Abstract
Brain spheroids or organoids derived from human pluripotent stem cells (hiPSCs) are still not capable of completely recapitulating in vivo human brain tissue, and one of the limitations is lack of microglia. To add built-in immune function, coculture of the dorsal forebrain spheroids with isogenic microglia-like cells (D-MG) was performed in our study. The three-dimensional D-MG spheroids were analyzed for their transcriptome and compared with isogenic microglia-like cells (MG). Cortical spheroids containing microglia-like cells displayed different metabolic programming, which may affect the associated phenotype. The expression of genes related to glycolysis and hypoxia signaling was increased in cocultured D-MG spheroids, indicating the metabolic shift to aerobic glycolysis, which is in favor of M1 polarization of microglia-like cells. In addition, the metabolic pathways and the signaling pathways involved in cell proliferation, cell death, PIK3/AKT/mTOR signaling, eukaryotic initiation factor 2 pathway, and Wnt and Notch pathways were analyzed. The results demonstrate the activation of mTOR and p53 signaling, increased expression of Notch ligands, and the repression of NF-κB and canonical Wnt pathways, as well as the lower expression of cell cycle genes in the cocultured D-MG spheroids. This analysis indicates that physiological 3-D microenvironment may reshape the immunity of in vitro cortical spheroids and better recapitulate in vivo brain tissue function for disease modeling and drug screening.
1. Introduction
Understanding the models established by human induced pluripotent stem cells (hiPSCs) requires genome-wide mapping to elucidate gene regulatory networks [1, 2]. Therefore, transcriptome analysis has been used to compare hiPSC-derived lineage-specific cells with somatic counterparts [3]. Recently, forebrain spheroids or organoids were derived from hiPSCs for disease modeling and as potential platforms for drug screening [4–7]. These spheroids need to contain critical components of the human brain, such as vascular cells and microglia, for proper function. Our previous study characterized microglia-like cells differentiated from hiPSCs and introduced isogenic microglia-like cells into forebrain spheroids [8]. The microglia-like cells were cocultured with isogenic dorsal cortical spheroids in order to build immune function within the spheroids. While extensive phenotypic characterizations were performed in our previous study, the fundamental metabolic pathways and signaling pathways in different culture systems were not analyzed yet. It is postulated that the microglia-like cells inside the spheroids retain more in vivo-like metabolic program and the associated phenotype due to the environmental change in intimate cell-cell and cell-matrix interactions compared to 2-D culture.
Metabolic programming was found to play important roles in homeostasis, tissue repair, immune function, epigenetic change, and cellular phenotype. For hiPSC reprogramming, the somatic cells with oxidative phosphorylation (OXPHOS) gain the glycolysis metabolism when they gain pluripotency [9, 10]. For human mesenchymal stem cell (MSC) aggregates, metabolic reconfiguration towards glycolysis supports reacquisition of the primitive stem cell phenotype [11]. Similarly, for microglia, limited data suggest that polarization to an M1 phenotype (proinflammatory) may be accompanied by a metabolic shift from oxidative phosphorylation to aerobic glycolysis for energy production [12, 13]. It has been suggested that default polarization of resident microglia uses OXPHOS (mitochondrial respiration, i.e., anti-inflammatory M2 phenotype) for the functions involved in tissue repair, while the metabolic program shifts towards glycolysis as well as nitric oxide and citrulline production for M1 polarization [12]. In addition, the metabolic environment of microglia can affect brain region-dependent gene regulation [13]. Therefore, modifications of immune response by the physiological 3-D microenvironment may reshape the innate immunity that gains the “memory” and “trained” inflammation response [12].
Based on the literature for metabolic programming of 3-D HepG2/C3A spheroids [14], this present study analyzed the genomics data for hiPSC-derived microglia-like cells (i.e., single cells in semiadherent culture) and the isogenic 3-D dorsal spheroids containing microglia-like cells. The focus is on changes in metabolic pathways and the signaling pathways that are involved in cell proliferation, cell death, inflammation, PIK3/AKT/mTOR signaling, and Wnt and Notch pathways in 3-D cortical spheroids derived from hiPSCs. This study provides global genomic details on the impact of 3-D culturing on the metabolic phenotype of cells inside the cortical spheroids. The analysis in this study can also help to establish better coculturing strategies for mimicking the in vivo structure and functions of the central nervous system in vitro.
2. Materials and Methods
2.1. Microglia-Like Cells Derived from iPSCs
Our lab has derived microglia-like cells from human iPSK3 cells free of feeders using a modified protocol [15]. Mesoderm induction was performed using activin A, BMP4, SCF, and VEGF, followed by SCF, Flt3L, IL-3, and GM-CSF treatment. Nonadherent cells were replated to tissue culture-treated plates in the presence of IL-3 and M-CSF. The day 23 cells expressed 48-59% CD11b and 52-64% CD45. Day 28 cells expressed 70 ± 6% CD11b and 80 ± 5% IBA-1. Day 30-33 cells expressed 68 ± 12% P2RY12 and 51 ± 10% CX3CR1 (Figure 1(a)) [8].
Figure 1.
Illustration of the MG group and D-MG group. (a) Derivation of microglia-like cells (MG) from hiPSCs. (b) Incorporation of MG into the isogenic dorsal spheroids (DMG). (c) Morphology of the cells in microglial differentiation and the D-MG spheroids. Day 30 MGs were labeled with CellTracker Red. White scale bar: 100 μm. Black scale bar: 200 μm.
2.2. Generation of Isogenic Dorsal Cortical Spheroids and Coculturing with Microglia-Like Cells
Neural differentiation was induced using dual SMAD inhibitors LDN193189 (LDN) and SB431542 (SB) on human iPSK3 cells in low attachment 96-well plates. The neural progenitor spheres were treated with patterning factors cyclopamine (a sonic hedgehog inhibitor) and fibroblast growth factor- (FGF-) 2 [7, 16]. The dorsal identity is defined by TBR1, PAX6, BRN2, and SATB2.
Dorsal spheroids (day 30) were cocultured with isogenic microglia-like cells at a 4 : 1 ratio (8 × 105 neurons to 2 × 105 microglia-like cells, i.e., 20%) in 50% DMEM/10% FBS and 50% neural differentiation medium composed of DMEM/F12 plus 2% B27 (Figures 1(b) and 1(c)). The CellTracker Red-labeled MGs were observed to move into the D-MG spheroids (Figure 1(c)). After three days of coculture (day 33), the dorsal spheroids containing microglia-like cells (D-MG group) and the microglia cells only (MG group) were harvested for RNA isolation and RNA-sequencing.
2.3. RNA-Sequencing
mRNA was isolated from the total RNA using the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs). cDNA libraries were generated from the isolated mRNA using an NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs), with a unique 6-nucleotide index primer (NEBNext Multiplex Oligos for Illumina), using Beckman Biomek 4000. The multiplexed sample was quantified with qPCR (Kapa Biosystems) specific for Illumina sequencing primers, and the average fragment size was determined with a Bioanalyzer High-Sensitivity DNA Chip (Agilent Technologies). The sequencing was performed on Illumina HiSeq 2500 at the Translational Science Laboratory of Florida State University. The pooled data were demultiplexed into individual sample data, and adapter primer sequences were removed [17].
2.4. RNA-Sequencing Data Analysis
The sequencing reads were analyzed using RNA-Seq Alignment version 1.1.1 (Illumina BaseSpace application). The reads were aligned with TopHat 2 [18] to the human genome (genome release GRCh38) using default parameters, and counts for each gene were generated. This workflow uses Cufflinks to generate FPKM (fragments per kilobase per million reads) normalized values [19]. DESeq2 was used to determine statistically significant differentially expressed genes (a false discovery rate (FDR) of <0.05 was used). 15,585 genes were considered to be expressed in this study by the DESeq2 software [20]. The top 500 genes that were upregulated and downregulated (1000 total genes) in the microglial culture versus the D-MG group were assessed for GO, KEGG pathway, and phenotype pathway analysis using WebGestalt [21, 22]. Significant enrichment was determined in WebGestalt using the hypergeometric test and the Benjamini-Hochberg FDR method [23] for multiple testing adjustment.
2.5. Reverse Transcription Polymerase Chain Reaction (RT-PCR) Analysis
Total RNA was isolated from different cell samples using the RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol followed by the treatment with a DNA-Free RNA Kit (Zymo, Irvine, CA) [24]. Reverse transcription was carried out using 2 g of total RNA, anchored oligo-dT primers (Operon, Huntsville, AL), and Superscript III (Invitrogen, Carlsbad, CA) (according to the protocol of the manufacturer). Primers specific for target genes (Supplementary ) were designed using the software Oligo Explorer 1.2 (GeneLink, Hawthorne, NY). The gene β-actin was used as an endogenous control for normalization of expression levels. Real-time RT-PCR reactions were performed on an ABI7500 instrument (Applied Biosystems, Foster City, CA), using SYBR1 Green PCR Master Mix (Applied Biosystems). The amplification reactions were performed as follows: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 95°C for 15 sec and 55°C for 30 sec and 68°C for 30 sec. Fold variation in gene expression was quantified by means of the comparative Ct method: 2−(ΔCt treatment − ΔCt control), which is based on the comparison of expression of the target gene (normalized to the endogenous control β-actin) between the compared samples.
3. Results and Discussions
Previously, our study derived and characterized microglia-like cells from the healthy iPSK3 cells and Ep-iPSC cells [8]. The cells were cocultured and integrated with isogenic dorsal cortical spheroids (Supplementary ). The whole-cell transcriptome analysis was performed for the microglial phenotype, neural inflammation, and the genes involved in Alzheimer's disease. Our current study focuses on different attributes of the 3-D dorsal cortical spheroids containing isogenic microglia-like cells (D-MG group) vs. microglia-like cells (MG group), including metabolic pathways and the signaling pathways that are involved in cell proliferation, cell death, inflammation, PIK3/AKT/mTOR signaling, EIF2 pathway, and Wnt and Notch pathways, which were not reported previously.
3.1. Central Metabolic Pathway
The 3-D spheroid culture usually is associated with slow growth rates, cytoskeleton reorganization, enhanced tight junctions and polarity, and the relocated membrane transporters [25–27]. In aerobic glycolysis, glucose is consumed through the glycolytic pathway to produce lactic acid, nucleotides, amino acids, and other metabolites, while glutamine is converted through glutaminolysis to citrate for cholesterol and lipid production. In oxidative phosphorylation, the generated pyruvate is oxidized to CO2 and water through the tricarboxylic (TCA) cycle, which produces ATP and coverts NADH to NADPH [28–30].
The central metabolic pathways are shown in Figure 2. The values were calculated using log2 (DMG/MG). Relative enzyme expression levels indicate an increase in glycolytic and pentose phosphate pathways in the D-MG group as well as the increased amino acid synthesis (Figure 2 and Supplementary ) [31]. The increased glycolysis is permissive to trigger microbiocidal activity of microglia and allows the cells to survive in the 3-D spheroids. However, enzymes in the glutaminolytic pathway, hexosamine, nucleotide, lipid synthesis, and TCA cycle have small differences among the two groups. Genes related to ATP synthesis and mitochondrial complexes I, III, and IV are shown in Supplementary and Supplementary . The differences between the two groups are small (within ±0.5 except NDUFA13 at -0.68). But the values are all negative for genes related to ATP synthesis and mitochondrial complexes I and III, indicating lower expression in the D-MG group. Together, these results indicate the different energetic/metabolic phenotypes between the D-MG and the MG groups.
Figure 2.
Ratios of gene abundance of central metabolic pathways in the D-MG group in comparison to the MG group. The FPKM (fragments per kilobase per million reads) normalized values for these genes are listed for both samples. The numbers are the log2 values of ratios of D-MG to MG. Negative values indicate that the genes are present in higher amounts in the MG group, while positive values indicate that the genes are present in higher amounts in the D-MG group. (a) Central metabolic pathways. (b) Log2 values of glycolysis genes of D-MG/MG ratios.
Metabolic shift towards glycolysis usually results in the elevated dependence on glutaminolysis or fatty acid synthesis in cell metabolism [32]. However, in this study, the cells did not show a significant increase in glutaminolysis in the spheroids (Figure 2 and Supplementary ). Cytoplasmic isocitrate dehydrogenase 1 (IDH-1) is involved in the reaction of isocitrate production from α-ketoglutarate in the cytosol. IDH-1 expression is decreased in the D-MG spheroids, which may be associated with NF-κB activation in a hypoxia-inducible factor- (HIF-1α-) dependent manner [33].
Similarly, 3-D culture of iPSC-derived endothelial cells also showed the glycolysis-dominated metabolism compared to ones in 2-D culture [34]. Moreover, human MSCs treated with interferon gamma exert an immunosuppressive phenotype by secreting PGE2/IDO and reconfigure the metabolic phenotype to aerobic glycolysis [35]. It was also suggested that activation of the AKT/mTOR signaling pathway is required for metabolic shift under this immune polarization.
The 3-D cultures may not always enhance the glycolysis, which is dependent on a specific cell type. For example, it was reported that 3-D cultures of hiPSC-derived cardiomyocytes displayed downregulation of genes involved in glycolysis and lipid biosynthesis and upregulation of genes involved in oxidative phosphorylation. Accordingly, the 3-D cultures showed lower fluxes for fatty acid synthesis and the increased TCA cycle activity, which improved both cell purity and metabolic maturation [36].
3.2. Hypoxia (Glycolysis and Oxidative Phosphorylation)
3.2.1. HIF-1α Pathway
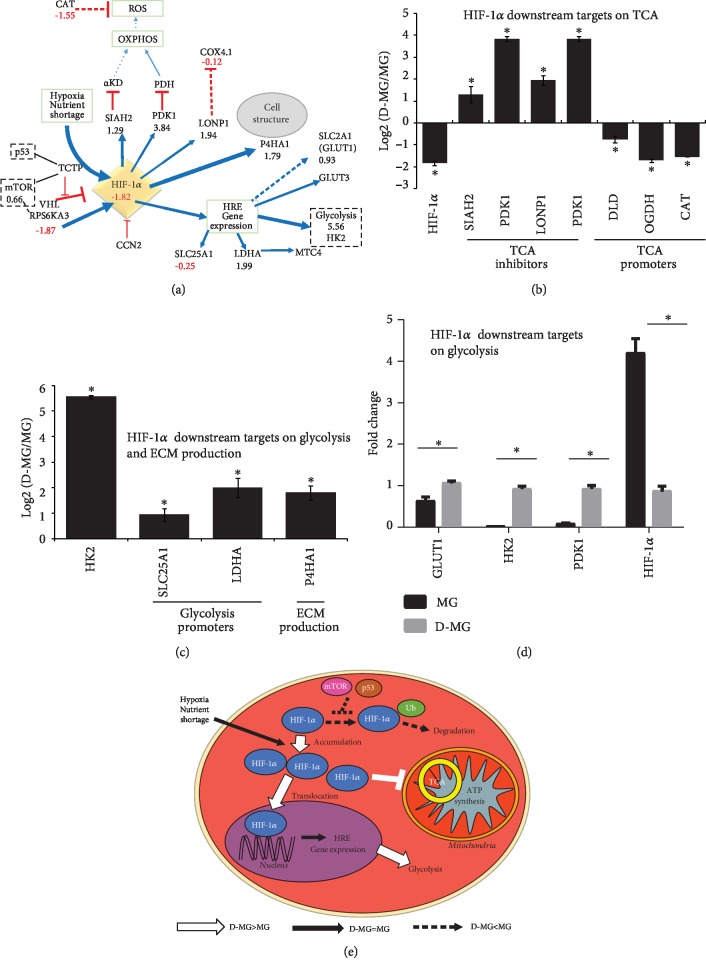
In 3-D spheroid culture, the inside of the spheroids is thought to be more hypoxic than the surface due to mass transfer limitation of oxygen [37], while this has been challenged by other studies as nonhypoxia-stabilized HIF expression [25]. Hypoxia is an important factor in regulating stem cell metabolism and phenotype [38]. When oxygen concentrations decrease, the oxygen-dependent prolyl hydroxylase domain proteins are inactivated and the HIF-1α protein is accumulated, which promotes HIF-1α translocation to the nucleus and its binding to hypoxia response elements, such as glucose transporters and glycolytic enzymes [39, 40]. Our results do not show the higher HIF-1α gene expression in the D-MG group but demonstrate the increased expression of HIF-1α pathway downstream genes, including SIAH2 (1.29), PDK1 (3.84), LDHA (1.99), LONP1 (1.94), and P4HA1 (1.79) (Figures 3(a)–3(c)). These results may indicate the nonhypoxia-stabilized HIF expression in the D-MG group. The downregulated HIF-1α gene expression in the D-MG group was also validated using RT-PCR, along with the upregulated glycolytic gene expression in the D-MG group (Figures 3(d) and 3(e)).
Figure 3.
HIF-1α-related pathway. (a) RNA-sequencing results for glycolytic genes and HIF-1α; relative transcript expression between D-MG and MG for HIF-1α and its downstream targets (b) on tricarboxylic cycle (TCA) and (c) on glycolysis and extracellular matrix (ECM) production. ∗ indicates p < 0.05 (n = 3). (d) Validation of glycolytic genes using RT-PCR. ∗ indicates p < 0.05 (n = 3). (e) Schematic diagram showing the major changes in D-MG versus MG for HIF-1α signaling: D-MG shows enhanced HIF-1α activities that reduce TCA and ATP production, increasing glycolysis mediated by HIF-1α.
HIF-1α induces pyruvate dehydrogenase kinase 1 (PDK1) expression, which inhibits mitochondrial pyruvate dehydrogenase (PDH) [38, 41]. This reduces pyruvate flux into the TCA cycle and lowers the mitochondrial oxygen requirements. The lactate production and secretion would be increased, as observed by Sart et al. [9]. HIF-1α also induces E3-ubiquitin ligase SIAH2 synthesis, which mediates the proteasomal degradation of the OGDH subunit of α-ketoglutarate dehydrogenase (α-KD) and forms a part of the feedback control of HIF-1α signaling. A modest reduction of the α-KD enzyme complex was observed in the D-MG group (DLD, -0.75, OGDH, -1.71) (Table 1), which may slow down TCA cycle activity. Interestingly, the citrate transportation into the cytoplasm by citrate transporter protein (SLC25A1, -0.25) was relatively comparable between these two groups. The extracellular matrix remodeling via collagen hydroxylases (P4HA1, 1.79) was upregulated in the D-MG group. The amounts of anabolic rate-limiting enzymes were increased or remained similar, while catabolic enzymes were unchanged between the two groups (Table 2). The rate-limiting glycolytic pathway steps (HK2, PKM, and PFKL) are the mostly increased enzymes of the pathway. These findings are consistent with the metabolic shift to aerobic glycolysis. In addition, high PFK level may also indicate high levels of AMP, since the spheroids usually have high ATP amounts [14].
Table 1.
Glycolysis vs. oxidative phosphorylation.
| Gene name | MG-1 | MG-2 | MG-3 | D-MG-1 | D-MG-2 | D-MG-3 | MG ave | DMG ave | DMG/MG | LOG2 (DMG/MG) |
|---|---|---|---|---|---|---|---|---|---|---|
| P4HA1 | 140.340 | 135.633 | 141.039 | 389.230 | 546.723 | 505.399 | 139.004 | 480.451 | 3.456 | 1.79 |
| SLC2A1 (GLUT1) | 579.1676 | 469.9516 | 562.8377 | 912.218 | 1098.52 | 1056.15 | 537.319 | 1022.293 | 1.903 | 0.93 |
| DLST | 17.881 | 18.114 | 17.362 | 26.470 | 26.133 | 24.754 | 17.786 | 25.786 | 1.450 | 0.54 |
| SLC16A3 | 304.016 | 219.663 | 293.687 | 255.760 | 244.563 | 255.972 | 272.455 | 252.098 | 0.925 | -0.11 |
| COX4I1 | 238.629 | 185.922 | 239.199 | 186.813 | 225.728 | 199.922 | 221.250 | 204.154 | 0.923 | -0.12 |
| SLC25A1 | 72.139 | 59.151 | 72.118 | 54.289 | 60.972 | 56.058 | 67.803 | 57.107 | 0.842 | -0.25 |
| DLD | 22.476 | 21.521 | 21.854 | 12.507 | 12.003 | 14.700 | 21.950 | 13.070 | 0.595 | -0.75 |
| OGDH | 28.692 | 23.353 | 27.126 | 8.233 | 7.421 | 8.492 | 26.390 | 8.049 | 0.305 | -1.71 |
The FPKM (fragments per kilobase per million reads) normalized values for these genes are listed for both samples. The numbers are the log2 values of ratios of D-MG to MG. Negative values indicate that the genes are present in higher amounts in the MG group, while positive values indicate that the genes are present in higher amounts in the D-MG group. The value 1 indicates two-fold change.
Table 2.
Rate-limiting enzymes for central metabolic pathways and the ratios of their expression in the D-MG group compared to the MG group.
| Pathway | Gene name | MG-1 | MG-2 | MG-3 | D-MG-1 | D-MG-2 | D-MG-3 | MG ave | DMG ave | LOG2 (DMG/MG) |
|---|---|---|---|---|---|---|---|---|---|---|
| Glucose phosphorylation | HK2 | 8.057 | 7.382 | 8.514 | 373.333 | 367.184 | 391.036 | 7.984 | 377.184 | 5.56 |
| Glycolysis | PKM | 144.193 | 130.476 | 142.160 | 1071.419 | 1269.244 | 1061.022 | 138.943 | 1133.895 | 3.03 |
| Glycine synthesis | SHMT2 | 34.121 | 29.636 | 33.060 | 146.550 | 175.435 | 141.673 | 32.272 | 154.553 | 2.26 |
| Asparagine synthesis | ASNS | 28.779 | 28.424 | 28.878 | 93.603 | 122.579 | 101.086 | 28.694 | 105.756 | 1.88 |
| Glycogenolysis | PYGB | 25.047 | 22.544 | 24.888 | 34.525 | 34.766 | 30.397 | 24.159 | 33.229 | 0.46 |
| Pyrimidine synthesis | CAD | 8.199 | 9.554 | 8.489 | 13.220 | 12.638 | 10.185 | 8.747 | 12.015 | 0.46 |
| Glycolysis | PFKL | 62.436 | 52.127 | 63.963 | 83.298 | 79.339 | 73.566 | 59.509 | 78.734 | 0.40 |
| Aspartate synthesis | GOT1 | 16.089 | 14.826 | 14.507 | 18.869 | 20.440 | 19.010 | 15.141 | 19.439 | 0.36 |
| Fatty acid synthesis | FASN | 28.814 | 22.644 | 29.725 | 36.264 | 34.796 | 31.673 | 27.061 | 34.244 | 0.34 |
| Purine synthesis | PRPS1 | 23.085 | 20.259 | 22.740 | 24.847 | 28.698 | 27.976 | 22.028 | 27.173 | 0.30 |
| Pentose phosphate | G6PD | 15.355 | 12.245 | 15.422 | 12.237 | 14.129 | 12.097 | 14.341 | 12.821 | -0.16 |
| Fatty acid synthesis | ACAA1 | 21.219 | 19.891 | 21.847 | 17.973 | 18.352 | 15.681 | 20.986 | 17.336 | -0.28 |
| Glutamine-glutamate conversion | GLUD1 | 49.375 | 44.127 | 51.590 | 35.530 | 41.837 | 38.198 | 48.364 | 38.522 | -0.33 |
| Methionine synthesis | MTR | 7.799 | 9.359 | 8.511 | 7.162 | 5.777 | 6.920 | 8.556 | 6.620 | -0.37 |
| TCA cycle | IDH2 | 103.753 | 84.981 | 101.212 | 66.326 | 79.889 | 75.170 | 96.649 | 73.795 | -0.39 |
| Serine synthesis | PHGDH | 141.285 | 116.771 | 138.838 | 92.636 | 114.237 | 92.781 | 132.298 | 99.884 | -0.41 |
| Hexose | GFPT1 | 46.208 | 45.599 | 46.304 | 26.099 | 32.086 | 29.340 | 46.037 | 29.175 | -0.66 |
| Fatty acid oxidation | CRAT | 23.955 | 20.886 | 25.945 | 14.810 | 14.697 | 14.392 | 23.596 | 14.633 | -0.69 |
| Proline synthesis | PYCR1 | 102.329 | 84.127 | 98.191 | 46.173 | 65.047 | 53.249 | 94.883 | 54.823 | -0.79 |
| Urea synthesis | CPS1 | 7.949 | 7.541 | 8.583 | 4.041 | 3.943 | 4.177 | 8.024 | 4.054 | -0.99 |
| Tyrosine synthesis | PAH | 0.441 | 0.551 | 0.468 | 0.171 | 0.184 | 0.224 | 0.487 | 0.193 | -1.33 |
| Cysteine synthesis | MAT1A | 5.443 | 4.897 | 5.280 | 0.793 | 0.724 | 0.529 | 5.207 | 0.682 | -2.93 |
The FPKM (fragments per kilobase per million reads) normalized values for these genes are listed for both samples. The numbers are the log2 values of ratios of D-MG to MG. Negative values indicate that the genes are present in higher amounts in the MG group, while positive values indicate that the genes are present in higher amounts in the D-MG group. The value 1 indicates two-fold change.
HIF-1α signaling can induce the expression of the mitochondrial protease LONP1 (1.94) (Figure 3). LONP1 degrades cytochrome C oxidase 4 subunit 1 (COX4-1) through electron transport chain complex IV, allowing the replacement of COX4-1 by COX4-2 [42], which is more efficient in enzymatic reaction. LONP1 is an essential central regulator of mitochondrial activity and is overexpressed during oncogenesis [43]. Although LONP1 was increased (1.94) based on our results, there was little change in the level of COX4-1 (-0.12). Reduced mitochondrial respiration normally results in fewer reactive oxygen species (ROS), correlated with the reduced level of catalase (CAT, -1.55). The reduced oxidative stress results in the diminished hydrogen peroxide damage and less oxidized proteins [14].
3.2.2. Glutamine Metabolism and Hexosamine Pathway
While the D-MG group mainly uses glycolysis as its major energetic metabolism, our results did not show an increased reliance on glutamine metabolism (Figure 2 and Supplementary ). Intracellular glutamine levels are regulated by plasma membrane transporters SLC38A2 and SLC1A5 [14]. Endoplasmic reticulum stress would induce the degradation of transporters and ultimately autophagy and cell death [14]. In D-MG spheroids of this study, both SLC38A2 (-0.53) and SLC1A5 (-1.86) were decreased, which may suggest enhanced autophagy in D-MG spheroids.
(1) Conversion of Glutamine into Glutamate. The glutamate demand of the cells is indicated by the expression of GFPT1 (-0.66). GFPT1 is the first and rate-limiting step of the hexosamine pathway and catalyzes the conversion of fructose 6-phosphate and glutamine to glucosamine 6-phosphate and glutamate. Consistently in this study, several enzymes in polysaccharide, proteoglycan, and glycosylation synthetic pathways were downregulated in the D-MG spheroids, including UDP-glucose pyrophosphorylase UGP2 (-0.51), UDP-glucose 6-dehydrogenase UGDH (-1.18), and UDP-glucose 4-epimerase GALE (-1.31).
(2) α-Ketoglutarate. Glutamate can be converted to α-ketoglutarate by the mitochondrial GLUD1 (-0.33) or by cytoplasmic alanine or aspartate aminotransferases [44]. The cytoplasmic enzymes were slightly upregulated (GOT1, 0.36, GOT2, 0.69) in the D-MG group. There are three possible routes by which α-ketoglutarate can be converted to citrate (Figure 2) [1]. It can be converted through the TCA cycle [2]. It could be converted from isocitrate to citrate (by IDH2, -0.39, and ACO2, 0.01), which affects the normal TCA cycle flux by reductive glutamate metabolism [32]. In this study, IDH3B was slightly increased (0.36) and it can only catalyze the “forward” reaction [3]. Cytoplasmic α-ketoglutarate, produced through GFPT1 (-0.66) and GOT1 (0.36), can be converted by IDH1 (-1.01) and ACO1 (-0.73), which are both downregulated in the D-MG group compared to the MG group.
(3) NADH. NADH/NAD+ and NADPH/NADP+ conversions are critical in the central metabolic pathways (Figure 2 and Supplementary ). The reduction in the mitochondrial MDH1 (-0.49) and the downregulation of NAD(P) transhydrogenase (NNT, -1.24) suggest that the conversions in mitochondria may have some significance. MDH1 was decreased (-0.49) but lacked a malate source (e.g., the SLC25A11 transporter was essentially unchanged (0.25)). The conversion of cytoplasmic pyruvate to lactate would consume the produced NADH. The richest source of NADPH is the pentose phosphate pathway, where G6PD (-0.16) was essentially unchanged between the two groups. These results indicate the reliance on mitochondrial NAD+/NADH in the D-MG group.
(3) Citrate. Citrate can be used for fatty acid synthesis. ATP-citrate synthase uses citrate to generate cytosolic acetyl-CoA. Acetyl-CoA is used for (1) histone acetylation by acetyl-CoA acyltransferase (ACAA1, -0.28), (2) fatty acid synthesis (FASN, 0.34), and (3) synthesis of cholesterol, steroid hormones, and other biomolecules. Glutamine is as important as glucose in metabolism, and it can block glutamate-dependent cellular pathways at either IDH1 or ACL step [14].
3.3. Signaling Pathways Involved in Metabolic Reprogramming
The analysis so far shows the metabolic shift to aerobic glycolysis in the 3-D spheroids, which may be driven by diffusion gradients [30, 37]. In order to investigate how metabolic reprogramming is orchestrated, the status of critical pathways for cellular function, including PI3K/AKT/mTOR, Myc, p53, NF-κB, EIF2, Wnt, and Notch, was analyzed.
3.3.1. PIK3/AKT/mTOR Signaling
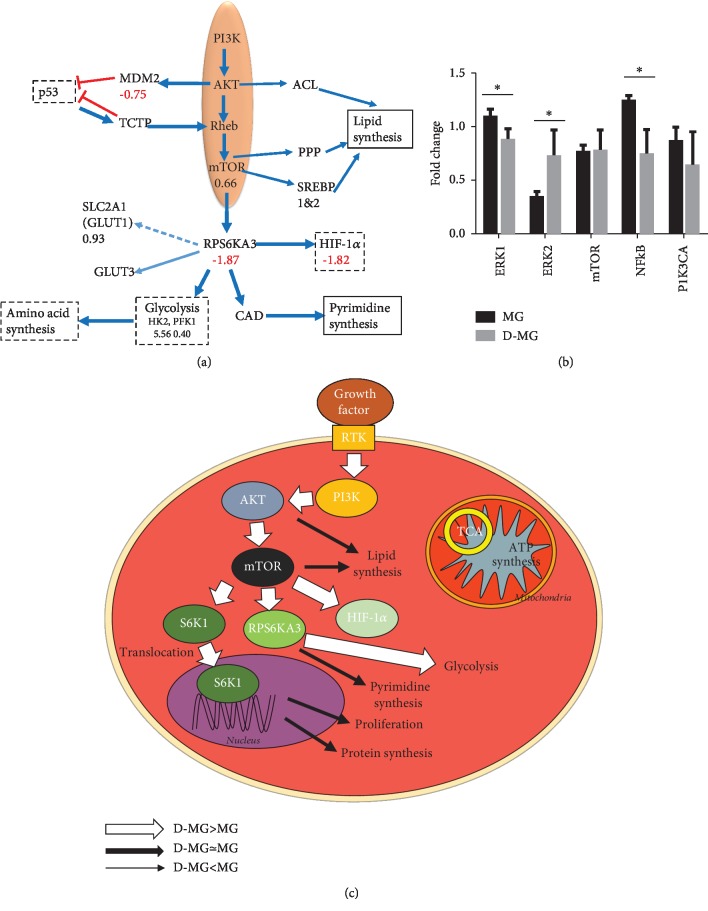
PI3K (a class I phosphoinositide 3-kinase) is one of the key signaling enzymes that are activated in 3-D spheroid cultures [45]. And the PIK3 pathway is directly related to cellular metabolism and growth. When cells consume the nutrients and growth factors, they generate amino acids, glucose, oxygen, and energy (i.e., ATP) and use these products to activate GTPase, such as Rheb, through PI3K. One key regulator of metabolism and growth-activated downstream of PI3K signaling is the mechanistic target of rapamycin (mTOR) [46]. Our results showed that the expression of mTOR (0.66) was slightly elevated in the D-MG spheroids and the downstream effectors were inhibited (e.g., ribosomal protein s6 kinase RPS6KA3, -1.87) (Figure 4(a)). mTOR stimulates pyrimidine synthesis via the RPS6KA-mediated phosphorylation of CAD (0.46), whereas AKT can phosphorylate ACL, enhancing its lipogenic activities and mTOR signaling. The expression of ACLY (-0.13) indicates that the activity of the AKT pathway was similar in the two groups. mTOR regulates several anabolic and catabolic pathways through fatty acid and cholesterol biosynthetic genes via the SREBP family (SREBP1 and SREBP2) [47].
Figure 4.
mTOR signaling in the D-MG group. (a) RNA-sequencing results. (b) Validation of ERK genes using RT-PCR. ∗ indicates p < 0.05 (n = 3). (c) Schematic diagram showing the major changes in D-MG versus MG condition for mTOR signaling. D-MG shows enhanced mTOR activities that reduce tricarboxylic cycle (TCA), and D-MG shows increased glycolysis mediated by mTOR.
mTOR signaling was reported to increase the translation of HIF-1α, glucose transporters, and glycolytic enzymes and promote metabolic reprogramming. The activation of the mTOR protein complex mTORC1 leads to the induction of genes encoding the enzymes of glycolysis, the pentose phosphate pathway, and lipid and sterol biosynthesis [35, 48]. mTOR was also reported to coordinate the activation of cell growth machinery together with amino acids in the presence of growth signals [49]. However, in the RT-PCR validation, mTOR gene expression was statistically insignificant, possibly due to assay variations (Figures 4(b) and 4(c)). In addition, ERK1 was downregulated while ERK2 was upregulated in the D-MG group.
3.3.2. Myc Signaling
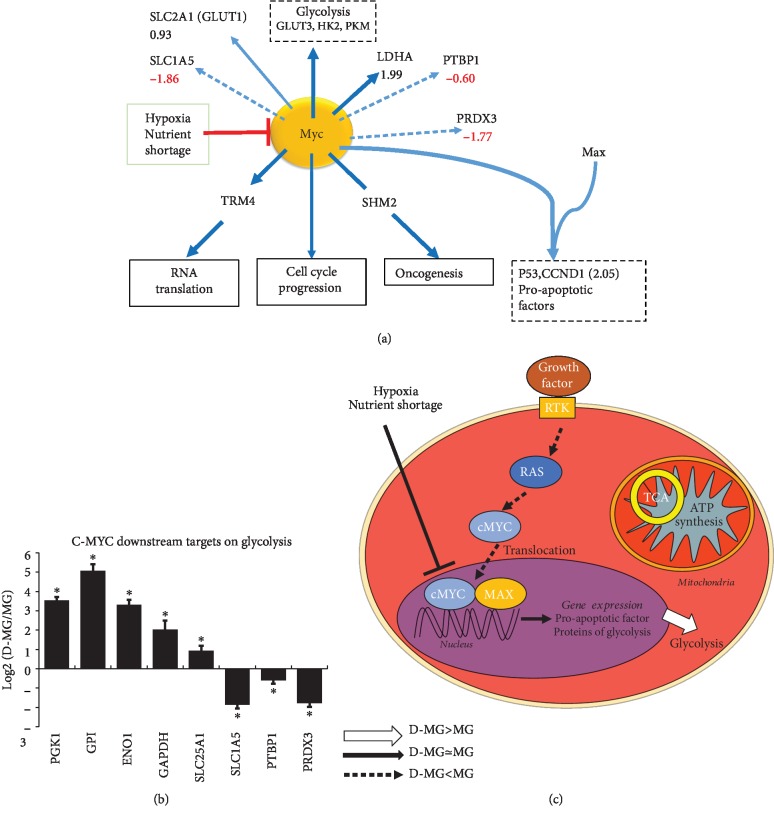
Myc is a transcription factor that dimerizes with MAX to bind to DNA and regulate gene expression involved in metabolism (glycolysis and glutaminolysis) and biosynthesis (nucleotide and lipid synthesis) [50]. Myc supports specific mRNA splice variants, such as glycolysis-affecting PKM2 over PKM1. Myc targets glucose membrane transporters such as GLUT1 (or SLC2A1) and glutamine transporter SLC1A5, which are important in cell proliferation [50]. Myc transactivates the gene expression of PFK, ENO, and LDHA (1.99) and indirectly increases GAPDH and PGK1, based on our genomics analysis (PGK1, 3.54; GPI, 5.07; ENO1, 3.30; and GAPDH, 2.02), indicating the relation between Myc and glycolysis (Figure 5). The gene expression of Myc-related proteins was downregulated in the D-MG spheroids including PTBP1 (-0.60), SLC1A5 (-1.86), and PRDX3 (-1.77) (Figure 5). The low Myc activity inside the D-MG spheroids might indicate the slow proliferation of cells.
Figure 5.
c-Myc signaling in the D-MG group. (a) RNA-sequencing results related to Myc. (b) Relative transcript expression between D-MG and MG conditions for c-Myc and its downstream targets; ∗ indicates p < 0.05 (N = 3). (c) Schematic diagram showing the major changes in D-MG versus MG for c-Myc signaling: D-MG shows enhanced c-Myc that mediates increased glycolysis.
3.3.3. p53 Signaling
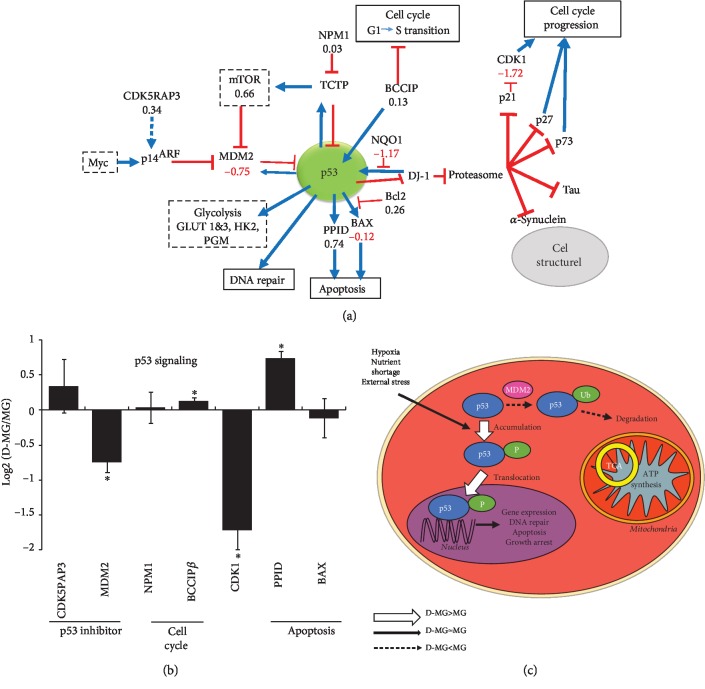
In addition, the expression of the transcriptional suppressor CDK5RAP3 is comparable between the D-MG group and the MG group (0.34). CDK5RAP3 is a novel activator of PAK4 and processes important prometastatic function [51]. It has been reported that CDK5RAP3 knockdown upregulated the tumor suppressor p14ARF at protein and mRNA levels, and ectopic expression of CDK5RAP3 repressed the transcription of p14ARF [51]. Therefore, the CDK5RAP3 expression may allow for the synthesis of p14ARF and its binding to MDM2. The MDM2 (-0.75) expression was decreased in the D-MG spheroids (Figure 6). The MDM2 expression can release p53 from inhibition and thereby lead to the stabilization, accumulation, and activation of p53. Also, the negative regulators of p53 are subjected to tight feedback regulation. For example, BCCIPβ overexpression delays the G1 to S progression and results in an elevated p21 expression, which would inhibit CDK1 induction of cell cycle progression. Though the D-MG spheroids did not show a significant increase in BCCIPβ expression (0.13) compared to the MG group (Figure 6), the expression of CDK1 was decreased (-1.72) in the D-MG group, which indicated a potential cell cycle arrest.
Figure 6.
p53 signaling in the D-MG group. (a) RNA-sequencing results related to p53. (b) Relative transcript expression between D-MG and MG for p53 and its downstream targets; ∗ indicates p < 0.05 (N = 3). (c) Schematic diagram showing the major changes in D-MG versus MG for p53 signaling: D-MG shows the increased p53 activities, which results in the decreased expression of proteins related to cell proliferation.
3.3.4. Wnt Pathway
In the canonical Wnt pathway, Wnt interacts with the Frizzled receptor that results in the inhibition of glycogen synthase kinase-3β (GSK-3β) [52]. Due to the inhibition, GSK-3β would not be able to phosphorylate β-catenin, leading to the nuclear entry of β-catenin. In the nucleus, β-catenin interacts with members of TCF transcriptional factors (i.e., T cell factor/lymphoid enhancer factor) and modulates target gene expression [53]. In our D-MG spheroids, the expression of β-catenin was slightly reduced compared to that in the MG group (CTNNB1, -0.66) (Figure 7). Wnt 3 (-1.06) and Wnt 5B (-1.55) were also decreased. The histidine triad nucleotide-binding protein 1 (HINT1, 0.23), which keeps the Wnt/β-catenin pathway inactive [54], and protein phosphorylase 2A (PPP2RA1, 0.28) were comparable in the two groups. Our previous studies evaluated the influence of the canonical Wnt pathway on neural patterning of hiPSCs [55, 56]. The reduced canonical Wnt/β-catenin signaling at a late stage of neural differentiation of hiPSCs can enrich cells with rostral forebrain identity [57]. In the noncanonical pathway, Wnt binds to the Frizzled receptor which activates Dishevelled and forms a complex with RAC1, mediating profilin binding to actin [58]. In the current study, the noncanonical Wnt pathway activity was comparable for the D-MG spheroids and MG group (RAC1, 0.27).
Figure 7.
Wnt signaling in the D-MG group. (a) RNA-sequencing results related to Wnt. (b) Relative transcript expression between D-MG and MG for Wnt and its downstream targets. ∗ indicates p < 0.05 (N = 3). (c) Schematic diagram showing the major changes in D-MG versus MG conditions for Wnt signaling; i.e., D-MG shows the decreased canonical Wnt signaling while noncanonical Wnt is increased, which results in the increased cytoskeletal rearrangement.
3.3.5. NF-κB Pathway
NF-κB was found to be involved in cellular responses to various stimuli such as stress, cytokines, free radicals, and viral antigens [59, 60]. The two classes of NF-κB proteins (p50/p52 and ReIA/ReIB) form heterodimers to function as transcriptional activators. In an inactive state, NF-κB dimers are sequestered in the cytoplasm by IkB inhibitors. They mask the nuclear localization signals of NF-κB proteins and keep NF-κB signaling in the inactive state. When active, NF-κB proteins enter the nucleus and turn on the IkBα repressor. It has been reported that hypoxia upregulated PRMT1 which asymmetrically methylated ReIA, inhibiting the binding of ReIA to DNA and further repressing NF-κB [61]. NF-κB signaling was found to be dysregulated in TGFβR3 epicardial cells, which also showed the impaired cell invasion and revealed the role of NF-κB signaling in TGFβR3 activation [62]. In our D-MG spheroids, the lower expression of TGFβR3 (-2.61) and TGFB1 (-0.60) indicates the less activated NF-κB pathway (Supplementary and Figure 4(b)). TGFBI expression is essentially similar (0.03), and PRMT1 expression (0.63) is slightly upregulated.
Microglial M1 immune response usually involves NF-κB signaling [59]. Knocking down of LRP1 in primary microglia led to the activation of both c-Jun N-terminal kinase and NF-κB pathways. The sensitivity to lipopolysaccharide (LPS) stimulation in the production of proinflammatory cytokines was also enhanced. The NF-κB inhibitor was shown not only to suppress the production of cytokines induced by the knockdown of LRP1 but also to restore the downregulated expression of LRP1 due to LPS stimulation.
3.3.6. Notch Pathway
Most Notch receptors were upregulated in the D-MG group, including DLL1 (5.85), DLL3 (2.84), DLL4 (1.49), JAG1 (2.55), JAG2 (1.70), HES1 (0.62), HES4 (2.44), and HES5 (7.93) (Figure 8(a)), indicating the enhanced cell-cell communications in 3-D culture. Our previous study also showed that Notch-1 expression was upregulated in the hybrid stem cell spheroids [63]. In addition, Notch signaling regulates the balance between the progenitor pool and the neuron pool, preventing premature neurogenesis [64]. Global deletion of Notch signaling leads to the accelerated differentiation into neurons. The activation of Notch by DLL1 causes neural stem cells to irreversibly commit to a glial fate and prevents the stem cells from adopting a neuronal fate [65]. When a Notch receptor interacts with its ligands, Delta and Serrate, it activates the membrane-tethered transcription factor and leads to the release of intracellular domain (NICD) and its translocation to the nucleus. Then, NICD interacts with CSL family regulators and transcript Hes1 and Hes5 expression. Notch signaling also promotes Wnt expression in Drosophila development [66]. Conversely, Wnt protein can promote the expression of Notch ligands DLLs, forming a positive feedback loop to maintain Notch signaling and Wnt protein expression.
Figure 8.
Log2 expression levels of the D-MG/MG ratios for genes about cell-cell interactions, cell death, and cell cycle. (a) Notch signaling. (b) Cell death. (c) Cell cycle and cell proliferation. (d) EIF2 signaling as a part of integrated stress response (ISR).
3.3.7. Cell Death and Cell Cycle
It is important to evaluate cell death pathways (necrosis or apoptosis) in the 3-D spheroids. The major changes associated with cell death are free radical damage, swelling, rupture, and cytolysis. Under stress, the cells sense DNA damage by the serine/threonine kinase ATM and p53 [67]. p53 accumulates in the mitochondrial matrix and triggers mitochondrial permeability transition (MPT) pore (PTP) opening. Then, p53 physically interacts with anti- and proapoptotic Bcl-2 and BAX family members to inhibit or activate their respective functions, leading to mitochondrial outer membrane permeabilization (MOMP) and apoptosis [68]. From our results, PPID (0.74), the essential component of the MPT pore located in the mitochondrial matrix, is slightly increased in the D-MG group (Figure 8(b)).
The exact molecular composition of the MOMP complex is assumed to contain hexokinase, voltage-dependent anion channel (VDAC, on the outer membrane), the adenine nucleotide translocase (ANT, in the inner membrane), and cyclophilin D (a peptidyl-prolyl isomerase in the matrix). When PTP opens, it leads to matrix swelling and depolarization of the membrane potential, causing subsequent rupture of the outer membrane and consequently apoptosis [68]. Based on our results, VDACs were increased (VDAC1, 1.84; VDAC3, 0.90) in the D-MG group (Figure 8(b)). MPT-driven necrosis works not only by the dissipation of mitochondrial transmembrane potential but also by the latent chromatinolytic activity of AIFM1 (apoptosis-inducing factor mitochondrion-associated 1) [69]. The slightly decreased AIFM1 (-0.38) in the D-MG spheroids indicates that necrosis is favored over apoptosis.
The mitochondrial HINT2 promotes angiogenesis via p53 and BAX. The activated NF-κB pathway also leads to the increased expression of antiapoptotic proteins, including Bcl-XL-binding protein v68 (PGAM5, 1.09) and Bcl-2-associated transcription factor 1 (BCLAF1, 0.96) in the D-MG spheroids (Figure 8(b)). The defender against apoptotic cell death (DAD1, 0.11) and Bcl-2 inhibitor of transcription 1 (PTRH2, 0.07) were unchanged. These antiapoptotic proteins can bind to and inactivate proapoptotic proteins. The proapoptotic protein BAX, Bcl-2-like protein 4, was unchanged (-0.02), but Bcl-2-associated athanogene 2 (BAG2, -2.90) was decreased in the D-MG spheroids. Consistent with higher expression of death genes in the D-MG group, the cell cycle-related genes were downregulated in the D-MG spheroids but highly expressed in the MG group (Figure 8(c)).
The sirtuin signaling proteins SIRT1 (0.48) and SIRT6 (1.01) were upregulated in the D-MG spheroids (Table 3), which regulate DNA damage and oxidative stress. The autophagy-related genes were essentially similar between the two groups. Poly (ADP-ribose) polymerase (PARP) is a family of proteins involved in a number of cellular processes such as DNA repair, genomic stability, and programmed cell death [70]. PARP6 (1.32), PARP3 (1.12), and PARP2 (0.97) were upregulated while PARP4 (-1.34), PARP14 (-2.44), PARP12 (-2.54), and PARP9 (-2.76) were downregulated in the D-MG group (Table 4). PARP1 (-0.01) was essentially unchanged between the two groups.
Table 3.
Sirtuin signaling and autophagy.
| Gene name | MG-1 | MG-2 | MG-3 | D-MG-1 | D-MG-2 | D-MG-3 | MG ave | DMG ave | DMG/MG | LOG2 (D-MG/MG) |
|---|---|---|---|---|---|---|---|---|---|---|
| Sirtuin signaling | ||||||||||
| SIRT6 | 11.268 | 9.088 | 9.289 | 20.250 | 21.520 | 17.741 | 9.882 | 19.837 | 2.007 | 1.01 |
| SIRT1 | 8.625 | 8.714 | 9.529 | 12.048 | 12.060 | 13.258 | 8.956 | 12.455 | 1.391 | 0.48 |
| SIRT4 | 3.945 | 5.113 | 3.877 | 5.457 | 4.254 | 5.735 | 4.312 | 5.148 | 1.194 | 0.26 |
| SIRT7 | 7.837 | 8.696 | 7.100 | 8.127 | 7.686 | 7.362 | 7.877 | 7.725 | 0.981 | -0.03 |
| SIRT5 | 1.763 | 2.161 | 1.591 | 1.743 | 1.551 | 1.505 | 1.838 | 1.599 | 0.870 | -0.20 |
| SIRT3 | 13.160 | 13.397 | 12.523 | 8.864 | 7.755 | 8.300 | 13.027 | 8.306 | 0.638 | -0.65 |
| SIRT2 | 50.715 | 42.689 | 48.365 | 27.236 | 31.360 | 27.618 | 47.256 | 28.738 | 0.608 | -0.72 |
| Autophagy | ||||||||||
| TFEB | 5.357 | 4.069 | 5.025 | 4.723 | 5.425 | 5.416 | 4.817 | 5.188 | 1.077 | 0.11 |
| BECN1 | 13.675 | 11.764 | 13.135 | 12.298 | 12.239 | 12.236 | 12.858 | 12.257 | 0.953 | -0.07 |
| LAMP1 | 57.074 | 48.954 | 58.426 | 46.114 | 51.499 | 47.309 | 54.818 | 48.307 | 0.881 | -0.72 |
The FPKM (fragments per kilobase per million reads) normalized values for these genes are listed for both samples. The numbers are the log2 values of ratios of D-MG to MG. Negative values indicate that the genes are present in higher amounts in the MG group, while positive values indicate that the genes are present in higher amounts in the D-MG group. The value 1 indicates two-fold change.
Table 4.
Gene expression of poly (ADP-ribose) polymerase (PARP) family proteins.
| Gene name | MG-1 | MG-2 | MG-3 | D-MG-1 | D-MG-2 | D-MG-3 | MG ave | DMG ave | DMG/MG | LOG2 (D-MG/MG) |
|---|---|---|---|---|---|---|---|---|---|---|
| PARP6 | 19.908 | 24.159 | 20.683 | 55.689 | 59.154 | 54.018 | 21.583 | 56.287 | 2.608 | 1.38 |
| PARP3 | 3.171 | 3.020 | 3.221 | 6.723 | 7.323 | 6.366 | 3.137 | 6.804 | 2.169 | 1.12 |
| PARP2 | 17.659 | 22.634 | 16.142 | 35.570 | 39.811 | 34.828 | 18.812 | 36.736 | 1.953 | 0.97 |
| PARP11 | 3.485 | 3.789 | 3.403 | 6.320 | 6.097 | 6.702 | 3.559 | 6.373 | 1.791 | 0.84 |
| PARP15 | 0.163 | 0.124 | 0.127 | 0.286 | 0.263 | 0.179 | 0.138 | 0.243 | 1.759 | 0.81 |
| PARP8 | 2.142 | 2.526 | 2.428 | 3.305 | 3.204 | 3.578 | 2.365 | 3.362 | 1.422 | 0.51 |
| PARP1 | 42.914 | 40.611 | 42.447 | 41.423 | 43.048 | 40.686 | 41.990 | 41.719 | 0.994 | -0.01 |
| PARP10 | 1.451 | 1.234 | 1.709 | 1.518 | 1.198 | 1.385 | 1.465 | 1.367 | 0.933 | -0.10 |
| PARP16 | 12.871 | 12.281 | 11.812 | 7.840 | 8.730 | 9.558 | 12.321 | 8.709 | 0.707 | -0.50 |
| PARP4 | 8.746 | 9.413 | 8.348 | 3.874 | 3.104 | 3.526 | 8.836 | 3.501 | 0.396 | -1.34 |
| PARP14 | 7.616 | 10.770 | 8.298 | 1.482 | 1.613 | 1.818 | 8.895 | 1.638 | 0.184 | -2.44 |
| PARP12 | 1.519 | 1.168 | 1.288 | 0.182 | 0.274 | 0.228 | 1.325 | 0.228 | 0.172 | -2.54 |
| PARP9 | 3.633 | 3.688 | 3.397 | 0.533 | 0.460 | 0.586 | 3.573 | 0.526 | 0.147 | -2.76 |
The FPKM (fragments per kilobase per million reads) normalized values for these genes are listed for both samples. The numbers are the log2 values of ratios of D-MG to MG. Negative values indicate that the genes are present in higher amounts in the MG group, while positive values indicate that the genes are present in higher amounts in the D-MG group. The value 1 indicates two-fold change.
3.3.8. Eukaryotic Initiation Factor 2 (eIF2) Pathway
eIF2 is one of the critical translation G proteins that are tightly regulated in the integrated stress response. In response to stress stimuli, eIF2α undergoes phosphorylation via different kinases, such as PKR-like endoplasmic reticulum kinase (PERK), which results in the formation of eIF2B. This blocks the p-eIF2 from its active GTP-bound state, causing a reduction of the translation of most mRNAs [71]. In the operative nervous systems, the eIF pathway was found to support the connectivity between neurons. The physiological extracellular cue Sema3A can trigger rapid and transient phosphorylation of eIF2a in axons for the axonal translational changes [72]. The D-MG group displayed the slightly elevated expression of eIF proteins, such as eIF-2α/EIF2S1 (0.220), eIF2β/EIF2S2 (0.274), and eIF2B proteins, such as EIF2B1 (0.43), EIF2B2 (0.47), EIF2B3 (0.45), and EIF2B5 (0.66) (Figure 8(d)), indicating potential stress such as hypoxia or insufficient nutrient uptake in the D-MG group. Also, the expression of EIF2AK3 (0.64), a stress sensing protein kinase that phosphorylates the alpha subunit of eIF2, was slightly elevated in the D-MG group. DDIT3 (0.58), which plays an essential role in inducing cell cycle arrest and apoptosis in response to ER stress, was also slightly elevated in the D-MG group. PPP1R15A (1.17) dephosphorylates the eIF-2α/EIF2S1, thereby reversing the shutoff of protein synthesis initiated by stress-inducible kinases and then facilitating cell recovery from stress.
3.3.9. Extracellular Matrix
The extracellular matrix- (ECM-) related genes were found to be differentially expressed in the two groups (Supplementary ) [1]. The D-MG group had higher expression of some collagen genes including COL7A1, COL20A1, COL9A1, and COL11A2. The MG group expressed high levels of different types of collagens, including COL23A1, COL6A1, COL4A1, COL13A1, COL5A2, COL5A1, COL14A1, and COL4A1 [2]. The D-MG group mainly enriched laminin LAMB4, while the MG group enriched LAMA3, LAMB1, LAMC1, LAMA2, LAMA1, etc. [3]. The D-MG group expressed higher levels of specific integrins, including ITGA7, ITGB8, ITGA2, and ITGAB3, while the MG group enriched ITGA6, ITGA11, ITGA8, ITGB6, ITGA1, ITGB5, etc. [4]. For ECM remodeling genes, the D-MG group enriched proteases MMP17, MMP10, MMP24, MMP7, and MMP1; the MG group had higher expression of proteases MMP23, MMP2, MMP14, TIMP1, MMP28, MMP26, and MMP15, etc.
Other pathways of our interests are shown in Supplementary , which include the AMPK pathway and PDL1/PD1 pathway (Figure 9 shows the relationship between miRNAs and PDL1/PD1 interactions) (Figure 9) [73]. Microglia, the main antigen-presenting cells in the human brain, maintain the equilibrium with T cells through the PD1 pathway as reported [74].
Figure 9.
Illustration of microglia with PDL1/PD1 interactions. The numbers in the illustration are from the normal DMG/MG ratios. The arrows show the ways of tumor cell and T cell interactions. MGs act as antigen-presenting cells (APC).
4. Conclusions
The genomics data reveal that the D-MG spheroids have higher expression of genes for glycolysis and hypoxia signaling, showing the metabolic shift to aerobic glycolysis, consistent with M1 polarization of microglia. However, glutamine conversion is not activated. The signaling pathway activities (activation of mTOR and p53, repression of NF-κB and canonical Wnt) are consistent with the slower proliferation rate and the accumulation of differentiated cells. The additional NADPH needed for citrate and lipid synthesis is mainly generated by pentose phosphate pathway activation. The reduction in the proliferation rate allows the cells to achieve higher ATP levels in the spheroids. The D-MG group enriches genes for NOTCH signaling, but not canonical Wnt signaling. The MG group has higher expression of genes related to cell cycle and proliferation. These results can help to establish better coculturing methods for efficiently mimicking the in vivo structure of the central nervous system.
Acknowledgments
The authors thank Dr. Brian K. Washburn in the FSU Department of Biological Sciences for his help with RNA-sequencing and RT-PCR analysis and Dr. Cynthia Vied in the FSU Translational Science Laboratory for the initial genomics analysis. This work is supported by the FSU Bridge Grant, NSF Career Award (No. 1652992 to YL), and by NIH R03EB020770 (YL).
Abbreviations
- 6PD:
6-Phosphate dehydrogenase
- ACAA1:
Acetyl-CoA acyltransferase
- ACL:
ATP citrate lyase
- ACO2:
Aconitase 2
- ADP:
Adenosine diphosphate
- AIFM1:
Apoptosis-inducing factor mitochondrion-associated 1
- AKT:
Protein kinase B
- AMP:
Adenosine monophosphate
- AMPK:
AMP-activated protein kinase
- ANT:
Adenine nucleotide translocase
- APC:
Antigen-presenting cells
- ARF:
ADP ribosylation factor
- ASNS:
Asparagine synthetase
- ATP:
Adenosine triphosphate
- BAG2:
Bcl-2-associated athanogene 2
- BAX:
Bcl-2-associated X
- BCCIPβ:
BRCA2 and CDKN1A-interacting protein isoform β
- Bcl-2:
B-cell lymphoma 2
- BCLAF1:
Bcl-2-associated transcription factor 1
- Bcl-XL:
B-cell lymphoma-extra large
- BECN1:
Beclin 1
- BMP4:
Bone morphogenetic protein 4
- BRN2:
POU class 3 homeobox 2
- CAD:
Carbamoyl-phosphate synthetase 2
- CAT:
Catalase
- CD11b:
Integrin alpha M
- CD45:
Protein tyrosine phosphatase receptor type C
- CDK:
Cyclin-dependent kinase
- CDK5RAP3:
CDK5 regulatory subunit-associated protein 3
- cDNA:
Complementary DNA
- CM:
Cardiomyocytes
- COL:
Collagen
- COX4-1:
Cytochrome C oxidase 4 subunit 1
- CPS1:
Carbamoyl-phosphate synthase 1
- CRAT:
Carnitine O-acetyltransferase
- CSL:
Recombination signal binding protein for immunoglobulin kappa J region
- CX3CR1:
C-X3-C motif chemokine receptor 1
- DAD1:
Defender against apoptotic cell death
- DDIT3:
DNA damage inducible transcript 3
- DLD:
Dihydrolipoamide dehydrogenase
- DLL:
Delta-like canonical Notch ligand
- DLST:
Dihydrolipoyllysine-residue succinyltransferase
- DMEM:
Dulbecco's modified Eagle's medium
- DMG/D-MG:
Coculture of the dorsal forebrain spheroids with isogenic microglia-like cells
- EC:
Endothelial cells
- ECM:
Extracellular matrix
- eIF2:
Eukaryotic initiation factor 2
- ENO:
Enolase
- Ep-iPSC:
Episomal-induced pluripotent stem cells
- ER:
Endoplasmic reticulum
- ERK:
Extracellular signal-regulated kinases
- FASN:
Fatty acid synthase
- FBS:
Fetal bovine serum
- FDR:
False discovery rate
- FGF:
Fibroblast growth factor
- FLT3L:
FMS-like tyrosine kinase 3 ligand
- FPKM:
Fragments per kilobase per million reads
- G6PD:
Glucose-6-phosphate dehydrogenase
- GALE:
UDP-glucose 4-epimerase
- GAPDH:
Glyceraldehyde-3-phosphate dehydrogenase
- GFPT1:
Glucosamine-fructose-6-phosphate aminotransferase isomerizing 1
- GLUD1:
Glutamate dehydrogenase 1
- GLUT1:
Glucose transporter 1
- GM-CSF:
Granulocyte-macrophage colony-stimulating factor
- GO:
Gene Ontology
- GOT:
Glutamic-oxaloacetic transaminase
- GPI:
Phosphor-glucose isomerase
- GSK-3β:
Glycogen synthase kinase-3β
- GTP:
Guanosine triphosphate
- HES:
Hairy and enhancer of split 1
- HIF-1α:
Hypoxia-inducible-factor 1α
- HINT:
Histidine triad nucleotide-binding protein
- hiPSCs:
Human induced pluripotent stem cells
- HK2:
Hexokinase 2
- IBA-1:
Allograft inflammatory factor 1
- IDH:
Cytoplasmic isocitrate dehydrogenase
- IDO:
Indoleamine-pyrrole 2,3-dioxygenase
- IkB:
Nuclear factor of kappa light polypeptide gene enhancer in B-cell inhibitor
- IL:
Interleukin
- ISR:
Integrated stress response
- ITG:
Integrins
- JAG:
Jagged
- KEGG:
Kyoto Encyclopedia of Genes and Genomes
- LAM:
Laminin
- LAMP1:
Lysosomal-associated membrane protein 1
- LDHA:
Lactate dehydrogenase A
- LDN:
SMAD inhibitor LDN193189
- LONP1:
Lon protease homolog, mitochondrial
- LPS:
Lipopolysaccharide
- LRP1:
Low-density lipoprotein receptor-related protein 1
- MAT1A:
Methionine adenosyltransferase 1A
- MAX:
Myc-associated factor X
- MDH1:
Malate dehydrogenase 1
- MDM2:
Mouse double minute 2 protooncogene
- MG:
Microglia-like cells
- MMP:
Matrix metalloproteinase
- MOMP:
Mitochondrial outer membrane permeabilization
- MPT:
Mitochondrial permeability transition
- MSC:
Human mesenchymal stem cell
- mTOR:
Mechanistic target of rapamycin
- MTR:
5-Methyltetrahydrofolate-homocysteine methyltransferase
- Mxi-1:
MAX-interacting protein 1
- Myc:
Myelocytomatosis
- NADH:
Nicotinamide adenine dinucleotide
- NADPH:
Nicotinamide adenine dinucleotide phosphate
- NANS:
Sialic acid synthase
- NDUFA13:
NADH dehydrogenase 1 alpha subcomplex subunit 13
- NF-κB:
Nuclear factor kappa-B
- NICD:
Notch intracellular domain
- NNT:
NAD(P) transhydrogenase
- OGDH:
Oxoglutarate dehydrogenase
- OXPHOS:
Oxidative phosphorylation
- P4HA1:
Prolyl 4-hydroxylase subunit alpha 1
- PAH:
Phenylalanine hydroxylase
- PAK4:
p21-activated kinase 4
- PARP:
Poly (ADP-ribose) polymerase
- PD1:
Programmed cell death protein 1
- PDH:
Mitochondrial pyruvate dehydrogenase
- PDK1:
Pyruvate dehydrogenase kinase 1
- PDL1:
Programmed death-ligand 1
- PERK:
PKR-like endoplasmic reticulum kinase
- PFK:
Phosphofructokinase
- PFKL:
6-Phosphofructokinase, liver type
- PGAM5:
Bcl-XL-binding protein v68
- PGE2:
Prostaglandin E2
- PGK1:
Phosphoglycerate kinase
- PHD:
Prolyl hydroxylase domain
- PHGDH:
Phosphoglycerate dehydrogenase
- PIK3:
Phosphoinositide 3-kinase
- PKM:
Pyruvate kinase isozymes M
- PPID:
Peptidyl-prolyl isomerase D
- PPP1R15A:
Protein phosphatase 1 regulatory subunit 15A
- PPP2RA1:
Protein phosphorylase 2A
- PRDX3:
Peroxiredoxin 3
- PRMT1:
Protein arginine methyltransferase 1
- PRPS1:
Phosphoribosyl pyrophosphate synthetase 1
- PTBP1:
Polypyrimidine tract-binding protein 1
- PTP:
Mitochondrial permeability transition pore
- PTRH2:
Bcl-2 inhibitor of transcription 1
- PYCR1:
Pyrroline-5-carboxylate reductase 1
- PYGB:
Glycogen phosphorylase B
- qPCR:
Quantitative polymerase chain reaction
- RAC1:
Ras-related C3 botulinum toxin substrate 1
- Rheb:
Ras homolog enriched in brain
- ROS:
Reactive oxygen species
- RPS6KA:
Ribosomal protein S6 kinase alpha
- RT-PCR:
Reverse transcription polymerase chain reaction
- SATB2:
Special AT-rich sequence-binding protein 2
- SB:
SMAD inhibitor SB431542
- SCF:
Stem cell factor
- Sema3A:
Semaphorin-3A
- SHMT2:
Serine hydroxymethyltransferase 2
- SIAH2:
Seven in absentia homolog 2
- SIRT:
Sirtuin
- SLC16A3:
Solute carrier family 16 member 3 (similar for SLC1A5, SLC25A1, and SLC38A2)
- SREBP:
Sterol regulatory element-binding protein
- TBR1:
T-box, brain 1
- TCA:
Tricarboxylic cycle
- TCF:
T cell factor/lymphoid enhancer factor
- TFEB:
Transcription factor EB
- TGFBI:
Transforming growth factor beta induced
- TGFβR3:
Transforming growth factor beta receptor 3
- TIMP:
Tissue inhibitor of metalloproteinases
- UDP:
Uridine diphosphate
- UGDH:
UDP-glucose 6-dehydrogenase
- UGP2:
UDP-glucose pyrophosphorylase
- VDAC:
Voltage-dependent anion channel
- VEGF:
Vascular endothelial growth factor
- α-KD:
α-Ketoglutarate dehydrogenase.
Data Availability
Data is available by contacting the corresponding author.
Additional Points
Significance. Most current forebrain spheroids or organoids lack the components of microglia. This study uses the genomics tool to analyze various pathways of dorsal forebrain spheroids/organoids integrated with isogenic microglia-like cells derived from human induced pluripotent stem cells. The metabolic shift to aerobic glycolysis, consistent with M1 polarization of microglia, in the cocultured spheroids was observed. This study enhances our understanding of 3-D coculturing impacts on the metabolic programming of cells inside the forebrain spheroids. The results can be used to establish better coculturing methods for efficiently mimicking in vivo structure of the central nervous system.
Disclosure
Liqing Song's current address is College of Medicine, Vanderbilt University, Nashville, Tennessee, USA. Julie Bejoy's current address is Department of Chemical Engineering, Carnegie Mellon University, Pittsburgh, Pennsylvania, USA. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Part of the results of this study will be presented in 2019 Annual Meeting of American Institute of Chemical Engineers (AIChE), Nov 10-15, 2019, Orlando, FL.
Conflicts of Interest
No competing financial interests exist.
Authors' Contributions
JB wrote the first version of the manuscript, did some analysis, and prepared most figures. XY did critical reviews and prepared some figures. LS performed the differentiation procedure and prepared genomics samples. TH and QS helped in microglial differentiation. RJ helped in data analysis and manuscript review. SS reanalyzed the results for figures and validated the statistical analysis. YL conceived the projects and wrote and revised the manuscript.
Supplementary Materials
Supplementary Table S1: primer sequence for target genes, Supplementary Table S2: genes related to ATP synthesis and mitochondrial complexes I, III, and IV. Supplementary Table S3: genes related to the NF-κB pathway. Supplementary Table S4: AMPK pathway and PDL1/PD1 pathway. Supplementary Figure S1: microglial differentiation using additional human iPSC line: Ep-iPSC. Supplementary Figure S2: regulation of central metabolism in D-MG. Supplementary Figure S3: log2 gene expression levels of D-MG/MG ratios for ECM-related genes.
References
- 1.Lau E., Paik D. T., Wu J. C. Systems-wide approaches in induced pluripotent stem cell models. Annual Review of Pathology. 2019;14:395–419. doi: 10.1146/annurev-pathmechdis-012418-013046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandenberger R., Wei H., Zhang S., et al. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nature Biotechnology. 2004;22(6):707–716. doi: 10.1038/nbt971. [DOI] [PubMed] [Google Scholar]
- 3.Muffat J., Li Y., Yuan B., et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nature Medicine. 2016;22(11):1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birey F., Andersen J., Makinson C. D., et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545(7652):54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasca S. P. The rise of three-dimensional human brain cultures. Nature. 2018;553(7689):437–445. doi: 10.1038/nature25032. [DOI] [PubMed] [Google Scholar]
- 6.Yan Y., Song L., Bejoy J., et al. Modeling neurodegenerative microenvironment using cortical organoids derived from human stem cells. Tissue Engineering Part A. 2018;24(13-14):1125–1137. doi: 10.1089/ten.tea.2017.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan Y., Song L., Madinya J., Ma T., Li Y. Derivation of cortical spheroids from human induced pluripotent stem cells in a suspension bioreactor. Tissue Engineering Part A. 2018;24(5-6):418–431. doi: 10.1089/ten.tea.2016.0400. [DOI] [PubMed] [Google Scholar]
- 8.Song L., Yuan X., Jones Z., et al. Functionalization of brain region-specific spheroids with isogenic microglia-like cells. Scientific Reports. 2019;9(1):11055–11072. doi: 10.1038/s41598-019-47444-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sart S., Agathos S. N., Li Y. Process engineering of stem cell metabolism for large scale expansion and differentiation in bioreactors. Biochemical Engineering Journal. 2014;84:74–82. doi: 10.1016/j.bej.2014.01.005. [DOI] [Google Scholar]
- 10.Perestrelo T., Correia M., Ramalho-Santos J., Wirtz D. Metabolic and mechanical cues regulating pluripotent stem cell fate. Trends in Cell Biology. 2018;28(12):1014–1029. doi: 10.1016/j.tcb.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y., Munoz N., Tsai A. C., Logan T. M., Ma T. Metabolic reconfiguration supports reacquisition of primitive phenotype in human mesenchymal stem cell aggregates. Stem Cells. 2017;35(2):398–410. doi: 10.1002/stem.2510. [DOI] [PubMed] [Google Scholar]
- 12.Orihuela R., McPherson C. A., Harry G. J. Microglial M1/M2 polarization and metabolic states. British Journal of Pharmacology. 2016;173(4):649–665. doi: 10.1111/bph.13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grabert K., Michoel T., Karavolos M. H., et al. Microglial brain region−dependent diversity and selective regional sensitivities to aging. Nature Neuroscience. 2016;19(3):504–516. doi: 10.1038/nn.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wrzesinski K., Fey S. J. Metabolic reprogramming and the recovery of physiological functionality in 3D cultures in micro-bioreactors. Bioengineering. 2018;5(1):p. 22. doi: 10.3390/bioengineering5010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pandya H., Shen M. J., Ichikawa D. M., et al. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nature Neuroscience. 2017;20(5):753–759. doi: 10.1038/nn.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan Y., Bejoy J., Xia J., Guan J., Zhou Y., Li Y. Neural patterning of human induced pluripotent stem cells in 3-D cultures for studying biomolecule-directed differential cellular responses. Acta Biomaterialia. 2016;42:114–126. doi: 10.1016/j.actbio.2016.06.027. [DOI] [PubMed] [Google Scholar]
- 17.Vied C., Ray S., Badger C. D., Bundy J. L., Arbeitman M. N., Nowakowski R. S. Transcriptomic analysis of the hippocampus from six inbred strains of mice suggests a basis for sex‐specific susceptibility and severity of neurological disorders. The Journal of Comparative Neurology. 2016;524(13):2696–2710. doi: 10.1002/cne.23989. [DOI] [PubMed] [Google Scholar]
- 18.Trapnell C., Pachter L., Salzberg S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C., Williams B. A., Pertea G., et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love M. I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):p. 550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Duncan D., Shi Z., Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Research. 2013;41(Web Server issue):W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang B., Kirov S., Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Research. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological) 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 24.Song L., Wang K., Li Y., Yang Y. Nanotopography promoted neuronal differentiation of human induced pluripotent stem cells. Colloids and Surfaces B: Biointerfaces. 2016;148:49–58. doi: 10.1016/j.colsurfb.2016.08.041. [DOI] [PubMed] [Google Scholar]
- 25.Bijonowski B. M., Daraiseh S. I., Yuan X., Ma T. Size-dependent cortical compaction induces metabolic adaptation in mesenchymal stem cell aggregates. Tissue Engineering Part A. 2019;25(7-8):575–587. doi: 10.1089/ten.tea.2018.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sart S., Tsai A.-C., Li Y., Ma T. Three-dimensional aggregates of mesenchymal stem cells: cellular mechanisms, biological properties, and applications. Tissue Engineering Part B: Reviews. 2014;20(5):365–380. doi: 10.1089/ten.teb.2013.0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sart S., Bejoy J., Li Y. Characterization of 3D pluripotent stem cell aggregates and the impact of their properties on bioprocessing. Process Biochemistry. 2017;59:276–288. doi: 10.1016/j.procbio.2016.05.024. [DOI] [Google Scholar]
- 28.Varum S., Rodrigues A. S., Moura M. B., et al. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One. 2011;6(6, article e20914) doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J., Nuebel E., Wisidagama D. R. R., et al. Measuring energy metabolism in cultured cells, including human pluripotent stem cells and differentiated cells. Nature Protocols. 2012;7(6):1068–1085. doi: 10.1038/nprot.2012.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMurtrey R. J. Analytic models of oxygen and nutrient diffusion, metabolism dynamics, and architecture optimization in three-dimensional tissue constructs with applications and insights in cerebral organoids. Tissue Engineering Part C: Methods. 2016;22(3):221–249. doi: 10.1089/ten.TEC.2015.0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang L., Shestov A. A., Swain P., et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532(7598):255–258. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metallo C. M., Gameiro P. A., Bell E. L., et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481(7381):380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang G., Sai K., Gong F., Yang Q., Chen F., Lin J. Mutation of isocitrate dehydrogenase 1 induces glioma cell proliferation via nuclear factor-κB activation in a hypoxia-inducible factor 1-α dependent manner. Molecular Medicine Reports. 2014;9(5):1799–1805. doi: 10.3892/mmr.2014.2052. [DOI] [PubMed] [Google Scholar]
- 34.Lin H., Du Q., Li Q., et al. A scalable and efficient bioprocess for manufacturing human pluripotent stem cell-derived endothelial cells. Stem Cell Reports. 2018;11(2):454–469. doi: 10.1016/j.stemcr.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y., Yuan X., Munoz N., Logan T. M., Ma T. Commitment to aerobic glycolysis sustains immunosuppression of human mesenchymal stem cells. Stem Cells Translational Medicine. 2019;8(1):93–106. doi: 10.1002/sctm.18-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Correia C., Koshkin A., Duarte P., et al. 3D aggregate culture improves metabolic maturation of human pluripotent stem cell derived cardiomyocytes. Biotechnology and Bioengineering. 2018;115(3):630–644. doi: 10.1002/bit.26504. [DOI] [PubMed] [Google Scholar]
- 37.Van Winkle A. P., Gates I. D., Kallos M. S. Mass transfer limitations in embryoid bodies during human embryonic stem cell differentiation. Cells, Tissues, Organs. 2012;196(1):34–47. doi: 10.1159/000330691. [DOI] [PubMed] [Google Scholar]
- 38.Ma T., Grayson W. L., Frohlich M., Vunjak-Novakovic G. Hypoxia and stem cell‐based engineering of mesenchymal tissues. Biotechnology Progress. 2009;25(1):32–42. doi: 10.1002/btpr.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathieu J., Zhou W., Xing Y., et al. Hypoxia-inducible factors have distinct and stage-specific roles during reprogramming of human cells to pluripotency. Cell Stem Cell. 2014;14(5):592–605. doi: 10.1016/j.stem.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mutoh T., Sanosaka T., Ito K., Nakashima K. Oxygen levels epigenetically regulate fate switching of neural precursor cells via hypoxia-inducible factor 1α-Notch signal interaction in the developing brain. Stem Cells. 2012;30(3):561–569. doi: 10.1002/stem.1019. [DOI] [PubMed] [Google Scholar]
- 41.Grayson W. L., Zhao F., Izadpanah R., Bunnell B., Ma T. Effects of hypoxia on human mesenchymal stem cell expansion and plasticity in 3D constructs. Journal of Cellular Physiology. 2006;207(2):331–339. doi: 10.1002/jcp.20571. [DOI] [PubMed] [Google Scholar]
- 42.Fukuda R., Zhang H., Kim J. W., Shimoda L., Dang C. V., Semenza G. L. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129(1):111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 43.Quirós P. M., Español Y., Acín-Pérez R., et al. ATP-dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Reports. 2014;8(2):542–556. doi: 10.1016/j.celrep.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 44.Weinberg F., Hamanaka R., Wheaton W. W., et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(19):8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J., Crawford R., Chen C., Xiao Y. The key regulatory roles of the PI3K/Akt signaling pathway in the functionalities of mesenchymal stem cells and applications in tissue regeneration. Tissue Engineering Part B: Reviews. 2013;19(6):516–528. doi: 10.1089/ten.TEB.2012.0672. [DOI] [PubMed] [Google Scholar]
- 46.Zhou J., Su P., Wang L., et al. mTOR supports long-term self-renewal and suppresses mesoderm and endoderm activities of human embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(19):7840–7845. doi: 10.1073/pnas.0901854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peterson T. R., Sengupta S. S., Harris T. E., et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146(3):408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Düvel K., Yecies J. L., Menon S., et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Molecular Cell. 2010;39(2):171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tee A. R. The target of rapamycin and mechanisms of cell growth. International Journal of Molecular Sciences. 2018;19(3):p. 880. doi: 10.3390/ijms19030880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stine Z. E., Walton Z. E., Altman B. J., Hsieh A. L., Dang C. V. MYC, metabolism, and cancer. Cancer Discovery. 2015;5(10):1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mak G. W.-Y., Lai W.-L., Zhou Y., Li M., Ng I. O.-L., Ching Y.-P. CDK5RAP3 is a novel repressor of p14ARF in hepatocellular carcinoma cells. PLoS One. 2012;7(7, article e42210) doi: 10.1371/journal.pone.0042210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X., Peterson K. A., Liu X. S., McMahon A. P., Ohba S. Gene regulatory networks mediating canonical Wnt signal directed control of pluripotency and differentiation in embryo stem cells. Stem Cells. 2013;31(12):2667–2679. doi: 10.1002/stem.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Azzolin L., Panciera T., Soligo S., et al. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158(1):157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 54.Wang L., Li H., Zhang Y., Santella R. M., Weinstein I. B. HINT1 inhibits β‐catenin/TCF4, USF2 and NFκB activity in human hepatoma cells. International Journal of Cancer. 2009;124(7):1526–1534. doi: 10.1002/ijc.24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bejoy J., Song L., Zhou Y., Li Y. Wnt/Yes-Associated protein interactions during neural tissue patterning of human induced pluripotent stem cells. Tissue Engineering Part A. 2018;24(7-8):546–558. doi: 10.1089/ten.tea.2017.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bejoy J., Wang Z., Bijonowski B., et al. Differential effects of heparin and hyaluronic acid on neural patterning of human induced pluripotent stem cells. ACS Biomaterials Science & Engineering. 2018;4(12):4354–4366. doi: 10.1021/acsbiomaterials.8b01142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moya N., Cutts J., Gaasterland T., Willert K., Brafman D. A. Endogenous WNT signaling regulates hPSC-derived neural progenitor cell heterogeneity and specifies their regional identity. Stem Cell Reports. 2014;3(6):1015–1028. doi: 10.1016/j.stemcr.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cohen E. D., Tian Y., Morrisey E. E. Wnt signaling: an essential regulator of cardiovascular differentiation, morphogenesis and progenitor self-renewal. Development. 2008;135(5):789–798. doi: 10.1242/dev.016865. [DOI] [PubMed] [Google Scholar]
- 59.Yang L., Liu C. C., Zheng H., et al. LRP1 modulates the microglial immune response via regulation of JNK and NF-κB signaling pathways. Journal of Neuroinflammation. 2016;13(1):p. 304. doi: 10.1186/s12974-016-0772-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y., Liu J., Yao S., et al. Nuclear factor kappa B signaling initiates early differentiation of neural stem cells. Stem Cells. 2012;30(3):510–524. doi: 10.1002/stem.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wrzesinski K., Rogowska-Wrzesinska A., Kanlaya R., et al. The cultural divide: exponential growth in classical 2D and metabolic equilibrium in 3D environments. PLoS One. 2014;9(9, article e106973) doi: 10.1371/journal.pone.0106973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clark C. R., Robinson J. Y., Sanchez N. S., et al. Common pathways regulate type III TGFβ receptor-dependent cell invasion in epicardial and endocardial cells. Cellular Signalling. 2016;28(6):688–698. doi: 10.1016/j.cellsig.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song L., Yuan X., Jones Z., et al. Assembly of human stem cell-derived cortical spheroids and vascular spheroids to model 3-D brain-like tissues. Scientific Reports. 2019;9(1, article 5977) doi: 10.1038/s41598-019-42439-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hatakeyama J., Wakamatsu Y., Nagafuchi A., Kageyama R., Shigemoto R., Shimamura K. Cadherin-based adhesions in the apical endfoot are required for active Notch signaling to control neurogenesis in vertebrates. Development. 2014;141(8):1671–1682. doi: 10.1242/dev.102988. [DOI] [PubMed] [Google Scholar]
- 65.Grandbarbe L., Bouissac J., Rand M., Hrabé de Angelis M., Artavanis-Tsakonas S., Mohier E. Delta-Notch signaling controls the generation of neurons/glia from neural stem cells in a stepwise process. Development. 2003;130(7):1391–1402. doi: 10.1242/dev.00374. [DOI] [PubMed] [Google Scholar]
- 66.Hayward P., Kalmar T., Arias A. M. Wnt/Notch signalling and information processing during development. Development. 2008;135(3):411–424. doi: 10.1242/dev.000505. [DOI] [PubMed] [Google Scholar]
- 67.Galluzzi L., Bravo-San Pedro J. M., Kepp O., Kroemer G. Regulated cell death and adaptive stress responses. Cellular and Molecular Life Sciences. 2016;73(11-12):2405–2410. doi: 10.1007/s00018-016-2209-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang C., Youle R. J. The role of mitochondria in apoptosis. Annual Review of Genetics. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Galluzzi L., Bravo-San Pedro J. M., Kroemer G. Organelle-specific initiation of cell death. Nature Cell Biology. 2014;16(8):728–736. doi: 10.1038/ncb3005. [DOI] [PubMed] [Google Scholar]
- 70.Beneke S., Burkle A. Poly(ADP-ribosyl)ation in mammalian ageing. Nucleic Acids Research. 2007;35(22):7456–7465. doi: 10.1093/nar/gkm735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hodgson R. E., Varanda B. A., Ashe M. P., Allen K. E., Campbell S. G. Cellular eIF2B subunit localization: implications for the integrated stress response and its control by small molecule drugs. Molecular Biology of the Cell. 2019;30(8):942–958. doi: 10.1091/mbc.E18-08-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cagnetta R., Wong H. H. W., Frese C. K., Mallucci G. R., Krijgsveld J., Holt C. E. Noncanonical modulation of the eIF2 pathway controls an increase in local translation during neural wiring. Molecular Cell. 2019;73(3):474–489.e5. doi: 10.1016/j.molcel.2018.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smolle M. A., Calin H. N., Pichler M., Calin G. A. Noncoding RNAs and immune checkpoints-clinical implications as cancer therapeutics. The FEBS Journal. 2017;284(13):1952–1966. doi: 10.1111/febs.14030. [DOI] [PubMed] [Google Scholar]
- 74.Schetters S. T. T., Gomez-Nicola D., Garcia-Vallejo J. J., Van Kooyk Y. Neuroinflammation: microglia and T cells get ready to tango. Frontiers in Immunology. 2018;8 doi: 10.3389/fimmu.2017.01905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: primer sequence for target genes, Supplementary Table S2: genes related to ATP synthesis and mitochondrial complexes I, III, and IV. Supplementary Table S3: genes related to the NF-κB pathway. Supplementary Table S4: AMPK pathway and PDL1/PD1 pathway. Supplementary Figure S1: microglial differentiation using additional human iPSC line: Ep-iPSC. Supplementary Figure S2: regulation of central metabolism in D-MG. Supplementary Figure S3: log2 gene expression levels of D-MG/MG ratios for ECM-related genes.
Data Availability Statement
Data is available by contacting the corresponding author.