Abstract
Pathogenic bacteria invade plant tissues and proliferate in the extracellular space. Plants have evolved the immune system to recognize and limit the growth of pathogens. Despite substantial progress in the study of plant immunity, the mechanism by which plants limit pathogen growth remains unclear. Here, we show that lignin accumulates in Arabidopsis leaves in response to incompatible interactions with bacterial pathogens in a manner dependent on Casparian strip membrane domain protein (CASP)‐like proteins (CASPLs). CASPs are known to be the organizers of the lignin‐based Casparian strip, which functions as a diffusion barrier in roots. The spread of invading avirulent pathogens is prevented by spatial restriction, which is disturbed by defects in lignin deposition. Moreover, the motility of pathogenic bacteria is negatively affected by lignin accumulation. These results suggest that the lignin‐deposited structure functions as a physical barrier similar to the Casparian strip, trapping pathogens and thereby terminating their growth.
Keywords: Arabidopsis, Casparian strip, CASPL, lignin, plant immunity
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Plant Biology
Lignin deposition is required for innate immune defense during incompatible plant‐pathogen interactions in a manner dependent on Casparian strip organizer proteins.
Introduction
During the course of the evolutionary plant–pathogen arms race, plants have developed multilayered defense mechanisms to detect invading pathogens and stop their growth (Jones & Dangl, 2006). Pattern‐triggered immunity (PTI), which relies on the recognition of pathogen‐associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs), provides basal resistance in plants (Zipfel & Felix, 2005; Boller & He, 2009; Zhang & Zhou, 2010). Adapted pathogens have overcome PTI by secreting effectors, which are recognized by plant resistance (R) proteins, leading to effector‐triggered immunity (ETI; Cui et al, 2015). ETI is often associated with programmed cell death (PCD), called the hypersensitive response (HR), which is readily induced at the infection site (Greenberg et al, 1994; Chisholm et al, 2006; Dodds & Rathjen, 2010; Coll et al, 2011). PTI and ETI responses overlap considerably, and they include oxidative bursts, transcriptional reprogramming, and the deposition of phenolic compounds such as lignin (Nicholson & Hammerschmidt, 1992; Yu et al, 1998; Torres, 2010; König et al, 2014). Effector‐triggered immunity is often regarded as a more robust and prolonged PTI response.
Lignin is a major phenolic polymer that constitutes the secondary cell wall in vascular plants (Vanholme et al, 2010). Lignin provides strength and imperviousness to cell walls, thus enabling long‐distance water transport in vascular tissues (Barros et al, 2015). The main building blocks of the lignin polymer are monolignols, p‐coumaryl, coniferyl, and sinapyl alcohol, giving rise to p‐hydroxyphenyl (H), guaiacyl (G), and syringyl (S) lignin units, respectively (Boerjan et al, 2003; Ralph et al, 2004). Monolignols are synthesized from phenylalanine via the phenylpropanoid pathway, which involves numerous enzyme reactions (Whetten & Sederoff, 1995; Bonawitz & Chapple, 2010). Knockout or knockdown mutations in genes encoding monolignol biosynthetic enzymes result in defective phenotypes, such as dwarfing and male sterility, implying that lignin is critical for plant growth and development (Schilmiller et al, 2009; Huang et al, 2010; Bonawitz et al, 2014). After their synthesis, monolignols are transported from the cytosol into the apoplast, where they are oxidatively polymerized to lignin units by the actions of laccases and peroxidases (Boerjan et al, 2003; Berthet et al, 2011; Lee et al, 2013). Monolignol composition and lignin content vary among plant species and tissues as well as during plant development (Voxeur et al, 2015). Angiosperms contain mostly G and S lignin, whereas gymnosperm lignin is predominantly composed of G units with trace amounts of H units.
In addition to its roles in growth and development, lignin has been suggested to be a physical barrier against pathogens (Nicholson & Hammerschmidt, 1992; Sattler & Funnell‐Harris, 2013; Malinovsky et al, 2014; Cesarino, 2019). Expression of lignin biosynthetic genes and lignin levels increased upon pathogen infection (Bhuiyan et al, 2008; Miedes et al, 2014). Analyses of plants in which monolignol biosynthetic genes were silenced or overexpressed have revealed that lignin formation is important for disease resistance. Arabidopsis knockout mutants for phenylalanine ammonia‐lyase (PAL), caffeic acid O‐methyltransferase (COMT), and cinnamyl alcohol dehydrogenase (CAD) showed reduction in basal resistance and/or effector‐triggered resistance against a number of microbial pathogens, including virulent and avirulent strains of the hemibiotrophic bacterial pathogen Pseudomonas syringae, the necrotrophic fungal pathogens Alternaria brassicicola and Botrytis cinerea, and the biotrophic fungal pathogen Blumeria graminis (Quentin et al, 2009; Huang et al, 2010; Tronchet et al, 2010). In wheat, tobacco, and flax, suppression of PAL, caffeoyl‐CoA O‐methyltransferase (CCoAOMT), COMT, and CAD increased susceptibility to fungal pathogens (Maher et al, 1994; Wróbel‐Kwiatkowska et al, 2007; Bhuiyan et al, 2008). In several contrasting cases, disruption of monolignol biosynthesis led to increased resistance to pathogens; however, this was correlated with changes in phenylpropanoid metabolism and accumulation or decrease of certain metabolites. Tobacco plants suppressed in OMT (COMT and CCoAOMT) showed enhanced resistance to Agrobacterium tumefaciens, which coincided with lower levels of vir inducers such as acetosyringone (Maury et al, 2010). Alfalfa suppressed in hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase (HCT) exhibited constitutive defense responses and was less susceptible to fungal infection, which resulted from increased production of defense hormones (Gallego‐Giraldo et al, 2011). Although previous studies support a role of lignin in plant immunity, many details remain unknown, for example, the temporal and spatial patterns of lignin polymerization, and what, if any, other factors are involved.
In this study, we report that lignin plays an essential regulatory role in plant immunity. We demonstrate that lignin deposition readily occurs during incompatible plant–pathogen interactions and this process requires Casparian strip membrane domain protein (CASP)‐like proteins (CASPLs), CASPL1D1 and CASPL4D1. We show that the spread of bacterial pathogens is unimpeded and disease resistance is decreased by defects in lignin deposition, that is, by treatment with a lignin biosynthesis inhibitor and by suppression of CASPL1D1, CASPL4D1, and lignin biosynthetic genes. Based on our results, we propose that lignin, together with CASPL proteins, forms a mechanical barrier similar to the Casparian strip in root endodermal cells and traps pathogens at the infection site, thereby conferring disease resistance in plants.
Results
Lignification is important for plant innate immunity
In a recent study, lignification has been implicated in basal immunity, and treatment with the bacterial PAMP flg22 markedly increased lignin accumulation in Arabidopsis seedlings (Chezem et al, 2017). In this study, we further examined pathogen‐induced lignification by challenging Arabidopsis seedlings (12‐day‐old) and adult plants (6‐week‐old) with virulent P. syringae pv. tomato (Pst) DC3000, avirulent Pst DC3000 (AvrRpm1) and Pst DC3000 (AvrRpt2), and the type III secretion‐defective Pst DC3000 hrcC − mutant that strongly activates the PTI response (Brooks et al, 2004). Lignin deposition was monitored by phloroglucinol staining and quantitatively measured by the acetyl bromide assay. In mock‐treated seedlings and leaves, staining was mostly seen in hypocotyls and leaf veins, and this pattern little changed upon Pst DC3000 treatment (Fig 1A). However, PTI‐activating Pst DC3000 hrcC − induced uniform staining and increased lignin content in cotyledons and the inoculated region of leaves, whereas ETI‐activating Pst DC3000 (AvrRpm1) and Pst DC3000 (AvrRpt2) displayed this response to a greater extent (Fig 1A–D). In adult plants, as an example, Pst DC3000 marginally increased the lignin levels following inoculation (11% and 15% increase at 1 and 2 days post‐inoculation (dpi), respectively). Unlike virulent bacteria, both PTI‐ and ETI‐activating pathogens led to a large increase in lignin content over days, for example, 22% and 51% increase at 1 and 2 dpi, respectively, for Pst DC3000 hrcC −, and 56–66% and 78–110% increase at 1 and 2 dpi, respectively, for Pst DC3000 (AvrRpm1) and Pst DC3000 (AvrRpt2) (Fig 1B and D). These results suggest that lignification is a defense response.
Figure 1. Lignification is an innate immune response.
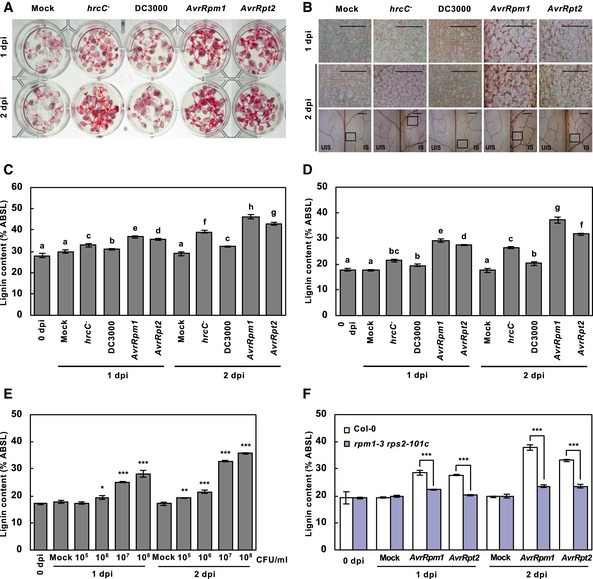
- Phloroglucinol staining of wild‐type seedlings inoculated with PTI‐inducing Pst DC3000 hrcC − , virulent Pst DC3000, and avirulent Pst DC3000 (AvrRpm1) and Pst DC3000 (AvrRpt2).
- Phloroglucinol staining of wild‐type adult leaves inoculated with PTI‐inducing Pst DC3000 hrcC − , virulent Pst DC3000, and avirulent Pst DC3000 (AvrRpm1) and Pst DC3000 (AvrRpt2). The upper images are enlarged ones of the lower boxes at 2 dpi. Scale bars, 100 μm.
- Quantification of lignin content in pathogen‐treated seedlings as in (A). Data are shown as means ± SD (n = 4; 20–30 seedlings each).
- Quantification of lignin content in pathogen‐treated wild‐type leaves as in (B). Data are shown as means ± SD (n = 4; 3–9 leaves each).
- Quantification of lignin content in wild‐type leaves treated with different titers of Pst DC3000 (AvrRpm1). Data are shown as means ± SD (n = 4; 3–9 leaves each).
- Quantification of lignin content in wild‐type and rpm1‐3 rps2‐101c leaves treated with avirulent bacteria. Data are shown as means ± SD (n = 4; 3–9 leaves each).
With the notion of AvrRpm1/AvrRpt2‐induced intense lignin accumulation, we focused on lignification during the ETI response. When treated with different titers of Pst DC3000 (AvrRpm1), plants accumulated lignin in a titer‐dependent manner (Fig 1E). We further determined lignin formation in rpm1‐3 rps2‐101c mutant lacking the respective R genes, RPM1 and RPS2 (Mindrinos et al, 1994). Avirulent pathogen‐induced lignification was abolished in rpm1‐3 rps2‐101c mutant (Fig 1F), verifying that it is an R gene‐mediated ETI response.
Lignin enhances disease resistance and prevents the spread of HR PCD
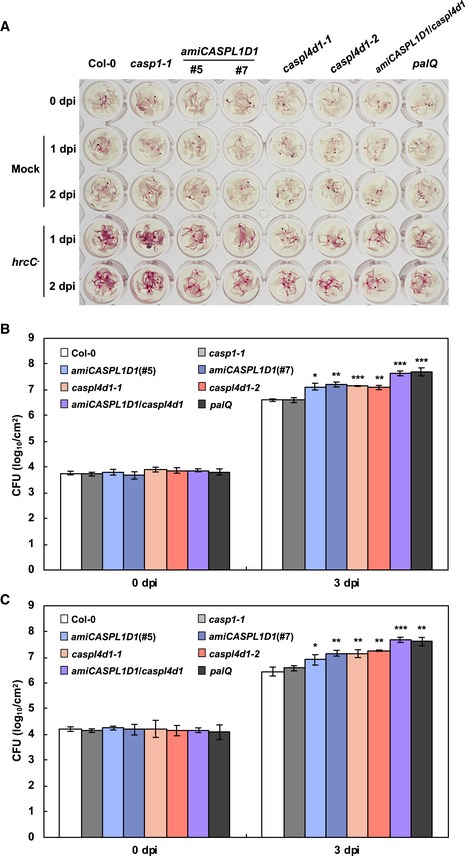
Phenylalanine ammonia‐lyase catalyzes the conversion of phenylalanine to cinnamic acid, the first committed step in lignin biosynthesis or the phenylpropanoid pathway, and Arabidopsis has four PAL genes (PAL1‐4) (Huang et al, 2010). The pal1/2/3/4 quadruple knockout mutant (hereafter designated as palQ; Huang et al, 2010) was used to investigate the role of lignification in plant immunity (Fig 2A). The importance of lignification in immune responses was additionally evaluated by treating plants with piperonylic acid (PA; Schalk et al, 1998; Lee et al, 2013), a monolignol biosynthesis inhibitor inactivating cinnamate 4‐hydroxylase (C4H), and coniferyl alcohol (CA), the monolignol giving rise to G lignin (Fig 2A). Disease resistance has been attributed mainly to G lignin in Arabidopsis (Miedes et al, 2014; Chezem et al, 2017).
Figure 2. Lignin enhances disease resistance and prevents the spread of HR PCD.
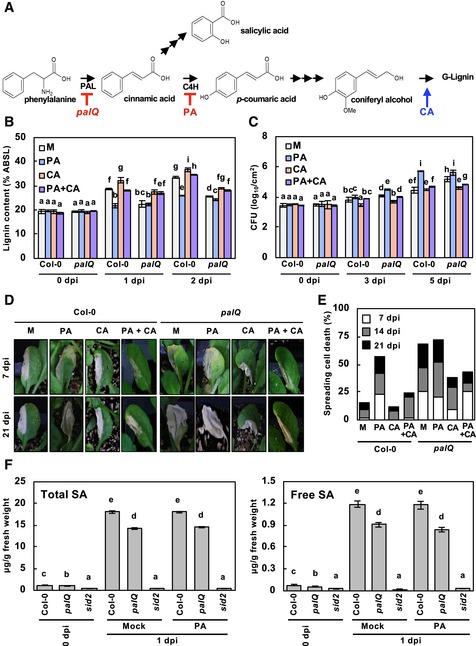
- Simplified lignin biosynthetic pathway.
- Quantification of lignin content in palQ leaves after the indicated treatments and pathogen infection. Data are shown as means ± SD (n = 4; 3–9 leaves each).
- Measurements of Pst DC3000 (AvrRpm1) growth. Data are shown as means ± SD (n = 3).
- Cell death phenotypes of leaves inoculated with Pst DC3000 (AvrRpm1).
- Quantification of leaves (n ≥ 30) with spreading cell death as in (D).
- Quantification of total and free SA in Col‐0 and palQ leaves after the indicated treatments and pathogen infection. Data are shown as means ± SD (n = 5; 6 leaves each).
Upon challenging with Pst DC3000 (AvrRpm1), palQ plants showed significantly lower lignin deposition than wild‐type plants (Fig 2B). Disease resistance was concomitantly decreased in palQ plants (Fig 2C). It was noted that HR PCD gradually spread across the entire infected leaves of palQ plants, whereas it was restricted to the infection site in wild‐type leaves (Fig 2D and E). PA‐treated wild‐type plants also accumulated less lignin and had the same defects in immune responses to pathogen challenge as palQ plants (Fig 2B–E). The phenylpropanoid pathway leads to the production of a number of phenolic compounds (Huang et al, 2010). Therefore, to clarify the effects of lignin, we externally supplied leaves with CA, which restored lignin deposition, disease resistance, and localized HR cell death in palQ and PA‐treated wild‐type plants (Fig 2B–E). These results suggest that lignification is required for defense against bacterial pathogens and for restricting HR cell death.
Salicylic acid (SA) is an essential defense hormone, and its biosynthesis is partially PAL‐dependent (Fig 2A). The bulk of SA is reportedly synthesized via the shikimate pathway through the reaction catalyzed by isochorismate synthase (ICS), also known as SA induction‐deficient 2 (SID2) in Arabidopsis (Wildermuth et al, 2001; Métraux, 2002; Huang et al, 2010). We could not rule out the possibility that the immune defects observed might be due to SA shortage, and we therefore measured SA content to assess whether PAL disruption and PA treatment affected SA accumulation. Total and free SA levels in palQ plants were still about 80% of wild‐type levels after infection with Pst DC3000 (AvrRpm1) and were not influenced by PA treatment (Fig 2F). In sid2 mutant, SA levels were only 2–3% of those in wild‐type plants. These results demonstrate that the defective immune responses, that is, increased susceptibility and uncontrolled HR cell death, in palQ and PA‐treated wild‐type plants, were due to monolignol and lignin deficiency, but not due to SA reduction. The possibility that PA and CA acted directly on bacteria was also tested by treating P. syringae with these compounds in media. The growth of bacteria was not influenced by treatments with PA and CA, even at concentrations 1,000‐fold higher than those used in plants, indicating that PA and CA had no direct effect on bacteria (Appendix Fig S1).
Lignin spatially restricts pathogens and limits their motility
Given the role for lignin as a physical barrier and in restricting HR cell death, we reasoned that the spread of invading pathogens may be additionally prevented by lignin deposition, thus enhancing pathogen resistance. To test our hypothesis, we infiltrated wild‐type leaves with green fluorescent protein (GFP)‐labeled Pst DC3000 and Pst DC3000 (AvrRpt2) (Melotto et al, 2006) and observed their localization. Whereas both virulent and avirulent bacteria colonized the apoplast, bacterial colonies were detected in nearby uninfected sites for Pst DC3000, but not for Pst DC3000 (AvrRpt2) (Fig 3A).
Figure 3. Lignin spatially restricts pathogens and limits their motility.
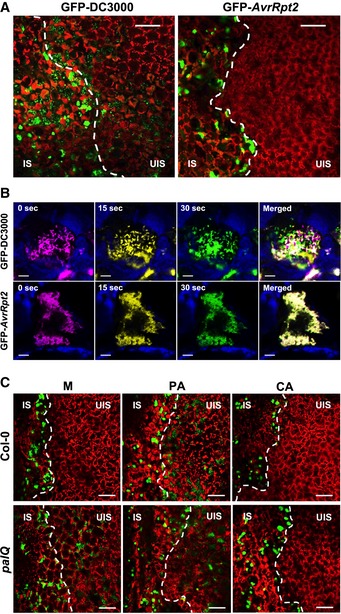
- Colonization patterns of GFP‐Pst DC3000 and Pst DC3000 (AvrRpt2) in wild‐type plants.
- Time‐lapse images of GFP‐Pst DC3000 and Pst DC3000 (AvrRpt2) captured at 0 (magenta), 15, (yellow), and 30 (green) s in Movies EV1 and EV2. Time in seconds in the movies is shown at the top of images. Blue color was used for chlorophyll autofluorescence.
- Colonization patterns of GFP‐Pst DC3000 (AvrRpt2) in wild‐type and palQ plants after PA and CA pretreatments.
The infection behavior led to the speculation that spatial restriction may disturb pathogen motility, especially as pathogens proliferate and fill the confined space. Thus, we assessed pathogen motility using in vivo real‐time imaging. Pst DC3000 was highly motile and moved across the intercellular space (Movie EV1). In sharp contrast, Pst DC3000 (AvrRpt2) appeared static and restricted in movement (Movie EV2). This was evident from the observation that in the merged time‐lapse images of movies at 0, 15, and 30 s, Pst DC3000 did not show substantial overlap, whereas Pst DC3000 (AvrRpt2) did (Fig 3B). However, when Pst DC3000 (AvrRpt2) motility was monitored at an earlier time point when cell density was lower, the bacteria were fairly active (Movie EV3), supporting our contention that spatial limitation contributes at least partially to the decreased motility. To check for this, Pst DC3000 and Pst DC3000 (AvrRpt2) bacteria were extracted from inoculated plants over days, and their movement in media was observed. Virulent and avirulent bacteria that greatly differed in motility inside plants both exhibited considerable motility in media (Movie EV4). These results demonstrate that increased population density in the confined space negatively affects mobility of Pst DC3000 (AvrRpt2) in plants.
Next, wild‐type leaves were treated with PA and challenged with Pst DC3000 (AvrRpt2). Compared to the mock control, PA pretreatment enabled pathogens to vigorously move around and migrate to the uninfected area (Fig 3C, Movie EV5). In lignin‐deficient palQ plants, invading Pst DC3000 (AvrRpt2) had high motility and spreading activity, which were disturbed by CA treatment with its incipient lignification (Fig 3C, Movies EV6 and EV7). Direct treatments of Pst DC3000 (AvrRpt2) with PA and CA in media did not affect pathogen motility (Movies EV8 and EV9). These results suggest that Pst DC3000 (AvrRpt2) is spatially confined in the apoplast, and this is primarily attributed to lignin during the ETI process.
Lignin deposits and encompasses pathogens in the extracellular space
In a previous study, fluorescence‐tagged monolignols that were synthesized to visualize the lignin polymerization process were successfully incorporated into lignin polymers (Tobimatsu et al, 2011, 2013). In the present study, nitrobenzofuran (NBD)‐tagged CA (CANBD) and dimethylaminocoumarin (DMAC)‐tagged CA (CADMAC) were used to monitor pathogen‐induced lignin formation. Following infiltration of an entire leaf with either CA and CANBD or CA and CADMAC mixtures, one half of the leaf was inoculated with Pst DC3000 (AvrRpm1). CANBD green or CADMAC blue fluorescence was detected solely in the pathogen‐infiltrated, lignin‐deposited site, as indicated by phloroglucinol staining, but not in the uninfected site of the inoculated leaf (Fig 4A). This indicates that fluorescence‐tagged CAs are compatible with pathogen‐induced lignin polymerization.
Figure 4. Lignin deposits and encompasses pathogens in the extracellular space.
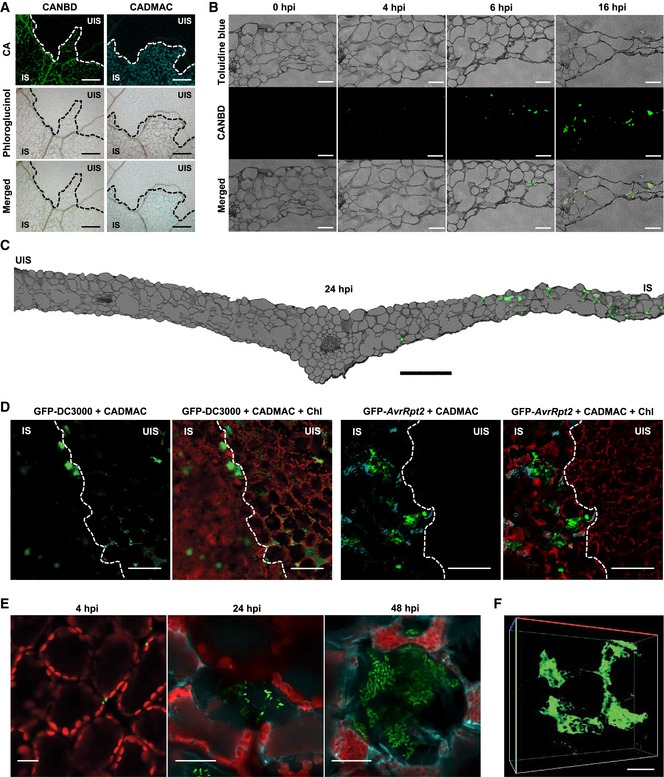
- CANBD‐ and CADMAC‐incorporated lignin polymerization. CANBD (green) and CADMAC (blue) fluorescence was observed in the lignified infection site at 2 dpi, as revealed by phloroglucinol staining.
- Cross‐sections of CANBD‐ and Pst DC3000 (AvrRpm1)‐treated leaves.
- Cross‐section of the entire leaf with one half leaf infected.
- CADMAC‐incorporated lignin deposition in the GFP‐Pst DC3000 (AvrRpt2)‐infected site at 2 dpi.
- Cellular observation of CADMAC‐incorporated lignin formation against infecting GFP‐Pst DC3000 (AvrRpt2).
- 3D image of CANBD‐polymerized structure. Images were taken at 24 hpi as in (C).
The time course of lignification was evaluated in cross‐sections of CA/CANBD‐pretreated and Pst DC3000 (AvrRpm1)‐infiltrated leaf tissues. The fluorescence signal of lignin was initially sporadic but eventually expanded in the area of dead cells during the course of pathogen infection (Fig 4B and C). Next, we examined how lignin is spatially constructed against pathogens by infiltrating CA/CADMAC‐pretreated leaves with GFP‐Pst DC3000 (AvrRpt2). Lignin polymerization was induced near bacterial colonies at the infected site of the leaf (Fig 4D). At the cellular level, only a few bacteria were initially observed, and as the number of bacteria increased, the surrounding cells experienced progressive lignification and simultaneous cell death (Fig 4E). Lignification largely increased, and cell death and shrinkage intensified at 48‐h post‐inoculation. As a result, the shrunken, lignified cells encompassed pathogens, confining them in space. When the infected leaf was chemically cleared, the wide, mesh‐like lignin structure was revealed in a 3D image (Fig 4F, Movie EV10).
CASPL genes are induced in response to avirulent bacterial treatment
A crucial question to address was whether lignin is associated with a defined structure. A search for known barrier structures led to the identification of the Casparian strip (CS), which functions as a diffusion barrier and prevents the movement of water and solutes in the root endodermis (Roppolo et al, 2011). Intriguingly, the CS is a lignin‐based structure and requires Casparian strip membrane domain proteins (CASPs; CASP1 to CASP5) that mediate lignin deposition and CS formation (Naseer et al, 2012). We postulated that a CS‐like structure (CSL) might be generated during the ETI response, implying that proteins similar to CASPs would be involved in CSL formation.
Evolutionary analysis of CASP genes revealed a large CASPL family, composed of 34 CASP‐like (CASPL) proteins in addition to CASP1 to CASP5 in Arabidopsis (Yang et al, 2015). Microarray data available at Genevestigator (http://www.genevestigator.ethz.ch/) indicated that, among CASP and CASPL genes, CASPL1D1 and CASPL4D1 transcripts accumulate to high levels after challenging with Pst DC3000 (AvrRpm1) (Fig 5A). The expression of CASPL1D1 and CASPL4D1 as well as CASP1 to CASP5 was evaluated in Pst DC3000 (AvrRpm1)‐challenged wild‐type plants. Consistently, the transcript levels of CASPL1D1 and CASPL4D1 increased upon Pst DC3000 (AvrRpm1) inoculation (Appendix Fig S2). CASP1 expression was slightly induced at 24‐h post‐inoculation (hpi), which differed from the Genevestigator data.
Figure 5. CASPLs are required for pathogen‐induced lignification.
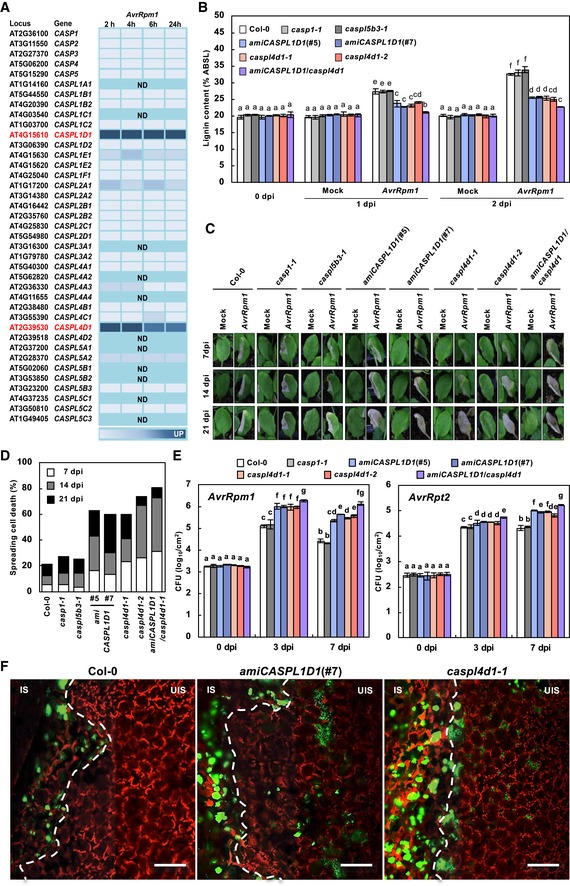
- Time course expression profiles of CASPLs in response to Pst DC3000 (AvrRpm1) treatment. Analysis was based on the Genevestigator database (http://www.genevestigator.ethz.ch/). The darker color corresponds to stronger expression. ND, not determined.
- Quantification of lignin content in wild‐type and caspl mutant leaves after Pst DC3000 (AvrRpm1) inoculation. Data are shown as means ± SD (n = 4; 3–9 leaves each).
- Cell death phenotypes of leaves inoculated with Pst DC3000 (AvrRpm1).
- Quantification of leaves (n ≥ 30) with spreading cell death after Pst DC3000 (AvrRpm1) inoculation.
- Measurements of Pst DC3000 (AvrRpm1) and Pst DC3000 (AvrRpt2) growth. Data are shown as means ± SD (n = 3).
- Colonization patterns of GFP‐Pst DC3000 (AvrRpt2) in caspl mutant plants. IS, infected site; UIS, uninfected site. White dash lines indicate the boundary between IS and UIS. Scale bars, 100 μm.
CASPL1D1 and CASPL4D1 are required for pathogen‐induced lignification
Based on the expression data, CASPL1D1 and CASPL4D1 were subjected to functional analysis, and knockout mutants caspl4d1‐1 (SALK_201606) and caspl4d1‐2 (SALK_078525) were obtained (Appendix Fig S3A–C). As no T‐DNA insertion mutants were available for CASPL1D1, CASPL1D1 knockdown lines (amiCASPL1D1) overexpressing artificial miRNAs were prepared (Appendix Fig S3D). In addition, amiCASPL1D1 caspl4d1 double mutants were generated by crossing amiCASPL1D1 (#7) with caspl4d1‐1. In addition, casp1‐1 (SAIL_265_H05) (Roppolo et al, 2011) and, as a negative control, caspl5b3‐1 (SALK_017299) mutants were included in the analysis (Appendix Fig S3E–G).
AvrRpm1‐induced lignification was largely reduced in amiCASPL1D1 and caspl4d1 plants but was not affected in casp1‐1 and caspl5b3‐1 plants (Fig 5B, Appendix Fig S4). In line with the immune responses of palQ and PA‐treated wild‐type plants, HR cell death spread beyond the infiltrated area in amiCASPL1D1 and caspl4d1 plants (Fig 5C and D). The restricted growth of Pst DC3000 (AvrRpm1) and Pst DC3000 (AvrRpt2) in wild‐type plants was alleviated in amiCASPL1D1 and caspl4d1 plants (Fig 5E and F). This was probably because bacteria lacked nutrients over time due to spatial limitation in wild‐type plants, whereas they were capable of obtaining nutrients from the surroundings in amiCASPL1D1 and caspl4d1 plants. Lignin deposition was reduced even further in amiCASPL1D1 caspl4d1 double mutants, but not completely abolished, suggesting the involvement of other factors besides CASPL1D1 and CASPL4D1 (Fig 5B). The spread of HR cell death and avirulent bacterial growth were also more increased in amiCASPL1D1 caspl4d1 plants (Fig 5C–E). GFP‐labeled Pst DC3000 (AvrRpt2) bacteria spread over the infection site and were vigorously motile in amiCASPL1D1 and caspl4d1 leaves (Fig 5F, Movies EV11 and EV12). None of these defects in immune responses were rescued by CA pretreatment (Fig EV1, Movies EV13 and EV14). These results together suggest that CASPL1D1 and CASPL4D1 are required for pathogen‐induced lignin formation and ETI, and act in a cooperative manner. CASPL1D1 and CASPL4D1 are phylogenetically distant, supporting their non‐redundant functions (Appendix Fig S5).
Figure EV1. Pathogen‐induced lignification depends on CASPL1D1 and CASPL4D1.
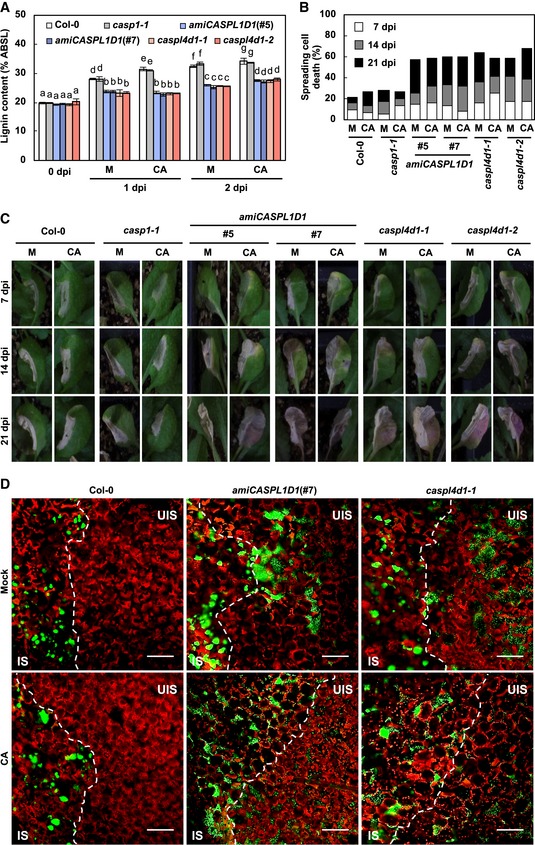
- Quantification of lignin content in CA‐pretreated wild‐type and caspl leaves after Pst DC3000 (AvrRpm1) inoculation. Data are shown as means ± SD (n = 4; 3–9 leaves each). Different letters indicate significant differences (Tukey's HSD test; P < 0.05).
- Quantification of leaves (n ≥ 30) with spreading cell death after Pst DC3000 (AvrRpm1) inoculation.
- Cell death phenotypes of leaves inoculated with Pst DC3000 (AvrRpm1).
- Colonization patterns of GFP‐Pst DC3000 (AvrRpt2). Bacterial inoculum was at 108 cfu/ml. IS, infected site; UIS, uninfected site. White dash lines indicate the boundary between IS and UIS. Scale bars, 100 μm.
In Chezem et al (2017), lignin biosynthetic mutants were more susceptible to virulent Pst DC3000 than wild‐type plants, which suggested that lignification contributes to basal immunity. Here, we asked whether CASPL1D1 and CASPL4D1 function in PTI‐associated lignification and basal immunity. Upon Pst DC3000 hrcC inoculation, lignin accumulation decreased in palQ and amiRCASPL1D1/caspl4d1 lines (Fig EV2A), indicating that CASPL1D1 and CASPL4D1 are required for lignification during PTI responses. At the same time, palQ and amiRCASPL1D1/caspl4d1 lines showed increased susceptibility to Pst DC3000 (Fig EV2B). These results suggest that CASPL1D1 and CASPL4D1 are involved in both PTI and ETI.
Figure EV2. CASPLs are implicated in basal immunity.
-
APhloroglucinol staining of wild‐type, caspl, and palQ seedlings flood‐inoculated with Pst DC3000 hrcC − at 108 cfu/ml.
-
B, CMeasurements of Pst DC3000 growth in wild‐type, caspl, and palQ plants syringe‐inoculated at 105 cfu/ml (B) and spray‐inoculated at 109 cfu/ml (C). Data are shown as means ± SD (n = 3). Asterisks indicate significant differences from the respective Col‐0 (t‐test; *P < 0.05; **P < 0.01; ***P < 0.001).
Stomatal closure is an essential PTI response, referred to as stomatal immunity (Melotto et al, 2006). Stomatal closure may also be part of ETI (Melotto et al, 2006; Freeman and Beattie, 2009). To test for a role of lignin in pathogen‐triggered stomatal closure, bacterial populations were additionally estimated in surface‐inoculated palQ and amiRCASPL1D1/caspl4d1 plants. Growth patterns of Pst DC3000 and Pst DC3000 (AvrRpm1) were little distinct between syringe‐ and surface‐inoculated plants (Figs 5E and EV2B and C, Appendix Fig S6), suggesting that lignin is not implicated in the stomatal immune response.
Subcellular localization of CASPL and lignin is correlated in pathogen‐infected cells
Next, we examined the temporal and spatial localization of CASPLs and lignin in pCASPL4D1::CASPL4D1‐mCherry transgenic plants that express CASPL4D1‐mCherry under the control of the CASPL4D1 promoter (Fig 6). pCASPL4D1::CASPL4D1‐mCherry plants were pretreated with CA/CADMAC and then infiltrated with Pst DC3000 (AvrRpm1). CASPL4D1‐mCherry proteins were first detected at the plasma membrane (2 hpi), after which CADMAC‐lignin formed around the cell membrane (4 hpi) where CASPL proteins were located (Fig 6A–C). Cells readily shrank and suffered severe deformation (8 and 24 hpi) indicative of HR cell death, resulting in strong signals of CASPL proteins and lignin interwoven over the dead cells. CADMAC‐lignin embraced CASPL4D1‐mCherry signals, manifesting the disengagement of the plasma membrane from the cell wall, and was conspicuous between cells (Fig 6B, arrows). In the complementation test, caspl4d1‐1 mutant was crossed with pCASPL4D1::CASPL4D1‐mCherry plants, and the resulting caspl4d1‐1 pCASPL4D1::CASPL4D1‐mCherry plants restored AvrRpm1‐induced lignin deposition (Appendix Fig S7), indicating that pCASPL4D1::CASPL4D1‐mCherry is functional. According to microscopic observations, we propose that lignin deposition serves to seal the apoplast and trap pathogens in the extracellular space of mesophyll cells (Figs 6D and 7).
Figure 6. Temporal and spatial localization of CASPL4D1 and lignin polymers.
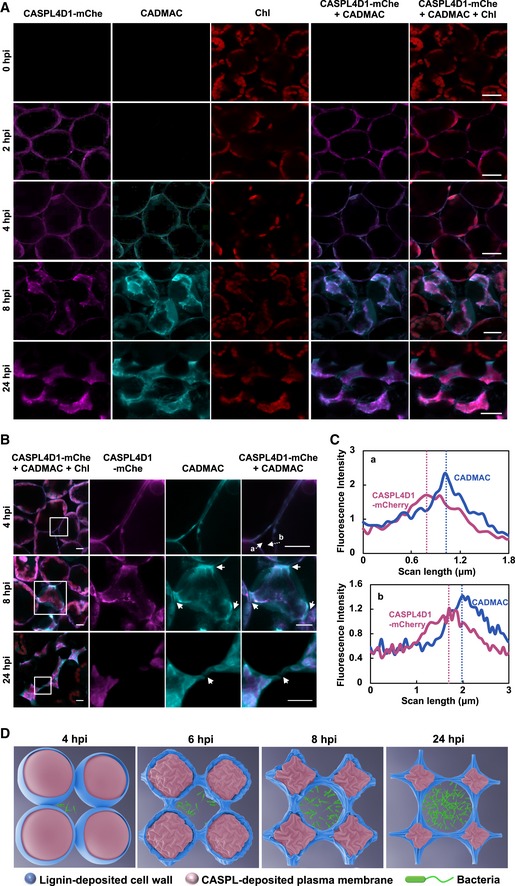
- Visualization of CASPL4D1 proteins and lignin by mCherry and CADMAC fluorescence.
- Magnified views of CASPL4D1 and lignin localization. Images in boxes were enlarged in the 2nd to 4th columns. Arrows indicate lignin deposition between cells.
- Relative fluorescence intensity profiles of CASPL1D1 (mCherry) and lignin (CADMAC) across transects indicated by dotted lines and letters (a, b) in (B).
- A model showing morphological changes of CASPL‐ and lignin‐deposited cells during the HR response.
Figure 7. A model for the role of the lignin‐deposited structure in plant immunity.
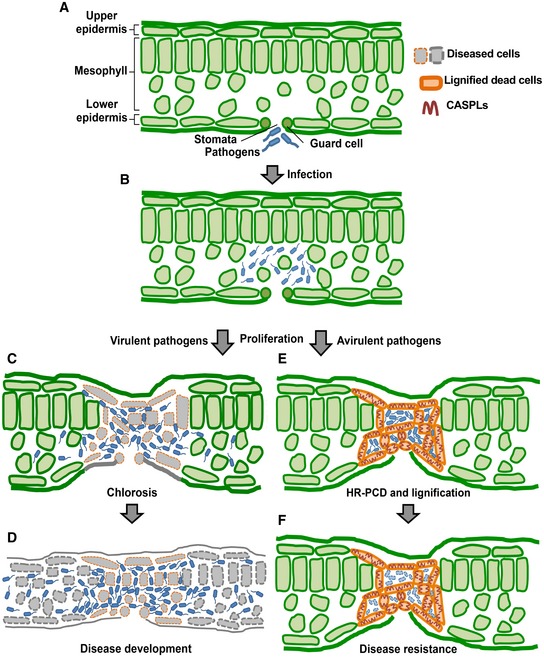
-
ABacterial pathogens invade leaf tissues through stomata.
-
BPathogens proliferate in the extracellular space or the apoplast.
-
C, DVirulent pathogens spread over the infection site (C) and develop disease (D).
-
E, FAvirulent pathogens are restricted to the infection site as a result of the construction of the CASPL‐ and lignin‐deposited structure (E) and are eventually eliminated from the infection site of the leaf (F).
Discussion
The phenylpropanoid pathway leads to the synthesis of lignin and many other phenolic compounds, including phytoalexins, stilbenes, coumarins, and flavonoids, which are implicated in disease resistance in plants (Nicholson & Hammerschmidt, 1992; Kuc, 1995; Dicko et al, 2005; König et al, 2014; Zernova et al, 2014). Accumulation of phenolic compounds has been associated with the HR and is described as a feature of HR PCD (Brisson et al, 1994; Bennett et al, 1996; Skalamera & Heath, 1998; Lee et al, 2001; Mellersh et al, 2002; Daniel & Guest, 2006). In particular, there are studies on the causal relationship between lignin and hypersensitive resistance of wheat to the stem rust fungus Puccinia graminis f.sp. tritici (Moerschbacher et al, 1990; Menden et al, 2007). Treatments of wheat with PAL and CAD inhibitors interfered with the development of lignified hypersensitive cell death and fungal growth (Moerschbacher et al, 1990). Analysis of phenolic compounds in resistant and susceptible wheat cultivars indicated the deposition of S‐rich lignin units, but no other phenolics, as a resistance‐specific reaction (Menden et al, 2007). These previous studies support a role of lignin in plant immune responses.
In this study, lignification was induced during incompatible plant–pathogen interactions. When lignin deposition was inhibited by PAL mutations and PA treatment, plants failed to confine avirulent bacterial pathogens to the infection site and limit pathogen growth. Intriguingly, HR cell death was readily induced but spread beyond the infection site in plants defective in lignin deposition. CA treatment of palQ and PA‐treated wild‐type plants rescued lignification and overcame the defects in immune responses, that is, spatial restriction of pathogens and cell death, and disease resistance, verifying the essential roles of lignin in ETI. However, the mechanism by which lignin localizes HR cell death is currently unclear. Whether the spread of HR cell death results from defects in lignification or is due to spatially unrestricted bacterial pathogens remains to be investigated.
HR PCD has long been regarded as a mechanism to prevent pathogens from spreading and to limit pathogen growth, as it is a rapid, localized reaction at the infection site. However, many lines of evidence contradict the proposed role of HR PCD in innate immunity. There have been reports of R genes or Arabidopsis mutants that exhibit disease resistance without the activation of HR cell death (Yu et al, 1998; Bendahmane et al, 1999; Clough et al, 2000; Bulgarelli et al, 2010; Coll et al, 2010). Conversely, ndr1 (for non‐race‐specific disease resistance) mutant plants were susceptible to avirulent P. syringae, while still retaining the HR cell death phenotype (Century et al, 1995). Previous observations and our results raise the concern as to whether HR PCD is essential for disease resistance in plants, and detailed investigation is required to determine the role of HR cell death.
Previous studies have shown that lignification is induced during basal resistance or PTI responses (Robertsen, 1986; Kawano & Shimamoto, 2013; Chezem et al, 2017). In a recent study, the SG2‐type R2R3‐MYB transcription factor MYB15 directly bound to gene promoters required for G‐lignin biosynthesis and activated lignification in response to flg22 (Chezem et al, 2017). Given that PTI and ETI responses, including lignin biosynthesis, qualitatively overlap (Tsuda et al, 2008; Thomma et al, 2011), it raises the question on whether MYB15 or other transcription factors control ETI‐related expression of monolignol biosynthetic genes. On the other hand, MYB36 was identified as a regulator for the expression of CS‐associated genes, such as CASPs and peroxidase 64 (PER64), leading to CS formation in root endodermal cells (Kamiya et al, 2015; Liberman et al, 2015).
Lignin is a structural component embedded into cell walls and has been implicated in plant growth and development, such as tracheary element formation, anther dehiscence, and pod shattering (Fukuda, 1997; Dawson et al, 1999; Liljegren et al, 2000). In this work, we provide compelling evidence suggesting that the lignin‐deposited CSL is formed during immune responses and this functions as a physical barrier against pathogens, similar to the endodermal CS. CS mainly consists of lignin and functions as a paracellular diffusion barrier in the root endodermis (Roppolo et al, 2011; Naseer et al, 2012; Lee et al, 2013). Lignin would provide rigidity and hydrophobicity to CS and CSL, which then renders these structures suitable for sealing the extracellular space of juxtaposed cells. The role of lignin as a mechanical barrier has additionally been demonstrated in a recent study, in which lignin accumulated in secession cells of the abscission zone and blocked the diffusion of cell wall enzymes for precise organ separation (Lee et al, 2018). Lignin polymerization occurs via monolignol oxidation, which implicate NADPH oxidases or respiratory burst oxidase homologs (RBOHs) for reactive oxygen species (ROS) production (Kärkönen & Koutaniemi, 2010). In fact, RBOHD/F are essential regulators for biotic interactions (Torres et al, 2002; Torres & Dangl, 2005; Morales et al, 2016) and for lignin biosynthesis associated with CS formation, organ abscission, and cell wall integrity signaling (Denness et al, 2011; Lee et al, 2013, 2018). It will be worthwhile to determine whether pathogen‐induced lignification is one of lignifying processes regulated by RBOHD/F.
Casparian strip membrane domain proteins are specifically expressed in the root endodermis, where they direct lignin deposition and therefore CS formation by scaffolding lignin polymerization enzymes such as PER64 (Lee et al, 2013). Previous studies and ours reveal that 34 Arabidopsis CASPLs are expressed in tissue‐ and condition‐specific manners, for example, in the abscission zone (CASPL1D1, CASPL1D2, CASPL4B1, CASPL4D1, and CASPL4D2; Roppolo et al, 2014; Lee et al, 2018) and the anther wall (CASPL1F1; Roppolo et al, 2014), and in response to cold stress (CASPL4C1; Yang et al, 2015) and biotic stress (CASPL1D1 and CASPL4D1; in this study). When ectopically expressed in the endodermis, many CASPLs behaved like CASPs, accumulating at the CS membrane domain (Roppolo et al, 2014). These results suggest that CASPs and CASPLs share a common function as scaffolding proteins, probably recruiting lignin polymerizing and/or other cell wall modifying enzymes, but are distinguished via their specific expression in cells, tissues, developmental stages, and stress responses.
In microscopic analyses, pathogens were encompassed by the lignified structure, and their motility decreased as bacteria proliferated but recovered when extracted from plants and resuspended in media. This lends support to the idea that reduced motility is attributed primarily to increased density due to pathogen growth in a limited space. On the other hand, there are reports suggesting that cells evoking the HR synthesize and secrete antimicrobial phenolics, phytoalexins, and pathogenesis‐related (PR) proteins, such as β‐glucanases and chitinases, leading to the inhibition of pathogen growth (Nicholson & Hammerschmidt, 1992; van Loon et al, 2006). The closed space may provide for a high local concentration of the released antimicrobial compounds and thus enable the suppression of pathogens more effectively. The lack of nutrients due to spatial closure, combined with attack by antimicrobials, would eventually lead to the elimination of pathogens.
Neutrophils, the immune phagocytes in animals, are armed with large, extracellular, web‐like structures, called neutrophil extracellular traps (NETs), that are formed primarily through a cell death process and are composed of DNA and DNA‐associated antimicrobial proteins (Brinkmann et al, 2004; Papayannopoulos, 2018). NETs trap and kill microbes extracellularly and are thought to serve as physical barriers that prevent the further spread of pathogens. It is interesting to note these similarities between the plant lignin barrier and animal NETs. In plants, extracellular DNA has been suggested to function in defense of the root tip against fungal infection (Wen et al, 2009), although more evidence is needed to clarify the exact role of extracellular DNA in plant defense.
Together, our findings underscore the importance of the lignin‐impregnated structure in plant immunity. We propose a model in which CASPLs are first positioned at the plasma membrane to guide lignin polymerization and together construct the CSL against invading pathogens (Fig 7). Whereas both CS and CSL commonly act as a diffusion barrier, they form structures of different patterns. In contrast to the former's continuous ring structure in the center of root endodermis cells (Roppolo et al, 2011), the latter displays a wide structure embracing dead cells. Previous studies have reported that the site of CS deposition is marked by CASP localization (Roppolo et al, 2011). In this context, CASPLs are uniformly distributed along the cell membrane, thus determining the CSL pattern. We cannot exclude the possibility that CASPLs localize to distinct membrane nanodomains where the lignin deposition machinery, equivalent to that formed in CS development, may be assembled, as observed for RBOHD and receptor kinases, flagellin sensing 2 (FLS2) and brassinosteroid insensitive 1 (BRI1) (Hao et al, 2014; Bücherl et al, 2017). The discrete CSL pattern may be visible at higher microscopic resolution. A leucine‐rich repeat receptor‐like kinase schengen3/gassho1 (SGN3/GSO1) (Pfister et al, 2014), a receptor‐like cytoplasmic kinase SGN1 (Alassimone et al, 2016), and CS integrity factors (CIF1/2) (Doblas et al, 2017; Nakayama et al, 2017) have also been identified as major players of CS formation. Further research is required to explore additional regulatory factors and mechanisms that regulate the distribution of CASPLs, and thus the organization of CSL.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana (ecotype Columbia, Col‐0) plants were grown at 23°C under long‐day conditions (16‐h light/8‐h dark cycle) for general growth and reproduction, and under short‐day conditions (8‐h light/16‐h dark cycle) for pathogen infection. The mutant lines used in this study are casp1‐1 (SAIL_265_H05), caspl4d1‐1 (SALK_201606), caspl4d1‐2 (SALK_078525), caspl5b3‐1 (SALK_017299), rpm1‐3 rps2‐101C (Mindrinos et al, 1994), palQ (Huang et al, 2010), and sid2 (Wildermuth et al, 2001). T‐DNA insertion sites were verified by sequencing using gene‐specific primers (Appendix Table S1), and homozygous lines were selected. To generate CASPL1D1 knockdown plants, an artificial microRNA (nucleotides 223–242) targeting CASPL1D1 was designed using the Web microRNA Designer (WMD) interface (Ossowski et al, 2008; Lee et al, 2013). The artificial microRNA sequence was incorporated into the miR319a precursor by overlap PCR using miR319a‐ and artificial microRNA‐specific primers (Appendix Table S1). The PCR products were cloned into the binary vector pCAMBIA 1300 to be expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The constructs were transformed into Agrobacterium tumefaciens GV3101 and then introduced into Col‐0 plants using the floral dip method (Clough & Bent, 1998).
Plant treatments
For bacterial pathogen inoculation, bacterial strains were grown at 28°C on King's B agar medium containing 50 μg/ml kanamycin and 100 μg/ml rifampicin for Pst DC3000, Pst DC3000 (AvrRpm1), Pst DC3000 (AvrRpt2), GFP‐Pst DC3000, and GFP‐Pst DC3000 (AvrRpt2), or 100 μg/ml rifampicin for Pst DC3000 hrcC − (King et al, 1954). Bacteria were resuspended in 10 mM MgCl2 for syringe and spray inoculation or in water for flood inoculation. Silwet L‐77 (0.025%) was included in bacterial suspensions for spray/flood inoculation. Six‐week‐old leaves grown in soil were syringe‐infiltrated at 105 colony‐forming units (cfu)/ml (OD600 = 0.0001) or spray‐inoculated at 109 cfu/ml (OD600 = 1) for bacterial growth and at 108 cfu/ml (OD600 = 0.1) for all other experiments. Twelve‐day‐old seedlings grown on 1/2 Murashige and Skoog (MS)‐sucrose (1%) agar media were flood‐inoculated for 2 min with bacterial inoculum at 108 cfu/ml (OD600 = 0.1) (Ishiga et al, 2011; Chezem et al, 2017). For all experiments, 10 mM MgCl2 was used as a mock treatment. For chemical treatments, coniferyl alcohol (Tobimatsu et al, 2011), piperonylic acid (Lee et al, 2013), CANBD (Tobimatsu et al, 2011), and CADMAC (Tobimatsu et al, 2011) were dissolved in 100% dimethyl sulfoxide (DMSO; Duchefa) to make stock solutions at concentrations of 50 mM for CA and PA and 1 mM for CANBD and CADMAC. CA and PA were diluted with distilled water to a final concentration of 50 μM. For the preparation of CA and CANBD or CA and CADMAC mixtures, CA and CANBD/CADMAC stocks were diluted to final concentrations of 100 μM and 1 μM, respectively. All treatments were carried out by syringe infiltration. Chemical‐treated leaves were air‐dried before pathogen inoculation.
Bacterial growth assay
Bacterial growth was determined as previously described (Shindo et al, 2012; Kwon et al, 2013). Leaves of 6‐week‐old plants were syringe‐inoculated with bacterial suspensions (OD600 = 0.0001) or spray‐inoculated with bacterial suspensions (OD600 = 1). Chemical pretreatments were carried out 1 h before pathogen infiltration. Two leaf disks (5 mm in diameter) per leaf were taken and pooled as one replicate. The pooled leaf disks were ground in 10 mM MgCl2, and cfu was determined by serial dilution plating on King's B agar medium containing 50 μg/ml kanamycin and 100 μg/ml rifampicin. Experiments were repeated 3–5 times with biologically independent samples.
Gene expression analysis
Total RNAs were extracted using TRIzol reagent and reverse‐transcribed into cDNAs using PrimeScript™ RT Reagent Kit (TaKaRa). Quantitative real‐time PCR was performed using KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems) with gene‐specific primers (Appendix Table S1) on a LightCycler 480 system (Roche) according to the manufacturer's protocol. For transcript normalization, Actin1 was used as a reference gene. Data were analyzed using LC480Conversion and LinRegPCR software (Heart Failure Research Center). Experiments were repeated at least three times with biologically independent samples.
Phloroglucinol staining
Phloroglucinol staining was performed at room temperature as previously described (Mitra & Loqué, 2014). Leaves and seedlings were dehydrated in 100% ethanol overnight, rehydrated in a graded series of ethanol (75%, 50%, and 25%) and water for 30 min each, and then stained with 3% phloroglucinol (Sigma‐Aldrich) dissolved in 30% HCl for 1 min. The stained leaves and seedlings were observed under an optical microscope (Leica EZ4E) or photographed using the camera (Canon).
Lignin quantification
Lignin content was determined by the acetyl bromide‐based method as previously described (Chang et al, 2008). Pathogen‐inoculated leaves and seedlings were frozen and ground to a fine powder in liquid nitrogen. The dried samples (3 ± 1 mg, accurately weighed) were washed serially with 70% ethanol, chloroform/methanol (1:1 v/v), and acetone. The washed pellets were completely dried at 45°C and digested with 1 ml of 25% acetyl bromide in acetic acid at 70°C for 1 h by vortexing every 10 min. Samples were cooled on ice and centrifuged for 5 min at 16,000 g. The supernatants (100 μl) were transferred to glass tubes and mixed with 2 M NaOH (400 μl), 0.5 M hydroxylamine hydrochloride (70 μl), and acetic acid (430 μl). The prepared solutions were transferred to 96‐well microplates, and the absorbance was measured using Microplate Reader (Molecular Devices) at 280 nm. The content of acetyl bromide soluble lignin (% ABSL) was calculated using Beer's Law (Kapp et al, 2015). The extinction coefficient used for Arabidopsis was 15.69 l/g cm (Foster et al, 2010).
SA quantification
Total and free SA were quantified as previously described (Kim et al, 2013; Quentin et al, 2016). Leaf samples (150 mg), together with o‐anisic acid (50 ng) as an internal standard, were ground in liquid nitrogen and extracted with 90% methanol. For total SA, the methanol‐evaporated samples were further hydrolyzed with 4 M HCl at 80°C for 1 h. The samples (for both total and free SA) were subjected to phase separation in ethyl acetate/cyclohexane (1:1 v/v). Following evaporation of the organic phase, the dry residues were solubilized in 10% methanol containing 0.1% TFA. Chromatography analysis was performed on HPLC 1290 system (Agilent) using ZORBAX Extend‐C18 column (2.1 × 150 mm, 5 μm; Agilent) combined with a guard column (UHPLC C18; Phenomenex) at 30°C. Mobile phases A and B were 0.1% TFA and 0.1% TFA in acetonitrile, respectively. The gradient profile was 10–60% (B) for 15 min followed by equilibration for 5 min, and the flow rate was 0.4 ml/min. Fluorescence was recorded with excitation/emission wavelengths of 305/407 nm for o‐anisic acid and SA.
Microscopic analysis of lignin
For histochemical analysis, CA (100 μM)/CANBD (1 μM)‐ and pathogen‐infiltrated leaves were fixed in 50 mM phosphate‐buffered saline (pH 7.0) containing 2.5% (v/v) glutaraldehyde and 4% (v/v) paraformaldehyde for 24 h at 4°C. Samples were then dehydrated in a graded series of ethanol (50, 70, 90, 95, and 100%) and embedded in LR White resin (Ted Pella). The resin was polymerized for 2 days at 55°C. Sections (7 μm) were prepared using a microtome (Leica RM 2255) and stained with 1% (w/v) toluidine blue O for 1 min. The samples were immersed in mineral oil and observed under a fluorescence microscope (Zeiss Axio Imager.A2). For 3D image analysis, CA (100 μM)/CANBD (1 μM)‐ and pathogen‐infiltrated leaves were dehydrated in 100% ethanol at 100°C for 1 min. After rinsing with distilled water and 10% glycerol, CANBD signals were detected using a confocal microscope (Zeiss LSM 700) at 488/530 nm excitation and emission. In total, 32 images were acquired along the z‐axis (approximately 31 μm) and converted into a 3D structure using the Zen 2.3 Blue imaging software (Zeiss).
Time‐lapse imaging of bacterial pathogens
Leaves were infiltrated with GFP‐Pst DC3000 or Pst DC3000 (AvrRpt2) and incubated for 1–2 days. Images for bacterial motility observed in vivo and in vitro were taken every 3 s for 5 min and every 1.8 s for 3 min, respectively, using a confocal microscope (Zeiss LSM 700), and converted to a video played at 3.3 frames/s for 30 s and at 10 frames/s for 10 s, respectively, using the Adobe Photoshop CS6 software (Adobe Systems).
Subcellular localization of CASPL4D1 and lignin polymers
Leaves were obtained from CA (100 μM)/CADMAC (1 μM)‐ and pathogen‐infiltrated pCASPL4D1::CASPL4D1‐mCherry leaves, fixed in 4% (v/v) paraformaldehyde for 24 h at 4°C, and washed with distilled water. Samples were observed under a confocal microscope (Zeiss LSM 700). Excitation and emission were set at 405/490 nm for CADMAC, 555/630 nm for mCherry, and 639/710 nm for chlorophyll. Confocal images were analyzed and processed using the Zen 2.1 Black imaging software (Zeiss).
Antimicrobial activity test
Sensitivity of bacterial pathogens to CA and PA was tested as previously described (Elleuch et al, 2010). King's B agar plates were covered with 5 ml of top agar media (0.7% agar) containing bacterial suspensions (OD600 = 0.02). Filter paper disks (6.5 mm in diameter) impregnated with the indicated chemicals and antibiotics were laid on the agar surface. The prepared plates were incubated for 2 days at 28°C. A clear zone indicates bacterial growth inhibition and antimicrobial activity.
Statistical analysis
Statistical analyses were performed using GraphPad Prism (v. 8.0). Significant differences between experimental groups were analyzed by one‐way ANOVA with Tukey's HSD test or unpaired Student's t‐test for multiple comparisons or single comparisons, respectively. Detailed information about statistical analysis is described in the figure legends. Statistical significance was set at P < 0.05. All experiments were repeated 3–5 times with similar results.
Author contributions
M‐HL, HSJ, and OKP designed the research. M‐HL and HSJ performed most of the experiments. SHK and HJC participated in lignin staining and quantification analysis. H‐JL analyzed cross‐section images. DR generated the pCASPL4D1::CASPL4D1‐mCherry line. JHC performed SA quantification experiments. YT and JR synthesized the CANBD and CADMAC. M‐HL, HSJ, and OKP analyzed the data and wrote the article. All authors contributed to the reviewing and editing of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Movie EV8
Movie EV9
Movie EV10
Movie EV11
Movie EV12
Movie EV13
Movie EV14
Review Process File
Acknowledgments
We thank Sheng Yang He for GFP‐Pst DC3000 and Pst DC3000 (AvrRpt2), Kee Hoon Sohn for rpm1‐3 rps2‐101c seeds, and Zhixiang Chen for palQ seeds. We also thank Eunjeong Jang for technical assistance and Kyoungho Kim for computer simulation and modeling. We are grateful to Niko Geldner and Yuree Lee for technical advice and critical reading of the manuscript. This work was supported by a Korea University grant, a Next‐Generation BioGreen 21 Program (SSAC, PJ013202) from the Rural Development Administration, and National Research Foundation of Korea (NRF) grants (2019R1A2C2003810; 2018R1A5A1023599, SRC) from the Korean government (MSIP). J.R. and Y.T. acknowledge support from the DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE‐FC02‐07ER64494 and DE‐SC0018409), Stanford's Global Climate and Energy Program (GCEP), DOE BER grant DOE ER65174‐1038248‐0017549, and the Japan Society for the Promotion of Science (KAKENHI grant no. 16H06198).
The EMBO Journal (2019) 38: e101948
References
- Alassimone J, Fujita S, Doblas VG, van Dop M, Barberon M, Kalmbach L, Vermeer JE, Rojas‐Murcia N, Santuari L, Hardtke CS et al (2016) Polarly localized kinase SGN1 is required for Casparian strip integrity and positioning. Nat Plants 2: 16113 [DOI] [PubMed] [Google Scholar]
- Barros J, Serk H, Granlund I, Pesquet E (2015) The cell biology of lignification in higher plants. Ann Bot 115: 1053–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A, Kanyuka K, Baulcombe DC (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11: 781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M, Gallagher M, Fagg J, Bestwick C, Paul T, Beale M, Mansfield J (1996) The hypersensitive reaction, membrane damage and accumulation of autofluorescent phenolics in lettuce cells challenged by Bremia lactucae . Plant J 9: 851–865 [Google Scholar]
- Berthet S, Demont‐Caulet N, Pollet B, Bidzinski P, Cezard L, Le Bris P, Borrega N, Herve J, Blondet E, Balzergue S et al (2011) Disruption of LACCASE4 and 17 results in tissue‐specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan NH, Selvaraj G, Wei Y, King J (2008) Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J Exp Bot 60: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Boller T, He SY (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44: 337–363 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Im Kim J, Tobimatsu Y, Ciesielski PN, Anderson NA, Ximenes E, Maeda J, Ralph J, Donohoe BS, Ladisch M (2014) Disruption of Mediator rescues the stunted growth of a lignin‐deficient Arabidopsis mutant. Nature 509: 376–380 [DOI] [PubMed] [Google Scholar]
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A (2004) Neutrophil extracellular traps kill bacteria. Science 303: 1532–1535 [DOI] [PubMed] [Google Scholar]
- Brisson LF, Tenhaken R, Lamb C (1994) Function of oxidative cross‐linking of cell wall structural proteins in plant disease resistance. Plant Cell 6: 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DM, Hernández‐Guzmán G, Kloek AP, Alarcón‐Chaidez F, Sreedharan A, Rangaswamy V, Peñaloza‐Vázquez A, Bender CL, Kunkel BN (2004) Identification and characterization of a well‐defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 17: 162–174 [DOI] [PubMed] [Google Scholar]
- Bücherl CA, Jarsch IK, Schudoma C, Segonzac C, Mbengue M, Robatzek S, MacLean D, Ott T, Zipfel C (2017) Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. Elife 6: e25114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Biselli C, Collins NC, Consonni G, Stanca AM, Schulze‐Lefert P, Valè G (2010) The CC‐NB‐LRR‐type Rdg2a resistance gene confers immunity to the seed‐borne barley leaf stripe pathogen in the absence of hypersensitive cell death. PLoS One 5: e12599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century KS, Holub EB, Staskawicz BJ (1995) NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc Natl Acad Sci USA 92: 6597–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarino I (2019) Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr Opin Biotechnol 56: 209–214 [DOI] [PubMed] [Google Scholar]
- Chang XF, Chandra R, Berleth T, Beatson RP (2008) Rapid, microscale, acetyl bromide‐based method for high‐throughput determination of lignin content in Arabidopsis thaliana . J Agric Food Chem 56: 6825–6834 [DOI] [PubMed] [Google Scholar]
- Chezem WR, Memon A, Li F‐S, Weng J‐K, Clay NK (2017) SG2‐type R2R3‐MYB transcription factor MYB15 controls defense‐induced lignification and basal immunity in Arabidopsis . Plant Cell 29: 1907–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host‐microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Fengler KA, Yu I‐C, Lippok B, Smith RK, Bent AF (2000) The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide‐gated ion channel. Proc Natl Acad Sci USA 97: 9323–9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Vercammen D, Smidler A, Clover C, Van Breusegem F, Dangl JL, Epple P (2010) Arabidopsis type I metacaspases control cell death. Science 330: 1393–1397 [DOI] [PubMed] [Google Scholar]
- Coll N, Epple P, Dangl J (2011) Programmed cell death in the plant immune system. Cell Death Differ 18: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE (2015) Effector‐triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66: 487–511 [DOI] [PubMed] [Google Scholar]
- Daniel R, Guest D (2006) Defence responses induced by potassium phosphonate in Phytophthora palmivora‐challenged Arabidopsis thaliana . Physiol Mol Plant Pathol 67: 194–201 [Google Scholar]
- Dawson J, Sözen E, Vizir I, Van Waeyenberge S, Wilson Z, Mulligan B (1999) Characterization and genetic mapping of a mutation (ms35) which prevents anther dehiscence in Arabidopsis thaliana by affecting secondary wall thickening in the endothecium. New Phytol 144: 213–222 [Google Scholar]
- Denness L, McKenna JF, Segonzac C, Wormit A, Madhou P, Bennett M, Mansfield J, Zipfel C, Hamann T (2011) Cell wall damage‐induced lignin biosynthesis is regulated by a reactive oxygen species‐and jasmonic acid‐dependent process in Arabidopsis . Plant Physiol 156: 1364–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicko MH, Gruppen H, Barro C, Traoré AS, van Berkel WJ, Voragen AG (2005) Impact of phenolic compounds and related enzymes in sorghum varieties for resistance and susceptibility to biotic and abiotic stresses. J Chem Ecol 31: 2671–2688 [DOI] [PubMed] [Google Scholar]
- Doblas VG, Smakowska‐Luzan E, Fujita S, Alassimone J, Barberon M, Madalinski M, Belkhadir Y, Geldner N (2017) Root diffusion barrier control by a vasculature‐derived peptide binding to the SGN3 receptor. Science 355: 280–284 [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Elleuch L, Shaaban M, Smaoui S, Mellouli L, Karray‐Rebai I, Fguira LF‐B, Shaaban KA, Laatsch H (2010) Bioactive secondary metabolites from a new terrestrial Streptomyces sp. TN262. Appl Biochem Biotechnol 162: 579–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M (2010) Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part I: lignin. J Vis Exp 37: e1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Beattie GA (2009) Bacterial growth restriction during host resistance to Pseudomonas syringae is associated with leaf water loss and localized cessation of vascular activity in Arabidopsis thaliana . Mol Plant Microbe Interact 22: 857–867 [DOI] [PubMed] [Google Scholar]
- Fukuda H (1997) Tracheary element differentiation. Plant Cell 9: 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego‐Giraldo L, Jikumaru Y, Kamiya Y, Tang Y, Dixon RA (2011) Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol 190: 627–639 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Guo A, Klessig DF, Ausubel FM (1994) Programmed cell death in plants: a pathogen‐triggered response activated coordinately with multiple defense functions. Cell 77: 551–563 [DOI] [PubMed] [Google Scholar]
- Hao H, Fan L, Chen T, Li R, Li X, He Q, Botella MA, Lin J (2014) Clathrin and membrane microdomains cooperatively regulate RbohD dynamics and activity in Arabidopsis . Plant Cell 26: 1729–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou Y‐H, Yu J‐Q, Chen Z (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153: 1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga Y, Ishiga T, Uppalapati SR, Mysore KS (2011) Arabidopsis seedling flood‐inoculation technique: a rapid and reliable assay for studying plant‐bacterial interactions. Plant Methods 7: 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kamiya T, Borghi M, Wang P, Danku JM, Kalmbach L, Hosmani PS, Naseer S, Fujiwara T, Geldner N, Salt DE (2015) The MYB36 transcription factor orchestrates Casparian strip formation. Proc Natl Acad Sci USA 112: 10533–10538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp N, Barnes WJ, Richard TL, Anderson CT (2015) Imaging with the fluorogenic dye Basic Fuchsin reveals subcellular patterning and ecotype variation of lignification in Brachypodium distachyon. J Exp Bot 66: 4295–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärkönen A, Koutaniemi S (2010) Lignin biosynthesis studies in plant tissue cultures. J Integr Plant Biol 52: 176–185 [DOI] [PubMed] [Google Scholar]
- Kawano Y, Shimamoto K (2013) Early signaling network in rice PRR‐mediated and R‐mediated immunity. Curr Opin Plant Biol 16: 496–504 [DOI] [PubMed] [Google Scholar]
- Kim HG, Kwon SJ, Jang YJ, Nam MH, Chung JH, Na Y‐C, Guo H, Park OK (2013) GDSL LIPASE1 modulates plant immunity through feedback regulation of ethylene signaling. Plant Physiol 163: 1776–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44: 301–307 [PubMed] [Google Scholar]
- König S, Feussner K, Kaever A, Landesfeind M, Thurow C, Karlovsky P, Gatz C, Polle A, Feussner I (2014) Soluble phenylpropanoids are involved in the defense response of Arabidopsis against Verticillium longisporum . New Phytol 202: 823–837 [DOI] [PubMed] [Google Scholar]
- Kuc J (1995) Phytoalexins, stress metabolism, and disease resistance in plants. Annu Rev Phytopathol 33: 275–297 [DOI] [PubMed] [Google Scholar]
- Kwon SI, Cho HJ, Kim SR, Park OK (2013) The Rab GTPase RabG3b positively regulates autophagy and immunity‐associated hypersensitive cell death in Arabidopsis . Plant Physiol 161: 1722–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sharma Y, Lee TK, Chang M, Davis KR (2001) Lignification induced by pseudomonads harboring avirulent genes on Arabidopsis . Mol Cells 12: 25–31 [PubMed] [Google Scholar]
- Lee Y, Rubio MC, Alassimone J, Geldner N (2013) A mechanism for localized lignin deposition in the endodermis. Cell 153: 402–412 [DOI] [PubMed] [Google Scholar]
- Lee Y, Yoon TH, Lee J, Jeon SY, Lee JH, Lee MK, Chen H, Yun J, Oh SY, Wen X (2018) A lignin molecular brace controls precision processing of cell walls critical for surface integrity in Arabidopsis . Cell 173: 1468–1480 [DOI] [PubMed] [Google Scholar]
- Liberman LM, Sparks EE, Moreno‐Risueno MA, Petricka JJ, Benfey PN (2015) MYB36 regulates the transition from proliferation to differentiation in the Arabidopsis root. Proc Natl Acad Sci USA 112: 12099–12104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF (2000) SHATTERPROOF MADS‐box genes control seed dispersal in Arabidopsis . Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CM (2006) Significance of inducible defense‐related proteins in infected plants. Annu Rev Phytopathol 44: 135–162 [DOI] [PubMed] [Google Scholar]
- Maher EA, Bate NJ, Ni W, Elkind Y, Dixon RA, Lamb CJ (1994) Increased disease susceptibility of transgenic tobacco plants with suppressed levels of preformed phenylpropanoid products. Proc Natl Acad Sci USA 91: 7802–7806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovsky FG, Fangel JU, Willats WG (2014) The role of the cell wall in plant immunity. Front Plant Sci 5: 178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury S, Delaunay A, Mesnard F, Cronier D, Chabbert B, Geoffroy P, Legrand M (2010) O‐methyltransferase (s)‐suppressed plants produce lower amounts of phenolic vir inducers and are less susceptible to Agrobacterium tumefaciens infection. Planta 232: 975–986 [DOI] [PubMed] [Google Scholar]
- Mellersh DG, Foulds IV, Higgins VJ, Heath MC (2002) H2O2 plays different roles in determining penetration failure in three diverse plant–fungal interactions. Plant J 29: 257–268 [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Menden B, Kohlhoff M, Moerschbacher BM (2007) Wheat cells accumulate a syringyl‐rich lignin during the hypersensitive resistance response. Phytochemistry 68: 513–520 [DOI] [PubMed] [Google Scholar]
- Métraux JP (2002) Recent breakthroughs in the study of salicylic acid biosynthesis. Trends Plant Sci 7: 332–334 [DOI] [PubMed] [Google Scholar]
- Miedes E, Vanholme R, Boerjan W, Molina A (2014) The role of the secondary cell wall in plant resistance to pathogens. Front Plant Sci 5: 358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu G‐L, Ausubel FM (1994) The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide‐binding site and leucine‐rich repeats. Cell 78: 1089–1099 [DOI] [PubMed] [Google Scholar]
- Mitra PP, Loqué D (2014) Histochemical staining of Arabidopsis thaliana secondary cell wall elements. J Vis Exp 87: e51381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerschbacher BM, Noll U, Gorrichon L, Reisener H‐J (1990) Specific inhibition of lignification breaks hypersensitive resistance of wheat to stem rust. Plant Physiol 93: 465–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales J, Kadota Y, Zipfel C, Molina A, Torres M‐A (2016) The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J Exp Bot 67: 1663–1676 [DOI] [PubMed] [Google Scholar]
- Nakayama T, Shinohara H, Tanaka M, Baba K, Ogawa‐Ohnishi M, Matsubayashi Y (2017) A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355: 284–286 [DOI] [PubMed] [Google Scholar]
- Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N (2012) Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci USA 109: 10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson RL, Hammerschmidt R (1992) Phenolic compounds and their role in disease resistance. Annu Rev Phytopathol 30: 369–389 [Google Scholar]
- Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V (2018) Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol 18: 134–147 [DOI] [PubMed] [Google Scholar]
- Pfister A, Barberon M, Alassimone J, Kalmbach L, Lee Y, Vermeer JE, Yamazaki M, Li G, Maurel C, Takano J (2014) A receptor‐like kinase mutant with absent endodermal diffusion barrier displays selective nutrient homeostasis defects. Elife 3: e03115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin M, Allasia V, Pegard A, Allais F, Ducrot P‐H, Favery B, Levis C, Martinet S, Masur C, Ponchet M (2009) Imbalanced lignin biosynthesis promotes the sexual reproduction of homothallic oomycete pathogens. PLoS Pathog 5: e1000264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin M, Baurès I, Hoefle C, Caillaud M‐C, Allasia V, Panabières F, Abad P, Hückelhoven R, Keller H, Favery B (2016) The Arabidopsis microtubule‐associated protein MAP65‐3 supports infection by filamentous biotrophic pathogens by down‐regulating salicylic acid‐dependent defenses. J Exp Bot 67: 1731–1743 [DOI] [PubMed] [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH et al (2004) Lignins: natural polymers from oxidative coupling of 4‐hydroxyphenyl‐propanoids. Phytochem Rev 3: 29–60 [Google Scholar]
- Robertsen B (1986) Elicitors of the production of lignin‐like compounds in cucumber hypocotyls. Physiol Mol Plant Pathol 28: 137–148 [Google Scholar]
- Roppolo D, De Rybel B, Tendon VD, Pfister A, Alassimone J, Vermeer JE, Yamazaki M, Stierhof Y‐D, Beeckman T, Geldner N (2011) A novel protein family mediates Casparian strip formation in the endodermis. Nature 473: 380–383 [DOI] [PubMed] [Google Scholar]
- Roppolo D, Boeckmann B, Pfister A, Boutet E, Rubio MC, Dénervaud‐Tendon V, Vermeer JE, Gheyselinck J, Xenarios I, Geldner N (2014) Functional and evolutionary analysis of the CASPARIAN STRIP MEMBRANE DOMAIN PROTEIN family. Plant Physiol 165: 1709–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler S, Funnell‐Harris D (2013) Modifying lignin to improve bioenergy feedstocks: strengthening the barrier against pathogens? Front Plant Sci 4: 70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalk M, Cabello‐Hurtado F, Pierrel M‐A, Atanassova R, Saindrenan P, Werck‐Reichhart D (1998) Piperonylic acid, a selective, mechanism‐based inactivator of the trans‐cinnamate 4‐hydroxylase: a new tool to control the flux of metabolites in the phenylpropanoid pathway. Plant Physiol 118: 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Stout J, Weng JK, Humphreys J, Ruegger MO, Chapple C (2009) Mutations in the cinnamate 4‐hydroxylase gene impact metabolism, growth and development in Arabidopsis . Plant J 60: 771–782 [DOI] [PubMed] [Google Scholar]
- Shindo T, Misas‐Villamil JC, Hörger AC, Song J, van der Hoorn RA (2012) A role in immunity for Arabidopsis cysteine protease RD21, the ortholog of the tomato immune protease C14. PLoS One 7: e29317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalamera D, Heath MC (1998) Changes in the cytoskeleton accompanying infection‐induced nuclear movements and the hypersensitive response in plant cells invaded by rust fungi. Plant J 16: 191–200 [DOI] [PubMed] [Google Scholar]
- Thomma BP, Nürnberger T, Joosten MH (2011) Of PAMPs and effectors: the blurred PTI‐ETI dichotomy. Plant Cell 23: 4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobimatsu Y, Davidson CL, Grabber JH, Ralph J (2011) Fluorescence‐tagged monolignols: synthesis, and application to studying in vitro lignification. Biomacromol 12: 1752–1761 [DOI] [PubMed] [Google Scholar]
- Tobimatsu Y, Wagner A, Donaldson L, Mitra P, Niculaes C, Dima O, Kim JI, Anderson N, Loque D, Boerjan W et al (2013) Visualization of plant cell wall lignification using fluorescence‐tagged monolignols. Plant J 76: 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JD (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Dangl JL (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol 8: 397–403 [DOI] [PubMed] [Google Scholar]
- Torres MA (2010) ROS in biotic interactions. Physiol Plant 138: 414–429 [DOI] [PubMed] [Google Scholar]
- Tronchet M, Balague C, Kroj T, Jouanin L, Roby D (2010) Cinnamyl alcohol dehydrogenases‐C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis . Mol Plant Pathol 11: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Glazebrook J, Cohen JD, Katagiri F (2008) Interplay between MAMP‐triggered and SA‐mediated defense responses. Plant J 53: 763–775 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voxeur A, Wang Y, Sibout R (2015) Lignification: different mechanisms for a versatile polymer. Curr Opin Plant Biol 23: 83–90 [DOI] [PubMed] [Google Scholar]
- Wen F, White GJ, VanEtten HD, Xiong Z, Hawes MC (2009) Extracellular DNA is required for root tip resistance to fungal infection. Plant Physiol 151: 820–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetten R, Sederoff R (1995) Lignin biosynthesis. Plant Cell 7: 1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Wróbel‐Kwiatkowska M, Starzycki M, Zebrowski J, Oszmiański J, Szopa J (2007) Lignin deficiency in transgenic flax resulted in plants with improved mechanical properties. J Biotechnol 128: 919–934 [DOI] [PubMed] [Google Scholar]
- Yang J, Ding C, Xu B, Chen C, Narsai R, Whelan J, Hu Z, Zhang M (2015) A Casparian strip domain‐like gene, CASPL, negatively alters growth and cold tolerance. Sci Rep 5: 14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I‐c, Parker J, Bent AF (1998) Gene‐for‐gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci USA 95: 7819–7824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernova OV, Lygin AV, Pawlowski ML, Hill CB, Hartman GL, Widholm JM, Lozovaya VV (2014) Regulation of plant immunity through modulation of phytoalexin synthesis. Molecules 19: 7480–7496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhou J‐M (2010) Plant immunity triggered by microbial molecular signatures. Mol Plant 3: 783–793 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Felix G (2005) Plants and animals: a different taste for microbes? Curr Opin Plant Biol 8: 353–360 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Movie EV8
Movie EV9
Movie EV10
Movie EV11
Movie EV12
Movie EV13
Movie EV14
Review Process File


