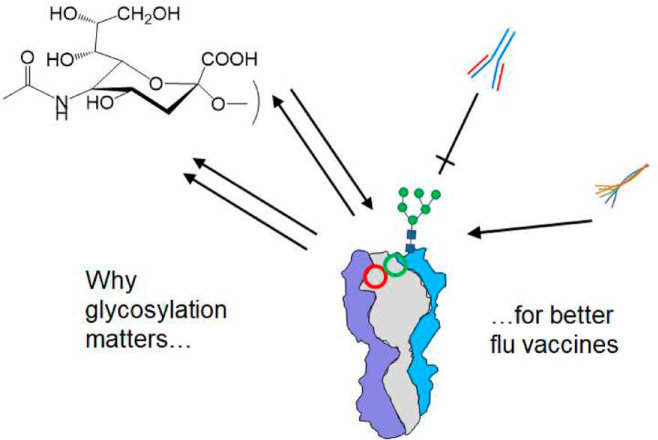
Immunodominant influenza A virus (IAV) antigens mutate rapidly, allowing the virus to escape host antibodies. The question remains how to design vaccines that recognize conserved but subdominant IAV antigens for broader immune protection. Glycosylation is a mechanism whereby IAV evades the innate and adaptive immune systems. However, its influence on immunodominance remains poorly understood. Although mass spectrometry methods for identifying glycopeptides are maturing, quantifying glycosylation variation among sets of IAV mutants remains a technical challenge.
Keywords: Glycoproteomics, glycoprotein structure, viruses, glycoproteins, clinical proteomics, influenza A virus
Graphical Abstract
Highlights
Glycosylation is not currently considered in flu vaccine design.
Glycosylation influences on immunodominance are not well understood.
Identification of site-specific glycosylation using mass spectrometry has matured.
New methods are needed to quantify site-specific glycosylation for vaccine design.
Abstract
Low vaccine efficacy against seasonal influenza A virus (IAV) stems from the ability of the virus to evade existing immunity while maintaining fitness. Although most potent neutralizing antibodies bind antigenic sites on the globular head domain of the IAV envelope glycoprotein hemagglutinin (HA), the error-prone IAV polymerase enables rapid evolution of key antigenic sites, resulting in immune escape. Significantly, the appearance of new N-glycosylation consensus sequences (sequons, NXT/NXS, rarely NXC) on the HA globular domain occurs among the more prevalent mutations as an IAV strain undergoes antigenic drift. The appearance of new glycosylation shields underlying amino acid residues from antibody contact, tunes receptor specificity, and balances receptor avidity with virion escape, all of which help maintain viral propagation through seasonal mutations. The World Health Organization selects seasonal vaccine strains based on information from surveillance, laboratory, and clinical observations. Although the genetic sequences are known, mature glycosylated structures of circulating strains are not defined. In this review, we summarize mass spectrometric methods for quantifying site-specific glycosylation in IAV strains and compare the evolution of IAV glycosylation to that of human immunodeficiency virus. We argue that the determination of site-specific glycosylation of IAV glycoproteins would enable development of vaccines that take advantage of glycosylation-dependent mechanisms whereby virus glycoproteins are processed by antigen presenting cells.
Viruses replicate by causing infected cells to produce virions that infect other cells. As has been observed by Fodor et al., virion composition determines virus stability, transmissibility, tropism, and immunogenicity (1). In the case of influenza A virus (IAV)1, viral hijacking of the host cell machinery results in error-prone replication. Some host cell proteins are taken up to construct the new virions, which are pleiomorphic in structure. Viral protein mutations caused by error-prone replication result in antigenic drift, which allows the virus to evade neutralization by immune system molecules. Hence, generating new vaccines annually is a critical effort in public health.
There are 18 known IAV hemagglutinin (HA), divided into two groups, and 11 known neuraminidase (NA) subtypes (2). Among the possible HA and NA combinations, only H1N1, H2N2 and H3N2 have caused pandemics. Today H1N1 and H3N2 circulate seasonally in humans. As shown in Fig. 1, IAV is a negative-sense, single-stranded RNA virus with 8 gene segments that code for at least 17 proteins (3). The trimeric HA glycoprotein binds sialylated glycans on the surface of host cells and facilitates subsequent endosomal membrane fusion. The NA glycoprotein cleaves sialic acids, allowing newly formed virions to escape the cell surface. The host adaptive immune system responds to IAV infection by generating antibodies that neutralize HA binding to host receptors.
Fig. 1.
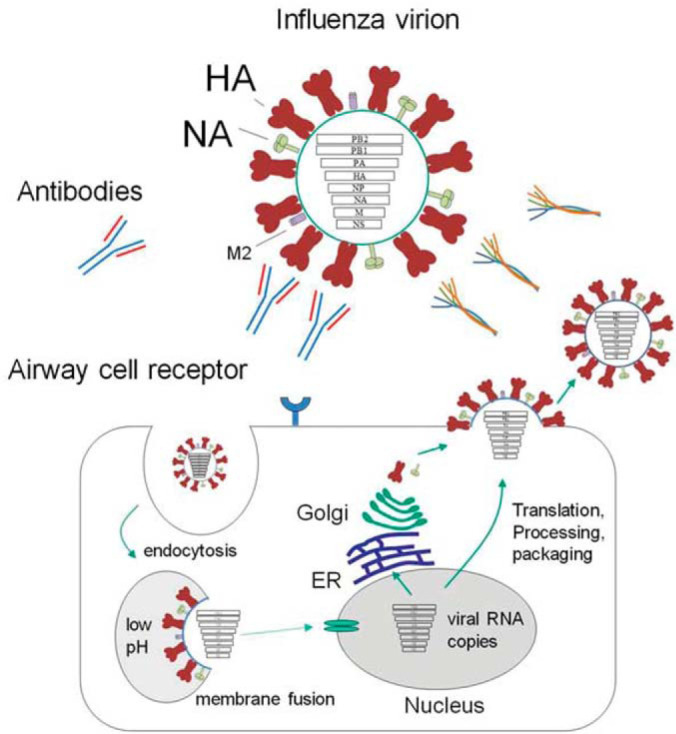
Influenza A viral cycle (127). Virion binding to airway cell receptors can be neutralized by antibody binding to the hemagglutinin (HA) head group or by innate immune system collectin binding to high mannose N-glycans on the HA head group. If neutralization does not occur, virion HA molecules bind airway cell sialic acid receptors and the virion is endocytosed. Low endosome pH causes virion membrane fusion and uncoating of ribonucleoprotein. Viral proteins are transported into the nucleus by host cell machinery. The negative sense RNA viral genome is converted to positive sense RNA that serves as a template for viral RNA production. Viral mRNA is exported from the nucleus and translated by cytoplasmic ribosomes. Viral proteins are imported to the nucleus where viral ribonucleoprotein complex assembly occurs. Viral ribonucleoproteins are transported to the plasma membrane. The HA and NA proteins pass through the secretory pathway where they are glycosylated and assembled into trimers. Viral progeny are assembled at the plasma membrane. Enveloped virions bud from the cell surface with the aid of NA cleavage of cell surface sialic acid residues.
The Need for a Better Flu Vaccine
Vaccines are injected intramuscularly or subcutaneously and must travel to the lymph nodes in order to elicit an antibody response. By contrast, a natural IAV infection occurs in human airways. The acute host infection immune reaction cascade includes cytokine release, influx of white blood cells, and cellular activation, resulting in clinical symptoms (4). This initial innate immune response limits the initial viral load and activates the adaptive immune system. The IAV infection induces systemic and local antibody responses (known as humoral immunity) and cytotoxic T cell responses (known as cellular immunity). In lymph nodes, naive B cells, through their surface antibodies, bind cognate antigens, become activated, and transition from IgM to IgG production. They then differentiate into memory B cells as they increase their immunoglobin specificity. Immunoglobins of the IgA class transported across the upper airway mucosal epithelium neutralize and clear infection. The lower airway is protected primarily by IgG.
The World Health Organization recommends the compositions of influenza vaccines based on surveillance, and laboratory and clinical observations (5). Despite this considerable effort, the effectiveness of the seasonal influenza vaccine remains unacceptably low, ranging from 10–60% (6). Thus far, neither the widespread accessibility of deep gene sequencing for efficient characterization of circulating strains, nor the availability of crystal coordinates for HA has led to improvement of vaccine efficacy. To address concern about future pandemics, the National Institute of Allergy and Infectious Diseases (NIAID) has released a strategic plan to address this problem (7).
Licensed vaccine classes include inactivated virus, live attenuated virus, and recombinant HAs (3). The majority of licensed inactivated or live attenuated vaccines are expressed in embryonated chicken eggs. Egg-based vaccine production has been practiced for 70 years; however, the many drawbacks of this system have spurred the exploration of new expression systems. It has been recognized for many years that IAV adapts to the cell in which it is propagated (8). Thus, IAV passaged in embryonated chicken eggs undergoes selection of strains adapted to growth in chicken cells. HA from such egg-adapted IAV display amino acid mutations near the receptor-binding site (9–12) and typically stimulate lower antibody titers. Mammalian tissue culture-grown IAV have shown superior vaccine protection in animal models compared with corresponding egg adapted IAV (13), and there now exists one inactivated vaccine expressed in mammalian cells, Flucelvax, approved by the FDA. Investigators have also used insect and plant cells for expression of HA (13–15). FluBlok is a licensed recombinant vaccine containing HA grown in insect cells from baculovirus vectors. Unlike egg-based expression, these alternative expression systems do not depend on a large supply of pathogen-free eggs for vaccine production, making them economical choices as well as ways to avoid the lowered immunogenicity that arises from egg adaptation.
The Number of HA Sequons Increases As IAV Circulates Seasonally in Humans
Glycosylation in the HA stalk region, at or near residues 15, 26, 289, 483, and 542, occurs in all HA forms (see (16) and references therein). These glycans may interact with glycan binding chaperones in the endoplasmic reticulum and appear to play roles in HA trimer assembly (17, 18). In contrast to seasonal strains, pandemic IAV, newly introduced to the human population, evades host antibody and innate immune defenses, and penetrates into the deep lung to infect bronchiolar, alveolar epithelial cells, and alveolar macrophages (19). To circulate in humans, these IAV must evade antibody recognition (20, 21). Thus, amino acid residues of the HA globular domain mutate rapidly under evolutionary pressure to avoid antibody recognition (22). Newly emerging pandemic IAV typically begin with a low degree of glycosylation of the HA globular domains, but the number of N-glycosylation consensus sites increases as the strains circulate seasonally. The amino acids shielded by N-glycosylation appear not to mutate at a high rate, relative to those that are exposed to antibody binding (20, 23).
The number of sequons on the HA globular head domain increases with the amount of time the IAV sub-type circulates seasonally in humans (21, 24–26). Genetic studies show evolutionary changes in the number of N-glycosylation sequons in the IAV protein sequences (16, 24). The number of N-glycosylation sequons for human-circulating H3N2 and H1N1 has increased over time and the pattern has changed (27). All of these findings suggest that amino acid mutations and increased glycosylation affect how HA interacts with immunity. Indeed, Bajic et al. showed that engineered hyperglycosylated HA restricted the resulting antibody repertoire to a subdominant epitope and that such antibodies protected against viral challenge (28). However, little is known about the types and structures of HA glycans and how these influence antibody responses.
Glycosylation Impacts IAV Antigenicity, Immunogenicity, and Immunodominance
As pointed out by Yewdell et al., among several respiratory RNA viruses with similar mutation rates and antigenic escape frequencies, only IAV undergoes genetic drift (29). Although the understanding of the reasons for IAV drift remains incomplete, the fact that immune responses in humans are focused on antigenic sites on the HA protein means that single point mutations have large impacts on viral escape.
Antigenicity refers to the capacity of a chemical structure (antigen) to bind antibodies or T cell receptors. Immunogenicity, by contrast, refers to the capacity of the antigen to induce an adaptive immune response. The immune system responds to complex antigens in a hierarchical manner, a concept known as immunodominance (30). Thus, immunodominant antigens may suppress immune responses to subdominant antigens. Broadly protective antibody responses appear to target subdominant conserved epitopes that have low variability due to the need to maintain viral function and display restricted gene usage (28).
Immune responses to viral antigens are governed by factors including antigen structure (concentration, conformation, and location), B cell immunodominance, and T-cell immunodominance. Aspects of T cell immunodominance have been reviewed (31). Briefly, because CD8+ T cells help clear viral infections, researchers have tried to develop vaccines that exploit these responses. CD8+ T cells recognize viral peptides processed by cellular proteasomes and presented by the major histocompatibility complex (MHC) class I molecules. Most T cell responses are generated against immunodominant viral peptides, which make up only a small fraction of the thousands of processed viral peptides. T Cell populations that recognize glycopeptides presented by MHC-I and MHC-II have been identified (32, 33), indicating the significance of glycosylation on acquired immune responses (34).
Immunodominance reflects many factors, including antigen presentation and T cell activation (35). Immunodominant antigens are recognized by large T cell populations, relative to those of subdominant antigens. This hierarchy is a reproducible pattern among individuals. Immunodominance largely results from the fact that only a small percentage of peptides bind MHC molecules with affinity enough for stable presentation to activate CD8+ T cells, a pattern that shows high evolutionary conservation (36). At present, the apparent inability of memory CD8+ T cells to protect against IAV has driven renewed focus on vaccines that elicit an antibody response (29).
Among IAV proteins, the order of antibody immunodominance is approximately HA > NA ⋙ nucleoprotein (36). It is therefore of interest that the number of HA (and NA) sequons increases during seasonal circulation in humans. In order to evaluate the influence of IAV glycosylation on binding of strain antibodies, researchers currently rely on genetic sequences to predict sequons. It is known that the number of sequons on the HA globular head domain increases with the amount of time the IAV sub-type circulates seasonally in humans (21, 24–26). Genetic information has been used to model HA glycosylation by inserting the generic N-glycosylation chitobiose core structure onto HA crystal structure coordinates (26, 37–39). However, information about the size and composition of glycans at individual sites and their impact on antigenic integrity is available for only a few strains.
The current understanding of the adaptive immune system derives from peptide antigens (40). Synthetic glycopeptides containing oligosaccharides generated reduced CD4+ T cell responses to IAV strains with glycosylation at the corresponding position (41). Avci et al. have described a mechanism whereby B cells take up glycoconjugates through a carbohydrate-recognizing B cell receptor and process the antigens in the endosome. The peptide portion of the resulting glycan-peptide is presented to carbohydrate-recognizing T cells, the stimulation of which results in a carbohydrate-specific adaptive immune response (40). There are only a few examples of natural glycopeptides inducing T cell responses (42). Nonetheless, the influence of site-specific glycosylation structure on recognition of IAV antigens by antigen presenting cells for subsequent T cell stimulation have not been defined clearly.
Comparing How Glycosylation Is Used to Evade the Human Immune System, in IAV and Human Immunodeficiency Virus 1 (HIV-1)
Although IAV and HIV-1 differ in their replication mechanisms, both exhibit sufficient antigenic variation in their surface proteins over time in the human population to evade the protection conferred by standard vaccine strategies. In both cases, the ability of the virus to evolve reduces the efficacy of vaccines. Glycosylation of the HIV-1 envelope protein trimer, consisting of gp120 and gp41, corresponds to about half its mass (43). The underlying protein is highly mutative and evolves constantly to evade host antibodies. The high density of N-glycosylation limits the accessibility of glycan biosynthetic processing enzymes, resulting in a shield of primarily high mannose N-glycans that facilitate viral escape by interfering with proteolytic processing of envelope peptides for presentation by the major histocompatibility complex (44, 45). Although broadly neutralizing antibodies to envelope protein have been identified that either tolerate the dense glycan shield or bind to epitopes that contain glycans, it has not been possible to formulate a vaccine that elicits such responses (46). By contrast, glycosylation of IAV HA appears to interfere with receptor binding and/or membrane fusion if too many sequons are occupied on the head domain (47–49).
The simplest mechanism whereby IAV escapes neutralizing antibodies of the adaptive immune system occurs through mutation(s) that diminish antibody binding affinity (50). In some cases, mutations can cause allosteric effects that decrease the antibody access to the epitope (51). Mutations that increase the avidity of HA for the host sialic acid receptor may cause IAV to bind host cells more avidly than competing antibodies (52, 53). Amino acid mutations may create new N-glycosylation sites (sequons) in the HA globular domain. These mutated sequences are synthesized, extruded into the ER lumen of infected cells, and modified by N-glycans, which can, as shown in Fig. 2, subsequently block antibody binding and therefore impact antigenicity (20, 38). Depending on their accessibilities, immature high mannose N-glycans may be trimmed by mannosidases and subsequently extended by galactosyltransferases to form complex-type N-glycans.
Fig. 2.
Mechanisms whereby hemagglutinin glycosylation influences IAV fitness (58).
Most IAV antigenicity studies do not account for changes in HA site-specific glycosylation because adding glycosylation lowers receptor binding avidity, thereby complicating interpretation of hemagglutination inhibition (HI) assays used in most serological analyses (38). The micro-neutralization (MN) assay is an alternative method that can overcome non-antigenic effects caused by changes in receptor binding affinity (54–56).
IAV viral fitness, the ability of the virus to propagate, is maintained by balancing receptor binding and membrane fusion with the release of new virions from the cell surface. A mutation that significantly increases receptor avidity may have a negative impact on fitness if virions cannot escape the cell surface. Thus, mutations that strengthen NA activity may help balance such increases in HA receptor binding avidity (57). Head group glycans can increase viral fitness by shielding HA residues from antibody binding and tuning receptor specificity (58). In addition, the subsequent appearance of new glycosylations sites can balance the increased receptor binding avidity of an amino acid mutation as a mechanism for maintaining fitness (58). Increased glycosylation may also compromise viral fitness by enabling the binding of HA by lectins of the innate immune system (59–62), or by negatively impacting assembly of stable HA trimers in the ER (16, 24, 38, 49, 63–65). Lectins, including surfactant protein D (SP-D) and mannose binding lectin (MBL), neutralize IAV by binding to glycosylated HA. Although these interactions depend on the glycan structures present at each glycosite, they cannot be predicted from HA sequence information alone.
Glycosylation and Antigenic Cartography of Influenza Viruses
Antigenic cartography (66) is used to assess the antigenic distance among HA molecules from different IAV strains (67–69). Antigenic distances are calculated from hemagglutination inhibition and microneutralization assays (69). Wan et al. developed a 3D antigenic cartography construction and visualization resource to study strain candidates for vaccines (68). Given its roles in shielding underlying protein sequences from antibody binding, glycosylation is likely to impact antigenic cartography of a given IAV strain. Expanded knowledge of site-specific glycosylation in different IAV strains, including the range of glycoforms present at each site, would enable the correlation between antigenic distance calculation and HA glycosylation. This would be a boon to efforts in predicting the pandemic potential of zoonotic viruses. It would also facilitate vaccine planning by improving the ability to predict whether a given seasonally circulating virus will likely escape vaccines.
Toward a Broadly Neutralizing IAV Vaccine
The major HA antigenic sites in the head domain show high rates of mutation, including the addition of new sequons (70). At the same time, the evolution of receptor binding sites and the stem domain is much more limited to conserve their functions (71–73). Antibody escape mutants occur in five major head domain antigenic clusters (50). Neutralizing antibodies appear to target regions proximal to the receptor binding site and mutations responsible for antigenic drift tend to occur within these proximal regions (74–78). This may be related to the accumulation of N-glycosylation on the head group that shield underlying antigenic sites (79). Such glycosylation will disrupt binding of sialic acid residues if it occurs too close to the receptor binding site, thus leaving an opening for neutralizing antibodies to bind.
In principle, broadly neutralizing antibodies can target the conserved sites of the receptor binding and stalk regions, respectively, which are present across different IAV strains (79). However, most human antibodies against HA (and NA) bind hypervariable residues, not those conserved among IAV strains. Efforts to generate broadly neutralizing antibodies have focused on the receptor binding site and the highly conserved stalk region (80), for which escape mutants would disrupt key viral functions and therefore have a high fitness cost. For this to work, however, it is necessary to direct the immune system away from the immunodominant variable residues toward subdominant residues that are conserved among strains.
Although most neutralizing antibodies target residues proximal to the receptor binding site, some appear to mimic the sialic acid receptor itself and bind to conserved residues, thus offering the potential for a broadly neutralizing response (78). Some researchers have pointed to the lack of accessibility of the stem region for the lack of broadly neutralizing antibodies from vaccines (81), whereas others have noted that the stem region of HA in virions should be accessible to antibody binding based on structural studies (82). Improved understanding of the dynamics of IAV immunodominance in human populations will be necessary in order to design vaccine strategies that succeed in generating antibodies against conserved epitopes that confer broadly neutralizing projection against IAV (83).
Structural analysis of HAs from pandemic and seasonal IAV indicates that while the HA fold is conserved, the surface properties and glycosylation patterns differ significantly among subtypes (80). A large-scale in vitro mutational analysis of the H1 and H3 HA receptor binding site identified many replication-competent mutations not yet observed in nature, indicating that the receptor binding site can accommodate much more sequence diversity than previously believed (84). These researchers noted that many deleterious single mutations were viable when present in combination with other substitutions, demonstrating epistatic effects in evolution of the HA receptor binding site. Natural mutations to the receptor binding site become part of a network of epistatic modifications that prevent reversion of individual substitutions (70). The recent decline in effectiveness of IAV vaccines has been attributed in part to HA substitutions that arise during virus growth in chicken eggs that reduce binding and neutralization by a receptor binding site broadly neutralizing antibody by orders of magnitude (85). This work highlighted the fact that much about the receptor binding site of HA remains unknown, despite decades of effort.
For H3N2, the mode of receptor binding has shifted as the virus has circulated since 1968. Thus, mutations that increased H3N2 sialic acid binding in early years after 1968 in H3N2 and subsequent strains are inhibitory more recently because of other substitutions in the receptor binding site (70). This suggests that many residues proximal to the receptor binding site coordinate the receptor binding behavior of HA. Further, after 2003, H3N2 preference moved to binding extended, branched N-glycans, indicating the ability of the virus to evolve to bind a subset of airway glycans as a way of maintaining fitness.
Proteomics of IAV
Mass spectrometric analysis of viral glycoproteins has been summarized in a recent review (86). Downard et al. developed an approach for using accurate mass measurement of proteolytic peptides of IAV proteins, referred to as proteotyping, to identify HA and NA from circulating IAV types and subtypes. The accurate mass values constitute signatures for conserved regions of IAV proteins that enable virus typing (87, 88). The investigators used this approach to differentiate seasonal strains from pandemic H1N1 (89–91) and study the evolution of H5N1 strains (92) and NA subtypes (93). They developed computer algorithms to identify virus reassortants from whole virus digests (94). FluShuffle considers combination of viral protein identities that match the mass spectral data using Gibbs sampling. FluResort uses those identities to calculate the weighted distance of each across two or more phylogentic trees through viral protein sequence alignment. As an extension to this approach, the FluClass algorithm performs phylogenetic classification using MS data starting from DNA- or protein-based phylogenetic trees (95). The MassTree algorithm identifies and displays protein mutations and calculates mutational frequencies across phylogenetic trees for studies of IAV evolution (96–98).
IAV Glycoproteomics
As reviewed (99), mass spectrometry has been used in proteomics studies of IAV proteins and in mass profiling of tryptic peptides and glycopeptides. An early pioneering study characterized N-glycosylation on three IAV strains (100). An LC-MS method has been used to characterize glycoforms at specific sites using alternating high and low collision energy values (a data-independent acquisition experiment known as MSE) combined with multiple reaction monitoring assays as a means of comparing recombinant HA samples as vaccine candidates (14, 101). The investigators who developed the MSE method also used this approach to analyze site-specific glycosylation in a series of engineered H3N2 HA variants with added sequons that mirror those that appeared during seasonal circulation since 1968 (49, 102). They also characterized HA glycosylation in a series of engineered H5N7 as part of an effort to define glycosylation structure-function relationships in this avian IAV strain (103). In additional work, they also examined glycosylation in a set of reference HA antigens used in influenza vaccine potency testing (104). We have used site-specific glycosylation information to model interactions between HA and surfactant protein-D (60, 102, 105–107).
MS Workflows for Assigning Glycopeptides
Confident assignment of glycopeptides requires building a search space consisting of the glycosylation variants of a measured proteome. For IAV, the hypervariable proteome (108), combined with the presence of host proteins in the viral architecture (1), require special consideration when applying proteomics methods. In addition, other post-translational modifications, including phosphorylation, have been observed on IAV proteins (109). Because the proteomics search space must include both IAV and host proteins, the rate of false positive identifications becomes unacceptably high if random mutations are allowed. Considering this, we recommend defining a range of allowable mutations in a custom database that contains sequences for a given IAV strain plus host proteome.
Mass Spectrometry Methods for Assigning Site-specific Glycosylation
For detailed glycoproteomics reviews, see (110–116). As shown in Fig. 3, glycopeptide glycoforms elute from a reversed phase chromatography column over a narrow retention time window. Identification of site-specific glycosylation requires tailored analytical and bioinformatics methods. Proteomics workflows identify and quantify proteins based on prediction of peptide tandem mass spectra from genomic databases. Although small PTMs have single predictable mass shifts, glycosylation at a given site is heterogeneous, pushing confident site-specific assignment of glycosylation beyond the scope of conventional proteomics workflows. In order to assign site-specific glycosylation, one must generate an appropriate list of theoretical glycan and peptide compositions (known as the search space) and use this list to assign the most probable glycopeptide composition (117). The number of possible glycoforms at each glycosylation site multiplies the size of the search space and the difficulty in making confident assignments. Assumptions made about the purity or complexity of the sample can greatly affect the quality and confidence of the results. Using a too small search space by assuming incorrectly that a glycoprotein sample is pure may lead to unacceptably high numbers of contaminant glycopeptides incorrectly assigned to the target glycoprotein. Overestimating the size of the search space by including too many glycoproteins and glycoforms leads to decreasing ability to assign glycopeptides with acceptable confidence.
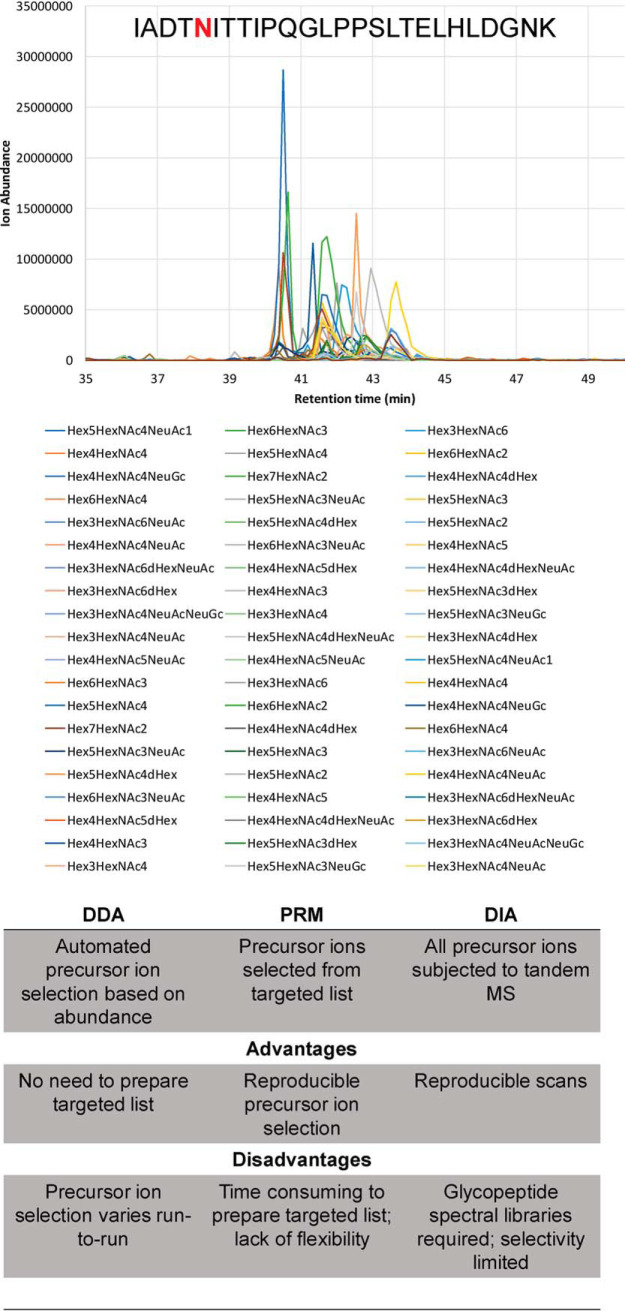
Fig. 3.
Comparison of acquisition methods for tandem MS of glycopeptides. Extracted ion chromatograms for glycopeptide IADTNITTIPQGLPPSLTELHLDGNK glycoforms are shown, illustrating that a large number of glycoforms elute over a narrow retention time range using reversed phase chromatography LC-MS as described (128). Automated precursor ion selection using data dependent acquisition, targeted precursor ion selection using parallel reaction monitoring (PRM), and data independent acquisition are compared.
The ability to distinguish HA glycosylation among different IAV strains or expression platforms with statistical significance depends on the quality of the mass spectral data. Typical top-N data-dependent acquisition (DDA) mass spectrometry methods, based on selection of the most abundant precursor ions, give rise to missing values that negatively impact the ability to quantify glycopeptides. In addition, glycosylation variants of a given peptide elute over a narrow retention time window using LC-MS. For this reason, DDA samples only the most abundant glycoforms. The under-sampling and reproducibility problems resulting from the stochastic nature of top-N DDA result in the inability to quantify a given precursor ion in a significant subset of a biological sample cohort. These missing values, the occurrence of which increases as the glycopeptide abundance decreases, necessitate the imputation of glycopeptide abundances, a step that reduces the resulting statistical rigor. The use of targeted quantification (118) solves the missing value problem but limits the number of precursor ions for which tandem mass spectra are available. This suggests the use of the MS1 dimension of a DDA or targeted quantification experiment to quantify glycopeptides. Thus, it would be important to develop metrics for assigning confidence of glycopeptides for which tandem mass spectra were not collected.
Data-independent analysis (DIA) eliminates the need to isolate precursor ions if peptides are chromatographically well resolved. In sequential window acquisition of all theoretical fragment ion spectra (SWATH)-MS DIA (119), for example, fragment ion spectra for all precursors are acquired within the specified m/z range and retention time window. However, there are limitations to DIA for the analysis of glycopeptides. First, one cannot construct a rigorous spectral library because of the lack of a comprehensive collection of synthetic glycopeptides. In addition, the glycopeptide oxonium ions and residue losses from the precursor ion do not provide peptide sequence information because these losses occur in all glycopeptides, irrespective of their glycan compositions. In order to be effective, DIA methods must consider the narrow retention time window over which glycopeptide glycoforms elute in typical reversed phase gradients. Further, if a large precursor window is fragmented, there may be problems determining which precursor peak produced the glycopeptide fragments. Despite these limitations, investigators have used low collision energy settings to produce Y-type ions for identification and quantification of IgG glycopeptides in a complex matrix of human plasma (120–122). A SWATH DIA method was used to quantify high mannose N-glycopeptides from yeast using manually created glycopeptide libraries (123, 124). Researchers developed a DIA strategy to quantify 25 N-glycopeptides from plasma using a search space of 161 glycoforms for a study of liver cirrhosis (121). Others have used DIA to produce comprehensive glycosylation maps of human serum IgM using extracted ion chromatograms of shared peptide-specific fragment ions to filter related glycoforms for a given glycosite (125). This approach allowed identification of glycopeptides with unexpected modifications. A targeted DIA method identified N-glycopeptides without predefined glycan compositions from 59 N-glycosylation sites from 41 glycoproteins, including 21 IgG glycopeptide glycoforms, from HILIC-enriched human blood plasma tryptic digest (126). Because of the statistical limitations of DDA, therefore, a targeted DIA approach may be necessary to produce a comprehensive catalogue of all HA glycoforms with sufficient confidence.
CONCLUSIONS
Because of the low efficacy of existing seasonal IAV vaccines, we need a paradigm change in IAV vaccine design. IAV strains are continually mutating under immune pressure, necessitating selection and manufacture of new vaccines every year. To mitigate the disease burden caused by IAV, we need to develop a broadly neutralizing vaccine capable of protecting against multiple strains. IAV researchers currently use genetics and structural biology studies in determining IAV antigenicity and immunogenicity, but as of today, glycosylation state of HA is largely ignored.
In the HIV vaccine field, researchers have moved toward mimicking the glycan shield in order to generate broadly neutralizing antibodies analogous to those that develop naturally in a subset of infected individuals. For IAV, glycosylation does not go to the extreme of forming a sterically restricted shield. There is a strong argument for the need to understand the influence of IAV glycosylation on transport to the lymph nodes and innate immune lectin mediated recognition and processing by dendritic cells and B cells toward generating an effective antibody response.
The technology to correlate glycopopulations of vaccine constructs with breadth of antibody response already exists. Exploitation of the knowledge of the most appropriate glycopopulations of HA, as well as the viral expression systems that can produce them, will mark a major step toward developing more effective IAV vaccines.
Footnotes
* This work is supported by NIH grant U01CA221234 and the Graduate Medical Sciences program at Boston University School of Medicine. No author has an actual or perceived conflict of interest with the contents of this article.
1 The abbreviations used are:
- IAV
- influenza A virus
- HA
- hemagglutinin
- NA
- neuraminidase
- NIAID
- National Institute of Allergy and Infectious Diseases
- MHC
- major histocompatibility complex.
REFERENCES
- 1. Hutchinson, E. C., Charles, P. D., Hester, S. S., Thomas, B., Trudgian, D., Martinez-Alonso, M., and Fodor, E. (2014) Conserved and host-specific features of influenza virion architecture. Nat. Commun. 5, 5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tong, S., Zhu, X., Li, Y., Shi, M., Zhang, J., Bourgeois, M., Yang, H., Chen, X., Recuenco, S., Gomez, J., Chen, L. M., Johnson, A., Tao, Y., Dreyfus, C., Yu, W., McBride, R., Carney, P. J., Gilbert, A. T., Chang, J., Guo, Z., Davis, C. T., Paulson, J. C., Stevens, J., Rupprecht, C. E., Holmes, E. C., Wilson, I. A., and Donis, R. O. (2013) New world bats harbor diverse influenza A viruses. PLoS Pathog. 9, e1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Paules, C., and Subbarao, K. (2017) Influenza. Lancet 390, 697–708 [DOI] [PubMed] [Google Scholar]
- 4. Behrens, G., and Stoll, M. (2006) Pathogenesis and Immunology. in Influenza Report (Kamps, B. S., Hoffman, C., and Preiser, W. eds.) pp 92–109, Flying Publisher, Paris [Google Scholar]
- 5. Houser, K., and Subbarao, K. (2015) Influenza vaccines: challenges and solutions. Cell Host Microbe 17, 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Centers for Disease Control and Prevention. Seasonal influenza vaccine effectiveness, 2005–2018. https://www.cdc.gov/flu/professionals/vaccination/effectiveness-studies.htm.
- 7. Erbelding, E. J., Post, D. J., Stemmy, E. J., Roberts, P. C., Augustine, A. D., Ferguson, S., Paules, C. I., Graham, B. S., and Fauci, A. S. (2018) A Universal Influenza Vaccine: The Strategic Plan for the National Institute of Allergy and Infectious Diseases. J. Infectious Dis. 218, 347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwarzer, J., Rapp, E., Hennig, R., Genzel, Y., Jordan, I., Sandig, V., and Reichl, U. (2009) Glycan analysis in cell culture-based influenza vaccine production: Influence of host cell line and virus strain on the glycosylation pattern of viral hemagglutinin. Vaccine 27, 4325–4336 [DOI] [PubMed] [Google Scholar]
- 9. Schild, G. C., Oxford, J. S., de Jong, J. C., and Webster, R. G. (1983) Evidence for host-cell selection of influenza virus antigenic variants. Nature 303, 706–709 [DOI] [PubMed] [Google Scholar]
- 10. Robertson, J. S., Bootman, J. S., Newman, R., Oxford, J. S., Daniels, R. S., Webster, R. G., and Schild, G. C. (1987) Structural changes in the haemagglutinin which accompany egg adaptation of an influenza A(H1N1) virus. Virology 160, 31–37 [DOI] [PubMed] [Google Scholar]
- 11. Katz, J. M., and Webster, R. G. (1988) Antigenic and structural characterization of multiple subpopulations of H3N2 influenza virus from an individual. Virology 165, 446–456 [DOI] [PubMed] [Google Scholar]
- 12. Wood, J. M., Oxford, J. S., Dunleavy, U., Newman, R. W., Major, D., and Robertson, J. S. (1989) Influenza A (H1N1) vaccine efficacy in animal models is influenced by two amino acid substitutions in the hemagglutinin molecule. Virology 171, 214–221 [DOI] [PubMed] [Google Scholar]
- 13. de Vries, R. P., Smit, C. H., de Bruin, E., Rigter, A., de Vries, E., Cornelissen, L. A., Eggink, D., Chung, N. P., Moore, J. P., Sanders, R. W., Hokke, C. H., Koopmans, M., Rottier, P. J., and de Haan, C. A. (2012) Glycan-dependent immunogenicity of recombinant soluble trimeric hemagglutinin. J. Virol. 86, 11735–11744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xie, H., Doneanu, C., Chen, W., Rininger, J., and Mazzeo, J. R. (2011) Characterization of a recombinant influenza vaccine candidate using complementary LC-MS methods. Curr Pharm. Biotechnol. 12, 1568–1579 [DOI] [PubMed] [Google Scholar]
- 15. Zhang, S., Sherwood, R. W., Yang, Y., Fish, T., Chen, W., McCardle, J. A., Jones, R. M., Yusibov, V., May, E. R., Rose, J. K., and Thannhauser, T. W. (2012) Comparative characterization of the glycosylation profiles of an influenza hemagglutinin produced in plant and insect hosts. Proteomics 12, 1269–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Das, S. R., Puigbo, P., Hensley, S. E., Hurt, D. E., Bennink, J. R., and Yewdell, J. W. (2010) Glycosylation focuses sequence variation in the influenza A virus H1 hemagglutinin globular domain. PLoS Pathog. 6, e1001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hebert, D. N., Foellmer, B., and Helenius, A. (1996) Calnexin and calreticulin promote folding, delay oligomerization and suppress degradation of influenza hemagglutinin in microsomes. EMBO J. 15, 2961–2968 [PMC free article] [PubMed] [Google Scholar]
- 18. Hebert, D. N., Zhang, J. X., Chen, W., Foellmer, B., and Helenius, A. (1997) The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J. Cell Biol. 139, 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gill, J. R., Sheng, Z. M., Ely, S. F., Guinee, D. G., Beasley, M. B., Suh, J., Deshpande, C., Mollura, D. J., Morens, D. M., Bray, M., Travis, W. D., and Taubenberger, J. K. (2010) Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch. Pathol. Lab. Med. 134, 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Skehel, J. J., Stevens, D. J., Daniels, R. S., Douglas, A. R., Knossow, M., Wilson, I. A., and Wiley, D. C. (1984) A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc. Natl. Acad. Sci. U.S.A. 81, 1779–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vigerust, D. J., and Shepherd, V. L. (2007) Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 15, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hensley, S. E., Das, S. R., Bailey, A. L., Schmidt, L. M., Hickman, H. D., Jayaraman, A., Viswanathan, K., Raman, R., Sasisekharan, R., Bennink, J. R., and Yewdell, J. W. (2009) Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 326, 734–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wanzeck, K., Boyd, K. L., and McCullers, J. A. (2011) Glycan shielding of the influenza virus hemagglutinin contributes to immunopathology in mice. Am. J. Respir. Crit. Care Med. 183, 767–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cherry, J. L., Lipman, D. J., Nikolskaya, A., and Wolf, Y. I. (2009) Evolutionary dynamics of N-glycosylation sites of influenza virus hemagglutinin. PLoS Curr. 1, RRN1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun, S., Wang, Q., Zhao, F., Chen, W., and Li, Z. (2011) Glycosylation site alteration in the evolution of influenza A (H1N1) viruses. PLoS ONE 6, e22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wei, C. J., Boyington, J. C., Dai, K., Houser, K. V., Pearce, M. B., Kong, W. P., Yang, Z. Y., Tumpey, T. M., and Nabel, G. J. (2010) Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci. Transl. Med. 2, 24ra21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tate, M. D., Brooks, A. G., and Reading, P. C. (2011) Specific sites of N-linked glycosylation on the hemagglutinin of H1N1 subtype influenza A virus determine sensitivity to inhibitors of the innate immune system and virulence in mice. J. Immunol. 187, 1884–1894 [DOI] [PubMed] [Google Scholar]
- 28. Petrova, V. N., and Russell, C. A. (2018) The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 16, 47–60 [DOI] [PubMed] [Google Scholar]
- 29. Altman, M. O., Angeletti, D., and Yewdell, J. W. (2018) Antibody immunodominance: the key to understanding influenza virus antigenic drift. Viral Immunol. 31, 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wrammert, J., Smith, K., Miller, J., Langley, W. A., Kokko, K., Larsen, C., Zheng, N. Y., Mays, I., Garman, L., Helms, C., James, J., Air, G. M., Capra, J. D., Ahmed, R., and Wilson, P. C. (2008) Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453, 667–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yewdell, J. W. (2006) Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity 25, 533–543 [DOI] [PubMed] [Google Scholar]
- 32. Dengjel, J., and Stevanovic, S. (2006) Naturally presented MHC ligands carrying glycans. Transfus. Med. Hemoth. 33, 38–44 [Google Scholar]
- 33. Dengjel, J., Rammensee, H. G., and Stevanovic, S. (2005) Glycan side chains on naturally presented MHC class II ligands. J. Mass Spectrom. 40, 100–104 [DOI] [PubMed] [Google Scholar]
- 34. Wolfert, M. A., and Boons, G. J. (2013) Adaptive immune activation: glycosylation does matter. Nat. Chem. Biol. 9, 776–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yewdell, J. W., and Bennink, J. R. (1999) Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 17, 51–88 [DOI] [PubMed] [Google Scholar]
- 36. Altman, M. O., Bennink, J. R., Yewdell, J. W., and Herrin, B. R. (2015) Lamprey VLRB response to influenza virus supports universal rules of immunogenicity and antigenicity. eLife 4, e07467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wilson, I. A., Ladner, R. C., Skehel, J. J., and Wiley, D. C. (1983) The structure and role of the carbohydrate moieties of influenza virus haemagglutinin. Biochem. Soc. Trans. 11, 145–147 [PubMed] [Google Scholar]
- 38. Das, S. R., Hensley, S. E., David, A., Schmidt, L., Gibbs, J. S., Puigbo, P., Ince, W. L., Bennink, J. R., and Yewdell, J. W. (2011) Fitness costs limit influenza A virus hemagglutinin glycosylation as an immune evasion strategy. Proc. Natl. Acad. Sci. U.S.A. 108, E1417–E1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tharakaraman, K., Raman, R., Stebbins, N. W., Viswanathan, K., Sasisekharan, V., and Sasisekharan, R. (2013) Antigenically intact hemagglutinin in circulating avian and swine influenza viruses and potential for H3N2 pandemic. Sci. Rep. 3, 1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Avci, F. Y., Li, X., Tsuji, M., and Kasper, D. L. (2013) Carbohydrates and T cells: a sweet twosome. Semin. Immunol. 25, 146–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jackson, D. C., Drummer, H. E., Urge, L., Otvos, L., and Brown, L. E. (1994) Glycosylation of a synthetic peptide representing a T-cell determinant of influenza virus hemagglutinin results in loss of recognition by CD4+ T-cell clones. Virology 199, 422–430 [DOI] [PubMed] [Google Scholar]
- 42. Sun, L., Middleton, D. R., Wantuch, P. L., Ozdilek, A., and Avci, F. Y. (2016) Carbohydrates as T-cell antigens with implications in health and disease. Glycobiology 26, 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lasky, L. A., Groopman, J. E., Fennie, C. W., Benz, P. M., Capon, D. J., Dowbenko, D. J., Nakamura, G. R., Nunes, W. M., Renz, M. E., and Berman, P. W. (1986) Neutralization of the AIDS retrovirus by antibodies to a recombinant envelope glycoprotein. Science 233, 209–212 [DOI] [PubMed] [Google Scholar]
- 44. Doe, B., Steimer, K. S., and Walker, C. M. (1994) Induction of HIV-1 envelope (gp120)-specific cytotoxic T lymphocyte responses in mice by recombinant CHO cell-derived gp120 is enhanced by enzymatic removal of N-linked glycans. Eur. J. Immunol. 24, 2369–2376 [DOI] [PubMed] [Google Scholar]
- 45. Li, H., Xu, C. F., Blais, S., Wan, Q., Zhang, H. T., Landry, S. J., and Hioe, C. E. (2009) Proximal glycans outside of the epitopes regulate the presentation of HIV-1 envelope gp120 helper epitopes. J. Immunol. 182, 6369–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Behrens, A. J., and Crispin, M. (2017) Structural principles controlling HIV envelope glycosylation. Curr. Opin. Struct. Biol. 44, 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vigerust, D. J., Ulett, K. B., Boyd, K. L., Madsen, J., Hawgood, S., and McCullers, J. A. (2007) N-linked glycosylation attenuates H3N2 influenza viruses. J. Virol. 81, 8593–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reichert, T., Chowell, G., Nishiura, H., Christensen, R. A., and McCullers, J. A. (2010) Does glycosylation as a modifier of original antigenic sin explain the case age distribution and unusual toxicity in pandemic novel H1N1 influenza? BMC Infect. Dis. 10, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Alymova, I. V., York, I. A., Air, G. M., Cipollo, J. F., Gulati, S., Baranovich, T., Kumar, A., Zeng, H., Gansebom, S., and McCullers, J. A. (2016) Glycosylation changes in the globular head of H3N2 influenza hemagglutinin modulate receptor binding without affecting virus virulence. Sci. Rep. 6, 36216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Caton, A. J., Brownlee, G. G., Yewdell, J. W., and Gerhard, W. (1982) The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31, 417–427 [DOI] [PubMed] [Google Scholar]
- 51. Yewdell, J. W., Taylor, A., Yellen, A., Caton, A., Gerhard, W., and Bachi, T. (1993) Mutations in or near the fusion peptide of the influenza virus hemagglutinin affect an antigenic site in the globular region. J. Virol. 67, 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yewdell, J. W., Caton, A. J., and Gerhard, W. (1986) Selection of influenza A virus adsorptive mutants by growth in the presence of a mixture of monoclonal antihemagglutinin antibodies. J. Virol. 57, 623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Temoltzin-Palacios, F., and Thomas, D. B. (1994) Modulation of immunodominant sites in influenza hemagglutinin compromise antigenic variation and select receptor-binding variant viruses. J. Exp. Med. 179, 1719–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Allen, J. D., and Ross, T. M. (2018) H3N2 influenza viruses in humans: Viral mechanisms, evolution, and evaluation. Hum. Vaccin. Immunother. 14, 1840–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Blackburne, B. P., Hay, A. J., and Goldstein, R. A. (2008) Changing selective pressure during antigenic changes in human influenza H3. PLoS Pathog. 4, e1000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sun, X., Jayaraman, A., Maniprasad, P., Raman, R., Houser, K. V., Pappas, C., Zeng, H., Sasisekharan, R., Katz, J. M., and Tumpey, T. M. (2013) N-linked glycosylation of the hemagglutinin protein influences virulence and antigenicity of the 1918 pandemic and seasonal H1N1 influenza A viruses. J. Virol. 87, 8756–8766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gaymard, A., Le Briand, N., Frobert, E., Lina, B., and Escuret, V. (2016) Functional balance between neuraminidase and haemagglutinin in influenza viruses. Clin. Microbiol. Infect. 22, 975–983 [DOI] [PubMed] [Google Scholar]
- 58. Kosik, I., Ince, W. L., Gentles, L. E., Oler, A. J., Kosikova, M., Angel, M., Magadán, J. G., Xie, H., Brooke, C. B., and Yewdell, J. W. (2018) Influenza A virus hemagglutinin glycosylation compensates for antibody escape fitness costs. PLoS Path. 14, e1006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. White, M. R., Boland, P., Tecle, T., Gantz, D., Sorenson, G., Tornoe, I., Holmskov, U., McDonald, B., Crouch, E. C., and Hartshorn, K. L. (2010) Enhancement of antiviral activity of collectin trimers through cross-linking and mutagenesis of the carbohydrate recognition domain. J. Innate Immun. 2, 267–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Crouch, E., Nikolaidis, N., McCormack, F., McDonald, B., Allen, K., Rynkiewicz, M., Cafarella, T., White, M., Lewnard, K., Leymarie, N., Zaia, J., Seaton, B., and Hartshorn, K. (2011) Mutagenesis of SP-D informed by evolution and xray crystallography enhances defenses against Influenza A Virus in vivo. J. Biol. Chem. 286, 40681–40692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Verma, A., White, M., Vathipadiekal, V., Tripathi, S., Mbianda, J., Ieong, M., Qi, L., Taubenberger, J. K., Takahashi, K., Jensenius, J. C., Thiel, S., and Hartshorn, K. L. (2012) Human H-ficolin inhibits replication of seasonal and pandemic influenza A viruses. J. Immunol. 189, 2478–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chang, W. C., White, M. R., Moyo, P., McClear, S., Thiel, S., Hartshorn, K. L., and Takahashi, K. (2010) Lack of the pattern recognition molecule mannose-binding lectin increases susceptibility to influenza A virus infection. BMC Immunol. 11, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schulze, I. T. (1997) Effects of glycosylation on the properties and functions of influenza virus hemagglutinin. J. Infect. Dis. 176, S24–S28 [DOI] [PubMed] [Google Scholar]
- 64. Chandrasekaran, A., Srinivasan, A., Raman, R., Viswanathan, K., Raguram, S., Tumpey, T. M., Sasisekharan, V., and Sasisekharan, R. (2008) Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat. Biotechnol. 26, 107–113 [DOI] [PubMed] [Google Scholar]
- 65. Tate, M. D., Job, E. R., Deng, Y. M., Gunalan, V., Maurer-Stroh, S., and Reading, P. C. (2014) Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 6, 1294–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Smith, D. J., Lapedes, A. S., de Jong, J. C., Bestebroer, T. M., Rimmelzwaan, G. F., Osterhaus, A. D. M. E., and Fouchier, R. A. M. (2004) Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376 [DOI] [PubMed] [Google Scholar]
- 67. Cai, Z., Zhang, T., and Wan, X. F. (2012) Antigenic distance measurements for seasonal influenza vaccine selection. Vaccine 30, 448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Barnett, J. L., Yang, J., Cai, Z., Zhang, T., and Wan, X. F. (2012) AntigenMap 3D: an online antigenic cartography resource. Bioinformatics 28, 1292–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cai, Z., Zhang, T., and Wan, X. F. (2011) Concepts and applications for influenza antigenic cartography. Influenza Other Respir. Viruses 5, 204–207 [PMC free article] [PubMed] [Google Scholar]
- 70. Wu, N. C., Thompson, A. J., Xie, J., Lin, C. W., Nycholat, C. M., Zhu, X., Lerner, R. A., Paulson, J. C., and Wilson, I. A. (2018) A complex epistatic network limits the mutational reversibility in the influenza hemagglutinin receptor-binding site. Nat. Commun. 9, 1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nobusawa, E., Aoyama, T., Kato, H., Suzuki, Y., Tateno, Y., and Nakajima, K. (1991) Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology 182, 475–485 [DOI] [PubMed] [Google Scholar]
- 72. Thyagarajan, B., and Bloom, J. D. (2014) The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin. eLife 3, e03300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wu, N. C., Young, A. P., Al-Mawsawi, L. Q., Olson, C. A., Feng, J., Qi, H., Chen, S. H., Lu, I. H., Lin, C. Y., Chin, R. G., Luan, H. H., Nguyen, N., Nelson, S. F., Li, X., Wu, T. T., and Sun, R. (2014) High-throughput profiling of influenza A virus hemagglutinin gene at single-nucleotide resolution. Sci. Rep. 4, 4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bush, R. M., Bender, C. A., Subbarao, K., Cox, N. J., and Fitch, W. M. (1999) Predicting the evolution of human influenza A. Science 286, 1921–1925 [DOI] [PubMed] [Google Scholar]
- 75. Wang, X., Ilyushina, N. A., Lugovtsev, V. Y., Bovin, N. V., Couzens, L. K., Gao, J., Donnelly, R. P., Eichelberger, M. C., and Wan, H. (2017) Amino acids in hemagglutinin antigenic site B determine antigenic and receptor binding differences between A(H3N2)v and ancestral seasonal H3N2 influenza viruses. J. Virol. 91, pii: e01512–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Popova, L., Smith, K., West, A. H., Wilson, P. C., James, J. A., Thompson, L. F., and Air, G. M. (2012) Immunodominance of antigenic site B over site A of hemagglutinin of recent H3N2 influenza viruses. PLoS ONE 7, e41895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Koel, B. F., Burke, D. F., Bestebroer, T. M., van der Vliet, S., Zondag, G. C., Vervaet, G., Skepner, E., Lewis, N. S., Spronken, M. I., Russell, C. A., Eropkin, M. Y., Hurt, A. C., Barr, I. G., de Jong, J. C., Rimmelzwaan, G. F., Osterhaus, A. D., Fouchier, R. A., and Smith, D. J. (2013) Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342, 976–979 [DOI] [PubMed] [Google Scholar]
- 78. Whittle, J. R., Zhang, R., Khurana, S., King, L. R., Manischewitz, J., Golding, H., Dormitzer, P. R., Haynes, B. F., Walter, E. B., Moody, M. A., Kepler, T. B., Liao, H. X., and Harrison, S. C. (2011) Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U.S.A. 108, 14216–14221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wu, N. C., and Wilson, I. A. (2017) A perspective on the structural and functional constraints for immune evasion: insights from influenza virus. J. Mol. Biol. 429, 2694–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wu, N. C., and Wilson, I. A. (2018) Structural insights into the design of novel anti-influenza therapies. Nat. Struct. Mol. Biol. 25, 115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Andrews, S. F., Huang, Y., Kaur, K., Popova, L. I., Ho, I. Y., Pauli, N. T., Henry Dunand, C. J., Taylor, W. M., Lim, S., Huang, M., Qu, X., Lee, J. H., Salgado-Ferrer, M., Krammer, F., Palese, P., Wrammert, J., Ahmed, R., and Wilson, P. C. (2015) Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 7, 316ra192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Harris, A. K., Meyerson, J. R., Matsuoka, Y., Kuybeda, O., Moran, A., Bliss, D., Das, S. R., Yewdell, J. W., Sapiro, G., Subbarao, K., and Subramaniam, S. (2013) Structure and accessibility of HA trimers on intact 2009 H1N1 pandemic influenza virus to stem region-specific neutralizing antibodies. Proc. Natl. Acad. Sci. U.S.A. 110, 4592–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zost, S. J., Wu, N. C., Hensley, S. E., and Wilson, I. A. (2019) Immunodominance and antigenic variation of influenza virus hemagglutinin: implications for design of universal vaccine immunogens. J. Infectious Dis. 219, S38–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wu, N. C., Xie, J., Zheng, T., Nycholat, C. M., Grande, G., Paulson, J. C., Lerner, R. A., and Wilson, I. A. (2017) Diversity of functionally permissive sequences in the receptor-binding site of influenza hemagglutinin. Cell Host Microbe 21, 742–753.e748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wu, N. C., Zost, S. J., Thompson, A. J., Oyen, D., Nycholat, C. M., McBride, R., Paulson, J. C., Hensley, S. E., and Wilson, I. A. (2017) A structural explanation for the low effectiveness of the seasonal influenza H3N2 vaccine. PLoS Pathog. 13, e1006682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Harvey, D. J. (2018) Mass spectrometric analysis of glycosylated viral proteins. Expert Rev. Proteomics 15, 391–412 [DOI] [PubMed] [Google Scholar]
- 87. Schwahn, A. B., Wong, J. W., and Downard, K. M. (2009) Subtyping of the influenza virus by high resolution mass spectrometry. Anal. Chem. 81, 3500–3506 [DOI] [PubMed] [Google Scholar]
- 88. Schwahn, A. B., and Downard, K. M. (2009) Antigenicity of a type A influenza virus through comparison of hemagglutination inhibition and mass spectrometry immunoassays. J. Immunoassay Immunochem. 30, 245–261 [DOI] [PubMed] [Google Scholar]
- 89. Schwahn, A. B., Wong, J. W., and Downard, K. M. (2010) Rapid differentiation of seasonal and pandemic H1N1 influenza through proteotyping of viral neuraminidase with mass spectrometry. Anal. Chem. 82, 4584–4590 [DOI] [PubMed] [Google Scholar]
- 90. Ha, J. W., Schwahn, A. B., and Downard, K. M. (2011) Proteotyping to establish gene origin within reassortant influenza viruses. PLoS ONE 6, e15771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Fernandes, N. D., and Downard, K. M. (2014) Origins of the reassortant 2009 pandemic influenza virus through proteotyping with mass spectrometry. J. Mass Spectrom. 49, 93–102 [DOI] [PubMed] [Google Scholar]
- 92. Ha, J. W., and Downard, K. M. (2011) Evolution of H5N1 influenza virus through proteotyping of hemagglutinin with high resolution mass spectrometry. Analyst 136, 3259–3267 [DOI] [PubMed] [Google Scholar]
- 93. Nguyen, A. P., and Downard, K. M. (2013) Subtyping of influenza neuraminidase using mass spectrometry. Analyst 138, 1787–1793 [DOI] [PubMed] [Google Scholar]
- 94. Lun, A. T., Wong, J. W., and Downard, K. M. (2012) FluShuffle and FluResort: new algorithms to identify reassorted strains of the influenza virus by mass spectrometry. BMC Bioinformatics 13, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ma, S., Downard, K. M., and Wong, J. W. (2015) FluClass: A novel algorithm and approach to score and visualize the phylogeny of the influenza virus using mass spectrometry. Anal. Chim. Acta 895, 54–61 [DOI] [PubMed] [Google Scholar]
- 96. Ma, S., Downard, K. M., and Wong, J. W. (2017) Phylogenetic Analysis Using Protein Mass Spectrometry. Methods Mol. Biol. 1549, 135–146 [DOI] [PubMed] [Google Scholar]
- 97. Akand, E. H., and Downard, K. M. (2017) Mutational analysis employing a phylogenetic mass tree approach in a study of the evolution of the influenza virus. Mol. Phylogenet. Evol. 112, 209–217 [DOI] [PubMed] [Google Scholar]
- 98. Akand, E. H., and Downard, K. M. (2018) Ancestral and compensatory mutations that promote antiviral resistance in influenza N1 neuraminidase revealed by a phylonumerics approach. J. Mol. Evol. 86, 546–553 [DOI] [PubMed] [Google Scholar]
- 99. Downard, K. M., Morrissey, B., and Schwahn, A. B. (2009) Mass spectrometry analysis of the influenza virus. Mass Spectrom. Rev. 28, 35–49 [DOI] [PubMed] [Google Scholar]
- 100. Mir-Shekari, S. Y., Ashford, D. A., Harvey, D. J., Dwek, R. A., and Schulze, I. T. (1997) The glycosylation of the influenza A virus hemagglutinin by mammalian cells. A site-specific study. J. Biol. Chem. 272, 4027–4036 [DOI] [PubMed] [Google Scholar]
- 101. An, Y., Rininger, J. A., Jarvis, D. L., Jing, X., Ye, Z., Aumiller, J. J., Eichelberger, M., and Cipollo, J. F. (2013) Comparative glycomics analysis of influenza Hemagglutinin (H5N1) produced in vaccine relevant cell platforms. J. Proteome Res. 12, 3707–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. An, Y., McCullers, J. A., Alymova, I., Parsons, L. M., and Cipollo, J. F. (2015) Glycosylation analysis of engineered H3N2 influenza A virus hemagglutinins with sequentially added historically relevant glycosylation sites. J. Proteome Res. 14, 3957–3969 [DOI] [PubMed] [Google Scholar]
- 103. Parsons, L. M., An, Y., de Vries, R. P., de Haan, C. A., and Cipollo, J. F. (2017) Glycosylation characterization of an influenza H5N7 hemagglutinin series with engineered glycosylation patterns: implications for structure-function relationships. J. Proteome Res. 16, 398–412 [DOI] [PubMed] [Google Scholar]
- 104. An, Y., Parsons, L. M., Jankowska, E., Melnyk, D., Joshi, M., and Cipollo, J. F. (2019) N-Glycosylation of Seasonal Influenza Vaccine Hemagglutinins: Implication for potency testing and immune processing. J. Virol. 92, e01693–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. van Eijk, M., Rynkiewicz, M. J., Khatri, K., Leymarie, N., Zaia, J., White, M. R., Hartshorn, K. L., Cafarella, T. R., van Die, I., Hessing, M., Seaton, B. A., and Haagsman, H. P. (2018) Lectin-mediated binding and sialoglycans of porcine surfactant protein D synergistically neutralize influenza A virus. J. Biol. Chem. 293, 10646–10662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Khatri, K., Klein, J. A., White, M. R., Grant, O. C., Leymarie, N., Woods, R. J., Hartshorn, K. L., and Zaia, J. (2016) Integrated Omics and Computational Glycobiology Reveal Structural Basis for Influenza A Virus Glycan Microheterogeneity and Host Interactions. Mol. Cell. Proteomics 15, 1895–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Cruz, E., Cain, J., Crossett, B., and Kayser, V. (2018) Site-specific glycosylation profile of influenza A (H1N1) hemagglutinin through tandem mass spectrometry. Hum. Vaccin. Immunother. 14, 508–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Vasin, A. V., Temkina, O. A., Egorov, V. V., Klotchenko, S. A., Plotnikova, M. A., and Kiselev, O. I. (2014) Molecular mechanisms enhancing the proteome of influenza A viruses: an overview of recently discovered proteins. Virus Res. 185, 53–63 [DOI] [PubMed] [Google Scholar]
- 109. Hutchinson, E. C., Denham, E. M., Thomas, B., Trudgian, D. C., Hester, S. S., Ridlova, G., York, A., Turrell, L., and Fodor, E. (2012) Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry. PLoS Path. 8, e1002993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Desaire, H., and Hua, D. (2009) When can glycopeptides be assigned based solely on high-resolution mass spectrometry data? Int. J. Mass Spectrom. 287, 21–26 [Google Scholar]
- 111. Dallas, D. C., Martin, W. F., Hua, S., and German, J. B. (2013) Automated glycopeptide analysis–review of current state and future directions. Briefings Bioinformatics 14, 361–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Mechref, Y. (2012) Use of CID/ETD mass spectrometry to analyze glycopeptides. Curr. Protoc. Protein Sci. Chapter 12, Unit 12.11.1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Hinneburg, H., Stavenhagen, K., Schweiger-Hufnagel, U., Pengelley, S., Jabs, W., Seeberger, P. H., Silva, D. V., Wuhrer, M., and Kolarich, D. (2016) The art of destruction: optimizing collision energies in quadrupole-time of flight (Q-TOF) instruments for glycopeptide-based glycoproteomics. J. Am. Soc. Mass Spectrom. 27, 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Hu, H., Khatri, K., Klein, J., Leymarie, N., and Zaia, J. (2016) A review of methods for interpretation of glycopeptide tandem mass spectral data. Glycoconj. J. 33, 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Leymarie, N., and Zaia, J. (2012) Effective use of mass spectrometry for glycan and glycopeptide structural analysis. Anal. Chem. 84, 3040–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Thaysen-Andersen, M., Packer, N. H., and Schulz, B. L. (2016) Maturing Glycoproteomics Technologies Provide Unique Structural Insights into the N-glycoproteome and Its Regulation in Health and Disease. Mol. Cell. Proteomics 15, 1773–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Khatri, K., Klein, J. A., and Zaia, J. (2017) Use of an informed search space maximizes confidence of site-specific assignment of glycoprotein glycosylation. Anal. Bioanal. Chem. 409, 607–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Gillette, M. A., and Carr, S. A. (2013) Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat. Methods 10, 28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Gillet, L. C., Navarro, P., Tate, S., Rost, H., Selevsek, N., Reiter, L., Bonner, R., and Aebersold, R. (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Sanda, M., and Goldman, R. (2016) Data independent analysis of IgG glycoforms in samples of unfractionated human plasma. Anal. Chem. 88, 10118–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Sanda, M., Zhang, L., Edwards, N. J., and Goldman, R. (2017) Site-specific analysis of changes in the glycosylation of proteins in liver cirrhosis using data-independent workflow with soft fragmentation. Anal. Bioanal. Chem. 409, 619–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Chapman, J. D., Goodlett, D. R., and Masselon, C. D. (2014) Multiplexed and data-independent tandem mass spectrometry for global proteome profiling. Mass Spectrom. Rev. 33, 452–470 [DOI] [PubMed] [Google Scholar]
- 123. Zacchi, L. F., and Schulz, B. L. (2016) SWATH-MS glycoproteomics reveals consequences of defects in the glycosylation machinery. Mol. Cell. Proteomics 15, 2435–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Medus, M. L., Gomez, G. E., Zacchi, L. F., Couto, P. M., Labriola, C. A., Labanda, M. S., Bielsa, R. C., Clerico, E. M., Schulz, B. L., and Caramelo, J. J. (2017) N-glycosylation triggers a dual selection pressure in eukaryotic secretory proteins. Sci. Rep. 7, 8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Pan, K. T., Chen, C. C., Urlaub, H., and Khoo, K. H. (2017) Adapting data-independent acquisition for mass spectrometry-based protein site-specific N-glycosylation analysis. Anal. Chem. 89, 4532–4539 [DOI] [PubMed] [Google Scholar]
- 126. Lin, C.-H., Krisp, C., Packer, N. H., and Molloy, M. P. (2018) Development of a data independent acquisition mass spectrometry workflow to enable glycopeptide analysis without predefined glycan compositional knowledge. J. Proteomics 172, 68–75 [DOI] [PubMed] [Google Scholar]
- 127. Matsuoka, Y., Matsumae, H., Katoh, M., Eisfeld, A. J., Neumann, G., Hase, T., Ghosh, S., Shoemaker, J. E., Lopes, T. J. S., Watanabe, T., Watanabe, S., Fukuyama, S., Kitano, H., and Kawaoka, Y. (2013) A comprehensive map of the influenza A virus replication cycle. BMC Systems Biol. 7, 97–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Klein, J. A., Meng, L., and Zaia, J. (2018) Deep sequencing of complex proteoglycans: a novel strategy for high coverage and site-specific identification of glycosaminoglycan-linked peptides. Mol. Cell. Proteomics 17, 1578–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]