ABSTRACT
Background
Three-dimensional optical (3DO) body scanning has been proposed for automatic anthropometry. However, conventional measurements fail to capture detailed body shape. More sophisticated shape features could better indicate health status.
Objectives
The objectives were to predict DXA total and regional body composition, serum lipid and diabetes markers, and functional strength from 3DO body scans using statistical shape modeling.
Methods
Healthy adults underwent whole-body 3DO and DXA scans, blood tests, and strength assessments in the Shape Up! Adults cross-sectional observational study. Principal component analysis was performed on registered 3DO scans. Stepwise linear regressions were performed to estimate body composition, serum biomarkers, and strength using 3DO principal components (PCs). 3DO model accuracy was compared with simple anthropometric models and precision was compared with DXA.
Results
This analysis included 407 subjects. Eleven PCs for each sex captured 95% of body shape variance. 3DO body composition accuracy to DXA was: fat mass R2 = 0.88 male, 0.93 female; visceral fat mass R2 = 0.67 male, 0.75 female. 3DO body fat test-retest precision was: root mean squared error = 0.81 kg male, 0.66 kg female. 3DO visceral fat was as precise (%CV = 7.4 for males, 6.8 for females) as DXA (%CV = 6.8 for males, 7.4 for females). Multiple 3DO PCs were significantly correlated with serum HDL cholesterol, triglycerides, glucose, insulin, and HOMA-IR, independent of simple anthropometrics. 3DO PCs improved prediction of isometric knee strength (combined model R2 = 0.67 male, 0.59 female; anthropometrics-only model R2 = 0.34 male, 0.24 female).
Conclusions
3DO body shape PCs predict body composition with good accuracy and precision comparable to existing methods. 3DO PCs improve prediction of serum lipid and diabetes markers, and functional strength measurements. The safety and accessibility of 3DO scanning make it appropriate for monitoring individual body composition, and metabolic health and functional strength in epidemiological settings.
This trial was registered at clinicaltrials.gov as NCT03637855.
Keywords: imaging, obesity, body composition, diabetes, strength, principal component analysis
Introduction
Excess adiposity plays a central role in the development of type 2 diabetes, cardiovascular disease, and several cancers (1–3). Although obesity is defined using BMI, a shape index, anthropometric and regional body composition measurements such as waist circumference (WC), waist-to-hip ratio, and visceral adipose tissue have been shown to be better predictors of metabolic disease and mortality risk (4, 5). These findings indicate that more detailed descriptors of body shape and composition might offer more accurate predictors of metabolic and disease risk factors.
Clinicians and investigators have long used various manual anthropometric measurements to estimate body fat mass and fat-free mass (FFM) as predictors of health risks. A typical assessment might include manually measured height and weight; waist, trunk, and extremity circumferences; and multiple skin-fold thicknesses (6). In practice, however, conventional manual anthropometric assessment requires well-trained technicians to produce precise results. Further, these methods are limited to linear and circumferential measurements that fail to capture most details of a person's shape.
Recently, 3-dimensional (3D) laser and optical (3DO) scanners have been investigated as an alternative to manual anthropometry for health assessment (7). 3DO scanners are becoming widely available in clinics, recreational facilities, and even in home settings. Lengths, girths, and circumferences can be extracted automatically from 3DO scans, allowing direct comparison with the manual measurements traditionally used to quantify human body shape and size. Several studies have investigated the use of 3DO body shape to estimate body composition (7–12). 3DO scanners capture high-resolution 3D body shape details, but the practical simplification of 3DO body scan data into a few coarse anthropometrics comparable to tape measurements still ignores in-depth shape information. We hypothesized that more detailed 3D body shape features from advanced statistical shape models of 3DO scans could provide superior estimates of metabolic risk. Statistical shape modeling using principal component analysis (PCA), a technique for dimensionality reduction, can capture complex shape features of highly detailed 3D meshes using a limited number of principal components (PCs). This technique is ideal for describing nuanced 3D body shape in a low number of features.
The primary aim of this study was to derive PC features of body shape from 3DO scans of a diverse sample of adults in terms of sex, age, BMI, and race/ethnicity, and evaluate the association of these shape measures to total and regional body composition, blood biomarkers, and functional strength. We hypothesized that PC-derived 3D body shape features would more accurately predict these somatic, biochemical, and strength disease-risk markers than conventional models using conventional anthropometric measurements. A secondary aim of this study was to quantify the test-retest precision of body composition estimates from 3DO PCA compared with DXA.
Methods
Experimental design
This analysis was part of Shape Up! Adults, an ongoing stratified cross-sectional observational study (NIH R01 DK109008, clinicaltrials.gov ID NCT03637855). Participants underwent whole-body 3DO scans, DXA for body composition assessment, blood serum tests for diabetes and cardiovascular disease biomarkers, as well as handgrip and thigh strength tests. Statistical body shape models were built from the 3DO scans. 3D shape features from these models were then used to predict body composition, serum biomarkers, and strength measurements. The approaches used to test the main hypothesis and secondary aim are presented in detail in the following sections.
Participants
Shape Up! Adults study participants were stratified by age (18–40 y, 40–60 y, >60 y), ethnicity (non-Hispanic white, non-Hispanic black, Hispanic, Asian, and native Hawaiian or Pacific Islander), sex, BMI [(in kg/m3) <18, 18–25, 25–30, >30], and geographic location (San Francisco, CA; Baton Rouge, LA; or Honolulu, HI). Participants in Shape Up! Adults were excluded if they could not stand without aid for 2 min, could not lie flat for 10 min without movement, had metal objects in their body, or had had significant body shape–altering procedures (e.g., liposuction, amputations, breast augmentation or reduction). Female participants were also excluded if pregnant or breastfeeding. All participants were examined at either the University of California, San Francisco (UCSF), Clinical and Translational Science Institute, the Pennington Biomedical Research Center (PBRC), or the University of Hawaii Cancer Center (UHCC) Body Composition Laboratory. The study protocol was approved by the institutional review boards at each site. Participants included in the present sample were recruited between October 2015 and September 2018. All participants gave written informed consent.
DXA scans
Each participant underwent 2 whole-body DXA scans, with repositioning, on either a Horizon/A system (Hologic, Inc) at UCSF or a Discovery/A system (Hologic, Inc) at PBRC or UHCC. Participants were scanned according to the manufacturer's guidelines. All DXA scans were analyzed at UHCC by a single certified technologist using Hologic Apex version 5.6 with the National Health and Nutrition Examination Survey Body Composition Analysis calibration option disabled. As per International Society for Clinical Densitometry guidelines, offset scanning was performed for subjects too wide to fit in the DXA scan field. DXA systems were calibrated according to standard Hologic procedures (13). In addition, Hologic spine and whole-body phantoms were circulated between study sites and it was determined that no correction factors were needed between the densitometers used in the study. Body composition measurements from DXA included total body mass, total and regional (trunk, arms, legs) fat mass, bone mineral content, and FFM.
Whole-body 3DO scanning
Each participant underwent two 3DO whole-body surface scans, with repositioning, on a Fit3D ProScanner (Fit3D, Inc). Participants followed a standard positioning protocol and wore skin-tight undergarments to minimize the effects of clothing on observed body shape. The ProScanner uses ≥1 light-coding depth sensors to capture 3D shape as the participant rotates 360° on the scanner platform. Each scan took approximately 40 s to complete. The iterative closest point algorithm was used to spatially align point clouds captured by the sensor as the subject rotated (14). The final point cloud was converted to a triangle mesh with approximately 300,000 vertices and 900,000 faces representing the human body shape. All 3DO scan data were transferred from the measurement sites and stored securely at UHCC prior to statistical analysis. All downstream analyses were performed on the reconstructed 3D meshes provided by Fit3D in Wavefront .obj format.
Blood serum biomarkers
A 40-mL whole blood sample was collected from each participant after an 8-h overnight fast, except for water and prescription medicines. Blood samples were placed on ice and processed within 4 h into plasma, serum, whole blood, and buffy coat components, following which they were stored at −80°C at each study site until analysis. Biochemical analysis of all blood samples was performed at PBRC. Serum chemistry panels were assayed through the use of a DXC600 instrument (Beckman Coulter, Inc). LDL cholesterol was calculated as:
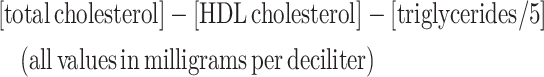 |
(1) |
as described by Friedewald et al. (15). Insulin was measured by immunoassay on an Immulite 2000 platform (Siemens Corp). Measurements of fasting glucose, glycated hemoglobin (HbA1c), total cholesterol, LDL and HDL cholesterol, serum triglycerides, and insulin were used in the present analysis.
Strength assessments
Isokinetic and isometric right leg strength were measured using a Biodex System 4 (Biodex Medical Systems Inc) or HUMAC NORM (Computer Sports Medicine Inc) dynamometer. Before measurements, participants walked on a treadmill to warm up for ≤5 min. They were then fastened into the dynamometer system with a seatbelt for measurement of right leg strength through knee extension and flexion. The participants then practised at an endurance of 50% of maximal effort for isokinetic and isometric testing. For isometric measurements the dynamometer was fixed at 60° from straight (full extension). For the isokinetic measurement, resistance was set at 60°/s. After practising each measurement, participants performed a set of 5 repetitions at maximal effort. Peak torque was recorded as the maximum torque (in newton-meters) achieved during the repetitions.
Handgrip strength for the right and left arms was measured with a handgrip dynamometer (JAMAR 5030J1; Sammons Preston Rolyan). Participants positioned their elbow at a 90° angle and were asked to squeeze the dynamometer as hard as they could, then encouraged to squeeze even harder. The strength of each hand was measured in kilograms, and the average of 3 measurements was taken.
Statistical shape modeling
Statistical shape models of 3DO body shape were constructed using a standardized 60,000-vertex body template that was warped to fit each participant's 3DO scan using the methods of Allen et al. (16). This registration process ensures vertex correspondence to specific anatomical locations across the dataset, enabling direct 3DO body shape comparison across the whole sample. To initialize registration, 75 markers were manually placed on physiological landmarks defined by the Civilian American and European Surface Anthropometry Resource Project (17). Marker placement was performed by an exercise physiologist using MeshLab version 1.3.2 (Consiglio Nazionale delle Ricerche). The template marker locations were transformed to align with the participant mesh markers, then the remaining template mesh vertices were warped to the participant mesh surface using a minimization process that preserved overall smoothness (16). After registration, PCA was performed to produce a statistical shape model that captured >95% of shape variation in the 60,000-vertex template space with only a small number of PCs. Each detailed 3DO body scan could then be represented in the 3DO PC space as a short vector of weights.
Statistical analysis
The average male and female body shapes in the sample were generated and visualized. Pearson correlation coefficients were calculated between body composition, blood marker measurements, strength, and the PCs that capture 95% of shape variance in each of the male and female models. Body shapes representing high (+3 SDs) and low (−3 SDs) states for each of these PCs were generated to illustrate the shape changes captured by each component. P values <0.05 were considered statistically significant with a secondary threshold at P < 0.001.
Stepwise linear regressions were performed to derive linear models for each of the outcome body composition, blood marker, and strength variables. Separate models were derived for each sex. Four different types of models were created: 1) “3D PC-only” models that included the first fifteen 3D body shape model PCs as candidate variables; 2) “Anthro-only” models that included height, weight, and linear circumference measurements extracted by the 3DO scanner software along with ethnicity and age as candidate variables; 3) “3D PC + Anthro” models that included both 3D PC and anthropometric candidate variables; and 4) “Simple Anthro” models for body composition only that used the same variables as described by Ng et al. (10). Variable selection was performed subject to minimization of the Schwarz–Bayesian information criterion. Fivefold cross-validation was performed to protect against overfitting. Model accuracies were assessed using coefficient of determinations (R2) and root mean squared errors (RMSEs). Differences in model accuracy by ethnic group were tested for significance using ANOVA tests on residuals calculated across the whole sample.
Measurement precision was quantified using RMSE and CV expressed as a percentage.
PCA and mesh processing were performed using Python version 3.6 (Python Software Foundation) and R version 3.3.3 (R Core Team). Regression analysis was performed in SAS version 9.4 (SAS Institute).
Results
Participants
Four hundred and fifty-six healthy adults of diverse race and ethnicity had completed the study at the time of this analysis. Three participants dropped out of the study during data collection. Nineteen participants were excluded for invalid DXA scans (9 with body parts off the scan field, 9 with high-density artifacts in the scan area, and 1 with movement artifacts). Twenty-seven participants were excluded for invalid 3DO scans (14 did not wear appropriate attire, 8 manufacturer errors, 3 template fitting errors, and 2 had movement artifacts). After these exclusions, 407 participants were included for analysis. Summary characteristics and counts of the sample participants are presented in Table 1. Subject counts by ethnicity are presented in Table 2. A CONSORT flow diagram detailing study enrollment is provided as Supplemental Figure 1.
TABLE 1.
Subject characteristics1
| Males | Females | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | Units | n | Mean | SD | Min. | Max. | n | Mean | SD | Min. | Max. |
| Age | y | 177 | 43.5 | 16.5 | 18.0 | 79.0 | 230 | 46.1 | 16.4 | 18.0 | 75.0 |
| Height | cm | 177 | 175.3 | 6.9 | 154.7 | 190.2 | 229 | 162.2 | 6.9 | 144.2 | 180.5 |
| Weight | kg | 177 | 85.5 | 20.3 | 40.6 | 173.5 | 228 | 70.3 | 19.3 | 35.4 | 152.7 |
| BMI | kg/m2 | 177 | 27.7 | 5.9 | 17.0 | 52.6 | 228 | 26.7 | 7.0 | 14.2 | 51.9 |
| Waist circ. | cm | 170 | 92.1 | 14.4 | 59.5 | 145.9 | 222 | 85.2 | 15.4 | 55.0 | 150.0 |
| Hip circ. | cm | 170 | 103.3 | 11.9 | 79.0 | 155.9 | 221 | 104.8 | 13.3 | 75.9 | 158.0 |
| Arm circ. | cm | 175 | 34.9 | 5.0 | 24.6 | 50.8 | 222 | 31.5 | 5.4 | 19.9 | 53.0 |
| Thigh circ. | cm | 175 | 58.9 | 6.1 | 44.3 | 82.2 | 222 | 60.0 | 7.6 | 40.3 | 85.3 |
| Waist-to-hip ratio | 170 | 0.89 | 0.07 | 0.74 | 1.03 | 221 | 0.81 | 0.08 | 0.66 | 1.06 | |
| Waist-to-height ratio | 170 | 0.53 | 0.08 | 0.39 | 0.81 | 222 | 0.53 | 0.10 | 0.35 | 0.88 | |
| Fat mass | kg | 177 | 19.8 | 9.8 | 6.1 | 66.4 | 230 | 24.8 | 11.3 | 6.3 | 72.7 |
| Lean mass | kg | 177 | 65.8 | 12.8 | 33.7 | 108.2 | 230 | 45.5 | 9.2 | 28.6 | 80.4 |
| Percentage fat | % | 177 | 22.2 | 6.5 | 9.4 | 48.2 | 230 | 33.9 | 7.4 | 12.9 | 48.6 |
| FMI | kg/m2 | 177 | 6.4 | 3.1 | 2.0 | 21.0 | 230 | 9.4 | 4.3 | 2.0 | 24.7 |
| FFMI | kg/m2 | 177 | 21.3 | 3.5 | 14.1 | 35.8 | 230 | 17.3 | 3.2 | 10.9 | 29.1 |
| Visceral fat mass | kg | 177 | 0.47 | 0.28 | 0.17 | 1.62 | 230 | 0.43 | 0.28 | 0.04 | 1.45 |
| HbA1c | % | 164 | 5.4 | 0.6 | 4.3 | 8.9 | 215 | 5.4 | 0.5 | 4.1 | 8.9 |
| Total cholesterol | mg/dL | 165 | 179.4 | 41.1 | 8.9 | 316.0 | 217 | 193.8 | 39.2 | 107.0 | 315.0 |
| LDL cholesterol | mg/dL | 163 | 106.2 | 31.7 | 44.2 | 212.4 | 215 | 112.8 | 32.1 | 37.1 | 209.8 |
| HDL cholesterol | mg/dL | 165 | 54.6 | 13.8 | 17.6 | 99.1 | 217 | 63.8 | 15.8 | 32.3 | 118.4 |
| Triglycerides | mg/dL | 163 | 93.2 | 58.6 | 29.0 | 383.0 | 215 | 86.2 | 50.1 | 23.0 | 360.0 |
| Glucose | mg/dL | 164 | 93.3 | 14.7 | 19.0 | 172.0 | 215 | 91.9 | 15.5 | 65.0 | 187.0 |
| Insulin | mIU/L | 144 | 12.0 | 9.9 | 2.0 | 47.7 | 191 | 10.1 | 6.7 | 2.1 | 29.8 |
| HOMA-IR | G × I | 144 | 2.9 | 2.8 | 0.4 | 17.5 | 191 | 2.4 | 1.9 | 0.4 | 10.2 |
| Handgrip left | kg | 142 | 40.8 | 11.7 | 11.7 | 79.7 | 189 | 22.6 | 7.7 | 2.7 | 52.7 |
| Handgrip right | kg | 149 | 40.8 | 11.4 | 12.0 | 79.7 | 193 | 23.3 | 7.5 | 6.0 | 46.0 |
| Isometric knee ext. | N m | 144 | 155.2 | 66.4 | 24.8 | 351.7 | 162 | 87.9 | 35.3 | 26.4 | 193.0 |
| Isokinetic knee ext. | N m | 141 | 160.4 | 70.6 | 2.9 | 339.2 | 187 | 88.8 | 36.5 | 8.7 | 222.7 |
Circ., circumference; ext., extension; FMI, fat mass index; FFMI, fat-free mass index; G, glucose; HbA1c, glycated hemoglobin; I, insulin; max., maximum; min., minimum.
TABLE 2.
Subject ethnicity distribution
| Males (n = 177) | Females (n = 230) | |||
|---|---|---|---|---|
| Ethnicity | Count | % | Count | % |
| Asian | 41 | 23.2 | 54 | 23.5 |
| Black | 44 | 24.9 | 57 | 24.8 |
| Hispanic | 15 | 8.5 | 17 | 7.4 |
| NHOPI1 | 4 | 2.3 | 5 | 2.2 |
| White | 73 | 41.2 | 97 | 42.2 |
NHOPI, native Hawaiian or Pacific Islander.
Shape models
PCA on registered 3DO scans produced unique statistical shape models for males and females. Eleven components captured 95% of the variance in each of the male and female body shape models. The mean body shapes of males and females in the sample are shown in Figure 1. Visualizations of the PC modes of body shape variation (±3 SDs for each from the average shape) are shown in Figures 2 and 3 for males and females, respectively. Univariate correlations of PCs to body composition, blood marker, and strength measurements are presented in Tables 3 and 4 for males and females, respectively.
FIGURE 1.
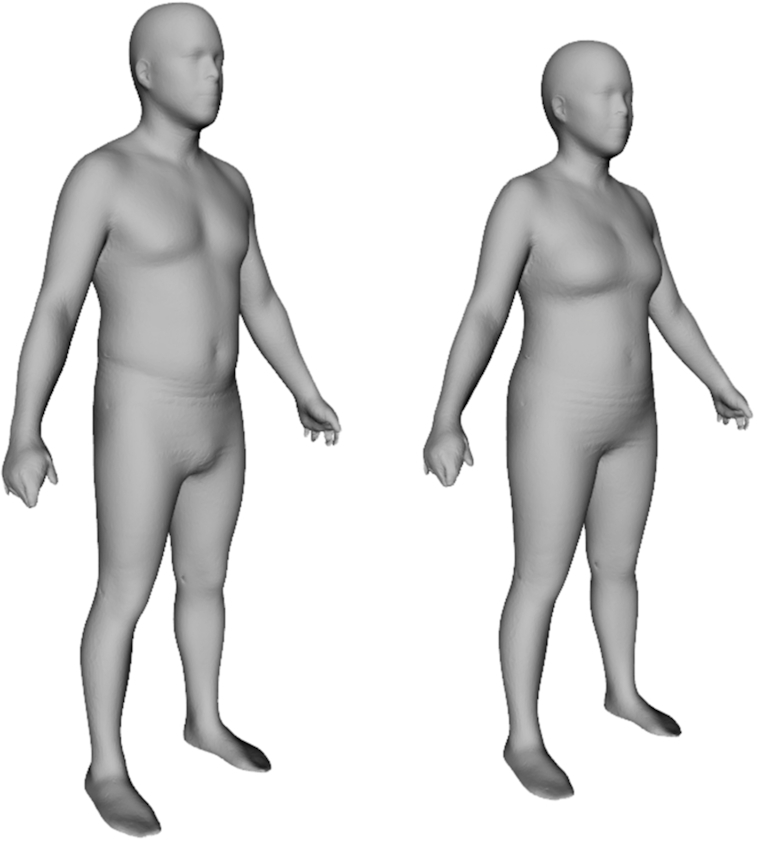
The average body shape of males and females in the sample. These 60,000-vertex 3-dimensional meshes are the sex-specific means of the registered meshes fitted to participant scans.
FIGURE 2.
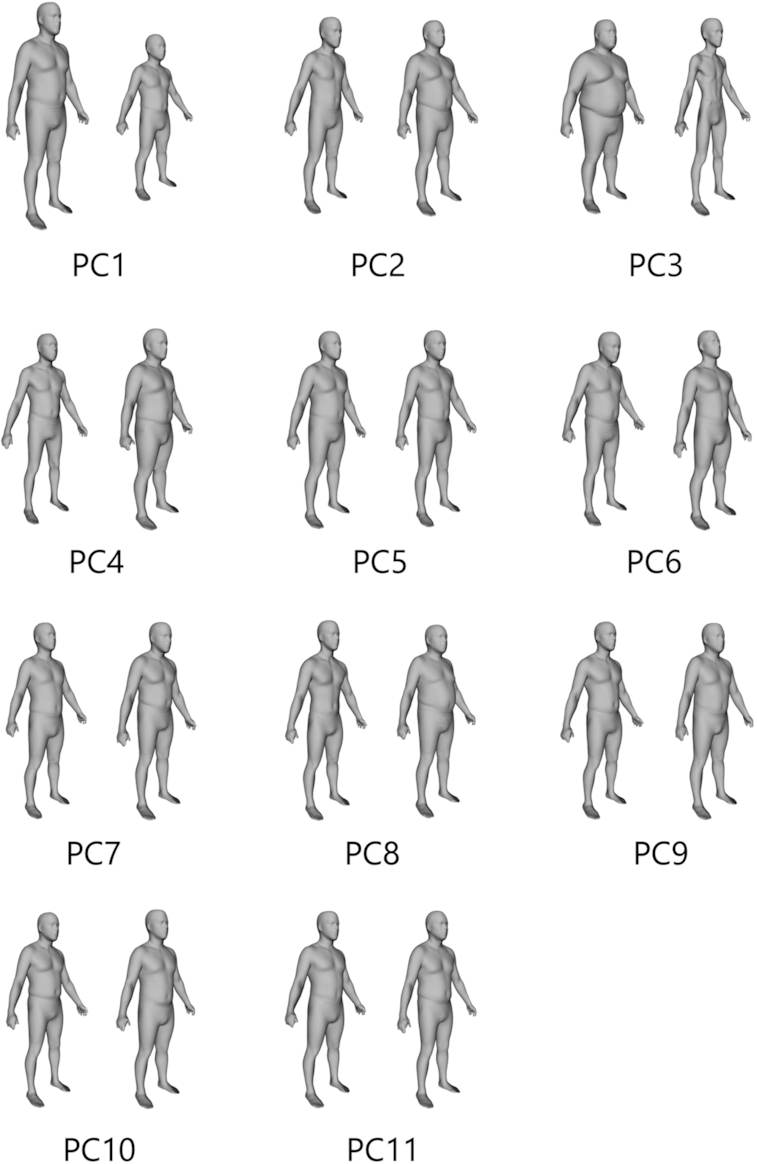
The first 11 principal components (PCs) that captured 95% of shape variance in males. Each component displays −3 SD to the left and +3 SD to the right.
FIGURE 3.
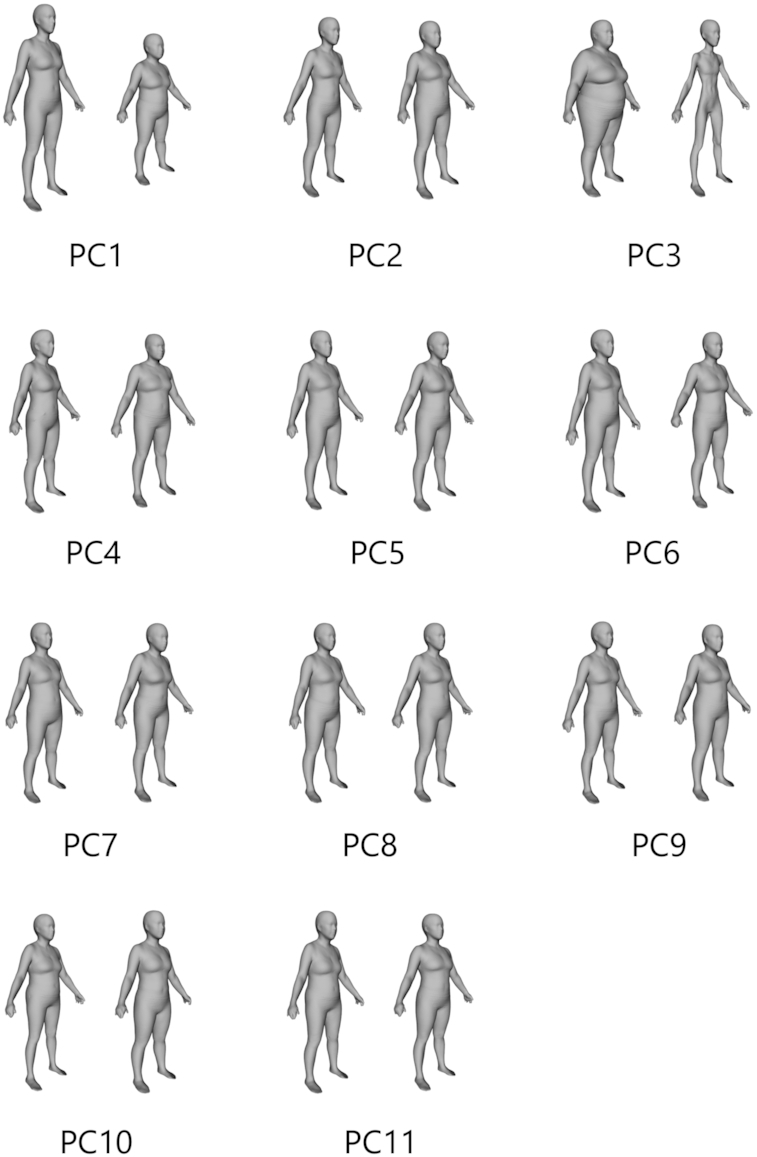
The first 11 principal components (PCs) that captured 95% of shape variance in females. Each component displays −3 SD to the left and +3 SD to the right.
TABLE 3.
Correlation of male body shape principal components (PCs) to anthropometric, body composition, blood, and strength metrics (n = 177)1
| Parameter | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 | PC9 | PC10 | PC11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | –0.19 | –0.25* | 0.32* | –0.20 | |||||||
| Height | –0.95* | ||||||||||
| Weight | –0.47* | 0.22 | –0.79* | 0.22 | |||||||
| BMI | –0.20 | 0.27* | –0.88* | 0.23 | |||||||
| Waist circ. | –0.32* | 0.24 | –0.84* | 0.17 | 0.20 | ||||||
| Hip circ. | –0.34* | 0.22 | –0.79* | 0.19 | –0.17 | ||||||
| Arm circ. | –0.33* | 0.24 | –0.72* | 0.27* | |||||||
| Thigh circ. | –0.36* | 0.16 | –0.65* | 0.31* | |||||||
| WHR | –0.52* | –0.21 | 0.22 | ||||||||
| WHtR | 0.28* | –0.87* | 0.16 | 0.23 | |||||||
| Fat mass | –0.23 | 0.20 | –0.77* | 0.20 | –0.17 | 0.21 | 0.19 | ||||
| Lean mass | –0.57* | 0.19 | –0.65* | 0.20 | |||||||
| Percentage fat | 0.16 | –0.58* | 0.32* | 0.28* | |||||||
| FMI | 0.21 | –0.79* | 0.19 | –0.16 | 0.24 | 0.20 | |||||
| FFMI | –0.25* | 0.26* | –0.77* | 0.22 | |||||||
| Visceral fat | 0.19 | –0.55* | –0.18 | 0.27* | 0.16 | ||||||
| Trunk fat | –0.20 | 0.24 | –0.79* | 0.16 | 0.23 | 0.18 | |||||
| Trunk lean | –0.55* | 0.20 | –0.70* | 0.20 | |||||||
| Arm fat | –0.23 | 0.20 | –0.75* | 0.22 | 0.19 | 0.19 | |||||
| Arm lean | –0.50* | 0.22 | –0.54* | 0.17 | 0.17 | ||||||
| Leg fat | –0.24 | –0.66* | 0.22 | –0.20 | 0.16 | 0.19 | |||||
| Leg lean | –0.58* | –0.58* | 0.21 | ||||||||
| HbA1c | 0.16 | –0.32* | 0.20 | ||||||||
| Total cholesterol | |||||||||||
| LDL cholesterol | –0.16 | ||||||||||
| HDL cholesterol | 0.36* | 0.16 | |||||||||
| Triglycerides | –0.16 | –0.20 | –0.19 | ||||||||
| Glucose | –0.21 | –0.20 | 0.17 | ||||||||
| Insulin | 0.31* | –0.53* | |||||||||
| HOMA-IR | 0.25 | –0.52* | |||||||||
| Handgrip left | –0.34* | –0.27 | –0.33* | –0.20 | |||||||
| Handgrip right | –0.31* | –0.26 | –0.26 | ||||||||
| Isometric knee extension | –0.50* | –0.39* | –0.31* | –0.23 | –0.19 | ||||||
| Isokinetic knee extension | –0.38* | –0.21 | 0.33* | 0.21 | –0.26 | –0.25 |
Only significant correlations are shown (P < 0.05 or *P < 0.001). Circ., circumference; FFM, fat-free mass; FFMI, fat-free mass index; HbA1c, glycated hemoglobin; WHR, waist-to-hip ratio; WHtR, waist-to-height ratio.
TABLE 4.
Correlation of female body shape principal components (PCs) to anthropometric, body composition, blood, and strength metrics (n = 230)1
| Parameter | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 | PC9 | PC10 | PC11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | 0.16 | –0.18 | –0.30* | –0.16 | |||||||
| Height | –0.95* | 0.14 | |||||||||
| Weight | –0.20 | –0.94* | 0.13 | ||||||||
| BMI | 0.14 | –0.96* | 0.15 | ||||||||
| Waist circ. | 0.19 | –0.93* | 0.18 | ||||||||
| Hip circ. | –0.15 | –0.91* | |||||||||
| Arm circ. | –0.92* | ||||||||||
| Thigh circ. | –0.16 | –0.85* | |||||||||
| WHR | 0.24* | 0.23* | –0.50* | 0.18 | |||||||
| WHtR | 0.23* | 0.20 | –0.90* | 0.19 | |||||||
| Fat mass | –0.94* | ||||||||||
| Lean mass | –0.36* | –0.82* | 0.14 | 0.15 | |||||||
| Percentage fat | 0.17 | –0.75* | 0.17 | ||||||||
| FMI | –0.93* | ||||||||||
| FFMI | 0.14 | –0.87* | 0.17 | ||||||||
| Visceral fat | 0.17 | –0.76* | |||||||||
| Trunk fat | 0.18 | –0.93* | 0.15 | ||||||||
| Trunk lean | –0.34* | –0.83* | –0.14 | ||||||||
| Arm fat | 0.13 | –0.82* | |||||||||
| Arm lean | –0.30* | –0.71* | 0.17 | ||||||||
| Leg fat | –0.14 | –0.84* | |||||||||
| Leg lean | –0.38* | –0.76* | 0.13 | 0.19 | |||||||
| HbA1c | –0.23* | –0.19 | –0.22 | ||||||||
| Total cholesterol | |||||||||||
| LDL cholesterol | |||||||||||
| HDL cholesterol | 0.33* | –0.16 | –0.14 | ||||||||
| Triglycerides | 0.14 | –0.21 | 0.16 | ||||||||
| Glucose | –0.29* | 0.14 | –0.14 | ||||||||
| Insulin | –0.55* | ||||||||||
| HOMA-IR | 0.16 | –0.52* | |||||||||
| Handgrip left | –0.34* | –0.24* | 0.26* | –0.32* | |||||||
| Handgrip right | –0.43* | –0.24* | 0.17 | 0.24* | –0.20 | ||||||
| Isometric knee extension | -0.44* | -0.27* | 0.33* | 0.35* | -0.35* | 0.21 | |||||
| Isokinetic knee extension | -0.34* | -0.30* | 0.19 | 0.21 | -0.16 | 0.21 |
Only significant correlations are shown (P < 0.05 or *P ≤ 0.001). Circ., circumference; FFM, fat-free mass; FFMI, fat-free mass index; HbA1c, glycated hemoglobin; WHR, waist-to-hip ratio; WHtR, waist-to-height ratio.
For men, PC3 was significantly correlated (P < 0.001) with all 12 body composition measures presented as well as the glucose and lipid metabolism biomarkers. PC8 and PC9 significantly correlated (P < 0.05) with 7 of the 12 body composition measures. Similarly, PC3 for women correlated strongly (P < 0.001) with all 12 body composition measures and several of the serum glucose and lipid metabolism biomarkers. PC1 generally captured overall body size, and PC3 captured thinness/thickness independent of height in both males and females as shown in Figures 2 and 3.
Prediction equations
Linear models selected from the stepwise regressions are shown in Table 5. The variables selected in each 3D PC-only model are provided. For body composition variables of total body, we directly regressed to predict fat mass, then used that equation along with scale weight and measured height to produce estimates of FFM, fat mass index, and FFM index. R2 and RMSE metrics for each model are shown. Full equations for the Anthro-only models are provided in Supplemental Tables 1 and 2.
TABLE 5.
Predictions of body composition from 3D optical scans1
| Model type | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3D PC-only; 5-fold CV | Anthro-only; 5-fold CV | 3D PC + Anthro; 5-fold CV | Simple Anthro (10); no CV | ||||||||
| Output variable | Sex | n | PC-only model | R 2 | RMSE | R 2 | RMSE | R 2 | RMSE | R 2 | RMSE |
| Fat mass, kg | M | 177 | PC1, 2, 3, 4, 5, 8, 9, 10, 13, 15 | 0.88 | 3.38 | 0.88 | 3.51 | 0.91 | 3.07 | 0.82 | 3.88 |
| F | 230 | PC1, 2, 3, 4, 8, 9, 11 | 0.93 | 2.96 | 0.95 | 2.66 | 0.95 | 2.63 | 0.94 | 2.84 | |
| FFM, kg | M | 177 | (scale weight) − (predicted FM) | 0.93 | 3.38 | 0.93 | 3.51 | 0.95 | 3.07 | 0.91 | 3.73 |
| F | 230 | 0.90 | 2.95 | 0.92 | 2.66 | 0.92 | 2.63 | 0.90 | 2.73 | ||
| Percentage fat, % | M | 177 | [(predicted FM)/(scale weight)] ×100 | 0.65 | 3.83 | 0.62 | 4.03 | 0.70 | 3.55 | 0.42 | 3.36 |
| F | 230 | 0.70 | 4.10 | 0.71 | 3.99 | 0.72 | 3.88 | 0.68 | 3.47 | ||
| FMI, kg/m2 | M | 177 | (predicted FM)/(height)2 | 0.87 | 1.11 | 0.87 | 1.14 | 0.90 | 1.01 | 0.81 | 1.13 |
| F | 230 | 0.93 | 1.13 | 0.95 | 1.01 | 0.95 | 0.99 | 0.94 | 1.04 | ||
| FFMI, kg/m2 | M | 177 | (predicted FFM)/(height)2 | 0.90 | 1.11 | 0.90 | 1.15 | 0.92 | 1.01 | 0.86 | 1.25 |
| F | 230 | 0.88 | 1.12 | 0.90 | 1.01 | 0.90 | 1.00 | 0.89 | 1.10 | ||
| Visceral fat mass, kg | M | 177 | PC2, 3, 6, 8, 9, 12, 13, 15 | 0.67 | 0.16 | 0.73 | 0.15 | 0.74 | 0.14 | 0.60 | 0.17 |
| F | 230 | PC1, 2, 3, 7, 10, 13, 14, 15 | 0.75 | 0.14 | 0.77 | 0.14 | 0.80 | 0.13 | 0.70 | 0.16 | |
| Trunk fat mass, kg | M | 177 | PC1, 2, 3, 4, 5, 6, 8, 9, 10, 11, 13, 15 | 0.91 | 1.68 | 0.92 | 1.68 | 0.94 | 1.45 | 0.82 | 2.37 |
| F | 230 | PC2, 3, 4, 8, 9, 11, 13, 14, 15 | 0.94 | 1.43 | 0.94 | 1.45 | 0.95 | 1.34 | 0.92 | 1.75 | |
| Trunk FFM, kg | M | 177 | PC1, 2, 3, 4, 5, 6, 11, 15 | 0.90 | 1.94 | 0.91 | 1.90 | 0.93* | 1.72* | 0.84 | 2.54 |
| F | 230 | PC1, 2, 3, 4, 5, 7, 8, 10 | 0.87 | 1.72 | 0.89 | 1.57 | 0.90 | 1.47 | 0.83 | 1.97 | |
| Arms fat mass, kg | M | 177 | PC1, 2, 3, 4, 5, 8, 9, 10, 13 | 0.84 | 0.26 | 0.81 | 0.28 | 0.86 | 0.24 | 0.80 | 0.56 |
| F | 230 | PC1, 2, 3, 9, 14, 15 | 0.70 | 0.58 | 0.75 | 0.54 | 0.77 | 0.51 | 0.88 | 0.60 | |
| Arms FFM, kg | M | 177 | PC1, 2, 3, 4, 6, 7, 9, 11, 13, 15 | 0.76 | 0.52 | 0.87 | 0.39 | 0.88 | 0.38 | 0.70 | 1.17 |
| F | 230 | PC1, 2, 3, 4, 11, 15 | 0.67 | 0.33 | 0.75 | 0.29 | 0.76 | 0.28 | 0.59 | 0.72 | |
| Legs fat mass, kg | M | 177 | PC1, 3, 4, 5, 8, 9, 10, 12, 13 | 0.71 | 0.87 | 0.70 | 0.86 | 0.75 | 0.82 | 0.70 | 1.59 |
| F | 230 | PC1, 3, 9, 11, 14, 15 | 0.83 | 0.86 | 0.84 | 0.81 | 0.87 | 0.75 | 0.81 | 1.80 | |
| Legs FFM, kg | M | 177 | PC1, 2, 3, 4, 5, 6, 7, 11, 13, 15 | 0.89 | 0.76 | 0.91 | 0.70 | 0.93 | 0.61 | 0.80 | 2.06 |
| F | 230 | PC1, 2, 3, 4, 8, 10, 14, 15 | 0.83 | 0.71 | 0.90 | 0.53 | 0.91 | 0.52 | 0.78 | 1.64 | |
| HbA1c, % | M | 164 | PC3, 15 | 0.21 | 0.56 | 0.29 | 0.53 | 0.31 | 0.52 | — | — |
| F | 215 | PC3, 6, 10 | 0.14 | 0.45 | 0.27 | 0.41 | 0.30 | 0.40 | — | — | |
| Total cholesterol, mg/dL | M | 165 | — | 0.00 | 41.10 | 0.00 | 41.10 | 0.00 | 41.10 | — | — |
| F | 217 | — | 0.00 | 39.32 | 0.07 | 37.84 | 0.07 | 37.84 | — | — | |
| LDL cholesterol, mg/dL | M | 163 | — | 0.00 | 31.66 | 0.00 | 31.76 | 0.00 | 31.76 | — | — |
| F | 215 | — | 0.00 | 32.09 | 0.05 | 31.38 | 0.05 | 31.38 | — | — | |
| HDL cholesterol, mg/dL | M | 165 | PC3 | 0.13 | 12.89 | 0.27 | 11.85 | 0.27 | 11.85 | — | — |
| F | 217 | PC3, 4 | 0.13 | 14.81 | 0.20 | 14.35 | 0.21 | 14.22 | — | — | |
| Triglycerides, mg/dL | M | 163 | PC3, 6, 10 | 0.07 | 56.64 | 0.21 | 52.96 | 0.23 | 52.23 | — | — |
| F | 215 | PC3, 4, 15 | 0.14 | 46.47 | 0.24 | 44.51 | 0.27 | 43.56 | — | — | |
| Glucose, mg/dL | M | 164 | PC3, 7, 15 | 0.20 | 13.19 | 0.14 | 13.46 | 0.25 | 12.61 | — | — |
| F | 215 | PC3, 15 | 0.11 | 14.65 | 0.16 | 14.37 | 0.18 | 14.13 | — | — | |
| Insulin, mIU/L | M | 144 | PC2, 3, 14 | 0.40 | 7.71 | 0.43 | 7.45 | 0.47 | 7.18 | — | — |
| F | 191 | PC1, 2, 3, 6, 15 | 0.43 | 5.08 | 0.38 | 5.37 | 0.50 | 4.84 | — | — | |
| HOMA-IR | M | 144 | PC2, 3, 15 | 0.36 | 2.23 | 0.43 | 2.12 | 0.47 | 2.05 | — | — |
| F | 191 | PC1, 2, 3, 15 | 0.41 | 1.43 | 0.44 | 1.41 | 0.50 | 1.34 | — | — | |
| Handgrip left, kg | M | 142 | PC1, 6, 8, 9, 10, 13, 15 | 0.51 | 8.26 | 0.37 | 9.45 | 0.54 | 8.10 | — | — |
| F | 189 | PC1, 3, 4, 5, 9, 15 | 0.37 | 6.15 | 0.20 | 6.96 | 0.40 | 6.05 | — | — | |
| Handgrip right, kg | M | 149 | PC1, 6, 8, 9, 13, 15 | 0.38 | 8.99 | 0.33 | 9.41 | 0.44* | 8.62* | — | — |
| F | 193 | PC1, 3, 4, 5, 9, 12, 15 | 0.42 | 5.68 | 0.37 | 6.00 | 0.45 | 5.65 | — | — | |
| Isometric knee extension, N m | M | 144 | PC1, 2, 3, 8, 9, 10, 13, 15 | 0.63 | 40.74 | 0.34 | 54.51 | 0.67 | 38.19 | — | — |
| F | 162 | PC1, 3, 4, 5, 9, 11 | 0.56 | 23.54 | 0.24 | 31.28 | 0.59 | 22.97 | — | — | |
| Isokinetic knee extension, N m | M | 141 | PC1, 3, 4, 7, 8, 11, 13, 15 | 0.50* | 49.95* | 0.57 | 46.45 | 0.64* | 42.73* | — | — |
| F | 187 | PC1, 3, 5, 7, 9, 11, 15 | 0.38 | 28.76 | 0.39 | 28.99 | 0.49 | 26.11 | — | — | |
Linear models were trained using 5-fold cross-validation. Models that exhibited statistically significant ( P < 0.05) differences in accuracy across ethnicities denoted by an asterisk (*). Anthro, anthropometric; FFM, fat-free mass; FFMI, fat-free mass index; FMI, fat mass index; HbA1c, glycated hemoglobin; PC, principal component; RMSE, root mean squared error; 3D, 3-dimensional.
Body composition estimation models using 3D PCs only, anthropometric measurements only, or a combination of the 2 variable types exhibited largely comparable R2 values, though RMSE values were generally lowest for the combined models. R2 values for the 3D PC-only models ranged from 0.65 for male percentage fat to 0.93 for male FFM and female fat mass. By comparison, the corresponding values from the Ng 2016 conventional anthropometric model had R2 values of 0.42, 0.91, and 0.94, respectively. Interestingly, across all model types, R2 values for prediction of fat mass were higher for females than for males whereas those for prediction of FFM were higher for males than females. This observation suggests that on average lean components of body mass are stronger contributors to body shape in males than in females, and that fat mass is a stronger contributor to body shape in females than males. 3D PC and anthropometric models for visceral fat notably outperformed (higher R2 and lower RMSE) the Ng 2016 conventional anthropometric models for visceral fat in both males and females. Similar trends were observed for prediction of fat and FFM in the arm and leg regions.
Blood serum biomarkers were predicted with moderate strength by 3D PCs and anthropometric measurements. For both male and female shape models, PC3 was selected in regression models for serum HbA1c, HDL cholesterol, triglycerides, glucose, insulin, and HOMA-IR. Body shape PCs and anthropometric estimates did not significantly predict LDL cholesterol. Insulin and HOMA-IR models achieved R2 values between 0.36 and 0.43 with 3–5 PCs of body shape selected as input variables.
Handgrip and leg strength measurements were most accurately estimated by combined 3D PC + Anthro models. Models for isometric knee extension strength achieved an R2 value of 0.67 for males and 0.59 for females. By comparison, the equivalent Anthro-only models achieved an R2 value of 0.34 for males and 0.24 for females. The 3D PC + Anthro models for right handgrip strength achieved an R2 of 0.44 for males and 0.45 for females, whereas equivalent Anthro-only models achieved an R2 of 0.33 for males and 0.37 for females.
Residual analysis to detect significant differences in model performance across ethnicities identified one 3D PC-only model (isokinetic knee extension strength in males, P = 0.02) and three 3D PC + Anthro models (trunk lean mass in males, P = 0.04; handgrip right in males, P = 0.05; isokinetic knee extension strength in males, P = 0.002).
Repeatability
Test-retest precision was calculated for body composition measurements from duplicate DXA scans and duplicate 3DO scans using the 3D PC-only models from Table 5. Subjects were excluded from this analysis if any of their 2 DXA scans or two 3DO scans were flagged to have artifacts. Precision results for male and female models are presented in Table 6. RMSE in total body percentage fat was 1.01 percentage units and 0.99 percentage units for males and females, respectively. Across both sexes, FFM measurements had lower CVs than corresponding fat mass measurements at both total body and regional levels, owing to the greater quantity of FFM compared with fat mass in the body. 3DO PCA measurement precision metrics were generally about 1 to 3 times the magnitude of corresponding DXA precision metrics.
TABLE 6.
Test-retest precision of body composition predictions from 3DO scans and DXA scans1
| 3DO PCA | DXA | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Male | Female | |||||||||
| n | %CV | RMSE | n | %CV | RMSE | n | %CV | RMSE | n | %CV | RMSE | |
| FMI, kg/m2 | 119 | 4.37 | 0.266 | 162 | 2.68 | 0.250 | 119 | 1.37 | 0.082 | 162 | 0.91 | 0.085 |
| FFMI, kg/m2 | 119 | 1.26 | 0.266 | 162 | 1.45 | 0.250 | 119 | 0.50 | 0.105 | 162 | 0.58 | 0.099 |
| Fat mass, kg | 119 | 4.30 | 0.809 | 162 | 2.67 | 0.655 | 119 | 1.36 | 0.251 | 162 | 0.91 | 0.223 |
| FFM, kg | 119 | 1.24 | 0.809 | 162 | 1.45 | 0.655 | 119 | 0.51 | 0.332 | 162 | 0.57 | 0.259 |
| Percentage fat, % | 119 | 4.69 | 1.010 | 162 | 2.92 | 0.987 | 119 | — | 0.277 | 162 | — | 0.308 |
| Visceral fat mass, kg | 119 | 7.40 | 0.033 | 162 | 6.78 | 0.028 | 119 | 6.80 | 0.030 | 162 | 7.40 | 0.031 |
| Trunk fat mass, kg | 119 | 5.14 | 0.466 | 165 | 2.88 | 0.321 | 119 | 2.66 | 0.239 | 165 | 2.00 | 0.222 |
| Trunk FFM, kg | 119 | 1.37 | 0.426 | 165 | 1.61 | 0.361 | 119 | 0.97 | 0.302 | 165 | 1.10 | 0.245 |
| Arms fat mass, kg | 119 | 4.76 | 0.054 | 161 | 3.45 | 0.055 | 119 | 3.02 | 0.034 | 161 | 2.73 | 0.043 |
| Arms FFM, kg | 119 | 3.11 | 0.135 | 162 | 2.45 | 0.059 | 119 | 1.64 | 0.072 | 162 | 1.82 | 0.043 |
| Legs fat mass, kg | 121 | 6.41 | 0.207 | 161 | 6.53 | 0.299 | 121 | 2.36 | 0.074 | 161 | 1.38 | 0.064 |
| Legs FFM, kg | 119 | 2.43 | 0.260 | 162 | 2.10 | 0.155 | 119 | 1.15 | 0.124 | 162 | 1.12 | 0.082 |
Repeat 3DO and DXA scans were each performed with repositioning. FFM, fat-free mass; FFMI, fat-free mass index; FMI, fat mass index; PCA, principal component analysis; RMSE, root mean squared error; 3DO, 3-dimensional optical.
Discussion
In this work we used advanced statistical shape modeling to derive PCs that compactly describe detailed body shape with data acquired using a 3DO whole-body scanner. We sought to determine whether these PCs could be used to better predict body composition, disease risk, and functional strength markers over existing conventional anthropometric models.
Across whole-body and regional body composition measurements, statistical shape analysis met or outperformed the previous best method of body composition assessment from 3DO scans that uses conventional anthropometrics-based prediction models. Coefficients of determination for the 3D PC fat mass index prediction model (R2: 0.87 males; R2: 0.93 females) compare favorably with both skinfold anthropometry (SFA) and bioelectrical impedance analysis (BIA), as described in a 2013 study by Hronek et al. (18) comparing DXA with SFA (R2: 0.86) and DXA with BIA (R2: 0.88). No significant differences in accuracy across ethnicities were found for any 3D PC-only body composition or blood biomarker models. This suggests that 3D PC features could be useful for estimating metabolic risk without the need for ethnicity-specific adjustments.
Test-retest precision metrics of body composition estimates from this technique are about 1 to 3 times greater than those of DXA. This indicates that 3DO estimates of body composition can be somewhat less precise than DXA, but the high safety and accessibility of 3D body scanning make this technology amenable to repeat measurements that can reduce measurement error. Notably, precision of visceral fat measurements from 3DO shape models (CV: 7.4% for males, 6.8% for females) was comparable to that of DXA (CV: 6.8% for males, 7.4% for females). The DXA visceral fat precision in the present study using Hologic densitometers was similar to the precision of CV: 7.3% reported by Ergun et al. (19) on a GE iDXA.
Several 3D body shape PCs were significantly correlated with HDL cholesterol, triglycerides, glucose, insulin, and HOMA-IR. This suggests significant associations between body shape and diabetes risk and metabolic state. It is known that WC significantly outperforms BMI for prediction of hypertension, dyslipidemia, and metabolic syndrome (20), and that waist-to-height ratio further outperforms both BMI and WC for several cardiometabolic risk factors (21). These findings indicate that more detailed descriptors of body shape can offer additional information about metabolic risk. Detailed 3D body shape PCs could be useful for direct risk classification in epidemiological studies, and for future investigation to identify more sophisticated shape phenotypes associated with metabolic disease.
LDL cholesterol was significantly correlated with only 1 male 3D body shape PC and with no female shape PCs. This result agrees with prior studies showing a general lack of significant correlation between LDL cholesterol concentrations and simple body shape metrics such as BMI or WC (22, 23).
3DO PCs significantly improved the accuracy of leg strength prediction compared with models using anthropometrics alone. This indicates that detailed body shape features capture information about functional ability and quality of body mass beyond conventional anthropometric measurements. It can be seen in Figure 3 that female PC11, selected in the regression models for both leg strength measurements, captures information about body posture. The −3SD image for this PC appears slouched whereas the +3SD image has a more erect posture. This is an example of a recognizable body shape cue associated with functional ability. Ethnicity-specific strength models could be warranted in males, because 3D PC + Anthro model performance for isokinetic knee extension and right handgrip strength varied significantly with ethnicity.
Previous studies on clinical application of 3DO body scanners report precise measurements of body shape and composition. Lee et al. (8) reported accurate prediction of total fat mass (training R2: 0.95; RMSE: 3.39 kg) using 1 length and 2 volumes from a custom-built 3DO scanner, along with sex. Ng et al. (10) demonstrated accurate prediction of total body composition (fat mass validation R2: 0.76; RMSE: 3.72 kg; FFM validation R2: 0.85; RMSE: 3.14 kg) using a combination of 4 length and volume measurements from a commercial 3DO scanner, along with sex as a covariate. These previous studies have been limited to linear, circumferential, volumetric, and surface area measurements extracted from scans using scanner manufacturers’ proprietary, often nonstandard, algorithms. This limitation can lead to incompatibility between datasets and fragile statistical models that apply only to specific scanner hardware/software combinations. By contrast, our study used the entire space of detailed 3D human shape to create PCA-based body shape models. This technique eliminates the need for inconsistent anthropometric measures for prediction of health status. With proper preprocessing and registration, 3DO body shape models can be agnostic to scanning technology, allowing comparison across a wide array of 3DO body scanning systems. Further studies could use more advanced supervised statistical shape modeling techniques to investigate health predictions that require more localized or fine-grained shape characteristics not captured by PCA, such as those caused by edema or cellulite.
This study had several limitations. The study population was restricted to adults, in whom body shape is relatively stable over time. Children, by contrast, undergo skeletal and overall body shape changes as they grow and thus the adult shape models derived here might not accurately interpret the unique body shapes and shape dynamics of children. Shape variation of children is an active area of research, and child-specific body shape models are currently under development in the Shape Up! Kids study (NIH R01DK111698). Furthermore, the adults in this cross-sectional study were recruited as healthy. It is not known if these models would apply to individuals with conditions that impact body composition. Some examples include cancer-related cachexia, wasting, sarcopenia, heart failure and associated edemas, and skeletal deformities related to arthritis and osteoporosis. Follow-up studies to assess the scalability of these models to special populations, and cohort studies to determine the efficacy of longitudinal disease risk estimation, are warranted.
Another limitation of this study was the requirement for manual landmark placement on points difficult to identify without palpation. This is a laborious, time-consuming process that could have introduced noise in the statistical shape models. We are addressing this limitation by investigating automated template registration techniques as well as the use of physical landmarks on the body at poor precision points. We also did not remove differences in pose that introduce unwanted variance in the body shape models. We are investigating deformable 3D body pose models to remove pose differences in future models (24–26). Validation of a robust landmark-free registration method would automate the analysis process and enable broad implementation of these modeling techniques. 3D body scans could be captured and analyzed using these rich shape features automatically in minutes with minimal need for manual data review, thereby greatly expanding accessibility of these models.
In conclusion, PCA of 3DO scans provides compact shape features that describe detailed individual body shape. These 3DO PCs can be used to predict body composition with greater accuracy than traditional anthropometric modeling approaches and have good repeatability. In addition, these 3DO PCs are passive, noninvasive measurements that provide improved prediction of blood lipid and diabetes markers, as well as functional strength. 3DO body scanning is safe, accessible, and increasingly affordable. Consequently, this technology is uniquely attractive for both longitudinal body shape and composition monitoring at the individual level, and metabolic health and functional assessment in epidemiological settings.
Supplementary Material
Acknowledgments
We thank all the members of the Shape Up! study team at University of Hawaii Cancer Center, Pennington Biomedical Research Center, University of California San Francisco, and University of Washington for their diverse and complementary contributions.
The authors’ responsibilities were as follows—BKN, MJS, BB, SBH, and JAS: designed research; NK, YEL, SK, BB, MCW, YN, PH, AKG, and DC: conducted research; CV and BC: provided essential materials; BKN, IP, BF, YN, MCW, and PH: analyzed data and performed statistical analysis; BKN, MJS, SBH, and JAS: wrote the manuscript and had primary responsibility for final content; and all authors: reviewed and approved the final manuscript. SBH reports membership of the Tanita Corporation Medical Advisory Board. The remaining authors had no conflicts of interest.
Notes
This work was partially supported by National Institutes of Health NORC Center Grants (P30DK072476, Pennington/Louisiana and P30DK040561, Harvard) and the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (R01DK109008 and R01DK111698).
Supplemental Figure 1 and Supplemental Tables 1 and 2 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
Abbreviations used: BIA, bioelectrical impedance analysis; FFM, fat-free mass; FM, fat mass; HbA1c, glycated hemoglobin; PBRC, Pennington Biomedical Research Center; PC, principal component; PCA, principal component analysis; RMSE, root mean squared error; SFA, skinfold anthropometry; UCSF, University of California, San Francisco; UHCC, University of Hawaii Cancer Center; WC, waist circumference; 3D, 3-dimensional; 3DO, 3-dimensional optical.
References
- 1. Zhang C, Rexrode KM, van Dam RM, Li TY, Hu FB. Abdominal obesity and the risk of all-cause, cardiovascular, and cancer mortality. Circulation. 2008;117(13):1658–67. [DOI] [PubMed] [Google Scholar]
- 2. Eckel RH, Kahn SE, Ferrannini E, Goldfine AB, Nathan DM, Schwartz MW, Smith RJ, Smith SR. Obesity and type 2 diabetes: what can be unified and what needs to be individualized?. J Clin Endocrinol Metab. 2011;96(6):1654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4(8):579–91. [DOI] [PubMed] [Google Scholar]
- 4. Price GM, Uauy R, Breeze E, Bulpitt CJ, Fletcher AE. Weight, shape, and mortality risk in older persons: elevated waist-hip ratio, not high body mass index, is associated with a greater risk of death. Am J Clin Nutr. 2006;84(2):449–60. [DOI] [PubMed] [Google Scholar]
- 5. Kuk JL, Katzmarzyk PT, Nichaman MZ, Church TS, Blair SN, Ross R. Visceral fat is an independent predictor of all‐cause mortality in men. Obesity. 2012;14(2):336–41. [DOI] [PubMed] [Google Scholar]
- 6. Heymsfield SB, Stevens J. Anthropometry: continued refinements and new developments of an ancient method. Am J Clin Nutr. 2017;105(1):1–2. [DOI] [PubMed] [Google Scholar]
- 7. Bourgeois B, Ng BK, Latimer D, Stannard CR, Romeo L, Li X, Shepherd JA, Heymsfield SB. Clinically applicable optical imaging technology for body size and shape analysis: comparison of systems differing in design. Eur J Clin Nutr. 2017;71(11):1329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee JJ, Freeland-Graves JH, Pepper MR, Stanforth PR, Xu B. Prediction of android and gynoid body adiposity via a three-dimensional stereovision body imaging system and dual-energy X-ray absorptiometry. J Am Coll Nutr. 2015;34(5):367–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee JJ, Freeland-Graves JH, Pepper MR, Yao M, Xu B. Predictive equations for central obesity via anthropometrics, stereovision imaging and MRI in adults. Obesity. 2014;22(3):852–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ng BK, Hinton BJ, Fan B, Kanaya AM, Shepherd JA. Clinical anthropometrics and body composition from 3D whole-body surface scans. Eur J Clin Nutr. 2016;70(11):1265–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Treleaven P, Wells J. 3D body scanning and healthcare applications. Computer. 2007;40(7):28–34. [Google Scholar]
- 12. Lin J-D, Chiou W-K, Weng H-F, Tsai Y-H, Liu T-H. Comparison of three-dimensional anthropometric body surface scanning to waist-hip ratio and body mass index in correlation with metabolic risk factors. J Clin Epidemiol. 2002;55(8):757–66. [DOI] [PubMed] [Google Scholar]
- 13. Lu Y, Mathur AK, Blunt BA, Gluer CC, Will AS, Fuerst TP, Jergas MD, Andriano KN, Cummings SR, Genant HK. Dual X‐ray absorptiometry quality control: comparison of visual examination and process‐control charts. J Bone Miner Res. 1996;11(5):626–37. [DOI] [PubMed] [Google Scholar]
- 14. Besl PJ, McKay ND. A method for registration of 3-D shapes. IEEE Trans Pattern Anal Mach Intell. 1992;14(2):239–56. [Google Scholar]
- 15. Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18(6):499–502. [PubMed] [Google Scholar]
- 16. Allen B, Curless B, Popović Z. The space of human body shapes: reconstruction and parameterization from range scans. In: ACM SIGGRAPH 2003 papers. New York, NY: ACM; 2003:587–94. [Google Scholar]
- 17. Robinette KM, Blackwell S, Daanen H, Boehmer M, Fleming S. Civilian American and European surface anthropometry resource (CAESAR), final report. Vol 1 Summary US Airforce Research Laboratory; 2002. [Google Scholar]
- 18. Hronek M, Kovarik M, Aimova P, Koblizek V, Pavlikova L, Salajka F, Zadak Z. Skinfold anthropometry—the accurate method for fat free mass measurement in COPD. Chronic Obstr Pulm Dis. 2013;10(5):597–603. [DOI] [PubMed] [Google Scholar]
- 19. Ergun DL, Rothney MP, Oates MK, Xia Y, Wacker WK, Binkley NC. Visceral adipose tissue quantification using lunar prodigy. J Clin Densitom. 2013;16(1):75–8. [DOI] [PubMed] [Google Scholar]
- 20. Janssen I, Katzmarzyk PT, Ross R. Waist circumference and not body mass index explains obesity-related health risk. Am J Clin Nutr. 2004;79(3):379–84. [DOI] [PubMed] [Google Scholar]
- 21. Ashwell M, Gunn P, Gibson S. Waist‐to‐height ratio is a better screening tool than waist circumference and BMI for adult cardiometabolic risk factors: systematic review and meta‐analysis. Obes Rev. 2012;13(3):275–86. [DOI] [PubMed] [Google Scholar]
- 22. Laclaustra M, Lopez-Garcia E, Civeira F, Garcia-Esquinas E, Graciani A, Guallar-Castillon P, Banegas JR, Rodriguez-Artalejo F. LDL cholesterol rises with BMI only in lean individuals: cross-sectional U.S. and Spanish representative data. Diabetes Care. 2018;41(10):2195–201. [DOI] [PubMed] [Google Scholar]
- 23. Shamai L, Lurix E, Shen M, Novaro GM, Szomstein S, Rosenthal R, Hernandez AV, Asher CR. Association of body mass index and lipid profiles: evaluation of a broad spectrum of body mass index patients including the morbidly obese. Obes Surg. 2011;21(1):42–7. [DOI] [PubMed] [Google Scholar]
- 24. Bogo F, Romero J, Loper M, Black MJ. FAUST: dataset and evaluation for 3D mesh registration. In: 2014 IEEE conference on computer vision and pattern recognition (CVPR 2014). Washington, DC: IEEE Computer Society; 2014:3794–801. [Google Scholar]
- 25. Zuffi S, Black MJ. The stitched puppet: a graphical model of 3D human shape and pose. In: 2015 IEEE conference on computer vision and pattern recognition (CVPR 2015). IEEE; 2016:3537–46. [Google Scholar]
- 26. Myronenko A, Song X. Point set registration: coherent point drifts. IEEE Trans Pattern Anal Mach Intell. 2010;32(12):2262–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


