Abstract
Background
We aimed to determine the promoter methylation status of the retinoic acid receptor-beta 2 (RARβ2) gene among breast cancer patients and to review relevant studies in this field in various populations.
Methods
We analyzed 400 samples which comprised blood specimens from 102 breast cancer patients, 102 first-degree female relatives of patients, 100 cancer-free females, 48 breast cancer tissues, and 48 adjacent normal breast tissues from the same patients. The RARβ2 methylation status was determined using methylation-specific polymerase chain reaction (MSP) and DNA sequencing methods.
Results
The presence of combined partially methylated (MU) and fully methylated (MM) forms of the RARβ2 gene (MU+MM) in the blood of patients was associated with susceptibility to breast cancer (odds ratio = 4.7, p = 0.05). A significantly higher frequency of the MM genotype was observed in cancer tissue (10.4%) compared to matched adjacent normal breast tissue (0%) (p = 0.02).
Conclusion
We found a higher frequency of RARβ2 gene methylation in the blood and cancer tissues of patients compared to the blood of controls and adjacent normal breast tissues. The survey of studies on various populations demonstrated a higher RARβ2 methylation frequency in breast cancer patients compared to normal individuals, and many reports suggest a significant association between hypermethylation of the gene and susceptibility to breast cancer.
Keywords: Breast cancer, RARβ2, Hypermethylation, Epigenetic, MSP, Iranian population
Introduction
Breast cancer is a heterogeneous malignancy in women associated with high morbidity and mortality [1,2]. Due to the considerably increased incidence rate of breast cancer, it could be considered as one of the most common health problems across different populations [3,4]. In developing countries including Iran, patients diagnosed with breast cancer are 10 years younger compared to those in developed countries [3]. Both genetic and epigenetic changes are involved in the individual susceptibility to breast cancer [5]. Epigenetic modifications alter gene expression through DNA hypermethylation, histone modification, and microRNA mechanisms [6]. In tumor cells, hypermethylation of CpG islands of tumor-related genes can effectively inactivate the gene, whereas in normal cells, CpG islands are not methylated which may lead to gene transcription [7].
Retinoic acid (RA) as a main signaling biomolecule derived from vitamin A is implicated in the control of main developmental processes such as differentiation and cell growth. Based on antiproliferative and differentiation effects of retinoids, RA is considered as the suppressor of cancer cell growth [7,8]. RA binds and activates 2 types of nuclear receptor heterodimers of the retinoic acid receptor (RAR) and the retinoid X receptor (RXR). Among them, the retinoic acid receptor-beta 2 (RARβ2) has a critical role in the chemopreventive effects of retinoids. The tumor suppressor gene RARβ2 is located on chromosome 3p24 and its loss is associated with RA resistance [7,8].
Reduced expression of RARB2 by genetic or epigenetic mechanisms is linked to many human cancers such as cervical [9,10], head and neck [11], non-small cell lung [12], prostate [13], bladder, brain, and breast cancers [4,14,15,16]. Knowledge of the DNA methylation status in the promoter regions of some candidate genes such as RARB2 may be useful for early detection, prognosis, and drug response prediction in breast cancer patients [6,17]. The aim of the present study was to evaluate the methylation status of the RARβ2 gene in the CpG islands of its promoter and to detect whether it is associated with the risk of breast cancer in an Iranian population with Kurdish ethnic background. Furthermore, we reviewed studies that have previously investigated the methylation status of RARβ2 in breast cancer patients.
Materials and Methods
Sample Collection
In the present case-control study, 400 samples including blood specimens from 102 female patients with breast cancer, 102 first-degree female relatives of patients (mother, sister, or daughter), 100 cancer-free females, 48 breast cancer tissues, and 48 adjacent normal breast tissues from the same patients were investigated. The study was approved by the Ethics Committee of Kermanshah University of Medical Sciences, Iran. All individuals agreed to participate in the study and signed informed consent in accordance with the principles of the Helsinki II declaration.
Genomic DNA Extraction
DNA was extracted from EDTA-treated whole blood and from paraffin-embedded tissue blocks using the phenol-chloroform method [21].
Methylation-Specific Polymerase Chain Reaction
The methylation status of RARβ2 was detected using methylation-specific polymerase chain reaction (MSP) as previously described [4]. The specific band intensity of the MSP product (170 bp) was determined using TotalLab TL120 software (TotalLab Ltd., Newcastle upon Tyne, UK). Based on TotalLab TL120 software analysis, only bands with an intensity higher than 50,000 values were considered. 8 samples from patients that were identified as MM (fully methylated) or UU (unmethylated) genotypes using the MSP method were selected for DNA sequencing (Macrogen Inc., Seoul, South Korea).
Statistical Analysis
The significance of differences in the frequencies of genotypes between the 3 study groups and between breast cancer tissues and adjacent normal breast tissues was determined using the chi-square test. Odds ratios (OR) and 95% confidence intervals (95% CI) were measured using SPSS 16.0 software (IBM Corp., Armonk, NY, USA). Statistical significance was assumed at the p < 0.05 level [1,18].
Literature Study and Data Extraction
We included all relevant studies published in the English language related to DNA methylation of the RARβ2 gene in breast cancer patients, obtained from searching the databases MEDLINE/PubMed and Scopus up to March 2016. Using the keywords ‘DNA methylation’, ‘RARBβ2’, and ‘breast cancer’, 21 eligible original articles were identified and included in the systematic review.
Results
The mean age of the patients was 49.8 ± 10.3 years (range 29-79 years). 10 (10.2%) and 28 (28.6%) patients were aged below 30 and 40 years, respectively. The result of direct genomic DNA sequencing from 1 sample is shown in figure 1. The frequencies of RARβ2 combined methylated genotype (MU+MM) in the blood of patients, relatives of patients, and healthy individuals were 8.8, 2.9, and 2%, respectively (χ2 = 6.3, p = 0.042) (table 1). The presence of RARβ2 MU+MM was associated with susceptibility to breast cancer (OR = 4.7, 95% CI 0.99-22.5, p = 0.05) (table 1). A higher frequency of fully methylated RARβ2 was observed in cancer tissue (10.4%) compared to matched adjacent normal breast tissue (0%) (p = 0.02) (table 2). As indicated in table 2, we did not detect the same frequency of RARβ MU+MM in the tumor tissue and white blood cells (WBC) of patients. Our results indicated that the MU+MM genotype of the RARβ2 gene was not correlated with breast cancer prognostic parameters in our population.
Fig. 1.
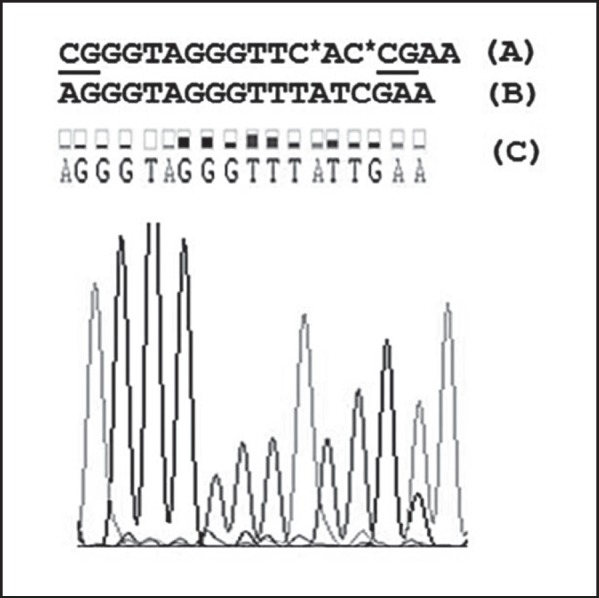
Analysis of a fragment of the RARβ2 gene in the promoter region using a DNA sequencing apparatus (Macrogen Inc., Seoul, South Korea). A Promoter sequence of the RARβ2 gene in the NCBI database; B obtained sequences from MethPrimer software; C sequences obtained by DNA sequencing using a sodium bisulfite-treated sample. Underlined CG represents the CpG islands that have not been modified by sodium bisulfite treatment and remained intact as CG, whereas those C* nucleotides that were not followed by a G nucleotide were modified to T after sodium bisulfite modification (B).
Table 1.
RARß2 methylation status in the blood of healthy individuals, relatives of patients, and patients
| RARß2 genotype | Patients, n (%) (n = 102) | Relatives of patients, n (%) (n = 102) | Healthy individuals, n (%) (n = 100) | χ2, p value | Odds ratio, 95% confidence interval, p |
|---|---|---|---|---|---|
| UU | 93 (91.2) | 99 (97.1) | 98 (98) | ||
| MU | 7 (6.9) | 3 (2.9) | 2 (2) | ||
| MM | 2 (1.9) | 0 (0) | 0 (0) | ||
| 7.67, 0.1a | |||||
| MU+MM | 9 (8.8) | 3 (2.9) | 2 (2) | 4.56, 0.033b 0.18, 0.66c |
4.7, 0.99–22.5, 0.05b |
Overall χ2 comparing 3 genotypes between 3 groups was 7.67, p = 0.1; overall χ2 comparing MU+MM versus UU genotype between 3 groups was 6.3, p = 0.042.
Comparing patients and healthy individuals.
Comparing relatives of patients and healthy individuals.
UU = Unmethylated; MU = partially methylated; MM = fully methylated.
Table 2.
RARß2 methylation status in cancer and adjacent normal breast tissues and blood of patients
| RARß2 genotypes | Cancer tissue, n (%) (n = 48) | Normal tissue, n (%) (n = 48) | Blood of patients, n (%) (n = 48) |
|---|---|---|---|
| UU | 39 (81.3) | 44 (91.7) | 42 (87.5) |
| MU | 4 (8.3) | 4 (8.3) | 4 (8.3) |
| MM | 5 (10.4) | 0 (0) | 2 (4.2) overall χ2 = 5.7, p = 0.22a |
| MU+MM | 9 (18.8) | 4 (8.3) | 6 (12.5) |
| overall χ2 = 2.3, p = 0.3a | |||
χ2 = 5.3, p = 0.07 comparing 3 genotypes between breast cancer tissue and adjacent normal breast tissue; χ2 = 5.3, p = 0.02 comparing MM versus UU between breast cancer tissue and adjacent normal breast tissue; χ2 = 1.39, p = 0.49 comparing 3 genotypes between cancer tissue and blood of patients; χ2 = 2.05, p = 0.35 comparing 3 genotypes between normal breast tissue and blood of patients.
UU = Unmethylated; MU = partially methylated; MM = fully methylated.
Discussion
MSP is a rapid, most widely used, and cost-effective method for gene methylation analysis [19]; however, it is not a quantitative method [20]. Hence, the results obtained in our study could be confirmed using a quantitative MSP method. However, a number of studies have suggested that MSP has a high sensitivity and specificity for the identification of breast cancer cells [21,22]. A frequency of 10-42% for the methylation of RARβ2 in the promoter region of breast cancer tissue has been reported [23]. Among our studied population, the frequency of methylated RARβ2 was found to be 18.8% in cancer tissues, which is lower than that reported for the population of Tehran (36.4%), the capital and a cosmopolitan city of Iran [24]. However, similarity between the frequency of RARβ2 methylation in breast cancer tissue in the present study (18.8%) and some Asian countries such as India (15%) [25] could be attributed to genetic admixture since the Kermanshah province is historically known as the gate of Asia through which passed the important silk road [26]. Also, the type of examined sample (blood or tissue) could be a reason for discrepancy in the reported methylation rates of RARβ2 among various populations. The presence of discordance in the methylation status of the RARβ2 promoter between whole blood samples and tumor tissue has been reported. Further, the frequency of hypermethylation of the gene might be ethnic-dependent. In this study, we identified a low methylation frequency of the RARβ2 gene (2.9%) among first-degree relatives of the patients similar to that among healthy individuals (2%). We detected a significantly higher frequency of fully methylated RARβ2 in cancer tissue (10.4%) compared to matched adjacent normal breast tissue (0%) (p = 0.02) and also blood of breast cancer patients (8.8%) compared to healthy individuals (2%), and this was associated with susceptibility to breast cancer (OR = 4.7, 95% CI 0.99-22.5, p = 0.05).
There are controversial reports regarding the methylation rate of the RARβ2 gene and its association with susceptibility to breast cancer among different populations (table 3). In many populations, including American [27,28,29,30], Chinese [31], Indian [25], Portuguese [32], and Belgian [33], a significant association between the methylation status of the RARβ2 gene and breast cancer incidence has been observed. Further, a meta-analysis by Fang et al. [34] reported that the methylation rate of RARβ2 is significantly increased among breast cancer patients compared to cancer-free controls. In a report from Russia, methylated RARβ2 was found to be 3 times more frequent in breast cancer patients than in fibroadenoma patients [35]. In contrast, a lack of association between the methylation status of RARβ2 and breast cancer risk has been reported in Russian [21], American [35], Turkish [36], and Senegalese [37] populations. Also, some studies found a lack of concordance between RARβ2 methylation in tumor tissue and paired WBC, suggesting that there might not be a direct link between WBC gene methylation and development of breast cancer (table 3).
Table 3.
RARß2 methylation status and breast cancer in different populations
| Population | Year of publication | Methylation method | Specimen | Methylation, % | Main finding | Reference |
|---|---|---|---|---|---|---|
| American | 2001 | MSP | breast cancer tissue white blood cells normal breast tissue |
37.5 3.6 0 |
high percent of methylated RARß2 in carcinomas (p < 0.01) | [28] |
| American | 2003 | MSP | invasive carcinomas in situ carcinomas benign breast tissue |
64 60 33 |
high frequency of methylated RARß2 in both benign lesions and invasive carcinomas (p = 0.05) | [38] |
| American | 2003 | MSP | breast cancer tissue normal breast tissue |
46 0 |
frequent hypermethylation in breast cancer tissue compared to normal tissue (p = 0.01) | [27] |
| American | 2005 | MSP | breast cancer tissue normal tissue of controls |
32 9 |
methylation of RARß2 was associated with increased breast cancer risk (p = 0.002) | [29] |
| American | 2005 | MSP | primary breast tumor sentinel lymph node tumor metastasis |
24 48 |
association of RARß2 hypermethylation with macroscopic SLN metastasis (p < 0.001) | [39] |
| American | 2009 | quantitative MSP |
breast cancer tissue | 27.5 | hypermethylation of RARß2 was inversely associated with histological grade of breast cancer (p = 0.13) | [40] |
| American | 2010 | quantitative MSP |
blood of breast cancer patients blood of women with benign breast disease blood from healthy controls |
6.7 2.3 1.1 |
absence of association of RARß2 methylation with breast cancer risk (p > 0.05) | [41] |
| American | 2011 | quantitative MSP |
breast cancer tissue | 27.5 | absence of association between RARß2 methylation status with intake of vitamin B complex (p > 0.05) | [42] |
| American | 2015 | MSP | blood of breast cancer patients breast cancer tissue blood of healthy individuals |
1.5 44.5 1.5 |
association between WBC DNA methylation of the RARß2 and breast cancer risk (OR: 0.67; 95% CI: 0.55–0.81) | [30] |
| Australian | 2006 | MSP | breast cancer tissue | 26 | RARß2 methylation is linked to poor histological tumor differentiation (p = 0.03) | [23] |
| Belgian | 2009 | quantitative MSP |
breast cancer tissue benign breast tissue |
29 0 |
significantly higher frequency of methylated RARß2 in inflammatory (53%) than in non-inflammatory breast cancer (23%) (p = 0.006) | [33] |
| Chinese | 2010 | MSP | blood of breast cancer patients blood of healthy individuals |
42 4 |
association of methylation status of RARß2 with sporadic breast cancer (p < 0.05) | [31] |
| Egyptian | 2015 | MSP | blood of breast cancer patients blood of breast benign lesion cases blood of healthy subjects |
95.9 14.5 0 |
association of RARß2 methylation with breast cancer risk but not with clinicopathological factors (p < 0.0001) | [22] |
| Japanese | 2012 | OS-MSP | breast cancer tissue normal breast tissue |
78 | high percentage of methylated RARß2 in carcinomas (p value not reported) | [43] |
| Indian | 2008 | MSP | breast cancer tissue normal breast tissues |
15 0 |
association of RARß2 methylation with older age at onset of disease (p = 0.04) | [25] |
| Indian | 2012 | MSP | blood of patients blood of healthy women breast carcinoma tissue adjacent normal tissues |
200 26 6.6 |
absence of association between RAR-ß2 hypermethylation and loss of protein expression (p = 0.70) | [44] |
| Iranian | 2010 | MSP | breast cancer tissue | 36.5 | association of hypermethylated RARß2 with younger age at diagnosis and negative family history of breast cancer (p = 0.033) | [24] |
| Present study- | MSP | breast cancer tissue normal breast tissue blood of breast cancer patients blood of relatives of patients blood of healthy women |
16.3 10.2 8.7 2.9 2 |
significantly higher frequency of methylated RARß2 in tissues and blood of patients than in the blood of 2 control groups (p < 0.05) | ||
| Italian | 2004 | MSP | invasive breast tissue benign breast tissue |
20 0 |
methylated status of RARß2 and its inverse correlation with the BRCA1 promoter (p < 0.03) | [45] |
| Portuguese | 2008 | quantitative MSP |
breast cancer tissue breast normal tissue |
53 25 |
significantly higher RARß2 methylation percentage in breast cancer tissue than in normal tissue (p < 0.01) | [32] |
| Russian | 2006 | MSP | blood of breast cancer patients blood of healthy individuals |
15 0 |
RARß2 methylation in circulating DNA is useful for the detection of malignant tumors (p value not reported) | [21] |
| Russian | 2008 | MSRA | breast cancer tissue breast normal tissue |
46 4 |
absence of association of RARß2 methylation with breast cancer risk (p > 0.05) | [46] |
| Senegalese | 2006 | quantitative MSP |
blood of breast cancer patients blood of healthy individuals |
26 8 |
absence of association of RARß2 methylation with breast cancer risk (p = 0.09) | [37] |
| Tunisian | 2010 | MSP | breast cancer tissue | 66.6 | frequent RARß2 methylation in breast cancer tissue (p = 0.043) | [35] |
| Turkish | 2010 | MethyLight assay |
breast cancer tissue adjacent normal tissue blood of breast cancer patients |
25 25.9 10 |
frequent RARß2 hypermethylation in tumor and adjacent tissues (p < 0.05) | [36] |
MSP = Methylation-specific polymerase chain reaction; OS-MSP = one-step MSP; MSRA = enzyme-linked methylation-sensitive restriction analysis; SLN = sentinel lymph node; WBC = white blood cells.
Conclusion
Our study results indicated that the presence of hypermethylation of the RARβ2 gene in the blood of breast cancer patients was associated with susceptibility to breast cancer. This suggests that RARβ2 hypermethylation in cancer tissue might be an epigenetic marker for susceptibility to breast cancer. The strength of the present research was that the RARβ2 methylation status was studied in both the blood and tissues of patients and that methylation was further determined in relatives of the patients. Despite this strength, the low sample size of studied tissues represents a limitation of our study. Among various populations, higher frequencies of RARβ2 methylation in breast cancer patients compared to cancer-free individuals have been reported, and in many studies, a significant association between hypermethylation of the gene and susceptibility to breast cancer has been observed. Some studies reported an association between the gene methylation rate and histological differentiation and tumor metastasis. The presence of inconsistent findings might be due to sample size, type of studied specimen (blood or tissue), breast cancer subtype, and ethnicity.
Contribution
K.Y. performed experiments and wrote the first draft of the manuscript. Z.R. designed the study and critically revised the manuscript.
Disclosure Statement
The authors declare no potential or actual conflicts of interest in relation to the current article.
Acknowledgement
This work was performed in partial fulfillment of the requirements for PhD by Research by Mr. Kheirolah Yari, and was financially supported by a grant from the Kermanshah University of Medical Sciences Office of Vice Chancellor for Research, Kermanshah, Iran.
References
- 1.Rahimi Z, Yari K, Rahimi Z. Matrix metalloproteinase-9-1562T allele and its combination with MMP-2-735 C allele are risk factors for breast cancer. Asian Pac J Cancer Prev. 2014;16:1175–1179. doi: 10.7314/apjcp.2015.16.3.1175. [DOI] [PubMed] [Google Scholar]
- 2.Yari K, Rahimi Z, Payandeh M, Rahimi Z. MMP-7 A-181G polymorphism in breast cancer patients from Western Iran. Breast Care (Basel) 2015;10:398–402. doi: 10.1159/000442231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pirouzpanah S, Taleban F-A, Mehdipour P, Atri M. Association of folate and other one-carbon related nutrients with hypermethylation status and expression of RARB, BRCA1, and RASSF1A genes in breast cancer patients. J Mol Med (Berl) 2015;93:917–934. doi: 10.1007/s00109-015-1268-0. [DOI] [PubMed] [Google Scholar]
- 4.Yari K, Payandeh M, Rahimi Z. Association of the hypermethylation status of PTEN tumor suppressor gene with the risk of breast cancer among Kurdish population from Western Iran. Tumour Biol. 2016;37:8145–8152. doi: 10.1007/s13277-015-4731-1. [DOI] [PubMed] [Google Scholar]
- 5.Marzese DM, Hoon DS, Chong KK, et al. DNA methylation index and methylation profile of invasive ductal breast tumors. J Mol Diagn. 2012;14:613–622. doi: 10.1016/j.jmoldx.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Lubecka-Pietruszewska K, Kaufman-Szymczyk A, Stefanska B, Fabianowska-Majewska K. Folic acid enforces DNA methylation-mediated transcriptional silencing of PTEN, APC and RARbeta2 tumour suppressor genes in breast cancer. Biochem Biophys Res Commun. 2013;430:623–628. doi: 10.1016/j.bbrc.2012.11.103. [DOI] [PubMed] [Google Scholar]
- 7.Moison C, Senamaud-Beaufort C, Fourrière L, et al. DNA methylation associated with polycomb repression in retinoic acid receptor β silencing. FASEB J. 2013;27:1468–1478. doi: 10.1096/fj.12-210971. [DOI] [PubMed] [Google Scholar]
- 8.Sirchia SM, Ferguson AT, Sironi E, et al. Evidence of epigenetic changes affecting the chromatin state of the retinoic acid receptor beta2 promoter in breast cancer cells. Oncogene. 2000;19:1556–1563. doi: 10.1038/sj.onc.1203456. [DOI] [PubMed] [Google Scholar]
- 9.Ivanova T, Petrenko A, Gritsko T, et al. Methylation and silencing of the retinoic acid receptor-β2 gene in cervical cancer. BMC Cancer. 2002;2:4. doi: 10.1186/1471-2407-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wongwarangkana C, Wanlapakorn N, Chansaenroj J, Poovorawan Y. Retinoic acid receptor beta promoter methylation and risk of cervical cancer. World J Virol. 2018;7:1. doi: 10.5501/wjv.v7.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu X-C, Ro JY, Lee JS, et al. Differential expression of nuclear retinoid receptors in normal, premalignant, and malignant head and neck tissues. Cancer Res. 1994;54:3580–3587. [PubMed] [Google Scholar]
- 12.Li Y, Lu DG, Ma YM, Liu H. Association between retinoic acid receptor-β hypermethylation and NSCLC risk: a meta-analysis and literature review. Oncotarget. 2017;8:5814. doi: 10.18632/oncotarget.14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dou M, Zhou X, Fan Z, et al. Clinical significance of retinoic acid receptor beta promoter methylation in prostate cancer: a meta-analysis. Cell Physiol Biochem. 2018;45:2497–2505. doi: 10.1159/000488268. [DOI] [PubMed] [Google Scholar]
- 14.Klajic J, Fleischer T, Dejeux E, et al. Quantitative DNA methylation analyses reveal stage dependent DNA methylation and association to clinico-pathological factors in breast tumors. BMC Cancer. 2013;13:456. doi: 10.1186/1471-2407-13-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadeq V, Isar N, Manoochehr T. Association of sporadic breast cancer with PTEN/MMAC1/TEP1 promoter hypermethylation. Med Oncol. 2011;28:420–423. doi: 10.1007/s12032-010-9473-8. [DOI] [PubMed] [Google Scholar]
- 16.Hashemi M, Rezaei H, Eskandari-Nasab E, Kaykhaei M-A, Taheri M. Association of promoter methylation and 32-bp deletion of the PTEN gene with susceptibility to metabolic syndrome. Mol Med Rep. 2013;7:342–346. doi: 10.3892/mmr.2012.1174. [DOI] [PubMed] [Google Scholar]
- 17.Conway K, Edmiston SN, May R, et al. DNA methylation profiling in the Carolina Breast Cancer Study defines cancer subclasses differing in clinicopathologic characteristics and survival. Breast Cancer Res. 2014;16:450. doi: 10.1186/s13058-014-0450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yari K, Rahimi Z, Moradi MT, Rahimi Z. The MMP-2-35 C allele is a risk factor for susceptibility to breast cancer. Asian Pac J Cancer Prev. 2013;15:6199–6203. doi: 10.7314/apjcp.2014.15.15.6199. [DOI] [PubMed] [Google Scholar]
- 19.Licchesi JD, Herman JG. Methylation-specific PCR. In: Jörg Tost, DNA Methylation , editor. Berlin: Springer; 2005. pp. pp 305–323. [Google Scholar]
- 20.Shen L, Waterland RA. Methods of DNA methylation analysis. Curr Opin Clin Nutr Metab Care. 2007;10:576–581. doi: 10.1097/MCO.0b013e3282bf6f43. [DOI] [PubMed] [Google Scholar]
- 21.Skvortsova T, Rykova E, Tamkovich S, et al. Cell-free and cell-bound circulating DNA in breast tumours: DNA quantification and analysis of tumour-related gene methylation. Brit J Cancer. 2006;94:1492–1495. doi: 10.1038/sj.bjc.6603117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swellam M, Abdelmaksoud MD, Sayed Mahmoud M, et al. Aberrant methylation of APC and RARβ2 genes in breast cancer patients. IUBMB Life. 2015;67:61–68. doi: 10.1002/iub.1346. [DOI] [PubMed] [Google Scholar]
- 23.Li S, Rong M, Iacopetta B. DNA hypermethylation in breast cancer and its association with clinicopathological features. Cancer Lett. 2006;237:272–280. doi: 10.1016/j.canlet.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Pirouzpanah S, Taleban FA, Atri M, Abadi A-R, Mehdipour P. The effect of modifiable potentials on hypermethylation status of retinoic acid receptor-beta2 and estrogen receptor-alpha genes in primary breast cancer. Cancer Causes Control. 2010;21:2101–2111. doi: 10.1007/s10552-010-9629-z. [DOI] [PubMed] [Google Scholar]
- 25.Bagadi SAR, Prasad CP, Kaur J, et al. Clinical significance of promoter hypermethylation of RASSF1A, RARβ2, BRCA1 and HOXA5 in breast cancers of Indian patients. Life Sci. 2008;82:1288–1292. doi: 10.1016/j.lfs.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 26.Rahimi Z, Akramipour R, Nagel RL, et al. The β-globin gene haplotypes associated with Hb D-Los Angeles (β121 (GH4) Glu→Gln) in western Iran. Hemoglobin. 2006;30:39–44. doi: 10.1080/03630260500454105. [DOI] [PubMed] [Google Scholar]
- 27.Fackler MJ, McVeigh M, Evron E, et al. DNA methylation of RASSF1A, HIN-1, RAR-β, Cyclin D2 and Twist in in situ and invasive lobular breast carcinoma. Int J Cancer. 2003;107:970–975. doi: 10.1002/ijc.11508. [DOI] [PubMed] [Google Scholar]
- 28.Evron E, Dooley WC, Umbricht CB, et al. Detection of breast cancer cells in ductal lavage fluid by methylation-specific PCR. Lancet. 2001;357:1335–1336. doi: 10.1016/s0140-6736(00)04501-3. [DOI] [PubMed] [Google Scholar]
- 29.Lewis CM, Cler LR, Bu D-W, et al. Promoter hypermethylation in benign breast epithelium in relation to predicted breast cancer risk. Clin Cancer Res. 2005;11:166–172. [PubMed] [Google Scholar]
- 30.Cho YH, McCullough LE, Gammon MD, et al. Promoter hypermethylation in white blood cell DNA and breast cancer risk. J Cancer. 2015;6:819. doi: 10.7150/jca.12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jing F, Yuping W, Yong C, et al. CpG island methylator phenotype of multigene in serum of sporadic breast carcinoma. Tumor Biol. 2010;31:321–331. doi: 10.1007/s13277-010-0040-x. [DOI] [PubMed] [Google Scholar]
- 32.Jeronimo C, Monteiro P, Henrique R, et al. Quantitative hypermethylation of a small panel of genes augments the diagnostic accuracy in fine-needle aspirate washings of breast lesions. Breast Cancer Res Treat. 2008;109:27–34. doi: 10.1007/s10549-007-9620-x. [DOI] [PubMed] [Google Scholar]
- 33.Van der Auwera I, Bovie C, Svensson C, et al. Quantitative assessment of DNA hypermethylation in the inflammatory and non-inflammatory breast cancer phenotypes. Cancer Biol Ther. 2009;8:2252–2259. doi: 10.4161/cbt.8.23.10133. [DOI] [PubMed] [Google Scholar]
- 34.Fang C, Jian Z-Y, Shen X-F, et al. Promoter methylation of the retinoic acid receptor beta2 (RARβ2) is associated with increased risk of breast cancer: a PRISMA compliant meta-analysis. PLoS One. 2015;10:e0140329. doi: 10.1371/journal.pone.0140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karray-Chouayekh S, Trifa F, Khabir A, et al. Aberrant methylation of RASSF1A is associated with poor survival in Tunisian breast cancer patients. J Cancer Res Clin Oncol. 2010;136:203–210. doi: 10.1007/s00432-009-0649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho YH, Yazici H, Wu H-C, et al. Aberrant promoter hypermethylation and genomic hypomethylation in tumor, adjacent normal tissues and blood from breast cancer patients. Anticancer Res. 2010;30:2489–2496. [PMC free article] [PubMed] [Google Scholar]
- 37.Hoque MO, Feng Q, Toure P, et al. Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. J Clin Oncol. 2006;24:4262–4269. doi: 10.1200/JCO.2005.01.3516. [DOI] [PubMed] [Google Scholar]
- 38.Pu RT, Laitala LE, Alli PM, et al. Methylation profiling of benign and malignant breast lesions and its application to cytopathology. Modern Pathol. 2003;16:1095–1101. doi: 10.1097/01.MP.0000095782.79895.E2. [DOI] [PubMed] [Google Scholar]
- 39.Shinozaki M, Hoon DS, Giuliano AE, et al. Distinct hypermethylation profile of primary breast cancer is associated with sentinel lymph node metastasis. Clin Cancer Res. 2005;11:2156–2162. doi: 10.1158/1078-0432.CCR-04-1810. [DOI] [PubMed] [Google Scholar]
- 40.Tao MH, Shields PG, Nie J, et al. DNA hypermethylation and clinicopathological features in breast cancer: the Western New York Exposures and Breast Cancer (WEB) Study. Breast Cancer Res Treat. 2009;114:559–568. doi: 10.1007/s10549-008-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brooks JD, Cairns P, Shore RE, et al. DNA methylation in pre-diagnostic serum samples of breast cancer cases: results of a nested case-control study. Cancer Epidemiol. 2010;34:717–723. doi: 10.1016/j.canep.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tao M-H, Mason JB, Marian C, et al. Promoter methylation of E-cadherin, p16, and RAR-β2 genes in breast tumors and dietary intake of nutrients important in one-carbon metabolism. Nutr Cancer. 2011;63:1143–1150. doi: 10.1080/01635581.2011.605982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamoto N, Nakayama T, Kajita M, et al. Detection of aberrant promoter methylation of GSTP1, RASSF1A, and RARβ2 in serum DNA of patients with breast cancer by a newly established one-step methylation-specific PCR assay. Breast Cancer Res Treat. 2012;132:165–173. doi: 10.1007/s10549-011-1575-2. [DOI] [PubMed] [Google Scholar]
- 44.Mirza S, Sharma G, Parshad R, et al. Clinical significance of Stratifin, ERα and PR promoter methylation in tumor and serum DNA in Indian breast cancer patients. Clin Biochem. 2010;43:380–386. doi: 10.1016/j.clinbiochem.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 45.Parrella P, Poeta ML, Gallo AP, et al. Nonrandom distribution of aberrant promoter methylation of cancer-related genes in sporadic breast tumors. Clin Cancer Res. 2004;10:5349–5354. doi: 10.1158/1078-0432.CCR-04-0555. [DOI] [PubMed] [Google Scholar]
- 46.Khodyrev D, Loginov V, Pronina I, et al. Methylation of promoter region of RAR-β2 gene in renal cell, breast, and ovarian carcinomas. Russ J Genet. 2008;44:983–988. [PubMed] [Google Scholar]


