Abstract
Objective
This study aimed to identify the changes of miRNAs in colorectal cancer (CRC) complicated with diabetes mellitus (DM) (CRC + DM) tissues and their potential effects.
Methods
The changes of miRNAs in CRC + DM tissues were determined by miRNA microarray. The expression levels of miR-99a in 40 clinical specimens and 6 CRC cell lines were determined by qRT-PCR. The capacity for miR-99a to induce cell proliferation and invasion was examined with miR-99a-overexpressing HCT-116 cells. The relative mTOR mRNA and protein levels were determined by qRT-PCR and Western blotting, respectively, in HCT-116 cells transfected with miR-99a. The dual luciferase assay was performed to confirm the direct regulation of miR-99a on mTOR 3′-UTR. The HCT-116 cells were treated with 100 mg/L advanced glycation end products (AGEs); then, the mTOR expression levels were determined by qRT-PCR, Western blotting, and immunohistochemistry.
Results
Seventeen miRNAs were found to be differentially expressed among normal tissue, CRC tissue, and CRC with DM tissue, including 15 upregulated and 2 downregulated with fold changs of more than 2 times. qRT-PCR confirmed that miR-99a was downregulated in CRC and CRC + DM tissues. In addition, miR-99a overexpression remarkably impaired CRC cell proliferation and metastasis, and negatively regulated mTOR signaling through direct binding to the 3′-UTR of mTOR. AGEs could suppress miR-99a and stimulate mTOR signaling in CRC cells. Increased mTOR was also identified in CRC with DM tissues.
Conclusion
Our findings indicate that miR-99a is a potential marker and therapeutic target of CRC complicated with DM, and that AGEs impair miR-99a-overactivated mTOR signaling in CRC with DM patients, which promotes CRC development.
Keywords: miR-99a, colorectal cancer, diabetes mellitus, mTOR
Introduction
Colorectal cancer (CRC) is the fifth most common cancer in China in men and the fourth in women,1 while being the third most common in men globally and the second in women.2 Approximately 746,300 men and 614,300 women were newly diagnosed with CRC in 2012 worldwide, and 693,900 died from this disease.2 Diabetes mellitus (DM), a major health problem globally, is a metabolic disorder characterized by hyperglycemia. In recent years, gene expression-based subtyping has been widely accepted as a valid approach for CRC stratification. There are many different classification systems, and it is considered that different subtypes of CRC have unique biological characteristics, clinical stratifications, and targeted interventions.3–6 CRC complicated with DM may be a special subtype of CRC. Multiple epidemiological studies have suggested that DM may significantly elevate the risk of cancers.4–6 Coughlin et al examined the relationship between DM and cancers in a large, prospective US cohort of 467,922 men and 588,321 women who had no reported history of cancer at enrollment.6 Their results suggested that DM may be an independent risk factor for death from cancers of the colon, liver, pancreas, and female breast, and be predictive of mortality from CRC. Recently, a series of studies and meta-analyses also indicated that DM is associated with increased risk of CRC in both women and men.7–11 The association remained after controlling for obesity, smoking, and physical exercise, which are important potential confounders.11 While there are limited studies related to the mechanism underlying the relationship between DM and CRC, it is hypothesized that hyperinsulinemia, high insulin levels, decreased bowel transit time, and increased fecal concentration of bile acids in DM may contribute to tumor growth.9
MicroRNAs (miRNAs) are noncoding, single-stranded RNA of about 21–23 nucleotides in length that inhibit protein expression in a post-transcriptional pattern via the interaction with complementary sites of the target mRNA.12 miRNAs play an important role in cell growth, proliferation, differentiation, motility, autophagy, and apoptosis.13,14 Deregulated expression of miRNAs is observed in various diseases,15–18 including CRC16,19,20 and DM.17,21–23
The mammalian target of rapamycin (mTOR) pathway controls protein synthesis in response to a large number of signals including nutrients such as glucose and amino acids, and growth factors such as insulin and IGF-1.24,25 This pathway is involved in many major cellular processes and plays a key role in a great number of pathological conditions, including cancer, obesity, diabetes, and neurodegeneration.24–26 Recent studies have revealed that the mTOR pathway is abnormally activated in CRC, which may contribute to the growth, metastasis, and progression of tumors.27 Moreover, the chronic activation of mTOR has been indicated to participate in the development of insulin resistance and type 2 diabetes.28,29 The high rate of protein synthesis associated with hyperactivation of mTOR has been considered to induce insulin resistance by promoting ER stress and the unfolded protein response.30 Many studies have shown that mTOR is highly active in the tissues of obese and high-fat-fed rodents.31 Overactivation of mTOR promotes adipogenesis, and the expansion of adipose tissue that characterizes the obese state represents the main risk factor for the development of insulin resistance and type 2 diabetes.32,33 It was demonstrated that insulin could regulate glycolytic activities via decreasing miR-99a and increasing mTOR in hepatocellular carcinoma.34 Meanwhile, previous study has shown that miR-99a could decrease cell proliferation, migration and possibly invasion of colorectal cancer cell lines by inhibiting the amount of phosphorylated mTOR protein.35 However, there is little information whether miRNA is involved in regulated mTOR in CRC complicated with DM. And the molecular mechanisms underlying the potential crosstalk are still nuclear.
In the present study, microarray analysis was used to estimate miRNA expression in tumor specimens from CRC patients and CRC with DM patients, along with normal colorectal mucosa samples. We further evaluated the expression of miR-99a in tumor tissues from 20 patients with CRC complicated with DM, 20 with CRC but not DM, along with 20 samples of normal tissues using quantitative polymerase chain reaction (qRT-PCR). Moreover, we explored the role of miR-99a in CRC with DM and revealed the relationship between miR-99a and mTOR in this clinical context.
Materials and methods
Patients and tumor samples
Forty specimens were obtained from CRC patients at the Sixth Affiliated Hospital of Sun Yat-Sen University in 2018. Blinded review of the original pathology slides was used to confirm the histopathological characteristics of CRC tissue specimens. Of the 40 patients, 20 were diagnosed with colorectal cancer with DM (CRC + DM) and 20 with colorectal cancer but not diabetes mellitus (CRC + nonDM). The diagnosis of DM was made in accordance with World Health Organization criteria: patients were classified as having diabetes if fasting blood glucose was ≥126 mg/dL (7 mmol/L). All of the tissue samples were extracted from surgical specimens or endoscopic biopsies, and none of the patients had undergone either chemotherapy or radiotherapy before the surgery (Table S1). The normal mucosa was collected in the bowel of nontumorous patients. Samples were subsequently frozen in liquid nitrogen and stored at −80°C. The study was performed in accordance with the Helsinki Declaration and approved by Sun Yat-sen University. All subjects were informed of the investigational nature of the study and provided their written informed consent.
Table S1.
Clinicopathological characteristics of patients
| Group | Patient ID | Gender | Age (years) | Tumor differentiation | Tumor location |
|---|---|---|---|---|---|
| CRC + DM | 0000180382 | Male | 52 | Moderately | Colon |
| CRC + DM | 0000107510 | Female | 81 | Undifferentiated | Colon |
| CRC + DM | 0000180657 | Male | 74 | Well | Colon |
| CRC + DM | 0000180766 | Male | 49 | Moderately | Colon |
| CRC + DM | 0000181317 | Female | 67 | Moderately | Colon |
| CRC + DM | 0000181002 | Female | 79 | Well/Moderately | Colon |
| CRC + DM | 0000178776 | Female | 79 | Moderately | Colon |
| CRC + DM | 0000178908 | Female | 50 | Moderately | Rectum |
| CRC + DM | 0000179669 | Male | 41 | Moderately/Poorly | Colon |
| CRC + DM | 0000180508 | Male | 50 | Moderately | Colon |
| CRC + DM | 0000179907 | Male | 60 | Moderately | Rectum |
| CRC + DM | 0000176460 | Male | 50 | Moderately | Rectum |
| CRC + DM | 0000176403 | Female | 65 | Moderately | Colon |
| CRC + DM | 0000175540 | Male | 70 | Moderately | Rectum |
| CRC + DM | 0000174817 | Female | 64 | Moderately/Poorly | Colon |
| CRC + DM | 0000174783 | Male | 72 | Well/Moderately | Colon |
| CRC + DM | 0000174234 | Female | 54 | Moderately | Colon |
| CRC + DM | 0000173801 | Male | 53 | Moderately | Rectum |
| CRC + DM | 0000173030 | Male | 30 | Well/Moderately | Colon |
| CRC + DM | 0000173719 | Male | 62 | Moderately | Colon |
| CRC | 0000180519 | Female | 72 | Moderately | Colon |
| CRC | 0000179083 | Male | 70 | Moderately | Rectum |
| CRC | 0000179656 | Female | 60 | Moderately | Colon |
| CRC | 0000179685 | Female | 48 | Moderately | Rectum |
| CRC | 0000176489 | Male | 40 | Undifferentiated | Rectum |
| CRC | 0000176509 | Male | 66 | Moderately | Rectum |
| CRC | 0000176701 | Male | 44 | Moderately | Rectum |
| CRC | 0000177107 | Male | 46 | Moderately | Rectum |
| CRC | 00000V3832 | Male | 65 | Moderately | Colon |
| CRC | 0000177712 | Female | 56 | Moderately | Rectum |
| CRC | 0000175532 | Male | 61 | Moderately | Colon |
| CRC | 0000176746 | Male | 59 | Moderately | Rectum |
| CRC | 0000177540 | Male | 70 | Moderately | Colon |
| CRC | 0000177733 | Male | 69 | Moderately | Rectum |
| CRC | 0000178244 | Female | 60 | Well | Colon |
| CRC | 0000177128 | Male | 52 | Moderately | Colon |
| CRC | 0000178421 | Female | 51 | Moderately | Colon |
| CRC | 0000178279 | Male | 66 | Moderately | Colon |
| CRC | 0000178551 | Male | 54 | Moderately | Colon |
| CRC | 0000178539 | Male | 85 | Moderately | Rectum |
miRNA microarray processing
Total RNA was extracted from specimens using Trizol reagent (Invitrogen, MA, USA), in accordance with the manufacturer’s protocol. The quantity and quality of RNA were evaluated by an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). Agilent Human miRNA (8*60K) arrays from Shanghai Biotechnology Corporation were used. Microarray hybridization, scanning, and analysis were also performed by Shanghai Biotechnology Corporation. Briefly, 2 µg of sample RNA was directly labeled with biotinylated signaling molecule using FlashTag Biotin HSR RNA Labeling kits (Affymetrix, USA). Hybridization and washing of the chips were performed in accordance with the manufacturer’s protocol. Scanning was performed using GeneChip Scanner 3000. GeneChip Operating Software was used to analyze the data. Average log2 ratios were calculated from the normalized data of each miRNA.
Cell culture and transfection
Human CRC cell lines HCT-15, HCT-116, HCT-8, SW480, and LOVO and normal human colon mucosal epithelial cell line NCM460 were purchased from the Cell Bank of Chinese Scientific Academy (Shanghai, China). The cells were routinely cultured in DMEM (Gibco, NY, USA) with 10% heat-inactivated fetal bovine serum (FBS; Gibco, NY, USA) and 1% penicillin–streptomycin (Gibco, NY, USA) in a humidified incubator at 37 °C with 5% CO2. miR-99a mimic was purchased from GenePharma (Shanghai, China). Cells were seeded into plates 12 h prior to transfection. Moreover, 100 nM miR-99a mimic or negative control (miR-NC) was transfected into cells using Lipofectamine 2000 (ThermoFisher, MA, USA), in accordance with the manufacturer’s instructions.
Quantitative real-time PCR (qRT-PCR)
Total RNA samples of cells and tissues were reverse-transcribed into cDNA using the PrimeScriptTM RT Reagent Kit (Takara, Tokyo, Japan) and Taqman MicroRNA Reverse Transcription Kit (ThermoFisher, MA, USA). The qRT-PCR was performed on a MiniOpticon™ real-time PCR detection instrument (Bio-Rad, CA, USA) with GoTaq® qPCR Master Mix (Promega, WI, USA) following standard procedures. GAPDH or U6 was used as an internal control. Primers used in this study were as follows: mTOR: 5′-ATTCCGACCTTCTGCCTTCA-3′ and 5′-AGTGAAGACTGTGGCATCCA-3′, GAPDH: 5′-GAGTCAACGGATTTGGTCGT-3′ and 5′-GACAAGCTTCCCGTTCTCAG-3′, and miR-99a: 5′-AAACCCGTAGATCCGATCT-3′.
Western blot analysis
The treated cells or tissues were washed with ice-cold PBS, and then lysed with RIPA containing proteinase and phosphatase inhibitors (Sigma-Aldrich, MO, USA). Protein concentration was quantified with Pierce™ BCA Protein Assay Kit (ThermoFisher, MA, USA), and then separated by 10% SDS-PAGE and transferred to PVDF membranes. The membranes were blocked with TBST containing 5% BSA for 1 h, and then incubated with anti-mTOR (Abcam, MA, USA) and anti-GAPDH (Abcam, MA, USA) overnight at 4 °C. The membranes were washed before incubation with specific secondary antibodies. Finally, the blots were visualized using chemiluminescence (ECL; Forevergen Biosciences Center, Guangzhou, China).
MTS assay
To estimate cell viability, the treated cells were seeded into 96-well plates at 1×103 cells per well 24 h before the MTS assay. The cells were incubated with MTS reagent for 2 h. Then, the OD value of each well at a wavelength of 490 nm was measured with a microplate reader (Diatek). All experiments were performed in triplicate.
Transwell assay
To evaluate cell mobility, the treated cells were suspended in serum-free medium and seeded in the upper compartment of modified Boyden chambers (BD Biosciences, CA, USA) at 2×104 cells/well. Medium containing 20% FBS was added to the lower compartment. The cells in the upper compartment migrated to the lower compartment through an 8-µm-pore-sized filter (for the migration) or a Matrigel-coated filter (for the invasion assay). After 48 h, the migrated/invasive cells were fixed in methanol and stained with crystal violet, and then photographed for counting. All experiments were performed in triplicate.
3′-UTR reporter assay for miRNA target validation
The whole sequence of the 3′-UTR of mTOR was amplified from the HCT-116 cDNA and cloned into the pmirGLO vector (Promega, WI, USA). The luciferase reporter plasmid with mutant mTOR 3′-UTR was constructed with MutanBest Kit (Takara, Tokyo, Japan), in accordance with the manufacturer’s protocol. All of the constructs were identified by sequencing. To confirm the direct interaction between miR-99a and mTOR, HCT-116 cells at 80% confluence were co-transfected with luciferase reporter plasmid and 100 nM miR-99a mimic or NC (miR-NC). At 48 h post-transfection, Firefly and Renilla luciferase activities were measured using Dual-Luciferase Reporter Assay (Promega, WI, USA). The final results are expressed as relative luciferase activity (Firefly LUC/Renilla LUC). All experiments were performed in triplicate.
Immunohistochemistry
The tissues were fixed in 10% formalin and then embedded in paraffin. The blocks were cut into 4-mm sections, deparaffinized, and rehydrated. After blocking with 3% hydrogen peroxide, the sections were incubated with mTOR primary antibody (Abcam, MA, USA) at 4 °C overnight. After washing with phosphate-buffered saline, the sections were incubated with the secondary antibody (Novus Biologicals, Shanghai, China) for 1 h at RT. Immunostaining was performed using 3,3′-diaminobenzidine tetrahydrochloride (DAB) (Sigma-Aldrich, MO, USA). Subsequently, sections were counterstained with hematoxylin.
Statistical analysis
All experiments were repeated at least three times. Data are expressed as mean ± standard error (SEM). Statistical analysis was performed with GraphPad Prism 7 (GraphPad Software Inc., CA, USA). .Statistical analysis for comparison of two groups was performed using two-tailed unpaired Student’s t-test. For comparison of more than two groups, one-way analysis of variance (ANOVA) followed by Tukey post hoc test was performed. P-values <0.05 were considered significant.
Results
Differential expression of miRNA among normal tissue, CRC with DM, and CRC without DM
To identify miRNA differentially expressed in CRC, we collected three groups of clinical tissue samples: normal (n=1), CRC without DM (CRC, n=1), and CRC with DM (CRC + DM, n=1). Then, we extracted miRNAs from the three samples and used the Agilent Human miRNA (8*60K) array to analyze the expression levels of 735 human miRNAs. The results showed that, compared with the normal group, 82 miRNAs were upregulated and 134 were downregulated with fold changs of more than 2 times in the CRC group (p<0.05). Meanwhile, compared with the CRC group, 62 miRNAs were upregulated and 103 were downregulated with fold changs of more than 2 times in the CRC + DM group (p<0.05).
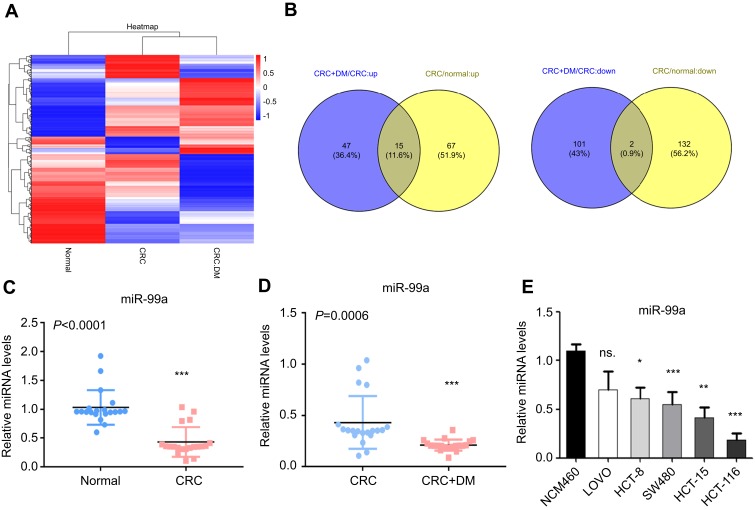
We further found that 17 miRNAs showed expression changes among all three groups with fold changs of more than 2 times. Among them, 15 miRNAs were sequentially upregulated from normal to CRC to CRC + DM groups, and two miRNAs were gradually downregulated in this order (Figure 1A and B). hsa-miR-99a-5p was the most significantly different miRNA in CRC + DM compared with the level in CRC + nonDM, and in CRC compared with the level in normal colon tissue, and the second downregulated miRNA was hsa-miR-214-3p (Figure 1B).
Figure 1.
MicroRNA expression patterns distinguish normal samples from CRC and CRC with DM. (A) Tissue miRNA microarray. Data are expressed as fold change of expression in CRC tissue versus normal tissue and CRC with DM versus normal tissue. (B) Venn diagram of differentially expressed miRNAs in CRC and CRC with DM samples. (C) The quantitative real-time PCR (qRT-PCR) analyses of miR-99a levels in normal (n=20) and CRC tissues (n=20). (D) The qRT-PCR analyses of miR-99a levels in CRC without DM (CRC + nonDM, n=20) and CRC with DM (CRC + DM, n=20). (E) The relative miR-99a expression levels in normal NCM460 and HCT-15, HCT-116, HCT-8, SW480, and LOVO tumor cells (*P<0.05, **P<0.01, ***P<0.001).
To further validate our results, the levels of miR-99a were estimated in 20 normal colon tissues and 40 CRC tissues using qRT-PCR. As shown in Figure 1C, miR-99a was dramatically downregulated (P<0.0001) in CRC tissues compared with that in normal colon tissues. To determine whether miR-99a is involved in CRC with DM, we examined it in 20 CRC and 20 CRC + DM tissue samples. The results indicated that CRC patients with DM exhibited markedly lower miR-99a expression (P=0.0006) than CRC patients without DM (Figure 1D). The expression levels of miR-99a were also evaluated in the normal human colon mucosal epithelial cell line NCM460 and the CRC cell lines HCT-15, HCT-116, HCT-8, SW480, and LOVO. As shown in Figure 1E, the qPCR results showed that miR-99a was suppressed in all analyzed CRC cells (Figure 1E). These results confirmed that the expression of miR-99a was significantly downregulated in clinical CRC tissues, CRC with DM tissues, and CRC cell lines, which is consistent with the microarray results.
miR-99a inhibits growth and mobility of CRC cells
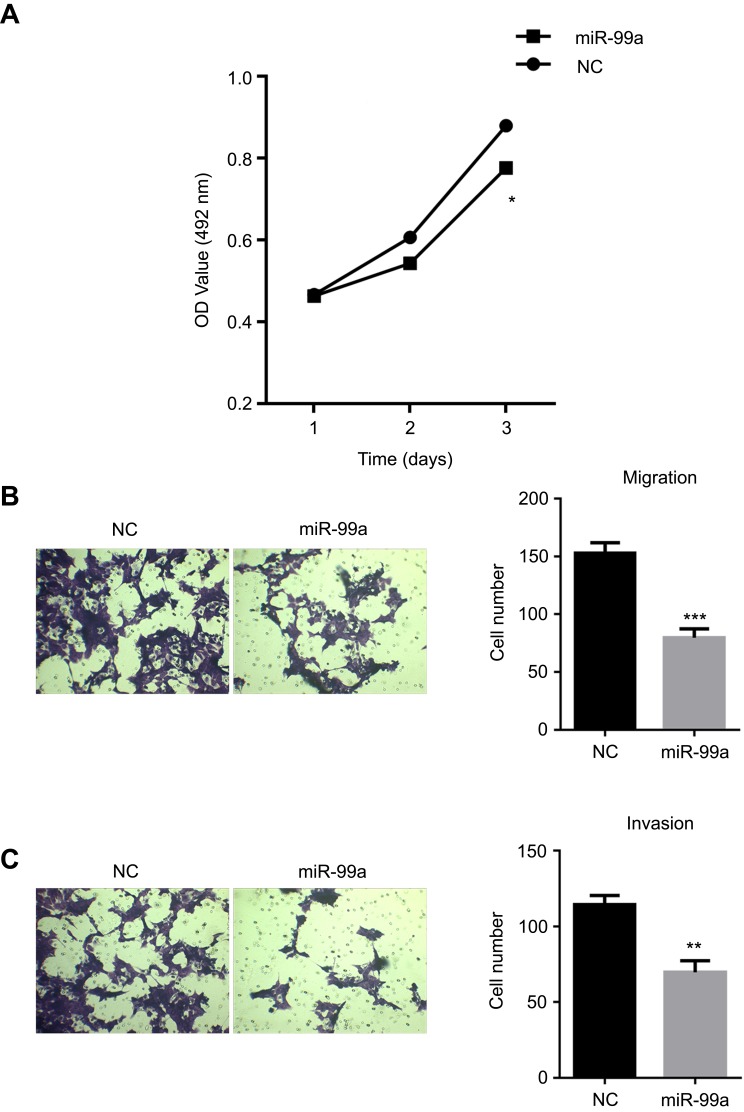
To investigate the role of miR-99a in CRC development and metastasis, we elevated the miR-99a level in HCT-116 using miR-99a mimic. Cell viability assays revealed that the ectopic expression of miR-99a impaired cell proliferation (Figure 2A). Furthermore, the role of miR-99a in CRC cell migration was estimated using Transwell assays. The cells transfected with miR-99a mimic exhibited lower migratory and invasive potential than the cells treated with miRNA mimic NC (Figure 2B and C). Taken together, these findings suggest that miR-99a is a negative regulator of growth and mobility in CRC cells.
Figure 2.
miR-99a suppressed proliferation and metastasis of CRC cells in vitro. HCT-116 cells were transiently transfected with 100 nM miR-99a mimic or negative control (NC) for 24 h. Cell viability was estimated by the MTS assay (A), while cell metastasis was determined by the Transwell migration (B) and invasion assay (C) (*P<0.05, **P<0.01, ***P<0.001).
miR-99a suppresses mTOR pathway through direct binding to 3′-UTR of mTOR
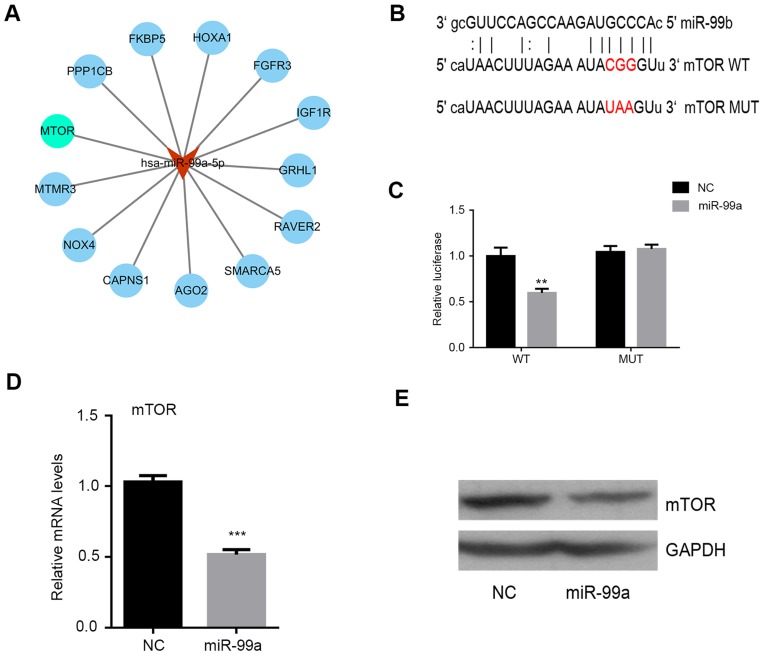
Mature miRNAs regulate gene expression by binding to the 3′-UTR of their target genes and blocking subsequent protein expression.We therefore searched for potential targets using the miRanda, TargetScan7.1, and PicTar, and only those targets detected by all programs were considered. And we found 13 potential target genes of miR-99a (Figure 3A). Among these target genes, mTOR with high degree formed local networks displayed their important roles in CRC, and a sequence of miR-99a complementary to the 3′-UTR of mTOR mRNA is shown in Figure 3B. Moreover, the results of luciferase reporter assay confirmed that miR-99a modulated mTOR levels through direct binding to the 3′-UTR of mTOR in CRC cells. Wild-type 3′-UTR of mTOR and mutant-type 3′-UTR (with a three-base mutation in the seed region sequence) fragments were inserted into pmirGLO luciferase reporter plasmid (Figure 3C). Exogenous overexpression of miR-99a suppressed relative luciferase activity in HCT-116 cells, while suppression was not observed in the cells transfected with mutant-type construct. Next, we evaluated the role of miR-99a in the mTOR pathway in CRC cells. As shown in Figure 3D and E, miR-99a mimic significantly downregulated the mRNA and protein levels of mTOR in the CRC cells. These findings suggest that miR-99a directly targeted mTOR in CRC cells.
Figure 3.
miR-99a targeting the mTOR gene in CRC cells. HCT-116 cells were transiently transfected with 100 nM miR-99a mimic or negative control (NC) for 48 h. The relative mTOR mRNA expression levels were determined by qRT-PCR (A). The mTOR protein expression levels were determined by Western blotting (B). The putative mir-99a binding site in mTOR 3′-UTR and an alteration of the mTOR 3′-UTR bearing mutant in the seed sequence are shown. The dual luciferase assay was performed to confirm the direct regulation of miR-99a on mTOR 3′-UTR (C). The relative mTOR mRNA expression levels were determined by qRT-PCR in the HCT-116 cells transfected with miR-99a mimic or NC (D). The mTOR protein levels were determined by western blotting in the HCT-116 cells transfected with miR-99a mimic or NC (E). (**P<0.01, ***P<0.001).
Advanced glycation end products (AGEs) stimulated mTOR through impairing miR-99a
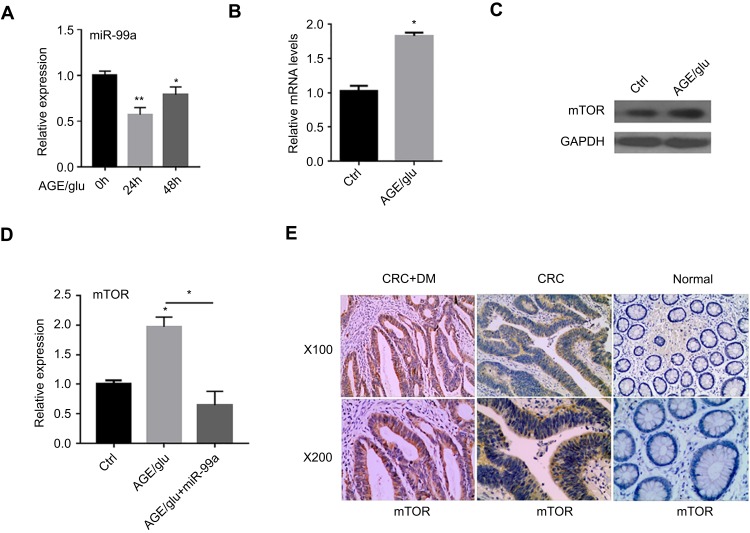
AGEs are formed from the nonenzymatic glycation of proteins and lipids with reducing sugars. To determine whether AGEs modulate the expression of miR-99a, HCT-116 cells were incubated with 100 mg/L AGEs or solvent control for 24h and 48 h, and then the levels of miR-99a were determined using qRT-PCR. The results showed that miR-99a was sharply decreased in AGE-treated cells compared with that in the control (Figure 4A). Moreover, the mRNA and protein expression levels of mTOR in AGE-treated cells at 48h were consistently elevated (Figure 4B and C), but the expression was inhibited after the miR-99a mimics treatment (Figure 4D). To further assess the role of miR-99a in CRC with DM patients, we determined the protein expression levels of mTOR in 20 CRC + DM and 20 CRC + nonDM tissues using IHC. As shown in Figure 4E, a remarkable increase in mTOR expression level was observed in diabetic CRC tissues compared with the level in nondiabetic CRC tissues. These results suggest that AGEs suppress miR-99a to overactivate the mTOR pathway, which in turn promotes CRC progression in DM patients.
Figure 4.
HCT-116 cells were treated with 100 mg/L AGEs or solvent control for 48 h. The relative miR-99a expression levels were determined by qRT-PCR (A). The mTOR expression levels were analyzed by qRT-PCR (B, D) and Western blotting (C). The relative mTOR expression levels in CRC without DM (CRC + nonDM, n=20) and CRC with DM (CRC + DM, n=20) were determined by immunohistochemistry (E) (*P<0.05, **P<0.01).
Discussion
Diabetes and cancer are leading causes of mortality and morbidity in the general population globally.36 In certain circumstances, DM and cancer are intricately linked to each other, while in other situations, they may independently act to reduce patients’ quality of life.37,38 With the development and application of genetic detection methods, many different molecular subtypes of CRC have been established by different classification systems. CRC complicated with DM may be a special subtype of CRC, and there is an urgent need to reveal the mechanistic link between them. Numerous epidemiological studies have implied that there is a relationship between DM and CRC,6,7,10,11,37 but only a few studies have attempted to link DM and CRC at the molecular level. miRNAs are broadly associated with the development of many pathological processes, and the expression of miRNAs is broadly altered in cancer and diabetes.17,18,20,21,39 In this context, the question arises of whether miRNAs exhibit crosstalk between CRC and DM. We hypothesized that the alteration of specific miRNAs may function as a common biological factor between DM and CRC. In the present study, microarray assay revealed distinctions in miRNA expression between CRC versus normal and CRC + DM versus CRC + nonDM tissues, and showed that miR-99a is commonly the most significantly downregulated miRNA in CRC with DM. We further determined the alteration of miR-99a of normal (n=20) versus CRC (n=40) and CRC + DM (n=20) versus CRC + nonDM (n=20) tissues. Our results showed that the expression of miR-99a decreased 50% in CRC compared with that in normal tissues, and 50% in CRC + DM compared with that in the CRC + nonDM group.
miR-99a, located on chromosome 21, has been reported to be a tumor suppressor in a variety of human cancers, including breast, oral, and hepatocellular cancers.40–43 Numerous studies have suggested that miR-99a participates in the regulation of epithelial-to-mesenchymal transition (EMT).43,44 EMT correlates with the increased migration and invasion of cancer cells.45,46 Exogenous overexpression of miR-99a was also shown to significantly inhibit the metastasis of oral cancer, non-small cell lung cancer, and cervical cancer cells.44,47,48 Moreover, miR-99a-3p is one of the significant predictors of chemotherapy response in patients with advanced colorectal cancer.49 Here, we confirmed that miR-99a was downregulated in human CRC tissues and cells and could remarkably suppress the metastasis and viability of CRC cells. miRNAs bind to the 3′-UTR of their target mRNA and interfere with translation.12 Therefore, we integrated bioinformatics-based predictions and the interaction networks of each target gene and found that mTOR was the most important potential target gene of miR-99a. Previous studies reported that miR-99a can inhibit mTOR expression by directly targeting its 3′-UTR in numerous human cancers including prostate cancer cells, childhood adrenocortical tumor cells, and breast cancer cells.46,50,51 We extended these studies and confirmed the relationship between miR-99a and mTOR in CRC cells. In addition, we demonstrated that miR-99a mimic was able to decrease the expression of mTOR at mRNA and protein levels in HCT116 cells. Moreover, we constructed luciferase reporter plasmids containing wild-type 3′-UTR of mTOR and found that miR-99a mimic could significantly reduce the relative luciferase activity. The repression of relative luciferase activity by miR-99a mimic was not observed in the cells transfected with constructs containing mutant-type 3′-UTR of mTOR. All of these results indicate that miR-99a can directly modulate mTOR gene expression through binding to the 3′-UTRs of mTOR mRNA.
mTOR, a serine-threonine protein kinase, belongs to the phosphoinositide 3-kinase (PI3K)-related kinase family.24 mTOR activity is controlled by the tumor suppressor complex TSC1/2. Growth factors and nutrients, such as glucose insulin and amino acids, activate PI3K-PKB and mitigate TSC1/2 activity, which causes mTOR activation.25 Activated mTOR regulates the translation initiation machinery by modulating S6 kinase and eIF4E binding protein 1 (4EBP1) phosphorylation, leading to increased protein synthesis and cell growth.32 A substantial number of studies support the importance of the mTOR pathway in cancer pathogenesis.27,52,53 The deregulation of protein synthesis caused by overactivation of mTOR plays a central role in tumor formation. Zhang et al53 reported that the activity of mTOR signaling is much higher in CRC than in normal noncancerous mucosa, and that the overactivation of mTOR occurs as an early event in the process of tumorigenesis and is involved in the progression from normal cells to hyperplasia to a neoplastic phenotype.27
Conclusion
Here, we focused on the role of miRNAs in CRC complicated with DM and provided evidence that decreased miR-99a and deregulation of mTOR signaling are involved in CRC + DM. In this study, we confirmed the tumor suppressor role of miR-99a in CRC, which is probably mediated by the targeting of mTOR. Our results suggest that a high concentration of AGEs in DM patients impairs miR-99a and increases the expression of mTOR. Moreover, it is reported here for the first time that the expression levels of mTOR are elevated in CRC complicated with DM tissues compared with that in CRC without DM tissue. This finding suggests that miR-99a is a potential link between CRC and DM, and may be a specific biomarker and therapeutic target of the subtype of CRC complicated with DM.
Supplementary material
Acknowledgments
We gratefully thank Forevergen Biosciences for assistance with the experiments. This work was supported by the grants of Guangzhou Tianhe District Science and Technology Program (grant number 201504KW045) and Natural Science Foundation of Guangdong Province (grant number 2017A030310229).
Disclosure
The authors declare no potential conflicts of interest in this work.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi: 10.3322/caac.21395 [DOI] [PubMed] [Google Scholar]
- 3.van de Poll-Franse LV, Houterman S, Janssen-Heijnen ML, Dercksen MW, Coebergh JW, Haak HR. Less aggressive treatment and worse overall survival in cancer patients with diabetes: a large population based analysis. Int J Cancer. 2007;120(9):1986–1992. doi: 10.1002/ijc.22532 [DOI] [PubMed] [Google Scholar]
- 4.Tudzarova S, Osman MA. The double trouble of metabolic diseases: the diabetes-cancer link. Mol Biol Cell. 2015;26(18):3129–3139. doi: 10.1091/mbc.E14-11-1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins KK. The diabetes-cancer link. Diabetes Spectr. 2014;27(4):276–280. doi: 10.2337/diaspect.27.4.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coughlin SS, Calle EE, Teras LR, Petrelli J, Thun MJ. Diabetes mellitus as a predictor of cancer mortality in a large cohort of US adults. Am J Epidemiol. 2004;159(12):1160–1167. doi: 10.1093/aje/kwh161 [DOI] [PubMed] [Google Scholar]
- 7.Woo H, Lee J, Lee J, et al. Diabetes mellitus and site-specific colorectal cancer risk in Korea: a case-control study. J Prev Med Public Health. 2016;49(1):45–52. doi: 10.3961/jpmph.15.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuhara H, Steinmaus C, Cohen SE, Corley DA, Tei Y, Buffler PA. Is diabetes mellitus an independent risk factor for colon cancer and rectal cancer? Am J Gastroenterol. 2011;106(11):1911–1921; quiz 1922. doi: 10.1038/ajg.2011.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Will JC, Galuska DA, Vinicor F, Calle EE. Colorectal cancer: another complication of diabetes mellitus? Am J Epidemiol. 1998;147(9):816–825. doi: 10.1093/oxfordjournals.aje.a009534 [DOI] [PubMed] [Google Scholar]
- 10.Zhu B, Wu X, Wu B, Pei D, Zhang L, Wei L. The relationship between diabetes and colorectal cancer prognosis: a meta-analysis based on the cohort studies. PLoS One. 2017;12(4):e0176068. doi: 10.1371/journal.pone.0176068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.La Vecchia C, Negri E, Decarli A, Franceschi S. Diabetes mellitus and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 1997;6(12):1007–1010. [PubMed] [Google Scholar]
- 12.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 13.Wust S, Drose S, Heidler J, et al. Metabolic maturation during muscle stem cell differentiation is achieved by miR-1/133a-mediated inhibition of the Dlk1-Dio3 mega gene cluster. Cell Metab. 2018;27(5):1026–39.e6. doi: 10.1016/j.cmet.2018.02.022 [DOI] [PubMed] [Google Scholar]
- 14.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33(4):1290–1297. doi: 10.1093/nar/gki200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patane S. Value of serial ST2 measurements in acute heart failure: miRNA regulation and genetic factors. J Am Coll Cardiol. 2018;71(12):1397. doi: 10.1016/j.jacc.2017.11.075 [DOI] [PubMed] [Google Scholar]
- 16.Yunqi H, Fangrui Y, Yongyan Y, et al. MiR-455 functions as a tumor suppressor through targeting GATA6 in colorectal cancer. Oncol Res. 2019;27(3):311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pheiffer C, Dias S, Rheeder P, Adam S. Decreased expression of circulating miR-20a-5p in South African women with gestational diabetes mellitus. Mol Diagn Ther. 2018;22(3):345–352. doi: 10.1007/s40291-018-0325-0 [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Pogue AI, Lukiw WJ. MicroRNA (miRNA) signaling in the human CNS in sporadic Alzheimer’s Disease (AD)-novel and unique pathological features. Int J Mol Sci. 2015;16(12):30105–30116. doi: 10.3390/ijms161226223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diaz T, Moreno I, Monzo M. miR-200 family in CRC primary tumors and metastases. J Surg Oncol. 2014;110(4):486. doi: 10.1002/jso.23683 [DOI] [PubMed] [Google Scholar]
- 20.Ghanbari R, Rezasoltani S, Hashemi J, et al. Expression analysis of previously verified fecal and plasma dow-regulated MicroRNAs (miR-4478, 1295-3p, 142-3p and 26a-5p), in FFPE tissue samples of CRC patients. Arch Iran Med. 2017;20(2):92–95. [PubMed] [Google Scholar]
- 21.Deng X, Qin S, Chen Y, et al. B-RCA revealed circulating miR-33a/b associates with serum cholesterol in type 2 diabetes patients at high risk of ASCVD. Diabetes Res Clin Pract. 2018;140:191–199. doi: 10.1016/j.diabres.2018.03.024 [DOI] [PubMed] [Google Scholar]
- 22.Cai F, Wu F, Cao J, Chen X. MicroRNA-146b-3p regulates the development and progression of cerebral infarction with diabetes through RAF1/P38MAPK/COX-2 signaling pathway. Am J Transl Res. 2018;10(2):618–628. [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao D, Wang NS, Chen F, Li ZB, Li XT, Zhu XX. Intravenous injection of miR-34a inhibitor alleviates diabetes mellitus-induced vascular endothelial dysfunction by targeting NOTCH1. Exp Clin Endocrinol Diabetes. 2019;127(4):255–262. [DOI] [PubMed] [Google Scholar]
- 24.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tee AR, Blenis J. mTOR, translational control and human disease. Semin Cell Dev Biol. 2005;16(1):29–37. doi: 10.1016/j.semcdb.2004.11.005 [DOI] [PubMed] [Google Scholar]
- 26.Tuo Y, Xiang M. mTOR: A double-edged sword for diabetes. J Leukoc Biol. 2018. doi: 10.1002/JLB.3MR0317-095RR [DOI] [PubMed] [Google Scholar]
- 27.Johnson SM, Gulhati P, Rampy BA, et al. Novel expression patterns of PI3K/Akt/mTOR signaling pathway components in colorectal cancer. J Am Coll Surg. 2010;210(5):767–76, 76–88. doi: 10.1016/j.jamcollsurg.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin X, Xu Z, Zhang Z, et al. Association of PI3K/AKT/mTOR pathway genetic variants with type 2 diabetes mellitus in Chinese. Diabetes Res Clin Pract. 2017;128:127–135. doi: 10.1016/j.diabres.2017.04.002 [DOI] [PubMed] [Google Scholar]
- 29.Suhara T, Baba Y, Shimada BK, Higa JK, Matsui T. The mTOR signaling pathway in myocardial dysfunction in type 2 diabetes mellitus. Curr Diab Rep. 2017;17(6):38. doi: 10.1007/s11892-017-0902-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tremblay F, Marette A. Amino acid and insulin signaling via the mTOR/p70 S6 kinase pathway. A negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J Biol Chem. 2001;276(41):38052–38060. [DOI] [PubMed] [Google Scholar]
- 31.Kim S, Jwa H, Yanagawa Y, Park T. Extract from Dioscorea batatas ameliorates insulin resistance in mice fed a high-fat diet. J Med Food. 2012;15(6):527–534. doi: 10.1089/jmf.2011.2008 [DOI] [PubMed] [Google Scholar]
- 32.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caron A, Richard D, Laplante M. The roles of mTOR complexes in lipid metabolism. Annu Rev Nutr. 2015;35:321–348. [DOI] [PubMed] [Google Scholar]
- 34.Li W, Wang J, Chen QD, et al. Insulin promotes glucose consumption via regulation of miR-99a/mTOR/PKM2 pathway. PLoS One. 2013;8(6):e64924. doi: 10.1371/journal.pone.0064924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jonathan R. The influence of miR-99a on mTOR signaling regulation in colorectal cancer cell lines [Electronic Theses and Dissertations]. 2016:2412. [Google Scholar]
- 36.Morrish NJ, Wang SL, Stevens LK, Fuller JH, Keen H. Mortality and causes of death in the WHO multinational study of vascular disease in diabetes. Diabetologia. 2001;44 Suppl 2:S14–S21. [DOI] [PubMed] [Google Scholar]
- 37.Zanders MM, Vissers PA, Haak HR, van de Poll-Franse LV. Colorectal cancer, diabetes and survival: epidemiological insights. Diabetes Metab. 2014;40(2):120–127. doi: 10.1016/j.diabet.2013.12.007 [DOI] [PubMed] [Google Scholar]
- 38.Payton S. Kidney cancer: diabetes and low serum cholesterol are risk factors for RCC. Nat Rev Urol. 2014;11(8):422. doi: 10.1038/nrurol.2014.157 [DOI] [PubMed] [Google Scholar]
- 39.Petrie JR, Guzik TJ, Touyz RM. Diabetes, hypertension, and cardiovascular disease: clinical insights and vascular mechanisms. Can J Cardiol. 2018;34(5):575–584. doi: 10.1016/j.cjca.2017.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Truini A, Coco S, Nadal E, et al. Downregulation of miR-99a/let-7c/miR-125b miRNA cluster predicts clinical outcome in patients with unresected malignant pleural mesothelioma. Oncotarget. 2017;8(40):68627–68640. doi: 10.18632/oncotarget.19800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen H, Tian Y, Yao X, Liu W, Zhang Y, Yang Z. MiR-99a inhibits keratinocyte proliferation by targeting Frizzled-5 (FZD5)/FZD8 through beta-catenin signaling in psoriasis. Pharmazie. 2017;72(8):461–467. doi: 10.1691/ph.2017.7018 [DOI] [PubMed] [Google Scholar]
- 42.Feliciano A, Garcia-Mayea Y, Jubierre L, et al. miR-99a reveals two novel oncogenic proteins E2F2 and EMR2 and represses stemness in lung cancer. Cell Death Dis. 2017;8(10):e3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mei LL, Qiu YT, Huang MB, Wang WJ, Bai J, Shi ZZ. MiR-99a suppresses proliferation, migration and invasion of esophageal squamous cell carcinoma cells through inhibiting the IGF1R signaling pathway. Cancer Biomark. 2017;20(4):527–537. doi: 10.3233/CBM-170345 [DOI] [PubMed] [Google Scholar]
- 44.Yu SH, Zhang CL, Dong FS, Zhang YM. miR-99a suppresses the metastasis of human non-small cell lung cancer cells by targeting AKT1 signaling pathway. J Cell Biochem. 2015;116(2):268–276. doi: 10.1002/jcb.24965 [DOI] [PubMed] [Google Scholar]
- 45.Turcatel G, Rubin N, El-Hashash A, Warburton D. MIR-99a and MIR-99b modulate TGF-beta induced epithelial to mesenchymal plasticity in normal murine mammary gland cells. PLoS One. 2012;7(1):e31032. doi: 10.1371/journal.pone.0031032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Z, Han Y, Cheng K, Zhang G, Wang X. miR-99a directly targets the mTOR signalling pathway in breast cancer side population cells. Cell Prolif. 2014;47(6):587–595. doi: 10.1111/cpr.12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Chang L, Li Z, et al. miR-99a and −99b inhibit cervical cancer cell proliferation and invasion by targeting mTOR signaling pathway. Med Oncol. 2014;31(5):934. doi: 10.1007/s12032-014-0374-0 [DOI] [PubMed] [Google Scholar]
- 48.Kuo YZ, Tai YH, Lo HI, et al. MiR-99a exerts anti-metastasis through inhibiting myotubularin-related protein 3 expression in oral cancer. Oral Dis. 2014;20(3):e65–e75. doi: 10.1111/odi.12133 [DOI] [PubMed] [Google Scholar]
- 49.Molina-Pinelo S, Carnero A, Rivera F, et al. MiR-107 and miR-99a-3p predict chemotherapy response in patients with advanced colorectal cancer. BMC Cancer. 2014;14:656. doi: 10.1186/1471-2407-14-656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun D, Lee YS, Malhotra A, et al. miR-99 family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res. 2011;71(4):1313–1324. doi: 10.1158/0008-5472.CAN-10-1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doghman M, El Wakil A, Cardinaud B, et al. Regulation of insulin-like growth factor-mammalian target of rapamycin signaling by microRNA in childhood adrenocortical tumors. Cancer Res. 2010;70(11):4666–4675. doi: 10.1158/0008-5472.CAN-09-3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang F, Wang L, Zhang S, et al. CD147 modulates autophagy through the PI3K/Akt/mTOR pathway in human prostate cancer PC-3 cells. Oncol Lett. 2015;9(3):1439–1443. doi: 10.3892/ol.2015.2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang YJ, Dai Q, Sun DF, et al. mTOR signaling pathway is a target for the treatment of colorectal cancer. Ann Surg Oncol. 2009;16(9):2617–2628. doi: 10.1245/s10434-009-0555-9 [DOI] [PubMed] [Google Scholar]