Abstract
BACKGROUND
Long noncoding RNAs (lncRNAs) are aberrant and play critical roles in gastric cancer (GC) progression and metastasis. Searching for coexpressed lncRNA clusters or representative biomarkers related to malignant phenotypes of GC may help to elucidate the mechanism of tumor development and predict the prognosis of GC.
AIM
To investigate the prognostic value of NOTCH1 associated with lncRNA in T cell acute lymphoblastic leukemia 1 (NALT1) in GC and the mechanism of its involvement in GC invasion and metastasis.
METHODS
RNA sequencing and corresponding clinical data were downloaded from The Cancer Genome Atlas database. The significance module was studied by weighted gene coexpression network analysis. A total of 336 clinical samples were included in the study. Gene silencing, reverse transcription polymerase chain reaction, western blotting, scrape motility assay, and Transwell migration assay were used to assess the function of hub-lncRNAs.
RESULTS
At the transcriptome level, 3339 differentially expressed lncRNAs were obtained. weighted gene coexpression network analysis was used to obtain 15 lncRNA clusters and observe their coexpression. Pearson’s correlation showed that blue module was correlated with tumor grade and survival. NALT1 was the hub-lncRNA of blue module and was an independent risk factor for GC prognosis. NALT1 was overexpressed in GC and its expression was closely related to invasion and metastasis. The mechanism may involve NALT1 regulation of NOTCH1, which is associated with lncRNA in T cell acute lymphoblastic leukemia, through cis regulation, thereby affecting the expression of the NOTCH signaling pathway.
CONCLUSION
NALT1 is overexpressed and promotes invasion and metastasis of GC. The mechanism may be related to regulation of NOTCH1 by NALT1 and its effect on NOTCH signaling pathway expression.
Keywords: Gastric cancer, Weighted gene coexpression network analysis, NALT1, NOTCH1, Cancer survival
Core tip: Analysis of the Cancer Genome Atlas and clinical data revealed that the long noncoding RNA (lncRNA) NOTCH1 associated with lncRNA in T cell acute lymphoblastic leukemia 1 (NALT1) was associated with poor prognosis of gastric cancer (GC). We investigated the possible mechanism of this association. We downloaded the Cancer Genome Atlas-Stomach Adenocarcinoma RNA-seq data and identified the differentially expressed lncRNAs. Weighted gene coexpression network analysis was used to clarify the connection between modules and clinical information. We then chose lncRNA NALT1 as the hub-lncRNA of blue-module. NALT1 was overexpressed in GC and promoted invasion and metastasis of GC. The over-expression of NALT1 was linked to poor prognosis in GC. The mechanism may be related to the regulation of NOTCH1 by NALT1 and its effect on the expression of NOTCH signaling pathway.
INTRODUCTION
Gastric cancer (GC) is one of the most common digestive system malignant tumors and poses a serious threat to human health[1]. Long-term survival of GC is still poor due to the high recurrence and distal metastasis rates, despite advances in diagnosis and treatment[2]. Therefore, it is necessary to explore new biomarkers and clarify relevant molecular mechanisms for the formulation of GC treatment strategies. With the completion of Human Genome Project and the parallel development of next-generation sequencing, human transcriptome analysis has revealed that > 98% of the transcriptional output encodes noncoding RNAs (ncRNAs), which have little or even no protein-coding capability[3]. Long noncoding RNAs (lncRNAs), with a length not less than 200 nucleotides, are dominant in the ncRNA family[4]. Accumulated studies have indicated that dysregulation of lncRNAs plays a crucial role in regulation of oncogenes or tumor suppressor genes[5]. LncRNAs are involved in cellular biological processes at multiple levels, including epigenetics, transcription, and post-transcriptional regulation[6]. Therefore, it is believed that the complex regulatory network between mRNA and lncRNAs may mediate the malignant phenotype of GC. For example, lncRNA GClnc1 acts as a “scaffold” to recruit the WDR5 and KAT2A complex and thus induces tumorigenesis, metastasis, and poor prognosis in GC[7]. HOXC-AS3 activated by abnormal histone modification may play an important role in regulation of GC cell proliferation and migration[8].
As a large multiplatform database of molecular maps and clinicopathological annotated data, The Cancer Genome Atlas (TCGA) facilitates the analysis of molecules associated with various clinicopathological parameters of cancer[9]. Using weighted gene coexpression network analysis (WGCNA), coexpression modules or networks of genes can be established to explore the correlation between different gene clusters and clinicopathological parameters[10]. In this investigation, lncRNAs with abnormal expression in GC were searched using transcriptome data of GC in TCGA database [TCGA-stomach adenocarcinoma (STAD)]. WGCNA was used to construct coexpression networks of differentially expressed lncRNAs (DELs) and to analyze the correlation between lncRNA clusters and clinicopathological parameters. In addition, it was revealed that lncRNA NOTCH1 associated with lncRNA in T cell acute lymphoblastic leukemia 1 (NALT1) was the hub-RNA of blue cluster and overexpressed in both GC tissues and cells. Mechanistic analyses revealed that NALT1 was involved in invasion and metastasis of GC, which was through the NOTCH signaling pathway. We studied the role of NALT1 in GC prognosis because the blue module was closely related to tumor differentiation and survival.
MATERIALS AND METHODS
Data download and DELs analysis
The RNA sequencing dataset and corresponding clinical records of GC were downloaded from TCGA (TCGA-STAD, https://portal.gdc.cancer.gov/). The RNA expression profile raw data were download by RTCGA Toolbox package in R platform. There were 406 samples, with 375 GC tissues and 31 normal tissues. Clinical data of 375 tumor tissue specimens were complete. Package edgeR was used to identify DELs. P < 0.05 and |log2FC| ≥ 1 were set as the cutoff criteria.
Construction of WGCNA
WGCNA package was used to construct a coexpression network of DELs[11]. The power function, which was dependent on soft-thresholding β, was used to create the weighted adjacency matrix. We transformed the adjacency matrix to topological overlap matrix (TOM). We performed average linkage hierarchical clustering based on per TOM dissimilarity measurement. The minimum size of the gene group for the genes dendrogram was 30, was the deep split, and 0.15 was the cutoff for module dendrogram and merged modules. Pearson’s correlation was used to search for biologically meaningful clusters and to evaluate the correlation between clusters and clinicopathological parameters. Age, tumor stage, grade, and overall survival (OS) were recorded. The module with the highest correlation was selected as the meaningful research cluster. Cytoscape 3.6.1 software (http://www.cytoscape.org/) was used to construct the network[12]. Molecular Complex Detection (MCODE) was used to filter the network module.
Patients and ethics statement
The study was approved by the Ethics Committee, Liaoning Cancer Hospital & Institute. The patients gave signed informed consent before surgery. There were 336 patients who underwent D2 lymph node dissected gastrectomy at Liaoning Cancer Hospital between January 2011 and March 2014, including 236 (70.2%) men and 100 (29.8%) women, with a mean age of 57 years. Patients treated with preoperative chemotherapy or radiotherapy were excluded. Adenosquamous carcinoma or neuroendocrine carcinoma patients were excluded. Tumor staging was based on the 8th edition of the tumor node metastasis (TNM) staging manual (American Joint Committee on Cancer/Union for International Cancer Control, 2017).
Patients with stage IIA and worse were routinely treated with postoperative adjuvant chemotherapy. Table 1 describes the clinicopathological parameters involved in the analysis. Follow-up was conducted every 3-6 mo within 5 years after the operation, including physical examination, pulmonary, abdominal, and pelvic computed tomography, blood count, endoscopic examination, and liver function examination. OS was the time between surgery and final follow-up or death. Disease-free survival (DFS) was the time between the date of resection and first diagnosis of recurrence. The final investigation date was 1 March 2019.
Table 1.
Patients characteristics and univariate analysis, n = 336
| Characteristics | n (%) |
DFS |
OS |
||||
| Mo | P value | F | Mo | P value | F | ||
| Age, median, yr | 0.280 | 1.172 | 0.641 | 0.218 | |||
| ≥ 60 | 178 | 37.73 | 44.80 | ||||
| < 60 | 158 | 40.77 | 45.95 | ||||
| Gender | 0.085 | 2.982 | 0.216 | 1.537 | |||
| Male | 236 (70.2) | 37.59 | 44.35 | ||||
| Female | 100 (29.8) | 42.86 | 47.68 | ||||
| Bormann type | 0.000 | 55.616 | 0.000 | 49.755 | |||
| I | 25 | 64.00 | 64.00 | ||||
| II | 147 | 45.92 | 51.50 | ||||
| III | 159 | 29.62 | 37.20 | ||||
| IV | 5 | 19.60 | 29.80 | ||||
| Tumor size | 0.000 | 26.829 | 0.000 | 20.674 | |||
| ≥ 5 cm | 142 | 30.99 | 38.99 | ||||
| < 5 cm | 194 | 45.13 | 49.99 | ||||
| Location | 0.142 | 3.899 | 0.325 | 2.247 | |||
| Up | 75 | 34.15 | 41.80 | ||||
| Middle | 90 | 42.04 | 47.60 | ||||
| Low | 171 | 39.84 | 45.70 | ||||
| Tumor histological morphology | 0.788 | 0.476 | 0.870 | 0.277 | |||
| Adenocarcinoma | 260 | 39.30 | 45.33 | ||||
| Mixed carcinoma | 75 | 38.35 | 45.13 | ||||
| Absolute signet ring cell carcinoma | 1 | 35.00 | 45.00 | ||||
| Lauren type | 0.298 | 2.420 | 0.309 | 2.349 | |||
| Intestinal | 156 | 38.79 | 44.88 | ||||
| Mixed carcinoma | 75 | 35.15 | 41.99 | ||||
| Diffuse | 105 | 42.57 | 48.42 | ||||
| Tumor differentiation | 0.000 | 20.700 | 0.000 | 17.649 | |||
| Moderate and high | 167 | 45.38 | 50.41 | ||||
| Poor | 169 | 33.01 | 40.33 | ||||
| Vessel invasion | 0.000 | 19.213 | 0.000 | 15.243 | |||
| Yes | 98 | 29.85 | 37.40 | ||||
| No | 238 | 42.99 | 48.61 | ||||
| Perineural invasion | 0.000 | 17.809 | 0.000 | 16.686 | |||
| Yes | 84 | 28.45 | 36.83 | ||||
| No | 252 | 42.73 | 48.17 | ||||
| T category | 0.000 | 105.672 | 0.000 | 91.002 | |||
| T1 | 60 | 65.95 | 65.98 | ||||
| T2 | 22 | 58.82 | 61.32 | ||||
| T3 | 12 | 60.25 | 62.00 | ||||
| T4 | 242 | 29.68 | 37.94 | ||||
| N category | 0.000 | 232.168 | 0.000 | 217.417 | |||
| N0 | 139 | 62.74 | 63.47 | ||||
| N1 | 58 | 36.12 | 48.07 | ||||
| N2 | 65 | 23.88 | 33.55 | ||||
| N3 | 74 | 10.66 | 18.16 | ||||
| TNM stage | 0.000 | 218.328 | 0.000 | 204.741 | |||
| I | 69 | 64.52 | 64.55 | ||||
| II | 81 | 63.59 | 66.40 | ||||
| III | 14 | 19.26 | 29.27 | ||||
| IV | 2 | 5.00 | 8.50 | ||||
| NALT1 | 0.000 | 20.645 | 0.000 | 18.035 | |||
| Weak-expression | 159 | 45.24 | 50.30 | ||||
| Over-expression | 177 | 33.69 | 40.88 |
DFS: Disease-free survival; OS: Overall survival.
Cell culture
Human normal gastric epithelial cell line GES-1 and human gastric cancer cell lines SGC-7901 and BGC-823 were obtained from China Medical University (Shenyang, China). Cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, United States), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen) at 37 °C under 5% CO2 and 1% O2. All experiments were repeated three times independently.
Real-time reverse transcription polymerase chain reaction
Total RNA was isolated from cells and tissues with TRIzol cell separation reagent (Invitrogen). Promega cDNA core kit (Madison, WI, United States) was used to generate cDNA from 500 ng total RNA. SYBR Master Mix (Takara Bio, Kusatsu, Japan) was used to perform real-time reverse transcription polymerase chain reaction (RT-PCR) (LightCycler 480; Roche AG, Basel, Switzerland). Each sample was analyzed three times. U6 was the loading control. Fold changes in mRNA expression in different cells were determined by 2–ΔΔCT normalization. Each sample was analyzed in triplicate. The sequence information is listed in Supplementary table 1.
Western blotting
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (10%-15%) and polyvinylidene difluoride membranes were used to separate and transfer protein (20 μg) from cells. TBS-Tween buffer (20 mmol/L Tris–HCl, 5% nonfat milk, 150 mmol/L NaCl, and 0.05% Tween-20, pH 7.5) was used to block membranes for 1 h at 21 °C after blotting. The membranes were incubated with primary antibodies overnight at 4 °C (NOTCH1, SNAI1, 1: 200, Boster Biological Technology; SLUG, MMP9, 1:150, Boster Biological Technology; and β-actin, 1: 4000, Santa Cruz Biotechnology, Santa Cruz, CA, United States). Finally, membranes were incubated with secondary antibody (1: 5000, Santa Cruz Biotechnology). β-Actin acted as a control. The gray value of proteins was measured by ImageJ (NIH, Bethesda, MD, United States). Averages of three independent data were presented as the final results.
Lentiviral vector system, plasmids, and cell transfection
Lentiviruses carrying siRNA sequences targeting human NALT1 were obtained from GeneChem (Shanghai, China). Full-length human NOTCH1 cDNA was subcloned into the pcDNA 3.1 vector. Lipofectamine 2000 Reagent (Invitrogen) was used to transfect the GC cells.
Scrape motility and Transwell invasion assays
Scrape motility assay was used to evaluate cell migration. GC cells were plated into culture inserts (Ibidi, Regensburg, Germany). After 24 h incubation, we removed the inserts. An inverted microscope (XDS-100; Shanghai Caikon Optical Instrument, Shanghai, China) was used to capture the wound monolayers images at 0 and 24 h post-wounding.
Transwell assay was performed to determine cell invasion. Transwell upper chambers coated with gelatin were used to plate GC cells. The lower chambers were coated with 600 μL fetal bovine serum (30%, Costar, Lowell, MA, United States). Methanol, hematoxylin, and eosin were used to fix and stain cells after incubation for 24 h (Sigma–Aldrich, St. Louis, MO, United States). We removed the upper chambers; the cells on the surface of the lower chambers were migrated cells that were counted and captured by microscopy at 100 × magnification in five fields. The average cell number per field represented the migrated cells.
Statistical analysis
Statistical analysis was performed using SPSS version 23.0 (IBM, Armonk, NY, United States). All data are presented as mean ± standard deviation. Data were compared by Student’s t test when homogeneity of variance was satisfied between groups; otherwise, the Wilcoxon-signed rank test was used. Multiple groups were compared using one-way analysis of variance. χ2 test or Fisher’s exact test reflected the correlation between clinicopathological parameters and biomarkers. Kaplan-Meier survival curves were generated from the survival data. Univariate survival analysis was performed by log-rank test. Cox proportional hazard model was used to evaluate the clinical significance of the biomarkers. P < 0.05 was considered to be statistically significant.
RESULTS
DELs in GC
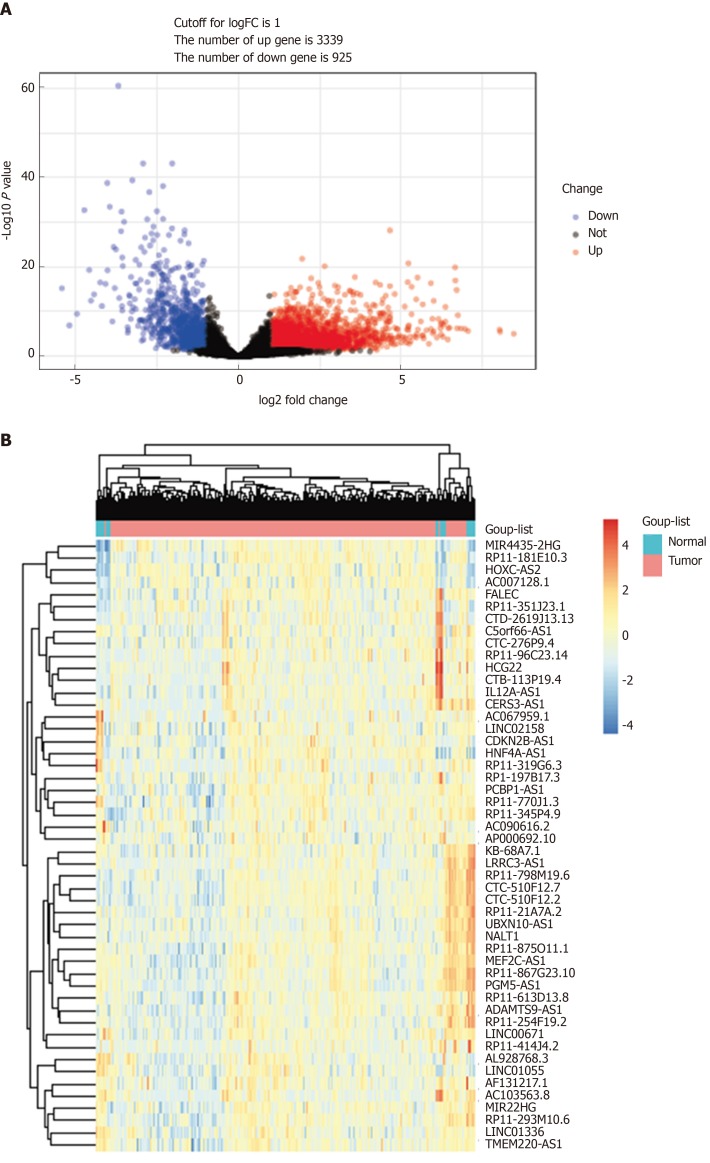
TCGA-STAD RNA sequence dataset included 375 GC samples and 31 para-carcinoma tissues. EdgeR was applied to identify the DELs (fold change ≥ 2, P < 0.05). A total of 4264 DELs were screened from the expression profile data, which included 14449 lncRNAs. Among them, 3339 were upregulated DELs (uDELs) and 925 were downregulated DELs (dDELs) (Figure 1A). All these were further investigated by WGCNA. The volcano showed the top 25 uDELs and dDELs (Figure 1B).
Figure 1.
DELs in gastric cancer. A: Volcano plots of the expression levels of DELs screened by limma; B: Heatmaps of the expression levels of top 50 DELs. DELs: Differentially expressed lncRNAs; lncRNAs: Long noncoding RNAs.
WCGNA and hub-lncRNA identification
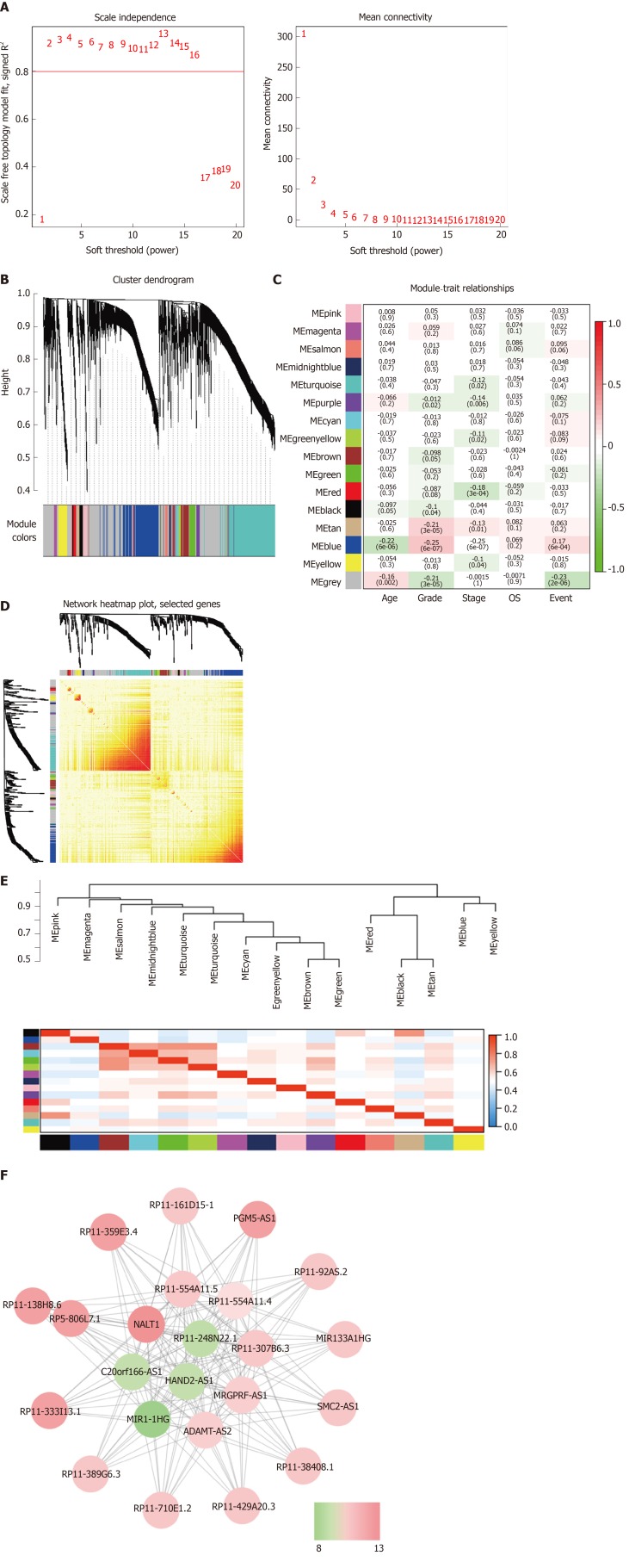
WGCNA was performed to identify the functional RNA cluster of GC patients. The scale-free network was constructed based on 4264 DELs with a soft threshold β = 2 (Figure 2A). A total of 15 RNA clusters were obtained that contained 36–729 DELs (Figure 2B). Correlation analysis showed that the blue module had the most significant correlation with clinical features. It was significantly correlated with grade (r = 0.25, P = 4 × 10–7; Figure 2C) and prognosis (r = 0.17, P = 6 × 10–4; Figure 2C). TOM plot was generated from the TOM plot function to verify module correlation (Figure 2D). Eigengenes were treated as representative profiles to quantify module similarity correlation. The dendrogram and heatmap showed the correlation and eigengene adjacency between modules, respectively (Figure 2E).
Figure 2.
Weighted gene coexpression network analysis identify function module. A: The soft-thresholding powers (β) of scale-free fit index and mean connectivity; B: Cluster dendrogram obtained by hierarchical clustering of adjacency-based dissimilarity; C: Heatmap of the correlation between module and gastric cancer clinical traits; D: Topological overlap matrix plot; E: Hierarchical clustering and heatmap of eigengene adjacency. The color bars on left and below indicate the module of each row or column; F: Weighted coexpression network for blue-module lncRNAs (Node colors are according to the molecular complex detection score). LncRNAs: Long noncoding RNAs.
The gene significance in the blue module was imported into Cytoscape software to construct a WGCN. MCODE was applied to filter the network module and select hub-lncRNAs. Weight > 0.4 and MCODE score > 8 combined were selected as significant. NALT1 had the highest score for follow-up (Figure 2F).
Clinical significance of NALT1
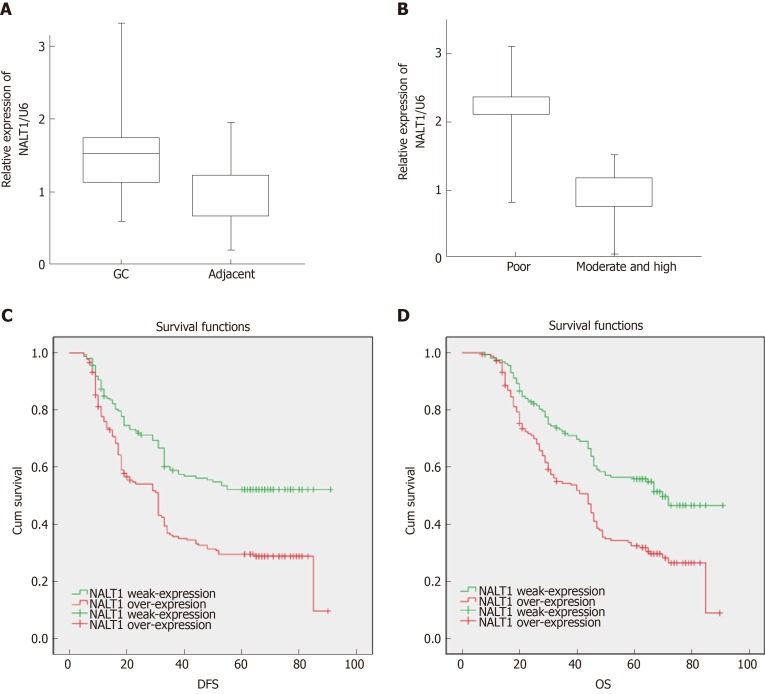
RT-PCR was used to detect expression of NALT1 in 35 paired samples (GC vs adjacent noncancerous samples). It was obvious that NALT1 was overexpressed in GC samples (Figure 3A). As predicted by bioinformatics, expression of NALT1 was closely related to tumor differentiation in 336 GC specimens (Figure 3B).
Figure 3.
The expression of NALT1 in GC tissue and its effect on survival. A: NALT1 expression in GC tissue compare to paired adjacent tissue in 35 patients; B: The relationship between NALT1 expression and tumor differentiation; C: Kaplan–Meier analysis of the correlation between NALT1 expression levels and DFS in 336 GC patients; D: Kaplan–Meier analysis of the correlation between NALT1 expression levels and OS in 336 GC patients. NALT1: NOTCH1 associated with lncRNA in T cell acute lymphoblastic leukemia 1; GC: Gastric cancer; DFS: Disease-free survival; OS: Overall survival.
To explore further the relationship between NALT1 and clinicopathological parameters, we grouped them according to the expression level of NALT1. The median value of NALT1 expression was set as the cutoff value in 336 samples. One hundred and seventy-seven (52.7%) samples were assigned to the overexpressed group and 159 (47.3%) to the weak expression group. Table 2 summarizes the correlation between clinicopathological parameters and NALT1 expression levels. Not surprisingly, expression of NALT1 was closely related to tumor differentiation (P = 0.000, χ2 = 247.392) and TNM stage (P = 0.001, χ2 = 17.625).
Table 2.
NALT1 expressions and clinicopathologic parameters
| Characteristics |
NALT1 |
|||
| Low | High | P value | χ2 | |
| 159 (47.3) | 177 (52.7) | |||
| Age | 0.444 | 0.680 | ||
| ≥ 60 | 88 (49.4) | 90 (50.6) | ||
| < 60 | 71 (44.9) | 87 (55.1) | ||
| Gender | 0.551 | 0.410 | ||
| Male | 109 (46.2) | 127 (53.8) | ||
| Female | 50 (50.0) | 50 (50.0) | ||
| Tumor size | 0.659 | 0.236 | ||
| ≥ 5 cm | 65 (45.8) | 77 (54.2) | ||
| < 5 cm | 94 (48.5) | 100 (51.5) | ||
| Location | 0.589 | 1.058 | ||
| Up | 32 (42.7) | 43 (57.3) | ||
| Middle | 42 (46.7) | 48 (53.3) | ||
| Low | 85 (49.7) | 86 (50.3) | ||
| Bormann type | 0.208 | 4.549 | ||
| I | 13 (52.0) | 12 (48.0) | ||
| II | 77(52.4) | 70 (47.6) | ||
| III | 68 (42.8) | 91 (57.2) | ||
| V | 1 (20.0) | 4 (80.0) | ||
| Tumor histological morphology | 0.569 | 1.126 | ||
| Adenocarcinoma | 123 (47.3) | 137 (52.7) | ||
| Absolute signet ring cell carcinoma | 35 (46.7) | 40 (53.3) | ||
| Mixed carcinoma | 1 (100) | 0 (0) | ||
| Lauren type | 0.190 | 3.326 | ||
| Intestinal | 71 (45.5) | 85 (54.5) | ||
| Mixed carcinoma | 31 (41.3) | 44 (58.7) | ||
| Diffuse | 57 (54.3) | 48 (45.7) | ||
| Tumor differentiation | 0.000 | 247.392 | ||
| Moderate and high | 151 (90.4) | 16 (9.6) | ||
| Poor | 8 (4.7) | 161 (95.3) | ||
| Vessel invasion | 0.471 | 0.658 | ||
| Yes | 43 (43.9) | 55 (56.1) | ||
| No | 116 (48.7) | 122 (51.3) | ||
| Perineural invasion | 0.706 | 0.195 | ||
| Yes | 38 (45.2) | 46 (54.8) | ||
| No | 121 (48.0) | 131 (52.0) | ||
| T category | 0.035 | 8.619 | ||
| T1 | 37 (61.7) | 23 (38.3) | ||
| T2 | 10 (45.5) | 12 (54.5) | ||
| T3 | 8 (66.7) | 4 (33.3) | ||
| T4 | 104 (43.0) | 138 (57.0) | ||
| N category | 0.000 | 20.693 | ||
| N0 | 86 (61.9) | 53 (38.1) | ||
| N1 | 22 (37.9) | 36 (62.1) | ||
| N2 | 26 (40.0) | 39 (60.0) | ||
| N3 | 25 (33.8) | 49 (66.2) | ||
| TNM stage | 0.001 | 17.625 | ||
| I | 41 (59.4) | 28 (40.6) | ||
| II | 49 (60.5) | 32 (39.5) | ||
| III | 68 (37.0) | 116 (63.0) | ||
| IV | 1 (50.0) | 1 (50.0) | ||
NALT1: NOTCH1 associated with lncRNA in T cell acute lymphoblastic leukemia 1; TNM: Tumor node metastasis.
Of the 336 patients, 319 had complete follow-up data, and 196 (58.3%) died before the end of follow-up. DFS was 5-91 mo, with a median of 33 mo, and OS was 7-91 mo, with a median of 45.5 mo. Patients with overexpression of NALT1 showed poorer DFS (45.24 vs 33.69 mo, P < 0.01) and OS (50.30 vs 40.88 mo, P < 0.01) (Table 1, Figure 3C, 3D). Multiple factor analysis included the clinicopathological parameters with P < 0.05 in univariate analysis. A backward stepwise method was applied in the Cox proportional hazards model. TNM stage and expression of NALT1 served as independent prognostic factors for prediction of DFS [TNM: P < 0.01, hazard ratio (HR) = 125.49, 95% confidence interval (CI): 42.47–370.76; NALT1: P = 0.02, HR = 0.71, 95%CI: 0.53-0.96; Table 3) and OS (TNM: P < 0.01, HR = 87.98, 95%CI: 37.29-207.38; NALT1: P = 0.02, HR = 0.71, 95%CI: 0.52-0.95; Table 3).
Table 3.
Multivariate analysis of significant prognostic factor for survival in gastric cancer patients
| Variables |
DFS |
OS |
||||
| P value | HR | 95%CI | P value | HR | 95%CI | |
| Bormann | 0.44 | 1.11 | 0.86-1.43 | 0.53 | 1.09 | 0.84-1.41 |
| Tumor size | 0.54 | 0.91 | 0.68-1.23 | 0.72 | 0.95 | 0.70-1.27 |
| Tumor differentiation | 0.81 | 0.91 | 0.43-1.98 | 0.74 | 0.88 | 0.41-1.88 |
| Vessel invasion | 0.45 | 1.34 | 0.63-2.85 | 0.50 | 1.30 | 0.61-2.80 |
| Perineural invasion | 0.18 | 0.82 | 0.60-1.10 | 0.25 | 0.84 | 0.62-1.14 |
| TNM stage | 0.00 | 125.49 | 42.47-370.76 | 0.00 | 87.98 | 37.29-207.38 |
| NALT1 expression | 0.02 | 0.71 | 0.53-0.96 | 0.02 | 0.71 | 0.52-0.95 |
HR: Hazard ratio; 95%CI: 95% confidence interval.
NALT1 was overexpressed and associated with invasion and metastasis of GC
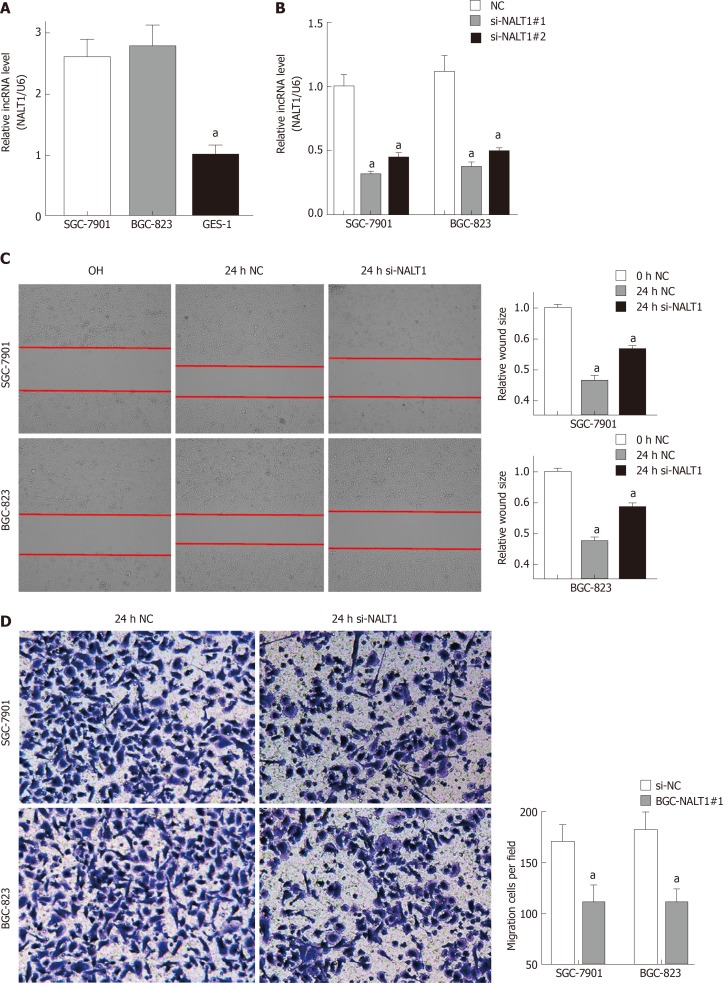
Similar to overexpression in GC tissues, NALT1 was also overexpressed in GC (SGC-7901 and BGC-823) cells (Figure 4A). To evaluate the biology of NALT1 in GC, NALT1 siRNAs were transfected into GC cell lines. Two siRNAs targeting NALT1 were detected (si-NALT1#1, si-NALT1#2), among which si-NALT1#1 was the most effective siRNA to remove NALT1 expression and was selected for subsequent research (Figure 4B). Scrape and Transwell assays were performed to evaluate the effect of NALT1 on cell invasion and metastasis. Decrease of NALT1 expression significantly decreased cell migration (Figure 4C and D).
Figure 4.
NALT1 induced GC invasion and metastasis. A: NALT1 expression in GC cells compare to GES-1 cells; B: The expression of NALT1 in GC cells before and after transfection si-NALT1 sequence; C: Scrape motility assays were monitored for 24 h in NALT1-knockdown GC cells; D: Transwell assays were used to evaluate the role of NALT1 in invasion in NALT1-knockdown. U6 was used as a loading control in RT-PCR; In all figures, 100 × magnification was used, n = 3, aP < 0.05. NALT1: NOTCH1 associated with lncRNA in T cell acute lymphoblastic leukemia 1; GC: Gastric cancer; GES-1: Human normal gastric epithelial cell line.
NALT1 and NOTCH1 affect invasion and metastasis synergistically
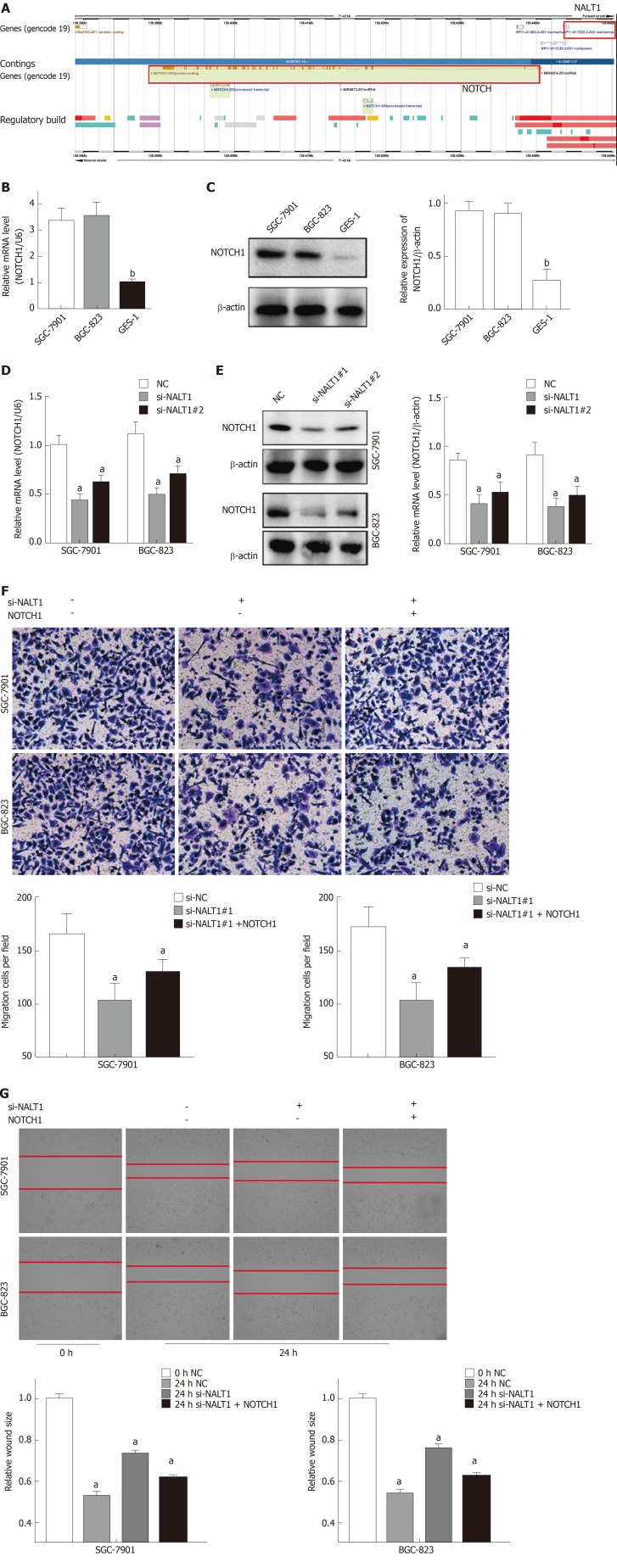
Bioinformatics analysis showed that NALT1 was 10 kb upstream of NOTCH1 (Figure 5A, http://asia.ensembl.org). Expression of NOTCH1 in GC was verified by western blotting and RT-PCR. It showed that compared with control GES-1 cells, NOTCH1 was overexpressed in BGC-823 and SGC-7901 cells at mRNA and protein levels (Figure 5B, 5C). With the downregulation of NALT1 expression, expression of NOTCH1 also decreased. Moreover, both the mRNA (Figure 5D) and protein (Figure 5E) levels of NOTCH1 were all significantly decreased. NALT1 may regulate NOTCH1 at the transcriptional level. As mentioned above, we found that downregulating expression of NALT1 significantly inhibited the invasiveness of GC cells, but this decrease was partially abolished by overexpression of NOTCH1 (Figure 5F and G). With the increase in NOTCH1 expression, the invasion and metastasis of cells were enhanced: the number (Figure 5F) and migratory distance (Figure 5G) of the cells increased significantly.
Figure 5.
NALT1 influence NOTCH1 expression. A: Schematic figure of NALT1 and NOTCH1 in chromosome 9. They were highlight with red box; B: NOTCH1 was overexpression in GC cells by RT-PCR; C: NOTCH1 was overexpression in GC cells by western-blot; D: The expression of NOTCH1 mRNA was effected by NALT1; E: The expression of protein was effected by NALT1; F: Transwell assay was used to evaluate the association of NALT1 and NOTCH1 in invasion in NALT1-knockdown and NALT1-knockdown + NOTCH1-overexpression GC cells; G: Scrape motility assays were monitored for 24 h in NALT1-knockdown and NALT1-knockdown + NOTCH1-overexpression GC cells. U6 and β-actin were used as loading control in RT-PCR and western-blot, respectively. In all figures, 100 × magnification was used. n = 3, aP < 0.05, bP < 0.01. NALT1: NOTCH1 associated with lncRNA in T cell acute lymphoblastic leukemia 1; GC: Gastric cancer.
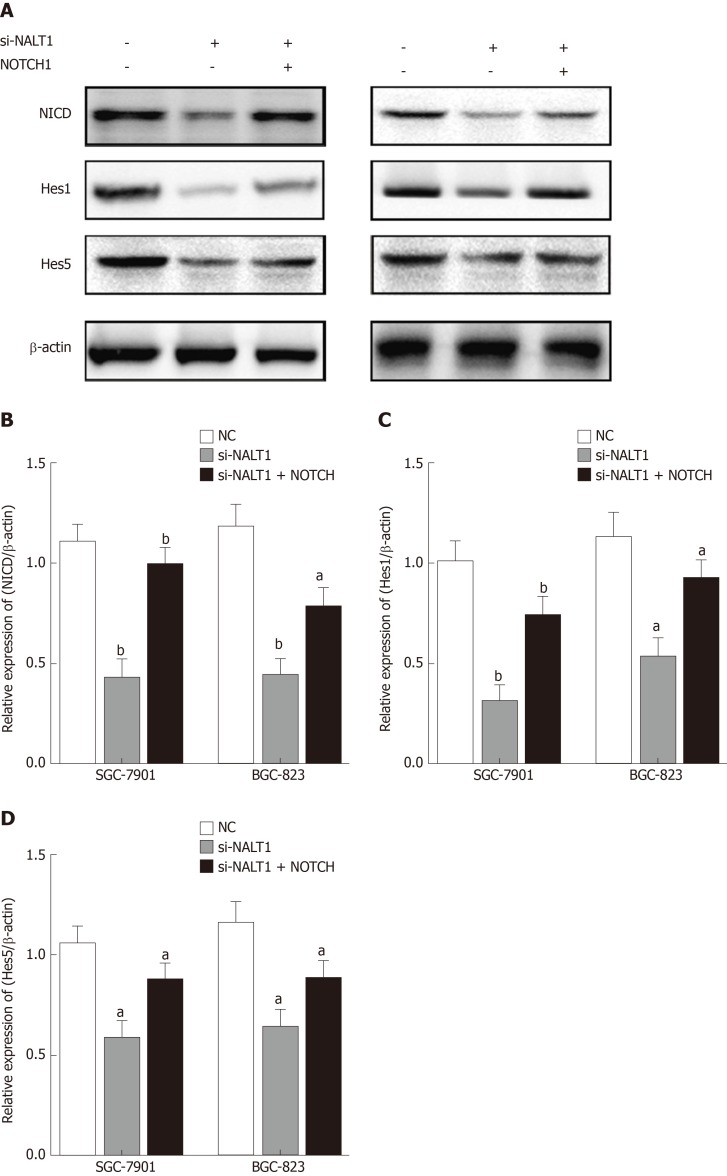
We studied the effect of NALT1 on NOTCH pathway components. We evaluated the expression levels of NOTCH intracellular cytoplasmic domain (NICD), HES1, and HES5 (downstream target genes of notch signaling pathway) in the cells treated with NALT1 siRNA. With knockdown of NALT1, expression of these proteins was decreased. However, the downregulation could partially be reversed by overexpression of NOTCH1 (Figure 6).
Figure 6.
Decreased NALT1 suppressed the NOTCH signaling pathway. A: Western blot was used to detect whether the loss of NALT1 led to the abnormal expression of NOTCH signaling pathway. NICD, Hes1, and Hes5 were selected; B: The relative expression of NICD; C: The relative expression of Hes1; D: The relative expression of Hes5. β-actin was used as a loading control; n = 3, aP < 0.05, bP < 0.01. NALT1: NOTCH1 associated with lncRNA in T cell acute lymphoblastic leukemia 1.
DISCUSSION
With the development of next-generation sequencing technology and molecular biology, thousands of genes have been proved to play an important role in the development of malignant tumors[13,14]. Moreover, the significance of ncRNA in gene expression regulation is being recognized[15,16]. LncRNAs can regulate gene expression at multiple levels: transcription and post-transcriptional processing as well as chromatin modification[17]. Correspondingly, lncRNAs can also bind various molecules (DNA, RNA, and protein) to perform biological functions. Therefore, it is of utmost importance to search for lncRNA that is closely related to the occurrence and development of GC and explore its functions and mechanisms. In this study, we downloaded transcriptome and clinical data from TCGA-STAD and attempted to explore GC-related lncRNAs through bioinformatics analysis. WGCNA is a scale-free network algorithm that is widely used in big data mining and analysis[11]. WGCNA can be used to study the differential coexpression between GC and normal tissue samples as a systematic biological method to explain the relationship between clinical characteristics and RNAs[10]. Different from other clustering algorithms, which use geometric distance between data for clustering, WGCNA clustering criteria focus on biological significance. These clustering modules are based on differences between RNA expression profiles and hub RNAs, which lead to changes in expression of cell signaling pathways[18]. Thus, the problem can be simplified to an equation between modular hub RNAs and clinical features to reveal the main causes of GC progression.
We found that the blue module was closely related to GC grade and survival through differential expression analysis and WGCNA. NALT1 is the hub-RNA of the blue module. NALT1, also named LINC01573, was first found in 2015 and contributes to the proliferation of T cell acute lymphoblastic leukemia[19]. However, its biological role in solid tumors has not been reported. In this study, we found that NALT1 was overexpressed in GC tissues and cells and was involved in tumor invasion and metastasis. Unfortunately, NALT1 overexpression is strongly associated with poor prognosis of GC. To explore further the role of NLAT1 in GC malignant phenotype, we studied its localization in the genome. As a neighbor, the well-known NOTCH1 is located 400 bp downstream of NALT1. Flank 10 k theory suggests that lncRNA can cis regulate adjacent genes within the range of 10000 bp on the chromosome location[20,21]. Therefore, we supposed that the influence of NALT1 on GC invasion and metastasis was realized by NOTCH1. The NOTCH signaling pathway regulates cell proliferation and differentiation in various gastrointestinal tissues, including the stomach[22]. The interaction between receptor and ligand initiates NOTCH signaling pathway, which activates NICD, thereby promoting transcription of downstream genes of the NOTCH pathway. The NOTCH family consists of four receptors and five ligands (Jagged-1, Jagged-2, Delta-like-1, Delta-like-3, and Delta-like-4). Mediated by NOTCH1 and NOTCH2 receptors, NOTCH is involved in the proliferation of gastric stem cells[23]. In addition, Xiao et al[24] proposed that inhibiting the activity of NOTCH signaling pathway could fully inhibit the invasion, metastasis, and proliferation of GC.
Unsurprisingly, NOTCH1 was also highly expressed in GC tissues and cells in the present study. Expression of NOTCH1 was decreased at mRNA and protein levels with knockdown of NALT1. This indicates that NALT1 regulates NOTCH1 expression at the transcriptional level, which is consistent with our hypothesis. In subsequent experiments, we found that downregulation of NALT1 inhibited the invasion and metastasis of GC, which could be partially restored by upregulation of NOTCH1. Subsequently, we detected the biomarkers (NICD, HES1, and HES5) of the NOTCH signaling pathway. Similarly, inhibited expression of NALT1 can downregulate the corresponding biomarkers. The upregulated NOTCH1 can partially abolished this effect. This further proved that the biological role of NALT1 in GC is achieved by NOTCH1 and NOTCH signaling pathway.
RNA sequencing and clinical data of TCGA-STAD patients were studied by WGCNA and other methods. NALT1 is one of the hub RNAs, which is closely related to poor prognosis of GC. Moreover, NALT1 is closely related to GC invasion and metastasis, which is achieved through the NOTCH signaling pathway.
Although we speculated about the important role of NALT1 in GC by bioinformatics analysis and verified it by molecular biological methods, there were some limitations to this study. First, the study lacked experimental evidence for a direct mechanism. Second, the expression and function of NALT1 in GC need to be verified by in vivo experiments.
ARTICLE HIGHLIGHTS
Research background
Gastric cancer (GC) is the most common and aggressive tumor of the digestive system and poses a serious threat to human health. Long noncoding RNAs (lncRNAs) are aberrant and play critical roles in GC. Since genes do not work alone, our aim was to elucidate the potential relationship between mRNA and noncoding RNA in this study.
Research motivation
Searching for coexpressed lncRNA clusters may help to elucidate the mechanism of tumor development and predict the prognosis of GC.
Research objectives
To explore the prognostic value of NOTCH1 associated with lncRNA in T cell acute lymphoblastic leukemia 1 (NALT1) in GC and the mechanism of its involvement in gastric cancer invasion and metastasis.
Research methods
Based on the TCGA database, we obtained differentially expressed lncRNAs. The significance module was studied by weighted gene coexpression network analysis. The function of NALT1 was assessed by reverse transcription polymerase chain reaction, western blotting, scrape motility assay, and Transwell migration assay.
Research results
Fifteen coexpression modules were constructed based on 3339 differentially expressed lncRNAs and weighted gene coexpression network analysis. The blue module was correlated with tumor grade and survival, and the hub-lncRNA of blue NALT1 was an independent risk factor for GC prognosis. Through cis regulation, NALT1 affected the expression of the NOTCH signaling pathway and was related to GC invasion and metastasis.
Research conclusions
NALT1 was overexpressed in GC and was an independent risk factor for GC prognosis. It affected invasion and metastasis of GC by regulating NOTCH1 and NOTCH signaling pathway.
Research perspectives
In future studies, we will verify the results of this study through in vivo experiments. The specific binding sites of NALT1 and NOTCH1 will be studied by chromatin immunoprecipitation and pull-down assays.
Footnotes
Institutional review board statement: The study was reviewed and approved by the Faculty of Science Ethics Committee at Liaoning Cancer Hospital & Institute (Cancer Hospital of China Medical University) (20150308-2).
Informed consent statement: All study participants provided informed written consent prior to their treatments and study enrollment.
Conflict-of-interest statement: The authors declare that there are no conflicts of interest related to this study.
ARRIVE guidelines statement: The authors have read the ARRIVE guidelines, and the manuscript was prepared and revised according to the ARRIVE guidelines.
Manuscript source: Unsolicited manuscript
Peer-review started: August 30, 2019
First decision: October 14, 2019
Article in press: November 13, 2019
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chuang SM S-Editor: Wang J L-Editor: Filipodia E-Editor: Ma YJ
Contributor Information
Hai-Yan Piao, Medical Oncology Department of Gastrointestinal Cancer, Liaoning Province Cancer Hospital & Institute (Cancer Hospital of China Medical University), Shenyang 110042, Liaoning Province, China.
Shuai Guo, Gastric Cancer Department, Liaoning Province Cancer Hospital & Institute (Cancer Hospital of China Medical University), Shenyang 110042, Liaoning Province, China.
Yue Wang, Gastric Cancer Department, Liaoning Province Cancer Hospital & Institute (Cancer Hospital of China Medical University), Shenyang 110042, Liaoning Province, China.
Jun Zhang, Gastric Cancer Department, Liaoning Province Cancer Hospital & Institute (Cancer Hospital of China Medical University), Shenyang 110042, Liaoning Province, China. zhangjun@cancerhosp-ln-cmu.com.
Data sharing statement
No data is shared.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Al-Batran SE, Homann N, Pauligk C, Goetze TO, Meiler J, Kasper S, Kopp HG, Mayer F, Haag GM, Luley K, Lindig U, Schmiegel W, Pohl M, Stoehlmacher J, Folprecht G, Probst S, Prasnikar N, Fischbach W, Mahlberg R, Trojan J, Koenigsmann M, Martens UM, Thuss-Patience P, Egger M, Block A, Heinemann V, Illerhaus G, Moehler M, Schenk M, Kullmann F, Behringer DM, Heike M, Pink D, Teschendorf C, Löhr C, Bernhard H, Schuch G, Rethwisch V, von Weikersthal LF, Hartmann JT, Kneba M, Daum S, Schulmann K, Weniger J, Belle S, Gaiser T, Oduncu FS, Güntner M, Hozaeel W, Reichart A, Jäger E, Kraus T, Mönig S, Bechstein WO, Schuler M, Schmalenberg H, Hofheinz RD FLOT4-AIO Investigators. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): a randomised, phase 2/3 trial. Lancet. 2019;393:1948–1957. doi: 10.1016/S0140-6736(18)32557-1. [DOI] [PubMed] [Google Scholar]
- 3.Liyanage KIP, Ganegoda GU. Therapeutic Approaches and Role of ncRNAs in Cardiovascular Disorders and Insulin Resistance. Biomed Res Int. 2017;2017:4078346. doi: 10.1155/2017/4078346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin C, Yang L. Long Noncoding RNA in Cancer: Wiring Signaling Circuitry. Trends Cell Biol. 2018;28:287–301. doi: 10.1016/j.tcb.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, Tang JY, Bao YJ, Hu Y, Lin Y, Sun D, Chen YX, Hong J, Chen H, Zou W, Fang JY. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016;6:784–801. doi: 10.1158/2159-8290.CD-15-0921. [DOI] [PubMed] [Google Scholar]
- 8.Zhang E, He X, Zhang C, Su J, Lu X, Si X, Chen J, Yin D, Han L, De W. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19:154. doi: 10.1186/s13059-018-1523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V Cancer Genome Atlas Research Network, Hu H. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018;173:400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Wu Y, Jin HY, Guo S, Dong Z, Zheng ZC, Wang Y, Zhao Y. Prognostic value of sorting nexin 10 weak expression in stomach adenocarcinoma revealed by weighted gene co-expression network analysis. World J Gastroenterol. 2018;24:4906–4919. doi: 10.3748/wjg.v24.i43.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aravanis AM, Lee M, Klausner RD. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell. 2017;168:571–574. doi: 10.1016/j.cell.2017.01.030. [DOI] [PubMed] [Google Scholar]
- 14.McDaniel AS, Stall JN, Hovelson DH, Cani AK, Liu CJ, Tomlins SA, Cho KR. Next-Generation Sequencing of Tubal Intraepithelial Carcinomas. JAMA Oncol. 2015;1:1128–1132. doi: 10.1001/jamaoncol.2015.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Q, Hao Q, Prasanth KV. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet. 2018;34:142–157. doi: 10.1016/j.tig.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Botía JA, Vandrovcova J, Forabosco P, Guelfi S, D'Sa K United Kingdom Brain Expression Consortium, Hardy J, Lewis CM, Ryten M, Weale ME. An additional k-means clustering step improves the biological features of WGCNA gene co-expression networks. BMC Syst Biol. 2017;11:47. doi: 10.1186/s12918-017-0420-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Wu P, Lin R, Rong L, Xue Y, Fang Y. LncRNA NALT interaction with NOTCH1 promoted cell proliferation in pediatric T cell acute lymphoblastic leukemia. Sci Rep. 2015;5:13749. doi: 10.1038/srep13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demitrack ES, Samuelson LC. Notch as a Driver of Gastric Epithelial Cell Proliferation. Cell Mol Gastroenterol Hepatol. 2017;3:323–330. doi: 10.1016/j.jcmgh.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demitrack ES, Gifford GB, Keeley TM, Horita N, Todisco A, Turgeon DK, Siebel CW, Samuelson LC. NOTCH1 and NOTCH2 regulate epithelial cell proliferation in mouse and human gastric corpus. Am J Physiol Gastrointest Liver Physiol. 2017;312:G133–G144. doi: 10.1152/ajpgi.00325.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao HJ, Ji Q, Yang L, Li RT, Zhang C, Hou JM. In vivo and in vitro effects of microRNA-124 on human gastric cancer by targeting JAG1 through the Notch signaling pathway. J Cell Biochem. 2018;119:2520–2534. doi: 10.1002/jcb.26413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data is shared.