Abstract
Tk-hefu is an artificial peptide designed based on the α-hairpinin scaffold, which selectively blocks voltage-gated potassium channels Kv1.3. Here we present its spatial structure resolved by NMR spectroscopy and analyze its interaction with channels using computer modeling. We apply protein surface topography to suggest mutations and increase Tk-hefu affinity to the Kv1.3 channel isoform. We redesign the functional surface of Tk-hefu to better match the respective surface of the channel pore vestibule. The resulting peptide Tk-hefu-2 retains Kv1.3 selectivity and displays ∼15 times greater activity compared with Tk-hefu. We verify the mode of Tk-hefu-2 binding to the channel outer vestibule experimentally by site-directed mutagenesis. We argue that scaffold engineering aided by protein surface topography represents a reliable tool for design and optimization of specific ion channel ligands.
Keywords: ion channel, molecular dynamics, neurotoxin, nuclear magnetic resonance (NMR), peptides, potassium channel, protein motif, protein structure, alpha-hairpinin, hefutoxin, pore blocker
Introduction
Ion channels are central to vital physiological processes ranging from regulation of cell osmotic pressure, motility, and transport of metabolites across the cell membrane to hormone secretion, muscle contraction, and brain functioning (1). There is an increasingly appreciated unmet demand in new selective ion channel ligands. This is not only due to the apparent need in selective molecular probes or markers for these proteins to be used in fundamental research but also because of the growing evidence of ion channel involvement in a great number of pathological processes. For instance, the number of known channelopathies, diseases associated with ion channel dysfunction, has increased dramatically in the past years and today includes several dozen if not hundred indications (2, 3). Ion channels are the second most privileged target of marketed drugs (∼20% of small-molecule drugs target ion channels) (4), and their importance is thought to increase (5).
Potassium channels in particular are one of the most diverse and widespread groups of membrane proteins found in all taxa of living organisms, from bacteria to humans (1). They are involved in physiological processes including ion transport, neural signaling, cell communication and proliferation, humoral regulation, and immune response (6, 7). In addition, many studies confirmed the involvement of these proteins in different pathologies, which makes them a promising target of pharmacological research (8, 9). For instance, the Kv1.3 channel isoform is a validated target in autoimmune diseases (10) and specific Kv1.3 blockers have shown a therapeutic effect in models of such diseases (11). This effect is thought to be due to the inhibition of proliferation of effector memory T cells, which rely on a high level of Kv1.3 expression (12). Thereby the search for highly specific ligands of potassium channels is a challenge for both fundamental science and drug discovery.
The current arsenal of potassium channel ligands has been compiled by using two basic approaches: (i) bioactivity-driven isolation from natural sources (13), and (ii) screening of compound libraries followed by structural optimization (14). Alternative strategies have been proposed but their net input is yet relatively small. One approach that is very often cited as the most promising in terms of future drug discovery is rational drug design based on the known 3D structures of protein targets (15). Although much effort has been invested in the development of rational design techniques, it is nowadays still far from becoming the mainstream stand-alone technique and usually supplements high-throughput screening, or is applied to optimize naturally occurring substances. In addition, small proteins or peptides are the most diverse class of potassium channel ligands (the principal source being scorpion venom), yet the methodology of rational design is underdeveloped for protein ligands compared with small molecules.
We argue, however, that the large amount of currently available theoretical data, coupled to the rapid development of computer simulation techniques allows the use of rational design for the discovery and optimization of artificial potassium channel blockers instead of the classical screening approach and exploitation of natural sources. In our recent work (16–18), we have used scaffold engineering to construct an artificial small protein potassium channel blocker based on a plant defense peptide Tk-AMP-X2 from wheat belonging to the recently recognized α-hairpinin family. This new blocker, named Tk-hefu, affects Kv1.3 with a half-maximal inhibitory concentration (IC50) of ∼35 μm. However, effective potassium channel blockers usually display activity at submicromolar concentrations (19, 20). In the present work, we succeed to significantly increase the binding affinity of Tk-hefu to Kv1.3 by using a set of computational techniques including the recently developed protein surface topography (PST)4 (21, 22).
Results
We used the following logic in our studies: (i) Tk-hefu spatial structure was studied by NMR; (ii) it was then used for computational analyses to determine the mode of binding to target channels (Kv1.3) and suggest modifications to increase the affinity; (iii) the proposed mutant peptide was synthesized and its activity was verified by electrophysiology. Table 1 lists amino acid sequences of peptides mentioned in this article.
Table 1.
Amino acid sequences of Tk-AMP-X2 and its derivatives
a Mutated residues are in bold type.
15N-Labeled Tk-hefu production
15N-Labeled recombinant peptide was produced in a bacterial expression system using the M9 medium. Tk-hefu was produced as part of a fusion protein, also containing thioredoxin (Trx), a His6 tag and cleavage site for enteropeptidase. The isolated fusion protein was treated with recombinant human enteropeptidase light chain (23), and 15N-Tk-hefu was purified by reversed-phase HPLC. The molecular mass of the labeled peptide was 3604.0 Da as determined by MALDI-MS (equal to the calculated value), indicating that all nitrogen atoms in the peptide were replaced by 15N. The final yield of 15N-Tk-hefu was ∼4 mg/1 liter of bacterial culture.
Three-dimensional structure of Tk-hefu in solution
The 3D structure of Tk-hefu was determined by NMR spectroscopy (PDB code 5LM0). 15N labeling allowed measuring three sets of J-couplings together with a number of NOESY distances. The statistics of the input data and parameters of the obtained structure are shown in Table 2. Structures from the set are characterized by low CYANA target function values and residual restraint violations, and a substantially low root mean square deviation (RMSD) value for backbone atoms indicating that the structure of Tk-hefu is defined accurately and precisely by experimental data in the region corresponding to residues 3–26.
Table 2.
Statistics for the 10 best NMR structures of Tk-hefu
| Parameter | Value |
|---|---|
| Distance and angle restraints | |
| Total NOEs | 179 |
| Intraresidual | 42 |
| Interresidual | 137 |
| Sequential (|I −J| = 1) | 60 |
| Medium range (2 ≤|I −J| ≤ 4) | 48 |
| Long range (|I −J| ≥ 5) | 29 |
| S-S bond restraints (upper/lower) | 6/6 |
| Angles | |
| ϕ | 27 |
| χ1 | 7 |
| Total restraints per residue | 8 |
| Statistics for the calculated set of structures | |
| CYANA target function (Å2) | 0.100 ± 0.005 |
| Restraint violations | |
| Distance (≥0.2 Å) | 0 |
| Angle (≥5°) | 0 |
| RMSD (Å) Elements of secondary structure | |
| Backbone | 0.5 ± 0.1 |
| All heavy atoms | 1.8 ± 0.2 |
| Ramachandran analysisa | |
| % Most favored regions | 100 |
| % Additional allowed regions | 0 |
| % Generally allowed regions | 0 |
| % Disallowed regions | 0 |
a Ramachandran analysis was performed with the PROCHECK tool at the RCSB validation server (http://deposit.rcsb.org/).
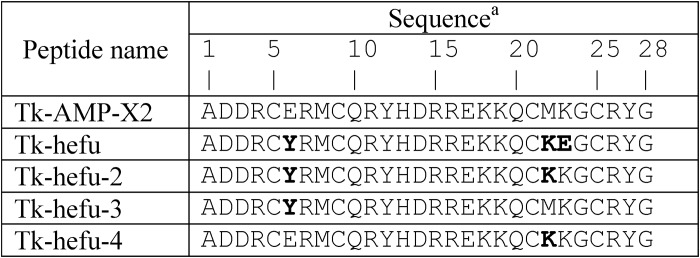
Fig. 1 shows NMR ensembles for Tk-hefu (Fig. 1A) and the parent peptide Tk-AMP-X2 (16) (Fig. 1B) as well as their ribbon representation (Fig. 1, C–F). Long-range distance restraints used for 3D structure calculation are plotted onto the ribbon structure of Tk-hefu in Fig. 1D and distance restraints distribution along the peptide sequence are shown in Fig. S1. Tk-hefu contains two antiparallel α-helices (Arg-4 through Cys-9 and Arg-15 through Arg-26) held together by two disulfide bonds (Cys-5 to Cys-25 and Cys-9 to Cys-21) conforming to the α-hairpinin-fold (17, 18). Some differences are noted between Tk-hefu and the parent peptide Tk-AMP-X2, where the first helix is one residue shorter and is followed by one turn of a 310 helix (Fig. 1F). This conclusion is confirmed by raw NMR data; the 3JHNHA coupling of Cys-9 is equal to 5.6 ± 0.2 Hz in Tk-hefu, which corresponds to α-helical conformation, and it is as large as 7.0 ± 0.2 Hz in Tk-AMP-X2. The second helix of Tk-hefu is also two residues longer at the C terminus than the corresponding helix of the parent peptide. Apart from the secondary structure, the orientation of helices is also different in the mutant peptide: the helices cross at 158 ± 3° (Fig. 1E), which is significantly greater than 133 ± 5° observed for Tk-AMP-X2 (Fig. 1F).
Figure 1.
Tk-hefu structure determined by NMR and its comparison with the parent peptide Tk-AMP-X2. A and B, ensembles of 10 independently derived NMR structures of Tk-hefu (A; PDB ID 5LM0) and Tk-AMP-X2 (B; PDB ID 2M6A) with the fewest restrain violations. N termini are labeled. Cystine side chains are in yellow. C, ribbon representation of Tk-hefu. The orientation of the molecule is as in A. The N-terminal α-helix is shown in green; C-terminal helix, sky blue; loops are presented as a gray string. S-S bonds are presented as yellow sticks, and other side chains are shown as lines; hydrophobic aliphatic and aromatic residues are orange; hydrophilic uncharged residues are magenta; positively charged residues, blue; and negatively charged, red. N and C termini and side chains are labeled. D, the same representation of Tk-hefu as in C but showing all atoms. Gray dashed lines denote the long-range NOESY connectivities between the protons of Tk-hefu. E and F, comparison of Tk-hefu (E) and Tk-AMP-X2 (F) structures. The color code is as in C, and the 310 helix in Tk-AMP-X2 is in smudge green. Side chains of residues discussed in the text are shown as lines and labeled. In C–F, the first structures from the set of 10 are presented (having the fewest restrain violations).
Analysis of NOESY data suggests that helices are more tightly packed in Tk-hefu, and the structure of this peptide is more stable: 29 long-range contacts are observed for the mutant versus 3 contacts found in the spectra of the parent peptide, and the conformation of both disulfide bridges is determined in Tk-hefu, whereas in Tk-AMP-X2 none of the disulfides exhibits a determined conformation. Detailed analysis of the obtained Tk-hefu structure reveals several stabilizing interactions that are introduced by the mutations. Namely, Tyr-6 forms π-cation contacts with Lys-22 and Lys-18, while Glu-23 forms a salt bridge with Lys-19. Thus, the three substitutions in the sequence of Tk-AMP-X2 affected significantly both the spatial structure and stability of the peptide.
Molecular modeling
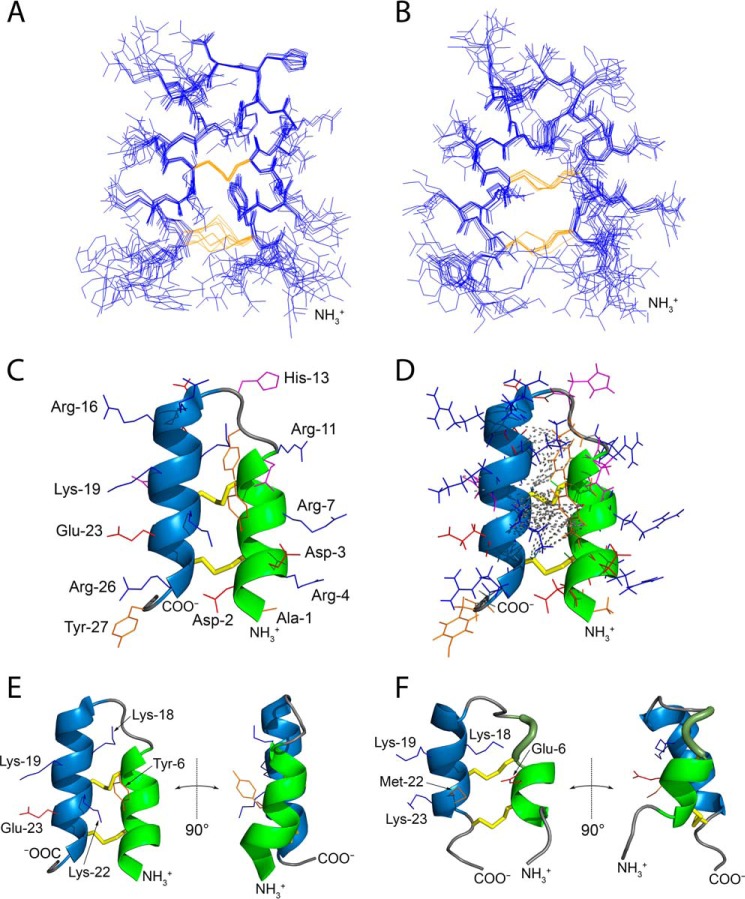
Having determined the NMR structure of Tk-hefu in solution, we were able to model its complex with the target channel. Kv1.3 was modeled by homology based on the available closely related Kv1.2 structure (24), and the Kv1.2/2.1 paddle chimera channel complex with charybdotoxin (ChTx) (25) was chosen as a template for the Kv1.3–Tk-hefu complex model. To date the most reliable approach to model Kv peptide blockers is the concept of the “functional dyad” (see “Discussion” below) (26), and our guide in such modeling was spatial superimposition of the Tk-hefu dyad comprising Tyr-6 and Lys-22 with the classical dyad of ChTx (Tyr-36 and Lys-27). This “manual” complex was then subjected to a series of optimizations including placement inside a hydrated lipid bilayer membrane and running a 200-ns molecular dynamics (MD) trajectory to capture a dynamic view of the channel-toxin interaction (Fig. 2).
Figure 2.
Molecular model of the Kv1.3–Tk-hefu complex suggests rational design of the Tk-hefu-2 high-affinity peptide. A, overall structure of the Kv1.3–Tk-hefu complex after 200-ns MD simulation inside a hydrated lipid bilayer membrane. Kv1.3 is in gray; the pore domain helices of the second protomer and voltage-sensing domain (VSD) of the fourth protomer, as well as extended extracellular loops of the VSDs are omitted for clarity. Lipids are shown in a space-filling representation; atoms are colored: oxygen, red; phosphorus, orange; nitrogen, blue; carbon of phospholipids (POPC and POPE), yellow; carbon of cholesterol, orange. Some lipids are omitted for clarity. Tk-hefu is presented in pink; the functional dyad residues Tyr-6 and Lys-22, as well as Glu-23 are shown as sticks. B, close-up view on Tk-hefu from A overlaid with Tk-hefu-2 for comparison. Asp-383 and Asp-399 of the channel are shown. Tk-hefu-2 is presented in semi-transparent blue, and lipids are omitted for clarity. C–E, protein surface topography maps showing ELP distribution in Kv1.3–Tk-hefu/Tk-hefu-2 complexes: C, Tk-hefu-2; D, Tk-hefu; E, Kv1.3. Semi-transparent green areas in C and D show contact areas with the Kv1.3 channel in complexes. Black boxes highlight the area, where ELP complementarity may be improved by the E23K substitution in Tk-hefu yielding Tk-hefu-2.
In our model of Tk-hefu complex with Kv1.3 (Fig. 2, A and B), Lys-22 sticks out to the selectivity filter, whereas Tyr-6 forms multiple van der Waals contacts with residues of the channel vestibule (Trp-386, Trp-387, Val-390, Val-395, Gly-396, Tyr-397, Gly-398, Asp-399, Met-400, His-401, Pro-402, and Val-403), in agreement with the classical functional dyad model.
We then applied the PST method (21) to the MD trajectory of the Kv1.3–Tk-hefu complex to reveal the fine details of intermolecular recognition. Vestibules of many cation channels carry an overall negative charge, and their ligands are often positively charged. Spherical projection maps of the electrostatic potential (ELP) vividly illustrate this case (Fig. 2, C–E): the Kv1.3 channel is characterized by a negatively charged surface interacting with the bound Tk-hefu (Fig. 2E). This is a result of the presence of a number of negatively charged residues in the channel entrance vestibule or turret (Glu-370, Asp-372, Asp-373, Asp-383, and Asp-399). Tk-hefu is highly positively charged due to a number of lysine and arginine residues (Arg-4, Arg-7, Arg-11, Arg-15, Arg-16, Lys-18, Lys-19, Lys-22, and Arg-26). These residues form an extended positively charged Kv1.3-interacting surface of the peptide (Fig. 2, C and D).
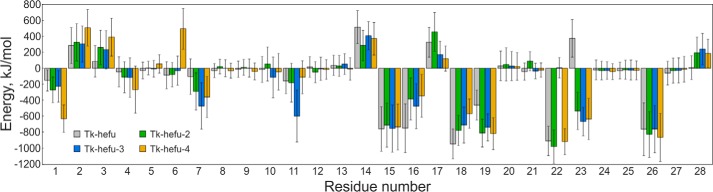
Black boxes in Fig. 2, C–E, highlight the area where ELP complementarity between Tk-hefu and Kv1.3 is not optimal due to the negatively charged Glu-23 in Tk-hefu. We performed computational analysis of the contributions of individual amino acid residues in Tk-hefu to the interaction energy. The resulting interaction energy profile (Fig. 3) demonstrates a negative impact of Glu-23 on the predicted binding affinity (positive energy contribution). We decided to produce Tk-hefu variants with an optimized ELP distribution. One of the peptides, which we named Tk-hefu-2, differs from the parent wheat defense peptide Tk-AMP-X2 by just two residues of the functional dyad, whereas Tk-hefu differs by three residues (Table 1). We built the Kv1.3–Tk-hefu-2 complex analogously to Kv1.3–Tk-hefu, and after MD simulation it indeed showed an optimized ELP distribution (Fig. 2C, black box) and favorable change in the interaction energy profile (Fig. 3) due to Lys-23 in Tk-hefu-2. Not only electrostatic forces drive intermolecular complex formation; in the case of Tk-hefu and Tk-hefu-2, however, they make the maximal contribution.
Figure 3.
Interaction energy profiles of Tk-hefu, Tk-hefu-2, -3, and -4 in complex with Kv1.3. Bar chart showing the contribution of amino acid residues to the interaction energy averaged over MD simulation. Error bars indicate standard deviations.
We also built models and performed MD simulation of Kv1.3 complexes with two single point mutants named Tk-hefu-3 and -4 containing the individual residues of the functional dyad (Table 1) to check their importance for channel binding. Predictably, in both cases computational analysis of the interaction energy showed unfavorable changes in the energy profiles due to the lack of one of the dyad residues (Fig. 3). Energy contribution of Met-22 in complex with Tk-hefu-3 is negligible, presumably because its side chain is too short to reach Kv1.3 pore, unlike Lys-22 in Tk-hefu-2.
Note that although a slight shift in the ligand orientation is observed when Kv1.3–Tk-hefu and Kv1.3–Tk-hefu-2 complexes are compared (Fig. 2B), the peptides retain very close positions and conformations. The modeled Tk-hefu-2 structure is generally stable in MD simulations in water (data not shown), although in a series of calculations we were able to observe two conformational states. One of them (more populated) is very similar to Tk-hefu, and the characteristic distance between the dyad residues (Tyr-6–Lys-22) is similar to that for Tk-hefu and ChTx. The other (less populated; less than ∼10% of simulation time) conformational state is similar to the “parent” Tk-AMP-X2 peptide with an increased distance in the dyad, which seems not optimal for the interaction with Kv channels. There were a number of transitions between these two states in MD runs, which involved a ∼30–40° axial rotation of the first α-helix, restructuring of the disulfides, and convergence of Tyr-6 and Lys-22.
Production of Tk-hefu derivatives
To verify our computational results we produced Tk-hefu-2, -3, and -4. We used the same bacterial expression system as previously for Tk-hefu (16). Synthetic genes coding for Tk-hefu-2, -3, and -4 were produced from oligonucleotides and cloned into the pET-32b expression vector. Trx was used as the fusion partner to ensure a high yield of the disulfide-containing peptides with native conformation (27). To enable selective proteolysis of the fusion proteins, a sequence encoding the enteropeptidase-cleavage site was introduced to the 5′ terminus of the synthetic genes. The resulting plasmids pET-32b–Tk-hefu-2, -3, and -4 were used to transform Escherichia coli BL21(DE3) cells. Target peptides were produced as the result of fusion proteins cleavage followed by separation using reversed-phase HPLC. Their primary structure was verified by Edman sequencing and MALDI-MS. The final yield of the peptides was ∼7 mg/1 liter of bacterial culture.
Electrophysiological recordings
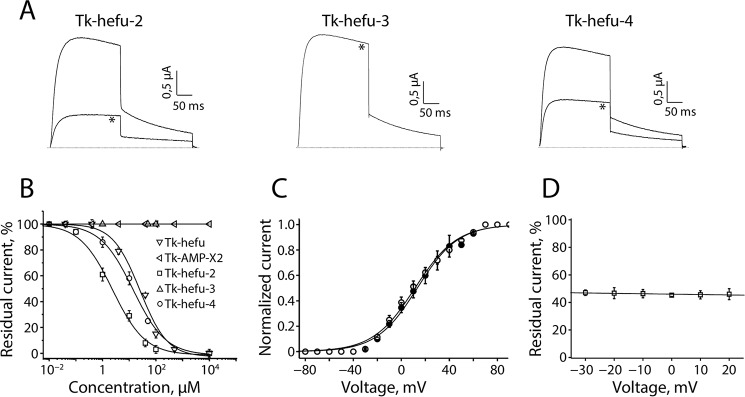
Because it was described previously that Tk-hefu could inhibit Kv1.1, Kv1.3, and Kv1.6 channels (16), Tk-hefu-2–4 were tested against Kv1.1–Kv1.3 and Kv1.6 channels (Fig. S2). At a concentration of 40 μm, Kv1.1 channels were not affected by Tk-hefu-2, whereas Kv1.2 and Kv1.6 channels were inhibited by 11.7 ± 1.9 and 17.4 ± 4.5%, respectively (n = 6). Interestingly, the same concentration of Tk-hefu-2 could inhibit the potassium current through Kv1.3 channels with 91.3 ± 1.4% (n = 8). Tk-hefu-3 could not inhibit any of the tested channels at concentrations up to 100 μm. 40 μm Tk-hefu-4 reduced the Kv1.3 currents by 71.8 ± 3.2% (n = 6). The same concentration of Tk-hefu-4 showed no activity on Kv1.1 and only minor activity on Kv1.2 and Kv1.6 (11.9 ± 1.2 and 4.9 ± 1.8%, respectively, n = 6; Fig. S2). The inhibitory effect of 10 μm Tk-hefu-2–4 on Kv1.3 channels is shown in Fig. 4A. Concentration-response curves were constructed to determine the concentrations at which half of Kv1.3 channels were blocked by Tk-hefu-2 and -4 compared with Tk-hefu. The IC50 values yielded 31.3 ± 5.1 μm for Tk-hefu (34.0 ± 2.8 μm reported previously (16)), 2.3 ± 0.4 μm for Tk-hefu-2, and 12.9 ± 1.8 μm for Tk-hefu-4 (n ≥ 6 for each point of the concentration-response curve; Fig. 4B). The parent peptide Tk-AMP-X2 showed no activity on Kv1.3 channels (n = 10).
Figure 4.
Tk-hefu-2, -3, and -4 block of human Kv1.3 channels studied by electrophysiology. A, activity of Tk-hefu mutants on Kv1.3 channels expressed in Xenopus laevis oocytes. Traces shown are representatives of at least six independent experiments (n ≥ 6). The dotted line indicates the zero current level. The asterisk (*) distinguishes the steady-state current after application of 10 μm peptide. B, concentration-response curves for Tk-AMP-X2, Tk-hefu, Tk-hefu-2, -3 and -4 on Kv1.3 channels obtained by plotting the percentage of remaining current as a function of increasing ligand concentrations. Error bars indicate the mean ± S.E. C, current-voltage relationship of Kv1.3 channels. Closed symbols, control condition; open symbols, after application of 2 μm Tk-hefu-2. D, percentage of inhibition upon application of 2 μm Tk-hefu-2 at a broad range of potentials is shown.
To investigate whether the observed current inhibition in Kv1.3 is attributed to the obstruction of the pore rather than to an altered channel gating upon ligand binding, the current-voltage relationship was investigated by constructing the current-voltage (I-V) curves (Fig. 4C). Application of 2 μm Tk-hefu-2 caused 48.2 ± 4.1% inhibition of the potassium current (n = 9). Tk-hefu-2 did not significantly alter the midpoint of activation because the V1/2 values yielded 15.8 ± 2.8 mV in control and 12.4 ± 1.9 mV after application of 2 μm peptide (n = 8). The inhibition of Kv1.3 channels induced by 2 μm Tk-hefu-2 was not voltage-dependent; no difference in the degree of block could be observed in the range of test potentials from −30 to +20 mV (Fig. 4D). Application of 2 μm Tk-hefu-2 resulted in a rapid decrease of the potassium current and binding was reversible upon washout (Fig. S3). Altogether, these experiments imply that current inhibition upon Tk-hefu-2 binding does not result from changes in the voltage dependence of channel gating. The inhibition of Kv1.3 channels occurred rapidly, and Tk-hefu-2 binding was reversible because the current recovered quickly and completely upon washout (data not shown).
Discussion
Small modifications to peptide sequence may bring about considerable structural change
Comparison of the parent peptide Tk-AMP-X2 (16) and Tk-hefu spatial structures established by NMR (Fig. 1) shows that small modification (three amino acid replacements; Table 1) led to a structural rearrangement. In particular, Tk-hefu is more tightly packed with α-helices running almost antiparallel to each other (∼160°), whereas Tk-AMP-X2 presents a less stabilized structure with crossed α-helices (∼130°). This rearrangement is easily explained by the new inter-helix contacts formed in Tk-hefu that act as staples in addition to the disulfide bonds: Tyr-6 from the N-terminal helix forms π-cation contacts with lysine residues from the C-terminal helix. The relative mobility of the α-hairpinin-fold should be kept in mind in future engineering experiments. This structural plasticity of α-hairpinins is not exceptional and many examples are known whereby small sequence changes bring about drastic impact on the 3D structure (28). For instance, subtle modifications to scorpion toxins affecting sodium channels are known to reassemble the structure of their “specificity modules” and switch taxon selectivity (29, 30).
Tk-hefu-2 mode of binding to Kv1.3
A large number of ion channel ligands are found in animal venoms. Perhaps the major source of potassium channel ligands is scorpion venoms, which may contain up to hundreds of components, mostly mini-proteins or peptides (19). Interestingly, the wide variety of potassium channel ligands from scorpion venom (KTx) are exceptionally pore blockers. Currently, ∼250 KTx are documented (20), and it is assumed that their variability is much greater. These peptides have different structural organization and can be grouped into five structural classes based on their fold: cysteine-stabilized α/β (CSαβ), cysteine-stabilized α/α with two disulfide bonds (2S-S CSαα), and CSαα with three disulfide bonds (3S-S CSαα), Kunitz, and inhibitor cystine knot. The 2S-S CSαα peptides are structurally similar to α-hairpinins and contain two antiparallel α-helices held together by two disulfide bonds.
For a large number of KTx, regardless of their structures, it is believed that a pair of amino acid residues (the functional dyad) is involved in ligand-receptor interaction and provides potassium channel blockage (26). This dyad is composed of two highly conserved residues, one of which is exclusively lysine, and the other may be tyrosine, phenylalanine, or leucine (31, 32). According to the functional dyad model, the conserved lysine side chain protrudes to the channel selectivity filter (the narrowest part of the outer side of the channel represented by rings of carbonyl oxygen atoms of the main chain; these atoms substitute the hydration shell oxygens surrounding potassium cations in aqueous solution), whereas the bulky tyrosine is needed to fix the toxin in the channel entrance vestibule; this was directly found in the complex between the classic CSαβ KTx ChTx and Kv1.2/2.1 paddle chimera channel (25).
Tk-hefu and Tk-hefu-2 peptides discussed here were designed based on the 2S-S CSαα KTx hefutoxin-1, which contains the classical functional dyad composed of Tyr-5 and Lys-19 (33). In our model of the Kv1.3–Tk-hefu complex (Fig. 2), Lys-22 interacts with the selectivity filter, whereas Tyr-6 fixes the ligand against the channel vestibule, as assumed in the functional dyad model. The important role of these two residues is supported by the computational analysis of their contribution to the interaction energy (Fig. 3) and confirmed by mutagenesis of Tk-hefu-2. Point mutation Y6E (yielding Tk-hefu-4) decreases the activity significantly, whereas the K22M mutation (as in Tk-hefu-3) results in a complete loss of function (Fig. 4). The relative importance of these two residues in Tk-hefu-2 is in accordance with data obtained for other KTx (26, 34).
To increase the binding affinity of Tk-hefu to Kv1.3 we inspected their interaction surfaces. Using the PST approach, we built Connolly maps of the concave and convex interacting surfaces of the channel and ligand approximating them to spherical projections and drawing the value of ELP in each point of the maps (Fig. 2). We then compared the produced maps and suggested a mutation to increase their complementarity. Quite to our surprise, this mutation reversed our initial modification to the parent Tk-AMP-X2 peptide used to produce Tk-hefu (K23E), and as a result the improved version Tk-hefu-2 actually differs by just two residues (the functional dyad) from the totally inactive wheat defense peptide Tk-AMP-X2.
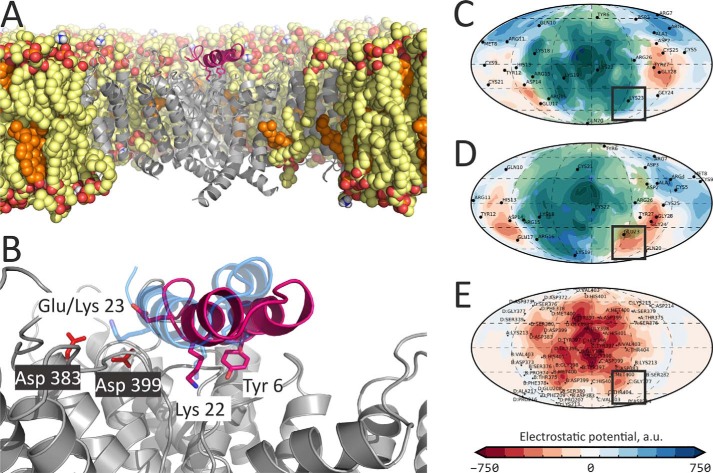
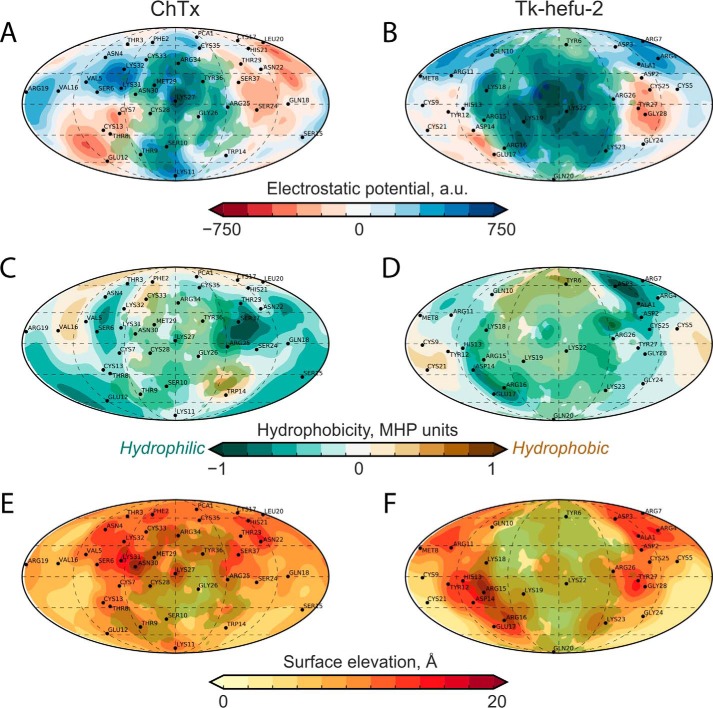
Further inspection of the ligand and channel interacting surfaces showed that the engineered complementarity brought about significant similarity of the resulting 2S-S CSαα Tk-hefu-2 peptide with the classic CSαβ KTx ChTx (Fig. 5). Although they exhibit drastically different folds, their functional surfaces interacting with the ion channel are quite similar as assessed by PST. Apparently, the α-hairpinin scaffold can be exploited to engineer ion channel ligands mimicking the classic channel blockers.
Figure 5.
Binding surface of Tk-hefu-2 is similar to ChTx. Protein surface topography permitted comparison of ChTx (left column; A, C, and E) and Tk-hefu-2 (right column; B, D, and F) surfaces with respect to ELP (A and B), molecular hydrophobicity potential (C and D), and relief (E and F). All map pairs are colored according to the scales below; panel B is the same as panel C in Fig. 2. Contact area with Kv1.3 is shaded green.
Outlook, protein surface topography as a tool to aid rational drug design
Perhaps because of Linus Pauling's (35) seminal contributions to the field of molecular recognition, the concept of complementarity has remained central to our understanding of how molecules form stable and selective complexes. Our recently proposed method of PST (21) is a powerful addition to the available toolkit to inspect complementarity in protein complexes. In this work, we relied upon ELP mapping, although it is widely recognized that other types of interactions play important roles in intermolecular recognition. The PST approach can be analogously used to map other properties, e.g. hydrophobicity, but it still remains just an elegant way to visualize and characterize peptides and proteins and their complexes, rather than an automated tool for prediction of favorable amino acid replacements. Here, we have demonstrated PST applicability to rational optimization of small protein ligands of ion channels. The proportion of biologics among marketed drugs is growing (36), but methods of medicinal chemistry, such as QSAR developed for small molecules (37), are underdeveloped for proteins. We argue that PST helps to overcome this deficit.
Experimental procedures
Expression vector construction
DNA sequences encoding Tk-hefu-2, -3, and -4 were constructed from a number of synthetic oligonucleotides (Table 3) by PCR. The previously obtained vector pET-32b–Tk-hefu (16) was used as a matrix. The target PCR fragments were amplified using a forward primer containing a KpnI restriction site and encoding an enteropeptidase cleavage site for fusion protein hydrolysis, and a reverse primer containing a BamHI restriction site and a stop codon. The PCR fragments were cloned into the expression vector pET-32b (Novagen) using respective restriction enzymes to produce pET-32b–Tk-hefu-2, -3, and -4.
Table 3.
Oligonucleotide primers used to synthesize genes encoding Tk-hefu derivatives
Restriction sites are underlined, stop codons are in italics, enteropeptidase cleavage site-encoding codons are in bold type, and the differing codons are shown on a gray background. The following pairs of primers were used to amplify the target sequences: Tk-hefu-2, f1 and rev1; Tk-hefu-3, f1 and rev2; and Tk-hefu-4, f2 and rev1.
Fusion protein expression and purification
E. coli BL21(DE3) cells were transformed using the expression vectors pET-32b–Tk-hefu-2, -3, and -4 and cultured at 37 °C in LB medium to the mid-log phase. Expression was then induced by 0.2 mm isopropyl β-d-thiogalactopyranoside. Cells were cultured at room temperature (24 °C) overnight (16 h) and harvested by centrifugation. The cell pellet was resuspended in 10 ml of 300 mm NaCl, 50 mm Tris-HCl buffer (pH 8.0) and ultrasonicated. The lysate was applied to a TALON Superflow resin (Clontech) and the fusion proteins Trx-Tk-hefu-2, -3, and -4 were purified according to the protocol supplied by the manufacturer.
To produce 15N-labeled Tk-hefu, M9 minimal medium with ISOGRO (Sigma) was used instead of LB medium. E. coli was first cultured in LB, harvested by centrifugation, and resuspended to A600 ∼ 0.1 in M9 medium with addition of ISOGRO (1 g/liter). Bacterial culture was then grown at 37 °C for ∼4 h to the mid-log phase (A600 ∼ 0.5). After induction the culture was kept at room temperature (24 °C) overnight (16 h).
Fusion protein cleavage and purification of the target peptides
Fusion proteins were dissolved in 50 mm Tris-HCl (pH 8.0) to a concentration of 1 mg/ml. Protein cleavage with human enteropeptidase light chain (1 unit of enzyme/1 mg of substrate) (23) was performed overnight (16 h) at 37 °C. Recombinant peptides were purified by reversed-phase HPLC on a Jupiter C5 column (10 × 250 mm; Phenomenex) in a linear gradient of acetonitrile concentration (0–60% in 60 min) in the presence of 0.1% TFA. The purity of the target peptides was checked by MS, N-terminal sequencing, and analytical chromatography on a Vydac 218TP54 C18 column (4.6 × 250 mm; Separations Group) in a shallow acetonitrile gradient.
Mass spectrometry
For molecular mass measurements an Ultraflex TOF-TOF (Bruker Daltonik) spectrometer was used as described previously (38). 2,5-Dihydroxybenzoic acid (Sigma) was used as a matrix. Measurements were performed in the linear mode with a mass-accuracy error not exceeding 100 ppm. Mass spectra were analyzed with the Data Analysis 4.3 and Data Analysis Viewer 4.3 software (Bruker).
NMR spectroscopy
To determine the spatial structure of Tk-hefu the 15N-labeled peptide was dissolved in either H2O/D2O (19:1) or 100% D2O. The concentration of the peptide was 1 mm, and pH was adjusted to 5.7. All NMR experiments were performed on an Avance 700 MHz spectrometer (Bruker Biospin) equipped with a cryoprobe at 30 °C. Unless otherwise stated, a relaxation delay of 1.4 s was used. The proton, nitrogen, and carbon carrier frequencies were adjusted to 4.7, 115.5, and 40 ppm, respectively. Spectral widths were equal to 10, 23, and 80 ppm in proton, nitrogen, and carbon dimensions. The Watergate technique (39) was applied to suppress strong solvent resonance in some spectra measured in H2O solution. 1H chemical shifts were measured relative to the protons of H2O, the chemical shift of their signal was chosen arbitrarily as 4.75. Chemical shifts of 13C and 15N were calculated from the respective gyromagnetic ratios. Proton and 15N resonance assignments for Tk-hefu were obtained by a standard procedure (40, 41) using 15N-HSQC (2 scans, 128 complex points), 3D 15N-TOCSY-HSQC (4 scans, 45 and 200 complex points), 3D 15N-NOESY-HSQC (8 scans, 45 and 150 complex points), and 13C-HSQC spectra (8 scans, 256 complex points) in the CARA software (42). The 3JHNHA coupling constants were determined from the intensity ratio of cross and diagonal peaks in the 3D HNHA spectrum (16 scans, 45 and 65 complex points) (43). The 3JHαHβ coupling constants were measured by a line shape analysis of Hα-Hβ cross-peaks in the DQF-COSY spectrum of Tk-hefu in D2O solution (relaxation delay of 3 s, 64 scans, 1024 complex points). 3JNHβ constants were calculated from the intensities of cross and diagonal peaks in the 3D HNHB experiment (8 scans, 40 and 65 complex points) (44).
NMR structure calculations
3D structure calculation was performed using the simulated annealing/molecular dynamics protocol as implemented in the CYANA software package version 3.0 (45). Upper interproton distance constraints were derived from NOESY (τm = 80 ms) cross-peaks via a 1/r6 calibration. Torsion angle restraints and stereospecific assignments were obtained from J coupling constants and NOE intensities. Hydrogen bonds were introduced based on temperature coefficients and water exchange rates of HN protons (protons with gradients less than 4.5 ppb/K were supposed to participate in hydrogen bonding). Temperature coefficients were measured in the range of 10–40 °C with the step of 5 degrees and the exchange rates were obtained using the CLEANEX experiment recorded with 20 ms mixing time and 64 complex points in the indirect dimension (46). The disulfide linkages were ascribed according to the structure of the parent peptide Tk-AMP-X2 (16, 47) and then confirmed by CYANA simulation. Visual analysis of the calculated structures and figure drawings were performed using the PyMOL software (48).
Kv1.3–Tk-hefu/Tk-hefu-2, -3, and -4 complexes modeling
Kv1.3 homology model was generated with Modeller 8.2 (49) using the Kv1.2 structure (PDB ID 3LUT) (24) as a template. Tk-hefu and Tk-hefu-2 complexes with Kv1.3 were modeled based on the crystal structure of the Kv1.2/2.1 paddle chimera in complex with ChTx (PDB ID 4JTA) (25). In this complex, the chimeric channel was replaced by our Kv1.3 model by structural alignment; furthermore, ChTx was replaced by either Tk-hefu or Tk-hefu-2, -3, and -4 by structural alignment of the appropriate dyads: Tyr-6 and Lys-22 of Tk-hefu onto the classical Tyr-36 and Lys-27 in ChTx with a slight manual adjustment. Tk-hefu-2, -3, and -4 structural model was obtained from the Tk-hefu NMR structure (reported in this paper) by in silico amino acid replacements E23K, K22M + E23K, and Y6E + E23K, respectively, introduced in PyMOL.
Molecular dynamics
The resulting Kv1.3–Tk-hefu/Tk-hefu-2, -3, and -4 complexes were placed inside a neuronal membrane-mimicking bilayer. We used an equilibrated fragment of bilayer (7 × 7 × 13.5 nm3; 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine/cholesterol, POPC/POPE/Ch, 100:50:50 molecules, respectively, solvated with 14,172 water molecules) that has been described in detail in our previous works (50, 51); some phospholipid and cholesterol molecules were removed to provide space for the protein. The TIP3P water model was used for resolvation; and the required number of Na+ ions to maintain electroneutrality was added. All systems were equilibrated (heated) during 100 ps of MD simulation. Positions of all Kv1.3 Cα atoms of residues, which are not involved in the channel pore vestibule, as well as the Nϵ atom of Lys-22 in Tk-hefu, Tk-hefu-2 and -4 (as well the thiomethyl carbon atom in Met-22 in Tk-hefu-3) were restrained during the equilibration. All simulations were performed with the GROMACS software (52) (version 5.1.2) using the AMBER99SB-ILDN parameters set. Simulations were carried out with a time step of 2 fs, imposing 3D periodic boundary conditions, in the isothermal-isobaric (NPT) ensemble with a semi-isotropic pressure of 1 bar, using the Berendsen pressure coupling algorithm, and at a constant temperature of 37 °C. The systems were then subjected to 110 ns of MD. The position of the Nϵ atom of Lys-22 (or the thiomethyl carbon atom of Met-22) in each complex was restrained inside the channel pore during the simulations.
Protein surface topography
A dynamic picture of Kv1.3–Tk-hefu/Tk-hefu-2 interaction with respect to ELP was assessed with the PST approach (21) implemented in our in-house IMPULSE software.5 Briefly, the 3D distribution of ELP on the target surface (here, the molecular surface of Tk-hefu or Tk-hefu-2) is transformed to 2D spherical projection maps, which are subjected to computational processing, e.g. MD-averaging. The resulting maps represent ELP distribution on the molecular surface of the peptide or its environment (here, the Kv1.3 channel). These maps suit well the visual and numerical assessment of matching physicochemical properties in complexes of interacting molecules, or the comparison of two or more similar molecules (e.g. bioactive peptides). In Fig. 2, C–E, such maps are provided. The axes of the projection were selected to place the pore-blocking residue of each peptide (Lys-22) in the center of the map; however, given that the residue marker is a projection of a single Lys atom from single MD frame, its position differs slightly from matching the exact center.
Interaction energy and intermolecular contacts
The Tk-hefu/Tk-hefu-2, -3, and -4 interaction energy profiles evaluation and intermolecular contacts identification within the modeled complexes in MD trajectories were performed using in-house software package IMPULSE.5 The AMBER99SB-ILDN parameters set and 1.5-nm cutoff distance for Lennard-Jones/electrostatic interactions were used during the intermolecular short-range nonbonded interaction energy estimation, the latter being the sum of the Lennard-Jones and electrostatic terms. Graphical representation of interaction energy profiles was performed using Python built-in libraries and the NumPy package.
Expression in Xenopus oocytes
For the expression of Kv genes (rat (r)Kv1.1 (GenBankTM accession number: NM173095), rKv1.2 (NM012970), human (h)Kv1.3 (L23499), and rKv1.6 (NM023954)) in Xenopus oocytes, linearized plasmids containing the respective genes were transcribed using the T7 or SP6 mMESSAGE-mMACHINE transcription kit (Ambion). Stage V–VI oocytes were harvested from an anesthetized female X. laevis frog. Oocytes were injected with 50 nl of cRNA at a concentration of 1 ng/nl using a micro-injector (Drummond Scientific). The oocytes were incubated in ND96 solution containing 96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 2 mm MgCl2, and 5 mm HEPES (pH 7.4), supplemented with 50 mg/liter of gentamycin sulfate.
Electrophysiological recordings
Two-electrode voltage-clamp recordings were performed at room temperature (18–22 °C) using a Geneclamp 500 amplifier (Molecular Devices) controlled by a pClamp data acquisition system (Axon Instruments) essentially as described (16). Whole-cell currents from oocytes were recorded 1–4 days after injection. Bath solution composition was ND96 or HK: 2 mm NaCl, 96 mm KCl, 1.8 mm CaCl2, 2 mm MgCl2, and 5 mm HEPES (pH 7.4). Voltage and current electrodes were filled with 3 m KCl. Resistances of both electrodes were kept between 0.7 and 1.5 mΩ.
The elicited currents were filtered at 0.5 kHz and sampled at 2 kHz using a four-pole low-pass Bessel filter. Leak subtraction was performed using a −P/4 protocol. Kv currents were evoked by a 250-ms depolarization to 0 mV followed by a 250-ms pulse to −50 mV, from a holding potential of −90 mV. To investigate the current-voltage relationship, currents were evoked by 10-mV depolarization steps from a holding potential of −90 mV. To assess the concentration dependence of the toxin-induced inhibitory effects, a concentration-response curve was constructed, in which the percentage of current inhibition was plotted as a function of toxin concentration. Data were fitted with the Hill equation: y = 100/[1 + (IC50/[toxin])h], where y is the amplitude of the toxin-induced effect, IC50 is toxin concentration providing 50% of block, [toxin] is toxin concentration, and h is the Hill coefficient. Comparison of two sample means was performed using a paired Student's t test (p < 0.05). All data represent at least 6 independent experiments (n ≥ 6) and are presented as mean ± S.E.
Author contributions
A. A. B., A. O. C., K. S. M., S. P., V. M. T., P. B. O., A. F. L., and E. V. N. investigation; A. A. B., A. O. C., K. S. M., S. P., V. M. T., P. B. O., and A. A. V. writing-original draft; A. O. C., K. S. M., and A. A. V. conceptualization; A. O. C., V. M. T., N. A. K., and R. G. E. software; A. O. C., K. S. M., V. M. T., and N. A. K. formal analysis; A. O. C., K. S. M., S. P., V. M. T., and A. A. V. validation; A. O. C., K. S. M., S. P., V. M. T., N. A. K., J. T., R. G. E., and A. A. V. methodology; A. O. C., J. T., R. G. E., and A. A. V. project administration; A. O. C. and A. A. V. writing-review and editing; K. S. M., A. S. A., E. V. G., J. T., R. G. E., and A. A. V. resources; K. S. M., S. P., V. M. T., and N. A. K. visualization; V. M. T. and N. A. K. data curation; A. S. A., E. V. G., J. T., R. G. E., and A. A. V. supervision; A. O. C., S. P., V. M. T., A. S. A., J. T., R. G. E., and A. A. V. funding acquisition.
Supplementary Material
Acknowledgments
Experiments were partially carried out using the equipment provided by the Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry core facility (CKP IBCH, supported by Russian Ministry of Education and Science Grant RFMEFI62117X0018). Computation of dynamic electrostatic maps and analysis of interaction energy were performed by VMT supported by Russian Science Foundation Grant 17-74-10172. We are grateful to O. Pongs for sharing the rKv1.1, rKv1.2, and rKv1.6 cDNA and M. L. Garcia for hKv1.3. We kindly thank Eugene A. Rogozhin from the Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry for protein sequencing. Access to computational facilities of the Supercomputer Center “Polytechnical” at the St. Petersburg Polytechnic University, Joint Supercomputer Center RAS (Moscow), and Shared Resource Center “Far Eastern Computing Resource” IACP FEB RAS is greatly appreciated.
This work was supported by grants from the Molecular and Cell Biology Program of the Russian Academy of Sciences (to A. A. V. and A. O. C.), Russian Academic Excellence Project “5–100” and the Basic Research Program at the National Research University Higher School of Economics (to R. G. E., A. O. C., and N. A. K.), F.W.O. Vlaanderen Grants GOC2319N and GOA4919N and CELSA/17/047 (BOF, KU Leuven) (to J. T.), and KU Leuven Grant PDM/19/164 (to S. P.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
The atomic coordinates and structure factors (code 5LM0) have been deposited in the Protein Data Bank (http://wwpdb.org/).
N. A. Krylov, unpublished data.
- PST
- protein surface topography
- CSαα
- cysteine-stabilized α/α-fold
- CSαβ
- cysteine-stabilized α/β-fold
- ChTx
- charybdotoxin
- ELP
- electrostatic potential
- KTx
- potassium channel ligand from scorpion venom
- Trx
- thioredoxin
- RMSD
- root mean square deviation
- PDB
- Protein Data Bank
- HSQC
- heteronuclear single quantum coherence.
References
- 1. Hille B. (2001) Ion channels of excitable membranes, 3rd Ed., Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 2. Jen J. C., Ashizawa T., Griggs R. C., and Waters M. F. (2016) Rare neurological channelopathies: networks to study patients, pathogenesis and treatment. Nat Rev Neurol. 12, 195–203 10.1038/nrneurol.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ptáćek L. J. (2015) Episodic disorders: channelopathies and beyond. Annu Rev Physiol. 77, 475–479 10.1146/annurev-physiol-021014-071922 [DOI] [PubMed] [Google Scholar]
- 4. Santos R., Ursu O., Gaulton A., Bento A. P., Donadi R. S., Bologa C. G., Karlsson A., Al-Lazikani B., Hersey A., Oprea T. I., and Overington J. P. (2017) A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16, 19–34 10.1038/nrd.2016.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaczorowski G. J., McManus O. B., Priest B. T., and Garcia M. L. (2008) Ion channels as drug targets: the next GPCRs. J. Gen. Physiol. 131, 399–405 10.1085/jgp.200709946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Urrego D., Tomczak A. P., Zahed F., Stühmer W., and Pardo L. A. (2014) Potassium channels in cell cycle and cell proliferation. Philos Trans. R. Soc. Lond. B Biol. Sci. 369, 20130094 10.1098/rstb.2013.0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feske S., Wulff H., and Skolnik E. Y. (2015) Ion channels in innate and adaptive immunity. Annu. Rev. Immunol. 33, 291–353 10.1146/annurev-immunol-032414-112212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Köhling R., and Wolfart J. (2016) Potassium channels in epilepsy. Cold Spring Harb. Perspect. Med. 6, a022871 10.1101/cshperspect.a022871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shah N. H., and Aizenman E. (2014) Voltage-gated potassium channels at the crossroads of neuronal function, ischemic tolerance, and neurodegeneration. Transl Stroke Res. 5, 38–58 10.1007/s12975-013-0297-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cahalan M. D., and Chandy K. G. (2009) The functional network of ion channels in T lymphocytes. Immunol. Rev. 231, 59–87 10.1111/j.1600-065X.2009.00816.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chandy K. G., and Norton R. S. (2017) Peptide blockers of Kv1.3 channels in T cells as therapeutics for autoimmune disease. Curr. Opin. Chem. Biol. 38, 97–107 10.1016/j.cbpa.2017.02.015 [DOI] [PubMed] [Google Scholar]
- 12. Beeton C., Wulff H., Standifer N. E., Azam P., Mullen K. M., Pennington M. W., Kolski-Andreaco A., Wei E., Grino A., Counts D. R., Wang P. H., LeeHealey C. J., S Andrews B., Sankaranarayanan A., et al. (2006) Kv1.3 channels are a therapeutic target for T cell-mediated autoimmune diseases. Proc. Natl. Acad. Sci. U.S.A. 103, 17414–17419 10.1073/pnas.0605136103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ortiz E., Gurrola G. B., Schwartz E. F., and Possani L. D. (2015) Scorpion venom components as potential candidates for drug development. Toxicon 93, 125–135 10.1016/j.toxicon.2014.11.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Obergrussberger A., Stölzle-Feix S., Becker N., Brüggemann A., Fertig N., and Möller C. (2015) Novel screening techniques for ion channel targeting drugs. Channels (Austin) 9, 367–375 10.1080/19336950.2015.1079675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lounnas V., Ritschel T., Kelder J., McGuire R., Bywater R. P., and Foloppe N. (2013) Current progress in structure-based rational drug design marks a new mindset in drug discovery. Comput. Struct. Biotechnol. J. 5, e201302011 10.5936/csbj.201302011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berkut A. A., Usmanova D. R., Peigneur S., Oparin P. B., Mineev K. S., Odintsova T. I., Tytgat J., Arseniev A. S., Grishin E. V., and Vassilevski A. A. (2014) Structural similarity between defense peptide from wheat and scorpion neurotoxin permits rational functional design. J. Biol. Chem. 289, 14331–14340 10.1074/jbc.M113.530477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oparin P. B., Mineev K. S., Dunaevsky Y. E., Arseniev A. S., Belozersky M. A., Grishin E. V., Egorov T. A., and Vassilevski A. A. (2012) Buckwheat trypsin inhibitor with helical hairpin structure belongs to a new family of plant defence peptides. Biochem. J. 446, 69–77 10.1042/BJ20120548 [DOI] [PubMed] [Google Scholar]
- 18. Nolde S. B., Vassilevski A. A., Rogozhin E. A., Barinov N. A., Balashova T. A., Samsonova O. V., Baranov Y. V., Feofanov A. V., Egorov T. A., Arseniev A. S., and Grishin E. V. (2011) Disulfide-stabilized helical hairpin structure and activity of a novel antifungal peptide EcAMP1 from seeds of barnyard grass (Echinochloa crus-galli). J. Biol. Chem. 286, 25145–25153 10.1074/jbc.M110.200378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuzmenkov A. I., Grishin E. V., and Vassilevski A. A. (2015) Diversity of potassium channel ligands: focus on scorpion toxins. Biochemistry (Mosc.) 80, 1764–1799 [DOI] [PubMed] [Google Scholar]
- 20. Kuzmenkov A. I., Krylov N. A., Chugunov A. O., Grishin E. V., and Vassilevski A. A. (2016) Kalium: a database of potassium channel toxins from scorpion venom. Database (Oxford) 2016, baw056 10.1093/database/baw056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koromyslova A. D., Chugunov A. O., and Efremov R. G. (2014) Deciphering fine molecular details of proteins' structure and function with a protein surface topography (PST) method. J. Chem. Inf. Model. 54, 1189–1199 10.1021/ci500158y [DOI] [PubMed] [Google Scholar]
- 22. Chugunov A. O., Koromyslova A. D., Berkut A. A., Peigneur S., Tytgat J., Polyansky A. A., Pentkovsky V. M., Vassilevski A. A., Grishin E. V., and Efremov R. G. (2013) Modular organization of α-toxins from scorpion venom mirrors domain structure of their targets, sodium channels. J. Biol. Chem. 288, 19014–19027 10.1074/jbc.M112.431650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gasparian M. E., Ostapchenko V. G., Schulga A. A., Dolgikh D. A., and Kirpichnikov M. P. (2003) Expression, purification, and characterization of human enteropeptidase catalytic subunit in Escherichia coli. Protein Expr. Purif. 31, 133–139 10.1016/S1046-5928(03)00159-1 [DOI] [PubMed] [Google Scholar]
- 24. Chen X., Wang Q., Ni F., and Ma J. (2010) Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement. Proc. Natl. Acad. Sci. U.S.A. 107, 11352–11357 10.1073/pnas.1000142107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Banerjee A., Lee A., Campbell E., and Mackinnon R. (2013) Structure of a pore-blocking toxin in complex with a eukaryotic voltage-dependent K+ channel. Elife 2, e00594 10.7554/eLife.00594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dauplais M., Lecoq A., Song J., Cotton J., Jamin N., Gilquin B., Roumestand C., Vita C., de Medeiros C. L., Rowan E. G., Harvey A. L., and Ménez A. (1997) On the convergent evolution of animal toxins: conservation of a diad of functional residues in potassium channel-blocking toxins with unrelated structures. J. Biol. Chem. 272, 4302–4309 10.1074/jbc.272.7.4302 [DOI] [PubMed] [Google Scholar]
- 27. McCoy J., and Lavallie E. (2001) Expression and purification of thioredoxin fusion proteins. Curr. Protoc. Mol. Biol. Chapter 16, Unit 6.7 [DOI] [PubMed] [Google Scholar]
- 28. Chi P. B., and Liberles D. A. (2016) Selection on protein structure, interaction, and sequence. Protein Sci. 25, 1168–1178 10.1002/pro.2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kahn R., Karbat I., Ilan N., Cohen L., Sokolov S., Catterall W. A., Gordon D., and Gurevitz M. (2009) Molecular requirements for recognition of brain voltage-gated sodium channels by scorpion alpha-toxins. J. Biol. Chem. 284, 20684–20691 10.1074/jbc.M109.021303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuldyushev N. A., Mineev K. S., Berkut A. A., Peigneur S., Arseniev A. S., Tytgat J., Grishin E. V., and Vassilevski A. A. (2018) Refined structure of BeM9 reveals arginine hand, an overlooked structural motif in scorpion toxins affecting sodium channels. Proteins 86, 1117–1122 10.1002/prot.25583 [DOI] [PubMed] [Google Scholar]
- 31. Shakkottai V. G., Regaya I., Wulff H., Fajloun Z., Tomita H., Fathallah M., Cahalan M. D., Gargus J. J., Sabatier J. M., and Chandy K. G. (2001) Design and characterization of a highly selective peptide inhibitor of the small conductance calcium-activated K+ channel, SkCa2. J. Biol. Chem. 276, 43145–43151 10.1074/jbc.M106981200 [DOI] [PubMed] [Google Scholar]
- 32. Gasparini S., Danse J. M., Lecoq A., Pinkasfeld S., Zinn-Justin S., Young L. C., de Medeiros C. C., Rowan E. G., Harvey A. L., and Ménez A. (1998) Delineation of the functional site of α-dendrotoxin: the functional topographies of dendrotoxins are different but share a conserved core with those of other Kv1 potassium channel-blocking toxins. J. Biol. Chem. 273, 25393–25403 10.1074/jbc.273.39.25393 [DOI] [PubMed] [Google Scholar]
- 33. Srinivasan K. N., Sivaraja V., Huys I., Sasaki T., Cheng B., Kumar T. K., Sato K., Tytgat J., Yu C., San B. C., Ranganathan S., Bowie H. J., Kini R. M., and Gopalakrishnakone P. (2002) κ-Hefutoxin1, a novel toxin from the scorpion Heterometrus fulvipes with unique structure and function: importance of the functional diad in potassium channel selectivity. J. Biol. Chem. 277, 30040–30047 10.1074/jbc.M111258200 [DOI] [PubMed] [Google Scholar]
- 34. Goldstein S. A., Pheasant D. J., and Miller C. (1994) The charybdotoxin receptor of a Shaker K+ channel: peptide and channel residues mediating molecular recognition. Neuron 12, 1377–1388 10.1016/0896-6273(94)90452-9 [DOI] [PubMed] [Google Scholar]
- 35. Pauling L. (1974) Molecular basis of biological specificity. Nature 248, 769–771 10.1038/248769a0 [DOI] [PubMed] [Google Scholar]
- 36. Morrison C. (2017) Fresh from the biotech pipeline-2016. Nat. Biotechnol. 35, 108–112 10.1038/nbt.3783 [DOI] [PubMed] [Google Scholar]
- 37. Cumming J. G., Davis A. M., Muresan S., Haeberlein M., and Chen H. (2013) Chemical predictive modelling to improve compound quality. Nat. Rev. Drug Discov. 12, 948–962 10.1038/nrd4128 [DOI] [PubMed] [Google Scholar]
- 38. Vassilevski A. A., Kozlov S. A., Samsonova O. V., Egorova N. S., Karpunin D. V., Pluzhnikov K. A., Feofanov A. V., and Grishin E. V. (2008) Cyto-insectotoxins, a novel class of cytolytic and insecticidal peptides from spider venom. Biochem. J. 411, 687–696 10.1042/BJ20071123 [DOI] [PubMed] [Google Scholar]
- 39. Piotto M., Saudek V., and Sklenár V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2, 661–665 10.1007/BF02192855 [DOI] [PubMed] [Google Scholar]
- 40. Wüthrich K. (1986) NMR of proteins and nucleic acids, Wiley, New York [Google Scholar]
- 41. Cavanagh J. (2007) Protein NMR spectroscopy: principles and practice, 2nd Ed., Academic Press, Amsterdam [Google Scholar]
- 42. Keller R. (2004) The Computer Aided Resonance Assignment Tutorial, CANTINA Verlag, Zurich [Google Scholar]
- 43. Vuister G. W., and Bax A. (1993) Quantitative J correlation: a new approach for measuring homonuclear three-bond J(HNH.alpha.) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc. 115, 7772–7777 10.1021/ja00070a024 [DOI] [Google Scholar]
- 44. Düx P., Whitehead B., Boelens R., Kaptein R., and Vuister G. W. (1997) Measurement of 15N-1H coupling constants in uniformly 15N-labeled proteins: application to the photoactive yellow protein. J. Biomol. NMR 10, 301–306 10.1023/A:1018393225804 [DOI] [PubMed] [Google Scholar]
- 45. Herrmann T., Güntert P., and Wüthrich K. (2002) Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR 24, 171–189 10.1023/A:1021614115432 [DOI] [PubMed] [Google Scholar]
- 46. Hwang T. L., van Zijl P. C., and Mori S. (1998) Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J. Biomol. NMR 11, 221–226 10.1023/A:1008276004875 [DOI] [PubMed] [Google Scholar]
- 47. Utkina L. L., Andreev Y. A., Rogozhin E. A., Korostyleva T. V., Slavokhotova A. A., Oparin P. B., Vassilevski A. A., Grishin E. V., Egorov T. A., and Odintsova T. I. (2013) Genes encoding 4-Cys antimicrobial peptides in wheat Triticum kiharae Dorof. et Migush.: multimodular structural organization, instraspecific variability, distribution and role in defence. FEBS J. 280, 3594–3608 10.1111/febs.12349 [DOI] [PubMed] [Google Scholar]
- 48. DeLano W. (2002) The PyMOL User's manual. DeLano Scientific, San Carlos, CA [Google Scholar]
- 49. Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., and Sali A. (2000) Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29, 291–325 10.1146/annurev.biophys.29.1.291 [DOI] [PubMed] [Google Scholar]
- 50. Lyukmanova E. N., Shenkarev Z. O., Shulepko M. A., Paramonov A. S., Chugunov A. O., Janickova H., Dolejsi E., Dolezal V., Utkin Y. N., Tsetlin V. I., Arseniev A. S., Efremov R. G., Dolgikh D. A., and Kirpichnikov M. P. (2015) Structural insight into specificity of interactions between nonconventional three-finger weak toxin from Naja kaouthia (WTX) and muscarinic acetylcholine receptors. J. Biol. Chem. 290, 23616–23630 10.1074/jbc.M115.656595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chugunov A. O., Volynsky P. E., Krylov N. A., Nolde D. E., and Efremov R. G. (2016) Temperature-sensitive gating of TRPV1 channel as probed by atomistic simulations of its trans- and juxtamembrane domains. Sci. Rep. 6, 33112 10.1038/srep33112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., and Berendsen H. J. (2005) GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.