Abstract
A heterobimetallic Mn/Fe cofactor is present in the R2 subunit of class Ic ribonucleotide reductases (R2c) and in R2-like ligand-binding oxidases (R2lox). Although the protein-derived metal ligands are the same in both groups of proteins, the connectivity of the two metal ions and the chemistry each cofactor performs are different: in R2c, a one-electron oxidant, the Mn/Fe dimer is linked by two oxygen bridges (μ-oxo/μ-hydroxo), whereas in R2lox, a two-electron oxidant, it is linked by a single oxygen bridge (μ-hydroxo) and a fatty acid ligand. Here, we identified a second coordination sphere residue that directs the divergent reactivity of the protein scaffold. We found that the residue that directly precedes the N-terminal carboxylate metal ligand is conserved as a glycine within the R2lox group but not in R2c. Substitution of the glycine with leucine converted the resting-state R2lox cofactor to an R2c-like cofactor, a μ-oxo/μ-hydroxo–bridged MnIII/FeIII dimer. This species has recently been observed as an intermediate of the oxygen activation reaction in WT R2lox, indicating that it is physiologically relevant. Cofactor maturation in R2c and R2lox therefore follows the same pathway, with structural and functional divergence of the two cofactor forms following oxygen activation. We also show that the leucine-substituted variant no longer functions as a two-electron oxidant. Our results reveal that the residue preceding the N-terminal metal ligand directs the cofactor's reactivity toward one- or two-electron redox chemistry, presumably by setting the protonation state of the bridging oxygens and thereby perturbing the redox potential of the Mn ion.
Keywords: ferritin, metalloprotein, ribonucleotide reductase, electron paramagnetic resonance (EPR), mass spectrometry (MS), X-ray crystallography, X-ray absorption spectroscopy, binuclear metal cofactor, R2-like ligand-binding oxidase, redox chemistry
Introduction
Three different native binuclear metal cofactors exist within the ferritin-like superfamily. In most groups, the cofactor consists of a diiron complex, whereas other groups contain a dimanganese complex or a mixed manganese/iron complex (1–7). The latter has been identified in two groups: the R2 subunits of subclass Ic ribonucleotide reductases (R2c)5 and the R2-like ligand-binding oxidases (R2lox) (4, 5, 7). Despite showing a high degree of similarity, these two proteins perform very different redox chemistry.
Ribonucleotide reductases (RNRs) catalyze the reduction of ribonucleotides to deoxyribonucleotides. In most subclasses of the aerobic class I RNRs, a binuclear metal cofactor in the R2 (NrdB) subunit generates a catalytic radical that is reversibly transferred to the active site in the R1 (NrdA) subunit (2, 3, 8). Subclass Ia contains a diiron cofactor that oxidizes a nearby tyrosine residue (2, 3, 8), whereas in subclass Ib, a tyrosyl radical is generated by a dimanganese complex (9–14). In subclass Ic (R2c), which contains a mixed manganese/iron complex (15–17), and probably also in the recently proposed dimanganese subclass Id (18, 19), the metal complex itself acts as the catalytic radical, with the Mn ion shuttling between the III and IV oxidation states. A metal-free subclass Ie was also recently discovered (20–22).
In contrast, the Mn/Fe cofactor in R2lox performs two-electron redox chemistry, catalyzing the formation of a tyrosine–valine ether cross-link in the protein scaffold during cofactor maturation (23, 24). The physiological function of R2lox is unknown. The name ligand-binding oxidase is derived from the observation that in R2lox proteins the R2 scaffold is remodeled to contain a hydrophobic channel leading from the protein surface to the active site in which a long-chain fatty acid ligand is bound that copurifies with the protein from heterologous expression hosts (23, 25). Although they are structurally more closely related to RNR R2 proteins (26), the function of R2lox proteins appears to resemble bacterial multicomponent monooxygenases, which catalyze two-electron oxidations on a variety of hydrocarbon substrates (1, 24).
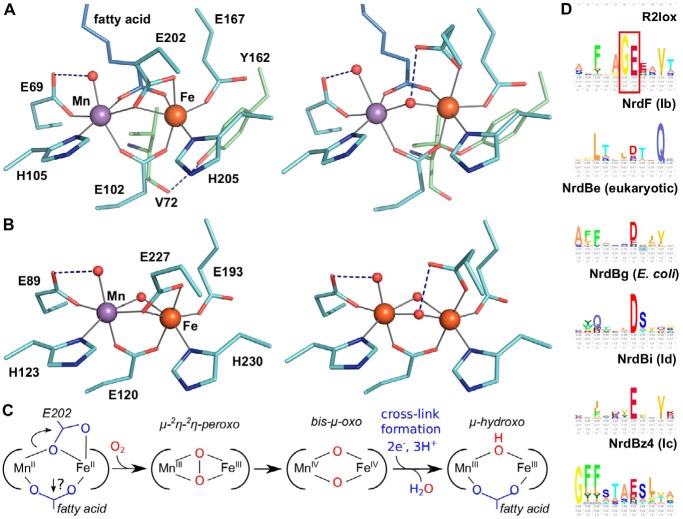
In both R2c and R2lox, cofactor assembly begins with the binding of MnII and FeII to the apoprotein scaffold, although each system achieves selective metallation by different strategies (27, 28). The two metal ions are bound in near-octahedral geometry, with the manganese ion occupying the N-terminal binding site (site 1) (Fig. 1, A and B) (23, 25, 29, 30). In the reduced state, each metal ion is coordinated by one histidine and one monodentate glutamate ligand, while two further glutamate residues bridge the metal ions (23, 24, 28). An oxo species, likely a water molecule, is bound at the open coordination site of the manganese ion. In R2lox, the fatty acid ligand also bridges both metal ions, whereas another oxo species, likely also a water molecule, occupies the corresponding position in R2c.
Figure 1.
A and B, active-site structures of R2lox (A) and R2c (B) in the nonactivated, reduced state (left) (PDB codes: R2lox, 4HR4; R2c, 4M1I) and oxidized resting state (right) (PDB codes: R2lox, 4HR0; R2c, 1SYY) (16, 23, 28). (Only a Fe/Fe-bound oxidized-state structure of R2c is available.) C, proposed activation pathway for R2lox. Both Glu-202 and the fatty acid need to shift from their bridging positions to allow O2 binding for R2lox, whereas for R2c only Glu-227 has to detach. D, sequence logos of R2lox and the biochemically characterized NrdB subclasses, centered on the N-terminal metal ligand following the conserved Gly in R2lox (marked with a red box). The biochemical R2 subclass Ia consists of several phylogenetically distinct subclasses and is represented here by subclasses NrdBe and NrdBg. The logos were generated from the RNRdb (http://rnrdb.pfitmap.org) (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.) HMMER (69) search profiles using the Skylign server (70).
Cofactor activation proceeds via reaction with O2 (Fig. 1C). For both systems, the initial bound O2 intermediate has been modeled as bridging the two metal ions as a side-on μ-2η-2η-peroxo species (31). To achieve side-on O2 binding, the C-terminal bridging/chelating glutamate, and in the case of R2lox also the lipid headgroup, need to shift away from their bridging coordination. Crystallographic data demonstrate that this does occur for the glutamate, which shifts to a monodentate coordination at the Fe ion following O2 activation in both R2lox and R2c (16, 23, 24). It is less clear that the lipid headgroup of R2lox is also displaced, as it is seen in the same position following activation. It is thus currently unclear whether cofactor activation in the two systems follows the same trajectory.
In R2c, O2 reduction generates a bis-μ-oxo MnIV/FeIV intermediate with a terminal hydroxo ligand on the Mn ion, and a similar intermediate is expected to be formed in R2lox (23, 31–34). Subsequent one- or two-electron/proton transfer events lead to the final, resting-state cofactor structure. In the case of R2c, the MnIV and FeIII ions are linked by two oxo (μ-oxo/μ-hydroxo) bridges with a terminal hydroxo ligand on the Mn ion, whereas in R2lox the two metal(III) ions are linked by a single oxygen (μ-hydroxo) bridge, with the fatty acid ligand occupying the position of one of the oxygen bridges of R2c and a terminal water ligand on the Mn ion (Fig. 1, A and B) (16, 23, 35–37). In R2lox, concomitant with cofactor activation, an ether cross-link is formed between the Cβ of a valine and the hydroxyl oxygen of a tyrosine close to the active site. Prior to cross-link formation, the tyrosine forms a hydrogen bond with the carbonyl oxygen of the valine (Fig. 1A) (23, 24). These two residues are conserved in the R2lox group but not conserved in R2c (5), which does not form such a covalent link in the protein scaffold.
Here, we introduced two different single point mutations into the R2lox scaffold to clarify whether cofactor activation in R2lox and R2c occurs by the same mechanism or at what point cofactor assembly diverges. These substitutions are targeted toward blocking the binding of the fatty acid ligand that copurifies with the protein, the only structural difference in the first coordination sphere between the two homologues. Substitution of one specific residue in the second coordination sphere, the glycine residue directly preceding the N-terminal glutamate ligand, Gly-68, was found to convert the activated R2lox cofactor into an R2c-like cofactor. This position is completely conserved as a glycine in the R2lox group, whereas it is not conserved in the characterized RNR R2 subclasses where it is generally occupied by a small hydrophobic residue (Fig. 1D). We examined the substitution of Gly-68 with a leucine or a phenylalanine (G68L/F). We found that the G68F substitution had no significant effects on cofactor reactivity, whereas the G68L variant stabilized an R2c-like cofactor and prevented cross-link formation. The EPR signature of the R2c-like cofactor matches that of an intermediate observed during oxygen activation of the WT protein (33), demonstrating that it is a physiologically relevant species.
Results and discussion
In analogy to our previous experiments with wt-R2lox (23), we crystallized the G68L/F-R2lox point mutants in metal-free form and soaked the crystals with equal concentrations of MnII and FeII in large excess under anoxic or aerobic conditions to obtain structures of the nonactivated reduced and oxidized resting states of the proteins, respectively. X-ray anomalous dispersion was used to quantify the relative amounts of Mn and Fe in each binding site in the crystals.
The G68L substitution blocks binding of the fatty acid ligand to the metal cofactor
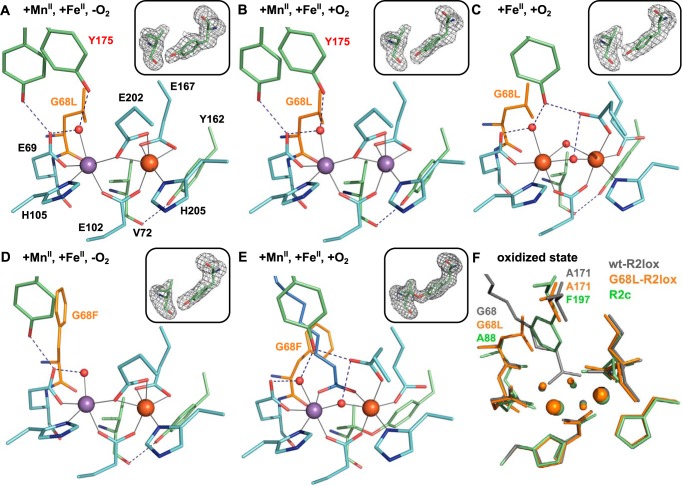
The G68L substitution excludes the fatty acid from the active site, although electron density for a hydrophobic ligand (modeled as caprylic acid in the crystal structures) is observed further up in the ligand-binding tunnel. Because the absence of the fatty acid ligand leaves an open coordination site on both metal ions, it causes some rearrangements in the reduced-state active site of G68L-R2lox as compared with the WT (Fig. 2A). Glu-167 acts as a bidentate ligand to Fe2 instead of its usual monodentate coordination, such that Fe2 remains near-octahedral. Mn1 retains a free coordination site, although this may be occupied by an additional water molecule that is not observed in the electron density (see X-ray absorption spectroscopy (XAS) characterization below). The second-sphere residue Tyr-175 is disordered but best modeled in two alternate conformations: its usual conformation as a hydrogen-bonding partner to the terminal water ligand of the site 1 metal ion and a rotated conformation in which it may form a hydrogen bond with the nonbinding carboxyl oxygen of Glu-69 instead, otherwise the other hydrogen-bonding partner of the water molecule (37). Tyr-175 is somewhat disordered in reduced-state wt-R2lox as well and may always be present in both conformations, just to different degrees (23, 24).
Figure 2.
A–E, active-site structures of G68L- (A–C) and G68F-R2lox (D and E) after soaking crystals with equal amounts of MnII and FeII under anoxic (A and D) or aerobic conditions (B and E) or with only FeII under aerobic conditions (C). All structures are shown in roughly the same orientation, with site 1 on the left. The substituted residue is highlighted in orange. Metal–ligand bonds are indicated by gray lines; hydrogen bonds are indicated by dashed blue lines. Residues in alternate conformations are labeled in red. The insets show mFo − DFc refined omit electron density contoured at 3.0 σ for residues Val-72 and Tyr-162. F, superposition of Mn/Fe–wt-R2lox (gray; PDB code 4HR0) (23), Fe/Fe–G68L-R2lox (orange), and Fe/Fe–R2c (green; PDB code 1SYY) (16) in the oxidized resting state, highlighting the similarity of the metal coordination and the residues that allow or block access of the fatty acid to the metal cofactor, Gly-68 and Ala-171 in R2lox, or Ala-88 and Phe-197 in R2c, respectively. The latter position is conserved as alanine in R2lox proteins and as phenylalanine in all RNR R2 proteins (5), whereas the former position is conserved as glycine in R2lox and most commonly occupied by an alanine in R2c (see Fig. 1D).
G68L-R2lox accumulates more Mn in site 2 than the WT
Following soaking of apoprotein crystals with equal concentrations of MnII and FeII under anoxic conditions, the metal distribution in G68L-R2lox was (within error) identical to the WT (23), with roughly 60% Mn in site 1 and 80% Fe in site 2 (Fig. S1). Soaking with MnII and FeII under aerobic conditions led to drastically different results, however. Under these conditions, wt-R2lox accumulates predominantly Mn in site 1 and Fe in site 2 (23), whereas aerobically soaked G68L-R2lox crystals contained mainly Mn in both sites (Fig. S1). The active site in aerobically soaked G68L-R2lox is in a reduced-state conformation that is practically identical to that obtained by soaking under anoxic conditions, and the Val-72–Tyr-162 ether cross-link is not observed (Fig. 2B). As in wt-R2lox (24), a Mn/Mn cofactor in G68L-R2lox therefore seems to be unable to react with O2 and consequently does not support cross-link formation.
In contrast, in solution reconstitution experiments, Mn/Fe cofactors were efficiently assembled in G68L-R2lox, although it also retained more Mn than the WT under the same conditions and, unlike wt-R2lox, assembled MnII2 complexes if only Mn was present, thus following an R2c-like metallation pathway (see supporting information S2 and S3.4 and Figs. S2 and S6) (27, 28). The large amount of Mn observed in site 2 in aerobically soaked crystals might therefore be caused by a combination of increased Mn affinity and the progressive depletion of bulk Fe by its reaction with O2, which would drive equilibrium protein metallation toward a Mn/Mn cofactor, as MnII does not react with O2 and thus remains available for protein binding. This would require the rate of cofactor maturation in G68L-R2lox to be slower than that of bound/free metal exchange. It has previously been inferred that these two rates are similar in wt-R2lox if a large excess of metal ions over protein is used, to rationalize the enrichment of Mn in site 1 that is observed in aerobic crystal soaking experiments but not in solution experiments in which near-stoichiometric amounts of metal ions are used (23, 27). Thus, the observation that in the presence of oxygen more Mn accumulates in both sites in the leucine-substituted variant suggests that cofactor maturation in G68L-R2lox is significantly slower than in the WT.
The G68L substitution does not prevent oxygen activation of the R2lox cofactor
Crystals were also soaked with an excess of FeII under aerobic conditions, forcing the protein to assemble a diiron cofactor. In this structure, Fe is bound in both sites, and the active site is in an oxidized-state conformation with two oxo-type species (i.e. bis-oxo or oxo/hydroxo) bridging the metal ions instead of the single μ-hydroxo bridge and the bridging fatty acid ligand seen in the WT (23). This arrangement leads to an active-site geometry that more closely resembles that of oxidized R2c than that of wt-R2lox (Fig. 2, C and F) (16). The G68L substitution thus does not prevent oxygen activation of the cofactor. However, electron density for the tyrosine–valine ether cross-link, although observed, was too weak to model the link, suggesting that the leucine substitution causes catalytic impairment.
Other second-sphere substitutions that block ligand binding do not cause any noticeable defects
In contrast to the G68L mutation, substitution of Gly-68 with a phenylalanine does not cause any visible defects in cofactor assembly, oxygen activation, or cross-link formation (Fig. 2E and supporting information S2). Phe-68 blocks the ligand-binding tunnel in the reduced state but does not exclude the fatty acid ligand from the metal site in the oxidized state where it adopts a different rotamer (Fig. 2, D and E). The active-site geometry is the same as in the WT in the oxidized state of G68F-R2lox, whereas in the reduced state the active site looks very similar to that in G68L-R2lox. It must be presumed that a fatty acid is bound higher up in the tunnel in G68F-R2lox in the reduced state as well, but electron density for it was not observed.
Another second-sphere substitution roughly opposite to Gly-68 in the ligand-binding tunnel, A171F (see Fig. 2F), demonstrates that the different effects of the G68L and G68F substitutions are not a consequence of the absence or presence of the fatty acid. The A171F substitution completely blocks ligand binding to the active site but has no discernible effect on cofactor assembly or cross-link formation (38), indicating that the phenotype of G68L-R2lox is caused specifically by this mutation. We further characterized the interesting effects of the G68L mutation using EPR and X-ray absorption spectroscopy as well as MS.
G68L-R2lox predominantly assembles an R2c-like MnIII/FeIII cofactor species in solution
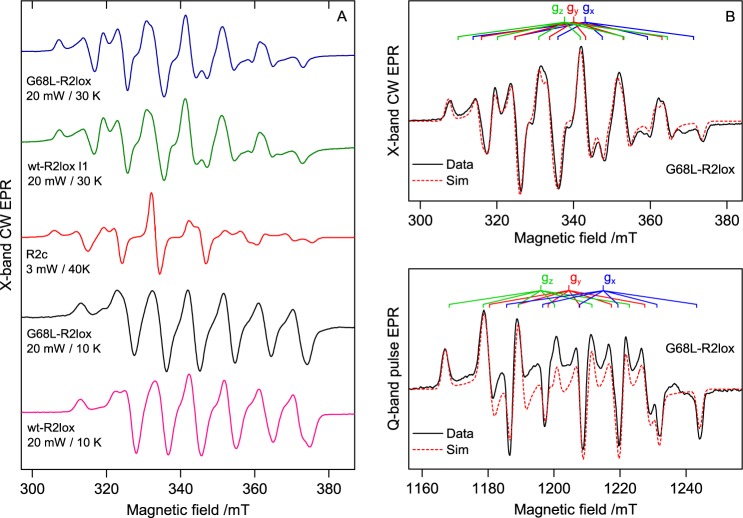
To elucidate the cofactor structure of the Gly-68 variants in solution, continuous-wave X-band EPR measurements were performed on aerobically Mn/Fe-reconstituted G68L- and G68F-R2lox. In Mn/Fe cofactors described in the literature, the two metal ions, in the III oxidation state, magnetically couple to form an effective spin ½ ground state, which gives rise to an EPR signal centered at approximately g = 2.0 (23, 36). The EPR spectrum is highly structured, a consequence of the hyperfine interaction of the 55Mn (I = 5/2) nucleus with the unpaired electron spin, which should split the EPR line into a sextet. The structure of the signal is a fingerprint of the two cofactor types. For R2lox, a simple six-line pattern is observed with an additional small line on the low-field edge, whereas for R2c a six-line pattern is observed, but several of the major lines display hyperfine structure (15, 17, 23, 27, 36).
Deviations from the six-line pattern come about because the cofactor's EPR signal changes depending upon how it is oriented in the applied magnetic field. As we measure samples in the frozen state, we probe all possible orientations the cofactor can take, with the resultant EPR spectrum thus representing the superposition of multiple hyperfine patterns (see Ref. 36 for more details).
The EPR line shape of G68L-R2lox was found to strongly depend on temperature and microwave power (Fig. S3). The dominant cofactor signal is best observed at higher temperatures (T = 30 K), similar to R2c (27). It also bears a strong resemblance to the cofactor signal of the R2c protein (Fig. 3A) (15, 17, 27, 36). The overall hyperfine structure matches that of the R2c cofactor signal, resolving approximately six lines with additional superstructure best resolved on the first, fifth, and sixth lines. The main difference between the two signals is that the homogeneous linewidth of the R2c spectrum is somewhat narrower than that of the G68L-R2lox spectrum. If instead the absorption line shapes are compared, the similarities between the two spectra are more easily observed (see supporting information S3.3 and Fig. S5).
Figure 3.
A, EPR spectra of the MnIII/FeIII cofactor of wt- and G68L-R2lox measured under different conditions. The EPR spectra of an O2 activation intermediate seen for wt-R2lox (I1) (33) and the MnIII/FeIII (one electron–reduced) state of the R2c cofactor (adapted from Voevodskaya et al. (17)) are shown for comparison. B, multifrequency simulations of the dominant G68L cofactor signal recorded at X- (T = 15 K) and Q-band (T = 19 K). Simulation parameters are given in Table 1. Signal processing and EPR simulations are described in supporting information S3.5–S3.7. CW, continuous-wave; mT, millitesla; mW, milliwatts.
In addition to this R2c-like cofactor signal, at lower temperatures and higher powers (T = 10 K, P = 20 milliwatts) a small wt-R2lox signal is observed in G68L-R2lox (Fig. 3A). Under these conditions, the R2c-like cofactor signal is completely suppressed due to slower relaxation properties, allowing the WT-like population to be quantified. It accounts for ∼30–40% of MnIII/FeIII centers (supporting information S5 and Table S3). At higher temperatures the WT-like contribution to the overall line shape is further suppressed due to a smaller exchange coupling compared with the R2c-like species, leading to thermal population of higher-spin states (see supporting information S4). In contrast, in G68F-R2lox only the WT-like cofactor form was observed (Fig. S4).
To uniquely constrain the set of field-dependent and field-independent spin Hamiltonian terms that describe the EPR spectrum of the R2c-like G68L-R2lox cofactor signal, higher-frequency (Q-band) measurements were performed (Fig. 3B). The fitted spin Hamiltonian parameters match those seen earlier for R2c (Table 1) (15, 17, 36). In particular, the fitted 55Mn hyperfine tensor is very similar, albeit 10% smaller in magnitude, to that previously reported for the R2c cofactor, demonstrating that the Mn coordination environment of both cofactors is similar. Likewise, the 57Fe (I = ½) hyperfine tensor, which can be interrogated by labeling the cofactor with 57Fe, demonstrates that the Fe coordination environment of both cofactors is also similar (Fig. S9). Taken together, these observations strongly suggest that the two cofactor forms are structurally equivalent, meaning that the G68L-R2lox protein harbors an R2c-like MnIII/FeIII cofactor.
Table 1.
Spin Hamiltonian parameters for EPR simulations of wt-, G68F-, and G68L-R2lox cofactor signals as compared with R2c (15, 17, 36)
Signal processing and EPR simulations are described in supporting information S3.5–S3.7.
|
g tensor |
55Mn hyperfine tensor (MHz) |
57Fe hyperfine tensor (MHz) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| gx | gy | gz | Ax | Ay | Az | Ax | Ay | Az | |
| wt-R2lox | 1.953 | 1.968 | 2.034 | 257 | 249 | 282 | 62.5 | 51.6 | 93.9 |
| G68Fa | 1.954 | 1.965 | 2.033 | 256 | 251 | 286 | |||
| G68L, producta | 1.955 | 1.968 | 2.033 | 258 | 253 | 286 | |||
| G68L, intermediateb | 2.007 | 2.025 | 2.039 | 281 | 365 | 246 | 59.2 | 60.1 | 65.9 |
| R2c | 2.009 | 2.015 | 2.024 | 314 | 392 | 269 | 50.2 | 48.4 | 44.5 |
a Based on single-frequency X-band simulations.
b For the dominant, high-temperature EPR signal of G68L, the g tensor and 55Mn A tensor are rotated relative to each other by the Euler angles (α, β, γ) = (−80.1°, 69.2°, 41.7°) (in the z′yz″ convention).
We note that the R2c-like cofactor species is also present in wt-R2lox in a fraction of centers. We had previously assigned this fraction to an incorrectly metallated population (36). However, the characteristics of this fraction match those seen for R2c, indicating that it represents a physiologically relevant cofactor state.
Additional cofactor signals could be observed for G68L-R2lox samples prepared with excess MnII and substoichiometric amounts of FeII following the initial incubation time of several hours. These additional signals decayed after ∼24 h of additional incubation at room temperature, leading to an increase in the EPR signals of both MnIII/FeIII cofactor signals (supporting information S6 and Figs. S11 and S12). The additional signal bears a strong resemblance to the recently postulated MnIV/MnIII cofactor that constitutes class Id RNR R2 proteins (18, 19) as well as to related MnIV/MnIII model complexes (39–43). It is important to note that the reduced MnII/MnII cofactor of class Ib and Id R2 proteins does not directly react with O2 (9, 13, 19). Instead, superoxide is thought to be the in vivo oxidant (14, 19, 44). In the G68L-R2lox system, Fenton-like chemistry of free FeII in solution could instead be the source of superoxide. Thus, the scaffold is able to assemble a MnIV/MnIII cofactor in a small fraction of centers under the condition of preloading both sites with Mn in addition to having a source of reactive oxygen species.
The EPR-visible form of the dimanganese cofactor (MnIV/MnIII) is unstable in G68L-R2lox (t½ < 12 h), presumably reflecting the slow rate of reduction of the cofactor to the III/III level, which is EPR-silent. Consistent with this notion is the progressive increase of the MnIII/FeIII signal in which we observe the product of the reduction, i.e. the slow reduction of the initially formed, EPR-silent MnIV/FeIII state to the EPR-observable MnIII/FeIII state. Thus, both results point to the G68L-R2lox cofactor having redox properties more similar to those of R2c, whose resting state is the IV/III oxidation level.
The R2c-like MnIII/FeIII cofactor species in G68L-R2lox contains two oxygen (μ-oxo/μ-hydroxo) bridges
The nature of the bridging ligand between the two metal sites strongly influences the magnetic properties of the cofactor and thus its EPR spectrum. The (super)exchange interaction (J) is mediated by the overlap of the orbitals of the bridging oxygen(s) with those of the metal ions. In the MnIII/FeIII cofactor of wt-R2lox, which has a single μ-hydroxo bridge and long metal ion separation, the superexchange interaction is small (J ≈ 10 cm−1) (27, 36), as also seen in related μ-hydroxo–bridged Mn model complexes (45). In contrast, the Mn and Fe model complexes, which additionally have an oxo bridge and shorter metal–metal distance, typically display a significantly larger superexchange interaction (43, 46–48). The superexchange interaction can be estimated from the temperature dependence of the EPR spectrum. For the G68L-R2lox cofactor, J coupling is estimated to be 33 cm−1 (supporting information S4 and Fig. S10), significantly larger than in wt-R2lox, suggesting that the G68L-R2lox cofactor has a more extensive oxygen-bridging network, i.e. at least one oxo bridge. The large exchange interaction probably precludes other connectivities such as a bridging peroxo, which is expected to have a smaller superexchange interaction because of the longer metal–metal distance and more extended through-bond superexchange pathway.
The structure of the EPR signal also indicates that the G68L-R2lox cofactor contains a μ-oxo bridge. The structure is primarily due to the 55Mn hyperfine interaction, as described above. The 55Mn hyperfine tensor provides detailed information on the local coordination of the Mn ion. In an earlier EPR and density functional theory analysis of the 55Mn hyperfine tensor of both the R2lox and the R2c cofactors, based on an extensive set of model complex data, it was shown that the local ligand field of the MnIII ion is different for the two cofactor forms (36). In R2lox, the MnIII site is tetragonally elongated, with the unique Jahn–Teller axis perpendicular to a vector connecting the Mn–Fe sites, whereas in R2c the MnIII site is instead tetragonally compressed, with the unique axis in a plane containing the Mn–Fe vector. Importantly, this rotation of the local ligand field axis is caused by the different protonation states of the bridging oxygen ligand in the two cofactors. Thus, the observation that the cofactor of G68L-R2lox displays approximately the same 55Mn hyperfine tensor as the R2c cofactor (Table 1) indicates that both cofactors contain a tetragonally compressed MnIII ion and therefore at least one oxo bridge (36).
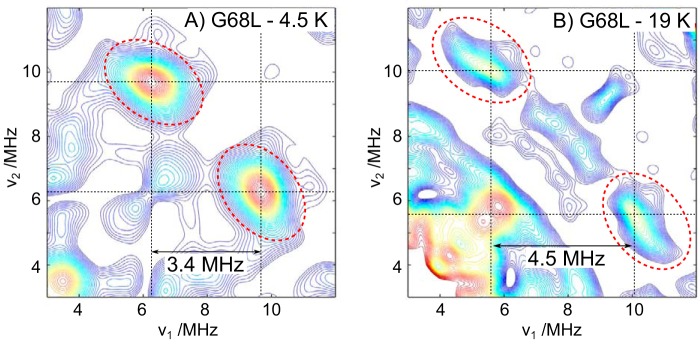
To test whether the G68L-R2lox cofactor also has a second, protonated bridge, double-resonance measurements were performed, i.e. 2H HYSCORE (hyperfine sublevel correlation) at Q-band (Fig. 4). In our previous study on wt-R2lox, we showed that this was the method of choice for detecting one or more protonated oxygen bridges. For the wt-R2lox cofactor, the single μ-OH bridge gives rise to a large 2H coupling of 3.4 MHz (36). This manifests in the HYSCORE spectrum as a doublet split by 3.4 MHz and centered about the 2H Larmor frequency (νN(2H) ≈ 8 MHz at 1200 milliteslas). For the R2c-like cofactor of G68L-R2lox, a new 2H coupling is observed at the expense of the WT signal. This coupling is also large (4.5 MHz), indicative of a μ-OH bridge, but clearly different from that of the WT.
Figure 4.
A and B, Q-band 2H HYSCORE measurements of the G68L-R2lox MnIII/FeIII cofactors at low temperature where the WT-like cofactor signal is dominant (A) and high temperature where the R2c-like cofactor signal is dominant (B). Red dashed circles indicate the cross-peaks of the μ-OH bridge signal for both cofactors.
In summary, multifrequency, multiresonance EPR data suggest that the dominant MnIII/FeIII cofactor seen in G68L-R2lox contains both a μ-oxo and a μ-hydroxo bridge. Its geometric structure most likely resembles that observed in the crystal structure of Fe only–soaked G68L-R2lox (Fig. 2C). We used XAS to further assess the cofactor species formed in G68L-R2lox.
X-ray absorption spectroscopy confirms that G68L-R2lox stabilizes a μ-O, μ-OH–bridged MnIII/FeIII cofactor
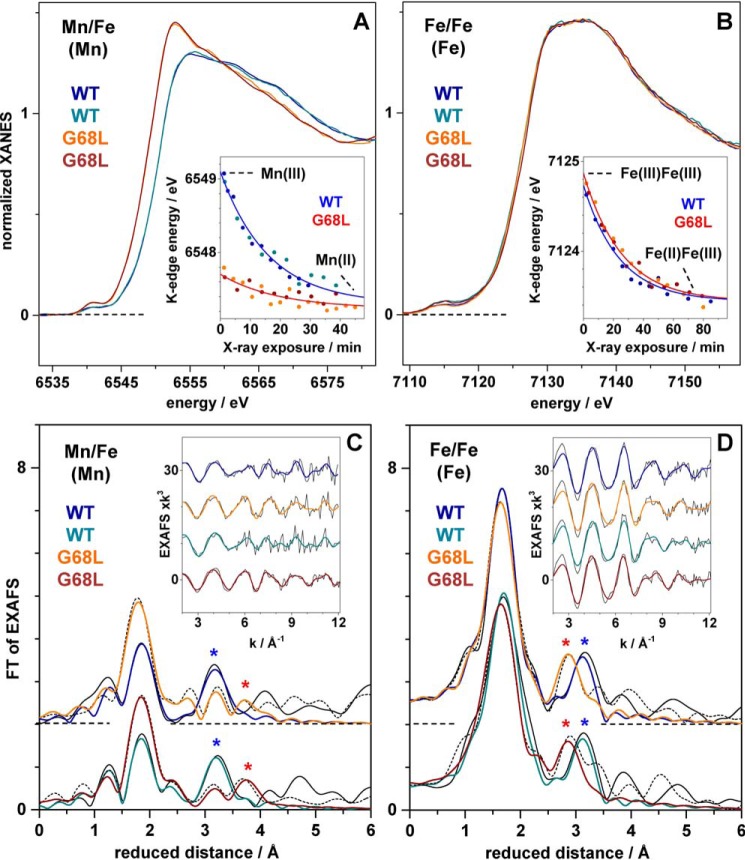
G68L-R2lox was aerobically reconstituted with either Mn and Fe or only Fe for XAS analysis. Both were examined because solution maturation of the R2lox protein in the presence of MnII and FeII leads to significant populations of both the MnIII/FeIII and FeIII/FeIII forms. Thus, Mn/Fe and Fe-only samples were characterized to derive complementary information on both cofactor types.
The Fe-only G68L samples show X-ray absorption near edge structure (XANES) spectra at the Fe K-edge that are very similar to the WT, suggesting a similar near-octahedral coordination of iron (Fig. 5A). The slightly larger pre-edge peak feature due to 1s → 3d electronic excitation transitions for G68L-R2lox compared with wt-R2lox implies lowered symmetry of the iron sites in this mutant. The K-edge energy of ∼7124.7 eV indicates the near-quantitative presence of FeIII (Table 2) (37), demonstrating that initial FeII2 centers activate oxygen to yield FeIII2 cofactors in both WT and G68L-R2lox.
Figure 5.
XAS analysis of WT and G68L variants of R2lox. Data for two preparations of each variant are shown. Left, spectra at the Mn K-edge of Mn/Fe-reconstituted variants. Right, spectra at the Fe K-edge of Fe only–reconstituted variants. XANES spectra in A and B and K-edge shifts due to X-ray photoreduction in the insets with fit curves for merged data sets (red and blue lines; time axes for data collected at SOLEIL (dark blue and orange data) were expanded by a factor of 1.2 versus data collected at Swiss Light Source (dark cyan and dark red data) for comparison; see Table 2). FTs in C and D of EXAFS spectra in the insets (vertically shifted for comparison; black lines, experimental data; colored lines, fit curves with parameters in Table 3). FTs were calculated for k-values of 3.8–12.3 Å−1 (cos-windows over 10% of both k-range ends) to emphasize metal–metal distances (asterisks in C and D).
Table 2.
K-edge energy and X-ray photoreduction rate in G68L-R2lox compared with wt-R2lox
Data represent mean values for two preparations of each R2lox variant (full parameter variation ranges are in parentheses). K-edge energies and X-ray photoreduction parameters are for proteins reconstituted with Mn and Fe (left, Mn data) or Fe only (right, Fe data).
| R2lox variant | Mn/Fe (Mn) | Fe/Fe (Fe) |
|---|---|---|
| EK-edge (eV) | ||
| WT | 6549.1 (1) | 7124.6 (1) |
| G68L | 6547.7 (1) | 7124.8 (1) |
| X-ray photoreduction rate (h−1)/ΔEK-edge (eV) | ||
| WT | 4.9 (2)/1.9 (2) | 2.7 (2)/1.3 (1) |
| G68L | 4.5 (3)/0.4 (1) | 2.1 (4)/1.4 (2) |
XANES spectra at the Mn K-edge collected for G68L-R2lox reconstituted with both Mn and Fe are more difficult to interpret due to the presence of both MnIII/FeIII cofactors and MnII2 cofactors in G68L-R2lox (see above and supporting information S2 and S3.4). For G68L-R2lox samples prepared for XAS, the MnII fraction was significant, as indicated by a ∼1.4-eV lower K-edge energy (Table 2), larger edge maximum, and increased pre-edge peak amplitude compared with wt-R2lox. Assuming that MnII and MnIII are present only in MnII2 or MnIII/FeIII cofactors, respectively, the results suggest that the Mn/Fe-reconstituted G68L-R2lox XAS samples contained ∼50% FeIII2, ∼20% MnIII/FeIII, and ∼30% MnII2 cofactors.
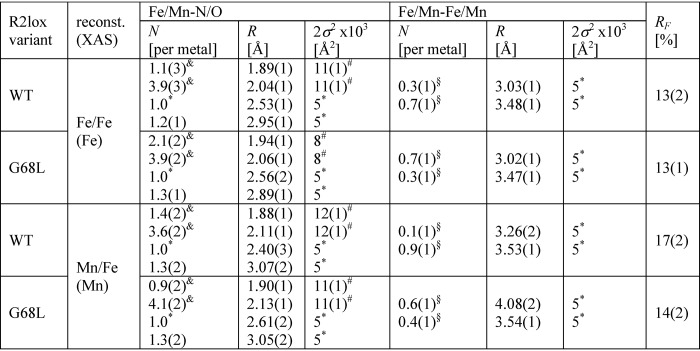
The Fe extended X-ray absorption fine structure (EXAFS) of G68L-R2lox reveals increased numbers of short iron–ligand bonds and the predominance of a ∼0.5-Å-shorter Fe–Fe distance than in wt-R2lox (∼3.05 Å) (Fig. 5D). The same short distance was observed in the fraction of Mn/Fe-reconstituted G68L-R2lox assigned to the MnIII/FeIII population (Table 3). The metal–metal distance of ∼3.05 Å in G68L-R2lox is almost identical to the distance in the (Mn/Fe)III/FeIII cofactors in R2c and mouse R2a proteins, for which such a distance has been assigned to the presence of a μ-O and a μ-OH bridge between the trivalent metal ions (49, 50). These findings suggest that an R2a/c-like μ-O, μ-OH–bridged (Mn/Fe)III/FeIII cofactor is formed in G68L-R2lox instead of the WT cofactor with a single μ-OH bridge, in agreement with the crystallographic and EPR data (Fig. S13).
Table 3.
EXAFS simulation parameters
Data correspond to spectra at the Fe or Mn K-edges in Fig. 5 of two Fe only–or Mn/Fe-reconstituted preparations each of the WT- and G68L-R2lox variants. Full parameter ranges from individual fitting of the two data sets are given in parentheses. N, coordination number; R, interatomic distance; 2σ2, Debye–Waller factor; RF, fit error sum calculated for reduced distances of 1–3.5 Å. Fit restraints: *,fixed parameters; #, coupled to yield the same 2σ2 value; &, coupled to yield a sum of 5; §, coupled to yield a sum of 1.
Mn EXAFS allows the MnII2 homodimer population to be characterized. It has overall ∼0.1-Å-longer manganese–ligand bonds typical for MnII and likely no short metal–ligand bonds compared with wt-R2lox as well as a larger metal–metal distance (∼4.1 Å), similar to metal(II) cofactors in R2 proteins lacking bridging oxides and holding a single carboxylate bridge (Fig. 5C and Table 3) (50, 51). In the crystal structure of aerobically Mn/Fe-soaked G68L-R2lox (Fig. 2B), Glu-202 adopts a semibridging position as in the reduced-state conformation, which may account for the ∼3.55-Å Mn–Mn distance also detectable by EXAFS, as supported by similar structures of R2 proteins (51). Mn/Mn–G68L-R2lox in solution may contain Glu-202 predominantly in a nonbridging position, which would explain the prevalence of the ∼4.1-Å distance in the EXAFS.
Finally, photoreduction of the metal ions was examined by extended X-ray exposure during successive monochromator scans on the same sample spot. This caused shifts of the Mn and Fe K-edge spectra to lower energies (37, 49, 52). The respective K-edge energies decreased exponentially due to MnIII → MnII or FeIII → FeII transitions (Fig. 5, A and B, insets). The magnitudes of the energy shifts reveal formation of FeII/FeIII centers after ∼80 min of X-ray exposure and about 2-fold faster formation of MnII (Table 2), as previously observed in wt-R2lox (24, 37). The much smaller K-edge energy shift in G68L-R2lox versus wt-R2lox corroborates that only ∼20% MnIII was initially present in the former variant. G68L-R2lox showed slightly (∼1.2-fold) slower reduction of MnIII and FeIII compared with wt-R2lox. The MnIII/FeIII and FeIII2 cofactors in G68L-R2lox may therefore possess a slightly less positive redox potential than the corresponding cofactor species in the WT (53).
The G68L substitution inhibits cross-link formation
To firmly establish whether the G68L substitution causes a catalytic impairment, we used MS to quantify the tyrosine–valine ether cross-link in Mn/Fe– and Fe/Fe–G68L-R2lox compared with the WT. We have recently shown that the cross-link is formed far more efficiently by a Mn/Fe cofactor than a diiron center in wt-R2lox (54). In agreement with our previous results, Mn/Fe–wt-R2lox contained roughly 5 times more cross-linked peptide than the iron-only form. In contrast, both Mn/Fe– and Fe/Fe–G68L-R2lox samples contained nearly 4 times less cross-link than Mn/Fe–wt-R2lox but slightly more than Fe/Fe–wt-R2lox (Fig. S14). Thus, although G68L-R2lox efficiently assembles Mn/Fe cofactors in solution that reduce O2, the leucine substitution does impair the catalytic process coupled to cofactor assembly, i.e. cross-link formation (38). It is not possible to determine the total amount of cross-link per protein, as only relative amounts of the same peptide across different samples can be determined. It appears likely, however, that the fraction of G68L-R2lox that forms the cross-link corresponds to the WT-like population observed by EPR, which would also suggest that the cross-link is formed near quantitatively in Mn/Fe–wt-R2lox.
The G68L variant stabilizes an intermediate of the R2lox maturation cycle
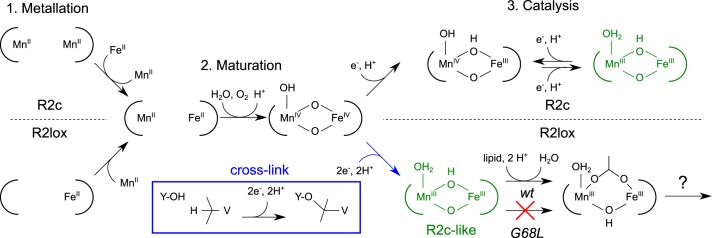
A recent study by Miller et al. (33) characterized a series of assembly and O2 activation intermediates of R2lox. Transient absorption spectroscopy identified relatively intense features in the visible region assigned as charge transfer bands associated with two early, bridging peroxo-type MnIII/FeIII cofactor intermediates, termed I1 and I2. The yield of these states was estimated to be very small (<5%), which would hamper their detection using other spectroscopies such as EPR. Interestingly, however, two EPR signals were observed in the same study and tentatively assigned to the same two intermediates, although the same kinetic information could not be obtained from the EPR experiment. The first EPR signal, assigned to the I1 state, matches the EPR signal reported here assigned as a μ-O, μ-OH–bridged MnIII/FeIII cofactor (Fig. 3A), demonstrating that the G68L variant stabilizes a physiologically relevant intermediate of the R2lox cofactor assembly pathway.
We would therefore propose that the two EPR intermediates observed in the study of Miller et al. (33) represent intermediates following O–O bond cleavage, as opposed to O2-binding/peroxo-type intermediates. The first is the R2c-like intermediate described in this study, whereas the second presumably represents an intermediate involved in the rebinding of the fatty acid to the cofactor, e.g. a μ-O, μ-OH2–bridged MnIII/FeIII cofactor (Fig. 6). We note that these two states are not expected to display an intense optical signature and would therefore not be observed in the transient absorption kinetics.
Figure 6.
Summary of cofactor maturation steps for R2lox and R2c. Although the two cofactors have divergent metallation (23, 27, 28) and catalytic chemistry (15–17, 23, 24), their maturation in the course of the reaction with O2 is likely to follow the same pathway (23, 31–37). The G68L-R2lox mutant traps an intermediate of the reaction following O–O bond cleavage (MnIV/FeIV) but preceding the loss of a bridging hydroxide and the rebinding of the lipid headgroup. Although cofactor maturation is coupled to cross-link formation in the WT, it is likely that redox equivalents are instead lost to the solution environment in G68L-R2lox.
Thus, our study, together with Miller et al. (33), suggests that lipid rebinding represents the rate-limiting step of maturation of the R2lox cofactor, with all preceding chemical steps, i.e. O–O bond cleavage and cross-link formation, occurring much faster. We suspect that the same process explains the very low yields of O2 activation intermediates seen in the Miller et al. (33) study because the detachment of the fatty acid from the reduced cofactor, which is needed for O2 binding to initiate cofactor activation (23, 24), is also a slow chemical event. This would then explain why R2lox cofactor maturation is inefficient in vitro, i.e. orders of magnitude slower than the kinetics measured for R2c (33, 34).
Conclusions
The results described here shed further light on the chemical activity of manganese/iron cofactors. Our data demonstrate that O2 activation and maturation of the R2c and R2lox cofactors proceed along the same pathway, only diverging following formation of the MnIV/FeIV state (Fig. 6). We further show that a small second-sphere perturbation of the protein scaffold can redirect their activity. Exchange of a single, uncharged, nonpolar amino acid for a larger hydrophobic residue is sufficient to convert one cofactor form into another, namely, to convert the R2lox cofactor into an R2c-like cofactor. This conversion is not limited to the structure the cofactor adopts but also extends to its chemical properties. Photoreduction kinetics, EPR parameters, and the observed inhibition of cross-link formation demonstrate that this single second-sphere mutation alters the redox potential of the R2lox cofactor, making it a less potent oxidant. In contrast, other changes to the second coordination sphere (G68F and A171F) affect the cofactor structure to varying degrees but not its reactivity (38).
When comparing the structures of the G68L and other R2lox variants with those of R2c, it appears that the leucine preceding the N-terminal glutamate ligand in G68L-R2lox exerts its effects on the structure and redox potential of the cofactor through a combination of increased hydrophobicity and decreased solvent accessibility of metal site 1 and steric exclusion of the fatty acid ligand (see Fig. 2). These structural changes explain the change in chemical properties of the two cofactor forms. We have previously shown that the different electronic structures of the R2lox and R2c cofactors are caused by the different protonation states of their bridging oxo ligands, with the ground electronic state (highest occupied molecular orbital) of the Mn ion switching from dz2 to dx2-y2, with concomitant reduction in its redox potential (36). The introduction of a leucine must therefore stabilize a different equilibrium bridge protonation state by remodeling the local H-bonding environment in the vicinity of the O2-derived bridge, presumably through solvent water exclusion.
In conclusion, we show that small perturbations of the second ligand sphere tune the structure and redox potential of Mn/Fe cofactors, opening up possibilities for the design of novel catalysts. The apparent unpredictability of the effects of these small perturbations suggests that site-saturation mutagenesis of the second sphere followed by screening is the most promising approach to identify cofactors with improved or novel reactivities.
Experimental procedures
Site-directed mutagenesis, protein production, and purification
The G68L and G68F point mutations were introduced into a construct encoding full-length Geobacillus kaustophilus R2loxI (accession number WP_011232245) inserted into pET-46 Ek/LIC (Novagen) (23) by site-directed mutagenesis using the QuikChange Lightning kit (Agilent) and verified by DNA sequencing. Both point mutants were produced and purified in metal-free form according to the same protocol as wt-R2lox (23). Briefly, protein was produced recombinantly in Escherichia coli BL21(DE3) (Novagen) grown in terrific broth (Formedium). To obtain metal-free protein, 0.5 mm EDTA was added to the cultures immediately before induction with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. Apoprotein was purified via heat denaturation of contaminating proteins and nickel-chelate affinity chromatography. Cells were disrupted by high-pressure homogenization in lysis buffer (25 mm HEPES-Na, pH 7.0, 300 mm NaCl, 20 mm imidazole, 0.5 mm EDTA). The lysate was cleared by centrifugation, incubated at 60 °C for 10 min, and again cleared by centrifugation. The supernatant was applied to a nickel-nitrilotriacetic acid-agarose (Protino, Macherey-Nagel) gravity-flow column. The beads were washed with lysis buffer containing 40 mm imidazole followed by the same buffer without EDTA. Protein was then eluted using lysis buffer containing 250 mm imidazole and without EDTA. The eluate was exchanged into storage buffer (25 mm HEPES-Na, pH 7.0, 50 mm NaCl) using a HiTrap Desalting column (GE Healthcare), concentrated to ∼1 mm, aliquoted, flash frozen in liquid nitrogen, and stored at −80 °C. Protein concentration was determined using the extinction coefficients at 280 nm determined for wt-R2lox, 47.8 and 50.6 mm−1 cm−1 for metal-free and metal-bound protein, respectively (36). The metal contents of purified apoprotein batches were quantified by total-reflection X-ray fluorescence (TXRF) (see below). The only metal that is generally present in significant amounts is Fe, occupying ≤10% of the cofactor sites.
Crystallization and diffraction data collection
Both R2lox variants were crystallized in metal-free form by vapor diffusion in hanging drops at 22 °C in the same condition as the WT protein, 25–32.5% (w/v) PEG 1500, 100 mm HEPES-Na, pH 7.2–7.5. To reconstitute the oxidized resting-state Mn/Fe cofactor, crystals of metal-free protein were removed from their drop and soaked in mother liquor additionally containing 5 mm each MnCl2 and (NH4)2Fe(SO4)2 for 1–2 h under aerobic conditions and then briefly washed in cryoprotectant solution (40% (w/v) PEG 1500, 100 mm HEPES-Na at the pH of the mother liquor) before flash cooling in liquid nitrogen. Crystals of G68L-R2lox were also soaked aerobically with only (NH4)2Fe(SO4)2 in the same way. To obtain the nonactivated reduced Mn/Fe cofactor, apoprotein crystals were soaked in 1 ml of 40% (w/v) PEG 1500, 100 mm HEPES-Na (at the pH of the mother liquor), 5 mm (NH4)2Fe(SO4)2, 5 mm MnCl2, 0.5% (w/v) sodium dithionite, 0.5 mm phenosafranin, 0.05% (v/v) Tween 20 for 1–2 h and flash-cooled directly without washing. Soaking solutions were always freshly prepared immediately before use using freshly dissolved (NH4)2Fe(SO4)2 and dithionite, to ensure that the Fe was ferrous and that oxygen was effectively removed from soaking solutions used to obtain reduced states, with phenosafranin serving as redox indicator. Data were collected at 100 K at beamlines X06SA and X10SA at the Swiss Light Source (Villigen, Switzerland) and I911-3 at Max II (Lund, Sweden). For the purpose of metal quantification, data collection proceeded in the order Fe edge–Mn edge on the same crystal. High-resolution “native” data were collected on separate crystals or after the anomalous data sets.
Structure determination, model building, and refinement
Diffraction data were processed with XDS (55). Both R2lox point mutants crystallized in the same space group as wt-R2lox, I222, with one molecule in the asymmetric unit (see Table S1). The structures were solved by Fourier synthesis using the structure of the WT protein in the same redox state (23) not containing any ligands as a starting model. Refinement was carried out with phenix.refine (56, 57) and iterated with rebuilding in Coot (58). Refinement included bulk solvent corrections, individual atomic coordinate and isotropic B factor refinement, and occupancy refinement for alternate conformations and metal ions bound on the protein surface but not the active-site metal ions. Metal–ligand bond lengths were restrained. Solvent molecules were added with phenix.refine and manually. Hydrogens were added to the models in the later stages of refinement. Structures were validated using MolProbity (59). Data and refinement statistics are given in Tables S1 and S2. All figures were prepared with PyMOL (version 1.8.6.2; Schrödinger, LLC).
Analysis of anomalous diffraction data
All anomalous data sets were integrated over the same resolution range (50.0–3.0 Å) with XDS (55) (Table S1). The Fe and Mn edge data sets from one crystal were placed on a common scale with XSCALE (55). Both scaled and unscaled data sets were analyzed. Anomalous difference maps were calculated with PHENIX (57) using the phases from a ligand-free model, and the relative amounts of Fe and Mn in each metal site were calculated as described previously (60). The intensities of the anomalous difference density peaks in spheres of 1.9-Å radius around the center of the peaks were integrated using MAPMAN (61). The relative amounts of Fe and Mn in each site were estimated from the integrated intensities at the Fe and Mn edges by taking into account the different contributions of both elements to the anomalous signal at the two wavelengths, assuming a total occupancy of each site of 1, as described (60). Because only relative amounts are calculated, the actual occupancy is irrelevant as long as it is high enough to yield a significant anomalous peak, which was the case in all data sets. The quantification results for scaled and unscaled data sets matched within 10%.
Sample preparation for XAS, EPR, and TXRF analyses
To obtain the oxidized resting state of the Mn/Fe cofactor for EPR analysis, 100–200 μm apoprotein (monomer concentration) was incubated with 2 eq of MnCl2 and 1 eq of (NH4)2Fe(SO4)2 in reconstitution buffer (100 mm HEPES-Na, pH 7.0, 50 mm NaCl) for 1 h at room temperature under aerobic conditions. To label the Mn/Fe cofactor with 57Fe, 57FeCl2 was used instead of natural abundance (NH4)2Fe(SO4)2. For XAS samples, 250–300 μm apoprotein was reconstituted in the same way at a protein:Mn:Fe ratio of 1:2:1 or 1:2.4:1.2. Fe only–loaded XAS samples were prepared using a 3- or 4-fold molar excess of (NH4)2Fe(SO4)2 over polypeptide chains. Excess metal ions were removed by passing the sample through a HiTrap Desalting column equilibrated in storage buffer. Mn/Fe-reconstituted EPR samples were exchanged into 50% 2H2O-based buffer (25 mm HEPES-Na, pH 7.0, 50 mm NaCl in 50% 2H2O) by four rounds of dilution and concentration. Mn only–loaded EPR samples were prepared analogously at a protein:Mn ratio of 1:1 or 1:2, both with and without removal of the excess MnII and following concentration. For EPR spectroscopy, the reconstituted protein was concentrated to 0.3–0.4 mm (X-band samples) or ∼1 mm (Q-band samples), and 50% (v/v) glycerol was added before transfer into EPR tubes and flash cooling in liquid nitrogen. For XAS samples, the reconstituted protein was concentrated to 1.5–3.5 mm. 10% (v/v) glycerol was added to one batch of samples, whereas the other was directly transferred into sample holders and flash-cooled in liquid nitrogen.
To assess the cofactor assembly mechanism of the R2lox variants using EPR and TXRF, 100 μm apoprotein (monomer concentration) was incubated with varying equivalents of MnCl2 and (NH4)2Fe(SO4)2 in reconstitution buffer for 1 h at room temperature under aerobic conditions. TXRF samples were prepared in duplicates. Fe titration series for EPR were prepared by adding 4 eq (per monomer) of MnCl2 to 100 μm apoprotein. Then, 0.2 eq of (NH4)2Fe(SO4)2 was added from a 10 mm stock solution. After 10-min incubation at room temperature, an aliquot was removed, and another 0.2 eq of (NH4)2Fe(SO4)2 was added to the remaining solution. These steps were repeated until 1 eq of (NH4)2Fe(SO4)2 had been added. For TXRF measurements, only three titration points were sampled, with final protein:Mn:Fe ratios of 1:4:0.2, 1:4:0.6, and 1:4:1. All samples were incubated at room temperature for 1 h after the last addition of (NH4)2Fe(SO4)2, i.e. for a total of 1 h 40 min. Following the reconstitution procedure, excess metal ions were removed as above, and the reconstituted protein was concentrated to 0.2–0.3 mm (EPR) or ∼0.5 mm (TXRF). 50% (v/v) glycerol was added to EPR samples before transfer into X-band tubes and flash cooling in liquid nitrogen.
XAS
XAS spectra at Mn and Fe K-edges were collected in fluorescence detection mode at the SuperXAS beamline of the Swiss Light Source and at the Samba beamline of SOLEIL (Paris, France) using standard setups as described previously (Si(111) or Si(220) double-crystal monochromator, energy-resolving five-element silicon-drift or 36-element Ge detector; samples held in a liquid-He cryostat at 20 K) (24, 37). XANES and EXAFS spectra were collected with scan durations of about 1 or 15 min (up to 20 scans on separate sample spots averaged for signal-to-noise ratio improvement; incident beam appropriately attenuated by Al filters). XAS spectra were processed using in-house software (62), and EXAFS spectra were simulated using phase functions calculated with FEFF9 (63).
TXRF
Metal contents of apoprotein preparations, metal titration, and selected EPR samples as well as X-ray spectroscopy samples were quantified using TXRF analysis on a Bruker PicoFox instrument (64). A gallium standard (Sigma) was added to the samples (v/v, 1:1) prior to the measurements. Technical duplicates were prepared of each sample. TXRF spectra were analyzed using the routines provided with the spectrometer.
EPR spectroscopy
X-band continuous-wave EPR measurements were performed in the temperature range of 10–30 K using a Bruker E500 spectrometer equipped with a Bruker ER 4116DM resonator, Oxford Instruments ESR 935 cryostat, and ITC503 temperature controller. Magnetic field modulation amplitude was 7.5 G, the microwave (mw) frequency was around 9.63 GHz. Q-band pulse EPR and HYSCORE measurements were performed between 4 and 20 K using a Bruker ELEXSYS E580 Q-band pulse EPR spectrometer equipped with a home-built TE011 microwave resonator (65) and cryogen-free variable temperature cryostat from Cryogenic Ltd. Field-swept EPR spectra were obtained via integration of the electron spin echo signal, produced by two microwave pulses of 156 ns with an interpulse delay of 348 ns and shot repetition time of 20 μs. Long mw pulses combined with a narrow integration window of 20 ns were used to isolate the S = 1/2 species from the S = 5/2 MnII hexaaquo and other background EPR signals (66). The mw frequency was around 34.12 GHz. HYSCORE spectra were measured using the pulse sequence tπ/2–τ–tπ/2–t1–tπ–t2–tπ–τ–echo with a shot repetition time of 500 μs. A pulse length of tπ/2 = 16 ns and interpulse delay time of τ = 240 ns were used. The interpulse distances t1 and t2 were sampled in the range of 200–1736 ns with an increment of 12 ns. Simulations of EPR spectra were performed using the EasySpin toolbox (67) for MATLAB®.
Mass spectrometric analysis of cross-link formation
Samples were kept in the dark during processing. Three samples each of 100 μm apo-wt-R2lox or apo-G68L-R2lox were incubated with either 4 eq (per monomer) of MnCl2 and 1 eq of (NH4)2Fe(SO4)2, with the Fe salt added in steps of 0.2 eq every 10 min to maximize Mn/Fe cofactor formation (27), or 3 eq of (NH4)2Fe(SO4)2 only in reconstitution buffer under aerobic conditions for 1 h at room temperature. Excess metal ions were removed by passing the samples through a HiTrap Desalting column equilibrated in storage buffer. The reconstituted protein was concentrated to 0.3–0.5 mm. From each assay replicate, 80 μg of protein were subjected to proteolytic digestion by Glu-C (Promega; enzyme:substrate ratio, 1:40) in 50 mm phosphate buffer, pH 7.6, using the SP3 sample preparation method (68). Glu-C in phosphate buffer shows a high rate of cleavage at the C-terminal side of glutamate and aspartate residues. From each replicate, 300 μl of digested sample were collected and acidified by addition of 100 μl of 10% formic acid (FA) prior to LC-MS. The autosampler of an HPLC 1200 system (Agilent Technologies) injected 1 μl (∼200 ng of peptides) into a C18 guard desalting column (Zorbax 300SB-C18, 5 × 0.3 mm, 5-μm bead size; Agilent). Then a 15-cm-long C18 PicoFrit column (100-μm internal diameter, 5-μm bead size; Nikkyo Technos Co., Tokyo, Japan) installed onto the nanoelectrospray ionization source was used. Solvent A was 97% water, 3% acetonitrile, 0.1% FA; and solvent B was 5% water, 95% acetonitrile, 0.1% FA. At a constant flow of 0.4 μl/min, a linear gradient went from 2% B up to 40% B in 45 min followed by a steep increase to 100% B in 5 min, plateau at 100% B for 5 min, and subsequent re-equilibration with 2% B. Online LC-MS was performed using an LTQ Orbitrap Velos Pro mass spectrometer (Thermo Scientific). Fourier transform (FT) MS master scans (automatic gain control target of 1e6) were acquired with a resolution of 30,000 and were followed by data-dependent MS/MS (automatic gain control target of 1e5) at a resolution of 7,500. In data-dependent MS/MS, the top two ions from the master scan were selected first for collision-induced dissociation (at 35% energy) and afterward for higher-energy collision dissociation (at 30% energy). Precursors were isolated with a 2 m/z window. Dynamic exclusion was used with 60-s duration. Each sample was analyzed in technical triplicates. MS raw files were then searched against a FASTA database containing only the R2lox protein sequence (but including both WT and G68L variants) using Sequest HT under Proteome Discoverer 1.4.0.288 (Thermo Scientific). No-enzyme was used as cleavage setting, oxidation of methionine was used as variable modification, and carbamidomethylation of cysteine was used as fixed modification (not relevant, however, because there are no cysteine residues in R2lox). Precursor mass tolerance was 15 ppm, and product mass tolerance was 0.02 Da. Peptide-spectrum matches with cross-correlation (Xcorr) >1.2 passed the fixed value peptide-spectrum match validator. Proteome Discoverer 1.4 can calculate protein areas based on the MS1 peak area integration of the three most abundant identified peptides. The MS1 peak area in each LC-MS run of m/z 689.038, z = 3 (which corresponds to the cross-linked peptide AVIRAATVYNMIVE–AVTLD), normalized to the protein area of the same run, was used as surrogate marker for the amount of cross-link in the samples. For an annotated spectrum of the cross-linked peptide, see Fig. S7 in Griese et al. (23).
Author contributions
Y. K., R. K., R. M. M. B., D. L., M. Haumann, and J. J. G. data curation; Y. K., R. K., R. M. M. B., D. L., M. Haumann, and J. J. G. formal analysis; Y. K., R. K., R. M. M. B., V. S., D. L., M. Haumann, and J. J. G. investigation; Y. K., R. K., R. M. M. B., D. L., and J. J. G. visualization; Y. K., R. K., R. M. M. B., D. L., M. Haumann, and J. J. G. methodology; Y. K., R. M. M. B., D. L., M. Haumann, M. Högbom, N. C., and J. J. G. writing-review and editing; R. M. M. B., M. Haumann, M. Högbom, and N. C. resources; M. Haumann, M. Högbom, and N. C. supervision; M. Haumann, M. Högbom, N. C., and J. J. G. funding acquisition; M. Högbom, N. C., and J. J. G. conceptualization; M. Högbom, N. C., and J. J. G. project administration; N. C. and J. J. G. writing-original draft; Y. K. collected and analyzed EPR data; R. K. collected and analyzed XAS and TXRF data; R. M. M. B. performed MS analysis of the cross-link; V. S. prepared samples for MS; D. L. performed sequence classification and analysis; M. Haumann collected and analyzed XAS and TXRF data; M. Högbom designed the study; N. C. designed the study and wrote the paper; J. J. G. performed protein production, purification and crystallization, collected and analyzed X-ray crystallographic data, prepared samples for EPR, XAS and TXRF, designed the study and wrote the paper.
Supplementary Material
Acknowledgments
We thank members of the Högbom lab and the staff at beamlines X06SA and X10SA/Swiss Light Source and I911-3/Max II for assistance with X-ray data collection as well as the beamline teams at SuperXAS/Swiss Light Source (M. Nachtegaal and coworkers) and Samba/SOLEIL (E. Fonda and coworkers) for technical support for XAS. We also thank Hannah S. Shafaat for providing the EPR spectrum of intermediate 1 of wt-R2lox Mn/Fe cofactor assembly.
This work was supported by Deutsche Forschungsgemeinschaft Grant Ha3265/6–1 and a Heisenberg Fellowship, German Bundesministerium für Bildung und Forschung Grant 05K14KE1 within the Röntgen-Ångström Cluster (to M. Haumann), Swedish Research Council Grants 2016-03770 (to J. J. G.) and 2017-04018 (to M. Högbom), European Research Council Grant HIGH-GEAR 724394, Knut and Alice Wallenberg Foundation Grant 2017.0275, the European Community's Seventh Framework Programme (FP7/2007–2013) under Grant Agreement 283570 (for BioStruct-X) (to M. Högbom), and Australian Research Council Future Fellowship FT140100834 (to N. C.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supporting information S1–S8, Figs. S1–S14, and Tables S1–S3.
The atomic coordinates and structure factors (codes 6I90, 6I92, 6I93, 6I94, and 6I95) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The mass spectrometric raw data and spectral libraries associated with this manuscript are available from ProteomeXchange with the accession number PXD015549.
- R2c
- class Ic RNR R2 protein
- RNR
- ribonucleotide reductase
- FA
- formic acid
- R2lox
- R2-like ligand-binding oxidase
- HYSCORE
- hyperfine sublevel correlation
- XANES
- X-ray absorption near edge structure
- EXAFS
- extended X-ray absorption fine structure
- TXRF
- total-reflection X-ray fluorescence
- mw
- microwave
- PDB
- Protein Data Bank
- FT
- Fourier transform
- XAS
- X-ray absorption spectroscopy.
References
- 1. Sazinsky M. H., and Lippard S. J. (2006) Correlating structure with function in bacterial multicomponent monooxygenases and related diiron proteins. Acc. Chem. Res. 39, 558–566 10.1021/ar030204v [DOI] [PubMed] [Google Scholar]
- 2. Hofer A., Crona M., Logan D. T., and Sjöberg B. M. (2012) DNA building blocks: keeping control of manufacture. Crit. Rev. Biochem. Mol. Biol. 47, 50–63 10.3109/10409238.2011.630372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nordlund P., and Reichard P. (2006) Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706 10.1146/annurev.biochem.75.103004.142443 [DOI] [PubMed] [Google Scholar]
- 4. Högbom M. (2011) Metal use in ribonucleotide reductase R2, di-iron, di-manganese and heterodinuclear—an intricate bioinorganic workaround to use different metals for the same reaction. Metallomics 3, 110–120 10.1039/c0mt00095g [DOI] [PubMed] [Google Scholar]
- 5. Högbom M. (2010) The manganese/iron-carboxylate proteins: what is what, where are they, and what can the sequences tell us? J. Biol. Inorg. Chem. 15, 339–349 10.1007/s00775-009-0606-5 [DOI] [PubMed] [Google Scholar]
- 6. Cotruvo J. A., and Stubbe J. (2011) Class I ribonucleotide reductases: metallocofactor assembly and repair in vitro and in vivo. Annu. Rev. Biochem. 80, 733–767 10.1146/annurev-biochem-061408-095817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Griese J. J., Srinivas V., and Högbom M. (2014) Assembly of nonheme Mn/Fe active sites in heterodinuclear metalloproteins. J. Biol. Inorg. Chem. 19, 759–774 10.1007/s00775-014-1140-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stubbe J. (2003) Di-iron-tyrosyl radical ribonucleotide reductases. Curr. Opin. Chem. Biol. 7, 183–188 10.1016/S1367-5931(03)00025-5 [DOI] [PubMed] [Google Scholar]
- 9. Cox N., Ogata H., Stolle P., Reijerse E., Auling G., and Lubitz W. (2010) A tyrosyl-dimanganese coupled spin system is the native metalloradical cofactor of the R2F subunit of the ribonucleotide reductase of Corynebacterium ammoniagenes. J. Am. Chem. Soc. 132, 11197–11213 10.1021/ja1036995 [DOI] [PubMed] [Google Scholar]
- 10. Stolle P., Barckhausen O., Oehlmann W., Knobbe N., Vogt C., Pierik A. J., Cox N., Schmidt P. P., Reijerse E. J., Lubitz W., and Auling G. (2010) Homologous expression of the nrdF gene of Corynebacterium ammoniagenes strain ATCC 6872 generates a manganese-metallocofactor (R2F) and a stable tyrosyl radical (Y) involved in ribonucleotide reduction. FEBS J. 277, 4849–4862 10.1111/j.1742-4658.2010.07885.x [DOI] [PubMed] [Google Scholar]
- 11. Cotruvo J. A., and Stubbe J. (2011) Escherichia coli class Ib ribonucleotide reductase contains a dimanganese(III)-tyrosyl radical cofactor in vivo. Biochemistry 50, 1672–1681 10.1021/bi101881d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Y., and Stubbe J. (2011) Bacillus subtilis class Ib ribonucleotide reductase is a dimanganese(III)-tyrosyl radical enzyme. Biochemistry 50, 5615–5623 10.1021/bi200348q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cotruvo J. A. Jr., and Stubbe J. (2010) An active dimanganese(III)-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Biochemistry 49, 1297–1309 10.1021/bi902106n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cotruvo J. A. Jr., Stich T. A., Britt R. D., and Stubbe J. (2013) Mechanism of assembly of the dimanganese-tyrosyl radical cofactor of class Ib ribonucleotide reductase: enzymatic generation of superoxide is required for tyrosine oxidation via a Mn(III)Mn(IV) intermediate. J. Am. Chem. Soc. 135, 4027–4039 10.1021/ja312457t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang W., Yun D., Saleh L., Barr E. W., Xing G., Hoffart L. M., Maslak M. A., Krebs C., and Bollinger J. M. Jr. (2007) A manganese(IV)/iron(III) cofactor in Chlamydia trachomatis ribonucleotide reductase. Science 316, 1188–1191 10.1126/science.1141179 [DOI] [PubMed] [Google Scholar]
- 16. Högbom M., Stenmark P., Voevodskaya N., McClarty G., Gräslund A., and Nordlund P. (2004) The radical site in chlamydial ribonucleotide reductase defines a new R2 subclass. Science 305, 245–248 10.1126/science.1098419 [DOI] [PubMed] [Google Scholar]
- 17. Voevodskaya N., Lendzian F., Ehrenberg A., and Gräslund A. (2007) High catalytic activity achieved with a mixed manganese-iron site in protein R2 of Chlamydia ribonucleotide reductase. FEBS Lett. 581, 3351–3355 10.1016/j.febslet.2007.06.023 [DOI] [PubMed] [Google Scholar]
- 18. Rozman Grinberg I., Lundin D., Hasan M., Crona M., Jonna V. R., Loderer C., Sahlin M., Markova N., Borovok I., Berggren G., Hofer A., Logan D. T., and Sjöberg B. M. (2018) Novel ATP-cone-driven allosteric regulation of ribonucleotide reductase via the radical-generating subunit. Elife 7, e31529 10.7554/eLife.31529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rose H. R., Ghosh M. K., Maggiolo A. O., Pollock C. J., Blaesi E. J., Hajj V., Wei Y., Rajakovich L. J., Chang W. C., Han Y., Hajj M., Krebs C., Silakov A., Pandelia M.-E., Bollinger J. M. Jr., et al. (2018) Structural basis for superoxide activation of Flavobacterium johnsoniae class I ribonucleotide reductase and for radical initiation by its dimanganese cofactor. Biochemistry 57, 2679–2693 10.1021/acs.biochem.8b00247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blaesi E. J., Palowitch G. M., Hu K., Kim A. J., Rose H. R., Alapati R., Lougee M. G., Kim H. J., Taguchi A. T., Tan K. O., Laremore T. N., Griffin R. G., Krebs C., Matthews M. L., Silakov A., et al. (2018) Metal-free class Ie ribonucleotide reductase from pathogens initiates catalysis with a tyrosine-derived dihydroxyphenylalanine radical. Proc. Natl. Acad. Sci. U.S.A. 115, 10022–10027 10.1073/pnas.1811993115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Srinivas V., Lebrette H., Lundin D., Kutin Y., Sahlin M., Lerche M., Eirich J., Branca R. M. M., Cox N., Sjöberg B.-M., and Högbom M. (2018) Metal-free ribonucleotide reduction powered by a DOPA radical in Mycoplasma pathogens. Nature 563, 416–420 10.1038/s41586-018-0653-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Srinivas V., Lebrette H., Lundin D., Kutin Y., Sahlin M., Lerche M., Eirich J., Branca R. M. M., Cox N., Sjöberg B.-M., and Högbom M. (2018) Metal-independent ribonucleotide reduction powered by a DOPA radical in Mycoplasma pathogens. BioRxiv 10.1101/348268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Griese J. J., Roos K., Cox N., Shafaat H. S., Branca R. M., Lehtiö J., Gräslund A., Lubitz W., Siegbahn P. E., and Högbom M. (2013) Direct observation of structurally encoded metal discrimination and ether bond formation in a heterodinuclear metalloprotein. Proc. Natl. Acad. Sci. U.S.A. 110, 17189–17194 10.1073/pnas.1304368110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Griese J. J., Kositzki R., Schrapers P., Branca R. M., Nordström A., Lehtiö J., Haumann M., and Högbom M. (2015) Structural basis for oxygen activation at a heterodinuclear manganese/iron cofactor. J. Biol. Chem. 290, 25254–25272 10.1074/jbc.M115.675223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andersson C. S., and Högbom M. (2009) A Mycobacterium tuberculosis ligand-binding Mn/Fe protein reveals a new cofactor in a remodeled R2-protein scaffold. Proc. Natl. Acad. Sci. U.S.A. 106, 5633–5638 10.1073/pnas.0812971106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lundin D., Poole A. M., Sjöberg B. M., and Högbom M. (2012) Use of structural phylogenetic networks for classification of the ferritin-like superfamily. J. Biol. Chem. 287, 20565–20575 10.1074/jbc.M112.367458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kutin Y., Srinivas V., Fritz M., Kositzki R., Shafaat H. S., Birrell J., Bill E., Haumann M., Lubitz W., Högbom M., Griese J. J., and Cox N. (2016) Divergent assembly mechanisms of the manganese/iron cofactors in R2lox and R2c proteins. J. Inorg. Biochem. 162, 164–177 10.1016/j.jinorgbio.2016.04.019 [DOI] [PubMed] [Google Scholar]
- 28. Dassama L. M., Krebs C., Bollinger J. M. Jr., Rosenzweig A. C., and Boal A. K. (2013) Structural basis for assembly of the Mn(IV)/Fe(III) cofactor in the class Ic ribonucleotide reductase from Chlamydia trachomatis. Biochemistry 52, 6424–6436 10.1021/bi400819x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dassama L. M., Boal A. K., Krebs C., Rosenzweig A. C., and Bollinger J. M. Jr., (2012) Evidence that the β subunit of Chlamydia trachomatis ribonucleotide reductase is active with the manganese ion of its manganese(IV)/iron(III) cofactor in site 1. J. Am. Chem. Soc. 134, 2520–2523 10.1021/ja211314p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Andersson C. S., Öhrström M., Popović-Bijelić A., Gräslund A., Stenmark P., and Högbom M. (2012) The manganese ion of the heterodinuclear Mn/Fe cofactor in Chlamydia trachomatis ribonucleotide reductase R2c is located at metal position 1. J. Am. Chem. Soc. 134, 123–125 10.1021/ja209678x [DOI] [PubMed] [Google Scholar]
- 31. Roos K., and Siegbahn P. E. (2011) Oxygen cleavage with manganese and iron in ribonucleotide reductase from Chlamydia trachomatis. J. Biol. Inorg. Chem. 16, 553–565 10.1007/s00775-011-0755-1 [DOI] [PubMed] [Google Scholar]
- 32. Martinie R. J., Blaesi E. J., Krebs C., Bollinger J. M. Jr., Silakov A., and Pollock C. J. (2017) Evidence for a di-μ-oxo diamond core in the Mn(IV)/Fe(IV) activation intermediate of ribonucleotide reductase from Chlamydia trachomatis. J. Am. Chem. Soc. 139, 1950–1957 10.1021/jacs.6b11563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miller E. K., Trivelas N. E., Maugeri P. T., Blaesi E. J., and Shafaat H. S. (2017) Time-resolved investigations of heterobimetallic cofactor assembly in R2lox reveal distinct Mn/Fe intermediates. Biochemistry 56, 3369–3379 10.1021/acs.biochem.7b00403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang W., Hoffart L. M., Krebs C., Bollinger J. M. Jr. (2007) A manganese(IV)/iron(IV) intermediate in assembly of the manganese(IV)/iron(III) cofactor of Chlamydia trachomatis ribonucleotide reductase. Biochemistry 46, 8709–8716 10.1021/bi700906g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kwak Y., Jiang W., Dassama L. M., Park K., Bell C. B. 3rd, Liu L. V., Wong S. D., Saito M., Kobayashi Y., Kitao S., Seto M., Yoda Y., Alp E. E., Zhao J., Bollinger J. M. Jr., et al. (2013) Geometric and electronic structure of the Mn(IV)Fe(III) cofactor in class Ic ribonucleotide reductase: correlation to the class Ia binuclear non-heme iron enzyme. J. Am. Chem. Soc. 135, 17573–17584 10.1021/ja409510d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shafaat H. S., Griese J. J., Pantazis D. A., Roos K., Andersson C. S., Popović-Bijelić A., Gräslund A., Siegbahn P. E., Neese F., Lubitz W., Högbom M., and Cox N. (2014) Electronic structural flexibility of heterobimetallic Mn/Fe cofactors: R2lox and R2c proteins. J. Am. Chem. Soc. 136, 13399–13409 10.1021/ja507435t [DOI] [PubMed] [Google Scholar]
- 37. Kositzki R., Mebs S., Marx J., Griese J. J., Schuth N., Högbom M., Schünemann V., and Haumann M. (2016) Protonation state of MnFe and FeFe cofactors in a ligand-binding oxidase revealed by X-ray absorption, emission, and vibrational spectroscopy and QM/MM calculations. Inorg. Chem. 55, 9869–9885 10.1021/acs.inorgchem.6b01752 [DOI] [PubMed] [Google Scholar]
- 38. Griese J. J., Kositzki R., Haumann M., and Högbom M. (2019) Assembly of a heterodinuclear Mn/Fe cofactor is coupled to tyrosine–valine ether cross-link formation in the R2-like ligand-binding oxidase. J. Biol. Inorg. Chem. 24, 211–221 10.1007/s00775-019-01639-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Teutloff C., Schäfer K. O., Sinnecker S., Barynin V., Bittl R., Wieghardt K., Lendzian F., and Lubitz W. (2005) High-field EPR investigations of Mn(III)Mn(IV) and Mn(II)Mn(III) states of dimanganese catalase and related model systems. Magn. Reson. Chem. 43, S51–64 10.1002/mrc.1685 [DOI] [PubMed] [Google Scholar]
- 40. Randall D. W., Sturgeon B. E., Ball J. A., Lorigan G. A., Chan M. K., Klein M. P., Armstrong W. H., and Britt R. D. (1995) 55Mn ESE-ENDOR of a mixed-valence Mn(III)Mn(IV) complex—comparison with the Mn cluster of the photosynthetic oxygen-evolving complex. J. Am. Chem. Soc. 117, 11780–11789 10.1021/ja00152a021 [DOI] [Google Scholar]
- 41. Randall D. W., Chan M. K., Armstrong W. H., and Britt R. D. (1998) Pulsed 1H and 55Mn ENDOR studies of dinuclear Mn(III)Mn(IV) model complexes. Mol. Phys. 95, 1283–1294 10.1080/00268979809483259 [DOI] [Google Scholar]
- 42. Peloquin J. M., Campbell K. A., Randall D. W., Evanchik M. A., Pecoraro V. L., Armstrong W. H., and Britt R. D. (2000) 55Mn ENDOR of the S2-state multiline EPR signal of photosystem II: implications on the structure of the tetranuclear Mn cluster. J. Am. Chem. Soc. 122, 10926–10942 10.1021/ja002104f [DOI] [Google Scholar]
- 43. Schäfer K. O., Bittl R., Lendzian F., Barynin V., Weyhermüller T., Wieghardt K., and Lubitz W. (2003) Multifrequency EPR investigation of dimanganese catalase and related Mn(III)Mn(IV) complexes. J. Phys. Chem. B 107, 1242–1250 10.1021/jp0259768 [DOI] [Google Scholar]
- 44. Berggren G., Duraffourg N., Sahlin M., and Sjöberg B. M. (2014) Semiquinone-induced maturation of Bacillus anthracis ribonucleotide reductase by a superoxide intermediate. J. Biol. Chem. 289, 31940–31949 10.1074/jbc.M114.592535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cox N., Ames W., Epel B., Kulik L. V., Rapatskiy L., Neese F., Messinger J., Wieghardt K., and Lubitz W. (2011) Electronic structure of a weakly antiferromagnetically coupled Mn(II)Mn(III) model relevant to manganese proteins: a combined EPR, 55Mn-ENDOR, and DFT study. Inorg. Chem. 50, 8238–8251 10.1021/ic200767e [DOI] [PubMed] [Google Scholar]
- 46. Schäfer K. O., Bittl R., Zweygart W., Lendzian F., Haselhorst G., Weyhermüller T., Wieghardt K., and Lubitz W. (1998) Electronic structure of antiferromagnetically coupled dinuclear manganese Mn(III)Mn(IV) complexes studied by magnetic resonance techniques. J. Am. Chem. Soc. 120, 13104–13120 10.1021/ja9827548 [DOI] [Google Scholar]
- 47. Wieghardt K., Bossek U., Nuber B., Weiss J., Bonvoisin J., Corbella M., Vitols S. E., and Girerd J. J. (1988) Synthesis, crystal structures, reactivity, and magnetochemistry of a series of binuclear complexes of manganese(II), -(III), and -(IV) of biological relevance. The crystal structure of [L′MnIV(μ-O)3MnIVL′](PF6)2·H2O containing an unprecedented short Mn···Mn distance of 2.296 Å. J. Am. Chem. Soc. 110, 7398–7411 10.1021/ja00230a021 [DOI] [Google Scholar]
- 48. Mukhopadhyay S., Mandal S. K., Bhaduri S., and Armstrong W. H. (2004) Manganese clusters with relevance to photosystem II. Chem. Rev. 104, 3981–4026 10.1021/cr0206014 [DOI] [PubMed] [Google Scholar]
- 49. Sigfridsson K. G., Chernev P., Leidel N., Popovic-Bijelic A., Gräslund A., and Haumann M. (2013) Rapid X-ray photoreduction of dimetal-oxygen cofactors in ribonucleotide reductase. J. Biol. Chem. 288, 9648–9661 10.1074/jbc.M112.438796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leidel N., Popović-Bijelić A., Havelius K. G., Chernev P., Voevodskaya N., Gräslund A., and Haumann M. (2012) High-valent [MnFe] and [FeFe] cofactors in ribonucleotide reductases. Biochim. Biophys. Acta 1817, 430–444 10.1016/j.bbabio.2011.12.008 [DOI] [PubMed] [Google Scholar]
- 51. Voevodskaya N., Lendzian F., Sanganas O., Grundmeier A., Gräslund A., and Haumann M. (2009) Redox intermediates of the Mn-Fe Site in subunit R2 of Chlamydia trachomatis ribonucleotide reductase: an X-ray absorption and EPR study. J. Biol. Chem. 284, 4555–4566 10.1074/jbc.M807190200 [DOI] [PubMed] [Google Scholar]
- 52. Griese J. J., and Högbom M. (2012) X-ray reduction correlates with soaking accessibility as judged from four non-crystallographically related diiron sites. Metallomics 4, 894–898 10.1039/c2mt20080e [DOI] [PubMed] [Google Scholar]
- 53. Haumann M., Müller C., Liebisch P., Iuzzolino L., Dittmer J., Grabolle M., Neisius T., Meyer-Klaucke W., and Dau H. (2005) Structural and oxidation state changes of the photosystem II manganese complex in four transitions of the water oxidation cycle (S0 → S1, S1 → S2, S2 → S 3, and S3,4 → S0) characterized by X-ray absorption spectroscopy at 20 K and room temperature. Biochemistry 44, 1894–1908 10.1021/bi048697e [DOI] [PubMed] [Google Scholar]
- 54. Griese J. J., Branca R. M. M., Srinivas V., and Högbom M. (2018) Ether cross-link formation in the R2-like ligand-binding oxidase. J. Biol. Inorg. Chem. 23, 879–886 10.1007/s00775-018-1583-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Griese J. J., and Högbom M. (2019) Location-specific quantification of protein-bound metal ions by X-ray anomalous dispersion: Q-XAD. Acta Crystallogr. D Struct. Biol. 75, 764–771 10.1107/S2059798319009926 [DOI] [PubMed] [Google Scholar]
- 61. Kleywegt G. J., and Jones T. A. (1996) xdlMAPMAN and xdlDATAMAN—programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr. D Biol. Crystallogr. 52, 826–828 10.1107/S0907444995014983 [DOI] [PubMed] [Google Scholar]
- 62. Dau H., Liebisch P., and Haumann M. (2003) X-ray absorption spectroscopy to analyze nuclear geometry and electronic structure of biological metal centers–potential and questions examined with special focus on the tetra-nuclear manganese complex of oxygenic photosynthesis. Anal. Bioanal. Chem. 376, 562–583 10.1007/s00216-003-1982-2 [DOI] [PubMed] [Google Scholar]
- 63. Rehr J. J., Kas J. J., Vila F. D., Prange M. P., and Jorissen K. (2010) Parameter-free calculations of X-ray spectra with FEFF9. Phys. Chem. Chem. Phys. 12, 5503–5513 10.1039/b926434e [DOI] [PubMed] [Google Scholar]
- 64. Klockenkämper R. (1997) Total-reflection X-ray Fluorescence Analysis, Wiley, New York [Google Scholar]
- 65. Reijerse E., Lendzian F., Isaacson R., and Lubitz W. (2012) A tunable general purpose Q-band resonator for CW and pulse EPR/ENDOR experiments with large sample access and optical excitation. J. Magn. Reson. 214, 237–243 10.1016/j.jmr.2011.11.011 [DOI] [PubMed] [Google Scholar]
- 66. Hofbauer W., and Bittl R. (2000) A novel approach to separating EPR lines arising from species with different transition moments. J. Magn. Reson. 147, 226–231 10.1006/jmre.2000.2202 [DOI] [PubMed] [Google Scholar]
- 67. Stoll S., and Schweiger A. (2006) EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 178, 42–55 10.1016/j.jmr.2005.08.013 [DOI] [PubMed] [Google Scholar]
- 68. Hughes C. S., Foehr S., Garfield D. A., Furlong E. E., Steinmetz L. M., and Krijgsveld J. (2014) Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 10, 757–757 10.15252/msb.20145625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Eddy S. R. (2011) Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wheeler T. J., Clements J., and Finn R. D. (2014) Skylign: a tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinformatics 15, 7 10.1186/1471-2105-15-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.