Abstract
Canonical Gremlin1 (GREM1) signaling involves binding to and sequestering bone morphogenetic proteins (BMPs) in the extracellular matrix, preventing the activation of cognate BMP receptor. Exquisite temporospatial control of the GREM1-BMP interaction is required during development, and perturbation of this balance leads to abnormal limb formation and defective kidney development. In addition to inhibition of BMP signaling, several other noncanonical signaling modalities of GREM1 have been postulated. Some literature reports have suggested that GREM1 can bind to and activate vascular endothelial growth factor receptor-2 (VEGFR2) in endothelial cells, human kidney epithelial cells, and others. These reports suggest that the GREM1 → VEGFR2 signaling can drive angiogenesis both in vitro and in vivo. We report here that, despite exhaustive attempts, we did not observe GREM1 activation of VEGFR2 in any of the cell lines reported by the above-mentioned studies. Incubation of endothelial colony–forming cells (ECFCs) or human umbilical vein endothelial cells (HUVECs) with recombinant VEGF triggered a robust increase in VEGFR2 tyrosine phosphorylation. In contrast, no VEGFR2 phosphorylation was detected when cells were incubated with recombinant GREM1 over a range of time points and concentrations. We also show that GREM1 does not interfere with VEGF-mediated VEGFR2 activation, suggesting that GREM1 does not bind with any great affinity to VEGFR2. Measurements of ECFC barrier integrity revealed that VEGF induces barrier function disruption, but recombinant human GREM1 had no effect in this assay. We believe that these results provide an important clarification of the potential interaction between GREM1 and VEGFR2 in mammalian cells.
Keywords: vascular endothelial growth factor (VEGF), bone morphogenetic protein (BMP), cell signaling, cell surface receptor, angiogenesis, vascular biology, protein phosphorylation, cancer biology, Gremlin1
Introduction
Bone morphogenetic proteins (BMPs)4 regulate a wide range of biological processes, such as lower-limb formation and kidney development (1, 2). The physiological action of BMPs is regulated by a number of secreted, protein antagonists, such as Gremlin1 (GREM1), Noggin, and Chordin, that bind to BMPs in the extracellular space (1, 3).This formation of a BMP-BMP antagonist complex prevents the activation of the BMPRI/II complex, preventing phosphorylation of R-SMAD1/5/9 and resulting SMAD-dependent gene expression (4). GREM1 binding to BMP4 has been shown to play a key role in mammalian kidney formation (5–7). In addition, GREM1 has been identified as a pathogenic mediator of diabetic kidney disease (8–10), pulmonary hypertension (11–13), pancreatitis (14), and a range of human cancers, including mesothelioma, glioma, and colorectal cancer (15–21). The mechanism of GREM1-mediated fibrosis or oncogenic signaling has not yet been definitively identified. Several groups have suggested noncanonical signaling mechanisms for GREM1 via Slit proteins (22), ROBO (23), and fibrillin (24). Two reports in Blood (25, 26) in 2007 and 2010 identified GREM1 as a novel agonist of the proangiogenic receptor VEGFR2. These reports suggest that exposure of human endothelial vein endothelial cells (HUVECs) to GREM1 (∼3 nm) induces phosphorylation of VEGFR2 on Tyr1175, the site associated with PLCγ/ERK activation (27). This group also published several follow-up reports on the requirement of αvβ3 integrins for GREM1-mediated VEGFR2 activation (28) and the ability of monomeric GREM1 to act as an antagonist of VEGFR2 (29). Sporadic reports in the literature have suggested similar GREM1 → VEGFR2 signaling in ARPE-19 retinal cells (30), HK-2 kidney tubule epithelial cells (31, 32), HaCaT skin keratinocytes, and primary skin fibroblasts (33). In contrast, another paper (46) suggests that GREM1 blocks VEGF signaling in the pulmonary microvascular endothelium. Given the wealth of new data implicating GREM1 signaling in human diseases such as cancer (21, 35), fibrosis of the kidney (36), and lung (11) as well as rheumatoid arthritis (37), we believe it is critical to define the precise signaling mechanisms of GREM1 signaling in mammalian cells. To that end, we report here that despite extensive effort, we failed to demonstrate GREM1-mediated activation of VEGFR2 in endothelial and other cells. Given the current model of GREM1 signaling in the literature, our data challenge the current dogma that GREM1 activates VEGFR2 phosphorylation and sheds new light on the likely signaling mechanisms engaged by GREM1 during development and disease.
Results
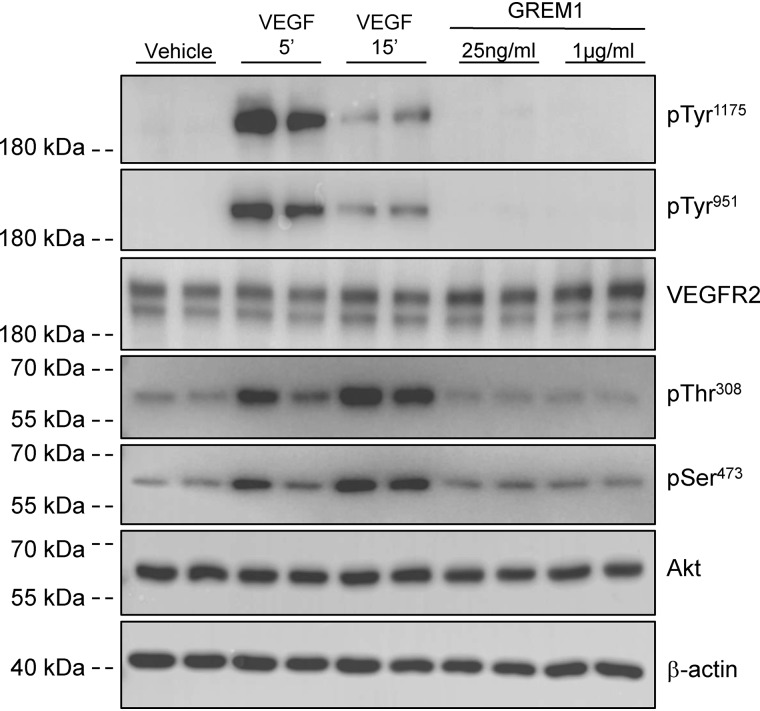
Endothelial colony–forming cells (ECFCs) are late outgrowth endothelial progenitor cells that can form new endothelial cells in vivo (34). ECFCs demonstrate a clear activation of VEGFR2 phosphorylation on both Tyr1175 (which binds Shc/Grb2/SOS, leading to PLCγ and ERK activation) and Tyr951 (which binds Shc/Grb2/Gab1, leading to PI3K/Akt activation) (27) in response to 25 ng/ml VEGF (Fig. 1). In contrast, GREM1 treatment for 15 min had no effect on phospho-VEGFR2 levels when 25 ng/ml (∼2.5 nm) or a supraphysiological concentration of 1 μg/ml (∼100 nm) was used (Fig. 1). A time course of GREM1 treatment using 5-, 10-, 15-, 30-, and 60-min and 16-h treatment also failed to demonstrate increased VEGFR2 phosphorylation (Fig. S1). Consistently, Akt phosphorylation was detected in ECFCs in response to VEGF after 15 min, with no increase evident in GREM1-treated cells (Fig. 1). No changes in total VEGFR2 or Akt were detected in any of the treatment groups. There was no difference in experimental outcome when ECFCs were grown on either collagen or fibrinogen to trigger co-activation of αvβ3, which was reported to enhance GREM1-mediated VEGFR2 phosphorylation (Figs. 1 and 2A) (28). Using immunofluorescence as a readout, increased phospho-VEGFR2 was detected at the plasma membrane of ECFCs (Fig. S2). Consistently, no increase was detected in GREM1-treated cells compared with vehicle (Fig. S2). These data suggest that in a VEGF-responsive cell where VEGFR2 phosphorylation is clearly evident, rhGREM1 cannot activate VEGFR2 phosphorylation despite the reports in the literature.
Figure 1.
Gremlin1 does not activate VEGFR2 phosphorylation in ECFCs. ECFCs isolated from human blood were seeded on fibrinogen (2 μg/ml in double-distilled H2O) and serum-reduced in 2% FBS EGM-2 medium overnight. On the day of the experiment, cells were serum-starved for 3 h prior to treatment with vehicle (PBS), rhVEGF (25 ng/ml; R&D Systems) for 5 and 15 min, or rhGREM1 (25 ng/ml and 1 μg/ml; R&D Systems) for 15 min. Protein lysates were run on SDS-PAGE and probed with the indicated antibodies reactive to pTyr1175 and pTyr951 on VEGFR2, total VEGFR2, pThr308 and pSer473 phospho-Akt antibodies, total Akt, and β-actin.
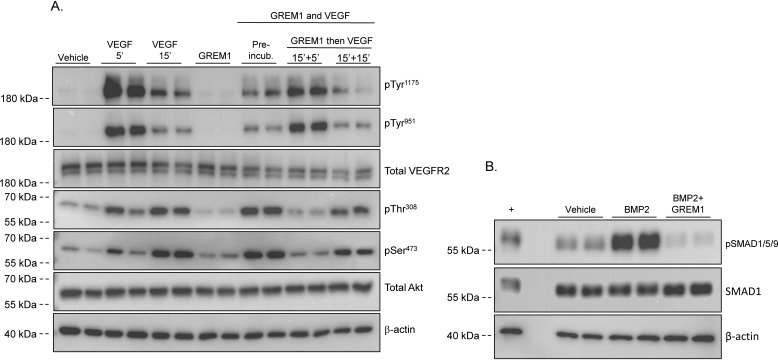
Figure 2.
Gremlin1 does not inhibit VEGF signaling via the VEGFR2 receptor. A, ECFCs isolated from human blood were seeded on fibrinogen and serum-reduced in 2% FBS EGM-2 medium overnight. On the day of the experiment, cells were serum-starved for 3 h prior to treatment with vehicle (PBS), rhVEGF (25 ng/ml) for 5 and 15 min or rhGREM1 (25 ng/ml; R&D Systems) for 15 min. rhGREM1 (25 ng/ml) and rhVEGF (25 ng/ml) were also preincubated for 15 min prior to the addition to cells for 15 min. In addition, rhGREM1 (25 ng/ml) was added to cells for 15 min prior to the addition of rhVEGF (25 ng/ml) for 5 and 15 min. Protein lysates were run on SDS-PAGE and probed with the indicated antibodies reactive to pTyr1175 and pTyr951 on VEGFR2, total VEGFR2, pThr308 and pSer473 phospho-Akt antibodies, total Akt, and β-actin as loading control. B, HEK293 cells were serum-reduced in 1% FBS overnight, followed by serum-free medium for 3 h. Cells were treated for 60 min with vehicle (4 mm HCl), rhBMP2 (5 ng/ml) or rhBMP2 (5 ng/ml), and rhGREM1 (25 ng/ml) that had been preincubated for 15 min at 37 °C. Protein lysates were run in SDS-PAGE and probed with the indicated antibodies reactive to pSmad1/5/9, total Smad1, and β-actin as loading control. +, positive control.
We tested the hypothesis that GREM1 may bind to and enhance or inhibit VEGF-mediated receptor activation. Preincubation of GREM1 with VEGF ligand did not inhibit VEGF-mediated VEGFR2 phosphorylation or Akt phosphorylation at the 15-min time point (Fig. 2A). We also explored the possibility that GREM1 may be acting as a partial agonist by binding and inhibiting VEGFR2 phosphorylation. Preincubation of ECFCs with GREM1, followed by VEGF, did not affect VEGF-mediated VEGFR2 or Akt phosphorylation (Fig. 2A). Similar data were obtained using three independent ECFC clones (data not shown). These data suggest that GREM1 is not acting as an antagonist or partial agonist at VEGFR2 and does not affect VEGF signaling in these cells. Importantly, the rhGREM1 used can completely block BMP2-mediated pSMAD1/5/9 phosphorylation in human embryonic kidney 293T (HEK293T) cells, confirming the integrity of the rhGREM1 used in our experiments (Fig. 2B). In addition to ECFCs, we have been unable to demonstrate GREM1-mediated activation of VEGFR2 in HUVECs (Fig. S3), human kidney epithelial cells (HK-2), HEK293 cells, or human retinal pigment epithelial ARPE-19 cells (as reported by other authors (38, 39)) (data not shown).
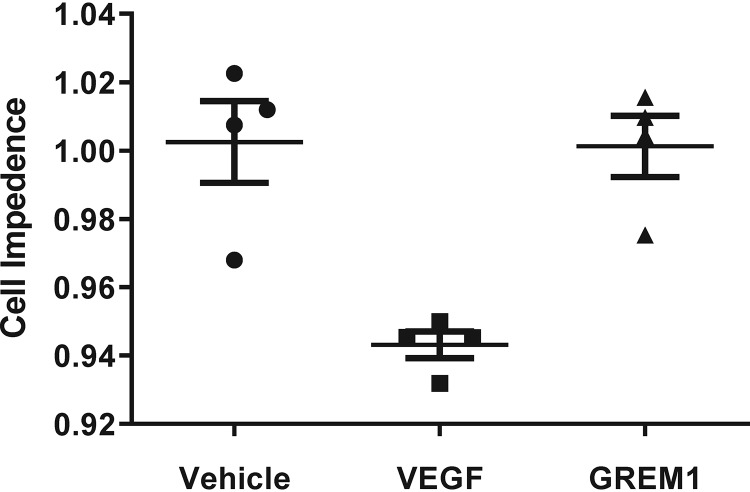
VEGF has previously been demonstrated to reduce barrier integrity in endothelial cells (40–44). We tested the ability of GREM1 to alter ECFC barrier function compared with VEGF as a functional readout of VEGFR signaling. Using the XCELLigence system, the ability of rhVEGF and rhGREM1 to reduce the integrity of the ECFC monolayer by decreasing cell impedance was assessed. The addition of rhVEGF (25 ng/ml) caused a transient reduction in ECFC barrier function within 30 min, which recovered after 60 min (Fig. 3). In contrast, rhGREM1 (100 ng/ml, ∼10 nm) had no effect on ECFC cell impedance and did not compromise ECFC monolayer integrity (Fig. 3). The addition of higher concentrations of rhGREM1 (1 μg/ml, ∼100 nm) also had no significant effect on ECFC cell impedance (data not shown). These data further support our conclusion that GREM1 does not signal via VEGFR2 or equivalent pathways that are engaged by VEGF in endothelial cells.
Figure 3.
GREM1 does not alter ECFC barrier function. ECFCs were plated in E-plates containing XCELLigence® electrodes as described under “Experimental procedures.” Cell impedance levels were normalized prior to the addition of vehicle (PBS; filled circles), rhVEGF (25 ng/ml; filled squares), or rhGREM1 (100 ng/ml; filled triangles). Changes in ECFC barrier function were measured for 60 min, and data are plotted as mean cell impedance ± S.D. (error bars). Cell index values were also taken after ligand-induced peak dip for at least 5 h. Data for an individual clone are shown and are representative of xCELLigence® data obtained from four independent ECFC clones tested in quadruplicate.
Discussion
In summary, we cannot demonstrate that GREM1 signals via the VEGFR2 in a range of cell lines that have been reported in the literature. We have considered the possibility that GREM1 treatment may be altering the expression levels of VEGFR2 in cells. However, quantitative PCR demonstrated no change in VEGFR2 mRNA levels in cells treated with GREM1 at 4, 8, and 16 h (Fig. S4). Our previous data demonstrating grossly normal development of the vasculature in retina and other tissues in Grem1−/− mice (36) also suggest that GREM1 signaling does not contribute a major physiological pro-angiogenic drive. We believe this is an important issue to resolve for the field, as GREM1 signaling via VEGR2 has been suggested as an important noncanonical signaling modality for GREM1 that may contribute to pathophysiological signaling in kidney fibrosis and cancer. We believe that our data shed important new light on this inconsistency and allow clarification in the field of GREM1/BMP signaling.
Experimental procedures
Cell culture
ECFCs were isolated by Dr. Reinhold Medina (Queen's University Belfast) and grown in endothelial cell growth medium 2 (EGM-2, PromoCell, Heidelberg, Germany) supplemented with 10% FBS and were grown in t75 cm2 flasks precoated with rat tail collagen type 1 (BD Biosciences) at 37 °C, 5% CO2, and 95% air. HEK293T cells were maintained in Dulbecco's modified Eagle's medium containing 1 g/liter glucose (Gibco) supplemented with 10% FBS and 100 μg/ml Primocin (Invitrogen). Cells were grown in t752 flasks at 37 °C, 5% CO2, and 95% air and plated on 60-mm dishes for treatments.
VEGFR2 activation assay
ECFCs were first seeded in 60-mm dishes coated with 2 μg/ml fibrinogen (Sigma-Aldrich) to achieve 70% confluence on the day of the experiment. Cells were serum-deprived in 2% FBS medium overnight, followed by a further 4 h in serum-free medium. Cells were treated with 25 ng/ml rhVEGF (R&D Systems) or 25 ng/ml rhGREM1 (R&D Systems) for 5 or 15 min. In parallel, cells were treated with 25 ng/ml rhVEGF and 25 ng/ml rhGREM1 preincubated at 37 °C for 15 min and then added to the cells for 15 min. Cells were also pretreated with 25 ng/ml rhGREM1 for 15 min, followed by 25 ng/ml rhVEGF for 5 or 15 min.
To confirm GREM1 activity, rhBMP2 treatment of HEK293T cells was also carried out. After serum deprivation, serum-free media containing either vehicle (4 mm HCl), 5 ng/ml rhBMP2 (R&D Systems), or 5 ng/ml rhBMP2 (R&D Systems) and 25 ng/ml rhGREM1 were preincubated for 15 min at 37 °C before adding to the cells for 60 min at 37 °C.
For protein extraction, cells were washed once with PBS and protein extracted with 150 μl of ice-cold radioimmunoprecipitation assay buffer (50 mm Tris-HCl (pH 7.4), 1% (v/v) Nonidet P-40, 0.5% (v/v) sodium deoxycholate, 150 mm sodium chloride (NaCl), and 1 mm EDTA) supplemented with 250 μm sodium orthovanadate (NaVO4; Sigma-Aldrich), 40 mm β-glycerolphosphate (Sigma-Aldrich), 1 mm sodium fluoride (NaF; Sigma-Aldrich), 2 μm microsystin-LR (Enzo® Life Sciences, New York), 1 mm phenylmethanesulfonyl fluoride (Sigma-Aldrich), and 1× protease inhibitor mixture (Sigma-Aldrich). Protein lysates were probed by Western blotting using antibodies (1:1000) reactive to pVEGFR2 (pTyr1175) (Cell Signaling), pVEGFR2 (pTyr951) (Cell Signaling), or total VEGFR2 (Cell Signaling) after blocking the membrane with 3% (w/v) BSA in TBS 0.1% (v/v) Tween 20 (TBS-T). Protein lysates were also probed with antibodies (1:1000) against pAkt (Thr308) (Cell Signaling), pAkt (Ser473) (Cell Signaling), total AKT (Cell Signaling), and β-actin (Cell Signaling) as the loading control in 5% (w/v) nonfat milk TBS-T. Reactive bands were visualized using anti-rabbit or mouse HRP using Immobilon Western Chemiluminescent HRP Substrate (Millipore, Watford, UK) and imaged on the G:BOX Chemi XX6 system (Syngene).
Barrier function analysis
To assess ECFC barrier function, we used the xCELLigence® DP Real-Time Cell Analyzer (ACEA Biosciences, Agilent, San Diego, CA). Briefly, E-plate 16 composed of gold-film electrodes positioned on the bottom surface of the well were coated with rat tail collagen type 1 (Corning) as described previously (45). Background measurements were recorded by filling wells with EBM-2 medium (Lonza). Cord blood–derived ECFCs suspended in EBM-2 were then added at a density of 20,000 cells/well and left at room temperature for 30 min to facilitate uniform attachment. Impedance/cell index measurements were then taken every 15 min. When a stable monolayer was formed, EBM-2 medium containing vehicle (PBS), rhVEGF (25 ng/ml; R&D Systems), or rhGREM1 (100 ng/ml (∼10 nm) or 1 μg/ml (∼100 nm); R&D Systems) was then added to the appropriate wells, and recordings were taken. Changes in cell impedance values were normalized prior to treatment and measured for at least 5 h post-treatment.
Author contributions
L. R. D. and D. P. B. data curation; L. R. D. formal analysis; L. R. D., C. L. O., and R. J. M. investigation; L. R. D. and D. P. B. writing-review and editing; C. L. O., R. J. M., and D. P. B. conceptualization; C. L. O. and R. J. M. methodology; D. P. B. funding acquisition; D. P. B. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Andriana Margariti (Queen's University Belfast) for supplying HUVECs and Dr. Philip Dunne, Prof. Mark Lawler, and colleagues in the Wellcome-Wolfson Institute for Experimental Medicine for advice and support.
The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4.
- BMP
- bone morphogenetic protein
- HUVEC
- human umbilical vein endothelial cell
- PLCγ
- phospholipase Cγ
- ERK
- extracellular signal-regulated kinase
- ECFC
- endothelial colony–forming cell
- PI3K
- phosphatidylinositol 3-kinase
- VEGF
- vascular endothelial growth factor
- EGM-2
- endothelial cell growth medium 2
- FBS
- fetal bovine serum
- HEK
- human embryonic kidney
- HRP
- horseradish peroxidase.
References
- 1. Rider C. C., and Mulloy B. (2010) Bone morphogenetic protein and growth differentiation factor cytokine families and their protein antagonists. Biochem. J. 429, 1–12 10.1042/BJ20100305 [DOI] [PubMed] [Google Scholar]
- 2. Brazil D. P., Church R. H., Surae S., Godson C., and Martin F. (2015) BMP signalling: agony and antagony in the family. Trends Cell Biol. 25, 249–264 10.1016/j.tcb.2014.12.004 [DOI] [PubMed] [Google Scholar]
- 3. Katagiri T., and Watabe T. (2016) Bone morphogenetic proteins. Cold Spring Harb. Perspect. Biol. 8, a021899 10.1101/cshperspect.a021899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. ten Dijke P. (2006) Bone morphogenetic protein signal transduction in bone. Curr. Med. Res. Opin. 22, S7–S11 10.1185/030079906X80576 [DOI] [PubMed] [Google Scholar]
- 5. Khokha M. K., Hsu D., Brunet L. J., Dionne M. S., and Harland R. M. (2003) Gremlin is the BMP antagonist required for maintenance of Shh and Fgf signals during limb patterning. Nat. Genet. 34, 303–307 10.1038/ng1178 [DOI] [PubMed] [Google Scholar]
- 6. Michos O., Panman L., Vintersten K., Beier K., Zeller R., and Zuniga A. (2004) Gremlin-mediated BMP antagonism induces theepithelial-mesenchymal feedback signaling controlling metanephric kidney and limb organogenesis. Development 131, 3401–3410 10.1242/dev.01251 [DOI] [PubMed] [Google Scholar]
- 7. Michos O., Gonçalves A., Lopez-Rios J., Tiecke E., Naillat F., Beier K., Galli A., Vainio S., and Zeller R. (2007) Reduction of BMP4 activity by gremlin 1 enables ureteric bud outgrowth and GDNF/WNT11 feedback signalling during kidney branching morphogenesis. Development 134, 2397–2405 10.1242/dev.02861 [DOI] [PubMed] [Google Scholar]
- 8. Dolan V., Murphy M., Sadlier D., Lappin D., Doran P., Godson C., Martin F., O'Meara Y., Schmid H., Henger A., Kretzler M., Droguett A., Mezzano S., and Brady H. R. (2005) Expression of gremlin, a bone morphogenetic protein antagonist, in human diabetic nephropathy. Am. J. Kidney Dis. 45, 1034–1039 10.1053/j.ajkd.2005.03.014 [DOI] [PubMed] [Google Scholar]
- 9. Walsh D. W., Roxburgh S. A., McGettigan P., Berthier C. C., Higgins D. G., Kretzler M., Cohen C. D., Mezzano S., Brazil D. P., and Martin F. (2008) Co-regulation of Gremlin and Notch signalling in diabetic nephropathy. Biochim. Biophys. Acta 1782, 10–21 10.1016/j.bbadis.2007.09.005 [DOI] [PubMed] [Google Scholar]
- 10. Zhang Y., and Zhang Q. (2009) Bone morphogenetic protein-7 and Gremlin: new emerging therapeutic targets for diabetic nephropathy. Biochem. Biophys. Res. Commun. 383, 1–3 10.1016/j.bbrc.2009.03.086 [DOI] [PubMed] [Google Scholar]
- 11. Cahill E., Costello C. M., Rowan S. C., Harkin S., Howell K., Leonard M. O., Southwood M., Cummins E. P., Fitzpatrick S. F., Taylor C. T., Morrell N. W., Martin F., and McLoughlin P. (2012) Gremlin plays a key role in the pathogenesis of pulmonary hypertension. Circulation 125, 920–930 10.1161/CIRCULATIONAHA.111.038125 [DOI] [PubMed] [Google Scholar]
- 12. Costello C. M., Howell K., Cahill E., McBryan J., Konigshoff M., Eickelberg O., Gaine S., Martin F., and McLoughlin P. (2008) Lung-selective gene responses to alveolar hypoxia: potential role for the bone morphogenetic antagonist gremlin in pulmonary hypertension. Am. J. Physiol. Lung Cell Mol. Physiol. 295, L272–L284 10.1152/ajplung.00358.2007 [DOI] [PubMed] [Google Scholar]
- 13. Ciuclan L., Sheppard K., Dong L., Sutton D., Duggan N., Hussey M., Simmons J., Morrell N. W., Jarai G., Edwards M., DuBois G., Thomas M., Van Heeke G., and England K. (2013) Treatment with anti-Gremlin 1 antibody ameliorates chronic hypoxia/SU5416–induced pulmonary arterial hypertension in mice. Am. J. Pathol. 183, 1461–1473 10.1016/j.ajpath.2013.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Staloch D., Gao X., Liu K., Xu M., Feng X., Aronson J. F., Falzon M., Greeley G. H., Rastellini C., Chao C., Hellmich M. R., Cao Y., and Ko T. C. (2015) Gremlin is a key pro-fibrogenic factor in chronic pancreatitis. J. Mol. Med. 93, 1085–1093 10.1007/s00109-015-1308-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang D.-J., Zhi X.-Y., Zhang S.-C., Jiang M., Liu P., Han X.-P., Li J., Chen Z., and Wang C.-L. (2012) The bone morphogenetic protein antagonist Gremlin is overexpressed in human malignant mesothelioma. Oncol. Rep. 27, 58–64 10.3892/or.2011.1463 [DOI] [PubMed] [Google Scholar]
- 16. Yin M., Tissari M., Tamminen J., Ylivinkka I., Rönty M., von Nandelstadh P., Lehti K., Hyytiäinen M., Myllärniemi M., and Koli K. (2017) Gremlin-1 is a key regulator of the invasive cell phenotype in mesothelioma. Oncotarget 8, 98280–98297 10.18632/oncotarget.21550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yan K., Wu Q., Yan D. H., Lee C. H., Rahim N., Tritschler I., DeVecchio J., Kalady M. F., Hjelmeland A. B., and Rich J. N. (2014) Glioma cancer stem cells secrete Gremlin1 to promote their maintenance within the tumor hierarchy. Genes Dev. 28, 1085–1100 10.1101/gad.235515.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karagiannis G. S., Berk A., Dimitromanolakis A., and Diamandis E. P. (2013) Enrichment map profiling of the cancer invasion front suggests regulation of colorectal cancer progression by the bone morphogenetic protein antagonist, gremlin-1. Mol. Oncol. 7, 826–839 10.1016/j.molonc.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lewis A., Freeman-Mills L., de la Calle-Mustienes E., Giráldez-Pérez R. M., Davis H., Jaeger E., Becker M., Hubner N. C., Nguyen L. N., Zeron-Medina J., Bond G., Stunnenberg H. G., Carvajal J. J., Gomez-Skarmeta J. L., Leedham S., and Tomlinson I. (2014) A polymorphic enhancer near GREM1 influences bowel cancer risk through differential CDX2 and TCF7L2 binding. Cell Rep. 8, 983–990 10.1016/j.celrep.2014.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karagiannis G. S., Musrap N., Saraon P., Treacy A., Schaeffer D. F., Kirsch R., Riddell R. H., and Diamandis E. P. (2015) Bone morphogenetic protein antagonist gremlin-1 regulates colon cancer progression. Biol. Chem. 396, 163–183 10.1515/hsz-2014-0221 [DOI] [PubMed] [Google Scholar]
- 21. Davis H., Irshad S., Bansal M., Rafferty H., Boitsova T., Bardella C., Jaeger E., Lewis A., Freeman-Mills L., Giner F. C., Rodenas-Cuadrado P., Mallappa S., Clark S., Thomas H., Jeffery R., et al. (2015) Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells outside the stem cell niche. Nat. Med. 21, 62–70 10.1038/nm.3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen B., Blair D. G., Plisov S., Vasiliev G., Perantoni A. O., Chen Q., Athanasiou M., Wu J. Y., Oppenheim J. J., and Yang D. (2004) Cutting edge: bone morphogenetic protein antagonists Drm/Gremlin and Dan interact with Slits and act as negative regulators of monocyte chemotaxis. J. Immunol. 173, 5914–5917 10.4049/jimmunol.173.10.5914 [DOI] [PubMed] [Google Scholar]
- 23. Tumelty K. E., Higginson-Scott N., Fan X., Bajaj P., Knowlton K. M., Shamashkin M., Coyle A. J., Lu W., and Berasi S. P. (2018) Identification of direct negative crosstalk between the SLIT2 and bone morphogenetic protein-Gremlin signaling pathways. J. Biol. Chem. 293, 3039–3055 10.1074/jbc.M117.804021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tamminen J. A., Parviainen V., Rönty M., Wohl A. P., Murray L., Joenväärä S., Varjosalo M., Leppäranta O., Ritvos O., Sengle G., Renkonen R., Myllärniemi M., and Koli K. (2013) Gremlin-1 associates with fibrillin microfibrils in vivo and regulates mesothelioma cell survival through transcription factor slug. Oncogenesis. 2, e66 10.1038/oncsis.2013.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stabile H., Mitola S., Moroni E., Belleri M., Nicoli S., Coltrini D., Peri F., Pessi A., Orsatti L., Talamo F., Castronovo V., Waltregny D., Cotelli F., Ribatti D., and Presta M. (2007) Bone morphogenic protein antagonist Drm/gremlin is a novel proangiogenic factor. Blood 109, 1834–1840 10.1182/blood-2006-06-032276 [DOI] [PubMed] [Google Scholar]
- 26. Mitola S., Ravelli C., Moroni E., Salvi V., Leali D., Ballmer-Hofer K., Zammataro L., and Presta M. (2010) Gremlin is a novel agonist of the major proangiogenic receptor VEGFR2. Blood 116, 3677–3680 10.1182/blood-2010-06-291930 [DOI] [PubMed] [Google Scholar]
- 27. Clegg L. W., and Mac Gabhann F. (2015) Site-specific phosphorylation of VEGFR2 is mediated by receptor trafficking: insights from a computational model. PLoS Comput. Biol. 11, e1004158 10.1371/journal.pcbi.1004158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ravelli C., Mitola S., Corsini M., and Presta M. (2013) Involvement of αvβ3 integrin in gremlin-induced angiogenesis. Angiogenesis 16, 235–243 10.1007/s10456-012-9309-6 [DOI] [PubMed] [Google Scholar]
- 29. Grillo E., Ravelli C., Corsini M., Ballmer-Hofer K., Zammataro L., Oreste P., Zoppetti G., Tobia C., Ronca R., Presta M., and Mitola S. (2016) Monomeric gremlin is a novel vascular endothelial growth factor receptor-2 antagonist. Oncotarget 7, 35353–35368 10.18632/oncotarget.9286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y., Chen Z., Cheng H., Chen J., and Qian J. (2017) Gremlin promotes retinal pigmentation epithelial (RPE) cell proliferation, migration and VEGF production via activating VEGFR2-Akt-mTORC2 signaling. Oncotarget 8, 979–987 10.18632/oncotarget.13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marquez-Exposito L., Lavoz C., Rodrigues-Diez R. R., Rayego-Mateos S., Orejudo M., Cantero-Navarro E., Ortiz A., Egido J., Selgas R., Mezzano S., and Ruiz-Ortega M. (2018) Gremlin regulates tubular epithelial to mesenchymal transition via VEGFR2: potential role in renal fibrosis. Front. Pharmacol. 9, 1195 10.3389/fphar.2018.01195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lavoz C., Poveda J., Marquez-Exposito L., Rayego-Mateos S., Rodrigues-Diez R. R., Ortiz A., Egido J., Mezzano S., and Ruiz-Ortega M. (2018) Gremlin activates the Notch pathway linked to renal inflammation. Clin. Sci. 132, 1097–1115 10.1042/CS20171553 [DOI] [PubMed] [Google Scholar]
- 33. Ji C., Huang J. W., Xu Q. Y., Zhang J., Lin M. T., Tu Y., He L., Bi Z. G., and Cheng B. (2016) Gremlin inhibits UV-induced skin cell damages via activating VEGFR2-Nrf2 signaling. Oncotarget 7, 84748–84757 10.18632/oncotarget.12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Medina R. J., Barber C. L., Sabatier F., Dignat-George F., Melero-Martin J. M., Khosrotehrani K., Ohneda O., Randi A. M., Chan J. K. Y., Yamaguchi T., Van Hinsbergh V. W. M., Yoder M. C., and Stitt A. W. (2017) Endothelial progenitors: a consensus statement on nomenclature. Stem Cells Transl. Med. 6, 1316–1320 10.1002/sctm.16-0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jang B. G., Kim H. S., Chang W. Y., Bae J. M., Oh H. J., Wen X., Jeong S., Cho N. Y., Kim W. H., and Kang G. H. (2017) Prognostic significance of stromal GREM1 expression in colorectal cancer. Hum. Pathol. 62, 56–65 10.1016/j.humpath.2016.12.018 [DOI] [PubMed] [Google Scholar]
- 36. Church R. H., Ali I., Tate M., Lavin D., Krishnakumar A., Kok H. M., Hombrebueno J. R., Dunne P. D., Bingham V., Goldschmeding R., Martin F., and Brazil D. P. (2017) Gremlin1 plays a key role in kidney development and renal fibrosis. Am. J. Physiol. Renal Physiol. 312, F1141–F1157 10.1152/ajprenal.00344.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Han E.-J., Yoo S.-A., Kim G.-M., Hwang D., Cho C.-S., You S., and Kim W.-U. (2016) GREM1 is a key regulator of synoviocyte hyperplasia and invasiveness. J. Rheumatol. 43, 474–485 10.3899/jrheum.150523 [DOI] [PubMed] [Google Scholar]
- 38. Li J., Liu H., Zou L., Ke J., Zhang Y., Zhu Y., Yang Y., Gong Y., Tian J., Zou D., Peng X., Gong J., Zhong R., Huang K., Chang J., and Miao X. (2017) A functional variant in GREM1 confers risk for colorectal cancer by disrupting a hsa-miR-185–3p binding site. Oncotarget 8, 61318–61326 10.18632/oncotarget.18095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rodrigues-Diez R., Rodrigues-Diez R. R., Lavoz C., Carvajal G., Droguett A., Garcia-Redondo A. B., Rodriguez I., Ortiz A., Egido J., Mezzano S., and Ruiz-Ortega M. (2014) Gremlin activates the Smad pathway linked to epithelial mesenchymal transdifferentiation in cultured tubular epithelial cells. Biomed. Res. Int. 2014, 802841 10.1155/2014/802841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weis S. M., and Cheresh D. A. (2005) Pathophysiological consequences of VEGF-induced vascular permeability. Nature 437, 497–504 10.1038/nature03987 [DOI] [PubMed] [Google Scholar]
- 41. Kevil C. G., Payne D. K., Mire E., and Alexander J. S. (1998) Vascular permeability factor/vascular endothelial cell growth factor-mediated permeability occurs through disorganization of endothelial junctional proteins. J. Biol. Chem. 273, 15099–15103 10.1074/jbc.273.24.15099 [DOI] [PubMed] [Google Scholar]
- 42. Behzadian M. A., Windsor L. J., Ghaly N., Liou G., Tsai N.-T., and Caldwell R. B. (2003) VEGF-induced paracellular permeability in cultured endothelial cells involves urokinase and its receptor. FASEB J. 17, 752–754 10.1096/fj.02-0484fje [DOI] [PubMed] [Google Scholar]
- 43. Satchell S. C., Anderson K. L., and Mathieson P. W. (2004) Angiopoietin 1 and vascular endothelial growth factor modulate human glomerular endothelial cell barrier properties. J. Am. Soc. Nephrol. 15, 566–574 10.1097/01.ASN.0000115397.22519.03 [DOI] [PubMed] [Google Scholar]
- 44. Chen X. L., Nam J.-O., Jean C., Lawson C., Walsh C. T., Goka E., Lim S.-T., Tomar A., Tancioni I., Uryu S., Guan J.-L., Acevedo L. M., Weis S. M., Cheresh D. A., and Schlaepfer D. D. (2012) VEGF-induced vascular permeability is mediated by FAK. Dev. Cell 22, 146–157 10.1016/j.devcel.2011.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Medina R. J., O'Neill C. L., Humphreys M. W., Gardiner T. A., and Stitt A. W. (2010) Outgrowth endothelial cells: characterization and their potential for reversing ischemic retinopathy. Invest. Ophthalmol. Vis. Sci. 51, 5906–5913 10.1167/iovs.09-4951 [DOI] [PubMed] [Google Scholar]
- 46. Rowan S. C., Piouceau L., Cornwell J., Li L., and McLoughlin P. (2018) Gremlin1 blocks vascular endothelial growth factor signalling in the pulmonary microvascular endothelium. Pulm. Circ. 10.1177/2045894018807205. 10.1177/2045894018807205 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.