See Smith and Stacey (doi:10.1093/brain/awz369) for a scientific commentary on this article.
Epilepsy surgery is currently guided by qualitative interpretation of neuroimaging and intracranial EEG, and leaves many patients with residual seizures. Kini et al. present a data-driven network-based method to accurately predict surgical outcome and identify resection targets in drug resistant epilepsy.
Keywords: seizures, electrocorticography, epilepsy surgery, network neuroscience, functional connectivity
Abstract
Patients with drug-resistant epilepsy often require surgery to become seizure-free. While laser ablation and implantable stimulation devices have lowered the morbidity of these procedures, seizure-free rates have not dramatically improved, particularly for patients without focal lesions. This is in part because it is often unclear where to intervene in these cases. To address this clinical need, several research groups have published methods to map epileptic networks but applying them to improve patient care remains a challenge. In this study we advance clinical translation of these methods by: (i) presenting and sharing a robust pipeline to rigorously quantify the boundaries of the resection zone and determining which intracranial EEG electrodes lie within it; (ii) validating a brain network model on a retrospective cohort of 28 patients with drug-resistant epilepsy implanted with intracranial electrodes prior to surgical resection; and (iii) sharing all neuroimaging, annotated electrophysiology, and clinical metadata to facilitate future collaboration. Our network methods accurately forecast whether patients are likely to benefit from surgical intervention based on synchronizability of intracranial EEG (area under the receiver operating characteristic curve of 0.89) and provide novel information that traditional electrographic features do not. We further report that removing synchronizing brain regions is associated with improved clinical outcome, and postulate that sparing desynchronizing regions may further be beneficial. Our findings suggest that data-driven network-based methods can identify patients likely to benefit from resective or ablative therapy, and perhaps prevent invasive interventions in those unlikely to do so.
Introduction
Epilepsy affects 65 million individuals, one-third of whom are resistant to antiepileptic medications. In these cases, surgery is often necessary to help reduce seizures (Kwan et al., 2011). Resections can be generous, with a significant amount of healthy tissue removed, frequently resulting in post-surgical side effects, including neuropsychological deficits and reduced quality of life (Kwan and Sperling, 2009). Unfortunately, some patients have seizure recurrence despite removal of assumed critical seizure generators, as mapped by extensive EEG, multimodal neuroimaging, and neuropsychological evaluation prior to surgery (Kwan et al., 2011). Because of the limitations of traditional epilepsy surgery, clinicians are turning to other less destructive therapeutic approaches. Specifically, responsive neurostimulation, deep brain stimulation, and targeted laser ablation techniques are increasingly being used to alleviate seizure burden and improve quality of life (Willie et al., 2014; Attiah et al., 2015; Thomas and Jobst, 2015). These interventions are hypothesized to act by disrupting connections and pathways involved in seizure spread (Thomas and Jobst, 2015). Identifying these important control regions is a critical step toward realizing the potential of these newer, less invasive techniques and for optimizing the use of established resective surgery.
While focal brain lesions have long been a target of epilepsy surgery with favourable success rates, patients without clear lesions may have seizures that arise from abnormal connectivity in broader networks that can be measured at the scale of electrocorticography (ECoG). Recent work supports the hypothesis that epilepsy can arise from disordered brain networks (Kramer and Cash, 2012). In epileptic networks, the seizure onset zone (SOZ) often not only drives seizure initiation and propagation, but also recruits regions that extend well beyond it to act as central hubs. These regions appear to strengthen in connectivity to each other, while weakening in connectivity to remaining regions (Khambhati et al., 2015; Besson et al., 2017). Because the epileptic network may be characterized by pathological foci embedded in this web of structural and functional connections (Burns et al., 2014), it is important to understand how aberrant cortical functioning drives seizure dynamics and manifests in the diverse roles of regions such as the epileptogenic, irritative, and propagation zones. Thus, a network approach that quantifies the complex synchronization and spread of neural activity is well suited to studying epilepsy in which changes in brain connectivity manifest across a wide range of spatiotemporal scales.
Recent efforts to translate and extend methods originally developed in network science have generated novel in silico approaches to model epileptic networks (Burns et al., 2014; Jirsa et al., 2017; Bassett et al., 2018) and identify important regions to target therapeutically with surgical (Goodfellow et al., 2016; Khambhati et al., 2016; Lopes et al., 2017; Sinha et al., 2017; Shah et al., 2019) or non-surgical (Taylor et al., 2015; Muldoon et al., 2018) interventions. While some of these models quantify brain dynamics through data-driven network models (Burns et al., 2014; Goodfellow et al., 2016; Khambhati et al., 2016; Shah et al., 2019), others integrate network architecture estimated by intracranial EEG or imaging with generative mathematical models that parameterize behaviour at each node (Lopes et al., 2017; Sinha et al., 2017). The virtual epileptic patient (VEP) (Jirsa et al., 2017) is one notable model that uses structural connectivity estimated from diffusion-weighted MRI to parameterize coupled ‘Epileptor’ oscillators (Jirsa et al., 2014), which predict seizure propagation and spread. The VEP framework is currently being studied in a prospective clinical trial to augment clinician decision-making in epilepsy surgery (US National Library of Medicine, 2018). Previous studies have established the potential of modelling to enhance our understanding of epileptic networks, but their translation to clinical care has been challenging. There are multiple potential reasons for this. Some approaches, such as the VEP, use models that generate synthetic seizures that look remarkably similar to clinical events, but because they are synthetic there is concern that they may not capture the complex interplay between brain regions with inherently different control properties. Many studies do not use expert clinical annotations from interictal, pre-ictal, and seizure epochs to evaluate the full spectrum of epileptic activity in each patient. Finally, validation has been challenging, particularly for studies that use clinical data. For a variety of reasons, most groups have not openly shared their methods and data so that other centres can reproduce and extend their studies. Specifically, we aim to overcome each of these shortcomings in this study using the virtual resection framework developed by our group.
Virtual resection (Khambhati et al., 2016) implements a brain network model in which the regions measured by individual intracranial electrode contacts are defined as nodes, and the statistical relationships between pairs of nodes known as functional connectivity are defined as edges (Bassett et al., 2018). This approach focuses on calculation of synchronizability, which describes the ease with which neural activity can propagate through the network. Virtual resection uses a push-pull framework in which synchronizing nodes dynamically oppose desynchronizers and thus the properties of the network are modulated as a function of time. By virtually removing nodes and recalculating synchronizability, we quantify each node or region’s control centrality (Khambhati et al., 2016) and thus estimate how well dynamic brain activity such as seizures would spread throughout the network if a given region were removed. While this framework has been shown to robustly characterize the spatiotemporal regulators of seizure dynamics (Khambhati et al., 2016), in our prior work we did not engage in any effort to predict surgical outcomes, nor did we investigate whether control centrality within the resection zone played a key role in accurately predicting that outcome. In the work presented here, we hypothesize that removing synchronizing versus desynchronizing brain regions as determined by the virtual resection method is predictive of good versus poor outcomes. In addition, we hypothesize that control centrality within the resection zone is predictive of epilepsy surgical outcomes.
In this multicentre retrospective cohort study, we seek to determine whether virtual resection network features can predict surgical outcome in 28 patients with drug-resistant epilepsy who underwent surgical therapy. Specifically, we ask: What is the spatiotemporal evolution of synchronizability during a seizure? Is there a relationship between network synchronizability and important clinical variables used to guide therapy? What is the accuracy of predicting surgical outcome from the control centrality of combined group nodes overlying resection cavities? We hypothesize that virtual resection will uncover important seizure dynamics as measured by network metrics that can separate patients based on post-surgical outcome and that network metrics will be sensitive to clinical variables such as seizure type.
Materials and methods
Patient dataset
All patients included in this study gave written informed consent in accordance with the Institutional Review Board of the University of Pennsylvania. All patients from both the Hospital of the University of Pennsylvania and the Mayo Clinic gave consent to have anonymized full-length electrophysiology recordings and brain MRI and CT scans available to the public on the open online portal IEEG.org (Kini et al., 2016a, b).
Twenty-eight patients undergoing surgical treatment for medically refractory epilepsy (Table 1) underwent implantation of subdural and depth electrodes to localize the SOZ after presurgical evaluation with scalp EEG recording of ictal epochs, MRI, fluorodeoxyglucose-PET (FDG-PET), and neuropsychological testing. The results of the presurgical studies suggested that focal cortical resection might be a therapeutic option. Patients were then implanted with intracranial electrodes including grid, strip and sparse depth electrodes, to define the epileptic network better. De-identified patient data were retrieved from the online International Epilepsy Electrophysiology Portal (IEEG Portal). Patients were excluded from the study if they (i) did not undergo resection; (ii) did not have complete post-resection imaging; or (iii) did not have complete electrophysiology data.
Table 1.
Summary of patient cohort demographics
| Good surgical outcome (Engel I or ILAE 1–2) | Poor surgical outcome (Engel II-IV or ILAE 3–6) | P-value | |
|---|---|---|---|
| Total number of subjects | 17 | 11 | |
| Age at surgery (HUP only)a | 0.99b | ||
| Mean ± SD | 36.2 ± 11.1 | 33.1 ± 18.6 | |
| Sex | 0.93c | ||
| Male | 9 | 6 | |
| Female | 8 | 5 | |
| Resected/ablated region | 0.11c | ||
| LTL | 3 | 6 | |
| RTL | 8 | 1 | |
| LFL/LPL/LFPL | 4 | 2 | |
| RTL/RFL/RFPL | 2 | 2 | |
| MRIa | 0.97b | ||
| Lesional | 7 | 4 | |
| Non-lesional | 9 | 5 | |
| Pathologya | 0.02b | ||
| HS/MTS | 6 | 2 | |
| Gliosis | 3 | 8 | |
| Malformations of cortical development | 5 | 0 | |
| Tumour/vascular/infection | 2 | 0 | |
| Seizure typea | 0.20b | ||
| Aura/focal aware | 14 | 3 | |
| Focal impaired awareness | 51 | 30 | |
| Focal with generalization | 31 | 11 | |
| Type of resection | 0.68c | ||
| Anterior temporal lobectomy and/or amygdalohippocampectomy | 3 (left), 7 (right) | 6 (left), 2 (right) | |
| Anterior temporal lobectomy+ | 1 (left) | None | |
| Partial resection/lesionectomy | 3 (left), 2 (right) | 2 (left), 1 (right) | |
| RF ablation | 1 (right) | None | |
| Volume of resection | |||
| Volume of tissue, cm3 | 0.83b | ||
| Mean ± SD | 19.3 ± 12.8 | 23.5 ± 18.9 | |
| Nodes removed, % | 0.72a | ||
| Mean ± SD | 19.3 ± 14.4 | 21.5 ± 10.8 |
Patients were grouped by surgical outcome. Second column shows patients who had a favourable surgical outcome. Third column shows patients who had a poor surgical outcome. FCD = focal cortical dysplasia; HS = hippocampal sclerosis; LFL = left frontal lobe; LFPL = left frontoparietal lobe; LPL = left parietal lobe; LTL = left temporal lobe; MTS = mesial temporal sclerosis; PNH = periventricular nodular heterotopia; RF = right frontal; RFL = right frontal lobe; RFPL = right frontoparietal lobe; RPL = right parietal lobe; RTL = right temporal lobe; SD = standard deviation; TSC = tuberous sclerosis complex.
aData for these fields were unknown in a minority of patients and thus were not included in the table.
bT-test; cPearson chi-square test.
ECoG signals for patients from the Hospital of the University of Pennsylvania were recorded and digitized at 500 Hz sampling rate and preprocessed to eliminate line noise. Cortical surface electrode configurations, determined by a multidisciplinary team of neurologists and neurosurgeons, consisted of linear and 2D arrays (2.3-mm diameter with 10-mm intercontact spacing) and sampled the neocortex and mesial cortex for epileptic foci. Signals were recorded using a referential montage with the reference electrode, chosen by the clinical team, distant to the site of seizure onset and spanned the duration of a patient’s stay in the epilepsy monitoring unit. All EEG recording systems and intracranial electrodes used were FDA approved and commercially available.
Clinical marking of seizure events
SOZ was marked on ECoG according to the standard clinical protocol in the Penn Epilepsy Center. Initial clinical markings are made on IEEG by board-certified staff epileptologist attendings (F.M., A.K., K.D., B.L.). These IEEG markings were made by clinicians blinded to surgical outcome but provided with available surgical conference notes that contained patient clinical data related to other multimodality testing, such as brain MRI, PET scan, neuropsychological testing, and ictal single-photon emission computed tomography (SPECT) scanning, used to finalize surgical approach and planning. The following seizure times were annotated for each seizure: (i) earliest electrographic change (EEC) (Litt et al., 2001); (ii) unequivocal electrographic onset (UEO); and (iii) termination of seizure (END). A pre-seizure state that spanned a period equal in duration from EEC to END was used to mark the baseline pre-ictal period for any given seizure. An epileptologist involved in the study (F.M.) examined the patient’s primary seizure type for the presence of localizing factors such as low voltage fast activity (LVFA), DC shift, and a clearly focal SOZ, which are known to predict outcome (Noe et al., 2013; Jin et al., 2016; Lagarde et al., 2016; Vakharia et al., 2018). If a disagreement regarding annotation arose, at least two epileptologists discussed the seizure in question until reaching consensus. All seizures were identified according to the current International League Against Epilepsy (ILAE) classification system, as focal aware seizures, focal impaired awareness seizures, or focal to bilateral tonic-clonic seizures. When clinical notes were unclear, the reviewing epileptologist made a decision on seizure type based on all available clinical data. In order to support the potential generalizability of our methods to any new patient, network measures were computed on all seizures, interpolated to fit into 10 sequential time bins spanning the pre-seizure and seizure epochs, and averaged within a patient’s group of seizures.
Surgical outcome
Surgical outcome was measured at a minimum of 1 year after surgery and determined based on medical records from the last available follow-up with a clinician. Patients who had surgical outcome of Engel I or ILAE 1–2 were marked as having favourable outcome and patients who had Engel II–IV or ILAE 3–6 were marked as having poor outcome (Wieser et al., 2008).
Image processing
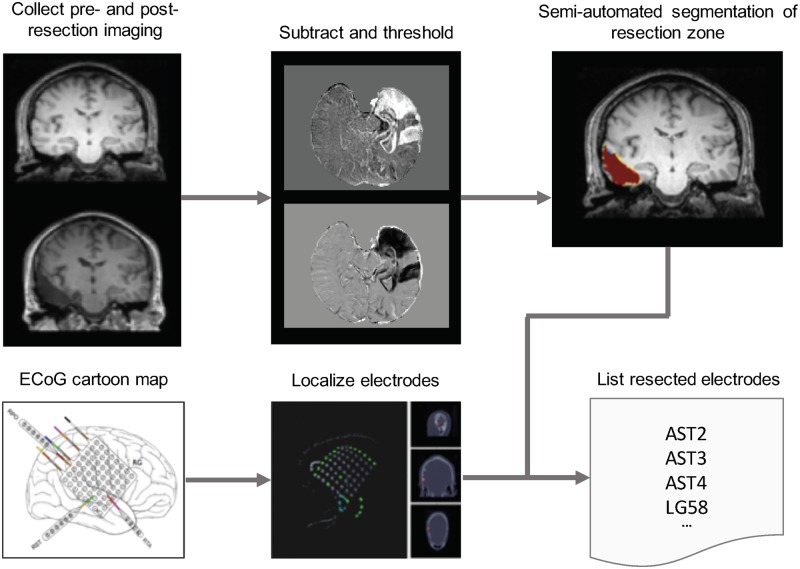
All patients, as part of their clinical neuroradiological work-up, underwent a clinical epilepsy neuroimaging protocol. Pre-implant T1-weighted MPRAGE MRI, post-implant T1-weighted MPRAGE MRI, and post-implant CT images were acquired to localize electrodes. In addition, patients underwent a post-resection imaging protocol acquired on average 6–8 months after implant and resection, which consisted of T1-weighted MRI and axial FLAIR MRI sequences. All images were stripped of headers, anonymized and registered to patient’s native pre-implant T1 MRI space for localization and segmentation (Fig. 1).
Figure 1.
Imaging pipeline for resection zone estimation. The imaging pipeline registers preoperative MRI, post-implant CT, and post-resection MRI together to allow for resection zone mapping and electrode localization. In-house software in addition to ITK-SNAP were used to map resection zones and localize electrodes for all patients using cartoon maps presented in surgical conference notes. Nodes that overlapped with the resection zone were virtually resected from the network to measure effect on synchronizability.
Electrode localization
In-house software (Azarion et al., 2014) was used to assist in localizing electrodes after registration of pre-implant and post-implant neuroimaging data. All electrode coordinates and labels were saved and matched with ECoG electrode names on IEEG.org. All electrode localizations were verified by a board-certified neuroradiologist (J.S.).
Resection zone segmentation
Pre-implant MRI was registered diffeomorphically using the Advanced Normalization Toolkit (ANTs) (Avants et al., 2011) to post-resection imaging to accurately segment the resection zone. Resection zones were estimated semi-automatically with the use of a random forest classifier and region-growing algorithm as part of the ITK-SNAP toolkit (Yushkevich et al., 2006). All resection estimates were confirmed by a board-certified neuroradiologist (J.S.).
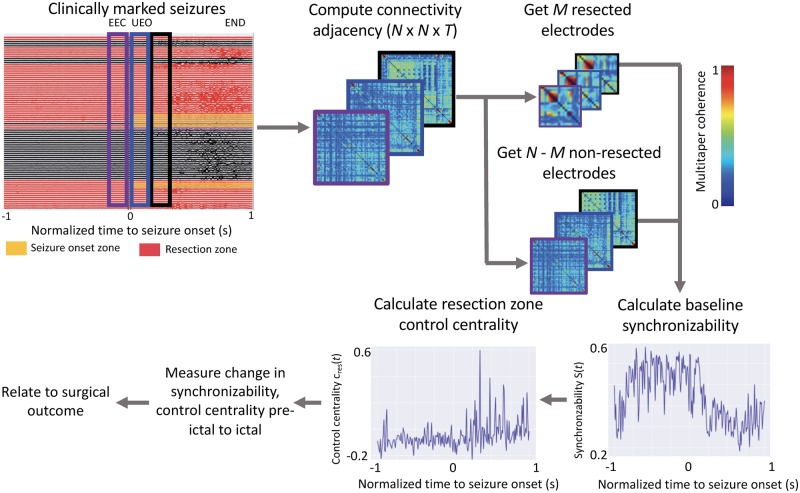
Network methods
The virtual resection method (Fig. 2) is described in greater detail in Khambhati et al. (2015). Electrodes in which the ECoG signal was obscured by artefact, as noted by an attending epileptologist, were removed from analysis to avoid biasing our results. A common average reference was applied to all neural signals by first computing a time-varying signal averaged across all electrodes and then by subtracting this signal from each electrode. All ECoG signals were notch filtered at 60 Hz to remove power line noise. We constructed functional brain networks in each time window using multitaper coherence estimation, which defines an edge between electrode pairs in terms of the correlation of the power spectra of signal activity over a specific frequency band. This procedure was done across different physiological frequency bands, namely: α/θ (5–15 Hz), β (15–25 Hz), low-γ (30–40 Hz), high-γ (95–105 Hz) and very high frequencies (>105 Hz). In addition, broadband cross-correlation was used to generate functional dynamic networks without regard to frequency specific information. In this study we compute functional networks directly from clinical recordings of intracranial EEG and not on modelled or simulated data. These networks were generated as N × N symmetric adjacency matrices, describing the network for all T time windows.
Figure 2.
Electrophysiology pipeline for epileptic network analysis. The electrophysiology pipeline uses coherence to construct adjacency matrices for each event, separately for seizure and pre-seizure data. Baseline synchronizability of the network and control centrality of each individual node is calculated. From the imaging pipeline, resected electrodes are determined, and control centrality of the resection zone is calculated. Metrics are used to generate predictions of surgical outcome. EEC = earliest electrographic change; UEO = unequivocal electrographic onset.
Because of its importance to seizure spread, we measured network synchronizability by first computing the Laplacian matrix of each adjacency matrix at time 1-s time windows. The Laplacian matrix can be interpreted as measuring the ease with which information diffuses between nodes in a network (Stam and Reijneveld, 2007). Next, at each time epoch t across all T epochs, we calculated the synchronizability measure, s(t) as the ratio of the second smallest eigenvalue to the largest eigenvalue of the Laplacian matrix, which quantifies the stability of the synchronous state (Stam and Reijneveld, 2007). To model the effects of resective surgery, we used the approach of virtual cortical resection, which quantifies control centrality as the contribution to synchronizability. Control centrality can be calculated either at each node or for the entirety of a region of interest by removing the node or nodes in question from the network and recalculating synchronizability. In this study, we remove the resection zone en bloc to calculate control centrality when comparing across patients, while for whole brain visualizations we calculate control centrality at the node level. This measure of change in synchronizability is referred to as the control centrality, or cres(t), and can be used to identify a region as (i) desynchronizing (removal of which increases post-resection network synchronizability) characterized by positive control centrality; or (ii) synchronizing (removal of which decreases post-resection network synchronizability) characterized by negative control centrality.
Statistical methods
All averages computed in this study use the median because it represents a better measure of centrality in skewed, non-normal distributions such as those seen in the distribution of network measures (Buzsaki and Mizuseki, 2014). We compared median time-varying network metrics between seizures in patients who experienced a favourable surgical outcome and patients who experienced a poor surgical outcome. We performed this comparison by normalizing each seizure event into 10 sequential time bins spanning the pre-seizure and seizure epochs, and using functional data analysis to test differences in temporal dynamics statistically between seizure types independently in each state (Ramsay and Silverman, 2010). Functional data analysis allowed us to test whether the area under the good outcome curves and poor outcome curves were significantly different by comparing the true area to the area expected in an appropriate permutation-based null model. The null model was created by reassigning surgical outcome to adjacency matrices uniformly at random up to 10 000 times and computing the median area under the resulting curves of functional network metrics.
The median network measures during pre-ictal epochs are compared to those of ictal epochs using Wilcoxon rank-sum test. The Δs(t) change from pre-ictal to ictal time periods was used as the feature for quantifying the ability to predict surgical outcome. We varied the threshold of Δs(t) to predict patients as having either good or poor surgical outcome, which generated a receiver operating characteristic (ROC) curve. We measured area under the ROC curve (AUC) as a marker for accuracy in predicting good versus poor surgical outcome; the ROC curve quantifies the trade-off between the true positive rate and the false positive rate for a binary classifier. We used the non-parametric DeLong test to compare ROC curves and determine whether any single predictive model derived from a specific frequency band performed significantly better than the others (DeLong et al., 1988). In the analysis of control centrality of the resection zone we present P-values noting that the significance threshold is 0.0083 to correct for six comparisons introduced by the separate frequency bands according to the Bonferroni method.
Data availability
Our codebase comprises the imaging and electrophysiology pipelines (https://github.com/ieeg-portal/EpiVR), and allows researchers to easily fetch data situated on the IEEG.org portal as well as to perform virtual cortical resection. The electrophysiology pipeline is dependent on Echobase, which can be found at https://github.com/akhambhati/Echobase. We have additionally made pre- and post-resection imaging as well as annotations of seizures along with their ECoG recordings from the entirety of their epilepsy monitoring unit (EMU) stay available to the public through IEEG.org. From this unique and powerful dataset we hope that other investigators may validate our methods or compare their performance to other virtual resection tools.
Results
Twenty-eight patients with drug-resistant epilepsy underwent implantation with ECoG electrodes to localize the SOZ preceding surgical resection. Patient-specific network models were constructed from clinically annotated (Supplementary Table 1) ECoG recordings stored on the cloud platform IEEG.org (Wagenaar et al., 2015; Kini et al., 2016a, b), and a quantitative pipeline using pre- and post-surgical imaging was used to determine which electrodes were resected.
Network synchronizability predicts surgical outcome
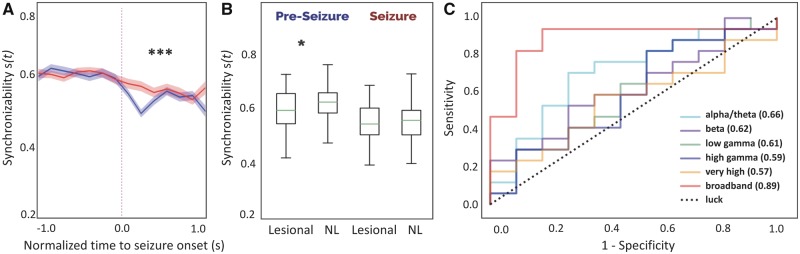
We began our virtual resection approach by examining the dynamic network changes in synchronizability s(t) that occur in the transition state from pre-ictal to ictal periods. Seizures were interpolated to be equal length across patients and onset times were aligned such that s(t) curves could be compared. Figure 3A shows the time course of median broadband synchronizability over pre-ictal and ictal periods between patients with good and poor outcome. These curves were different in the ictal period between outcome groups (functional data analysis curve area test, 10 000 permutations, P < 0.001) suggesting that there is predictive information in the magnitude and temporal evolution of s(t). Patients with favourable surgical outcomes had decreased synchronizability at the time of seizure onset across frequency bands while s(t) remained high in those with poor surgical outcomes. Synchronizability curves for other frequency bands can be found in Supplementary Fig. 1.
Figure 3.
Time-varying network synchronizability is predictive of surgical outcome. (A) Median base network synchronizability in good outcome patients (blue) and poor outcome patients (red) for broadband intracranial EEG (IEEG). ***P < 0.001. Shaded areas show 95% confidence intervals. (B) Patients with lesional MRI have higher pre-ictal synchronizability than non-lesional (NL) patients. *P < 0.05 (lesional, pre-ictal: min = 0.42, 25%ile = 0.55, median = 0.60, 75%ile = 0.67, max = 0.74; NL, pre-ictal: min = 0.33, 25%ile = 0.59, median = 0.63, 75%ile = 0.67, max = 0.85; lesional, ictal: min = 0.40, 25%ile = 0.51, median = 0.55, 75%ile = 0.61, max = 0.70; NL, ictal: min = 0.40, 25%ile = 0.51, median = 0.56, 75%ile = 0.60, max = 0.74). (C) ROC curves were constructed by calculating difference in s(t) from pre-ictal to ictal periods and sweeping the threshold for classification. Broadband IEEG predicts surgical outcome significantly better than other bands as assessed by the DeLong test, which statistically compares ROC curves generated from correlated data.
We also asked whether there is an association between synchronizability and clinical variables used to guide therapy such as lesion status on MRI. We found that patients who were non-lesional (n = 14) had higher pre-ictal synchronizability in broadband compared to patients with lesions (n = 11) (rank-sum statistic −1.984, P = 0.047) (Fig. 3B), even though there was no correlation with lesion status and outcome (chi-square test = 0.178, P = 0.67) or electrode number (rank-sum statistic 267, P = 0.79) in our study.
To quantify the ability of synchronizability to predict surgical outcome, we calculated median change in s(t) from pre-ictal to ictal periods, and performed a sweep of that feature to generate an ROC curve (Fig. 3C). Despite the small sample size, the Δs(t) derived from broadband ECoG data (AUC = 0.89, 95% CI 0.76–1.00) is predictive of surgical outcome. This ROC curve showed greater statistical significance than that observed in each of the other frequency bands. Selecting the point on the ROC that gave the greatest number of correct classifications, we choose a threshold of Δs(t) = 0.0279 to determine the performance of our predictive model based on broadband networks across all 28 patients in our cohort. We found an accuracy of 0.86 with a true positive rate of 0.88 and a true negative rate of 0.82 (Supplementary Table 2).
Virtual resection provides novel clinical insight
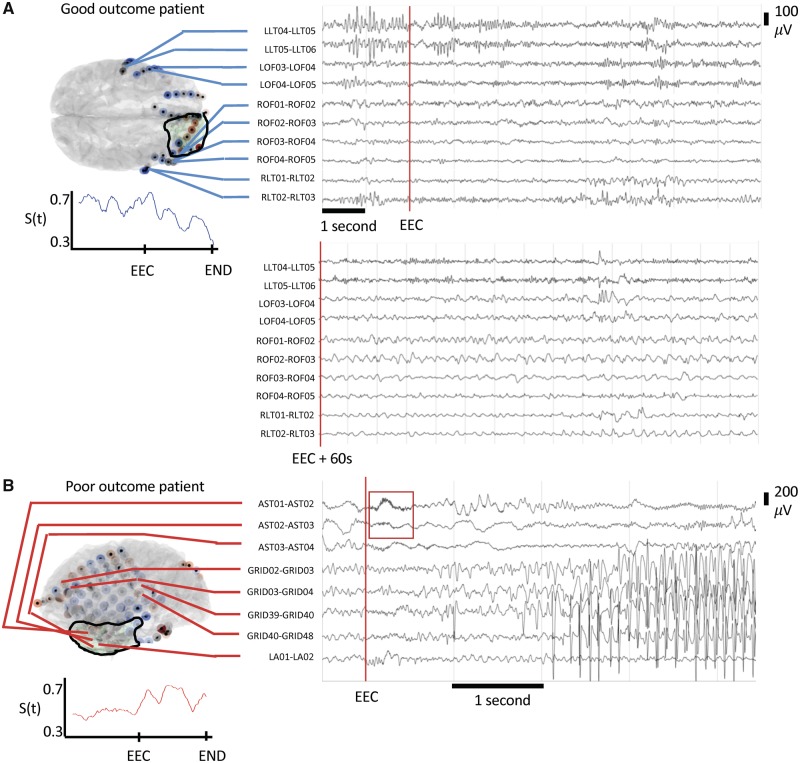
We sought to determine whether the virtual resection model provides additional information on patient prognosis that is not merely correlated with traditional electrographic findings such as DC shift, clear seizure focus, and low voltage fast activity. While a significant decrease in broadband synchronizability at seizure onset carries an odds ratio of 35 for good surgical outcome, having a focal SOZ, low voltage fast activity, or a DC shift do not perform as well with odds ratios of 4.3, 1.5 and 7, respectively (Supplementary Table 3). We present the EEG and synchronizability of two patients that our model predicted correctly (Fig. 4). In these cases, following electrographic features alone did not correlate with outcome and we found that virtual resection provides novel clinical information not captured by traditional clinical analysis.
Figure 4.
Synchronizability provides novel clinical information. (A) Our model correctly predicted HUP073 to be seizure-free after surgery. A board-certified epileptologist determined there is no DC shift, low voltage fast activity, or clearly focal seizure onset. This stereotyped clinical seizure results in arousal from sleep without evident EEG change. While the clinically marked seizure onset began with an arousal pattern in LLT04–06, here rhythmic activity begins in ROF1–3 60 s into seizure, later progressing to RLT1–3. Resection was performed in the right frontal region. Synchronizability decreases throughout the seizure in this patient, predicting seizure freedom after surgery. (B) Our model correctly predicted the poor outcome of Patient HUP080. In the displayed seizure, a board-certified epileptologist determined seizure onset electrodes of AST1–3 with the presence of low voltage fast activity (red box), and a clearly focal seizure onset but no DC shift. Synchronizability increases after earliest electrographic change (EEC) in this patient, correctly predicting the surgical outcome of this patient to be poor.
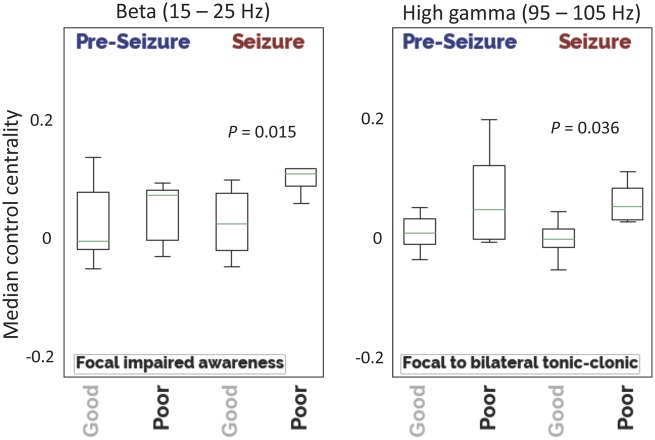
We then asked the following questions: How does control centrality cres(t) of the resection zone change before and during a seizure? How does it differ between patients who fare favourably and patients who fare poorly after surgery? We observed that median cres(t) was lower in good outcome patients compared to poor outcome patients during the ictal period. In focal impaired awareness seizures the β frequency analysis was strongest (rank-sum statistic –2.10, P = 0.036), while in focal to bilateral tonic-clonic seizures this effect was strongest in high-γ (rank-sum statistic –2.43, P = 0.015) (Fig. 5). We note that after adjusting the α-level to 0.0083 for multiple comparisons, these results no longer reached statistical significance. The effect was not significant during pre-ictal periods or in other frequency bands (Supplementary Fig. 2). Additionally, calculated cres(t) is robust to segmentation error at the resection zone margin (Supplementary Fig. 3). These findings align with our initial hypotheses as well as with the theoretical understanding of the virtual resection network analysis.
Figure 5.
Control centrality of resection zone. Median node-level control centrality—calculated for the entire group of nodes lying within the resection zone—are shown for different seizure types. Patients with a good outcome have regions that play a greater synchronizing role resected compared to poor outcome patients. This effect is greatest in the β-band for focal impaired awareness seizures (good: min = −0.044, 25%ile = −0.021, median = 0.017, 75%ile = 0.060, max = 0.220; poor: min = 0.046, 25%ile = 0.071, median = 0.088, 75%ile = 0.095, max = 0.183) and in the high-γ band for focal to bilateral tonic-clonic seizures (good: min = −0.050, 25%ile = −0.017, median = −0.006, 75%ile = 0.010, max = 0.034; poor: min = 0.019, 25%ile = 0.023, median = 0.042, 75%ile = 0.067, max = 0.090).
Virtual resection maps spatial anomalies in seizure networks
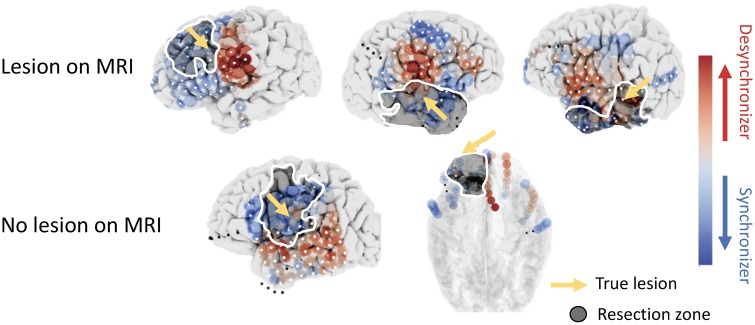
To examine the implications of virtual resection results on clinical management, we sought to elucidate the role that various clinical features can play on virtual resection features. We sought to study the spatial distribution of cres(t) in patients who had undergone focal resections with malformations of cortical development (MCD), such as focal cortical dysplasia. There were six patients with MCD, two of which were read as MRI-normal. Five of these patients had favourable outcome. Figure 6 shows patients with MCD on pathology and their respective mean control centrality across pre-ictal and ictal epochs derived using the broadband cross-correlation metric for functional connectivity. In high frequency bands, during pre-ictal periods, we observed that MCDs exhibit stronger synchronizers peri-lesionally (pre-seizure: rank-sum statistic –2.08, P = 0.04). We also found strongly desynchronizing nodes in the β-band during ictal periods peri-lesionally within the resection zone compared to non-lesional patients (seizure period: rank-sum statistic 1.71, P = 0.09). However, neither of these results achieved statistical significance after correcting for multiple comparisons. These closely localized desynchronizing nodes near lesions may be important controllers that act on nearby seizure-generating regions next to dysmorphic dysplastic tissue. As this finding was present even in the two non-lesional patients with MCD, spatial maps of control centrality may act as a biomarker to uncover hidden epileptogenic lesions that may not be easy to identify on standard clinical neuroimaging.
Figure 6.
Spatial patterns uncover MCDs in resected regions in five patients. Spatial maps of mean control centrality for each node are shown for all five patients: three lesional and two non-lesional on MRI, seen above. All patients had pathology-confirmed MCDs (yellow arrow). Two patients had non-lesional findings on MRI despite presenting a spatial pattern of node-level control centrality similar to those in other patients with MRI-positive MCDs. Specifically, strong desynchronizing regions (red) are seen in all resected regions (darkened zone with white outline), which contain MCDs (synchronizers, blue). Artefactual electrodes not included and analyses are denoted with black dots.
Discussion
In this study, we investigate the ability of virtual resection to accurately predict surgical outcome using functional networks derived from ECoG data, expertly annotated seizure markings, and rigorous co-registration of intracranial electrodes and resected brain regions. We determine that decreased synchronizability at the time of seizure onset is predictive of good outcome, performing better than traditional electrographic features. We further suggest that good outcome patients have brain regions removed with a relatively greater synchronizing effect on broader networks than those of poor outcome patients. Finally, we propose that our robust pipeline incorporating rigorous clinical marking and validation of ECoG, quantified resection zones on standardized MRI after surgery, and sharing of all code and data are novel contributions that make this study important.
In our validation of the virtual resection method, we uncover relationships between synchronizability and important clinical variables. Broadband synchronizability has a significant decrease at seizure onset in good but not poor outcome patients, and pre-ictal synchronizability is higher in non-lesional patients than those with a clear lesion on MRI. The notion that patients with a good outcome have a decrease in synchronizability at seizure onset is intriguing because it suggests a greater resistance to the propagation of oscillatory epileptic activity throughout the network early in seizures. For patients without this decrease in synchronizability, traditional resections may not provide a cure as the existing epileptic network may poorly constrain abnormal activity. Furthermore, heightened pre-ictal synchronizability in non-lesional patients could underlie differences in the pathophysiology of seizure generation. Furthermore, the robustness of our methods to predict both non-lesional and lesional patient outcomes equally well is exciting and a clear strength of this study as there is substantial literature supporting improved outcomes in lesional epilepsy (Noe et al., 2013; Vakharia et al., 2018). While our retrospective analysis is not designed to prove conclusively the mechanism by which either of these observations occur, future experimental studies may explore these concepts in vivo. As we continue to extend virtual resection and provide support of its utility, we further aim to initiate a prospective clinical trial in the future.
We used differences in synchronizability between patients with good and poor outcome to predict surgical outcome and show that it provides novel information not present in traditional electrographic features. Using the change in median s(t) from the pre-ictal to the ictal period, we predict surgical outcome with accuracies that compare favourably to other recently published in silico models of resective epilepsy surgery (Goodfellow et al., 2016; Sinha et al., 2017; Tomlinson et al., 2017; Taylor et al., 2018). We also show that examining synchronizability curves can uncover novel information about the epileptic network that is often not present in traditional electrographic localizing features of focal SOZ, DC shift, and LVFA. These findings support the use of our model as an adjunct to traditional EEG interpretation and may identify patients for whom surgery could eliminate their seizures as well as those unlikely to benefit from intervention. We envision clinicians interacting with our model in a similar approach to Fig. 4, where synchronizability, control centrality, and EEG are viewed alongside each other. A holistic assessment of the full clinical information such as electrographic features is always warranted; however, our findings support the use of synchronizability to support surgical decision-making.
Examining the contribution of the resection zone to overall network dynamics may be a powerful tool for assessing surgical intervention. While the results of this analysis are not statistically significant after adjusting for multiple comparisons, median control centrality of the resection zone is lower in good outcome patients (Supplementary Fig. 2) across all frequency bands that are correlated. We observe that patients with a good outcome may be more likely to have lower control centrality of their resection zones, meaning that the resected tissue plays more of a synchronizing role in the overall network. It follows that such patients would have good outcome as the topology of the resulting functional brain network would have decreased synchronizability. Performing a high resolution spatial mapping of individual synchronizing and desynchronizing nodes such as in Fig. 6 may also provide insights into these concepts in the context of specific pathologies. However, larger collaborative datasets are needed to robustly uncover disease-specific patterns as our results do not reach statistical significance after adjusting for multiple comparisons. It is tempting to infer that our finding suggests that these regions function as a macro-scale ‘inhibitory surround’, analogous to that seen in more controlled studies of seizure propagation in animal models and humans (Weiss et al., 2013; Bink et al., 2018), but this hypothesis would require further investigation.
Broadband cross-correlation may have higher predictive power compared to individual frequency bands because it provides the most general assessment of network connectivity without frequency-specific information. Given that broadband has high correlation with all other frequency bands (Supplementary Fig. 4), broadband may be the most generalizable as it would not be as affected by differences in individual seizure pathophysiology. Furthermore, previous network models of epilepsy have found broadband to be highly predictive (Tomlinson et al., 2017). On the other hand, when examining control centrality of the resection zone parsed by seizure types we found frequency differences that are not apparent in the broadband analysis. The finding that high-γ synchronization was most associated with good outcome in focal seizures that generalized whereas β synchronization was associated with good outcome in focal seizures that did not spread may be rooted in different mechanisms underlying these events.
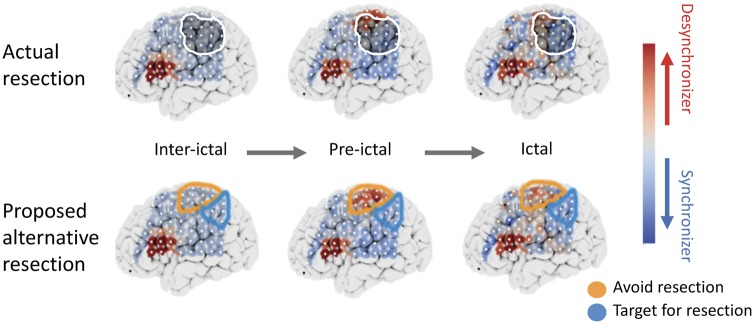
Recent work has attempted to study the spatiotemporal dynamics of seizures from initiation to termination (Kramer et al., 2008; Khambhati et al., 2016; Wang et al., 2017). Onset patterns are not determined by initiation of aberrant activity in the SOZ core alone, but additionally by how changes in excitability in surrounding healthy tissue cause the onset to become evident (Ray et al., 2008). These network state changes may serve as control mechanisms enabling desynchronous activity to disrupt seizures or to coalesce tightly bound and functionally cohesive network components (Kramer and Cash, 2012). As a result, some nodes may be seizure desynchronizers that should potentially be left intact in any resection plan (Fig. 7). In Fig. 7, the proposed resection zone is the group of synchronizing nodes that fell within the original resection zone, while nodes marked to avoid are desynchronizing nodes that were resected. The predictions of our model need to be validated in a prospective trial before translating them into patient care. Additionally, other nodes may act as strong synchronizers during seizure evolution and perhaps could be especially targeted for ablation, resection or stimulation. Studying these findings broadly across connected brain regions could identify potential targets for focal therapy outside of the seizure onset.
Figure 7.
Proposed framework for optimal resection targets using virtual resection. Median node-level control centralities during interictal, pre-ictal and ictal periods are shown for a sample patient who had poor outcome. The top row shows spatial maps with the resection zone (darkened region with white outline) and the bottom row shows regions that perhaps should be targeted for resection due to the presence of strong synchronizing nodes (blue), or should be avoided during resection because of the presence of strong desynchronizing nodes (red). Future experiments assessing the effects of stimulation of these nodes and subsequent changes in synchronizability will allow clinicians to better predict the effect of targeted resection of these nodes.
The virtual resection method allows for mapping of control mechanisms of epileptic networks outside of seizures as well. The model can be extended to interictal periods without the need for ictal markings because repetitive, stable topographical patterns in functional connectivity emerge across long interictal time periods (Wang et al., 2017). Additionally, subgraphs identified in interictal periods are similar topologically to those identified during seizures, allowing for generalization of the virtual resection method to interictal epochs (Wang et al., 2017). Interictal epileptiform discharges may also be incorporated into the virtual resection framework by studying the regions that generate spikes in terms of their interictal, pre-ictal and ictal control centrality. The flexibility of our methods to various modes of analysis is a clear strength of our study.
The natural next step in testing the idea that resection should target synchronizers and preserve desynchronizers is to relate the effects of local stimulation to regional control centrality. Brain stimulation performed either intraoperatively or in the epilepsy monitoring unit may provide a safe and effective way to experimentally test network hypotheses by determining whether activation of certain nodes results in the generation or interruption of epileptic activity. Relating stimulation to virtual resection measures may be useful to describe spatiotemporal dynamics fully, and would provide an avenue to formulate an algorithm to target resection. As recent technology allows intracranial EEG streaming to online cloud platforms (Baldassano et al., 2019), our virtual resection method and calculation of network metrics could also be implemented in real time during stimulus-based mapping. If successful, these methods could be extended to guide therapy with closed-loop neurostimulation devices such as the Neuropace RNS.
The methods and results of our model are derived from patient-specific ECoG and imaging data and compare favourably with previous studies of in silico models of resective epilepsy surgery. In particular, studies have often performed the identification of ictogenic nodes whose resection influences outcome by using neural mass models parameterized by functional connectivity. First, Sinha et al. (2017) identified nodes that caused the network to transition the fastest into seizure dynamics via a subcritical Hopf bifurcation (Kalitzin et al., 2010). Second, Goodfellow et al. (2016) used a more mechanistic model that identified nodes where removal would reduce epileptiform dynamics via saddle-nodes on a limit cycle bifurcation (Goodfellow et al., 2016, 2017). In contrast, our current virtual resection study uses a network framework to describe directly the node level and global dynamics of each patient’s seizures as they occur. Our approach is thus not constrained by simulated seizures whose dynamics may be at odds with those observed in clinical data, and instead allows clinicians to assess intracranial EEG network properties derived from the very same data on which they currently base clinical decisions. Furthermore, our quantitative imaging pipeline and high-quality dataset is larger than either of the previous studies, which bolsters the generalizability of our results. We also use data from each seizure and a corresponding preictal period for each patient in our study rather than just the first seizure (Goodfellow et al., 2016) or purely interictal data (Sinha et al., 2017). Each of these advantages of our study brings us closer to a clinical tool and demonstrates the significant novelty of our work in the field of personalized network models of epilepsy.
Our study has several important limitations. The small number of patients tested decreases the statistical power of this study, and before the technique can be applied clinically, it must be tested on a broader range of epilepsy types. It also must be validated on stereotactic EEG recordings, which are rapidly becoming the standard for many epilepsy centres worldwide as they use depth electrodes to sample broader brain networks yet require a less invasive implantation procedure. Another limitation of our network neuroscience approach is the sensor-level brain regions that we call ‘nodes’ are neither spatially discrete nor fixed in their locations across patients as intracranial electrodes are not implanted with connectivity studies in mind. One limitation of all studies using functional connectivity derived from ECoG is the sampling bias introduced by electrode placement, which may impact the reliability of summary statistics derived from network models. We have found that even upon 20–40% resampling of electrodes, network metrics including control centrality and synchronizability are reliable and perform in line with other metrics used in the field (Conrad et al., 2019). Furthermore, statistical methods such as jackknife resampling can be used to determine relative spatial confidence in network model results (Conrad et al., 2019). Unfortunately, we cannot validate whether the predictions regarding changes in synchronizability after resection result in an altered epileptic network that is less likely to manifest seizures. Direct brain recordings post-surgery would be an ideal method of model validation but these are not performed as part of clinical care at our institution.
A further nuance of our study is that all seizures were considered and analysed for every patient. Patients undergoing electrode implantation and monitoring are frequently observed to have aberrant seizures and discharges attributed to electrode trauma as well as events occurring in regions that do not give rise to the patient’s stereotyped clinical events (Sperling, 1997; Hudgins et al., 2016; Parvizi and Kastner, 2018). The true causes of these seizures and their significance are unknown. Additionally, in our synchronizability analysis we average all seizures within a patient, even for mixtures of focal impaired awareness and bilateral tonic-clonic seizures. This approach diminishes sensitivity in detecting dynamic changes associated with different seizure subtypes. However, we feel that ‘human filtering’ of data by selecting subtypes would add bias to our results and add a level of subjectivity to our methods that would make them quite difficult to translate to clinical practice, particularly at different medical centres using slightly different ECoG interpretation criteria. While this attests to the generalizability of the method, we need to refine spatiotemporal mapping further.
We present a method for rigorously mapping epileptic networks and predicting outcome based upon rigorously validated resection of network nodes. In spite of study limitations, our results suggest that these tools may have value in planning epilepsy surgeries, identifying patients in advance who are likely to have good outcomes, and also those who are less likely to benefit from surgical resection. We hope that this work may be a step in further standardizing invasive epilepsy procedures and treatment, and initiating multicentre clinical trials that reduce individual variation from centre to centre in this vital part of patient care.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Mr John Frommeyer for technical assistance with www.ieeg.org; Dr Steven Baldassano for technical discussion/suggestions regarding seizure annotations. The authors would like to additionally acknowledge Dr Jay Pathmanathan for discussion and suggestions regarding clinical annotations and analysis.
Glossary
Abbreviations
- ECoG
electrocorticography
- MCD
malformations of cortical development
- ROC
receiver operating characteristic
- SOZ
seizure onset zone
See Smith and Stacey (doi:10.1093/brain/awz369) for a scientific commentary on this article.
Funding
Virtual cortical resection from NINDS R01-NS099348–01 (Litt; Bassett). Additional grant funding provided by NIH 1-T32-NS-091006–01 (Training Program in Neuro-engineering and Medicine), NIH R25-NS065745, The Mirowski Family Foundation, Neil and Barbara Smit, and Jonathan Rothberg. D.S.B. would also like to acknowledge support from the Alfred P. Sloan Foundation, the John D. and Catherine T. MacArthur Foundation, and the ISI Foundation.
Competing interests
The authors report no competing interests.
References
- Attiah MA, Paulo DL, Danish SF, Stein SC, Mani R. Anterior temporal lobectomy compared with laser thermal hippocampectomy for mesial temporal epilepsy: a threshold analysis study. Epilepsy Res 2015; 115: 1–7. [DOI] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011; 54: 2033–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azarion AA, Wu J, Pearce A, Krish VT, Wagenaar J, Chen W, et al. An open-source automated platform for three-dimensional visualization of subdural electrodes using CT-MRI coregistration. Epilepsia 2014; 55: 2028–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassano S, Zhao X, Brinkmann BH, Kremen V, Bernabei J, Cook MJ, et al. Cloud computing for seizure detection in implanted neural devices. J Neural Eng 2019; 16: 026016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Zurn P, Gold GI. On the nature and use of models in network neuroscience. Nat Rev Neurosci 2018; 19: 566–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson P, Bandt SK, Proix T, Lagarde S, Jirsa VK, Ranjeva JP, et al. Anatomic consistencies across epilepsies: a stereotactic-EEG informed high-resolution structural connectivity study. Brain 2017; 1323: 152–160. [DOI] [PubMed] [Google Scholar]
- Bink H, Sedigh-Sarvestani M, Fernandez-Lamo I, Kini L, Ung H, Kuzum D, et al. Spatiotemporal evolution of focal epileptiform activity from surface and laminar field recordings in cat neocortex. J Neurophysiol 2018; 19: 2068–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns SP, Santaniello S, Yaffe RB, et al. Network dynamics of the brain and influence of the epileptic seizure onset zone. Proc Natl Acad Sci U S A 2014; 111: E5321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsaki G, Mizuseki K. The log-dynamic brain: how skewed distributions affect network operations. Nat Rev Neurosci 2014; 15: 264–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad EC, Bernabei JM, Kini LG, Shah P, Shinohara RT, Davis KA, et al. Sensitivity of functional connectivity to electrocorticography electrode resampling: Implications for personalized network models in drug-resistant epilepsy. bioRxiv 2019. [Google Scholar]
- De Long ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988; 44: 837–45. [PubMed] [Google Scholar]
- Goodfellow M, Rummel C, Abela E, Richardson MP, Schindler K, Terry JR. Estimation of brain network ictogenicity predicts outcome from epilepsy surgery. Sci Rep 2016; 6: 29215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow M, Rummel C, Abela E, Richardson MP, Schindler K, Terry JR. Computer models to inform epilepsy surgery strategies: prediction of postoperative outcome. Brain 2017; 140: e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudgins E, Brown MG, Litt B, Davis K, Richardson AG, Lucas T. Focal seizures induced by intracranial electroencephalogram grids. Cureus 2016; 8: e831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirsa VK, Proix T, Perdikis D, Woodman MM, Wang H, Gonzalez-Martinez J, et al. The Virtual Epileptic Patient: Individualized whole-brain models of epilepsy spread. Neuroimage 2017; 145: 377–88. [DOI] [PubMed] [Google Scholar]
- Jin B, So NK, Wang S. Advances of intracranial electroencephalography in localizing the epileptogenic zone. Neurosci Bull 2016; 32: 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirsa VK, Stacey WC, Quilichini PP, Ivanov AI, Bernard C. On the nature of seizure dynamics. Brain 2014; 137: 2210–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitzin SN, Velis DN, Lopes da Silva FH. Stimulation-based anticipation and control of state transitions in the epileptic brain. Epilepsy Behav 2010; 17: 310–23. [DOI] [PubMed] [Google Scholar]
- Khambhati AN, Davis KA, Oommen BS, Chen SH, Lucas TH, Litt B, et al. Dynamic Network Drivers of Seizure Generation, Propagation and Termination in Human Neocortical Epilepsy. Kramer M, ed. PLoS Comput Biol 2015; 11: e1004608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khambhati AN, Davis KA, Lucas TH, Litt B, Bassett DS. Virtual cortical resection reveals push-pull network control preceding seizure evolution. Neuron 2016; 91: 1170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kini LG, Davis KA, Wagenaar JB. Data integration: Combined imaging and electrophysiology data in the cloud. Neuroimage 2016a; 124: 1175–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kini LG, Gee JC, Litt B. Computational analysis in epilepsy neuroimaging: a survey of features and methods. NeuroImage Clin 2016b; 11: 515–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MA, Cash SS. Epilepsy as a disorder of cortical network organization. Neuroscientist 2012; 18: 360–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MA, Roopun AK, Carracedo LM, Traub RD, Whittington MA, Kopell NJ. Rhythm generation through period concatenation in rat somatosensory cortex. PLoS Comput Biol 2008; 4: e1000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan P, Schachter SC, Brodie MJ. Drug-Resistant Epilepsy. N Engl J Med 2011; 365: 919–26. [DOI] [PubMed] [Google Scholar]
- Kwan P, Sperling YR. Refractory seizures: Try additional antiepileptic drugs (after two have failed) or go directly to early surgery evaluation? Epilepsia 2009; 50: 57–62. [DOI] [PubMed] [Google Scholar]
- Lagarde S, Bonini F, McGonigal A, Chauvel P, Gavaret M, Scavarda D, et al. Seizure-onset patterns in focal cortical dysplasia and neurodevelopmental tumors: relationship with surgical prognosis and neuropathological subtypes. Epilepsia 2017; 57: 1426–35. [DOI] [PubMed] [Google Scholar]
- Litt B, Esteller R, Echauz J, D’Alessandro M, Shor R, Henry T, et al. Epileptic seizures may begin hours in advance of clinical onset: a report of five patients. Neuron 2001; 30: 51–64. [DOI] [PubMed] [Google Scholar]
- Lopes MA, Richardson MP, Abela E, Rummel C, Schindler K, Goodfellow M, et al. An optimal strategy for epilepsy surgery: Disruption of the rich-club? PLOS Comput Biol 2017; 13: e1005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muldoon SF, Constantini J, Webber WRS, Lesser R, Bassett DS. Locally stable brain states predict suppression of epileptic activity by enhanced cognitive effort. Neuroimage Clin 2018; 18: 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noe K, Sulc V, Wong-Kisiel L, Wirrell E, Van Gompel JJ, Wetjen N, et al. Long-term outcomes after nonlesional extratemporal epilepsy surgery. JAMA Neurol 2013; 70: 1003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvizi J, Kastner S. Promises and limitations of human intracranial electroencephalography. Nat Neurosci 2018; 21: 474–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay JO, Silverman BW. Functional data analysis. Berlin: Springer; 2010. [Google Scholar]
- Ray S, Crone NE, Niebur E, Franaszczuk PJ, Hsiao SS. Neural correlates of high-gamma oscillations (60–200 Hz) in Macaque local field potentials and their potential implications in electrocorticography. J Neurosci 2008; 28: 11526–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Bernabei JM, Kini L, Ashourvan A, Boccanfuso J, Archer R, et al. High interictal connectivity within the resection zone is associated with favorable post-surgical outcomes in focal epilepsy patients. NeuroImage: Clinical 2019; 23: 101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha N, Dauwels J, Kaiser M, Cash SS, Westover MB, Wang Y, et al. Predicting neurosurgical outcomes in focal epilepsy patients using computational modelling. Brain 2017; 140: 319–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling MR. Clinical challenges in invasive monitoring in epilepsy surgery. Epilepsia 1997; 36: S6–12. [DOI] [PubMed] [Google Scholar]
- Stam CJ, Reijneveld JC. Graph theoretical analysis of complex networks in the brain. Nonlinear Biomedical Physics 2007; 1: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor PN, Sinha N, Wang Y, Vos SB, de Tisi J, Miserocchi A, et al. The impact of epilepsy surgery on the structural connectome and its relation to outcome. Neuroimage Clin 2018; 18: 202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor PN, Thomas J, Sinha N, Dauwels J, Kaiser M, Thesen T, et al. Optimal control based seizure abatement using patient derived connectivity. Front Neurosci 2015; 9: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GP, Jobst BC. Critical review of the responsive neurostimulator system for epilepsy. Med Devices (Auckland) 2015; 8: 401–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson SB, Porter BE, Marsh ED. Interictal network synchrony and local heterogeneity predict epilepsy surgery outcome among pediatric patients. Epilepsia 2017; 58: 402–11. [DOI] [PubMed] [Google Scholar]
- US National Library of Medicine. Improving Epilepsy Surgery Management and prognosis Using Virtual Epileptic Patient Software (VEP) (EPINOV). https://clinicaltrials.gov/ct2/show/study/NCT03643016#contacts (22 August 2018, date last accessed).
- Vakharia VN, Duncan JS, Witt JA, Elger CE, Staba R, Engel J. Getting the best outcomes from epilepsy surgery. Ann Neurol 2018; 83: 676–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenaar JB, Worrell GA, Ives Z, Dumpelmann M, Litt B. Collaborating and sharing data in epilepsy research. J Clin Neurophysiol 2015; 32: 235–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Trevelyan AJ, Valentin A, Alarcon G, Taylor PN, Kaiser M. Mechanisms underlying different onset patterns of focal seizures. PLOS Comput Biol 2017; 13: e1005475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss SA, Banks GP, McKhann GM, Goodman RR, Emerson RG, Trevelyan AJ, et al. Ictal high frequency oscillations distinguish two types of seizure territories in humans. Brain 2013; 136: 3796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser HG, Blume WT, Fish D, Goldensohn A, Hufnagel A, King D, et al. Proposal for a new classification of outcome with respect to epileptic seizures following epilepsy surgery. Epilepsia 2008; 42: 282–86. [PubMed] [Google Scholar]
- Willie JT, Laxpati NG, Drane DL, Dowda A, Appin C, Hao C, et al. Real-time magnetic resonance-guided stereotactic laser amygdalohippocampotomy for mesial temporal lobe epilepsy. Neurosurgery 2014; 74: 569–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006; 31: 1116–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our codebase comprises the imaging and electrophysiology pipelines (https://github.com/ieeg-portal/EpiVR), and allows researchers to easily fetch data situated on the IEEG.org portal as well as to perform virtual cortical resection. The electrophysiology pipeline is dependent on Echobase, which can be found at https://github.com/akhambhati/Echobase. We have additionally made pre- and post-resection imaging as well as annotations of seizures along with their ECoG recordings from the entirety of their epilepsy monitoring unit (EMU) stay available to the public through IEEG.org. From this unique and powerful dataset we hope that other investigators may validate our methods or compare their performance to other virtual resection tools.