a attenuates cell proliferation and invasiveness and induces apoptosis (54). PANC-1 cells transfected with miR-663 exhibit decreased tumor growth in comparison to untransfected cells after subcutaneous implantation into the flanks of immuno-deficient mice (54). Elongation factor 1A2 (eEF1A2) has been identified as a direct target of miR-663 (54). eEF1A2 is critical for protein synthesis because it negatively regulates protein biosynthesis by binding to amino-acylated t-RNAs and facilitating their recruitment to the ribosome (55) (Figure 3).
Up-regulated miRs Mediating Metastasis in PDAC Xenografts
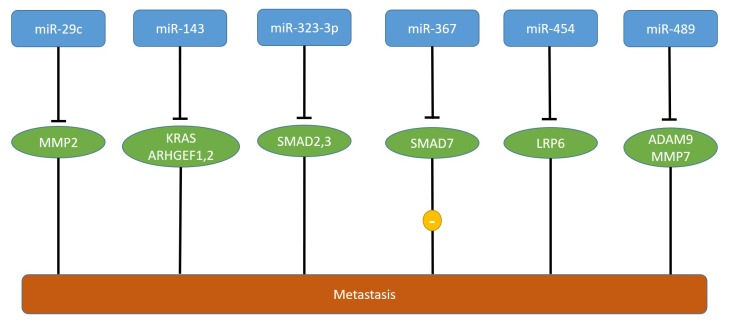
High levels of miR-367 correlate with poor prognosis in PDAC patients (56). miR-367 promotes migration and invasion of PANC-1 and BxPC-3 pancreatic carcinoma cells (56). miR-367 induces epithelial- mesenchymal transition (EMT) by increasing TGFβ-induced transcriptional activity (56). In vivo, PANC-1 cells transfected with miR-367 and injected into the spleen of nude mice give rise to increased numbers of liver metastatic colonies in comparison to non-transfected cells, whereas no effect on tumor growth was observed (56). SMAD family protein 7 (SMAD7) was identified as a direct target of miR-367 (56) (Figure 4). SMAD7 acts as an inhibitor of TGFβ signaling and as a metastasis suppressor in human cancer (57-59). In PDAC, TGFβ signaling contributes to cell invasion and metastasis by inducing EMT (60). Low levels of SMAD7 correlates with poor prognosis and lymph node metastasis in patients with pancreatic cancer (61). Nevertheless, one should be aware that TGFβ can have opposite effects on tumor progression and metastasis depending on the stage of carcinogenesis and the responsiveness of tumor cells (62,63). TCGA-derived steady state levels of miR-663a revealed very low expression in PDAC and normal pancreatic tissues (Figure 2).
Figure 4. miRs affecting metastasis of pancreatic ductal adenocarcinoma xenografts. miR-367 is up-regulated, whereas miRs -29c, -143, -323-3p, -367, -454-5p, and miR-489, are down-regulated in pancreatic tumors in comparison to corresponding normal pancreatic tissues models. The corresponding targets are shown. ADAM 9: A disintegrin and metalloproteinase 9; ARHGEF1,2: rho guanine nucleotide exchange factor 1,2; KRAS: kirsten rat sarcoma; LRP6: low-density lipoprotein-related protein C; MMP2,7: matrix metalloproteinase 2,7.
Down-regulated miRs Mediating Metastasis in PDAC Xenografts
Down-regulated expression of miR-29c and matrix metallo-protease 2 (MMP2) correlate with progression of PDAC (64). miR-29c suppresses migration and invasion of miR-29c transfected PANC-1 and BxPC-3 pancreatic carcinoma cells and targets MMP2 directly (64) (Figure 4). In vivo, miR-29c transfected Hs-766 T pancreatic carcinoma cells give rise to reduced liver metastasis in comparison to non-transfected cells after injection into the pancreas (64). MMP2 can act as a promoter of metastasis, but its functional role is context-dependent and clinical trials failed to validate MMP2 as an anti-metastatic target in several types of cancer (65-68).
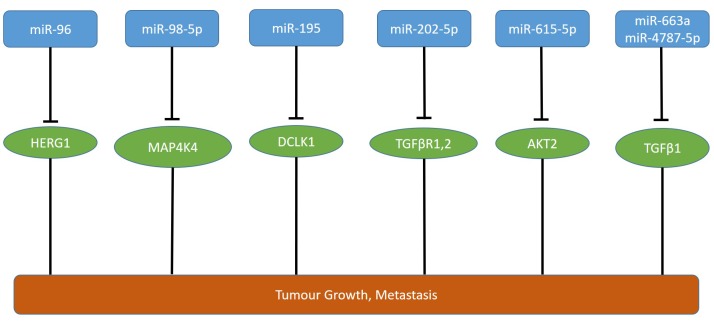
miR-96 decreases proliferation, migration and invasion of PANC-1 and CFPAC-1 pancreatic carcinoma cells in vitro (69). CFPAC-1 cells transfected with miR-96 exhibit reduced growth after subcutaneous injection into the flanks of nude mice and no metastatic nodules after injection into the pancreatic capsule in contrast to the untransfected cell line (69). Human ether-a-go-go 1 (HERG1) potassium channel was identified as a direct target of miR-96 (69,70) (Figure 5). HERG1 is often up-regulated in PDAC and functions as an oncogene (71). HERG1 is overexpressed in many types of cancers and has been shown to be involved in functions such as invasion, proliferation and apoptosis (72,73).
Figure 5. miRs affecting tumor growth and metastasis of pancreatic ductal adenocarcinoma xenografts. miR-96, -98-5p, -195, -202-5p, -615-5p, -663a and 4787-5p are down-regulated in pancreatic tumors in comparison to corresponding normal tissues. AKT2: Protein kinase β; DCLK1: doublecortin-like kinase 1; HERG1: human ether-a-go-go related gene product; MAP4K4: mitogen-activated protein kinase kinase kinase kinase 4; TGFβ1: transforming growth factor β1; TGFβR1,2: transforming growth factor β receptor 1 and 2.
miR-98-5p inhibits migration, invasion and proliferation of PDAC cell lines CFPAC-1 and MIA-PaCa-2 (74). Low levels of miR-98-5p correlate with tumor size and lymph node metastasis (74). In vivo, tumor volume and weight of miR-98-5p overexpressing pancreatic carcinoma cells was decreased, whereas silencing of miR-98-5p increased tumor growth in vivo (74). In an orthotopic pancreatic cancer in vivo model, liver nodules were elevated in MIA PaCa-2 cells with silenced expression of miR-98-5p in comparison to the control cell line (74). MAP4K4 was identified as a direct target of miR-98-5p (74) (Figure 5). It was shown that overexpression of MAP4K4 increases growth of hepatocellular carcinoma cell lines in vitro and in vivo (75). In gastric cancer cells, silencing of MAP4K4 by short hairpin RNA was shown to suppress proliferation and to induce G1 cell cycle arrest and apoptosis (76).
miR-143 inhibits migration and invasion of PANC-1 pancreatic carcinoma cells in vitro and liver metastasis of these cells in an orthotopic xenograft model (77). miR-143 targets directly KRAS and RHO guanine nucleotide exchange factors ARHGEF1 and ARHGEF2 (77) (Figure 4). miR-143 decreases RHO GTPase activity of ras-related C3 botulinum toxin substrate 1 (RAC1), cell division control protein 42 (CDC42) and RHO GTPase A (RHO A), inhibits MMP2 and MMP9 and elevates the protein level of E-cadherin (77). Guanine nucleotide exchange factors commonly activate RHO GTPases through catalysis of the exchange of GDP for GTP and are crucial for cell migration and invasion (78-80).
miR-195 inhibits proliferation, migration and invasion of PANC-1 and AsPC-1 pancreatic carcinoma cells in vitro and in vivo and low miR-195 expression indicates a poor clinical outcome (81). Overexpression of miR-195 in PANC-1 cells inhibits subcutaneous tumor growth and alleviates metastasis of PANC-1 cells transfected with miR-195 to the liver after injection into the spleen of nude mice (81). Doublecortin-like kinase 1 (DCLK1) was identified as a direct target of miR-195 (81) (Figure 5). miR-195 regulates pERK1/2, p38, p21, vimentin, E-cadherin, RHOA and MMP9 expression in pancreatic carcinoma cells by targeting DCLK1 (81). DCLK1 is a microtubule-binding member of the calmodulin-dependent kinase family, is overexpressed in intraepithelial lesions as well as in surgical resection specimen of PDAC and is a stem cell marker, inhibitor of proliferation and mediator of EMT in PDAC (82-85).
Expression of miR-202-5p in PANC-1 cells reduces tumor growth and liver metastasis in athymic mice after orthotopic implantation in comparison to the untransfected cell line (86). In an immuno-competent mouse model, making use of PAN 02 cells, derived from a carcinogen-induced mouse model of pancreatic carcinoma, miR-202-5p reduced tumor burden and liver metastasis after orthotopic implantation (86). miR-202-5p inhibits TGFβ1-induced EMT of PDAC cells (86). Also, miR-202-5p interferes with TGFβ-mediated activation of pancreatic stellar cells, accumulation of extracellular matrix (ECM) proteins and fibrotic stromal deposition (86). Transforming growth factor β receptor 1 and 2 (TGFRβ1, TGFRβ2) have been identified as direct targets of miR-202-5p (86) (Figure 5). TGFβ signaling is a mediator of EMT of pancreatic carcinoma cells (87-91). The TGFβ-TGFβR complex recruits downstream effectors such as SMAD2 and SMAD3 (R-SMADs), which form complexes with SMAD4 and these complexes translocate into the nucleus, resulting in activation of genes involved in EMT by recruitment of transcriptional coactivators or corepressors (87-91). TCGA derived steady-state levels of miR-202-5p did not reveal a tendeny of deregulation of its expression in PDAC samples in comparison to normal pancreatic tissues (Figure 2).
Down-regulation of miR-323-3p correlates with PDAC progression (92). miR-323-3p inhibits invasion and migration of PANC-1 and BxPC-3 pancreatic carcinoma cells (92). Metastatic lung foci were inhibited in PANC-1 cells transfected with miR-323-3p after tail vein injection in nude mice in comparison to control cells (92). miR-323 inactivates the TGFβ signaling pathway by directly targeting SMAD2 and SMAD3 which are functional effectors of the TGFβ pathway in PDAC cells (92-95) (Figure 4). Down-regulation of miR-323-3p, with increased expression of SMAD2 and SMAD3, contributes to a more aggressive phenotype of PDAC (92). Steady state levels for miR-323-3p did not show a tendeny of deregulation of its expression in PDAC in comparison to normal pancreatic tissues (Figure 2).
miR-454 inhibits invasion, proliferation and arrests PANC-1 and MIA PaCa-2 pancreatic carcinoma cells in the G2/M phase (96). Furthermore, miR-454 inhibits tube formation of endothelial cells (96). Tail vein injection of miR-454 transduced PANC-1 cells revealed reduced lung metastasis in comparison to the corresponding control cell line (96). As a direct target, low density lipoprotein receptor-related protein 6 (LRP6) has been identified (96) (Figure 4). LRP6 together with LRP5 and the family of frizzled receptors are activating components of the wingless/integrated (WNT) signaling pathway (97-99). Calcipotriol has been identifed as a WNT-pathway inhibitor targeting LRP6 (100). Targeting of WNT signaling in cancer is pursued by many ongoing drug discovery programmes (101-103).
miR-489 inhibits migration of PANC1 and BxPC3 pancreatic carcinoma cells (104). PANC-1 cells transfected with miR-489 exhibit reduced lung metastasis after tail vein injection in comparison to the untransfected cell line. Substantially reduced liver metastasis was observed after injection of these cells into the mouse spleen (104). Conversely, inhibition of miR-489 in BxPC-3 cells leads to increased metastasis to the lungs after tail vein injection and enhanced liver metastasis after injection into the spleen of immuno-deficient mice in comparison to the control cell line (104). miR-489 is down-regulated by the KRAS-NFĸB-YY1 axis in PDAC cells (104). KRAS and NFĸB are constitutively activated in the vast majority of PDACs and yin-yang (YY1) is a ubiquitously distributed transcription factor that binds to the promoter region of its target genes (105-107). A disintegrin and metalloproteinase 9 (ADAM9) and matrix metalloprotease 7 (MMP7) were identified as direct targets of miR-489 (104) (Figure 4). ADAM9 and MMP7 are metalloproteinases that remodel the ECM, faciltating cancer cells to form local or distant metastases (108-112). Expression of miR-489, YY1, ADAM9 and MMP7 correlated with progression of human PDAC (104).
High expression of miR-615-5p correlates with significantly higher survival time in PDAC patients (113). Overexpression of miR-615-5p inhibits MIA-PaCa-2 and PANC-1 cell migration and invasion in vitro (113). Tumor growth of MIA-PaCa-2 cell transfected with miR-615-5p was inhibited after subcutaneous implantation and fewer and smaller colonies in the liver were observed after tail vein injection in comparison to the control cell line (113). Serine-threonine kinase AKT2 was identified as a direct target of miR-615-5p (113) (Figure 5). AKT2 together with AKT1 are isoforms of AKT (114). Activation and amplification of AKT2 has been observed in human PDACs (115-117).
miR-663a and -4787-5p
miR-663a and -4787-5p suppress TGFβ1-induced EMT in PDAC cells MIA PaCa-2 and PANC-1 (118). Expression of both miRs in PANC-1 cells reduces tumor weights and volumes and the number of metastatic lesions in liver, spleen, lungs and kidneys after orthotopic implantation into the pancreas in comparison to the non-transfected cells (118). Both miRs target TGFβ1 directly (118) (Figure 5). In PDAC, TGFβ1 can act both as a tumor suppressor and promoter depending on tumor stage and cellular context (119,120).
Synopsis of Identified PDAC-related miRs
miRs modulating growth of PDAC xenografts. The identifed miRs of this sub-category modulate tumor growth of PDAC-related preclinical in vivo models. They might also be involved in metastasis, but corresponding preclinical in vivo experiments are pending. Up-regulated tumor growth promoting miRs such as miR-10b (31), miR-194 (34) and miR-451 (37) interfere with mRNAs of TS genes such as TIP30, DACH1 and CAB39. Down-regulated tumor growth inhibiting miRs, miR-96 (40), miR-130b (43), miR-141 (46), miR-148 (50) inhibit signal transducers such as KRAS and HERG1, STAT3, MAP4K4 and AMPKα1. miR-663a (54) interferes with translation by targeting elongation factor eEF1A2.
miRs modulating metastasis of PDAC xenografts. In the category metastasis-related miRs, examples of tumor growth and metastasis affecting miRs as well as miRs affecting metastasis only have been summarized. miR-367 (56), the only up-regulated miR in this category, targets SMAD7, a negative regulator of TGFβ signaling (57-59).
Down-regulation of miRs-29c (64), -143 (75), -323-3p (92), -367 (56), -454 (96), -489 (104) promote metastasis, but not tumor growth in preclinical in vivo models. miRs-96 (69), -98-5p (74), -195 (81), -202-5p (86), -615-5p (113), -663a (118) and -4787 (118) inhibit tumor growth as well as metastasis in corresponding preclinical in vivo models.
As outlined, miR-29c (64) and miR-489 (104) interfere with metalloproteinases MMP2, MMP7 and ADAM9. However, these targets have pro- and tumoral activities and clinical studies with small-molecule inhibitors did not reach the projected endpoints (66-68). Other metastasis-related miRs such as miR-96 (96), -98-5p (74), miR-454 (96) and miR-615p (113) affect signal transduction (113) through targets such as HERG1, MAP4K4, LRP6 and AKT2. miR-195 (81) inhibits DCLK1 which modifies microtubules and therefore has an impact on migration and metastasis. None of the latter targets has reached the status of a clinically validated target.
Noteworthy, 4 miRs such as miR-202-5p (86), -323-3p (92), -663a (118) and -4187 (118) are down-regulated and activate TGFβ signaling through up-regulation of targets such as TGFRβ1and TGFRβ2, TGFβ1, SMAD2, 3, emphasizing the role of activation of this pathway in the progression of PDAC. However, miR-367 is up-regulated and targets SMAD7 which is a negative regulator of pro-metastatic TGFβ signaling as outlined above (56-59) and therefore it is a candidate for functional inhibition. Galunisertib (LY2157299), a small molecule kinase inhibitor of TGFRβ1, is presently evaluated together with gemcitabine for first-line treatment in patients with unresectable PDAC (121). Furthermore, combination studies of galunisertib and checkpoint inhibitory antibody durvalumab (MEDI 4736) are underway (122). Further candidates for functional reconstitution are miR-96 (KRAS and HERG1), miR-130b (STAT3), miR-141 (MAP4K4), miR-148 (AMPKalpha1) and miR-663 (eEF1A2). In addition to miR-367 (SMAD7), further candidates for functional inhibition are miR-10b (TIP30), miR-194 (DACH1) and miR-451 (CAB39) which target TS genes.
Therapeutic Aspects
As outlined, 4 up-regulated and 16 down-regulated miRs have been identified as possible therapeutic candidates for treatment of PDAC. However, for miRs -96, -143, -195, -202-5p, -454, -663 and -4787-5p, data correlating expression status and clinical outcome are pending. Of note, miR-96 and -663 can be found in both categories: down-regulated miRs with growth inhibitory function and down-regulated miRs inhibiting metastasis. This is not a contradiction, because miR-96 and miR-663 modulate different targets in these categories. From a therapeutic point of view, up-regulated miRs are candidates for inhibition of function and down-regulated miRs are candidates for reconstitution of function. miRs-10b (31), -194 (34), -451 (37) and -367 (56) are candidates for inhibition of function. Among the miRs to be functionally reconstituted are two miRs targeting KRAS (miR-96 and -143) and four down-regulated miRs with an inhibitory function of TGFβ signaling (miRs-202-5p, -323-3p -663a and -4187). The latter are candidates for reconstitution therapy. An exception in this context is miR-367 which is up-regulated and inhibits SMAD7, an inhibitory component of TGFβ signaling and therefore is a candidate for functional inhibition. Analysis of the expression status of miRs and components of the TGFβ signaling pathway in clinical specimens will guide the selection of the appropriate therapeutic miR candidates. Inhibition of KRAS by reconstitution of miRs-96 and -143 is critical, because these miRs do not discriminate between wild-type and mutated KRAS due to targeting of the conserved 3’-UTRs of their corresponding mRNAs.
miR inhibitors are referred to as antagomirs and are single-stranded RNA molecules in the range of 22-25 nts complementary to a site of the mRNA which is the target of the miR under consideration, preventing binding of the corresponding miR to its target mRNA (21,22,123,124). Usually, chemical modifications such as 2’-methoxy groups and phosphothioates are incorporated into antagomirs to make them resistant to degradation (21,22). Another version of miR-inhibitors are miR sponges. They are artificial constructs with multiple reiterated miR-binding sites that compete with the natural mRNA target for binding of the corresponding miR and are usually delivered into mammalian cells by expression vectors (125,126). The alternative therapeutic option, functional reconstitution of miRs, can be performed making use of miR mimetics, small double-stranded synthetic RNA molecules designed to mimic endogenous mature miRs leading to cleavage, deadenylation or inhibition of translation of the corresponding mRNAs (126). They mimic the transient duplex product of DICER complex processing. The two strands of the synthetic mimic separate and the single-strand mature miR is incorporated into the RISC to down-regulate mRNA transcripts. Reconstitution of miR function by delivery of expression vectors for the corresponding miR is another option for therapeutic intervention (126).
After identification of the target, the miR-related agent has to be designed, its chemistry and delivery efficiency have to be optimized, therapeutic efficacy of the candidate miR-related agent has to be worked out in preclinical in vitro and in vivo studies and toxicity in rodents and non-human primates has to be assessed. Finally, issues such as dose-finding, toxicity and therapeutic efficacy have to be addressed in clinical studies (22,124).
There are many issues which are potential bottlenecks and have to be optimized for the therapeutic performance of miR-related therapeutic agents. One of the issues is the improvement of the delivery systems. Optimization of liposomal carriers, conjugation of nucleic-acid related agents to poly-ethylene glycole (PEG), use of biodegradable polymers, cyclodextrin and dendrimers, have resulted in significant progress (127-131). Conjugation of miR-related agents to N-acetyl-D-galactosamine has significantly improved delivery to the liver based on its high affinity to the asialo glycoprotein receptor expressed in liver (132). The size of the unconjugated RNA is another critical issue for delivery of miR-related agents. They have a size between 7 and 20 kD, are filtered by the kidneys and secreted which holds true for all complexes smaller than 50 kD (133). After internalisation into target cells, escaping the endosomal vesicles for release into the cytoplasm is a further bottleneck, because the delivery complexes are often trapped as phagolysosomes (134). Further critical issues are immunogenicity of the miR-related agents, removal from extracellular spaces by macrophages and monocytes and carrier toxicity such as hemolysis, thrombogenicity and complement activation (135-138). Toxicity due to hybridisation of the miR-related agents to non-target nucleic acids is another alerting issue.
Since PDAC has already metastasized to distant organs in the vast majority of patients after diagnosis, miR-related therapeutic agents should be able to eradicate metastasis, which is a significant hurdle. This senario is reflected in none of the preclinical in vivo models. miR-related therapeutic agents should exhibit anti-proliferative together with anti-invasive properties as a desired profile. In case the miR-related agent has only anti-invasive capacity, the desired profile can be probably achieved by combination with other agents.
In the near past, miR-based therapeutic agents have witnessed serious drawbacks in clinical studies (139). In 2018, Regulus Therapeutics has halted clinical studies of treatment of polycystic kidney disease with anti-miR agent RGLS4326 due to toxicity issues and paused enrollment for a phase II study of miR-21 inhibitor RG-012 in patients with Alpert syndrome, a rare genetic kidney disease (139). In 2016, Regulus has halted a phase I study of RG-101 for treatment of infection with hepatitis C virus with anti-miR-122 due to jaundice in two patients, despite excellent efficacy data (139). In the oncology area, Mirna Therapeutics has shut down a phase I study of MRX-34 for treatment of multiple types of cancers in 2016 (139). Presently, miRagen is clinically evaluating Cobomersen (MRG106) in several types of hematological cancers in phase I studies without reporting serious side effects (139). The clinical evaluation of second-generation miRs without the side-effects as outlined above will be soon underway and tell us more about the therapeutic utility of miRs in cancer therapy.
Conflicts of Interest
FB and AN are employees of Roche, UHW has been a former employee of Roche.
Authors’ Contributions
UHW and AN were jointly designing and writing the paper, FB has performed the bioinformatics analysis and was preparing the figures.
References
- 1.Munoz Martin AJ, Adeva J, Martinez-Galin J, Reina IJ, Hidalgo M. Pancreatic ductal adenocarcinoma: metastatic disease. Clin Transl Oncol. 2017;19:1423–1429. doi: 10.1007/s12094-017-1690-6. PMID: 28623515. DOI: 10.1007/s12094-017-1690-6. [DOI] [PubMed] [Google Scholar]
- 2.Sahin ICH, Icobuzio-Donahue CA, O’Reilly EM. Molecular signature of pancreatic adenocarcinoma: an insight from genotype to phenotype and challenges for targeted therapy. Exp Opin Ther Targets. 2016;20:341–359. doi: 10.1517/14728222.2016.1094057. PMID: 26439702. DOI: 10.1517/14728222.2016.1094057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohta T, Nagakawa T, Ueno K, Kayahara M, Mori K, Kobayashi H, Takeda T, Miyazaki I. The mode of lymphatic and local spread of pancreatic carcinomas less than 4.0 cm in size. Int Surg. 1993;78:208–212. PMID: 8276542. [PubMed] [Google Scholar]
- 4.Gall FP, Kessler H, Hermanek P. Surgical treatment of ductal pancreatic carcinoma. Eur J Surg Oncol. 1991;17:173–181. PMID: 2015922. [PubMed] [Google Scholar]
- 5.Manji GA, Olive KP, Saenger YM, Oberstein P. Current and emerging therapies in metastatic pancreatic cancer. Clin Cancer Res. 2017;23:1670–1678. doi: 10.1158/1078-0432.CCR-16-2319. PMID: 28373365. DOI: 10.1158/1078-0432.CCR-16-2319. [DOI] [PubMed] [Google Scholar]
- 6.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:2140–2141. doi: 10.1056/NEJMc1412266. PMID: 25427123. DOI: 10.1056/NEJMc1412266. [DOI] [PubMed] [Google Scholar]
- 7.Dreyer SB, Chang DK, Bailey P, Biankin AV. Pancreatic cancer genomes: implications for clinical management and therapeutic development. Clin Cancer Res. 2017;23:1638–1646. doi: 10.1158/1078-0432.CCR-16-2411. PMID: 28373362. DOI: 10.1158/1078-0432.CCR-16-2411. [DOI] [PubMed] [Google Scholar]
- 8.Borazanci E, Dang CV, Robey RW, Bates SE, Chabot JA, Von Hoff DD. Pancreatic cancer: “A riddle wrapped in a mystery inside an enigma”. Clin Cancer Res. 2017;23:1629–1637. doi: 10.1158/1078-0432.CCR-16-2070. PMID: 28373361. DOI: 10.1158/1078-0432.CCR-16-2070. [DOI] [PubMed] [Google Scholar]
- 9.Johnson BA 3rd, Yarchoan M, Lee V, Laheru DA, Jaffee EM. Strategies for increasing pancreatic tumor immunogenicity. Clin Cancer Res. 2017;23:1656–1669. doi: 10.1158/1078-0432.CCR-16-2318. PMID: 28373364. DOI: 10.1158/1078-0432.CCR-16-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evan GI, Hah N, Littlewood TD, Sodir NM, Campos T, Downer M, Evans RM. Re-engineering the pancreas tumor microenvironment: a regenerative program hacked. Clin Cancer Res. 2017;23:1647–1655. doi: 10.1158/1078-0432.CCR-16-3275. PMID: 28373363. DOI: 10.1158/1078-0432.CCR-16-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerra C, Barbacid M. Genetically engineered mouse models of pancreatic adenocarcinoma. Mol Oncol. 2013;7:232–247. doi: 10.1016/j.molonc.2013.02.002. PMID: 23506980. DOI: 10.1016/j.molonc.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang CI, Boj SF, Clevers H, Tuveson DA. Preclinical models of pancreatic ductal adenocarcinoma. J Pathol. 2016;238:197–204. doi: 10.1002/path.4651. PMID: 26419819. DOI: 10.1002/path.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lösch S, Buchholz M, Gress TM, Weidle UH. LKW, a putative dual-specificity kinase is down-regulated in several invasive systems. Cancer Genomics Proteomics. 2004;1:177–188. [PubMed] [Google Scholar]
- 14.Lösch S, Buchholz M, Gress TM, Weidle UH. Identification of a novel transmembrane protein (UKW): Association with invasive status of mammary carcinoma cell lines and expression in pancreatic carcinoma. Cancer Genomics Proteomics. 2004;1:263–274. [PubMed] [Google Scholar]
- 15.Costa-Silva B, Aiello NM, Ocean AJ, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, Xiang J, Zhang T, Theilen TM, Garcia-Santos G, Williams C, Ararso Y, Huang Y, Rodriques G, Shen TL, Labori KJ, Lothe IM, Kure EH, Hernandez J, Dussot A, Ebbesen SH, Grandgenett PM, Hollingsworth MA, Jain M, Mallva K, Batra SK, Jarnagin WR, Schwartz RE, Matei I, Peinado H, Stanger BZ, Bromberg J, Lyden D. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. PMID: 25985394. DOI: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamisawa T, Isawa T, Koike M, Tsuruta K, Okamoto A. Hematogenous metastases of pancreatic ductal carcinoma. Pancreas. 1995;11:345–349. doi: 10.1097/00006676-199511000-00005. PMID: 8532650. [DOI] [PubMed] [Google Scholar]
- 17.Disibio G, French SW. Metastatic patterns of cancers. Results from a large autopsy study. Arch Pathol Lab Med. 2008;132:931–939. doi: 10.5858/2008-132-931-MPOCRF. PMID: 18517275. DOI: 10.1043/15432165(2008)132[931:MPOCRF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 18.Deeb A, Haque SU, Olowokure O. Pulmonary metastases in pancreatic cancer, is there a survival influence. J Gastrointest Oncol. 2015;6:E48–51. doi: 10.3978/j.issn.2078-6891.2014.114. PMID: 26029466. DOI: 10.3978/j.issn.2078-6891.2014.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. Metazoan microRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. PMID: 29570994. DOI: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slotwinski R, Lech G, Slotwinska SM. microRNA in pancreatic diagnosis and therapy. Cent Eur J Immunol. 2018;43:314–324. doi: 10.5114/ceji.2018.80051. PMID: 30588176. DOI: 10.5114/ceji.2018.80051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Rana TM. Therapeutic targeting of microRNAs. Current status and future challenges. Nat Rev Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. PMID: 25011539. DOI: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 22.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. PMID: 28209991. DOI: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 23.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. PMID: 15965474. DOI: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 24.Peter ME. Targeting of mRNAs by multiple miRNAs: the next step. Oncogene. 2010;29:2161–2164. doi: 10.1038/onc.2010.59. PMID: 20190803. DOI: 10.1038/onc.2010.59. [DOI] [PubMed] [Google Scholar]
- 25.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Ann Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. PMID: 19630570. DOI: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 26.Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanese N, Garzon R, Ageilan RI, Alder H, Volinia S, Rassanti L, Liu X, Liu CG, Kipps TJ, Negrini M, Croce CM. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. PMID: 18362358. DOI: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calin GA, Dumitru CD, Shimizu M, Bich R, Zupo S, Noch E, Rattan S, Keating M, Rai K, Rassenti l, Kipps T, Negrini M, Bullrich R, Croce CM. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. PMID: 12434020. DOI: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weidle UH, Birzele F, Kollmorgen G, Nopara A. Potential microRNA-related targets for therapeutic intervention with ovarian cancer metastasis. Cancer Genomics Proteomics. 2018;15:1–15. doi: 10.21873/cgp.20061. PMID: 29275359. DOI: 10.21873/cgp.20061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weidle UH, Dickopf S, Hintermair C, Kollmorgen F, Birzele F, Brinkmann U. The role of microRNAs in breast cancer metastasis: preclinical validation and potential therapeutic targets. Cancer Genomics Proteomics. 2018;15:17–39. doi: 10.21873/cgp.20062. PMID: 29275360. DOI: 10.21873/cgp.20062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weidle UH, Epp A, Birzele F, Brinkmann U. The functional role of prostate cancer metastasis-related microRNAs. Cancer Genomics Proteomics. 2019;16:1–19. doi: 10.21873/cgp.20108. PMID: 30587496. DOI: 10.21873/cgp.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ouyang H, Gore J, Deitz S, Korc M. microRNA-10b enhances pancreatic cancer cell invasion by suppressing TIP30 expression and promoting EGF and TGF-β actions. Oncogene. 2014;33:4664–4674. doi: 10.1038/onc.2013.405. PMID: 24096486. DOI: 10.1038/onc.2013.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakata K, Ohuchida K, Mizumoto K, Kayashima T, Ikenaga N, Sakai H, Lin C, Fujita H, Otsuka T, Aishima S, Nagai E, Oda Y, Tanaka M. MicroRNA-10b is overexpressed in pancreatic cancer, promotes its invasiveness, and correlates with a poor prognosis. Surgery. 2011;150:916–922. doi: 10.1016/j.surg.2011.06.017. PMID: 22018284. DOI: 10.1016/j.surg.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 33.Zhang C, Li A, Zhang X, Xiao H. A novel TIP30 protein complex regulate EGF receptor signaling and endocytic degradation. J Biol Chem. 2011;286:9373–9381. doi: 10.1074/jbc.M110.207720. PMID: 21252234. DOI: 10.1074/jbc.M110.207720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Zhao CY, Zhang SH, Yu DH, Chen Y, Liu QH, Shi M, Ni CR, Zhu MH. Upregulation of miR-194 contributes to tumor growth and progression in pancreatic ductal adenocarcinoma. Oncol Rep. 2014;31:1157–1164. doi: 10.3892/or.2013.2960. PMID: 24398877. DOI: 10.3892/or.2013.2960. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Han N, Zhou S, Yuan X, Xu H, Zhang C, Yin T, Wu K. The DACH/EYA/SIX gene network and its role in tumor initiation and progression. Int J Cancer. 2016;138:1067–1075. doi: 10.1002/ijc.29560. PMID: 26096807. DOI: 10.1002/ijc.29560. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Zhou R, Yuan X, Han N, Zhou S, Xu H, Guo M, Yu S, Zhang C, Yin T, Wu K. Dach1 is a novel predictive and prognostic biomarker in hepatocellular carcinoma as a negative regulator of Wnt/β-catenin signaling. Oncotarget. 2015;6:8621–8634. doi: 10.18632/oncotarget.3281. PMID: 25940701. DOI: 10.18632/oncotarget.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo R, Gu J, Zhang Z, Wang Y, Gu C. MiR-451 promotes cell proliferation and metastasis in pancreatic cancer through targeting CAB39. Biomed Res Int. 2017;2017:2381482. doi: 10.1155/2017/2381482. PMID: 28197410. DOI: 10.1155/2017/2381482. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Kullmann L, Krahn MP. Controlling the master-upstream regulation of the tumor suppressor LKB1. Oncogene. 2018;37:3045–3057. doi: 10.1038/s41388-018-0145-z. PMID: 29540834. DOI: 10.1038/s41388-018-0145-z. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Wang N, Liu P, Xie X. AMPK and cancer. Exp Suppl. 2016;107:203–226. doi: 10.1007/978-3-319-43589-3_9. PMID: 27812982. DOI: 10.1007/978-3-319-43589-3_9. [DOI] [PubMed] [Google Scholar]
- 40.Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W, Liu J, Yu J, Chen J. miR-86 suppresses KRAS and functions as a tumor suppressor in pancreatic cancer. Cancer Res. 2010;70:6015–6025. doi: 10.1158/0008-5472.CAN-09-4531. PMID: 20610624. DOI: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- 41.Hall A. The cellular functions of small GTP-binding proteins. Science. 1990;249:635–640. doi: 10.1126/science.2116664. PMID: 2116664. DOI: 10.1126/science.2116664. [DOI] [PubMed] [Google Scholar]
- 42.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. PMID: 28666118. DOI: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B, Tian K, Deng SC, Li X, Zhu S, Gong Q, Niu Y, Wang CY. MiR-130 b is a prognostic marker and inhibits cell proliferation and invasion in pancreatic cancer through targeting STAT3. PLoS One. 2013;8:e73803. doi: 10.1371/journal.pone.0073803. PMID: 24040078. DOI: 10.1371/journal.pone.0073803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role of STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. PMID: 19851315. DOI: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee M, Hirpara JL, Eu JQ, Sethi G, Wang L, Goh BC, Wong AL. Targeting STAT3 and oxidative phosphorylation in oncogene-addicted tumors. Redox Biol. 2018:101073. doi: 10.1016/j.redox.2018.101073. PMID: 30594485. DOI: 10.1016/j.redox.2018.101073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao G, Wang B, Liu Y, Zhang JG, Deng SC, Quin A, Tian K, Li X, Zhu S, Niu Y, Gong Q, Wang CY. miRNA-141, downregulated in pancreatic cancer, inhibits cell proliferation and invasion by directly targeting MAP4K4. Mol Cancer Ther. 2013;12:2569–2580. doi: 10.1158/1535-7163.MCT-13-0296. PMID: 24013097. DOI: 10.1158/1535-7163.MCT-13-0296. [DOI] [PubMed] [Google Scholar]
- 47.Gao X, Gao C, Liu G, Hu J. MAP4K4: an emerging therapeutic target in cancer. Cell Biosci. 2016;6:56. doi: 10.1186/s13578-016-0121-7. PMID: 27800153. DOI:10.1186/s13578-016-0121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yue J, Xie M, Gou X, Lee P, Schneider MD, Wu X. Microtubules regulate focal adhesion dynamics through MAP4K4. Dev Cell. 2014;31:572–585. doi: 10.1016/j.devcel.2014.10.025. PMID: 25490267. DOI: 10.1016/j.devcel.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang JJ, Wang H, Rashid A, Tan TH, Hwang RF, Hamilton SR, Abbruzzese JL, Evans DB, Wang H. Expression of MAP4K4 is associated with worse prognosis in patients with stage II pancreatic ductal adenocarcinoma. Clin Cancer Res. 2008;14:7043–7049. doi: 10.1158/1078-0432.CCR-08-0381. PMID:18981001. DOI: 10.1158/1078-0432.CCR-08-0381. [DOI] [PubMed] [Google Scholar]
- 50.Zhao G, Zhang JG, Liu Y, Qin Q, Wang B, Tian K, Liu L, Li X, Niu Y, Deng SC, Wang CY. miR-148b functions as a tumor suppressor in pancreatic cancer by targeting AMPKα1. Mol Cancer Ther. 2013;12:83–93. doi: 10.1158/1535-7163.MCT-12-0534-T. PMID: 23171948. DOI: 10.1158/1535-7163.MCT-12-0534-T. [DOI] [PubMed] [Google Scholar]
- 51.Lee YK, Park SY, Kim YM, Kim DC, Öee WS, Surh YJ, Park OJ. Suppression of mTOR via Akt-dependent and –independent mechanisms in selenium-treated colon cancer cells. Carcinogenesis. 2010;31:1092–1099. doi: 10.1093/carcin/bgq040. PMID: 20164123. DOI: 10.1093/carcin/bgq040. [DOI] [PubMed] [Google Scholar]
- 52.Lee YK, Lee WS, Kim GS, Park OJ. Anthocyanins are novel AMPKα1 stimulators that suppress tumor growth by inhibiting mTOR phosphorylation. Oncol Rep. 2010;24:1471–1477. doi: 10.3892/or_00001007. PMID: 21042741. [DOI] [PubMed] [Google Scholar]
- 53.Stapleton D, Mitchelhill KI, Gao D, Widmer J, Mitchell BJ, Teh T, House CM, Fernandez CS, Cox T, Witters LA, Kemp BE. Mammalian AMP-activated protein kinase subfamily. J Biol Chem. 1996;271:611–614. doi: 10.1074/jbc.271.2.611. PMID: 8557660. DOI: 10.1074/jbc.271.2.611. [DOI] [PubMed] [Google Scholar]
- 54.Zang W, Wang Y, Wang T, Du Y, Chen X, Li M, Zhao G. miR-663 attenuates tumor growth and invasiveness by targeting eEF1A2 in pancreatic cancer. Mol Cancer. 2015;14:37. doi: 10.1186/s12943-015-0315-3. PMID: 25744894. DOI: 10.1186/s12943-015-0315-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem. 2002;269:5360–5368. doi: 10.1046/j.1432-1033.2002.03290.x. PMID: 12423334. DOI: 10.1046/j.14321033.2002.03290.x. [DOI] [PubMed] [Google Scholar]
- 56.Zhu Z, Xu Y, Zhao J, Liu Q, Feng W, Fan J, Wang P. miR-367 promotes epithelial-to-mesenchymal transition and invasion of pancreatic ductal adenocarcinoma cells by targeting the Smad7-TGF-β signaling pathway. Br J Cancer. 2015;112:1367–1375. doi: 10.1038/bjc.2015.102. PMID: 25867271. DOI: 10.1038/bjc.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi W, Sun C, He B, Xiong W, Shi X, Yao D, Cao X. GADD34-PP1c recruited by Smad7 dephosphorylates TGFbeta type I receptor. J Cell Biol. 2004;164:291–300. doi: 10.1083/jcb.200307151. PMID: 14718519. DOI: 10.1083/jcb.200307151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. PMID: 11278251. DOI: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 59.Luo L, Li N, Lv N, Huang D. SMAD7: a timer of tumor progression targeting TGF-β signaling. Tumor Biol. 2014;35:8379–8385. doi: 10.1007/s13277-014-2203-7. PMID: 24935472. DOI: 10.1007/s13277-014-2203-7. [DOI] [PubMed] [Google Scholar]
- 60.Nakajima S, Doi R, Toyoda E, Tsuji S, Wada M, Kozumi M, Tulachan SS, Ito D, Kami K, Mori T, Kawaguchi Y, Fujimoto K, Hosotani R, Imamura M. N-cadherin expression and epithelial-mesenchymal transition in pancreatic carcinoma. Clin Cancer Res. 2004;10:4125–4133. doi: 10.1158/1078-0432.CCR-0578-03. PMID: 15217949. DOI: 10.1158/1078-0432.CCR-0578-03. [DOI] [PubMed] [Google Scholar]
- 61.Wang P, Fan J, Chen Z, Meng ZQ, Luo JM, Lin JH, Zhou ZH, Chen H, Wang K, Xu ZD, Liu M. Low level expression of Smad7 correlates with lymph node metastasis and poor prognosis in patients with pancreatic cancer. Ann Surg Oncol. 2009;16:826–835. doi: 10.1245/s10434-008-0284-5. PMID: 19165547. DOI: 10.1245/s10434-008-0284-5. [DOI] [PubMed] [Google Scholar]
- 62.David CJ, Massague J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat Rev Mol Cell Biol. 2018;19:419–435. doi: 10.1038/s41580-018-0007-0. PMID: 29643418. DOI: 10.1038/s41580-018-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morikawa M, Derynck R, Miyazono K. Contextual determinants of TGFβ action in development, immunity and cancer. Nat Rev Mol Cell Biol. 2018;19:419–435. doi: 10.1038/s41580-018-0007-0. PMID: 29643418. DOI: 10.1038/s41580-018-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zou Y, Li J, Chen Z, Li X, Zhena S, Yi D, Zhona A, Chen J. miR-29c suppresses pancreatic cancer liver metastasis in an orthtopic implantation model in nude mice and affects survival in pancreatic cancer patients. Carcinogenesis. 2015;36:676–684. doi: 10.1093/carcin/bgv027. PMID: 25863127. DOI: 10.1093/carcin/bgv027. [DOI] [PubMed] [Google Scholar]
- 65.Overall CM, Dean RA. Degradomics: systems biology of the protease web. Pleiotropic roles of MMPs in cancer. Cancer Metastasis Rev. 2006;25:69–75. doi: 10.1007/s10555-006-7890-0. PMID: 16680573. DOI: 10.1007/s10555-006-7890-0. [DOI] [PubMed] [Google Scholar]
- 66.Conlon GA, Murray GI. Recent advances in understanding the roles of matrix metalloproteinases in tumor invasion and metastasis. J Pathol. 2019;247:629–640. doi: 10.1002/path.5225. PMID: 30582157. DOI: 10.1002/path.5225. [DOI] [PubMed] [Google Scholar]
- 67.Winer A, Adams S, Mignatti P. Matrix metalloproteinase inhitors in cancer therapy: turning past failures into future successes. Mol Cancer Ther. 2018;17:1147–1155. doi: 10.1158/1535-7163.MCT-17-0646. PMID: 29735645. DOI: 10.1158/1535-7163.MCT-17-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Piperigkou Z, Manou D, Karamanou K, Theocharis AD. Strategies to target matrix metalloproteinases as therapeutic approach in cancer. Meth Mol Biol. 2018;1731:325–348. doi: 10.1007/978-1-4939-7595-2_27. PMID: 29318564. DOI: 10.1007/978-1-4939-7595-2_27. [DOI] [PubMed] [Google Scholar]
- 69.Feng J, Yu J, Li Z, Chen Z, Zhang W, Wang B, Yang L, Xu H, Zhang G, Xu Z. HERG1 functions as an oncogene in pancreatic cancer and is downregulated by miR-96. Oncotarget. 2014;5:5832–5844. doi: 10.18632/oncotarget.2200. PMID: 25071021. DOI: 10.18632/oncotarget.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trudeau MC, Warmke JW, Ganetzky B, Robertson GA. HERG, a human inward rectifier in the voltage-gated potassium channel family. Science. 1995;269:92–95. doi: 10.1126/science.7604285. PMID: 7604285. DOI: 10.1126/science.7604285. [DOI] [PubMed] [Google Scholar]
- 71.Lastraioli E, Perrone G, Sette A, Fiore A, Crociani O, Manoli S, D’Amico M, Masselli M, Iorio J, Callea M, Borzomati D, Nappo G, Bartolozzi F, Santini D, Bencini L, Farsi M, Boni L, Di Costanzo F, Schwab A, Onetti Muda A, Coppola R, Arcangeli A. hERG1 channels drive tumor malignancy and may serve as prognostic factor in pancreatic ductal adenocarcinoma. Br J Cancer. 2015;112:1076–1087. doi: 10.1038/bjc.2015.28. PMID: 25719829. DOI: 10.1038/bjc.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang H, Zhang Y, Cao L, Han H, Wang J, Yang B, Nattel S, Wang Z. HERG K+ channel, a regulator of tumor cell apoptosis and proliferation. Cancer Res. 2002;62:4843–4848. PMID: 12208728. [PubMed] [Google Scholar]
- 73.Camacho J. Ether a go-go potassium channels in cancer. Cancer Lett. 2006;233:1–9. doi: 10.1016/j.canlet.2005.02.016. PMID: 16473665. DOI: 10.1016/j.canlet.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 74.Zhao G, Wang B, Liu Y, Zhang JG, Deng SC, Qin Q, Tian K, Li X, Zhu S, Niu Y, Gong Q, Wang CY. miRNA-141, downregulated in pancreatic cancer, inhibits proliferation and invasion by directly targeting MAP4K4. Mol Cancer Ther. 2013;12:3569–2580. doi: 10.1158/1535-7163.MCT-13-0296. PMID: 24013097. DOI: 10.1158/1535-7163.MCT-13-0296. [DOI] [PubMed] [Google Scholar]
- 75.Liu AW, Cai J, Zhao XL, Jiang TH, He TF, Fu HQ, Zhu MH, Zhang SH. ShRNA-targeted MAP4K4 inhibits hepatocellular carcinoma growth. Clin Cancer Res. 2011;17:710–720. doi: 10.1158/1078-0432.CCR-10-0331. PMID: 21196414. DOI: 10.1158/1078-0432.CCR-10-0331. [DOI] [PubMed] [Google Scholar]
- 76.Liu YR, Qu GQ, Lu YM, Kong WM, Liu Y, Chen WX, Liao XH. Silencing of MAP4K4 by short hairpin RNA suppresses proliferation, induces G1 cell cycle arrest and induces apoptosis in gastric cancer cells. Mol Med Rep. 2016;13:41–48. doi: 10.3892/mmr.2015.4510. PMID: 26549737. DOI: 10.3892/mmr.2015.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hu Y, Ou Y, Wu K, Chen Y, Sun W. miR-143 inhibits the metastasis of pancreatic cancer and an associated signaling pathway. Tumor Biol. 2012;33:1863–1870. doi: 10.1007/s13277-012-0446-8. PMID: 23070684. DOI: 10.1007/s13277-012-0446-8. [DOI] [PubMed] [Google Scholar]
- 78.Lazer G, Katzav S. Guanine nucleotide exchange factors for RhoGTPases: good therapeutic targets for cancer therapy. Cell Signal. 2011;23:969–979. doi: 10.1016/j.cellsig.2010.10.022. PMID: 21044680. DOI: 10.1016/j.cellsig.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 79.Fukata M, Kaibuchi K. Rho-family GTPases in cadherin-mediated cell-cell adhesion. Nat Rev Mol Cell Biol. 2001;2:887–897. doi: 10.1038/35103068. PMID: 11733768. DOI: 10.1038/35103068. [DOI] [PubMed] [Google Scholar]
- 80.Bustelo XR. RHO GTPases in cancer: known facts, open questions, and therapeutic challenges. Biochem Soc Trans. 2018;46:741–760. doi: 10.1042/BST20170531. PMID: 29871878. DOI: 10.1042/BST20170531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou B, Sun C, Hu X, Zhan H, Zou H, Feng Y, Qiu F, Zhang S, Wu L, Zhang B. MicroRNA-195 suppresses the progression of pancreatic cancer by targeting DCLK1. Cell Physiol Biochem. 2017;44:1867–1881. doi: 10.1159/000485876. PMID: 29224010. DOI: 10.1159/000485876. [DOI] [PubMed] [Google Scholar]
- 82.Bailey JM, Alsina J, Rasheed ZA, McAllister FM, Fu YY, Plentz R, Zhang H, Pasricha PJ, Bardeesy N, Matsui W, Maitra A, Leach SD. DCLK1 marks a morphologically distinct subpopulation of cells with stem cell properties in preinvasive pancreatic cancer. Gastroenterology. 2014;146:245–256. doi: 10.1053/j.gastro.2013.09.050. PMID: 24096005. DOI: 10.1053/j.gastro.2013.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sureban SM, May R, Lightfoot SA, Hoskins AB, Lerner M, Brackett DJ, Postier RG, Ramanujam R, Mohammed A, Rao CV, Wyche JH, Anant S, Houchen CW. DCAMKL-1 regulates epithelial-mesenchymal transition in human pancreatic cells through a miR-200a dependent mechasism. Cancer Res. 2011;71:2328–2338. doi: 10.1158/0008-5472.CAN-10-2738. PMID: 21285251. DOI: 10.1158/0008-5472.CAN-10-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sureban SM, May R, Ramalingam D, Natarajan G, Anant S, Houchen CV. Selective blockade of DCAMKL-1 results in tumor growth arrest by a let-7a micrRNA-dependent mechanism. Gastroenterology. 2009;137:649–659. doi: 10.1053/j.gastro.2009.05.004. PMID: 19445940. DOI: 10.1053/j.gastro.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Westphalen CB, Quante M, Wang TC. Functional implication of Dclk1 and Dclk1-expressing cells in cancer. Small GTPases. 2017;8:164–171. doi: 10.1080/21541248.2016.1208792. PMID: 27458755. DOI: 10.1080/21541248.2016.1208792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mody HR, Hung SW, Pathak RK, Griffin J, Cruz-Monserrate Z, Govidarajan R. miR-202 diminishes TGFβ receptors and attenuates TGFβ1-induced EMT in pancreatic cancer. Mol Cancer Res. 2017;15:1029–1039. doi: 10.1158/1541-7786.MCR-16-0327. PMID: 28373289. DOI: 10.1158/1541-7786.MCR-16-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Neuzillet C, de Gramont A, Tijeras-Raballand A, de Mestier L, Cros J, Faivre S, Raymond E. Perspectives of TGFβ inhibition in pancreatic and hepatocellular carcinomas. Oncotarget. 2014;5:78–94. doi: 10.18632/oncotarget.1569. PMID: 24393789. DOI: 10.18632/oncotarget.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Truty MJ, Urrutia R. Basics of TGFβ and pancreatic cancer. Pancreatology. 2007;7:423–435. doi: 10.1159/000108959. PMID: 17898532. DOI: 10.1159/000108959. [DOI] [PubMed] [Google Scholar]
- 89.Katsuno Y, Lamouille S, Derynck R. TGFβ signaling and epithelial-mesenchymal transition in cancer progression. Curr Opin Oncol. 2013;25:76–84. doi: 10.1097/CCO.0b013e32835b6371. PMID: 23197193. DOI: 10.1097/ CCO.0b013e32835b6371. [DOI] [PubMed] [Google Scholar]
- 90.Heldin CH, Vanlandewijck M, Moustakas A. Regulation of EMT by TGFβ in cancer. FEBS Lett. 2012;586:1959–1970. doi: 10.1016/j.febslet.2012.02.037. PMID: 22710176. DOI: 10.1016/j.febslet.2012.02.037. [DOI] [PubMed] [Google Scholar]
- 91.Tsubakihara Y, Moustakas A. Epithelial-mesenchymal transition and metastasis under the control of transforming growth factor β. Int J Mol Sci. 2018;19(11) doi: 10.3390/ijms19113672. PMID: 30463358. DOI: 10.3390/ijms19113672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang C, Liu P, Wu H, Cui P, Li Y, Liu Z, Gou S. MicroRNA-323-3p inhibits cell division and metastasis in pancreatic ductal adenocarcinoma via direct suppression of SMAD2 and SMAD3. Oncotarget. 2016;7:14912–14924. doi: 10.18632/oncotarget.7482. PMID: 26908446. DOI: 10.18632/oncotarget.7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Levy L, Hill CS. Alterations in components of the TGFβ superfamily pathways in human cancer. Cytokine Growth Factor Rev. 2006;17:41–58. doi: 10.1016/j.cytogfr.2005.09.009. PMID: 16310402. DOI: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 94.Wrighton KH, Lin X, Feng XH. Phospho-control of TGFβ superfamily signaling. Cell Res. 2009;19:8–20. doi: 10.1038/cr.2008.327. PMID: 19114991. DOI: 10.1038/cr.2008.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Massague J, Gomis RR. The logic of TGFβ signaling. FEBS Lett. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. PMID: 16678165. DOI: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 96.Fan Y, Shi C, Li T, Kuang T. microRNA-454 shows anti-angiogenic and anti-metastatic activity in pancreatic ductal adenocarcinoma by targeting LRP6. Am J Cancer Res. 2017;7:139–147. PMID: 28123855. [PMC free article] [PubMed] [Google Scholar]
- 97.Williams BO, Insogna KL. Where Wnts went: the exploding field of Lrp5 and Lrp6 signaling in bone. J Bone Min Res. 2009;24:171–178. doi: 10.1359/jbmr.081235. PMID: 19072724. DOI: 10.1359/jbmr.081235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu G, Bafico A, Harris VK, Aaronson SA. A novel mechanism for Wnt activation of canonical signaling through the LRP6 receptor. Mol Cell Biol. 2003;23:5825–5835. doi: 10.1128/MCB.23.16.5825-5835.2003. PMID: 12897152. DOI: 10.1128/MCB.23.16.5825-5835.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garg B, Giri B, Majumder K, Dudeja V, Banerjee S, Saluja A. Modulation of post-translational modifications in β-catenin and LRP6 inhibits Wnt signaling pathway in pancreatic cancer. Cancer Lett. 2017;388:64–72. doi: 10.1016/j.canlet.2016.11.026. PMID: 27919787. DOI: 10.1016/j.canlet.2016.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Arensman MD, Nguyen P, Kershaw KM, Lay AR, Ostertag-Hill CA, Sherman MH, Downes M, Liddle C, Evans RM, Dawson DW. Calcipotriol targets LRP6 to inhibit Wnt signaling in pancreatic cancer. Mol Cancer Res. 2015;13:1509–1519. doi: 10.1158/1541-7786.MCR-15-0204. PMID: 26224368. DOI: 10.1158/1541-7786.MCR-15-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Javadinia SA, Shahidsales S, Fabipakdel A, Joudi-Mashhad M, Mehramiz M, Talebian S, Maftouh M, Mardani R, Hassanian SM, Khazaei M, Ferns GA, Avan A. Therapeutic potential of targeting the Wnt/β-catenin pathway in the treatment of pancreatic cancer. J Cell Biochem. 2018 doi: 10.1002/jcb.27835. PMID: 30368889.DOI: 10.1002/jcb.27835. [DOI] [PubMed] [Google Scholar]
- 102.Kahn M. Can we safely target the WNT pathway. Nat Rev Drug Discov. 2014;13:513–532. doi: 10.1038/nrd4233. PMID: 24981364. DOI: 10.1038/nrd4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. PMID: 23258168. DOI: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 104.Yuan P, He XH, Rong YF, Cao J, Li Y, Hu YP, Li D, Lou W, Liu MF. KRAS/NF-kappaB/YY1/miR-489 signaling axis controls pancreatic cancer metastasis. Cancer Res. 2017;77:100–111. doi: 10.1158/0008-5472.CAN-16-1898. PMID: 27793842. DOI: 10.1158/0008-5472.CAN-16-1898. [DOI] [PubMed] [Google Scholar]
- 105.Wang H, Abbruzzese JL, Evans DB, Larry L, Cleary KR, Chiao PJ. The nuclear factor-kappa B RelA transcription factor is constitutively activated in haman pancreatic adenocarcinoma cells. Clin Cancer Res. 1999;5:119–127. PMID: 9918209. [PubMed] [Google Scholar]
- 106.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. PMID: 20303878. DOI: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: Structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. PMID: 16314846. DOI: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 108.Duffy MJ. The role of proteolytic enzymes in cancer invasion and metastasis. Clin Exp Metastasis. 1992;10:145–155. doi: 10.1007/BF00132746. PMID: 1582084. [DOI] [PubMed] [Google Scholar]
- 109.Grützmann R, Foerder M, Alldinger I, Staub E, Brümmendorf T, Röpke S, Li X, Kristiansen G, Jesenofsky R, Sipos B, Löhr M, Lüttges J, Ockert D, Klöppel G, Saeger HD, Pilarsky C. Gene expression profiles of microdissected pancreatic ductal adenocarcinoma. Virchows Arch. 2003;443:508–517. doi: 10.1007/s00428-003-0884-1. PMID: 12942322. DOI: 10.1007/s00428-003-0884-1. [DOI] [PubMed] [Google Scholar]
- 110.Jakubowska K, Pryczynicz A, Januszwewska J, Sidorkiewicz I, Kemona A, Niewinski A, Lewczuk L, Kedra B, Guzinka-Ustymowicz K. Expressions of metalloproteinases 2, 7, and 9 in carcinogenesis of pancreatic ductal adenocarcinoma. Dis Markers. 2016;2016:9895721. doi: 10.1155/2016/9895721. PMID: 27429508. DOI: 10.1155/2016/9895721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Oria VO, Lopatta P, Schilling O. The pleiotropic roles of ADAM 9 in the biology of solid tumors. Cell Mol Life Sci. 2018;75:2291–2301. doi: 10.1007/s00018-018-2796-x. PMID: 29550974. DOI: 10.1007/s00018-018-2796-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Murphy G. The ADAMs: signalling scissors in the tumor microenvironment. Nat Rev Cancer. 2008;8:929–941. doi: 10.1038/nrc2459. PMID: 19005493. DOI: 10.1038/nrc2459. [DOI] [PubMed] [Google Scholar]
- 113.Sun Y, Zhang T, Wang C, Jin X, Jia C, Yu S, Chen J. MiRNA-615-5p functions as a tumor suppressor in pancreatic ductal adenocarcinoma by targeting AKT2. PLoS One. 2015;10:e0119783. doi: 10.1371/journal.pone.0119783. PMID: 25856297. DOI: 10.1371/journal.pone.0119783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fortier AM, Asselin E, Cadrin M. Functional specificty of Akt isoforms in cancer progression. Biomol Concepts. 2011;2:1–11. doi: 10.1515/bmc.2011.003. PMID: 25962016. DOI: 10.1515/bmc.2011.003. [DOI] [PubMed] [Google Scholar]
- 115.Altomare DA, Tanno S, De Rienzo A, Klein-Szanto AJ, Tanno S, Skele KL, Hofman JP, Testa JR. Frequent activation of AKT2 kinase in human pancreatic carcinomas. J Cell Biochem. 2002;87:470–476. doi: 10.1002/jcb.10287. PMID: 14735903. [DOI] [PubMed] [Google Scholar]
- 116.Cheng JQ, Ruggeri B, Klein WM, Sonoda G, Altomare DA, Watson DK, Testa JR. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc Natl Acad Sci USA. 1996;93:3636–3641. doi: 10.1073/pnas.93.8.3636. PMID: 8622988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ruggeri BA, Huang L, Wood M, Cheng JQ, Testa JR. Amplification and overexpression of the AKT2 oncogene in a subset of human pancreatic ductal adenocarcinomas. Mol Carcinog. 1998;21:81–86. PMID: 9496912. DOI: https://doi.org/10.1002/(SICI)1098-2744(199802)21:2<121::AID-MC6>3.0.CO;2-Q. [PubMed] [Google Scholar]
- 118.Mody HR, Hung SW, Al Saggar M, Griffin J, Govindarajan R. Inhibition of S-Adenosylmethionine-dependent methyl-transferase attenuates TGFβ1- induced EMT and metastasis in pancreatic cancer: putative roles of miR-663a and miR-4787-5p. Mol Cancer Res. 2016;14:1124–1135. doi: 10.1158/1541-7786.MCR-16-0083. PMID: 27624777. DOI: 10.1158/1541-7786.MCR-16-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Javle M, Li Y, Tan D, Dong X, Chang P, Kar S, Li D. Biomarkers of TGF-β signaling pathway and prognosis of pancreatic cancer. PLoS One. 2014;9:e85942. doi: 10.1371/journal.pone.0085942. PMID: 24465802. DOI: 10.1371/journal.pone.0085942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Katz LH, Likhter M, Jogunoori W, Belkin M, Ohshiro K, Mishra L. TGF-β signaling in liver and gastrointestinal cancers. Cancer Lett. 2016;379:166–172. doi: 10.1016/j.canlet.2016.03.033. PMID: 27039259. DOI: 10.1016/j.canlet.2016.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Melisi D, Garcia-Carbonero R, Macarulla T, Pezet D, Deplanque G, Fuchs M, Trojan J, Oettle H, Kozloff M, Cleverly A, Smith C, Estrem ST, Gueorgieva I, Lahn MMF, Blunt A, Benhadji KA, Tabernero J. Galunisertib plus gemcitabine vs. Gemcitabine for first-line treatment of patients with unresectable pancreatic cancer. Br J Cancer. 2018;119:1208–1214. doi: 10.1038/s41416-018-0246-z. PMID: 30318515. DOI: 10.1038/s41416-018-0246-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Holmgaard RB, Schaer DA, Li Y, Castaneda SP, Murphy MY, Xu X, Inigo I, Dobkin J, Manro JR, Iversen DW, Surguladze D, Hall GE, Novosiadly RD, Benhadji KA, Plowman GD, Kalos M, Driscoll KE. Targeting the TGFβ pathway with galunisertib, a TGFβRI small molecule inhibitor, promotes anti-tumor immunnity leading to durable, complete responses, as monotherapy and in combination with checkpoint blockade. J Immunother Cancer. 2018;6:47. doi: 10.1186/s40425-018-0356-4. PMID: 29866156. DOI: 10.1186/s40425-018-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nguyen DD, Chang S. Development of novel therapeutic agents by inhibition of oncogenic microRNAs. Int J Mol Sci 19: 2017;pii:E65. doi: 10.3390/ijms19010065. PMID: 29280958. DOI: 10.3390/ijms19010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ling H, Fabbri M, Calin GA. microRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. PMID: 24172333. DOI: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ors-Kumoglu G, Gulce-Iz S, Biray-Avci C. Therapeutic microRNAs in human cancer. Cytotechnology. 2019;71:411–435. doi: 10.1007/s10616-018-0291-8. PMID: 30600466. DOI: 10.1007/s10616-018-0291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gambari R, Brognara E, Spandidos DA, Fabbri E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: New trends in the development of miRNA therapeutic strategies in oncology. Int J Oncol. 2016;49:5–32. doi: 10.3892/ijo.2016.3503. PMID: 27175518. DOI: 10.3892/ijo.2016.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shende P, Ture N, Gaud RS, Trotta F. Lipid- and polymer-based plexes as therapeutic carriers for bioactive molecules. Int J Pharm. 2019;558:250–260. doi: 10.1016/j.ijpharm.2018.12.085. PMID: 30641179. DOI: 10.1016/j.ijpharm.2018.12.085. [DOI] [PubMed] [Google Scholar]
- 128.Wang Y, Wu C. Site-specific conjugation of polymers to proteins. Biomacromolecules. 2018;19:1804–1825. doi: 10.1021/acs.biomac.8b00248. PMID: 29722971. DOI: 10.1021/acs.biomac.8b00248. [DOI] [PubMed] [Google Scholar]
- 129.Xu Z, Wang D, Cheng Y, Yang M, Wu LP. Polyester based nanovehicles for siRNA delivery. Mater Sci Eng Mater Biol Appl. 2018;92:1006–1015. doi: 10.1016/j.msec.2018.05.031. PMID: 30184724. DOI: 10.1016/j.msec.2018.05.031. [DOI] [PubMed] [Google Scholar]
- 130.Buddolla AL, Kim S. Recent insights into the development of nucleic acid-based nanoparticles for tumor-targeted drug delivery. Colloids Surf B Interfaces. 2018;172:315–322. doi: 10.1016/j.colsurfb.2018.08.057. PMID: 30176511. DOI:10.1016/j.colsurfb.2018.08.057. [DOI] [PubMed] [Google Scholar]
- 131.Ekladious I, Colson YL, Grinstaff MW. Polymer-drug conjugate therapeutics: advances, insights and prospects. Nat Rev Drug Discov. 2018;18 (4):273–294. doi: 10.1038/s41573-018-0005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Prakash TP, Graham MJ, Yu J, Carty R, Low A, Chappell A, Schmidt K, Zhao C, Aghajan M, Murray HF, Riney S, Booten SL, Murray SF, Gaus H, Crosby J, Lima WF, Guo S, Monia BP, Swayze EE, Seth PP. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10 fold in mice. Nucleic Acids Res. 2014;42:8796–87807. doi: 10.1093/nar/gku531. PMID: 24992960. DOI: 10.1093/nar/gku531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Broderick JA, Zamore PD. MicroRNA therapeutics. Gene Ther. 2011;18:1104–1110. doi: 10.1038/gt.2011.50. PMID: 21525952. DOI: 10.1038/gt.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologics. J Control Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. PMID: 21078351. DOI: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 135.Shen ZL, Xia YQ, Yang QS, Tian WD, Chen K, Ma YQ. Polymer-nucleic acid interactions. Polymer-nucleic acid interactions. Top Curr Chem. 2017;375:44. doi: 10.1007/s41061-017-0131-x. PMID: 28357711. DOI: 10.1007/s41061-017-0131-x. [DOI] [PubMed] [Google Scholar]
- 136.Majzoub RN, Ewert KK, Safinya CR. Cationic liposome-nucleic acid nanoparticle assemblies with applications in gene delivery and gene silencing. Philos Trans A Math Phys Eng Sci. 2016;374:pii 20150129. doi: 10.1098/rsta.2015.0129. PMID: 27298431. DOI: 10.1098/rsta.2015.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Beloor J, Zeller S, Choi CS, Lee SK, Kumar P. Cationic cell-penetrating peptides as vehicles for siRNA delivery. Ther Deliv. 2015;6:491–507. doi: 10.4155/tde.15.2. PMID: 25996046. DOI: 10.4155/tde.15.2. [DOI] [PubMed] [Google Scholar]
- 138.Javanmardi S, Aghamaali S, Tamaddon AM. Progress in microRNA delivery using synthetic nanovectors in cancer therapy. Curr Pharm Des. 2018;24:3678–3696. doi: 10.2174/1381612825666181120160316. PMID: 30465494. DOI: 10.2174/1381612825666181120160316. [DOI] [PubMed] [Google Scholar]
- 139.Jones D. Setbacks shadow microRNA therapies in the clinic. Nat Biotechnol. 2018;36:909–910. doi: 10.1038/nbt1018-909. PMID: 30307922. DOI: 10.1038/nbt1018-909. [DOI] [PubMed] [Google Scholar]