Abstract
tau is a microtubule (MT)-associated protein that promotes tubulin assembly and stabilizes MTs by binding longitudinally along the MT surface. tau can aberrantly aggregate into pathological inclusions that define Alzheimer's disease, frontotemporal dementias, and other tauopathies. A spectrum of missense mutations in the tau-encoding gene microtubule-associated protein tau (MAPT) can cause frontotemporal dementias. tau aggregation is postulated to spread by a prion-like mechanism. Using a cell-based inclusion seeding assay, we recently reported that only a few tau variants are intrinsically prone to this type of aggregation. Here, we extended these studies to additional tau mutants and investigated their MT binding properties in mammalian cell-based assays. A limited number of tau variants exhibited modest aggregation propensity in vivo, but most tau mutants did not aggregate. Reduced MT binding appeared to be the most common dysfunction for the majority of tau variants due to missense mutations, implying that MT-targeting therapies could potentially be effective in the management of tauopathies.
Keywords: aggregation, Alzheimer disease, microtubule, prion, tau protein (tau), tauopathy
Introduction
Pathogenic mutations in the MAPT gene that encodes the microtubule (MT)2-associated protein tau directly cause some forms of tauopathies, a heterogeneous group of disorders with brain-laden pathological tau inclusions that includes Alzheimer's disease (AD), Pick's disease, chronic traumatic encephalopathy, and other conditions (1). AD, the most common form of dementia, has two classical pathological lesions: Aβ senile plaques and neurofibrillary tangles composed of hyperphosphorylated, aggregated tau (2). Of these two pathological features, the presence of neurofibrillary tangles strongly correlates with symptoms such as cognitive decline and other dementia signs in patients with AD (3, 4). In addition, MAPT mutations are directly associated with frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17t) (5, 6).
As an MT-associated protein, tau binds longitudinally along the MT surface, providing MT stability and promoting tubulin assembly (7–10). Physiologically, tau is primarily expressed in neurons and is concentrated in the distal axon (11). In the human brain, tau protein is alternatively spliced into six major different isoforms based on inclusion or exclusion of exons 2, 3, and 10 (12, 13). Inclusion of one or two N-terminal domains generates 0N, 1N, and 2N isoforms due to alternative splicing of exons 2 and 3. Varied forms of tau also result from the presence of three (3R) or four (4R) MT-binding repeats of 31 or 32 amino acids due to alternative splicing of exon 10 (14, 15).
More than 50 pathogenic MAPT mutations have been identified (5, 6, 16). Of note, many of these are intronic and silent mutations that affect exon 10 splicing and thus the ratio of 3R/4R tau isoforms expressed. Missense mutations directly alter the primary protein sequence, but in some cases they can also affect exon 10 splicing (6, 16). Most tau missense mutations are clustered within the MT-binding domain (MTBD) (6, 17), suggesting that impairment of tau–MT interactions can be directly involved in pathogenesis. Loss of MT mass due to MT instability is a common feature of AD (18–21). tau–MT dysfunction can affect synaptic plasticity and impair axonal transport of vesicles and other molecules, causing cognitive deficits in learning and memory (22–25). Defects in tau can also activate MT-severing proteins such as katanin and cause degradation of MTs (26). Predominantly in vitro studies indicate that tau mutants can alter tubulin assembly and MT binding (17, 27). tau post-translational modifications such as phosphorylation can also decrease MT-binding activity (28–30).
Because of the progressive nature of tauopathies, tau aggregation has been hypothesized to propagate from neuron to neuron by a prion-like mechanism (31). In clinical staging of AD patients, it has been proposed that aggregated forms of tau may spread from the hippocampus to the entorhinal region, and eventually to the rest of the neocortex (32, 33). Experimentally, tau can transfer from cell to cell and can be seeded by preformed aggregated tau fibrils to induce aggregation (34–36). Transgenic mouse models of tau can be injected with tau seeds of recombinant proteins, mouse brain lysate, and even human brain lysate to induce neurofibrillary tangles with tau fibrillar aggregates (37–41). Although there is no clinical evidence of iatrogenic spread between AD patients (42), experimental studies support the hypothesis that tau can spread in a prion-like manner along anatomical connections to other neurons.
A previous study used a cell-based assay to examine prion-like seeding in 19 missense pathogenic tau mutants and revealed that only mutants at the Pro-301 position were uniquely prone to seed induced aggregation (43). Building from this unexpected finding, we investigated and characterized an extensive series of tau mutants for MT binding using a mammalian cell-based assay, and we extended the previous series of pathogenic tau mutants for prion-like seeding. These studies show that most tau mutants share a common mechanism of impaired MT binding with only heterogeneous potential for aggregation.
Results
tau variants with mutations at the Pro-301 position severely impaired MT binding compared with WT tau and several other tau mutants in the R1 and R2 repeats
Although tau–MT associations can be visualized in the cytoplasm by immunofluorescent labeling, the amount of tau that is directly bound to MTs cannot be quantified by this method (Fig. S1). A previously established cell-based MT-binding assay (44–46) was performed on diverse tau missense mutants (Fig. 1) to assess changes in MT binding associated with a spectrum of tau variants with missense tau mutations. Furthermore, most previous studies investigated tau mutants with an in vitro MT-binding assay that used recombinant tau expressed from bacteria and tubulin assembled from bovine or porcine sources (Table 1). This cell-based MT-binding assay is more physiologically relevant as it, at least partially, incorporates the effects of post-translational modifications such as phosphorylation (Fig. S2), differential tau folding in mammalian cells, and interactions with human MT isotypes in HEK293T cells. The 0N4R human tau isoform was used for all the assays, but all the tau mutations are numbered according to the 2N4R human tau isoform, which is the longest tau isoform expressed in the human brain.
Figure 1.
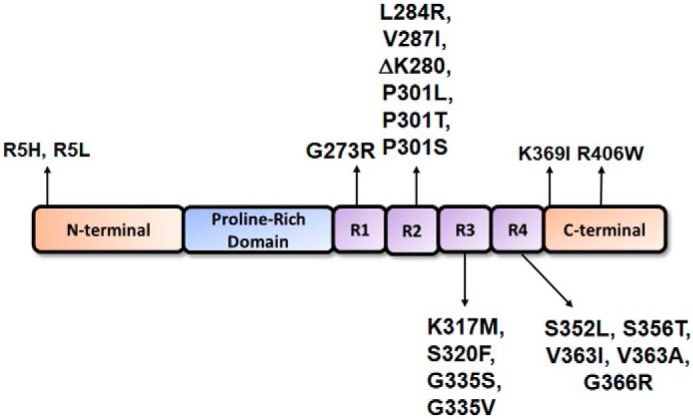
Schematic of 4R tau protein depicting major structural domains. tau is an MT-associated protein that consists of an N-terminal region, a proline-rich domain, an MTBD composed of four MT-binding repeats in the 4R isoforms, and a C-terminal region. Indicated are the locations of the pathogenic tau missense mutants that were investigated herein; they were numbered relative to the longest 2N4R tau isoform expressed in the human brain. Most tau missense mutants are clustered within the MTBD.
Table 1.
Summary of findings on tau aggregation and MT binding or polymerization for tau variants with pathogenic mutations investigated
The new mammalian cell-based MT binding and tau aggregation results shown in this study are identified by asterisks. Data from previous studies are included with the references. Impaired MT binding and MT dysfunction are the most common mechanisms for tau mutants. The aggregation propensity of tau mutants in vivo is more varied and only observed for a subset of tau mutants. ↑ = increase; ↓ = decrease; ↔ = unchanged.
| tau mutations | Cell-based MT binding assay | In vitro MT binding assay | In vitro MT assembly | In vitro tau aggregation | In vivo tau aggregation |
|---|---|---|---|---|---|
| R5H | ↑* | NA | ↓ (106) | ↑ (106) | ↔ (43) |
| R5L | ↑* | NA | ↓ (107, 108) | ↔ or ↑ (82, 107–109) | ↔ (43) |
| G273R | ↔* | NA | NA | NA | ↑* |
| L284R | ↓* | NA | NA | NA | ↔* |
| V287I | ↔* | NA | NA | NA | ↔* |
| ΔK280 | ↓* (45) | ↓ or ↔ (51, 52) | ↓ (51, 52, 110) | ↑ (52, 111) | ↑ or ↔ (43, 45) |
| P301L | ↓* or ↔ (45, 53) | ↓ or ↔ (49–52) | ↓ (50–52, 108, 110, 112) | ↑ (52, 82, 108, 109, 113, 114) | ↑* as reviewed in Ref. 17 |
| P301S | ↓* (55) | NA | ↓ (112, 115, 116) | ↑ (114, 116) | ↑ as reviewed in Ref. 17 |
| P301T | ↓* | NA | NA | NA | ↑ (43) |
| K317M | ↓* | NA | NA | NA | ↔* |
| S320F | ↔* | NA | ↓ (82) | ↑ (82) | ↑ (43) |
| G335S | ↓* | NA | ↓ (116) | ↔ (116) | ↔* |
| G335V | ↓* | NA | ↓ (116, 117) | ↑ (116, 117) | ↑* |
| S352L | ↓* | ↓ (118) | ↓ (82, 118) | ↑ (82, 118) | ↔* |
| S356T | ↓* | NA | NA | NA | ↑* |
| V363I | ↓* | NA | ↔ (119) | ↔ (119) | ↑* (120) |
| V363A | ↓* | NA | ↓ (119) | ↔ (119) | ↔* (120) |
| G366R | ↓* | NA | ↓ (90) | ↔ (90) | ↔* |
| K369I | ↓* | NA | ↓ (82, 89) | ↓ (82) | ↔* |
| R406W | ↓* or ↔ (45) | ↓ or ↔ (49, 50, 53) | ↓ (50, 108) | ↑ (108, 113) | ↔ (43) |
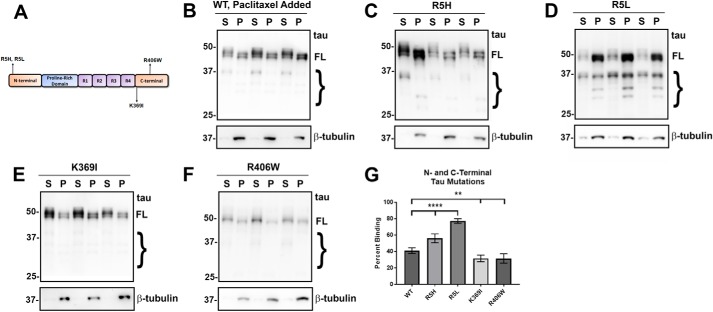
First, to validate the MT-binding assay, wildtype (WT) tau was expressed in HEK293T cells, and the assay was performed with or without paclitaxel, a drug that hyperstabilizes and promotes the formation of MTs (47, 48). Without paclitaxel, most of the tubulin is not polymerized and soluble (Fig. 2A); hence, the majority of tau (∼87%) is also soluble. When paclitaxel is present, tubulin polymerizes into MTs and shifts into the pellet fractions. In the presence of paclitaxel-stabilized MTs, ∼41% of WT tau is found in the MT pellet fraction (Fig. 2B). Therefore, detection of tubulin in the insoluble fraction was used to confirm that paclitaxel was active in all the experiments that followed. Notably, there are also several smaller tau bands observed by immunoblot in this assay, and these are likely due to some degradation from incubation at 37 °C required for these MT-binding assays. However, the major bands are likely full-length tau with other forms of post-translational modifications (Fig. S3).
Figure 2.
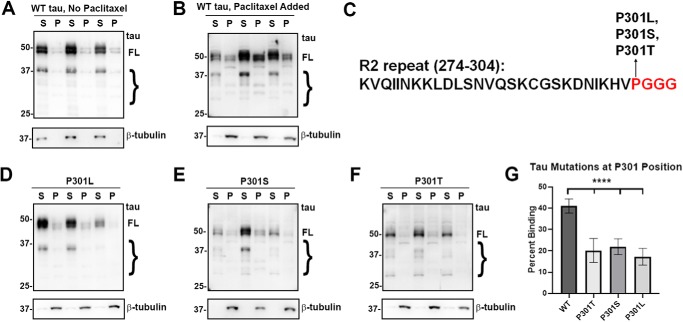
tau mutants at the Pro-301 residue significantly impaired MT binding. A and B, cell-based MT-binding assay performed with HEK293T cells transfected to express WT tau with or without the presence of paclitaxel as described under “Experimental procedures.” Antibody specific for β-tubulin (clone TUB 2.1) was used to assay the polymerization of tubulins. 3026 is a polyclonal antibody against total tau. S = supernatants; P = pellet fractions. Without paclitaxel, the majority of tubulin is not polymerized and soluble, whereas with paclitaxel, the majority of tubulin is polymerized as MTs in the pellet fraction. C, P301L, P301S, and P301T tau mutants are in the PGGG motif of the R2 repeat. In the presence of paclitaxel, P301L (D), P301S (E), and P301T (F) all demonstrate significantly decreased MT binding when compared with WT tau. The relative molecular masses of protein markers are indicated on the left. On the right, FL is for full-length tau, and the brace indicates degradative tau bands. G, one-way ANOVA with Dunnett's test was performed with n = 18 for WT tau and n = 3 for each of these tau mutants. ****, p < 0.0001.
Three tau mutants (P301L, P301S, and P301T) located at the same amino acid residue within the PGGG motif of the R2 repeat (Fig. 2C) were assessed, because previous in vitro and cell culture assays for P301L tau had shown significant decrease in MT binding (49–54). Likewise, studies using brain lysate of P301S tau transgenic mice have confirmed a decreased MT binding (55). In the cell-based assay, it was revealed that all these mutants at Pro-301 reduced MT binding similarly and significantly (Fig. 2, D–G). These findings show that the change of a proline residue to a leucine, serine, or threonine have a similar effect; thus, the Pro-301 position and disruption of this PGGG motif is likely more important than the specific amino acid change.
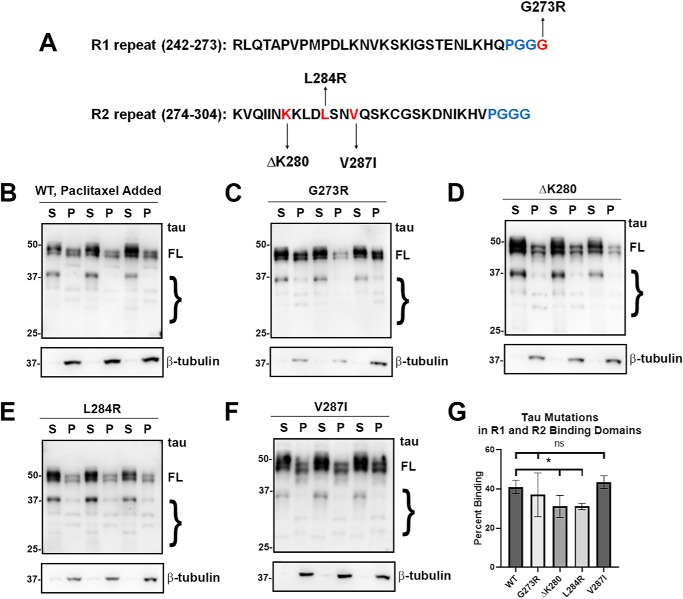
Mutants within the R1 and R2 repeats such as G273R, ΔK280, L284R, and V287I were assessed for altered MT binding (Fig. 3). Interestingly, G273R, a mutant in another PGGG motif but within the R1 repeat, did not significantly affect MT binding compared with WT tau (Fig. 3, C and G). Likewise, the V287I mutant did not affect MT binding (Fig. 3, F and G). We observe that both ΔK280 and L284R significantly decreased MT binding (Fig. 3, D, E, and G). The effects for ΔK280 are consistent with previous in vitro studies that showed varied results but generally indicated that ΔK280 decreased MT binding (51, 52, 56). Interestingly, G273R and V287I flank the other two mutations, suggesting that only a restricted region of the R2 repeat is responsible for most of the R2 binding activity. Overall, tau mutants in the R1 and R2 repeats show mixed results for MT binding (Fig. 3G).
Figure 3.
tau mutants in the R1 and R2 repeats had varied effects on MT binding. A, tau mutants G273R, ΔK280, L284R, and V287I are present within the R1 or R2 repeat as indicated. B–F, MT-binding assay was performed in the presence of paclitaxel as described under “Experimental procedures.” Relative to WT tau, ΔK280 and L284R tau demonstrated decreased MT binding, whereas G273R tau and V287I tau did not have a significant effect. Immunoblots were probed with anti-β-tubulin antibody TUB 2.1 or total tau antibody 3026 as indicated. S = supernatants; P = pellet fractions. The relative molecular masses of protein markers are indicated on the left. On the right, FL is for full-length tau, and the brace indicates degraded tau bands. G, one-way ANOVA with Dunnett's test was performed with n = 18 for WT tau and n = 3 for each of these tau mutants. *, p < 0.05; ns, no statistical significance.
Most tau variants with mutations in the R3 and R4 repeats impaired MT binding
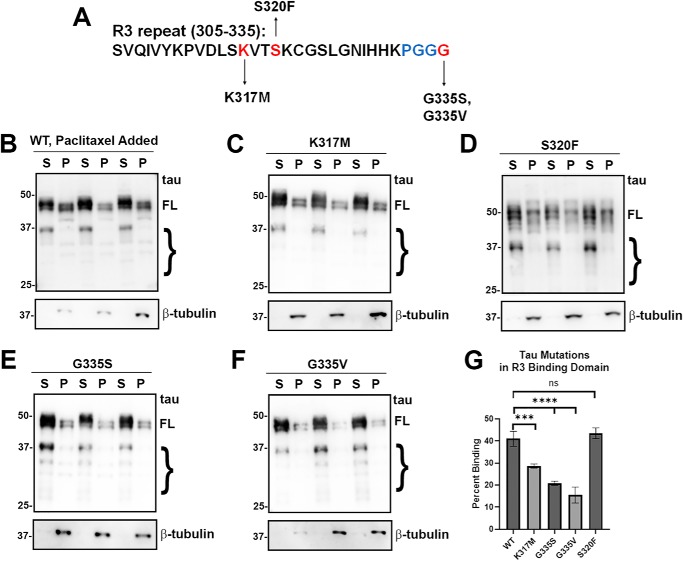
Many pathogenic tau variants with missense mutations are clustered within R3 and R4 repeats, a region that prominently contributes to tau MT binding (Fig. 1). Within the R3 repeat, tau mutations are scattered in multiple locations (Fig. 4A). K317M, a mutant near the center of the R3 repeat, significantly impaired MT binding (Fig. 4, C and G). Both G335S and G335V also decreased MT binding (Fig. 4, E–G). Surprisingly, S320F, a mutant at the center of R3, did not alter MT binding (Fig. 4, D and G). This mutant also appeared to have more degradative bands.
Figure 4.
Most tau mutants in the R3 repeat decreased MT binding. A, tau mutants K317M, S320F, G335S, and G335V are located within the R3 repeat. B–F, MT-binding assay was performed in the presence of paclitaxel as described under “Experimental procedures.” HEK293T cells were transfected with WT or the indicated tau mutations. K317M, G335S, and G335V tau mutants significantly decreased MT binding, whereas the S320F tau had no major change. Immunoblots were probed with anti-β-tubulin antibody TUB 2.1 or total tau antibody 3026 as indicated. S = supernatants; P = pellet fractions. The relative molecular masses of protein markers are indicated on the left. On the right, FL is for full-length tau, and the brace indicates degradative tau bands. G, one-way ANOVA with Dunnett's test was performed with n = 18 for WT tau and n = 3 for each tau mutant. ***, p < 0.001; ****, p < 0.0001; and ns, no statistical significance.
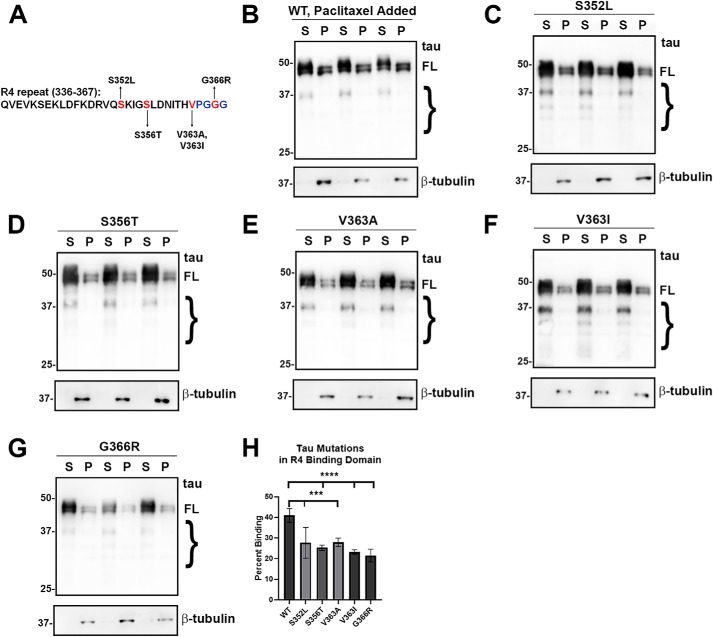
Similar to the majority of mutants in the R3 repeat, mutants such as S352L, S356T, V363A, V363I, and G366R in the R4 repeat decreased MT binding (Fig. 5). Thus, most mutants in the R3 and R4 repeats impaired MT binding, indicating that impairment of binding in R3 and R4 repeats is not well-compensated by the other repeats.
Figure 5.
Most of the tau variants mutants in the R4 repeat significantly reduced MT binding. A, tau mutants S352L, S356T, V363I, V363A, and G366R are located within the R4 repeat. B–G, MT-binding assay was performed in the presence of paclitaxel as described under “Experimental procedures.” HEK293T cells were transfected with WT or the indicated tau mutations. S352L, S356T, V363I, V363A, and G366R tau mutants significantly decreased MT binding. Immunoblots were probed with anti-β-tubulin antibody TUB 2.1 or total tau antibody 3026 as indicated. S = supernatants; P = pellet fractions. The relative molecular masses of protein markers are indicated on the left. On the right, FL is for full-length tau, and the brace indicates degradative tau bands. H, one-way ANOVA with Dunnett's test was performed with n = 18 for WT tau and n = 3 for each tau mutant. ***, p < 0.001; ****, p < 0.0001.
tau variants within the N- and C-terminal regions display varying effects on MT binding
In addition to mutants within the MTBD, we also tested several tau mutants clustered around the N- and C-terminal regions of tau (Fig. 6A). Surprisingly, R5L and R5H were the only mutants that depicted increased MT binding (Fig. 6, C and D), suggesting that these N-terminal tau mutations likely cause long-range conformational changes promoting the tau–MT interactions, at least in a proportion of tau molecules.
Figure 6.
tau mutants in the N and C termini had differential effects on MT binding. A, diagram shows N- and C-terminal tau mutants. B–F, MT-binding assay was performed in the presence of paclitaxel as described under “Experimental procedures.” HEK293T cells were transfected with WT or the indicated tau mutations. Both the R5H and R5L tau mutants in the N terminus significantly increased MT binding, whereas the mutants near the C terminus, K369I and R406W, decreased MT binding. Immunoblots were probed with anti-β-tubulin antibody TUB 2.1 or total tau antibody 3026 as indicated. S = supernatants; P = pellet fractions. The relative molecular masses of protein markers are indicated on the left. On the right, FL is for full-length tau, and the brace indicates degradative tau bands. G, one-way ANOVA with Dunnett's test was performed with n = 18 for WT and n = 3 for each tau mutant. **, p < 0.01; ****, p < 0.0001.
Both C-terminal region mutants K369I and R406W displayed decreased MT binding (Fig. 6, E and F). Although these tau mutations are not within the MTBD, they are both close to this region. This finding suggests that the area adjacent to the repeats also can contribute to MT binding or that these mutations induce conformational changes in the adjacent MTBD.
tau mutants in the PGGG motifs and the R4 repeat have varied aggregation propensities
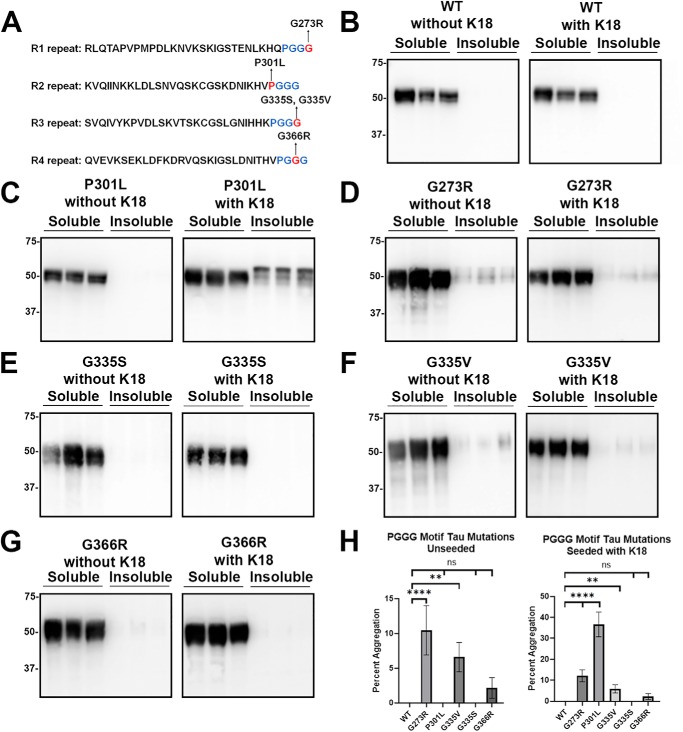
We previously studied the aggregation properties of 19 FTDP-17 tau missense mutants with or without exogenous K18 seeds and found that only S320F moderately self-aggregated, whereas all three mutants (P301L, P301S, and P301T) at Pro-301 within the R2 PGGG motif were robustly permissive to seed-induced aggregation with conformational templating (43). These studies were extended to include new tau mutants used for MT binding studies. First, additional mutants within the PGGG sequences were assessed for their propensity to aggregate: G273R in the R1 repeat; G335S and G335V in the R3 repeat; and G366R in the R4 repeat (Fig. 7A). As a baseline, WT tau did not aggregate with or without K18 seeding (Fig. 7, B and H). In contrast, P301L tau was used as a positive control that normally does not intrinsically aggregate but will readily aggregate when seeded with preformed exogenous K18 fibrils (Fig. 7, C and H). The G273R and G335V tau mutants self-aggregated at modest levels with or without seeding (Fig. 7, D, F, and H). G366R within the R4 repeat negligibly aggregated, but this was not statistically significant (Fig. 7, G and H). None of these mutants within the PGGG motif aggregated more when seeded with K18 preformed fibrils further highlighting the uniqueness of Pro-301 tau mutants (Fig. 7 and Fig. S4).
Figure 7.
tau mutants in the PGGG motif showed varied susceptibility to prion-like seeding. A, diagram depicts tau mutations in the PGGG motif within R1, R2, R3, and R4 repeats. HEK293T cells were transfected with WT or tau mutants and assessed for aggregation with or without exogenous K18 seeds. B, WT tau did not aggregate by itself or in the presence of K18 seeds. C, P301L did not aggregate without seeding but had robust induced aggregation in the presence of K18 seeds. D and F, G273R and G335V demonstrated modest levels of self-aggregation with or without exogenous K18 seeds. E, interestingly, G335S did not aggregate, despite being at the same location as G335V. G, G366R showed low levels of aggregation, but this was not statistically significant. Immunoblots were probed with total tau antibody 3026. The relative molecular masses of protein markers are indicated on the left. H, one-way ANOVA with Dunnett's test was performed with n = 9 for WT tau and n = 3 for each tau mutant. ****, p < 0.0001; **, p < 0.01; and ns, no statistical significance.
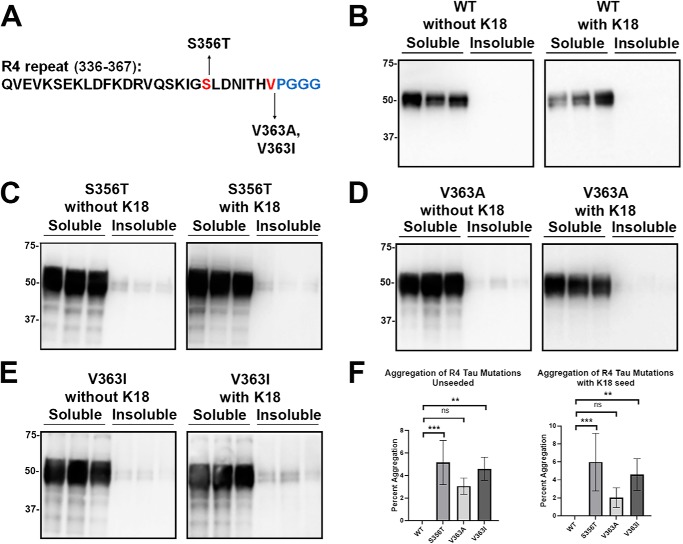
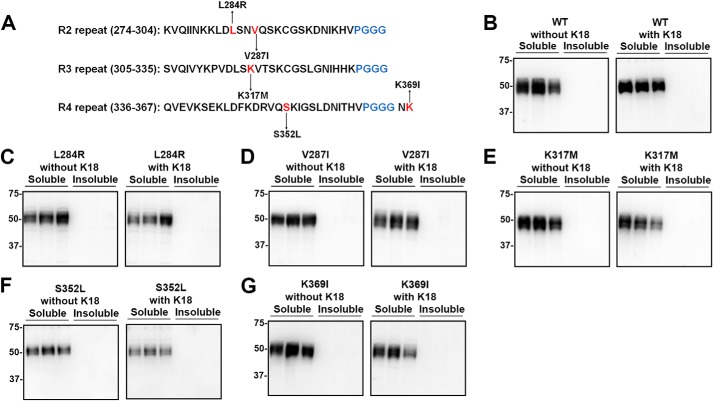
The tau mutants (S356T, V363A, and V363I) close to the R4 PGGG motifs within the R4 repeat were assessed for aggregation (Fig. 8A). S356T and V363I tau modestly aggregated even without seeding, but this did not increase with the presence of K18 seeds (Fig. 8, C and E). The V363A mutant in the same location as V363I might modestly aggregate but was not statistically significant (Fig. 8, D and F). Other tau variants with mutations in various repeats (L284R, V287I, K317M, S352L and K369I) did not aggregate with or without K18 seeding (Fig. 9).
Figure 8.
Some tau mutants in the R4 repeat modestly aggregated without exogenous seeding. A, tau mutants S356T, V363A, and V363I are located within the R4 repeat. HEK293T cells were transfected with WT or tau mutants indicated and assessed for aggregation with or without exogenous K18 fibrillar seeds. Compared with WT tau (B), S356T (C) and V363I (E) mutants modestly self-aggregated without and with seeding. D, V363A did not significantly aggregate with or without K18 seeds. Immunoblots were probed with total tau antibody 3026. The relative molecular masses of protein markers are indicated on the left. One-way ANOVA with Dunnett's test was performed with n = 9 for WT tau and n = 3 for each tau mutant (F). *, p < 0.05; **, p < 0.01; and ns, no statistical significance.
Figure 9.
Additional tau mutants did not show any propensity to aggregate. A, diagram depicts tau mutations L284R, V287I, K317M, S352L, and K369I within R2, R3, and R4 repeats. HEK293T cells were transfected with WT or tau mutants and were assessed for aggregation with or without exogenous K18 fibrillar seeds. Compared with WT tau (B), L284R (C), V287I (D), K317M (E), S352L (F), and K369I (G) did not aggregate with or without exogenous K18 seeds. Immunoblots were probed with total tau antibody 3026. The relative molecular masses of protein markers are indicated on the left.
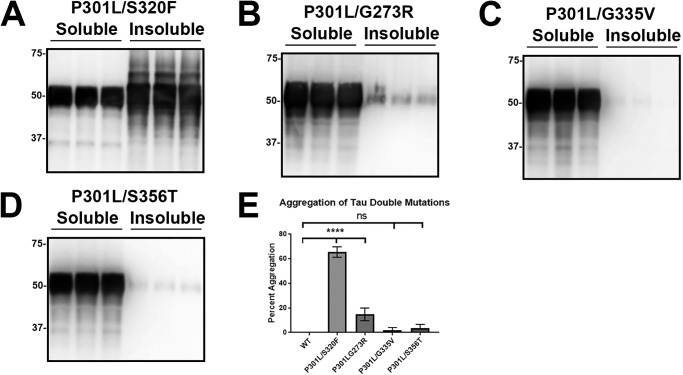
P301L/S320F tau uniquely self-aggregated compared with other tau double mutants
We previously discovered that the combined double tau mutant P301L/S320F robustly self-aggregates without exogenous seeding (43). From our additional screening, tau mutants with some baseline ability to self-aggregate were combined to P301L to create similar double mutants. Surprisingly, other double mutants did not exhibit the enhanced aggregation effect. P301L/G273R tau aggregated modestly compared with P301L/S320F tau (Fig. 10, A, B, and E). Other double mutants like P301L/G335V tau and P301L/S356T tau did not aggregate (Fig. 10, C, D, and E). In fact, the addition of P301L to these mutants did not change their ability to aggregate. This suggests that P301L/S320F tau may form a unique conformation and represent a different tau strain.
Figure 10.
P301L/S320F tau was uniquely capable of robust self-aggregation compared with other double tau mutants. HEK293T cells were transfected with different tau double mutations and were fractionated into Triton-soluble and Triton-insoluble fractions as described under “Experimental procedures.” A, P301L/S320F tau double mutant presented extensive intrinsic aggregation. B, P301L/G273R tau modestly aggregated but did not show an enhanced effect from the addition of P301L. P301L/G335V tau (C) and P301L/S356T tau (D) did not significantly aggregate. Immunoblots were probed with total tau antibody 3026. The relative molecular masses of protein markers are indicated on the left. E, one-way ANOVA with Dunnett's test was performed with n = 9 for WT tau and n = 3 for each tau mutant. ****, p < 0.0001; and ns, statistical significance.
Discussion
Aggregation of tau to form pathological inclusions is a hallmark of tauopathies and is universally observed in postmortem brains of patients with MAPT mutations, although variable brain regional distribution can be observed even between patients with the same MAPT mutation (57). In recent years, prion-like conformational templating of aggregated tau has gained significant attention as a mechanism in the progressive spread of tauopathies in many experimental models (31, 58–61). Recently, we investigated the aggregation propensity of 19 FTDP-17 tau mutants in a cell-based tau-seeding assay and demonstrated that except for three mutants at Pro-301 (i.e. P301L, P301S and P301T), WT and the other assayed tau mutants displayed relatively limited to no aggregation (43). Herein, we extend these studies to an additional 12 tau mutants and demonstrated that none of them were prone to seeded aggregation, although four mutants (G273R, G335V, S356T or V363I) were identified that modestly and intrinsically aggregated. Similarly, we had previously found that the S320F mutant had a modest tendency to aggregate but was not enhanced with exogenous K18 seeding (43). Combining the P301L and S320F mutants resulted in tau that robustly aggregated even without seeding. However, combining the P301L mutant with either G273R, G335V, or S356T did not have a synergistic effect of tau aggregation (Fig. 10). These findings further support the notion that tau aggregation is tightly regulated by global folding, such as the proposed paperclip-like structure that is not intrinsically permissive to aggregation (62), as well as key local molecule structures supported by recent cryo-EM studies (63, 64). The synergistic aggregation of the combined P301L/S320F tau double mutant is likely due to the unique relative distance and locations of these two mutations initiating and potentiating an amyloid-permissive stretch (43). In this structure, S320F enhances the hydrophobicity of an amyloid pocket, whereas the Pro-301 mutations disrupt a fold that would otherwise suppress amyloid formation (43, 63). The increased propensity of S320F tau to promote aggregation with or without seeding is consistent with the notion that it has greater property to aggregate with a lower nucleation threshold and a moderate ability to elongate as amyloid structures. P301L tau displays a greater potential for amyloid elongation but is limited by the lack of significant nucleation to initiate aggregation. Thus, the combination of both S320F and P301L yields an aggressive tau strain that can self-aggregate and is highly permissive to amyloid aggregation. Similar to this double mutant, other structural alterations such as post-translational modifications can result in unique tau strains permissive to aggregation as suggested by some experimental models (38, 65, 66).
One limitation of our seeding studies is that we used in vitro polymerized K18 tau as preformed fibrils to induce intracellular tau aggregation. Some studies have shown that tau aggregates isolated from brain lysates have higher-seeding potencies than recombinant tau fibrils (67). In addition, our in vitro generated tau fibrils likely do not comprise all of the misfolded tau strains that can occur in vivo (68, 69). Therefore, seeding of WT and other tau mutant could occur with more virulent seeds. Nevertheless, our data further highlight the uniqueness of Pro-301 tau mutants.
tau is an “intrinsically disordered” protein that can assume a variety of different conformations (6, 70, 71). When tau associates with MTs, it adopts a more stable conformation but transiently binds to MT (8, 9, 72, 73). The tau–MT interaction is very flexible and allows tau to readily move between the MT-bound and -unbound states (73, 74) and can be regulated by tau phosphorylation (29, 30, 75–78). This interaction allows tau to maintain MT homeostasis by stabilizing MTs, regulating their dynamic instability, and promoting tubulin assembly (6, 7, 10, 79). Different tau mutants also impair physiological dynamic instability and tubulin assembly (17, 49, 80–82), both of which are likely regulated by the MT-binding affinity of tau. Compared with the small subset of tau mutants that presented enhanced prion-like seeded aggregation, most tau mutants investigated so far display reduced activity of tubulin polymerization activity or MT binding (Table 1) (17). However, most studies investigating the impact of tau mutants on MT binding used in vitro MT binding with recombinant tau expressed from bacteria that lack the folding and post-translational modifications associated with mammalian expression. Herein, we conducted a survey of FTDP-17 tau mutants present in various regions of tau for effects on MT binding using a cell-based assay. When compared with previous in vitro studies of MT binding or tubulin polymerization assays, our cell-based findings for ΔK280 and P301L, P301S, G335S, G335V, S352L, V363A, G366R, K369I, and R406W consistently demonstrate reduced MT interactions (Table 1). Furthermore, we showed that L284R, P301T, K317M, and S356T also reduce MT binding, supporting the overarching notion that impaired MT binding is the most common altered property of pathogenic tau mutants due to missense mutations. Surprisingly, and at odds with in vitro MT assembly assays, both the R5H and R5L mutants increased MT interaction in the cell-based assay. The reasons for these differences are not clear, but mammalian expression of these tau mutations must confer long-range conformational differences allowing potentiated MT interaction.
A few missense mutants such as G273R and V287I did not appreciably affect tau MT binding. It is likely that these could primarily influence exon 10 splicing due to their close proximity to other splicing mutations that impact key RNA sequence elements that regulate splicing (83, 84). Many intronic, silent, and even missense MAPT mutations affect the splicing of exon 10, altering the normal ratio of 3R to 4R tau isoforms (6, 57, 83, 84). How this altered ratio of WT tau isoforms results in neurodegeneration is still unclear, but altered isoform-specific cellular localization and MT-binding properties could be involved. Generally, 3R tau isoforms bind MTs with lower affinity and promote MT polymerization less than 4R tau isoforms (14, 85, 86). As a result, 3R tau isoforms are less potent regulators of MT dynamics (80, 87). Furthermore, 3R and 4R tau appear to have different binding sites and interactions with MTs (81, 88). It is possible for some missense tau mutants to affect both MT binding and tau exon 10 splicing, resulting in a change in the 3R/4R ratio that causes dysregulation of MT dynamics (81).
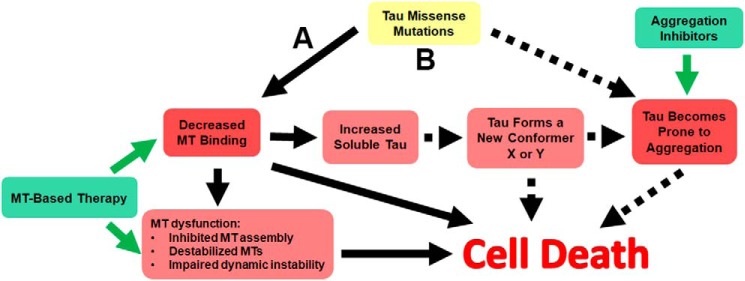
Based on our results and recent studies on tau prion-like seeding and propagation (17, 31, 43, 58–61), we propose an integrated paradigm of tau pathogenesis to highlight different but not mutually exclusive mechanisms that can lead to tauopathies (Fig. 11). The majority of tau missense mutants can affect MT binding and cause MT dysfunction by impairing MT stabilization, tubulin assembly, and regulation of dynamic instability (Fig. 11A). These changes in MT associations may also result in altered cellular localization. Although tau is mainly an axonal protein (11), tau pathology in AD occurs predominantly in somatodendritic compartments such as neurofibrillary tangles and neuropil threads with a paucity of white matter pathology (6, 32, 91), likely a result of improper cellular trafficking of pathogenic tau. For many forms of mutant tau, decreased MT binding leads to an increased pool of soluble tau, although tau phosphorylation at specific residues can also contribute to reduced MT binding, possibly in sporadic cases of AD and other tauopathies (29, 30, 75–78). Furthermore, in AD Aβ oligomers or aggregates may be the primary driver of tau toxicity by promoting hyperphosphorylation of different tau isoforms (92–94). For WT tau and tau mutants that are not intrinsically prone to aggregation, unbound tau may slowly convert into new conformers that aggregate over time. The combined events of impaired MT binding, MT dysfunction, soluble tau conformers, and eventual tau aggregates can all contribute to toxicity and cell death. Consistent with this model, several MT-based drugs have been shown to have some benefits in animal models of tauopathies (95–97).
Figure 11.
Proposed mechanisms of tau pathogenesis. A, model for the majority of tau variants due to missense mutations that predominantly impair MT binding, resulting in increased unbound tau as depicted by arrows with solid lines. These altered tau properties may result in aberrant tau cellular distribution, reduced MT assembly/stability, and MT dynamics. An increased pool of unbound tau can convert into new conformers/oligomers that can be toxic and/or permissive to aggregated pathological inclusions. For these mutants, therapies that directly target MT dysfunctions and restore function are more likely to be beneficial. B, model associated with tau variants due to missense mutations that robustly potentiate tau aggregation, but also impair MT interactions. These mutants can lead to neuronal demise through two independent pathways depicted by arrows with solid lines and dashed lines. Therapies that target MT dysfunctions and tau aggregation are likely to have an impact on disease course for these types of mutants, but it is still unclear which would be the most beneficial.
For tau mutants such as those involving the Pro-301 residue that have robust impacts on aggregation, the best approach is likely to be a combination of MT-based and aggregation inhibitors therapies (Fig. 11B). However, due to the unique robust aggregation-driven nature of models based on Pro-301 tau mutants, it would be prudent to cautiously assess the universality of experimental studies and translational therapies based solely on these models for AD and other tauopathies. Nevertheless, it is possible that some of the many post-translational modifications linked to pathogenic tau (6, 91, 98, 99) might mimic the impact of these mutations on tau aggregation propensity and prion-like conformational templating.
Although familial tau mutations are responsible for only a minority of cases with tauopathies, the wide spectrum of tau mutants provide information on the diverse pathomechanisms that can provide invaluable insight to understanding sporadic cases of tauopathies. Since numerous diverse tau missense mutants lead to a similar outcome of impaired MT interaction, other tau modifications in patients with sporadic tauopathies might also result in a similar aberrant outcome that leads to neurodegeneration. Our findings presented here in addition to the observed MT changes in AD (18–21) underscore that a greater emphasis on understanding and targeting MT changes in tauopathies should be considered. MT function is associated with numerous cellular functions that can result in many modes of cell injury and toxicity. It is also important to gain further insights into the relative involvement of MT dysfunction versus tau aggregation in the pathobiology of tauopathies. Even if several pathogenic mechanisms are involved, MT-based therapy will likely be important in treating Alzheimer's disease and other tauopathies.
Experimental procedures
tau protein purification
The tau protein fragment K18, which consists of the MT-binding repeats from Gln-244 to Glu-372 relative to the 2N4R human tau with an added methionine at the N terminus, was expressed from the bacterial plasmid pRK172 in BL21 (DE3)/RIL Escherichia coli (Agilent Technologies, Santa Clara, CA). K18 tau protein and K18 with tau mutations were purified as described previously (43, 100). Protein concentration was measured by the bicinchoninic acid assay with albumin as a standard.
Preparation of tau amyloid seeds
K18 tau protein or K18 with mutations were diluted in sterile PBS at 1 mg/ml with 50 μm heparin and shaking at 1050 rpm at 37 °C for at least 2 days. As described previously, amyloid formation was confirmed by K114 or thioflavin T assays (101). K18 fibrils were centrifuged and resuspended in sterile PBS to remove heparin. The purified K18 fibrils were placed in bath sonication for 60 min to produce short fibrils for seeding experiments, as described previously (43, 102).
Plasmids for mammalian expression and site-directed mutagenesis
The 0N4R human tau cDNA isoform was cloned into mammalian expression vector pcDNA3.1(+). The different missense MAPT mutations were created with QuikChange site-directed mutagenesis (Agilent Technologies, Santa Clara, CA) using mutation-specific oligonucleotides. All mutations and the entire cDNA tau sequence were confirmed by Sanger sequencing performed as a service by Genewiz (South Plainfield, NJ).
Cell culture and transfection
HEK293T cells were maintained in Dulbecco's modified Eagle's media with 10% fetal bovine serum and antibiotics (100 units/ml penicillin, 100 μg/ml streptomycin) at 37 °C and 5% CO2. HEK293T cells were transfected by calcium phosphate precipitation. Cells were plated at 20–40% confluence in 12-well plates. For each well, 1.5 μg of DNA was added to 18.75 μl of 0.25 m CaCl2. This DNA/CaCl2 mix was further added to an equal amount of 2× BES buffer (50 mm BES, 280 mm NaCl, 1.5 mm Na2HPO4, pH 6.96) and left at room temperature for 15–20 min. The final solution was added dropwise to each well. For cell-seeding studies, 1 μm K18 was added 1 h after transfection as described previously (102). At 16 h after transfection, cells were washed with PBS, and fresh Dulbecco's modified Eagle's medium in 3% FBS was added. Cells were harvested 48 h after media change or at a final time of 64 h.
Biochemical cellular aggregation assay
Cells were lysed in 200 μl of Triton Lysis Buffer (25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 20 mm NaF) with a mix of protease inhibitors. Cell lysates were centrifuged at 100,000 × g and 4 °C for 30 min (36, 43, 103). The soluble fractions were collected. The insoluble fraction was washed with additional Triton Lysis Buffer and centrifuged again at 100,000 × g and 4 °C for 30 min. After the wash step, the pellet was resuspended in Triton Lysis Buffer. Sample buffer (10 mm Tris, pH 6.8, 1 mm EDTA, 40 mm DTT, 0.005% bromphenol blue, 0.0025% pyronin yellow, 1% SDS, 10% sucrose) was added to both soluble and insoluble fractions and were boiled for 10 min. The insoluble fraction was probe-sonicated and boiled again for 10 min to ensure homogeneity.
MT-binding assay
Cells were lysed in 200 μl of PEM buffer (80 mm PIPES, pH 6.8, 1 mm EGTA, 1 mm MgCl2) supplemented with 0.1% Triton X-100, 2 mm GTP, 20 μm paclitaxel, and a mix of protease inhibitors as described previously (44–46). Cell lysates were incubated in a 37 °C water bath for 30 min and then centrifuged at 100,000 × g for 30 min to pellet MTs. Supernatant was transferred to a new tube, and the pellet (MT fraction with bound proteins) was resuspended in PEM buffer. The pellet fraction was homogenized, and SDS gel loading buffer was added to both fractions. Equivalent amounts of supernatant and pellet were loaded on SDS-polyacrylamide gels for Western blot analysis. Percent MT bound tau was calculated as pellet/(supernatant + pellet) × 100.
Antibodies
Anti-β-tubulin (clone TUB 2.1) is a mouse mAb (Sigma). tau 3026 is a rabbit polyclonal antibody against recombinant full-length 0N3R human tau (90). AT8 is an antibody specific for tau phosphorylated at Ser-202/Thr-205 (Invitrogen), and PHF-1 antibody is specific for tau phosphorylated at Ser-396/Ser-404 (generously provided by Peter Davies, Albert Einstein University, New York). T14 is an antibody against the N-terminal part of tau (83–120 amino acids), and T46 is an antibody against the C-terminal part of tau (404–441 amino acids) (104).
Western blot analysis
Equal proportions of each sample were loaded on 10% polyacrylamide gels and resolved by SDS-PAGE. After transfer, the membranes were blocked in 5% milk dissolved in TBS for 1 h. Membranes probed with phosphorylation-specific antibodies were blocked in 5% BSA dissolved in TBS for 1 h. The membranes were incubated in primary antibody overnight at 4 °C at dilutions of 1:1000 for 3026 tau antibody, β-tubulin antibody, and tau phosphorylation-specific antibodies AT8 and PHF-1. The membranes were washed in TBS and incubated in goat anti-rabbit or anti-mouse secondary antibodies conjugated to horseradish peroxidase (Jackson ImmunoResearch) at 1:4000 dilution for 1 h. After TBS washes, the membranes were exposed and imaged after adding Western Lightning Plus ECL reagents (PerkinElmer Life Sciences).
Statistical analysis
The specific signals in each lane were quantified based on densitometric analysis with ImageJ. Statistical comparison tests were performed on GraphPad Prism for one-way or two-way analysis of variance (ANOVA), with post hoc analysis using Dunnett's test to compare each group to the control.
Immunofluorescence of fixed cells
Double immunofluorescence was performed on cells similar to previous studies except for the fixation step (102, 105). Cell fixation was performed at −20 °C in 95% acetic acid and 5% methanol for 30 min. After PBS washes, the cells were blocked in PBS with 2% FBS and 0.1% Triton X-100 for 30 min. Primary antibody such as 3026 at 1:4000 and β-tubulin 1:1000 were incubated for 1 h. After the PBS wash, the slides were incubated in the dark for 1 h in Alexa-fluor 488- or 594-conjugated secondary antibodies (Invitrogen). Afterward, the slides were incubated in 4′,6-diamidino-2-phenylindole (Invitrogen) for 5 min to stain nuclei and mounted with Fluoromount-G (Southern Biotech, Birmingham, AL). Immunofluorescent images were captured with an Olympus BX51 fluorescence microscope Olympus, Center Valley, PA).
Author contributions
Y. X. and B. I. G. conceptualization; Y. X. formal analysis; Y. X., Z. A. S., J. D. K., K. H. S., and C. J. R. investigation; Y. X. and B. I. G. writing-original draft; Z. A. S. and K. H. S. writing-review and editing; B. I. G. supervision; B. I. G. funding acquisition.
Supplementary Material
This work was supported by National Institutes of Health NINDS Grant R01NS089022, Florida Department of Health Grants 7AZ25 and 9AZ17, and National Institutes of Health NIA Grant F30AG063446 (to Z. A. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4.
- MT
- microtubule
- AD
- Alzheimer's disease
- FTDP-17
- frontotemporal dementia with parkinsonism linked to chromosome 17
- HEK293T
- human embryonic kidney 293T
- MAPT
- microtubule associated protein tau
- MTBD
- microtubule binding domain
- 3R
- three repeat
- 4R
- four repeat
- FBS
- fetal bovine serum
- ANOVA
- analysis of variance
- BES
- N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid.
References
- 1. Kovacs G. G. (2018) Tauopathies. Handb. Clin. Neurol. 145, 355–368 10.1016/B978-0-12-802395-2.00025-0 [DOI] [PubMed] [Google Scholar]
- 2. Jack C. R. Jr., Bennett D. A., Blennow K., Carrillo M. C., Dunn B., Haeberlein S. B., Holtzman D. M., Jagust W., Jessen F., Karlawish J., Liu E., Molinuevo J. L., Montine T., Phelps C., Rankin K. P., et al. (2018) NIA-AA research framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement. 14, 535–562 10.1016/j.jalz.2018.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nelson P. T., Alafuzoff I., Bigio E. H., Bouras C., Braak H., Cairns N. J., Castellani R. J., Crain B. J., Davies P., Del Tredici K., Duyckaerts C., Frosch M. P., Haroutunian V., Hof P. R., Hulette C. M., et al. (2012) Correlation of Alzheimer disease neuropathologic changes with cognitive status: a review of the literature. J. Neuropathol. Exp. Neurol. 71, 362–381 10.1097/NEN.0b013e31825018f7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nelson P. T., Jicha G. A., Schmitt F. A., Liu H., Davis D. G., Mendiondo M. S., Abner E. L., and Markesbery W. R. (2007) Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort: neuritic plaques and neurofibrillary tangles “do count” when staging disease severity. J. Neuropathol. Exp. Neurol. 66, 1136–1146 10.1097/nen.0b013e31815c5efb [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hutton M., Lendon C. L., Rizzu P., Baker M., Froelich S., Houlden H., Pickering-Brown S., Chakraverty S., Isaacs A., Grover A., Hackett J., Adamson J., Lincoln S., Dickson D., Davies P., et al. (1998) Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393, 702–705 10.1038/31508 [DOI] [PubMed] [Google Scholar]
- 6. Wang Y., and Mandelkow E. (2016) Tau in physiology and pathology. Nat. Rev. Neurosci. 17, 5–21 10.1038/nrg.2015.11 [DOI] [PubMed] [Google Scholar]
- 7. Weingarten M. D., Lockwood A. H., Hwo S.-Y., and Kirschner M. W. (1975) A protein factor essential for microtubule assembly (tau factor/tubulin/electron microscopy/phosphocellulose). Proc. Natl. Acad. Sci. U.S.A. 72, 1858–1862 10.1073/pnas.72.5.1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Santarella R. A., Skiniotis G., Goldie K. N., Tittmann P., Gross H., Mandelkow E. M., Mandelkow E., and Hoenger A. (2004) Surface-decoration of microtubules by human tau. J. Mol. Biol. 339, 539–553 10.1016/j.jmb.2004.04.008 [DOI] [PubMed] [Google Scholar]
- 9. Al-Bassam J., Ozer R. S., Safer D., Halpain S., and Milligan R. A. (2002) MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J. Cell Biol. 157, 1187–1196 10.1083/jcb.200201048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kadavath H., Hofele R. V., Biernat J., Kumar S., Tepper K., Urlaub H., Mandelkow E., and Zweckstetter M. (2015) Tau stabilizes microtubules by binding at the interface between tubulin heterodimers. Proc. Natl. Acad. Sci. U.S.A. 112, 7501–7506 10.1073/pnas.1504081112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Black M. M., Slaughter T., Moshiach S., Obrocka M., and Fischer I. (1996) Tau is enriched on dynamic microtubules in the distal region of growing axons. J. Neurosci. 16, 3601–3619 10.1523/JNEUROSCI.16-11-03601.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goedert M., Spillantini M. G., Potier M. C., Ulrich J., and Crowther R. A. (1989) Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: differential expression of tau protein mRNAs in human brain. EMBO J. 8, 393–399 10.1002/j.1460-2075.1989.tb03390.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goedert M., Spillantini M. G., Jakes R., Rutherford D., and Crowther R. A. (1989) Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer's disease. Neuron 3, 519–526 10.1016/0896-6273(89)90210-9 [DOI] [PubMed] [Google Scholar]
- 14. Lee G., Neve R. L., and Kosik K. S. (1989) The microtubule binding domain of tau protein. Neuron 2, 1615–1624 10.1016/0896-6273(89)90050-0 [DOI] [PubMed] [Google Scholar]
- 15. Himmler A., Drechsel D., Kirschner M. W., and Martin D. W. (1989) Tau consists of a set of proteins with repeated C-terminal microtubule-binding domains and variable N-terminal domains. Mol. Cell. Biol. 9, 1381–1388 10.1128/MCB.9.4.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iqbal K., Liu F., and Gong C.-X. (2016) Tau and neurodegenerative disease: the story so far. Nat. Rev. Neurol. 12, 15–27 10.1038/nrneurol.2015.225 [DOI] [PubMed] [Google Scholar]
- 17. Strang K. H., Golde T. E., and Giasson B. I. (2019) MAPT mutations, tauopathy, and mechanisms of neurodegeneration. Lab. Invest. 99, 912–928 10.1038/s41374-019-0197-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Köpke E., Tung Y. C., Shaikh S., Alonso A. C., Iqbal K., and Grundke-Iqbal I. (1993) Microtubule-associated protein tau. Abnormal phosphorylation of a non-paired helical filament pool in Alzheimer disease. J. Biol. Chem. 268, 24374–24384 [PubMed] [Google Scholar]
- 19. Hempen B., and Brion J. P. (1996) Reduction of acetylated α-tubulin immunoreactivity in neurofibrillary tangle-bearing neurons in Alzheimer's disease. J. Neuropathol. Exp. Neurol. 55, 964–972 10.1097/00005072-199609000-00003 [DOI] [PubMed] [Google Scholar]
- 20. Zhang F., Su B., Wang C., Siedlak S. L., Mondragon-Rodriguez S., Lee H.-G., Wang X., Perry G., and Zhu X. (2015) Posttranslational modifications of α-tubulin in Alzheimer disease. Transl. Neurodegener. 4, 9 10.1186/s40035-015-0030-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cash A. D., Aliev G., Siedlak S. L., Nunomura A., Fujioka H., Zhu X., Raina A. K., Vinters H. V., Tabaton M., Johnson A. B., Paula-Barbosa M., Avíla J., Jones P. K., Castellani R. J., Smith M. A., and Perry G. (2003) Microtubule reduction in Alzheimer's disease and aging is independent of tau filament formation. Am. J. Pathol. 162, 1623–1627 10.1016/S0002-9440(10)64296-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Biundo F., Del Prete D., Zhang H., Arancio O., and D'Adamio L. (2018) A role for tau in learning, memory and synaptic plasticity. Sci. Rep. 8, 3184 10.1038/s41598-018-21596-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Z.-X., Tan L., and Yu J.-T. (2015) Axonal transport defects in Alzheimer's disease. Mol. Neurobiol. 51, 1309–1321 10.1007/s12035-014-8810-x [DOI] [PubMed] [Google Scholar]
- 24. Dejanovic B., Huntley M. A., De Mazière A., Meilandt W. J., Wu T., Srinivasan K., Jiang Z., Gandham V., Friedman B. A., Ngu H., Foreman O., Carano R. A. D., Chih B., Klumperman J., Bakalarski C., et al. (2018) Changes in the synaptic proteome in tauopathy and rescue of Tau-induced synapse loss by C1q antibodies. Neuron 100, 1322–1336.e7 10.1016/j.neuron.2018.10.014 [DOI] [PubMed] [Google Scholar]
- 25. Forner S., Baglietto-Vargas D., Martini A. C., Trujillo-Estrada L., and LaFerla F. M. (2017) Synaptic impairment in Alzheimer's disease: a dysregulated symphony. Trends Neurosci. 40, 347–357 10.1016/j.tins.2017.04.002 [DOI] [PubMed] [Google Scholar]
- 26. Qiang L., Yu W., Andreadis A., Luo M., and Baas P. W. (2006) Tau protects microtubules in the axon from severing by katanin. J. Neurosci. 26, 3120–3129 10.1523/JNEUROSCI.5392-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rossi G., and Tagliavini F. (2015) Frontotemporal lobar degeneration: old knowledge and new insight into the pathogenetic mechanisms of tau mutations. Front. Aging Neurosci. 7, 192 10.3389/fnagi.2015.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Biernat J., Gustke N., Drewes G., Mandelkow E. M., and Mandelkow E. (1993) Phosphorylation of Ser262 strongly reduces binding of tau to microtubules: Distinction between PHF-like immunoreactivity and microtubule binding. Neuron 11, 153–163 10.1016/0896-6273(93)90279-Z [DOI] [PubMed] [Google Scholar]
- 29. Sengupta A., Kabat J., Novak M., Wu Q., Grundke-Iqbal I., and Iqbal K. (1998) Phosphorylation of tau at both Thr 231 and Ser 262 is required for maximal inhibition of its binding to microtubules. Arch. Biochem. Biophys. 357, 299–309 10.1006/abbi.1998.0813 [DOI] [PubMed] [Google Scholar]
- 30. Fischer D., Mukrasch M. D., Biernat J., Bibow S., Blackledge M., Griesinger C., Mandelkow E., and Zweckstetter M. (2009) Conformational changes specific for pseudophosphorylation at serine 262 selectively impair binding of tau to microtubules. Biochemistry 48, 10047–10055 10.1021/bi901090m [DOI] [PubMed] [Google Scholar]
- 31. Ayers J. I., Giasson B. I., and Borchelt D. R. (2018) Prion-like spreading in tauopathies. Biol. Psychiatry 83, 337–346 10.1016/j.biopsych.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Braak H., and Braak E. (1991) Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 10.1007/BF00308809 [DOI] [PubMed] [Google Scholar]
- 33. Braak H., Alafuzoff I., Arzberger T., Kretzschmar H., and Del Tredici K. (2006) Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 112, 389–404 10.1007/s00401-006-0127-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Frost B., Jacks R. L., and Diamond M. I. (2009) Propagation of Tau misfolding from the outside to the inside of a cell. J. Biol. Chem. 284, 12845–12852 10.1074/jbc.M808759200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kfoury N., Holmes B. B., Jiang H., Holtzman D. M., and Diamond M. I. (2012) Trans-cellular propagation of Tau aggregation by fibrillar species. J. Biol. Chem. 287, 19440–19451 10.1074/jbc.M112.346072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guo J. L., and Lee V. M. (2011) Seeding of normal Tau by pathological Tau conformers drives pathogenesis of Alzheimer-like tangles. J. Biol. Chem. 286, 15317–15331 10.1074/jbc.M110.209296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Iba M., Guo J. L., McBride J. D., Zhang B., Trojanowski J. Q., and Lee V. M. (2013) Synthetic tau fibrils mediate transmission of neurofibrillary tangles in a transgenic mouse model of Alzheimer's-like tauopathy. J. Neurosci. 33, 1024–1037 10.1523/JNEUROSCI.2642-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sanders D. W., Kaufman S. K., DeVos S. L., Sharma A. M., Mirbaha H., Li A., Barker S. J., Foley A. C., Thorpe J. R., Serpell L. C., Miller T. M., Grinberg L. T., Seeley W. W., and Diamond M. I. (2014) Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82, 1271–1288 10.1016/j.neuron.2014.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaufman S. K., Thomas T. L., Del Tredici K., Braak H., and Diamond M. I. (2017) Characterization of tau prion seeding activity and strains from formaldehyde-fixed tissue. Acta Neuropathol. Commun. 5, 41 10.1186/s40478-017-0442-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Clavaguera F., Bolmont T., Crowther R. A., Abramowski D., Frank S., Probst A., Fraser G., Stalder A. K., Beibel M., Staufenbiel M., Jucker M., Goedert M., and Tolnay M. (2009) Transmission and spreading of tauopathy in transgenic mouse brain. Nat. Cell Biol. 11, 909–913 10.1038/ncb1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Clavaguera F., Akatsu H., Fraser G., Crowther R. A., Frank S., Hench J., Probst A., Winkler D. T., Reichwald J., Staufenbiel M., Ghetti B., Goedert M., and Tolnay M. (2013) Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc. Natl. Acad. Sci. U.S.A. 110, 9535–9540 10.1073/pnas.1301175110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Irwin D. J., Abrams J. Y., Schonberger L. B., Leschek E. W., Mills J. L., Lee V. M., and Trojanowski J. Q. (2013) Evaluation of potential infectivity of Alzheimer and Parkinson disease proteins in recipients of cadaver-derived human growth hormone. JAMA Neurol. 70, 462–468 10.1001/jamaneurol.2013.1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Strang K. H., Croft C. L., Sorrentino Z. A., Chakrabarty P., Golde T. E., and Giasson B. I. (2018) Distinct differences in prion-like seeding and aggregation between Tau protein variants provide mechanistic insights into tauopathies. J. Biol. Chem. 293, 2408–2421 10.1074/jbc.M117.815357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huda M. N., Erdene-Ochir E., and Pan C.-H. (2017) Assay for phosphorylation and microtubule binding along with localization of Tau protein in colorectal cancer cells. J. Vis. Exp. 2017, 10.3791/55932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vogelsberg-Ragaglia V., Bruce J., Richter-Landsberg C., Zhang B., Hong M., Trojanowski J. Q., and Lee V. M. (2000) Distinct FTDP-17 missense mutations in Tau produce Tau aggregates and other pathological phenotypes in transfected CHO cells. Mol. Biol. Cell. 11, 4093–4104 10.1091/mbc.11.12.4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alberico E. O., Duan A. R., and Goodson H. V. (2017) Measuring Tau–microtubule affinity through cosedimentation assays. Methods Cell Biol. 141, 115–134 10.1016/bs.mcb.2017.06.015 [DOI] [PubMed] [Google Scholar]
- 47. Vallee R. B. (1982) A taxol-dependent procedure for the isolation of microtubules and microtubule-associated proteins (MAPs). J. Cell Biol. 92, 435–442 10.1083/jcb.92.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hamel E., del Campo A. A., Lowe M. C., and Lin C. M. (1981) Interactions of taxol, microtubule-associated proteins, and guanine nucleotides in tubulin polymerization. J. Biol. Chem. 256, 11887–11894 [PubMed] [Google Scholar]
- 49. DeTure M., Ko L. W., Yen S., Nacharaju P., Easson C., Lewis J., van Slegtenhorst M., Hutton M., and Yen S. H. (2000) Missense tau mutations identified in FTDP-17 have a small effect on tau-microtubule interactions. Brain Res. 853, 5–14 10.1016/S0006-8993(99)02124-1 [DOI] [PubMed] [Google Scholar]
- 50. Hong M., Zhukareva V., Vogelsberg-Ragaglia V., Wszolek Z., Reed L., Miller B. I., Geschwind D. H., Bird T. D., McKeel D., Goate A., Morris J. C., Wilhelmsen K. C., Schellenberg G. D., Trojanowski J. Q., and Lee V. M. (1998) Mutation-specific functional impairments in distinct tau isoforms of hereditary FTDP-17. Science 282, 1914–1917 10.1126/science.282.5395.1914 [DOI] [PubMed] [Google Scholar]
- 51. D'Souza I., Poorkaj P., Hong M., Nochlin D., Lee V. M., Bird T. D., and Schellenberg G. D. (1999) Missense and silent tau gene mutations cause frontotemporal dementia with parkinsonism-chromosome 17 type, by affecting multiple alternative RNA splicing regulatory elements. Proc. Natl. Acad. Sci. U.S.A. 96, 5598–5603 10.1073/pnas.96.10.5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Barghorn S., Zheng-Fischhöfer Q., Ackmann M., Biernat J., von Bergen M., Mandelkow E. M., and Mandelkow E. (2000) Structure, microtubule interactions, and paired helical filament aggregation by tau mutants of frontotemporal dementias. Biochemistry 39, 11714–11721 10.1021/bi000850r [DOI] [PubMed] [Google Scholar]
- 53. Nagiec E. W., Sampson K. E., and Abraham I. (2001) Mutated tau binds less avidly to microtubules than wildtype tau in living cells. J. Neurosci. Res. 63, 268–275 [DOI] [PubMed] [Google Scholar]
- 54. Dayanandan R., Van Slegtenhorst M., Mack T. G., Ko L., Yen S. H., Leroy K., Brion J. P., Anderton B. H., Hutton M., and Lovestone S. (1999) Mutations in tau reduce its microtubule binding properties in intact cells and affect its phosphorylation. FEBS Lett. 446, 228–232 10.1016/S0014-5793(99)00222-7 [DOI] [PubMed] [Google Scholar]
- 55. Yoshiyama Y., Higuchi M., Zhang B., Huang S. M., Iwata N., Saido T. C., Maeda J., Suhara T., Trojanowski J. Q., and Lee V. M. (2007) Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 53, 337–351 10.1016/j.neuron.2007.01.010 [DOI] [PubMed] [Google Scholar]
- 56. Fischer D., Mukrasch M. D., von Bergen M., Klos-Witkowska A., Biernat J., Griesinger C., Mandelkow E., and Zweckstetter M. (2007) Structural and microtubule binding properties of tau mutants of frontotemporal dementias. Biochemistry 46, 2574–2582 10.1021/bi061318s [DOI] [PubMed] [Google Scholar]
- 57. Ghetti B., Oblak A. L., Boeve B. F., Johnson K. A., Dickerson B. C., and Goedert M. (2015) Invited review: frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: a chameleon for neuropathology and neuroimaging. Neuropathol. Appl. Neurobiol. 41, 24–46 10.1111/nan.12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Goedert M., Eisenberg D. S., and Crowther R. A. (2017) Propagation of Tau aggregates and neurodegeneration. Annu. Rev. Neurosci. 40, 189–210 10.1146/annurev-neuro-072116-031153 [DOI] [PubMed] [Google Scholar]
- 59. Lewis J., and Dickson D. W. (2016) Propagation of tau pathology: hypotheses, discoveries, and yet unresolved questions from experimental and human brain studies. Acta Neuropathol. 131, 27–48 10.1007/s00401-015-1507-z [DOI] [PubMed] [Google Scholar]
- 60. Goedert M., Masuda-Suzukake M., and Falcon B. (2017) Like prions: the propagation of aggregated tau and α-synuclein in neurodegeneration. Brain 140, 266–278 10.1093/brain/aww230 [DOI] [PubMed] [Google Scholar]
- 61. Jucker M., and Walker L. C. (2013) Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 501, 45–51 10.1038/nature12481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jeganathan S., von Bergen M., Brutlach H., Steinhoff H. J., and Mandelkow E. (2006) Global hairpin folding of tau in solution. Biochemistry 45, 2283–2293 10.1021/bi0521543 [DOI] [PubMed] [Google Scholar]
- 63. Fitzpatrick A. W. P., Falcon B., He S., Murzin A. G., Murshudov G., Garringer H. J., Crowther R. A., Ghetti B., Goedert M., and Scheres S. H. (2017) Cryo-EM structures of tau filaments from Alzheimer's disease. Nature 547, 185–190 10.1038/nature23002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Falcon B., Zhang W., Schweighauser M., Murzin A. G., Vidal R., Garringer H. J., Ghetti B., Scheres S. H. W., and Goedert M. (2018) Tau filaments from multiple cases of sporadic and inherited Alzheimer's disease adopt a common fold. Acta Neuropathol. 136, 699–708 10.1007/s00401-018-1914-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kaufman S. K., Sanders D. W., Thomas T. L., Ruchinskas A. J., Vaquer-Alicea J., Sharma A. M., Miller T. M., and Diamond M. I. (2016) Tau prion strains dictate patterns of cell pathology, progression rate, and regional vulnerability in vivo. Neuron 92, 796–812 10.1016/j.neuron.2016.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Guo J. L., Narasimhan S., Changolkar L., He Z., Stieber A., Zhang B., Gathagan R. J., Iba M., McBride J. D., Trojanowski J. Q., and Lee V. M. (2016) Unique pathological tau conformers from Alzheimer's brains transmit tau pathology in nontransgenic mice. J. Exp. Med. 213, 2635–2654 10.1084/jem.20160833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Falcon B., Cavallini A., Angers R., Glover S., Murray T. K., Barnham L., Jackson S., O'Neill M. J., Isaacs A. M., Hutton M. L., Szekeres P. G., Goedert M., and Bose S. (2015) Conformation determines the seeding potencies of native and recombinant Tau aggregates. J. Biol. Chem. 290, 1049–1065 10.1074/jbc.M114.589309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fichou Y., Vigers M., Goring A. K., Eschmann N. A., and Han S. (2018) Heparin-induced tau filaments are structurally heterogeneous and differ from Alzheimer's disease filaments. Chem. Commun. 54, 4573–4576 10.1039/C8CC01355A [DOI] [PubMed] [Google Scholar]
- 69. Zhang W., Falcon B., Murzin A. G., Fan J., Crowther R. A., Goedert M., and Scheres S. H. (2019) Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases. Elife 8, e43584 10.7554/eLife.43584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Manger L. H., Foote A. K., Wood S. L., Holden M. R., Heylman K. D., Margittai M., and Goldsmith R. H. (2017) Revealing conformational variants of solution-phase intrinsically disordered Tau protein at the single-molecule level. Angew. Chem. Int. Ed. Engl. 56, 15584–15588 10.1002/anie.201708242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Skrabana R., Sevcik J., and Novak M. (2006) Intrinsically disordered proteins in the neurodegenerative processes: formation of tau protein paired helical filaments and their analysis. Cell. Mol. Neurobiol. 26, 1085–1097 10.1007/s10571-006-9083-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Butner K. A., and Kirschner M. W. (1991) Tau protein binds to microtubules through a flexible array of distributed weak sites. J. Cell Biol. 115, 717–730 10.1083/jcb.115.3.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li X. H., Culver J. A., and Rhoades E. (2015) Tau binds to multiple tubulin dimers with helical structure. J. Am. Chem. Soc. 137, 9218–9221 10.1021/jacs.5b04561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Makrides V., Massie M. R., Feinstein S. C., and Lew J. (2004) Evidence for two distinct binding sites for Tau on microtubules. Proc. Natl. Acad. Sci. U.S.A. 101, 6746–6751 10.1073/pnas.0400992101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Alonso A. C., Zaidi T., Grundke-Iqbal I., and Iqbal K. (1994) Role of abnormally phosphorylated Tau in the breakdown of microtubules in Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 91, 5562–5566 10.1073/pnas.91.12.5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gustke N., Steiner B., Mandelkow E.-M., Biernat J., Meyer H. E., Goedert M., and Mandelkow E. (1992) The Alzheimer-like phosphorylation of tau protein reduces microtubule binding and involves Ser-Pro and Thr-Pro motifs. FEBS Lett. 307, 199–205 10.1016/0014-5793(92)80767-b [DOI] [PubMed] [Google Scholar]
- 77. Kiris E., Ventimiglia D., Sargin M. E., Gaylord M. R., Altinok A., Rose K., Manjunath B. S., Jordan M. A., Wilson L., and Feinstein S. C. (2011) Combinatorial Tau pseudophosphorylation: markedly different regulatory effects on microtubule assembly and dynamic instability than the sum of the individual parts. J. Biol. Chem. 286, 14257–14270 10.1074/jbc.M111.219311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Garver T. D., Lehman R. A., and Billingsley M. L. (1996) Microtubule assembly competence analysis of freshly-biopsied human tau, dephosphorylated Tau, and Alzheimer tau. J. Neurosci. Res. 44, 12–20 [DOI] [PubMed] [Google Scholar]
- 79. Breuzard G., Hubert P., Nouar R., De Bessa T., Devred F., Barbier P., Sturgis J. N., and Peyrot V. (2013) Molecular mechanisms of Tau binding to microtubules and its role in microtubule dynamics in live cells. J. Cell Sci. 126, 2810–2819 10.1242/jcs.120832 [DOI] [PubMed] [Google Scholar]
- 80. Bunker J. M., Kamath K., Wilson L., Jordan M. A., and Feinstein S. C. (2006) FTDP-17 mutations compromise the ability of Tau to regulate microtubule dynamics in cells. J. Biol. Chem. 281, 11856–11863 10.1074/jbc.M509420200 [DOI] [PubMed] [Google Scholar]
- 81. LeBoeuf A. C., Levy S. F., Gaylord M., Bhattacharya A., Singh A. K., Jordan M. A., Wilson L., and Feinstein S. C. (2008) FTDP-17 mutations in Tau alter the regulation of microtubule dynamics. J. Biol. Chem. 283, 36406–36415 10.1074/jbc.M803519200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Combs B., and Gamblin T. C. (2012) FTDP-17 Tau mutations induce distinct effects on aggregation and microtubule interactions. Biochemistry 51, 8597–8607 10.1021/bi3010818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Liu F., and Gong C. X. (2008) Tau exon 10 alternative splicing and tauopathies. Mol. Neurodegener. 3, 8 10.1186/1750-1326-3-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Qian W., and Liu F. (2014) Regulation of alternative splicing of tau exon 10. Neurosci. Bull. 30, 367–377 10.1007/s12264-013-1411-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Goode B. L., Chau M., Denis P. E., and Feinstein S. C. (2000) Structural and functional differences between 3-repeat and 4-repeat Tau isoforms: implications for normal tau function and the onset of neurodegenerative disease. J. Biol. Chem. 275, 38182–38189 10.1074/jbc.M007489200 [DOI] [PubMed] [Google Scholar]
- 86. Goedert M., and Jakes R. (1990) Expression of separate isoforms of human tau protein: correlation with the tau pattern in brain and effects on tubulin polymerization. EMBO J. 9, 4225–4230 10.1002/j.1460-2075.1990.tb07870.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Panda D., Samuel J. C., Massie M., Feinstein S. C., and Wilson L. (2003) Differential regulation of microtubule dynamics by three- and four-repeat tau: implications for the onset of neurodegenerative disease. Proc. Natl. Acad. Sci. U.S.A. 100, 9548–9553 10.1073/pnas.1633508100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Goode B. L., and Feinstein S. C. (1994) Identification of a novel microtubule binding and assembly domain in the developmentally regulated inter-repeat region of tau. J. Cell Biol. 124, 769–782 10.1083/jcb.124.5.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Neumann M., Schulz-Schaeffer W., Crowther R. A., Smith M. J., Spillantini M. G., Goedert M., and Kretzschmar H. A. (2001) Pick's disease associated with the novel Tau gene mutation K369I. Ann. Neurol. 50, 503–513 10.1002/ana.1223 [DOI] [PubMed] [Google Scholar]
- 90. Rossi G., Bastone A., Piccoli E., Mazzoleni G., Morbin M., Uggetti A., Giaccone G., Sperber S., Beeg M., Salmona M., and Tagliavini F. (2012) New mutations in MAPT gene causing frontotemporal lobar degeneration: biochemical and structural characterization. Neurobiol. Aging 33, 834 10.1016/j.neurobiolaging.2011.08.008 [DOI] [PubMed] [Google Scholar]
- 91. Guo T., Noble W., and Hanger D. P. (2017) Roles of tau protein in health and disease. Acta Neuropathol. 133, 665–704 10.1007/s00401-017-1707-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ma Q.-L., Yang F., Rosario E. R., Ubeda O. J., Beech W., Gant D. J., Chen P. P., Hudspeth B., Chen C., Zhao Y., Vinters H. V., Frautschy S. A., and Cole G. M. (2009) Amyloid oligomers induce phosphorylation of Tau and inactivation of insulin receptor substrate via c-Jun N-terminal kinase signaling: suppression by omega-3 fatty acids and curcumin. J. Neurosci. 29, 9078–9089 10.1523/JNEUROSCI.1071-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Busciglio J., Lorenzo A., Yeh J., and Yankner B. A. (1995) β-Amyloid fibrils induce tau phosphorylation and loss of microtubule binding. Neuron 14, 879–888 10.1016/0896-6273(95)90232-5 [DOI] [PubMed] [Google Scholar]
- 94. Zheng W.-H., Bastianetto S., Mennicken F., Ma W., and Kar S. (2002) Amyloid β peptide induces tau phosphorylation and loss of cholinergic neurons in rat primary septal cultures. Neuroscience 115, 201–211 10.1016/S0306-4522(02)00404-9 [DOI] [PubMed] [Google Scholar]
- 95. Makani V., Zhang B., Han H., Yao Y., Lassalas P., Lou K., Paterson I., Lee V. M., Trojanowski J. Q., Ballatore C., Smith A. B. 3rd, and Brunden K. R. (2016) Evaluation of the brain-penetrant microtubule-stabilizing agent, dictyostatin, in the PS19 tau transgenic mouse model of tauopathy. Acta Neuropathol. Commun. 4, 106 10.1186/s40478-016-0378-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Das V., and Miller J. H. (2012) Microtubule stabilization by peloruside A and paclitaxel rescues degenerating neurons from okadaic acid-induced tau phosphorylation. Eur. J. Neurosci. 35, 1705–1717 10.1111/j.1460-9568.2012.08084.x [DOI] [PubMed] [Google Scholar]
- 97. Zhang B., Carroll J., Trojanowski J. Q., Yao Y., Iba M., Potuzak J. S., Hogan A.-M. L., Xie S. X., Ballatore C., Smith A. B. 3rd., Lee V. M., and Brunden K. R. (2012) The microtubule-stabilizing agent, epothilone D, reduces axonal dysfunction, neurotoxicity, cognitive deficits, and Alzheimer-like pathology in an interventional study with aged Tau transgenic mice. J. Neurosci. 32, 3601–3611 10.1523/JNEUROSCI.4922-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kontaxi C., Piccardo P., and Gill A. C. (2017) Lysine-directed post-translational modifications of Tau protein in Alzheimer's disease and related tauopathies. Front. Mol. Biosci. 4, 56 10.3389/fmolb.2017.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Martin L., Latypova X., and Terro F. (2011) Post-translational modifications of tau protein: implications for Alzheimer's disease. Neurochem. Int. 58, 458–471 10.1016/j.neuint.2010.12.023 [DOI] [PubMed] [Google Scholar]
- 100. Giasson B. I., Forman M. S., Higuchi M., Golbe L. I., Graves C. L., Kotzbauer P. T., Trojanowski J. Q., and Lee V. M. (2003) Initiation and synergistic fibrillization of tau and α-synuclein. Science 300, 636–640 10.1126/science.1082324 [DOI] [PubMed] [Google Scholar]
- 101. Crystal A. S., Giasson B. I., Crowe A., Kung M. P., Zhuang Z. P., Trojanowski J. Q., and Lee V. M. (2003) A comparison of amyloid fibrillogenesis using the novel fluorescent compound K114. J. Neurochem. 86, 1359–1368 10.1046/j.1471-4159.2003.01949.x [DOI] [PubMed] [Google Scholar]
- 102. Waxman E. A., and Giasson B. I. (2011) Induction of intracellular Tau aggregation is promoted by α-synuclein seeds and provides novel insights into the hyperphosphorylation of Tau. J. Neurosci. 31, 7604–7618 10.1523/JNEUROSCI.0297-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sorrentino Z. A., Vijayaraghavan N., Gorion K.-M., Riffe C. J., Strang K. H., Caldwell J., and Giasson B. I. (2018) Physiological C-terminal truncation of α-synuclein potentiates the prion-like formation of pathological inclusions. J. Biol. Chem. 293, 18914–18932 10.1074/jbc.RA118.005603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kosik K. S., Orecchio L. D., Binder L., Trojanowski J. Q., Lee V. M., and Lee G. (1988) Epitopes that span the tau molecule are shared with paired helical filaments. Neuron. 1, 817–825 10.1016/0896-6273(88)90129-8 [DOI] [PubMed] [Google Scholar]
- 105. Waxman E. A., Covy J. P., Bukh I., Li X., Dawson T. M., and Giasson B. I. (2009) Leucine-rich repeat kinase 2 expression leads to aggresome formation that is not associated with α-synuclein inclusions. J. Neuropathol. Exp. Neurol. 68, 785–796 10.1097/NEN.0b013e3181aaf4fd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Hayashi S., Toyoshima Y., Hasegawa M., Umeda Y., Wakabayashi K., Tokiguchi S., Iwatsubo T., and Takahashi H. (2002) Late-onset frontotemporal dementia with a novel exon 1 (Arg5His) tau gene mutation. Ann. Neurol. 51, 525–530 10.1002/ana.10163 [DOI] [PubMed] [Google Scholar]
- 107. Poorkaj P., Muma N. A., Zhukareva V., Cochran E. J., Shannon K. M., Hurtig H., Koller W. C., Bird T. D., Trojanowski J. Q., Lee V. M., and Schellenberg G. D. (2002) An R5L τ mutation in a subject with a progressive supranuclear palsy phenotype. Ann. Neurol. 52, 511–516 10.1002/ana.10340 [DOI] [PubMed] [Google Scholar]
- 108. Mutreja Y., Combs B., and Gamblin T. C. (2019) FTDP-17 Mutations alter the aggregation and microtubule stabilization propensity of Tau in an isoform-specific fashion. Biochemistry 58, 742–754 10.1021/acs.biochem.8b01039 [DOI] [PubMed] [Google Scholar]
- 109. Chang E., Kim S., Yin H., Nagaraja H. N., and Kuret J. (2008) Pathogenic missense MAPT mutations differentially modulate tau aggregation propensity at nucleation and extension steps. J. Neurochem. 107, 1113–1123 10.1111/j.1471-4159.2008.05692.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rizzu P., Van Swieten J. C., Joosse M., Hasegawa M., Stevens M., Tibben A., Niermeijer M. F., Hillebrand M., Ravid R., Oostra B. A., Goedert M., van Duijn C. M., and Heutink P. (1999) High prevalence of mutations in the microtubule-associated protein Tau in a population study of frontotemporal dementia in the Netherlands. Am. J. Hum. Genet. 64, 414–421 10.1086/302256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. von Bergen M., Barghorn S., Li L., Marx A., Biernat J., Mandelkow E. M., and Mandelkow E. (2001) Mutations of Tau protein in frontotemporal dementia promote aggregation of paired helical filaments by enhancing local β-structure. J. Biol. Chem. 276, 48165–48174 10.1074/jbc.M105196200 [DOI] [PubMed] [Google Scholar]
- 112. Bugiani O., Murrell J. R., Giaccone G., Hasegawa M., Ghigo G., Tabaton M., Morbin M., Primavera A., Carella F., Solaro C., Grisoli M., Savoiardo M., Spillantini M. G., Tagliavini F., Goedert M., and Ghetti B. (1999) Frontotemporal dementia and corticobasal degeneration in a family with a P301S mutation in Tau. J. Neuropathol. Exp. Neurol. 58, 667–677 10.1097/00005072-199906000-00011 [DOI] [PubMed] [Google Scholar]
- 113. Nacharaju P., Lewis J., Easson C., Yen S., Hackett J., Hutton M., and Yen S. H. (1999) Accelerated filament formation from tau protein with specific FTDP-17 missense mutations. FEBS Lett. 447, 195–199 10.1016/S0014-5793(99)00294-X [DOI] [PubMed] [Google Scholar]
- 114. Goedert M., Jakes R., and Crowther R. A. (1999) Effects of frontotemporal dementia FTDP-17 mutations on heparin-induced assembly of Tau filaments. FEBS Lett. 450, 306–311 10.1016/S0014-5793(99)00508-6 [DOI] [PubMed] [Google Scholar]
- 115. Iovino M., Pfisterer U., Holton J. L., Lashley T., Swingler R. J., Calo L., Treacy R., Revesz T., Parmar M., Goedert M., Muqit M. M., and Spillantini M. G. (2014) The novel MAPT mutation K298E: mechanisms of mutant Tau toxicity, brain pathology and tau expression in induced fibroblast-derived neurons. Acta Neuropathol. 127, 283–295 10.1007/s00401-013-1219-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Spina S., Murrell J. R., Yoshida H., Ghetti B., Bermingham N., Sweeney B., Dlouhy S. R., Crowther R. A., Goedert M., and Keohane C. (2007) The novel Tau mutation G335S: clinical, neuropathological and molecular characterization. Acta Neuropathol. 113, 461–470 10.1007/s00401-006-0182-5 [DOI] [PubMed] [Google Scholar]
- 117. Neumann M., Diekmann S., Bertsch U., Vanmassenhove B., Bogerts B., and Kretzschmar H. A. (2005) Novel G335V mutation in the tau gene associated with early onset familial frontotemporal dementia. Neurogenetics 6, 91–95 10.1007/s10048-005-0210-y [DOI] [PubMed] [Google Scholar]
- 118. Nicholl D. J., Greenstone M. A., Clarke C. E., Rizzu P., Crooks D., Crowe A., Trojanowski J. Q., Lee V. M., and Heutink P. (2003) An English kindred with a novel recessive tauopathy and respiratory failure. Ann. Neurol. 54, 682–686 10.1002/ana.10747 [DOI] [PubMed] [Google Scholar]
- 119. Rossi G., Bastone A., Piccoli E., Morbin M., Mazzoleni G., Fugnanesi V., Beeg M., Del Favero E., Cantù L., Motta S., Salsano E., Pareyson D., Erbetta A., Elia A. E., Del Sorbo F., et al. (2014) Different mutations at V363 MAPT codon are associated with atypical clinical phenotypes and show unusual structural and functional features. Neurobiol. Aging 35, 408–417 10.1016/j.neurobiolaging.2013.08.004 [DOI] [PubMed] [Google Scholar]
- 120. Morelli F., Romeo M., Barzago M. M., Bolis M., Mattioni D., Rossi G., Tagliavini F., Bastone A., Salmona M., and Diomede L. (2018) V363I and V363A mutated tau affect aggregation and neuronal dysfunction differently in C. elegans. Neurobiol. Dis. 117, 226–234 10.1016/j.nbd.2018.06.018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.