Abstract
Salivary duct carcinoma (SDC) is a rare, aggressive salivary gland malignancy, which often presents at an advanced stage. A proportion of SDC are characterized by HER2 amplification and/or overexpression of androgen receptor (AR), which could be targeted in a subset of patients, but the presence of AR splice variant‐7 (AR‐V7) in some SDC cases could result in resistance to anti‐androgen therapy. We evaluated a cohort of 28 cases of SDC for potentially targetable biomarkers and pathways using immunohistochemistry (IHC) and next‐generation sequencing (DNA and RNA) assays. Pathogenic genetic aberrations were found in all but 1 case and affected TP53 (n = 19), HRAS (n = 7), PIK3CA, ERBB2 (HER2), and NF1 (n = 5 each); KMT2C (MLL3) and PTEN (n = 3 each); BRAF (p.V600E), KDM5C and NOTCH1 (n = 2 each). Androgen receptor was expressed in all cases and 13 of 27 harbored the AR‐V7 splice variant (including a case without any other detectable genetic alteration). HER2 IHC was expressed in 11 of 28 cases. The majority of SDC cases had no biomarkers predictive of immunotherapy response: 5 cases exhibited low (1%‐8%) programmed death ligand 1 (PD‐L1) expression in tumor cells, 2 cases exhibited elevated TMB, and no samples exhibited microsatellite instability. Notably, the pre‐treatment biopsies from 2 patients with metastatic disease, who demonstrated clinical responses to anti‐androgen therapy, showed AR expression and no AR splice variants. We conclude that comprehensive molecular profiling of SDCs can guide the selection of patients for targeted therapies involving AR, HER2, PD‐L1, mitogen‐activated protein kinase, and PIK3CA pathways.
Keywords: biomarkers, head and neck cancer, molecular genetics, next generation sequencing
Salivary duct carcinoma (SDC) is a genetically diverse malignancy associated with several potentially targetable molecular pathways that have been uncovered by comprehensive molecular profiling. Our study detected genetic aberrations in 27 of 28 cases of SDC, including targetable alterations involving PIK3CA/PTEN, BRAF, and HER2. In addition, androgen receptor splice variant AR‐V7 was found in approximately half of cases, which has implications in resistance to anti‐androgen therapy.
![]()
1. INTRODUCTION
Salivary duct carcinoma (SDC) is a rare, high‐grade salivary gland malignancy characterized by an apocrine phenotype and morphologic similarity to ductal carcinoma of the breast.1 Men over the age of 50 are most frequently affected, and the parotid gland is the most common site. SDC tends to present with an enlarging mass sometimes accompanied by facial nerve paralysis.2 With the exception of cases occurring as intracapsular SDC ex pleomorphic adenoma (PA), this tumor is highly aggressive. Patients often present with advanced stage disease including frequent nodal metastases, and overall survival is poor, with approximately 65% of patients dying from progressive disease within 48 months.2, 3 Standard management for SDC consists of surgical resection followed by radiation therapy; however, local recurrence and metastatic rates remain high, and treatment options for recurrent and metastatic SDC are limited.3 Therefore, there is an unmet need for targeted therapies to help better manage this aggressive malignancy.
Several characteristic findings in SDC have been exploited in the development of targeted therapy including the overexpression of HER2/neu (ERBB2) found in 37% of SDC ERBB2 gene amplification is found in 72% of overexpressing cases.2 Clinical responses to HER2‐targeted agents, alone or in combination with chemotherapy, have been reported in individual patients with HER2 + SDC.4, 5, 6, 7 Additionally, androgen receptor (AR) expression which is a defining feature of SDC, is found in up to 98% of cases.1, 8 Some argue that AR‐negative SDC is virtually non‐existent, as many such cases are either AR‐positive after repeat immunohistochemistry (IHC) testing, or are subsequently reclassified as a different entity with morphologic similarities to SDC.1 As a result, androgen deprivation therapy (ADT) has emerged as another potential targeted therapeutic strategy for SDC patients, potentially even in the adjuvant setting, and has shown promising results in several small studies.9, 10, 11, 12, 13
Despite some headway in targeted therapy for SDC, neither anti‐HER2 nor anti‐androgen therapies have been standardized or clinically validated. In addition, mechanisms of resistance to HER2‐based treatments and ADT may hinder its clinical benefit in some patients.14, 15 For instance, alterations in phosphoinositide 3‐kinase (PI3K) signaling, either by PIK3CA mutation or PTEN loss, may be involved in resistance to HER2‐targeted therapy.15 Additionally, the discovery of constitutively active AR splice variants, notably AR splice variant‐7 (AR‐V7), which are implicated in resistance to ADT in prostate cancer, has led to the discovery of these variants in SDC specimens as well.14, 16, 17, 18 A small study conducted by Cappelletti et al16 suggests that the presence of AR‐V7 in SDC cells may likewise affect response to ADT, but this requires more extensive investigation.
The treatment of numerous solid and hematologic malignancies (eg non‐small cell lung carcinoma, bladder carcinoma, melanoma, classical Hodgkin lymphoma) has been markedly improved due to therapy with immune check point inhibitors against programmed death 1 and its ligand PD‐L1 (immuno‐oncology [I‐O] treatment). Predictors of a response to these inhibitors include the expression of PD‐L1 in cancer or tumor‐associated immune cells (IC), tumor mutational burden (TMB), microsatellite instability (MSI) status, and the presence of tumor infiltrating lymphocytes.19, 20, 21 While immunotherapy has not yet been clinically validated for the treatment of salivary gland malignancies including SDC, several trials are underway. A study of Cohen et al22 revealed a modest therapeutic benefit (12% response rate) of pembrolizumab in the patients with PD‐L1 positive (≥1% positive cancer or IC) SDC, and it is thought that a combination treatment with chemotherapy may improve the clinical response rates (NCT03360890).
In this study, we report the findings from comprehensive molecular profiling focusing on identification of potential novel molecular targets and I‐O biomarkers in a cohort of SDC.
2. MATERIALS AND METHODS
2.1. Patients and samples selection
Twelve cases of primary SDC from the Department of Pathology at Thomas Jefferson University Hospital and 16 cases of SDC (6 primary cases and 10 recurrent/metastatic SDC cases, including one matched primary and metastatic tumor from the same patient) from Caris Life Sciences met the following inclusion criteria: Confirmed diagnosis of SDC and availability of sufficient formalin‐fixed paraffin‐embedded tissue from the primary and/or recurrent/metastatic tumor for molecular assays. All cases were re‐reviewed by a board‐certified pathologist to confirm the diagnosis and select appropriate slides for molecular profiling. A case was considered as SDC ex PA if histologic evidence of a PA was present in the same specimen, or if there was a clinical history of PA occurring previously in the same site. The Institutional Review Board of the Thomas Jefferson University Hospital approved the study (IRB #18D.142).
2.2. Immunohistochemistry
Quantification of AR staining (SP107 clone; Ventana) was performed with positivity defined as strong nuclear staining in ≥10% of tumor cells. Detection of the splice variant AR‐V7 was assessed at protein level by IHC (EPR15656; Abcam) (16 cases from Caris) and at mRNA level by anchored multiplex PCR for targeted RNA sequencing (ArcherDX) (12 cases from Thomas Jefferson University Hospital).
Positive HER2 IHC was defined as 3+ (strong membranous staining) in ≥10% of tumor cells. IHC for CD274 (PD‐L1) was performed using either 28‐8 (Agilent) or SP142 (Ventana) clones, and PD‐L1 positivity was defined as membranous expression in ≥1% cancer cells (TC).23 Additionally, PD‐L1 expression on the IC was recorded. All IHC assays were run with both positive and negative controls using fully automated staining platforms (Ventana‐Roche and DAKO‐Agilent). The assays were conducted in a CLIA/CAP/ISO15189 certified clinical laboratory (Caris Life Sciences).23
2.3. Next‐generation sequencing
Next‐generation sequencing (NGS) was performed using two commercially available platforms: the Caris Life Sciences (n = 22) and Foundation Medicine (n = 6). The Caris panel utilizes SureSelect XT biotinylated RNA probes from Agilent, to capture DNA fragments from the exons of 592 genes. Sequencing is performed using NextSeq instruments from Illumina. The complete list of the tested genes is available here: http://www.carismolecularintelligence.com/solid_tumors_international.24 The Foundation Medicine NGS platform was reported previously.25 For variant classification, variants of genes that were pre‐determined for their cancer related and clinical significance were interpreted by board‐certified clinical molecular geneticists at Caris and categorized as pathogenic, presumed pathogenic, variant of unknown significance, presumed benign, or benign according to American College of Medical Genetics and Genomics standards.
The TMB was assessed by counting the number of nonsynonymous missense mutations excluding common germline variants. TMB was considered high if ≥10 mutations/megabase were detected. This threshold was calculated based on Caris Life Sciences' cohort of 148 salivary gland carcinomas using the 80th percentile cutoff value as recently suggested by Samstein et al.26
Microsatellite instability status was explored in 18 cases by analyzing the microsatellite loci of the genes on the Caris panel, as reported previously.23 Gene copy number variations were assessed in 22 cases by comparing the depth of NGS sequence reads on the Caris panel to reads from a diploid control. Genes harboring with ≥6 copies were considered amplified.23, 27
ArcherDx FusionPlex Assay (ArcherDX) was used to search for gene fusions. Fifty‐three gene targets were analyzed in 12 SDCs (the panel is available here: https://www.carismolecularintelligence.com/tumor-profiling-menu/mi-profile-usa-excluding-new-york).28
3. RESULTS
3.1. Clinical characteristics of the cohort
Table 1 summarizes the cohort's characteristics; clinical follow‐up information was available for 12 patients (from Thomas Jefferson University Hospital). In accordance with known trends, the majority of the patients were elderly men with tumor in the parotid gland, and 12/12 patients with available clinical follow‐up presented with nodal (specifically stage pN2b) metastatic disease. In the cohort from Caris Life Sciences, 8 patients were with metastatic lesions (involving liver, lung, brain, skin, bone, and lymph nodes), 2 with local recurrences involving the skin, and 6 with primary tumors; clinical follow‐up was not available for this group.
Table 1.
Summary of clinicopathologic features including patient demographics, tumor characteristics, treatment, and follow‐up
| Clinicopathologic feature | N (%) |
|---|---|
| Age, mean (range) (y) | 66.25 (41‐91) |
| Sex | |
| Male | 24/27 (89%) |
| Female | 3/27 (11%) |
| Primary tumor site | |
| Parotid | 24/27 (89%) |
| Submandibular | 2/27 (7%) |
| Lip | 1/27 (4%) |
| Source of neoplastic tissue | |
| Primary tumor, de novo SDC | 13/28 (46%) |
| Primary tumor, SDC ex pleomorphic adenoma | 5/28 (18%) |
| Recurrent/metastatic SDC | 10/28 (36%) |
| pT stage | |
| 1 | 3/12 (25%) |
| 2 | 0/12 (0%) |
| 3 | 2/12 (17%) |
| 4 | 7/12 (58%) |
| pN stage | |
| 0 | 0/12 (0%) |
| 1 | 0/12 (0%) |
| 2 | 12/12 (100%) (all stage pN2b) |
| Treatment | |
| Chemoradiotherapy (CRT) | 4/12 (33%) |
| Radiotherapy | 7/12 (58%) |
| Anti‐androgen therapy | 2/12 (17%) (both combined with CRT) |
| No therapy (surveillance) | 1/12 (8%) |
| Follow‐up, mean (range) (mo) | 18.4 (4‐45) |
| Clinical outcome | |
| NED | 6/12 (50%) |
| AWD | 2/12 (17%) (1 DM + 1 LR) |
| DOD | 1/12 (8%) |
| DOC | 1/12 (8%) |
| LTF | 2/12 (17%) |
Abbreviations: AWD, alive with disease; DM, distant metastasis; DOC, died of another cause; DOD, died of disease; LR, locoregional recurrence; LTF, lost to follow‐up; NED, no evidence of disease; SDC, salivary duct carcinoma.
3.2. Treatment and clinical follow‐up
Following surgical resection, 7 of 12 received adjuvant radiation therapy, 4 of 12 received chemoradiotherapy, and 1 patient received no additional therapy.
Two patients (1 male, 1 female) with distant metastatic disease were treated with anti‐androgen therapy (one received bicalutamide and the other received enzalutamide), both in conjunction with chemoradiotherapy. One patient had lung metastases at the time of diagnosis, and she was alive with stable pulmonary metastatic disease at 42 months after initial diagnosis. The other patient demonstrated a total reduction in metastatic tumor burden of 28% after two scans, and then showed stable disease at the time of his third scan but passed away soon afterward. Neither of these patients had AR‐V7 detected in their tumor samples.
3.3. Molecular characteristics of SDCS
Results of molecular profiling assays are summarized in Table 2. HER2 was overexpressed in 39% (11/28) of the cases. All 28 cases expressed strong nuclear staining for AR by IHC (100%) (Figure 1D), and 13/27 (48%) harbored splice variant AR‐V7 detected by either variant‐specific antibody (showing nuclear localization) (Figure 2) or mRNA (Table 2). AR‐V7 positivity by IHC (n = 16) ranged from 10%‐95% (mean 53%) of tumor cells. A single case without any detected pathogenic mutation in our cohort had AR‐V7 present in the tumor.
Table 2.
Summary of molecular genetic features of salivary duct carcinomas
| Molecular genetic features | ||
|---|---|---|
| Androgen receptor (AR) | 100% positive (28/28) via IHC | 13/27 (48%) had AR‐V7 (9 primary, 4 recurrent/metastatic) |
| HER‐2/neu receptor | 11/28 (39%) positive via IHC | |
| Mutational profilea |
n = 19 (68%): TP53 n = 7 (25%): HRAS n = 5 (18%): PIK3CA, NF1 n = 3 (11%): MLL3, PTEN n = 2 (7%): BRAF, ERBB2, KDM5C, NOTCH1 n = 1 (4%): FGFR1, CDKN1B, CREBBP, MLL2, CHEK2, RB1, BAP1, AKT1, PBRM1, SF3B1, ARID1A, SMARCE1, KMT2C, FBXW7, TSC2, PIK3R1, ATRX, C176Y |
|
| Gene amplifications |
NOTCH1 (n = 2, both primary) ERBB2 (n = 4, 3 primary cases and 1 metastatic case) SDC4 (n = 1, primary case) EGFR (n = 1, metastatic case) MDM2, LGR5, KDM5A, ERC1, CDC73 (all in one metastatic case) |
|
| Archer fusion panel (n = 12) |
WASF2:FGR (n = 1) TBL1XR1:PIK3CA (n = 1) FGFR2:PAWR (n = 1) |
|
| Predictors to immune checkpoint inhibitors | ||
|---|---|---|
| PD‐L1 expressionb |
23/28 (82%) negative 5 cases (18%) with low PD‐L1 positivity (1%‐8% positive cancer cells)b |
One case with PD‐L1 amplification |
| Tumor mutational burden (TMB) |
Low (range 3‐6) in 20/22 (91%) High (14 and 16 mutations per Mb) in 2/22 (9%) |
|
| Microsatellite instability status | 100% (18/18) microsatellite stable | |
Abbreviations: AR‐V7, androgen receptor splice variant‐7; IHC, immunohistochemistry; PD‐L1, programmed death ligand 1.
Full sequencing was performed using two different platforms: the Caris Life Sciences (n = 22) and Foundation Medicine (n = 6).
PD‐L1 expression was assessed by 28‐8 (Agilent) and SP142 (Ventana) clones.
Figure 1.
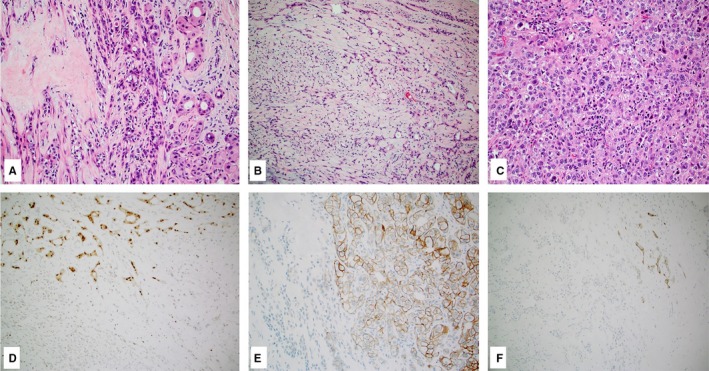
Case of salivary duct carcinoma ex pleomorphic adenoma (A‐C, hematoxylin and eosin, 10×) showing expression of androgen receptor (D, 10×), HER2 (E, 10×), and programmed death ligand 1 (F, 10×) restricted to the malignant component only
Figure 2.
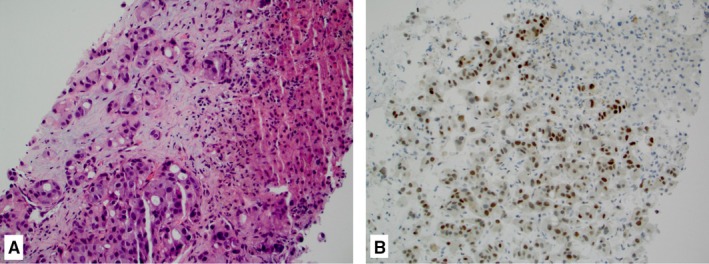
Case of metastatic salivary duct carcinoma in the liver (A, hematoxylin and eosin, 20×) showing diffuse nuclear expression of androgen receptor splice variant‐7 (B, 20×)
A case with matched primary and metastatic SDC harbored identical TP53 and ERBB2 mutations in both samples. A case of SDC ex PA harbored a TP53 mutation in the carcinoma component, while the PA component was devoid of any mutation. Additionally, in this case (Figure 1A‐C), HER2 was positive in the carcinoma but negative in the adenoma component (Figure 1E). Both PA and SDC were devoid of AR‐V7.
3.4. Genomic profile of SDCS
Pathogenic genetic alterations were detected in 27 of 28 cases, many of which harbored more than one alteration. The most frequently encountered were TP53 (n = 19), HRAS (n = 7), PIK3CA (n = 5), NF1 (n = 5), ERBB2 (n = 5) (3 cases with amplification alone, 1 [matched primary and metastatic case] with point mutation p.L755S alone, and 1 case with amplification and two point mutations: p.R678Q, p.V842I), KMT2C (n = 3), PTEN (n = 3), BRAF (p.V600E), KDM5C and NOTCH1 (n = 2 each). All other mutations were rare and affected single SDCs (Table 2). Co‐mutations of HRAS (p.Q61R) and PIK3CA (p. E542A, p.H1047R) genes were seen in 5 of 7 cases with HRAS mutations, all of which were de novo SDC with concomitant HER2 overexpression.
Gene fusion results, available for 12 cases from Caris, were rare and included WASF2:FGR, TBL1XR1:PIK3CA, and FGFR2:PAWR fusions in single cases (Table 2).
3.5. I‐O biomarkers in SDCS
Five cases had detectable (≥1% of tumor cells) PD‐L1 expression, and only 2 cases exhibited more than 5% tumor cell PD‐L1 expression. One case showed PD‐L1/JAK2 gene co‐amplification by NGS but was negative for PD‐L1 expression by IHC. Immune cells expression of PD‐L1 was detected in 5 cases (3 cases without detectable TC PD‐L1 expression) but was not scored due to the lack of uniform criteria.
All tested samples (n = 18) were microsatellite stable (MSS). Twenty of 22 tested cases had a low TMB (range 3‐6 mutations/Mb), while 2 cases (one primary and one recurrent SDC) had TMB > 10 per Mb (14 and 16 mutations per Mb). Both of these cases were PD‐L1 negative.
4. DISCUSSION
In this study, we explored the molecular genetic characteristics of a well‐defined cohort of patients with SDC. Our study revealed common mutations affecting the mitogen‐activated protein kinase (MAPK) and PIK3CA/AKT pathways along with consistent AR and frequent HER2 expression. The molecular genetic landscape of our SDC specimens is comparable to findings in other studies on SDC and closely resembles that of apocrine breast cancer.2, 29, 30
Half of the tumors studied harbored alterations in MAPK pathway (HRAS/BRAF/NF1) genes, which is in line with a recent study of Dalin et al30 HRAS gene mutations were particularly common (25%) indicating their significant oncogenic potential in SDCs. These mutations were not mutually exclusive with PIK3CA mutations as co‐mutations were present in 5 of 7 HRAS‐mutated cases. This observation was recently reported in SDC by Dalin et al30 and previously in other cancer types.31 In contrast to Dalin et al,30 who found co‐occurring PIK3CA mutations in all of their cases with HRAS p.G13R (n = 3) but none of their cases with HRAS p.Q61R (n = 4), all 5 of our PIK3CA/HRAS co‐mutated cases harbored the HRAS p.Q61R mutation. Therefore, both HRAS p.Q61R and p.G13R seem to interact with PIK3CA in promoting oncogenesis in SDC. Furthermore, combined HRAS/PIK3CA mutations were found in de novo SDC cases and none of our SDC ex PA cases, which is a similar finding to that of Chiosea et al.32 The finding of frequent PIK3CA/HRAS co‐mutations in SDC may be clinically relevant as a potential cause of anti‐HER2 resistance given that all 4 of our cases with PIK3CA/HRAS co‐mutations were also HER2‐positive. However, none of the 12 patients from our cohort has been treated with anti‐HER2 agents although previous data indicate a significant clinical benefit of trastuzumab‐based therapy among patients with HER2‐positive SDC.6, 30, 33, 34 More recent studies suggest that dual HER2 inhibition with trastuzumab plus pertuzumab, which is a HER2 dimerization inhibitor antibody, or trastuzumab plus chemotherapy, may lead to even better clinical outcomes when compared to trastuzumab alone.35, 36
While mutations in PIK3CA, HRAS, and BRAF seem to be more common in de novo SDC, SDC ex PA have been reported to more frequently overexpress HER family members such as HER2, HER3 (ERBB3), and EGFR.37 In our study, 4 of 5 SDC ex PA, 5 of 13 de novo SDC, and 2 of 10 recurrent/metastatic SDC were positive by HER2 IHC. In a large series of 151 SDCs, PIK3CA/HRAS/BRAF mutations and HER2 positivity were reported to be mutually exclusive.37 In contrast, in our 7 cases with PIK3CA/HRAS/BRAF mutations, 4 cases (all with PIK3CA/HRAS co‐mutations) showed HER2 expression by IHC. However, the 2 cases with HRAS mutation alone, along with 1 case with BRAF p.V600E, were negative for HER2 IHC. Additionally, the 2 cases with HER2 amplification did not show PIK3CA/HRAS/BRAF mutations, suggesting that perhaps HER2 amplification but not HER2 overexpression by IHC alone may be mutually exclusive with these mutations.
The presence of PIK3CA mutations in a subset of SDC represents a viable therapeutic target. A large series of patients with various tumor histologies showed that the presence of the PIK3CA p.H1047R mutation specifically was an independent predictor for tumor response to PI3K pathway inhibitors.38 All 4 of our patients with PIK3CA mutations had this particular mutation, and one had a concurrent PIK3CA p.E542A mutation. Besides PIK3CA mutations, loss of PTEN activates the PI3K pathway and thus may represent another potential target. Several studies have reported frequent loss of PTEN expression, detected via IHC or FISH analysis, in up to 51% of SDC.15, 37 In head and neck squamous cell carcinoma, tumor cells with PTEN loss were shown to be commonly resistant to pan‐PI3K inhibitors; however, in vitro studies have shown promising results for targeted PI3K therapy in SDC cells.15 Finally, the discovery of a TBL1XR1‐PIK3CA fusion, which has been reported to result in overexpression of the complete PIK3CA protein, in one of our cases further supports the important role of PIK3CA in the oncogenesis of SDC.39
The BRAF p.V600E mutation was detected in two of our cases, similarly to the previously reported studies.2, 32, 37 Although the overall percentage of SDCs with mutated BRAF may be low, there is a potential for targeted therapy for these patients, as BRAF inhibitors are used successfully in melanoma and other BRAF‐mutated tumors. A phase 2 study of vemurafenib in a variety of BRAF p.V600 mutation‐positive non‐melanoma cancers showed partial response in 1 patient with SDC.40 Additionally, a recent report describes a patient who presented with widely metastatic disease and achieved an excellent response to combination chemotherapy with dabrafenib (BRAF inhibitor) and trametinib (MEK inhibitor).41 Clearly, routine testing for BRAF mutations in SDC seems warranted to select patients for targeted therapy.
The presence of AR‐V7, a biomarker predictive of resistance to anti‐androgen therapy in prostate carcinoma, in 13/27 (48%) of cases of SDC indicates a potential role for refining patient selection for hormonal therapies. The AR‐V7 splice variant encodes a truncated AR protein that possesses only the transactivating N‐terminal domain without the C‐terminal ligand‐binding domain (which normally serves as the binding site for anti‐androgen agents), resulting in constitutive activation of AR.3, 8 With the use of RT‐PCR or RNA sequencing, AR‐V7 mRNA has been reported in up to 50% of SDC tumors.17 Other AR isoforms such as AR‐V3 and AR‐45 are found less frequently and with unknown clinical significance.8 The presence of AR‐V7 in prostate cancer tumor cells has been associated with resistance to ADT, though there are limited studies thus far regarding its effect in SDC response to anti‐androgen therapy.17, 42 Cappelletti et al16 report a single patient with SDC treated with ADT who developed resistance and was found to have AR‐V7 in circulating tumor cells. In our study, the 2 patients treated with ADT did not have AR‐V7 in their initial tumor samples, and they showed clinical response in the form of stabilization of the disease. Interestingly, the tumors with AR‐V7 in our study harbored this protein prior to initiation of any systemic therapies, as the tissue tested came from tumor samples from the initial surgical resection. Therefore, it seems that prior exposure to ADT is not necessary for the development of AR‐V7 in SDC. Kang et al17 report a similar finding, in that SDC specimens from 2 patients with no prior exposure to ADT demonstrated high signal for AR‐V7 with RNA in situ hybridization (RISH). Additionally, they found that only 1 patient of 3 treated with ADT had clinical benefit, and this patient was AR‐V7 negative by RISH.17
We also explored the previously established predictive biomarkers to immune checkpoint inhibitors including PD‐L1, TMB and MSI status. Five of our cases exhibited low level (1%‐8%) PD‐L1 expression in tumor cells. A recent study by Sato et al,43 which utilized the PD‐L1 clone E1L3N (Cell Signaling Technology), revealed higher rates of PD‐L1 expression in SDC tumor cells, with 4 of 18 cases showing high expression (defined as greater than 10%) and 5 of 18 cases showing low expression (1%‐9%); moreover high PD‐L1 expression strongly correlated with shorter overall survival. In our study, two different anti‐PD‐L1 assays (laboratory developed SP142 assay and 28‐8 complementary diagnostic assay used for head and neck squamous cell carcinoma) were used in 2 different groups of cases, respectively. We found 1/12 TC positive using 28‐8 and 4/16 TC positive using SP142 clones. Importantly, all tested cases were MSI stable (tissue site agnostic assay for the use of pembrolizumab), while the vast majority (20/22) exhibited low TMB. Based on the obtained data for predictive biomarkers, SDC patients are less likely to benefit from the immune checkpoint inhibitors. In a clinical trial of 26 patients with PD‐L1‐positive advanced salivary gland cancer of various histologies, the objective response rate for pembrolizumab monotherapy was only 12%.22 However, additional trials (NCT03360890) are underway with the hope of achieving improved clinical response rates using combination pembrolizumab plus chemotherapy.
In summary, SDC is a genetically diverse neoplasm associated with several potentially targetable molecular pathways, indicting the need for individual patient's tumor assessment. Some of the detected molecular alterations (eg AR‐V7, PIK3CA, HRAS) may undermine the therapeutic benefits of standardized treatments (anti‐AR and anti‐HER2). The predictive biomarkers to immune checkpoint inhibitors indicate a small potential for therapeutic benefit of these agents in some patients.
CONFLICT OF INTEREST
None declared.
Gargano SM, Senarathne W, Feldman R, et al. Novel therapeutic targets in salivary duct carcinoma uncovered by comprehensive molecular profiling. Cancer Med. 2019;8:7322–7329. 10.1002/cam4.2602
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Williams L, Thompson L, Seethala RR, et al. Salivary duct carcinoma: the predominance of apocrine morphology, prevalence of histologic variants, and androgen receptor expression. Am J Surg Pathol. 2015;39(5):705‐713. [DOI] [PubMed] [Google Scholar]
- 2. Nardi V, Sadow PM, Juric D, et al. Detection of novel actionable genetic changes in salivary duct carcinoma helps direct patient treatment. Clin Cancer Res. 2013;19(2):480‐490. [DOI] [PubMed] [Google Scholar]
- 3. Schmitt NC, Kang H, Sharma A. Salivary duct carcinoma: an aggressive salivary gland malignancy with opportunities for targeted therapy. Oral Oncol. 2017;74:40‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Corrêa TS, Matos G, Segura M, Dos Anjos CH. Second‐line treatment of HER2‐positive salivary gland tumor: ado‐trastuzumab emtansine (T‐DM1) after progression on trastuzumab. Case Rep Oncol. 2018;11(2):252‐257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaidar‐Person O, Billan S, Kuten A. Targeted therapy with trastuzumab for advanced salivary ductal carcinoma: case report and literature review. Med Oncol. 2012;29(2):704‐706. [DOI] [PubMed] [Google Scholar]
- 6. Limaye SA, Posner MR, Krane JF, et al. Trastuzumab for the treatment of salivary duct carcinoma. Oncologist. 2013;18(3):294‐300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Park JC, Ma TM, Rooper L, et al. Exceptional responses to pertuzumab, trastuzumab, and docetaxel in human epidermal growth factor receptor‐2 high expressing salivary duct carcinomas. Head Neck. 2018;40(12):E100‐E106. [DOI] [PubMed] [Google Scholar]
- 8. Dalin MG, Watson PA, Ho AL, Morris LG. Androgen receptor signaling in salivary gland cancer. Cancers (Basel). 2017;9(12):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boon E, van Boxtel W, Buter J, et al. Androgen deprivation therapy for androgen receptor‐positive advanced salivary duct carcinoma: a nationwide case series of 35 patients in The Netherlands. Head Neck. 2018;40(3):605‐613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jaspers H, Verbist BM, Schoffelen R, et al. Androgen receptor‐positive salivary duct carcinoma: a disease entity with promising new treatment options. J Clin Oncol. 2011;29(16):e473‐e476. [DOI] [PubMed] [Google Scholar]
- 11. Locati LD, Perrone F, Cortelazzi B, et al. Clinical activity of androgen deprivation therapy in patients with metastatic/relapsed androgen receptor‐positive salivary gland cancers. Head Neck. 2016;38(5):724‐731. [DOI] [PubMed] [Google Scholar]
- 12. van Boxtel W, Locati LD, van Engen‐van Grunsven A, et al. Adjuvant androgen deprivation therapy for poor‐risk, androgen receptor‐positive salivary duct carcinoma. Eur J Cancer. 2019;110:62‐70. [DOI] [PubMed] [Google Scholar]
- 13. Yamamoto N, Minami S, Fujii M. Clinicopathologic study of salivary duct carcinoma and the efficacy of androgen deprivation therapy. Am J Otolaryngol. 2014;35(6):731‐735. [DOI] [PubMed] [Google Scholar]
- 14. Mitani Y, Rao PH, Maity SN, et al. Alterations associated with androgen receptor gene activation in salivary duct carcinoma of both sexes: potential therapeutic ramifications. Clin Cancer Res. 2014;20(24):6570‐6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Saintigny P, Mitani Y, Pytynia KB, et al. Frequent PTEN loss and differential HER2/PI3K signaling pathway alterations in salivary duct carcinoma: implications for targeted therapy. Cancer. 2018;124(18):3693‐3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cappelletti V, Miodini P, Reduzzi C, et al. Tailoring treatment of salivary duct carcinoma by liquid biopsy: ARv7 expression in circulating tumor cells. Ann Oncol. 2018;29(7):1598‐1600. [DOI] [PubMed] [Google Scholar]
- 17. Kang H, Antonarakis ES, Luo J, et al. Detection of AR‐V7 transcript with RNA in situ hybridization in human salivary duct cancer. Oral Oncol. 2018;84:134‐136. [DOI] [PubMed] [Google Scholar]
- 18. Yang RK, Zhao P, Lu C, Luo J, Hu R. Expression pattern of androgen receptor and AR‐V7 in androgen deprivation therapy‐naive salivary duct carcinomas. Hum Pathol. 2019;84:173‐182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD‐1 blockade. Science. 2017;357(6349):409‐413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herbst RS, Soria J‐C, Kowanetz M, et al. Predictive correlates of response to the anti‐PD‐L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563‐567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Taube JM, Klein A, Brahmer JR, et al. Association of PD‐1, PD‐1 ligands, and other features of the tumor immune microenvironment with response to anti‐PD‐1 therapy. Clin Cancer Res. 2014;20(19):5064‐5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cohen RB, Delord JP, Doi T, et al. Pembrolizumab for the treatment of advanced salivary gland carcinoma: findings of the phase 1b KEYNOTE‐028 study. Am J Clin Oncol. 2018; 41(11): 1083‐1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gatalica Z, Xiu J, Swensen J, Vranic S. Comprehensive analysis of cancers of unknown primary for the biomarkers of response to immune check‐point blockade therapy. Eur J Cancer. 2018;94:179‐186. [DOI] [PubMed] [Google Scholar]
- 24. Caris Molecular Intelligence Profiling Menu. http://www.carismolecularintelligence.com/solid_tumors_international. 2019. Accessed May 16, 2019.
- 25. Technical Specifications, FoundationOne CDx. https://assets.ctfassets.net/vhribv12lmne/4ZHUEfEiI8iOCk2Q6saGcU/671b313cb6bb85bfe861f83e31c9716d/F1CDx_TechInfo_09-03.pdf. 2018. Accessed May 16, 2019.
- 26. Samstein RM, Lee C‐H, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vanderwalde A, Spetzler D, Xiao N, Gatalica Z, Marshall J. Microsatellite instability status determined by next‐generation sequencing and compared with PD‐L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018;7(3):746‐756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Caris Molecular Intelligence, MI Profile for U.S. (excluding New York). https://www.carismolecularintelligence.com/tumor-profiling-menu/mi-profile-usa-excluding-new-york. 2019. Accessed May 16, 2019.
- 29. Chiosea SI, Williams L, Griffith CC, et al. Molecular characterization of apocrine salivary duct carcinoma. Am J Surg Pathol. 2015;39(6):744‐752. [DOI] [PubMed] [Google Scholar]
- 30. Dalin MG, Desrichard A, Katabi N, et al. Comprehensive molecular characterization of salivary duct carcinoma reveals actionable targets and similarity to apocrine breast cancer. Clin Cancer Res. 2016;22(18):4623‐4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oda K, Okada J, Timmerman L, et al. PIK3CA cooperates with other phosphatidylinositol 3ʹ‐kinase pathway mutations to effect oncogenic transformation. Cancer Res. 2008;68(19):8127‐8136. [DOI] [PubMed] [Google Scholar]
- 32. Chiosea SI, Thompson LD, Weinreb I, et al. Subsets of salivary duct carcinoma defined by morphologic evidence of pleomorphic adenoma, PLAG1 or HMGA2 rearrangements, and common genetic alterations. Cancer. 2016;122(20):3136‐3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kadowaki S, Yatabe Y, Hirakawa H, et al. Complete response to trastuzumab‐based chemotherapy in a patient with human epidermal growth factor receptor‐2‐positive metastatic salivary duct carcinoma ex pleomorphic adenoma. Case Rep Oncol. 2013;6(3):450‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thorpe LM, Schrock AB, Erlich RL, et al. Significant and durable clinical benefit from trastuzumab in 2 patients with HER2‐amplified salivary gland cancer and a review of the literature. Head Neck. 2017;39(3):E40‐E44. [DOI] [PubMed] [Google Scholar]
- 35. van Boxtel W, Boon E, Weijs W, van den Hoogen F, Flucke UE, van Herpen C. Combination of docetaxel, trastuzumab and pertuzumab or treatment with trastuzumab‐emtansine for metastatic salivary duct carcinoma. Oral Oncol. 2017;72:198‐200. [DOI] [PubMed] [Google Scholar]
- 36. Kurzrock R, Meric‐Bernstam F, Hurwitz H, et al. Targeted therapy for advanced salivary cancer with HER2 or hedgehog alterations: interim data from MyPathway. J Clin Oncol. 2017;35(15_suppl):6086. [Google Scholar]
- 37. Shimura T, Tada Y, Hirai H, et al. Prognostic and histogenetic roles of gene alteration and the expression of key potentially actionable targets in salivary duct carcinomas. Oncotarget. 2018;9(2):1852‐1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Janku F, Wheler JJ, Naing A, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early‐phase clinical trials. Cancer Res. 2013;73(1):276‐284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. 2014;5:4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med. 2015;373(8):726‐736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin V, Nabell LM, Spencer SA, Carroll WR, Harada S, Yang ES. First‐line treatment of widely metastatic BRAF‐mutated salivary duct carcinoma with combined BRAF and MEK inhibition. J Natl Compr Canc Netw. 2018;16(10):1166‐1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Antonarakis ES, Lu C, Wang H, et al. AR‐V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371(11):1028‐1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sato F, Akiba J, Kawahara A, et al. The expression of programed death ligand‐1 could be related with unfavorable prognosis in salivary duct carcinoma. J Oral Pathol Med. 2018;47(7):683‐690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


