Abstract
Background
Breast cancer has a high mortality rate and is the most common cancer of women worldwide. Our gene co-expression network analysis identified the genes closely related to the pathological stage of breast cancer.
Material/Methods
We performed weighted gene co-expression network analysis (WGCNA) from the Gene Expression Omnibus (GEO) database, and performed pathway enrichment analysis on genes from significant modules.
Results
A non-metastatic sample (374) of breast cancer from GSE102484 was used to construct the gene co-expression network. All 49 hub genes have been shown to be upregulated, and 19 of the 49 hub genes are significantly upregulated in breast cancer tissue. The roles of the genes CASC5, CKAP2L, FAM83D, KIF18B, KIF23, SKA1, GINS1, CDCA5, and MCM6 in breast cancer are unclear, so in order to better reveal the staging of breast cancer markers, it is necessary to study those hub genes. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes indicated that 49 hub genes were enriched to sister chromatid cohesion, spindle midzone, microtubule motor activity, cell cycle, and something else. Additionally, there is an independent data set – GSE20685 – for module preservation analysis, survival analysis, and gene validation.
Conclusions
This study identified 49 hub genes that were associated with pathologic stage of breast cancer, 19 of which were significantly upregulated in breast cancer. Risk stratification, therapeutic decision making, and prognosis predication might be improved by our study results. This study provides new insights into biomarkers of breast cancer, which might influence the future direction of breast cancer research.
MeSH Keywords: Biological Markers; Genes, abl; Triple Negative Breast Neoplasms
Background
Breast cancer has a high mortality rate and is the most common cancer of women worldwide. A study published in 2013 used statistical methods to estimate that the number of new cancer cases in Europe in 2012 would be about 3.45 million, to which the largest contributor would be breast cancer. With the third-highest mortality [1]. The incidence of breast cancer in postmenopausal women is higher [2], and breast cancer is considered one of the leading causes of death in postmenopausal women, accounting for 23% of all cancer deaths [3]. In recent years, the morbidity rate of women under 50 years old has been increasing, and due to the female infertility induced by breast cancer treatments such as chemotherapy, women are caught in a dilemma of choosing between survival and fertility [4]. The occurrence of breast cancer is related to heredity [5]; therefore, the BRCA1/2 mutation has been studied for use in risk assessment in families with a high prevalence of breast cancer [6]. Many genetic tests such as the PAM50 ROR, Breast Cancer Index, and EndoPredict have been reported to predict the development of advanced recurrences [6]. There are many treatments for breast cancer, including chemotherapy, surgery, targeted therapy, hormone replacement therapy, radiation therapy, and combination therapy [3], but the large number of treatments available is a proof that each treatment is not fully effective. The poor prognosis of breast cancer is related to drug resistance, and inappropriate pathological stage may aggravate it. It has been shown that scientific and accurate pathological staging can improve individual prognosis.
Weighted gene co-expression network analysis (WGCNA) is a systematic and comprehensive biological method to explore the correlation between genes, which stands out among many biological methods. The WGCNA approach operates on 2 assumptions: that molecules with similar expression patterns may be involved in specific biological functions (co-regulation of genes), and scale-free distribution [7]. In simple terms, the gene distribution is more consistent with the scale-free network distribution by selecting soft threshold β to weight the correlation coefficient so as to maximize the use of information and avoid information loss [8]. To facilitate their use, a WGCNA R software package summarized and standardized its methods, including network construction, module detection, gene selection, topological property calculation, and visualization [9].
WGCNA builds modules by identifying potential links and correlations between high-throughput genes. Modules closely related to clinical features were used as hub modules for subsequent analysis until the discovery of hub genes tightly related to the disease. This method is not only used for the detection of specific biomarkers of normal and abnormal tissues (such as cancer screening and specific biomarkers of gene-related diseases), but also for the identification of hub genes between abnormal tissues (such as tumor staging, grading, and metastasis). Therefore, the purpose of the present study was to apply the differentially expressed genes (DEGs) co-expression network constructed by WGCNA to identify a series of hub genes related with breast cancer pathological stage. These hub genes as biomarkers may provide better diagnosis and more effective treatment for breast cancer patients, thus leading to earlier detection and better results. This study may contribute to the establishment of a complete biomarker system for the pathological staging of breast cancer.
Material and Methods
Data collection and Preprocessing
The breast cancer gene expression profile of dataset GSE102484 [10], downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), was raw and was based on the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array platform. In the 683 samples of its data set, we selected 374 samples of non-metastatic breast cancer, and all of our samples were female. Then, the preprocessing of 374 original expression data includes the normalization of data using a robust multi-array averaging (RMA) [11] algorithm and the filtering of the nsFilter algorithm. There were 374 samples and 10 093 probes used for subsequent analysis. Furthermore, an independent data set, GSE20685 [12], also downloaded from GEO for module preservation analysis and validation.
Weighted gene co-expression network construction
The WGCNA package in R software was used to construct the co-expression network. First, samples with a Z.K value <−2.5 were deleted as outliers and did not participate in the later analysis. For the construction of a co-expression network, we converted the correlation matrix with the removed outlier samples into the adjacency matrix based on value, and the specific calculation was aij=|cor(xi, xj)|β, and Xi and xj are the nodes i and j of the network. The β was determined by scale-free topology criterion and R2 >0.8 made the network approximately meet the scale-free network distribution generally. For details, refer to original authors Zhang and Horvath. Then, this study transformed the adjacency matrix into a topological overlap matrix (TOM) after a series of complex calculations. TOM provides a simplified diagram of the network, allowing the visualization of the network and facilitating the identification of network modules. Then, the TOM graph was analyzed by average linkage hierarchical clustering based on the phase dissimilarity (1-TOM). Finally, the dynamic shear method was used to obtain the original modules and all the unidentified genes were assigned to a module. The original modules that we obtained were screened and merged before moving on to the next analysis. According to the author’s recommendation, the number of genes in each module was 30 and above. At the same time, each module was marked with a different color for the convenience of research, and the unrecognized genes were grayed out.
Module preservation analysis
To verify the stability of the module, gene expression profiles of 327 samples from data set GSE20685 were used for module preservation analysis. We used the module preservation method in WGCNA R software to calculate the Z summary score (Z score) and medianRank [13], which assesses whether the module is preserved or not. Since Z score and medianRank have their own advantages and disadvantages, in order to treat each module equally, the analysis method with both of them is usually adopted. If the Z score is greater than 10, it is considered that the module is highly preserved, and the higher the Z score is, the higher the stability of the module is, and the more reliable the subsequent analysis will be. A medianRank of the modules close to zero indicates a high degree of module preservation.
Identifying clinically significant modules
The selection of hub modules for subsequent analysis was based on the calculation of the correlation between clinical information and gene modules and the similarity of module expression in samples, including modules eigengene (ME), gene significance (GS), and module significance (MS).
Hub genes identification, validation, and functional annotation
The hub gene is defined as the gene with the highest degree of connectivity in the hub module. Specifically, our study determined hub genes based on 2 values, and selected geneModuleMembership >0.8 and geneTraitSignificance >0.2 as the hub genes. This limitation leads to a high degree of modularity and clinical characteristics of the hub gene. Moreover, genes in hub modules were projected into a protein–protein interaction (PPI) network to further clarify the interaction and association between genes, which was one of the references for our analysis of genes and diseases and the evidence supporting the status of hub genes. The “limma” package is used to identify differentially expressed genes that are widely used in disease and gene research. We used “limma” to test our hub genes. If the P value of the gene is less than the selected significance level (0.05 or 0.01), the selection of the gene is considered statistically significant and it is considered to be validated. Verification makes our selection of hub genes more scientific and convincing.
To further clarify how hub genes influence related clinical characteristics of interesting modules, we used Enrichr (http://amp.pharm.mssm.edu/Enrichr/) database to Gene Ontology (GO) function module of the gene annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis [14].
Survival analysis
Survival analysis is used to determine the relationship between the expression profile of one or more genomes and survival time. Survival analysis of WGCNA is usually done by the non-parametric Kaplan-Meier method [15]. The patients were divided into 2 groups according to median expression value of hub genes by the Kaplan-Meier method, and the results were presented by drawing a survival curve. As a common way to compare survival curves, the log-rank test can conclude that there is no statistically significant difference between groups by analyzing the significance of differences between actual and theoretical values. To assess the relationship between hub genes and breast cancer patients, the Kaplan-Meier survival analysis and log-rank test using the “survival” package of R software were conducted.
Results
Weighted co-expression network construction
The 374 non-metastatic breast cancer samples without stage IV in the independent data set GSE102484 from GEO were filtered by RMA method normalization and nsFilter. Since there were 7 samples with Z.K value <−2.5 (GSM2738700, GSM2738857, GSM2738923, GSM2738999, GSM2739109, GSM2739182, and GSM2739239), these 7 samples were considered as outliers and were excluded from subsequent analysis (Figure 1). Therefore, the gene expression profile of 367 samples was used to construct the gene co-expression network by using the WGCNA package. First, we selected the soft threshold β=5 (scale-free R2=0.90) according to Supplementary Figure 1 as the weighting coefficient to ensure a scale-free network. Second, our study used average linkage hierarchical clustering, the TOM-based dissimilarity, dynamic tree clipping, and merging processing to identify modules, and it obtained 18 modules marked with different colors (Supplementary Figure 2).
Figure 1.
Sample dendrogram and trait heatmap. Seven samples with a Z.K value <−2.5 are outliers. The color intensity was proportional to older age as well as higher stage, Tstage, and Nstage.
Module preservation analysis
We used “modulePreservation” [13] to calculate the preservation statistics of 2 independent modules and to determine whether the modules were preserved according to Z summary score and medianRank. Figure 2 shows that the Z summary score of the brown, turquoise, and yellow modules were all above 50, and the highest Z summary score of the brown module indicated that this module was the best preserved. To compensate for the weakness that Z summary score is dependent on module size, we continued to analyze the relevant medianRank. Among the 3 modules of brown, turquoise, and yellow, the medianRank of the brown module was still prominent. Brown modules, whose Z summery score is more than 60 were considered to be the best preserved modules in this study.
Figure 2.
The medianRank and Zsummary statistics of the module preservation based on GSE20685. The medianRank of the modules close to zero indicates the high degree of module preservation, and the Zsummary of the modules close to zero indicates the low degree of module preservation.
Identifying hub modules
The selection of hub modules was based on the correlation between modules and traits. Therefore, brown modules were selected as the hub modules in this study according to the closest correlation between modules and stages (r=0.22, P=3e-05, Figure 3). At the same time, we also found that the brown module was also the most correlated with Tstage (r=0.18, P=4e-04, Figure 3), and the correlation between Nstage and the brown module was also high (r=0.14, P=0.008, Figure 3). Furthermore, the brown module showed high genetic significance and module membership (cor=0.49, P=4.4e-67, Supplementary Figure 3), which further confirmed the status of the brown module as a hub module.
Figure 3.
Heatmap of the correlation between module eigengenes and clinical traits of breast cancer. Each module is based on the pattern of their co-expression. Stage indicated bipolar disorder (breast cancer and non-breast cancer).
Hub genes identification, validation and functional annotation
There were 1093 genes in the brown module and 49 that meet our requirements (geneModuleMembership >0.8 and geneTraitSignificance >0.2), and the special data are shown in Table 1. The 2290 genes were shown to be associated with breast cancer in previous studies through Junior Doc (http://www.drwang.top/), and 21 of our 49 hub genes had not been reported (e.g., TPX2, MCM10, NCAPG, and KIF4A). The PPI network of brown modules showed significant connectivity, and the overall effect of the network graph was good (Supplementary Figure 4). In this network, the size and color depth of nodes are proportional to their degree of connection, which facilitated our qualitative observation. The node degree, betweenness, stress, closeness, and clustering coefficient of 49 hub genes in the brown module were quantitatively compared (Table 2). AURKA gene has the largest node and the darkest color, which was an important object for our subsequent discussion and research. Independent GSE20685 was used by us for gene validation, and the condition for our hub genes to pass the verification was P<0.05. Supplementary Figure 5 intuitively shows our verification results, in which the P values of 49 hub genes are less than 0.05, which indicates that the selection of our 49 hub genes was statistically significant, scientific, and convincing. Meanwhile, according to |log2(fold change)| ≥0.5, 19 of the 49 hub genes were significantly upregulated (Supplementary Figure 6).
Table 1.
The 49 hub genes most associated with breast cancer.
| Probe ID | Gene symbol | Entrez gene ID | Gene module membership | Gene trait significance |
|---|---|---|---|---|
| 209642_at | BUB1 | 699 | 0.924293226 | 0.235607323 |
| 209408_at | KIF2C | 11004 | 0.92218934 | 0.241907024 |
| 210052_s_at | TPX2 | 22974 | 0.914480792 | 0.223144303 |
| 224753_at | CDCA5 | 113130 | 0.91205407 | 0.259025302 |
| 220651_s_at | MCM10 | 55388 | 0.910563071 | 0.208487817 |
| 204822_at | TTK | 7272 | 0.907948229 | 0.219457711 |
| 204825_at | MELK | 9833 | 0.903514378 | 0.231607588 |
| 218726_at | HJURP | 55355 | 0.90328239 | 0.22698554 |
| 225687_at | FAM83D | 81610 | 0.899886619 | 0.203016176 |
| 218663_at | NCAPG | 64151 | 0.896054426 | 0.205216007 |
| 218755_at | KIF20A | 10112 | 0.895166941 | 0.226348808 |
| 202870_s_at | CDC20 | 991 | 0.892240716 | 0.239137318 |
| 221520_s_at | CDCA8 | 55143 | 0.89118746 | 0.244566866 |
| 206364_at | KIF14 | 9928 | 0.890950252 | 0.26234143 |
| 209464_at | AURKB | 9212 | 0.890552966 | 0.22142283 |
| 203755_at | BUB1B | 701 | 0.888672889 | 0.213901874 |
| 218355_at | KIF4A | 24137 | 0.887945991 | 0.228378944 |
| 202954_at | UBE2C | 11065 | 0.887818368 | 0.226938633 |
| 203554_x_at | PTTG1 | 9232 | 0.881155019 | 0.213686403 |
| 205046_at | CENPE | 1062 | 0.880069219 | 0.22209321 |
| 208079_s_at | AURKA | 6790 | 0.879152052 | 0.218061231 |
| 203358_s_at | EZH2 | 2146 | 0.875743388 | 0.207933283 |
| 222608_s_at | ANLN | 54443 | 0.871339701 | 0.234975449 |
| 205339_at | STIL | 6491 | 0.869107261 | 0.210922715 |
| 205024_s_at | RAD51 | 5888 | 0.867508527 | 0.213183274 |
| 228069_at | MTFR2 | 113115 | 0.864955684 | 0.211359463 |
| 228868_x_at | CDT1 | 81620 | 0.86478764 | 0.229292335 |
| 205733_at | BLM | 641 | 0.860785004 | 0.217032378 |
| 228323_at | CASC5 | 57082 | 0.857092896 | 0.202429723 |
| 201710_at | MYBL2 | 4605 | 0.85635078 | 0.236329323 |
| 206102_at | GINS1 | 9837 | 0.856311574 | 0.208385671 |
| 222077_s_at | RACGAP1 | 29127 | 0.853841921 | 0.256705857 |
| 204709_s_at | KIF23 | 9493 | 0.851379193 | 0.21561731 |
| 229610_at | CKAP2L | 150468 | 0.85011016 | 0.229596216 |
| 204033_at | TRIP13 | 9319 | 0.849593284 | 0.202381238 |
| 218009_s_at | PRC1 | 9055 | 0.849581571 | 0.221410912 |
| 207746_at | POLQ | 10721 | 0.848529181 | 0.247638427 |
| 214804_at | CENPI | 2491 | 0.846773238 | 0.207038989 |
| 217640_x_at | SKA1 | 220134 | 0.837739498 | 0.203041011 |
| 219650_at | ERCC6L | 54821 | 0.82821799 | 0.202549981 |
| 222039_at | KIF18B | 146909 | 0.825933796 | 0.203151907 |
| 207828_s_at | CENPF | 1063 | 0.818716418 | 0.230482778 |
| 228273_at | PRR11 | 55771 | 0.815331726 | 0.215057337 |
| 204023_at | RFC4 | 5984 | 0.815233799 | 0.213293858 |
| 213008_at | FANCI | 55215 | 0.810580105 | 0.2165155 |
| 204817_at | ESPL1 | 9700 | 0.808284083 | 0.211142978 |
| 202107_s_at | MCM2 | 4171 | 0.803926063 | 0.207204313 |
| 209680_s_at | KIFC1 | 3833 | 0.801065976 | 0.213650537 |
| 201930_at | MCM6 | 4175 | 0.800871266 | 0.214708557 |
Gene module membership represents the degree of linkage between hub genes and other genes, while gene trait significance represents the relationship between genes and clinical features.
Table 2.
The node degree, betweenness, stress, closeness, and clustering coefficient of 49 hub genes in brown module.
| Gene | Node degree | Betweenness | Stress | Closeness | Clustering coefficient |
|---|---|---|---|---|---|
| AURKA | 50 | 0.01994251 | 556 | 1 | 0.77306122 |
| TPX2 | 49 | 0.00885408 | 464 | 0.98039216 | 0.80272109 |
| BUB1 | 49 | 0.00885408 | 464 | 0.98039216 | 0.80272109 |
| BUB1B | 49 | 0.00885408 | 464 | 0.98039216 | 0.80272109 |
| CDCA8 | 49 | 0.00885408 | 464 | 0.98039216 | 0.80272109 |
| KIF2C | 49 | 0.01860824 | 508 | 0.98039216 | 0.78401361 |
| TTK | 49 | 0.00885408 | 464 | 0.98039216 | 0.80272109 |
| NCAPG | 49 | 0.00885408 | 464 | 0.98039216 | 0.80272109 |
| KIF20A | 48 | 0.0078717 | 418 | 0.96153846 | 0.81471631 |
| MELK | 48 | 0.0078717 | 418 | 0.96153846 | 0.81471631 |
| CDC20 | 47 | 0.00599724 | 352 | 0.94339623 | 0.83718779 |
| AURKB | 47 | 0.00603083 | 352 | 0.94339623 | 0.83718779 |
| MCM10 | 47 | 0.01567858 | 408 | 0.94339623 | 0.81128585 |
| CENPF | 47 | 0.00603083 | 352 | 0.94339623 | 0.83718779 |
| UBE2C | 47 | 0.00730838 | 386 | 0.94339623 | 0.82146161 |
| KIF4A | 47 | 0.00538752 | 336 | 0.94339623 | 0.84458834 |
| RACGAP1 | 46 | 0.00583064 | 326 | 0.92592593 | 0.84251208 |
| KIF23 | 46 | 0.00457007 | 294 | 0.92592593 | 0.85797101 |
| CENPE | 45 | 0.00420229 | 268 | 0.90909091 | 0.86464646 |
| PRC1 | 44 | 0.00395004 | 248 | 0.89285714 | 0.86892178 |
| CDCA5 | 44 | 0.00448217 | 252 | 0.89285714 | 0.86680761 |
| HJURP | 44 | 0.00449643 | 262 | 0.89285714 | 0.8615222 |
| TRIP13 | 43 | 0.00307517 | 204 | 0.87719298 | 0.88704319 |
| PTTG1 | 41 | 0.00319721 | 176 | 0.84745763 | 0.89268293 |
| ANLN | 41 | 0.00234005 | 160 | 0.84745763 | 0.90243902 |
| FANCI | 41 | 0.00240147 | 160 | 0.84745763 | 0.90243902 |
| CDT1 | 40 | 0.00249934 | 158 | 0.83333333 | 0.89871795 |
| MCM2 | 40 | 0.00238095 | 152 | 0.83333333 | 0.9025641 |
| ESPL1 | 40 | 0.00218884 | 142 | 0.83333333 | 0.90897436 |
| KIF14 | 40 | 0.00258938 | 152 | 0.83333333 | 0.9025641 |
| RAD51 | 38 | 0.00218296 | 138 | 0.80645161 | 0.90184922 |
| MCM6 | 36 | 0.00155343 | 98 | 0.78125 | 0.92222222 |
| CASC5 | 36 | 7.88E-04 | 58 | 0.78125 | 0.95396825 |
| SKA1 | 36 | 0.00155359 | 96 | 0.78125 | 0.92380952 |
| KIF18B | 36 | 0.00151695 | 98 | 0.78125 | 0.92222222 |
| KIFC1 | 36 | 0.00120015 | 80 | 0.78125 | 0.93650794 |
| ERCC6L | 36 | 0.00174436 | 110 | 0.78125 | 0.91269841 |
| CKAP2L | 35 | 0.00166605 | 94 | 0.76923077 | 0.9210084 |
| RFC4 | 34 | 0.00115046 | 74 | 0.75757576 | 0.93404635 |
| MYBL2 | 34 | 0.00150041 | 82 | 0.75757576 | 0.92691622 |
| POLQ | 33 | 0.00670463 | 108 | 0.74626866 | 0.89772727 |
| EZH2 | 33 | 8.32E-04 | 54 | 0.74626866 | 0.94886364 |
| CENPI | 32 | 4.99E-04 | 36 | 0.73529412 | 0.96370968 |
| FAM83D | 29 | 2.09E-04 | 16 | 0.70422535 | 0.98029557 |
| STIL | 29 | 2.24E-04 | 16 | 0.70422535 | 0.98029557 |
| BLM | 26 | 6.48E-04 | 44 | 0.67567568 | 0.93230769 |
| GINS1 | 26 | 3.55E-04 | 26 | 0.67567568 | 0.96 |
| PRR11 | 18 | 5.66E-05 | 4 | 0.6097561 | 0.9869281 |
| MTFR2 | 15 | 0 | 0 | 0.58823529 | 1 |
To determine the mechanism of action of the 49 hub genes in the brown module, the 49 hub genes were uploaded to Enichr for GO function annotation and KEGG enrichment analysis. GO function annotation indicated that the brown module was enriched to sister chromatid cohesion, spindle midzone, and microtubule motor activity (Table 3). KEGG enrichment analysis suggested the brown module was enriched in cell cycle (Table 4). Tables 3, 4 show that the crude and adjusted P value, Z score, the combined score of pathways, and genes were included in the pathway.
Table 3.
GO function annotation of 49 hub genes.
| Series | Name | P value | Adjusted P value | Z score | Combined score | Genes |
|---|---|---|---|---|---|---|
| GO Cellular Component | Spindle midzone (GO: 0051233) | 7.05E-22 | 9.03E-20 | −2.19 | 106.46 | KIF14; BUB1B; CDCA8; TTK; KIF23; AURKB; AURKA; CDC20; TPX2; CENPF; RACGAP1; PRC1; KIF20A |
| GO Cellular Component | Mitotic spindle (GO: 0072686) | 4.49E-18 | 2.37E-16 | −2.3 | 91.68 | CDC20; TPX2; CENPF; RACGAP1; ESPL1; CKAP2L; PRC1; TTK; KIF23; KIF20A; AURKB; AURKA |
| GO Cellular Component | Mitotic spindle midzone (GO: 1990023) | 5.56E-18 | 2.37E-16 | −2.06 | 81.88 | TPX2; CENPE; RACGAP1; ESPL1; CKAP2L; KIF14; BUB1B; CDCA8; KIF23; AURKB; AURKA |
| GO Biological Process | Sister chromatid cohesion (GO: 0007062) | 6.67E-18 | 1.35E-15 | −2.76 | 109.02 | CDC20; CENPE; CENPF; ERCC6L; CENPI; CDCA5; BUB1B; CDCA8; KIF2C; BUB1; AURKB; SKA1 |
| GO Cellular Component | Spindle microtubule (GO: 0005876) | 1.69E-17 | 5.42E-16 | −2.32 | 89.73 | KIF14; TTK; KIF23; AURKB; SKA1; AURKA; CDC20; TPX2; CENPE; CENPF; PRC1; KIF4A; KIF20A |
| GO Cellular Component | Spindle (GO: 0005819) | 2.60E-16 | 6.67E-15 | −2.45 | 88.06 | TTK; KIF23; AURKB; SKA1; AURKA; CDC20; TPX2; CENPE; CENPF; PRC1; KIF2C; KIF20A; MCM2 |
| GO Cellular Component | Mitotic spindle microtubule (GO: 1990498) | 1.87E-15 | 3.99E-14 | −2.1 | 71.08 | TPX2; RACGAP1; ESPL1; CKAP2L; PRC1; KIF4A; KIF23; AURKB; SKA1; AURKA |
| GO Biological Process | Mitotic cell cycle (GO: 0000278) | 9.03E-15 | 9.17E-13 | −2.82 | 91.31 | CDT1; TPX2; CENPE; CENPF; KIF18B; CDCA5; BUB1B; KIF2C; SKA1; AURKA |
| GO Cellular Component | Meiotic spindle (GO: 0072687) | 1.20E-14 | 2.20E-13 | −1.98 | 63.49 | CDC20; TPX2; CENPF; PRC1; TTK; KIF23; KIF20A; AURKB; AURKA |
| GO Biological Process | Microtubule-based movement (GO: 0007018) | 2.69E-14 | 1.82E-12 | −2.46 | 76.95 | CENPE; KIF18B; RACGAP1; KIFC1; KIF4A; KIF14; KIF2C; KIF23; KIF20A |
| GO Cellular Component | Spindle pole (GO: 0000922) | 6.45E-14 | 1.03E-12 | −2.19 | 66.53 | CDC20; TPX2; CENPF; PRC1; TTK; KIF23; AUNIP; KIF20A; AURKB; AURKA |
| GO Cellular Component | Kinesin complex (GO: 0005871) | 9.68E-14 | 1.13E-12 | −1.72 | 51.51 | CENPE; KIF18B; KIFC1; KIF4A; KIF14; KIF2C; KIF23; KIF20A |
| GO Cellular Component | Kinesin I complex (GO: 0016938) | 9.68E-14 | 1.13E-12 | −1.68 | 50.33 | CENPE; KIF18B; KIFC1; KIF4A; KIF14; KIF2C; KIF23; KIF20A |
| GO Molecular Function | ATP-dependent microtubule motor activity (GO: 1990939) | 5.81E-13 | 1.22E-10 | −1.91 | 53.71 | CENPE; KIF18B; KIFC1; KIF4A; KIF14; KIF2C; KIF23; KIF20A |
| GO Molecular Function | Microtubule motor activity (GO: 0003777) | 2.84E-12 | 2.98E-10 | −2.06 | 54.82 | CENPE; KIF18B; KIFC1; KIF4A; KIF14; KIF2C; KIF23; KIF20A |
| GO Molecular Function | ATP-dependent microtubule motor activity, plus-end-directed| (GO: 0008574) | 7.36E-11 | 5.15E-09 | −2.59 | 60.38 | CENPE; BLM; KIF18B; KIFC1; KIF4A; KIF14; KIF2C; KIF23; KIF20A |
| GO Biological Process | Mitotic metaphase plate congression (GO: 0007080) | 4.10E-10 | 2.08E-08 | −2.79 | 60.28 | CENPE; KIFC1; CDCA5; KIF14; CDCA8; KIF2C |
| GO Molecular Function | Protein-DNA unloading ATPase activity (GO: 0140083) | 1.11E-09 | 4.14E-08 | −2.51 | 51.79 | CENPE; BLM; KIF18B; KIFC1; KIF14; KIF2C; KIF23; KIF20A |
| GO Molecular Function | ATPase activity, uncoupled (GO: 0042624) | 1.18E-09 | 4.14E-08 | −2.56 | 52.69 | CENPE; BLM; KIF18B; KIFC1; KIF14; KIF2C; KIF23; KIF20A |
| GO Molecular Function | ATP-dependent microtubule motor activity, minus-end-directed (GO: 0008569) | 1.33E-09 | 4.14E-08 | −2.54 | 51.86 | CENPE; BLM; KIF18B; KIFC1; KIF14; KIF2C; KIF23; KIF20A |
| GO Molecular Function | ATPase activity, coupled (GO: 0042623) | 1.49E-09 | 4.14E-08 | −2.61 | 53.06 | CENPE; BLM; KIF18B; KIFC1; KIF14; KIF2C; KIF23; KIF20A |
| GO Molecular Function | ATPase activity (GO: 0016887) | 1.58E-09 | 4.14E-08 | −2.53 | 51.31 | CENPE; BLM; KIF18B; KIFC1; KIF14; KIF2C; KIF23; KIF20A |
| GO Molecular Function | Intracellular ATPase-gated chloride channel activity (GO: 0005260) | 5.56E-09 | 1.30E-07 | −2.63 | 50.06 | CENPE; BLM; KIF18B; KIFC1; KIF14; KIF2C; KIF23; KIF20A |
| GO Biological Process | Mitotic spindle midzone assembly (GO: 0051256) | 6.60E-09 | 2.68E-07 | −2.1 | 39.62 | RACGAP1; KIF4A; KIF23; AURKB |
| GO Biological Process | Metaphase plate congression (GO: 0051310) | 1.04E-08 | 3.51E-07 | −2.44 | 44.81 | CENPE; CENPF; KIF2C; FAM83D |
| GO Biological Process | Anaphase-promoting complex-dependent catabolic process (GO: 0031145) | 4.13E-08 | 0.000001197 | −2.51 | 42.73 | CDC20; PTTG1; UBE2C; BUB1B; AURKB; AURKA |
| GO Biological Process | Chromosome segregation (GO: 0007059) | 1.13E-07 | 0.000002705 | −2.33 | 37.27 | CDT1; CENPE; CENPF; HJURP; SKA1 |
| GO Biological Process | Spindle organization (GO: 0007051) | 1.20E-07 | 0.000002705 | −2.13 | 33.93 | TTK; AUNIP; AURKB; AURKA |
| GO Biological Process | Protein ubiquitination involved in ubiquitin-dependent protein catabolic process (GO: 0042787) | 5.14E-07 | 0.00001043 | −2.89 | 41.83 | CDC20; PTTG1; UBE2C; BUB1B; AURKB; AURKA |
| GO Molecular Function | Microtubule plus-end binding (GO: 0051010) | 7.81E-07 | 1.64E-05 | −2.32 | 32.66 | RACGAP1; KIF14; KIF2C; KIF23; FAM83D; SKA1 |
Table 4.
KEGG enrichment analysis of 49 hub genes.
| Name | P value | Adjusted P value | Z score | Combined score | Genes |
|---|---|---|---|---|---|
| Cell cycle | 6.34E-10 | 1.01E-08 | −5.14 | 108.9 | CDC20; PTTG1; ESPL1; BUB1B; TTK; MCM6; BUB1; MCM2 |
| Oocyte meiosis | 0.00001346 | 0.0001077 | −27.81 | 311.96 | CDC20; PTTG1; ESPL1; BUB1; AURKA |
| DNA replication | 0.00009322 | 0.0004972 | −49.47 | 459.08 | RFC4; MCM6; MCM2 |
| Fanconi anemia pathway | 0.0003139 | 0.001256 | −42.68 | 344.27 | FANCI; BLM; RAD51 |
| Human T-cell leukemia virus 1 infection | 0.002015 | 0.006448 | −19.4 | 120.41 | CDC20; PTTG1; ESPL1; BUB1B |
| Homologous recombination | 0.004537 | 0.0121 | −49.65 | 267.88 | BLM; RAD51 |
Survival analysis
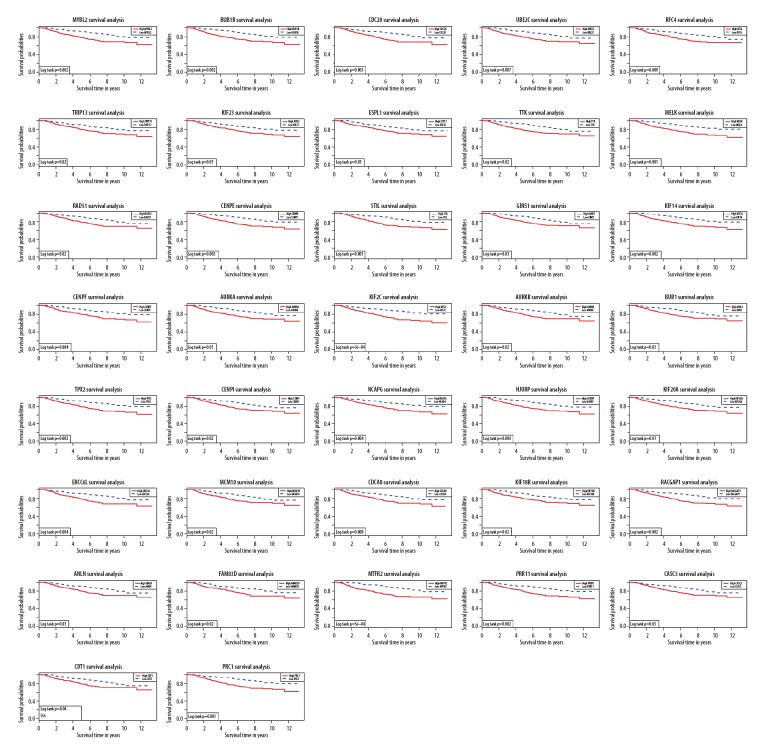
Breast cancer patients were divided into a high-expression group and a low-expression group according to the median expression value of each hub gene, and survival curves were plotted accordingly. The P value of 37 of 49 hub genes was less than 0.05, indicating that the 37 hub genes were statistically significant (Figure 4). The survival curve generally shows a downward trend with the increase of time. On the graph line, the slope is larger, which means lower survival rate. The image of the high-expression group of 49 hub genes was steeper than that of the low-expression group, indicating that the high expression of these hub genes was closely related to the poor prognosis of patients.
Figure 4.
Survival curves for patients in different groups. Red lines represent high expression of hub genes, while blue lines represent low expression of hub genes.
Hub gene mutation analysis
The independent data set GSE29044 from the GEO database was used for mutation analysis (Figure 5). All 49 hub genes were shown to be upregulated in the mutation analysis, suggesting that our hub genes are scientific and persuasive, and Supplementary Figure 6 shows that 19 of 49 hub genes were significantly upregulated.
Figure 5.
Mutation analysis of 49 genes was based on independent data set GSE29044.
Discussion
Breast cancer is an epithelial malignant tumor of ductal lobules at the end of the mammary gland. It is the most common cancer among women and has a poor prognosis, causing a large number of deaths worldwide every year. The research activity on breast cancer is in direct proportion to its harm to human beings, but quantity does not mean quality; the pathogenesis of breast cancer has not been fully elucidated, and the genetic standard of breast cancer staging is not perfect. Although some studies have used the WGCNA method to explore molecular markers related to the pathogenesis, diagnosis, treatment, and prognosis of breast cancer, the present study may contribute to the establishment of a more complete set of molecular markers for pathological staging of breast cancer. This may lead to better treatment regimens for different patients and better prognostic estimates.
In our study, we used 367 samples without stage IV to construct the co-expression network. After dynamic tree shearing, we identified 18 modules, among which the brown module showed the strongest correlation with pathological staging. The Z summery score and medianRank indicated that the brown module has good stability. Therefore, the brown module as a candidate module continues to identify candidate biomarkers. Finally, we identified the 49 hub genes that are candidate biomarkers for breast cancer pathological stage, and 21 hub genes that were not inquired about in Junior Doc. The PPI chart of 49 hub genes showed the ideal degree of connectivity, and it could be intuitively seen that the hub genes were interacting with each other. Both survival analysis and mutation analysis yielded satisfactory results: high expression of 49 hub genes was closely related to the poor prognosis of patients, and all 49 hub genes were shown to be upregulated in the mutation analysis. Moreover, to better illustrate how hub genes works, our study also carried out GO functional annotation and KEGG enrichment analysis for 49 hub genes. GO functional annotation of hub genes were suggested to focus on sister chromatid cohesion, mitotic cell cycle, spindle midzone, and microtubule motor activity. Similarly, hub genes identified by KEGG were enriched in the cell cycle and DNA replication. These hub pathways are basically all involved in cell division. The sister chromatid cohesion pathway gene members are CDC20, CENPE, CENPF, ERCC6L, CENPI, BUB1B, CDCA8, KIF2C, BUB1, AURKB, and SKA1. The sister chromatid cohesion pathway is a hub of mitotic chromosomes separation; this process is mediated by the cohesive element protein complexes. It has been reported in colorectal cancer [16], bladder cancer [17], head and neck squamous cell carcinoma [18], and other cancers. Previous studies on breast cancer found that sister chromatid cohesion can inhibit breast cancer cells and induce their apoptosis and autophagy [19], and the survival rate of breast cancer patients with defective sister chromatid cohesion expression is lower than those with higher sister chromatid cohesion expression [20]. In addition, studies have shown that cancer cells that recognize mutations in sister chromatid cohesion may suggest new therapeutic opportunities [21]. Cell cycle refers to the time needed for one cell to divide one time, and its members have CDC20, PTTG1, ESPL1, BUB1B, TTK, MCM6, BUB1, and MCM2. Many studies have shown that the stagnation of the cell cycle is a target for the treatment of breast cancer [22].
In terms of individual hub genes, KIF4A, MCM10, and TPX2 were also listed as the hub genes associated with poor prognosis of breast cancer in other studies that also used the WGCNA method to identify the pathological process of breast cancer [23]. In another study looking at biomarkers of prognosis in invasive breast cancer [24], only MELK in the 6 hub genes was the same as our hub gene, suggesting that our study could be complementary. A study using grade 1, 2, and 3 differentially expressed genes of cancer to construct a hierarchical specific molecular interaction network indicated that KIF2C and UBE2C are potential biomarkers for breast cancer diagnosis and prognosis [25]. The present study offers more new insights than the hub genes provided by the latest research on genetic markers for breast cancer [26]. Most of the hub genes associated with breast cancer are well understood. AURKA and AURKB also are leading predictors of poor prognosis [27]. AURKA has the highest degree of connectivity in the PPI network. Our research on this gene is relatively mature, and it has been reported in many studies that this gene is closely related to breast cancer. AURKA inhibitors have long been important drugs in the clinical treatment of breast cancer. The latest research shows that AURKA inhibitors combined with other inhibitors provide a new approach for the treatment of breast cancer [28]. BUB1B causes higher chromosomal instability in breast cancer cells [29], and BUB1 is associated with cancer stem cells [30]. CDT1, CDC20 CENPE CENPF, and CENPI expression in breast cancer cells are on the increase, and are significantly associated with shorter survival [31,32]. KIF14, KIF20A, KIF2C, KIF4A, and KIFC1 belonging to the kinesin family and have been proven to be potential biomarkers of breast cancer prognosis [33,34], but there have been few studies on KIF18B and KIF23. HJURP is a histone chaperone, a prognostic factor for disease-free survival and overall survival in breast cancer patients, and a predictive marker for radiotherapy sensitivity [35]. Among the hub genes in the MCM series, MCM2 and MCM10 have been extensively studied in the pathological process of breast cancer, but MCM6 has received relatively little attention. However, poor prognosis due to overexpression of MCM6 has been reported in lung cancer [36]. Additionally, the abnormal expression of TPX2 has basically become a universal biomarker for poor prognosis of cancer, which has been reported in gastric cancer, non-small cell lung cancer, liver cancer, and other cancers [37,38]. CASC5, also known as KNL1, is an important gene involved in chromosome separation and is expressed in various cancer cells. Inhibition of this gene expression can induce cell cycle arrest and inhibit cell proliferation and migration. KNL1 and BUB1 have similar effects and both function by activating the kinetochore-bound Mad1-Mad2 [39]. However, CASC5 has not been adequately investigated in relation to breast cancer, nor has BUB1. The protein encoded by the CKAP2L gene plays an important role in neuroprogenitor cell division, and mutations in this gene are associated with spindle tissue defects [40]. CKAP2L is reported to be the main cause of Filippi syndrome [40]. There have been few studies on the relationship between CKAP2L and cancer, and few studies have been reported so far: the deletion of this gene is associated with oral squamous cell carcinoma, and the latest study shows that the upregulated expression of this gene in lung adenocarcinoma may be closely correlated with the poor prognosis of patients. However, the exact role of CKAP2L in cancer progression, metastasis, and drug resistance remains unclear. In particular, research on the relationship between CKAP2L and breast cancer is scant. In addition, understanding of the molecular basis of CASC5, CKAP2L, FAM83D, KIF18B, KIF23, SKA1, GINS1, CDCA5, and MCM6 in breast cancer is poor, so in order to better reveal the staging of breast cancer markers, it is necessary to study the hub genes. Our understanding of certain genes is still incomplete, and our co-expression networks might provide new clues to the complex regulation of these different molecules. However, compared with other tumor databases with more tumor database samples, the data set samples that this study used are relatively small; there may be bias, and many related studies have been published. Furthermore, although it found that some new genes may be related to the pathological process of breast cancer, these new genes may not provide accurate information about the actual biological characteristics of the tumor.
Conclusions
We established a co-expression network to identify the hub genes related to the pathological staging of breast cancer, identifying 49 hub genes that were associated with the pathologic stage of breast cancer, 19 of which were significantly upregulated in breast cancer. Our results may provide new insights into biomarkers for breast cancer, but more research is needed to validate these findings
Supplementary Data
Analysis of network topology for various soft-theholding powers. The left panel showed the scale-free fit index, signed R2 (y-axis) and the soft threshold power (x-axis).
Clustering dendrogram of genes and modules identfied by weighted gene co-expression network analysis based on a dissimilarity measure (1-TOM).
Scatter diagram for module membership vs. gene significance of stage (breast cancer or non-breast cancer) in brown module.
The protein-protein network of the hub genes in brown module. Within the network, node sizes and color depth are proportional to their connectivity.
The P value of 49 hub genes for gene validation.
19 hub genes were significantly up-egulated in breast cancer that indicates the high expression of those genes is related to breast cancer.
Footnotes
Availability of data and material
All data and material are available in the GEO database.
Conflict of interests
None.
Source of support: Departmental sources
References
- 1.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, et al. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Cao J, Eshak ES, Liu K, et al. Television viewing time and breast cancer incidence for Japanese premenopausal and postmenopausal women: The JACC Study. Cancer Res Treat. :2019. doi: 10.4143/crt.2018.705. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akram M, Iqbal M, Daniyal M, et al. Awareness and current knowledge of breast cancer. Biol Res. 2017;50:33. doi: 10.1186/s40659-017-0140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghaemi SZ, Keshavarz Z, Tahmasebi S, et al. Conflicts women with breast cancer face with: A qualitative study. J Family Med Prim Care. 2019;8:27–36. doi: 10.4103/jfmpc.jfmpc_272_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plasterer C, Tsaih SW, Lemke A, et al. Identification of a rat mammary tumor risk locus that is syntenic with the commonly amplified 8q12.1 and 8q22.1 regions in human breast cancer patients. G3 (Bethesda) 2019 doi: 10.1534/g3.118.200873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duffy MJ, Walsh S, McDermott EW, et al. Biomarkers in breast cancer: Where are we and where are we going? Adv Clin Chem. 2015;71:1–23. doi: 10.1016/bs.acc.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 8.Zhao W, Langfelder P, Fuller T, et al. Weighted gene coexpression network analysis: State of the art. J Biopharm Stat. 2010;20:281–300. doi: 10.1080/10543400903572753. [DOI] [PubMed] [Google Scholar]
- 9.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng SH, Huang TT, Cheng YH, et al. Validation of the 18-gene classifier as a prognostic biomarker of distant metastasis in breast cancer. PLoS One. 2017;12:e0184372. doi: 10.1371/journal.pone.0184372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 12.Kao KJ, Chang KM, Hsu HC, et al. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: Implications for treatment optimization. BMC Cancer. 2011;11:143. doi: 10.1186/1471-2407-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langfelder P, Luo R, Oldham MC, et al. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen EY, Tan CM, Kou Y, et al. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goel MK, Khanna P, Kishore J. Understanding survival analysis: Kaplan-Meier estimate. Int J Ayurveda Res. 2010;1:274–78. doi: 10.4103/0974-7788.76794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi Y, Qi H, Liu Z, et al. Bioinformatics analysis of key genes and pathways in colorectal cancer. J Comput Biol. 2019;26:364–75. doi: 10.1089/cmb.2018.0237. [DOI] [PubMed] [Google Scholar]
- 17.Aquila L, Ohm J, Woloszynska-Read A. The role of STAG2 in bladder cancer. Pharmacol Res. 2018;131:143–49. doi: 10.1016/j.phrs.2018.02.025. [DOI] [PubMed] [Google Scholar]
- 18.Stoepker C, Ameziane N, van der Lelij P, et al. Defects in the fanconi anemia pathway and chromatid cohesion in head and neck cancer. Cancer Res. 2015;75:3543–53. doi: 10.1158/0008-5472.CAN-15-0528. [DOI] [PubMed] [Google Scholar]
- 19.Zhou H, Zheng L, Lu K, et al. Downregulation of cohesin loading factor nipped-B-like protein (NIPBL) induces cell cycle arrest, apoptosis, and autophagy of breast cancer cell lines. Med Sci Monit. 2017;23:4817–25. doi: 10.12659/MSM.906583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Repo H, Loyttyniemi E, Nykanen M, et al. The expression of cohesin subunit SA2 predicts breast cancer survival. Appl Immunohistochem Mol Morphol. 2016;24:615–21. doi: 10.1097/PAI.0000000000000240. [DOI] [PubMed] [Google Scholar]
- 21.De Koninck M, Losada A. Cohesin mutations in cancer. Cold Spring Harb Perspect Med. 2016;6 doi: 10.1101/cshperspect.a026476. pii: a026476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ajji PK, Binder MJ, Walder K, et al. Balsamin induces apoptosis in breast cancer cells via DNA fragmentation and cell cycle arrest. Mol Cell Biochem. 2017;432:189–98. doi: 10.1007/s11010-017-3009-x. [DOI] [PubMed] [Google Scholar]
- 23.Tang J, Kong D, Cui Q, et al. Prognostic genes of breast cancer identified by gene co-expression network analysis. Front Oncol. 2018;8:374. doi: 10.3389/fonc.2018.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu J, Du Z, Wang Y, et al. Weighted gene co-expression network analysis reveals modules and hub genes associated with the development of breast cancer. Medicine (Baltimore) 2019;98:e14345. doi: 10.1097/MD.0000000000014345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jayanthi V, Das AB, Saxena U. Grade-specific diagnostic and prognostic biomarkers in breast cancer. Genomics. :2019. doi: 10.1016/j.ygeno.2019.03.001. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 26.Cai Y, Mei J, Xiao Z, et al. Identification of five hub genes as monitoring biomarkers for breast cancer metastasis in silico. Hereditas. 2019;156:20. doi: 10.1186/s41065-019-0096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao Y, Liao Y, Li J, et al. Polymorphisms in AURKA and AURKB are associated with the survival of triple-negative breast cancer patients treated with taxane-based adjuvant chemotherapy. Cancer Manag Res. 2018;10:3801–8. doi: 10.2147/CMAR.S174735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korobeynikov V, Borakove M, Feng Y, et al. Combined inhibition of Aurora A and p21-activated kinase 1 as a new treatment strategy in breast cancer. Breast Cancer Res Treat. 2019;177(2):369–82. doi: 10.1007/s10549-019-05329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mansouri N, Movafagh A, Sayad A, et al. Targeting of BUB1b gene expression in sentinel lymph node biopsies of invasive breast cancer in Iranian female patients. Asian Pac J Cancer Prev. 2016;17:317–21. doi: 10.7314/apjcp.2016.17.s3.317. [DOI] [PubMed] [Google Scholar]
- 30.Han JY, Han YK, Park GY, et al. Bub1 is required for maintaining cancer stem cells in breast cancer cell lines. Sci Rep. 2015;5:15993. doi: 10.1038/srep15993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naorem LD, Muthaiyan M, Venkatesan A. Integrated network analysis and machine learning approach for the identification of key genes of triple-negative breast cancer. J Cell Biochem. 2019;120:6154–67. doi: 10.1002/jcb.27903. [DOI] [PubMed] [Google Scholar]
- 32.Thangavelu PU, Lin CY, Vaidyanathan S, et al. Overexpression of the E2F target gene CENPI promotes chromosome instability and predicts poor prognosis in estrogen receptor-positive breast cancer. Oncotarget. 2017;8:62167–82. doi: 10.18632/oncotarget.19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang K, Gao J, Luo M. Identification of key pathways and hub genes in basal-like breast cancer using bioinformatics analysis. Onco Targets Ther. 2019;12:1319–31. doi: 10.2147/OTT.S158619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu G, Xu Y, Chen W, et al. RNA interference of IQ motif containing GTPase-activating protein 3 (IQGAP3) inhibits cell proliferation and invasion in breast carcinoma cells. Oncol Res. 2016;24:455–61. doi: 10.3727/096504016X14685034103635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Z, Huang G, Sadanandam A, et al. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 2010;12:R18. doi: 10.1186/bcr2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu YZ, Wang BS, Jiang YY, et al. MCMs expression in lung cancer: Implication of prognostic significance. J Cancer. 2017;8:3641–47. doi: 10.7150/jca.20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomii C, Inokuchi M, Takagi Y, et al. TPX2 expression is associated with poor survival in gastric cancer. World J Surg Oncol. 2017;15:14. doi: 10.1186/s12957-016-1095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider MA, Christopoulos P, Muley T, et al. AURKA, DLGAP5, TPX2, KIF11 and CKAP5: Five specific mitosis-associated genes correlate with poor prognosis for non-small cell lung cancer patients. Int J Oncol. 2017;50:365–72. doi: 10.3892/ijo.2017.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez-Rodriguez JA, Lewis C, McKinley KL, et al. Distinct roles of RZZ and Bub1-KNL1 in mitotic checkpoint signaling and kinetochore expansion. Curr Biol. 2018;28:3422–29.e5. doi: 10.1016/j.cub.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hussain MS, Battaglia A, Szczepanski S, et al. Mutations in CKAP2L, the human homolog of the mouse Radmis gene, cause Filippi syndrome. Am J Hum Genet. 2014;95:622–32. doi: 10.1016/j.ajhg.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of network topology for various soft-theholding powers. The left panel showed the scale-free fit index, signed R2 (y-axis) and the soft threshold power (x-axis).
Clustering dendrogram of genes and modules identfied by weighted gene co-expression network analysis based on a dissimilarity measure (1-TOM).
Scatter diagram for module membership vs. gene significance of stage (breast cancer or non-breast cancer) in brown module.
The protein-protein network of the hub genes in brown module. Within the network, node sizes and color depth are proportional to their connectivity.
The P value of 49 hub genes for gene validation.
19 hub genes were significantly up-egulated in breast cancer that indicates the high expression of those genes is related to breast cancer.