Abstract
Endoplasmic reticulum protein of 29 kDa (ERp29) is a thioredoxin-homologous endoplasmic reticulum (ER) protein that regulates the biogenesis of cystic fibrosis transmembrane conductance regulator (CFTR) and the epithelial sodium channel (ENaC). ERp29 may promote ENaC cleavage and increased open probability by directing ENaC to the Golgi via coat complex II (COP II) during biogenesis. We hypothesized that ERp29's C-terminal KEEL ER retention motif, a KDEL variant that is associated with less robust ER retention, strongly influences its regulation of ENaC biogenesis. As predicted by our previous work, depletion of Sec24D, the cargo recognition component of COP II that we previously demonstrated to interact with ENaC, decreases ENaC functional expression without altering β-ENaC expression at the apical surface. We then tested the influence of KDEL ERp29, which should be more readily retrieved from the proximal Golgi by the KDEL receptor (KDEL-R), and a KEEL-deleted mutant (ΔKEEL ERp29), which should not interact with the KDEL-R. ENaC functional expression was decreased by ΔKEEL ERp29 overexpression, whereas KDEL ERp29 overexpression did not significantly alter ENaC functional expression. Again, β-ENaC expression at the apical surface was unaltered by either of these manipulations. Finally, we tested whether the KDEL-R itself has a role in ENaC forward trafficking and found that KDEL-R depletion decreases ENaC functional expression, again without altering β-ENaC expression at the apical surface. These results support the hypothesis that the KDEL-R plays a role in the biogenesis of ENaC and in its exit from the ER through its association with COP II. The cleavage of the extracellular loops of the epithelial sodium channel (ENaC) α and γ subunits increases the channel's open probability and function. During ENaC biogenesis, such cleavage is regulated by the novel 29-kDa chaperone of the ER, ERp29. Our data here are consistent with the hypothesis that ERp29 must interact with the KDEL receptor to exert its regulation of ENaC biogenesis. The classically described role of the KDEL receptor is to retrieve ER-retained species from the proximal Golgi and return them to the ER via coat complex I machinery. In contrast, our data suggest a novel and important role for the KDEL receptor in the biogenesis and forward trafficking of ENaC.
Keywords: epithelial sodium channel (ENaC), trafficking, chaperone, endoplasmic reticulum (ER), coat complex II (COPII), biogenesis, endoplasmic reticulum protein of 29 kDa (ERp29), KDEL receptor, KEEL motif
Introduction
The epithelial sodium channel (ENaC)3 is found in the apical membrane in a wide variety of epithelial cells (1, 2). In the airway, ENaC constitutes the rate-limiting step for Na+ absorption and is hypothesized to play a significant role in mucus hydration (3). Its functional overexpression has been shown to cause both decreased mucociliary clearance and increased morbidity and mortality in mouse models, which may mimic the cystic fibrosis airway. ENaC is likely a heterotrimer (4) composed of three similar subunits, α, β, and γ (5). Each subunit maintains its respective N and C termini in the cytoplasm, with large extracellular loops (6–8). Our group's recent data have suggested that nascent ENaC can exit the endoplasmic reticulum (ER) and reach the Golgi by interacting with the coat complex II (COP II) ER export machinery and that ENaC exiting the ER via this pathway may undergo cleavage of the luminal/extracellular loops of the α and γ subunits en route to the apical surface (9).
This cleavage of ENaC's α and γ subunits in their extracellular loops, which increases the channel's open probability (Po) is a fundamental and unique feature of ENaC biogenesis and regulation. Indeed, furin, a trans-Golgi resident pro-protein convertase, can cleave the luminal/extracellular loops of ENaC's α and γ subunits at two sites and one site, respectively, during biogenesis (10). An additional cleavage of the γ subunit at the plasma membrane is required for full activation of the channel (11). Especially interesting is that newly synthesized ENaC can also bypass furin proteolysis and reach the apical membrane in an uncleaved, low-open probability (Po) form that has immature (endoglycosidase H–sensitive) glycosyl side chains (12). These uncleaved, “nearly silent” channels can be proteolytically activated after delivery to the cell surface by either endogenous cell surface channel–activating proteases or exogenous proteases, such as trypsin or elastase (13–15). The mechanism(s) determining whether ENaC undergoes or bypasses furin cleavage during biogenesis is not known, but our recent work suggests that the ER chaperone ERp29 (ER protein of 29 kDa) is a critical determinant of whether ENaC undergoes cleavage during biogenesis (2).
ERp29 is an ER-luminal resident that is ubiquitously expressed and is especially abundant in epithelia (16). Interestingly, ERp29 is homologous to the thioredoxins but lacks the characteristic thioredoxin CXXC motif. Instead, ERp29 contains a single Cys residue at position 157 (18). Our group has previously demonstrated that ERp29 promotes biogenesis of the cystic fibrosis transmembrane conductance regulator (CFTR) (19); these were the first data demonstrating that an ER chaperone could promote CFTR biogenesis. Furthermore, our data also demonstrated that ERp29 is present at the cell surface and in the culture medium, suggesting that ERp29 itself could escape ER retention and be found in more distal secretory compartments (19).
CFTR and ENaC share similarities in their biogenesis and trafficking, so we hypothesized that ERp29 would also regulate ENaC biogenesis and function. Our recently published work has begun to demonstrate that ERp29 plays a role in directing ENaC's itinerary during biogenesis, specifically by increasing ENaC's interaction with the Sec24D cargo recognition component of COP II as well as by increasing ENaC's cleavage, presumably in the Golgi, during biogenesis (2). In considering which structural features or motifs in ERp29 may be critical for its functional role, we demonstrated that ERp29's single cysteine, Cys157, is a key determinant of ERp29's function. In this present work, we focus on ERp29's C-terminal KEEL motif, which presumably promotes retention of ERp29 in the ER through interaction with the KDEL receptor (KDEL-R) (20, 21). The KDEL-R cycles between the ER and Golgi and facilitates the retrieval of ER-resident proteins in and from the Golgi through recognition of their KDEL (or KEEL) retention motif (22–24). ERp29's KEEL motif is associated with less robust ER retention (27), which is consistent with our previous work demonstrating that ERp29 can be found at the surface of and is secreted by epithelial cells (19). In these experiments testing the hypothesis that ERp29's KEEL motif is a critical determinant of the regulation of ENaC biogenesis by ERp29, our data are consistent with both ERp29's C-terminal KEEL motif and the KDEL-R itself playing key roles in directing ENaC's itinerary during biogenesis.
Results
The cleavage state of ENaC at the apical surface regulates ENaC open probability
The Po of ENaC at the apical surface of epithelia is largely regulated by cleavage of its α and γ subunits in their extracellular loops (10); uncleaved channels are “nearly silent” and have a Po approximating 0, whereas fully cleaved channels have a Po approximating 1. We (2, 9, 28, 29) and others (12, 30) have previously demonstrated that both cleaved and uncleaved channels are present at the surface of epithelial cells and that the uncleaved channels can be acutely activated by treating the apical surface with exogenous trypsin.
In this paradigm (exemplified by Fig. 1C; see also Refs. 2 and 31), the amiloride-sensitive short-circuit current (Isc) at baseline represents apical surface channels that are already cleaved (Cut Fraction in Fig. 1C), whereas the trypsin-stimulated change in amiloride-sensitive Isc represents channels at the apical surface that are uncleaved and nearly silent at the beginning of the experiment and are acutely activated by trypsin cleavage (Uncut Fraction in Fig. 1C). The total amiloride-sensitive Isc is then the sum of the Isc of the cut and uncut fractions and is reflective of the total amount of ENaC at the apical surface (Total in Fig. 1C). Using this technique, our group has previously demonstrated that overexpression of both WT ERp29 and C157S ERp29, an ERp29 mutant where its single Cys157 was mutated to a serine, as well as specific depletion of ERp29 using siRNA, can modulate the fraction of uncleaved versus cleaved ENaC present at the surface of cultured epithelial cells (2).
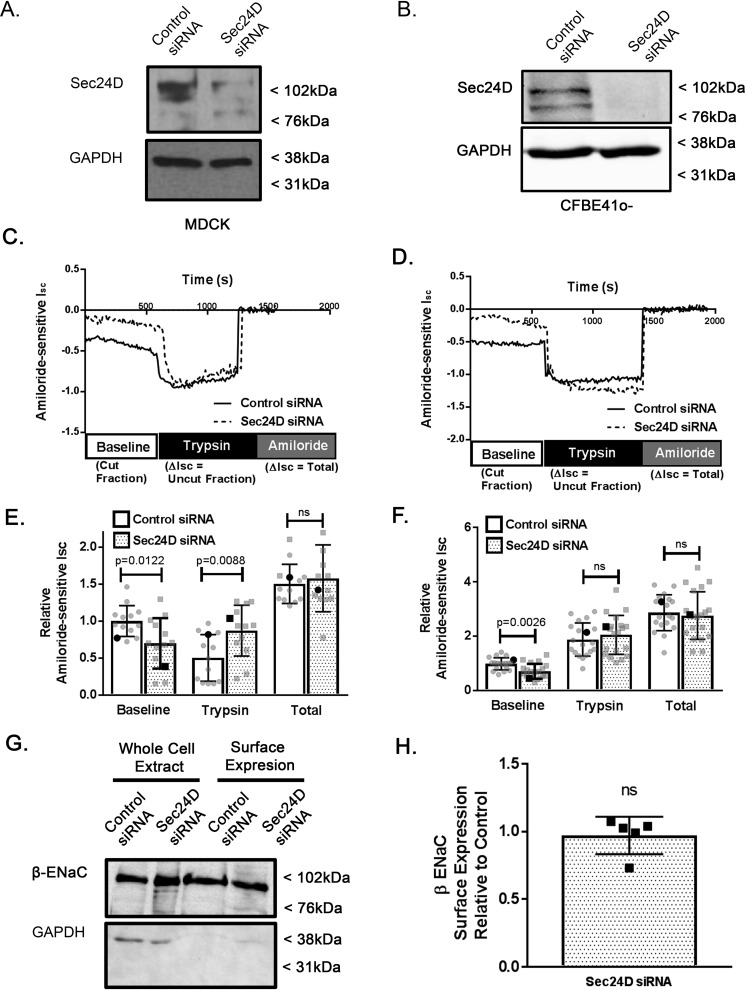
Figure 1.
Sec24D regulates ENaC cleavage in MDCK αβγ ENaC and CFBE41o− epithelial cells. MDCK αβγ-ENaC cells (A, C, E, G, and H) or CFBE41o− epithelial cells (B, D, and F) were transiently transfected with nontargeted (control) or Sec24D siRNA and grown as polarized monolayers. Cells were mounted in Ussing chambers for Isc measurements (A–F) or underwent surface biotinylation (G and H) as described under “Experimental procedures.” A and B, representative immunoblot analysis performed after completion of the Isc measurements confirming siRNA-mediated depletion of Sec24D in MDCK αβγ-ENaC cells (A) or CFBE41o− epithelial cells (B). Shown are representative Isc traces from experiments in MDCK αβγ-ENaC (C) or CFBE41o− (D) cells, with annotations depicting experimental protocol. E and F, summary of amiloride-sensitive Isc measurements (relative to control baseline) are presented as mean ± S.D. (error bars); individual data points are depicted in gray, whereas representative traces from C and D are shown in black in E and F, respectively. Baseline Isc represents ENaC that is at the membrane in a cleaved/active form. E, application of trypsin to the apical surface in MDCK αβγ-ENaC cells acutely activates uncleaved/nearly silent ENaC (n = 13, p = 0.0122 for baseline, p = 0.0088 for trypsin, p = ns for total). Boldface points in E denote the representative Isc traces from C. F, Isc measurements of Sec24D-depleted CFBE41o− epithelial cells demonstrating decreased baseline ENaC-mediated Isc (n = 19, p = 0.0026). Boldface points in F denote the representative Isc traces from D. G, representative experiment demonstrating unchanged apical surface expression of β-ENaC by surface biotinylation. F, densitometric quantification of β-ENaC surface expression relative to control (n = 5 independent experiments, p = ns by Wilcoxon signed-rank test).
Sec24D regulates the cleavage state of ENaC at the apical surface
Sec24D is a cargo recognition component of COP II, and our previous work has demonstrated that both the cytoplasmic chaperone Hsp70 (70-kDa heat shock protein) (9) and ERp29 (2) promote the interaction of Sec24D with β-ENaC. In contrast, overexpression of C157S ERp29 inhibits the association of β-ENaC with Sec24D (2). We therefore initially hypothesized that modulating Sec24D expression itself would alter ENaC trafficking and functional expression and tested this hypothesis in Ussing chambers after depletion of Sec24D expression using specific siRNA (representative immunoblot, Fig. 1A).
When Sec24D was depleted in Madin–Darby canine kidney (MDCK) αβγ-ENaC cells, there was a lower baseline ENaC-mediated Isc (representative Isc traces in Fig. 1C, summary data in Fig. 1E, baseline; n = 13, p = 0.0122) and greater increase in amiloride-sensitive Isc upon application of trypsin than in controls (Fig. 1E, trypsin; n = 13, p = 0.0088). Interestingly, and again consistent with our previous data regarding modulation of ERp29 expression and function, the total amiloride-sensitive Isc was similar for control and Sec24D-depleted cells (Fig. 1E, total n = 13, p = ns), suggesting that the number of fully activated epithelial sodium channels at the apical surface after trypsin was similar. Assuming that all apical surface ENaC is fully activated by trypsin in this protocol, these data suggest that Sec24D depletion does not alter ENaC surface expression, which was confirmed in surface biotinylation experiments (Fig. 1, G (representative experiment) and H (densitometry of n = 5 individual experiments), p = ns).
Similar experiments in CFBE41o− cells that endogenously express functional ENaC demonstrated that siRNA-mediated depletion of Sec24D (representative immunoblot, Fig. 1B) also resulted in decreased baseline ENaC Isc (representative Isc traces in Fig. 1D, summary data in Fig. 1F, n = 19, p = 0.0026), suggesting that these observations are neither species– or cell type–specific; nor are they an artifact of exogenous ENaC expression in MDCK cells. Taken together, these data support the hypothesis that Sec24D, like ERp29, is a determinant of ENaC functional expression by regulating its fractional cleavage state and therefore its Po.
ERp29's KEEL ER retention motif is a determinant in regulating ENaC cleavage in MDCK cells
We have previously demonstrated that ERp29 regulates the interaction of β-ENaC with Sec24D (2), and Fig. 1 further demonstrates the potential importance of this interaction with regard to the fractional cleavage and functional expression of ENaC at the surface of epithelial cells. We next tested the influence of mutating ERp29's KEEL ER retention motif on ENaC functional expression in Ussing chambers as well as the interaction of β-ENaC with Sec24D.
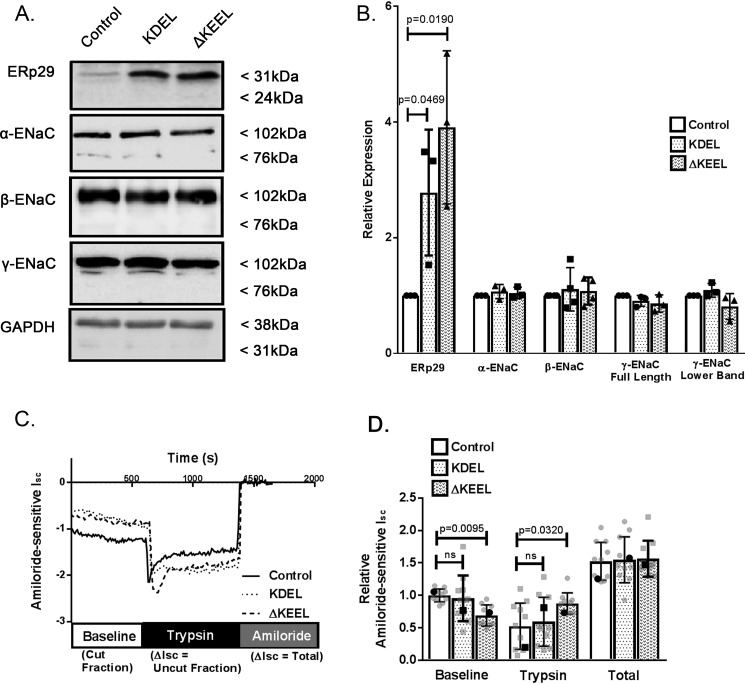
In this context, we constructed two ERp29 mutants: 1) a mutant ERp29 containing a KDEL retention motif (KDEL ERp29) that should more avidly interact with the KDEL-R and be better retained in the ER and/or returned to the ER from the proximal Golgi by the KDEL-R and 2) A KEEL-deleted mutant (ΔKEEL ERp29) that should interact less strongly with the KDEL-R. We tested whether overexpression of these mutants would regulate ENaC functional expression in αβγ-ENaC–expressing MDCK cells. Fig. 2A demonstrates successful overexpression of these mutant ERp29s as assessed by immunoblotting. We also assessed the whole-cell expression of the ENaC subunits in response to overexpression of these ERp29 mutants (Fig. 2A). ∼3.3-Fold and ∼2.8-fold increased expressions of KDEL and ΔKEEL ERp29, respectively, were obtained when αβγ-ENaC–expressing MDCK cells were transiently transfected with the appropriate plasmids, compared with cells transfected with control plasmid (Fig. 2, A (representative immunoblot) and D (densitometry of n = 3 experiments), p = 0.0469 and p = 0.0190, respectively). Overexpression of either KDEL or ΔKEEL ERp29 did not alter the whole-cell expression of α-, β-, or γ-ENaC (Fig. 2, A and B).
Figure 2.
ERp29's KEEL ER retention motif is determinant in regulating ENaC cleavage in MDCK cells. A, MDCK αβγ-ENaC cells were transfected with control plasmid (pSK−), or plasmids expressing either KDEL ERp29 or ΔKEEL Erp29. ERp29 or the individual ENaC subunits were detected by immunoblot analysis of whole-cell lysate proteins using anti-ERp29, anti-HA (α-ENaC), anti-V5 (β-ENaC), and anti-γ-ENaC. B, densitometric quantification of the relative expression of the ERp29 and the ENaC subunits for n = 3–4 independent experiments (p = 0.0469 ERp29-KDEL versus control, p = 0.0190 ERp29-ΔKEEL versus control by ANOVA). C and D, MDCK αβγ-ENaC cells transfected with control plasmid (pSK−) or plasmids expressing either KDEL ERp29 or ΔKEEL Erp29 were grown as polarized monolayers and mounted in Ussing chambers for Isc measurements. C, representative Isc experiment. D, summary data for amiloride-sensitive Isc measurements normalized to control amiloride-sensitive baseline Isc (individual data points in gray and mean ± S.D. (error bars), representative data from C in black), demonstrating decreased baseline and increased trypsin response ENaC-mediated Isc versus control in ΔKEEL ERp29–transfected cells (n = 11; p = 0.0095 ΔKEEL versus control for baseline, p = 0.0320 ΔKEEL versus control for trypsin by ANOVA). For KDEL ERp29 versus control (n = 11), all p values were ns. Boldface points in D denote the representative Isc traces from C.
Cells overexpressing KDEL ERp29 did not show any significant change in baseline Isc or in Isc change after trypsin addition (representative Isc traces in Fig. 2C, summary data in Fig. 2D, baseline and trypsin; n = 11, p = ns and p = ns, respectively), suggesting that increased avidity of ERp29 for the KDEL-R did not alter ENaC biogenesis. However, cells overexpressing ΔKEEL ERp29 had reduced baseline Isc and a greater increase in currents after trypsin addition (Fig. 2C, baseline and trypsin; n = 9, p = 0.0095 and p = 0.032, respectively), consistent with less cleaved/active ENaC at the surface. Finally, in both cases, total amiloride-sensitive Isc was similar to controls (Fig. 2D, total; p = ns), suggesting that surface expression of ENaC was not modulated by either KDEL ERp29 or ΔKEEL ERp29, as was similarly observed in our previous work with WT ERp29, ERp29 C157S, and ERp29 siRNA-mediated depletion (2).
Taken together, these data support the hypothesis that ERp29's KEEL motif is a determinant in ENaC functional expression by regulating its baseline cleavage state and therefore its Po. These data are also consistent with expression of these ERp29 mutants not altering ENaC surface expression and provide further support for a potential role of ERp29's interaction with the KDEL-R as a determinant of ENaC biogenesis.
ERp29's KEEL motif is a determinant in regulating the cleavage state of ENaC at the apical surface but not total ENaC surface expression
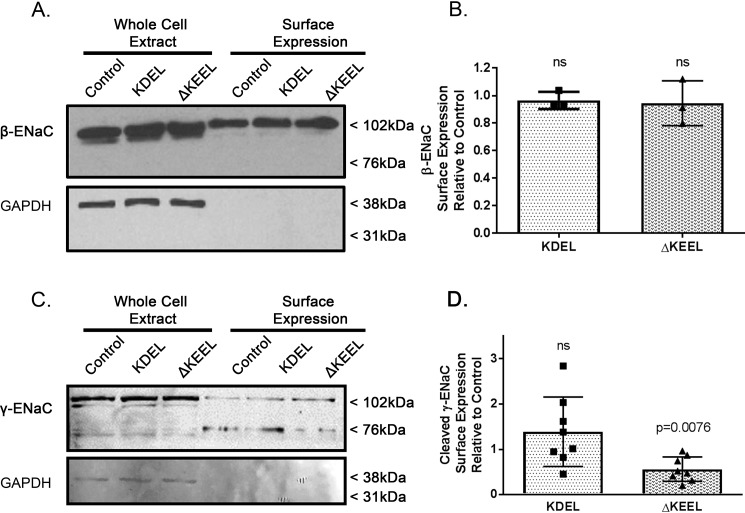
We performed surface biotinylation experiments to directly confirm that ERp29 does not modulate the total amount of ENaC at the apical surface (Fig. 3, A and B). As predicted by our data in Fig. 2, regarding ENaC functional expression in Ussing chambers, β-ENaC expression at the apical surface when either KDEL or ΔKEEL ERp29 was overexpressed was similar to each other and to controls (Fig. 3, A (representative experiment) and B (densitometry of n = 3 independent experiments), p = ns). These data thus suggest that ERp29's C-terminal KEEL motif specifically modulates ENaC functional expression by altering its cleavage state/Po and not its abundance at the surface.
Figure 3.
ERp29's KEEL motif is a determinant in regulating the cleavage state of ENaC at the apical surface but not total ENaC surface expression. MDCK αβγ-ENaC cells were transiently transfected with KDEL or ΔKEEL ERp29 and grown as polarized monolayers. Control cells were transfected with pSK−. Apical surface proteins were biotinylated and isolated by neutravidin precipitation, and β-ENaC (A) or γ-ENaC (C) was revealed by immunoblotting. In these representative immunoblots (A and C), the presence of GAPDH in the whole-cell lysates and absence of GAPDH immunoreactivity in the surface expression samples suggests that intracellular proteins were not labeled by the surface biotinylation reagent. B, densitometric quantification of β-ENaC surface expression relative to control (n = 3 independent experiments, p = ns by Wilcoxon signed-rank test). D, densitometric quantification of γ-ENaC surface expression for n = 8 independent experiments (control versus KDEL p = ns, control versus ΔKEEL ERp29 p = 0.0076, by Wilcoxon signed-rank test). Error bars, S.D.
To further test this hypothesis, the surface expression of the γ-ENaC subunit was analyzed (Fig. 3, C and D). Here we observed a significant decrease in cleaved γ-ENaC (∼72 kDa) at the surface when ΔKEEL ERp29 was overexpressed, but not when KDEL ERp29 was overexpressed (Fig. 3D, control versus ΔKEEL ERp29, p = 0.0076, n = 8). In these experiments, surface expression of uncleaved γ-ENaC (∼102 kDa) was not significantly different from control when either KDEL or ΔKEEL ERp29 was expressed (summary data not shown, p = ns).
Together, these data further support the hypothesis that (ΔKEEL) ERp29 alters ENaC functional expression by modulating the cleavage state of the γ-ENaC subunit, rather than through either modulation of whole-cell expression of the ENaC subunits or by modulation of total ENaC expression at the apical surface.
ERp29's KEEL motif modulates ENaC interaction with Sec24D
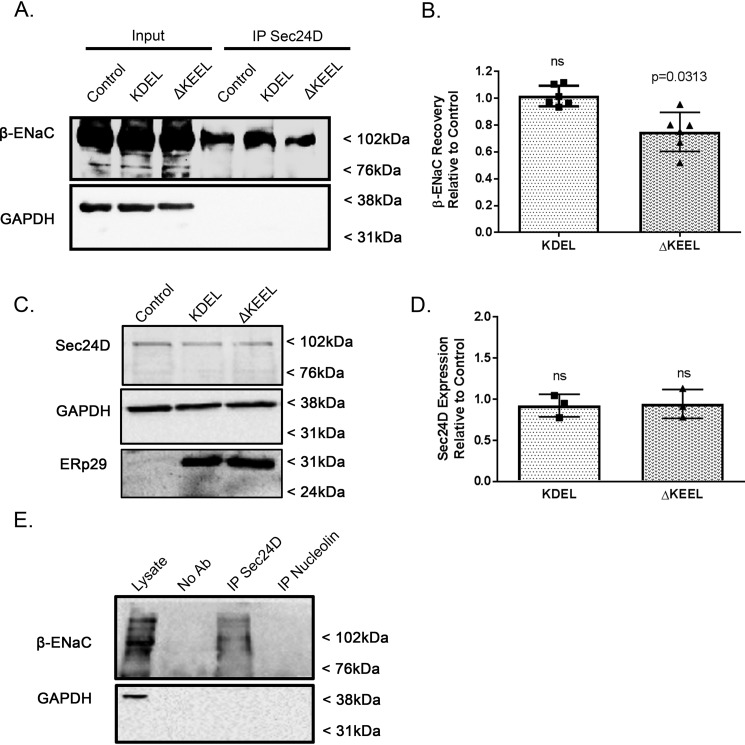
Using a co-immunoprecipitation approach (Fig. 4), we next confirmed our previous finding that β-ENaC interacts with Sec24D (2, 9) and found that the interaction of β-ENaC and Sec24D was not changed by overexpression of KDEL ERp29 (Fig. 4; n = 6, p = ns). In contrast, overexpression of ΔKEEL ERp29 caused a small but significant decrease in the association of β-ENaC with Sec24D (Fig. 4; n = 6, p = 0.0313 compared with control). Transfection of these ERp29 mutants did not alter the whole-cell expression of Sec24D (Fig. 4, C and D), suggesting that decreased Sec24D abundance was not the cause of decreased β-ENaC/Sec24D association. Fig. 4E presents control experiments demonstrating the specificity of the co-precipitation of β-ENaC with Sec24D and confirm our previously published control experiments in this regard (9). As we have previously demonstrated that ERp29 promotes the association of ENaC with Sec24D (2), these data suggest that ERp29's C-terminal KEEL motif plays an important role in this function of ERp29. These data also begin to suggest a potential role for the interaction of ERp29 and the KDEL-R in regulating ENaC's association with Sec24D and its movement from ER to Golgi via COP II.
Figure 4.
Deletion of ERp29's C-terminal KEEL motif decreases association of β-ENaC with Sec24D. MDCK αβγ-ENaC cells were transfected with either KDEL or ΔKEEL ERp29 or mock-transfected (Control). A and B, cell lysates were prepared under nondenaturing conditions and subject to immunoprecipitation (IP) with anti-Sec24D. β-ENaC was detected in immunoblots of the precipitated proteins and whole-cell lysates (Input) using anti-V5. GAPDH was detected in immunoblots of the lysates, but not the precipitated proteins, suggesting that there was not nonspecific co-precipitation of this housekeeping protein. A, representative experiment. Input lanes, 10% of the total protein subject to immunoprecipitation. B, densitometric quantification of co-precipitated β-ENaC (n = 6 independent experiments), suggesting that expression of ΔKEEL ERp29 decreases co-precipitation of β-ENaC with Sec24D relative to the amount of β-ENaC that co-precipitates with Sec24D under control conditions (p = 0.0313, by Wilcoxon signed-rank test). Expression of KDEL ERp29 did not alter the co-precipitation of β-ENaC with Sec24D compared with control (n = 6, p = ns). C and D, MDCK αβγ-ENaC cells were transfected with either KDEL- or ΔKEEL-ERp29 or mock-transfected (Control), and whole-cell lysates were prepared. Expression of Sec24D and ERp29 was revealed by immunoblot, with GAPDH immunoreactivity as a loading control. C, representative immunoblots. D, densitometric quantification of Sec24D in MDCK αβγ-ENaC cells that were transfected with either KDEL- or ΔKEEL-ERp29 relative to the amount of Sec24 in control (n = 3, p = ns). E, MDCK αβγ-ENaC cell lysate was prepared under nondenaturing conditions and was subject to immunoprecipitation without primary antibody (No Ab control), with anti-Sec24D, or with anti-nucleolin as a nonspecific antibody control. β-ENaC and GAPDH (as a nonspecific interaction control) in the precipitated proteins were revealed by immunoblotting. Lysate lane, 10% of the total protein subject to immunoprecipitation. Error bars, S.D.
The KDEL receptor regulates the cleavage state of ENaC at the apical surface
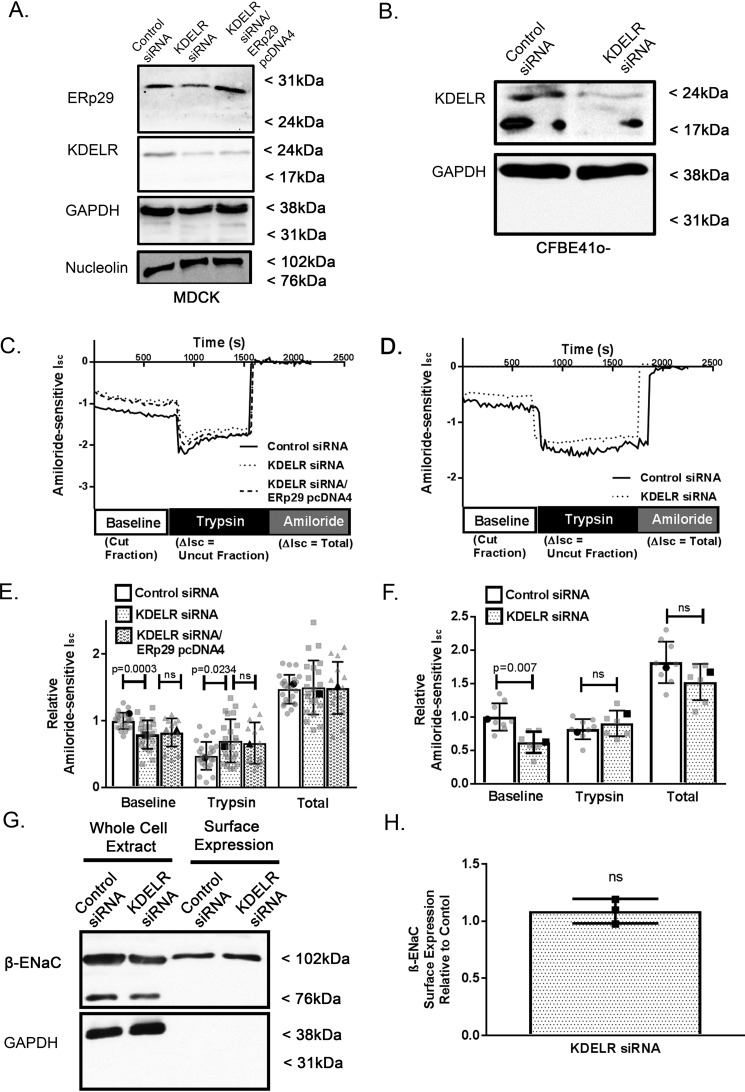
Because our data suggest that ERp29's KEEL motif plays a significant role in regulating ENaC cleavage state at the apical surface, we tested whether the KDEL receptor itself influences ENaC functional expression and cleavage state. MDCK αβγ-ENaC cells (Fig. 5, A, C, and E) or CFBE41o− cells (Fig. 5, B, D, and F) were transfected with nontargeting (control) or KDEL-R1–specific siRNA and grown as polarized monolayers. These monolayers were then mounted in Ussing chambers for Isc measurements (Fig. 5); siRNA-mediated depletion of KDEL-R expression was confirmed by immunoblotting of whole-cell lysates after cells had undergone Isc measurements (Fig. 5, A (MDCK αβγ-ENaC cells) and B (CFBE41o− cells)).
Figure 5.
The KDEL receptor regulates ENaC cleavage in MDCK αβγ-ENaC and CFBE41o− epithelial cells. A, C, and E, MDCK αβγ-ENaC cells were transiently co-transfected with pSK− plasmid and either nontargeting (control siRNA) or KDEL-R1 siRNA (KDELR siRNA) or co-transfected with both KDEL-R1 siRNA– and WT ERp29–expressing plasmid (KDELR siRNA/ERp29 pcDNA4). A, immunoblot analysis confirming decreased expression of KDEL-R and/or overexpression of ERp29 in whole-cell lysates (immunoblots for GAPDH and nucleolin serve as loading controls). C and E, depletion of the KDEL-R decreased baseline ENaC-mediated Isc (control, KDEL-R1 siRNA n = 24 baseline control versus KDEL-R siRNA p = 0.0003, trypsin control versus KDEL-R siRNA p = 0.0234 by ANOVA); however, overexpression of ERp29 did not rescue the decrease in baseline ENaC-medicated Isc (KDEL-R1/WT ERp29 n = 15, p = ns versus KDEL-R siRNA by ANOVA). Boldface points in C denote the representative Isc traces from E. B, D, and F, CFBE41o− cells were transfected with nontargeting (control) or KDEL-R1 siRNA. B, immunoblot analysis confirming depletion of KDEL-R expression. D and F, Isc measurements of KDEL-R–depleted CFBE41o− epithelial cells demonstrating decreased baseline ENaC-mediated Isc (n = 9, p = 0.0007). Boldface points in F denote the representative Isc traces from D. Shown are a representative surface biotinylation experiment (G) and densitometric quantification (H) of the relative expression of the β-ENaC subunits at the surface of transfected MDCK αβγ-ENaC, demonstrating that depletion of the KDEL-R does not alter β-ENaC surface expression (n = 3 independent experiments, p = ns by Wilcoxon signed-rank test). Error bars, S.D.
When KDEL-R1 was depleted in MDCK αβγ-ENaC cells, there was a significant decrease in baseline ENaC-mediated Isc (Fig. 5, E (baseline); n = 24, p = 0.0003) and a greater increase in amiloride-sensitive Isc than in controls after the application of trypsin (Fig. 5E, trypsin; n = 24, p = 0.0234). The total amiloride-sensitive Isc was again similar for control and KDEL-R1–depleted cells (Fig. 5E, total; n = 24, p = ns), suggesting that the number of fully activated epithelial sodium channels at the apical surface after trypsin was similar and therefore that depletion of KDEL-R1 alters the ENaC cleavage state without altering its surface expression.
To further assess whether ERp29 may act through or in concert with KDEL-R1 to regulate ENaC biogenesis, we tested whether overexpression of WT ERp29 could reverse or overcome the effect of KDEL-R1 depletion. MDCK αβγ-ENaC cells were transiently transfected with nontargeting (control) siRNA or KDEL-R1–specific siRNA or co-transfected with KDEL-R1-specific siRNA and a plasmid expressing WT ERp29 (ERp29 pcDNA4). Fig. 5A demonstrates successful depletion of KDEL-R1 and overexpression of WT ERp29 in these experiments. As shown in Fig. 5E, overexpression of ERp29 did not reverse this decrease in baseline ENaC-mediated Isc (n = 15, p = ns versus KDEL-R siRNA) and demonstrated no differences in the trypsin response (p = ns versus KDEL-R siRNA).
Surface biotinylation experiments were performed to directly confirm that total surface ENaC expression was not altered by KDEL-R1 depletion (Fig. 5, G (representative experiment) and H (densitometry of n = 3 independent experiments), p = ns).
To further confirm that this phenomenon was not MDCK αβγ-ENaC cell–specific, similar experiments were performed using siRNA to deplete expression of KDEL-R1 in CFBE41o− cells. A representative immunoblot analysis of whole-cell lysates shown in Fig. 5B demonstrates successful depletion of KDEL-R1 expression, and representative amiloride-sensitive Isc traces are shown in Fig. 5D. As summarized in Fig. 5E, depletion of KDEL-R1 caused a significant decrease in baseline ENaC-mediated Isc CFBE41o− cells (Fig. 5E, n = 9, p = 0.0007).
Taken together, these data suggest that the KDEL-R, most likely KDEL-R1, modulates ENaC functional expression by regulating its cleavage state and therefore its Po. These data therefore further support the hypothesis that the KDEL-R plays a role in the regulation of ENaC biogenesis and trafficking.
β-ENaC associates with KDEL-R and ERp29
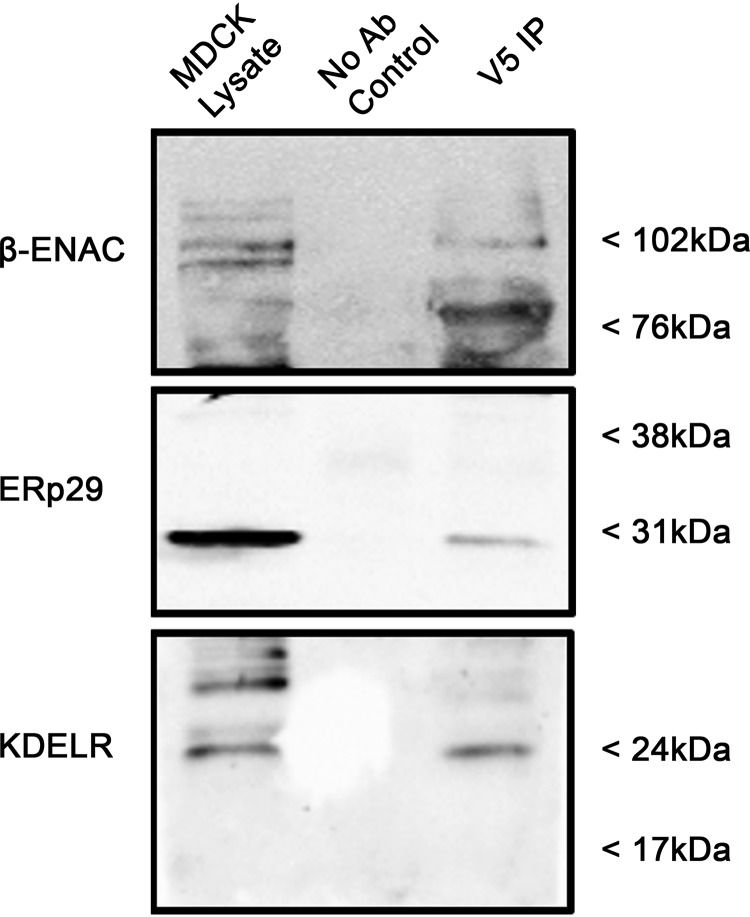
To demonstrate that both ERp29 and KDEL-R associate with ENaC, a co-immunoprecipitation experiment was performed (Fig. 6). Here, MDCK αβγ-ENaC lysates were prepared under nondenaturing conditions. Mouse anti-V5 antibody was used to precipitate (V5-tagged) β-ENaC and its interacting proteins. As shown in Fig. 6, both ERp29 and KDEL-R were readily detected in the proteins that co-precipitate with β-ENaC. Together, these data and the data demonstrating that the association of β-ENaC with Sec24D can be altered by expression of ΔKEEL ERp29 further support the hypothesis that the KDEL-R, by interacting with ERp29 and ENaC, may influence ENaC association with Sec24D and therefore ENaC biogenesis.
Figure 6.
β-ENaC associated with KDEL-R and ERp29. Whole-cell MDCK αβγ-ENaC lysates were prepared under nondenaturing conditions. β-ENaC and its interacting proteins were precipitated using mouse anti-V5 antibody. Anti-V5 was omitted from the precipitation depicted by the No Ab Control sample. β-ENaC (V5), ERp29, and KDLER were revealed by immunoblotting, with the lysate lane equal to 10% of the total lysate that was subjected to immunoprecipitation (IP). These data are representative of three independent experiments.
Discussion
ENaC represents the rate-limiting step of Na+ absorption across many epithelia (32) and plays a key role in the regulation of blood volume and blood pressure as well as airway liquid surface volume (33–35). Understanding ENaC regulation is therefore crucial in studying both hypertension and diseases of the airway, such as cystic fibrosis. Because of similarities in CFTR and ENaC biogenesis, we had previously tested the hypothesis that ERp29 regulates ENaC functional expression, and our data suggested that ERp29, by interacting with ENaC either directly or through a larger complex, directs ENaC for cleavage during biogenesis, which in turn increases ENaC functional expression (2). Data from this and other work from our group (2, 9) further suggested that the interaction of ENaC with the Sec24D cargo recognition component of COP II is a key step in this process. Here, we directly tested the hypothesis that modulating Sec24D expression would alter ENaC trafficking and functional expression and demonstrated that depletion of Sec24D decreased ENaC functional expression without altering expression of β-ENaC at the apical surface (Fig. 1). These data support the hypotheses that 1) ENaC association with Sec24D promotes ENaC cleavage and 2) uncleaved ENaC at the cell surface may have arrived there by a Sec24D/COP II-independent pathway.
Our previous work also demonstrated that either depleting ERp29 expression or mutating Cys157 of ERp29 could modulate both the interaction of ENaC with Sec24D and the fraction of uncleaved versus cleaved ENaC present at the surface of cultured epithelial cells (2). However, the mechanism by which ERp29 within the ER lumen promotes the association of ENaC with cytoplasmic Sec24D remained unclear. Therefore, we examined the role of ERp29's KEEL ER retention motif in ENaC's interaction with Sec24D and its biogenesis. As ERp29's C-terminal KEEL motif presumably promotes both the association of ERp29 with the organellar membranes (ER and Golgi) and the retention of ERp29 in the ER through its interaction with the KDEL-R, we interrogated whether both ERp29's C-terminal KEEL motif and the KDEL-R itself would also be crucial components of ENaC biogenesis. ERp29's KEEL ER retention motif is a KDEL variant that is associated with less robust ER retention (27); that ERp29's retention in the ER is “leaky” is consistent with our group's previously published data demonstrating that ERp29 was not confined to the ER and was also present both at the surface and in the conditioned medium of cultured epithelial cells (19). Interestingly, our data (Figs. 2–4) demonstrated that overexpression of KDEL ERp29, which should be more robustly retained in the ER, did not show any significant effects on ENaC biogenesis, suggesting that ERp29's role in ENaC biogenesis is not dependent on the strength of its association with KDEL-R beyond the proximal Golgi. In contrast, overexpression of ΔKEEL ERp29 had reduced baseline Isc and a greater increase in currents after the addition of trypsin (Fig. 2) as well as reduced cleaved γ-ENaC at the surface (Fig. 3, C and D). This difference in ENaC function was not due to differences in total ENaC (β-ENaC) surface expression (Fig. 3, A and B); nor were there changes in the whole-cell expression of any of the ENaC subunits (Fig. 2, A and B). These data mirror our previous data, where we interfered with ERp29 function by siRNA-mediated depletion or overexpression of ERp29 C157S (2), and therefore suggest that ERp29's KEEL ER retention motif is critical for ERp29's regulation of ENaC and promotion of ENaC biogenesis.
Additional evidence that ERp29's C-terminal KEEL is important in the regulation of ENaC biogenesis is found in Fig. 4, where we show that overexpression of the ΔKEEL ERp29 causes decreased β-ENaC association with Sec24D. Interestingly, overexpression of KDEL ERp29 did not significantly alter this β-ENaC/Sec24D association. Together, these data suggest that ERp29's C-terminal KEEL motif, or the similar KDEL motif, is crucial in modulating ENaC biogenesis and functional expression (Fig. 3) by promoting its interaction with Sec24D (Fig. 4). These data also suggested the hypothesis that the KDEL-R, the most likely interaction partner of ERp29's C-terminal ER retention domain, may also play a role in directing ENaC for cleavage during biogenesis. Considering the mechanism(s) underlying these effects, we interrogated whether the KDEL-R itself was playing a role in ENaC biogenesis and forward trafficking.
Much previous work has focused primarily on the role of the KDEL-R in retrograde trafficking of ER residents from the cis-Golgi back to the ER (36). However, the KDEL-R cycles between the ER and Golgi and is present in both COP II and COP I vesicles (37), and it has been shown to be highly concentrated at budding ER exit sites, where COP II vesicles form (38). We hypothesized that the KDEL-R plays a role in forward trafficking of ENaC via COP II vesicles. In support of this hypothesis, the depletion of KDEL-R (using siRNA targeted toward KDEL-R1) resulted in a significant decrease in baseline ENaC-mediated Isc (Fig. 5, C and E) and a greater increase in trypsin-stimulated, amiloride-sensitive Isc in MDCK cells (Fig. 5E), again with no significant difference in total ENaC at the surface (Fig. 5, E, G, and H). These data, which mirror those for the effects of interfering with ERp29 and Sec24D function, support the hypothesis that KDEL-R1 regulates ENaC functional expression by altering its cleavage state, perhaps by facilitating the process by which ENaC is directed to the Golgi for cleavage during biogenesis. It is important to note that in our experiments, we are inferring that we are depleting KDEL-R1 expression with specific siRNA by immunoblotting. A potential limitation of this interpretation is that the commercial antibody directed at KDEL-R1 that we used to probe for KDEL-R on these blots likely cross-reacts with KDEL-R2 (peptide antigen, 21 of 21 amino acids identical to KDEL-R1) and KDEL-R3 (peptide antigen, 20 of 21 amino acids identical to KDEL-R1). Future studies will be necessary to rigorously address the expression and function of the other KDEL-R isoforms (KDEL-R2 and KDEL-R3) in these model systems and the potential role of these other KDEL-R isoforms in ENaC biogenesis.
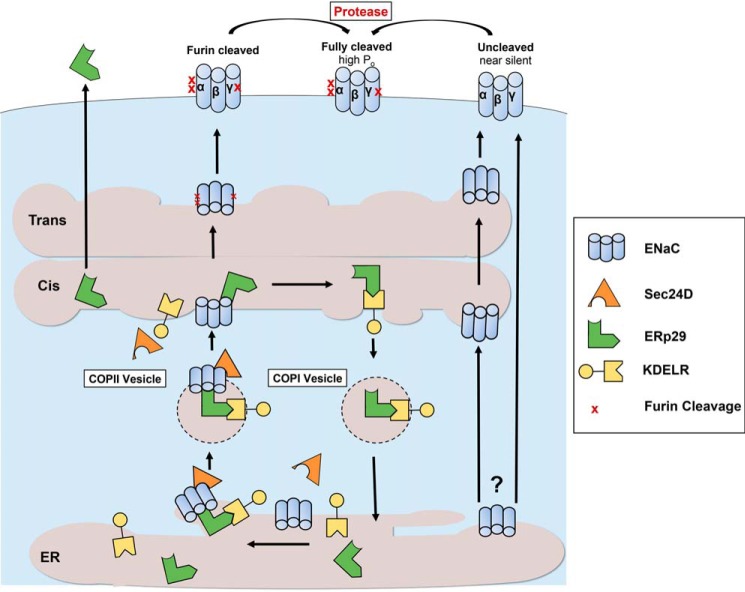
Together, these data and our previous work (2) further support a model of ENaC biogenesis outlined in Fig. 7, where ERp29 interacts with ENaC, and the ENaC/ERp29 complex subsequently interacts with the KDEL-R (likely KDEL-R1) via ERp29's KEEL motif. This promotes the association of ENaC with Sec24D and its transport to the Golgi via COP II vesicles. The cleavage of the luminal/extracellular loops of ENaC's α and γ subunits by furin then occurs in the trans-Golgi or later compartments (10), and such processing is disrupted when an ER exit signal is removed (39). Our data here and in our previous work (2) suggest that interference with any component of this model (ERp29, KDEL-R, or Sec24D) may cause ENaC to avoid furin processing and arrive at the plasma membrane in an uncleaved state.
Figure 7.
A model of KDEL-R and ERp29 in ENaC biogenesis. The KDEL-R works in concert with WT ERp29 to promote the association of ENaC with the Sec24D cargo recognition component of COP II at the ER membrane. This facilitates COP II–mediated transport of ENaC from the ER to the cis-Golgi. ENaC subsequently transverses the Golgi stacks to the trans-Golgi and later compartments, where it undergoes furin-mediated cleavage to its cleaved and higher-Po form. ERp29 that remains associated with the KDEL-R in the cis-Golgi returns to the ER via COP I vesicles to reinitiate the cycle. Without the KDEL-R, or when deleting ERp29's KDEL-R–binding motif (ΔKEEL), ENaC has decreased association with Sec24D/COP II and thus can bypass processing/cleavage in the Golgi to reach the surface in its uncleaved, low-Po (nearly silent) form.
To further test this model and to address whether ERp29 may act through or in concert with KDEL-R in a complex, we tested whether overexpression of WT ERp29 could reverse or overcome the effect of KDEL-R depletion. The decrease in baseline ENaC-mediated Isc resulting from the knockdown of KDEL-R1 in MDCK αβγ (Fig. 5E) could not be rescued by overexpression of ERp29. These data suggest that depletion of the KDEL-R blocks the influence of ERp29 on ENaC biogenesis and cleavage and therefore further support the hypothesis that the KDEL-R regulates ENaC biogenesis and cleavage downstream of ERp29.
Interestingly, our data here also begin to uncover factors that may alter the reciprocal relationship of decreasing ENaC cleavage and increasing uncleaved (trypsin-activatable) ENaC. Depletion of Sec24D (Fig. 1F) and KDEL-R in CFBE41o− cells (Fig. 5F) did not result in increased tryspin-stimulated Isc while baseline Isc was decreased. We have previously demonstrated that ERp29 expression is significantly greater in CFBE41o− cells than in MDCK αβγ-ENaC cells (2). These data support the hypothesis that increased ERp29 may have stabilized ENaC in the ER when ER exit machinery is made limiting and prevented ENaC from getting to the apical membrane in its uncleaved form through the proposed non-COP II pathway. In contrast, depletion or mutation of ERp29 allows ENaC to escape the ER through the proposed non-COP II pathway. These data also suggest the hypotheses that the ratio of ERp29 to KDEL-R and/or Sec24D/COPII may determine ENaC's trafficking itinerary and that COP II's overall capacity to carry proteins out of the ER may, in fact, be limiting under certain conditions. Future studies will test these hypotheses.
Experimental procedures
Cell culture
MDCK type I cells that stably express C-terminally epitope-tagged murine ENaC subunits (α-HA, β-V5, and γ-Myc, MDCK αβγ-ENaC) were a gift of Dr. T. Kleyman (University of Pittsburgh) and were maintained in 50:50 Dulbecco's modified Eagle's medium (Gibco) and Ham's F-12 (CellGro) supplemented with 10% fetal bovine serum (Gemini), streptomycin (100 μg/ml), and penicillin (100 units/ml) (Invitrogen). These cells were maintained under antibiotic selective pressure using hygromycin (Roche Applied Science), blasticidin (Invitrogen), and G418 (CellGro).
CFBE41o− parental cells (immortalized human cystic fibrosis bronchial epithelial cells) were previously obtained from Dr. J. P. Clancy (then at the University of Alabama (Birmingham, AL), now at Cincinnati Children's Hospital Medical Center) and cultured as detailed previously by our group (28). We have previously demonstrated that these cells endogenously express functional ENaC (28).
MDCK or CFBE41o− cells were grown on polarized monolayers on Snapwells (Costar, Corning Life Sciences) for ion transport assays or on Transwells for biochemical assays. Once cells had achieved resistances of ≥ 400 Ω·cm2, cells were treated with 1 μg/ml dexamethasone (Sigma-Aldrich) for 48 h prior to experimentation.
Transient overexpression of ERp29
pcDNA4 plasmids encoding WT, KDEL, or ΔKEEL ERp29 were generated by site-directed mutagenesis as described previously (17), and plasmid maxi-preps were prepared using the QIAfilter Plasmid Maxi-Prep kit (Qiagen). Sequences of mutant plasmids were confirmed by automated sequence analysis in the Children's Hospital of Philadelphia Nucleic Acid and Protein Core Facility. Transient transfections of αβγ-ENaC–expressing MDCK cells were performed with Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. For transfections of cells on tissue culture plates, 2 μg of plasmid was used, and for transfections of cells on Snapwells or Transwells, 0.1 μg/cm2 plasmid plus 0.5 μg/cm2 carrier DNA was used. Cells were subsequently assayed 48–72 h after transfection as described below.
Depletion of Sec24D or the KDEL-R by siRNA
Sec24D or KDEL-R1 expression was depleted using pools of four specific siRNAs (Dharmacon SMARTpool ON-TARGETplus Sec24D siRNA (L-008493-01) or Dharmacon SMARTpool ON-TARGETplus KDELR1 siRNA (L-019136-01), respectively). 20 pmol of the specific pooled siRNA or control siRNA (Dharmacon ON-TARGETplus Nontargeting Control siRNA Pool D-001810-10) was delivered to MDCK or CFBE41o− cells by transfection with Lipofectamine 2000 according to the manufacturer's protocol. Cells were subsequently assayed 48–72 h after transfection as described below.
Antibodies
Rabbit anti-ERp29 (ab11420) and mouse monoclonal anti-KDEL-R (ab69659) were purchased from Abcam (Cambridge, MA). Mouse monoclonal anti-KDEL-R (NPB2-12873) was purchased from Novus (Littleton, CO). (It should be noted that this antibody likely recognizes KDEL-R1, -R2 and -R3; however, in these experiments we focused on the 24 kDa band, which demonstrated decreased expression when cells were treated with the KDEL-R1–specific siRNA mentioned above.) Rabbit anti-α-ENaC (PA1-920A) and rabbit anti-γ-ENaC (PA1-922) were from Thermo Fisher Scientific. Rabbit anti-HA (to detect α-ENaC) was from BD Biosciences (catalog no. 631207). Rabbit anti-c-Myc (to detect γ-ENaC) was from Sigma–Aldrich (C3956). Mouse monoclonal anti-V5 (to detect β-ENaC) was from Invitrogen (46-0705). Mouse monoclonal anti-GAPDH was from Millipore (Billerica, MA). Rabbit Anti-nucleolin (ab22758) was from Abcam (Cambridge, MA). Horseradish peroxidase–conjugated secondary antibodies (anti-mouse (NA931V) and anti-rabbit (NA934V)) were from GE Healthcare.
Immunoblotting
Our general techniques for immunoblot analyses have been published previously (2). In brief, whole-cell lysates were prepared by incubating cells on ice for 30 min in RIPA buffer (150 mm NaCl, 50 mm Tris-HCl, pH 8, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) containing a 1:1000 dilution of protease inhibitor mixture (Sigma–Aldrich). The lysates were collected, passaged through a 21-gauge needle, and cleared by centrifugation (14,000 × g for 15 min at 4 °C). Protein content in the lysate supernatants was determined using DC protein assay reagents (Bio-Rad) and BSA as a standard. Samples were denaturated using 6× Laemmli sample buffer (125 mm Tris, pH 6.8, 4% SDS, 10% glycerol, 0.006% bromphenol blue, 1.8% 2-mercaptoethanol, final concentration 1–2×), and equal amounts of protein (typically 50 μg) were resolved using SDS-PAGE and transferred to nitrocellulose using semi-dry techniques (Bio-Rad). Nonspecific protein binding was diminished by incubating the membrane in 5% nonfat milk in TBS (10 mm Tris-HCl, pH 8, 150 mm NaCl) with 0.1% Tween 20. Primary antibodies and horseradish peroxidase–conjugated secondary antibodies were applied in TBS with 0.1% Tween 20 with 1% nonfat milk. Immunoreactivity was detected by chemiluminescence (SuperSignal West Pico or Femto; Thermo Fisher Scientific) and fluorography using either film (Hyblot ECL, Amersham Biosciences) or a ChemiDoc Touch Imaging System (version 5.2.1; Bio-Rad). Densitometry was performed using an Alpha-Imager 2200 system (AlphaInnotech, Santa Clara, CA) for film exposures or the Image Lab software (Bio-Rad) for images visualized by the ChemiDoc system.
Co-immunoprecipitation
For co-immunoprecipitation experiments, cells were lysed under nondenaturing conditions in RIPA buffer without SDS, and protein content was determined as above. Protein A-agarose beads (catalog no. 22811, Thermo Fisher Scientific) that had been preincubated with primary antibody for 1 h were then incubated overnight with cell lysate proteins (500 μg of total protein) at 4 °C. Precipitated proteins were released by heating the samples for 3.5 min at 90 °C in 2× Laemmli sample buffer, resolved by SDS-PAGE, and revealed by immunoblotting.
Surface biotinylation
MDCK αβγ-ENaC cells were transiently transfected with WT or mutant ERp29 or were treated with siRNA for Sec24D or KDEL-R1 and grown on Transwells until transepithelial resistance of ≥500 Ω·cm2 was achieved, or they were grown in a T25 flask. The cells were placed on ice for 30 min and washed with PBS containing Ca2+ and Mg2+, and their apical surface was exposed to 1 mg/ml Sulfo-NHS-SS-biotin (Thermo Fisher Scientific) in the biotinylation buffer (10 mm H3BO4, 137 mm NaCl, 1 mm CaCl2, pH 8.0) twice for 25 min on ice. The biotinylation reaction was terminated by washing the cells with a quenching buffer (192 mm glycine, 25 mm Tris-HCl, pH 8.3), followed by a 20-min incubation with this quenching buffer. After cell lysis in RIPA buffer, biotinylated proteins were precipitated using NeutrAvidin beads (catalog no. 29200, Thermo Fisher Scientific), resolved by SDS-PAGE, and revealed by immunoblotting. As in our previous work using this technique (2), we routinely assessed that GAPDH was present in whole-cell lysates but not in the neutravidin-precipitated proteins as a control for cellular integrity and lack of labeling of intracellular proteins in these experiments.
Transepithelial ion transport measurements in Ussing chambers
MDCK αβγ-ENaC or CFBE41o− cells were grown as polarized epithelial monolayers on Snapwells as described above. When transepithelial resistance was ≥300 Ω·cm2, as assessed by an epithelial volt-ohmmeter (World Precision Instruments, Sarasota, FL), cells were transfected with either control or specific siRNA, or with the indicated WT ERp29, KDEL ERp29, ΔKEEL ERp29, or control plasmid, or co-transfected with specific siRNA and WT ERp29 or control plasmid as described above and in the specific experiments. After 48 h and when transepithelial resistance was >700 Ω·cm2, cells were mounted in a vertical Ussing chamber (Physiologic Instruments, San Diego, CA) and underwent continuous voltage clamping for determination of Isc. The bath solutions were symmetric and contained 115 mm NaCl, 25 mm NaHCO3, 2.4 mm KH2PO4, 1.24 mm K2HPO4, 1.2 mm MgCl2, 1.2 mm CaCl2, 10 mm glucose, pH 7.4, at 37 °C. Isc was analyzed using Acquire & Analyze data acquisition software (Physiologic Instruments, San Diego, CA). Resistance was monitored and calculated by Ohm's law using 2-mV bidirectional pulses every 90 s. Apical application of 10 μm amiloride was used to define ENaC-mediated currents. 10 μg/ml trypsin (final concentration; Sigma-Aldrich) was added to the apical bath as indicated. Isc was determined at baseline and subsequently after trypsin and after amiloride were added to the apical surface of the same epithelium. Because of day-to-day variability in baseline Isc, data were normalized by the average amiloride-sensitive Isc at baseline in control cells for a given day's experiment prior to analysis (2).
Statistical analysis
Statistical significance was determined by a two-tailed Student's t test, a Mann–Whitney U or Wilcoxon rank-sum test (if data were not normally distributed), or one-way ANOVA techniques in the case of multiple comparisons, as appropriate. For immunoblotting data (including co-precipitation and surface biotinylation), densities of the experimental lanes are expressed relative to the densities of their respective controls, and a Wilcoxon signed-rank test was utilized to test for differences from the reference value of 1.0. Graphs of data were generated, and statistical analysis was performed using GraphPad Prism version 7.04; these graphs depict individual data points as well as means ± S.D. A p value of ≤0.05 was considered significant.
Author contributions
Y. B., J. V., L. S., and R. C. R. conceptualization; Y. B., J. V., M. N. O., L. B., M. B., and R. C. R. formal analysis; Y. B., J. V., M. N. O., L. B., M. B., J. L. J., D. G., L. S., and R. C. R. investigation; Y. B., J. V., M. N. O., L. B., M. B., J. L. J., D. G., L. S., and R. C. R. methodology; Y. B., J. V., and R. C. R. writing-original draft; Y. B., J. V., M. N. O., L. B., M. B., J. L. J., D. G., L. S., and R. C. R. writing-review and editing; R. C. R. resources; R. C. R. supervision; R. C. R. funding acquisition; R. C. R. visualization; R. C. R. project administration.
This work was supported by NHLBI, National Institutes of Health, Grant R01 HL135670 and Cystic Fibrosis Foundation Grants RUBENS15P0 and RUBENS16G0 (to R. C. R) as well as Cystic Fibrosis Foundation Post-doctoral Fellowship BIKARD16F0 (to Y. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ENaC
- epithelial sodium channel
- ER
- endoplasmic reticulum
- ERp29
- ER protein of 29 kDa
- KDEL-R
- KDEL receptor
- COP II
- coat complex II
- MDCK
- Madin–Darby canine kidney
- CFBE41o−
- cystic fibrosis bronchial epithelial
- Po
- open probability
- CFTR
- cystic fibrosis transmembrane conductance regulator
- ns
- not significant
- HA
- hemagglutinin
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- Ω
- ohms
- RIPA
- radioimmune precipitation assay
- ANOVA
- analysis of variance.
References
- 1. Duc C., Farman N., Canessa C. M., Bonvalet J. P., and Rossier B. C. (1994) Cell-specific expression of epithelial sodium channel α, β, and γ subunits in aldosterone-responsive epithelia from the rat: localization by in situ hybridization and immunocytochemistry. J. Cell Biol. 127, 1907–1921 10.1083/jcb.127.6.1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grumbach Y., Bikard Y., Suaud L., Chanoux R. A., and Rubenstein R. C. (2014) ERp29 regulates epithelial sodium channel functional expression by promoting channel cleavage. Am. J. Physiol. Cell Physiol. 307, C701–C709 10.1152/ajpcell.00134.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Althaus M. (2013) ENaC inhibitors and airway re-hydration in cystic fibrosis: state of the art. Curr. Mol. Pharmacol. 6, 3–12 10.2174/18744672112059990025 [DOI] [PubMed] [Google Scholar]
- 4. Debonneville C., Flores S. Y., Kamynina E., Plant P. J., Tauxe C., Thomas M. A., Münster C., Chraïbi A., Pratt J. H., Horisberger J. D., Pearce D., Loffing J., and Staub O. (2001) Phosphorylation of Nedd4–2 by Sgk1 regulates epithelial Na+ channel cell surface expression. EMBO J. 20, 7052–7059 10.1093/emboj/20.24.7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mall M., Grubb B. R., Harkema J. R., O'Neal W. K., and Boucher R. C. (2004) Increased airway epithelial Na+ absorption produces cystic fibrosis-like lung disease in mice. Nat. Med. 10, 487–493 10.1038/nm1028 [DOI] [PubMed] [Google Scholar]
- 6. Hansson J. H., Schild L., Lu Y., Wilson T. A., Gautschi I., Shimkets R., Nelson-Williams C., Rossier B. C., and Lifton R. P. (1995) A de novo missense mutation of the β subunit of the epithelial sodium channel causes hypertension and Liddle syndrome, identifying a proline-rich segment critical for regulation of channel activity. Proc. Natl. Acad. Sci. U.S.A. 92, 11495–11499 10.1073/pnas.92.25.11495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Staub O., Dho S., Henry P., Correa J., Ishikawa T., McGlade J., and Rotin D. (1996) WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 15, 2371–2380 10.1002/j.1460-2075.1996.tb00593.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Strautnieks S. S., Thompson R. J., Gardiner R. M., and Chung E. (1996) A novel splice-site mutation in the γ subunit of the epithelial sodium channel gene in three pseudohypoaldosteronism type 1 families. Nat. Genet. 13, 248–250 10.1038/ng0696-248 [DOI] [PubMed] [Google Scholar]
- 9. Chanoux R. A., Robay A., Shubin C. B., Kebler C., Suaud L., and Rubenstein R. C. (2012) Hsp70 promotes epithelial sodium channel functional expression by increasing its association with coat complex II and its exit from endoplasmic reticulum. J. Biol. Chem. 287, 19255–19265 10.1074/jbc.M112.357756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hughey R. P., Bruns J. B., Kinlough C. L., Harkleroad K. L., Tong Q., Carattino M. D., Johnson J. P., Stockand J. D., and Kleyman T. R. (2004) Epithelial sodium channels are activated by furin-dependent proteolysis. J. Biol. Chem. 279, 18111–18114 10.1074/jbc.C400080200 [DOI] [PubMed] [Google Scholar]
- 11. Adebamiro A., Cheng Y., Rao U. S., Danahay H., and Bridges R. J. (2007) A segment of γ ENaC mediates elastase activation of Na+ transport. J. Gen. Physiol. 130, 611–629 10.1085/jgp.200709781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hughey R. P., Bruns J. B., Kinlough C. L., and Kleyman T. R. (2004) Distinct pools of epithelial sodium channels are expressed at the plasma membrane. J. Biol. Chem. 279, 48491–48494 10.1074/jbc.C400460200 [DOI] [PubMed] [Google Scholar]
- 13. Adebamiro A., Cheng Y., Johnson J. P., and Bridges R. J. (2005) Endogenous protease activation of ENaC: effect of serine protease inhibition on ENaC single channel properties. J. Gen. Physiol. 126, 339–352 10.1085/jgp.200509285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bruns J. B., Carattino M. D., Sheng S., Maarouf A. B., Weisz O. A., Pilewski J. M., Hughey R. P., and Kleyman T. R. (2007) Epithelial Na+ channels are fully activated by furin- and prostasin-dependent release of an inhibitory peptide from the γ-subunit. J. Biol. Chem. 282, 6153–6160 10.1074/jbc.M610636200 [DOI] [PubMed] [Google Scholar]
- 15. Vuagniaux G., Vallet V., Jaeger N. F., Hummler E., and Rossier B. C. (2002) Synergistic activation of ENaC by three membrane-bound channel-activating serine proteases (mCAP1, mCAP2, and mCAP3) and serum- and glucocorticoid-regulated kinase (Sgk1) in Xenopus oocytes. J. Gen. Physiol. 120, 191–201 10.1085/jgp.20028598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shnyder S. D., and Hubbard M. J. (2002) ERp29 is a ubiquitous resident of the endoplasmic reticulum with a distinct role in secretory protein production. J. Histochem. Cytochem. 50, 557–566 10.1177/002215540205000413 [DOI] [PubMed] [Google Scholar]
- 17. Yan W., Samaha F. F., Ramkumar M., Kleyman T. R., and Rubenstein R. C. (2004) Cystic fibrosis transmembrane conductance regulator differentially regulates human and mouse epithelial sodium channels in Xenopus oocytes. J. Biol. Chem. 279, 23183–23192 10.1074/jbc.M402373200 [DOI] [PubMed] [Google Scholar]
- 18. Barak N. N., Neumann P., Sevvana M., Schutkowski M., Naumann K., Malesević M., Reichardt H., Fischer G., Stubbs M. T., and Ferrari D. M. (2009) Crystal structure and functional analysis of the protein disulfide isomerase-related protein ERp29. J. Mol. Biol. 385, 1630–1642 10.1016/j.jmb.2008.11.052 [DOI] [PubMed] [Google Scholar]
- 19. Suaud L., Miller K., Alvey L., Yan W., Robay A., Kebler C., Kreindler J. L., Guttentag S., Hubbard M. J., and Rubenstein R. C. (2011) ERp29 regulates ΔF508 and wild-type cystic fibrosis transmembrane conductance regulator (CFTR) trafficking to the plasma membrane in cystic fibrosis (CF) and non-CF epithelial cells. J. Biol. Chem. 286, 21239–21253 10.1074/jbc.M111.240267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hubbard M. J., McHugh N. J., and Carne D. L. (2000) Isolation of ERp29, a novel endoplasmic reticulum protein, from rat enamel cells: evidence for a unique role in secretory-protein synthesis. Eur. J. Biochem. 267, 1945–1957 10.1046/j.1432-1327.2000.01193.x [DOI] [PubMed] [Google Scholar]
- 21. Demmer J., Zhou C., and Hubbard M. J. (1997) Molecular cloning of ERp29, a novel and widely expressed resident of the endoplasmic reticulum. FEBS Lett. 402, 145–150 10.1016/S0014-5793(96)01513-X [DOI] [PubMed] [Google Scholar]
- 22. Tang B. L., Wong S. H., Qi X. L., Low S. H., and Hong W. (1993) Molecular cloning, characterization, subcellular localization and dynamics of p23, the mammalian KDEL receptor. J. Cell Biol. 120, 325–338 10.1083/jcb.120.2.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lewis M. J., and Pelham H. R. (1992) Ligand-induced redistribution of a human KDEL receptor from the Golgi complex to the endoplasmic reticulum. Cell 68, 353–364 10.1016/0092-8674(92)90476-S [DOI] [PubMed] [Google Scholar]
- 24. Munro S., and Pelham H. R. (1987) A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907 10.1016/0092-8674(87)90086-9 [DOI] [PubMed] [Google Scholar]
- 25. Deleted in proof.
- 26. Deleted in proof.
- 27. Sargsyan E., Baryshev M., Szekely L., Sharipo A., and Mkrtchian S. (2002) Identification of ERp29, an endoplasmic reticulum lumenal protein, as a new member of the thyroglobulin folding complex. J. Biol. Chem. 277, 17009–17015 10.1074/jbc.M200539200 [DOI] [PubMed] [Google Scholar]
- 28. Suaud L., Miller K., Panichelli A. E., Randell R. L., Marando C. M., and Rubenstein R. C. (2011) 4-Phenylbutyrate stimulates Hsp70 expression through the Elp2 component of elongator and STAT-3 in cystic fibrosis epithelial cells. J. Biol. Chem. 286, 45083–45092 10.1074/jbc.M111.293282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chanoux R. A., Shubin C. B., Robay A., Suaud L., and Rubenstein R. C. (2013) Hsc70 negatively regulates epithelial sodium channel trafficking at multiple sites in epithelial cells. Am. J. Physiol. Cell Physiol. 305, C776–C787 10.1152/ajpcell.00059.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hanwell D., Ishikawa T., Saleki R., and Rotin D. (2002) Trafficking and cell surface stability of the epithelial Na+ channel expressed in epithelial Madin-Darby canine kidney cells. J. Biol. Chem. 277, 9772–9779 10.1074/jbc.M110904200 [DOI] [PubMed] [Google Scholar]
- 31. Tong Z., Illek B., Bhagwandin V. J., Verghese G. M., and Caughey G. H. (2004) Prostasin, a membrane-anchored serine peptidase, regulates sodium currents in JME/CF15 cells, a cystic fibrosis airway epithelial cell line. Am. J. Physiol. Lung Cell Mol. Physiol. 287, L928–L935 10.1152/ajplung.00160.2004 [DOI] [PubMed] [Google Scholar]
- 32. Hanukoglu I., and Hanukoglu A. (2016) Epithelial sodium channel (ENaC) family: phylogeny, structure-function, tissue distribution, and associated inherited diseases. Gene 579, 95–132 10.1016/j.gene.2015.12.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rossier B. C. (2014) Epithelial sodium channel (ENaC) and the control of blood pressure. Curr. Opin. Pharmacol. 15, 33–46 10.1016/j.coph.2013.11.010 [DOI] [PubMed] [Google Scholar]
- 34. Büsst C. J. (2013) Blood pressure regulation via the epithelial sodium channel: from gene to kidney and beyond. Clin. Exp. Pharmacol. Physiol. 40, 495–503 10.1111/1440-1681.12124 [DOI] [PubMed] [Google Scholar]
- 35. Hobbs C. A., Da Tan C., and Tarran R. (2013) Does epithelial sodium channel hyperactivity contribute to cystic fibrosis lung disease? J. Physiol. 591, 4377–4387 10.1113/jphysiol.2012.240861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Capitani M., and Sallese M. (2009) The KDEL receptor: new functions for an old protein. FEBS Lett. 583, 3863–3871 10.1016/j.febslet.2009.10.053 [DOI] [PubMed] [Google Scholar]
- 37. Scales S. J., Pepperkok R., and Kreis T. E. (1997) Visualization of ER-to-Golgi transport in living cells reveals a sequential mode of action for COPII and COPI. Cell 90, 1137–1148 10.1016/S0092-8674(00)80379-7 [DOI] [PubMed] [Google Scholar]
- 38. Martínez-Menárguez J. A., Geuze H. J., Slot J. W., and Klumperman J. (1999) Vesicular tubular clusters between the ER and Golgi mediate concentration of soluble secretory proteins by exclusion from COPI-coated vesicles. Cell 98, 81–90 10.1016/S0092-8674(00)80608-X [DOI] [PubMed] [Google Scholar]
- 39. Mueller G. M., Kashlan O. B., Bruns J. B., Maarouf A. B., Aridor M., Kleyman T. R., and Hughey R. P. (2007) Epithelial sodium channel exit from the endoplasmic reticulum is regulated by a signal within the carboxyl cytoplasmic domain of the α subunit. J. Biol. Chem. 282, 33475–33483 10.1074/jbc.M707339200 [DOI] [PubMed] [Google Scholar]