Abstract
Cytosolic calcium (cCa2+) entry into mitochondria is facilitated by the mitochondrial membrane potential (ΔΨm), an electrochemical gradient generated by the electron transport chain (ETC). Is has been assumed that as long as mutations that affect the ETC do not affect the ΔΨm, the mitochondrial Ca2+ (mCa2+) homeostasis remains normal. We show that knockdown of NDUFAF3 and SDHB reduce ETC activity altering mCa2+ efflux and influx rates while ΔΨm remains intact. Shifting the equilibrium toward lower [Ca2+]m accumulation renders cells resistant to death. Our findings reveal an unexpected relationship between complex I and II with the mCa2+ homeostasis independent of ΔΨm.
Keywords: Calcium flux, mitochondrial membrane potential, cell death, migration
1. Introduction
The electron transport chain (ETC) consists of four protein complexes embedded in the inner mitochondrial membrane that sequentially transport electrons delivered by NADH (Marreiros et al., 2016). The free energy from redox reactions at complex I, III and IV is used to translocate protons across the inner mitochondrial membrane, forming an electrochemical gradient called the mitochondrial membrane potential (ΔΨm), which is used by the F0-F1-ATP synthetase to generate ATP (Saraste, 1999). In addition, ΔΨm allows the import and export of metabolites, proteins and ions to and from the mitochondrial matrix (Poburko and Demaurex, 2012; Zorova et al., 2017). In fact, the ΔΨm is considered to be the main driving force for the mitochondrial Ca2+ (mCa2+) uptake (Nicholls and Budd, 2000) and its maintenance is essential to prevent the onset of cell death (Duchen, 2000).
The influx of Ca2+ into the mitochondria promotes the activity of at least seven different mitochondrial proteins including three rate-limiting dehydrogenases of the TCA cycle (Glancy and Balaban, 2012). Thus, Ca2+ plays a strategic task in coordinating cellular workload and generation of ATP (Denton, 2009).
Dysfunctions either in complex I, the larger enzyme of the ETC composed by 44 subunits; 14 highly conserved core subunits that harbor bioenergetic functions, and 30 accessory subunits (Hirst, 2010) or complex II (succinate-ubiquinone oxidoreductase), the only member of the ETC that also participates in the TCA cycle and is comprised of four subunits (SDHA, SDHB, SDHC, and SDHD) (Bezawork-Geleta et al., 2017) have been associated with several diseases that show severe alterations in Ca2+ homeostasis such as cancer (Bustos et al., 2017; Kluckova et al., 2013; Urra et al., 2017) and neurodegenerative conditions such as Parkinson’s (Cieri et al., 2017; Gatt et al., 2016; Haelterman et al., 2014; Keeney et al., 2006), Alzheimer’s disease (Kim et al., 2001; Muller et al., 2018; Zhang et al., 2015) and Leigh syndrome (Quinlan et al., 2012; Wasniewska et al., 2001). However, the relation between complex I or complex II dysfunction and Ca2+ dyshomeostasis has been poorly explored and only associated with loss of ΔΨm (Mbaya et al., 2010; Visch et al., 2004), which occurs in specific scenarios, while in most the ΔΨm remains constant thanks to the reversal of ATP-synthase activity (Chinopoulos, 2011).
Here, we show that knockdown of both complex I assembly factor NDUFAF3 and complex II catalytic subunit SDHA reduce mitochondrial Ca2+ fluxes and mitochondrial respiration without changing the ΔΨm. Moreover, we demonstrate that the reduction in mCa2+ flux protects these cells from cell death induced by mitochondrial Ca2+ overload. Our findings reveal that complex I and complex II play a role in Ca2+ homeostasis beyond their role in the maintenance of the ΔΨm.
2. Materials and Methods
2.1. Materials
Antibodies: From Santa Cruz Biotechnology®: NDUFAF3 (sc-99317), β-actin (sc-47778). Abcam®: SDHA (ab14715), SDHB (ab14714), ATP5A (ab14748) NDUFB8, (ab110242). Reagents and inhibitors were purchased from Sigma-Aldrich unless specifically stated.
2.2. Cell culture and transfection.
MCF7 cell line, derived from human breast cancer, was purchased from ATCC (ATCC® Number: HTB-22™) and cultured in DMEM HG supplemented with 10% FBS. For NDUFAF3 knockdown (NDUFAF3-KD), MCF7 cells where transduced with lentiviral particles (MISSION® Lentiviral Transduction Particles – Sigma-Aldrich SHCLNV-NM_199069) and 24h after transduction selected with puromycin. For SDHA knockdown (SDHA-KD) cells, shRNA construct in retroviral untagged vector was used (Origene™ TR315539). Transfection was done with FuGENE® in reduced serum media for 24 hours and selected with puromycin.
2.3. Western Blotting.
Cell extracts were prepared with ice-cold 1X RIPA buffer (EMD Millipore®) supplemented with protease and phosphatase inhibitors (Complete EDTA-free, PhosSTOP, Roche). Equals amount of protein extracts were separated in NuPAGE 4–12% Bis-Tris protein gels (ThermoFisher Scientific) and transferred to PDVF membranes.
2.4. Immunopurification of complex I and II.
MCF7 Scramble, MCF7 NDUFAF3-KD and MCF7 SDHA-KD cells were lysed in lysis buffer [150 mM Tris-HCl, 150 mM NaCl (Merck), 1 mM EDTA (Chemix, 5 mM NaF (Sigma-Aldrich), 0.5% v/v NP-40 (Sigma-Aldrich), pH 7.4] containing protease inhibitors 1 mM phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich) and protease inhibitor cocktail (PIC; Cytoskeleton, Inc.). The lysates were centrifuged at 11,000 g at 4 °C for 10 min. The supernatants were incubated with Complex I or Complex II immunocapture KIT (Abcam, ab109711 and ab109799, respectively) for 16 h at 4°C. After incubation, immunoprecipitation reaction products were washed ten times in buffer (150 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl, pH 7.4, 5 mM NaF, 0.5% v/v NP-40). Immunocomplexes were eluted by boiling the samples in reducing sample buffer for 5 min and then, size-fractionated on SDS–PAGE.
2.5. Mitochondrial complex I assay.
The activity of mitochondrial complex I was determined using the complex I enzymatic activity assay kit (ab109721) as recommended by the manufacture. Briefly, MCF7 neg shRNA and -NDUFAF3-KD cells were lysated as described in section 2.4 and the complex I activity, define as the oxidation of NADH to NAD+, determine spectrophotometrically at 450 nm.
2.6. Mitochondrial Complex II assay.
The activity of mitochondrial complex II was determined using the complex II enzymatic activity assay kit (ab109908) as recommended by the manufacture. Briefly, MCF7 neg shRNA and MCF7 SDHA-KD cells were lysed as described in section 2.4 and the complex II immunocaptured within the wells of the microplate. The activity of complex II, was determine by the reduction of the dye DCPIP (2,6-dichlorophenolindophenol) during the production of ubiquinol spectrophotometrically at 600 nm.
2.7. Extracellular flux analysis.
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were assessed in an extracellular flux analyzer XFe96 (Agilent Technologies™, CA, USA) as described previously [1]. Briefly, MCF7 cell lines generated for this study were seeded on XFe96-well plates (15,000 cells per well) and pre-incubated overnight at 37°C in 5% CO2 atmosphere. The following day, the culture medium was replaced with assay media (unbuffered DMEM supplemented with glutaMAX®, 10 mM glucose and 1 mM pyruvate, pH 7.4) 1 hour prior to the assay and left for the duration of the experiment. After establishing the baseline OCR, cells were sequentially challenged with oligomycin (1 μM), FCCP (125 nM) and rotenone + Antimycin A (both 1 μM) to reveal basal, maximal, ATP-coupled respiration and proton leak. For ECAR, the same assay media was used and cells were sequentially challenged with glucose (10 mM), oligomycin (1 μM) and the hexokinase inhibitor 2-deoxyglucose (2-DG, 100 mM) to reveal the glycolytic capacity of the cells. The data were normalized by protein content.
2.8. Mitochondrial Ca2+ dynamics in permeabilized cells.
MCF7 cell lines generated for this study (7.5×106 cells) were permeabilized using digitonin (40 μM) containing intracellular-like medium [ICM; permeabilization buffer: 120 mM KCl; 10 mM NaCl; 1 mM KH2PO4; 20 mM HEPES-Tris, pH 7.2; 2 μM thapsigargin, and protease inhibitors (Roche Applied Science, Minneapolis, MN, USA) supplemented with succinate (3 mM)]. Fura-2FF (ThermoFisher Scientific®) was added to measure extramitochondrial Ca2+ using a fluorimeter (Photon Technology International). After baseline recording, 10 μM Ca2+ was added to cells at 350s to measure mitochondrial Ca2+ uptake, followed by the injection of the MCU inhibitor Ru360 (1 μM) at 550s, the inhibitor of the Na+/Ca2+ exchanger CGP37157 (10 μM) at 610s, and FCCP (2 μM) at 750s.
2.9. Cell proliferation assay.
Cell lines were incubated for 0, 24, 48 and 72 h. At each time point, culture medium was removed, and cells were carefully washed twice with PBS. After washing, cells were incubated at room temperature for 20 minutes in 0.5% crystal violet 20% methanol staining solution. Next, the plate was washed carefully four times in a gentle stream of tap water, inverted on a filter paper to remove the remaining liquid and dried at room temperature for at least 1 hour. Finally, crystal violet was solubilized adding 200 μL of methanol per well. OD was measured at 570 nm.
2.10. Cell migration assay.
MCF7 cell lines with density of 1 × 106 cells/well were seeded in 6-well plates and grown until a confluent monolayer of cells was achieved. Then, a cross-shaped scratch over the well using a sterile 200 μl tip was made. Wells were washed with sterile PBS to remove detached cells and debris and then cultured in 2 ml of DMEM high glucose media supplemented with 25 mM HEPES and 1% fetal bovine serum to avoid cell proliferation. Wells were photographed in multiple locations with 20x magnification in an Olympus CKX41 microscope using a Qimaging micropublisher 3.3 RTV camera. Images were analyzed with Image J and data expressed as percentage (%) of gap closure as previously described (Liang et al., 2007).
2.11. Cell viability assay.
MCF7 cell lines were seed in 24-well plates and incubated for 24 h. Then, the cells were either exposed to DMSO (control), 5 μM ionomycin, 40 μM C2-ceramide or 30 μM thapsigargin. After 24h of exposure, cells were trypsinized, washed with PBS, and re-suspended in PBS solution with 5 mg/mL of propidium iodide (PI, ThermoFisher Scientific). Incorporation of PI in dead cells was detected using a BD FACSAria III flow cytometer.
2.12. Measurement of Mitochondrial Superoxide
Mitochondrial superoxide was measured by using the mitochondrial oxygen free radical indicator MitoSOX Red (molecular probes; Invitrogen) as previously described (Dong et al., 2017). Briefly, cells were grown on coverslips and loaded with 5 μM MitoSOX Red for 30 min. Coverslips were mounted in a confocal microscope (510 Meta; Carl Zeiss, Inc.) and images were obtained at 561 nm excitation using a 63x oil objective. Images were analyzed, and MitoSOX Red fluorescence was quantified using ImageJ software (NIH). For flow cytometry experiments, 3 × 104 cells were trypsinized and re-suspended in fresh medium without FBS and MitoSOX fluorescence detected using a BD FACSCanto flow cytometer.
2.13. Mitochondrial membrane potential.
MCF7 cell lines were loaded with tetramethylrhodamine methyl ester (TMRM, 5 nM, Life Technologies) for 30 min at 37°C and 5% CO2. Hoechst 33342 (Life Technologies) was used as nuclear counterstain. Images were acquired using a Nikon C2+ spectral confocal microscope using the same settings for all experiments. TMRM fluorescence was quantified with ImageJ software.
2.14. Statistics.
All data are summarized as mean ± SEM; significance of differences was assessed using unpaired t-tests. Differences were accepted as significant at the 95% level (p < 0.05).
3. Results
3.1. Knockdown of NDUFAF3 and SDHA reduces mitochondrial oxygen consumption and increases glycolysis in MCF7 cells.
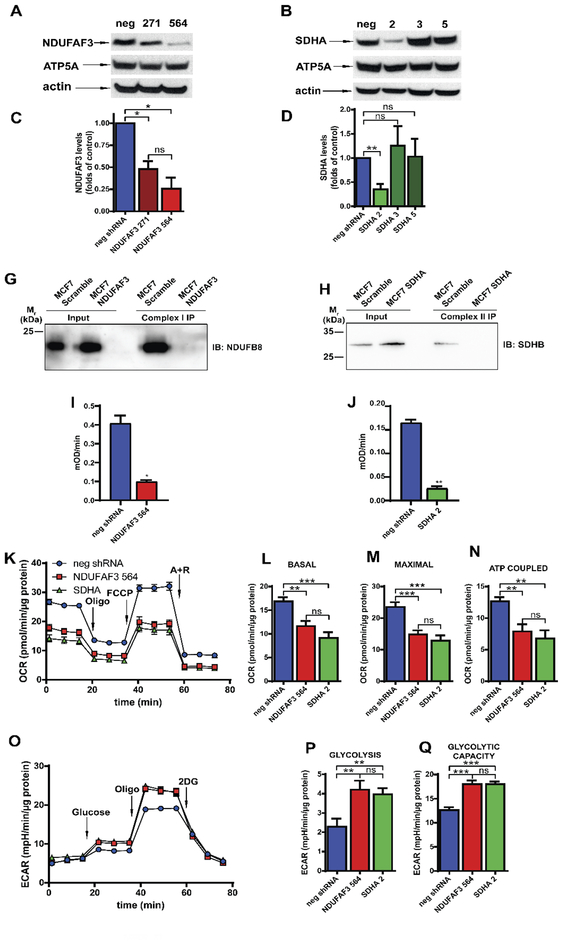
NDUFAF3 and SDHA are essential for the assembly of complex I and complex II (van den Ecker et al., 2012; Zurita Rendon and Shoubridge, 2012) and their absence is presumed to disrupt mitochondrial respiration. Here, we experimentally determined for the first time the effect of knocking down NDUFAF3 and SDHA on oxygen consumption in MCF7 cells. Using two different shRNA against NDUFAF3 and three against SDHA we generated stable knockdown MCF7 cell lines. As observed in Figure 1A and 1B, NDUFAF3–271 and NDUFAF3–564 cell lines show a decrease in NDUFAF3 protein expression of 52% and 74.5% respectively (Fig. 1A and C). On the other hand, from the stable cell lines generated to knockdown SDHA, only SDHA-2 showed a significant reduction in the expression of SDHA (65%) (Fig. 1B, D). The level of complex V subunit ATP5A, which was used as a mitochondrial loading control, remained unchanged (Fig. 1A and B) similar to what is observed with the housekeeping protein actin. Given these results, we continued our studies in cell lines NDUFAF3–564 and SDHA-2 which hereafter we refer to as NDUFAF3 KD and SDHA KD respectively. To explore the assembly status of complex I and complex II in these cells, we determine whether subunits NDUFB8 and SDHB co-immunoprecipitated with complex I and complex II respectively. As expected, when complex I is immunoprecipitated in the control cell line (neg shRNA) the complex I structural subunit NDUFB8 is present indicating assembly of complex I. However, this subunit is not seen when complex I is immunoprecipitated in NDUFAF3 KD cells, confirming that NDUFAF3 assembly factor is required for complex I assembly (Barbi de Moura et al., 2012) (Fig. 1E). Similarly, the complex II structural subunit SDHB co-immunoprecipitated with complex II in the control cell line, but not in the SDHA KD cell line (Fig. 1F) confirming the absence of the SDHA catalytic subunit impairs complex II assembly (Saxena et al., 2016). Activity assays confirmed the reduced function of complex I and complex II in NDUFAF3 KD (Fig. 1G) and SDHA KD (Fig. 1H) cells, respectively.
Figure 1. Knockdown of NDUFAF3 and SDHA subunits affects global cellular bioenergetics.
A. Representative Western blots of NDUFAF3, ATP5A in total lysates from MCF7 cells stable expressing either a non-target shRNA or a shRNA against NDUFAF3 (clones 271 and 564). B. Representative Western blots of SDHA, and ATP5A in total lysate from MCF7 cells stable expressing either a non-target shRNA (neg) or a shRNA against SDHA (clones 2, 3 and 5). C. Analysis of NDUFAF3 expression normalized over mitochondrial marker ATP5A and expressed as average fold increase over control cells (neg). D. Analysis of SDHA levels normalized over mitochondrial marker ATP5A and expressed as average fold increase over control cells. E. Co-immunoprecipitation of complex I and NDUFB8. F. Co-immunoprecipitation of complex II and SDHB. G. Determination of complex I activity at 450 nm over time. At the straight portion of the progression curve, slope (mOD/min) was calculated. H. Determination of complex II activity at 600 nm over time. At the straight portion of the progression curve, slope (mOD/min) was calculated. I. Representative traces of oxygen consumption rate (OCR) in neg shRNA, NDUFAF3–564 and SDHA-2 MCF7 cells exposed sequentially to oligomycin (oligo, 1 μM), FCCP (0.125 μM) and antimycin A plus rotenone (A+R, 1 μM each). J. Basal respiration calculated by subtracting the non-mitochondrial OCR (after A+R) from the OCR measurement before oligomycin. K. Maximal respiration calculated by subtracting non-mitochondrial OCR (after A+R) to the OCR after FCCP. L. ATP coupled OCR calculated by subtracting OCR after and before oligomycin. M. Representative traces of extracellular acidification rate (ECAR) in cells exposed sequentially to glucose (10mM), oligomycin (oligo, 1 μM) and 2-deoxyglucose (2DG, 100 mM). N. Glycolysis calculated by subtracting ECAR measured before glucose from ECAR after 2-DG. O. Glycolytic capacity calculated by subtracting ECAR measured before 2-DG from ECAR measured after oligomycin. All data shown represent mean ± SEM of at least three independent experiments. * = p<0.05, ** = p <0.01, *** = p < 0.001, ns = not significant.
Next, to determine whether NDUFAF3 KD and SDHA KD cell lines have impaired mitochondrial respiration, we performed oxygen consumption measurements. Both NDUFAF3 KD and SDHA KD cell lines showed lower levels of basal and maximal oxygen consumption rate (OCR) than the control cell line (neg shRNA) (Fig. 1I, J and K), confirming a defect in the electron transport chain. In addition, the respiration coupled to the ATP generation, calculated as the difference in OCR before and after oligomycin injection was also significantly diminished (Fig. 1L).
It is well documented that mitochondrial electron transport chain inhibition induces a compensatory rise of glycolysis (Hill et al., 2012; Saxena et al., 2016). Thus, by measuring the extracellular acidification rate (ECAR) we determined that NDUFAF3 KD and SDHA KD cells present higher glycolytic flux than the control cells (Fig. 1M and N). In addition, by inhibiting ATP synthase with oligomycin, we were able to determine that NDUFAF3 and SDHA KD cells demonstrated a higher glycolytic capacity, suggesting a metabolic remodeling toward glycolysis (Fig. 1O).
3.2. Intact complex I or complex II are necessary to maintain mCa2+ homeostasis.
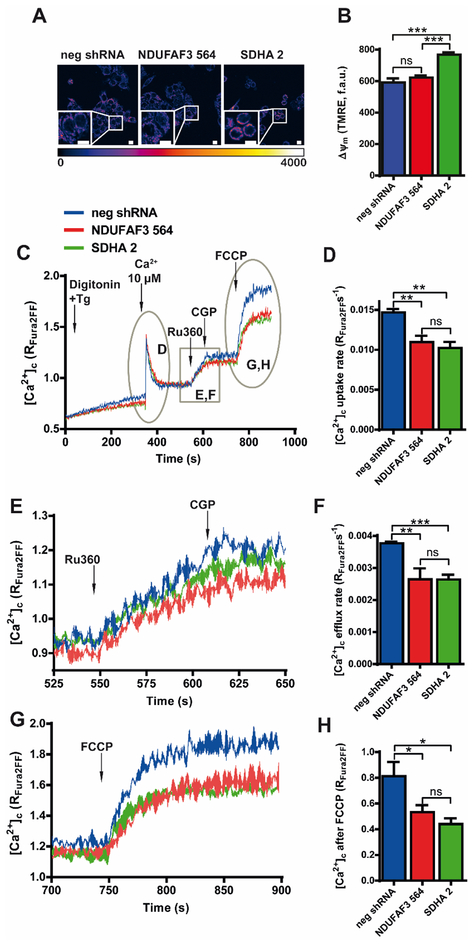
Maintenance of the ΔΨm is essential for cellular Ca2+ homeostasis (Contreras et al., 2010). Inhibition of complex I activity has been associated with both increased and decreased ΔΨm (Abramov et al., 2010; Distelmaier et al., 2009a; Distelmaier et al., 2009b; Koopman et al., 2005; Valsecchi et al., 2012), however, in NDUFAF3 KD cells the ΔΨm remained unaffected (Figure 2A and B). On the other hand, complex II plays an important role in maintaining ΔΨm in normoxic and hypoxic conditions (Hawkins et al., 2010). Surprisingly, SDHA knockdown induced a small, but significant increase in ΔΨm (Figure 2A and B) rather than a decrease. Under these conditions we hypothesized that mitochondrial Ca2+ flux should remain unchanged or increased in the case of the SDHA knockdown cells. Thus, we examined cytosolic clearance, mitochondrial Ca2+ efflux and release of total mitochondrial Ca2+ by assessing free extramitochondrial Ca2+ concentration in permeabilized cells as described before (Doonan et al., 2014). First, we triggered Ca2+ uptake, by adding a pulse of Ca2+ to permeabilized neg-shRNA, NDUFAF3, and SDHA KD cells in presence of the Ca2+ indicator, Fura-2FF. Surprisingly, the Ca2+ uptake rate was diminished in NDUFAF3 and SDHA KD cells (Fig. 2C and D). Then, in order to visualize [Ca2+]m efflux, we blocked mitochondrial Ca2+ uptake by adding Ru360, which revealed that both NDUFAF3 and SDHA KD cells have reduced efflux rate, suggesting that [Ca2+]m steady state was lower in the KD cells (Fig. 2E and F). Mitochondrial efflux was stopped with the addition of NCLX inhibitor CGP-37157. Finally, to determine the total levels of mitochondrial Ca2+, the mitochondrial membrane potential was dissipated by adding the uncoupler carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP), which revealed that NDUFAF3 and SDHA KD cells have a significantly lower [Ca2+]m content compared with control cells (Fig. 2G and H). Altogether, these results confirm that the absence of either complex I or complex II activity and/or levels, reduce [Ca2+]m efflux and influx rate, shifting the equilibrium of mitochondrial Ca2+ towards a lower [Ca2+]m accumulation.
Figure 2. Effects of NDUFAF3 and SDHA knockdown on mitochondrial membrane potential and Ca2+ influx and efflux rates.
A. Representative confocal images of neg shRNA, NDUFAF3–564 and SDHA-2 MCF7 cell lines loaded with mitochondrial membrane potential indicator TMRE. B. Quantification of TMRE fluorescence C. Representative Ca2+ traces of neg shRNA, NDUFAF3–564 and SDHA-2 MCF7 cell lines permeabilized with 40 μg/mL of digitonin and loaded with the ratiometric Ca2+ indicator Fura 2-FF. A pulse of 10 μM Ca2+ was added at 350 s to measure mitochondrial Ca2+ uptake, followed by the addition of 1 μM Ru360 at 550 s, 10 μM CGP37157 at 610 s, and 2 μM uncoupler FCCP, at 750 s. D. Quantification of Ca2+ uptake rate. E. Magnification of Ca2+ efflux from panel A (between 525 and 650 s). F. Quantification of Ca2+ efflux rate after addition of RU360. G. Magnification of Ca2+ release after FCCP addition from panel A (between 700 and 900 s). H. Quantification of FCCP-induced release of mitochondrial Ca2+. All data shown represent mean ± SEM of three independent experiments. *p<0.05, **p<0.01, ***p<0.001. ns = not significant.
3.3. Effect of NDUFAF3 and SDHA knockdown in cellular proliferation and migration.
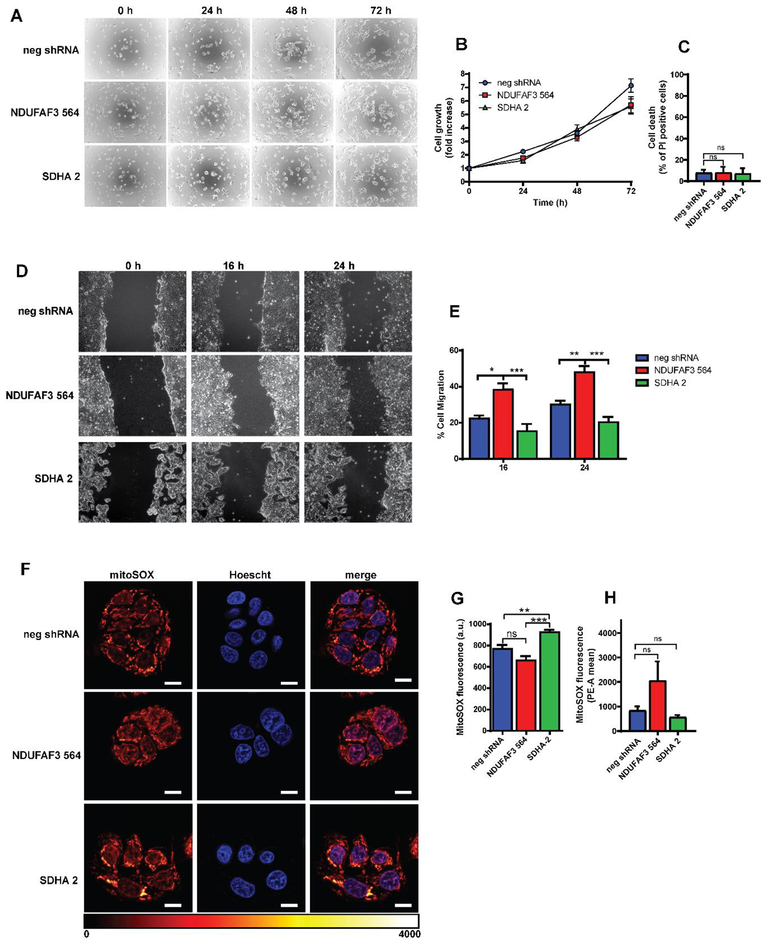
Inhibition of Ca2+ transfer to mitochondria lowers mitochondrial respiration reducing the clonogenic capacity of breast cancer cells (Cardenas et al., 2016). As the lack of complex I or complex II activity decreases respiration and the flux of Ca2+ to mitochondria, we assessed proliferation in NDUFAF3 and SDHA KD cells and no differences were found after 72h (Fig. 3A and B). Accordingly, no increase in cell death was observed in the KD cells compared to the control (Fig. 3C). Cellular migration can also be affected by a reduction of mitochondrial Ca2+ content (Tosatto et al., 2016), thus, we studied migrating behavior in NDUFAF3 and SDHA KD cells. Remarkably, NDUFAF3 KD cells migrate faster than control cells (3D and E), while SDHA KD cells behave as controls, suggesting that another mechanism independent of mitochondrial Ca2+ homeostasis operates during migration in complex I KD cells. Reactive oxygen species (ROS), which are known to increase with mutations in mitochondrial or nuclear DNA-encoded ETC proteins that diminish complex I or II (Sabharwal and Schumacker, 2014; Sullivan and Chandel, 2014), play an important role in migration and cell adhesion (Hurd et al., 2012). We therefore determined the levels of superoxide in both NDUFAF3 and SDHA KD cells by confocal microscopy, as done before by our group (Mallilankaraman et al., 2012b), and by flow cytometry. Surprisingly, mitochondrial superoxide levels in NDUFAF3 KD cells were similar to controls with both techniques (Fig. 3F–H), while SDHA KD cells show a small, but significant rise in mitochondrial superoxide levels by confocal microscopy than is not seen by flow cytometry (Fig. 3F–H).
Figure 3. Effects of NDUFAF3 and SDHA knockdown on proliferation, migration and mitochondrial ROS generation.
A. Representative images of proliferating cells stained with crystal violet at 0, 24, 48 and 72 hrs. B. Cell proliferation rates were determined by absorbance of crystal violet stain. C. Cell death determine as the percentage of cells that incorporated propidium iodide (PI). D. Representative images of a wound healing assay of neg shRNA, NDUFAF3–564 and SDHA-2 cells. E. Migration was determined as the percentage of cells covering the scratched area. F. Representative images of neg shRNA, NDUFAF3–564 and SDHA-2 cells stained with MITOSOX red. Scale bar represents 10 μm. G. Quantitation of MITOSOX red fluorescence. H. quantification of MITOSOX red fluorescence by flow cytometry. All data shown represent the mean ± SEM of three independent experiments. *p<0.05, **p<0.01, ***p<0.001.
3.4. The absence of complex I and II prevents cell death induced by mCa2+ overload.
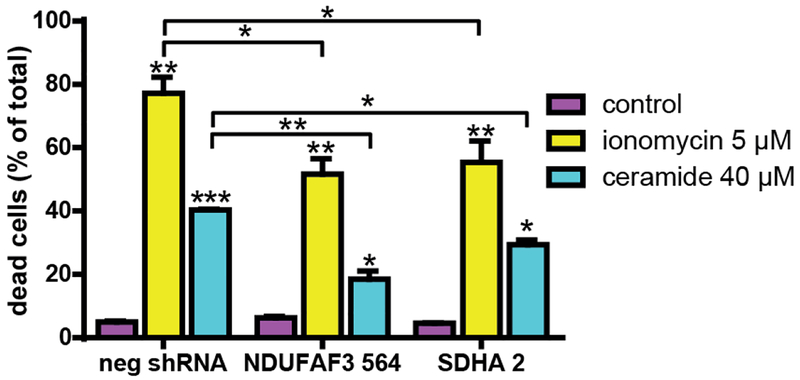
Given the shifting of the mCa2+ equilibrium towards lower [Ca2+]m accumulation in NDUFAF3 and SDHA KD cells, we determined whether these cells were more resistant to cell death induced by mitochondrial Ca2+ overload. Accordingly, we treated all cells with 5 μM ionomycin or 40 μM ceramide C2 for 24h and measured cell death using propidium iodide and flow cytometry. As shown in Figure 4, complex I and complex II KD cells were significantly more resistant to these challenges than control cells.
Figure 4. Effects of NDUFAF3 and SDHA knockdown on mitochondrial Ca2+ overload-induced cell death.
Quantification of dead cells was determined by propidium iodide incorporation measured with flow cytometry (10,000 events) in cells treated either with ionomycin or ceramide for 24 h. All data shown represent the mean ± SEM of three independent experiments. *p<0.05, **p<0.01, ***p<0.001.
4. Discussion
Although the regulation of the ETC complex and its influence on cell physiology has been studied at length (Guo et al., 2018; Lobo-Jarne and Ugalde, 2018), there are still aspects that have received little attention. For instance, Ca2+, a pivotal regulator of mitochondrial function (Cardenas et al., 2010), and key determinant of cell fate (Berridge et al., 2003) has been assumed to be regulated by the ETC indirectly through the ΔΨm. Here, we show that either complex I or II regulate mCa2+ homeostasis independently of the ΔΨm. Downregulation of NDUFAF3, an early assembly factor of complex I, decreased basal, ATP-coupled and maximal oxygen consumption and the mitochondrial Ca2+ fluxes in the human cancer cell line MCF7. The ΔΨm was conserved to a similar extent as in control cells. The same characteristics were observed during complex II subunit SDHA knockdown. SDHA contains the prosthetic group that oxidizes succinate to fumarate. Mitochondrial internal membrane damage can cause low ETC activity (Brand and Nicholls, 2011), however, this possibility was discarded in our system since proton leak showed no difference between knockdown and control cells (data not shown), indicating that the internal mitochondrial membrane was intact.
The lack of ΔΨm depolarization observed in NDUFAF3 and SDHA KD cells was expected as it has been established that in either absence or low activity of complex I, II, III and IV, the ΔΨm is maintained by complex V (F1F0-ATP synthase) reverse activity, which pumps protons towards the mitochondrial intermembrane space hydrolyzing ATP as the energy source (Chinopoulos, 2011; Jonckheere et al., 2012). The maintenance of the ΔΨm is essential to avoid the activation of the intrinsic branch of apoptosis (Gottlieb et al., 2003). For example, in neurons carrying either a severe mutant form of complex I that reduced its activity to less than 10%, or a mutant form of complex IV that reduced its activity to 40%, the ΔΨm remained stable (Abramov et al., 2010). Similarly, primary isocortical neurons and astrocytes derived from a complex I deficient Ndufs4 knockout mouse showed no alteration in ΔΨm (Bird et al., 2014). However, in both cases the ΔΨm collapsed in response to oligomycin, a well-known inhibitor of complex V activity (Hong and Pedersen, 2008), demonstrating that the ΔΨm was maintained by complex V in reverse mode. Supporting the notion that the ATP synthase reverse activity could maintain the ΔΨm in NDUFAF3 and SDHA KD cells, both cell lines exhibited an increased glycolytic flux. Glycolysis and OXPHOS are tightly coupled by intermediary metabolites, as has beenrevealed in cancer cells (Vander Heiden et al., 2009). Complex I uses NADH as a source of electrons for proton pumping and for ubiquinone reduction. When complex I is inhibited for example by rotenone, NADH concentration increases, diminishing NAD+/NADH ratio, which in turn, increases the conversion of pyruvate to lactate. Therefore, increasing NADH concentration, increases glycolytic flux as well. Also, when OXPHOS activity is decreased due to low SDH activity, AMPK is activated, upregulating lactate dehydrogenase activity (Hou et al., 2018), which replenishes ATP levels necessary to maintain Complex V working in reverse and maintaining ΔΨm.
Strikingly, the ΔΨm in SDHA knockdown cells is even higher than in control cells. Inhibition of complex I can induce a mitochondrial hyperpolarization due to low activity of complex II, III, IV and V (Forkink et al., 2014), and therefore a similar mechanism could be expected to operate in these cells. Since ΔΨm is the main driving force for Ca2+ uptake into mitochondria, no changes were expected in the mitochondrial Ca2+ flow in NDUFAF3 and SDHA KD cells. Unexpectedly, we found that the Ca2+ influx to the mitochondrial matrix as well as the mitochondrial Ca2+ efflux was lower in both knockdown cell lines (Fig. 2), causing a lower overall mitochondrial Ca2+ content. As far as we know, this is the first time that a correlation between either complex I or II and mitochondrial Ca2+ homeostasis independent of ΔΨm has been established. In a previous report, fibroblasts from patients carrying isolated mutations in complex I exhibited a reduced mCa2+ uptake/accumulation that impaired ATP production, but this occurred alongside a reduction in the ΔΨm (Visch et al., 2004). We, on the other hand, show that a reduction of either complex I or complex II in absence of an altered ΔΨm, establishes a new Ca2+ “set-point” (Mallilankaraman et al., 2012b; Nicholls, 2017). The entrance of Ca2+ to mitochondria is mainly through the mitochondrial calcium uniporter (MCU) complex, which is immersed in the inner mitochondrial membrane. It is composed by several functional and regulatory subunits, MCU being the pore-forming subunit (Baughman et al., 2011; Csordás et al., 2013; De Stefani et al., 2011; Mallilankaraman et al., 2012a; Mallilankaraman et al., 2012b; Patron et al., 2014; Perocchi et al., 2010; Plovanich et al., 2013; Sancak et al., 2013). The intake of Ca2+ through MCU is regulated by the MCUb (Raffaello et al., 2013) MICU1 (Patron et al., 2014) (Hoffman et al., 2013), MICU2 (Plovanich et al., 2013), MCUR1 (Mallilankaraman et al., 2012a) and EMRE (Sancak et al., 2013). MICU1 and MICU2 inhibit the pore-conformation of the complex, while MCUR1, regulates positively the pore activity (Woods et al., 2019). Since a decrease in MCU protein levels was not detected in our KD cell lines (data not shown), the possibility of the existence of a spatial and/or functional interaction between ETC proteins and MCU-regulatory subunits should be not dismissed. Additionally, the MCU complex is post-transcriptionally regulated by phosphorylation and chemical modification such S-glutathionylation (Pallafacchina et al., 2018), which may explain the phenomenon observed in NDUFAF3 and SDHA KD cells. New experiments are necessary to determine the exact mechanism responsible for the reduction of mCa2+ flux observed in these cells.
In many cellular contexts, a reduction in mCa2+ uptake/accumulation allows cells to resist insults that would otherwise cause cell death mediated by activation of the mitochondrial permeability transition (MPT) (Angelova et al., 2019; Kon et al., 2017; Luongo et al., 2015; Oropeza-Almazan et al., 2017). Therefore, it is not surprising that NDUFAF3 and SDHA KD cells show resistance to death upon stimulation with known inductors of cell death mediated by mCa2+ overload such as ionomycin (Nemani et al., 2018) and ceramide (Parra et al., 2013). Interestingly, a decrease of either complex I or II activity due to subunit mutations can generate cancer mainly by increasing resistance to apoptosis (Kluckova et al., 2013; Urra et al., 2017). Therefore, in the context of cancer mediated by either complex I or II impaired activity, it is possible to speculate that at least the resistance to apoptosis could be the result of low mCa2+ uptake/accumulation.
The lack of mitochondrial respiration does not affect proliferation, as shown here for both NDUFAF3 and SDHA KD cell lines, as long the ΔΨm is sustained by an upregulation of glycolysis and the reverse activity of the F0F1-ATP synthetase. Martinez-Reyes et al., 2016, elegantly demonstrated that restoring the reverse activity of the F0F1-ATP synthetase by knocking out its endogenous physiological inhibitor ATPIF1, was sufficient to restore the ΔΨm and proliferative capability of cells lacking mtDNA. On the other hand, restoring TCA cycle oxidative metabolites by ectopic expression of S. cerevisae NDI1 and C. intestinalis, which does not recover the ΔΨm, was not sufficient to maintain proliferation (Martinez-Reyes et al., 2016), highlighting the importance of the ΔΨm in proliferation. Cell migration is a pivotal step in metastasis (Yilmaz and Christofori, 2010), and is responsible for most patient deaths from solid tumors (Steeg, 2016). Several reports show that down-modulation of certain complex I subunits by genetic or pharmacologic means generates an enhanced migratory behavior of cancer cells and metastasis (He et al., 2013; Ishikawa et al., 2008; Li et al., 2015; Yuan et al., 2015), through a ROS-mediated mechanism that involves modulation of extracellular matrix proteins. In NDUFAF3 KD cells we observed an increase in cell migration, however, this was independent of ROS generation. On the other hand, germline complex II mutations that cause pheochromocytoma/paraganglioma syndrome type 4 (PGL4), which is characterized by the presence of pheochromocytoma and paraganglioma, two types of cancer that very rarely metastasize (Angelousi et al., 2015), and gastrointestinal stromal tumors and renal tumors, which have the potential to metastasize, act through a mechanism mediated by ROS (Shanmugasundaram and Block, 2016; Shi et al., 2017). However, SDHA knockdown cell lines did not show any increase in their migrative behavior compared with control cells. These results that seem controversial remind us there are many factors that regulate complex cellular phenomena such as cellular migration.
5. Conclusions
Overall, our data demonstrate that decreasing complex I and II levels affects Ca2+ homeostasis, leading to a decrease in the mCa2+ set-point independent of ΔΨm. This could have some interesting implications, as until now, the mitochondrial Ca2+ homeostasis has been overlooked as related to the array of diseases that involve mutations of the ETC that impair mitochondrial function without loss of the ΔΨm. Thus, the modulation of the mCa2+ offers a new window of intervention in cancer and other diseases related to either complex I or II downregulation activity.
HIGHLIGHTS.
Knockdown of NDUFAF3 and SDHB altered mitochondrial Ca2+ efflux and influx rates in absence of ΔΨm loss.
Shifting the equilibrium toward lower concentration of mitochondrial Ca2+ accumulation renders cells resistant to death.
Our findings reveal an unexpected relationship between complex I and II with the mitochondrial calcium homeostasis independent of ΔΨm.
6. Acknowledgments
This work was supported by FONDECYT #1160332 (CC), #11170291 (FJ), #1160518 (OC), Universidad de Aysén Semilla grant (FJ), Universidad de Chile-Universidad de Aysén collaboration grant (JR), FONDECYT postdoctoral grant #3170813 (FU), #3150623 (CBA), CONICYT PhD fellowship #21180306 (PC), FONDAP program grant #15150012 (CC) and NIH P30NS047243 (AL), R01GM109882, R01HL086699, R01HL142673, and 1S10RR027327 (MM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abramov AY, Smulders-Srinivasan TK, Kirby DM, Acin-Perez R, Enriquez JA, Lightowlers RN, Duchen MR, Turnbull DM, 2010. Mechanism of neurodegeneration of neurons with mitochondrial DNA mutations. Brain 133, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelousi A, Kassi E, Zografos G, Kaltsas G, 2015. Metastatic pheochromocytoma and paraganglioma. Eur J Clin Invest 45, 986–997. [DOI] [PubMed] [Google Scholar]
- Angelova PR, Vinogradova D, Neganova ME, Serkova TP, Sokolov VV, Bachurin SO, Shevtsova EF, Abramov AY, 2019. Pharmacological Sequestration of Mitochondrial Calcium Uptake Protects Neurons Against Glutamate Excitotoxicity. Mol Neurobiol 56, 2244–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbi de Moura M, Vincent G, Fayewicz SL, Bateman NW, Hood BL, Sun M, Suhan J, Duensing S, Yin Y, Sander C, Kirkwood JM, Becker D, Conrads TP, Van Houten B, Moschos SJ, 2012. Mitochondrial respiration--an important therapeutic target in melanoma. PLoS One 7, e40690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL, Koteliansky V, Mootha VK, 2011. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476, 341–U111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ, Bootman MD, Roderick HL, 2003. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol 4, 517–529. [DOI] [PubMed] [Google Scholar]
- Bezawork-Geleta A, Rohlena J, Dong L, Pacak K, Neuzil J, 2017. Mitochondrial Complex II: At the Crossroads. Trends Biochem Sci 42, 312–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird MJ, Wijeyeratne XW, Komen JC, Laskowski A, Ryan MT, Thorburn DR, Frazier AE, 2014. Neuronal and astrocyte dysfunction diverges from embryonic fibroblasts in the Ndufs4fky/fky mouse. Biosci Rep 34, e00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand MD, Nicholls DG, 2011. Assessing mitochondrial dysfunction in cells. Biochemical Journal 435, 297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos G, Cruz P, Lovy A, Cardenas C, 2017. Endoplasmic Reticulum-Mitochondria Calcium Communication and the Regulation of Mitochondrial Metabolism in Cancer: A Novel Potential Target. Front Oncol 7, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas C, Miller RA, Smith I, Bui T, Molgo J, Muller M, Vais H, Cheung KH, Yang J, Parker I, Thompson CB, Birnbaum MJ, Hallows KR, Foskett JK, 2010. Essential Regulation of Cell Bioenergetics by Constitutive InsP(3) Receptor Ca2+ Transfer to Mitochondria. Cell 142, 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas C, Muller M, McNeal A, Lovy A, Jana F, Bustos G, Urra F, Smith N, Molgo J, Diehl JA, Ridky TW, Foskett JK, 2016. Selective Vulnerability of Cancer Cells by Inhibition of Ca Transfer from Endoplasmic Reticulum to Mitochondria. Cell Rep 15, 219–220. [DOI] [PubMed] [Google Scholar]
- Chinopoulos C, 2011. Mitochondrial consumption of cytosolic ATP: not so fast. FEBS Lett 585, 1255–1259. [DOI] [PubMed] [Google Scholar]
- Cieri D, Brini M, Cali T, 2017. Emerging (and converging) pathways in Parkinson’s disease: keeping mitochondrial wellness. Biochem Biophys Res Commun 483, 1020–1030. [DOI] [PubMed] [Google Scholar]
- Contreras L, Drago I, Zampese E, Pozzan T, 2010. Mitochondria: the calcium connection. Biochim Biophys Acta 1797, 607–618. [DOI] [PubMed] [Google Scholar]
- Csordás G, Golenár T, Seifert EL, Kamer KJ, Sancak Y, Perocchi F, Moffat C, Weaver D, de la Fuente Perez S, Bogorad R, Koteliansky V, Adijanto J, Mootha VK, Hajnóczky G, 2013. MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca2+ uniporter. Cell metabolism 17, 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R, 2011. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476, 336–U104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton RM, 2009. Regulation of mitochondrial dehydrogenases by calcium ions. Biochim Biophys Acta 1787, 1309–1316. [DOI] [PubMed] [Google Scholar]
- Distelmaier F, Koopman WJ, van den Heuvel LP, Rodenburg RJ, Mayatepek E, Willems PH, Smeitink JA, 2009a. Mitochondrial complex I deficiency: from organelle dysfunction to clinical disease. Brain 132, 833–842. [DOI] [PubMed] [Google Scholar]
- Distelmaier F, Visch HJ, Smeitink JA, Mayatepek E, Koopman WJ, Willems PH, 2009b. The antioxidant Trolox restores mitochondrial membrane potential and Ca2+ -stimulated ATP production in human complex I deficiency. J Mol Med (Berl) 87, 515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Shanmughapriya S, Tomar D, Siddiqui N, Lynch S, Nemani N, Breves SL, Zhang X, Tripathi A, Palaniappan P, Riitano MF, Worth AM, Seelam A, Carvalho E, Subbiah R, Jana F, Soboloff J, Peng Y, Cheung JY, Joseph SK, Caplan J, Rajan S, Stathopulos PB, Madesh M, 2017. Mitochondrial Ca2+ Uniporter Is a Mitochondrial Luminal Redox Sensor that Augments MCU Channel Activity. Mol Cell 65, 1014–1028 e1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan PJ, Chandramoorthy HC, Hoffman NE, Zhang X, Cardenas C, Shanmughapriya S, Rajan S, Vallem S, Chen X, Foskett JK, Cheung JY, Houser SR, Madesh M, 2014. LETM1-dependent mitochondrial Ca2+ flux modulates cellular bioenergetics and proliferation. FASEB J 28, 4936–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen MR, 2000. Mitochondria and calcium: from cell signalling to cell death. J Physiol 529 Pt 1, 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forkink M, Manjeri GR, Liemburg-Apers DC, Nibbeling E, Blanchard M, Wojtala A, Smeitink JA, Wieckowski MR, Willems PH, Koopman WJ, 2014. Mitochondrial hyperpolarization during chronic complex I inhibition is sustained by low activity of complex II, III, IV and V. Biochim Biophys Acta 1837, 1247–1256. [DOI] [PubMed] [Google Scholar]
- Gatt AP, Duncan OF, Attems J, Francis PT, Ballard CG, Bateman JM, 2016. Dementia in Parkinson’s disease is associated with enhanced mitochondrial complex I deficiency. Mov Disord 31, 352–359. [DOI] [PubMed] [Google Scholar]
- Glancy B, Balaban RS, 2012. Role of Mitochondrial Ca2+ in the Regulation of Cellular Energetics. Biochemistry 51, 2959–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E, Armour SM, Harris MH, Thompson CB, 2003. Mitochondrial membrane potential regulates matrix configuration and cytochrome c release during apoptosis. Cell Death Differ 10, 709–717. [DOI] [PubMed] [Google Scholar]
- Guo R, Gu J, Zong S, Wu M, Yang M, 2018. Structure and mechanism of mitochondrial electron transport chain. Biomed J 41, 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haelterman NA, Yoon WH, Sandoval H, Jaiswal M, Shulman JM, Bellen HJ, 2014. A mitocentric view of Parkinson’s disease. Annu Rev Neurosci 37, 137–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins BJ, Levin MD, Doonan PJ, Petrenko NB, Davis CW, Patel VV, Madesh M, 2010. Mitochondrial complex II prevents hypoxic but not calcium- and proapoptotic Bcl-2 protein-induced mitochondrial membrane potential loss. J Biol Chem 285, 26494–26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XL, Zhou AF, Lu H, Chen Y, Huang GC, Yue X, Zhao PW, Wu YX, 2013. Suppression of Mitochondrial Complex I Influences Cell Metastatic Properties. Plos One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill BG, Benavides GA, Lancaster JR Jr., Ballinger S, Dell’Italia L, Jianhua Z, Darley-Usmar VM, 2012. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol Chem 393, 1485–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, 2010. Towards the molecular mechanism of respiratory complex I. Biochemical Journal 425, 327–339. [DOI] [PubMed] [Google Scholar]
- Hoffman NE, Chandramoorthy HC, Shamugapriya S, Zhang X, Rajan S, Mallilankaraman K, Gandhirajan RK, Vagnozzi RJ, Ferrer LM, Sreekrishnanilayam K, Natarajaseenivasan K, Vallem S, Force T, Choi ET, Cheung JY, Madesh M, 2013. MICU1 motifs define mitochondrial calcium uniporter binding and activity. Cell Rep 5, 1576–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou WL, Yin J, Alimujiang M, Yu XY, Ai LG, Bao YQ, Liu F, Jia WP, 2018. Inhibition of mitochondrial complex I improves glucose metabolism independently of AMPK activation. J Cell Mol Med 22, 1316–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd TR, DeGennaro M, Lehmann R, 2012. Redox regulation of cell migration and adhesion. Trends Cell Biol 22, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J, 2008. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science 320, 661–664. [DOI] [PubMed] [Google Scholar]
- Jonckheere AI, Smeitink JA, Rodenburg RJ, 2012. Mitochondrial ATP synthase: architecture, function and pathology. J Inherit Metab Dis 35, 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney PM, Xie J, Capaldi RA, Bennett JP Jr., 2006. Parkinson’s disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J Neurosci 26, 5256–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Vlkolinsky R, Cairns N, Fountoulakis M, Lubec G, 2001. The reduction of NADH ubiquinone oxidoreductase 24- and 75-kDa subunits in brains of patients with Down syndrome and Alzheimer’s disease. Life Sci 68, 2741–2750. [DOI] [PubMed] [Google Scholar]
- Kluckova K, Bezawork-Geleta A, Rohlena J, Dong L, Neuzil J, 2013. Mitochondrial complex II, a novel target for anti-cancer agents. Biochim Biophys Acta 1827, 552–564. [DOI] [PubMed] [Google Scholar]
- Kon N, Satoh A, Miyoshi N, 2017. A small-molecule DS44170716 inhibits Ca(2+)-induced mitochondrial permeability transition. Sci Rep 7, 3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman WJ, Verkaart S, Visch HJ, van der Westhuizen FH, Murphy MP, van den Heuvel LW, Smeitink JA, Willems PH, 2005. Inhibition of complex I of the electron transport chain causes O2-. -mediated mitochondrial outgrowth. Am J Physiol Cell Physiol 288, C1440–1450. [DOI] [PubMed] [Google Scholar]
- Li LD, Sun HF, Liu XX, Gao SP, Jiang HL, Hu X, Jin W, 2015. Down-Regulation of NDUFB9 Promotes Breast Cancer Cell Proliferation, Metastasis by Mediating Mitochondrial Metabolism. PLoS One 10, e0144441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CC, Park AY, Guan JL, 2007. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc 2, 329–333. [DOI] [PubMed] [Google Scholar]
- Lobo-Jarne T, Ugalde C, 2018. Respiratory chain supercomplexes: Structures, function and biogenesis. Semin Cell Dev Biol 76, 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luongo TS, Lambert JP, Yuan A, Zhang X, Gross P, Song J, Shanmughapriya S, Gao E, Jain M, Houser SR, Koch WJ, Cheung JY, Madesh M, Elrod JW, 2015. The Mitochondrial Calcium Uniporter Matches Energetic Supply with Cardiac Workload during Stress and Modulates Permeability Transition. Cell Rep 12, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K, Cárdenas C, Doonan PJ, Chandramoorthy HC, Irrinki KM, Golenár T, Csordás G, Madireddi P, Yang J, Müller M, Miller R, Kolesar JE, Molgó J, Kaufman B, Hajnóczky G, Foskett JK, Madesh M, 2012a. MCUR1 is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism. Nature Cell Biology 14, 1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K, Doonan P, Cardenas C, Chandramoorthy HC, Muller M, Miller R, Hoffman NE, Gandhirajan RK, Molgo J, Birnbaum MJ, Rothberg BS, Mak DO, Foskett JK, Madesh M, 2012b. MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca(2+) uptake that regulates cell survival. Cell 151, 630–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marreiros BC, Calisto F, Castro PJ, Duarte AM, Sena FV, Silva AF, Sousa FM, Teixeira M, Refojo PN, Pereira MM, 2016. Exploring membrane respiratory chains. Biochim Biophys Acta 1857, 1039–1067. [DOI] [PubMed] [Google Scholar]
- Martinez-Reyes I, Diebold LP, Kong H, Schieber M, Huang H, Hensley CT, Mehta MM, Wang T, Santos JH, Woychik R, Dufour E, Spelbrink JN, Weinberg SE, Zhao Y, DeBerardinis RJ, Chandel NS, 2016. TCA Cycle and Mitochondrial Membrane Potential Are Necessary for Diverse Biological Functions. Mol Cell 61, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbaya E, Oules B, Caspersen C, Tacine R, Massinet H, Pennuto M, Chretien D, Munnich A, Rotig A, Rizzuto R, Rutter GA, Paterlini-Brechot P, Chami M, 2010. Calcium signalling-dependent mitochondrial dysfunction and bioenergetics regulation in respiratory chain Complex II deficiency. Cell Death and Differentiation 17, 1855–1866. [DOI] [PubMed] [Google Scholar]
- Muller M, Ahumada-Castro U, Sanhueza M, Gonzalez-Billault C, Court FA, Cardenas C, 2018. Mitochondria and Calcium Regulation as Basis of Neurodegeneration Associated With Aging. Front Neurosci 12, 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemani N, Carvalho E, Tomar D, Dong Z, Ketschek A, Breves SL, Jana F, Worth AM, Heffler J, Palaniappan P, Tripathi A, Subbiah R, Riitano MF, Seelam A, Manfred T, Itoh K, Meng S, Sesaki H, Craigen WJ, Rajan S, Shanmughapriya S, Caplan J, Prosser BL, Gill DL, Stathopulos PB, Gallo G, Chan DC, Mishra P, Madesh M, 2018. MIRO-1 Determines Mitochondrial Shape Transition upon GPCR Activation and Ca(2+) Stress. Cell reports 23, 1005–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls DG, 2017. Brain mitochondrial calcium transport: Origins of the set-point concept and its application to physiology and pathology. Neurochem Int 109, 5–12. [DOI] [PubMed] [Google Scholar]
- Nicholls DG, Budd SL, 2000. Mitochondria and neuronal survival. Physiol Rev 80, 315–360. [DOI] [PubMed] [Google Scholar]
- Oropeza-Almazan Y, Vazquez-Garza E, Chapoy-Villanueva H, Torre-Amione G, Garcia-Rivas G, 2017. Small Interfering RNA Targeting Mitochondrial Calcium Uniporter Improves Cardiomyocyte Cell Viability in Hypoxia/Reoxygenation Injury by Reducing Calcium Overload. Oxid Med Cell Longev 2017, 5750897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallafacchina G, Zanin S, Rizzuto R, 2018. Recent advances in the molecular mechanism of mitochondrial calcium uptake. F1000Res 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra V, Moraga F, Kuzmicic J, Lopez-Crisosto C, Troncoso R, Torrealba N, Criollo A, Diaz-Elizondo J, Rothermel BA, Quest AF, Lavandero S, 2013. Calcium and mitochondrial metabolism in ceramide-induced cardiomyocyte death. Biochim Biophys Acta 1832, 1334–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron M, Checchetto V, Raffaello A, Teardo E, Vecellio Reane D, Mantoan M, Granatiero V, Szabo I, De Stefani D, Rizzuto R, 2014. MICU1 and MICU2 finely tune the mitochondrial Ca2+ uniporter by exerting opposite effects on MCU activity. Mol Cell 53, 726–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perocchi F, Gohil VM, Girgis HS, Bao XR, McCombs JE, Palmer AE, Mootha VK, 2010. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature 467, 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plovanich M, Bogorad RL, Sancak Y, Kamer KJ, Strittmatter L, Li AA, Girgis HS, Kuchimanchi S, De Groot J, Speciner L, Taneja N, Oshea J, Koteliansky V, Mootha VK, 2013. MICU2, a Paralog of MICU1, Resides within the Mitochondrial Uniporter Complex to Regulate Calcium Handling. Plos One 8, e55785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poburko D, Demaurex N, 2012. Regulation of the mitochondrial proton gradient by cytosolic Ca(2)(+) signals. Pflugers Arch 464, 19–26. [DOI] [PubMed] [Google Scholar]
- Quinlan CL, Orr AL, Perevoshchikova IV, Treberg JR, Ackrell BA, Brand MD, 2012. Mitochondrial Complex II Can Generate Reactive Oxygen Species at High Rates in Both the Forward and Reverse Reactions. Journal of Biological Chemistry 287, 27255–27264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaello A, De Stefani D, Sabbadin D, Teardo E, Merli G, Picard A, Checchetto V, Moro S, Szabo I, Rizzuto R, 2013. The mitochondrial calcium uniporter is a multimer that can include a dominant-negative pore-forming subunit. EMBO J 32, 2362–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabharwal SS, Schumacker PT, 2014. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat Rev Cancer 14, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Markhard AL, Kitami T, Kovács-Bogdán E, Kamer KJ, Udeshi ND, Carr SA, Chaudhuri D, Clapham DE, Li AA, Calvo SE, Goldberger O, Mootha VK, 2013. EMRE Is an Essential Component of the Mitochondrial Calcium Uniporter Complex. Science 342, 1379–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M, 1999. Oxidative phosphorylation at the fin de siecle. Science 283, 1488–1493. [DOI] [PubMed] [Google Scholar]
- Saxena N, Maio N, Crooks DR, Ricketts CJ, Yang YF, Wei MH, Fan TWM, Lane AN, Sourbier C, Singh A, Killian JK, Meltzer PS, Vocke CD, Rouault TA, Linehan WM, 2016. SDHB-Deficient Cancers: The Role of Mutations That Impair Iron Sulfur Cluster Delivery. Jnci-Journal of the National Cancer Institute 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugasundaram K, Block K, 2016. Renal Carcinogenesis, Tumor Heterogeneity, and Reactive Oxygen Species: Tactics Evolved. Antioxid Redox Signal 25, 685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YN, Li Y, Wang LP, Wang ZH, Liang XB, Liang H, Zhang L, Li B, Fan LQ, Zhao Q, Ma ZX, Zhao XF, Zhang ZD, Liu Y, Tan BB, Wang D, Wang LL, Hao YJ, Jia N, 2017. Gastrointestinal stromal tumor (GIST) with liver metastases: An 18-year experience from the GIST cooperation group in North China. Medicine (Baltimore) 96, e8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeg PS, 2016. Targeting metastasis. Nat Rev Cancer 16, 201–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Chandel NS, 2014. Mitochondrial reactive oxygen species and cancer. Cancer Metab 2, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosatto A, Sommaggio R, Kummerow C, Bentham RB, Blacker TS, Berecz T, Duchen MR, Rosato A, Bogeski I, Szabadkai G, Rizzuto R, Mammucari C, 2016. The mitochondrial calcium uniporter regulates breast cancer progression via HIF-1alpha. EMBO Mol Med 8, 569–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urra FA, Munoz F, Lovy A, Cardenas C, 2017. The Mitochondrial Complex(I)ty of Cancer. Front Oncol 7, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsecchi F, Monge C, Forkink M, de Groof AJ, Benard G, Rossignol R, Swarts HG, van Emst-de Vries SE, Rodenburg RJ, Calvaruso MA, Nijtmans LG, Heeman B, Roestenberg P, Wieringa B, Smeitink JA, Koopman WJ, Willems PH, 2012. Metabolic consequences of NDUFS4 gene deletion in immortalized mouse embryonic fibroblasts. Biochim Biophys Acta 1817, 1925–1936. [DOI] [PubMed] [Google Scholar]
- van den Ecker D, van den Brand MA, Ariaans G, Hoffmann M, Bossinger O, Mayatepek E, Nijtmans LG, Distelmaier F, 2012. Identification and functional analysis of mitochondrial complex I assembly factor homologues in C. elegans. Mitochondrion 12, 399–405. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB, 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visch HJ, Rutter GA, Koopman WJ, Koenderink JB, Verkaart S, de Groot T, Varadi A, Mitchell KJ, van den Heuvel LP, Smeitink JA, Willems PH, 2004. Inhibition of mitochondrial Na+-Ca2+ exchange restores agonist-induced ATP production and Ca2+ handling in human complex I deficiency. J Biol Chem 279, 40328–40336. [DOI] [PubMed] [Google Scholar]
- Wasniewska M, Karczmarewicz E, Pronicki M, Piekutowska-Abramczuk D, Zablocki K, Popowska E, Pronicka E, Duszynski J, 2001. Abnormal calcium homeostasis in fibroblasts from patients with Leigh disease. Biochem Biophys Res Commun 283, 687–693. [DOI] [PubMed] [Google Scholar]
- Woods JJ, Nemani N, Shanmughapriya S, Kumar A, Zhang M, Nathan SR, Thomas M, Carvalho E, Ramachandran K, Srikantan S, Stathopulos PB, Wilson JJ, Madesh M, 2019. A Selective and Cell-Permeable Mitochondrial Calcium Uniporter (MCU) Inhibitor Preserves Mitochondrial Bioenergetics after Hypoxia/Reoxygenation Injury. ACS Cent Sci 5, 153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz M, Christofori G, 2010. Mechanisms of motility in metastasizing cells. Mol Cancer Res 8, 629–642. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wang W, Li H, Yu Y, Tao J, Huang S, Zeng Z, 2015. Nonsense and missense mutation of mitochondrial ND6 gene promotes cell migration and invasion in human lung adenocarcinoma. BMC Cancer 15, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang S, Maezawa I, Trushin S, Minhas P, Pinto M, Jin LW, Prasain K, Nguyen TD, Yamazaki Y, Kanekiyo T, Bu G, Gateno B, Chang KO, Nath KA, Nemutlu E, Dzeja P, Pang YP, Hua DH, Trushina E, 2015. Modulation of mitochondrial complex I activity averts cognitive decline in multiple animal models of familial Alzheimer’s Disease. EBioMedicine 2, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorova LD, Popkov VA, Plotnikov EY, Silachev DN, Pevzner IB, Jankauskas SS, Babenko VA, Zorov SD, Balakireva AV, Juhaszova M, Sollott SJ, Zorov DB, 2017. Mitochondrial membrane potential. Anal Biochem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita Rendon O, Shoubridge EA, 2012. Early complex I assembly defects result in rapid turnover of the ND1 subunit. Hum Mol Genet 21, 3815–3824. [DOI] [PMC free article] [PubMed] [Google Scholar]