Abstract
Background/Aim: Proteomics of invasiveness opens a window on the complexity of the metastasis-engaged mechanisms. The extend and types of this complexity require elucidation. Materials and Methods: Proteomics, immunohistochemistry, immunoblotting, network analysis and systems cancer biology were used to analyse acquisition of invasiveness by human breast adenocarcinoma cells. Results: We report here that invasiveness network highlighted the involvement of hallmarks such as cell proliferation, migration, cell death, genome stability, immune system regulation and metabolism. Identified involvement of cell-virus interaction and gene silencing are potentially novel cancer mechanisms. Identified 6,113 nodes with 11,055 edges affecting 1,085 biological processes show extensive re-arrangements in cell physiology. These high numbers are in line with a similar broadness of networks built with diagnostic signatures approved for clinical use. Conclusion: Our data emphasize a broad systemic regulation of invasiveness, and describe the network of this regulation.
Keywords: Breast cancer, invasiveness, proteomics, systems biology
Systemic analysis of omics data has proven mature in delivering novel insights into mechanisms of carcinogenesis (1-4). It is accepted that the status of cancer networks is more informative for understanding tumor growth than the status of individual genes or proteins (1-4). Even relatively small changes in the activity or expression of many individual proteins and genes may result in a significant cumulative effect on cell physiology. This has led to development of diagnostic signature which has entered the clinic (5,6). Examples of approved clinical applications of diagnostic signatures are Mammaprint Dx and OncoType Dx diagnostic panels (7-9).
The success of diagnostic panels is further developed by implementation of a network analysis (1,3,10). The network analysis allows a comprehensive overview of all engaged components and mechanisms, e.g. proteins, genes, functional and physical interactions. Proteomics, genomics, transcriptomics, metabolomics and electronic health records provide rich sources of data. An understanding that tumorigenesis is the result of coordinated action of many regulatory processes promotes development of tools for systemic analysis, which enhances quality and biological relevance of conclusions (11,12). Tumorigenesis involves hundreds of components, and is not anymore a chain of changes in few tumor suppressors or/and oncogenes (6,13,14). Reported networks of thousands of components and connections represent steps of the tumorigenic transformation of cells or responses to cancer regulators (15). Complexity of the cancer networks raises a question regarding how much of the cellular physiology has to change, when cells acquire invasiveness.
Metastasis is the main cause of lethality in breast cancer. Invasion of malignant cells from the site of a primary tumor into surrounding tissue is the first step toward a metastatic disease (13,16,17). Development of markers to predict transformation of cancer from a localized into a spread disease has been an area of intensive research. Many markers and panels have been identified, including reports of clinical applicability of some of them (8,9,18-24). These reports provide valuable insights into breast carcinogenesis, with description of specific pathways. However, a comprehensive analysis of all regulatory processes engaged in invasiveness has not been reported. Prediction of a large complexity of regulatory mechanisms engaged in acquisition of invasiveness comes from reports that more than one classical hallmark may be affected in one step of carcinogenesis (25-27).
Proteomics is the only technology allowing a comprehensive and simultaneous analysis of thousands of proteins (28,29). Proteome profiles have been reported for human breast epithelial cells at different steps of the carcinogenic transformation and anti-cancer drug treatments (25-28). Proteome profiling of tumors and normal tissues have also been reported (21-24). However, a comprehensive coverage of all cellular proteins is still a challenge. Top-down and bottom-up proteomics are two main approaches. Separation in a two-dimensional gel (2-DE) or ionization in a mass spectrometer allow identification of intact proteins in the top-down approach, whereas LC-MS/MS uses peptides of digested proteins as analytes in the bottom-up approach (30,31). Separation of intact proteins allows detection of protein forms as they are in a cell, and therefore is preferable for representing a proteome, as compared to detection of peptides by LC-MS/MS. Because of technical limitations none of the two proteomic approaches deliver a full and comprehensive coverage of the proteome (30,31). To compensate the lack of full coverage, proteomics may use systems biology to extract regulatory components and mechanisms reflected by the identified proteins. Integration of proteomics with different omics and targeted studies by systems biology has been widely employed (10,25,28,29,32-35).
Introduction of diagnostic signatures unveiled complexity of engaged mechanisms, and calls for their systemic analysis. The question remains about the description of these systemic mechanisms. Which regulatory processes are involved? What are the relations between these regulatory processes? Do they have a clinical impact? We report here a proteome profiling and systemic analysis of acquisition of invasiveness by human breast adenocarcinoma MCF7 cells and comparison with aggressive breast adenocarcinoma MDA-MB-231 cells. We show that the invasiveness is associated with mechanisms of relevance to established and two potentially novel cancer hallmarks. The invasiveness network complexity is high, but it is comparable to networks associated with other carcinogenesis mechanisms and diagnostic signatures. This is a significant broadening of the number and types of the invasiveness-related regulatory processes.
Materials and Methods
Cells and reagents. Human breast adenocarcinoma MCF7 and MDA-MB-231 cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). MCF7 are tumorigenic but not metastatic cells, while MDA-MB-231 are aggressive and metastatic cells. Cells were regularly tested for contaminations, e.g. mycoplasma. Antibodies to HNF4α (sc-6556), BRMS1 (sc-101219) and actin (sc-376421) were obtained from Santa Cruz Biotechnology (Dallas, TX, USA). Antibodies to cyclin G1 (PA5-36050) and β-catenin-like protein 1 (PA5-21112) were obtained from Invitrogen (by Sedeer Medical, Doha, Qatar).
Generation of an isogenic model of invasiveness. We used collagen gel invasion assay to select invasive isogenic clones of MCF7 cells. Cells invading a collagen type I layer were selected. Collagen type 1 from rat tails was obtained from Sigma-Aldrich (C3867; Darmstadt, Germany). Non-invasive cells were removed by washing with cell culture medium. Collagen-invading cells were collected and expanded as single-cell clones in culture plates under substrate-anchored conditions, and passed again through the collagen invasiveness-assay until highly invasive cell clones were generated. Two cycles of selection of collagen-invading cells were performed.
Proteome profiling. For proteome profiling, two-dimensional gel electrophoresis, gel image analysis and MALDI TOF mass spectrometry were used, as described earlier (36). In brief, cells were solubilized in urea-containing buffer for isoelectrofocusing. The first dimension isoelectrofocusing was performed in IPGDry strips, linear, pH 3-10, 18 cm in an IPGPhor instrument (Amersham Biosciences, Uppsala, Sweden). The second dimension SDS-PAGE was performed in Dalt Six (Amersham Biosciences). We generated seven 10% SDS-PAGE large-size gels for each MCF7 and MCFc46 and six large-size gels for MDA-MB-231 cells, and stained them with silver to detect proteins. Protein spots were analyzed using dedicated software (Image Master Platinum v6.0, GE Healthcare, Uppsala, Sweden). Statistical significance of reproducibility of spot expression in 2D gels and differences in expression were evaluated by using the ImageMaster 2D Platinum Version 6.0 software. Proteins whose expression was changed by more than 50% up or down between MCF7, MCF7c46 and MDA-MB-231 were considered for identification. Student’s t-test was used to ensure the statistical significance of the observed changes in expression (p<0.05).
Protein identification. Protein spots were excised from the gels, destained and subjected to in-gel digestion with trypsin (modified, sequence grade porcine, Promega, Madison, WI, USA), as described earlier (36). Tryptic peptides were concentrated and desalted by C18 ZipTip’s. Peptides were eluted with 65% acetonitrile, containing the matrix α-cyano-4-hydroxycinnamic acid, applied directly onto the metal target and analyzed by MALDI TOF MS on a MALDI R instrument (Micromass/Waters, Manchester, UK). Embedded software (MassLinx) was used to collect and process mass spectra. Peptide spectra were internally calibrated using autolytic peptides from trypsin (842.51, 1045.56, and 2211.10 Da). To identify proteins, we performed searches in the NCBInr sequence database using the ProFound search engine (http://65.219.84.5/service/prowl/profound.html). One miscut, alkylation and partial oxidation of methionine were allowed. Search parameters were set to no limitations of pI and “mammalian” was selected for species search. Significance of the identification was evaluated according to the probability value, Z value and sequence coverage.
Systemic analysis. Protein names were translated into gene ontology (GO) terms. Functional and pathway analysis was performed using Cytoscape (cytoscape.org) (37). BioGrid database was used for building networks, and BiNGO was used for building of networks of biological processes. We used tools for union and intersection of networks, available in the Cytoscape platform as plug-in applications. Fischer’s exact test was used to calculate a p-value determining the network connectivity. The p-values for each network can be retrieved from the file deposited at BioModels at ebi.ac.uk, identifier MODEL1904060001.
For analysis of clinically relevant observations, we used open-source clinical datasets and tools, e.g. The Cancer Proteome Atlas (https://www.tcpaportal.org), Genomics Data Commons of the National Cancer Institute (https://portal.gdc.cancer.gov/) and NCBI databases relevant to proteins, genetics and genome (https://www.ncbi.nlm.nih.gov/search).
Immunoblotting. For immunoblotting, cell lysates were resolved on SDS polyacrylamide gels and transferred onto Hybond P membranes (Amersham Biosciences, Piscataway, NJ, USA). Membranes were blocked with 5% (v/v) BSA for one hour and then incubated with a primary antibody against target proteins with dilutions, as recommended by the manufacturer, and followed by incubation with an HRP-conjugated secondary antibody (GE Healthcare, Uppsala, Sweden). The following antibodies were used: anti-HNF4α (c-19, sc-6556), anti-BRMS1 (4H7, sc-101219) and anti-actin (H-6, sc-376421) all from Santa Cruz biotechnology Inc. The proteins were visualized using Western Blotting Luminol Reagents (Santa Cruz Biotechnology Inc).
Invasiveness assay. Membrane was covered with matrigel, and 1,000 cells were seeded in wells of the 96 well-plate ChemoTx® chemotaxis system (cat. no. #116-8; NeuroProbe, Gaithersburg, MD, USA). After 24 hours, the membrane was fixed with 4% paraformaldehyde. The non-invaded cells were removed by cotton swab from the upper chamber of the well. Membrane was stained with 0.5% crystal violet and cells were counted under light microscope.
Immunohistochemistry. Breast cancer BC081120d tissue microarrays (TMA) containing 99 cases of invasive ductal carcinoma, 1 intraductal carcinoma, 9 adjacent normal breast tissue, 1 cancer adjacent breast tissue, single core per case were obtained from US Biomax (Rockville, MD, USA). Sections were deparaffinized, and antigen unmasking solution citrate-based solution (H-3300) was used for antigen retrieval. Vectastain ABC kit (PK-6200) and DAB peroxidase substrate kit (SK-4100) were used for staining. Primary antibodies to cyclin G1 (PA5-36050) and β-catenin-like protein 1 (PA5-21112) were obtained from Invitrogen. After staining, TMA were mounted with a cover oil and cover glasses. Images were taken in a microscope. Intensity of staining, frequency of staining of tumor cells and stromal elements, and histological structure of the samples, including histology of tumor cells were evaluated. Immunohistochemistry staining data were analyzed in relation to the clinical information about the studied cases, e.g. TNM.
Results
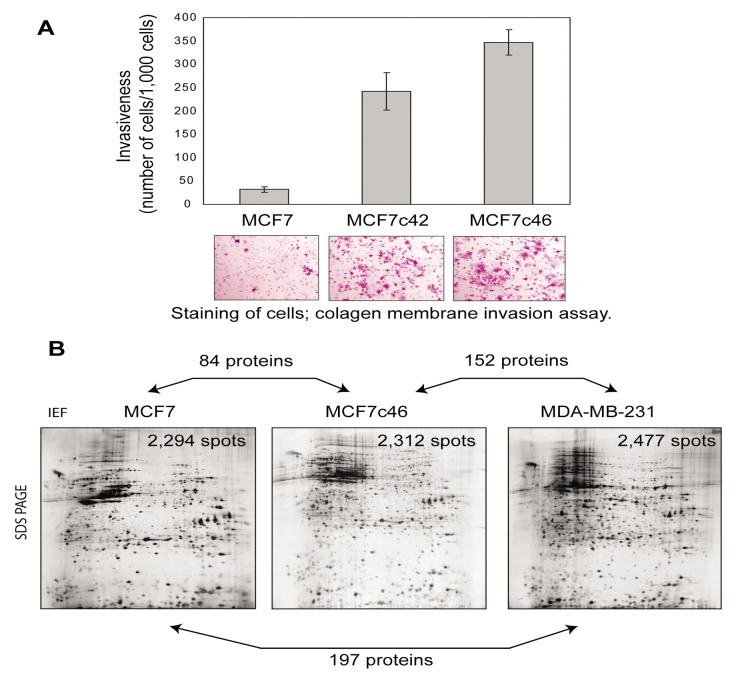
Generation of clones of invasive MCF7 cells. Collagen invasiveness assay was used to collect clones of MCF7 cells, which acquired enhanced invasiveness into a collagen layer. Two cycles of selection were performed. The first cycle consisted of collection of the collagen-invading cells and their expansion. We collected 10 highly invasive clones for further expansion; this number was due to sufficiency of no more than 5 clones for the second cycle of selection. After expansion of the selected clones, the second cycle of selection of highly collagen-invasive cells was performed with the cells collected after the first cycle. After the second cycle, we collected another 10 highly invasive clones for a validation study using a membrane invasiveness assay. For our proteomics study, we selected the clone MCF7c46, as this clone was among the most invasive and stable in maintaining its invasive phenotype during the long-term culturing and experiments, i.e. more than 5 months of the monitoring time before freezing the cells (Figure 1A). MDA-MB-231 cells were also used for our proteomics study, as these cells are metastatic and have reported rates of invasiveness 300-500 cells/1,000 cells, comparable to the rate of MCF7c46 clone shown in Figure 1A.
Figure 1. Generation of proteome profile of acquisition of invasiveness by MCF7 human breast epithelial cells. A) Generation of invasive MCF7c46 cells. The clones MCF7c42 and MCF7c46 cells show higher invasive activity, compared to their parental MCF7 cells. Quantification of invading cells is shown in the graph. Images of membranes with stained cells (purple) are shown. Parental cells (MCF7) and clones (MCF7c42 and MCF7c46) are indicated. B) Images of representative 2D gels with indication of numbers of identified proteins. Representative 2D gels generated with non-invasive MCF7, invasive MCF7c46 and metastatic MDA-MB-231 cells are shown. Numbers of protein spots reproducibly (with p<0.05) detected in 2D gels are indicated in the gel images. Directions of isoelectric focusing (IEF) and SDS-PAGE are indicated on the top and on the side of the left gel image. Numbers of proteins identified as differentially expressed between the annotated by double arrows pairs are shown on the double arrows. List of identified proteins are presented in Supplementary Tables I-SIII.
Proteome profiling and validation. 2D gels were generated for MCF7, MCF7c46 and MDA-MB-231 cells. More than 2,000 protein spots were reproducibly detected in 2D gels for each tested cell line. Significance of reproducible detection was at p<0.05, with analysis of 7 large-size gels each for MCF7 and MCF7c46, and 6 large-size gels for MDA-MB-231 cells (Figure 1B). The 2D gels had size of 18cm strip for the first dimension and up to 20 cm separation in the second dimension. This size is sufficient for reproducible separation of more than 5,000 intact proteins, and detection of 2,000 – 2,500 spots is well within the saturation limit of separation (Figure 1B).
The overall patterns of protein separations in 2D gels of the tested cells were similar, which is in line with the same tissue origin of the cells, i.e. from breast epithelium (Figure 1B). Gel image analysis was performed to detect protein spots differentially expressed between MCF7c46 and MCF7, MCF7 and MDA-MB-231, and MCF7c46 and MDA-MB-231. These three combinations allowed extraction of proteins which are invasiveness-specific, and exclude proteins differentially expressed due to differences between the cell lines and due to invasiveness-unrelated changes upon selection of the MCF7c46 clone.
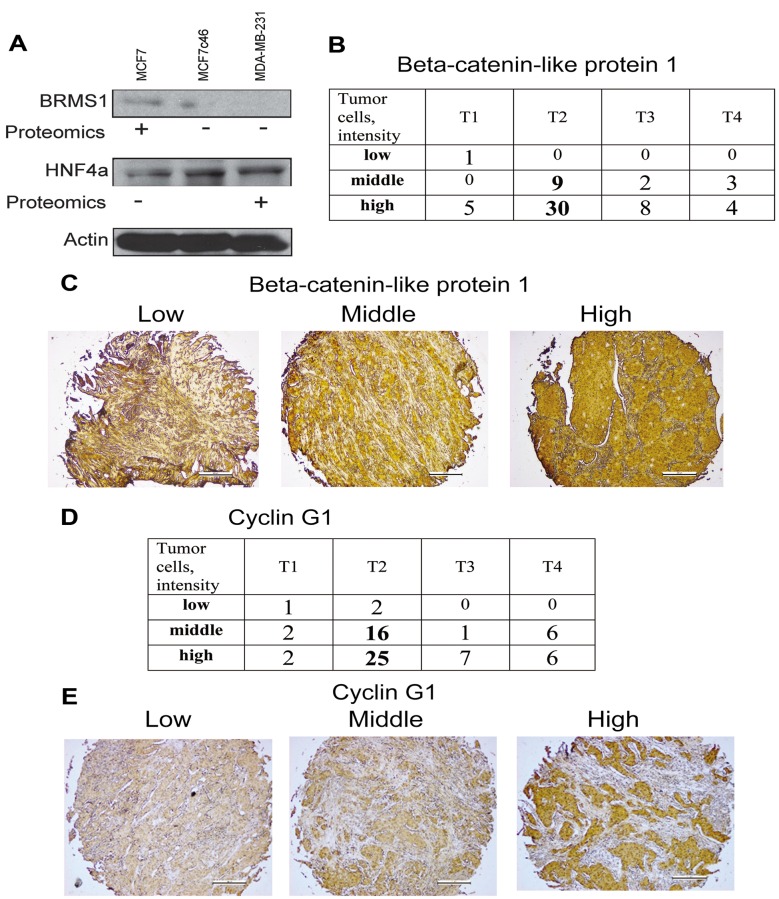
We identified 84 spots corresponding to proteins whose expression changed upon acquisition of the invasive phenotype by MCF7 cells (parental MCF7 vs. invasive MCF7c46), 152 proteins with different expression between invasive MCF7c46 and metastatic MDA-MB-231, and 197 proteins with different expression between non-invasive parental MCF7 and MDA-MB-231 cells (https://www.researchgate.net/publication/33523 4512_SupplInf_Mousa_etal_CGP) To verify the proteomic data, we monitored expression of HNF4α and BRMS1 in MCF7, MCF7c46 and MDA-MB-231 cells, and observed correlations of proteomic data and immunoblotting results (Figure 2A).
Figure 2. Validation studies confirmed the proteomic data. A) Expression of BRMS1 and HNF4α in MCF7, MCF7c46 and MDA-MB-231 cells. Immunoblotting confirmed expression pattern of these proteins detected in the proteome profiling. For proteomics entries for BRMS1 see #64 in Supplementary Table I and #157 in the Supplementary Table II, and for HNF4α see #51. (+) indicates increased expression. Actin immunoblotting was performed for control of loading. B) Immunohistochemistry of β-catenin-like protein 1. Tissue microarray of 62 cases of invasive ductal carcinomas was used for evaluation of β-catenin-like protein 1 expression. Intensity of staining of tumor cells (low, middle and high) in relation to T1, T2, T3 and T4 of TNM classification of cases is presented as numbers of cases. C) Representative images of low (left), middle (middle) and high (right) staining of tissues for expression of β-catenin-like protein 1 are shown. D) Immunohistochemistry of cyclin G1. Tissue microarray of 68 cases of invasive ductal carcinomas was used for evaluation of cyclin G1 expression. Intensity of staining of tumor cells (low, middle and high) in relation to T1, T2, T3 and T4 of TNM classification of cases is presented as numbers of cases. E) Representative images of low (left), middle (middle) and high (right) staining of tissues for expression of cyclin G1 are shown.
To validate our data with clinical samples, we performed an immunohistochemistry study of tissue microarrays containing invasive ductal carcinoma histological sections. We explored expression of 2 of the identified proteins, cyclin G1 and β-catenin-like protein 1 (Figure 2B-E). A significant (predominantly high) expression of β-catenin-like protein 1 (CTNNBL1) and cyclin G1 was observed in invasive ductal carcinomas of breast (Figure 2B-E). Expression levels of CTNNBL1in 62 cases and cyclin G1 in 68 cases, and examples of immunohistochemistry staining are shown in Figure 2B-E. Proteomic data show that both proteins were upregulated upon acquisition of invasiveness. Predominantly high expression of these proteins in clinical samples of invasive ductal carcinomas confirmed the proteomics results.
The approach used here with proteome profiling of non-invasive MCF7, invasive clone MCF7c46, and metastatic MDA-MB-231 cells ensured high probability of identification of proteins associated with acquisition of the invasive phenotype by the cells, and excluded most of the invasiveness-unrelated changes. The identified proteins were subjected to a systemic analysis by building networks for the three sets of identified proteins (Figure 1B), followed by the network analysis, as described in the next section. The validation study (Figure 2) confirmed proteomics data for tested proteins.
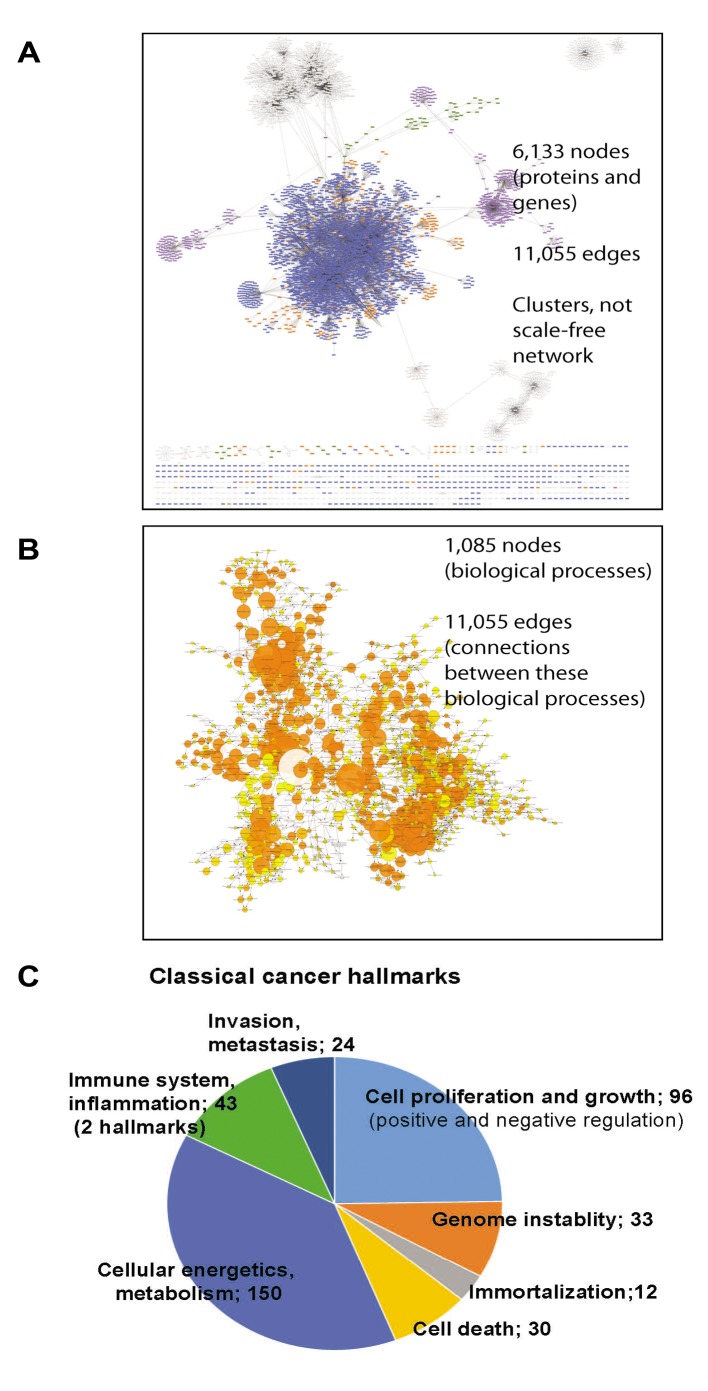
Systemic analysis of invasiveness-specific proteome signature. To identify invasiveness-specific proteome changes, we built networks with the 3 datasets, i.e. MCF7 vs. MCF7c46, MCF7 vs. MDA-MB-231 and MCF7c46 vs. MDA-MB-231 pairs (https://www.researchgate.net/publication/335234512_SupplInf_Mousa_etal_CGP; file deposited at BioModels at ebi.ac.uk, identifier MODEL1904060001). The networks were built in Cytoscape using the BioGrid database. These networks were used to detect an intersection between MCF7 vs. MCF7c46 and MCF7 vs. MDA-MB-231 networks. This intersection would deliver nodes affected upon acquisition of invasiveness and metastatic phenotypes and reflect cell type differences. From this intersection network we subtracted nodes of the MCF7c46 vs. MDA-MB-231 network. The resulting network would have nodes and connections (species and edges) representing invasiveness, and not cell origin (Figure 3A). The network showed clustered nodes and a number of nodes without direct connections to the large single network. The 6,113 nodes and 11,055 edges of the network represent 1,085 biological processes connected by 1,883 edges (Figure 3B; see MODEL1906040001 at ebi.ac.uk BioModels). This large size of the network is expected, as the identified by us invasiveness-related proteins would reflect regulatory processes retrieved by Cytoscape from an extensive database of physical interactions and functional dependencies of proteins and genes, discovered, validated and deposited by other researchers. The size of the network shows that the acquisition of invasiveness is not confined to few pathways only but engages many regulatory mechanisms.
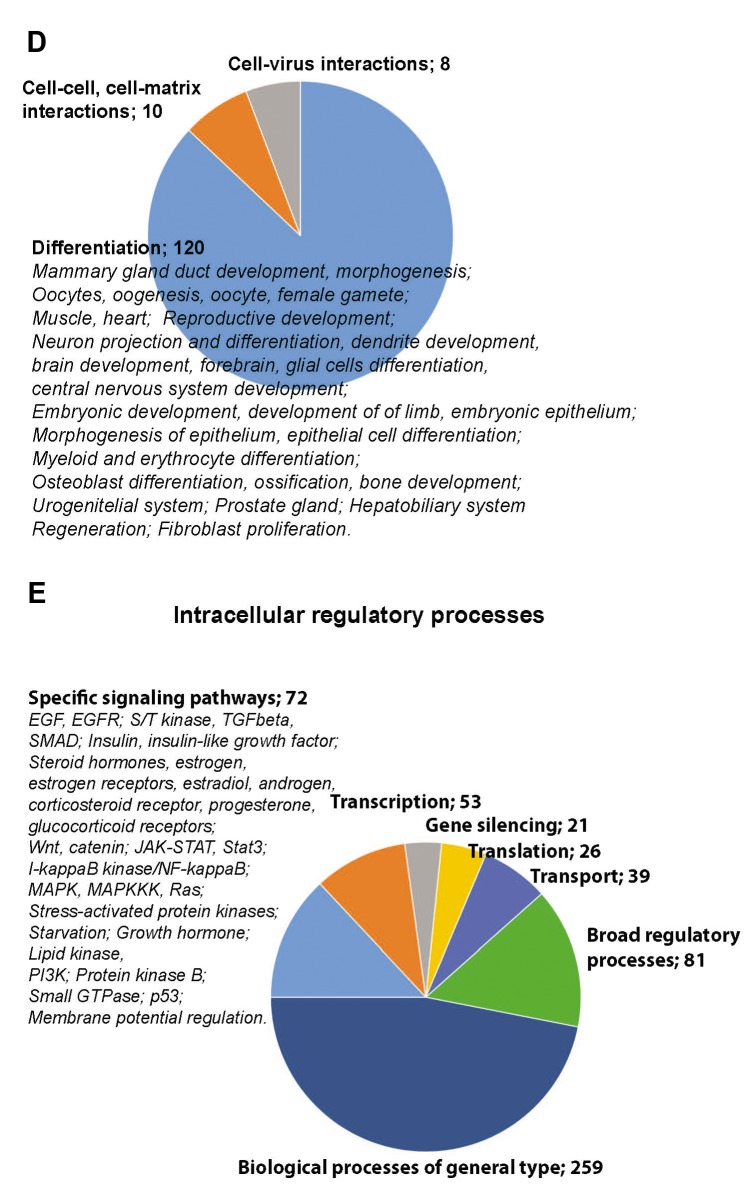
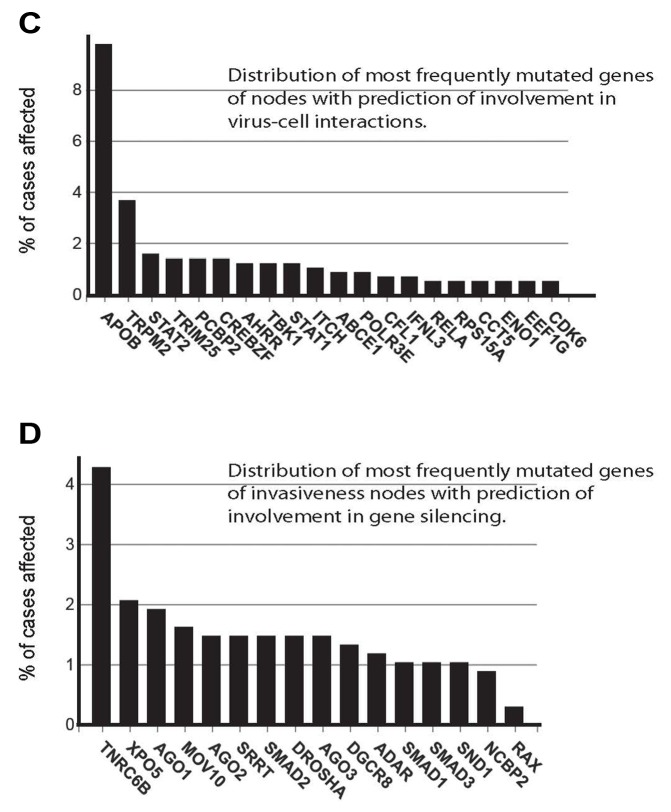
Figure 3. Systemic analysis of the proteome profiling. A) Network of species affected by the identified invasiveness-relevant proteins shows presence of clusters and a large number of involved nodes, e.g. 6,113 nodes. B) Biological processes represented in the invasiveness network are shown. Relations between these processes are presented as a network. Size of the nodes correlates with the number of species engaged in that biological process. Original data of the networks shown in (A) and (B) are in files deposited at BioModels at ebi.ac.uk, identifier MODEL1906040001, with statistic values for each process/node. Distribution of biological processes of relevance to currently accepted cancer hallmarks (C), biological processes not yet included in the cancer hallmarks (D), and relevant to intracellular regulatory mechanisms (E) are shown.
Analysis of 1,085 biological processes and nodes in these processes allowed an evaluation of the complexity of engaged regulatory mechanisms. The established cancer hallmarks were represented in the network, e.g. immortalization, genome instability, cell proliferation, cell death, immune system, cellular energetics and metabolism, and invasion and metastasis (Figure 3C). A representation of regulation of angiogenesis was observed via detection of angiogenesis-regulating signaling pathways and developmental processes engaging angiogenesis (Figure 3D and E). Observation of cell-virus interactions suggests that virus integration and virus defense mechanisms are components of the invasiveness regulatory mechanisms (Figure 3D). Role of oncoviruses in human tumorigenesis is well-documented (38).
Analysis of intracellular regulatory processes showed the involvement of transcription, translation, transport and gene silencing mechanisms (Figure 3E). Of these mechanisms, gene silencing by short RNAs is gaining a recognition as cancer-regulating mechanism (39).
More than the half of the processes represent biological functions on the levels of inter-cellular regulations or processes involving different signaling pathways. List of these processes is the source for further in silico analysis, and is available as an original .cys file of a Cytoscape session at BioModels, identifier MODEL1904060001. If transcription, transport and translation are expected to be affected, mechanisms of gene silencing and virus-host interaction emerge as novel ways to regulate carcinogenesis.
Representation of invasiveness nodes in clinical observations. Our systemic analysis predicts involvement of many proteins and genes (Figure 3). If these predictions are correct, we should be able to observe that the identified nodes are affected in cancer. Our network analysis suggests that the changes of the nodes may be on the level of protein or gene expression, mutations and activities. We evaluated the potential clinical impact of the identified invasiveness nodes by using open-source clinical datasets, e.g. The Cancer Proteome Atlas (https://www.tcpaportal.org), Genomics Data Commons of the National Cancer Institute (https://portal.gdc.cancer.gov/) and NCBI databases relevant to proteins, genetics and genome (https://www.ncbi.nlm.nih.gov/search). These databases contain data about mutations and expression of genes and proteins in different cancers. As a control, we generated random gene lists using http://molbiotools.com/randomgeneset generator.html tool, and run it in parallel with our list of nodes. If identified by us invasiveness nodes would be retrieved as affected in breast cancer, that would support the clinical relevance of these nodes.
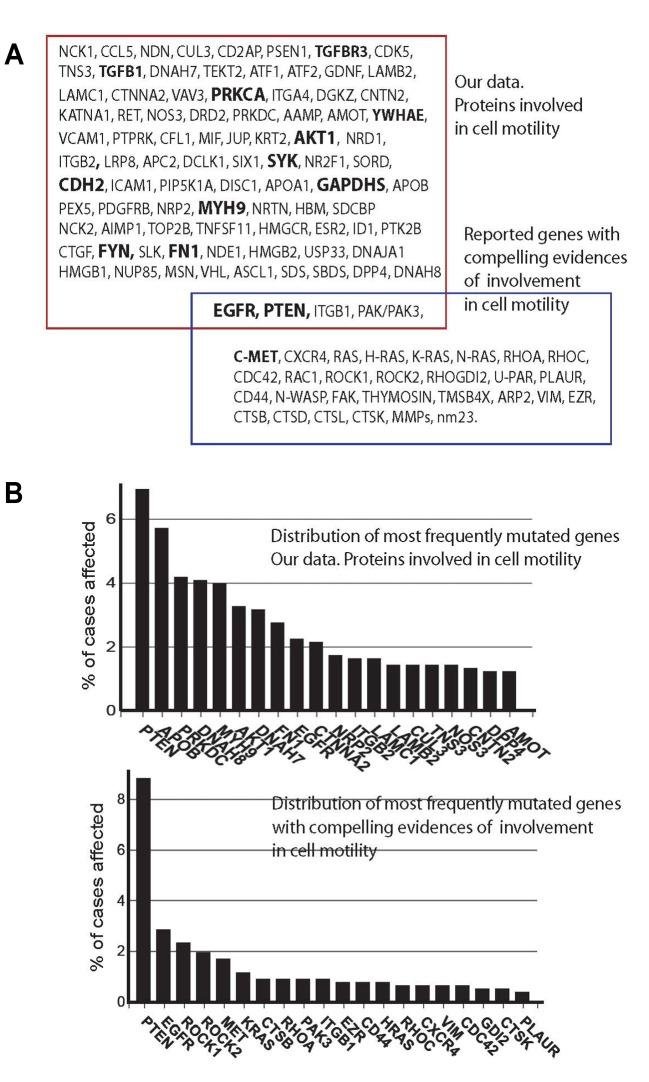
We focused here on nodes involved in cell motility, as a validation example. The validation example would show whether our approach delivers clinically relevant data. We observed that the motility nodes have a strong record of publications linking them to breast cancer (https://www.research gate.net/publication/335234512_SupplInf_Mousa_etal_CGP), confirming relevance of the nodes to breast tumorigenesis. When we compared name-by-name our motility nodes to the motility genes reported earlier, we observed only 4 common nodes/genes (Figure 4A). However, the similarity was in many times higher when affected biological processes were compared with processes represented by the diagnostic signatures (Figure 5). The reason for this difference may be in the ways the lists were produced. Our list was generated from the invasiveness-related network, and has a strong component of a functional impact, whereas the motility signature-genes (Supplementary Table IV) were selected as individual species in a correlation study without taking into account their functional activities. Similar differences in low overlaps in name-by-name comparisons and large overlaps of represented functional processes were observed for reported diagnostic signatures (25). This suggests that system biology approach is more informative in analysis of data as compared to comparing lists name-by-name.
Figure 4. Nodes engaged in regulation of cells motility and correlations with clinical outputs. A) Intersection of the nodes involved in cell motility and identified in this work with the set of genes with compelling evidences of involvement in cell motility is shown (40). GO terms in bold annotate nodes which showed correlations with clinical data on survival of breast cancer patients. Survival data were retrieved from the TCPA portal (https://www.tcpaportal.org/), and contain data of 436 cases. B) Frequencies of mutations of genes of the motility nodes (upper graph) and previously reported motility genes (lower graph) (40) are shown. Top 20 mutated nodes are annotated. C) Frequencies of mutations of genes of the cell-virus interaction nodes. Top 20 nodes are annotated. D) Frequencies of mutations of genes of the gene silencing nodes. Top 16 nodes are annotated. Data for B, C and D were retrieved from GDC database (https://portal.gdc.cancer.gov/). The top nodes with highest mutation frequencies are shown.
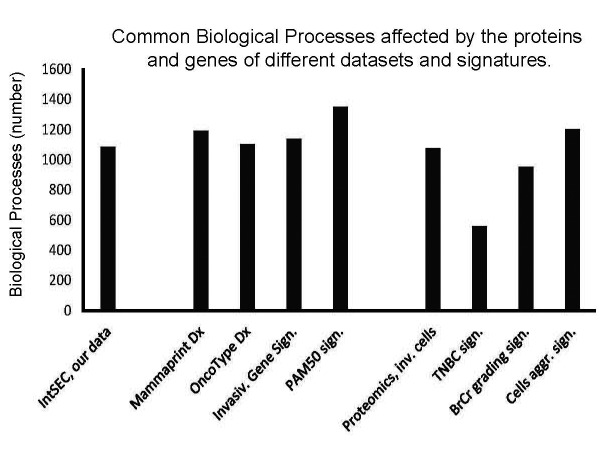
Figure 5. Invasiveness-network shows engagement of biological processes at a scale comparable to the numbers of biological processes engaged by the protein and gene diagnostic signatures. Numbers of biological processes reported here: invasiveness network (1,085 processes), diagnostic gene-signatures Mammaprint Dx (1,194 processes), OncoType Dx (1,103 processes), Invasiveness Gene Signature (1,140 processes), PAM50 (1,353 processes), and proteomics signatures of invasive cells (152 proteins; 1,076 processes), triple-negative breast cancer (11 proteins; 559 processes), grading signature (49 proteins; 954 processes), and aggressiveness signature (44 proteins; 1,203 processes). Note engagement of large number of biological processes by the signatures, identified by systemic analysis (see file of the networks, which is deposited ta BioModels at ebi.ac.uk, identifier MODEL1904060001).
Validation of predicted by systems biology activities may be performed in many ways. One way is an experimental interrogation of each node in a network. However, with thousands of nodes, it is an unrealistic task. Accepted and realistic is a validation study of selected nodes. We did a validation study for 4 nodes, i.e. BRMS1, HNF4α, CTNNBL1 and CCNG1, and confirmed our proteomic data (Figure 2). Another way of validation is the use of large and rich resources deposited in various databases, which may describe relations of a node of interest to breast cancer. We performed searches for representation of the motility sub-set of invasiveness nodes with 1) published reports of involvement in breast cancer, and 2) frequencies of mutations of genes in breast cancer.
Search of publications confirmed that the identified nodes are relevant to breast cancer (Supplementary Tables V-VIII). The majority of motility nodes have been broadly studied in the context of breast cancer. Comparison with a similar search for established gene signature of cell motility (Supplementary Table VI) showed 32 genes (94% of all genes) with more than 5 references. Out of the reported by us motility nodes, 61 (67%) have more than 5 references. Only 5 nodes were not retrieved with searches for “breast” & “cancer”. However, these nodes were retrieved with PubMed searches for links to “cancer”, and showed relevance to testicular, lung, prostate, lung, pancreatic, ovarian cancers. Thus, these motility nodes do have records linking them to carcinogenesis.
Searches of publications reflecting breast cancer engagement of cell-virus interaction and gene silencing nodes (Supplementary Table VII and Table VIII) support involvement of these nodes in breast cancer. Of 35 cell-virus interaction nodes, 31 were reported as relevant to breast cancer. The 4 other nodes were reported as involved in regulations of hepatitis C virus, Epstein-Barr virus, p53-dependent and virus-dependent transcription. Therefore, all 35 cell-virus interaction nodes relevant to invasiveness regulatory network have also been reported as cancer-relevant. The similar observation was made for the gene silencing invasiveness-relevant nodes. Twenty-seven nodes have been reported in the context of breast cancer. Other eight nodes were retrieved as involved in cancer-relevant regulatory processes, e.g. processing of Epstein-Barr and hepatitis C viruses, hepatocellular carcinoma, leukemia, glioblastoma, stem cells differentiation and leiomyoma. Thus, retrieved publications support involvement of reported here nodes in cancer.
In addition to reviewing publications, a search for cancer-relevant mutations in the genes of invasiveness nodes, and modulations of expression as genes and/or proteins may support clinical relevance. A search of the Cancer Proteome Atlas database with the motility nodes retrieved 13 nodes with recorded correlations between expression of the corresponding proteins and survival of breast cancer patients (Figure 4A, nodes in bold). We also searched a breast cancer database CGD for mutations of the nodes genes. Mutation rates of our motility nodes were found to be similar to the rates of the confirmed motility genes (Figure 4B). Comparison to the mutation rates of random lists of genes showed up to 2x higher mutation frequency for the motility nodes as compared to the random gene list. Search for mutation rates of genes of nodes involved in cell-virus interactions and gene silencing sub-sets of the invasiveness nodes showed that most of the rates were below 2% (Figure 4C and D). The mutation rates for all sets of nodes (Figure 4B, C and D) were not sufficiently high to claim a strong contribution of mutations to carcinogenesis. Literature reports, on the other side, support involvement of these nodes in cancer. For example, the mutation rate of CDK6 gene is below 1.0%, but there are more than 300 publications linking this node to breast cancer, including activity of CDK6 (Figure 4C; Supplementary Table VII). Therefore, the identified by us nodes may be involved in breast tumorigenesis on different levels, as genes or proteins. Systems biology and network analysis allow combination different types of data, e.g. mutagenesis, expression and activities. This merging of different data is crucial for supporting involvement of identified nodes in tumorigenesis, including proteins identified by proteomics and nodes identified by the network analysis.
The network effect of observed invasiveness nodes is supported by the comparison of biological processes affected by the invasiveness nodes and the reported gene and protein signatures (Figure 5). For this comparison, we built networks for diagnostic signatures (file deposited at BioModels at ebi.ac.uk, identifier MODEL1904060001). Mammaprint Dx, OncoType Dx, Invasiveness Gene Signature and PAM50 signature are approved for use in diagnosis of breast cancer (8,9,19,20). There are also reported proteome signatures relevant to metastasis and aggressiveness of breast cancer (21-24). We built networks with the signatures’ genes or proteins and extracted biological processes affected by these genes or proteins. Then, we compared these biological processes with the processes associated with our invasiveness network. We observed a similarity in quantity and types of affected biological processes (Figure 5). Our network shows engagement of 1,085 biological processes. The networks of the diagnostic gene-signatures indicated involvement of more than 1,000 biological processes for gene-based signatures (from 1,103 to 1,353). The networks of reported proteomic signatures showed engagement in the range of 1,000 processes for proteomic signatures of 152, 49 and 44 proteins. Only TNBC 11-protein signature resulted in 559 processes (Figure 5). Regulation of proliferation, cell differentiation, motility, cell death, various metabolic and signaling pathways are examples of similar processes (file deposited at BioModels at ebi.ac.uk, identifier MODEL1904060001). This analysis of our data and reported earlier signatures suggest that the high complexity of engaged mechanisms is a general phenomenon associated with tumorigenesis. This makes cancer from a disease of few oncogenes and tumor suppressors into a systemic disease involving hundreds of components.
Discussion
Cell signaling is a network. The size and shape of this network may include hundreds to thousands of components (1-4,10,12,15,16). When cells acquire an invasive phenotype, it is expected that the affected regulatory processes would be complex. After all, the cells may have to modify cell-cell and cell-substrate interactions, proliferation, death, migration and differentiation status (13-16). All these activities constitute established cancer hallmarks. The reported herein proteome profiling and systemic analysis describes this complexity of invasiveness for human breast adenocarcinoma cells. We observed that this complexity covers all established cancer hallmarks and adds two more processes, i.e. gene silencing and mechanisms engaged in cell-virus interactions.
Engaging cancer hallmarks in metastasis is expected, but the scale of this engagement has been under investigation (5,40). Our data show that the cellular transformation into an invasive phenotype is a large-scale process. The number of engaged biological processes is 1,085 (Figure 4), showing that the inputs to these processes are multiple. The multiplicity of inputs is a feature of a robust signaling. When there are many triggers, the probability that the cells would respond is higher, as compared to a single input which may be compensated by the network signaling (12-14,41). As an example, we observed 24 biological processes directly linked to invasiveness and metastasis (Figure 3C). Ten more biological processes were involved in the regulation of cell-cell and cell-matrix interactions (Figure 3D). These 34 processes ensure a robust response.
Our data do not contest validity of oncogenes and tumor suppressors as strong drivers of carcinogenesis. We suggest that the regulatory mechanisms are not passive passengers, but are active participants affecting cancer drivers. These mechanisms are often identical to normal regulatory processes, but are tricked to support malignant growth (1-3,41). The described here invasiveness-relevant mechanisms have been reported as strong regulators of cellular physiology. For example, the nodes regulating cell motility represent potent regulators of transcription, (de)differentiation, cell cycle and cytoskeletal regulation (Figure 4A).
Viruses are known to affect tumorigenesis (38). It is expected that tumor cells would develop mechanisms to interact with viruses. Gene silencing by microRNAs is another regulatory mechanism engaged in carcinogenesis (39,42-44). The observations of virus-cell interaction and gene silencing mechanisms as part of a cell motility relevant to invasiveness (Figure 4C and D), adds another layer of complexity to the regulation of invasiveness.
Reported diagnostic protein and gene signatures provide an opportunity to cross-validate our results. When we validated our data with networks built with Mammaprint Dx, OncoType Dx, IGS and PAM50 genes signatures and with 4 proteomics signatures, the overlap in biological processes was both quantitative (Figure 5) and qualitative (files of signatures deposited at BioModels at ebi.ac.uk, identifier MODEL 1904060001). The signatures contain 70 (Mammaprint Dx,; 6,366 nodes), 21 (OncoTarget Dx,; 3,259 nodes), 186 (IGS; 11,047 nodes), and 50 (PAM50; 2,402 nodes) genes, and 152 (9,190 nodes), 49 (4,810 nodes), 11 (TNBC signature; 845 nodes) and 44 (4,315 nodes) proteins (Figure 5; see also the file deposited at BioModels at ebi.ac.uk, identifier MODEL 1904060001) (8,9,19-24). The reported signatures show engagement of a high number of biological processes (Figure 5). This is a strong indication that each step of tumorigenesis affects profoundly cellular physiology, where thousands of cancer “drivers”, “modulators” and “passengers” are equally important for transforming a normal cell into a cancerous one. Our results illustrate the case of acquisition of invasiveness by breast adenocarcinoma cells, by describing the network of the invasiveness, e.g. nodes and connections.
Supplementary Information
Supplementary Tables I-VIII can be accessed online at https://www.researchgate.net/publication/335234512_SupplInf_Mousa_etal_CGP and the file of the networks is deposited at BioModels at ebi.ac.uk (identifier MODEL1904060001).
Conflicts of Interest
The Authors declared no conflict of interest regarding this study.
Authors’ Contributions
HM, ME, RGM, NS and SS performed analysis of data. HM, ME, RGM, KWL performed validation studies. KWL generated the model and performed proteome profiling. KWL and NS did mass spectrometry. SS designed and supervised the project, analyzed data, wrote the manuscript. All Authors participated in reviewing the manuscript.
Acknowledgements
This work was supported in part by the NPRP9-453-3-089, QUCG-CMED-2018/2019-2, IRGC-05-SI-18-307 and MRC-03-19-039 grants.
References
- 1.Chakraborty S, Hosen MI, Ahmed M, Shekhar HU. Onco-Multi-OMICS Approach: A New Frontier in Cancer Research. Biomed Res Int. 2018;2018:9836256. doi: 10.1155/2018/9836256. PMID: 30402498. DOI: 10.1155/2018/9836256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang E, Zaman N, Mcgee S, Milanese JS, Masoudi-Nejad A, O’Connor-McCourt M. Predictive genomics: a cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data. Semin Cancer Biol. 2014;30:4–12. doi: 10.1016/j.semcancer.2014.04.002. PMID: 24747696. DOI: 10.1016/j.semcancer.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Clarke R. Introduction: Cancer Gene Networks. Methods Mol Biol. 2017;13:1–9. doi: 10.1007/978-1-4939-6539-7_1. PMID: 27807826. DOI: 10.1007/978-1-4939-6539-7_1. [DOI] [PubMed] [Google Scholar]
- 4.Mueller C, Haymond A, Davis JB, Williams A, Espina V. Protein biomarkers for subtyping breast cancer and implications for future research. Expert Rev Proteomics. 2018;15(2):131–152. doi: 10.1080/14789450.2018.1421071. PMID: 29271260. DOI: 10.1080/14789450.2018.1421071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai X, Xiang L, Li T, Bai Z. Cancer Hallmarks, biomarkers and breast cancer molecular subtypes. J Cancer. 2016;7:1281–1294. doi: 10.7150/jca.13141. PMID: 27390604. DOI: 10.7150/jca.13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ali M, Aittokallio T. Machine learning and feature selection for drug response prediction in precision oncology applications. Biophys Rev. 2019;11(1):31–39. doi: 10.1007/s12551-018-0446-z. PMID: 30097794. DOI: 10.1007/s12551-018-0446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu F, Quan F, Xu J, Zhang Y, Xie Y, Zhang J, Lan Y, Yuan H, Zhang H, Cheng S, Xiao Y, Li X. Breast cancer prognosis signature: linking risk stratification to disease subtypes. Brief Bioinform. 2018;3 doi: 10.1093/bib/bby073. PMID: 30184043. DOI: 10.1093/bib/bby073. [DOI] [PubMed] [Google Scholar]
- 8.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. PMID: 12490681. DOI: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 9.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. PMID: 15591335. DOI: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 10.Korcsmaros T, Schneider MV, Superti-Furga G. Next generation of network medicine: interdisciplinary signaling approaches. Integr Biol (Camb) 2017;9(2):97–108. doi: 10.1039/c6ib00215c. PMID: 28106223. DOI: 10.1039/c6ib00215c. [DOI] [PubMed] [Google Scholar]
- 11.Yoo BC, Kim KH, Woo SM, Myung JK. Clinical multi-omics strategies for the effective cancer management. J Proteomics. 2018;88:97–106. doi: 10.1016/j.jprot.2017.08.010. PMID: 28821459. DOI: 10.1016/j.jprot.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Kann BH, Thompson R, Thomas CR, Dicker A, Aneja S. Artificial intelligence in oncology: current applications and future directions. Oncology. 2019;33(2):1–20. PMID: 30784028. [PubMed] [Google Scholar]
- 13.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. PMID: 22000009. DOI: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Souchelnytskyi S. Bridging proteomics and systems biology: what are the roads to be traveled. Proteomics. 2005;5(16):4123–4137. doi: 10.1002/pmic.200500135. PMID: 16196099. DOI: 10.1002/pmic.200500135. [DOI] [PubMed] [Google Scholar]
- 15.Kuperstein I, Bonnet E, Nguyen HA, Cohen D, Viara E, Grieco L, Fourquet S, Calzone L, Russo C, Kondratova M, Dutreix M, Barillot E, Zinovyev A. Atlas of Cancer Signalling Network: a systems biology resource for integrative analysis of cancerdata with Google Maps. Oncogenesis. 2015;4:e160. doi: 10.1038/oncsis.2015.19. PMID: 26192618. DOI: 10.1038/oncsis.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redig AJ, McAllister SS. Breast cancer as a systemic disease: a view of metastasis. J Intern Med. 2013;274(2):113–126. doi: 10.1111/joim.12084. PMID: 23844915. DOI: 10.1111/joim.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steeg PS. Targeting metastasis. Nat Rev Cancer. 2016;16(4):201–218. doi: 10.1038/nrc.2016.25. PMID: 27009393. DOI: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCart Reed AE, Kalita-De Croft P, Kutasovic JR, Saunus JM, Lakhani SR. Recent advances in breast cancer research impacting clinical diagnostic practice. J Pathol. 2019;247(5):552–562. doi: 10.1002/path.5199. PMID: 30426489. DOI: 10.1002/path.5199. [DOI] [PubMed] [Google Scholar]
- 19.Wallden B, Storhoff J, Nielsen T, Dowidar N, Schaper C, Ferree S, Liu S, Leung S, Geiss G, Snider J, Vickery T, Davies SR, Mardis ER, Gnant M, Sestak I, Ellis MJ, Perou CM, Bernard PS, Parker JS. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med Genomics. 2015;8:54. doi: 10.1186/s12920-015-0129-6. PMID: 26297356. DOI: 10.1186/s12920-015-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356(3):217–226. doi: 10.1056/NEJMoa063994. PMID: 17229949. DOI: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 21.Dun MD, Chalkley RJ, Faulkner S, Keene S, Avery-Kiejda KA, Scott RJ, Falkenby LG, Cairns MJ, Larsen MR, Bradshaw RA, Hondermarck H. Proteotranscriptomic profiling of 231-BR breast cancer cells: Identification of potential biomarkers and therapeutic targets for brain metastasis. Mol Cell Proteomics. 2015;14(9):2316–2330. doi: 10.1074/mcp.M114.046110. PMID: 26041846. DOI: 10.1074/mcp.M114.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsson N, Carlsson P, James P, Hansson K, Waldemarson S, Malmström P, Fernö M, Ryden L, Wingren C, Borrebaeck CA. Grading breast cancer tissues using molecular portraits. Mol Cell Proteomics. 2013;12(12):3612–3623. doi: 10.1074/mcp.M113.030379. PMID: 23982162. DOI: 10.1074/mcp.M113.030379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu NQ, Stingl C, Look MP, Smid M, Braakman RB, De Marchi T, Sieuwerts AM, Span PN, Sweep FC, Linderholm BK, Mangia A, Paradiso A, Dirix LY, Van Laere SJ, Luider TM, Martens JW, Foekens JA, Umar A. Comparative proteome analysis revealing an 11-protein signature for aggressive triple-negative breast cancer. J Natl Cancer Inst. 2014;106(2):djt376. doi: 10.1093/jnci/djt376. PMID: 24399849. DOI: 10.1093/jnci/djt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terp MG, Lund RR, Jensen ON, Leth-Larsen R, Ditzel HJ. Identification of markers associated with highly aggressive metastatic phenotypes using quantitative comparative proteomics. Cancer Genomics Proteomics. 2012;9(5):265–273. PMID: 22990106. [PubMed] [Google Scholar]
- 25.Souchelnytskyi S. Intersection between genes controlling vascularization and angiogenesis in renal cell carcinomas. Exp Oncol. 2018;40(2):140–143. PMID: 29949527. [PubMed] [Google Scholar]
- 26.Tian S, Roepman P, Van’t Veer LJ, Bernards R, de Snoo F, Glas AM. Biological functions of the genes in the mammaprint breast cancer profile reflect the hallmarks of cancer. Biomark Insights. 2010;5:129–138. doi: 10.4137/BMI.S6184. PMID: 21151591. DOI: 10.4137/BMI.S6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolf DM, Lenburg ME, Yau C, Boudreau A, van ‘t Veer LJ. Gene co-expression modules as clinically relevant hallmarks of breast cancer diversity. PLoS One. 2014;9(2):e88309. doi: 10.1371/journal.pone.0088309. PMID: 24516633. DOI: 10.1371/journal.pone.0088309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanash S, Taguchi A. The grand challenge to decipher the cancer proteome. Nat Rev Cancer. 2010;10(9):652–660. doi: 10.1038/nrc2918. PMID: 20733593. DOI: 10.1038/nrc2918. [DOI] [PubMed] [Google Scholar]
- 29.Hondermarck H, Vercoutter-Edouart AS, Révillion F, Lemoine J, el-Yazidi-Belkoura I, Nurcombe V, Peyrat JP. Proteomics of breast cancer for marker discovery and signal pathway profiling. Proteomics. 2001;1:1216–1232. doi: 10.1002/1615-9861(200110)1:10<1216::AID-PROT1216>3.0.CO;2-P. PMID: 11721634. DOI: 10.1002/1615-9861(200110)1:10<1216::AID-PROT1216>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 30.Kim YI, Cho JY. Gel-based proteomics in disease research: Is it still valuable. Biochim Biophys Acta Proteins Proteom. 2019;1867(1):9–16. doi: 10.1016/j.bbapap.2018.08.001. PMID: 30392562. DOI: 10.1016/j.bbapap.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Toby TK, Fornelli L, Kelleher NL. Progress in top-down proteomics and the analysis of proteoforms. Annu Rev Anal Chem (Palo Alto Calif) 2016;9(1):499–519. doi: 10.1146/annurev-anchem-071015-041550. PMID: 27306313. DOI: 10.1146/annurev-anchem-071015-041550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreira JM, Cabezón T, Gromova I, Gromov P, Timmermans-Wielenga V, Machado I, Llombart-Bosch A, Kroman N, Rank F, Celis JE. Tissue proteomics of the human mammary gland: towards an abridged definition of the molecular phenotypes underlying epithelial normalcy. Mol Oncol. 2010;4(6):539–561. doi: 10.1016/j.molonc.2010.09.005. PMID: 21036680. DOI: 10.1016/j.molonc.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suman S, Basak T, Gupta P, Mishra S, Kumar V3, Sengupta S, Shukla Y. Quantitative proteomics revealed novel proteins associated with molecular subtypes of breast cancer. J Proteomics. 2016;148:183–193. doi: 10.1016/j.jprot.2016.07.033. PMID: 27498393. DOI: 10.1016/j.jprot.2016.07.033. [DOI] [PubMed] [Google Scholar]
- 34.Zakharchenko O, Greenwood C, Lewandowska A, Hellman U, Alldridge L, Souchelnytskyi S. Meta-data analysis as a strategy to evaluate individual and common features of proteomic changes in breast cancer. Cancer Genomics Proteomics. 2011;8(1):1–14. PMID: 21289332. [PubMed] [Google Scholar]
- 35.Low SK, Zembutsu H, Nakamura Y. Breast cancer: The translation of big genomic data to cancer precision medicine. Cancer Sci. 2018;109(3):497–506. doi: 10.1111/cas.13463. PMID: 29215763. DOI: 10.1111/cas.13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saini RK, Attarha S, da Silva Santos C, Kolakowska J, Funa K, Souchelnytskyi S. Proteomics of dedifferentiation of SK-N-BE2 neuroblastoma cells. Biochem Biophys Res Commun. 2014;454(1):202–209. doi: 10.1016/j.bbrc.2014.10.065. PMID: 25450381. DOI: 10.1016/j.bbrc.2014.10.065. [DOI] [PubMed] [Google Scholar]
- 37.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. PMID:14597658. DOI: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang Y, Moore PS, Weiss RA. Human oncogenic viruses: nature and discovery. Philos Trans R Soc Lond B Biol Sci. 2017;372(1732):pii 20160264. doi: 10.1098/rstb.2016.0264. PMID: 28893931. DOI: 10.1098/rstb.2016.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. PMID: 19167326. DOI: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sahai E. Mechanisms of cancer cell invasion. Curr Opin Genet Dev. 2005;15:87–96. doi: 10.1016/j.gde.2004.12.002. PMID: 15661538. DOI: 10.1016/j.gde.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Alberghina L, Westerhoff HV, editors. Springer-Verlag, Berlin Heidelberg. 2005. Systems Biology: Definitions and Perspectives. ISBN: 978-3-540-22968-1. DOI: 10.1007/b95175. [Google Scholar]
- 42.Petrovic N, Ergün S, Isenovic ER. Levels of microRNA heterogeneity in cancer biology. Mol Diagn Ther. 2017;21(5):511–523. doi: 10.1007/s40291-017-0285-9. PMID: 28620889. DOI: 10.1007/s40291-017-0285-9. [DOI] [PubMed] [Google Scholar]
- 43.Volinia S, Galasso M, Croce CM. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc Natl Acad Sci USA. 2012;109(8):3024–3029. doi: 10.1073/pnas.1200010109. PMID: 22315424. DOI: 10.1073/pnas.1200010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weidle UH, Dickopf S, Hintermair C, Kollmorgen G, Birzele F, Brinkmann U. The role of micro RNAs in breast cancer metastasis: preclinical validation and potential therapeutic targets. Cancer Genomics Proteomics. 2018;15(1):17–39. doi: 10.21873/cgp.20062. PMID: 29275360. DOI: 10.21873/cgp.20062. [DOI] [PMC free article] [PubMed] [Google Scholar]