Abstract
Genetic variations in miRNAs binding site might participate in cancer risk. This study aimed to systematically review the association between miRNA‐binding site polymorphisms and colorectal cancer (CRC). Electronic literature search was carried out on PubMed, Web of Science (WOS), Scopus, and Embase. All types of observational studies till 30 November 2018 were included. Overall 85 studies (21 SNPs) from two systematic searches were included analysis. The results showed that in the Middle East population, the minor allele of rs731236 was associated with decreased risk of CRC (heterozygote model: 0.76 [0.61‐0.95]). The minor allele of rs3025039 was related to increased risk of CRC in East Asian population (allelic model: 1.25 [1.01‐1.54]). Results for rs3212986 were significant in overall and subgroup analysis (P < .05). For rs1801157 in subgroup analysis the association was significant in Asian populations (including allelic model: 2.28 [1.11‐4.69]). For rs712, subgroup analysis revealed a significant (allelic model: 1.41 [1.23‐1.61]) and borderline (allelic model: 0.92 [0.84‐1.00]) association in Chinese and Czech populations, respectively. The minor allele of rs17281995 increased risk of CRC in different genetic models (P < .05). Finally, rs5275, rs4648298, and rs61764370 did not show significant associations. In conclusion, minor allele of rs3025039, rs3212986, and rs712 polymorphisms increases the risk of CRC in the East Asian population, and heterozygote model of rs731236 polymorphism shows protective effect in the Middle East population. In Europeans, the minor allele of rs17281995 may increase the risk of CRC, while rs712 may have a protective effect. Further analysis based on population stratifications should be considered in future studies.
Keywords: colorectal cancer, meta‐analysis, microRNAs, polymorphism
MicroRNA‐binding site polymorphisms in 3'UTR of target genes could play critical roles on cancer genes regulation by affecting miRNA:mRNA interactions, but no comprehensive study has been considered on association between cancers and the mentioned polymorphisms. In this study, the authors carried out two comprehensive systematic searches and a meta‐analysis on 21 included polymorphisms on colorectal cancer (CRC). The results have shown that these polymorphisms can serve as genetic biomarkers of CRC, and their roles were exclusively related to population stratifications. It is strongly recommended that further studies should be performed to display the effect of population stratifications.
1. INTRODUCTION
Colorectal cancer is one of the most serious illnesses in both sexes. It has been recognized as the second and third common cancers in females and males, respectively.1, 2, 3 Incidence and mortality of colorectal cancer (CRC)was about 6.1% of new cancer cases and was around 9.2% of cancer death based on Global Cancer Statistics 2018.4 Its incidence is three times higher in developed countries than developing counters.4 CRC imposes enormous global burden which could be related to aging and population growth, socioeconomic status, diet, life styles, and habits including smoking, western diet, and physical activity.5, 6, 7 Early diagnosis of CRC leads to lesser treatment cost besides better survival and prognosis.8 Early prognosis or diagnosis of CRC is also important in cancer survival. Nine of 10 people with CRC would have more than 5 years of survival, if the diagnosis is performed at the stage one while diagnosis in the last stage leads to merely 1 year of survival. For this purpose, finding novel biomarkers for noninvasive early diagnosis of CRC will be crucial in disease treatment.
Some risk factors of CRC including diet and smoking could be modified in contrast to genetic factors.9, 10, 11 MicroRNAs (miRNAs) are important genetic factors which are regulating around 60% of human protein‐coding genes.12 It is believed that miRNAs play an important role in the pathogenesis of CRC.13 miRNA polymorphisms might participate in cancer prognosis through their effect on miRNA gene transcription, processing, expression, and target selection.14, 15, 16 A meta‐analysis in 2016 has been implemented on the association between miR‐27a rs895819 in the loop of pre‐miRNA and shows that this SNP may be a risk factor for CRC (for instance in allelic model OR = 1.21 [1.11‐1.31]).13 A systematic review and meta‐analysis has been published in 2014 based on the role of two polymorphisms in miR‐146a and in miR‐196a2 on the susceptibility towards CRC. The results revealed that miR‐196a2 polymorphism rs11614913 is associated with the risk of CRC.17 Another review paper in 2015 described the association of miRNA variants (in miR‐146a, hsa‐miR‐149, and hsa‐miR‐196a2) and CRC and showed that rs2910164 (1.24 [1.03‐1.49]) and rs2292832 (1.18 [1.08‐1.38]) may increase the risk of CRC, and rs11614913 and rs3746444 (0.57 [0.34‐0.95]) may decrease the risk of CRC.18 In 2017, a review article was published on the risk of CRC and polymorphisms in microRNA gene. Based on these results let‐7, miR‐149, miR‐603, miR‐34b/c, and miR‐146a gene SNPs were associated with CRC.19
Polymorphisms in miRNA‐binding sites may also alter the risk and survival of a variety of human complex diseases including CRC.20, 21, 22 miRNA‐binding sites are conserved through evolution and contain lesser polymorphisms.23 Polymorphisms in these sites can affect miRNA:mRNA interactions and target mRNA expression.24, 25 In one study, the association between let‐7 miRNA‐binding site polymorphisms and CRC outcome has been described, based on one miRNA, one database (PubMed), and also CRC risk was not investigated.26 miRNAs’ target site polymorphisms may potentially play a role in the interaction between miRNAs and their target mRNA, which is dependent on the effect of polymorphism on miRNA:mRNA interactions. There was also a meta‐analysis on 3'UTR polymorphisms and the risk of cancers,27 but the results were only for two polymorphisms and were not specific for CRC or miRNA‐binding sites. To the best of our knowledge, there is no previous systematic review on the association between miRNA‐binding site polymorphisms and CRC. Therefore, the lack of a comprehensive systematic review focusing on miRNA‐binding site polymorphisms and CRC is obvious.
Because of importance and economic burden of CRC, and regarding the significant role of miRNA‐binding site polymorphisms on CRC according to the previous studies besides lack of a systematic review on this subject, the necessity of such study on association between miRNA‐binding site polymorphisms and CRC, as prognostic markers, is quite clear. For this purpose, the main objective of the current systematic review was to explore and reveal the association of 3'UTR and miRNA‐binding site polymorphisms with the risk of CRC. The secondary specific objective was to determine the effect of ethnicity on these associations.
2. METHODS AND ANALYSIS
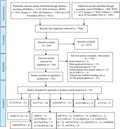
The methods of this study have been developed according to the PRISMA‐P 2015 checklist.28 PRISMA 2009 flow diagram,29 used to display the flow of document number through the different phases of the study (Figure 1). The protocol of this systematic review is registered in International Prospective Register for Systematic Reviews (PROSPERO) on January 11, 2018 (Registration ID = CRD42018084094).
Figure 1.
Flow diagram for systematic review
2.1. Eligible studies and participants
This study imposed a restriction on the study design. Observational studies (case‐control, cohort, and cross‐sectional), describing the association between miRNA‐binding site polymorphisms and CRC, were eligible for inclusion. Primary documents will be screened according to the PECO criteria (Participants, Exposure, Comparisons, and Outcomes) and objectives of this study. Studies with deviation from Hardy‐Weinberg equilibrium30 (HWE) and with the lack of required primary data or data for estimating genotype numbers were excluded. This study also applied a restriction on publication date. Only documents published from January 1, 1992 to November 30, 2018 were searched. This restriction was based on two reasons; first: miRNA discovery date, and second: most recent publications were relevant to our study subject. There was no restriction about the language of documents related to the topic of this study. Non‐English languages articles were translated by free language translation services or by a translator. There was also no limitation on age, gender, ethnicity, and method of genotyping. The study did not impose a restriction on colorectal cancer stages (I, II, III, and IV). Colorectal polyps and family‐based case‐control studies were not considered for inclusion.
2.2. MicroRNAs binding site polymorphism
Polymorphisms in miRNA‐binding sites have been reported to be associated with cancers.31, 32 These SNPs are conserved through evolution.23 These sites act as diagnostic and prognostic biomarkers associated with cancer risk and outcome.33 Their association with susceptibility, outcome, treatment, prognosis, and progression of CRC has also been reported.20, 34, 35, 36 In this systematic review, studies that evaluated the relationship between miRNA‐binding site polymorphisms and CRC were included and the primary outcome of this review was finding association between miRNA‐binding site polymorphisms and CRC susceptibility. Moreover, subgroup analysis for ethnicity was carried out on association of CRC risk with microRNA‐binding site polymorphisms.
2.3. Search methods for studies identification
In order to identify the relevant papers on miRNA‐binding site polymorphisms and colorectal cancer, online systematic search (electronic searches) of literature was performed in PubMed, Embase, Scopus, and Web of Science. We developed PubMed search syntax, as the main database, this syntax was adapted to other database. PubMed search syntax was performed by combined medical subject headings (MeSH), Emtree terms, keywords of related papers, also free text words. Key search terms were “colorectal neoplasms,” “miRNA,” “Polymorphism, Single Nucleotide,” and their equivalents (Table S1). To identify more results, we also manually checked references from included primary articles and relevant reviews, conference papers, gray literature, as well as contact with corresponding authors for missing data.
2.4. Data collection
2.4.1. Screening for eligible studies
Screening and eligibility checking was performed in three following steps. First, duplicate documents were removed. Second, for screening, two reviewers independently scrutinize remaining documents by checking title and/or abstract. Third, full texts' eligibility was independently scrutinized by two reviewers. Any disagreements between two reviewers were resolved by consensus strategy and third‐person strategy.
2.4.2. Data extraction and management
A data extraction form was created and then piloted by two reviewers. This form included the following data: the name of first author, country of study, year of publication, study design, age, gender, ethnicity, names of 3'UTR or binding site SNPs, genotyping methods, minor allele frequency (MAF), HWE, sample size, matching criteria (such as age and sex), source of controls (HB, hospital base or PB, population base), odds ratio (OR), confidence interval (95% CIs), and other related raw data. In the next step, two reviewers independently extracted data based on the extraction form. Disagreements were resolved by strategies listed above.
2.5. Analysis
2.5.1. Meta‐analysis
Meta‐analysis was performed by using R (3.5.2). Odds ratio and 95% CI were used to investigate the associations between each polymorphism in miRNA‐binding site and CRC. The meta‐analysis was performed based on different genetic models (allelic model (A vs a), homozygous model (AA vs aa), heterozygote model (Aa vs aa), AA vs Aa model, dominant model (AA + Aa vs aa), recessive model (AA vs Aa + aa), and overdominant model (Aa vs AA + aa)). All included studies were at the risk of various types of heterogeneity. For exploring possible sources of heterogeneity, included studies were divided according to the type of polymorphisms. For each polymorphism, if sufficient studies were included, subgroup analysis (based on ethnicity) was applied. Odds ratios were estimated by fixed effects model (FEM) or random effects model (REM), according to the heterogeneity level. Level of heterogeneity between primary studies was obtained by the Cochran's Q test (P < .05 is statistically significant) and the I 2 statistic in forest plots. We used the following guide to interpret the amount of heterogeneity: I 2 < 25% = low heterogeneity; 25 ≥ I 2 < 50% = moderate heterogeneity; 50 ≥ I 2 < 75% = sever heterogeneity; 75% ≥ I 2 = highly sever heterogeneity.
2.5.2. Reporting biases and sensitivity analysis
We used Begg's test and Egger's regression method to assess the potential publication bias in primary studies. Main results were depicted by funnel plots (for visual assessment). Sensitivity analysis was performed by the leave‐one‐out method.
3. RESULTS
In the systematic search, at the first stage we found 9221 documents, with 222 polymorphisms in 3′UTR and miRNA‐binding site of genes that were studied for the risk of CRC. Among them we included main polymorphisms in second search for meta‐analysis (these polymorphisms were selected because the meta‐analysis for all included polymorphisms was not possible, also in order to decrease the false positive prediction of miRNA‐binding sites polymorphisms, only polymorphisms that were mentioned in two studies or more were included, one of these studies should report polymorphism in miRNA‐binding site). Twenty‐five polymorphisms were included (rs10082466, rs10434, rs8176318, rs17281995, rs3212986, rs1368439, rs1131445, rs5275, rs61764370, rs712, rs108621, rs696, rs3135500, rs8679, rs16870224, rs731236, rs3025039, rs3025040, rs3025053, rs4648298, rs1801157, rs3742330, rs4846049, rs854551, and rs9138). Second search strategy applied for these polymorphisms, which contained 5170 documents. Finally, we included 54 studies on the role of 3′UTR polymorphisms and 52 studies on the role of miRNA‐binding site polymorphisms and risk of CRC for all the selected polymorphisms (Tables 1 and 2). Finally, 21 polymorphisms with two or more than two included studies were eligible for final analysis (these studies are shown in detail in Tables 3 and 4). For rs17281995 polymorphism, the pooled analysis based on three included articles showed significant increased risk of CRC in different genetic models, including homozygote model 2.29 (1.25‐4.19). Seven of 21 included polymorphisms in our meta‐analysis were polymorphisms with more than four included articles (rs731236, rs3025039, rs3212986, rs712, rs5275, rs4648298, and rs1801157). The basic characteristics of studies included in the meta‐analysis are shown following (Table 4).
Table 1.
miRNA‐binding sites polymorphisms and colorectal cancer risk (included from first search strategy)
| References | Study design | rsID (target miRNA) |
|---|---|---|
| 37 | Case‐control | rs10082466 (miR‐27a) |
| 38 | Case‐control | rs11466537 (miR‐1193) |
| 39 | Case‐control | rs12904 (miR‐200 family: miR‐200c, miR‐429, and miR‐200b) |
| 40 | Case‐control | rs12915554 (miR‐185‐3p) |
| 41 | Case‐control | rs141178472 (miR‐520a) |
| 42 | Case‐control | rs16917496 (miR‐502) |
| 43 | Case‐control | rs1710 (miRNA‐binding site polymorphisma) |
| 44 | Case‐control | rs2015 (miR‐376a‐5p) |
| 45 | Case‐control | rs2737 (miR‐379) |
| 46 | Case‐control | rs3135500 (miR‐158, miR‐215, miR‐98, miR‐573) |
| 47 | Case‐control | rs11169571 (miR‐1283, miR‐520d‐5p) |
| 48 | Case‐control | rs34149860 (miR‐29b) |
| 49 | Case‐control | rs4648298 (miR‐21, miR590) |
| 50 | Case‐control | rs3814058 (miR‐129‐5p) |
| 51 | Case‐control | rs4245739 (miR‐191) |
| 52 | Case‐control | rs4804800 (miR‐622, miR‐1238) |
| 53 | Case‐control | rs4939827 (miR‐375) |
| 54 | Case‐control | rs5275 (miR‐542‐3p) |
| 55 | Case‐control | rs61764370 (let‐7) |
| 56 | Case‐control | rs61764370 (let‐7) |
| 57 | Case‐control | rs696 (miR449a) |
| 58 | Case‐control | rs696 (miR‐449a, miR‐34b) |
| 36 | Case‐control | rs712 (let‐7) |
| 59 | Case‐control | rs712 (miR‐200b, miR‐429, miR‐200c, miR‐193b) |
| 60 | Case‐control | rs8679 (miR‐145) |
| 61 | Case‐control | rs12997 (miR‐330‐3p), rs1043784 (miR‐584), rs10038999 (miR‐629), rs1129976 (miR‐150) |
| 62 | Case‐control | rs712 (let‐7), rs61764370 (let‐7) |
| 63 | Case‐control | rs17468, rs2317676 (miRNA‐binding site polymorphisms) |
| 64 | Case‐control | rs3135500, rs1368439 (miRNA ‐binding site polymorphisms) |
| 65 | Case‐control | rs13347 (miR‐509‐3p), rs10836347, rs11821102 (miRNA‐binding site polymorphisms) |
| 66 | Case‐control | rs5186 (miR‐155), rs710100 (miR‐155), rs411103 (miR‐27b) |
| 67 | Case‐control | rs847 (miR‐98, let‐7i/f/g), rs848 (miR‐558, miR‐621, let‐7i), rs1295685 (miR‐621) |
| 68 | Case‐control | rs7930 (miR‐4273‐5p), rs8117825 (miR‐3126‐5p, miR‐337‐3p), rs16853287 (miR‐128‐3p, miR‐140‐3p) |
| 69 | Case‐control | rs1590 (miR‐532‐5p, miR‐768‐3p), rs1434536, rs17023107 (miRNA‐binding site polymorphisms) |
| 70 | Case‐control | rs4143815 (miR‐570), rs1059293, rs27194, rs43216 (miRNA‐binding site polymorphisms) |
| 71 | Case‐control | rs1062044 (miR‐423‐5p), rs17477864 (miR‐186‐5p), rs3824998 (miR‐221‐3p), rs4768914 (miR‐200c‐3P), rs1046165 (miR‐451a) |
| 72 | Case‐control | rs108621 (miR‐193a‐3p, miR‐338‐3p), rs3212986 (miR‐15a) |
| 73 | Case‐control | rs3660, rs1044129, rs1053667, rs4901706, rs11337 (miRNA‐binding site polymorphisms) |
| 74 | Case‐control | rs1131445 (miR‐135a/135b), rs1051208 (miR‐213), rs743554, rs16870224, rs11515 (miRNA‐binding site polymorphisms) |
| 75 | Case‐control | rs1126547 (hsa‐miR‐141, hsa‐miR‐200a), rs2229090 (miR‐1225‐3p, miR‐3123, miR‐3619), rs9914073 (miR‐548c‐3p, miR‐605), rs17339395 (miR‐4299), rs7356 (miR‐3149,miR‐1183), rs1803541 (miR‐568, miR‐802), rs4596 (miR‐518a‐5p, miR‐527, miR‐1205), rs4781563 (miR‐2355‐3p, miR‐4288), rs45522131 (miR‐26a/b, miR‐374a) |
| 76 | Case‐control | rs61764370 (let‐7), rs8679 (miR‐145‐3p), rs1804197, rs41116, rs397768, rs4585, rs712, rs16950113 (miRNA‐binding site polymorphisms) |
| 22 | Case‐control | rs17281995 (miR‐337, miR‐582, miR‐200a*, miR‐184, miR‐212), rs3135500 (miR‐158, miR‐215, miR‐98, miR‐573), rs1131445 (miR‐135a, miR‐135b, miR‐143, miR‐18, miR‐18a), rs1368439 (miR‐513, miR‐210, miR‐27b, miR‐27a), rs916055 (miR‐588, miR‐183), rs11677 (miR‐187, miR‐638, miR‐154, miR‐453, miR‐296), rs16870224 (miR‐9, miR‐30a‐3p, miR‐30e‐3p), rs1051690 (miR‐618, miR‐612) |
| 77 | Case‐control | rs2147578 (miR‐128‐3p,216a‐3p,3681‐3p), rs112462125 (miR‐197‐3p), rs7844527 (miR‐146a‐5p,146b‐5p), rs7814028 (miR‐5001‐3p,miR‐6819‐3p), rs12677572 (miR‐891a‐5p), rs60719452 (miR‐548‐5p,548ab,548ak,548au‐5p,548ay‐5p,548b‐5p,548d‐5p,548i,548y), rs61095617 (miR‐1307‐5p), rs75511849 (miR‐100‐3p) |
| 78 | Case‐control | rs88640,3 (miR‐4647, miR‐588, miR‐125, let‐7), rs4077531, rs3733492, rs12732, rs1532602, rs4071, rs17552409, rs17243454, rs4729655, rs7631009, rs6782006, rs974034, rs7372 (miRNA‐binding site polymorphisms) |
| 79 | Case‐control | rs712 (miR‐200b, miR‐429, miR‐200c, miR‐193b), rs709805 (miR‐324‐3p), rs2289965 (miR‐142‐3p, miR‐324‐5p), rs3012518 (miR‐299‐3p), rs2839629 (miR‐18a, miR‐18b), rs904960 (miR‐32, miR‐25, miR‐367, miR‐363), rs3734279 (miR‐203), rs354476 (miR‐125a, miR‐125b), rs495714 (miR‐324‐3p, miR‐196b, miR‐196a), rs1048650 (miR‐22), rs496550 (miR‐363), rs473351 (miR‐182) |
| 80 | Case‐control | rs2233921 (miR‐3925‐3p, miR‐3140‐3p, miR‐1825, miR‐1825, miR‐3925‐3p, miR‐3140‐3p), rs971 (miR‐4744, miR‐3154, miR‐610, miR‐4744, miR‐3154, hsa‐miR‐610), rs6997097 (miR‐3605‐5p, miR‐3545‐3p, miR‐3605‐5p, miR‐3545‐3p), rs8191670, rs2740439, rs4639, rs1043180, rs1055678, rs1052536 rs2307285, rs2307294, rs1534862, (miRNA‐binding site polymorphisms) |
| 34 | Case‐control | rs2279398 miR‐370, rs1047854, rs11206394, rs1128287, rs1131445, rs12462695, rs15049, rs17111100, rs2275085, rs2283606, rs2839531, rs3135499, rs3757417, rs3803098, rs747343, rs9118 (miRNA‐binding site polymorphisms) |
| 81 | Case‐control | rs2155209 (miR‐1296, miR‐296‐5p), rs11226 (miR‐296‐5p, miR‐1296), rs1051669 rs11571475, rs7963551, rs12593359, rs7180135, rs45507396, rs8176318, rs13447749, rs9995, rs14448,rs300171, rs300170, rs3218547, rs10131, rs1051685, rs2440, rs1051677, rs897477, rs2035990 (miRNA‐binding site polymorphisms) |
miRNA‐binding site polymorphism: the polymorphism located in miRNA‐binding sites (according to the referenced article).
Table 2.
3ʹUTR polymorphisms and colorectal cancer risk (included from first search strategy)
| Reference | Study design | rsID |
|---|---|---|
| 82 | Case‐control | rs1058881 |
| 83 | Case‐control | rs1059234 |
| 84 | Case‐control | rs731236 |
| 85 | Case‐control | rs108621 |
| 86 | Case‐control | rs142559064 |
| 40 | Case‐control | rs146588909 |
| 87 | Case‐control | rs17281995 |
| 88 | Case‐control | rs1801157 |
| 89 | Case‐control | rs1801157 |
| 90 | Case‐control | rs1801157 |
| 91 | Case‐control | rs2075786 |
| 44 | Case‐control | rs2241703 |
| 92 | Case‐control | rs3025039 |
| 93 | Case‐control | rs3025039 |
| 94 | Case‐control | rs3025039 |
| 95 | Case‐control | rs3025039 |
| 96 | Case‐control | rs3212986 |
| 50 | Case‐control | rs3732360 |
| 97 | Case‐control | rs3742330 |
| 98 | Nested case‐cohort | rs5275 |
| 99 | Case‐control | rs78378222 |
| 100 | Case‐control | rs5275 |
| 101 | Case‐control | rs5275 |
| 102 | Case‐control | rs57898959 |
| 103 | Case‐control | rs8176318 |
| 104 | Case‐control | rs696 |
| 105 | Case‐control | rs713041 |
| 106 | Case‐control | rs7579 |
| 107 | Case‐control | rs8878 |
| 108 | Case‐control | rs9138 |
| 109 | Case‐control | rs9138 |
| 110 | Case‐control | CDX2‐G1312T |
| 111 | Case‐control | rs868, rs7591 |
| 112 | Case‐control | rs5275, rs4648298 |
| 113 | Case‐control | rs67085638, rs77628730 |
| 114 | Case‐control | rs4648298, rs5276, rs13306035 |
| 115 | Case‐control | rs1205, rs3093075 |
| 116 | Case‐control | rs7975232, rs1544410 |
| 117 | Case‐control | rs16930073, rs8491, rs854551 |
| 118 | Case‐control | rs11875, rs1042669, rs4149206 |
| 119 | Case‐control | rs3025040, rs10434, rs3025053 |
| 72 | Case‐control | rs735482, rs2336219, rs1052133 |
| 62 | Case‐control | rs12245, rs12587, rs9266, rs1137282 |
| 120 | Case‐control | rs3742330, rs10719, rs14035, rs11077 |
| 121 | Case‐control | rs334348, rs334349, rs1590, rs868, rs420549 |
| 122 | Case‐control | rs11708581, rs12163565, rs390802, rs123598 |
| 37 | Case‐control | rs2120132, rs2099902, rs10450310, rs10082466 |
| 123 | Case‐control | rs4846049, rs1537514, rs3737967, rs4846048 |
| 124 | Case‐control | rs1137188, rs3025039, rs3025040, rs3025053, rs10434 |
| 125 | Nested case‐cohort | rs11168267, rs11574113, rs731236, rs3847987, rs11574143 |
| 66 | Case‐control | rs12009, rs700082, rs1057035, rs10404, rs1939861, rs3757261 |
| 52 | Case‐control | rs7248637, rs11465421, rs10824792, rs2083771, rs1052972 |
| 43 | Case‐control | rs1707, rs17179101, rs17179108, rs1063320, rs9380142, rs1610696 |
| 68 | Case‐control | rs4985036, rs9970671, rs11861556, rs17500814, rs12678, rs9129, rs2561819 |
| 126 | Case‐control | rs2302821, rs45544737, rs34337770, rs7730368, rs16870224, rs4957343, rs9312555 |
| 127 | Case‐control | rs10849, rs10890324, rs293796, rs7641176, rs293782, rs293783, rs6809452, rs6544991, rs6720549, rs6713506, rs2537742 |
| 128 | Case‐control | rs2298753, rs706209, rs13420827, rs6058896, rs3827869, rs1832683, rs4846049, rs9282787, rs9332, rs854571, rs1544468, rs10418, rs757158, rs854551, rs3917577 |
Table 3.
Genotyping and analysis results of polymorphism with less than four eligible studies
| Gene | rsID | Case | Control | References | Sig. in genetic models | ||||
|---|---|---|---|---|---|---|---|---|---|
| CC | GC | GG | CC | GC | GG | Yesa | |||
| CD86 | rs17281995 | 7 | 48 | 137 | 0 | 55 | 164 | 87 | |
| 24 | 161 | 475 | 8 | 114 | 434 | 22 | |||
| 12 | 75 | 217 | 7 | 67 | 181 | 129 | |||
| CC | TC | TT | CC | TC | TT | ||||
| PARP1 | rs8679 | 53 | 335 | 687 | 66 | 482 | 873 | 76 | No |
| 12 | 60 | 111 | 14 | 86 | 90 | 60 | |||
| AA | GA | GG | AA | GA | GG | ||||
| VEGF | rs10434 | 8 | 57 | 214 | 9 | 83 | 213 | 119 | No |
| 19 | 143 | 209 | 11 | 93 | 142 | 124 | |||
| CC | TC | TT | CC | TC | TT | ||||
| MLH3 | rs108621 | 219 | 562 | 311 | 300 | 665 | 428 | 85 | No |
| 14 | 62 | 124 | 9 | 59 | 132 | 72 | |||
| CC | CT | TT | CC | CT | TT | ||||
| IL‐16 | rs1131445 | 36 | 110 | 103 | 34 | 159 | 201 | 74 | No |
| 65 | 287 | 308 | 53 | 240 | 251 | 22 | |||
| GG | TG | TT | GG | TG | TT | ||||
| IL12B | rs1368439 | 2 | 29 | 61 | 2 | 35 | 68 | 64 | No |
| 21 | 188 | 465 | 15 | 164 | 388 | 22 | |||
| AA | GA | GG | AA | GA | GG | ||||
| PTGER4 | rs16870224 | 11 | 130 | 523 | 4 | 116 | 439 | 22 | No |
| 2 | 68 | 179 | 14 | 109 | 271 | 74 | |||
| AA | CA | CC | AA | CA | CC | ||||
| BRCA1 | rs8176318 | 127 | 504 | 484 | 109 | 504 | 560 | 103 | No |
| 119 | 445 | 509 | 144 | 634 | 640 | 81 | |||
| AA | GA | GG | AA | GA | GG | ||||
| VEGF | rs3025053 | 0 | 36 | 243 | 0 | 27 | 278 | 119 | No |
| 6 | 91 | 274 | 4 | 67 | 175 | 124 | |||
| AA | CA | CC | AA | CA | CC | ||||
| MTHFR | rs4846049 | 79 | 344 | 373 | 83 | 351 | 371 | 123 | No |
| 17 | 157 | 276 | 9 | 113 | 278 | 128 | |||
| AA | AC | CC | AA | AC | CC | Yesb | |||
| SPP1 | rs9138 | 31 | 138 | 99 | 20 | 102 | 152 | 108 | |
| 20 | 42 | 38 | 19 | 43 | 50 | 109 | |||
| AA | GA | GG | AA | GA | GG | ||||
| NOD2 | rs3135500 | 15 | 37 | 40 | 19 | 48 | 38 | 64 | Yesc |
| 31 | 42 | 15 | 10 | 43 | 35 | 46 | |||
| 120 | 303 | 243 | 81 | 265 | 209 | 22 | |||
| GG | TG | TT | GG | TG | TT | ||||
| KRAS | rs61764370 | 0 | 66 | 375 | 2 | 35 | 202 | 130 | No |
| 1 | 45 | 151 | 2 | 68 | 288 | 56 | |||
| 6 | 167 | 916 | 10 | 215 | 1200 | 76 | |||
| AA | AG | GG | AA | AG | GG | ||||
| NFKBIA | rs696 | 55 | 181 | 118 | 155 | 480 | 380 | 104 | No |
| 233 | 460 | 308 | 212 | 531 | 262 | 58 | |||
| 57 | 58 | 28 | 22 | 62 | 53 | 57 |
VEGF, vascular endothelial growth factor.
Allelic model, OR: 1.28, 95% CI (1.08‐1.52); Recessive model, OR: 2.23, 95% CI (1.22‐4.07); Dominant model, OR: 1.23, 95% CI (1.01‐1.49); Homozygote, OR: 2.29, 95% CI (1.25‐4.19); Heterozygote CC vs GC OR: 2.06, 95% CI (1.10‐3.83).
Overdominant model, OR: 1.59, 95% CI (1.19‐2.12).
AA vs AG OR: 2.50, 95% CI (1.12‐5.57).
Table 4.
The basic characteristic of included studies (polymorphisms with at least four eligible studies were included)
| SNPs | First author | Year | Country | Population subgroupa | Case | Study design | Gender | Age | Sample size (case‐control) | Genotyping method | Quality score | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs731236 | Budhathoki | 2016 | Japan | East Asian | CRC | Nested case‐control | F/M | 40‐69 | 356/708 | TaqMan | 8 | 125 |
| Takeshige | 2015 | Japan | East Asian | CRC | Case‐control | F/M | 20‐74 | 685/778 | PCR‐RFLP | 9 | 131 | |
| Park | 2006 | Korea | East Asian | CRC | Case‐control | F/M | 23‐81 | 190/318 | PCR‐RFLP | 6 | 132 | |
| Hughes | 2011 | Czech Republic | European | CRC | Case‐control | F/M | >29 | 717/615 | KASPar | 8 | 133 | |
| Bentley | 2012 | New Zealand | European | CRC | Case‐control | F/M | — | 199/182 | TaqMan | 7 | 134 | |
| Gromowski | 2016 | Poland | European | CRC | Case‐control | — | — | 195/390 | TaqMan | 4 | 135 | |
| Laczmanska | 2014 | Poland | European | CRC | Case‐control | F/M | 32‐87 | 157/175 | SNaPshot Multiplex Kit | 6 | 84 | |
| Flügge | 2007 | Russia | European | CRC | Case‐control | F/M | 29‐85 | 256/256 | PCR‐RFLP | 6 | 136 | |
| Mahmoudi | 2010 | Iran | Middle East | CRC | Case‐control | F/M | 14‐90 | 160/180 | PCR‐RFLP | 6 | 137 | |
| Moossavi | 2017 | Iran | Middle East | CRC | Case‐control | F/M | — | 100/100 | PCR‐RFLP | 6 | 138 | |
| Safaei | 2012 | Iran | Middle East | CRC | Case‐control | F/M | — | 112/112 | PCR‐RFLP | 6 | 139 | |
| Atoum | 2014 | Jordan | Middle East | CRC | Case‐control | F/M | — | 93/102 | PCR‐RFLP | 6 | 140 | |
| Alkhayal | 2016 | Saudi Arabia | Middle East | CRC | Case‐control | F/M | 21‐89 | 100/100 | Sequencing | 5 | 141 | |
| Gunduz | 2012 | Turkey | Middle East | CRC | Case‐control | F/M | — | 43/42 | PCR‐RFLP | 6 | 142 | |
| Yaylım‐Eraltan | 2007 | Turkey | Middle East | CRC | Case‐control | — | — | 26/52 | PCR‐RFLP | 4 | 143 | |
| Dilmec | 2009 | Turkey | Middle East | CRC | Case‐control | F/M | — | 56/169 | PCR‐RFLP | 4 | 144 | |
| Kupfer | 2011 | USA | African | CRC | Case‐control | F/M | — | 938/811 | Sequenom MassARRAY | 7 | 145 | |
| Slattery | 2001 | USA | Caucasian, African, Hispanic | CRC | Case‐control | F/M | 30‐79 | 427/366 | PCR‐RFLP | 9 | 146 | |
| Ochs‐Balcom | 2008 | USA | Caucasian | CRC | Case‐control | F/M | ≥40 | 250/246 | TaqMan | 8 | 147 | |
| Yamaji | 2011 | Japan | East Asian | Adenoma | Case‐control | F/M | 40‐79 | 684/640 | TaqMan | 7 | 148 | |
| Peters | 2004 | USA | European | Adenoma | Nested Case‐control | F/M | 55‐74 | 716/727 | PCR‐RFLP | 7 | 149 | |
| Peters | 2004 | USA | African | Adenoma | Nested Case‐control | F/M | 55‐74 | 763/774 | PCR‐RFLP | 7 | 149 | |
| rs30259039 | Hofmann | 2008 | Austria | Caucasian | CRC | Case‐control | F/M | 29‐83 | 427/427 | TaqMan | 7 | 150 |
| Wu | 2009 | Germany | Caucasian | CRC | Case‐control | F/M | 33‐91 | 157/117 | PCR‐RFLP | 5 | 151 | |
| Ungerback | 2009 | Sweden | Caucasian | CRC | Case‐control | — | — | 302/336 | MegaBACE™ SNuPe™ Genotyping Kit | 5 | 95 | |
| Bayhan | 2014 | Turkey | Caucasian | CRC | Case‐control | — | — | 43/44 | PCR‐RFLP | 4 | 152 | |
| Jannuzzi | 2015 | Turkey | Caucasian | CRC | Case‐control | F/M | — | 103/129 | PCR‐RFLP | 8 | 153 | |
| Yang | 2017 | China | East Asian | CRC | Case‐control | F/M | 20‐83 | 371/246 | iMLDR method | 7 | 124 | |
| Bae | 2008 | Korea | East Asian | CRC | Case‐control | F/M | 18‐95 | 262/229 | PCR‐RFLP | 5 | 154 | |
| Chae | 2008 | Korea | East Asian | CRC | Case‐control | F/M | 21‐89 | 465/413 | PCR/DHPLC | 4 | 141 | |
| Jang | 2013 | Korea | East Asian | CRC | Case‐control | F/M | — | 390/492 | PCR‐RFLP | 6 | 155 | |
| Lau | 2014 | Malaysia | South Asian | CRC | Case‐control | — | 40‐90 | 130/212 | TaqMan | 5 | 156 | |
| Credidio | 2011 | Brazil | Caucasian, African | CRC | Case‐control | F/M | 25‐97 | 261/261 | PCR‐RFLP | 4 | 157 | |
| Wu | 2011 | China | East Asian | Adenoma | Case‐control | F/M | 18‐75 | 224/200 | TaqMan | 8 | 158 | |
| rs3212986 | Hou | 2014 | China | East Asian | CRC | Case‐control | F/M | — | 204/204 | MALDI‐MS | 7 | 159 |
| Moreno | 2006 | Spain | _ | CRC | Case‐control | F/M | — | 349/300 | APEX | 7 | 160 | |
| Ni | 2014 | China | East Asian | CRC | Case‐control | F/M | — | 213/240 | TaqMan | 8 | 161 | |
| Yueh | 2017 | Taiwan | East Asian | CRC | Case‐control | F/M | — | 362/362 | PCR‐RFLP | 7 | 162 | |
| Zhang | 2018 | China | East Asian | CRC | Case‐control | F/M | — | 200/200 | TaqMan | 5 | 72 | |
| rs712 | Dai | 2016 | China | Chinese | CRC | Case‐control | F/M | 36‐75 | 430/430 | iMLDR | 7 | 62 |
| Jiang | 2015 | China | Chinese | CRC | Case‐control | F/M | — | 586/476 | PCR‐RFLP | 5 | 36 | |
| Landi | 2012 | Czech Republic | Czechs | CRC | Case‐control | F/M | — | 717/1171 | KASPar | 7 | 79 | |
| Pan | 2014 | China | Chinese | CRC | Case‐control | F/M | — | 339/313 | PCR‐RFLP | 7 | 59 | |
| Schneiderova | 2017 | Czech Republic | Czechs | CRC | Case‐control | F/M | 21‐78 | 1057/1405 | KASPar | 6 | 76 | |
| rs5275 | Makar (DALS) | 2013 | USA | Caucasian | CRC | Case‐control | F/M | 30‐79 | 2003/2549 | Illumina™ GoldenGate assay | 6 | 163 |
| Pereira | 2010 | Portugal | Caucasian | CRC | Case‐control | F/M | 50‐75 | 115/256 | PCR‐RFLP | 5 | 100 | |
| Siezen (PPHV) | 2006 | Netherlands | Caucasian | CRC | Nested Case‐control | F/M | — | 200/388 | PCR‐RFLP | 7 | 164 | |
| Siezen (DOM) | 2006 | Netherlands | Caucasian | CRC | Nested Case‐control | F/M | — | 442/693 | PCR‐RFLP | 6 | 164 | |
| Vogel | 2014 | Norway | Caucasian | CRC | Case‐control | F/M | 50‐64 | 189/399 | KBioscience | 8 | 165 | |
| Zhang | 2012 | China | East Asian | CRC | F/M | 93‐30 | 343/340 | 6 | 101 | |||
| Cox | 2004 | Spain | Caucasian | CRC | Case‐control | F/M | 24‐92 | 290/271 | TaqMan | 6 | 166 | |
| Andersen | 2013 | Denmark | Caucasian | CRC |
Case‐Cohort Study |
F/M | 50‐64 | 931/1738 | KASPar | 9 | 167 | |
| Thompson | 2009 | USA | Caucasian, African, Other | CRC | Case‐control | F/M | — | 421/480 | TaqMan | 9 | 168 | |
| Gunter | 2006 | USA | _ | Adenoma | Case‐control | F/M | 43‐74 | 210/197 | TaqMan | 8 | 169 | |
| Pereira | 2016 | Portugal | Caucasian | Adenoma | Case‐control | F/M | 50‐75 | 191/474 | — | 6 | 170 | |
| Siezen | 2006 | Netherlands | Caucasian | Adenoma | Case‐control | F/M | — | 378/396 | TaqMan | 7 | 171 | |
| Vogel | 2014 | Norway | Caucasian | Adenoma | Case‐control | F/M | 50‐64 | 983/399 | KBioscience | 8 | 165 | |
| Gong | 2009 | USA | _ | Adenoma | Case‐control | F/M | 30‐74 | 162/211 | PCR‐RFLP | 8 | 112 | |
| Ali | 2005 | USA | Caucasian | Adenoma | Nested Case‐control | F/M | 55‐74 | 749/756 | TaqMan | 7 | 172 | |
| Ashktorab | 2008 | USA | African | Adenoma | Case‐control | F/M | — | 70/136 | TaqMan | 7 | 173 | |
| rs4648298 | Iglesias | 2009 | Spain | Caucasian | CRC | Case‐control | F/M | — | 284/123 | PCR‐RFLP | 7 | 114 |
| Mosallaei | 2018 | Iran | Caucasian | CRC | Case‐control | F/M | — | 88/88 | PCR‐RFLP | 5 | 49 | |
| Ueda | 2008 | Japan | East Asian | Adenoma | Case‐control | M | 47‐59 | 455/1051 | PCR‐RFLP | 5 | 174 | |
| Gong | 2009 | USA | _ | Adenoma | Case‐control | F/M | 30‐74 | 162/211 | PCR‐RFLP | 8 | 112 | |
| rs1801157 | Ramzi | 2014 | Malaysia | Asian | CRC | Case‐control | F/M | >18 | 124/173 | Illumina's BeadArray | 7 | 175 |
| Razmkhah | 2013 | Iran | Caucasian | CRC | Case‐control | — | — | 109/262 | PCR‐RFLP | 4 | 176 | |
| Amara | 2015 | Tunis | African | CRC | Case‐control | F/M | — | 80/80 | PCR‐RFLP | 5 | 177 | |
| Dimberg | 2007 | Sweden | Caucasian | CRC | Case‐control | F/M | 29‐103 | 258/300 | PCR‐RFLP | 5 | 88 | |
| Hidalgo‐Pascual | 2007 | Spain | Caucasian | CRC | Case‐control | F/M | 35‐87 | 151/141 | FRET | 4 | 89 | |
| Shi | 2013 | Taiwan | Asian | CRC | Case‐control | F/M | >30 | 349/516 | PCR‐DHPLC | 6 | 90 |
Different classifications for population subgroup were used for each polymorphism.
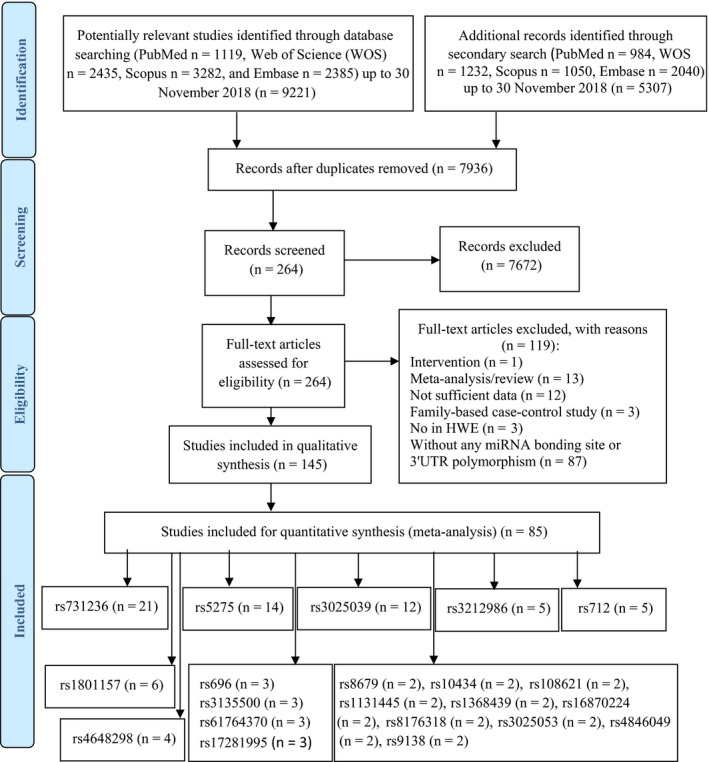
For rs731236 in overall meta‐analysis (based on minor allele; t) no significant result for the risk of CRC was observed, but in subgroup analysis in Middle East population the results were significant in heterozygote (Tt vs TT) (0.76 [0.61‐0.95]) and overdominant models (Tt vs TT + tt) (0.75 [0.61‐0.92]), and borderline significance was observed in dominant model (tt + Tt vs TT) (0.81 [0.66‐1.00]) (Figure 2, Figure S2).
Figure 2.
Forest plot related to rs731236 and risk of CRC. A, Heterozygote model. B, Overdominant model
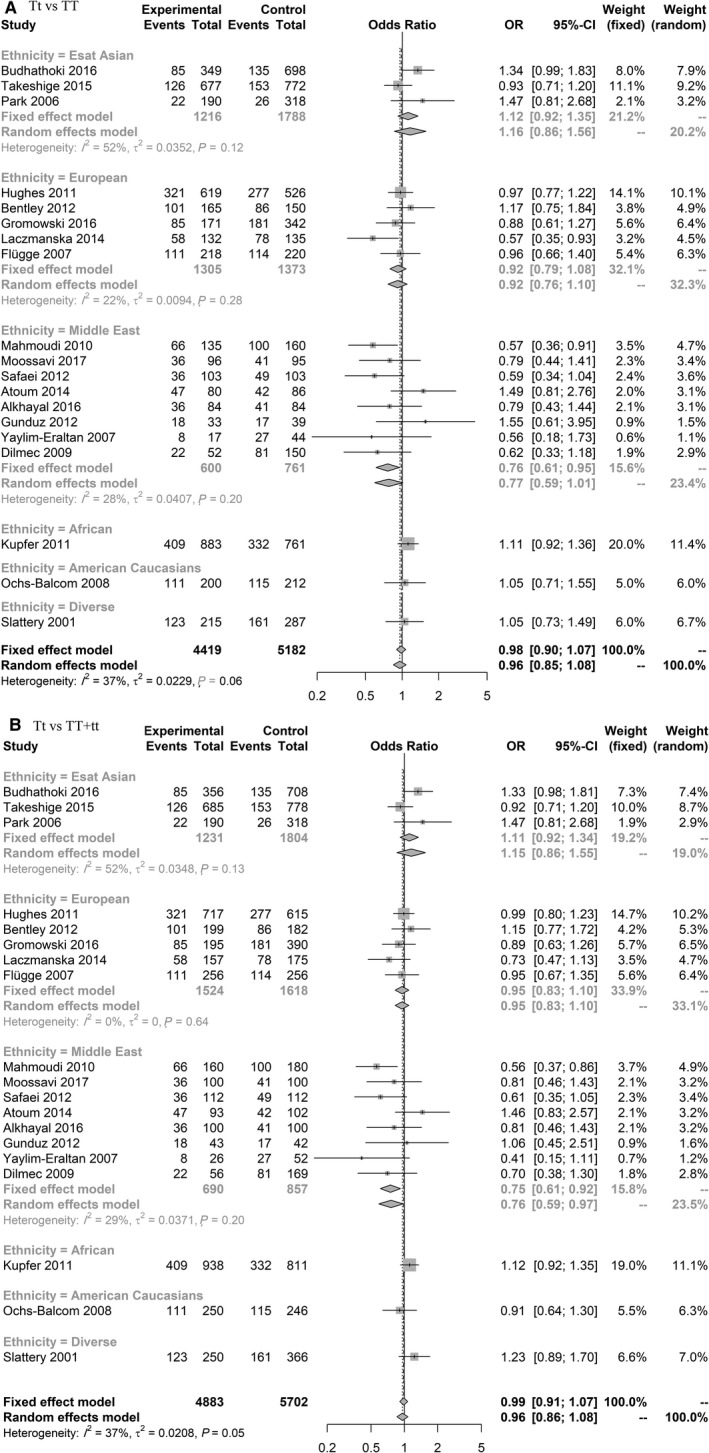
For rs3025039 in overall, there was no significant association, but subgroup analysis revealed significant results (based on minor allele; T). In East Asian population, the allelic model (T vs C) (1.25 [1.01‐1.54]) significantly increased the risk of CRC and in dominant model (TT + TC vs CC) (1.29 [1.00‐1.66]) there was a trend towards significance (Figure 3, Figure S3).
Figure 3.
Forest plot related to rs3025039 and risk of CRC. A, Allelic model. B, Dominant model
In meta‐analysis for rs3212986, there were significant results in both overall and subgroup analysis in different genetic models (based on minor allele; T), including homozygote model (TT vs GG) 1.76 (1.08‐2.86) (Figure 4, Figure S4).
Figure 4.
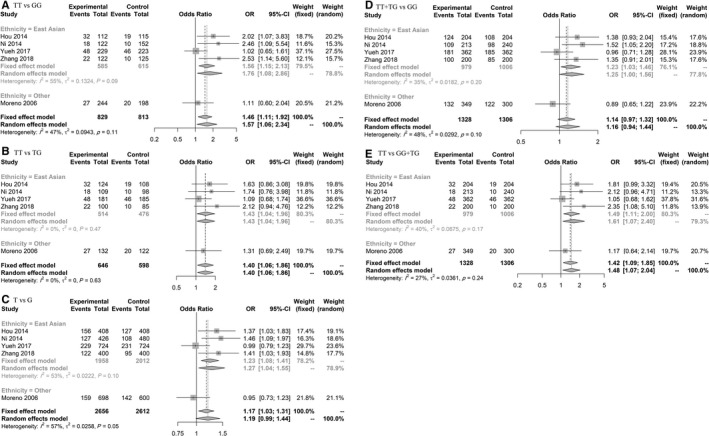
Forest plot related to rs3212986 and risk of CRC. A, Homozygote model. B, TT vs TG model. C, Allelic model. D, Dominant model. E, Recessive model
Although we did not find any significant result for rs712 in overall models, subgroup analysis revealed significant and borderline association in Chinese and Czech populations, respectively, on six genetic models (based on minor allele; T), including homozygote model (TT vs GG) in Chinese 2.51 (1.70‐3.69) and in Czech 0.85 (0.72‐1.01) populations (Figure 5, Figure S5).
Figure 5.
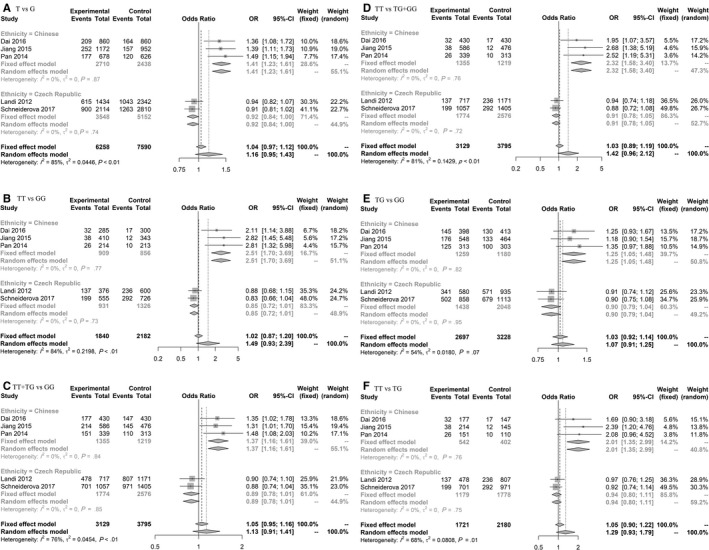
Forest plot related to rs712 and risk of CRC. A, Allelic model. B, Homozygote model. C, Dominant model. D, Recessive model. E, Heterozygote model. F, TT vs TG model
The allele (A) of rs1801157 polymorphism increased risk of CRC in Asian population, while we did not find any significant results in Caucasian populations (Table 5).
Table 5.
Meta‐analysis of association between rs1801157 and risk of CRC
| Classification | Allelic | Dominant | Recessive | Overdominant | ||||
|---|---|---|---|---|---|---|---|---|
| OR [95% CI] |
Q test P value |
OR [95% CI] |
Q test P value |
OR [95% CI] |
Q test P value |
OR [95% CI] |
Q test P value |
|
| Caucasian (n = 3) | 0.98 [0.82‐1.17] | .89 | 1.03 [0.83‐1.27] | .90 | 0.75 [0.44‐1.26] | .45 | 1.09 [0.88‐1.35] | .76 |
| Asian (n = 2) | 2.28 [1.11‐4.69] | .02 | 2.20 [0.66‐7.30] | <.01 | 4.94 [1.69‐14.42] | .58 | 1.57 [0.28‐8.88] | <.01 |
| Overall (n = 6) | 1.56 [0.97‐2.50] | <.01 | 1.59 [0.93‐2.70] | <.01 | 2.03 [0.73‐5.63] | <.01 | 1.24 [0.78‐2.00] | <.01 |
| Classification | Homozygote | AA vs AG | Heterozygote (AG vs GG) | |||
|---|---|---|---|---|---|---|
| OR [95% CI] |
Q test P value |
OR [95% CI] |
Q test P value |
OR [95% CI] |
Q test P value |
|
| Caucasian (n = 3) | 0.75 [0.44‐1.29] | .50 | 0.72 [0.42‐1.25] | .39 | 1.07 [0.86‐1.33] | .83 |
| Asian (n = 2) | 4.86 [1.63‐14.50] | .39 | 4.96 [1.59‐15.45] | .90 | 1.78 [0.38‐8.39] | <.01 |
| Overall (n = 6) | 2.31 [0.73‐7.27] | <.01 | 1.75 [0.69‐4.40] | <.01 | 1.43 [0.87‐2.35] | <.01 |
The bold values are statistically significant.
Finally for rs5275 (based on minor allele; C) and rs4648298 (based on minor allele; G), we performed meta‐analysis according to three different subgroup analyses (CRC cases, adenoma, and overall). The results in all different genetic models were not significant except dominant model (0.82 [0.70‐0.97]) in adenoma for rs5275, also the allelic model (C vs T) showed borderline association 0.92 (0.85‐1.00) (Tables 6). For rs4648298 recessive, homozygote, and heterozygote (CG vs GG) models the analysis was not possible, because of zero number in GG genotype in all included studies (Table 7).
Table 6.
Meta‐analysis of association between rs5275 and risk of CRC (n = 9) and adenoma (n = 7)
| Classification | Allelic | Dominant | Recessive | Overdominant | ||||
|---|---|---|---|---|---|---|---|---|
| OR [95% CI] |
Q test P value |
OR [95% CI] |
Q test P value |
OR [95% CI] |
Q test P value |
OR [95% CI] |
Q test P value |
|
| CRC | 1.03 [0.98‐1.09] | .16 | 1.03 [0.92‐1.16] | .18 | 1.04 [0.97‐1.12] | .38 | 0.97 [0.90‐1.04] | .70 |
| Adenoma | 0.92 [0.85‐1.00] | .78 | 0.82 [0.70‐0.97] | .19 | 0.94 [0.83‐1.05] | .07 | 0.90 [0.71‐1.15] | <.01 |
| Overall | 1.00 [0.95‐1.04] | .16 | 0.96 [0.87‐1.05] | .05 | 1.01 [0.95‐1.08] | .09 | 0.95 [0.86‐1.04] | .01 |
| Classification | Homozygote | CC vs CT | Heterozygote (CT vs TT) | |||
|---|---|---|---|---|---|---|
| OR [95% CI] |
Q test P value |
OR [95% CI] |
Q test P value |
OR [95% CI] |
Q test P value |
|
| CRC | 1.05 [0.93‐1.18] | .13 | 1.04 [0.96‐1.13] | .59 | 1.01 [0.90‐1.15] | 33 |
| Adenoma | 0.85 [0.71‐1.02] | .38 | 1.06 [0.83‐1.36] | <.01 | 0.79 [0.59‐1.06] | <.01 |
| Overall | 0.98 [0.89‐1.09] | .10 | 1.03 [0.93‐1.14] | .03 | 0.88 [0.76‐1.03] | .02 |
The bold values are statistically significant.
Table 7.
Meta‐analysis of association between rs4648298 and risk of CRC (n = 2) and adenoma (n = 2)
| Classification | Allelic | Dominant/Overdominant/Heterozygotea | ||
|---|---|---|---|---|
| OR [95% CI] |
Q test P value |
OR [95% CI] |
Q test P value |
|
| CRC | 1.93 [0.21‐17.52] | <.01 | 0.47 [0.04‐5.39] | <.01 |
| Adenoma | 1.02 [0.48‐2.18] | .99 | 0.98 [0.46‐2.11] | .99 |
| Overall | 1.41 [0.49‐4.05] | <.01 | 1.47 [0.47‐4.63] | <.01 |
These models had similar results, because of zero number in GG genotype.
4. DISCUSSION
This study aimed to investigate miRNA‐binding site polymorphisms and risk of CRC, which may potentially play roles in various conditions. The effects shown for these polymorphisms associated with miRNA:mRNA interactions. Polymorphisms in miRNA‐binding site can negatively or positively influence these interactions by different mechanisms such as effect of hybrid stability, target sites accessibility, local RNA secondary structure, and structural accessibility. Among 222 included polymorphisms, 25 were eligible for inclusion in our secondary search strategy. Fourteen polymorphisms, with less than four eligible studies, were included in the pooled analysis. The rs17281995 polymorphism is located in 3'UTR of CD86 gene and binding site of miR‐337 and miR‐582.22 The minor allele (C) of rs17281995 polymorphism increased the risk of CRC in different genetic models. Although the results are based on limited number of studies but the strong association is noteworthy. This was also observed in the previous review based on two included articles.129 The nonsignificant results are not conclusive and cannot rule out the association between these polymorphisms and the risk of CRC, because of limited number of included studies and also ethnic differences in studied populations. Further studies need to confirm these results. In addition, seven polymorphisms, with more than four eligible studies, were included in the final meta‐analysis.
The rs731236 polymorphism is located in 3'UTR of vitamin D receptor gene. Its downregulation is related to cancer progression.178 There are several previous meta‐analyses on the role of rs731236 on CRC risk. Most of the previous meta‐analyses179, 180, 181, 182, 183 found no significant association between the risk of CRC and rs731236. While Serrano et al in their meta‐analysis184 found significant results based on analyzing both of colorectal cancer and adenoma. Therefore, all previous meta‐analysis results were according to fewer included studies, the overall CRC population and no subgroup analysis were carried out and in some studies adenoma was also included for calculating the risk of CRC. In our study, we carried out subgroup analysis based on different ethnicity and found that the results were different after stratification according to ethnicity. While in overall analysis our results are in line with the previous meta‐analysis, showing no relation between the risk of CRC and rs731236 polymorphism. In Middle East population we observed a significant association between this polymorphism and CRC. This result was not reported previously. We also found a heterozygote advantage for the risk of CRC with heterozygote (Tt) showing protective effects compared with homozygotes (TT, tt). Similarly, in a study on pediatric solid tumor, the heterozygote model decreased the risk of CRC compared to homozygote model. The survival rate of subjects with CRC was significantly decreased in heterozygote model compared to homozygote model.185 More studies are needed to specify the reason for our interesting observation.
In overall analysis, based on 11 included studies, rs3025039 was not related to the risk of CRC, but is showing association in Caucasian and East Asian populations. Based on subgroup analysis, minor allele in East Asian was related to an increased risk of CRC. This SNP is located in 3'UTR of vascular endothelial growth factor gene which may affect hsa‐miR‐591 target sites.186 This gene affects angiogenesis, tumor growth, and metastasis.187 It is also related to CRC outcomes and treatment.124 Thus the association between rs3025039 and CRC risk may be related to the effect of this SNP on miRNA:mRNA interactions. However, in the previous meta‐analysis with five included studies, no significant association was found between this polymorphism and risk of CRC.188 This might be due to heterogeneity of their data in different populations requiring further subgroup analysis.
According to the results based on five included studies, rs3212986 increased the risk of CRC in all genetic models, which was similar to previous meta‐analysis,189 we also found to the same results in East Asian population. This polymorphism is located in binding site of miR‐15a in 3'UTR of ERCC1.72 The polymorphisms and mRNA level of this gene had previously been investigated in CRC.190
For rs1801157 minor allele (A) increased risk of CRC was observed in Asian population. This result is similar to previous meta‐analysis by Xu,191 which found significant association in non‐Caucasian populations. This polymorphism is located in 3'UTR of CXCL12 in a putative miRNA‐binding site for miR‐941.192 The effect of CXCL12 polymorphisms on CRC was previously observed in different studies. The CXCL12 binds to CXCR4 and affects different clinical features of cancers such as progression, angiogenesis, and metastasis.193 Thus the observed association for rs1801157 A allele and CRC may be related to its effect on miRNA:mRNA interactions and CXCL12 expression.
We also found no significant association between rs712 and risk of CRC, in the overall meta‐analysis of five included studies. However, subgroup analysis revealed remarkable and completely different results in Chinese and Czech Republic populations. In Chinese, we observed a strong risk while in Czech population a protective effect was shown in all various models. There is one study similar to our results which confirm the increase risk of this polymorphism in Chinese population.194 In two other meta‐analyses it has been reported that this polymorphism may increases the overall risk of different types of cancers in the Chinese population.195, 196 This variant is within let‐7 KRAS binding site. KRAS, is an important oncogene, which has been previously described to be associated with different types of cancers. This gene influence cancer cells differentiation and proliferation, and is highly mutated in many type of cancers such as CRC.197, 198 Based on our results differences between populations should be considered for the effect of this binding site polymorphism in future studies.
In addition, our results (based on 10 eligible studies) showed that rs5275 was not related to the risk of CRC. While the minor allele of rs5275 may have a protective effect on the risk of adenoma. This polymorphism is located in COX‐2 gene at miR‐542‐3p target site. COX‐2 is usually overexpressed in colorectal adenoma patients,199 and has effect on pro‐inflammatory prostaglandins and links between inflammation and cancer progression.200 Therefore, the minor allele of rs5275 may be associated with a decreased risk of colorectal adenoma by downregulating COX‐2 expression.
4.1. Strength and limitations
Our study had several advantages: First, this is the first systematic review for evaluating the role of miRNA‐binding site polymorphisms on CRC susceptibility, and 25 polymorphisms were included in our pooled analysis. Second, to reduce the publication biases and include all relevant documents we carried out a systematic search on four common databases, as well as other sources such as references of relevant reviews. Third, there was no language bias, we included all relevant documents without any language restriction. Fourth, our study has high power and strength reliability because of our comprehensive and double search strategies and subgroup analyzing based on different ethnicity. Fifth, to reduce binding site false positive prediction, related to bioinformatics tools, we only included polymorphisms located in miRNA‐binding site or 3'UTR (stated at least in two of the included documents).
There are also some limitations in our study. First, based on insufficient data, it was mandatory to exclude some relevant documents. Second, some polymorphisms had two or three included article. Third, CRC is a multifactorial disease and we only included genetic effect.
5. CONCLUSION
miRNA‐binding site polymorphisms in this meta‐analysis showed significant association with CRC in different populations. Interestingly, rs731236 polymorphism showed a significant association with CRC in Middle East population with a heterozygote advantage. The minor allele in the East Asian populations for rs3025039, rs3212986, and rs712, and also in Asian population for rs1801157, increased the risk of CRC. The minor allele of rs712 may have a protective effect on the risk of CRC in Czech populations, while rs17281995 showed risk effect in the European population. Finally, it can be concluded that these miRNA‐binding site polymorphisms play different roles on the risk of CRC in various populations which should be considered in data analysis and interpretation in the future studies.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
Supporting information
ACKNOWLEDGMENTS
This study was supported by Endocrinology and Metabolism Research Institute, Tehran University of Medical Sciences (Grant No. 1395‐02‐105‐2087).
Gholami M, Larijani B, Sharifi F, et al. MicroRNA‐binding site polymorphisms and risk of colorectal cancer: A systematic review and meta‐analysis. Cancer Med. 2019;8:7477–7499. 10.1002/cam4.2600
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87‐108. [DOI] [PubMed] [Google Scholar]
- 2. American Cancer Society . Global Cancer Facts & Figures, 3rd edn Atlanta: American Cancer Society; 2015. [Google Scholar]
- 3. Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D. F B. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. [DOI] [PubMed]
- 4. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 5. Bray F, Møller B. Predicting the future burden of cancer. Nat Rev Cancer. 2006;6:63. [DOI] [PubMed] [Google Scholar]
- 6. Nowatzki J, Moller B, Demers A. Projection of future cancer incidence and new cancer cases in Manitoba, 2006–2025. Chronic Dis Injuries Can. 2011;31:71‐78. [PubMed] [Google Scholar]
- 7. Mahan LK, Raymond JL. Krause's Food & the Nutrition Care Process‐E‐Booked. Winnipeg, Canada: Elsevier Health Sciences; 2016. [Google Scholar]
- 8. Incisive Health . Saving Lives, Averting Costs: An Analysis of the Financial Implications of Achieving Earlier Diagnosis of Colorectal, Lung and Ovarian Cancer. London: Incisive Health/Cancer Research UK; 2014. [Google Scholar]
- 9. Ferlay J, Parkin D, Steliarova‐Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer. 2010;46:765‐781. [DOI] [PubMed] [Google Scholar]
- 10. Azzoni C, Bottarelli L, Campanini N, et al. Distinct molecular patterns based on proximal and distal sporadic colorectal cancer: arguments for different mechanisms in the tumorigenesis. Int J Colorectal Dis. 2007;22:115‐126. [DOI] [PubMed] [Google Scholar]
- 11. Bognar G, Ledniczky G, Istvan G, Ondrejka P. Molecular mechanisms in development of colorectal cancer metastasis. Magyar Sebeszet. 2006;59:342‐349. [PubMed] [Google Scholar]
- 12. Friedman RC, Farh KK‐H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92‐105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu F, Dear K, Huang L, et al. Association between microRNA‐27a rs895819 polymorphism and risk of colorectal cancer: a meta‐analysis. Cancer Genetics. 2016;209:388‐394. [DOI] [PubMed] [Google Scholar]
- 14. Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gong J, Tong Y, Zhang HM, et al. Genome‐wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum Mutat. 2012;33:254‐263. [DOI] [PubMed] [Google Scholar]
- 16. Wu M, Jolicoeur N, Li Z, et al. Genetic variations of microRNAs in human cancer and their effects on the expression of miRNAs. Carcinogenesis. 2008;29:1710‐1716. [DOI] [PubMed] [Google Scholar]
- 17. Xu L, Tang W. Associations of polymorphisms in mir‐196a2, mir‐146a and mir‐149 with colorectal cancer risk: a meta‐analysis. Pathol Oncol Res. 2016;22:261‐267. [DOI] [PubMed] [Google Scholar]
- 18. Liu X‐X, Wang M, Xu D, et al. Quantitative assessment of the association between genetic variants in microRNAs and colorectal cancer risk. Biomed Res Int. 2015;2015:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rong G‐Q, Zhang X‐M, Chen B, Yang X‐D, Wu H‐R, Gong W. MicroRNA gene polymorphisms and the risk of colorectal cancer. Oncol Lett. 2017;13:3617‐3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim JG, Chae YS, Lee SJ, et al. Genetic variation in microRNA‐binding site and prognosis of patients with colorectal cancer. J Cancer Res Clin Oncol. 2015;141:35‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Esquela‐Kerscher A, Slack FJ. Oncomirs–microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259. [DOI] [PubMed] [Google Scholar]
- 22. Landi D, Gemignani F, Naccarati A, et al. Polymorphisms within micro‐RNA‐binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579‐584. [DOI] [PubMed] [Google Scholar]
- 23. Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38:1452. [DOI] [PubMed] [Google Scholar]
- 24. Vaishnavi V, Manikandan M, Munirajan AK. Mining the 3′ UTR of autism‐implicated genes for SNPs perturbing microRNA regulation. Genomics, Proteomics Bioinformatics. 2014;12:92‐104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gopalakrishnan C, Kamaraj B, Purohit R. Mutations in microRNA binding sites of CEP genes involved in cancer. Cell Biochem Biophys. 2014;70:1933‐1942. [DOI] [PubMed] [Google Scholar]
- 26. Langevin SM, Christensen BC. Let‐7 microRNA‐binding‐site polymorphism in the 3′ UTR of KRAS and colorectal cancer outcome: a systematic review and meta‐analysis. Cancer Med. 2014;3:1385‐1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li J, Li Z, Kan Q, Sun S, Li Y, Wang S. Association of p21 3′ UTR gene polymorphism with cancer risk: evidence from a meta‐analysis. Sci Rep. 2015;5:13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moher D, Shamseer L, Clarke M, et al. Preferred reporting items for systematic review and meta‐analysis protocols (PRISMA‐P) 2015 statement. Systematic Rev. 2015;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta‐analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gholami M, Amoli MM, Sharifi F. Letter to the Editor: Comments on “Association between the ICAM‐1 gene polymorphism and coronary heart disease risk: a meta‐analysis”. Biosci Rep. 2019;39:BSR20190554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xiong F, Wu C, Chang J, et al. Genetic variation in an miRNA‐1827 binding site in MYCL1 alters susceptibility to small‐cell lung cancer. Cancer Res. 2011;71:5175‐5181. [DOI] [PubMed] [Google Scholar]
- 32. Zhang L, Liu Y, Song F, et al. Functional SNP in the microRNA‐367 binding site in the 3′ UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc Natl Acad Sci. 2011;108:13653‐13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salzman DW, Weidhaas JB. SNPing cancer in the bud: microRNA and microRNA‐target site polymorphisms as diagnostic and prognostic biomarkers in cancer. Pharmacol Ther. 2013;137:55‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kang BW, Jeon H‐S, Chae YS, et al. Impact of genetic variation in MicroRNA‐binding site on susceptibility to colorectal cancer. Anticancer Res. 2016;36:3353‐3361. [PubMed] [Google Scholar]
- 35. Saridaki Z, Weidhaas JB, Lenz H‐J, et al. A let‐7 microRNA‐binding site polymorphism in KRAS predicts improved outcome in patients with metastatic colorectal cancer treated with salvage cetuximab/panitumumab monotherapy. Clin Cancer Res. 2014;20:4499‐4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jiang Q‐H, Peng H‐X, Zhang Y, Tian P, Xi Z‐L, Chen H. rs712 Polymorphism within let‐7 microRNA‐binding site might be involved in the initiation and progression of colorectal cancer in Chinese population. OncoTargets Ther. 2015;8:3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zanetti KA, Haznadar M, Welsh JA, et al. 3′‐UTR and functional secretor haplotypes in mannose‐binding lectin 2 are associated with increased colon cancer risk in African Americans. Cancer Res. 2012;72:1467‐1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He H, Lei L, Chen E, et al. The screening of the functional microRNA binding site SNPs in sporadic colorectal cancer genes. Cancer Biol Ther. 2017;18:407‐413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mao Y‐Y, Jing F‐Y, Jin M‐J, et al. rs12904 polymorphism in the 3UTR of EFNA1 is associated with colorectal cancer susceptibility in a Chinese population. Asian Pac J Cancer Prev. 2013;14:5037‐5041. [DOI] [PubMed] [Google Scholar]
- 40. Li J, Liu H, Zou LI, et al. A functional variant in GREM1 confers risk for colorectal cancer by disrupting a hsa‐miR‐185‐3p binding site. Oncotarget. 2017;8:61318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ding L, Jiang Z, Chen Q, Qin R, Fang Y, Li H. A functional variant at miR‐520a binding site in PIK3CA alters susceptibility to colorectal cancer in a Chinese Han population. Biomed Res Int. 2015;2015:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mosallayi M, Simonian M, Khosravi S, et al. Polymorphism (rs16917496) at the miR‐502 binding site of the lysine methyltransferase 5A (SET8) and its correlation with colorectal cancer in Iranians. Adv Biomed Res. 2017;6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garziera M, Catamo E, Crovella S, et al. Association of the HLA‐G 3′ UTR polymorphisms with colorectal cancer in Italy: a first insight. Int J Immunogenet. 2016;43:32‐39. [DOI] [PubMed] [Google Scholar]
- 44. Yang Y, Ding J, Gao Z‐G, Wang Z‐J. A variant in SIRT2 gene 3′‐UTR is associated with susceptibility to colorectal cancer. Oncotarget. 2017;8:41021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen J, Shi YI, Li Z, et al. A functional variant of IC53 correlates with the late onset of colorectal cancer. Mol Med. 2011;17:607‐618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ahangari F, Salehi R, Salehi M, Khanahmad H. A miRNA‐binding site single nucleotide polymorphism in the 3′‐UTR region of the NOD2 gene is associated with colorectal cancer. Med Oncol. 2014;31:173. [DOI] [PubMed] [Google Scholar]
- 47. Chang J, Tian J, Yang Y, et al. A rare missense variant in TCF7L2 associates with colorectal cancer risk by interacting with a GWAS‐identified regulatory variant in the MYC enhancer. Cancer Res. 2018;78:5164‐5172. [DOI] [PubMed] [Google Scholar]
- 48. Datta S, Sherva RM, De La Cruz M, et al. Single nucleotide polymorphism facilitated down‐regulation of the cohesin stromal antigen‐1: implications for colorectal cancer racial disparities. Neoplasia. 2018;20:289‐294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mosallaei M, Simonian M, Ahangari F, et al. Single nucleotide polymorphism rs4648298 in miRNAs hsa‐miR21 and hsa‐miR590 binding site of COX gene is a strong colorectal cancer determinant. J Gastrointestinal Oncol. 2018;9:448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ni H, Su B, Pan L, Li X, Zhu X, Chen X. Functional variants inPXRare associated with colorectal cancer susceptibility in Chinese populations. Cancer Epidemiol. 2015;39:972‐977. [DOI] [PubMed] [Google Scholar]
- 51. Gansmo LB, Romundstad P, Birkeland E, et al. MDM4 SNP34091 (rs4245739) and its effect on breast‐, colon‐, lung‐, and prostate cancer risk. Cancer Med. 2015;4:1901‐1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lu S, Bevier M, Huhn S, et al. Genetic variants in C‐type lectin genes are associated with colorectal cancer susceptibility and clinical outcome. Int J Cancer. 2013;133:2325‐2333. [DOI] [PubMed] [Google Scholar]
- 53. Shaker OG, Mohammed SR, Mohammed AM, Mahmoud Z. Impact of micro RNA‐375 and its target gene SMAD‐7 polymorphism on susceptibility of colorectal cancer. J Clin Lab Anal. 2018;32:e22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ross J, Lockett L, Brookes D, et al. An association between the PTGS2 rs5275 polymorphism and colorectal cancer risk in families with inherited non‐syndromic predisposition. Eur J Hum Genet. 2013;21:1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nejat D, Zubeyde Y, Orkun G. The K‐ras let‐7 miRNA binding site variant and K‐ras mutations in colon cancer. [abstract]. In: Proceedings of the AACR Special Conference on RAS Oncogenes: From Biology to Therapy; Feb 24–27, 2014; Lake Buena Vista, FL. Philadelphia (PA): AACR. Mol Cancer Res 2014;12(12 Suppl): Abstract nr A48. [Google Scholar]
- 56. Kjersem JB, Ikdahl T, Guren T, et al. Let‐7 miRNA‐binding site polymorphism in the KRAS 3′ UTR; colorectal cancer screening population prevalence and influence on clinical outcome in patients with metastatic colorectal cancer treated with 5‐fluorouracil and oxaliplatin+/− cetuximab. BMC Cancer. 2012;12:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Simonian M, Mosallayi M, Miraghajani M, et al. Single nucleotide polymorphism rs696 in miR449a binding site of NFKBIA gene is correlated with risk of colorectal cancer. Gastroenterol Hepatol Bed Bench. 2018;11:48. [PMC free article] [PubMed] [Google Scholar]
- 58. Song S, Chen D, Lu J, et al. NFκB1 and NFκBIA polymorphisms are associated with increased risk for sporadic colorectal cancer in a southern Chinese population. PLoS ONE. 2011;6:e21726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pan X‐M, Sun R‐F, Li Z‐H, et al. A let‐7 KRAS rs712 polymorphism increases colorectal cancer risk. Tumor Biol. 2014;35:831‐835. [DOI] [PubMed] [Google Scholar]
- 60. Alhadheq AM, Purusottapatnam Shaik J, Alamri A, et al. The effect of poly (ADP‐Ribose) polymerase‐1 gene 3′ untranslated region polymorphism in colorectal cancer risk among Saudi cohort. Dis Markers. 2016;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gong J, Shen N, Zhang H‐M, et al. A genetic variant in microRNA target site of TGF‐β signaling pathway increases the risk of colorectal cancer in a Chinese population. Tumor Biol. 2014;35:4301‐4306. [DOI] [PubMed] [Google Scholar]
- 62. Dai Q, Wei HL, Huang J, Zhou TJ, Chai L, Yang Z‐H. KRAS polymorphisms are associated with survival of CRC in Chinese population. Tumor Biol. 2016;37:4727‐4734. [DOI] [PubMed] [Google Scholar]
- 63. Ye P, Li Z, Jiang H, Liu T. SNPs in microRNA‐binding sites in the ITGB1 and ITGB3 3′‐UTR increase colorectal cancer risk. Cell Biochem Biophys. 2014;70:601‐607. [DOI] [PubMed] [Google Scholar]
- 64. Chaleshi V, Tajali R, Savabkar S, et al. Lack of association between NOD2 rs3135500 and IL12B rs1368439 microRNA binding site SNPs and colorectal cancer susceptibility in an Iranian population. Microrna. 2016;5:152‐156. [DOI] [PubMed] [Google Scholar]
- 65. Wu X‐M, Yang H‐G, Zheng B‐A, Cao H‐F, Hu Z‐M, Wu W‐D. Functional genetic variations at the microRNA binding‐site in the CD44 gene are associated with risk of colorectal cancer in Chinese populations. PLoS ONE. 2015;10:e0127557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Slaby O, Sachlova M, Brezkova V, et al. Identification of microRNAs regulated by isothiocyanates and association of polymorphisms inside their target sites with risk of sporadic colorectal cancer. Nutr Cancer. 2013;65:247‐254. [DOI] [PubMed] [Google Scholar]
- 67. Yu Y, Zhou J, Gong C, et al. Dietary factors and microRNA‐binding site polymorphisms in the IL13 gene: risk and prognosis analysis of colorectal cancer. Oncotarget. 2017;8:47379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee A‐R, Park J, Jung KJ, Jee SH, Kim‐Yoon S. Genetic variation rs7930 in the miR‐4273‐5p target site is associated with a risk of colorectal cancer. OncoTargets Ther. 2016;9:6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gu D, Li S, Du M, et al. A genetic variant located in the miR‐532‐5p‐binding site of TGFBR1 is associated with the colorectal cancer risk. J Gastroenterol. 2019;54:141‐148. [DOI] [PubMed] [Google Scholar]
- 70. Catalano C, da Silva Filho MI, Frank C, et al. Investigation of single and synergic effects of NLRC5 and PD‐L1 variants on the risk of colorectal cancer. PLoS ONE. 2018;13:e0192385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ke J, Tian J, Li J, et al. Identification of a functional polymorphism affecting microRNA binding in the susceptibility locus 1q25. 3 for colorectal cancer. Mol Carcinog. 2017;56:2014‐2021. [DOI] [PubMed] [Google Scholar]
- 72. Zhang Q, Zheng X, Li X, et al. The polymorphisms of mi RNA‐binding site in MLH 3 and ERCC 1 were linked to the risk of colorectal cancer in a case–control study. Cancer Med. 2018;7:1264‐1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhao Y, Yang H, Luo X, Wang C, Zhang R, Guo Z. Single nucleotide polymorphisms at the microRNA‐binding site of KIAA0423 are associated with colorectal cancer. Biotechnol Biotechnol Equip. 2016;30:1163‐1167. [Google Scholar]
- 74. Azimzadeh P, Romani S, Mohebbi SR, et al. Association of polymorphisms in microRNA‐binding sites and colorectal cancer in an Iranian population. Cancer Genet. 2012;205:501‐507. [DOI] [PubMed] [Google Scholar]
- 75. Naccarati A, Pardini B, Stefano L, et al. Polymorphisms in miRNA‐binding sites of nucleotide excision repair genes and colorectal cancer risk. Carcinogenesis. 2012;33:1346‐1351. [DOI] [PubMed] [Google Scholar]
- 76. Schneiderova M, Naccarati A, Pardini B, et al. MicroRNA‐binding site polymorphisms in genes involved in colorectal cancer etiopathogenesis and their impact on disease prognosis. Mutagenesis. 2017;32:533‐542. [DOI] [PubMed] [Google Scholar]
- 77. Gong J, Tian J, Lou J, et al. A functional polymorphism in lnc‐LAMC2‐1: 1 confers risk of colorectal cancer by affecting miRNA binding. Carcinogenesis. 2016;37:443‐451. [DOI] [PubMed] [Google Scholar]
- 78. Vymetalkova V, Pardini B, Rosa F, et al. Polymorphisms in microRNA binding sites of mucin genes as predictors of clinical outcome in colorectal cancer patients. Carcinogenesis. 2017;38:28‐39. [DOI] [PubMed] [Google Scholar]
- 79. Landi D, Gemignani F, Pardini B, et al. Identification of candidate genes carrying polymorphisms associated with the risk of colorectal cancer by analyzing the colorectal mutome and microRNAome. Cancer. 2012;118:4670‐4680. [DOI] [PubMed] [Google Scholar]
- 80. Pardini B, Rosa F, Barone E, et al. Variation within 3′‐UTRs of base excision repair genes and response to therapy in colorectal cancer patients: a potential modulation of microRNAs binding. Clin Cancer Res. 2013;19:6044‐6056. [DOI] [PubMed] [Google Scholar]
- 81. Naccarati A, Rosa F, Vymetalkova V, et al. Double‐strand break repair and colorectal cancer: gene variants within 3′ UTRs and microRNAs binding as modulators of cancer risk and clinical outcome. Oncotarget. 2016;7:23156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Naumov I, Lisiansky V, Kazanov D, et al. Polymorphisms in the CD24 gene play a role in the risk for colorectal neoplasia. Gastroenterology. 2011;140:S‐98. [Google Scholar]
- 83. Liu B, Zhang Y, Jin M, et al. Association of selected polymorphisms of CCND1, p21, and caspase8 with colorectal cancer risk. Mol Carcinog. 2010;49:75‐84. [DOI] [PubMed] [Google Scholar]
- 84. Laczmanska I, Laczmanski L, Bebenek M, et al. Vitamin D receptor gene polymorphisms in relation to the risk of colorectal cancer in the Polish population. Tumor Biol. 2014;35:12397‐12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Vymetalkova V, Pardini B, Rosa F, et al. Variations in mismatch repair genes and colorectal cancer risk and clinical outcome. Mutagenesis. 2014;29:259‐265. [DOI] [PubMed] [Google Scholar]
- 86. Timofeeva MN, Kinnersley B, Farrington SM, et al. Recurrent coding sequence variation explains only a small fraction of the genetic architecture of colorectal cancer. Sci Rep. 2015;5:16286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pazienza P, Actis G, Borghesio E, et al. CD86 genetic variants are associated with colorectal cancer (CRC) and ulcerative colitis, but not adenoma, risk: a clue to a different pathogenetic mechanism of CRC? Digestive Liver Dis. 2011;43:S196. [Google Scholar]
- 88. Dimberg J, Hugander A, Löfgren S, Wågsäter D. Polymorphism and circulating levels of the chemokine CXCL12 in colorectal cancer patients. Int J Mol Med. 2007;19:11‐15. [PubMed] [Google Scholar]
- 89. Hidalgo‐Pascual M, Galan J, Chaves‐Conde M, et al. Analysis of CXCL12 3'UTR G> A polymorphism in colorectal cancer. Oncol Rep. 2007;18:1583‐1587. [DOI] [PubMed] [Google Scholar]
- 90. Shi M‐D, Chen J‐H, Sung H‐T, Lee J‐S, Tsai L‐Y, Lin H‐H. CXCL12‐G801A polymorphism modulates risk of colorectal cancer in Taiwan. Arch Med Sci. 2013;9:999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bellido F, Guinó E, Jagmohan‐Changur S, et al. Genetic variant in the telomerase gene modifies cancer risk in Lynch syndrome. Eur J Hum Genet. 2013;21:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kang M‐J, Jung S‐A, Jung JM, et al. Associations between single nucleotide polymorphisms of MMP2, VEGF, and HIF1A genes and the risk of developing colorectal cancer. Anticancer Res. 2011;31:575‐584. [PubMed] [Google Scholar]
- 93. Savabkar S, Chaleshi V, Farahbakhsh FB, et al. VEGF gene+ 936C/T polymorphism decreases the risk of colorectal cancer. Eur J Oncol. 2015;20:88‐93. [Google Scholar]
- 94. Bae SJ, Kim JW, Kang H, Hwang SG, Oh D, Kim NK. Gender‐specific association between polymorphism of vascular endothelial growth factor (VEGF 936 C> T) gene and colon cancer in Korea. Anticancer Res. 2008;28:1271‐1276. [PubMed] [Google Scholar]
- 95. Ungerbäck J, Elander N, Dimberg J, Söderkvist P. Analysis of VEGF polymorphisms, tumor expression of VEGF mRNA and colorectal cancer susceptibility in a Swedish population. Mol Med Rep. 2009;2:435‐439. [DOI] [PubMed] [Google Scholar]
- 96. Joshi AD, Corral R, Siegmund KD, et al. Red meat and poultry intake, polymorphisms in the nucleotide excision repair and mismatch repair pathways and colorectal cancer risk. Carcinogenesis. 2008;30:472‐479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhao Y, Du Y, Zhao S, Guo Z. Single‐nucleotide polymorphisms of microRNA processing machinery genes and risk of colorectal cancer. OncoTargets Ther. 2015;8:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Andersen V, Østergaard M, Christensen J, Overvad K, Tjønneland A, Vogel U. Polymorphisms in the xenobiotic transporter Multidrug Resistance 1 (MDR1) and interaction with meat intake in relation to risk of colorectal cancer in a Danish prospective case‐cohort study. BMC Cancer. 2009;9:407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Stacey SN, Sulem P, Jonasdottir A, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43:1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Pereira C, Pimentel‐Nunes P, Brandao C, Moreira‐Dias L, Medeiros R, Dinis‐Ribeiro M. COX‐2 polymorphisms and colorectal cancer risk: a strategy for chemoprevention. Eur J Gastro Hepatol. 2010;22:607‐613. [DOI] [PubMed] [Google Scholar]
- 101. Ying Z, Chang‐Ming L, Jian‐Zhi Z, Xiao‐Qin C. Relationship between polymorphisms in the promoter region of the COX‐2 gene and susceptibility to colorectal cancer. World Chinese J Digestol. 2012;20:1579‐1584. [Google Scholar]
- 102. Mohd Shafi'i MS, Shahpudin S, Mustapha MA, et al. The genetic variation A> G at 3'UTR of nuclear factor kappa B 1 A (NFkB1A) influences susceptibility of sporadic colorectal cancer in Malaysian population. Int Med J. 2012;19:98‐101. [Google Scholar]
- 103. Mullany LE, Wolff RK, Herrick JS, Buas MF, Slattery ML. SNP regulation of microRNA expression and subsequent colon cancer risk. PLoS ONE. 2015;10:e0143894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Gao J, Pfeifer D, He L‐J, et al. Association of NFKBIA polymorphism with colorectal cancer risk and prognosis in Swedish and Chinese populations. Scand J Gastroenterol. 2007;42:345‐350. [DOI] [PubMed] [Google Scholar]
- 105. Bermano G, Pagmantidis V, Holloway N, et al. Evidence that a polymorphism within the 3′ UTR of glutathione peroxidase 4 is functional and is associated with susceptibility to colorectal cancer. Genes Nutrition. 2007;2:225‐232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Méplan C, Hesketh J. Genetic polymorphisms in selenoprotein P gene affect colorectal, prostate and breast cancer risk. Proc Nutrition Soc. 2013;72. [Google Scholar]
- 107. Dimberg J, Skarstedt M, Löfgren S, Zar N, Matussek A. Protein expression and gene polymorphism of CXCL10 in patients with colorectal cancer. Biomed Rep. 2014;2:340‐343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Fan Y, Zhang X, Yang Z‐H, et al. The polymorphisms of osteopontin gene and plasma osteopontin protein levels with susceptibility to colorectal carcinoma. DNA Cell Biol. 2013;32:594‐600. [DOI] [PubMed] [Google Scholar]
- 109. Kamal A, Darwish RK, Saad S, et al. Association of osteopontin gene polymorphisms with colorectal cancer. Cancer Invest. 2017;35:71‐77. [DOI] [PubMed] [Google Scholar]
- 110. Xia X, Xu E, Quan S, Huang Q, Lai M. No association between the polymorphisms in CDX2 coding regions and colorectal cancer in Chinese. Mol Cell Biochem. 2009;331:27. [DOI] [PubMed] [Google Scholar]
- 111. Xicola RM, Bontu S, Doyle BJ, et al. Association of a let‐7 miRNA binding region of TGFBR1 with hereditary mismatch repair proficient colorectal cancer (MSS HNPCC). Carcinogenesis. 2016;37:751‐758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Gong Z, Bostick RM, Xie D, et al. Genetic polymorphisms in the cyclooxygenase‐1 and cyclooxygenase‐2 genes and risk of colorectal adenoma. Int J Colorectal Dis. 2009;24:647‐654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Li Y, Jing F, Ding Y, He Q, Zhong Y, Fan C. Long noncoding RNA CCAT1 polymorphisms are associated with the risk of colorectal cancer. Cancer Genet. 2018;222:13‐19. [DOI] [PubMed] [Google Scholar]
- 114. Iglesias D, Nejda N, Azcoita MM, Schwartz S, González‐Aguilera JJ. Effect of COX2‐765G> C and c. 3618A> G polymorphisms on the risk and survival of sporadic colorectal cancer. Cancer Causes Control. 2009;20:1421‐1429. [DOI] [PubMed] [Google Scholar]
- 115. Slattery ML, Curtin K, Poole EM, et al. Genetic variation in C‐reactive protein in relation to colon and rectal cancer risk and survival. Int J Cancer. 2011;128:2726‐2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Rasool S, Kadla SA, Rasool V, et al. Role of the VDR Bsm I and Apa I polymorphisms in the risk of colorectal cancer in Kashmir. Oncol Res Treatment. 2014;37:345‐349. [DOI] [PubMed] [Google Scholar]
- 117. Cheng T‐YD, Makar KW, Neuhouser ML, et al. Interaction between genetic variants in one‐carbon metabolism and folate biomarkers on colorectal cancer risk: The Women's Health Initiative observational cohort [abstract]. In: Proceedings of the 105th Annual Meeting of the American Association for Cancer Research; 2014 Apr 5–9; San Diego, CA. Philadelphia (PA): AACR; Caner Research. 2014;74(19 Suppl): Abstract nr 2197. [Google Scholar]
- 118. Zhang H, Liao L‐H, Liu S‐M, et al. Microsomal glutathione S‐transferase gene polymorphisms and colorectal cancer risk in a Han Chinese population. Int J Colorectal Dis. 2007;22:1185‐1194. [DOI] [PubMed] [Google Scholar]
- 119. Jeon YJ, Kim JW, Park HM, et al. Interplay between 3′‐UTR polymorphisms in the vascular endothelial growth factor (VEGF) gene and metabolic syndrome in determining the risk of colorectal cancer in Koreans. BMC Cancer. 2014;14:881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Cho SH, Ko JJ, Kim JO, et al. 3’‐Utr polymorphisms in the mirna machinery genes drosha, dicer1, ran, and xpo5 are associated with colorectal cancer risk in a Korean population. PLoS ONE. 2015;10:e0131125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Tomsic J, Guda K, Liyanarachchi S, et al. Allele‐specific expression of TGFBR1 in colon cancer patients. Carcinogenesis. 2010;31:1800‐1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Lin M, Zhang L, Hildebrandt MA, Huang M, Wu X, Ye Y. Common, germline genetic variations in the novel tumor suppressor BAP1 and risk of developing different types of cancer. Oncotarget. 2017;8:74936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Jeon YJ, Kim JW, Park HM, et al. Genetic variants in 3′‐UTRs of methylenetetrahydrofolate reductase (MTHFR) predict colorectal cancer susceptibility in Koreans. Sci Rep. 2015;5:11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Yang M, Xiao X, Xing X, Li X, Xia T, Long H. KRAS and VEGF gene 3'‐UTR single nucleotide polymorphisms predicted susceptibility in colorectal cancer. PLoS ONE. 2017;12:e0174140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Budhathoki S, Yamaji T, Iwasaki M, et al. Vitamin D receptor gene polymorphism and the risk of colorectal cancer: a nested case‐control study. PLoS ONE. 2016;11:e0164648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Poole EM, Hsu L, Xiao L, et al. Genetic variation in prostaglandin E2 synthesis and signaling, prostaglandin dehydrogenase, and the risk of colorectal adenoma. Cancer Epidemiol Biomarkers Prev. 2010;19:547‐557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Schafmayer C, Buch S, Egberts JH, et al. Genetic investigation of DNA‐repair pathway genes PMS2, MLH1, MSH2, MSH6, MUTYH, OGG1 and MTH1 in sporadic colon cancer. Int J Cancer. 2007;121:555‐558. [DOI] [PubMed] [Google Scholar]
- 128. Cheng T‐Y, Makar KW, Neuhouser ML, et al. Folate‐mediated one‐carbon metabolism genes and interactions with nutritional factors on colorectal cancer risk: Women's Health Initiative Observational Study. Cancer. 2015;121:3684‐3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Landi D, Moreno V, Guino E, et al. Polymorphisms affecting micro‐RNA regulation and associated with the risk of dietary‐related cancers: a review from the literature and new evidence for a functional role of rs17281995 (CD86) and rs1051690 (INSR), previously associated with colorectal cancer. Mutat Res. 2011;717:109‐115. [DOI] [PubMed] [Google Scholar]
- 130. Ryan BM, Robles AI, Harris CC. KRAS‐LCS6 genotype as a prognostic marker in early‐stage CRC–letter. Clin Cancer Res. 2012;18:3487‐3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Takeshige N, Yin G, Ohnaka K, et al. Associations between vitamin D receptor (VDR) gene polymorphisms and colorectal cancer risk and effect modifications of dietary calcium and vitamin D in a Japanese population. Asian Pac J Cancer Prev. 2015;16:2019‐2026. [DOI] [PubMed] [Google Scholar]
- 132. Park K, Woo M, Nam J, Kim JC. Start codon polymorphisms in the vitamin D receptor and colorectal cancer risk. Cancer Lett. 2006;237:199‐206. [DOI] [PubMed] [Google Scholar]
- 133. Hughes DJ, Hlavatá I, Soucek P, et al. Variation in the vitamin D receptor gene is not associated with risk of colorectal cancer in the Czech Republic. J Gastrointest Cancer. 2011;42:149‐154. [DOI] [PubMed] [Google Scholar]
- 134. Bentley RW, Keown DA, Gearry RB, et al. Vitamin D receptor polymorphisms in colorectal cancer in New Zealand: an association study. NZ Med J. 2012;125:47‐51. [PubMed] [Google Scholar]
- 135. Gromowski T, Kąklewski K, Marciniak W, et al. Vitamin D concentration and frequent variants of VDR gene as a markers of detection probability of breast, lung, prostate and colorectal cancers. (PhD theses). International Conference "Clinical Genetics of Cancer"; 2016.
- 136. Flügge J, Krusekopf S, Goldammer M, et al. Vitamin D receptor haplotypes protect against development of colorectal cancer. Eur J Clin Pharmacol. 2007;63:997‐1005. [DOI] [PubMed] [Google Scholar]
- 137. Mahmoudi T, Mohebbi SR, Pourhoseingholi MA, Fatemi SR, Zali MR. Vitamin D receptor gene ApaI polymorphism is associated with susceptibility to colorectal cancer. Dig Dis Sci. 2010;55:2008‐2013. [DOI] [PubMed] [Google Scholar]
- 138. Moossavi M, Parsamanesh N, Mohammadoo‐Khorasani M, et al. Positive correlation between vitamin D receptor gene FokI polymorphism and colorectal cancer susceptibility in South‐Khorasan of Iran. J Cell Biochem. 2018;119:8190‐8194. [DOI] [PubMed] [Google Scholar]
- 139. Safaei A, Rostami F, Karimi AM, Kh AE, Khorshidi F. Association of vitamin D receptor polymorphism (VDR rs 2238136) with colorectal cancer. J Kerman Univ Med Sci. 2012;19:1‐8. [Google Scholar]
- 140. Atoum MF, Tchoporyan MN. Association between circulating vitamin D, the Taq1 vitamin D receptor gene polymorphism and colorectal cancer risk among Jordanians. Asian Pac J Cancer Prev. 2014;15:7337‐7341. [DOI] [PubMed] [Google Scholar]
- 141. Chae YS, Kim JG, Sohn SK, et al. Association of vascular endothelial growth factor gene polymorphisms with susceptibility and clinicopathologic characteristics of colorectal cancer. J Korean Med Sci. 2008;23:421‐427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Gündüz M, Cacına C, Toptaş B, Yaylım‐Eraltan İ, Tekand Y, İsbir T. Association of vitamin D receptor gene polymorphisms with colon cancer. Genetic Testing Mol Biomarkers. 2012;16:1058‐1061. [DOI] [PubMed] [Google Scholar]
- 143. Yaylım‐Eraltan İ, Arzu Ergen H, Arıkan S, et al. Investigation of the VDR gene polymorphisms association with susceptibility to colorectal cancer. Cell Biochem Funct. 2007;25:731‐737. [DOI] [PubMed] [Google Scholar]
- 144. Dilmec F, Özgönül A, Akkafa F, Uzunkoy A, van Kuilenburg AB. Determination of ApaI and TaqI Polymorphisms of VDR gene in a group of Turkish patients with colorectal cancer. Int J Hematol Oncol. 2009;28:18‐22. [Google Scholar]
- 145. Kupfer SS, Anderson JR, Ludvik AE, et al. Genetic associations in the vitamin D receptor and colorectal cancer in African Americans and Caucasians. PLoS ONE. 2011;6:e26123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Slattery ML, Yakumo K, Hoffman M, Neuhausen S. Variants of the VDR gene and risk of colon cancer (United States). Cancer Causes Control. 2001;12:359‐364. [DOI] [PubMed] [Google Scholar]
- 147. Ochs‐Balcom HM, Cicek MS, Thompson CL, et al. Association of vitamin D receptor gene variants, adiposity and colon cancer. Carcinogenesis. 2008;29:1788‐1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Yamaji T, Iwasaki M, Sasazuki S, Sakamoto H, Yoshida T, Tsugane S. Association between plasma 25‐hydroxyvitamin D and colorectal adenoma according to dietary calcium intake and vitamin D receptor polymorphism. Am J Epidemiol. 2011;175:236‐244. [DOI] [PubMed] [Google Scholar]
- 149. Peters U, Hayes RB, Chatterjee N, et al. Circulating vitamin D metabolites, polymorphism in vitamin D receptor, and colorectal adenoma risk. Cancer Epidemiol Prev Biomarkers. 2004;13:546‐552. [PubMed] [Google Scholar]
- 150. Hofmann G, Langsenlehner U, Renner W, et al. Common single nucleotide polymorphisms in the vascular endothelial growth factor gene and colorectal cancer risk. J Cancer Res Clin Oncol. 2008;134:591‐595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Wu G‐Y, Hasenberg T, Magdeburg R, Bönninghoff R, Sturm JW, Keese M. Association between EGF, TGF‐β1, VEGF gene polymorphism and colorectal cancer. World J Surg. 2009;33:124‐129. [DOI] [PubMed] [Google Scholar]
- 152. Bayhan Z, Simşek T, Ergül E, Utkan NZ, Canturk NZ, Cekmen M. Serum cytokine levels in patients with colorectal cancers according to tumor stages and VEGF gene polymorphism. Hepatogastroenterology. 2014;61:1889‐1894. [PubMed] [Google Scholar]
- 153. Jannuzzi AT, Özhan G, Yanar HT, Alpertunga B. VEGF gene polymorphisms and susceptibility to colorectal cancer. Genetic Test Mol Biomarkers. 2015;19:133‐137. [DOI] [PubMed] [Google Scholar]
- 154. Su C, Li D, Li N, et al. Studying the mechanism of PLAGL2 overexpression and its carcinogenic characteristics based on 3'‐untranslated region in colorectal cancer. Int J Oncol. 2018;52:1479‐1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Jang MJ, Jeon YJ, Kim JW, et al. Association of VEGF and KDR single nucleotide polymorphisms with colorectal cancer susceptibility in Koreans. Mol Carcinog. 2013;52:60‐69. [DOI] [PubMed] [Google Scholar]
- 156. Lau T, Roslani A, Lian L, et al. Association between EGF and VEGF functional polymorphisms and sporadic colorectal cancer in the Malaysian population. Genet Mol Res. 2014;13:5555‐5561. [DOI] [PubMed] [Google Scholar]
- 157. Credidio L, Lima C, Leal R, et al. C936T polymorphism of the VEGF gene in relation to the risk and the clinical and biological characteristics of sporadic colorectal adenocarcinoma. BMC Res Notes. 2014;7:768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Wu X, Li D, Liu Z, et al. Vascular endothelial growth factor 1498C/T, 936C/T polymorphisms associated with increased risk of colorectal adenoma: a Chinese case–control study. Mol Biol Rep. 2011;38:1949‐1955. [DOI] [PubMed] [Google Scholar]
- 159. Hou R, Liu Y, Feng Y, et al. Association of single nucleotide polymorphisms of ERCC1 and XPF with colorectal cancer risk and interaction with tobacco use. Gene. 2014;548:1‐5. [DOI] [PubMed] [Google Scholar]
- 160. Moreno V, Gemignani F, Landi S, et al. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12:2101‐2108. [DOI] [PubMed] [Google Scholar]
- 161. Ni M, Zhang W‐Z, Qiu J‐R, et al. Association of ERCC1 and ERCC2 polymorphisms with colorectal cancer risk in a Chinese population. Sci Rep. 2014;4:4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Yueh T‐C, Chou A‐K, Gong C‐L, et al. The contribution of excision repair cross‐complementing group 1 genotypes to colorectal cancer susceptibility in Taiwan. Anticancer Res. 2017;37:2307‐2313. [DOI] [PubMed] [Google Scholar]
- 163. Makar KW, Poole EM, Resler AJ, et al. COX‐1 (PTGS1) and COX‐2 (PTGS2) polymorphisms, NSAID interactions, and risk of colon and rectal cancers in two independent populations. Cancer Causes Control. 2013;24:2059‐2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Siezen CL, Bueno‐de‐Mesquita HB, Peeters PH, Kram NR, van Doeselaar M, van Kranen HJ. Polymorphisms in the genes involved in the arachidonic acid‐pathway, fish consumption and the risk of colorectal cancer. Int J Cancer. 2006;119:297‐303. [DOI] [PubMed] [Google Scholar]
- 165. Vogel LK, Sæbø M, Høyer H, et al. Intestinal PTGS2 mRNA levels, PTGS2 gene polymorphisms, and colorectal carcinogenesis. PLoS ONE. 2014;9:e105254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Cox D, Pontes C, Guinó E, et al. Polymorphisms in prostaglandin synthase 2/cyclooxygenase 2 (PTGS2/COX2) and risk of colorectal cancer. Br J Cancer. 2004;91:339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Andersen V, Holst R, Kopp TI, Tjønneland A, Vogel U. Interactions between diet, lifestyle and IL10, IL1B, and PTGS2/COX‐2 gene polymorphisms in relation to risk of colorectal cancer in a prospective Danish case‐cohort study. PLoS ONE. 2013;8:e78366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Thompson CL, Plummer SJ, Merkulova A, et al. No association between cyclooxygenase‐2 and uridine diphosphate glucuronosyltransferase 1A6 genetic polymorphisms and colon cancer risk. World J Gastroenterol. 2009;15:2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Gunter MJ, Canzian F, Landi S, Chanock SJ, Sinha R, Rothman N. Inflammation‐related gene polymorphisms and colorectal adenoma. Cancer Epidemiol Prev Biomarkers. 2006;15:1126‐1131. [DOI] [PubMed] [Google Scholar]
- 170. Pereira C, Queirós S, Galaghar A, et al. Influence of genetic polymorphisms in prostaglandin E2 pathway (COX‐2/HPGD/SLCO2A1/ABCC4) on the risk for colorectal adenoma development and recurrence after polypectomy. Clin Transl Gastroenterol. 2016;7:e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Siezen CL, Tijhuis MJ, Kram NR, et al. Protective effect of nonsteroidal anti‐inflammatory drugs on colorectal adenomas is modified by a polymorphism in peroxisome proliferator‐activated receptor δ. Pharmacogenet Genomics. 2006;16:43‐50. [DOI] [PubMed] [Google Scholar]
- 172. Ali I, Luke B, Dean M, Greenwald P. Allellic variants in regulatory regions of cyclooxygenase‐2: association with advanced colorectal adenoma. Br J Cancer. 2005;93:953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Ashktorab H, Tsang S, Luke B, et al. Protective effect of Cox‐2 allelic variants on risk of colorectal adenoma development in African Americans. Anticancer Res. 2008;28:3119‐3123. [PMC free article] [PubMed] [Google Scholar]
- 174. Ueda N, Maehara Y, Tajima O, Tabata S, Wakabayashi K, Kono S. Genetic polymorphisms of cyclooxygenase‐2 and colorectal adenoma risk: The Self Defense Forces Health Study. Cancer Sci. 2008;99:576‐581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Ramzi NH, Chahil JK, Lye SH, et al. Role of genetic & environment risk factors in the aetiology of colorectal cancer in Malaysia. Indian J Med Res. 2014;139:873. [PMC free article] [PubMed] [Google Scholar]
- 176. Razmkhah M, Ghaderi A. SDF‐1alpha G801A polymorphism in Southern Iranian patients with colorectal and gastric cancers. Indian J Gastroenterol. 2013;32:28‐31. [DOI] [PubMed] [Google Scholar]
- 177. Amara S, Chaar I, Khiari M, et al. Relationship between SDF‐1G801A polymorphism and its expression in Tunisian patients with colorectal cancer. J Immunoassay Immunochem. 2015;36:182‐194. [DOI] [PubMed] [Google Scholar]
- 178. Larriba MJ, Bonilla F, Muñoz A. The transcription factors Snail1 and Snail2 repress vitamin D receptor during colon cancer progression. J Steroid Biochem Mol Biol. 2010;121:106‐109. [DOI] [PubMed] [Google Scholar]
- 179. Touvier M, Chan DS, Lau R, et al. Meta‐analyses of vitamin D intake, 25‐hydroxyvitamin D status, vitamin D receptor polymorphisms, and colorectal cancer risk. Cancer Epidemiol Prev Biomarkers. 2011;20:1003‐1016. [DOI] [PubMed] [Google Scholar]
- 180. Bai Y‐H, Lu H, Hong D, Lin C‐C, Yu Z, Chen B‐C. Vitamin D receptor gene polymorphisms and colorectal cancer risk: a systematic meta‐analysis. World J Gastroenterol. 2012;18:1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Xu Y, He B, Pan Y, et al. Systematic review and meta‐analysis on vitamin D receptor polymorphisms and cancer risk. Tumor Biol. 2014;35:4153‐4169. [DOI] [PubMed] [Google Scholar]
- 182. Sheng S, Chen Y, Shen Z. Correlation between polymorphism of vitamin D receptor TaqI and susceptibility to colorectal cancer: a meta‐analysis. Medicine. 2017;96:e7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Pan Z, Chen M, Hu X, et al. Associations between VDR gene polymorphisms and colorectal cancer susceptibility: an updated meta‐analysis based on 39 case‐control studies. Oncotarget. 2018;9:13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Serrano D, Gnagnarella P, Raimondi S, Gandini S. Meta‐analysis on vitamin D receptor and cancer risk: focus on the role of TaqI, ApaI, and Cdx2 polymorphisms. Eur J Cancer Prev. 2016;25:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Bienertová‐Vašků J, Drábová K, Zlámal F, et al. Pre‐treatment VD levels and VDR receptors as potential predictors of occurrence and overall survival in paediatric patients with solid tumours—A single institution pilot study. Tumor Biol. 2016;37:9209‐9219. [DOI] [PubMed] [Google Scholar]
- 186. Buroker NE, Ning X‐H, Zhou Z‐N, et al. SNPs, linkage disequilibrium, and chronic mountain sickness in Tibetan Chinese. Hypoxia. 2017;5:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Sun W. Angiogenesis in metastatic colorectal cancer and the benefits of targeted therapy. J Hematol Oncol. 2012;5:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Zhou L, Luan H, Dong X, Jin G, Man D, Shang H. Vascular endothelial growth factor gene polymorphisms and colorectal cancer risk: a meta‐analysis. Genet Mol Res. 2011;10:3674‐3688. [DOI] [PubMed] [Google Scholar]
- 189. Chen J, Sun N, Hu G, et al. Association of ERCC1 polymorphisms with the risk of colorectal cancer: a meta‐analysis. Crit Rev Eukaryot Gene Expr. 2017;27(3):267–275. [DOI] [PubMed] [Google Scholar]
- 190. Sæbø M, Skjelbred CF, Nexø BA, et al. Increased mRNA expression levels of ERCC1, OGG1 and RAI in colorectal adenomas and carcinomas. BMC Cancer. 2006;6:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Xu K, Dai H, Wang S, Zhang J, Liu T. The cXcl12 rs1801157 polymorphism and risk of colorectal cancer: a meta‐analysis. OncoTargets Ther. 2018;11:2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Schulz M, Karpova D, Spohn G, et al. Variant rs1801157 in the 3’UTR of SDF‐1ss does not explain variability of healthy‐donor G‐CSF responsiveness. PLoS ONE. 2015;10:e0121859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Teicher BA, Fricker SP. CXCL12 (SDF‐1)/CXCR4 pathway in cancer. Clin Cancer Res. 2010;16:2927‐2931. [DOI] [PubMed] [Google Scholar]
- 194. Du X‐Y, Hu Y‐Y, Xie C, et al. Significant association between Let‐7‐KRAS rs712 G> T polymorphism and cancer risk in the Chinese population: a meta‐analysis. Oncotarget. 2017;8:13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Ying H‐Q, Wang F, He B‐S, et al. The involvement of Kras gene 3′‐UTR polymorphisms in risk of cancer and influence on patient response to anti‐EGFR therapy in metastatic colorectal cancer: a meta‐analysis. OncoTargets Ther. 2014;7:1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Zhao W‐H, Qu X‐F, Xing Z‐G, Zhao L‐Q, Qin L, Lv C. Association of rs712 polymorphism in Kras gene 3’‐luntranslated region and cancer risk: a meta‐analysis. J BUON. 2015;20:309‐316. [PubMed] [Google Scholar]
- 197. Oczko‐Wojciechowska M, Pfeifer A, Rusinek D, et al. The prevalence of somatic RAS mutations in medullary thyroid cancer—a Polish population study. Endokrynologia Polska. 2015;66:121‐125. [DOI] [PubMed] [Google Scholar]
- 198. Smith G, Bounds R, Wolf H, Steele R, Carey F, Wolf C. Activating K‐Ras mutations outwith ‘hotspot'codons in sporadic colorectal tumours–implications for personalised cancer medicine. Br J Cancer. 2010;102:693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Eberhart CE, Coffey RJ, Radhika A, Giardiello FM, Ferrenbach S, Dubois RN. Up‐regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology. 1994;107:1183‐1188. [DOI] [PubMed] [Google Scholar]
- 200. Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


