Abstract
Keshan disease (KD) is an endemic cardiomyopathy, which mainly occurs in China. Selenium deficiency is believed to play an important role in the pathogenesis of KD, but the molecular mechanism of selenium-induced damage remains unclear. To identify the key genes involved in selenium-induced damage, we compared the expression profiles of selenium-related genes between patients with KD and normal controls. Total RNA was isolated, amplified, labeled, and hybridized to Agilent human 4 × 44 K whole genome microarrays. Selenium-related genes were screened using the Comparative Toxicogenomics Database. The microarray data were subjected to single-gene and gene ontology (GO) expression analysis using R Studio and Gene Set Enrichment Analysis (GSEA) software. Quantitative real-time PCR was conducted to validate the microarray results. We identified 16 upregulated and 11 downregulated selenium-related genes in patients. These genes are involved in apoptosis, metabolism, transcription regulation, ion transport, and growth and development. Of the significantly enriched GO categories in KD patients, we identified four apoptosis-related, two metabolism-related, four growth and development-related, and four ion transport-related GOs. Based on our results, we suggest that selenium might contribute to the development of KD through dysfunction of selenium-related genes involved in apoptosis, metabolism, ion transport, and growth and development in the myocardium.
1. Introduction
Keshan disease (KD) is an endemic cardiomyopathy with a high mortality rate. It was first reported in Keshan County in northeast China in 1935, while similar cases were found in Nagano Prefecture in Japan and the northern mountains of North Korea in the 1950s. It is characterized by acute heart failure, congestive heart failure, and cardiac arrhythmia. It can be classified into acute, sub-acute, chronic, and latent KD. Currently, acute and sub-acute cases are almost nonexistent, but chronic and latent cases are still scattered in many areas. Most patients are distributed among 2953 townships in 16 provinces from the northeast to the southwest of China, mainly in remote rural areas [1]. Although the incidence of KD has declined in recent years, records still indicate 379,800 chronic KD patients and 1,295,700 latent KD patients, with a total prevalence of 0.13% in China [1].
The etiology of KD has been widely discussed, but it is still unclear. Currently, KD is considered as a gene-environment interaction disease linked to selenium deficiency [2]. Selenium, an essential micronutrient, exerts its function through selenoproteins or small selenium metabolites in the human body [3]. It has been shown that all selenoproteins that have enzymic functions are selenium-dependent, generally with selenocysteine at the active site as a redox center to reduce oxidative damage to biomolecules. In cardiovascular disease, one hypothesis is that selenium deficiency may increase the production of oxidized low-density lipoprotein, thereby promoting the incidence of heart disease [4]. Previous evidence has supported selenium deficiency as a key environmental risk factor for KD. Selenium levels in soil and food are clearly lower in endemic areas than in nonendemic areas, and selenium levels in the blood and hair are significantly lower in KD patients than normal controls [5]. Moreover, it has been reported that the incidence of KD dramatically declined after selenium supplementation [6]. Molecular work has implicated genes related to selenium activity in KD, for example, the selenium-related proteins, glutathione peroxidase 1 (GPX1) and thioredoxin reductase 1 (TrxR1) were found to be decreased significantly in the myocardium of patients with KD compared to controls [7, 8]. However, information is still lacking on expression changes of global selenium-related genes in KD patients. Such data may contribute to understanding of the development of KD.
Blood gene expression profiling has been applied to patients with heart diseases including heart failure and idiopathic dilated cardiomyopathy, and linked to pathological changes as surrogates for disease predication and research [9, 10]. It has been reported that some gene expression signatures in blood can act as valuable diagnostic and prognostic tools for monitoring of disease progression. Moreover, one important pathological change observed in hearts from 308 autopsy cases of KD between 1959 and 1983 is that the tributaries of the intramural coronary system of the left ventricle are closely related to the distribution of necrotic KD foci. Multiple miliary foci usually develop around the branching type of coronary arteries, and the cross section of the terminal arteriole is usually the necrotic focus [11]. In addition to the above, PBMCs appear to hold a great advantage because they are easily obtained for further study or early detection compared to heart tissue. Thus, in this study, we compared the expression profiles of 1517 selenium-related genes in peripheral blood mononuclear cells (PBMCs) of patients with KD and normal controls to identify the key genes that contribute to selenium-induced damage in KD. Both single-gene and gene-set expression analyses were conducted to identify differentially expressed genes and gene ontology (GO) terms, which may provide clues to the pathological mechanism of KD.
2. Methods
All studies were approved by the Institutional Review Board (IRB) of Xi'an Jiaotong University. Informed-consent documents were signed by all participants.
2.1. Groups and Diagnostic Criteria
Inclusion criteria for patients with KD were the following: (1) KD diagnosis according to “The Diagnostic Standard of the KD in China” (WS/T 210 - 2011); and (2) living in KD areas for more than six months. Chronic KD was diagnosed based on slow onset, cardiac function grade higher than I as per the New York Heart Association, and cardiothoracic ratio higher than 0.55. Latent KD was diagnosed based on insidious onset, cardiac function grade I or normal, and cardiothoracic ratio less than 0.55. Inclusion criteria for normal controls were the following: (1) good health; (2) residence in an endemic area for more than six months; (3) match by age and area of origin with the patients. Patients with other heart diseases and severe illnesses, such as congenital heart disease, diabetes, and hyperlipidemia, were excluded from the study. Based on the inclusion and exclusion criteria, 16 patients and 16 normal controls were selected for the microarray experiment. All were from Xunyi and Huangling counties, Shaanxi Province, China, where the crude prevalence of KD was 1.13% in 2012 [1]. All participants underwent electrocardiography, chest X-ray, and echocardiography. Details of patients and controls are given in Table 1.
Table 1.
Characteristics of KD patient-control sample pairs.
| Chip1 (P-C) a | Chip2 (P-C) | Chip3 (P-C) | Chip4 (P-C) | ||
|---|---|---|---|---|---|
| N | 4–4 | 4–4 | 4–4 | 4–4 | |
| Gender (male/female) | 4/0-4/0 | 0/4–0/4 | 0/4–0/4 | 0/4–0/4 | |
| Average age (year) | 44.75–52.00 | 41.25–39.74 | 49.75–48.25 | 57.75–58.75 | |
| N (latent/chronic) | 2/2 | 2/2 | 2/2 | 2/2 | |
| Heart rate (bpm) | 75, 71/54, 62 | 66, 66/85, 76 | 82, 75/63, 85 | 95, 87/62, 64 | |
| Cardiothoracic ratio | 0.61,0.58/0.46,0.45 | 0.57,0.58/0.53,0.52 | 0.63,0.59/0.50,0.50 | 0.87,0.60/0.47,0.44 | |
| NYHA classb | Ⅲ,Ⅱ /Ⅰ,Ⅰ | Ⅱ,Ⅲ /Ⅰ,Ⅰ | Ⅱ,Ⅲ /Ⅰ,Ⅰ | Ⅲ,Ⅲ /Ⅰ,Ⅰ | |
| Electrocardiograph | RBBB,ST-T/LBBB, RBBB | ST-T,ST-T/ST-T, ST-T | VPB,RBBB/ST-T, ST-T | AF,AF/RBBB, RBBB | |
| LVEFc | 0.62,0.74/0.46,0.47 | 0.48,0.62/0.49,0.72 | 0.60,0.62/0.77,0.74 | 0.68,0.65/0.54,0.77 |
aP—C, patient-control.
bNYHA class, New York Heart Association class.
cLeft ventricular ejection fraction.
2.2. Blood Collection and PBMC Isolation
We collected two milliliters peripheral blood from each subject into heparinized vacutainer tubes (Becton Dickenson, San Jose, CA, USA). Blood mixtures were centrifuged at 800 g for 30 min with Lympholyte-H (Cedarlane Laboratories, ON, Canada) to isolate PBMCs, and then rinsed twice using PBS. Cells were stored at −80°C until total RNA isolation.
2.3. Extraction of Total RNA from PBMCs
Total RNA was isolated from PBMCs using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Concentration and purity detection was performed by an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, California, USA) and gel electrophoresis containing formaldehyde.
2.4. Microarray Hybridization
Isolated total RNA from PBMCs of each patient-control pair was first transcribed into complementary DNA (cDNA), and then reverse transcribed into cRNA and labeled with CyDye using an amino allyl MessageAmp aRNA Kit (Ambion). Before reverse transcription, RNase-free DNase I was used to remove residual genomic DNA. For each patient-control pair, 0.5 μg labeled cRNA was purified separately and mixed together with hybridization buffer before microarray hybridization. Agilent Human 4 × 44 K Whole Genome microarray (G4112F), which consists of 44290 oligonucleotide probes representing 41675 human genes, was employed for microarray hybridization. Hybridization signals were scanned by Gene-Pix 4000B (Axon Instruments Inc., Foster City, CA, USA), Feature Extraction 9.3 software (Agilent Technologies), and Spotfire 8.0 (Spotfire Inc., Cambridge, MA, USA) software were used to record and analyze microarray data. Fluorescent spots that failed to pass the quality control procedures were excluded. Data files were imported into Excel 2010 prior to statistical analysis.
2.5. Quantitative RT-PCR Validation
We selected seven differentially expressed genes for validation by quantitative RT-PCR: ACACB, APOA1, BCL2L1, BNIP3L, CYB5A, PGC-1alpha, and HBA2. GAPDH was selected as an internal reference gene. Total RNA samples were prepared in the same way as in the microarray experiment and further converted to cDNA using an RT reagent kit (TAKARA, Takara Biotechnology, CHN). Quantitative RT-PCRs were conducted using SYBR premix Ex Taq™ II (TAKARA, Dalian, CHN) and iQ5™ Real Time PCR Systems (Bio-Rad, CA, USA). We used 2-△△C(t) to calculate relative expression of each individual gene [12]. Paired samples t-tests were conducted to compare quantitative RT-PCR data between controls and KD patients using SPSS version 19.0 software, and P < 0.05 was considered a statistically significantly difference.
2.6. Gene Expression Analysis
To investigate the expression levels of selenium-related genes in PBMCs of patients with KD, we downloaded 1660 selenium-related genes from the Comparative Toxicogenomics Database (CTD, http://ctdbase.org/). After merging these with the normalized microarray data mentioned above, 1517 selenium-related genes (see Supplemental Data ) were obtained and analyzed by R Studio software. CTD is a publicly available database and provides data as well as detailed steps on “what genes/protein interact with a chemical?”. It provides manually curated chemical-gene interactions in vertebrates and invertebrates from the published literature. Significantly differentially expressed selenium-related genes were defined by a fold change (FC) cut-off of >1.5, a P-value < 0.05, and a false discovery rate (FDR) <0.05 [13].
To further identify differently expressed GO terms between patients and controls, we used gene set enrichment analysis (GSEA) software. It provides a normalized enrichment score (NES), which reflects the degrees of over-expression of gene sets, with positive and negative NES values, respectively, indicating the up- and downregulation of gene sets in patients compared to controls. The false discovery rate (FDR) is the estimated probability that a gene set with a specific NES represents a false positive finding. The nominal P-value estimates the statistical significance of the enrichment score for a single-gene set. In this study, we choseFDR < 0.25 and P < 0.05 to determine significantly differentially expressed GOs. Gene ontology database 6.0 containing 5,917 GO categories was downloaded from the GSEA website (http://www.broadinstitute.org/gsea/index.jsp) for use in this study.
3. Results
3.1. Quantitative RT-PCR Analysis
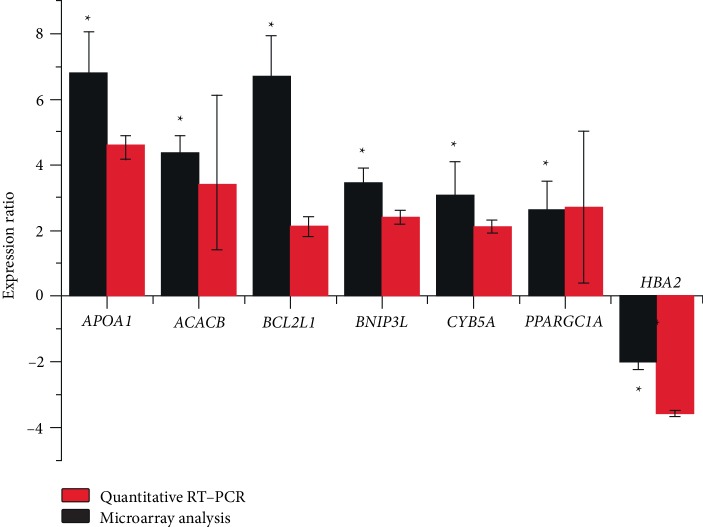
Quantitative RT-PCR results revealed the increased levels of the following six transcripts: ACACB, APOA1, BCL2L1, BNIP3L, CYB5A, PGC-1α, and a decreased level of HBA2, in KD patients compared to controls (P < 0.05). The differences in the relative expression of these genes were consistent with the results of the microarray analysis (Figure 1).
Figure 1.
Histogram showing expression levels of seven selected genes as measured by oligonucleotide microarrays and Quantitative RT-PCR.
3.2. Single-Gene Expression Analysis
We identified 27 differentially expressed selenium-related genes (16 up- and 11 downregulated) in KD patients compared to controls. The 16 upregulated genes were involved in various biological processes, including apoptosis, transcription regulation, metabolism, cytoskeleton and movement, growth and development, and ion transport. The 11 downregulated genes were involved in signal transduction, protein synthesis and modification, cytoskeleton and movement, growth and development, ion channels, and transport proteins (Table 2).
Table 2.
Differently expressed selenium related genes in patients compared to controls.
| Gene Name | Pubic ID | Function | logFCa | P-value | FDR | |
|---|---|---|---|---|---|---|
| Up-regulated Genes | ||||||
| OPTN | NM_001008211 | Transcription regulator | 1.056 | <0.001 | 0.002 | |
| WASF2 | NM_006990 | Cytoskeleton and cell movement | 1.180 | <0.001 | 0.005 | |
| PRSS8 | NM_002773 | Metabolism | 1.260 | <0.001 | 0.006 | |
| TNFAIP6 | NM_007115 | Apoptosis | 1.199 | <0.001 | 0.006 | |
| SELENBP1 | NM_003944 | Cell cycle | 1.004 | <0.001 | 0.006 | |
| IL1RL2 | NM_003854 | Interleukin | 1.432 | <0.001 | 0.013 | |
| BAG1 | NM_004323 | Oncogene related | 0.898 | <0.001 | 0.019 | |
| OSBP2 | NM_030758 | Ion channel and transport protein | 1.057 | <0.001 | 0.020 | |
| TMC5 | NM_024780 | Other | 0.977 | <0.001 | 0.028 | |
| SLC25A37 | AY032628 | Ion channel and transport protein | 1.121 | <0.001 | 0.030 | |
| FECH | NM_001012515 | Metabolism | 0.958 | <0.001 | 0.039 | |
| GCLC | NM_001498 | Metabolism | 0.994 | <0.001 | 0.033 | |
| LTBP3 | NM_021070 | Growth factor related | 0.976 | <0.001 | 0.038 | |
| ARG1 | NM_000045 | Metabolism | 0.938 | <0.001 | 0.046 | |
| CSF1 | NM_005211 | Cytokine | 1.203 | <0.001 | 0.044 | |
| BCL2L1 | NM_138578 | Apoptosis | 1.205 | <0.001 | 0.048 | |
| Down-regulated Genes | ||||||
| LEP | NM_017526 | Receptor | −2.013 | <0.001 | <0.001 | |
| HBA2 | NM_000517 | Ion channel and transport protein | -−.698 | <0.001 | <0.001 | |
| SLC29A1 | NM_004955 | Ion channel and transport protein | −1.607 | <0.001 | <0.001 | |
| CSNK2A1 | NM_177559 | Signal transduction related | −1.668 | <0.001 | <0.001 | |
| TGFβ1 | NM_015927 | Growth factor related | −1.278 | <0.001 | 0.006 | |
| RHOBTB3 | NM_014899 | Signal transduction related | −1.531 | <0.001 | 0.006 | |
| ATP6V0D1 | NM_004691 | Ion channel and transport protein | −1.127 | <0.001 | 0.019 | |
| SPTAN1 | NM_003127 | Cytoskeleton and cell movement | −1.118 | <0.001 | 0.021 | |
| SLC3A2 | NM_002394 | Ion channel and transport protein | −1.092 | <0.001 | 0.030 | |
| CAPNS1 | NM_001749 | Cytokine | −1.061 | <0.001 | 0.033 | |
| COPE | NM_007263 | Protein synthesis and modification | −1.048 | <0.001 | 0.036 | |
aFC, Fold Change.
3.3. Gene Set Expression Analysis
GSEA detected significant enrichment of multiple GO categories in patients with KD compared to controls. Of these, four were apoptosis-related, two were metabolism-related, four were ion transport-related, and four were development-related (Table 3). The enrichment GO categories with the highest normalized enrichment score (NES) are shown in Figure 2.
Table 3.
Significant enrichment of Gene Ontology categories in KD patients compared to controls.
| Gene ontology category | Function | NESa | P-value |
|---|---|---|---|
| Negative_regulation_of_neuron_apoptotic_process | Apoptosis | 1.650 | <0.001 |
| Regulation_of_extrinsic_apoptotic_signaling_pathway | Apoptosis | 1.665 | <0.001 |
| Apoptotic_mitochondrial_changes | Apoptosis | 1.534 | <0.001 |
| Apoptotic_signaling_pathway | Apoptosis | 1.566 | <0.001 |
| Cellular_modified_amino_acid_metabolic_process | Metabolism | 1.701 | <0.001 |
| Alpha_amino_acid_metabolic_process | Metabolism | 1.494 | <0.001 |
| Epidermal_cell_differentiation | Growth and development | 2.084 | <0.001 |
| Negative_regulation_of_epithelial_cell_proliferation | Growth and development | 1.992 | <0.001 |
| Positive_regulation_of_dendrite_development | Growth and development | 1.629 | <0.001 |
| Gland_morphogenesis | Growth and development | 1.615 | <0.001 |
| Metal_ion_transport | Ion transport | 1.794 | <0.001 |
| Cation_transport | Ion transport | 1.588 | <0.001 |
| Inorganic_cation_transmembrane_transporter_activity | Ion transport | 1.527 | <0.001 |
| Monovalent_inorganic_cation_transport | Ion transport | 1.519 | <0.001 |
aDenotes normalized enrichment score (NES) calculated by GSEA. Positive NES indicate gene ontology up-regulated in patients compared to controls.
Figure 2.
Enrichment plots with the highest normalized enrichment score (NES) for the up-regulated and downregulated genes data.
4. Discussion
In this study, we compared expression of selenium-related genes between patients with KD and normal controls, and identified a set of differentially expressed selenium-related genes and GO categories. Based on the results of this and previous studies, we suggest that selenium deficiency might contribute to KD via effects on the expression and biological function of selenium related-genes involved in apoptosis, metabolism, growth and development, and ion transport.
4.1. Apoptosis
Myocardial apoptosis is one of the primary pathological changes seen in KD [14], but the molecular mechanisms underlying this dysfunction, have not been fully elucidated. In this study, we found that the apoptosis-related gene BCL2-like-1 (BCL2L1) was significantly upregulated in patients. GO analysis further indicated four enriched apoptosis-related GO categories in patients compared to controls. This result is consistent with a previous study [15, 16], which suggested that apoptosis may play a key role in the mechanism of selenium-related damage in KD.
The outer mitochondrial membrane protein, BCL2L1 belongs to the anti-apoptotic B cell lymphoma/leukemia 2 (BCL2) family. BCL2L1 could prevent the release of mitochondrial apoptogenic factors including cytochrome C into the cytoplasm to attenuate apoptosis by regulating the opening of the voltage-dependent anion channels (VDAC). Previous reports have implicated BCL2L1 and related genes in KD. Evaluation of apoptotic percentage in myocardial tissues from 30 patients with KD (10 acute, 10 sub-acute, and 10 chronic), and five healthy controls, showed that the TUNEL positivity of patients was higher (acute: 2/10; sub-acute: 8/10; chronic: 7/10) than that of the controls [16]. Using Agilent microarray analysis, Xiang et al. identified six apoptosis-related genes that were differentially expressed in KD patients, but BCL2L1 was downregulated in patients with chronic KD [17]. There is no obvious explanation for this inconsistency, given that the cumulative data indicate the possible roles of apoptosis in the pathological process of KD. In addition to the evidence noted above, selenium deficiency has been associated with upregulated mRNA levels of BCL-2 and BCL-2-associated X Protein (Bax) (P < 0.05) [18].
4.2. Metabolism
We also found that the metabolism-related genes arginase 1 (ARG1), glutamate-cysteine ligase catalytic subunit (GCLC), Protease serine 8 (PRSS8), and ferrochelatase (FECH) were significantly upregulated in patients. GO expression analysis further indicated enrichment of two metabolism-related GO categories in patients compared to controls.
ARG1—which is predominantly expressed in the cytoplasm of the liver, endothelial cells and vascular smooth muscle cells—hydrolyzes L-arginine into L-ornithine and urea. Previous studies have shown that plasma arginase I levels were significantly higher in patients with heart failure than controls [19], and serum arginase activity was found to be significantly higher in 33 KD patients compared with 56 controls [20]. Increased arginase activity in blood of these patients may arise from enhanced spillover of enzyme from damaged myocytes [21]. These were consistent with our results. Increased cardiomyocyte arginase activity in cardiovascular disorders may negatively influence the regulation of contractility, endothelial dysfunction, and heart failure by reduction of NO bioavailability [22, 23]. Additionally, some studies on selenium and arginase activity have been provided. Selenium has no effect on rat liver arginase activity when added to liver in vitro [24]. While another study showed that ARG1 activity was significantly increased when IL-4-treated macrophages were supplemented with selenium and increased activity may contribute to the resolution of inflammation [25]. Thus, it is difficult to explain the mechanisms of dietary intake of selenium on arginase activity in patients with KD using the present data, but it seems that patients with KD supplemented with selenium should not be responsible for overexpression of ARG1 in patients with KD.
GCLC is a subunit of the first rate-limiting enzyme of glutathione synthesis. Promoter polymorphisms of GCLC have been associated with increased risk of ischemic heart disease in the Kazakhstani population [26]. Uthus and Ross showed that the expression of GCLC was upregulated by selenium deprivation in both rat and mouse liver [27], while increased levels of glutamate-cysteine ligase (GCL) may drive glutathione (GSH) synthesis in response to oxidants, providing a protective mechanism against oxidative stress [28].
4.3. Growth & Development
Another two enriched categories in KD was that of the growth and development-related genes. These included upregulated expression of latent transforming growth factor binding protein 3 (LTBP3) and selenium binding protein 1 (SELENBP1) and downregulated expression of transforming growth factor beta 1 (TGFβ1) in patients with KD compared to normal controls.
There is compelling evidence that TGFβ1 plays a critical role in various cardiac pathologies involved in heart failure [29]; however, in contrast to decreased TGFβ1 levels in KD, increased TGFβ1 levels have been found in the myocardium of human patients with idiopathic hypertrophic cardiomyopathy [30], atherosclerotic, and restenotic lesions [31]. LTBP3 forms a complex with TGFβ1 protein, and may be involved in the TGFβ1/SMAD3 pathway as a TGFβ1 target gene [32]. Decreased TGFβ1 and increased LTBP3 in patients with KD may suggest a dysregulated TGFβ signaling pathway, which would be a novel component of the mechanism underlying myocardial injury in KD. Further supporting this, Reddi and Bollineni showed that selenium deficiency increases oxidative stress via TGFβ1 [33]. Thus, selenium deficiency in patients with KD may be an inducing factor for oxidative injury of the myocardium via the TGFβ signaling pathway.
Human SELENBP1, a member of the selenium-binding protein family, has been shown to mediate the intracellular transport of selenium [34]. SELENBP1 is expressed in a variety of tissue types, including heart tissue. Some evidence has shown that decreased SELENBP1 in lung and colorectal cancer is related to poor prognosis and increased expression level in zebrafish heart, and can be used as a biomarker of aging [35]. Other previous studies may provide clues to understanding the effect of increased SELENBP1 in KD. Fang et al. found that increasing SELENBP1 in either human colorectal or breast cancer cells resulted in reduced GPX1 activity, while selenium deficiency was associated with increased SELENBP1 expression [36].
4.4. Ion Transport
The final enriched category we identified in KD was that of ion transport, including ion channel and transport-protein-related genes. Oxysterol binding protein 2(OSBP2) and solute carrier family 25 member 37 (SLC25A37) were significantly upregulated, while human hemoglobin A2 (HBA2), solute carrier family 29 member 1 (SLC29A1), ATPase H+ transporting V0 subunit d1 (ATP6V0D1), and solute carrier family 3 member 2 (SLC3A2) were significantly decreased in patients with KD compared to normal controls. GO expression analysis results showed that four ion transport-related categories were significantly enriched in patients.
HBA2 is a tetramer of α and β globin chains. Previous work has shown that the selenium content of red blood cells was significantly lowered in KD, and the activity of methemoglobin reductase was lower in rats fed with grain from KD areas than nonKD areas [6]. SLC25A37 localizes in the mitochondrial inner membrane. The solute carrier family 25 (SLC25) have been shown to transport the molecules include ATP/ADP, amino acids (glutamate, aspartate, lysine, histidine, and arginine), malate, ornithine, and citruline [37]. ATP6V0D1 encodes a component of vacuolar ATPase. Our analysis revealed a significant change in the expression of three members of a solute carrier family including SLC25A37, SLC29A1 and SLC3A2, and ATP6V0D1 may involve in energy transfer in mitochondria [38]. Obvious increases in the numbers of enlarged and swollen mitochondria and significantly decreased levels of oxidative phosphorylation were observed in heart tissue from patients with KD. This might further help to explain the role of energy metabolism disorders in the mechanism of KD.
In conclusion, we identified a set of differently expressed selenium-related genes and GO categories between patients with KD and normal controls. We suggest that selenium might contribute to the pathogenesis of KD through dysfunction of selenium-related genes involved in apoptosis, metabolism, ion transport, and growth and development in the myocardium. These results contribute to increased understanding of the molecular mechanisms underlying KD.
Acknowledgments
This work was supported by the National Natural Scientific Foundation of China (81602809 and 81273008).
Data Availability
All relevant data are contained within the paper. Additional information can be obtained by contacting Dr. Shulan He (hesl@nxmu.edu.cn).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Xiaojuan Liu and Shulan He share co-senior authorship as the first authors. Xiong Guo and Wuhong Tan share co-senior authorship as the correspondence authors.
Supplementary Materials
Selenium related genes in gene expression microarray data from Keshan Disease patients and healthy controls.
References
- 1.Li Q., Liu M., Hou J., Jiang C., Li S., Wang T. The prevalence of Keshan disease in China. International Journal of Cardiology. 2013;168(2):1121–1126. doi: 10.1016/j.ijcard.2012.11.046. [DOI] [PubMed] [Google Scholar]
- 2.Lei C., Niu X., Wei J., Zhu J., Zhu Y. Interaction of glutathione peroxidase-1 and selenium in endemic dilated cardiomyopathy. Clinica Chimica Acta. 2009;399(1-2):102–108. doi: 10.1016/j.cca.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 3.Hori E., Yoshida S., Fuchigami T., Haratake M., Nakayama M. Cardiac myoglobin participates in the metabolic pathway of selenium in rats. Metallomics. 2018;10(4):614–622. doi: 10.1039/C8MT00011E. [DOI] [PubMed] [Google Scholar]
- 4.Tinggi U. Selenium: its role as antioxidant in human health. Environmental Health and Preventive Medicine. 2008;13(2):102–108. doi: 10.1007/s12199-007-0019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X. H., Xiang Y. Z., Qu F. R., Song S. L., Wang L., Guan S. F. Study on the relationship between selenium level and the incidence of keshan disease. Journal of Control of Endemic Diseases. 2005;6:351–353. [Google Scholar]
- 6.Loscalzo J. Keshan disease, selenium deficiency, and the selenoproteome. New England Journal of Medicine. 2014;370(18):1756–1760. doi: 10.1056/NEJMcibr1402199. [DOI] [PubMed] [Google Scholar]
- 7.Pei J., Fu W., Yang L., Zhang Z., Liu Y. Oxidative stress is involved in the pathogenesis of keshan disease (an endemic dilated cardiomyopathy) in China. Oxidative Medicine and Cellular Longevity. 2013;2013:1–5. doi: 10.1155/2013/474203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pei J. R., Liu M. F., Liu Y., et al. Expression of 8-hydroxydeoxyguanylic acid, thioredoxin reductase 1 and glutathione peroxidase 1 in myocardial tissue of patients with keshan disease at autopsy. Chinese Journal of Endemiology. 2012;31:631–634. [Google Scholar]
- 9.Vanburen P., Ma J., Chao S., Mueller E., Schneider D. J., Liew C. C. Blood gene expression signatures associate with heart failure outcomes. Physiological Genomics. 2011;43(8):392–397. doi: 10.1152/physiolgenomics.00175.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szmit S., Jank M., Maciejewski H., et al. Gene expression profiling in peripheral blood nuclear cells in patients with refractory ischaemic end-stage heart failure. Journal of Applied Genetics. 2010;51(3):353–368. doi: 10.1007/bf03208866. [DOI] [PubMed] [Google Scholar]
- 11.Li G. S., Wang F., Kang D., Li C. Keshan disease: an endemic cardiomyopathy in China. Human Pathology. 1985;16(6):602–609. doi: 10.1016/S0046-8177(85)80110-6. [DOI] [PubMed] [Google Scholar]
- 12.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Dalman M. R. Fold change and p-value cutoffs significantly alter microarray interpretations. BMC Bioinformatics. 2012;13(2) doi: 10.1186/1471-2105-13-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Y., Gao C., Wang X., Liu Y. Preliminary quantitative proteomics analysis in chronic and latent Keshan disease by iTRAQ labeling approach. Oncotarget. 2017;8(62):105761–105774. doi: 10.18632/oncotarget.2239722397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xin L., Zhu Y. H., Zhang C. Y. The relationship between low-selenium and the mechanisms of oxygenated lesion of myocardial cells and the expression of apoptosis-related genes. Foreign Medical Sciences. 2011 [Google Scholar]
- 16.Zhong X., Bo Y., Zhang L. Myocardial injury of keshan disease and apoptosis. Chinese Jouranl of Cndemiology. 1997 [Google Scholar]
- 17.Xiang Y. Z., He S. L., Wang X. H., Liu Y., Zhang W. M., Huang G. Y. Differentially expressed genes and apoptosis-related pathways in Keshan disease. Chinese Journal of Endemiology. 2017;36:859–865. [Google Scholar]
- 18.Yang J., Zhang Y., Hamid S., et al. Interplay between autophagy and apoptosis in selenium deficient cardiomyocytes in chicken. Journal of Inorganic Biochemistry. 2017;170:17–25. doi: 10.1016/j.jinorgbio.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Quitter F., Figulla H. R., Ferrari M., Pernow J., Jung C. Increased arginase levels in heart failure represent a therapeutic target to rescue microvascular perfusion. Clinical Hemorheol Microcirculation. 2013;54(1):75–85. doi: 10.3233/CH-2012-1617. [DOI] [PubMed] [Google Scholar]
- 20.Wang M. Detection and analysis of serum arginase activity in patients with Keshan disease. Endemic disease bulletin. 1999;14:18–19. [Google Scholar]
- 21.Maxwell A. J. Mechanisms of dysfunction of the nitric oxide pathway in vascular diseases. Nitric Oxide. 2002;6(2):101–124. doi: 10.1006/niox.2001.0394. [DOI] [PubMed] [Google Scholar]
- 22.Post H., Pieske B. Arginase: a modulator of myocardial function. American Journal of Physiology-Heart and Circulatory Physiology. 2006;290(5):H1747–H1748. doi: 10.1152/ajpheart.00056.2006. [DOI] [PubMed] [Google Scholar]
- 23.Jung A. S., Kubo H., Wilson R., Houser S. R., Margulies K. B. Modulation of contractility by myocyte-derived arginase in normal and hypertrophied feline myocardium. Am J Physiol Heart Circ Physiol. 2006;290(5):H1756–762. doi: 10.1152/ajpheart.01104.2005. [DOI] [PubMed] [Google Scholar]
- 24.Wright C. L. Effect of selenium on urease and arginase. The Journal of Pharmacology and Experimental Therapeutics. 1940;68:220–230. [Google Scholar]
- 25.Nelson S. M., Lei X., Prabhu K. S. Selenium levels affect the IL-4-induced expression of alternative activation markers in murine macrophages. The Journal of Nutrition. 2011;141(9):1754–1761. doi: 10.3945/jn.111.141176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skvortsova L., Perfelyeva A., Khussainova E., Mansharipova A., Forman H. J., Djansugurova L. Association of gclm -588c/t and gclc -129t/c promoter polymorphisms of genes coding the subunits of glutamate cysteine ligase with ischemic heart disease development in kazakhstan population. Disease Markers. 2017;2017:8. doi: 10.1155/2017/4209257.4209257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uthus E. O., Ross S. A. Dietary selenium affects homocysteine metabolism differently in fisher-344 rats and CD-1 mice. The Journal of Nutrition. 2007;137(5):1132–1136. doi: 10.1093/jn/137.5.1132. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi T., Watanabe Y., Saito Y., et al. Mice lacking the glutamate-cysteine ligase modifier subunit are susceptible to myocardial ischaemia-reperfusion injury. Cardiovascular Research. 2010;85(4):785–795. doi: 10.1093/cvr/cvp342. [DOI] [PubMed] [Google Scholar]
- 29.Song W., Wang X. The role of TGFβ1 and LRG1 in cardiac remodelling and heart failure. Biophysical Reviews. 2015;7(1):91–104. doi: 10.1007/s12551-014-0158-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li G., Li R.-K., Mickle D. A. G., et al. Elevated insulin-like growth factor-I and transforming growth factor-beta 1 and their receptors in patients with idiopathic hypertrophic obstructive cardiomyopathy, A possible mechanism. Circulation. 1998;98(19 Suppl):p. I144–9.I149–5010 [PubMed] [Google Scholar]
- 31.O'Brien E. R., Bennett K. L., Garvin M. R., et al. Beta ig-h3, a transforming growth factor-beta-inducible gene, is overexpressed in atherosclerotic and restenotic human vascular lesions. Arterioscler Thrombosis Vascular Biology. 1996;16(4):576–584. doi: 10.1161/01.atv.16.4.576. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y., Handley D., Kaplan T., et al. High throughput determination of TGFbeta1/SMAD3 targets in A549 lung epithelial cells. Plos One. 2011;6:e20319. doi: 10.1371/journal.pone.0020319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddi A. S., Bollineni J. S. Selenium-deficient diet induces renal oxidative stress and injury via TGF-beta1 in normal and diabetic rats. Kidney International. 2001;59(4):1342–1353. doi: 10.1046/j.1523-1755.2001.0590041342.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang S., Li F., Younes M., Liu H., Chen C., Yao Q. Reduced selenium-binding protein 1 in breast cancer correlates with poor survival and resistance to the anti-proliferative effects of selenium. PLoS One. 2013;8:p. e63702. doi: 10.1371/journal.pone.0063702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng S., Wang S., Wang Y., Yang Q., Wang D., Li H. Identification and expression of carbonic anhydrase 2, myosin regulatory light chain 2 and selenium-binding protein 1 in zebrafish Danio rerio: Implication for age-related biomarkers. Gene Expression Patterns. 2018;29:47–58. doi: 10.1016/j.gep.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 36.Fang W., Goldberg M. L., Pohl N. M., et al. Functional and physical interaction between the selenium-binding protein 1 (SBP1) and the glutathione peroxidase 1 selenoprotein. Carcinogenesis. 2010;31(8):1360–1366. doi: 10.1093/carcin/bgq114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmieri F. The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Archives. 2004;447:689–709. doi: 10.1007/s00424-003-1099-7. [DOI] [PubMed] [Google Scholar]
- 38.Aquila H., Link T. A., Klingenberg M. Solute carriers involved in energy transfer of mitochondria form a homologous protein family. FEBS Letters. 1987;212(1):1–9. doi: 10.1016/0014-5793(87)81546-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Selenium related genes in gene expression microarray data from Keshan Disease patients and healthy controls.
Data Availability Statement
All relevant data are contained within the paper. Additional information can be obtained by contacting Dr. Shulan He (hesl@nxmu.edu.cn).