Abstract
Introduction: Recently, volumetric absorptive microsampling (VAMS) has been used for accurate sampling of a fixed peripheral blood volume (10 µL) on a volumetric swab, and long-term sample storage. The mPlex-Flu assay is a novel, high-throughput assay that simultaneously measures the concentration of antibodies against the hemagglutinin (HA) proteins from multiple influenza virus strains with ≤5 µL of serum. Here we describe combining these two methods to measure multidimensional anti-influenza IgG activity in whole blood samples collected by a finger stick and VAMS, with correction for serum volume based on simultaneous hemoglobin measurement. Methods: We compared capillary blood samples obtained from a finger stick using a VAMS device with serum samples collected by traditional phlebotomy from 20 subjects, with the influenza antibody profiles measured by the mPlex-Flu assay. Results: We found that results with the two sampling methods were highly correlated within subjects and across all influenza strains (mean R2 = 0.9470). Adjustment for serum volume, based on hemaglobin measurement, was used to estimate serum volume of samples and improved the accuracy. IgG measurements were stable over 3 weeks when VAMS samples were stored at room temperature or transported using a variety of shipping methods. Additionally, when volunteers performed finger-stick VAMS at-home by themselves, the comparison results of anti-HA antibody concentrations were highly consistent with sampling performed by study personnel on-site (R2 = 0.9496). Conclusions: This novel approach can provide a simple, accurate, and low-cost means for monitoring the IgG anti-influenza HA antibody responses in large population studies and clinical trials.
Keywords: Volumetric absorptive microsampling (VAMS), mPlex-Flu assay, influenza A virus antibodies, immunity assay, clinical influenza vaccine studies
Introduction
Both seasonal and emerging influenza virus infection are among the largest reoccurring global public health threats [1], and vaccination is the major method of prevention [2]. Flu vaccines are currently designed to elicit antibodies against hemagglutinin (HA), the most abundant glycoprotein on the viral surface [3]. Protective antibodies block the ability of HA to bind to sialic acid on target cells, or enhance viral clearance, preventing infection [4]. Measuring antibody-mediated immunity is critical to evaluate vaccine efficacy and immunity to seasonal and emerging influenza viruses. However, a major translational barrier is obtaining serum samples to measure antibody-mediated influenza immunity, which is resource-intensive, time-consuming, and expensive [5]. This limits our ability to conduct large-scale influenza vaccine clinical trials, measure population immunity, and assess the mismatch between circulating influenza strains and the seasonal influenza vaccine in real time. Solving this translational barrier would greatly improve our ability to conduct high-quality clinical trials of influenza vaccines, perform large-scale assessments of population immunity to influenza, and greatly decrease the resource intensity of clinical and translational influenza research.
Several factors contribute to this translational barrier. Most assays of antibody-mediated influenza immunity, such as hemagluttinin inhibition (HAI), enzyme-linked immunosorbent assay (ELISA), and microneutralization (MN) assays all require at the minimum 0.1–0.2 mL of serum to perform with appropriate replicates. Such quantities of serum are usually obtained by venipuncture phlebotomy performed by a healthcare professional, requiring subject travel to the research facility or collection point. After collection, blood samples require post-phlebotomy processing including serum separation, aliquotting, and storage. Thus, developing a translational research solution would require addressing two barriers: (1) developing and validating a simple method for in-the-field collection of small amounts of peripheral blood or serum, and (2) coupling microsampling to an assay measuring anti-influenza IgG that uses very small sample volumes (5–20 µL). Here we describe and validate such a system, using a combination of volumetric absorptive microsampling (VAMS) [6] coupled with a Luminex-based assay (mPlex-Flu) [7–12] to quantitatively measure IgG antibodies against 33 strains of influenza HA.
One approach to simplifying sample collection is to perform a finger or heal stick to draw a drop of blood, using a disposable lancet as is done for diabetic blood glucose monitoring. The blood drop, generally 50–200 µL, is adsorbed onto filter paper and dried. Samples are then eluted and analyzed at a later date. This microsampling dried blood spot (DBS) method was first introduced in 1963 [13]. It has been used to assess the HIV-1 antibodies in newborns, in population-based surveys for more than 25 years [14–16], and for analysis of anti-drug antibodies in FDA clinical trials. DBS is safer and simpler than venipuncture. It enables self-sampling at-home and can greatly reduce costs for clinical- or population-based studies. In addition, IgG and IgM antibodies in dried blood sample are known to be stable at room temperature for weeks and at −20°C for years. A significant drawback of DBS, however, is the high variability of sample volumes. This makes calculation of a concentration problematic and limits its use for quantitative measures of antibody abundance. In contrast, VAMS devices adsorb a consistent volume of blood from a finger stick, generally 10 or 20 µL, and have been recently used to collect samples for antibody testing in many fields (Reviewed in Ref. [6]). This new technique overcomes the issue of inconsistent blood volumes between sample blood spots in the DBS method. VAMS allows accurate and precise sampling with standard deviation ≤0.4 µL with 10 µL blood samples [17].
To address the translational barrier of measuring anti-influenza antibody-mediated immunity, we have previously developed a Luminex-based multiplex assay (mPlex-Flu assay) that can simultaneously measure absolute antibody concentrations (IgG, IgM, or IgA) against up to 50 influenza strains using ≤5 µL of serum [7,8,11,12]. The mPlex-Flu assay has a continuous linear readout over four logs, with low Type-I (false positives, specificity) and Type-II (false negatives, sensitivity) errors [10]. It provides absolute concentrations for strain-specific anti-influenza IgG antibody levels, as opposed to 8–12 discrete titer levels for other assays (e.g. HAI, MN), with extremely low inter- and intra-subject variance [9]. Notably, the mPlex-Flu assay also has a very high correlation with both standard HAI and MN assays, with several added advantages, including simultaneous measurement of absolute anti-HA IgG levels for a large number of influenza strains [7,8,11,12], greater precision of clinical trial group statistical comparisons [9,10], and a low per-sample cost.
Unlike traditional DBS sampling, VAMS also permits whole blood IgG measurements to be adjusted for serum or plasma volume. Whole blood is composed of a cellular component, primarily erythrocytes, and a noncellular component, plasma (when anticoagulant is used) or serum (when clotting is allowed to occur). The hematocrit (HCT) is the fraction of whole blood occupied by cells; IgG is only present in the noncellular serum fraction (∼45–75%). Thus IgG concentrations, as traditionally measured in plasma, (IgGs) will be higher than those measured in whole blood (IgGBL) such that [IgG]BL = [IgG]S(1 – HCT) [18]. This factor could lead to systemic underestimation of anti-HA IgG levels with VAMS. HCT cannot be measured directly in a DBS or VAMS sample. However, the concentration of hemoglobin (Hgb), an iron-binding molecule contained by red blood cells, can be measured. There is a tight correlation between whole blood Hgb levels and HCT, generally HCTBL = 3.0[Hgb]BL. While some exceptions apply (e.g. sickle cell disease, β thallasemia) [19], such estimates of HCT from Hgb generally have only a modest error under normal conditions [20]. As blood sample volume is known in VAMS [21], [Hgb]BL can be used to estimate IgGS, facilitating comparison of VAMS measured anti-HA IgG concentrations with more traditional direct plasma measurement methods.
Development of a simple method to measure anti-HA IgG levels using small volume blood samples remotely collected by study subjects would greatly improve our ability to conduct more robust clinical trials, population immunity surveys, and augment current influenza surveillance efforts. For example, most influenza vaccine clinical trials have measured anti-HA IgG titers in peripheral blood samples pre-vaccination (day 0), and at days 7 and 28 post-vaccination [22], while others have collected samples at more distant time points [23]. A substantial expense in these trials is sample collection by trial personnel. For the same reasons (i.e. cost and inconvenience of phlebotomy for sample collection), large-scale surveys of population antibody-mediated immunity to influenza are rare. We are not aware of any current studies assessing IgG-mediated immunity to multiple (>30) influenza strains with over 1000 subjects. Finally, the the United States Center for Disease Control and the World Health Organization (WHO) both conduct extensive influenza virus field surveillance programs [24], collecting viral samples by nasal swab to isolate and sequence influenza strains in people with influenza-like illnesses. Yet, these programs generally do not collect serum to assess antibody-mediated influenza immunity, likely due to the cost and time needed for phlebotomy and sample processing. In these cases, a simple method to collect and analyze samples for anti-influenza IgG concentrations would decrease the barriers to multiple sample collection (cost, inconvenience, sample processing), and improve scientific knowledge of influenza immunity.
Here we describe using a combination of VAMS with the mPlex-Flu assay and Hgb measurement to quantitatively estimate serum IgG antibody concentrations against 33 strains of influenza virus HAs. Whole blood anti-influenza IgG concentrations are adjusted for serum volume using measured Hgb and used to estimate serum anti-influenza IgG to facilitate comparison with standard methods. This study validates the accuracy, reproducibility, and sample stability of this novel assay combination. Overall, we show that the combination of VAMS with the mPlex-Flu assay could be a powerful tool for large sample size analysis of multidimensional influenza antibody-mediated immunity for use in influenza vaccine and population immunity studies.
Methods
Human Subjects Ethics Statement
This study was approved by the Research Subjects Review Board at the University of Rochester Medical Center (RSRB approval number RSRB00070463), and informed consent was obtained from all participants. Research data were coded such that subjects could not be identified, either directly or through linked identifiers. Subject identification numbers were re-encoded for publication.
Participants and Sample Collection
Twenty-one healthy volunteers 18–65 years of age were recruited for this study. Subjects who were taking immunosuppressive medications were excluded. All subjects had samples collected by both venous phlebotomy and VAMS (Mitra Collection Kit; Neoteryx, CA, USA). For venous phlebotomy, standard venipuncture was performed and 3–4 mL of blood was collected in a serum collection tube (BD, NJ, USA), centrifuged (3000 RPM, 4°C, 12 min), and sera were aliquotted into 100 µL cryo-vials and stored at −20°C until analysis.
Study Design, VAMS Sample Collection and Storage
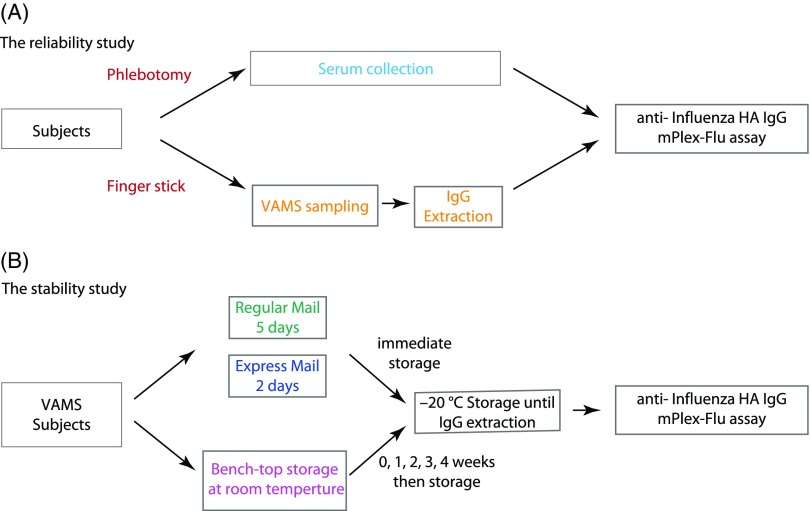
The study was designed to assess both variability between standard venipuncture for serum and VAMS sampling, and reproducibility of results when subjects performed VAMS sampling remotely after instruction. At the initial study visit, each volunteer donated one venous blood sample by phlebotomy and one VAMS blood sample by finger stick. Both samples were collected by study coordinators on-site at the University of Rochester Clinical Research Center. Study subjects were then trained to perform a finger stick with the lancet device and collect the VAMS sample. After training, one VAMS kit was sent home with the volunteer. Three days later, the volunteer self-collected a second VAMS sample and returned it in sealed packaging to the Research Center for analysis (Fig. 1).
Fig. 1.
Experimental design. HA = hemagglutinin; VAMS = volumetric microsampling.
VAMS blood samples were collected using the manufacturer’s 10 µL collection kit (Neoteryx, CA, USA). After alcohol swabbing, the lateral portion of the participant’s finger was punctured using a contact-activated lancet. Gentle pressure was applied to the finger to allow a drop of blood to collect at the skin surface. A porous, hydrophilic VAMS tip was held against the blood drop until completely filled with blood. Each tip absorbed 10 µL of blood, and two tips were present in each collection kit, for a total of 20 µL of blood per collection. Tips were allowed to dry for 1–2 h at room temperature in protective cassettes. For the stability experiment, 14 VAMS blood samples from one donor were collected at the same time. All VAMS tips were placed in separate and sealed containers with silica desiccant packets and stored at −20°C until analysis.
Extraction of Antibodies and Hemoglobin (Hgb) from VAMS Samples
The absorbent tips from each VAMS collection kit (containing 10 µL blood sample) were soaked in 200 µL extraction buffer (PBS + 1% BSA + 0.5% Tween) in 1 mL 96-well plates (Masterblock, GBO, Austria) and shaken overnight to extract the antibodies as described previously [16]. Hgb was extracted along with IgG by this method and quantified as described below.
mPlex-Flu Assay
The mPlex-Flu assay was performed as described previously [7,8,12]. Briefly, venous phlebotomy serum samples were diluted 1:5000 with PBS, while the 200 µL extractions from VAMS device (1:20) were further diluted 1:250, to yield a final 1:5000 dilution of the VAMS samples. For both serum and VAMS samples, 200 µL of diluted sample was used for analysis and added to a black, clear-bottom 96-well plate (Microplate, GBO, Austria). Standard serum (STD01) was made in our laboratory [7,12], and the standard curve for each influenza virus strain was generated by 1:4 serially diluting STD01 for each batch of samples. Fifty microliter of the diluted sample was added into each reaction well. All samples were run in duplicate.
The influenza HA bead panel used in this study is shown in Table 1, comprising 30 separate recombinant HAs. Fifty microliter of beads mix was added to each well of the plate as previously described [7,8,12]. Plates were incubated with gentle shaking for 2 h at room temperature and then washed (PBS + 1% Bris + 0.1% BSA). A magnet placed under the plate immobilized the beads during washes. After three washes, a goat anti-human IgG-PE secondary antibody (Southern Biotech, Cat No:2040–09) was added, and plates were incubated for another 2 h. After three more washes, beads were resuspended in drive fluid (Luminex Co., TX) and the beads were analyzed using MAGPIX Multiplex Reader (Luminex Co., TX). The calculation of IgG antibody concentration against each individual influenza virus strain rHA was performed by Bio-Plex Manager 6.2 software (Bio-Rad Co., CA).
Table 1.
The panel of influenza virus recombinant hemagglutinins (rHAs) in mPlex-Flu assay
| Influenza type | Subtype | Full strain name | Abbreviation | Genbank accession # |
|---|---|---|---|---|
| A | H1 | A/South Carolina/1/18 | SC18 | AF117241.1 |
| A/Puerto Rico/8/1934 | PR8 | CY148243.1 | ||
| A/USSR/90/1977 | USSR77 | DQ508897.1 | ||
| A/New Caledonia/20/1999 | NewCal99 | DQ508889.1 | ||
| A/Texas/36/1991 | Tex91 | CY125100.1 | ||
| A/California/07/2009 | Cali09 | FJ966974.1 | ||
| A/Michigan/45/2015 | Mic15 | KY117023.1 | ||
| H2 | A/Japan/305/1957 | Jap57 | L20407.1 | |
| H3 | A/Port Chalmers/1/1973 | PC73 | CY112249.1 | |
| A/Hong Kong/1/1968 | HK68 | CY009348.1 | ||
| A/Perth/16/2009 | Per09 | GQ293081.1 | ||
| A/Victoria/361/2011 | Vic11 | KM821347 | ||
| A/Texas/50/2012 | Tex12 | KC892248.1 | ||
| A/Switzerland/9715293/2013 | Swi13 | EPI537866 | ||
| A/Hong Kong/4801/2014 | HK14 | EPI653201 | ||
| A/Singapore/INFIMH-16-0019/2016 | Sin16 | EPI1164036 | ||
| H5 | A/Viet Nam/1203/2004 | Viet04 | EF541403 | |
| A/Beijing/01/2003 | BJ03 | EF587277 | ||
| H6 | A/chicken/Taiwan/67/2013 | chTW13 | KJ162860.1 | |
| H7 | A/mallard/Netherlands/12/2000 | malNert00 | EF470586 | |
| A/rhea/North Carolina/39482/1993 | rheaNC93 | KF695239 | ||
| A/Shanghai/1/2013 | SH13 | KF021597.1 | ||
| H9 | A/guinea fowl/Hong Kong/WF10/1999 | gfHK99 | AY206676.1 | |
| B | B/Brisbane/60/2008 | B/Bris08 | CY115343 | |
| B/Massachusetts/2/2012 | B/Mass12 | KF752446.1 | ||
| B/Phuket/3027/2013 | B/Phu13 | EPI540673 | ||
| Chimeric HA | Group 1 Stalk | A/Indonesia/5/05 head, A/California/07/2009 stalk | cH5/1 | |
| A/gf/HK/WF10/1999 head, A/California/07/2009 stalk | cH9/1 | |||
| Group 2 Stalk | A/Indonesia/5/05 head, A/Victoria/361/2011 stalk | cH5/3 | ||
| A/duck/Czech/1956 head, A/Shanghai/1/2013 stalk | cH4/7 |
Measurement of Hemoglobin (Hgb) and Hematocrit (HCT)
We measured Hgb concentrations using a Hemoglobin colormetric Assay Kit (Abcam, Cat No:ab234046, MA, USA). Briefly, 20 µL of blood or extracted samples were incubated with 180 µL of Hgb detector buffer at room temperature for 15 min in 96-well plates. Negative and positive controls for Hgb at concentrations of 0, 50, 100, 150, 200, and 250 mg/dL six standards were used to construct a standard curve. The absorbency at 575 nm (OD 575) was measured using a Synergy Microplate reader (BioTek, VT, US). The concentration of Hgb was then calculated based on the OD575 of samples with the standard curve. The HCT of venous whole blood samples taken by venipuncture was measured using an AcT Diff Coulter Counter (Beckman Coulter, CA) following manufacture instructions.
Estimation and Adjustment of the Effects of HCT on IgG Concentration in VAMS Saples
To create a standard curve to estimate serum IgG concentration, we collected heparinezed whole blood by phlebotomy (Ref No:366480, BD Vacutainer), and separated the cellular and plasma portions from each subject by centrifugation. We next created a range of HCTs, (∼10%, 20%, 30%, 40%, 50%, and 60%) by manually mixing the plasma and red blood cells in a 2-mL total volume. As the volume of red cells might vary even with accurate pipetting, we measured the HCT and Hgb using the Coulter Counter, as described above, for all control blood mixtures, and used the resulting values for further calculation. We next collected VAMS samples in triplicate from the standards. HCT and Hgb were measured for all VAMS samples, and anti-influenza HA IgG antibody concentrations were measured from the corresponding venipuncture serum samples and the VAMS sample extractions.
To obtain the estimated serum concentration ([IgG
S]) from the VAMS blood concentration ([IgG
VAMS]), we first built a relationship between the estimated ratio of 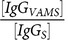 and the measured HCT values from five subjects using a generalized estimating equation (GEE) model with identity link function [25]. A compound symmetry variance–covariance matrix [26] was used to model the within-subject correlation. The GEE model gives us the following relationship between the estimated ratio of
and the measured HCT values from five subjects using a generalized estimating equation (GEE) model with identity link function [25]. A compound symmetry variance–covariance matrix [26] was used to model the within-subject correlation. The GEE model gives us the following relationship between the estimated ratio of 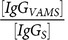 and the measured real HCT values.
and the measured real HCT values.
| (1) |
Next, we obtained the relationship between HCT values and Hgb values using measured HCT and Hgb data from five subjects through the GEE model with the identity link function and compound symmetry variance–covariance matrix:
| (2) |
Combining equations 1 and 2 results in the following relationship between the estimated ratio of  and the measured real Hgb values.
and the measured real Hgb values.
| (3) |
which can be used to estimate the serum anti-influenza HA IgG concentration from the hemaglobin measurement:
| (4) |
VAMS Sample Stability Analysis
To assess the stability of VAMS samples at room temperature over time, 14 VAMS samples were collected from the same subject. An initial two VAMS samples were stored at −20°C after drying for 2 h. A further eight VAMS samples were left on the lab bench at room temperature (22–25°C, controlled, but not monitored). Two of these were then moved into 20°C storage at days 7, 14, 21, and 28 after the initial sampling. The remaining four VAMS samples were mailed back to the lab using the United States Postal Service (USPS) and 2-day overnight delivery. The samples sent by express delivery service returned in 2 days and those sent by the standard post returned in 5 days (Fig. 1). The VAMS samples were stored at −20°C upon arrival in the laboratory.
Statistical Analysis
Spearman’s correlation coefficient [27] with the Benjamini–Hochberg multiple testing correction method [28] was used to measure the reliability of mPlex-Flu results from VAMS versus conventional venous phlebotomy samples, the reproduciblity of mPlex-Flu results from VAMS collected by volunteers at-home versus VAMS collected by study coordinators on-site, and the stability of mPlex-Flu results from VAMS samples stored at room temperature over time or after shipping. For calculation of correlation coefficients, measurements from the mPlex-Flu assay using various VAMS samples were either combined across multiple influenza virus types or separated by influenza virus type and subject.
Subject demographic differences were analyzed using the binomial exact test. Because the sample size is small and the data were not normally distributed, we used GEE models with identity link functions [29] to compare the mean measurements from the mPlex-Flu assay results obtained with VAMS versus conventional serum sampling under different room temperature storage times and shipping methods. GEE models with identity link functions were also used to build the relationship between the estimated serum concentration ([IgG S]) and the VAMS blood concentration ([IgG VAMS]. The within-subject correlations were accounted for using compound symmetry variance–covariance matrix.
Statistical analysis software SAS v9.4 (SAS Institute, Inc., Cary, NC) and R version 3.5.1 were used for all the data analysis. The significance level for all tests was set at P = 0.05.
Results
Subject Demographics
Twenty-one healthy volunteers were recruited for this study and their demographics are shown in Supplementary Table 1. More female subjects took part in this study (71%) than male (P = 0.0784), with majority of volunteers being Caucasian (90%; P = 0.002). The distribution of age groups is relatively uniform with fewer volunteers ≤20 years of age.
mPlex-Flu Assay Results from VAMS and Serum Sampling are Highly Correlated
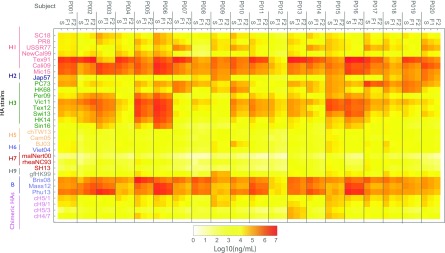
In order to compare the variability of mPlex-Flu results between capillary blood VAMS versus venous serum sampling, we measured anti-influenza IgG concentrations using the mPlex-Flu assay on simultaneous VAMS finger stick and serum from venipuncture samples (n = 20 subjects). The results are shown in heatmap form in Fig. 2.
Fig. 2.
Anti-HA IgG antibody concentration against 30 influenza virus strains assessed by mPlex-Flu assay. The blood samples of 20 subjects were collected by phlebotomy serum sampling (S), VAMS sampling on-site (F1), and VAMS sampling at-home (F2) were tested by mPlex-Flu assay with a 30 influenza virus HA panel in the same 1:5,000 dilution. The IgG concentrations of samples were calculated based on a standard curve for individual virus strain generated by standard serum with Bio-Plex Manager 6.2 software. The mean concentration of duplicates are shown in the heatmap. HA = hemagglutinin; VAMS = volumetric absorptive microsampling. Influenza strain full names are provided in Table 1.
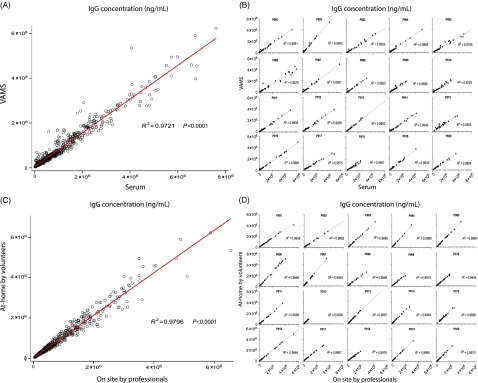
To compare the anti-HA IgG concentrations from mPlex-Flu in samples obtained by VAMS versus traditional phlebotomy, we used the Spearman’s correlation coefficient [27] with the Benjamini–Hockberg multiple testing correction method [28]. We found a high overall correlation of the mPlex-Flu results between the two sampling methods (r = 0.9721;P < 0.001) (Fig. 3A) with n = 20 subjects assayed for anti-HA IgG against 30 strains of influenza (total n = 620 data points).
Fig. 3.
Correlation analysis. (A and B) The correlation of concentration of influenza virus IgG antibodies against 30 strains of influenza virus by mPlex-Flu assay using VAMS sampling versus venous serum sampling. (A) The overall correlation (n = 620); (B) the analysis separated by individual subject (n = 31). (C and D) The correlation of concentration of anti-influenza virus IgG antibodies against 30 strains of influenza virus by mPlex-Flu assay with VAMS finger stick from on-site professionals with that from volunteers at-home. (C) The overall correlation (n = 620). (D) The correlation of concentration of influenza virus antibodies separated by individual subject (n = 31). HA = hemagglutinin; VAMS = volumetric absorptive microsampling.
We also found high correlations when we calculated the (r) values separated by individual influenza virus strains for 20 subjects. All results are listed in Table 2, with the mean r = 0.9470. When we calculated the correlation (r) values separated by individual subject for 30 influenza virus strains, we found the even higher correlation (r) values. The results are shown in Fig. 3B, with the mean r = 0.9836, which is likely due to smaller within-subject variability than between-subject variability. All the above results suggest that mPlex-Flu results from VAMS are highly correlated with that of serum sampling when assessing individual strain-specific anti-HA IgG for individual human subjects.
Table 2.
Correlation between mPlex-Flu anti-HA IgG results: paired samples comparing VAMS versus serum sampling, and on-site versus remote VMAS sampling
| Influenza virus type | Sub types | Abbreviation | VAMS versus Serum | F1 versus F2 * | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Ratio(%) † | SCC | Ratio(%) ‡ | SCC | |||||||
| Mean | SD | r | P corr | Mean | SD | r | P corr | ||||
| A | H1 | SC18 | 20 | 76.21 | 14.40 | 0.9324 | <0.0001 | 110.09 | 17.41 | 0.8787 | <0.0001 |
| PR8 | 20 | 78.15 | 14.79 | 0.9625 | <0.0001 | 110.01 | 15.58 | 0.9214 | <0.0001 | ||
| USSR77 | 20 | 79.46 | 15.56 | 0.9684 | <0.0001 | 109.51 | 15.68 | 0.9423 | <0.0001 | ||
| NewCal99 | 20 | 76.45 | 13.98 | 0.9532 | <0.0001 | 110.01 | 16.71 | 0.9418 | <0.0001 | ||
| Tex91 | 20 | 78.81 | 15.25 | 0.9526 | <0.0001 | 109.49 | 19.42 | 0.9331 | <0.0001 | ||
| Cali09 | 20 | 77.27 | 14.91 | 0.9542 | <0.0001 | 110.10 | 16.23 | 0.9275 | <0.0001 | ||
| Mic15 | 20 | 82.92 | 14.18 | 0.9688 | <0.0001 | 110.39 | 13.43 | 0.9532 | <0.0001 | ||
| H2 | Jap57 | 20 | 87.86 | 22.41 | 0.9684 | <0.0001 | 109.03 | 12.42 | 0.9629 | <0.0001 | |
| H3 | PC73 | 20 | 84.23 | 14.57 | 0.9484 | <0.0001 | 111.18 | 18.55 | 0.9514 | <0.0001 | |
| HK68 | 20 | 77.65 | 13.86 | 0.9685 | <0.0001 | 109.96 | 15.72 | 0.9651 | <0.0001 | ||
| Per09 | 20 | 76.95 | 13.93 | 0.9586 | <0.0001 | 110.35 | 16.18 | 0.9701 | <0.0001 | ||
| Vic11 | 20 | 77.84 | 14.01 | 0.9532 | <0.0001 | 110.85 | 17.14 | 0.9664 | <0.0001 | ||
| Tex12 | 20 | 77.65 | 13.23 | 0.9761 | <0.0001 | 110.16 | 17.39 | 0.9703 | <0.0001 | ||
| Swi13 | 20 | 74.40 | 14.45 | 0.9788 | <0.0001 | 110.79 | 18.98 | 0.9763 | <0.0001 | ||
| HK14 | 20 | 76.18 | 13.87 | 0.9792 | <0.0001 | 110.85 | 17.37 | 0.9790 | <0.0001 | ||
| Sin16 | 20 | 74.88 | 13.88 | 0.9866 | <0.0001 | 110.62 | 16.05 | 0.9907 | <0.0001 | ||
| H5 | Viet04 | 20 | 81.39 | 16.03 | 0.8874 | <0.0001 | 109.00 | 15.90 | 0.9660 | <0.0001 | |
| BJ03 | 20 | 82.20 | 17.97 | 0.9492 | <0.0001 | 109.10 | 14.41 | 0.9571 | <0.0001 | ||
| H6 | chTW13 | 20 | 81.61 | 17.82 | 0.9684 | <0.0001 | 109.89 | 15.40 | 0.9028 | <0.0001 | |
| H7 | malNert00 | 20 | 80.43 | 16.83 | 0.9416 | <0.0001 | 109.99 | 15.89 | 0.9503 | <0.0001 | |
| rheaNC93 | 20 | 95.81 | 30.95 | 0.9502 | <0.0001 | 110.56 | 12.87 | 0.9674 | <0.0001 | ||
| SH13 | 20 | 82.29 | 20.00 | 0.9501 | <0.0001 | 112.42 | 15.79 | 0.9765 | <0.0001 | ||
| H9 | gfHK99 | 20 | 95.60 | 28.62 | 0.9641 | <0.0001 | 109.96 | 14.31 | 0.9711 | <0.0001 | |
| B | B/Bris08 | 20 | 84.54 | 21.35 | 0.9604 | <0.0001 | 109.30 | 16.42 | 0.9052 | <0.0001 | |
| B/Mass12 | 20 | 86.05 | 14.80 | 0.9547 | <0.0001 | 109.25 | 14.70 | 0.9515 | <0.0001 | ||
| B/Phu13 | 20 | 96.35 | 19.67 | 0.9679 | <0.0001 | 111.80 | 17.08 | 0.9630 | <0.0001 | ||
| Chimeric HA | Group 1 | cH5/1 | 20 | 77.91 | 16.23 | 0.9164 | <0.0001 | 108.44 | 15.34 | 0.9030 | <0.0001 |
| Stalk | cH9/1 | 20 | 84.92 | 19.84 | 0.9142 | <0.0001 | 109.41 | 14.85 | 0.9072 | <0.0001 | |
| Group 2 | cH5/3 | 20 | 79.70 | 26.15 | 0.9679 | <0.0001 | 111.65 | 15.59 | 0.9817 | <0.0001 | |
| Stalk | cH4/7 | 20 | 83.10 | 19.90 | 0.9451 | <0.0001 | 110.12 | 15.25 | 0.9589 | <0.0001 | |
Influenza strain full names are provided in Table 1.
HA = hemagglutinin; VAMS = volumetric absorptive microsampling; SCC = Spearman’s correlation coefficient.
F1: the finger-stick VAMS done by study coordinators on-site; F2:the finger-stick VAMS done by volunteers at-home.
The ratio of anti-influenza virus HA IgG of VAMS sampling to that of serum sampling, expressed as a percentage (%).
The ratio of anti-influenza virus HA IgG concentration F1 (on-site) to that of F2 (remote), expressed as a percentage (%).
Adjustment of mPlex-Flu Results for Estimated Serum Volume
To compare the IgG concentrations measured by mPlex-Flu in VAMS versus traditional phlebotomy sampling, we calculated the ratio of mPlex-Flu values from VAMS sampling as a fraction of that from serum for each strain:
| (5) |
where the subscript V denotes a VAMS sample and S denotes a serum sample. We found that the mean of ratio for all anti-HA IgG measurements of R V/S was 81.63% (Table 2), which is <1.0. This result is consistent with the fact that the denominator used to calculate [IgGS] is serum volume, which is a fraction the whole blood volume in the VAMS sample. This would result in lower [IgGVAMS] compared to [IgGS]. To adjust the estimate of [IgGVAMS] for the actual serum volume in the VAMS sample, we next estimated the serum fraction (i.e. (1 – HCT)) using the hemaglobin concentration of VAMS samples (see Methods). As expected, we found that Hgb is highly correlated with HCT (Fig. 4A), and that the RV/S is inversely proportional to Hgb, as expected (Fig. 4B). These data were then used to construct a GEE for [IgGS] as a function of [IgGVAMS] and HCT (see Methods).
Fig. 4.
Relationship between sample Hgb, HCT, and correction of the IgGVAMS concentration. (A) The correlation of HCT (determined using an automated counter) with Hgb measured from experimental whole blood samples. (B) The correlation between the ratio of anti-HA IgG from VAMS versus serum samples with Hgb concentration (n = 5 subjects). HA = hemagglutinin; VAMS = volumetric absorptive microsampling.
To validate this model, we calculated the estimated values [IgGES] for the VAMS samples of the 20 subjects with measured [IgGVAMS] and [Hgb], then compared the estimates with the measured [IgGS] values. The adjusted ratios RV/S of 30 HA influenza strain antibody concentration improved from range 74.40–95.35% (mean 81.63%) to range 82.93–109.3% (mean 90.22%) (see Table 3). [IgGES] values were highly correlated with the actual [IgGS] values and correlation coefficients R are shown in Table 3. Overall, the results demonstrate that the estimated [IgGES] is closer to the measured [IgGS] value than the uncorrected [IgGVAMS].
Table 3.
VAMS and serum measurement ratio comparisons pre- and post-adjustment for hematocrit and correlations between predicted and measured serum IgG concentrations
| Influenza virus type | Subtypes | Abbreviation | N | Pre-adjustment | Post-adjustment | |||
|---|---|---|---|---|---|---|---|---|
| RV/S(%) | RV/S(%) | SCC | ||||||
| Mean | SD | Mean | SD | r | ||||
| A | H1 | SC18 | 20 | 76.21 | 14.40 | 84.42 | 13.10 | 0.8481 |
| PR8 | 20 | 78.15 | 14.79 | 86.42 | 13.17 | 0.9188 | ||
| USSR77 | 20 | 79.46 | 15.56 | 88.10 | 14.07 | 0.9744 | ||
| NewCall99 | 20 | 76.45 | 13.98 | 84.87 | 13.09 | 0.9444 | ||
| Tex91 | 20 | 78.81 | 15.25 | 87.39 | 13.04 | 0.9669 | ||
| Cali09 | 20 | 77.27 | 14.91 | 86.05 | 13.97 | 0.9383 | ||
| Mic15 | 20 | 82.92 | 14.18 | 92.16 | 12.77 | 0.9534 | ||
| H2 | Jap57 | 20 | 87.86 | 22.41 | 96.73 | 19.13 | 0.9774 | |
| H3 | PC73 | 20 | 84.23 | 14.57 | 94.09 | 13.94 | 0.9579 | |
| HK68 | 20 | 77.65 | 13.86 | 86.26 | 13.14 | 0.9820 | ||
| Per09 | 20 | 76.95 | 13.93 | 85.49 | 14.13 | 0.9774 | ||
| Vic11 | 20 | 77.84 | 14.01 | 86.46 | 13.23 | 0.9398 | ||
| Tex12 | 20 | 77.65 | 13.23 | 85.94 | 13.29 | 0.9669 | ||
| Swi13 | 20 | 74.40 | 14.45 | 82.61 | 13.71 | 0.9459 | ||
| HK14 | 20 | 76.18 | 13.87 | 84.48 | 13.19 | 0.9549 | ||
| Sin16 | 20 | 74.88 | 13.88 | 82.93 | 12.73 | 0.9744 | ||
| H5 | Viet04 | 20 | 81.39 | 16.03 | 90.88 | 14.41 | 0.9489 | |
| BJ03 | 20 | 82.20 | 17.97 | 89.79 | 16.23 | 0.9504 | ||
| H6 | chTW13 | 20 | 81.61 | 17.82 | 90.16 | 15.09 | 0.9323 | |
| H7 | malNert00 | 20 | 80.43 | 16.83 | 88.43 | 15.12 | 0.9278 | |
| rheaNC93 | 20 | 95.81 | 30.95 | 103.89 | 28.55 | 0.9579 | ||
| SH13 | 20 | 82.29 | 20.00 | 90.03 | 17.16 | 0.9143 | ||
| H9 | gfHK99 | 20 | 95.60 | 28.62 | 105.37 | 27.75 | 0.9429 | |
| B | B/Bris08 | 20 | 84.54 | 21.35 | 94.12 | 19.94 | 0.9098 | |
| B/Mass12 | 20 | 86.05 | 14.80 | 95.24 | 13.05 | 0.9729 | ||
| B/Phu13 | 20 | 96.35 | 19.67 | 107.4 | 20.63 | 0.9549 | ||
| Chimeric HA | Group 1 | cH5/1 | 20 | 77.91 | 16.23 | 85.39 | 13.93 | 0.9278 |
| Stalk | cH9/1 | 20 | 84.92 | 19.84 | 93.79 | 19.08 | 0.9128 | |
| Group 2 | cH5/3 | 20 | 79.70 | 26.15 | 86.75 | 22.13 | 0.9068 | |
| Stalk | cH4/7 | 20 | 83.10 | 19.90 | 90.97 | 17.85 | 0.8737 | |
| Average | 81.63 | 90.22 | ||||||
Influenza strain full names are provided in Table 1.
HA = hemagglutinin; VAMS = volumetric absorptive microsampling; SCC = Spearman’s correlation coefficient.
RV/S = the ratio of anti-influenza virus HA IgG concentration of VAMS sampling to that of serum sampling, expressed as a percentage (%).
VAMS is a Highly Reproducible Method When Performed at-Home
One advantage of the VAMS method is the safety and simplicity of the process. It is easy for study volunteers to learn and perform at-home. Previously published data have shown that the volume of blood captured in the 10 µL VAMS device varies <0.4 µL [17]. But no study has shown the reputability of VAMS sampling by participants at-home compared to on-site by a nurse in the mPlex-Flu assay to evaluate influenza virus antibodies. To estimate this correlation, a second finger-stick collection was performed by the 20 subjects at-home 3 days later. These samples were then hand-delivered back to the laboratory in a provided envelope.
Using the same analytic approach, we calculated the correlation of at-home (F2) and on-site (F1) sampling for measurement of IgG-mediated immunity across multiple influenza virus strains, grouped by strain and subject and adjusted using simultaneous Hgb measurements. The results are shown in Table 2, and Figs. 3C and 3D. We found no statistically significant difference between the results obtained with on-site versus at-home VAMS sampling. These data suggest that VAMS sampling could be preformed at-home by the study subjects, as the anti-HA antibody concentrations are highly consistent with sampling performed by study personnel on-site. These results support the consistency of VAMS sampling for future influenza vaccine or infection immunity studies.
Stability of Anti-Influenza HA Antibodies in VAMS Samples
We next examined the stability of anti-influenza virus HA antibodies in samples stored in VAMS device at room temperature, and during transport (e.g. postal service, 2-day express mail). This is an essential aspect of quality control that needs to be addressed for future applications of VAMS. Prior studies have shown that antibodies on DBS filter paper are stable for more than 20 years when stored at 4 or −20°C [16]. To determine the stability of antibodies in the VAMS device over time at room temperature, we used the mPlex-Flu assay to compare the antibody activity of VAMS tips stored at −20°C immediately after drying (control) with other VAMS tips left for 7, 14, 21, or 28 days at room temperature. The results are shown in Fig. 5A. We found no detectable antibody activity decrease at room temperature from storing the VAMS devices at room temperature environment, and antibody measurements still kept 94.5% of the control level up to 21 days. After storing VAMS devices at room temperature to 28 days, the antibody activity level was significantly decreased to 80% of the control levels from the control devices stored at −20°C (statistical results were shown in the table in Fig. 5B).
Fig. 5.
The stability of multiple dimensional IgG antibodies collected with VAMS finger stick stored at room temperature or after shipping. The mean concentration of influenza virus IgG antibodies against 30 strains of influenza virus HA by mPlex-Flu assay is shown in the heatmap (n = 4). USPS = United States Postal Service; FEDEX = Federal Express Overnight Shipping; SD = Standard Deviation; HA = hemagglutinin; VAMS = volumetric absorptive microsampling. Influenza strain full names are provided in Table 1.
To confirm the stability of the antibodies during the shipping process, we compared two commonly used shipping methods: standard first-class mail and 2-day commercial shipping (Federal Express, FedEx) to send two duplicate groups of the VAMS devices to our lab in New York State in August. USPS first-class mail took 5 days and commercial shipping took 2 days. After the samples were received back at the lab, the anti-HA IgG antibody levels were evaluated by mPlex-Flu assay (Fig. 5A). No statistically significant difference was detected between results from samples transported via the two shipping methods (Fig. 5B), suggesting that the VAMS samples are stable during shipping process (2–5 days) even during the summer time, when temperatures may be elevated.
Discussion
In this report, we have demonstrated the utility of capillary VAMS sampling, combined with the mPlex-Flu assay, for measuring anti-influenza HA IgG antibody levels. This combination addresses a significant translational barrier in influenza research and population health research: how to measure antibody-mediated influenza immunity in a large number of subjects at modest expense using a minimally invasive method for sample collection. The VAMS sampling method is inexpensive, can be used remotely by study volunteers, requires only a finger stick, and yields consistent results compared to serum samples obtained with standard phlebotomy. VAMS samples are stable at room temperature for up to 21 days, and during standard 2–5 day shipping at ambient temperatures. Combined with the mPlex-Flu method, we are able to measure IgG reactivity against more than 30 influenza strains simultaneously from a 10-µL VAMS finger-stick sample [8].
We have also demonstrated that the VAMS-mPlex Flu method overcomes an issue with the use of traditional DBS methods for estimating the concentration of analytes distributed only in the serum compartment [30], such as IgG. Serum typically accounts for ∼45–80% of total blood volume, and thus whole blood concentrations of IgG will be lower than serum estimates, and vary by test subject HCT. This could lead to underestimation of vaccine infection-induced changes in IgG in clinical trials or research studies. We have shown here that simultaneous measurement of Hgb content in VAMS samples can be used to estimate serum volume and to improve estimates of serum [IgG]. One minor caveat is that even with this adjustment, the ratio of IgG concentrations of samples obtained by VAMS versus serum was still RV/S ≠ 1.0. This could be due to a variety of factors, including variation in microcapillary sampling by gender, Hgb retained on the VAMS device out of proportion to IgG, or other factors. Thus, further work could be done to improve the estimating equations in larger population studies. Nevertheless, the improved correlation and prediction of [IgGS] from [IgGV] suggests that this relatively simple method may be adequate for many studies.
It is also important to note that serum volume correction may not be needed when comparing the ratio of pre- to post-vaccine anti-HA IgG concentrations over a short time period, and in the same subject, when plasma volume is unlikely to change significantly. This is likely the case for measurements within 30 days of vaccination, which constitute a large fraction of the samples collected in current influenza vaccine research studies. However, conditions in which the red cell volume is increased (erythrocytosis) or markedly decreased (sickle cell disease, β -thallasemia, blood loss anemia) may affect between subject comparisons [19,31]. Thus studies with large population comparisons between subjects, and over long time periods in diverse populations, may benefit from this adjustment to avoid underestimating absolute anti-HA IgG concentrations. Finally, whether such differences in how IgG concentrations are expressed have clinical relevance in vaccine immunity studies is currently unclear.
We suggest that this method may improve future studies of the longitudinal persistence of IgG-mediated influenza immunity. It is known that IgG-mediated influenza immunity can decrease after vaccination over months, but vaccine trials and research studies rarely sample beyond 90 days post-vaccination [22]. A major translational issue in vaccine trials is the expense and difficulty of having study subjects come to a study center for phlebotomy to monitor vaccine responses. A combined VAMS + mPlex-Flu methodology could allow for longer term remote sample collection from study subjects, along with assay of IgG-mediated anti-influenza antibody levels. This would greatly lower the cost and difficulty of such monitoring, and aid in current research initiatives to develop a universal influenza vaccine [32], to understand the persistence of vaccine-mediated immunity [33], and to predict who will respond to influenza vaccines long term [34].
The combination of VAMS sampling and mPlex-Flu analysis also has the potential to reduce translational barriers to large-scale population studies of antibody-mediated influenza immunity [2,24,35]. This method would enable remote subject enrollment, consent, and sample collection across large populations and disparate geographic areas. In addition, this method also addresses a significant translational barrier to determining the true efficacy of the seasonal influenza vaccine, which is based on data collected by influenza surveillance field teams. Currently, WHO and CDC field surveillance teams only collect vaccine history and swabs for influenza strain genotyping. Providing WHO and CDC teams with VAMS collection kits when surveilling influenza-like illness cases in the field, especially in remote areas, would allow public health agencies to determine if infected subjects actually had, or lacked, antibody-mediated immunity against the circulating influenza strains [36]. This is not currently done on any large scale and would provide much more accurate information regarding vaccine efficacy. Finally, the mPlex-Flu assay provides rich, multidimensional data regarding IgG anti-HA reactivity. Informatics methods can be used to calculate the antigenic distance between different influenza strain HAs using sequence comparisons. When combined with anti-HA IgG levels from the study of very large populations, such data could be used to create large-scale HA antigenic landscapes [37,38] for future influenza immunity research.
Finally, we also would like to note that this approach is not pathogen-specific, but could be adapted to estimating antibody-mediated immunity after vaccination for any other viral pathogens, where the key antibody targets are known and broad populations need to be surveyed. In addition, other biologic data could easily be obtained from a second VAMS device (another 10 µL of blood), including DNA samples for genomic sequencing of the host or blood-borne pathogens. Such methods are likely to be increasingly used in clinical trials in remote and rural areas, as well as newer “siteless” clinical trials [39,40].
Acknowledgments
We would like to thank all the study volunteers without whom this work would not be possible. We also are very grateful for the outstanding study coordination by Susanne Heininger, the excellent technical assistance from the research coordinators at the University of Rochester Clinical Research Center, and Jonathan Kasper and Eric Mendelson for their excellent technical assistance with the hemaglobin assays. In addition, we would also like to thank the reviewer who suggested correcting the IgG measurements from VAMS specimens for serum volume. This greatly improved both the manuscript and the utility of the method.
Footnotes
Financial Support
This work was supported by the National Institutes of Health Institute of Allergy, Immunology and Infectious Diseases grant R21 AI138500 (MZ, JW), and the University of Rochester Clinical and Translational Science Award UL1 TR002001 from the National Center for Advancing Translational Sciences of the National Institutes of Health (JW, DL, MZ). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. None of the above funders had any role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/cts.2019.410.
click here to view supplementary material
Data Availability
The primary data generated by this study are available from https://figshare.com/s/7f8992762e960e0aa023
Author Contributions
JW, SPH, and MSZ conceived of the investigational plan, designed the experiments and analytic methods. JW, AW, and JE carried out the experiments. JW, DL, JE, and MSZ analyzed the data. JW, DL, AW, JE, SPH, and MSZ wrote and edited the manuscript.
Disclosures
The authors declare no conflicts of interest.
References
- 1. Jayasundara K, et al. Natural attack rate of influenza in unvaccinated children and adults: a meta-regression analysis. BMC Infectious Diseases 2014; 14: 670. doi: 10.1186/s12879-014-0670-5 Available from: https://www.ncbi.nlm.nih.gov/pubmed/25495228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flannery B, et al. Early estimates of seasonal influenza vaccine effectiveness - United States, January 2015. Morbidity and Mortality Weekly Report 2015; 64(1): 10–5. Available from: https://www.ncbi.nlm.nih.gov/pubmed/25590680. [PMC free article] [PubMed] [Google Scholar]
- 3. Ohmit SE, et al. Influenza hemagglutination-inhibition antibody titer as a correlate of vaccine-induced protection. Journal of Infectious Diseases 2011; 204(12): 1879–1885. doi: 10.1093/infdis/jir661 Available from: https://www.ncbi.nlm.nih.gov/pubmed/21998477. [DOI] [PubMed] [Google Scholar]
- 4. Gamblin SJ, Skehel JJ. Influenza hemagglutinin and neuraminidase membrane glycoproteins. Journal of Biological Chemistry 2010; 285(37): 28403–28409. doi: 10.1074/jbc.R110.129809 Available from: https://www.ncbi.nlm.nih.gov/pubmed/20538598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lim MD. Dried blood spots for global health diagnostics and surveillance: opportunities and challenges. The American Journal of Tropical Medicine and Hygiene 2018; 99(2): 256–265. doi: 10.4269/ajtmh.17-0889 Available from: http://www.ajtmh.org/content/journals/10.4269/ajtmh.17-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kok MGM, Fillet M. Volumetric absorptive microsampling: current advances and applications. Journal of Pharmaceuticals and Biomedical Analysis 2018; 147: 288–296. doi: 10.1016/j.jpba.2017.07.029 Available from: https://www.ncbi.nlm.nih.gov/pubmed/28803682. [DOI] [PubMed] [Google Scholar]
- 7. Wang J, et al. Multi-dimensional measurement of antibody-mediated heterosubtypic immunity to influenza. PLoS One 2015; 10(6): e0129858. doi: 10.1371/journal.pone.0129858 Available from: https://www.ncbi.nlm.nih.gov/pubmed/26103163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang J, et al. Broad cross-reactive IgG responses elicited by adjuvanted vaccination with recombinant influenza hemagglutinin (rHA) in ferrets and mice. PLoS One 2018; 13(4): e0193680. doi: 10.1371/journal.pone.0193680 Available from: https://www.ncbi.nlm.nih.gov/pubmed/29641537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li D, et al. Continuous readout versus titer based assays of influenza vaccine trials: sensitivity, specificity and false discovery rates. Computational and Mathematical Methods in Medicine 2019; 2019: 9287120. doi: 10.1155/2019/9287120 Available from: https://www.ncbi.nlm.nih.gov/pubmed/31205481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li D, et al. Improved specificity and false discovery rates for multiplex analysis of changes in strain specific anti-influenza IgG. Computational and Mathematical Methods in Medicine 2019; 2019: 2019:3053869. doi: 10.1155/2019/3053869 Available from: https://www.ncbi.nlm.nih.gov/pubmed/31178920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang J, et al. IL-15 enhances cross-reactive antibody recall responses to seasonal H3 influenza viruses in vitro. F1000Res 2017; 6: 2015. doi: 10.12688/f1000research.12999.1 Available from: https://www.ncbi.nlm.nih.gov/pubmed/29479423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tesini BL, et al. Broad hemagglutinin-specific memory B cell expansion by seasonal influenza virus infection reflects early-life imprinting and adaptation to the infecting virus. Journal of Virology 2019; 93(8). doi: 10.1128/JVI.00169-19 Available from: https://www.ncbi.nlm.nih.gov/pubmed/30728266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guthrie RK, Roach AW, Ferguson JK. Serology of aerobic aquatic actinomycetes. Ii. Effects of growth medium and culture age on antigen levels. Journal of Bacteriology 1963; 86: 173–178. Available from: https://www.ncbi.nlm.nih.gov/pubmed/14058938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hannon WH, Lewis DS, Powell MK. A quality assurance program for human immunodeficiency virus seropositivity screening of dried-blood spot specimens. Infection Control & Hospital Epidemiology 1988; 9(8): 361 Available from: https://www.ncbi.nlm.nih.gov/pubmed/3171134. [DOI] [PubMed] [Google Scholar]
- 15. Nicoll A, et al. The public health applications of unlinked anonymous seroprevalence monitoring for HIV in the United Kingdom. International Journal of Epidemiology 2000; 29(1): 1–10. Available from: https://www.ncbi.nlm.nih.gov/pubmed/10750597. [DOI] [PubMed] [Google Scholar]
- 16. Williams D, et al. Long-term stability of HIV-1 antibody in dried blood spot samples and eluates. AIDS 2014; 28(12): 1850–1851. doi: 10.1097/QAD.0000000000000348 Available from: https://www.ncbi.nlm.nih.gov/pubmed/25006831. [DOI] [PubMed] [Google Scholar]
- 17. Spooner N, et al. A device for dried blood microsampling in quantitative bioanalysis: overcoming the issues associated blood hematocrit. Bioanalysis 2015; 7(6): 653–659. doi: 10.4155/bio.14.310 Available from: https://www.ncbi.nlm.nih.gov/pubmed/25514576. [DOI] [PubMed] [Google Scholar]
- 18. Lewis SM, Bain B, Bates I. Dacie and Lewis Practical Haematology. Churchill Livingstone, Philadelphia, Penns., 2006. doi: 10.1016/B0-443-06660-4/X5001-6 [DOI] [Google Scholar]
- 19. Quint L, et al. Relationship between haemoglobin and haematocrit in the definition of anaemia. Tropical Medicine & International Health 2006; 11(8): 1295–1302. doi: 10.1111/j.1365-3156.2006.01679.x Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1365-3156.2006.01679.x. [DOI] [PubMed] [Google Scholar]
- 20. Ziemann M, et al. Reliability of capillary hemoglobin screening under routine conditions. Transfusion 2011; 51(12): 2714–2719. doi: 10.1111/j.1537-2995.2011.03183.x Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/j.1537-2995.2011.03183.x. [DOI] [PubMed] [Google Scholar]
- 21. Spooner N, et al. A device for dried blood microsampling in quantitative bioanalysis: overcoming the issues associated blood hematocrit. Bioanalysis 2015; 7(6): 653–659. doi: 10.4155/bio.14.310 Available from: https://www.ncbi.nlm.nih.gov/pubmed/25514576. [DOI] [PubMed] [Google Scholar]
- 22. Osterholm MT, et al. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. The Lancet Infectious Diseases 2012; 12(1): 36–44. doi: 10.1016/S1473-3099(11)70295-X Available from: https://www.ncbi.nlm.nih.gov/pubmed/22032844. [DOI] [PubMed] [Google Scholar]
- 23. Nayak JL, et al. Effect of influenza A(H5N1) vaccine prepandemic priming on CD4+ T-cell responses. The Journal of Infectious Diseases 2015; 211(9): 1408–1417. doi: 10.1093/infdis/jiu616 Available from: https://www.ncbi.nlm.nih.gov/pubmed/25378637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Layne SP. Human influenza surveillance: the demand to expand. Emerging Infectious Diseases 2006; 12(4): 562–568. doi: 10.3201/eid1204.051198 Available from: https://www.ncbi.nlm.nih.gov/pubmed/16704802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hardin JW, Hilbe JM. Generalized Estimating Equations. Boca Raton, FL: Chapman Hall/CRC; 2003. Available from: http://www.loc.gov/catdir/enhancements/fy0646/2002067404-d.html.
- 26. Wolfinger RD. Heterogeneous variance: covariance structures for repeated measures. Journal of Agricultural, Biological, and Environmental Statistics 1996; 1: 205. doi: 10.2307/1400366 [DOI] [Google Scholar]
- 27. Spearman C. The proof and measurement of association between two things. The American Journal of Psychology 1904; 15(1): 72–101. Available from: http://www.jstor.org/stable/1412159. [PubMed] [Google Scholar]
- 28. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological) 1995; 57(1): 289–300. [Google Scholar]
- 29. Dobson A, et al. An Introduction to Generalized Linear Models. Chapman and Hall/CRC Press, New York, 2008. doi: 10.1201/9781584889519. [DOI] [Google Scholar]
- 30. Cable RG, et al. The difference between fingerstick and venous hemoglobin and hematocrit varies by sex and iron stores. Transfusion 2012; 52(5): 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. De Kesel PM, et al. Hemato-critical issues in quantitative analysis of dried blood spots: challenges and solutions. Bioanalysis 2013; 5(16): 2023–2041. [DOI] [PubMed] [Google Scholar]
- 32. Paules CI, Fauci AS. A universal flu vaccine is vital. Scientific American 2018; 318(2): 8. [DOI] [PubMed] [Google Scholar]
- 33. Foppa IM, et al. Vaccination history as a confounder of studies of influenza vaccine effectiveness. Vaccine: X 2019; 1: 100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Devarajan P, Swain SL. Original antigenic sin: friend or foe in developing a broadly cross-reactive vaccine to influenza? Cell Host & Microbe 2019; 25(3): 354–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jester B, et al. Mapping of the US domestic influenza virologic surveillance landscape. Emerging Infectious Diseases 2018; 24(7): 1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hay AJ, McCauley JW. The WHO global influenza surveillance and response system (GISRS) A future perspective. Influenza and Other Respiratory Viruses 2018; 12(5): 551–557. doi: 10.1111/irv.12565 Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/irv.12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Smith DJ, et al. Mapping the antigenic and genetic evolution of influenza virus. Science 2004; 305(5682): 371–376. doi: 10.1126/science.1097211 Available from: https://www.ncbi.nlm.nih.gov/pubmed/15218094. [DOI] [PubMed] [Google Scholar]
- 38. Fonville JM, et al. Antibody landscapes after influenza virus infection or vaccination. Science 2014; 346(6212): 996–1000. doi: 10.1126/science.1256427 Available from: https://www.ncbi.nlm.nih.gov/pubmed/25414313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cutts FT, et al. Operational challenges in large clinical trials: examples and lessons learned from the Gambia pneumococcal vaccine trial. PLoS Clinical Trials 2006; 1(3): e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hirsch IB, et al. Incorporating site-less clinical trials into drug development: a framework for action. Clinical Therapeutics 2017; 39(5): 1064–1076. doi: 10.1016/j.clinthera.2017.03.018 Available from: http://www.sciencedirect.com/science/article/pii/S014929181730200X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/cts.2019.410.
click here to view supplementary material
Data Availability Statement
The primary data generated by this study are available from https://figshare.com/s/7f8992762e960e0aa023