Abstract
Background & objectives:
Duchenne muscular dystrophy (DMD) is an X-linked recessive disorder and is caused mainly by deletion, duplication and point mutations in the DMD gene. Diagnosis of DMD has been a challenge as the mutations in the DMD gene are heterogeneous and require more than one diagnostic strategy for the validation of the mutation. This study was planned to evaluate the targeted next-generation sequencing (NGS) as a single platform to detect all types of mutations in the DMD gene, thereby reducing the time and costs compared to conventional sequential testing and also provide precise genetic information for emerging gene therapies.
Methods:
The study included 20 unrelated families and 22 patients from an Indian population who were screened for DMD based on phenotypes such as scoliosis, toe walking and loss of ambulation. Peripheral blood DNA was isolated and subjected to multiplex ligation-dependent probe amplification (MLPA) and targeted NGS of the DMD gene to identify the nature of the mutation.
Results:
In the study patients, 77 per cent of large deletion mutations and 23 per cent single-nucleotide variations (SNVs) were identified. Novel mutations were also identified along with reported deletions, point mutations and partial deletions within the exon of the DMD gene.
Interpretation & conclusions:
Our findings showed the importance of NGS in the routine diagnostic practice in the identification of DMD mutations over sequential testing. It may be used as a single-point diagnostic strategy irrespective of the mutation type, thereby reducing the turnaround time and cost for multiple diagnostic tests such as MLPA and Sanger sequencing. Though MLPA is a sensitive technique and is the first line of a diagnostic test, the targeted NGS of the DMD gene may have an advantage of having a single diagnostic test. A study on a larger number of patients is needed to highlight NGS as a single, comprehensive platform for the diagnosis of DMD.
Keywords: Deletions, Duchenne muscular dystrophy, multiplex ligation-dependent probe amplification, next-generation sequencing, partial deletion, single-nucleotide variation
Duchenne muscular dystrophy (DMD) is an X-linked recessive neuromuscular disorder characterized by progressive muscle degeneration and weakness1, affecting only males. Clinically, the DMD phenotype includes reduced motor functions, Gowers' sign2, increased creatine kinase, scoliosis, and toe walking3.
Genetically, DMD results from mutations in the DMD gene, which is one of the longest genes (OMIM: 310200; ~2.5 Mbp) located on chromosome Xp.21.2. It has 79 exons encoding a 14 kbp transcript and protein product of about 427 kDa4. It is expressed in skeletal, cardiac and smooth muscles, forming a major part of the dystrophin-glycoprotein complex linking the muscle contractile cytoskeleton to the extracellular matrix, thereby maintaining a structural integrity of the muscle tissue5. Mutations in the DMD gene generally result in disruption of the reading frame eventually resulting in the synthesis of a truncated/dysfunctional protein leading to protein degradation6. The frequency of occurrence of mutations in the DMD gene, however, is not entirely clear due to lack of a well-defined epidemiological survey. However, from the existing reports on the spectrum of DMD gene mutations, an intragenic deletion mutation of one or more exons accounts for 60 per cent of mutations and is the most common type reported among DMD patients7. Point mutations account for approximately 15-20 per cent of DMD cases, majorly resulting in premature stop codons8, and duplications account for approximately 11 per cent of DMD cases9.
Most commonly used diagnostic techniques such as multiplex ligation probe amplification (MLPA)10 allow detection of mutations such as deletions/duplication in patients11. However, small-nucleotide variations such as single-nucleotide variations (SNVs) and insertion/deletion variants9 are not detected by MLPA unless the detection probe is situated at the mutation locus12. The undetected samples then undergo Sanger sequencing of the exons, which increases the cost and time taken to arrive at a conclusion13. The next most comprehensive and accurate method of detection to completely sequence the entire DMD gene is next-generation sequencing (NGS), which can detect deletions/duplications and point mutations14. NGS might serve as a single, comprehensive platform for the diagnosis and detection of mutations in DMD. Moreover, the sequence information provided by NGS can easily be applied to treatment initiatives, such as exon skipping, which is one of the most promising forms of therapeutic strategies requiring precise mutational information.
The present study was planned for the identification of novel mutations in patients with DMD using both MLPA and targeted NGS of the DMD gene. Further, the advantages of using NGS in the diagnosis of DMD, mainly in the identification of point mutations and partial deletions of exons were compared with sequential conventional testing.
Material & Methods
This study was approved by the Institutional Ethics Committee of Dystrophy Annihilation Research Trust (DART), Bengaluru, India, and written informed consent was obtained from the patients and/or their families. The study was conducted between February 2016 and March 2017, and consisted of 20 families and 22 unrelated patients who volunteered based on suspected symptoms unique to DMD such as scoliosis, toe walking and inability to walk or being wheelchair-bound and who were referred by a clinician for the diagnosis of DMD by MLPA and NGS.
Sample collection genomic DNA isolation: Blood samples (2 ml) from the DMD patients were collected in a hospital setup using an EDTA Vacutainer (Becton Dickinson, USA). Genomic DNA was isolated from 200 μl of peripheral blood using a QIAamp blood DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
MLPA and targeted NGS of the DMD gene: All DMD patients were initially diagnosed using an MLPA kit (SALSA MLPA KIT P034/035 DMD, MRC-Holland, Amsterdam, The Netherlands) as the first line of diagnosis for the identification of one or more deletions or duplications. All samples irrespective of the MLPA result were subjected to NGS of the DMD gene. The FASTQ files of all the 22 patients included in this study were uploaded on to a repository (http://basil.strandls.com/clinicaldata/sls-DART/). Each sample had a pair of R1 and R2 FASTQ files, for paired-end forward and reverse reads (read 1 and read 2). The TruSight Inherited Disease panel (Illumina, CA, USA) that contains 460 genes associated with severe recessively inherited Mendelian diseases, including the DMD gene, was used for targeted NGS. An analytical validation of the panel showed a sensitivity of 97.82 per cent, a specificity of 99.9 per cent and a reproducibility of greater than 98 per cent. The gene coverage analysis on this panel revealed that exonic and flanking intronic regions of the DMD gene (NM_004006) showed a coverage of greater than 99 per cent (at ≥20 reads). Genomic DNA was used for library preparation15 for NGS and sequenced on the NextSeq platform (Illumina, CA, USA), according to the manufacturer's instructions.
Library preparation and sequencing: Genomic DNA was quantified and 50-60 ng of the purified genomic DNA from each patient was used for library preparation for NGS as described15. DNA was fragmented and tagged simultaneously using Nextera® technology (Illumina, CA, USA), and using a limited-cycle polymerase chain reaction (PCR) approach16. The adapters were incorporated followed by sample-specific barcodes from which NGS libraries were prepared.
For target enrichment, approximately 500 ng of an individual sample library was pooled with three other libraries having unique indices. These were subjected to enrichment that involved two successive hybridization steps with target-specific biotinylated probes. These probes were targeted at the exons and exon-intron junctions of the 460 genes. The target-specific probes were used to pull down the regions of interest. The target libraries were amplified using limited-cycle PCR steps. The tagged and amplified sample libraries were checked for quality. These were quantified using a BioAnalyzer (Agilent, CA, USA). About 6-10 pM of the pooled library was loaded and sequenced on the MiSeq platform (Illumina, CA, USA), according to the manufacturer's instructions.
Analysis of NGS data: The trimmed FASTQ files were generated using a MiSeq Reporter from Illumina. The reads were aligned against the whole-genome build hg19 using Strand NGS v2.6 (Strand Life Sciences, Bengaluru)15. The Strand NGS variant caller was used to detect variants at locations in the target regions covered by a minimum of 10 reads with at least two variant reads. Reads that failed the quality control metrics, such as reads with average quality <20, and reads with ambiguous characters were filtered.
Variants were imported into StrandOmics v3.4 (https://strandls.com/bioinformatics/). Annotation and prioritization of variants with suggestive American College of Medical Genetics and Genomics (ACMG) labels were done by automated pipelines in StrandOmics17. StrandOmics is a clinical interpretation and reporting platform that combines knowledge from internally curated literature content and various publically available data sources (such as Uniprot, OMIM, HGMD, ClinVar, ARUP, dbSNP, 1000 Genomes, Exome Variant Server and Exome Aggregation Consortium). In addition to databases, bioinformatics prediction tools (such as SIFT, PolyPhen HVAR/HDIV, Mutation Taster, Mutation Assessor, FATHMM, LRT for missense variants and NNSPLICE and ASSP tools for variants in essential splice sites and exon-intron boundaries) have also been integrated to assess the pathogenicity of the variants. This integrated knowledge is used to prioritize automatically a list of variants based on the ACMG guidelines17, the inheritance model, disease phenotype, sequence conservation across various species and allelic frequency in our laboratory's internal patient-pooled database. A variant was labelled 'novel' if it had not been previously reported in the literature or in any public database.
The identified variants were labelled according to the ACMG recommended standards for the interpretation and reporting of sequence variations. Accordingly, the variants were classified into five categories: (i) pathogenic, (ii) likely pathogenic, (iii) variant of uncertain significance (VUS), (iv) likely benign, and (v) benign. 'Pathogenic', 'likely pathogenic' and 'variants of uncertain significance' were considered for reporting.
In addition to SNVs and indels, copy number analysis was performed to identify large deletions or insertions ranging from a single exon to multiple exon deletions in DMD18. In addition, split-read analysis for the identification of breakpoints was done as previously described18,19.
Results
All DMD patients were initially diagnosed using MLPA, and subsequently irrespective of the results obtained from the MLPA, targeted NGS of the DMD gene was carried out.
Mutation frequency among the study participants: The Table shows the mutations identified using the MLPA and NGS analyses of both deletions and SNV and status of the mutation as either previously reported or novel. Nearly 77 per cent of large deletion mutations and 23 per cent of SNVs/small deletion mutations accounted for the distribution of mutations in the DMD gene (Table). The results of deletions identified by MLPA were related with the deletions identified by NGS with respect to large deletions except for non-detection of point mutations in three patients (DMD18-21) and partial deletion of exon 33 in one patient (DMD22), which was identified as a complete deletion of exons 33-45 by MLPA.
Table.
List of Duchenne muscular dystrophy (DMD) patients with the mutations as diagnosed by multiplex ligation-dependent probe amplification (MLPA) and next-generation sequencing (NGS)
| DMD ID | Ambulation status | Age at onset (yr) | Age at present (yr) | MLPA_EXON Deletion | NGS_EXON Deletion/SNV | cDNA | Type of mutation | DMD database |
|---|---|---|---|---|---|---|---|---|
| DMD1 | Ambulant | 3 | 9 | 46-49 | 46-49 | c.(6614+1_6615-1)_(7200+1_7201-1) del | Deletion | Reported |
| DMD2 | Non-ambulant | 4 | 13 | 46-55 | 46-55 | c.(6614+1_6615-1)_(8217+1_8818-1) del | Deletion | Reported |
| DMD3 | Non-ambulant | 5 | 8 | 46-48 | 46-48 | c.(6614+1_6615-1)_(7098+1_7099-1) del | Deletion | Reported |
| DMD4 | Ambulant | 6 | 11 | 45-52 | 45-52 | c.(6438+1_6439-1)_ 7660+1_7661-1) del | Deletion | Reported |
| DMD5 | Non-ambulant | 3 | 10 | 8, 9 | 8, 9 | c.(649+1_650-1)_ (960+1_961-1) del | Deletion | Reported |
| DMD6 | Ambulant | 3 | 10 | 18-29 | 18-29 | c.(2168+1_2169-1)_(4071+1_4072-1) del | Deletion | Novel |
| DMD7 | Non-ambulant | 10 | 16 | 45-52 | 45-52 | c.(6438+1_6439-1)_ 7660+1_7661-1) del | Deletion | Reported |
| DMD8 | Non-ambulant | 6 | 12 | 46-47 | 46-47 | c.(6614+1_6615-1)_(6912+1_6913-1) del | Deletion | Reported |
| DMD9 | Non-ambulant | 3 | 8 | 49-50 | 49-50 | c.(7098+1_7099-1)_(7309+1_7310-1) del | Deletion | Reported |
| DMD10 | Ambulant | 2 | 7 | 49-50 | 49-50 | c.(7098+1_7099-1)_(7309+1_7310-1) del | Deletion | Reported |
| DMD11 | Ambulant | 4 | 6 | 44 | 44 | c.(6290+1_6291-1)_(6438+1_6439-1) del | Deletion | Reported |
| DMD12 | Ambulant | 3 | 17 | 48-52 | 48-52 | c.(6912+1_6913-1)_(7660+1_7661-1) del | Deletion | Reported |
| DMD13 | Non-ambulant | 5 | 13 | 46-50 | 46-50 | c.(6614+1_6615-1)_(7309+1_7310-1) del | Deletion | Reported |
| DMD14 | Ambulant | 4 | 10 | 51 | 51 | c.(7309+1_7310-1)_(7542+1_7543-1) del | Deletion | Reported |
| DMD15 | Non-ambulant | 6 | 17 | 51 | 51 | c.(7309+1_7310-1)_(7542+1_7543-1) del | Deletion | Reported |
| DMD16 | Non-ambulant | 4 | 15 | 51 | 51 | c.(7309+1_7310-1)_(7542+1_7543-1) del | Deletion | Reported |
| DMD17 | Non-ambulant | 3 | 11 | ND | 21 | c. 2661_2662delinsA (p.Glu888*) | Nonsense | Novel |
| DMD18 | Ambulant | 4 | 7 | ND | 46 | c. 6622G>T (pGlu2208*) | Nonsense | Novel |
| DMD19 | Ambulant | 7 | 11 | ND | 8 | c. 724C>T (p.Gln242*) | Nonsense | Reported |
| DMD20 | Non-ambulant | 2 | 16 | ND | 51 | c. 7348dupG (p.Val2450*) | Frameshift | Reported |
| DMD21 | Non-ambulant | 3 | 10 | ND | 7 | c. 583C>T (p.Arg195*) | Nonsense | Reported |
| DMD22 | Non-ambulant | 8 | 16 | 33-45 | 33 (PD) 34-45 | c. 4525_(6615-738) del | Deletion | Novel |
ND, non-detected; PD, partial deletion; SNV, single-nucleotide variation; cDNA, complementary DNA
Large deletions: A total of 13 different large deletions were identified among 16 DMD patients; large deletions (deletion of more than 1 exon) were higher among the 22 patients studied, which accounted for 77 per cent of them. Among the large deletions and point mutations identified, 21 patients had the mutation in the region spanning the rod domain from exon 8-55. Exon 8 is placed in the hinge region between the actin-binding domain (ABD) and the central rod domain. DMD5 had a deletion of exon 8 spanning the hinge region and exon 9 from the central rod domain. Patient DMD22 had a deletion mutation of exons 33-45 [c.4525_ (6615-738) del], which was not reported previously (Table). Exon 33 was found to be partially deleted, whereas the rest of the exons 34-35 were completely deleted. Among the large deletions, patient DMD6 had deletion of exons 18-29 again novel deletion, not reported previously.
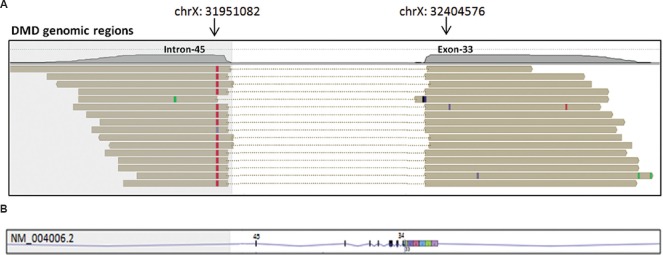
Partial deletion of exon 33 in DMD22: In patient DMD22, there was a deletion of 149 nucleotides from the 3' end of exon 33 and complete deletion of exons 34-45 of the DMD gene (Figure). The deletion was identified by a split-read analysis tool, available in the Strand NGS software, which allowed us to determine the breakpoint(s) of a deletion, if it occurred within the coding region. For this, reads that did not align with an alignment score of greater than 95 per cent were subjected to split-read alignment18, as described previously19. In brief, the input reads were split into two segments, each segment was mapped independently to the reference genome and the split segments that were required to align uniquely were investigated, with an alignment score of at least 97 per cent. Based on these split-read alignment scores, a structural variant caller integrated into Strand NGS was used to call out large deletions, insertions, inversions and translocation events.
Figure.
(A) Split-read alignment: In split-read alignment, input sequence reads split into two segments, mapping each segment to a different location on the reference genome as represented by the horizontal brown bars. Mismatch with respect to the reference genome in each sequence reads are shown as red, green and black bars. As seen in this Figure, each read aligns to exon 33 and intron 45 of the DMD gene. The deletion breakpoints are represented with arrows, (B) Lower panel shows the corresponding transcript region of the DMD transcript (NM_004006.2) in reverse strand, to which the split-reads map. Vertical bars represent exons. Based on this, it was accurately determined that the deletion encompassed 149 bp from the 3' end of exon 33 and complete deletion of exons 34-45 of the DMD gene. DMD, Duchenne muscular dystrophy.
Single-nucleotide variation (SNV): SNVs accounted for about 23 per cent of the total patients. There were no specific hotspots found to be associated with the point mutations as these were distributed across different exons among the study patients. Among the patients displaying point mutations as shown in the Table, DMD17 presented a point mutation in exon 21 (c.2661_2662delinsA; protein change p. Glu888Asn*) and DMD18 presented a nonsense mutation in exon 46 (c.6622G>T; protein change p.GLu2208*), which were not reported previously. Patient DMD19 had a previously reported point mutation in exon 8 (c.724C>T; protein change p. Glm242*), whereas a frameshift mutation was observed in patient DMD20 in exon 51 (c.7348dupG; protein change p. Val2450*) and DMD21 displayed a point mutation in exon 7 (c.583C>T; protein change p. Arg195*), which was not previously reported in the Leiden Muscular Dystrophy Database8. Among the point mutations presented in the study group, point mutations in patients DMD17, 18 and 20 were found in the central rod domain and DMD19 and 21 were localized in the ABD of dystrophin.
Ambulatory status, present age, and type of mutation: Ambulatory status of the study patients was examined to analyze the effect of the mutation type with respect to age at which the patients would become wheelchair-bound. The Table shows the ambulatory status, age at loss of ambulation, age at onset, present age and the mutation type of the patients in the study group. There was no association among the ambulatory status, age at loss of ambulation and the type of mutation among the patients.
Discussion
An accurate and comprehensive one-point diagnosis has been a challenge in the case of DMD due to the variability in mutation type and availability of more than one technology such as mPCR20, MLPA and NGS14,21, each with a different sensitivity in identifying the type of mutation22,23. The precise location of the point mutation is necessary for designing and implementing therapeutic strategies such as exon skipping24. MLPA is limited to the precise identification of deletions/duplications, whereas SNVs are undetected; there is, therefore, the need for additional techniques such as Sanger sequencing and NGS. Hence, it is imperative to adapt to a single, comprehensive platform that is robust and a sensitive technology to illuminate all possible information on mutations in the DMD gene. Moreover, employing multiple testing strategies is time-consuming and expensive.
In this study, of the 22 DMD patients, large deletions were found in 16 (DMD1-16), by MLPA, and the same was validated, displaying exactly the same mutations, by NGS. However, SNVs from patients DMD17-21 were not detected using the conventional MLPA technique.
Large deletions were found between exons 8 and 55 in 17 patients and SNVs in five. The deletion of the exons ranged from a minimum of a single exon to a maximum of 11 exons. The deletions were found to be situated majorly in the rod domain of the dystrophin protein. The SNVs in the study group were found to be in the rod domain of dystrophin, while patient DMD21 had a previously reported nonsense mutation in exon 7 (c.583C>T; p. Arg195*)25, situated in the ABD2 within the rod domain where dystrophin binds with the actin filaments26,27.
Previously, methods of detection of DMD included MLPA and array comparative genomic hybridization28, methods that are limited in their capacity to detect large deletions and duplications and do not detect SNVs, thus making NGS the strategy of choice13 as a single-point detection technique especially for DMD. Novel mutations that were not previously reported in large deletions in patient DMD 6 with deletion of exons 18-29 were identified. In patient DMD22 where a large deletion of exons 33-45 by MLPA was identified, NGS showed previously not reported partial deletion of exon 33 and complete deletion of exons 34-45. Partial deletions of an exon in the proband and carrier have been reported13, suggesting a probability of the presence of breakpoints within exons.
Novel variant identified in exons 33-45 of patients DMD22, are predicted to cause premature termination of the protein. Due to the introduction of a premature stop codon, the aberrant transcript will likely be targeted by the nonsense-mediated mRNA decay mechanism, which might result in loss of function. A novel variant identified in DMD6 and DMD22 results in multi-exon deletions involving exons 18-29 and exons 33-45, respectively. Exonic deletion spanning these exons of the DMD gene has been reported in patients with a clinical diagnosis of DMD29,30 and, thus, is likely to be disease-causing.
Exon skipping is one of the most promising strategies in the clinical trial phase30 and used in other strategies such as the premature termination suppression drugs for example, ataluren (PTC124)31. In the case of complete deletion of exons, strategies such as exon skipping can be employed to skip one or more exons to restore the reading frame32. However, clear genetic information on the mutation in the DMD gene is a prerequisite before the patient can benefit from the exon-skipping strategy33. One such example in our study was the partial deletion of exon 33 and complete deletion of exons 34-45. Breakpoints in the DMD gene were previously thought to be within introns until it was reported that breakpoints are also present within exons9. The presence of internal exonic breakpoints results in the partial deletion of exons, which was not identified using MLPA. In our study, a partial deletion in exon 33 in patient DMD22 was identified with NGS, which was not identified using MLPA. An in-frame mutation, which is supposed to present a milder phenotype, could actually be pathogenic in the case of partial deletion within exons. A targeted NGS approach gives a precise location that could identify such breakpoints. Moreover, partial deletions may also change the effect of exon skipping if the antisense oligonucleotides (AONs) are not designed based on the breakpoints within exons13.
One of the limitations of the study was the sample size and it would be appropriate to conduct similar studies using a larger sample size to arrive at a conclusion in using NGS as a single platform in the diagnosis of DMD.
In conclusion, our findings showed NGS as a confirmatory platform for the detection of DMD irrespective of the mutation detected by MLPA. This would have a significant implication in saving on time and cost when compared to sequential testing. Also NGS is important in the context of non-detectable mutation by MLPA. With emerging therapeutic strategies such as exon skipping, NGS would provide more information on the design of AONs for mutations including point mutations and partial exon deletions in DMD.
Acknowledgment
Authorsthank Dr V. Viswanathan, pediatric neurologist, Child Trust Hospital, Chennai, for support and advice for the study, Shri Ravdeep Singh Anand, founder, Dystrophy Annihilation Research Trust (DART), for his contribution toward the research facility and infrastructure; Dr Ashwin Dalal, Centre for DNA Finger-printing and Diagnostics, for his support with MLPA and Anish Joseph Chacko, for proofreading the final manuscript.
Footnotes
Financial support & sponsorship: The study was funded by a 2015 Strand Life Foundation grant, Bengaluru.
Conflicts of Interest: None.
References
- 1.Moser H. Duchenne muscular dystrophy: Pathogenetic aspects and genetic prevention. Hum Genet. 1984;66:17–40. doi: 10.1007/BF00275183. [DOI] [PubMed] [Google Scholar]
- 2.Bushby K, Finkel R, Birnkrant DJ, Case LE, Clemens PR, Cripe L, et al. Diagnosis and management of duchenne muscular dystrophy, part 1: Diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 2010;9:77–93. doi: 10.1016/S1474-4422(09)70271-6. [DOI] [PubMed] [Google Scholar]
- 3.Koumbourlis AC. Scoliosis and the respiratory system. Paediatr Respir Rev. 2006;7:152–60. doi: 10.1016/j.prrv.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Koenig M, Monaco AP, Kunkel LM. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988;53:219–28. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- 5.Chevron MP, Girard F, Claustres M, Demaille J. Expression and subcellular localization of dystrophin in skeletal, cardiac and smooth muscles during the human development. Neuromuscul Disord. 1994;4:419–32. doi: 10.1016/0960-8966(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 6.Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–5. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 7.Koenig M, Beggs AH, Moyer M, Scherpf S, Heindrich K, Bettecken T, et al. The molecular basis for duchenne versus Becker muscular dystrophy: Correlation of severity with type of deletion. Am J Hum Genet. 1989;45:498–506. [PMC free article] [PubMed] [Google Scholar]
- 8.Aartsma-Rus A, Van Deutekom JCT, Fokkema IF, Van Ommen GJ, Den Dunnen JT. Entries in the Leiden duchenne muscular dystrophy mutation database: An overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–44. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- 9.Aartsma-Rus A, Ginjaar IB, Bushby K. The importance of genetic diagnosis for duchenne muscular dystrophy. J Med Genet. 2016;53:145–51. doi: 10.1136/jmedgenet-2015-103387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai KKS, Lo IFM, Tong TMF, Cheng LYL, Lam STS. Detecting exon deletions and duplications of the DMD gene using multiplex ligation-dependent probe amplification (MLPA) Clin Biochem. 2006;39:367–72. doi: 10.1016/j.clinbiochem.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Santos R, Gonçalves A, Oliveira J, Vieira E, Vieira JP, Evangelista T, et al. New variants, challenges and pitfalls in DMD genotyping: Implications in diagnosis, prognosis and therapy. J Hum Genet. 2014;59:454–64. doi: 10.1038/jhg.2014.54. [DOI] [PubMed] [Google Scholar]
- 13.Wei X, Dai Y, Yu P, Qu N, Lan Z, Hong X, et al. Targeted next-generation sequencing as a comprehensive test for patients with and female carriers of DMD/BMD: A multi-population diagnostic study. Eur J Hum Genet. 2014;22:110–8. doi: 10.1038/ejhg.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim BC, Lee S, Shin JY, Kim JI, Hwang H, Kim KJ, et al. Genetic diagnosis of duchenne and Becker muscular dystrophy using next-generation sequencing technology: Comprehensive mutational search in a single platform. J Med Genet. 2011;48:731–6. doi: 10.1136/jmedgenet-2011-100133. [DOI] [PubMed] [Google Scholar]
- 15.Mannan AU, Singh J, Lakshmikeshava R, Thota N, Singh S, Sowmya TS, et al. Detection of high frequency of mutations in a breast and/or ovarian cancer cohort: Implications of embracing a multi-gene panel in molecular diagnosis in India. J Hum Genet. 2016;61:515–22. doi: 10.1038/jhg.2016.4. [DOI] [PubMed] [Google Scholar]
- 16.Fraz S, Haiying G, Nicholas C. Next-generation sequencing library preparation: simultaneous fragmentation and tagging using in vitro transposition. Nat Methods. 2009;6:856. [Google Scholar]
- 17.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh J, Mishra A, Pandian AJ, Mallipatna AC, Khetan V, Sripriya S, et al. Next-generation sequencing-based method shows increased mutation detection sensitivity in an Indian retinoblastoma cohort. Mol Vis. 2016;22:1036–47. [PMC free article] [PubMed] [Google Scholar]
- 19.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–76. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beggs AH, Kunkel LM. Improved diagnosis of duchenne/Becker muscular dystrophy. J Clin Invest. 1990;85:613–9. doi: 10.1172/JCI114482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Yang Y, Liu J, Chen XC, Liu X, Wang CZ, et al. Whole dystrophin gene analysis by next-generation sequencing: A comprehensive genetic diagnosis of duchenne and Becker muscular dystrophy. Mol Genet Genomics. 2014;289:1013–21. doi: 10.1007/s00438-014-0847-z. [DOI] [PubMed] [Google Scholar]
- 22.Janssen B, Hartmann C, Scholz V, Jauch A, Zschocke J. MLPA analysis for the detection of deletions, duplications and complex rearrangements in the dystrophin gene: Potential and pitfalls. Neurogenetics. 2005;6:29–35. doi: 10.1007/s10048-004-0204-1. [DOI] [PubMed] [Google Scholar]
- 23.Kohli S, Saxena R, Thomas E, Singh J, Verma IC. Gene changes in duchenne muscular dystrophy: Comparison of multiplex PCR and multiplex ligation-dependent probe amplification techniques. Neurol India. 2010;58:852–6. doi: 10.4103/0028-3886.73744. [DOI] [PubMed] [Google Scholar]
- 24.Abbs S, Tuffery-Giraud S, Bakker E, Ferlini A, Sejersen T, Mueller CR. Best practice guidelines on molecular diagnostics in duchenne/Becker muscular dystrophies. Neuromuscul Disord. 2010;20:422–7. doi: 10.1016/j.nmd.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Cunniff C, Andrews J, Meaney FJ, Mathews KD, Matthews D, Ciafaloni E, et al. Mutation analysis in a population-based cohort of boys with duchenne or Becker muscular dystrophy. J Child Neurol. 2009;24:425–30. doi: 10.1177/0883073808324770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norwood FL, Sutherland-Smith AJ, Keep NH, Kendrick-Jones J. The structure of the N-terminal actin-binding domain of human dystrophin and how mutations in this domain may cause duchenne or Becker muscular dystrophy. Structure. 2000;8:481–91. doi: 10.1016/s0969-2126(00)00132-5. [DOI] [PubMed] [Google Scholar]
- 27.Juan-Mateu J, Gonzalez-Quereda L, Rodriguez MJ, Baena M, Verdura E, Nascimento A, et al. DMD mutations in 576 dystrophinopathy families: A step forward in genotype-phenotype correlations. PLoS One. 2015;10:e0135189. doi: 10.1371/journal.pone.0135189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.del Gaudio D, Yang Y, Boggs BA, Schmitt ES, Lee JA, Sahoo T, et al. Molecular diagnosis of duchenne/Becker muscular dystrophy: Enhanced detection of dystrophin gene rearrangements by oligonucleotide array-comparative genomic hybridization. Hum Mutat. 2008;29:1100–7. doi: 10.1002/humu.20841. [DOI] [PubMed] [Google Scholar]
- 29.Florentin L, Mavrou A, Kekou K, Metaxotou C. Deletion patterns of Duchenne and Becker muscular dystrophies in Greece. J Med Genet. 1995;32:48–51. doi: 10.1136/jmg.32.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murugan S, Chandramohan A, Lakshmi BR. Use of multiplex ligation-dependent probe amplification (MLPA) for Duchenne muscular dystrophy (DMD) gene mutation analysis. Indian J Med Res. 2010;132:303–11. [PubMed] [Google Scholar]
- 31.Pichavant C, Aartsma-Rus A, Clemens PR, Davies KE, Dickson G, Takeda S, et al. Current status of pharmaceutical and genetic therapeutic approaches to treat DMD. Mol Ther. 2011;19:830–40. doi: 10.1038/mt.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 33.Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar I, van Deutekom J, van Ommen GJ, et al. Theoretic applicability of antisense-mediated exon skipping for duchenne muscular dystrophy mutations. Hum Mutat. 2009;30:293–9. doi: 10.1002/humu.20918. [DOI] [PubMed] [Google Scholar]