Abstract
Apoptosis-linked gene-2 (ALG-2), also known as programmed cell death 6 (PDCD6), has recently been reported to be aberrantly expressed in various tumors and required for tumor cell viability. The aim of the present study was to investigate whether ALG-2 plays a crucial role in tumor cell proliferation, migration and tumorigenicity. In this study, we examined the expression of PDCD6 in glioblastoma cell lines and found that ALG-2 was generally expressed in glioblastoma cell lines. We also performed an analysis of an online database and found that high expression of ALG-2 was associated with poor prognosis (p = 0.039). We found that over-expression of ALG2 in glioblastoma could inhibit cell proliferation and, conversely, that down-regulation of ALG2 could promote cell proliferation. Further studies showed that over-expression of ALG2 inhibited the migration of tumor cells, whereas down-regulation of ALG2 promoted tumor cell migration. Finally, in vitro and in vivo studies showed that over-expression of ALG2 inhibited the tumorigenic ability of tumor cells, while down-regulation of ALG2 promoted tumor cell tumorigenic ability. In conclusion, ALG2 has a tumor suppressive role in glioblastoma and might be a potential target for the treatment of glioblastoma.
Keywords: ALG-2, PDCD6, Proliferation, Migration, Glioblastoma
1. Introduction
Glioblastoma (GBM) is the most common malignant intracranial tumor and is mainly characterized by high proliferation, invasion, vascularization and high mortality [1]. Despite the increasing clinical practice in recent years, the standard procedure for the treatment of GBM remains surgical excision followed by chemoradiotherapy. Additionally, the survival of GBM patients is still poor, with a median survival rate of only approximately 15 months [2]. Therefore, the exploration of new potential biomarkers is urgently needed for the current treatment of GBM.
The calcium-binding protein apoptosis-linked gene-2 (ALG-2), also known as programmed cell death 6 (PDCD6), is associated with cell proliferation and death. It was originally identified as a novel gene linked to apoptosis [3] and is a ubiquitously expressed protein containing five serially repetitive EF-hand structures [4]. Previous studies have reported that ALG2 participates in T-cell receptor-Fas and glucocorticoid-induced apoptosis [5,6], as well as endoplasmic reticulum stress induced apoptosis during organ formation [7,8]. In addition, many tumor studies have analyzed the expression of ALG2 in clinical tumor tissues or cell lines and found that ALG2 has opposing effects in different tumors. ALG2 is up-regulated in hepatomas, lung cancer tissue and metastatic ovarian cancer cells [9,10]; moreover, ALG2 positively regulates cell migration and invasion in metastatic ovarian cancer cells [10]. In contrast, decreased ALG2 expression was observed in non-small cell lung cancer (NSCLC) and gastric cancer [11,12]. ALG2 has not yet been defined as an oncogene or tumor suppressor. Thus, we examined the role of ALG2 in GBM.
In the current study, we focus on the function of ALG2 in glioblastoma cell proliferation, migration and tumorigenicity. We characterized ALG2 as a novel tumor suppressor gene that inhibits the proliferation, migration and tumorigenicity of GBM cells. In conclusion, our results suggested that ALG2 is a significant prognostic biomarker in GBM.
2. Materials and methods
2.1. Cell lines and culture
The human glioblastoma cell lines A172, LN229, U87 and U251 and the human embryonic renal cell line 293FT were purchased from ATCC (Manassas, VA, United States). Glioblastoma cells were cultured in DMEM (Life Technologies, United States) supplemented with 10% fetal bovine serum (Invitrogen) and 1% penicillin /streptomycin (Life Technologies, United States). 293FT cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin, 0.5 mg/ml G418, 4 mM L-glutamine, 0.1 mm non-essential amino acid and 1 mm sodium pyruvate. All cells were maintained in a 5% CO2 humidified incubator at 37 °C.
2.2. BrdU staining
Cells (2 × 104) were plated in 24-well plates and incubated with 10 μg/ml BrdU for 30 min at 37 °C. The cells were fixed in 4% paraformaldehyde, permeabilized with 0.3% Triton X-100 and treated with 1 mol/L HCL. The cells were blocked with 10% goat serum in PBS for 1 h, incubated with a primary rat antibody against BrdU (1:200, ab6326; Abcam) overnight at 4 °C and then incubated a secondary antibody. For the nuclear staining, 300 nM DAPI was used.
2.3. Western blotting
Cells were lysed, and then, the protein was separated on a 10% SDS-PAGE gel and transferred onto a polyvinylidene fluoride membrane. The polyvinylidene fluoride membrane was then incubated with 5% milk for 1 h and probed with 1:1000 rabbit polyclonal ALG2 antibody (ab133326, Abcam) or 1:2000 α-tubulin mouse polyclonal antibody (AT819, Beyotime). The secondary antibodies utilized were HRP-labeled goat anti-mouse (A0216, Beyotime) and goat anti-rabbit IgG (A0208, Beyotime). Appropriate secondary antibodies (HRP-labeled goat anti-mouse or anti-rabbit IgG) were used at a dilution of 1:500.
2.4. Cell viability assay
Transfected cells (1 × 103) were seeded in 96-well plates and maintained in an incubator containing 5% CO2 at 37 °C. At the indicated time points, 10 μl of CCK-8 was added to each well and incubated at 37 °C for 2 h, after which the absorbance at 450 nm was measured. All experiments were independently repeated three times.
2.5. Migration and wound healing assay
Glioblastoma cells were seeded in 6-well plates and grown to near confluence. The confluent cell monolayer was scratched with a white micropipette tip to create an artificial ‘wound.’ The media was removed, and the cultures were washed twice with PBS to remove the floating cells and then refreshed with fresh serum-free medium. Images were obtained at 36 h using an inverted phase-contrast microscope (Olympus Inc., Shinjuku-ku, Tokyo, Japan). All tests were performed in triplicate. For the transwell assay, a transwell system with permeable filters was used for the migration assays. Briefly, 105 cells were added to the upper chamber containing 1% FBS, and 10% FBS was added to the lower chamber in accordance with the manufacturer’s protocol. The cells were incubated for 4 h, and migrated cells were fixed in methanol and then stained with 0.1% crystal violet for 20 min. After images were obtained, the stained cells were solubilized with 33% acetic acid and quantified on a microplate reader at a wavelength of 560 nm.
2.6. Plasmids and transfection
ALG2si-1#, ALG2si-2# and GFPsi specific sequences were synthesized at BGI (Beijing, China) and cloned into the pLKO.1 vector. Flag-tagged full-length ALG2 sequences were cloned by PCR-based amplification and ligated into the pCDH-CMV-EF1-copGFP vector. Recombinant transfection was performed with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). The target plasmid was transfected into 293FT cells with packaging plasmids (pLP1, pLP2, and pLP/VSVG) for 48 h. The supernatant was collected and filtered and then used to infect the target cells (U87 and LN229) in the presence of polybrene.
2.7. RNA purification and reverse transcription-polymerase chain reaction
Total RNA was extracted from the cells using TRIzola® reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The methods for reverse transcription and quantitative real-time PCR reaction have been previously described [13].
2.8. Patient data analysis
Kaplan–Meier analysis and survival curves were downloaded from the online database R2: microarray analysis and visualization platform (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi).
2.9. Animal studies
Four-week-old NOD/SCID female mice were used for the xenograft assays. For the subcutaneous injections, 1 × 106 cells suspended in PBS were injected into both flanks of NOD/SCID mice. The diameter of each xenograft tumor was measured with a caliper when the tumor was visible, and a tumor growth curve was created. Eight days after tumor growth, the tumors were removed and weighed. For the orthotopic transplant test, 1 × 105 cells suspended in 10 μl of PBS were injected into the brains of NOD/SCID mice. The mice were maintained in an SPF room, and the survival time was recorded and plotted using GraphPad Prism. All studies were approved by the Purdue Animal Care and Use Committee of Southwestern University.
2.10. IHC assay
The immunohistochemistry (IHC) staining method has been previously described [14]. Ki-67 positive cells were counted in randomly chosen microscopic fields.
2.11. Statistical analysis
All data were collected from at least three independent experiments, and the results are presented as the mean ± S.D. P-values were calculated via Student’s t-test using GraphPad Prism (version 6.0), and P < 0.05 was considered significant.
3. Results
3.1. Low ALG2 expression is indicative of poor prognosis in glioblastoma patients
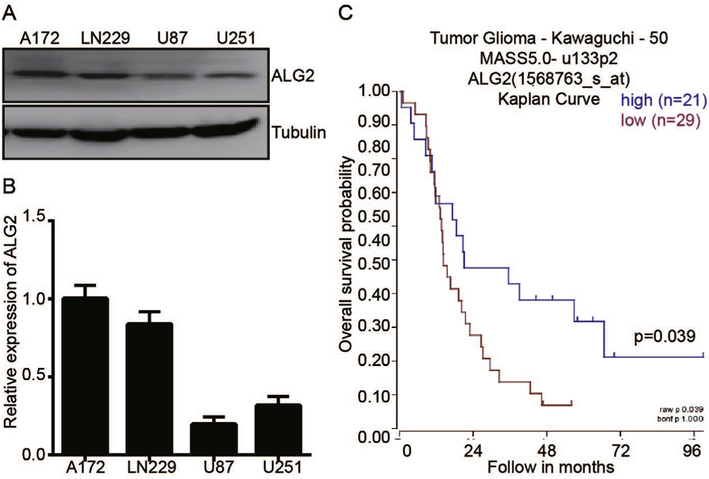
Several reports have revealed that ALG2 is abnormally expressed in a variety of tumors [10,15–18]; however, no results have been reported in GBM patients. To determine the role of ALG2 in GBM, we performed a Western blot and an qRT–PCR assay in 4 GBM cell lines. We found that ALG2 is expressed in all 4 cell lines, with the A172 and LN229 cell lines exhibiting high levels of ALG2 and U87 and U251 exhibiting low levels of ALG2 (Fig. 1A and Fig. 1B). The A172 cell line, which exhibits high levels of ALG2, has no tumorigenic capability in immunosuppressed mice, indicating that ALG2 may play a tumor-suppressor activity in GBM. To confirm this result, we assessed the correlation between the ALG2 expression level and the GBM patient survival state using an online database (Tumor Glioma-Kawaguchi- 50). A Kaplan-Meier analysis of overall survival shows that low ALG2 expression is associated with a poor outcome, while high ALG2 expression is associated with a better outcome, p = 0.039 (Fig. 1C). These data suggest that ALG2 may act as a tumor-suppressor gene in GBM. To elucidate the role of ALG2 in GBM, we selected the U87 and LN229 cell lines for subsequent experiments in this study.
Fig. 1.
Low expression of ALG2 is indicative of poor survival in GBM. The ALG2 expression level in four GBM cell lines was analyzed by Western blot (A) and qRT–PCR (B). (C) Kaplan-Meier analysis of progression-free survival using the online dataset from the Tumor Glioma Kawaguchi database; log-rank test p-values were indicated.
3.2. ALG2 is required for cell proliferation in GBM cells
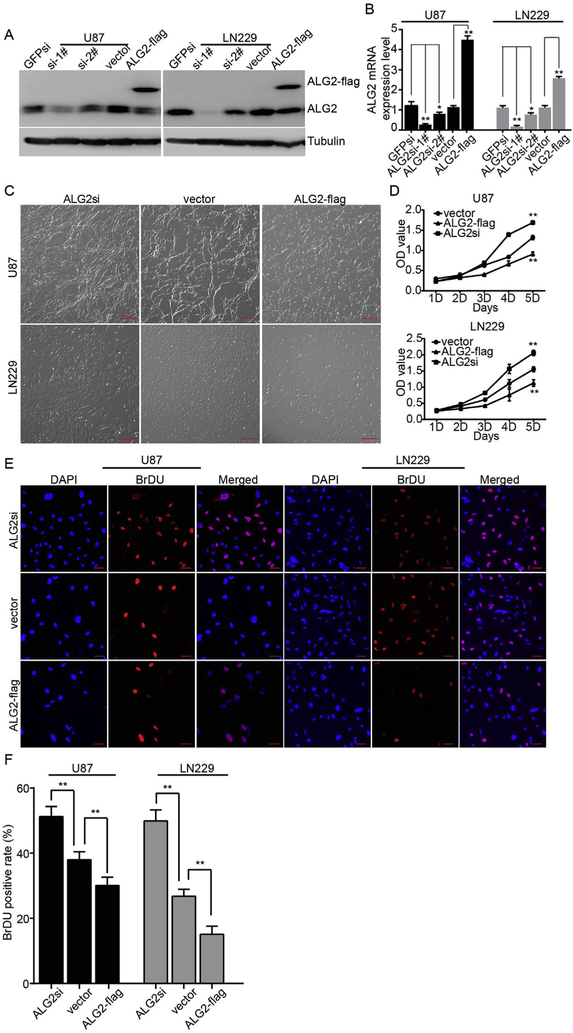
To evaluate the requirement of ALG2 in GBM cell proliferation, we knocked down and over-expressed ALG2 in U87 and LN229 cells using lentiviral expression systems. For the knock-down experiments, we designed two pairs of different interfering primers (si-1# and si-2#) against different regions of the ALG2 sequence and used GFPsi as a control. Western blotting and qRT–PCR assays showed that ALG2si-1# more effectively down-regulated ALG2 expression in U87 and LN229 cells than ALG2si-2#. We therefore used the ALG2si-1# plasmid for subsequent experiments (Fig. 2A and B). In addition, we also over-expressed ALG2 fused with GFP in U87 and LN229 cells. As expected, the expression of the fusion proteins resulted in a significant up-regulation of ALG2 in these two cell lines (Fig. 2A and B). Both GFPsi and empty vector were used as negative controls and did not affect ALG2 expression; thus, the empty vector was used as a negative control for the subsequent experiments. Visualization of the cells under a microscope revealed a significant change in cell number compared to that observed in the vector group. Additionally, ALG2si increased cell proliferation, while ALG2-flag inhibited cell proliferation (Fig. 2C and D). These data suggest that ALG2 negatively regulates GBM cell proliferation. To confirm this finding, we performed a CCK-8 assay for 5 days and obtained results similar to those shown in Fig. 2C and D. Accordingly, a BrDU assay showed that over-expression of ALG2 significantly decreased the number of BrDU-positive cells and that down-regulation of ALG2 increased the number of BrDU-positive cells (Fig. 2E and F). These results suggested that ALG2 negatively regulates GBM cell proliferation.
Fig. 2.
ALG2 inhibits GBM cell proliferation. Over-expression or knock down of ALG2 in U87 and LN229 cells was determined by Western blot (A) and qRT–PCR (B) to detect ALG2 expression levels. (C) Morphologic examination of GBM cells in which ALG2 was over-expressed or knocked down; scale bar is 100 μm. (D) A CCK-8 assay was performed to determine the effect of over-expression or knock down of ALG2 on the proliferation of U87 and LN229 cells. (E) A BrdU assay was performed; positive cells are indicated in the fluorescent images, and the quantification of BrdU positive ratios is shown in (F), *p < 0.05, **p < 0.01.
3.3. ALG2 is involved in GBM cell migration
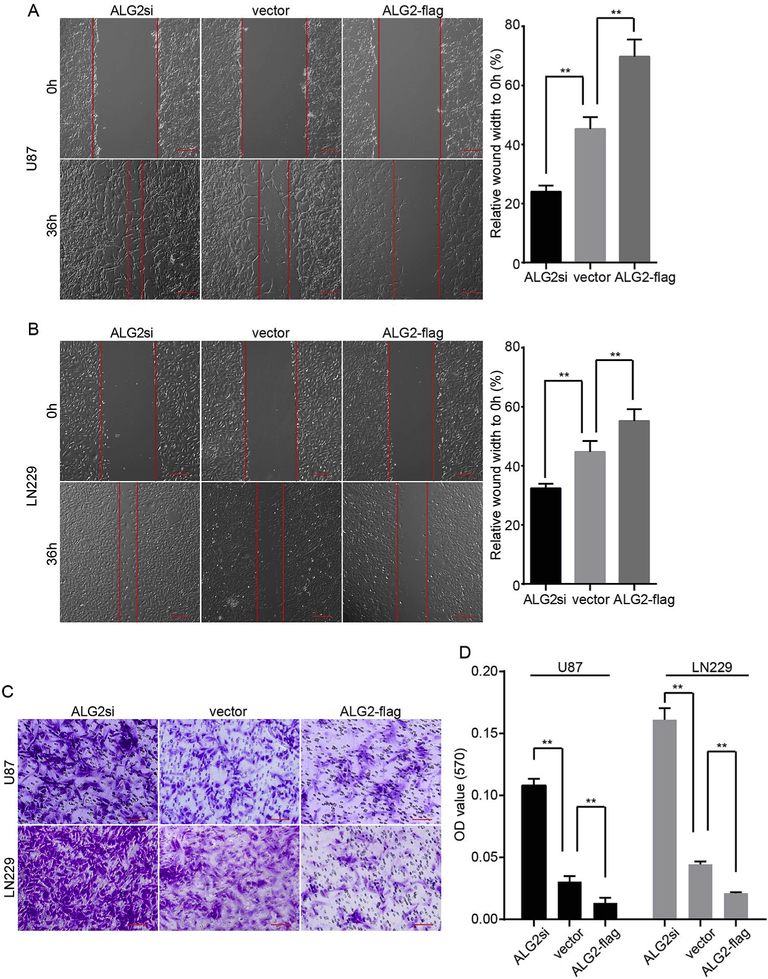
Next, we performed wound healing and transwell assays to determine if the migration ability of GBM cells was affected by over-expression or knock down of ALG2. The wound healing assays revealed that the wounds were significantly wider in the ALG2-flag group than in the vector group. Conversely the wounds within the ALG2si group were narrower than those in the vector group after 36 h (Fig. 3A and B). Consistently, the transwell migration assays revealed a significant decrease in the migration of cells expressing ALG2-flag as compared to that observed in the vector group, while the migration rate of the ALG2si group was higher than those of the vector group (Fig. 3C and D). These results suggested that knock down of ALG2 increases the migration ability of GBM cells, while over-expression of ALG2 inhibits GBM cell migration ability. Together, these findings suggest that ALG2 plays an essential role in GBM cell migration.
Fig. 3.
ALG2 inhibits the migration capacity of GBM cells. Would healing and transwell assays were performed to determine whether ALG2 participates in GBM cell migration. (A) and (B) show the migration rate of U87 (A) and LN229 (B) cells in which ALG2 was over-expressed or knocked down; quantification of the relative migration rate is shown in the right panel. (C) U87 and LN229 cells from the transwell assay were stained with crystal violet and representative images are presented. (D) The quantification of the migration rate. Statistics were calculated using a two-tailed Student’s t-test, **p < 0.01.
3.4. ALG2 is involved in GBM cell self-renewal and tumorigenesis in vitro and in vivo
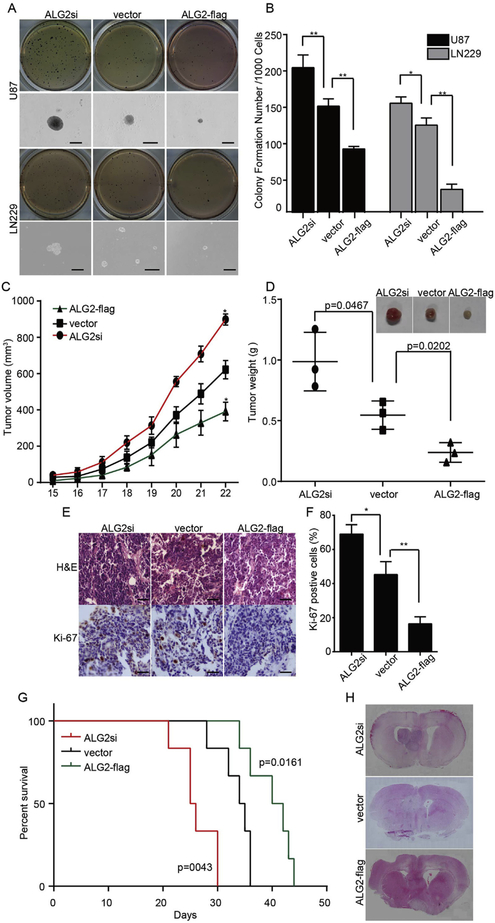
We next evaluated the role of ALG2 in the self-renewal and tumorigenesis abilities of GBM cells. Soft agar assays were performed to evaluate self-renewal ability in vitro. Over-expression of ALG2 results in a decrease in the number and size of colonies compared to those in the vector group. Additionally, cells from the ALGsi group showed an increase in the number and size of colonies compared to those in the vector group (Fig. 4A and B). The subcutaneous xenograft experiment revealed that tumors formed by ALG2si-U87 cells grew more quickly than cells in the vector control group, while cells expressing ALG2-flag grew much more slowly than cells in the vector group (Fig. 4C). After 3 weeks, the mice were sacrificed, and the tumors were excised, weighted and photographed. Accordingly, compared to tumors from the vector group, tumors formed by ALG2si-U87 cells were the heaviest, and tumors from the ALG2-flag group were the lightest (Fig. 4D). The tumor tissues were also subjected to IHC staining for Ki-67 (a cell proliferation marker), which showed that ALG2 inhibited cell proliferation in vivo. These results suggested that ALG2 inhibits GBM cell self-renewal and tumorigenesis. To further confirm these conclusions, we performed an orthotopic transplant experiment using U87 cells. The survival time of animals injected with cells expressing ALG2-flag was significantly longer than the survival time of animals in the vector group, while the survival time of the ALG2si group was significantly decreased (Fig. 4G). H&E staining revealed a significant increase in the tumor size of the ALG2si group compared to vector group and a significant decrease in tumor size in the ALG2-flag group. Taken together, these results confirm that ALG2 inhibits the self-renewal and tumorigenesis abilities of GBM cells.
Fig. 4.
ALG2 plays an important role in the tumorigenicity of GBM cells in vitro and in vivo. (A) Representative images of soft agar colony-formation assays; colonies were stained with MTT and scored (B). The growth curve (C), images and weight (D) of tumors formed by U87 cells in which ALG2 was over-expressed or knocked down; p-values are indicated. (E) Hematoxylin and eosin (H&E) and IHC staining of Ki-67 in corresponding tumors. (F) The quantification of Ki-67 positive cells, calculated by counting cells from 5 random fields. Statistics were calculated using a two-tailed Student’s t-test, *p < 0.05, **p < 0.01. (G) The survival times of mice bearing U87 GBM tumors in which ALG2 was over-expressed or knocked down and of the control group; n = 6, p-values were determined by the log-rank test and are indicated. (H) H&E staining of mouse brains.
4. Discussion
In this study, we identified ALG2 as a novel tumor suppressor gene in GBM. High levels of ALG2 expression indicated a good outcome, whereas low ALG2 expression levels indicated a poor outcome as determined by the analysis of an online database, suggesting that ALG2 may play an important role in the pathogenesis of glioblastoma. The apoptosis-linked gene-2 (ALG2), also known as programmed cell death 6 (PDCD6), is dysregulated in tumors of various origins. Here, we found that ALG2 controls cell proliferation and migration; down-regulation of ALG2 increased GBM cell proliferation and migration, and up-regulation of ALG2 attenuated cell proliferation and migration. The potential for ALG2 to act as a tumor suppressor gene has been reported. Over-expression of ALG2 inhibited VEGF-induced proliferation and invasion by targeting the PI3K/mTOR/p70S6K signaling pathway [19]. Yan Huang et al. reported that ALG2 participates in FSH-stimulated ovarian cancer cell proliferation and inhibits cell apoptosis through the PI3K/AKT and SAPK/JNK signal transduction pathways [16]. Moreover, Kazuho Suzuki et al. reported ALG2 as a novel p53-responsive gene that accumulates in the nucleus in response to DNA damage-induced apoptosis [20]. Nevertheless, numerous reports revealed that ALG2 is up-regulated in a variety of human tumors. Berchtold et al. reported the up-regulation of ALG2 in hepatomas and lung cancer tissue and the contribution of ALG2 to cancer cell viability [9,21]. Similar results were reported by Won Sang Park et al., which revealed ALG2 as a prognostic biomarker for advanced gastric cancer [15]. In addition, ALG2 is highly expressed in metastatic ovarian cancer cells and positively regulates cell migration and invasion [10]. PDCD6 has also been identified as a potential prognostic biomarker for early stage adenocarcinoma and advanced-stage oral cancer [22,23]. The opposing effects of ALG2 in these tumors suggest that its function may be species or cell-type specific. Hence, there is an urgent need to address the question of whether ALG2 plays an important role in GBM cells. Our study is the first report suggesting that ALG2 is an important tumor-suppressor gene in GBM. In conclusion, our results demonstrated that ALG2 inhibits the proliferation, migration, self-renewal and tumorigenic ability of GBM cells. Nevertheless, the molecular mechanism by which ALG2 regulates GBM cell proliferation, migration, self-renewal and tumorigenesis require further study.
Acknowledgments
Our work was supported by the China Postdoctoral Science Foundation (2016M592624,2016M590851), Chongqing Postdoctoral Science Foundation (xm2015026, Xm2016005, Xm2016087), the Natural Science Foundation of Chongqing(cstc2015jcyjA10120) and the Chongqing University Innovation Team Building Special Fund (CXTDX201601010).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2017.03.032.
References
- [1].Olar A, Aldape KD, Using the molecular classification of glioblastoma to inform personalized treatment, J. Pathol. 232 (2) (2014) 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Meyer MA, Malignant gliomas in adults, N. Engl. J. Med 359 (17) (2008) 1850 author reply 1850. [DOI] [PubMed] [Google Scholar]
- [3].Vito P, Interfering with apoptosis: Ca2+-binding protein ALG-2 and Alzheimer’s disease gene ALG-3 (vol 271, pg 521, 1996), Science 271 (5256) (1996), 1655–1655. [DOI] [PubMed] [Google Scholar]
- [4].Lo KWH, et al. , Apoptosis-linked gene product ALG-2 is a new member of the calpain small subunit subfamily of Ca2þ-binding proteins, Biochemistry 38 (23) (1999) 7498–7508. [DOI] [PubMed] [Google Scholar]
- [5].Shibata H, et al. , The penta-EF-hand protein ALG-2 interacts with a region containing PxY repeats in Alix/AIP1, which is required for the subcellular punctate distribution of the amino-terminal truncation form of Alix/AIP1,J. Biochem 135 (1) (2004) 117–128. [DOI] [PubMed] [Google Scholar]
- [6].Jung YS, et al. , Apoptosis-linked gene 2 binds to the death domain of Fas and dissociates from Fas during Fas-mediated apoptosis in Jurkat cells, Biochem. Biophys. Res. Commun 288 (2) (2001) 420–426. [DOI] [PubMed] [Google Scholar]
- [7].Rao RV, et al. , Molecular components of a cell death pathway activated by endoplasmic reticulum stress, J. Biol. Chem 279 (1) (2004) 177–187. [DOI] [PubMed] [Google Scholar]
- [8].Mahul-Mellier AL, et al. , Alix, making a link between apoptosis-linked gene-2, the endosomal sorting complexes required for transport, and neuronal death in vivo, J. Neurosci 26 (2) (2006) 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].la Cour JM, et al. , Up-regulation of ALG-2 in hepatomas and lung cancer tissue, Am. J. Pathol 163 (1) (2003) 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Su D, et al. , PDCD6 is an independent predictor of progression free survival in epithelial ovarian cancer, J. Transl. Med (2012) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].NishioHe K, Y.Q., et al. , Genetic variation in PDCD6 and susceptibility to lung cancer, Asian Pac. J. Cancer Prev. 13 (9) (2012) 4689–4693. [DOI] [PubMed] [Google Scholar]
- [12].Yamada Y, et al. , Identification of prognostic biomarkers in gastric cancer using endoscopic biopsy samples, Cancer Sci 99 (11) (2008) 2193–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ke XX, et al. , Inhibition of H3K9 methyltransferase G9a repressed cell proliferation and induced autophagy in neuroblastoma cells, PLoS One 9 (9) (2014) e106962.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hou J, et al. , CSN6 controls the proliferation and metastasis of glioblastoma by CHIP-mediated degradation of EGFR, Oncogene (2017) 1134–1144. [DOI] [PubMed] [Google Scholar]
- [15].Yoon JH, et al. , Programmed cell death 6 (PDCD6) as a prognostic marker for gastric cancers, Tumor Biol. 33 (2) (2012) 485–494. [DOI] [PubMed] [Google Scholar]
- [16].Huang Y, et al. , FSH inhibits ovarian cancer cell apoptosis by up-regulating survivin and down-regulating PDCD6 and DR5, Endocrine-Related Cancer 18 (1) (2011) 13–26. [DOI] [PubMed] [Google Scholar]
- [17].Shi SQ, et al. , Association between two single nucleotide polymorphisms of PDCD6 gene and increased endometriosis risk, Hum. Immunol 74 (2) (2013) 215–218. [DOI] [PubMed] [Google Scholar]
- [18].Su D, et al. , PDCD6 and ovarian cancer metastasis, findings of a proteomic study, Ejc Suppl. 7 (2) (2009), 453–453. [Google Scholar]
- [19].Rho SB, et al. , Programmed cell death 6 (PDCD6) inhibits angiogenesis through PI3K/mTOR/p70S6K pathway by interacting of VEGFR-2, Cell. Signal 24 (1) (2012) 131–139. [DOI] [PubMed] [Google Scholar]
- [20].Suzuki K, et al. , Programmed cell death 6, a novel p53-responsive gene, targets to the nucleus in the apoptotic response to DNA damage, Cancer Sci. 103(10) (2012) 1788–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].la Cour JM, et al. , The apoptosis linked gene ALG-2 is dysregulated in tumors of various origin and contributes to cancer cell viability, Mol. Oncol 1 (4) (2008) 431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Aviel-Ronen S, et al. , Genomic markers for malignant progression in pulmonary adenocarcinoma with bronchioloalveolar features, Proc. Natl. Acad. Sci. U. S. A 105 (29) (2008) 10155–10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ambatipudi S, et al. , Genomic profiling of advanced-stage oral cancers reveals chromosome 11q alterations as markers of poor clinical outcome, PLoS One(2) (2011) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]