Abstract
Multidrug‐resistant Staphylococcus aureus infections place a huge burden on the healthcare sector and the wider community. An increasing rate of infections caused by methicillin‐resistant Staphylococcus aureus (MRSA) has necessitated the development of alternative agents. We previously reported that usnic acid (UA) has activity against MRSA; here, we report the effect of UA in combination with norfloxacin on the drug resistance of MRSA clinical isolates. We observed that the combination of UA–norfloxacin significantly reduces the bacterial burden in mouse models infected with S. aureus, without causing any detectable associated toxicity. Proteomic analysis indicated that UA–norfloxacin induces oxidative stress within cells, which leads to membrane damage and inhibits metabolic activity and biosynthesis of peptidoglycan and fatty acids. Collectively, this study provides evidence that UA in combination with norfloxacin may be a potential candidate for development into a resistance‐modifying agent for the treatment of invasive MRSA infections.
Keywords: drug resistance reversal, MRSA, phytomolecules, Staphylococcus aureus, synergy, usnic acid
This is the first report on the drug resistance‐modifying potential of usnic acid (UA) through inhibition of the multidrug resistance (MDR) efflux pump and down‐regulation of proteins involved in peptidoglycan and fatty acid biosynthesis. This compound may be helpful in the management of infection caused by MRSA through (a) lowering the prescribed amount of antibiotics, (b) decreasing MDR generation, and (c) intensifying the efficacy of antibiotics against MRSA/VRSA under both in vitro and in vivo conditions. These results may be helpful in the development of anti‐MRSA drug combinations from economical and non‐toxic natural products.
Abbreviations
- CFU
colony forming units
- CLSI
Clinical and Laboratory Standards Institute
- CTC
5‐cyano‐2,3‐di‐(p‐tolyl) tetrazolium chloride
- EtBr
ethidium bromide
- GAPDH
glyceraldehyde 3‐phosphate dehydrogenase
- IPG
immobilized pH gradient
- MALDI‐TOF/TOF
matrix‐assisted laser desorption/ionization time‐of‐flight/time‐of‐flight
- MDR
multidrug resistance
- MIC
minimum inhibitory concentration
- MRSA
methicillin‐resistant Staphylococcus aureus
- NO
nitric oxide
- PI
propidium iodide
- ROS
reactive oxygen species
- UA
usnic acid
- VRSA
vancomycin‐resistant Staphylococcus aureus
The infections caused by multidrug‐resistant pathogens pose a serious global public health challenge. The Centers for Disease Control and Prevention (CDC) has outlined the top 18 drug‐resistant threats, including methicillin‐resistant Staphylococcus aureus (MRSA), which require urgent attention to prevent the spread of infections 1. Recent reports from hospital and community surveillance studies together with the Infectious Diseases Society of America (ISDA) have termed the nosocomial infections the ‘ESKAPE pathogens’ (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species) 2, 3. Staphylococcus aureus, a Gram‐positive bacterium, is a ubiquitous member of the human microbiota and was recognized as a major pathogen in the late 19th century. It is the causative agent for a wide range of infections including skin infections, sepsis, toxic shock syndrome, endocarditis and osteomyelitis 4. This pathogen has gained wide attention due to the emergence of MRSA, vancomycin‐resistant Staphylococcus aureus (VRSA) and multidrug resistance (MDR) strains which have acquired resistance to agents of last resort including vancomycin and linezolid. This scenario presents an urgent need to develop alternative approaches to combat such infections, which include modulating drug plasma concentration within the therapeutic window 5 and use of immune modulation therapy and probiotics 6.
Plant‐derived natural products have gained global attention as novel antimicrobial compounds due to their being recognized as safe and their use in traditional medicine systems 7, 8. It has been well established that plant secondary metabolites possess antibacterial and resistance‐modifying properties with a possible novel mechanism of action 9, 10, 11 and simultaneously minimize the side effects associated with conventional antibiotics 12. Combination therapy has been successful in the treatment of drug‐resistant tuberculosis and extended‐spectrum β‐lactam‐producing Gram‐negative bacterial infections 13, 14, 15. Combination therapy includes two antibiotics to which the bacterial pathogens are susceptible in order to improve the clinical outcome with respect to monotherapy by broadening the antibacterial spectrum, synergistic effects and reduced risk of resistance development. Several studies have reported the synergistic interactions of many plant bioactive compounds with clinically used antibiotics to minimize the infection and also result in reduced rates of emergence of resistance 16, 17.
The bioactivity‐guided fraction of Usnea subfloridana led to the identification of usnic acid (UA), which has potent antibacterial activity against multidrug‐resistant clinical isolates of S. aureus with membrane damaging potential 18. The present study deals with the investigation of the synergistic interaction of UA with norfloxacin against clinical isolates of MRSA through in vitro and in vivo assays. A gel‐based proteomic approach was further employed to understand the possible mechanism of synergy.
Materials and methods
Bacterial strains and growth media
The details of clinical isolates used in the present study are presented in Table S1. All the bacterial strains were grown and maintained on Muller Hinton Agar (MHA)/Broth (MHB) at 37 °C. The inoculum was prepared in 0.85% NaCl and adjusted at 0.5 McFarland standard to obtain approximately 108 colony forming units (CFU)·mL−1.
PCR amplification of mecA gene
All the eight clinical isolates of S. aureus were checked for the presence of the mecA gene, a marker for MRSA that impart resistance to β‐lactam antibiotics using PCR 19.
Checkerboard assay for interaction study
The fractional inhibitory concentration (FIC) of UA with different antibiotics was quantified through a modified broth checkerboard method 20. The synergy between the antibiotics and UA was estimated using the fold reduction and fractional inhibitory concentration index (FICI) 21. The FIC was calculated as follows (where MIC is minimum inhibitory concentration):
Resistance studies
The propensity of bacteria to develop resistance and the mutation prevention concentration of the UA and norfloxacin combination was evaluated following methodology describe earlier 22, 23 using reference strain MTCC‐96 of S. aureus. First, the MIC of UA alone and in combination with norfloxacin was determined in MTCC‐96 as per the broth microdilution assay, in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines. Following this, serial passaging was carried out by transferring the bacterial cells growing at sub‐MIC of the compound/antibiotic and subjecting them to another MIC assay. After 24 h of incubation, cells at sub‐MIC of the test compound/antibiotic were further transferred and assayed for MIC determination. The process was carried out for 20 passages and the fold increase in MIC was plotted against the number of passages to evaluate the propensity of the bacteria to develop resistance. The post‐antibiotic effect of UA alone and in combination with norfloxacin against the clinical isolate MRSA‐2071 was measured using a previously described method 24.
Time–kill studies
To assess the bactericidal potential of UA and norfloxacin against MRSA‐2071, a time–kill kinetic study was performed at different concentrations alone and in combination as per the CLSI methods 25. Each analysis was carried out in triplicate and compared to untreated control. The criterion for a bactericidal effect was ≥ 3‐log10 decrease in CFU count at a specified time while a decline of less than 3‐log10 CFU·mL−1 was interpreted as bacteriostatic.
In vivo efficacy
Swiss Albino mice (male, 5–6 weeks old, 22–25 g, n = 5 per group) were infected with 106 CFU·mL−1 of S. aureus (MTCC‐96) through an intravenous route to evaluate therapeutic efficacy. The treatment groups comprised combinations (norfloxacin 8 mg·kg−1 + UA 1 mg·kg−1; norfloxacin 4 mg·kg−1 + UA 0.5 mg·kg−1) along with norfloxacin alone (1, 5, 10 and 16 mg·kg−1 body weight) and UA alone (0.5, 1 and 4 mg·kg−1 body weight). Oral administration of treatments once daily was initiated 16 h after infection and continued till day 7. Blood was collected from retro‐orbital plexus 24 h after the last dose for toxicity assessment and spleen to check the bacterial load 18. The bacterial count was finally expressed as CFU·g−1 tissue and presented as the mean ± SEM. The reduction in the bacterial load within the tissues was assessed by comparing the load in the treatment groups with respect to infection control.
In vivo toxicity determination
In vivo toxicity in Swiss Albino mice was studied in accordance with the Organization for Economic Co‐operation and Development test guideline no. 423 with additional modifications 26. The animals were observed for body weight changes in addition to morbidity and mortality. In addition, serum from the blood was analyzed for various biochemical parameters pertaining to hematology, liver, and kidney functioning along with the lipid profile.
Ethidium bromide accumulation and efflux
In order to evaluate the efflux pump inhibitory/modulatory potential of UA, spectrofluorometric determination of ethidium bromide (EtBr) efflux was performed as per the earlier reported method 27. Efflux inhibition was assessed by measuring the decline in fluorescence intensity of EtBr over a time of 30 min. Reserpine, an efflux pump inhibitor, was used as positive control. The experiments were performed thrice and the data were expressed as mean ± SEM. An EtBr accumulation assay was carried out using a flow cytometer (LSR II, BD Bioscience, San Diego, CA, USA) following the method described earlier 28. Further, the expression analysis of five different membrane‐associated efflux pump genes was evaluated upon exposure to UA and norfloxacin, alone and in combination.
Membrane permeabilization assay
The effect of the UA–norfloxacin combination on membrane permeability in MRSA‐2071 was assessed following propidium iodide (PI) uptake method described earlier and the fluorescence was measured at excitation and emission of 544 nm and 620 nm, respectively 29 (FLUOStar Omega; BMG Labtech, Ortenberg, Germany). A negative control contained untreated MRSA‐2071 bacterial cells and PI with Milli‐Q water while cells treated with 2.8% formaldehyde and 0.04% glutaraldehyde served as positive control. Untreated cells which possess intact membrane and do not allow PI to intercalate with DNA and fluoresce served as a negative control. Formaldehyde and glutaraldehyde used as fixatives which destroy the integrity of the bacterial membrane and thus allow PI to bind to DNA served as a positive control.
Cytoplasmic membrane depolarization assay
Membrane depolarization was studied in MRSA‐2071 using membrane‐potential‐sensitive cyanine dye diSC3‐5, according to the earlier reported method 30. MRSA‐2071 cells with mid‐logarithmic phase (D 600 = 0.6) were collected and washed once with buffer (5 mm HEPES, pH 7.2, 5 mm glucose) and resuspended in same buffer to achieve the D 600 = 0.05. The cell suspension was incubated with 0.4 μm diSC3‐5 until dye uptake was maximal (approximately 90%), after which 100 mm KCl was added to equilibrate the cytoplasmic and external K+ concentration. Staphylococcus aureus cells (D 600 = 0.3) were treated with phytomolecules and carbonyl cyanide m‐chlorophenylhydrazone as a positive control at 37 °C. Culture without treatment served as a negative control. The fluorescence reading was monitored with a microplate reader (FLUOStar Omega) at an excitation wavelength of 622 nm and an emission wavelength of 670 nm. The blank with only cells and the dye was used to deduct the background. The experiments were performed thrice and data expressed as mean values ± SEM (**P < 0.01 vs control; P > 0.05 represents non‐significant: Dunnett's test).
Proteomic studies to investigate the mechanism of action of UA–norfloxacin combination
The mode of anti‐staphylococcal activity of the UA–norfloxacin combination was determined through a gel‐based proteomic approach. Protein extraction was carried out from bacterial cells (MRSA‐2071) exposed to sub‐lethal concentrations of UA, norfloxacin and UA–norfloxacin combination using a TRIzol extraction protocol 31. Untreated MRSA‐2071 cells grown to the same attenuance served as control. Protein samples (225 μg) were used for passive rehydration of 17 cm immobilized pH gradient (IPG) strips (linear pH 4–7; Bio‐Rad, Hercules, CA, USA) overnight. A gradient of voltage starting from 200 V for 4 h (step and hold), 500 V for 1 h (step and hold), 1000 V for 1 h (step and hold), 8000 V for 13 500 Volt‐hour (gradient) and 8000 V for 8 h (step and hold), 500 V for 4 h (step and hold) and 200 V for 2 h (step and hold) resulting in a total of approximately 93 kV for isoelectric focussing (IEF) was used followed by resolution on 12.5% SDS/PAGE in triplicate. Coomassie brilliant blue stained images were imported to ht analyzer 2d (Genomic Solution, Ann Arbor, MI, USA) software for comparative analysis. Spots present in all the gels were considered for detailed multivariate analysis and differentially expressed spots were excised and stored at 4 °C for subsequent mass spectrometric analysis.
In‐gel digestion and protein identification by MALDI‐TOF/TOF
In‐gel digestion of the excised spots was performed following the earlier described protocol 31, 32. The protein identification was performed with matrix‐assisted laser desorption/ionization time‐of‐flight/time‐of‐flight (MALDI‐TOF/TOF) mass spectrometry (UltrafleXtreme™ mass spectrometer; Bruker Daltonics Inc., Bremen, Germany). A combined MS and LIFT tandem mass spectrometry was performed using biotools 3.0 software (Bruker Daltonics Inc.). The TOF spectra were recorded in positive ion reflector mode with a mass range from 700 to 3500 Da. Only proteins matched by a minimum of two unique peptide sequences were included in the results list. Sequence coverage was also considered to evaluate protein identification.
Functional pathway and network analysis
The differentially expressed proteins identified in two‐dimensional electrophoresis were subjected to DAVID (Database for Annotation, Visualization, and Integrated Discovery) database version 6.7 (https://david.ncifcrf.gov/) 33. The list of UniProt IDs of the proteins was further imported into KOBAS 3.0 (KEGG Orthology‐Based Annotation System) database and the pathways were mapped against S. aureus using default settings 34. In addition, a protein–protein interaction analysis was also carried out using the STRING v10.0 database (https://string-db.org/) with default parameters and S. aureus subsp. aureus as the organism of interest 35.
Measurement of reactive oxygen species
The analysis of intracellular levels of reactive oxygen species (ROS) was carried out using the ROS‐sensitive probe CM‐H2DCFDA (Invitrogen, Carlsbad, CA, USA) as per the method described by Wang and Joseph 36. The ROS level was measured in terms of reduced fluorescent compound dichlorofluorescein (DCF) with excitation and emission wavelengths of 485‐10 nm and 520 nm, respectively, using a microplate reader (FLUOStar Omega). Ciprofloxacin served as a positive control, while untreated MRSA‐2071 cells served as a negative control in the study.
Measurement of nitrite level
The nitrite level within the MRSA‐2071 cells exposed to UA and norfloxacin alone and in combination was quantified by spontaneous oxidation of nitric oxide (NO) 37 using a Griess reagent kit according to the manufacturer's protocol (Life Technologies, Camarillo, CA, USA). Sodium nitroprusside, a standard NO producer, was used as a positive control, while a reference sample was prepared by mixing Griess reagent and deionized water. The absorbance of the nitrite was measured at 548 nm using a microplate reader (FLUOStar Omega). The experiment was performed in triplicate and data expressed are mean values ± SEM (**P < 0.01 vs control: Dunnett's test).
Metabolic activity using resazurin
A resazurin assay was performed in MRSA‐2071 cells exposed to UA and norfloxacin alone and in combination at different concentrations to check the cell viability and metabolic activity 38. Untreated MRSA‐2071 actively growing cells served as a positive control in the study. Resazurin dye, at a concentration 10 μg·mL−1, was used and the fluorescence intensity was measured at 590 nm for 30 min at 15 s intervals in a microplate reader (FLUOStar Omega). The experiment was performed in triplicate and data expressed are mean values ± SEM (**P < 0.01 vs control: Dunnett's test).
Assessment of bacterial respiratory activity using 5‐cyano‐2,3‐di‐(p‐tolyl) tetrazolium chloride
Respiratory activity assay using 5‐cyano‐2,3‐di‐(p‐tolyl) tetrazolium chloride (CTC) was performed as described previously 39. MRSA‐2071 cells exposed to UA and norfloxacin alone and in combination at MIC concentrations for 4 h were used for the assay. CTC at the final concentration of 5 mm was added, followed by fixing the cells with 2.8% formaldehyde and 0.04% glutaraldehyde and counterstaining with 4′6‐diamidino‐2‐phenylindole. A negative control was prepared by disrupting the membrane prior to addition of CTC. Further, the samples were analyzed through flow cytometer using the FL‐1 channel (488 nm; blue argon laser) and cell quest 3.3 analysis software (LSR II, Becton‐Dickinson, San Diego, CA, USA).
Western blot analysis
The overnight grown culture of MRSA‐2071 in MHB at 37 °C with agitation was diluted in MHB and again grown at 37 °C to a D of 0.6 with or without treatments. The cells were harvested, the pellet was washed and resuspended in TE buffer (10 mm Tris, pH 7.5, 1 mm EDTA) followed by sonication, and cell debris was removed by centrifugation at 11 269 g for 10 min at 4 °C. Following the estimation of proteins using the Bradford assay, aliquots of protein (80 μg) were loaded on 12.5% SDS/PAGE gels and blot transferred onto polyvinylidene difluoride membrane. After blocking with 5% skimmed milk, MurA was probed with anti‐MurA antibody at 1 : 2000 dilution, followed by incubation with secondary alkaline phosphatase conjugated goat anti‐rabbit antibody at 1 : 5000 dilution. The blot was further developed using nitro blue tetrazolium/5‐bromo‐4‐chloro‐3‐indolyl‐phosphate substrate with glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) as an endogenous control.
qRT‐PCR analysis
The qRT‐PCR analysis of identified genes was analyzed in MRSA‐2071 cells in the presence of UA and norfloxacin alone and in combination. The list of all the primers is presented in Table S4. Cells were grown to mid‐log phase in the presence of different treatments at sub‐lethal concentrations and real‐time quantification of RNA transcript was analyzed by SYBR GreenER qPCR mix (Invitrogen) using the 7900HT fast real‐time PCR system (Applied Biosystems, Foster City, CA, USA). Observations were recorded in terms of log RQ after normalization with GAPDH, an endogenous control.
Ethical clearance
The protocols (CIMAP/IAEC/2016‐19/06 and CIMAP/IAEC/2016‐19/01) were duly approved by Institutional Animal Ethics Committee on 29 February 2016.
Statistical analysis
The data were subjected to a one‐way ANOVA to analyze the mean values obtained for the treatment and control. Dunnett's test was used to compare the treatment and control and statistical significance was set at *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001, and P > 0.05 was considered to be non‐significant.
Results and Discussion
The extensive use of antibiotics has raised serious health concerns throughout the world due to the development and emergence of MRSA, Vancomycin Intermediate Staphylococcus aureus/ Vancomycin Resistant Staphylococcus aureus (VISA/VRSA) and MDR S. aureus, urging the need to identify new agents. Plant secondary metabolites have displayed a potential as antibacterial agents individually and are known to potentiate the activity of other antibiotics through different mechanisms and thus they could be employed for the treatment of resistant pathogens 40, 41, 42, 43, 44. UA, a dibenzofuran derivative from lichens, has already been established as having antibacterial potential against MRSA with the probable mode of action being its membrane disruptive property 18, 45. However, no detailed reports are available for the resistance‐modifying potential of UA in combination with conventional antibiotics against MRSA. The present comprehensive study describes the synergistic interaction of UA with norfloxacin against clinical isolates of S. aureus (MRSA), which were found to be multidrug resistant and harbor mecA gene (Table S2 and Fig. S1). In order to assess the utility of UA (particularly in combination) as a therapeutic agent, the propensity of bacteria (MRSA‐2071) to acquire resistance against UA alone as well as in combination with norfloxacin was evaluated (Fig. S2a). The results indicate that clinical strains of S. aureus (MRSA‐2071) were not able to develop resistance against UA alone as well as in combination with norfloxacin even after 20 subsequent passages. A 2‐ to 4‐fold increase in the MIC of UA alone was observed after 12–20 subsequent passages. However, in the case of norfloxacin alone, more than 900‐fold increase in MIC was observed during the same period. The prolongation of the post‐antibiotic effect of norfloxacin in the presence of UA represents a continued suppression of growth after a short exposure to the antimicrobial agents. Also, UA not only increased the inherent susceptibility of S. aureus to norfloxacin, but also simultaneously reduced the emergence of norfloxacin resistance (Table S3, Fig. S2b). All these results collectively suggest the suitability of UA in combination with norfloxacin. A similar observation has been made using an isoliquiritigenin derivative (IMRG4), a natural plant flavonoid 46, suggesting the therapeutic potential of the UA–norfloxacin combination for treating MDR infections with delayed resistance.
In vitro combination study
Synergy and the resistance‐modifying potential of UA with clinically used antibiotics were investigated and are presented in terms of fold reduction and FICI (Table 1). In combination with oxacillin and tetracycline, UA displayed synergy with 4–16‐fold reduction in MIC against all the tested clinical isolates. However, the UA–vancomycin combination showed synergistic interaction against four of the eight isolates (FICI 0.374–0.49) and the remaining four exhibited additive interaction with FICI 0.5–0.75. As evident from the results, UA could cause significant reduction of up to 16‐fold in the MIC of norfloxacin with FICI ≤ 0.50 and thus it was selected for further studies. The clinical breakout point for the susceptibility of norfloxacin is ≤ 10 mg·L−1 47, and its MIC in combination with UA was observed to reach close to the susceptible limit as defined in the European Committee on Antimicrobial Susceptibility Testing guidelines (31.25 mg·L−1).
Table 1.
In vitro anti‐staphylococcal activity of UA with different antibiotics against methicillin‐resistant clinical isolates of Staphylococcus aureus. FICI, fractional inhibitory concentration index; FR, fold reduction in MIC of antibiotics; MIC, minimum inhibitory concentration; NOR, norfloxacin; OXA, oxacillin; TET, tetracycline; UA, usnic acid; VAN, vancomycin
| Strains | MIC of UA alone (μg·mL−1) | MIC of antibiotics alone (μg·mL−1) | MIC of OXA in presence of UA (μg·mL−1) | MIC of NOR in presence of UA (μg·mL−1) | MIC of VAN in presence of UA (μg·mL−1) | MIC of TET in presence of UA (μg·mL−1) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OXA | NOR | VAN | TET | MIC | FICI (FR) | MIC | FICI (FR) | MIC | FICI (FR) | MIC | FICI (FR) | ||
| SA‐2071 | 25 | 1000 | 500 | 3.125 | 25 | 15.6 | 0.265 (64) | 31.25 | 0.31 (16) | 0.78 | 0.49 (4) | 3.125 | 0.25 (8) |
| SA‐1745 | 25 | 1000 | 500 | 6.125 | 50 | 31.25 | 0.281 (32) | 62.5 | 0.375 (8) | 1.56 | 0.379 (4) | 6.25 | 0.375 (8) |
| SA‐5944 | 25 | 1000 | 500 | 1.56 | 12.5 | 31.25 | 0.28 (32) | 125 | 0.375 (4) | 0.39 | 0.5 (4) | 3.125 | 0.37 (4) |
| SA‐4627 | 25 | 500 | 250 | 1.56 | 50 | 62.5 | 0.25 (8) | 62.5 | 0.312 (8) | 0.39 | 0.5 (4) | 12.5 | 0.31 (4) |
| SA‐4423 | 50 | 500 | 250 | 1.56 | 25 | 62.5 | 0.375 (8) | 62.5 | 0.37 (8) | 0.39 | 0.375 (4) | 6.25 | 0.3 (4) |
| SA‐4620 | 25 | 1000 | 250 | 1.56 | 25 | 62.5 | 0.31 (16) | 31.25 | 0.375 (8) | 0.78 | 0.75 (2) | 3.125 | 0.375 (8) |
| SA‐3151 | 25 | 500 | 500 | 1.56 | 25 | 31.25 | 0.31 (16) | 62.5 | 0.325 (8) | 0.195 | 0.625 (8) | 6.25 | 0.31 (4) |
| SA‐10760 | 50 | 250 | 500 | 3.125 | 50 | 62.5 | 0.375 (4) | 125 | 0.49 (4) | 0.78 | 0.374 (4) | 6.25 | 0.312 (8) |
In‐vivo anti‐staphylococcal activity and toxicity
The bacterial load in spleen tissue of Swiss Albino mice was determined to evaluate the in vivo efficacy of the UA–norfloxacin combination (Fig. 1). A dose‐dependent significant reduction was observed where 25% of the effective dose (ED25; 0.5 mg·kg−1) of UA in combination with ED25 (4 mg·kg−1) of norfloxacin could reduce the bacterial load up to 50% (P < 0.01), whereas up to 80% (P < 0.001) reduction was recorded when the UA–norfloxacin combination was administered at 50% of the effective dose (ED50; Nor 8 mg·kg−1 and UA 1 mg·kg−1). Toxicity studies were also performed to evaluate the effect of the UA–norfloxacin combination on healthy animals and animals infected with MTCC‐96. No lethal effects were recorded post‐treatment with the combination in the biochemical parameters compared to vehicle control, with and without infection (Table 2). The results further provide evidence for the low toxicity and acceptable safety profiles in mice models for therapeutic use. These are in accordance with previous reports describing the efficiency of plant molecules in combination with conventional antibiotics in reducing the bacterial load in animal models 48, 49.
Figure 1.
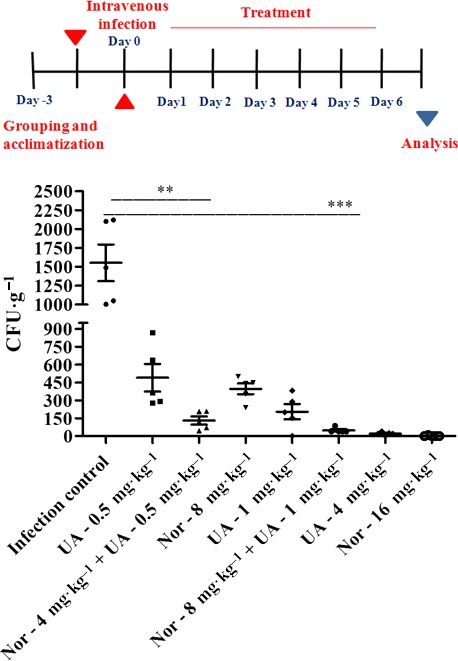
In vivo anti‐staphylococcal efficacy of UA, norfloxacin (Nor) and UA–norfloxacin combination at various doses in terms of reduction of bacterial burden (Staphylococcus aureus MTCC‐96) in spleen tissue. The infection (1 × 106 CFU·mL−1 in 200 μL) was induced through the intravenous route. The bacterial load (CFU·g−1) of spleen tissue was expressed as mean ± SEM. (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, Dunnett's test.)
Table 2.
Hematological and biochemical changes recorded during the toxicity study in Swiss Albino mice. Values are mean ± SD. ALKP, alkaline phosphatase; RBC, red blood cells; SGOT, serum glutamate–oxaloacetate transaminase; SGPT, serum glutamate–pyruvate transaminase; WBC, white blood cells.
| Parameters | Uninfected | Infected with MTCC‐96 | ||
|---|---|---|---|---|
| Control | UA + Nor | Control | UA + Nor | |
| Change in body weight (g) | 3.64 ± 0.38 | 4.36 ± 0.44 | −3.33 ± 0.31 | −2.26 ± 0.27 |
| Relative organ weight | ||||
| Liver | 1.453 ± 0.19 | 2.115 ± 0.24 | 1.414 ± 0.27 | 1.178 ± 0.28 |
| Spleen | 0.556 ± 0.42 | 1.152 ± 0.30 | 1.257 ± 0.22 | 1.097 ± 0.5 |
| SGPT (U·L−1) | 7.15 ± 0.54 | 6.95 ± 0.63 | 6.835 ± 0.49 | 6.874 ± 0.31 |
| SGOT (U·L−1) | 10.96 ± 0.66 | 11.08 ± 0.38 | 12.36 ± 0.74 | 13.05 ± 0.63 |
| Serum creatinine (mg·dL−1) | 0.14 ± 0.08 | 0.17 ± 0.27 | 0.15 ± 0.18 | 0.16 ± 0.11 |
| Serum ALKP (U·L−1) | 118.04 ± 3.18 | 120.27 ± 2.21 | 114.39 ± 1.76 | 117.64 ± 2.91 |
| Serum total cholesterol (mg·dL−1) | 61.42 ± 2.09 | 57.92 ± 2.16 | 63.37 ± 1.88 | 65.69 ± 3.13 |
| Serum bilirubin (mg·dL−1) | 0.07 ± 0.017 | 0.06 ± 0.005 | 0.07 ± 0.004 | 0.08 ± 0.013 |
| Serum triglycerides (mg·dL−1) | 28.74 ± 3.74 | 31.36 ± 2.58 | 29.49 ± 2.14 | 31.33 ± 1.76 |
| RBC (millions·mm−3) | 4.532 ± 0.56 | 5.971 ± 0.29 | 6.059 ± 0.4 | 6.114 ± 0.28 |
| WBC (millions·mm−3) | 8.15 ± 0.56 | 12.22 ± 0.49 | 11.4 ± 0.27 | 12.23 ± 0.53 |
| Hemoglobin (g·dL−1) | 12.96 ± 0.35 | 12.14 ± 0.28 | 11.175 ± 1.04 | 11.54 ± 0.82 |
Killing kinetics
A dose‐ and time‐dependent decline in viable MRSA‐2071 cells was observed upon exposure to the UA–norfloxacin combination. The bactericidal effect of norfloxacin was achieved at a very low concentration in combination with UA (Fig. S3). Similar observations have also been reported earlier wherein plant molecules in combination with conventional antibiotics have been found to reduce the dose of partner antibiotics by many fold irrespective of their own activity 50.
EtBr accumulation and efflux
Increased efflux is one of the major mechanisms which prevents antimicrobial agents from reaching their targets and contributes to the MDR phenotypes of pathogens 27. The efflux pump inhibitory potential of UA was evaluated using a spectrofluorometric assay, displaying a decline in EtBr efflux over a time duration of 30 min in MRSA‐2071 cells exposed to UA as compared to untreated cells. UA demonstrated an efflux pump inhibitory potential in MRSA‐2071 cells, comparable to that of reserpine (Fig. 2A). Flow cytometric analysis displayed a 1.8‐fold shift in the intensity of red fluorescence inside the cells in the presence of UA as compared to untreated control. In the presence of reserpine, however, a 1.2‐fold reduction in the intensity of red fluorescence was observed (Fig. 2B). The overexpression of efflux pump genes norA, norB, norC, mepA and mdeA, which are implicated in conferring multidrug resistance 51, upon norfloxacin exposure obtained in this study are in accordance with previous reports 52, 53. However, the down‐regulation of these in the presence of UA alone and further reduced expression in case of the UA–norfloxacin combination clearly demonstrate the efflux pump inhibitory potential of UA in MRSA (Fig. 2C). Collectively, these results indicate that UA modulates the activity of the MDR efflux pump of MRSA‐2071.
Figure 2.
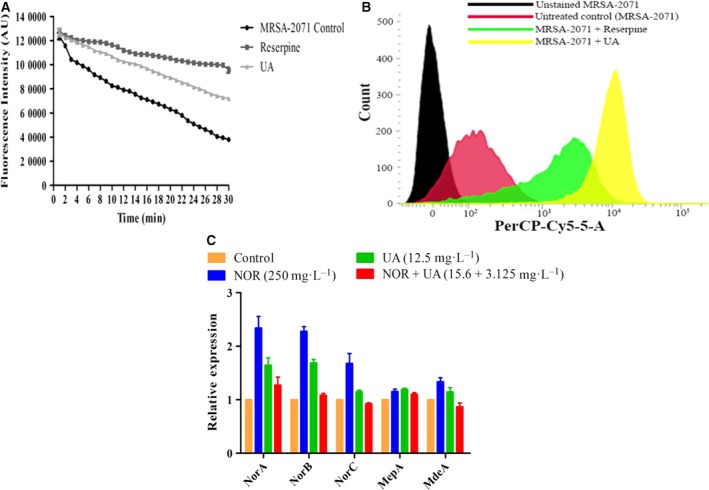
(A,B) Efflux pump modulatory potential of UA in clinical isolate MRSA‐2071 using EtBr as marker, determined through spectrofluorometric analysis (A) and flow cytometric analysis (B). Reserpine, an efflux pump inhibitor, was used as positive control in both the experiments. (C) Expression analysis of efflux pump genes in the presence of UA alone and in combination with norfloxacin.
Alteration of membrane permeability and potential
The effect of the UA–norfloxacin combination on membrane permeability and potential was determined using spectroscopic techniques. Notably, MRSA‐2071 cells exposed to UA alone and the UA–norfloxacin combination displayed an increase in PI uptake as compared to untreated control (Fig. 3A) indicating altered membrane permeability. DiSC3‐5 is well known to intercalate into the cytoplasmic membrane of energized cells leading to quenching of the initial fluorescence, and increased fluorescence highlights the disruption in the membrane potential 54, 55. The UA–norfloxacin combination was found to dissipate the membrane potential to a much higher extent in comparison to UA and norfloxacin alone (Fig. 3B). All these results collectively demonstrate the membrane‐damaging ability of the UA–norfloxacin combination in MRSA‐2071.
Figure 3.
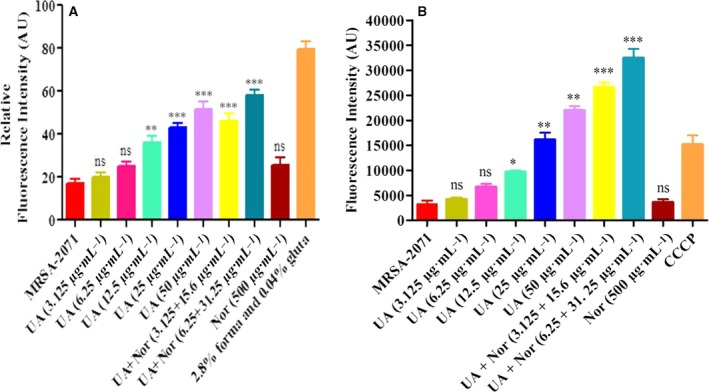
Prospecting membrane disruptive property of UA–norfloxacin combination against clinical isolate MRSA‐2071. (A) Membrane permeabilization using PI through spectrofluorimetry. Data represent mean ± SEM of three independent experiments (**P ≤ 0.01, ***P ≤ 0.001, Dunnett's test). (B) Dissipation of membrane potential by increase in the fluorescence of DiSC 3‐(5) with the treatment of UA and norfloxacin alone as well as in combination at different concentrations (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, Dunnett’s test). Forma; Formaldehyde; Gluta; Glutaraldehyde.
Modulation of pathways and networks in MRSA‐2071 due to UA–norfloxacin exposure revealed by gel‐based proteomic analysis
Multivariate analysis between the proteome of MRSA‐2071 control and the UA–norfloxacin combination (0.5 MIC – 3.125 + 15.6 mg·L−1) indicated the presence of around 1200 spots among which 34 spots exhibited differential expression (P ≤ 0.05). The gel image with identified proteins is presented in Fig. S4. The protein identification details obtained from the MALDI‐TOF/TOF analysis are presented in Table 3. DAVID, PANTHER and STRING databases were used to classify and identify the potential function of altered proteins within the cells (Figs S5, S6). A cluster of proteins involved in oxidative stress responses was found to be up‐regulated in response to the UA–norfloxacin combination in MRSA‐2071 cells, which is in accordance with the earlier report where similar results were obtained in MRSA cells exposed to sub‐inhibitory concentrations of UA 44. Oxidants such as ROS and NO affect the viability of cells and hinder the process of pathogenesis 56 by irreversible oxidative damage to DNA, lipids and proteins 57, 58 and bring the tissues to homeostasis 59, 60. The treatment further decreased the expression of some of the proteins involved in peptidoglycan and fatty acid biosynthesis. In particular, a 6.64‐fold decline was observed in the expression level of UDP‐N‐acetylglucosamine 1‐carboxyvinyltransferase (MurA), which catalyzes the first step in peptidoglycan biosynthesis. In addition, four proteins which were uniquely identified in the cells exposed to the UA–norfloxacin combination at sub‐inhibitory concentrations were mainly involved in transcriptional regulation, namely DEAD‐box ATP‐dependent RNA helicase CshA, transcriptional regulatory protein WalR, elongation factor Tu and UPF0355 protein MRSA252. Moreover, sequence level in‐depth analysis of the uncharacterized protein UPF0355 identified uniquely in the combination‐treated cells displayed a putative role in modulating the operon regulating the downstream expression of genes involved in oxidative stress responses. Another remarkable observation was a 0.95‐fold down‐regulation of d‐lactate dehydrogenase, encoded by ldhD, which has been implicated in imparting vancomycin resistance in S. aureus by incorporating d‐lactate at the terminal residue of peptidoglycan precursors 61, 62. Another key observation in cells exposed to the UA–norfloxacin combination was the down‐regulation of tetracycline resistance protein tetM from transposon Tn5251 (TetM), implicated in imparting resistance to tetracycline. The above observations clearly point towards a resistance‐modifying ability of the UA–norfloxacin combination. Fatty acid biosynthesis is the first stage of membrane lipid biogenesis, playing a vital role in bacterial physiology 63, 64. FabI, unique to prokaryotes and a key regulator in controlling the elongation of the acyl chain for saturated fatty acids 65, 66, has been identified as a promising drug target 61. In the present study, the UA–norfloxacin combination was observed to down‐regulate the expression of enoyl‐(acyl‐carrier‐protein) reductase (NADPH) (FabI) as well as the initiation condensation enzyme 3‐oxoacyl‐(acyl‐carrier‐protein) synthase 3 encoded by fabH which may lead to inhibition of fatty acid biosynthesis.
Table 3.
List of differentially expressed proteins in methicillin resistant Staphylococcus aureus SA‐2071 upon UA–norfloxacin exposure at sub‐lethal concentrations obtained from two‐dimensional electrophoresis analysis. D, down‐regulated; U, up‐regulated
| Spot no. | Name of protein | Uniprot ID | Gene | M (Da) | No. of peptides | Fold change | Score | P‐value |
|---|---|---|---|---|---|---|---|---|
| U1 | Superoxide dismutase (Mn/Fe) | P99098 | sodA | 22 711 | 12 | 0.27 | 243 | 0.004 |
| U2 | Alkyl hydroperoxide reductase subunit C | P99074 | ahpC | 20 977 | 8 | 0.33 | 217 | 0.005 |
| U3 | Enolase | P99088 | eno | 47 117 | 23 | 0.29 | 332 | 0.008 |
| U4 | Catalase | Q7A5T2 | katA | 58 380 | 27 | 0.23 | 207 | 0.009 |
| U5 | Probable quinol oxidase subunit 2 | Q7A698 | qoxA | 41 777 | 24 | 0.31 | 199 | 0.016 |
| U6 | Probable thiol peroxidase | P99146 | tpx | 18 019 | 6 | 0.41 | 151 | 0.024 |
| U7 | Bacterial non‐heme ferritin | Q7A4R2 | ftnA | 19 589 | 5 | 0.49 | 148 | 0.027 |
| U8 | Protein GrpE | P99086 | grpE | 24 008 | 10 | 0.61 | 97 | 0.001 |
| U9 | Iron–sulfur cluster repair protein ScdA | Q7A7U6 | scdA | 25 485 | 17 | 0.35 | 226 | 0.019 |
| U10 | Transketolase | P99161 | tkt | 72 317 | 22 | 0.24 | 316 | 0.013 |
| U11 | Chaperone Protein DnaK | P99110 | dnaK | 66 361 | 20 | 0.35 | 208 | 0.024 |
| U12 | Peroxide responsive repressor (PerR) | Q7A4T8 | perR | 17 183 | 4 | 0.37 | 114 | 0.003 |
| U13 | UPF0413 protein SAUSA300_0983 | Q2FI75 | SAUSA300_0903 | 31 364 | 12 | 0.29 | 328 | 0.004 |
| U14 | Putative 2‐hydroxyacid dehydrogenase SAR2389 | Q6GEC9 | SAR2389 | 34 675 | 15 | 0.15 | 175 | 0.004 |
| D1 | Fructose‐bisphosphate aldolase Class I | P99117 | fda | 32 913 | 13 | 1.37 | 266 | 0.043 |
| D2 | d‐Lactate dehydrogenase | P99116 | ldhD | 36 756 | 18 | 0.95 | 254 | 0.036 |
| D3 | LipidII: glycine glycyltransferase | Q7A447 | femX | 48 513 | 21 | 1.48 | 310 | 0.001 |
| D4 | Putative antiporter subunit nnhG2 | Q7A722 | mnhG2 | 16 371 | 9 | 1.22 | 163 | 0.027 |
| D5 | DNA translocase FtsK | P64165 | ftsK | 90 682 | 26 | 1.06 | 336 | 0.013 |
| D6 | Trigger factor | P99080 | tig | 48 609 | 15 | 1.69 | 154 | 0.032 |
| D7 | UDP‐N‐acetylmuramoyl tripeptide–d‐alanyl‐d‐alanine ligase | A0A0H3JMW3 | murF | 50 065 | 8 | 3.53 | 186 | 0.007 |
| D8 | Enoyl‐(acyl‐carrier‐protein) reductase (NADPH) | A0A0H3JLH9 | fabI | 27 992 | 4 | 1.98 | 119 | 0.004 |
| D9 | 3‐Oxoacyl‐(acyl‐carrier‐protein) synthase 3 | P99159 | fabH | 33 879 | 11 | 1.96 | 176 | 0.023 |
| D10 | 30S Ribosomal protein S12 | P0A0G8 | rpsL | 15 287 | 9 | 1.88 | 143 | 0.018 |
| D11 | Uncharacterized lipoprotein SAUSA300_0411 | Q2FJK3 | SAUSA300_0411 | 30 917 | 10 | 1.52 | 281 | 0.047 |
| D12 | Tetracycline resistance protein tetM from transposon Tn5251 | Q54807 | tetM(5251) | 72 556 | 16 | 2.15 | 294 | 0.024 |
| D13 | Deoxyribose phosphate aldolase 2 | P99174 | deoC2 | 23 218 | 14 | 1.36 | 173 | 0.022 |
| D14 | Formate acetyltransferase | Q7A7X6 | pflB | 84 862 | 23 | 1.73 | 304 | 0.021 |
| D15 | Adenine phosphoribosyltransferase | P68779 | apt | 19 117 | 12 | 1.84 | 265 | 0.03 |
| D16 | Fructose‐bisphosphate aldolase Class I | P99117 | fda | 32 913 | 16 | 1.34 | 208 | 0.005 |
| D17 | Fructose‐bisphosphate aldolase Class II | P99075 | fba | 30 836 | 18 | 1.42 | 314 | 0.014 |
| D18 | ATP synthase subunit c | Q7A4E6 | atpE | 6979 | 3 | 1.26 | 101 | 0.029 |
| D19 | HTH‐type transcriptional regulator SarR | Q7A425 | sarR | 13 669 | 5 | 1.9 | 151 | 0.006 |
| D20 | UDP‐N‐acetylglucosamine 1‐carboxyvinyltransferase | P84058 | murA | 45 054 | 15 | 6.64 | 163 | .012 |
| 1a | 60 kDa chaperonin | P99083 | groEL | 57 630 | 13 | n/a | 138 | 0.034 |
| 2a | General stress protein | W8U554 | yugI_1 | 14 723 | 7 | n/a | 127 | 0.005 |
| 1b | 50S ribosomal protein L17 | Q7A469 | rplQ | 13 529 | 5 | n/a | 134 | 0.014 |
| 1c | DEAD‐box ATP‐dependent RNA helicase CshA | Q7A4G0 | cshA | 56 942 | 20 | 0 | 193 | 0.023 |
| 2c | Transcriptional regulatory protein WalR | Q7A8E1 | walR | 27 192 | 13 | 0 | 269 | 0.016 |
| 3c | Elongation factor Tu | P99152 | tuf | 43 104 | 15 | 0 | 338 | 0.018 |
| 4c | UPF0355 protein MRSA252 | Q6GJR0 | SAR0405 | 15 051 | 6 | 0 | 193 | 0.026 |
a Uniquely expressed in only UA‐treated cells. b Uniquely identified in norfloxacin‐exposed cells. c Unique in UA–norfloxacin‐treated cells).
Validation of proteomic signatures through cell‐based assays
Upon the exposure to the UA–norfloxacin combination, a considerable increase in H2O2 and NO levels was observed which was significantly higher compared to UA alone (Fig. 4). The present study for the first time reports the ROS‐mediated anti‐staphylococcal activity and resistance‐modifying potential of the UA–norfloxacin combination against a clinical isolate MRSA‐2071 (Indian origin). The up‐regulation of genes involved in mitigation of oxidative stress indicates that the UA–norfloxacin combination induces oxidative stress which could be detrimental to the cell and thus provides the basis for proposing it as an antibacterial and resistance‐modifying agent (see Fig. 6C). Functional assays clearly indicated decreased metabolic and respiratory activity upon exposure to the UA–norfloxacin combination (Fig. 5). These results also validate the proteomic data where enzymes involved in metabolic processes were found to be down‐regulated. Additionally, the down‐regulation of MurA, which is also linked to rest of the Mur proteins involved in successive steps of peptidoglycan biosynthesis and cell division (Fig. 6A), was further validated using immunoblotting and RT‐PCR, which clearly indicate the downregulation of MurA in UA–norfloxacin‐exposed cells (Fig. 6B). The RT‐PCR results were also in accordance with the proteomics data (Fig. 6C,D) where the murA transcripts followed a similar pattern of down‐regulation in the cells exposed to the UA–norfloxacin combination. The altered membrane potential and permeability, decreased expression of fatty acid biosynthesizing enzymes and disruption in cellular respiration collectively validate the membrane‐damaging potential of UA in combination with norfloxacin.
Figure 4.
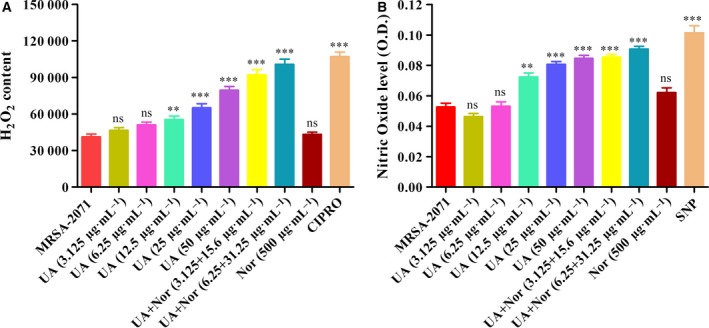
Effect of UA–norfloxacin combination on generation of ROS (H2O2 and NO). (A) Measurement of intracellular H2O2 levels in MRSA‐2071 exposed to different concentrations of UA and norfloxacin alone as well as in combination. Ciprofloxacin was used as a positive control in the study. (B) Effect of UA–norfloxacin combination at variable concentrations on generation of NO levels. Sodium nitroprusside was included in the study as positive control. The tests were performed thrice and data are expressed as mean ± SEM (ns, not significant; **P < 0.01, ***P < 0.001, Dunnett's test).
Figure 5.
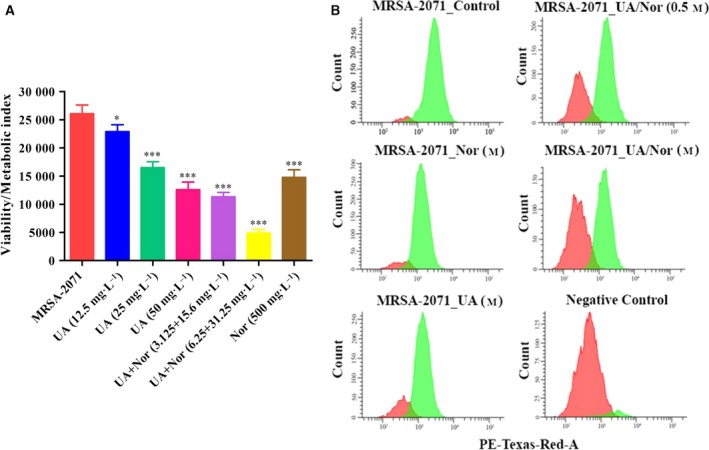
(A) Metabolic activity assay using resazurin. A gradual decline in metabolic activity of MRSA‐2071 cells was recorded with various concentrations of UA alone and the decrease was most prominent in the cells exposed to MIC concentrations of UA–norfloxacin combinations. The results are mean ± SEM derived from three independent experiments (*P ≤ 0.05, ***P ≤ 0.001 Dunnett's test). (B) Flow cytometric analysis for respiratory activity. Graphical representation of cell count vs CTC mean intensity (phycoerythrin‐Texas‐Red‐A) obtained in fluorescence‐activated cell sorting analysis of MRSA‐2071 control and treated samples. A dose‐dependent significant reduction in mean CTC intensity was observed compared to untreated control (n = 3). Negative control was prepared by fixing the cells with formaldehyde and glutaraldehyde prior to CTC staining.
Figure 6.
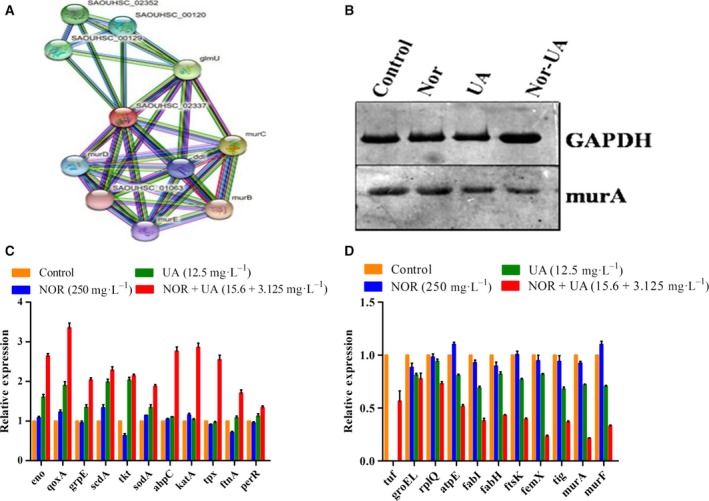
(A) Interactions of MurA with other proteins identified using STRING v.10.0 database. MurA, involved in catalyzing the first step in peptidoglycan biosynthesis, displayed strong interactions with proteins regulating peptidoglycan and cell‐wall biogenesis. (B) Western blot analysis of MurA. The alteration of MurA protein in response to UA–norfloxacin treatment was validated using western blotting, which confirmed the down‐regulation of MurA, indicated by lowered band width and intensity. GAPDH, an endogenous control, was used as loading control in the experiment. (C,D) Quantitative real‐time PCR analysis relative mRNA expression levels of genes up‐regulated in MRSA‐2071 (C) and down‐regulated (D) under UA–norfloxacin treatment alone and in combination using RT‐PCR. The relative expression analysis of each gene was carried out by normalizing the data with GAPDH as an endogenous control and the error bars represent standard deviation (n = 3).
The experiments were carried out in other clinical isolates as well to ensure the reproducibility of the results obtained with MRSA‐2071 (Figs S7–S10) .
In summary, this is the first comprehensive study describing the resistance‐modifying potential of UA in clinical isolate MRSA‐2071. The initial assays showed that UA acts synergistically with norfloxacin and inhibits the efflux pumps, one of the major processes involved in multidrug resistance. Further, the detailed mechanism of action was explored, which revealed that the UA–norfloxacin combination acts by perturbing multiple vital pathways essential for the bacteria to survive and persist. The efficacy and safety profile in the murine mouse model provide evidence the combination as a therapeutic agent against MRSA infections. The detailed analysis evidently demonstrates the effect of UA on inhibition of efflux pumps, followed by induction of oxidative stress, down‐regulation of peptidoglycan and fatty acid biosynthesis, altered membrane potential, perturbed respiration and metabolic activity. The UA–norfloxacin combination was found to further augment these observations as compared to UA alone.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
SS and MPD conceived and designed the study. SS, VG, PK and RJ performed the experiments. SS, RK, AP and MPD analyzed the data. SS, AP and MPD wrote the manuscript.
Supporting information
Table S1. Source of clinical isolates used in the present study.
Table S2. Antibiotic sensitivity/resistance profiling of clinical isolates of S. aureus.
Table S3. Mutation frequency of S. aureus (MTCC‐96) with UA and norfloxacin.
Table S4. List of primers used in the present study.
Fig. S1. PCR amplification of mecA gene from the clinical isolates of S. aureus. (Lane 1: SA‐2071, Lane 2: SA‐1745, Lane 3: SA‐5944, Lane 4: SA‐4627, Lane 5: SA‐3151, Lane 6: SA‐4423, Lane 7: SA‐10760, Lane 8: SA‐4620, Lane 9: MTCC SA‐96, Lane 10: Ladder.)
Fig. S2. (a) PAE of norfloxacin alone as well as in combination with UA against clinical isolate MRSA‐2071. (b) Propensity of development of resistance of S. aureus against UA and norfloxacin combination compared to norfloxacin alone.
Fig. S3. Time kill kinetics of MRSA‐2071 at different concentrations of UA and norfloxacin alone as well as in combination. These data represent mean ± SEM of three independent experiments.
Fig. S4. (a) Protein expression profiles of soluble proteins from MRSA‐2071 cells exposed to sub‐lethal concentrations of UA and norfloxacin alone as well as in combination. Proteins were resolved on 17 cm pH 4–7 IPG strips and 12.5% 20 cm SDS/PAGE gels. (½ MIC UA‐ 12.5 mg·L−1; ½ MIC Nor‐ 250 mg·L−1; ½ MIC UA/Nor‐ 3.125 + 15.6 mg·L−1). (b) Temporal proteome changes of MRSA‐2071 under UA–norfloxacin treatment identified using classical two‐dimensional electrophoresis. Proteins were separated in first dimension isoelectric focussing on 17 cm IPG strips of pH 4–7 range followed by second dimension separation on 12.5% SDS/PAGE. Multivariate analysis was performed to identify the differentially expressing protein spots in response to UA–norfloxacin treatment compared to untreated control and UA and norfloxacin alone, all at sub‐inhibitory concentrations (0.5 MIC). Green arrows indicate up‐regulation, red indicate down‐regulation, blue indicate unique to UA exposed cells, brown indicate unique in Nor exposed cells and yellow indicate proteins unique in cells exposed to UA–norfloxacin combination.
Fig. S5. Functional clustering and biological pathways using DAVID and PANTHER tools associated with the differentially expressed proteins identified in UA–norfloxacin treated MRSA‐2071 cells. Pie chart showing the protein classes (A), biological processes (B) and molecular functions (C).
Fig. S6. Protein–protein interaction network of differentially expressed proteins. The differentially expressed proteins in MRSA‐2071 cells in response to UA–norfloxacin combination were searched for their protein–protein interactions using the web resource STRING v.10.0 (http://www.string-db.org/). Protein–protein interactions are shown in evidence view and proteins are linked based on neighborhood, gene fusion, co‐occurrence, co‐expression, experimental evidences, existing databases and text mining. Network analysis was set at high stringency (STRING score ≥ 0.7).
Fig. S7. Time kill assay of different MRSA strains alone as well as in combination in three different clinical isolates namely MRSA‐1745, MRSA‐4627 and MRSA‐5944. These data represent mean ± SEM of three independent experiments.
Fig. S8. Efflux pump modulatory potential of UA in clinical isolates MRSA‐1745, MRSA‐4627 and MRSA‐5944 using EtBr as marker, determined through spectrofluorometric analysis. Reserpine, an efflux pump inhibitor, was used as positive control in the experiment.
Fig. S9. Prospecting membrane disruptive property of UA–norfloxacin combination against clinical isolates MRSA‐1745, MRSA‐4627 and MRSA‐5944 using PI through spectrofluorimetry. These data represent mean ± SEM of three independent experiments (**P ≤ 0.01, ***P ≤ 0.001, Dunnett's test).
Fig. S10. Membrane depolarization assay of UA–norfloxacin combination using diSC3‐5 against clinical isolates MRSA‐1745, MRSA‐4627 and MRSA‐5944 via spectrofluorimetry. These data represent mean ± SEM of three independent experiments (**P ≤ 0.01, ***P ≤ 0.001, Dunnett's test).
Acknowledgements
The authors are grateful to Prof. K. N. Prasad, Sanjay Gandhi Post Graduate Institute of Medical Sciences (SGPGIMS), Lucknow, India for providing the clinical isolates of methicillin‐resistant S. aureus. The DST‐INSPIRE fellowship from Department of Science and Technology, Govt. of India to SS is dully acknowledged. This work was part of In‐house project MLP‐02 and CSIR Network Project BSC‐0203 of CSIR‐CIMAP, Lucknow, India. The facility and encouragement provided by Director, CSIR‐CIMAP and Director, IHBT Palampur are gratefully acknowledged.
References
- 1. Centers for Disease Control and Prevention (CDC) (2013) Antibiotic resistance threats in the United States, 2013. (https://www.cdc.gov/drugresistance/biggest_threats.html)
- 2. Bush K and Jacoby GA (2010) Updated functional classification of β‐lactamases. Antimicrob Agents Chemother 54, 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rice LB (2010) Progress and challenges in implementing the research on ESKAPE pathogens. Infect Control Hosp Epidemiol 31 (Suppl. 1), S7–S10. [DOI] [PubMed] [Google Scholar]
- 4. Tong SY, Davis JS, Eichenberger E, Holland TL and Fowler VG (2015) Staphylococcus aureus infections: pathophysiology, clinical manifestations and management. Clin Microbiol Rev 28, 603–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abdul‐Aziz MH, Lipman J, Mouton JW, Hope WW and Roberts JA (2015) Applying pharmacokinetic/pharmacodynamic principles in critically ill patients: optimizing efficacy and reducing resistance development. Semin Respir Crit Care Med 36, 136–153. [DOI] [PubMed] [Google Scholar]
- 6. Hrvatin V and Vancouver BC (2017) Combating antibiotic resistance: New drugs or alternative therapies? CMAJ 189 (37), E‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zeng WC, He Q, Sun Q, Zhong K and Gao H (2012) Antibacterial activity of water‐soluble extract from pine needles of Cedrus deodara . I J Food Micro 153, 78–84. [DOI] [PubMed] [Google Scholar]
- 8. Tiwari BK, Valdramidi VP, O'Donnell CP, Muthukumarappan K, Bourke P and Cullen PJ (2009) Application of natural antimicrobials for food preservation. J Agric Food Chem 57, 5987–6000. [DOI] [PubMed] [Google Scholar]
- 9. Garcia SR, Ng C, Anderson H, Chao JD, Zheng X, Pfeifer T, Gay YA, Roberge M and Thompson CJ (2011) Synergistic drug combinations for tuberculosis therapy identified by a novel high‐throughput screen. Antimicrob Agents Chemother 55, 3861–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cottarel G and Wierzbowski J (2007) Combination drugs, an emerging option for antibacterial therapy. Trends Biotechnol 25, 547–555. [DOI] [PubMed] [Google Scholar]
- 11. Price RN and Douglas NM (2009) Artemisinin combination therapy for malaria: beyond good efficacy. Clin Infect Dis 49, 1638–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ekambaram SP, Perumal SS, Balakrishnan A, Marappan N, Gajendran SS and Viswanathan V (2016) Antibacterial synergy between rosmarinic acid and antibiotics against methicillin‐resistant Staphylococcus aureus . J Intercult Ethnopharmacol 5 (4), 358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chung PY, Navaratnam P and Chung LY (2011) Synergistic antimicrobial activity between pentacyclic triterpenoids and antibiotics against Staphylococcus aureus strains. Annals of Clin Microbio Antimicrob 10, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eja ME, Arikpo GE, Enyi‐Idoh KH and Ikpeme EM (2011) An evaluation of the antimicrobial synergy of Garlic (Allium sativum) and Utazi (Gongronema ratifolium) on Escherichia coli and Staphylococcus aureus Malaysian . J Microbiol 7, 49–53. [Google Scholar]
- 15. Tohidpour A, Sattari M, Omidbaigi R, Yadegar A and Nazemi J (2010) Antibacterial effect of essential oils from two medicinal plants against methicillin‐resistant Staphylococcus aureus (MRSA). Phytomedicine 17, 142–145. [DOI] [PubMed] [Google Scholar]
- 16. Chen Y, Liu T, Wang K, Hou C, Cai S, Huang Y, Du Z, Huang H, Kong J and Chen Y (2016) Baicalein inhibits Staphylococcus aureus biofilm formation and the quorum sensing system in vitro. PLoS ONE 11, e0153468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhardwaj M, Singh BR, Singh DK, Kumar V, Prasanna VOR, Varan Singh S, Nirupama KR, Pruthvishree and Archana Saraf BS (2016) Potential of herbal drug and antibiotic combination therapy: a new approach to treat multidrug resistant bacteria. Pharma Anal Acta 7, 523. [Google Scholar]
- 18. Gupta VK, Verma S, Gupta S, Singh A, Pal A, Srivastava SK, Srivastava PK, Singh PC and Darokar MP (2012) Membrane damaging potential of natural L‐(‐)‐usnic acid in Staphylococcus aureus . Eur J Clin Microbiol Infect Dis 31, 3375–3383. [DOI] [PubMed] [Google Scholar]
- 19. Pouranajaf A, Ardebili A, Goudarzi L, Khodabandeh M, Narimani T and Abbaszadeh H (2014) PCR‐based identification of methicillin‐resistant Staphylococcus aureus strains and their antibiotic resistance profiles. Asian Pac J Trop Biomed 4 (Suppl. 1), S293–S297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Novy P, Urban J, Leuner O, Vadlejch J and Kokoska L (2011) In vitro synergistic effects of baicalin with oxytetracycline and tetracycline against Staphylococcus aureus . J Antimicrob Chemother 66, 1298–1300. [DOI] [PubMed] [Google Scholar]
- 21. Odds FC (2003) Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 52, 1. [DOI] [PubMed] [Google Scholar]
- 22. Konai MM, Ghosh C, Yarlagadda V, Samaddar S and Haldar J (2014) Membrane active phenylalanine conjugated lipophilic norspermidine derivatives with selective antibacterial activity. J Med Chem 57 (22), 9409–9423. [DOI] [PubMed] [Google Scholar]
- 23. Heisig P and Tschorny R (1994) Characterization of fluoroquinolone resistant mutants of Escherichia coli selected In vitro. Antimicrob Agents Chemother 38 (6), 1284–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Craig WA and Gudmundsson S (1991) The Postantibiotic effect. Antibiotics in laboratory medicine, Baltimore. Williams and Wilkins 3, 403–431. [Google Scholar]
- 25. Clinical and Laboratory Standards Institute (1999) Methods for determining bactericidal activity of antimicrobial agents. Approved guidelines M26‐A. CLSI. , Wayne, PA, USA. [Google Scholar]
- 26. Saikia D, Parihar S, Ojha CS, Kumar JK, Chanotiya CS, Shanker K and Negi AS (2010) Antitubercular potential of some semisynthetic analogues of phytol. Bioorganic Med Chem Lett 20, 508–512. [DOI] [PubMed] [Google Scholar]
- 27. Viveiros M, Martins M, Couto I, Rodrigues L, Spengler G, Martins A, Kristiansen JE, Molnar J and Amaral L (2008) New methods for the identification of efflux mediated MDR bacteria, genetic assessment of regulators and efflux pump constituents, characterization of efflux systems and screening of inhibitors of efflux pumps. Curr Drug Targets 9, 760–778. [DOI] [PubMed] [Google Scholar]
- 28. Kalle AM and Rizvi A (2011) Inhibition of bacterial multidrug resistance by Celecoxib, a cyclooxygenase‐2 inhibitor. Antimicrob Agents Chemother 55, 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tyagi P, Singh M, Kumari H, Kumari A and Mukhopadhyay K (2015) Bactericidal activity of curcumin I is associated with damaging of bacterial membrane. PLoS ONE 10 (3), e0121313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Silverman JA, Perlmutter NG and Shapiro HM (2003) Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus . Antimicrob Agents Chemother 47, 2538–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reddy PJ, Rao AA, Malhotra D, Sharma S, Kumar R, Jain R, Gollapalli K, Pendharkar N, Srikanth R and Srivastava S (2013) A simple protein extraction method for proteomic analysis of diverse biological specimens. Curr Proteomics 10, 298–311. [Google Scholar]
- 32. Shevchenko A, Tomas H, Havlis J, Olsen JV and Mann M (2007) In‐gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1, 2856–2860. [DOI] [PubMed] [Google Scholar]
- 33. Huang DW, Sherman BT and Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY and Wei L (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39, W316–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, Mering C et al (2013) STRING v9.1: Protein‐protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41, D808–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang H and Joseph JA (1999) Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic Biol Med 27 (5/6), 612–616. [DOI] [PubMed] [Google Scholar]
- 37. Murrell GA, Doland MM, Jang D, Szabo C, Warren RF and Hannafin JA (1996) Nitric oxide: an important articular free radical. J Bone Joint Surg Am 78 (2), 265–274. [PubMed] [Google Scholar]
- 38. Mariscal A, Rosa M, Lopez G, Manuel CV and Crehuet JF (2009) Fluorescent assay based on resazurin for detection of activity of disinfectants against bacterial biofilm. Appl Microbiol Biotechnol 82, 773–783. [DOI] [PubMed] [Google Scholar]
- 39. Rodriguez GG, Phipps D, Ishiguro K and Ridgway HF (1992) Use of a fluorescent redox probe for direct visualization of actively respiring bacteria. Appl Environ Microbiol 58 (6), 1801–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gibbons S, Oluwatuyi M, Veitch NC and Gray AI (2003) Bacterial resistance modifying agents from Lycopus europaeus . Phytochemistry 62, 83–87. [DOI] [PubMed] [Google Scholar]
- 41. Stapleton PD, Shah S, Anderson JC, Hara Y, Hamilton‐Miller JMT and Taylor PW (2004) Modulation of β‐lactam resistance in Staphylococcus aureus by catechins and gallates. Int J Antimicrob Agents 23, 462–467. [DOI] [PubMed] [Google Scholar]
- 42. Oluwatnyi M, Kaatz GW and Gibbons S (2004) Antibacterial and resistance modifying activity of Rosmarinus officinalis . Phytochemistry 65, 3249–3254. [DOI] [PubMed] [Google Scholar]
- 43. Shibata H, Kondo K, Katsuyama R, Kawazoe K, Sato Y, Murakami K, Takaishi Y, Arakaki N and Higuti T (2005) Alkyl gallates, intensifiers of β‐lactam susceptibility in methicillin‐resistant Staphylococcus aureus . Antimicrob Agents Chemother 49, 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pompilio A, Riviello A, Crocetta V, Giuseppe FD, Pomponio S, Sulpizio M, Carmine D, Angelucci S, Barone L, Giulio AD et al (2016) Evaluation of antibacterial and antibiofilm mechanisms by usnic acid against methicillin‐resistant Staphylococcus aureus . Future Microbiol 11, 1315–1338. [DOI] [PubMed] [Google Scholar]
- 45. Gupta VK, Gaur R, Sharma A, Akther J, Saini M, Bhakuni RS and Pathania R (2018) A novel bi‐functional chalcone inhibits multi‐drug resistant Staphylococcus aureus and potentiates the activity of fluoroquinolones. Bioorg Chem 83, 214–225. [DOI] [PubMed] [Google Scholar]
- 46. The European Committee on Antimicrobial Susceptibility Testing . Breakpoint tables for interpretation of MICs and zone diameters. Version 7.1, 2017 (http://www.eucast.org)
- 47. Uppu DSSM, Konai MM, Sarkar P, Samaddar S, Fensterseifer ICM, Farias‐Junior C, Krishnamoorthy P, Shome BR, Franco OL and Haldar J (2017) Membrane‐active macromolecules kill antibiotic‐tolerant bacteria and potentiate antibiotics towards Gram‐negative bacteria. PLoS ONE 12, e0183263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ali F, Sangwan PL, Koul S, Pandey A, Bani S, Abdullah ST, Sharma PR, Kitchlu S and Khan IA (2012) 4‐epi‐Pimaric acid: a phytomolecule as potent antibacterial and antibiofilm agent for oral cavity pathogens. Eur J Clin Mirobiol Infect Dis 31, 149–159. [DOI] [PubMed] [Google Scholar]
- 49. Lu X, Yang X, Li X, Lu Y, Ren Z, Zhao L, Hu X, Jiang J and You X (2013) In vitro activity of sodium new houttuyfonate alone and in combination with oxacillin or netilmicin against methicillin‐resistant Staphylococcus aureus . PLoS ONE 8, e68053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hassan KA, Skurray RA and Brown MH (2007) Active export proteins mediating drug resistance in staphylococci. J Mol Microbiol Biotechnol 12, 180–196. [DOI] [PubMed] [Google Scholar]
- 51. Kaatz GW, McAleese F and Seo SM (2005) Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob Agents Chemother 49, 1857–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Velamakanni S, Yao Y, Gutmann DA and van Veen HW (2008) Multidrug transport by the ABC transporter Sav 1866 from Staphylococcus aureus . Biochemistry 47, 9300–9308. [DOI] [PubMed] [Google Scholar]
- 53. Strahl H and Hamoen LW (2010) Membrane potential id important for bacterial cell division. Proc Natl Acad Sci USA 107, 12281–12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang L, Dhillon P, Yan H, Farmer S and Hancock REW (2000) Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa . Antimicrob Agents Chemother 44, 3317–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miller RA and Britigan BE (1997) Role of oxidants in microbial pathophysiology. Clin Microbiol Rev 10, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Singh V, Pal A and Darokar MP (2015) A polyphenolic flavonoid glabridin: oxidative stress response in multidrug resistant Staphylococcus aureus . Free Radic Biol Med 87, 48–57. [DOI] [PubMed] [Google Scholar]
- 57. Rahal A, Kumar A, Singh V, Yadav B, Tiwari R, Chakraborty S and Dhama K (2014) Oxidative stress, prooxidants and antioxidants: the interplay. Biomed Res Int 2014, 761264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Laurent M, Lepoivre M and Tenu PJ (1996) Kinetic modeling of the nitric oxide gradient generated in vitro by adherent cells expressing inducible nitric oxide synthase. Biochem J 314 (Pt 1), 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wink DA and Mitchell JB (1998) Chemical biology of nitric oxide: Insights into regulatory, cytotoxic and cytoprotective mechanisms of nitric oxide. Free Radic Biol Med 25 (4–5), 434–456. [DOI] [PubMed] [Google Scholar]
- 60. Milewski WM, Boyle‐Vavra S, Moreira B, Ebert CC and Daum RS (1996) Overproduction of a 37‐kilodalton protein homologous to NAD+‐linked‐D‐Lactate dehydrogenase associated with vancomycin resistance in Staphylococcus aureus . Antimicrob Agents Chemother 40 (1), 166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bugg TD, Wright GD, Dutka‐Malen S, Arthur M, Courvalin P and Walsh CT (1991) Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 30, 10408–10415. [DOI] [PubMed] [Google Scholar]
- 62. Heath RJ, White SW and Rock CO (2002) Inhibitors of fatty acid synthesis as antimicrobial chemotherapeutics. Appl Microbiol Biotechnol 58, 695–703. [DOI] [PubMed] [Google Scholar]
- 63. Campbell JW and Cronan JE Jr (2001) Bacterial fatty acid biosynthesis: targets for antibacterial drug discovery. Annu Rev Microbiol 55, 305–332. [DOI] [PubMed] [Google Scholar]
- 64. Zhang YM, White SW and Rock CO (2006) Inhibiting bacterial fatty acid synthesis. J Biol Chem 281, 17541–17544. [DOI] [PubMed] [Google Scholar]
- 65. Zhang YM and Rock CO (2008) Membrane lipid homeostasis in bacteria. Nat Rev Microbio 6, 222–233. [DOI] [PubMed] [Google Scholar]
- 66. Zheng CJ, Sohn MJ and Kim WG (2009) Vinaxanthone, a new FabI inhibitor from Penicillium sp. J Antimicrob Chemother 63, 949–953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Source of clinical isolates used in the present study.
Table S2. Antibiotic sensitivity/resistance profiling of clinical isolates of S. aureus.
Table S3. Mutation frequency of S. aureus (MTCC‐96) with UA and norfloxacin.
Table S4. List of primers used in the present study.
Fig. S1. PCR amplification of mecA gene from the clinical isolates of S. aureus. (Lane 1: SA‐2071, Lane 2: SA‐1745, Lane 3: SA‐5944, Lane 4: SA‐4627, Lane 5: SA‐3151, Lane 6: SA‐4423, Lane 7: SA‐10760, Lane 8: SA‐4620, Lane 9: MTCC SA‐96, Lane 10: Ladder.)
Fig. S2. (a) PAE of norfloxacin alone as well as in combination with UA against clinical isolate MRSA‐2071. (b) Propensity of development of resistance of S. aureus against UA and norfloxacin combination compared to norfloxacin alone.
Fig. S3. Time kill kinetics of MRSA‐2071 at different concentrations of UA and norfloxacin alone as well as in combination. These data represent mean ± SEM of three independent experiments.
Fig. S4. (a) Protein expression profiles of soluble proteins from MRSA‐2071 cells exposed to sub‐lethal concentrations of UA and norfloxacin alone as well as in combination. Proteins were resolved on 17 cm pH 4–7 IPG strips and 12.5% 20 cm SDS/PAGE gels. (½ MIC UA‐ 12.5 mg·L−1; ½ MIC Nor‐ 250 mg·L−1; ½ MIC UA/Nor‐ 3.125 + 15.6 mg·L−1). (b) Temporal proteome changes of MRSA‐2071 under UA–norfloxacin treatment identified using classical two‐dimensional electrophoresis. Proteins were separated in first dimension isoelectric focussing on 17 cm IPG strips of pH 4–7 range followed by second dimension separation on 12.5% SDS/PAGE. Multivariate analysis was performed to identify the differentially expressing protein spots in response to UA–norfloxacin treatment compared to untreated control and UA and norfloxacin alone, all at sub‐inhibitory concentrations (0.5 MIC). Green arrows indicate up‐regulation, red indicate down‐regulation, blue indicate unique to UA exposed cells, brown indicate unique in Nor exposed cells and yellow indicate proteins unique in cells exposed to UA–norfloxacin combination.
Fig. S5. Functional clustering and biological pathways using DAVID and PANTHER tools associated with the differentially expressed proteins identified in UA–norfloxacin treated MRSA‐2071 cells. Pie chart showing the protein classes (A), biological processes (B) and molecular functions (C).
Fig. S6. Protein–protein interaction network of differentially expressed proteins. The differentially expressed proteins in MRSA‐2071 cells in response to UA–norfloxacin combination were searched for their protein–protein interactions using the web resource STRING v.10.0 (http://www.string-db.org/). Protein–protein interactions are shown in evidence view and proteins are linked based on neighborhood, gene fusion, co‐occurrence, co‐expression, experimental evidences, existing databases and text mining. Network analysis was set at high stringency (STRING score ≥ 0.7).
Fig. S7. Time kill assay of different MRSA strains alone as well as in combination in three different clinical isolates namely MRSA‐1745, MRSA‐4627 and MRSA‐5944. These data represent mean ± SEM of three independent experiments.
Fig. S8. Efflux pump modulatory potential of UA in clinical isolates MRSA‐1745, MRSA‐4627 and MRSA‐5944 using EtBr as marker, determined through spectrofluorometric analysis. Reserpine, an efflux pump inhibitor, was used as positive control in the experiment.
Fig. S9. Prospecting membrane disruptive property of UA–norfloxacin combination against clinical isolates MRSA‐1745, MRSA‐4627 and MRSA‐5944 using PI through spectrofluorimetry. These data represent mean ± SEM of three independent experiments (**P ≤ 0.01, ***P ≤ 0.001, Dunnett's test).
Fig. S10. Membrane depolarization assay of UA–norfloxacin combination using diSC3‐5 against clinical isolates MRSA‐1745, MRSA‐4627 and MRSA‐5944 via spectrofluorimetry. These data represent mean ± SEM of three independent experiments (**P ≤ 0.01, ***P ≤ 0.001, Dunnett's test).


