Abstract
Precise modification and processing of rRNAs are required for the production of ribosomes and accurate translation of proteins. Small nucleolar ribonucleoproteins (snoRNPs) guide the folding, modification, and processing of rRNAs and are thus critical for all eukaryotic cells. Bcd1, an essential zinc finger HIT protein functionally conserved in eukaryotes, has been implicated as an early regulator for biogenesis of box C/D snoRNPs and controls steady-state levels of box C/D snoRNAs through an unknown mechanism. Using a combination of genetic and biochemical approaches, here we found a conserved N-terminal motif in Bcd1 from Saccharomyces cerevisiae that is required for interactions with box C/D snoRNAs and the core snoRNP protein, Snu13. We show that both the Bcd1–snoRNA and Bcd1–Snu13 interactions are critical for snoRNP assembly and ribosome biogenesis. Our results provide mechanistic insight into Bcd1 interactions that likely control the early steps of snoRNP maturation and contribute to the essential role of this protein in maintaining the steady-state levels of snoRNAs in the cell.
Keywords: ribosome assembly, RNA, RNA binding protein, RNA modification, RNA methylation, small nucleolar RNA (snoRNA), ribonuclear protein (RNP), assembly factor, box C/D snoRNA, snoRNP assembly, Bcd1, Snu13, small nucleolar ribonucleoprotein (snoRNP) assembly, zinc finger HIT-type containing 6 (ZNHIT6), ribosome biogenesis, posttranscriptional regulation, rRNA processing
Introduction
Ribosomes are essential and conserved macromolecular machines that catalyze the production of proteins in all cells. Ribosome integrity is extensively monitored through its biogenesis in a highly regulated process that involves nearly 200 assembly factors (1, 2). A critically important class of assembly factors for ribosome biogenesis is small nucleolar ribonucleoproteins (snoRNPs)3 that ensure the proper folding, modification, and processing of ribosomal RNAs for the coordinated binding of ribosomal proteins (3, 4). snoRNPs fall into two major groups based on their conserved RNA elements: box H/ACA snoRNPs direct the isomerization of uridines into pseudouridines, whereas box C/D snoRNPs guide the 2′-O-methylation of target RNAs. In these complexes, a small nucleolar RNA (snoRNA) binds to a set of essential core proteins to form the catalytically active snoRNP (5).
Eukaryotic box C/D snoRNPs contain four core proteins: Snu13, Nop56, Nop58, and the methyltransferase Nop1 in yeast (SNU13, NOP56, NOP58, and FBL/fibrillarin in humans). These proteins are proposed to assemble on the snoRNA in a hierarchal manner (6–9), in a process that is regulated by the action of several assembly factors (10). In yeast, these assembly factors include the Hsp90/R2TP (Rvb1, Rvb2, Tah1, and Pih1) chaperone/co-chaperone system and the proteins Rsa1, Hit1, and Bcd1 (11–20). However, how these assembly factors drive the biogenesis of snoRNPs is not understood.
Among the assembly factors linked to box C/D snoRNP biogenesis, Bcd1 is essential and specific to this process, whereas others are implicated in the biogenesis of other RNPs (16, 21–23). Examples are the involvement of Rsa1 in assembly of the large ribosomal subunits (24) and the participation of R2TP in diverse cellular processes including transcription, chromatin remodeling, phosphatidylinositol-3 kinase-related protein kinase (PIKK) signaling, mitotic spindle assembly, and apoptosis (25–27).
The importance of Bcd1 as a factor required for box C/D snoRNA accumulation was first revealed using a tetO7 shutoff allele of the essential BCD1 gene in a microarray screen that was designed to monitor the abundance and processing of noncoding RNAs (22, 23). Later, RNAi-mediated depletion of the human homologue of BCD1, ZNHIT6 indicated the conservation of Bcd1/ZNHIT6 function for maintaining box C/D snoRNA levels in human cells (11). Several more recent studies have identified the network of binding proteins for Bcd1 and ZNHIT6 (11–13, 21) including Rvb2, Pih1, Rsa1, Nop58, and Snu13 in yeast cells (11, 12) and RUVBL1/RUVBL2, PIH1, NUFIP, ZNHIT3, NOP58, and SNU13 in human cells (13). However, the mechanism of these interactions and the chronology of the binding events remain largely unclear.
Bcd1/ZNHIT6 are thought to be involved at early steps during the assembly of box C/D snoRNPs (13, 21). Using quantitative proteomics in human cell lines, ZNHIT6 was identified as part of an early protein-only complex together with assembly factors RUVBL1/2, NUFIP, ZNHIT3, and the core proteins SNU13 and NOP58 (13). More recently, depletion of Bcd1 was shown to result in a significant loss of interactions between Rsa1 and Nop58, suggesting that Bcd1 participates in loading of Nop58 into an early pre-snoRNP complex (21). Furthermore, Bcd1 controls the interaction of several snoRNP-related proteins with C/D snoRNAs (21). Although depletion of BCD1 was shown to have minimal effect on the amount of Snu13 and Nop56 associated with box C/D snoRNAs, it significantly reduced the snoRNA association of assembly factors Rvb2, Pih1 and Rsa1, and the core proteins Nop58 and Nop1 (21). It is unclear how Bcd1 coordinates all these protein-protein and protein-RNA interactions and regulates the steady-state level of snoRNAs. Intriguingly, Bcd1 is reported to interact with RNAs directly in a nonspecific manner, and to bind to longer RNAs with higher affinity (16). Together, how Bcd1 gains specificity for assembly of box C/D snoRNPs and the significance of Bcd1-RNA interaction for maintaining snoRNA levels remain unclear.
Here we define the domain organization of Bcd1/ZNHIT6 and use a combination of genetic and biochemical approaches to show that Bcd1 interacts with snoRNAs and Snu13 via a conserved element near its N-terminal region. Further, we demonstrate that perturbation of these interactions leads to a significant decrease in steady-state levels of box C/D snoRNAs as well as defects in ribosome biogenesis. Together, our data provide insights into Bcd1-mediated interactions with box C/D snoRNAs and reveal their significance for snoRNP production and ribosome biogenesis.
Results
Bcd1 and ZNHIT6 share similar domains that are arranged differently
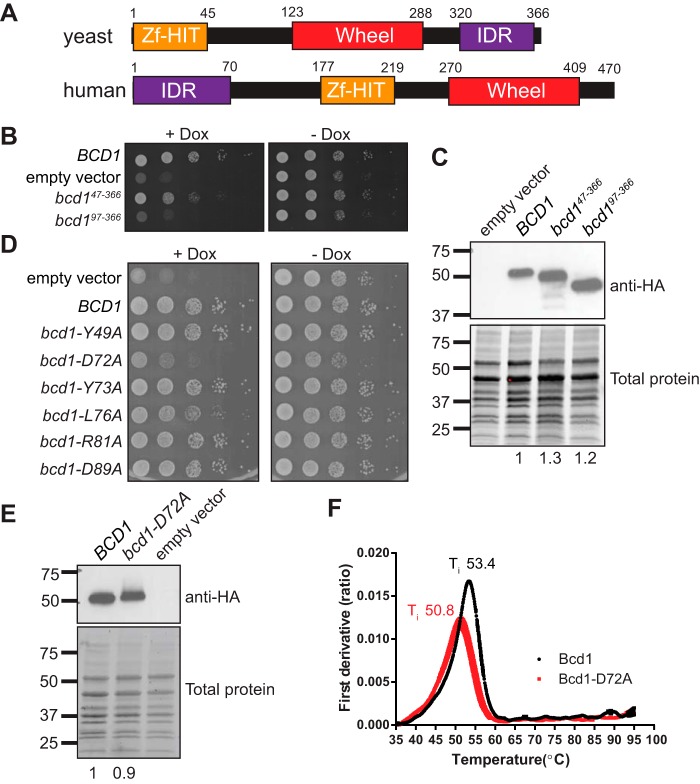
Yeast Bcd1 is an essential protein (22, 28) with a zinc finger HIT domain (Zf-HIT) at its N terminus (residues 1–45) which has high structural homology to the Zf-HIT domain of Hit1 (16). The Zf-HIT domain is important for the stability of Bcd1 (16) and mediates the interaction of ZNHIT6 with RUVBL1/2 (29). We reasoned that because deletion of Zf-HIT domain of Bcd1 is not lethal in yeast (16), additional elements must contribute to the essential function of the protein. To identify the important domains of Bcd1, we first performed a sequence alignment across eukaryotes using Clustal Omega, followed by a domain analysis using IUPred and Phyre2 (30–32). Bcd1 proteins fall into two groups (Fig. 1A): the first group, exemplified by human Bcd1 (ZNHIT6), contains an intrinsically disordered region (IDR) at its N terminus (residues 1 to ∼70 of ZNHIT6), a Zf-HIT domain in the middle (residues 215–258), and a wheel domain (residues 304–452). The wheel domain consists of a twisted five-stranded β sheet surrounded by several α helices and was predicted based on homology to a newly identified domain in Cns1, an essential Hsp90 co-chaperone (33). The second group, exemplified by yeast Bcd1 (Fig. 1A), contains a Zf-HIT domain at its N terminus (residues 1–45) (16), followed by a wheel domain (123–288) and an intrinsically disordered region at the C terminus (310–366). Thus, Bcd1 and ZNHIT6 share similar domains that are organized differently.
Figure 1.
A conserved residue in the N-terminal region of Bcd1 is important for cell growth. A, domain organization of Bcd1 and ZNHIT6. B, serial dilution growth assay of yeast expressing BCD1 under a doxycycline (Dox) repressible promoter (tetO7::BCD1), and supplemented with the indicated plasmids. C, Western blot analysis of Bcd1 and its variants. Total protein was resolved on a Mini-PROTEAN TGX Stain-Free gel (Bio-Rad) and imaged prior to the transfer to serves as loading control. Anti-HA intensity is normalized to the total protein and reported under each lane. D, serial dilution growth assay of tetO7::BCD1 cells supplemented with the indicated plasmids. E, Western blot analysis of Bcd1 and Bcd1-D72A. The anti-HA levels relative to the loading control is reported under each lane. F, the inflection temperature (Ti) of Bcd1 and Bcd1-D72A as a function of increasing temperature. The peaks in the first derivative view correspond to the Ti of the proteins.
A conserved region in the N terminus of Bcd1 is essential for yeast viability
Previous studies showed that expression of the Zf-HIT domain of Bcd1 (residues 1–45) alone does not support the growth of yeast depleted of endogenous BCD1 (16). However, a fragment encompassing residues 1–96 of Bcd1 is sufficient to maintain cell viability in the absence of BCD1 (21). To investigate the reason for this growth rescue, we first checked the expression of Bcd1 fragments lacking either the Zf-HIT domain (bcd147–366) or residues 1–97 (bcd197–366) in cells depleted of the endogenous BCD1 via a doxycycline-repressible promoter (tetO7::BCD1). As expected, the expression of bcd147–366 partially rescued the lethality of BCD1-depleted cells. Cells expressing bcd197–366 grew similarly to cells expressing an empty vector, implicating that Bcd1 residues 47–97 are important for cell viability (Fig. 1B). To confirm that the observed growth defects are not because of lack of protein expression, we assayed the expression of the two variant proteins (Bcd147–366 and Bcd197–366) by Western blotting (Fig. 1C). These data indicated that the two Bcd1 variants were expressed and their levels were similar to WT Bcd1.
To identify which Bcd1 residues are critical for function, we individually mutated highly conserved amino acids within this region to alanine (Fig. S1) and checked the effect of each mutation on the growth of yeast cells depleted of their endogenous BCD1 and supplemented with a plasmid expressing the variant protein. Of all the conserved residues tested, the alanine substitution of Asp-72 (bcd1-D72A) showed a strong growth phenotype that resembled the growth defect observed in cells expressing vector only (Fig. 1D). Mutation of Leu-76 to alanine (L76A) also conferred a growth phenotype, albeit to a much lesser extent than D72A, indicating the overall importance of the region-spanning residues 72–76 (Fig. 1D). It is noteworthy that mutating the highly conserved residue Tyr-73 within this region did not show any effect on cell growth (Fig. 1D), indicating the specific importance of Asp-72, and to a lesser extent, Leu-76 for Bcd1 function. Because the effect from D72A mutation on cell growth was much greater than L76A, we focused on this residue for further analyses.
To check whether the observed phenotype in cells expressing Bcd1-D72A was a result of protein destabilization because of the mutation, we compared the expression levels of WT and D72A proteins by Western blotting. These analyses confirmed that the protein variant was expressed to similar levels as WT Bcd1 (Fig. 1E). To compare the stability of the D72A variant and WT Bcd1 proteins, we overexpressed and purified each protein and measured their thermal unfolding (Fig. 1F). Bcd1 and Bcd1-D72A show similar apparent melting temperatures (ΔTi 2.6 °C, Fig. 1F), suggesting the D72A mutation does not lead to a global destabilization of the protein and instead likely affects Bcd1 function.
D72A disrupts the Bcd1 binding to the core snoRNP protein, Snu13
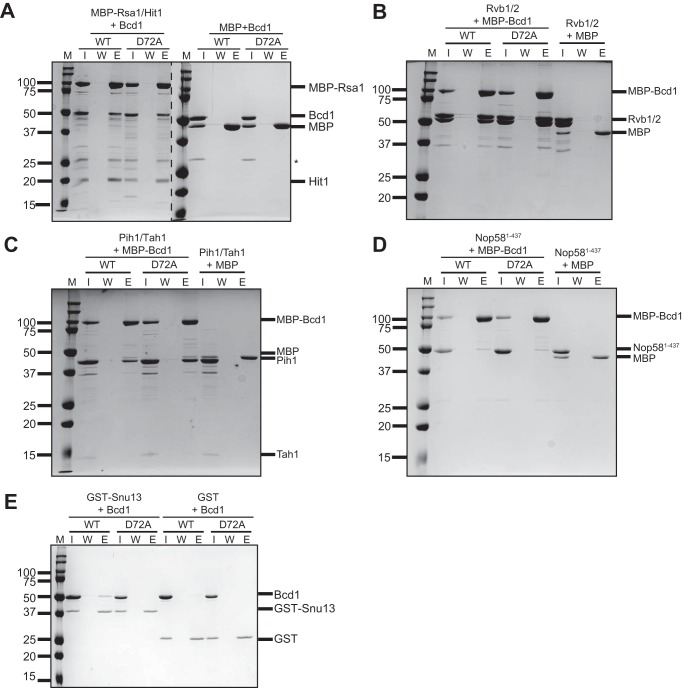
Bcd1 interacts with core box C/D snoRNP proteins Nop58 and Snu13 and assembly factors Rvb2, Pih1, and Rsa1 (11, 12). The interaction network of Bcd1/ZNHIT6 has been established using in vitro pulldown experiments and quantitative proteomic studies in cell culture (13). However, the coordination of the events remains largely unknown (10). We reasoned that if Bcd1 interactions with its binding partners are essential, the growth defect observed upon expression of Bcd1-D72A may arise from disruption of these important interactions. To test this, we performed pulldown assays to compare the binding interaction of WT and D72A variant of Bcd1 to its known binding partners (Fig. 2). We purified the core proteins individually and co-purified Rvb1 with Rvb2, Tah1 with Pih1, and MBP-tagged Rsa1 with Hit1 (removal of the MBP moiety renders the Rsa1/Hit1 complex unstable). Because the C terminus of Nop58 is highly charged and intrinsically disordered, we could not purify the protein to high purity and yield and therefore used a Nop58 variant that lacks the last 73 amino acids of the protein (Nop581–437).
Figure 2.
In vitro protein-binding assays testing the interaction network of Bcd1 with its binding partners. A, MBP-Rsa1/Hit1 immobilized on amylose resin can pull down WT and D72A variant Bcd1. B–D, MBP-Bcd1 (WT) or MBP-Bcd1-D72A (D72A) bound to amylose resin pulldowns Rvb1/2 (B), Nop58 (C), and Tah1/Pih1 (D). E, GST-Snu13 immobilized on GSH Sepharose resin can pull down WT but fails to interact with Bcd1-D72A. M, molecular weight marker (kDa); I, input; W, last wash; E, eluate; *, nonspecific contaminant.
First, the complex of MBP-Rsa1/Hit1 was immobilized on amylose resin and incubated with excess of either WT or D72A variant of Bcd1. After extensive washing of the resin, the retained proteins were eluted and analyzed on SDS-PAGE. We did not observe any significant difference in the binding of the D72A variant versus WT Bcd1 to MBP-Rsa1/Hit1 (Fig. 2A). We next immobilized MBP-tagged Bcd1 or Bcd1-D72A on amylose resin and incubated it with either Rvb1/Rvb2 complex (Fig. 2B), Tah1/Pih1 complex (Fig. 2C), or Nop581–438 (Fig. 2D). Rvb1/Rvb2 and Tah1/Pih1 demonstrated robust binding to either MBP-Bcd1 WT or D72A (Fig. 2, B and C). In contrast, Nop581–437 bound weakly to both (Fig. 2D). To confirm these findings, we performed the reciprocal pulldown by immobilizing GST-Nop581–437 or GST-Tah1/Pih1 or GST alone on GSH Sepharose beads and incubated them WT and D72A variant of Bcd1. As expected, both WT and D72A variant of Bcd1 were pulled down by GST-Nop581–437 or GST-Tah1/Pih1 but not by GST alone (Fig. S2, A and B). We did not perform the reciprocal pulldown with Rvb1/2 as tagging these proteins can interfere with their oligomeric state (34). Strikingly, when Bcd1 or Bcd1-D72A were tested for their interaction with GST-Snu13, no binding was observed for Bcd1-D72A (Fig. 2E). In the reciprocal pulldown, untagged Snu13 did not bind to either WT MBP-Bcd1 or D72A variant (data not shown). It is likely that the N-terminal region of Bcd1 is responsible for establishing its transient interaction with Snu13 and hence a large tag such as MBP could hinder this interaction. Collectively, these in vitro binding assays indicate that the Bcd1-D72A variant abolishes binding to Snu13, but does not impact interactions with Rvb1/2, Tah1/Pih1, or Nop58.
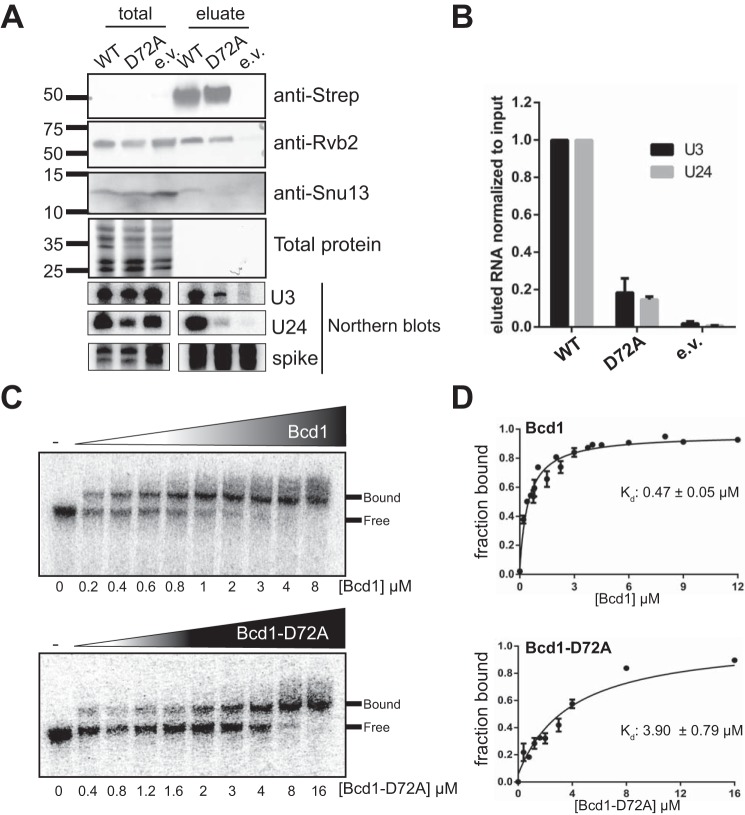
To confirm these results further, we expressed N-terminally Strep-tagged BCD1 or Bcd1-D72A in yeast cells and purified the protein from mid-log phase yeast total lysates and analyzed the proteins that co-eluted using Western blotting. Although WT Bcd1 co-purified both Rvb2 and Snu13 (Fig. 3A), the Bcd1-D72A variant bound to Rvb2 but failed to co-purify Snu13 (Fig. 3A), further corroborating the results of pulldown assays performed using recombinant proteins (Fig. 2E). To further support this finding, we performed the reciprocal pulldown using an N-terminally Strep-tagged Snu13 overexpressed in cells expressing HA-tagged WT or D72A variant of Bcd1. Analysis of the eluted proteins by Western blotting revealed that Strep-Snu13 can pull down Bcd1 from WT, but not bcd1-D72A cells (Fig. S2C). Taken together, these data confirm the results of in vitro binding assays that D72A variant fails to interact with Snu13.
Figure 3.
Mutation of a conserved residue of Bcd1 hampers the ability of the protein to interact with RNA. A, strep-tagged Bcd1 (WT) or Bcd1-D72A was used to pull down snoRNPs from yeast cells. Protein and RNA from input and eluates were analyzed by Western and Northern blotting. B, quantification of the eluted RNA relative to input is shown in A. Data from two independent experiments were averaged. Error bars indicate mean ± S.E. C, EMSA shows that Bcd1 binds to snR51 (a box C/D snoRNA), albeit weakly, whereas Bcd1-D72A binding to the snoRNA is weaker. D, quantification of the data shown in C. Data show averages from four independent experiments. Errors bars indicate mean ± S.E.
Bcd1-D72A variant is impaired in binding to RNA
Bcd1 interacts with RNAs in a nonspecific manner (16), yet it contributes specifically to the maintenance of box C/D snoRNA levels in the cell (22, 23). To test if the severe growth defect observed upon expression of Bcd1-D72A may partially arise from the loss of interaction of Bcd1 with RNA, we compared the RNAs co-purified with WT Strep-tagged Bcd1 and D72A variants (see above) using Northern blotting. Although the levels of purified WT Bcd1 and Bcd1-D72A are similar, significantly less U3 and U24 snoRNAs were bound to Bcd1-D72A as compared with Bcd1-WT (Fig. 3, A and B). To corroborate this in vivo finding, we in vitro transcribed a box C/D snoRNA (snR51) and compared its affinity for Bcd1 and Bcd1-D72A using EMSAs. Although WT Bcd1 binds to snR51 with a Kd of 0.47 ± 0.05 μm, the D72A variant binds more weakly (Kd 3.90 ± 0.79 μm) (Fig. 3, C and D). Taken together, these data suggest that Bcd1 Asp-72 affects the ability of the protein to interact with snoRNAs.
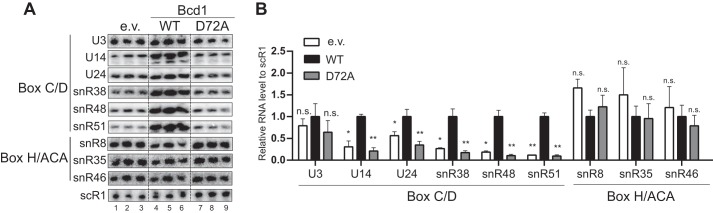
Bcd1D72A abolishes production of box C/D snoRNAs
Bcd1 is essential for the maintenance of box C/D snoRNA levels (22, 23). We therefore reasoned that the severe growth phenotype observed upon expressing Bcd1-D72A may be a result of perturbing box C/D snoRNA levels. To test this hypothesis, we used Northern blotting to analyze the steady-state levels of six box C/D snoRNAs in mid-log phase cells expressing WT BCD1, bcd1-D72A, or an empty vector (Fig. 4, A and B). These were compared with levels of scR1, the RNA component of the signal recognition particle, and three box H/ACA snoRNAs as controls (Fig. 4). As previously observed in the cells depleted of BCD1 (21–23), lower levels of the tested box C/D snoRNAs, and unchanged levels of H/ACA snoRNAs and scR1 RNA were observed when an empty vector was expressed (Fig. 4). Similarly, in the cells expressing Bcd1-D72A, the levels of the tested box C/D snoRNAs decreased (Fig. 4). This change was unique to box C/D snoRNAs as the three box H/ACA snoRNAs tested did not show significant variations in cells expressing Bcd1-D72A. Because the expression level of Bcd1 and Bcd1-D72A is similar (Fig. 1E), the significant drop in the level of box C/D snoRNAs strongly suggests that the conserved Bcd1 Asp72 is critical for maintaining the cellular levels of box C/D snoRNAs.
Figure 4.
Bcd1-D72A variant abrogates the production of box C/D snoRNAs. A, Northern blotting analyses of steady-state expression levels of box C/D and box H/ACA snoRNAs in tetO7::BCD1 cells treated with doxycycline and containing an empty vector (lanes 1–3), or a vector expressing Bcd1 (lanes 4–6), or Bcd1-D72A (lanes 7–9). B, quantification of data shown in A as normalized to scR1. Lane 6 was excluded from data analyses. Graph bars represent the mean and S.D. from at least two biological replicates. Significance is relative to the WT and was determined using an unpaired t test. n.s., nonsignificant; p > 0.05; *, p < 0.05; **, p < 0.01.
Dysregulation of Bcd1 impairs ribosome biogenesis
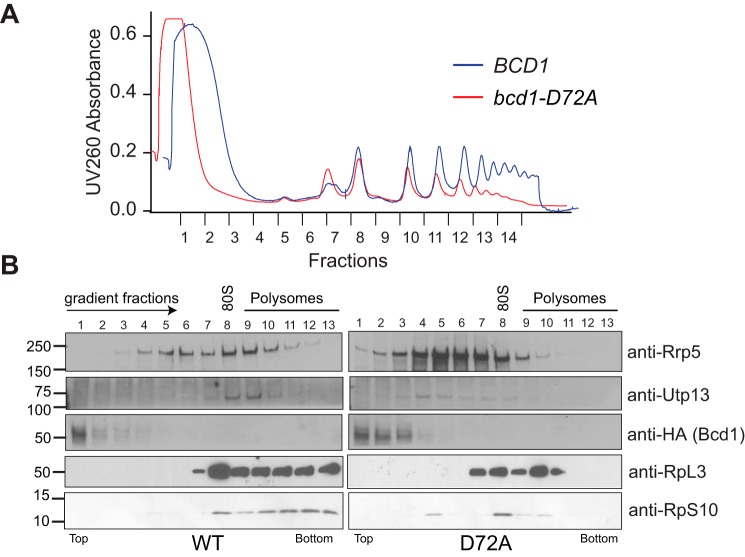
Box C/D snoRNAs are active players in the assembly of ribosomes and direct the folding, processing, and 2′-O-methylation of rRNAs during transcription and ribosomal protein binding (5). Because of their critical role in ribosome assembly, alteration of snoRNA levels can have profound effects on cell viability, especially in rapidly dividing cells such as cancer cells (35, 36). To assess the significance of Bcd1 interactions with Snu13 and snoRNAs for snoRNP biogenesis and ribosome production, we grew cells expressing Bcd1 or the Bcd1-D72A variant to mid-log phase and stalled translation by addition of cycloheximide. Polysome profiles on 10–50% sucrose gradients revealed a significant decrease in polysomes and an increase in the amount of free 60S subunit in cells expressing Bcd1-D72A compared with WT (Fig. 5A). Western blot analysis of gradient fractions (Fig. 5B) revealed that in WT cells, RpS10, a small ribosomal subunit (SSU) protein, was mainly in fractions corresponding to the monosomes and the polysomes. Rpl3, a large ribosomal subunit protein, was also found mostly in fractions corresponding to the monosomes and the polysomes. Utp13, a ribosome assembly factor and a subunit of the UTPB complex, which is involved in the biogenesis of SSU processome (37) was found in fractions corresponding to 90S. Rrp5, a protein involved in the assembly of both 40S and 60S subunits (38), was found in fractions corresponding to maturing 60S as well as SSU processome. However, in bcd1-D72A cells RpS10 was mainly found in 40S and 80S fractions (Fig. 5B). There was also a significant decrease in RpS10 in the polysomes, indicating a drop in the pool of translating ribosomes. Similarly, RpL3 was mostly found in 60S and 80S fractions. Some Rpl3 was also detected in early polysome fractions, albeit to a much lesser extent than WT. Interestingly, both Utp13 and Rrp5 shifted toward lighter fractions in cells expressing Bcd1-D72A, indicating a defect in the formation of SSU processome and perturbation of ribosome assembly.
Figure 5.
Ribosome biogenesis is impaired in cells expressing Bcd1-D72A. A, total lysate gradients of cells expressing either Bcd1 (WT) or Bcd1-D72A (D72A). B, Western blot analysis of gradient fractions shown in A.
To reveal how this mutation impairs ribosome biogenesis, we analyzed rRNA processing in WT and bcd1-D72A cells. To avoid possible effects from the addition of doxycycline on rRNA processing, we used CRISPR-Cas9 genome engineering to introduce bcd1-D72A mutation into BY4741 yeast strain. We isolated total RNA from BY4741 or bcd1-D72A cells in the log phase and subjected them to Northern blotting.
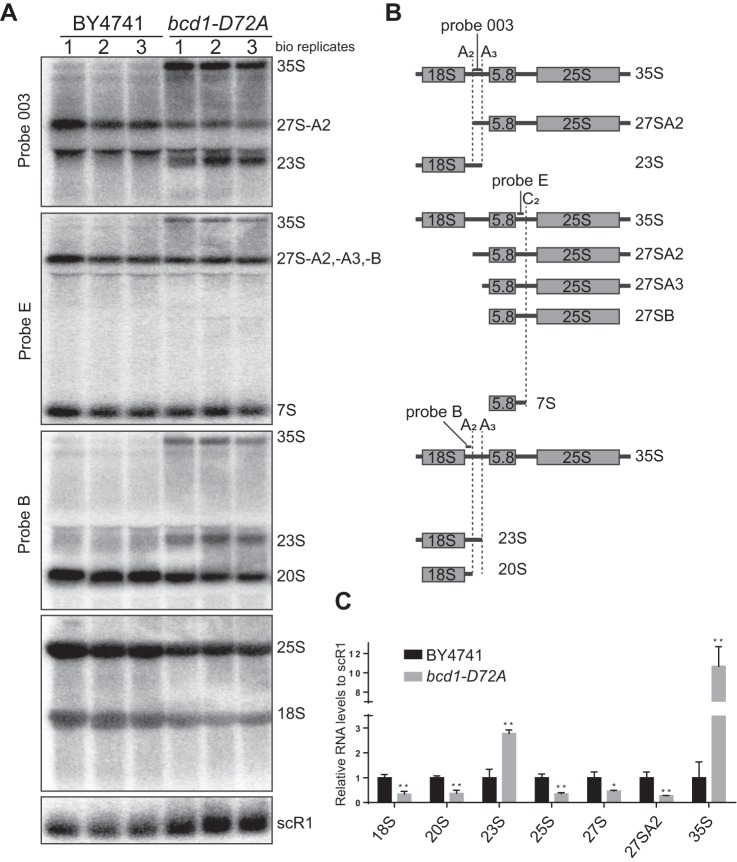
In yeast the majority of rRNA processing occurs co-transcriptionally (39, 40), where A0 and A1 cleavages in 5′-ETS are followed by the A2 cleavage in ITS1, resulting in the formation of 20S as the major 18S precursor (reviewed in Ref. 41). In an alternative pathway, shown to happen under unfavorable growth conditions, 35S is cleaved further downstream of the A2 site at the so-called A3 site, resulting in the formation of 23S (40–47). Our analysis revealed a reduction in the levels of both 18S and 25S rRNAs in bcd1-D72A cells (Fig. 6), explaining the decrease in their polysome levels (Fig. 5). The levels of 20S and 27S A2, the products of the A2 cleavage in ITS1 were also lowered in bcd1-D72A cells compared with WT. This reduction was accompanied by an increase in the unprocessed 35S as well as 23S suggesting a switch from co-transcriptional A2 processing to posttranscriptional A3 processing pathway.
Figure 6.
Alteration of rRNA processing in Bcd1-D72A cells. A, Northern blot analysis of the rRNA processing in BY4741 and bcd1-D72A cells. B, schematic of the binding sites for the probes and the intermediates they detect. The positions of the cleavage sites are marked by dashed lines. C, quantification of data shown in A. Band intensities were normalized to scR1. Graph bars show averages from three biological replicates shown in A. Significance is relative to the WT and was determined using an unpaired t test. n.s., nonsignificant; p > 0.05; *, p < 0.05; **, p < 0.01.
Collectively, these data indicate the critical role of Bcd1-mediated interactions during snoRNP assembly and their impact on ribosome biogenesis.
Discussion
snoRNAs regulate the biogenesis and modification of ribosomes and are thus critically important for control of the cellular translational output. Despite their importance, little is currently known about how snoRNA levels are controlled posttranscriptionally. This is an important question because snoRNA level changes can lead to a decrease in ribosome biogenesis as well as the alteration of the rRNA modification pattern, which impacts translation fidelity and the choice of mRNAs translated (48–51). Indeed, alteration of snoRNA levels is commonly observed in various human cancers, including breast, brain, lung, and prostate cancers (35).
The abundance of snoRNAs is controlled by the binding to a set of core proteins for the formation of catalytically active snoRNPs. Complex formation protects snoRNAs from degradation by nucleases, thus regulating their turnover rate (10, 52). Assembly factors assist in binding of the core proteins to snoRNAs, but their mechanism of function is poorly understood (10). Here, we show that the interactions of the assembly factor Bcd1 with the core protein Snu13 and snoRNAs are critical for maintaining box C/D snoRNA levels. Furthermore, our mutational analyses revealed a critical region of Bcd1 (residues 72–76) which is important for the essential function of the protein for interaction with Snu13 and snoRNAs.
Based on previous proteomic experiments in human cell lines, a model for snoRNP assembly was suggested (13). In this model, ZNHIT6 associates with an early protein-only pre-snoRNP complex and is proposed to stay bound during maturation to be part of a complex that recruits the box C/D snoRNAs. ZNHIT6 is thought to leave the maturing pre-snoRNP concomitantly with the binding of fibrillarin and Nop56 (13). Whether ZNHIT6 presence is required for snoRNA recruitment and how it controls the steady-state level of box C/D snoRNAs has so far remained elusive.
In yeast, an N-terminal fragment (residues 1–97) of Bcd1 is sufficient for growth and maintenance of steady-state expression levels of box C/D snoRNAs, although it associates poorly with snoRNAs (21). In vitro, full-length Bcd1 and a viable N-terminally truncated form of Bcd1 that lacks the Zf-HIT domain (residues 1–45) interact with snoRNAs but their binding can be competed out in presence of longer RNAs (16). These data suggested that the key function of Bcd1 is independent of its ability to bind to RNA (16, 21). Our mutational screening approach enabled us to define the effect of single amino acid substitutions in the region of Bcd1 that is associated with its essential function (residues 45–97) and to reassess the ability of the protein to interact with RNAs in its full-length form. We show that the D72A mutation significantly weakens the binding of Bcd1 to RNA and Snu13, causing a severe growth defect in yeast cells and altering the steady-state levels of snoRNAs. Our data strongly suggest that the ability of Bcd1 to interact with Snu13 and snoRNAs is important for its essential function in snoRNP assembly and for maintenance of box C/D snoRNA steady-state levels.
Secondary structure prediction of Bcd1-D72A using PSIPRED and its comparison to the WT protein indicates that Asp-72 is located in a short loop between two helices (Fig. S1). The D72A mutation could result in the disappearance of the short loop and formation of a long helix by combining the two helices. Thus, we suggest that the Bcd1-D72A mutation weakens the interaction of the protein with Snu13 and RNA by a conformational change between the Zf-HIT and wheel domains of the protein. Structural analysis of Bcd1 will be essential to clarify its interactions that are critically important for its role in snoRNP biogenesis.
snoRNAs are produced as larger precursors that are trimmed by exonucleases for maturation in a process that is linked to the assembly of the snoRNP core proteins (52). The conserved box C/D RNA elements, which form Kink(K)-turns, are essential for snoRNP assembly and serve as the binding site for the recruitment of Snu13 that is followed by ordered recruitment of other core proteins (9, 53–55). Thus, the association of Snu13 with pre-snoRNAs is required for establishing the hierarchy of binding of the other core proteins and is thought to be the first step in commitment of snoRNAs to assembly (10). Similarly, Bcd1 is found to associate with pre-snoRNAs at an early stage, and likely co-transcriptionally (21). We therefore anticipate that our identified Bcd1-mediated contacts with Snu13 and snoRNAs will be essential and critical for early engagement of the snoRNP assembly machinery to protect snoRNAs from rapid turnover.
Eukaryotes transcribe their major ribosomal RNAs (18S, 5.8S, and 25S/28S) as a large polycistronic precursor (35S) (1). In yeast, this precursor can undergo processing either co-transcriptionally (∼70%) or posttranscriptionally (∼30%) (39). The hallmark of the co-transcriptional route is the cleavage at site A2 within ITS1, resulting in the formation of 20S and 27SA2, whereas the posttranscriptional route is marked by the production of 35S precursor and the A3 cleavage, resulting in the formation of 23S and 27SA3. It was shown that under stress, yeast cells switch their rRNA processing toward posttranscriptional route (40, 46, 47). This route is considered for most part to be nonproductive (46). However, there is evidence of 23S processing to make new 18S rRNA (47). The bcd1-D72A cells, which are impaired in snoRNP biogenesis, show the features of posttranscriptional rRNA processing: accumulation of 35S accompanied by an increase in 23S and a drop in 20S levels. They grow and make new ribosomes, albeit at a lower rate. It is therefore tempting to speculate that hypomethylation of the rRNA pushes the balance between co-transcriptional and posttranscriptional rRNA processing toward the latter. It remains to be examined whether or not the new ribosomes made in these cells are the products of the A3 cleavage pathway or the co-transcriptional pathway.
Bcd1 is conserved from yeast to human. Because the Bcd1 mutation analyzed in this study is conserved in ZNHIT6, we used the cBioPortal (56, 57) to investigate the common cancer mutations in ZNHIT6 (Fig. S3). Interestingly, the most common form of mutation observed in ZNHIT6 is a truncation at residue Arg-258, which likely results in the production of a truncated form of the protein that lacks the conserved region encompassing Asp-277, and thus fails to interact with Snu13 and snoRNAs. Thus, modulation of the Bcd1/ZNHIT6 essential interactions by a single point mutation may be exploited by cancer cells and effectively contribute to the alteration of snoRNA levels and perturbation of ribosome biogenesis. This may be an effective means to unbalance the level of snoRNAs in cancer cells, or perhaps modulate the cellular translational program under certain stress conditions. Confirmation of this awaits the future analyses of ZNHIT6 function in human cells.
Materials and methods
Yeast strains and plasmids
S. cerevisiae strains used in this study are listed in Table S1. Oligonucleotides and plasmids used in this study are listed in Table S2 and Table S3, respectively.
Making bcd1-D72A yeast strain using CRISPR-Cas9
Genome editing was carried out as described previously (58). A primer was designed for guide recognition sequence mutagenesis (Table S2) and used to amplify the pCAS9 vector (Addgene 60847) (59). A double-stranded 160-mer repair DNA was generated using oligos listed in Table S2. The D72A CRISPR was carried out by transforming pCAS9-Bcd1 and the PCR product into the WT BY4741 strain. Transformants were plated onto G418 and allowed to grow for 72 h at 37 °C. Individual colonies were selected and restreaked onto YPD. Mutations in individual colonies were confirmed by PCR and sequencing.
Yeast growth assays
tetO7::BCD1 cells containing ZNHIT6 or Bcd1 expression plasmids were grown to saturation in synthetic glucose liquid culture lacking histidine (His−). Cultures were diluted in sterile water to a final concentration of 107 cells/ml, followed by four successive cascade dilutions in a 1:10 ratio. Dilutions were spotted onto synthetic His− medium plates with and without doxycycline and grown at 30 °C.
Protein expression and purification
All proteins were expressed in Escherichia coli Rosetta2 (DE3) cells (Novagen). Cells were grown at 37 °C to A600 of 0.6 in 2× YT media supplemented with the appropriate antibiotics and then transferred to 18 °C. Protein expression was induced by addition of 0.3 mm or 1 mm isopropyl β-D-1-thiogalactopyranoside (IPTG) for pGEX-6-P2 or pET23/pSV272 harboring cells, respectively. Cultures were harvested 18 h after induction.
Bcd1 and Bcd1-D72A were expressed as His-tagged proteins and purified on Ni-NTA resin in buffer A. The protein was eluted with 250 mm imidazole. The tag was cleaved overnight and the protein was further purified over a MonoS ion exchange column in buffer B and eluted with a linear gradient of 150 mm to 1 m NaCl over 20 column volumes. Final purification was performed on a Superdex S-200 gel filtration column (GE Healthcare) equilibrated in 150 mm NaCl, 20 mm Tris/HCl, pH 7.5, 5% glycerol, 40 μm ZnCl2, and 1 mm DTT. His-MBP–tagged Bcd1 and Bcd1-D72A were purified on Ni-NTA in buffer A and eluted as above. The proteins were buffer exchanged overnight in buffer B and further purified over MonoQ and Superdex 200 columns and stored in 150 mm NaCl, 20 mm Tris/HCl, pH 7.5, 5% glycerol and 40 μm ZnCl2 and 1 mm DTT. Rvb1/2 were expressed and purified as published previously (60). MBP-Rsa1/GST-Hit1 were co-expressed and purified using Ni-NTA in 200 mm NaCl, 50 mm HEPES/NaOH, pH 7.5, 5% glycerol. The complex was eluted by increasing the concentration of imidazole to 200 mm, and further purified on a GSH Sepharose column (GE Healthcare). The GST tag of Hit1 was removed overnight using PreScission protease (GE Healthcare) in buffer B. The GST tag and the protease were removed using a MonoQ ion exchange column. The complex was further polished using a Superdex 200 gel filtration column equilibrated in 200 mm NaCl, 20 mm HEPES/NaOH, 5% glycerol, and 1 mm DTT. GST-Snu13 was purified using GSH Sepharose in buffer C. The protein was further purified using a Superdex 75 gel filtration column in buffer D. Nop581–437 was purified as a GST-tagged protein in buffer C. The GST-tag was cleaved off overnight using PreScission protease in 150 mm NaCl, 30 mm Tris/HCl, pH 7.5, 5% glycerol, and 2 mm 2-mercaptoethanol. Both GST-tagged and untagged proteins were further purified using MonoQ and Superdex 200 and stored in buffer D. Tah1 and His-tagged Pih1 were co-expressed and purified on Ni-NTA resin in 100 mm NaCl, 25 mm sodium phosphate, 5% glycerol, and 30 mm imidazole. The complex was eluted by increasing the concentration of imidazole to 200 mm. The His tag of Pih1 was cleaved overnight using tobacco etch virus protease in 100 mm NaCl, 25 mm sodium phosphate, and 10% glycerol. The uncleaved His-Pih1 and the tobacco etch virus protease were removed by a second step of Ni-NTA. The complex was further purified using a Superdex 200 gel filtration column equilibrated in 100 mm NaCl, 25 mm sodium phosphate, 5% glycerol, and 1 mm DTT. GST-tagged Tah1 and His-MBP–tagged Pih1 were co-expressed and purified as above with the exception that after the first Ni-NTA, the complex was further purified on GSH Sepharose. His-MBP tag was cleaved overnight. The complex was polished on S-200 gel filtration column.
In vitro interaction studies
5 μm of the untagged proteins were mixed with 3 μm tagged protein (or control) in 150 mm NaCl and 20 mm Tris/HCl, pH 7.5, 5% glycerol, and 40 μm ZnCl2, and preincubated on ice for 15 min. 10% of the reaction was set aside as the input control before addition of 25 μl of equilibrated amylose resin (New England Biolabs) or GSH Sepharose. The mixture was incubated for 30 min at 4 °C, washed, and eluted with binding buffer supplemented with either 20 mm maltose (amylose resin) or reduced glutathione (GSH Sepharose). Input, final wash, and the eluate from each binding study were analyzed on a SDS-PAGE.
In vitro RNA-protein–binding assay
In vitro transcribed RNA was labeled using [γ-32P]ATP. Trace amount of labeled RNA was folded and mixed with appropriate protein and incubated for 20 min in 20 mm Tris, pH 7.5, 100 mm KCl, 5 mm MgCl2 at 30 °C before mixing with the heparin loading dye (1 mg/ml final concentration) and loading on a running 8% acrylamide/TBE gel at 4 °C. Protein-bound and unbound fractions were quantified using Quantity One (Bio-Rad), and data were fit to a single binding isotherm using Prism (GraphPad).
Measuring the thermal stability of Bcd1 and its variant
3 μm of Bcd1-WT or D72A variant were loaded as triplicates into glass capillaries (NanoTemper Technologies) and intrinsic protein fluorescence at 330 and 350 nm was monitored between 35 and 95 °C in the Tycho instrument (NanoTemper Technologies). Temperature inflection values (Ti) were obtained by automated data analysis using the accompanying software.
Analysis of the steady-state levels of snoRNAs
Cells were grown in selective media in the presence of doxycycline, to A600 ∼ 0.6. Total RNA from three biological replicates of each strain was isolated using the hot phenol method. snoRNAs were separated on 10% acrylamide/urea gels, transferred to Hybond nylon membrane (GE Healthcare) and probed as indicated. Bands were quantified using Image Lab software (Bio-Rad).
Analysis of the steady-state levels of pre-rRNAs
BY4741 or bcd1-D72A cells were grown in YPD to mid-log phase. Total RNAs from three biological samples of each yeast strain were isolated using the hot phenol method. rRNAs were separated on a 1% agarose/formaldehyde gel and transferred by capillarity overnight in 20× saline sodium citrate on a nylon membrane (GE Healthcare). Membranes were probed as indicated. Band were quantified using Image Lab software (Bio-Rad).
Pulldown assay from yeast cells
tetO7::BCD1 yeast cells were grown for 16 h in YPD while constitutively expressing a twin-Strep–tagged Bcd1 from a TEF promoter. Strep-tagged protein was purified using Strep-Tactin (IBA Lifesciences) in 150 mm NaCl, 50 mm Tris/HCl, pH 7.5, 5% glycerol, and 5 mm MgCl2, and eluted in the same buffer supplemented with 2.5 mm desthiobiotin. Eluted proteins were analyzed by Western blotting. Eluted RNA was precipitated and analyzed by Northern blotting as described above with the exception that 1 fmol of an in vitro transcribed random RNA was spiked into each tube to serve as the control for precipitation efficiency.
Twin-Strep–Snu13 was expressed from GPD promoter in tetO7::BCD1 cells expressing endogenous Bcd1 as well as HA-tagged WT or D72A variant of Bcd1. Strep purification and data analysis were performed as described above.
Sucrose density gradient analysis
Yeast cells were grown to mid log phase in YPD supplemented with 10 mg/liter doxycycline and harvested after addition of 0.1 mg/ml cycloheximide, washed, and lysed in ice-cold gradient buffer (20 mm HEPES, pH 7.4, 5 mm MgCl2, 100 mm NaCl, and 3 mm DTT) supplemented with 0.1 mg/ml cycloheximide and cOmplete Protease Inhibitor mixture (Roche). Lysate was cleared by 10 min of centrifugation at 10,000 × g, applied to 10–50% sucrose gradients in gradient buffer, and centrifuged for 2 h at 40,000 rpm in a SW 41 Ti rotor. Gradients were fractionated and scanned by UV 260 nm absorbance. Fractions were analyzed for their protein content by Western blotting.
Antibodies
HRP-conjugated anti-rabbit and anti-mouse secondary antibodies were obtained from Rockland Immunochemicals. The Utp13, Rrp5, and Rps10 antibodies (a gift from K. Karbstein, Scripps, FL) and the Rvb2 (a gift from W. Houry) were raised in rabbits. Bcd1 was detected using an anti-HA antibody obtained from BioLegend or HRP-conjugated anti-Strep (IBA Bioscience). Rpl3 was detected by an antibody obtained from the Developmental Studies Hybridoma Bank, The University of Iowa, deposited by J.R. Warner.
Author contributions
S. K. and H. G. conceptualization; S. K., R. E. D., T. F. R. H., and H. G. data curation; S. K., R. E. D., and H. G. formal analysis; S. K., R. E. D., and H. G. validation; S. K., R. E. D., T. F. R. H., and H. G. investigation; S. K., R. E. D., and H. G. visualization; S. K. and H. G. methodology; S. K. and H. G. writing-original draft; S. K. and H. G. project administration; S. K. and H. G. writing-review and editing; H. G. resources; H. G. supervision; H. G. funding acquisition.
Supplementary Material
Acknowledgments
We thank J. A. Ball for purification of Nop581–437 and GST-Snu13, and K. Araki for help with gradient fractionation. We also thank K. P. Hopfner for the Rvb1/2 expression plasmid; W. Houry and K. Karbstein for antibodies; and G. L. Conn, C. M. Dunham, D. Reines, and members of the Ghalei lab for comments on the manuscript.
This work was supported by Department of Biochemistry at Emory School of Medicine startup funds (to H. G.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3 and Tables S1–S3.
- snoRNP
- small nucleolar ribonucleoprotein
- snoRNA
- small nucleolar RNA
- Zf-HIT
- zinc finger HIT
- SSU
- small subunit
- Ni-NTA
- nickel-nitrilotriacetic acid
- Ti
- inflection temperature
- IDR
- intrinsically disordered region.
References
- 1. Woolford J. L. Jr., and Baserga S. J. (2013) Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195, 643–681 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klinge S., and Woolford J. L. Jr. (2019) Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 20, 116–131 10.1038/s41580-018-0078-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sloan K. E., Warda A. S., Sharma S., Entian K. D., Lafontaine D. L. J., and Bohnsack M. T. (2017) Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14, 1138–1152 10.1080/15476286.2016.1259781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Decatur W. A., and Fournier M. J. (2003) RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278, 695–698 10.1074/jbc.R200023200 [DOI] [PubMed] [Google Scholar]
- 5. Watkins N. J., and Bohnsack M. T. (2012) The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 3, 397–414 10.1002/wrna.117 [DOI] [PubMed] [Google Scholar]
- 6. Watkins N. J., Dickmanns A., and Lührmann R. (2002) Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 22, 8342–8352 10.1128/mcb.22.23.8342-8352.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Omer A. D., Ziesche S., Ebhardt H., and Dennis P. P. (2002) In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. U.S.A. 99, 5289–5294 10.1073/pnas.082101999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tran E. J., Zhang X., and Maxwell E. S. (2003) Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J. 22, 3930–3940 10.1093/emboj/cdg368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Watkins N. J., Segault V., Charpentier B., Nottrott S., Fabrizio P., Bachi A., Wilm M., Rosbash M., Branlant C., and Luhrmann R. (2000) A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103, 457–466 10.1016/s0092-8674(00)00137-9 [DOI] [PubMed] [Google Scholar]
- 10. Massenet S., Bertrand E., and Verheggen C. (2017) Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 14, 680–692 10.1080/15476286.2016.1243646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McKeegan K. S., Debieux C. M., Boulon S., Bertrand E., and Watkins N. J. (2007) A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol. Cell. Biol. 27, 6782–6793 10.1128/MCB.01097-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McKeegan K. S., Debieux C. M., and Watkins N. J. (2009) Evidence that the AAA+ proteins TIP48 and TIP49 bridge interactions between 15.5K and the related NOP56 and NOP58 proteins during box C/D snoRNP biogenesis. Mol. Cell. Biol. 29, 4971–4981 10.1128/MCB.00752-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bizarro J., Charron C., Boulon S., Westman B., Pradet-Balade B., Vandermoere F., Chagot M. E., Hallais M., Ahmad Y., Leonhardt H., Lamond A., Manival X., Branlant C., Charpentier B., Verheggen C., and Bertrand E. (2014) Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control. J. Cell Biol. 207, 463–480 10.1083/jcb.201404160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boulon S., Marmier-Gourrier N., Pradet-Balade B., Wurth L., Verheggen C., Jády B. E., Rothé B., Pescia C., Robert M. C., Kiss T., Bardoni B., Krol A., Branlant C., Allmang C., Bertrand E., and Charpentier B. (2008) The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 180, 579–595 10.1083/jcb.200708110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Back R., Dominguez C., Rothé B., Bobo C., Beaufils C., Morera S., Meyer P., Charpentier B., Branlant C., Allain F. H., and Manival X. (2013) High-resolution structural analysis shows how Tah1 tethers Hsp90 to the R2TP complex. Structure 21, 1834–1847 10.1016/j.str.2013.07.024 [DOI] [PubMed] [Google Scholar]
- 16. Bragantini B., Tiotiu D., Rothé B., Saliou J. M., Marty H., Cianférani S., Charpentier B., Quinternet M., and Manival X. (2016) Functional and structural insights of the zinc-finger HIT protein family members involved in box C/D snoRNP biogenesis. J. Mol. Biol. 428, 2488–2506 10.1016/j.jmb.2016.04.028 [DOI] [PubMed] [Google Scholar]
- 17. Rothé B., Saliou J. M., Quinternet M., Back R., Tiotiu D., Jacquemin C., Loegler C., Schlotter F., Peña V., Eckert K., Moréra S., Dorsselaer A. V., Branlant C., Massenet S., Sanglier-Cianférani S., Manival X., and Charpentier B. (2014) Protein Hit1, a novel box C/D snoRNP assembly factor, controls cellular concentration of the scaffolding protein Rsa1 by direct interaction. Nucleic Acids Res. 42, 10731–10747 10.1093/nar/gku612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kakihara Y., Makhnevych T., Zhao L., Tang W., and Houry W. A. (2014) Nutritional status modulates box C/D snoRNP biogenesis by regulated subcellular relocalization of the R2TP complex. Genome Biology 15, 404 10.1186/s13059-014-0404-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao R., Kakihara Y., Gribun A., Huen J., Yang G., Khanna M., Costanzo M., Brost R. L., Boone C., Hughes T. R., Yip C. M., and Houry W. A. (2008) Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J. Cell Biol. 180, 563–578 10.1083/jcb.200709061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Prieto M. B., Georg R. C., Gonzales-Zubiate F. A., Luz J. S., and Oliveira C. C. (2015) Nop17 is a key R2TP factor for the assembly and maturation of box C/D snoRNP complex. BMC Mol. Biol. 16, 7 10.1186/s12867-015-0037-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paul A., Tiotiu D., Bragantini B., Marty H., Charpentier B., Massenet S., and Labialle S. (2019) Bcd1p controls RNA loading of the core protein Nop58 during C/D box snoRNP biogenesis. RNA 25, 496–506 10.1261/rna.067967.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peng W. T., Robinson M. D., Mnaimneh S., Krogan N. J., Cagney G., Morris Q., Davierwala A. P., Grigull J., Yang X., Zhang W., Mitsakakis N., Ryan O. W., Datta N., Jojic V., Pal C., et al. (2003) A panoramic view of yeast noncoding RNA processing. Cell 113, 919–933 10.1016/s0092-8674(03)00466-5 [DOI] [PubMed] [Google Scholar]
- 23. Hiley S. L., Babak T., and Hughes T. R. (2005) Global analysis of yeast RNA processing identifies new targets of RNase III and uncovers a link between tRNA 5′ end processing and tRNA splicing. Nucleic Acids Res. 33, 3048–3056 10.1093/nar/gki608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kressler D., Doère M., Rojo M., and Linder P. (1999) Synthetic lethality with conditional dbp6 alleles identifies rsa1p, a nucleoplasmic protein involved in the assembly of 60S ribosomal subunits. Mol. Cell. Biol. 19, 8633–8645 10.1128/mcb.19.12.8633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kakihara Y., and Houry W. A. (2012) The R2TP complex: Discovery and functions. Biochim. Biophys. Acta 1823, 101–107 10.1016/j.bbamcr.2011.08.016 [DOI] [PubMed] [Google Scholar]
- 26. Nano N., and Houry W. A. (2013) Chaperone-like activity of the AAA+ proteins Rvb1 and Rvb2 in the assembly of various complexes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20110399 10.1098/rstb.2011.0399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Snider J., Thibault G., and Houry W. A. (2008) The AAA+ superfamily of functionally diverse proteins. Genome Biol. 9, 216 10.1186/gb-2008-9-4-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., Arkin A. P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., et al. (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- 29. Cloutier P., Poitras C., Durand M., Hekmat O., Fiola-Masson E., Bouchard A., Faubert D., Chabot B., and Coulombe B. (2017) R2TP/Prefoldin-like component RUVBL1/RUVBL2 directly interacts with ZNHIT2 to regulate assembly of U5 small nuclear ribonucleoprotein. Nat. Commun. 8, 15615 10.1038/ncomms15615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dosztanyi Z., Csizmok V., Tompa P., and Simon I. (2005) IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 10.1093/bioinformatics/bti541 [DOI] [PubMed] [Google Scholar]
- 32. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. E. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schopf F. H., Huber E. M., Dodt C., Lopez A., Biebl M. M., Rutz D. A., Muhlhofer M., Richter G., Madl T., Sattler M., Groll M., and Buchner J. (2019) The co-chaperone Cns1 and the recruiter protein Hgh1 link Hsp90 to translation elongation via chaperoning elongation factor 2. Mol. Cell 74, 73–87.e78 10.1016/j.molcel.2019.02.011 [DOI] [PubMed] [Google Scholar]
- 34. Cheung K. L., Huen J., Kakihara Y., Houry W. A., and Ortega J. (2010) Alternative oligomeric states of the yeast Rvb1/Rvb2 complex induced by histidine tags. J. Mol. Biol. 404, 478–492 10.1016/j.jmb.2010.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gong J., Li Y., Liu C. J., Xiang Y., Li C., Ye Y., Zhang Z., Hawke D. H., Park P. K., Diao L., Putkey J. A., Yang L., Guo A. Y., Lin C., and Han L. (2017) A pan-cancer analysis of the expression and clinical relevance of small nucleolar RNAs in human cancer. Cell Rep. 21, 1968–1981 10.1016/j.celrep.2017.10.070 [DOI] [PubMed] [Google Scholar]
- 36. Monaco P. L., Marcel V., Diaz J. J., and Catez F. (2018) 2′-O-methylation of ribosomal RNA: Towards an epitranscriptomic control of translation? Biomolecules 8, E106 10.3390/biom8040106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barandun J., Hunziker M., and Klinge S. (2018) Assembly and structure of the SSU processome—a nucleolar precursor of the small ribosomal subunit. Curr. Opin. Struct. Biol. 49, 85–93 10.1016/j.sbi.2018.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Venema J., and Tollervey D. (1996) RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 15, 5701–5714 [PMC free article] [PubMed] [Google Scholar]
- 39. Kos M., and Tollervey D. (2010) Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell 37, 809–820 10.1016/j.molcel.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Osheim Y. N., French S. L., Keck K. M., Champion E. A., Spasov K., Dragon F., Baserga S. J., and Beyer A. L. (2004) Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell 16, 943–954 10.1016/j.molcel.2004.11.031 [DOI] [PubMed] [Google Scholar]
- 41. Henras A. K., Plisson-Chastang C., O'Donohue M. F., Chakraborty A., and Gleizes P. E. (2015) An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 6, 225–242 10.1002/wrna.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dunbar D. A., Wormsley S., Agentis T. M., and Baserga S. J. (1997) Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell. Biol. 17, 5803–5812 10.1128/mcb.17.10.5803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Venema J., and Tollervey D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33, 261–311 10.1146/annurev.genet.33.1.261 [DOI] [PubMed] [Google Scholar]
- 44. Mullineux S. T., and Lafontaine D. L. (2012) Mapping the cleavage sites on mammalian pre-rRNAs: Where do we stand? Biochimie 94, 1521–1532 10.1016/j.biochi.2012.02.001 [DOI] [PubMed] [Google Scholar]
- 45. Kos-Braun I. C., and Kos M. (2017) Post-transcriptional regulation of ribosome biogenesis in yeast. Microbial Cell 4, 179–181 10.15698/mic2017.05.575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kos-Braun I. C., Jung I., and Kos M. (2017) Tor1 and CK2 kinases control a switch between alternative ribosome biogenesis pathways in a growth-dependent manner. PLoS Biol. 15, e2000245 10.1371/journal.pbio.2000245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Talkish J., Biedka S., Jakovljevic J., Zhang J., Tang L., Strahler J. R., Andrews P. C., Maddock J. R., and Woolford J. L. Jr. (2016) Disruption of ribosome assembly in yeast blocks cotranscriptional pre-rRNA processing and affects the global hierarchy of ribosome biogenesis. RNA 22, 852–866 10.1261/rna.055780.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Erales J., Marchand V., Panthu B., Gillot S., Belin S., Ghayad S. E., Garcia M., Laforets F., Marcel V., Baudin-Baillieu A., Bertin P., Coute Y., Adrait A., Meyer M., Therizols G., et al. (2017) Evidence for rRNA 2′-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. U.S.A. 114, 12934–12939 10.1073/pnas.1707674114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baudin-Baillieu A., Fabret C., Liang X. H., Piekna-Przybylska D., Fournier M. J., and Rousset J. P. (2009) Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic Acids Res. 37, 7665–7677 10.1093/nar/gkp816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krogh N., Jansson M. D., Hafner S. J., Tehler D., Birkedal U., Christensen-Dalsgaard M., Lund A. H., and Nielsen H. (2016) Profiling of 2′-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res. 44, 7884–7895 10.1093/nar/gkw482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marcel V., Ghayad S. E., Belin S., Therizols G., Morel A. P., Solano-Gonzalez E., Vendrell J. A., Hacot S., Mertani H. C., Albaret M. A., Bourdon J. C., Jordan L., Thompson A., Tafer Y., Cong R., et al. (2013) p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 24, 318–330 10.1016/j.ccr.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kufel J., and Grzechnik P. (2019) Small nucleolar RNAs tell a different tale. Trends Genet. 35, 104–117 10.1016/j.tig.2018.11.005 [DOI] [PubMed] [Google Scholar]
- 53. Szewczak L. B., DeGregorio S. J., Strobel S. A., and Steitz J. A. (2002) Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 9, 1095–1107 [DOI] [PubMed] [Google Scholar]
- 54. Schultz A., Nottrott S., Watkins N. J., and Lührmann R. (2006) Protein-protein and protein-RNA contacts both contribute to the 15.5K-mediated assembly of the U4/U6 snRNP and the box C/D snoRNPs. Mol. Cell. Biol. 26, 5146–5154 10.1128/MCB.02374-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cahill N. M., Friend K., Speckmann W., Li Z. H., Terns R. M., Terns M. P., and Steitz J. A. (2002) Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 21, 3816–3828 10.1093/emboj/cdf376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., and Schultz N. (2012) The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., and Schultz N. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ryan O. W., Poddar S., and Cate J. H. (2016) CRISPR-Cas9 genome engineering in Saccharomyces cerevisiae cells. Cold Spring Harb. Protoc. 2016, pdb-prot086827 10.1101/pdb.prot086827 [DOI] [PubMed] [Google Scholar]
- 59. Ryan O. W., Skerker J. M., Maurer M. J., Li X., Tsai J. C., Poddar S., Lee M. E., DeLoache W., Dueber J. E., Arkin A. P., and Cate J. H. (2014) Selection of chromosomal DNA libraries using a multiplex CRISPR system. eLife 3, e03703 10.7554/eLife.03703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhou C. Y., Stoddard C. I., Johnston J. B., Trnka M. J., Echeverria I., Palovcak E., Sali A., Burlingame A. L., Cheng Y., and Narlikar G. J. (2017) Regulation of Rvb1/Rvb2 by a domain within the INO80 chromatin remodeling complex implicates the yeast Rvbs as protein assembly chaperones. Cell Rep. 19, 2033–2044 10.1016/j.celrep.2017.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.