Abstract
A highly specialized cytoskeletal protein, keratin 75 (K75), expressed primarily in hair follicles, nail beds, and lingual papillae, was recently discovered in dental enamel, the most highly mineralized hard tissue in the human body. Among many questions this discovery poses, the fundamental question regarding the trafficking and secretion of this protein, which lacks a signal peptide, is of an utmost importance. Here, we present evidence that K75 is expressed during the secretory stage of enamel formation and is present in the forming enamel matrix. We further show that K75 is secreted together with major enamel matrix proteins amelogenin and ameloblastin, and it was detected in Golgi and the endoplasmic reticulum (ER)–Golgi intermediate compartment (ERGIC) but not in rough ER (rER). Inhibition of ER–Golgi transport by brefeldin A did not affect the association of K75 with Golgi, whereas ameloblastin accumulated in rER, and its transport from rER into Golgi was disrupted. Together, these results indicate that K75, a cytosolic protein lacking a signal sequence, is secreted into the forming enamel matrix utilizing portions of the conventional ER–Golgi secretory pathway. To the best of our knowledge, this is the first study providing insights into mechanisms of keratin secretion.
Keywords: biomineralization, secretion, keratin, Golgi, endoplasmic reticulum (ER), ameloblast, ameloblastin, amelogenin
Introduction
Keratins comprise a large and diverse family of epithelial cytoskeletal proteins that evolved from two ancestral genes during the expansion of tetrapods into the land environments (1). There are 54 keratins in humans, divided into two major groups, type I (acidic) and type II (basic), which assemble into heterodimers (2). Based on their location and function, keratins are further divided into epithelial, hair follicle, and hair keratins (2). Keratins organize into heavily cross-linked networks of intermediate filaments, which provide mechanical rigidity to the cells and play important roles in cell–cell contacts as components of the desmosomal complexes (3, 4). In addition to the major role in the cytoskeleton, recent studies suggest that keratins can have other important functions. Specifically, keratin 17 was detected in cell nuclei where it was shown to regulate gene expression (5). Hair follicle type II keratin 75 (K752; also known as K6hf) is expressed in ectodermal appendages of amniotes, such as hair and feather follicles, nail beds, and lingual papillae (6–8). A single amino acid substitution, A161T, in this protein causes a common skin disorder, pseudofolliculitis barbae, colloquially called barber's itch (9). We recently demonstrated that K75 is present in dental enamel–forming cells, ameloblasts, and enamel (10). Moreover, it was found that dental enamel in individuals carrying the pseudofolliculitis barbae–causing polymorphism has altered structure and mechanical properties and is more susceptible to caries (10). Recently, a number of other epithelial keratins were discovered in enamel, and an association between mutations in these keratins and a higher susceptibility to caries has been established (11). Furthermore, in our recent study, K75 was identified in forming porcine enamel by Western blotting and mass spectrometry (MS) (12). Importantly, unlike other transient enamel matrix proteins (EMPs), K75 was not degraded by resident enamel proteases KLK4 and MMP20, strongly suggesting that it incorporates into mature extracellular enamel matrix (EEM).
Although hairs and teeth evolved independently, they share a number of regulatory pathways involved in their morphogenesis (13, 14). For example, expression of K75 and several other keratins in ameloblasts, enamel-forming cells, is regulated by DLX3 transcription factor (10), which plays a major role in organogenesis of hairs and teeth (15–17). Mutations in DLX3 cause tricho-dento-osseus syndrome, manifested by severe hair and tooth defects. Specifically, enamel in tricho-dento-osseus syndrome patients lacks the rod pattern, which is the hallmark of mammalian enamel (18).
Mature enamel is the hardest tissue in the body; it comprises 95% carbonated hydroxyapatite mineral and ∼1% organic matrix. The basic building blocks of enamel, enamel rods, are arrays of elongated crystals aligned with their crystallographic c axes (19–21). The rods are woven into a very complex 3D structure, which is responsible for the unique mechanical resilience of this material (20, 22–25). Although mature enamel is a highly mineralized material, it starts as a hydrogel containing arrays of tiny crystallites (26–29). Enamel formation (amelogenesis) is carried out by specialized epithelial cells, ameloblasts, in a highly coordinated multistep process (30). Enamel matrix deposition begins at the surface of forming dentin by so-called presecretory ameloblasts. They differentiate into secretory ameloblasts, polarized cells with the nuclei located at the basal (proximal) pole, and the Tomes' processes, a specialized cellular secretory apparatus responsible for enamel rod formation, at the apical (distal) pole (31–33). The secretory EEM mainly consists of three proteins, amelogenin (AMELX) (34) comprising ∼90% of the matrix, ameloblastin (AMBN) (35), and enamelin (ENAM) (36), and a metalloproteinase, MMP20 (37), which play important roles in the regulation of mineralization (38–45). All EMPs possess a signal sequence and are secreted via the conventional ER–Golgi secretory pathway (46, 47). Interestingly, several articles reported interactions between EMPs and cytosolic keratins, suggesting the presence of novel trafficking mechanisms in ameloblasts (48, 49). Once the full thickness of enamel is deposited, the maturation stage begins, at which the majority of EMPs are removed and replaced by thickening enamel crystals (43). Only a small fraction of a heavily cross-linked fibrous material remains in the mature enamel (28). This material comprises rod sheaths, enveloping enamel rods, and enamel tufts, protein-rich structures in the inner enamel (50–52) that contain K75 and other keratins (10, 11).
Our discovery of K75 in enamel (10, 12) raises a number of questions regarding its roles in enamel formation and its functional properties. One question, with a potential impact beyond enamel research, is of particular interest, namely when and how this cytosolic protein lacking a lead sequence enters the extracellular space. There are only a few reports in the literature describing keratins outside cells (53, 54), but these studies do not address the secretory pathway question. It is possible that K75 ends up in the enamel matrix as a part of cellular debris embedded in the matrix during enamel secretion (55). Alternatively, cytosolic proteins can be secreted via unconventional protein secretion mechanisms (56–58), including Golgi bypass (56, 59), secretory autophagy (60, 61), secretory lysosomes (62), and endosomes (63). Conversely, studies show that a number of proteins lacking the signal sequence can enter the classical secretory pathway via posttranslational translocation into rough ER (rER) (64, 65).
To gain insights into the mechanisms of K75 trafficking and secretion, we carried out a number of experiments to characterize the expression pattern and trafficking pathway of this protein during enamel formation. Importantly, unlike the majority of trafficking studies, we performed our experiments in vivo because ameloblasts, highly specialized polarized secretory cells, cannot maintain their differentiated phenotype in vitro.
Results
K75 is expressed by secretory ameloblasts and is present in forming enamel
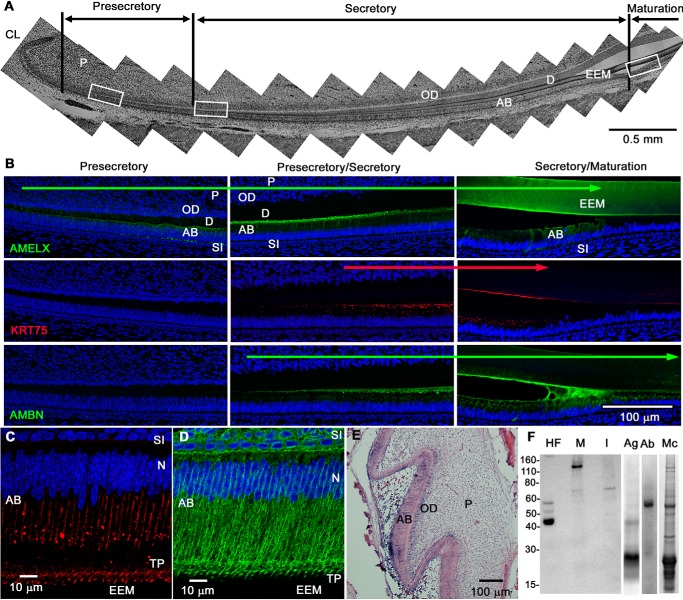
Taking advantage of the constantly growing murine incisors, containing all stages of enamel development (Fig. 1A), we were able to observe K75 expression pattern during amelogenesis and to compare it with AMELX and AMBN, two major EMPs with well-established expression profiles (46, 66, 67). Fig. 1B shows immunofluorescence (IF) images taken from the three boxed areas in Fig. 1A. The IF results indicate that only AMELX is expressed at the presecretory stage of amelogenesis, and all three proteins are expressed during the secretory stage (Fig. 1B). At the transition between secretory and maturation stages, AMELX and K75 expression cease, whereas AMBN expression persists through the maturation stage (Fig. 1B). The expression profiles of AMELX and AMBN observed in this study are consistent with earlier findings (46, 66, 67). Importantly, our results clearly demonstrate that K75 protein is exclusively expressed during the secretory stage of amelogenesis. Higher magnification imaging of secretory-stage ameloblasts revealed that the K75 distribution in secretory ameloblasts was remarkably different from other typical cytokeratins, such as K14 (Fig. 1, C and D). Although IF of K14 revealed a network of filaments throughout the ameloblast, K75 was found in large granular bodies located in the distal parts of ameloblasts and a distinct signal organized into a band in the central portion of the cytoplasm. Strong K75 signal was also present in Tomes' processes. In situ hybridization (ISH) studies revealed that Krt75 has a unique expression pattern in the craniofacial region. Its expression is confined to cells of hair follicles, lingual papillae, and enamel organ (Figs. 1E and S1, A and B). The presence of K75 in forming molar and incisal enamel from 14-day-old rats was also detected by Western blotting (WB) (Fig. 1F). A band around 70 kDa was observed in both molar and incisal enamel, and in the case of molar enamel, a higher-molecular-mass band between 110 and 160 kDa, likely corresponding to a K75 dimer with an acidic keratin, was detected (Fig. 1F). The fact that K75 is observed at a higher than nominal K75 molecular mass of 59 kDa suggests that it undergoes posttranslational modifications, potentially being glycosylated in forming enamel (68). The 70-kDa band was further analyzed using MS, which identified with 100% confidence five K75 fragments covering 8% of its sequence (Fig. S1C). Importantly no enamel matrix proteins were identified in this band. Overall, these results corroborate our earlier observation of K75 in enamel (10, 12) and revealed that K75 expression is associated with secretory ameloblasts.
Figure 1.
Characterization of K75 expression in developing enamel tissue. A, sagittal section of an apical portion of rat incisor in bright-field. Presecretory, secretory, and maturation stages are identified based on the morphological changes of ameloblasts. The three white boxes indicate areas shown in B. B, AMELX, K75, and AMBN expression at the presecretory, secretory, and late secretory–early maturation stages of amelogenesis. AMELX was found early in the presecretory and secretory ameloblasts; it ceased at the secretory–maturation transition. K75 and AMBN were expressed throughout the secretory stage. K75 expression ceased at secretory–maturation transition, whereas AMBN expression continued into the maturation stage. C and D, high-magnification IF images of K75 (C) and K14 (D) in the mouse secretory ameloblasts. K14 showed a typical fibrillar network of tonofilaments, whereas K75 was present in the form of granules mainly located in the distal half of the cell body and in the form of long bands located in the central cytoplasm. E, in situ hybridization of Krt75 in mouse first molar at P1. Krt75 mRNA signal was present in ameloblasts, stratum intermedium, and odontoblasts. F, Western blot of hair follicle extract (HF), molar (M), and incisal (I) secretory enamel from 14-day-old rat using guinea pig anti-K75 antibodies, which were used for IF studies. Ag and Ab, molar secretory enamel lane stained with AMELX and AMBN antibodies, respectively; Mc, molar secretory enamel lane stained with Coomassie Brilliant Blue. The scale on the left represents estimated molecular mass in kDa. AB, ameloblast; D, dentin; N, nucleus; OD, odontoblast; P, pulp; SI, stratum intermedium; TP, Tomes' process; CL, cervical loop.
K75 is present in membrane-delineated vesicles in secretory ameloblasts
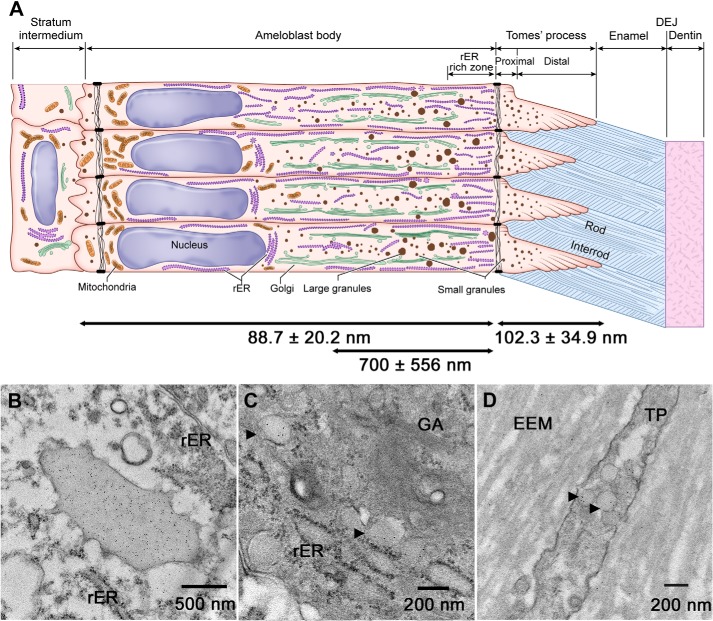
To better assess the intracellular localization of K75 in the secretory ameloblasts, immunogold transmission EM (IG-TEM) studies were carried out. Ameloblasts are polarized secretory cells with extensive rER and Golgi networks throughout the cell body (Figs. 2A and S2–S4). In addition to typical perinuclear rER and Golgi structures, a large cylindrical Golgi complex aligned with the cell axis exists in the midportion of an ameloblast (31, 33) (Figs. S2 and S3). The cis face of this central Golgi complex is mainly oriented toward the plasma membrane, and the trans face is mainly oriented toward the core of the cell, which is rich in vesicles. The space between the cis Golgi and plasma membrane is lined with the rER network. The very distal end of the ameloblast cell body, bordering the Tomes' process, is free of Golgi and rich in rER and secretory vesicles (33) (Fig. S4). Tomes' processes are free of rER and Golgi cisternae but are filled with small secretory granules and contain a tubulovesicular network (69, 70) (Figs. 2D and S4). The IG-TEM analysis revealed K75 signal in the membrane-delineated vesicles containing electron-dense material in the cell bodies and Tomes' processes (Figs. 2, B–D, S3, and S4). Two distinct groups of vesicles were identified based on their sizes. Large vesicles of various shapes (700 ± 556 nm) were observed primarily in the apical region of the cell body (Figs. 2, A and B, and S3). Smaller circular vesicles were observed throughout the ameloblast body (89 ± 20 nm) (Fig. 2, A and C, and S3) and in Tomes' process (102 ± 35 nm) (Fig. 2, A and D). The differences between the two groups were not statistically significant, suggesting that they belong to the same population. Our observations indicate that the small vesicles are covered with one single membrane bilayer, and no membrane-delineated compartments were found within the vesicles, suggesting that these are not multivesicular bodies or autophagosomes. K75 signal was also detected in the EEM surrounding the Tomes' processes (Figs. 2D and 3, G and J). Our IG-TEM studies failed to identify association of K75 with tonofilaments, whereas other keratins, e.g. K14, were clearly associated with them (Fig. S5A). Furthermore, we were not able to identify this protein in multivesicular bodies. Collectively, these results suggest that K75 is translocated into vesicles in the cell bodies of ameloblasts and is exocytosed via Tomes' processes. To the best of our knowledge, this is the first observation of a keratin, a cytosolic protein lacking a signal sequence, in secretory vesicles.
Figure 2.
Distribution of K75 in secretory ameloblasts by IG-TEM. A, schematic depiction of secretory ameloblasts. The double arrows represent the regions in which different populations of vesicles were detected. The numbers represent mean diameters of the vesicles for each population, and the regions of the cell in which these vesicles exist are indicated by double arrows. Note that the sizes of the small vesicles in the cell body and in the Tomes' processes are not statistically significantly different. For size measurements, five ameloblasts were selected, and 10–20 vesicles per group per ameloblast were measured. B, large membrane-delineated vesicle positive for K75. C, small-sized K75-positive vesicles in the cell body. D, small K75-positive vesicles in the Tomes' process. Arrowheads point toward the small vesicles containing K75. Note that one of the vesicles is undergoing exocytosis. The size of the gold particles is 6 nm. GA, Golgi apparatus; TP, Tomes' process; DEJ, dentinoenamel junction.
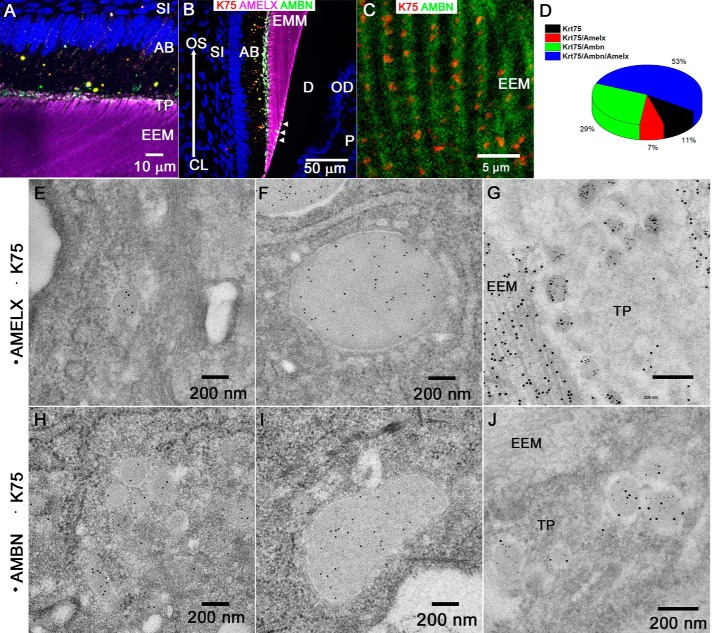
Figure 3.
A and B, triple IF labeling of K75, AMELX, and AMBN visualized by laser-scanning confocal microscopy of secretory ameloblasts and enamel in incisors (A) and molars (B) from 4-week-old rat. Note the bright overlap structures at the dentinoenamel junction resembling the enamel tufts (white arrowheads). The large arrow identifies the sequence of enamel deposition. C, high-magnification image showing colocalization of K75 and AMBN in secretory enamel matrix. Note that the K75 signal consists of arrays of compact clusters and overlaps with the AMBN signal. D, pie chart showing the extent of the overlap of K75 with AMELX and AMBN based on the quantitative analysis of the incisal secretory ameloblast bodies from three 4-week-old rats. E–G, colocalization of K75 (6 nm) with AMELX (12 nm) and AMBN (12 nm) in secretory ameloblasts of 4-week-old rats by IG-TEM. E and H, small vesicles in the cell bodies. F and I, large vesicles in the cell bodies. G and J, small vesicles in Tomes' processes. AB, ameloblast; CL, cervical loop; D, dentin; P, pulp; OD, odontoblast; N, nucleus; OS, occlusal surface; SI, stratum intermedium; TP, Tomes' process. Data from three animals were used for IF and IG-TEM colocalization analyses.
K75 colocalized with two major EMPs, AMBN and AMELX, in secretory ameloblasts
The discovery of K75 in membrane-delineated secretory vesicles prompted us to carry out colocalization studies of K75 with AMBN and AMELX. IF revealed significant levels of colocalization of K75 with the EMPs in cell bodies and Tomes' processes (Fig. 3, A and B) as well as discrete structures, resembling strings of beads colocalizing with AMBN in the enamel rod sheaths (Fig. 3C). Importantly, all three proteins are present in structures resembling enamel tufts at the dentinoenamel junction in forming molars (Fig. 3B, white arrowheads).
A quantitative IF colocalization analysis in the bodies of secretory ameloblasts revealed that close to ∼90% of K75 colocalizes with AMBN or AMELX, and ∼50% of K75 overlaps with both EMPs (Fig. 3D). In contrast, only about 30% of either AMBN or AMELX signal overlapped with K75 (Fig. S5, B, C, and F).
In addition to IF colocalization studies, a high-resolution IG-TEM analysis was also carried out. The results revealed that K75 colocalizes with AMBN and AMELX in small and large vesicles in the bodies of ameloblasts and in the Tomes' processes (Figs. 3, E–J, S3, and S4). The quantitative analysis of small secretory vesicles in the Tomes' processes revealed that the colocalization of K75 with AMBN as well as with AMELX is greater than 90%. Similarly, more than 90% of the vesicles containing either AMBN or AMELX contained K75 (Fig. S5, D, E, and G). Overall, the results of the IF and IG-TEM colocalization studies strongly indicate that K75, despite the lack of a signal sequence, enters the main secretory pathway and is cotransported with the EMPs.
K75 colocalizes with ERGIC and Golgi markers
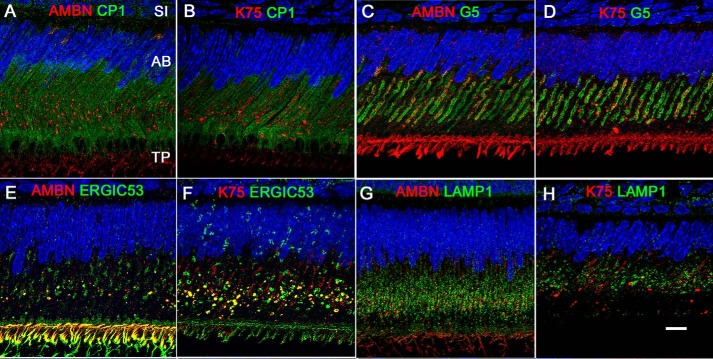
To investigate the trafficking route of K75 in secretory ameloblasts, IF colocalization of K75 and markers for rER (calreticulin peptide 1 (CP1)), ERGIC (ERGIC53; also known as LMAN1), Golgi (Golga 5 (G5)), and lysosomes (LAMP1) was studied. In parallel, colocalization experiments with the same markers were conducted with AMBN, which is secreted via the classical secretory mechanism (47). K75 and AMBN had low levels of colocalization with rER (Fig. 4, A and B). However, a significant degree of overlap for both AMBN and K75 was observed with ERGIC and Golgi (Figs. 4, E and F, and S6). In the ameloblasts cell bodies, ERGIC signal was present in large granules, which were often attached to the cis Golgi face (Figs. S3 and S6). Interestingly, a strong ERGIC signal overlapping with K75 and AMBN was present in the apical portions of ameloblasts and in the Tomes' processes (Fig. 4, E and F).
Figure 4.
IF colocalization of AMBN (A, C, E, and G) and K75 (B, D, F, and H) with different cellular organelle markers in the secretory ameloblasts from 4 weeks old mice. A and B, colocalization with rER marker CP1. Note that the colocalization for both proteins is very low. C and D, overlap with Golgi marker G5. The G5 signal appears as two well-organized long strips roughly parallel to each other, flanking the core of the ameloblast body. Both AMBN and K75 signals organized into bands adjacent to the two strips of Golgi complexes in the ameloblast core, partially overlapping with G5. Furthermore, large AMBN- and K75-positive granules were closely associated with the Golgi cisternae, potentially translocating their contents into Golgi for further processing. E and F, overlap with ERGIC marker ERGIC53. The ERGIC53 signal was associated with large granules in the ameloblast cell bodies where it colocalized with AMBN and K75. The ERGIC53 signal was also found in the distal portions of ameloblasts and Tomes' processes. Prominent overlap of K75 and AMBN with ERGIC53 was evident. G and H, overlap with lysosome marker LAMP1. LAMP1 signal was present throughout ameloblast cell bodies in the form of granules of different shapes and sizes. There was a very limited overlap of K75 with LAMP1, whereas AMBN was more prominently associated with LAMP1. Scale bar, 10 μm. AB, ameloblast; SI, stratum intermedium; TP, Tomes' process.
ER–Golgi trafficking inhibition using brefeldin A (BFA) does not affect colocalization of K75 with Golgi but significantly reduces its colocalization with ERGIC
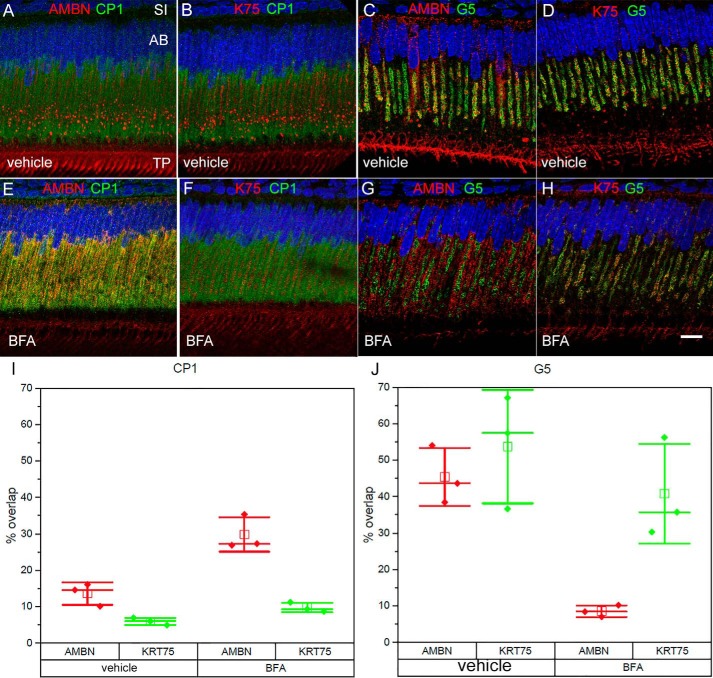
To further assess the details of the K75 secretory route, we conducted trafficking inhibition experiments using BFA. BFA, a potent inhibitor of rER–Golgi transport, is widely used in studies of protein trafficking (46, 71). Mice were injected with BFA dissolved in ethanol. No obvious changes were found between the 1-h ethanol vehicle group, used as controls, when compared with the nontreated specimens (Fig. 5, A–D, versus Fig. 4, A–D). A quantitative colocalization analysis of secretory ameloblasts in the vehicle group showed that 14 ± 2.5% of AMBN and 6 ± 1% of K75 colocalized with rER marker CP1 (Fig. 5, A, B, and I). Although no significant overlap between K75 and CP1 is expected because K75 lacks the signal peptide, low colocalization levels of AMBN and CP1 can be due to a very short residence time of AMBN molecules in rER as previously reported (46). In contrast, substantial levels of colocalization of K75 (54 ± 16%) and AMBN (45.3 ± 8%) with Golgi marker G5 were found (Fig. 5, C, D, and J). In the BFA-treated group, 1 h after injection, the colocalization pattern of AMBN changed dramatically. Specifically, AMBN signal associated with the central Golgi complex significantly decreased (8.5 ± 2%; p < 0.01), whereas its overlap with the rER significantly increased (30 ± 5%; p < 0.01) (Fig. 5, E, G, I, and J). These results demonstrate that the disruption of rER–Golgi transport by BFA leads to the accumulation of AMBN in rER and its depletion from Golgi, which is anticipated for an extracellular protein secreted via the classical rER–Golgi-dependent pathway (57, 72). The treatment with BFA, however, had a more limited effect on the localization of K75 in ER and Golgi. Colocalization of K75 and CP1 in the treatment group remained at low levels comparable with the control (10 ± 1%) (Fig. 5, F and I), whereas the quantitative analysis revealed no significant changes in colocalization of K75 and G5 between the control and treatment groups (41 ± 14 versus 54 ± 16%; p = 0.34) (Fig. 5, H and J). Colocalization studies using antibody against another Golgi marker, GM130, yielded similar results; namely the BFA treatment led to a dramatic decrease in overlap of AMBN and GM130, whereas the colocalization of K75 and GM130 remained essentially unchanged (Fig. S7).
Figure 5.
IF analysis of secretory ameloblasts from 4-week old mouse incisors after BFA treatment. A–D, vehicle controls 1 h after vehicle injection. E–H, corresponding colabeling images after 1-h BFA treatment. E, colocalization of AMBN with CP1. Considerable overlap between AMBN and CP1 can be seen, indicating that the classic secretory protein AMBN accumulated in ER after BFA treatment. F, colocalization of K75 with CP1 was not substantially affected by BFA, although K75-labeled large granular vesicles could no longer be detected. G, colocalization of AMBN with G5. After BFA treatment, overlap of AMBN with Golgi complex dramatically decreased, indicating that trafficking from ER to Golgi was blocked by BFA. H, colocalization of K75 with G5. The K75 signal was still associated with the central Golgi complex, indicating that K75 potentially utilized an unconventional pathway to translocate into Golgi apparatus. I and J, box graphs showing the overlap of AMBN and K75 with CP1 (I) and G5 (J) in the BFA treatment groups and vehicle controls. Filled diamonds represent the values of the overlap (%), open squares represent mean, central cross-bars represent median values, and end cross-bars represent S.D. Scale bar, 10 μm. AB, ameloblast; SI, stratum intermedium; TP, Tomes' process.
The distribution of ERGIC53 and its colocalization with K75 and AMBN in the vehicle controls were similar to untreated samples (Figs. 4, E and F, and 6, A and B). After 1-h treatment with BFA, large granules positive for ERGIC53 disappeared, and instead a diffuse ERGIC53 signal was detected throughout the ameloblast body (Figs. 4, E and F, and 6, A and B, versus Figs. 6, C and D, and S8). The treatment also led to a significant decrease of ERGIC53 signal in the Tomes' processes. These observations prompted us to conduct quantitative analysis of ERGIC53 colocalization with K75 and AMBN after BFA treatment. The quantitative colocalization analysis revealed significant reduction in K75 colocalization with ERGIC53 (29.5 ± 5.8 versus 9.9 ± 1.6%; p = 0.003) (Fig. 6E). At the same time, the colocalization of AMBN and ERGIC53 remained unchanged (21.8 ± 1.1% in vehicle control versus 26.5 ± 6.3% for BFA treatment; p = 0.5) (Fig. 6E). To rule out potential effects of the carrier (ethanol), we conducted similar experiments using PBS as the carrier, which lead to similar results but in a shorter time frame, possibly due to a slower degradation of BFA when using ethanol as a solvent (Fig. S9).
Figure 6.
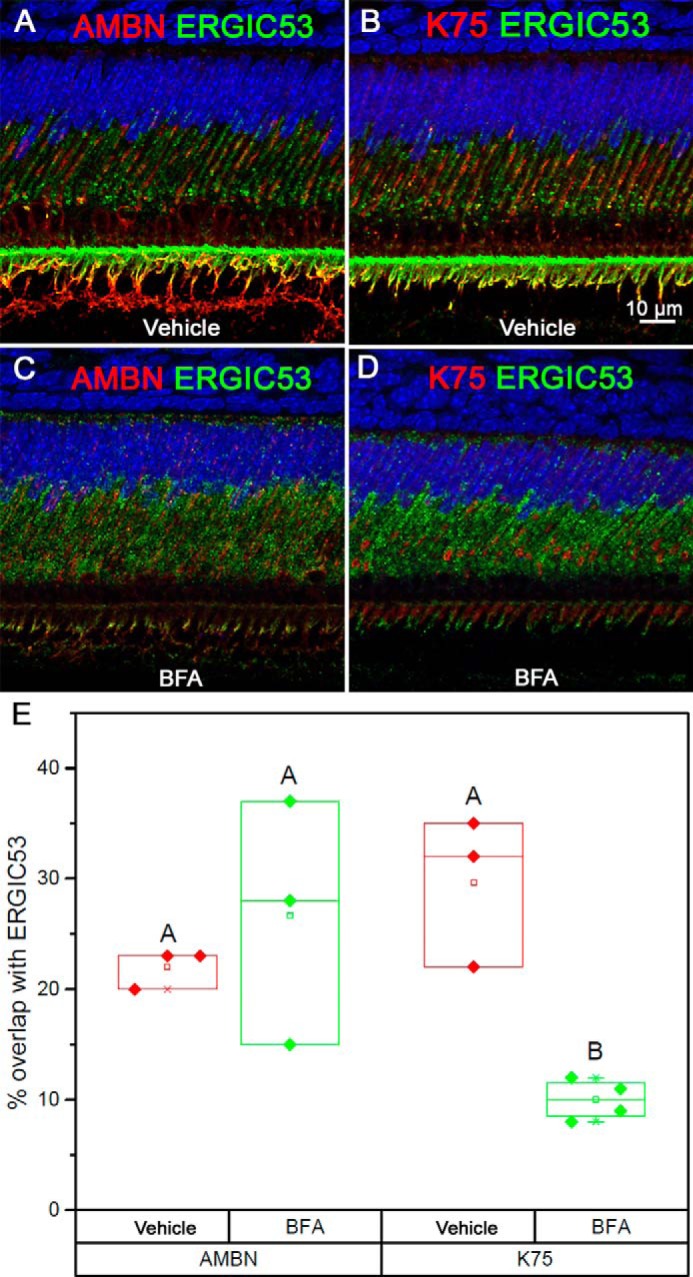
IF analysis of secretory ameloblasts from 4-week old mouse incisors after BFA treatment. A and B, vehicle controls 1 h after vehicle injection. C and D, BFA treatment 1 h after BFA injection. E, box graphs show the overlap of AMBN and K75 with ERGIC53 in the BFA treatment groups and vehicle controls. Filled diamonds represent the values of the overlap (%), open squares represent mean, central cross-bars represent median values, and end cross-bars represent S.D. Scale bar, 10 μm.
TEM analysis of the animals treated with BFA in ethanol revealed dilation of the rER, further confirming disruption of rER–Golgi trafficking (Fig. S10). IG-TEM of the control group detected AMBN in small vesicles, larger compartments of irregular shapes, potentially ERGIC, and Golgi apparatus, whereas no AMBN signal was detected in the rER (Fig. S10). After the treatment with BFA, we observed the accumulation of AMBN signal in rER; however, no signal was observed in the Golgi apparatus (Fig. S10), consistent with the results of IF studies. Overall, the results of BFA inhibition experiments indicate that although AMBN trafficking from rER to Golgi was blocked, as anticipated for a typical secreted protein, the BFA treatment did not affect association of K75 with Golgi.
Discussion
The presence of heavily cross-linked keratin matrix in enamel was first proposed in the late 19th century, and a number of more recent papers suggested the presence of keratins in enamel (52). However, unequivocal evidence demonstrating the presence of keratin in enamel was lacking. Our recent studies, for the first time, demonstrate the presence of several keratins, including K75, in enamel matrix and that mutations in these genes cause structural and functional defects of enamel (10–12, 73). The present results further corroborate our earlier observations of K75 in enamel (10, 12) and revealed that K75 expression is associated with secretory-stage ameloblasts.
One of the arguments against the presence of keratins in enamel matrix was the fact that these are cytosolic proteins that lack a signal sequence and therefore cannot be secreted. There are several potential scenarios of translocation of cytosolic proteins into the extracellular space: they can be dislodged into the matrix as a part of cellular debris (55), or they can be secreted via the canonical ER–Golgi pathway (64, 65) or by a number unconventional protein secretion mechanisms (56). Our colocalization studies demonstrate that secretion of K75, at least in part, utilizes the canonical secretory apparatus. Specifically, K75 is present in secretory vesicles together with two major EMPs, AMELX and AMBN, which possess a signal sequence and are secreted via the classical secretory pathway (46, 47). K75 was also found in ERGIC and Golgi apparatus, cellular compartments associated with the classical secretory pathway. To the best of our knowledge, there are only a few other examples of cytosolic proteins lacking a signal peptide that utilize the conventional secretory pathway (64, 65, 74, 75). Although there were a couple of reports showing keratins extracellularly (53, 54), the mechanisms by which these keratin proteins enter the extracellular space were not previously studied. Our study is the first to demonstrate that K75 is cosecreted with AMBN and AMELX and that it utilizes, at least partially, the canonical ER–Golgi secretory pathway.
The fact that K75 colocalizes with AMBN and AMELX in secretory vesicles and the enamel matrix opens the possibility of interactions between K75 and EMPs. Ravindranath et al. (48, 49) reported interactions between AMELX and K5 and K14. Further studies are needed to clarify this question. In another study using a yeast-two hybrid system, it was found that tuftelin, another protein expressed by ameloblasts, interacts with K6 and K5 (76).
ER–Golgi trafficking inhibition experiments using BFA provide further insights into the secretory pathway of K75 in ameloblasts. Although BFA treatment, which disrupts rER–Golgi transport, led to the accumulation of AMBN in dilated rER and its disappearance from Golgi as described previously (46), BFA treatment did not significantly affect the colocalization of K75 with Golgi. At the same time, even though ERGIC53 signal distribution in the cells changed, the degree of AMBN colocalization with ERGIC53 remained unchanged, whereas colocalization of K75 and ERGIC53 decreased significantly. Hence, although K75 utilizes portions of the canonical ER–Golgi secretory pathway, it behaves differently than typical secretory proteins containing a signal sequence. According to our model, K75 enters the classical secretory pathway not via rER, as do most secreted proteins, but is translocated via ERGIC. Once in the ERGIC it is cotransported with other secretory cargo through the Golgi apparatus and secreted from the Tomes' processes. At the same time, the fact that the BFA treatment, which leads to breakdown of ERGIC, does not significantly affect K75 colocalization with Golgi apparatus suggests that either BFA treatment prevents K75 from clearing Golgi or there is another ERGIC-independent mechanism of K75 translocation into the Golgi. To the best of our knowledge, this is the first demonstration of a trafficking route for a cytosolic protein that utilizes portions of the conventional rER–Golgi secretory pathway, and it is very different from other unconventional pathways utilized by cytosolic proteins (57, 58).
Our observation of ERGIC53 in association with EMPs, in a distal rER-rich region of ameloblast and in the Tomes' processes, the ameloblast secretory apparatus, raises the possibility that some elements of the ERGIC system can be involved in a secretory pathway that bypasses Golgi. Earlier studies of ameloblasts revealed an extensive tubulovesicular system in the distal portions of ameloblasts and the Tomes' processes (69, 70) that is potentially a part of ERGIC. One possible scenario is that some of these ERGIC structures are produced in the distal rER-rich zone (Figs. S4 and S6) and transported into the Tomes' process directly. There are several examples of Golgi bypass (56, 57, 59); however, it is possible that, in ameloblasts, secreted proteins can simultaneously be transported via the conventional rER–Golgi pathway and through the Golgi bypass mechanism (59) involving ERGIC.
It is important to emphasize that ameloblasts are highly specialized supersecretory cells, and their secretory pathways are best studied in vivo. The fact that in tissue cultures ameloblast-derived cells never attain their secretory phenotype presents a major challenge for studies of ameloblast cell biology. This constraint potentially applies to other cell types. The vast majority of cell trafficking and secretion studies are conducted in tissue culture systems, and often the data obtained in vitro are not in a good agreement with in vivo observations (57), potentially because cells in vitro are not able to differentiate properly. We anticipate that the mechanisms of secretion revealed in this study are not unique to ameloblasts but are much more universal, and other cell types with high secretory activity might utilize them as well.
Conclusions
Overall, these results further support our previous observations of K75 in ameloblasts and enamel and demonstrate that it is expressed during the secretory stage of amelogenesis. Furthermore, our data demonstrate that the majority of K75 is cotransported together with EMPs and is secreted from the Tomes' processes. Using cell compartment markers, we detected K75 in ERGIC and Golgi apparatus. We further conducted experiments with the ER–Golgi trafficking inhibitor BFA, which revealed that although the treatment disrupted colocalization of K75 with ERGIC, its association with Golgi remained unchanged. Collectively, these results demonstrate that K75, a cytosolic protein lacking a signal sequence, utilizes portions of the classical secretory pathway. Our results also suggest that some of the secreted proteins might utilize the Golgi bypass mechanism via ERGIC-associated vesicles. Together, these results provide novel insights into the K75 secretion processes and cellular trafficking mechanisms in ameloblasts and potentially other systems.
Experimental procedures
All methods were performed in accordance with the relevant guidelines and regulations. Four-week-old Sprague-Dawley rats (Charles River Laboratories, Wilmington, MA) and 4-week-old mice (C57BL/6J, The Jackson Laboratory, Bar Harbor, ME) were used in the study. All animal procedures described were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Tissue preparation for ISH and IF and IG TEM studies
Four-week-old Sprague-Dawley rats (Charles River Laboratories) and 4-week old mice (C57BL/6J, The Jackson Laboratory) were euthanized with CO2. Mandibles were dissected and immediately submersed in 50-ml volumes of 4% paraformaldehyde in 10 mm PBS for IF or TEM or Karnovsky fixative (2% glutaraldehyde, 2% formaldehyde in 10 mm PBS) for TEM at 4 °C in less than 3 min after euthanasia. After 24–48 h of fixation at 4 °C, samples were placed into demineralization solution containing 0.1 m EDTA (pH 7.2–7.4) for 1–2 weeks. The demineralization solution was changed every other day.
For IG-TEM experiments, animals were anesthetized with isoflurane and perfused through the left ventricle first with cold PBS for 30 s and then with cold 4% paraformaldehyde in 0.1 m phosphate buffer or cold 1% glutaraldehyde in 0.1 m phosphate buffer for 15–20 min. The mandibles were dissected and further fixed in the same fixative solution for another 8–12 h at 4 °C followed by demineralization as described above. For IF, whole demineralized jaws were dehydrated using a Leica ASP 300S automatic processer (Leica Biosystems, Buffalo Grove, IL) and paraffin-embedded using a standard protocol (77).
For TEM studies, after demineralization mandibular bone around the incisors was trimmed, and the molars were removed. The distal fragments of the jaws containing apical portions of the incisors were further cross-sectioned into 1–1.5-mm-thick pieces, processed, and embedded in LR White or Embed 812 (catalog numbers 14381 and 14120, Electron Microscopy Sciences, Hatfield, PA) according to published protocols (47). In brief, for Embed 812 processing, incisor pieces were postfixed in 1% ferrocyanide reduced osmium tetroxide for 1 h, washed in PBS, dehydrated in graded ethanol, and infiltrated with propylene oxide. The samples were embedded in Embed 812 and cured at 65 °C for 2 days. For LR White processing, some of the samples were postfixed in osmium tetroxide, whereas others were not as osmication reduces the immunoreactivity of the samples. Incisor pieces were washed in PBS, dehydrated in graded ethanol, embedded in LR White, and cured at 60 °C for 1–2 days.
For ISH, mice at postnatal day 1 were euthanized with CO2, and samples were fixed with 4% paraformaldehyde in 10 mm PBS for 24 h and embedded in paraffin as described above. For IF and ISH, the paraffin blocks were sectioned using a Leica RM 2225 microtome (Leica Biosystems, Buffalo, IL) into 10-μm-thick sections using a stainless steel microtome knife (Leica 818, Leica, Germany). The sections were mounted on 3-triethoxysilylpropylamine (440140, Millipore-Sigma)–coated glass slides. For TEM, the resin blocks were sectioned into 70-nm-thick sections using a Leica EM UC7 ultramicrotome (Leica Biosystems) equipped with a diamond knife (Electron Microscopy Sciences). The sections were mounted on carbon coated Ni grids (Electron Microscopy Sciences).
IF studies
An optimized IF procedure (77, 78) developed in our laboratory, aimed at reducing false positives, was used. Briefly, after deparaffinization, sections underwent trypsin treatment for 5–30 min at 37 °C or heat treatment in 10 mm sodium citrate buffer (pH 6.0) for 10–20 min for antigen retrieval. The sections were blocked in 10% serum from the secondary Ab host animal in Tris-buffered saline (TBS) for 1 h at room temperature and incubated with primary Abs at 4 °C overnight followed by incubation with secondary Abs for 45 min at room temperature. The sections were washed with TBS, incubated with 1.5% Sudan Black B (199664, Sigma) in 70% ethanol to minimize autofluorescence, and counterstained with 4′,6-diamidino-2-phenylindole (DAPI).
Primary Abs used include rabbit anti-AMELX at 1:100 (ABT260, Abcam, Cambridge, MA), goat anti-AMBN at 1:100 (sc-33102, Santa Cruz Biotechnology, Dallas, TX), rabbit anti-AMBN at 1:100 (sc-50534, Santa Cruz Biotechnology), rabbit anti-G5 at 1:100 (NBP1-83352, Novus Biologicals, Centennial, CO), rabbit anti-GM130 at 1:100 (Ab52649, Abcam), rabbit anti-ERGIC53 at 1:100 (sc-66880, Santa Cruz Biotechnology), monoclonal rabbit anti-CP1 at 4 μg/ml (CPTC-CALR-1-s, Developmental Studies Hybridoma Bank, Iowa City, IA), rat anti-LAMP1 at 4 μg/ml (1D4B-s, Developmental Studies Hybridoma Bank), rabbit anti-K5 at 1:2000 (905501, BioLegend, San Diego, CA), rabbit anti-K14 at 1:1000 (905301, BioLegend); and guinea pig anti-K75, an antibody widely used in IF studies (6, 10, 79), at 1:100 (20R-2647, Fitzgerald Industries International, Acton, MA). Guinea pig serum (88R-1015, Fitzgerald Industries International), other normal sera, or IgGs purchased from Santa Cruz Biotechnology or Jackson ImmunoResearch Laboratories were used as isotype controls. Secondary Abs used included Alexa Flour 555/Cy3/Cy5–conjugated donkey anti-primary Ab host IgG (heavy + light), diluted 1:500, purchased from Jackson ImmunoResearch Laboratories. Mounted slides were analyzed using a Nikon A1 confocal microscope system (Nikon Instruments Inc., Melville, NY) in the Center for Biologic Imaging at the University of Pittsburgh. Colocalization analysis was performed using NIS-Elements software (Nikon Instruments Inc.) provided with the microscope. K75, AMBN, and AMELX colocalization analysis was performed on the samples collected from three rats per group. Statistical analysis of the colocalization data was performed using Microsoft Excel and Origin software packages (OriginLab Corp., Northampton, MA).
In situ hybridization
ISH was performed following a published protocol (80, 81). Briefly, the sections of P1 mice were deparaffinized, rehydrated, postfixed, and subjected to prehybridization on the 1st day. Digoxigenin-labeled RNA probes to the sense and antisense strands of mouse Krt75 partial cDNA (421 bp) corresponding to exon 9, including the C-terminal tail domain and part of the 3′ noncoding region (1494–1996 bp of NCBI Reference Sequence NM_133357.3), were hybridized to the sections. The sections were blocked in 10% sheep serum for 2 h, incubated with 1:2000 anti-digoxigenin alkaline phosphatase Ab in 1% sheep serum mixture overnight at 4 °C on the 2nd day, and washed and incubated in developing solution (1681451, Roche Applied Science) on the 3rd day. The reaction was stopped by washing in PBS. The slides were mounted in toluene (4112, Thermo Fisher) and observed under a light microscope in bright-field mode.
Western blotting
The enamel matrix was collected from apical portions of continually growing incisors and unerupted first molars from 14-day-old Wistar rats. For incisal enamel matrix preparation, mandibles of 14-day-old Wistar rats were dissected, and the basal bone was removed to expose unerupted portions of incisors under a dissecting microscope. The cellular components of enamel organ, including the ameloblast layer, were carefully removed, and the forming enamel tissue was scraped from the dentin surface. The matrix was transferred to TBS (Thermo Scientific) with proteinase inhibitor mixture (Thermo Scientific) at a 1:100 ratio. Molar enamel preparation was carried out according to published procedure(82). Maxillary and mandibular unerupted first molars were extracted, and pulp tissue was removed under a dissecting microscope. The crowns were cleaned of surrounding tissues, and enamel matrix proteins were extracted using a published method (82) with slight modifications. Briefly, the dissected molars were incubated with 0.17 m HCl, 0.98% formic acid for 2 h at 4 °C, and the supernatant was concentrated using 3000-molecular-weight-cutoff centrifugal filtering unit (Millipore). Whiskers were processed according to a published procedure (83). Briefly, whiskers were plucked from the rat snout, and the hair follicles were collected in 200 μl of protein extraction buffer with proteinase inhibitor mixture. The hair follicles were vortexed and incubated at 37 °C overnight. For WB, the samples were centrifuged, and the supernatant was collected for analysis. The concentration of the proteins in the samples was detected using a BCA assay. Ten micrograms of proteins were loaded in a Bis-Tris 4–12% gel (Life Technologies) and subjected to SDS-PAGE. The protein bands were transferred from the gel to a polyvinylidene difluoride membrane (Life Technologies), and bands containing proteins of interest were detected using ECL substrate (catalog number 32106, Thermo Scientific). Primary Abs used were anti-K75 guinea pig IgG antibodies at 1:1000 (20R-2647, Fitzgerald Industries International), rabbit anti-AMELX at 1:1000 (ABT260, Abcam), and rabbit anti-AMBN at 1:1000 (sc-50534, Santa Cruz Biotechnology). Secondary Abs used were donkey anti-guinea pig IgG conjugated with HRP at 1:2000 (706-035-148, Jackson ImmunoResearch Laboratories) and goat anti-rabbit IgG conjugated with HRP at 1:2000 (catalog number 32460, Thermo Scientific).
Mass spectrometry
For MS, the bands of interest were dissected from the gels and submitted to the University of Pittsburgh Biomedical Mass Spectrometry Center for analysis.
IG-TEM studies
The IG-TEM studies were performed on 70-nm-thick sections mounted on carbon-coated Ni grids. Immunolabeling was performed according to published procedures (46, 47). Briefly, grids were blocked in the same blocking solution as in the IF protocol described above for 1 h, incubated with primary antibody for 1 h, washed with TBS, then incubated with secondary antibody for 1 h, washed, and counterstained with 2% uranyl acetate for 10 min. Primary Ab concentrations 10 times higher than in the IF assays described above were used. Six-nanometer colloidal gold–conjugated donkey anti-guinea pig IgG at 1:30 (706-195-148, Jackson ImmunoResearch Laboratories) and 12-nm colloidal gold–conjugated donkey anti-rabbit IgG at 1:30 (711-205152, Jackson ImmunoResearch Laboratories) were used as secondary Abs. Guinea pig serum (88R-1015, Fitzgerald Industries International) and rabbit serum (011-000-001, Jackson ImmunoResearch Laboratories) were used as isotype controls. All grids were observed with JEM 1400Plus TEM (JEOL USA Inc.) at 80 kV, located in the Center for Biologic Imaging at University of Pittsburgh. To assess the distribution of K75 in vesicles of different sizes in the ameloblast cell bodies and the Tomes' processes, sections from three rats were labeled with anti-K75 Abs and analyzed. The vesicles containing at least one gold particle were counted and measured, and at least 50 vesicles per animal were analyzed. For IG-TEM colocalization of K75 with EMP in Tomes' processes, LR White sections from three rats were double-labeled for K75 and either AMBN or AMELX. Vesicles with at least one gold particle were counted, and more than 400 vesicles were analyzed.
Brefeldin A experiments
BFA powder (B-8500, LC Laboratories, Woburn, MA) dissolved in DMSO at 25 mg/ml concentration was used as a stock solution. BFA stock solution was further dissolved in 30% ethanol or PBS to a final concentration of 0.5 mg/ml prior to the experiments. Four-week-old mice were used in this study; all animals were anesthetized by isoflurane. Three mice received intraperitoneal injection of 0.5 mg of BFA in ethanol, and three mice injected with ethanol only were used as vehicle control. The mice were euthanized 1 h after the injection. In another set of experiments, three mice were injected with 0.5 mg of BFA in PBS. These animals were euthanized 15, 30, and 60 min after the injection. One mouse was injected with the vehicle (control) and euthanized 60 min after the injection. The animals were euthanized in CO2 1 h after the injection, and samples were fixed in 4% paraformaldehyde according to the IF or TEM procedures as described above. An independent two-sample t test was used to analyze the colocalization differences between the vehicle control and BFA/ethanol-treated mice (n = 3).
Quantitative IF colocalization analysis
Quantitative colocalization analysis was performed using a well-established methodology (84, 85). The colocalization analysis was conducted using proprietary NIS-Elements software. The quantitative colocalization analysis was conducted on sections from three animals. Colocalization was calculated based on the number of pixels in confocal slices from a z-stack containing two channels. The colocalization was assessed according to established procedures (84–86) using Pearson correlation analysis and Manders' coefficients, which represent a ratio of overlapping pixels to the total number of pixels in each channel. For background adjustment, adjacent sections were treated with naïve sera (isotype control), and signal intensity from these sections was used to determine the background fluorescence. The colocalization analysis was conducted using proprietary NIS-Elements software. The quantitative colocalization analysis was conducted on sections from three animals. The data were analyzed using a t test assuming unequal variance in the OriginPro 2015 software package (OriginLab Corp.).
Author contributions
X. Y., O. D., M. I. M., and E. B. conceptualization; X. Y. and H. Y. formal analysis; X. Y. and E. B. supervision; X. Y., H. Y., Y. Y., and E. B. investigation; X. Y. and H. Y. visualization; X. Y., H. Y., and Y. Y. methodology; X. Y., H. Y., O. D., M. I. M., and E. B. writing-original draft; X. Y. and E. B. project administration; X. Y., H. Y., and E. B. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Donna Beer Stolz, Mara Sullivan, Ming Sun, Jonathan Franks, Morgan Jessup, and Callen Wallace at the Center for Biological Imaging at the University of Pittsburgh for expert support of our imaging and TEM studies. We also acknowledge Heather Szabo-Rogers and Yong Wan (Department of Oral Biology, University of Pittsburgh) for help with ISH and Miu Okubo and Risako Chiba (School of Dental Medicine, Tsurumi University, Japan) and Sana Khalid (Department of Oral Biology, University of Pittsburgh) for help with WB. We are thankful to Dr. Meir Aridor (Department of Cell Biology, University of Pittsburgh) for advice and helpful discussions.
This work was supported by the University of Pittsburgh and by NIAMS, National Institutes of Health, Intramural Research Program Grants AR041171 and AR041124 (to M. I. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S10.
- K
- keratin
- ER
- endoplasmic reticulum
- ERGIC
- ER–Golgi intermediate compartment
- rER
- rough ER
- EMP
- enamel matrix protein
- EEM
- extracellular enamel matrix
- AMELX
- amelogenin
- AMBN
- ameloblastin
- ENAM
- enamelin
- MMP
- matrix metalloproteinase
- IF
- immunofluorescence
- ISH
- in situ hybridization
- WB
- Western blotting
- IG-TEM
- immunogold transmission EM
- G5
- Golga 5
- BFA
- brefeldin A
- Ab
- antibody
- TBS
- Tris-buffered saline
- CP1
- calreticulin peptide 1
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- HRP
- horseradish peroxidase
- P1
- postnatal day 1.
References
- 1. Vandebergh W., and Bossuyt F. (2012) Radiation and functional diversification of α keratins during early vertebrate evolution. Mol. Biol. Evol. 29, 995–1004 10.1093/molbev/msr269 [DOI] [PubMed] [Google Scholar]
- 2. Moll R., Divo M., and Langbein L. (2008) The human keratins: biology and pathology. Histochem. Cell Biol. 129, 705–733 10.1007/s00418-008-0435-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fuchs E., and Cleveland D. W. (1998) A structural scaffolding of intermediate filaments in health and disease. Science 279, 514–519 10.1126/science.279.5350.514 [DOI] [PubMed] [Google Scholar]
- 4. Herrmann H., and Aebi U. (2004) Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular scaffolds. Annu. Rev. Biochem. 73, 749–789 10.1146/annurev.biochem.73.011303.073823 [DOI] [PubMed] [Google Scholar]
- 5. Hobbs R. P., Jacob J. T., and Coulombe P. A. (2016) Keratins are going nuclear. Dev. Cell 38, 227–233 10.1016/j.devcel.2016.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Winter H., Langbein L., Praetzel S., Jacobs M., Rogers M. A., Leigh I. M., Tidman N., and Schweizer J. (1998) A novel human type II cytokeratin, K6hf, specifically expressed in the companion layer of the hair follicle. J. Invest. Dermatol. 111, 955–962 10.1046/j.1523-1747.1998.00456.x [DOI] [PubMed] [Google Scholar]
- 7. Wang Z., Wong P., Langbein L., Schweizer J., and Coulombe P. A. (2003) Type II epithelial keratin 6hf (K6hf) is expressed in the companion layer, matrix, and medulla in anagen-stage hair follicles. J. Invest. Dermatol. 121, 1276–1282 10.1111/j.1523-1747.2003.12644.x [DOI] [PubMed] [Google Scholar]
- 8. Lin S. J., Wideliz R. B., Yue Z., Li A., Wu X., Jiang T. X., Wu P., and Chuong C. M. (2013) Feather regeneration as a model for organogenesis. Dev. Growth Differ. 55, 139–148 10.1111/dgd.12024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Winter H., Schissel D., Parry D. A., Smith T. A., Liovic M., Lane E. B., Edler L., Langbein L., Jave-Suarez L. F., Rogers M. A., Wilde J., Peters G., and Schweizer J. (2004) An unusual Ala12Thr polymorphism in the 1A α-helical segment of the companion layer-specific keratin K6hf: evidence for a risk factor in the etiology of the common hair disorder pseudofolliculitis barbae. J. Invest. Dermatol. 122, 652–657 10.1111/j.0022-202X.2004.22309.x [DOI] [PubMed] [Google Scholar]
- 10. Duverger O., Ohara T., Shaffer J. R., Donahue D., Zerfas P., Dullnig A., Crecelius C., Beniash E., Marazita M. L., and Morasso M. I. (2014) Hair keratin mutations in tooth enamel increase dental decay risk. J. Clin. Investig. 124, 5219–5224 10.1172/JCI78272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duverger O., Carlson J. C., Karacz C. M., Schwartz M. E., Cross M. A., Marazita M. L., Shaffer J. R., and Morasso M. I. (2018) Genetic variants in pachyonychia congenita-associated keratins increase susceptibility to tooth decay. PLoS Genet. 14, e1007168 10.1371/journal.pgen.1007168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chiba R., Okubo M., Yamamoto R., Saito M. M., Kobayashi S., Beniash E., and Yamakoshi Y. (2019) Porcine keratin 75 in developing enamel. J. Oral Biosci. 61, 163–172 10.1016/j.job.2019.06.002 [DOI] [PubMed] [Google Scholar]
- 13. Biggs L. C., and Mikkola M. L. (2014) Early inductive events in ectodermal appendage morphogenesis. Semin. Cell Dev. Biol. 25, 11–21 10.1016/j.semcdb.2014.01.007 [DOI] [PubMed] [Google Scholar]
- 14. Sharpe P. T. (2001) Fish scale development: hair today, teeth and scales yesterday? Curr. Biol. 11, R751–R752 10.1016/S0960-9822(01)00438-9 [DOI] [PubMed] [Google Scholar]
- 15. Price J. A., Bowden D. W., Wright J. T., Pettenati M. J., and Hart T. C. (1998) Identification of a mutation in DLX3 associated with tricho-dento-osseous (TDO) syndrome. Hum. Mol. Genet. 7, 563–569 10.1093/hmg/7.3.563 [DOI] [PubMed] [Google Scholar]
- 16. Duverger O., Lee D., Hassan M. Q., Chen S. X., Jaisser F., Lian J. B., and Morasso M. I. (2008) Molecular consequences of a frameshifted DLX3 mutant leading to tricho-dento-osseous syndrome. J. Biol. Chem. 283, 20198–20208 10.1074/jbc.M709562200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duverger O., Zah A., Isaac J., Sun H. W., Bartels A. K., Lian J. B., Berdal A., Hwang J., and Morasso M. I. (2012) Neural crest deletion of Dlx3 leads to major dentin defects through down-regulation of Dspp. J. Biol. Chem. 287, 12230–12240 10.1074/jbc.M111.326900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y., Han D., Zhang H., Liu H., Wong S., Zhao N., Qiu L., and Feng H. (2015) Morphological analyses and a novel de novo DLX3 mutation associated with tricho-dento-osseous syndrome in a Chinese family. Eur. J. Oral Sci. 123, 228–234 10.1111/eos.12197 [DOI] [PubMed] [Google Scholar]
- 19. Nanci A. (2007) Ten Cate's Oral Histology: Development, Structure, and Function, Elsevier Health Sciences, New York [Google Scholar]
- 20. Stifler C. A., Wittig N. K., Sassi M., Sun C.-Y., Marcus M. A., Birkedal H., Beniash E., Rosso K. M., and Gilbert P. U. P. A. (2018) X-ray linear dichroism in apatite. J. Am. Chem. Soc. 140, 11698–11704 10.1021/jacs.8b05547 [DOI] [PubMed] [Google Scholar]
- 21. Glimcher M. J., Daniel E. J., Travis D. F., and Kamhi S. (1965) Electron optical and X-ray diffraction studies of the organization of the inorganic crystals in embryonic bovine enamel. J. Ultrastruct. Res. 50, Suppl. 7, 1–77 [PubMed] [Google Scholar]
- 22. Bajaj D., and Arola D. D. (2009) On the R-curve behavior of human tooth enamel. Biomaterials 30, 4037–4046 10.1016/j.biomaterials.2009.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baldassarri M., Margolis H. C., and Beniash E. (2008) Compositional determinants of mechanical properties of enamel. J. Dent. Res. 87, 645–649 10.1177/154405910808700711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chai H., Lee J. J., Constantino P. J., Lucas P. W., and Lawn B. R. (2009) Remarkable resilience of teeth. Proc. Natl. Acad. Sci. U.S.A. 106, 7289–7293 10.1073/pnas.0902466106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Macho G. A., Jiang Y., and Spears I. R. (2003) Enamel microstructure—a truly three-dimensional structure. J. Hum. Evol. 45, 81–90 10.1016/S0047-2484(03)00083-6 [DOI] [PubMed] [Google Scholar]
- 26. Hu Y., Smith C. E., Cai Z., Donnelly L. A., Yang J., Hu J. C., and Simmer J. P. (2016) Enamel ribbons, surface nodules, and octacalcium phosphate in C57BL/6 Amelx−/− mice and Amelx+/− lyonization. Mol. Genet. Genomic Med. 4, 641–661 10.1002/mgg3.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Daculsi G., and Kerebel B. (1978) High-resolution electron-microscope study of human enamel crystallites—size, shape, and growth. J. Ultrastruct. Res. 65, 163–172 10.1016/S0022-5320(78)90053-9 [DOI] [PubMed] [Google Scholar]
- 28. Travis D. F., and Glimcher M. J. (1964) The structure and organization of, and the relationship between the organic matrix and the inorganic crystals of embryonic bovine enamel. J. Cell Biol. 23, 447–497 10.1083/jcb.23.3.447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fincham A. G., Moradian-Oldak J., Diekwisch T. G., Lyaruu D. M., Wright J. T., Bringas P. Jr., and Slavkin H. C. (1995) Evidence for amelogenin “nanospheres” as functional components of secretory-stage enamel matrix. J. Struct. Biol. 115, 50–59 10.1006/jsbi.1995.1029 [DOI] [PubMed] [Google Scholar]
- 30. Lacruz R. S., Habelitz S., Wright J. T., and Paine M. L. (2017) Dental enamel formation and implications for oral health and disease. Physiol. Rev. 97, 939–993 10.1152/physrev.00030.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garant P. R., and Nalbandian J. (1968) Observations on the ultrastructure of ameloblasts with special reference to the Golgi complex and related components. J. Ultrastruct. Res. 23, 427–443 10.1016/S0022-5320(68)80108-X [DOI] [PubMed] [Google Scholar]
- 32. Katchburian E., and Holt S. J. (1972) Studies on the development of ameloblasts. J. Cell Sci. 11, 415–447 [DOI] [PubMed] [Google Scholar]
- 33. Warshawsky H. (1968) The fine structure of secretory ameloblasts in rat incisors. Anat. Rec. 161, 211–229 10.1002/ar.1091610207 [DOI] [PubMed] [Google Scholar]
- 34. Snead M. L., Lau E. C., Zeichner-David M., Fincham A. G., Woo S. L., and Slavkin H. C. (1985) DNA sequence for cloned cDNA for murine amelogenin reveal the amino acid sequence for enamel-specific protein. Biochem. Biophys. Res. Commun. 129, 812–818 10.1016/0006-291X(85)91964-3 [DOI] [PubMed] [Google Scholar]
- 35. Krebsbach P. H., Lee S. K., Matsuki Y., Kozak C. A., Yamada K. M., and Yamada Y. (1996) Full-length sequence, localization, and chromosomal mapping of ameloblastin—a novel tooth-specific gene. J. Biol. Chem. 271, 4431–4435 10.1074/jbc.271.8.4431 [DOI] [PubMed] [Google Scholar]
- 36. Hu C.-C., Fukae M., Uchida T., Qian Q., Zhang C. H., Ryu O. H., Tanabe T., Yamakoshi Y., Murakami C., Dohi N., Shimizu M., and Simmer J. P. (1997) Cloning and characterization of porcine enamelin mRNAs. J. Dent. Res. 76, 1720–1729 10.1177/00220345970760110201 [DOI] [PubMed] [Google Scholar]
- 37. Bartlett J. D., Simmer J. P., Xue J., Margolis H. C., and Moreno E. C. (1996) Molecular cloning and mRNA tissue distribution of a novel matrix metalloproteinase isolated from porcine enamel organ. Gene 183, 123–128 10.1016/S0378-1119(96)00525-2 [DOI] [PubMed] [Google Scholar]
- 38. Beniash E., Simmer J. P., and Margolis H. C. (2005) The effect of recombinant mouse amelogenins on the formation and organization of hydroxyapatite crystals in vitro. J. Struct. Biol. 149, 182–190 10.1016/j.jsb.2004.11.001 [DOI] [PubMed] [Google Scholar]
- 39. Fang P. A., Conway J. F., Margolis H. C., Simmer J. P., and Beniash E. (2011) Hierarchical self-assembly of amelogenin and the regulation of biomineralization at the nanoscale. Proc. Natl. Acad. Sci. U.S.A. 108, 14097–14102 10.1073/pnas.1106228108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Margolis H. C., Beniash E., and Fowler C. E. (2006) Role of macromolecular assembly of enamel matrix proteins in enamel formation. J. Dent. Res. 85, 775–793 10.1177/154405910608500902 [DOI] [PubMed] [Google Scholar]
- 41. Yamazaki H., Tran B., Beniash E., Kwak S. Y., and Margolis H. C. (2019) Proteolysis by MMP20 prevents aberrant mineralization in secretory enamel. J. Dent. Res. 98, 468–475 10.1177/0022034518823537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fincham A. G., Moradian-Oldak J., and Simmer J. P. (1999) The structural biology of the developing dental enamel matrix. J. Struct. Biol. 126, 270–299 10.1006/jsbi.1999.4130 [DOI] [PubMed] [Google Scholar]
- 43. Simmer J. P., and Hu J. C. (2002) Expression, structure, and function of enamel proteinases. Connect. Tissue Res. 43, 441–449 10.1080/03008200290001159 [DOI] [PubMed] [Google Scholar]
- 44. Paine M. L., Zhu D.-H., Luo W., Bringas P. Jr., Goldberg M., White S. N., Lei Y.-P., Sarikaya M., Fong H. K., and Snead M. L. (2000) Enamel biomineralization defects result from alterations to amelogenin self-assembly. J. Struct. Biol. 132, 191–200 10.1006/jsbi.2000.4324 [DOI] [PubMed] [Google Scholar]
- 45. Tao J., Fijneman A., Wan J., Prajapati S., Mukherjee K., Fernandez-Martinez A., Moradian-Oldak J., and De Yoreo J. J. (2018) Control of calcium phosphate nucleation and transformation through interactions of enamelin and amelogenin exhibits the “Goldilocks effect”. Cryst. Growth Design 18, 7391–7400 10.1021/acs.cgd.8b01066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nanci A., Zalzal S., Lavoie P., Kunikata M., Chen W., Krebsbach P. H., Yamada Y., Hammarström L., Simmer J. P., Fincham A. G., Snead M. L., and Smith C. E. (1998) Comparative immunochemical analyses of the developmental expression and distribution of ameloblastin and amelogenin in rat incisors. J. Histochem. Cytochem. 46, 911–934 10.1177/002215549804600806 [DOI] [PubMed] [Google Scholar]
- 47. Zalzal S. F., Smith C. E., and Nanci A. (2008) Ameloblastin and amelogenin share a common secretory pathway and are co-secreted during enamel formation. Matrix Biol. 27, 352–359 10.1016/j.matbio.2008.01.003 [DOI] [PubMed] [Google Scholar]
- 48. Ravindranath R. M., Basilrose R. M. Sr., Ravindranath N. H., and Vaitheesvaran B. (2003) Amelogenin interacts with cytokeratin-5 in ameloblasts during enamel growth. J. Biol. Chem. 278, 20293–20302 10.1074/jbc.M211184200 [DOI] [PubMed] [Google Scholar]
- 49. Ravindranath R. M., Tam W. Y., Bringas P. Jr., Santos V., and Fincham A. G. (2001) Amelogenin-cytokeratin 14 interaction in ameloblasts during enamel formation. J. Biol. Chem. 276, 36586–36597 10.1074/jbc.M104656200 [DOI] [PubMed] [Google Scholar]
- 50. Robinson C., and Hudson J. (2011) Tuft protein: protein cross-linking in enamel development. Eur. J. Oral Sci. 119, Suppl. 1, 50–54 10.1111/j.1600-0722.2011.00906.x [DOI] [PubMed] [Google Scholar]
- 51. Robinson C., Fuchs P., and Weatherell J. A. (1977) Fate of matrix proteins during development of dental enamel. Calcif. Tissue Res. 22, 185–190 [DOI] [PubMed] [Google Scholar]
- 52. Duverger O., Beniash E., and Morasso M. I. (2016) Keratins as components of the enamel organic matrix. Matrix Biol. 52–54, 260–265 10.1016/j.matbio.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Alix-Panabières C., Vendrell J.-P., Slijper M., Pellé O., Barbotte E., Mercier G., Jacot W., Fabbro M., and Pantel K. (2009) Full-length cytokeratin-19 is released by human tumor cells: a potential role in metastatic progression of breast cancer. Breast Cancer Res. 11, R39 10.1186/bcr2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chan R., Rossitto P. V., Edwards B. F., and Cardiff R. D. (1986) Presence of proteolytically processed keratins in the culture-medium of mcf-7 cells. Cancer Res. 46, 6353–6359 [PubMed] [Google Scholar]
- 55. Goldberg M., Vermelin L., Mostermans P., Lécolle S., Septier D., Godeau G., and Legeros R. Z. (1998) Fragmentation of the distal portion of Tomes' processes of secretory ameloblasts in the forming enamel of rat incisors. Connect. Tissue Res. 38, 159–169; discussion 201–205 10.3109/03008209809017033 [DOI] [PubMed] [Google Scholar]
- 56. Rabouille C. (2017) Pathways of unconventional protein secretion. Trends Cell Biol. 27, 230–240 10.1016/j.tcb.2016.11.007 [DOI] [PubMed] [Google Scholar]
- 57. Nickel W., and Rabouille C. (2009) Mechanisms of regulated unconventional protein secretion. Nat. Rev. Mol. Cell Biol. 10, 148–155 10.1038/nrm2617 [DOI] [PubMed] [Google Scholar]
- 58. Rabouille C., Malhotra V., and Nickel W. (2012) Diversity in unconventional protein secretion. J. Cell Sci. 125, 5251–5255 10.1242/jcs.103630 [DOI] [PubMed] [Google Scholar]
- 59. Grieve A. G., and Rabouille C. (2011) Golgi bypass: skirting around the heart of classical secretion. Cold Spring Harb. Perspect. Biol. 3, a005298 10.1101/cshperspect.a005298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ponpuak M., Mandell M. A., Kimura T., Chauhan S., Cleyrat C., and Deretic V. (2015) Secretory autophagy. Curr. Opin. Cell Biol. 35, 106–116 10.1016/j.ceb.2015.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Davis S., Wang J., and Ferro-Novick S. (2017) Crosstalk between the secretory and autophagy pathways regulates autophagosome formation. Dev. Cell 41, 23–32 10.1016/j.devcel.2017.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Blott E. J., and Griffiths G. M. (2002) Secretory lysosomes. Nat. Rev. Mol. Cell Biol. 3, 122–131 10.1038/nrm732 [DOI] [PubMed] [Google Scholar]
- 63. Simons M., and Raposo G. (2009) Exosomes—vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21, 575–581 10.1016/j.ceb.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 64. Stefanovic S., and Hegde R. S. (2007) Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128, 1147–1159 10.1016/j.cell.2007.01.036 [DOI] [PubMed] [Google Scholar]
- 65. Shao S., and Hegde R. S. (2011) Membrane protein insertion at the endoplasmic reticulum. Annu. Rev. Cell Dev. Biol. 27, 25–56 10.1146/annurev-cellbio-092910-154125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bègue-Kirn C., Krebsbach P. H., Bartlett J. D., and Butler W. T. (1998) Dentin sialoprotein, dentin phosphoprotein, enamelysin and ameloblastin: tooth-specific molecules that are distinctively expressed during murine dental differentiation. Eur. J. Oral Sci. 106, 963–970 10.1046/j.0909-8836.1998.eos106510.x [DOI] [PubMed] [Google Scholar]
- 67. Bleicher F., Couble M. L., Farges J. C., Couble P., and Magloire H. (1999) Sequential expression of matrix protein genes in developing rat teeth. Matrix Biol. 18, 133–143 10.1016/S0945-053X(99)00007-4 [DOI] [PubMed] [Google Scholar]
- 68. Snider N. T., and Omary M. B. (2014) Post-translational modifications of intermediate filament proteins: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 15, 163–177 10.1038/nrm3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Uchida T., and Warshawsky H. (1992) Zinc iodide-osmium tetroxide impregnation of the “tubulo-vesicular system” in Tomes' process of the rat incisor ameloblast. Anat. Rec. 232, 325–339 10.1002/ar.1092320302 [DOI] [PubMed] [Google Scholar]
- 70. Yoshida H., Inoue M., Inoue S., and Akisaka T. (1996) Ultrastructure of kitten molar secretory ameloblasts after quick-freezing and freeze-substitution. J. Electron Microsc. 45, 483–490 10.1093/oxfordjournals.jmicro.a023472 [DOI] [Google Scholar]
- 71. Klausner R. D., Donaldson J. G., and Lippincott-Schwartz J. (1992) Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116, 1071–1080 10.1083/jcb.116.5.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nickel W. (2003) The mystery of nonclassical protein secretion. Eur. J. Biochem. 270, 2109–2119 10.1046/j.1432-1033.2003.03577.x [DOI] [PubMed] [Google Scholar]
- 73. Duverger O., Cross M. A., Smith F. J. D., and Morasso M. I. (2019) Enamel anomalies in a pachyonychia congenita patient with a mutation in KRT16. J. Invest. Dermatol. 139, 238–241 10.1016/j.jid.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hayoun D., Kapp T., Edri-Brami M., Ventura T., Cohen M., Avidan A., and Lichtenstein R. G. (2012) HSP60 is transported through the secretory pathway of 3-MCA-induced fibrosarcoma tumour cells and undergoes N-glycosylation. FEBS J. 279, 2083–2095 10.1111/j.1742-4658.2012.08594.x [DOI] [PubMed] [Google Scholar]
- 75. Merendino A. M., Bucchieri F., Campanella C., Marcianò V., Ribbene A., David S., Zummo G., Burgio G., Corona D. F., Conway de Macario E., Macario A. J., and Cappello F. (2010) Hsp60 is actively secreted by human tumor cells. PLoS One 5, e9247 10.1371/journal.pone.0009247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Paine C. T., Paine M. L., and Snead M. L. (1998) Identification of tuftelin- and amelogenin-interacting proteins using the yeast two-hybrid system. Connect. Tissue Res. 38, 257–267; discussion 295–303 10.3109/03008209809017046 [DOI] [PubMed] [Google Scholar]
- 77. Yang X., and Beniash E. (2019) Immunofluorescence procedures for developing enamel tissues. Methods Mol. Biol. 1922, 191–196 10.1007/978-1-4939-9012-2_19 [DOI] [PubMed] [Google Scholar]
- 78. Yang X., Vidunas A. J., and Beniash E. (2017) Optimizing immunostaining of enamel matrix: application of Sudan Black B and minimization of false positives from normal sera and IgGs. Front. Physiol. 8, 239 10.3389/fphys.2017.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Langbein L., Yoshida H., Praetzel-Wunder S., Parry D. A., and Schweizer J. (2010) The keratins of the human beard hair medulla: the riddle in the middle. J. Invest. Dermatol. 130, 55–73 10.1038/jid.2009.192 [DOI] [PubMed] [Google Scholar]
- 80. Verdelis K., Szabo-Rogers H. L., Xu Y., Chong R., Kang R., Cusack B. J., Jani P., Boskey A. L., Qin C., and Beniash E. (2016) Accelerated enamel mineralization in Dspp mutant mice. Matrix Biol. 52–54, 246–259 10.1016/j.matbio.2016.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Szabo-Rogers H. L., Geetha-Loganathan P., Whiting C. J., Nimmagadda S., Fu K., and Richman J. M. (2009) Novel skeletogenic patterning roles for the olfactory pit. Development 136, 219–229 10.1242/dev.023978 [DOI] [PubMed] [Google Scholar]
- 82. Yamakoshi Y., Richardson A. S., Nunez S. M., Yamakoshi F., Milkovich R. N., Hu J. C., Bartlett J. D., and Simmer J. P. (2011) Enamel proteins and proteases in Mmp20 and Klk4 null and double-null mice. Eur. J. Oral Sci. 119, 206–216 10.1111/j.1600-0722.2011.00866.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Winter H., Hofmann I., Langbein L., Rogers M. A., and Schweizer J. (1997) A splice site mutation in the gene of the human type I hair keratin hHa1 results in the expression of a tailless keratin isoform. J. Biol. Chem. 272, 32345–32352 10.1074/jbc.272.51.32345 [DOI] [PubMed] [Google Scholar]
- 84. Dunn K. W., Kamocka M. M., and McDonald J. H. (2011) A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 300, C723–C742 10.1152/ajpcell.00462.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bolte S., and Cordelières F. P. (2006) A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- 86. Manders E. M. M., Verbeek F. J., and Aten J. A. (1993) Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169, 375–382 10.1111/j.1365-2818.1993.tb03313.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.