SUMMARY
Chronic inflammatory diseases are associated with altered hematopoiesis that could result in neutrophilia and anemia. Here we report that genetic or chemical manipulation of different inflammasome components altered the differentiation of hematopoietic stem and progenitor cells (HSPC) in zebrafish. Although the inflammasome was dispensable for the emergence of HSPC, it was intrinsically required for their myeloid differentiation. In addition, Gata1 transcript and protein amounts increased in inflammasome-deficient larvae, enforcing erythropoiesis and inhibiting myelopoiesis. This mechanism is evolutionarily conserved, since pharmacological inhibition of the inflammasome altered erythroid differentiation of human erythroleukemic K562 cells. In addition, caspase-1 inhibition rapidly upregulated GATA1 protein in mouse HSPC promoting their erythroid differentiation. Importantly, pharmacological inhibition of the inflammasome rescued zebrafish disease models of neutrophilic inflammation and anemia. These results indicate that the inflammasome plays a major role in the pathogenesis of neutrophilia and anemia of chronic diseases and reveal druggable targets for therapeutic interventions.
In Brief
Chronic inflammatory diseases are associated to altered hematopoiesis that could result in neutrophilia and anemia. In this issue of Immunity, Tyrkalska et al. (2019) provide evidence of an evolutionary conserved mechanism by which inflammasome regulates the erythroid/myeloid decision in HSC, which might contribute to the hematopoietic bias of these diseases.
Graphical Abstract
INTRODUCTION
Hematopoiesis is the process of blood cell formation that occurs during embryonic development and across adulthood to produce the blood system (Jagannathan-Bogdan and Zon, 2013). In vertebrates, blood development involves two main waves of hematopoiesis: the primitive one during early embryonic development and the definitive one, which occurs in later stages (Gore et al., 2018). Definitive hematopoiesis engages multipotent hematopoietic stem cells (HSCs), which migrate eventually to the bone marrow, or kidney marrowin zebrafish, and give rise to all blood lineages (Birbrair and Frenette, 2016; Cumano and Godin, 2007). HSC maturation involves the diversification of the lymphoid (T, B, and NK cells) and myeloid and erythroid cell lineages (megakaryocytes, erythrocytes, granulocytes, and macrophages) (Kondo, 2010; Kondo et al., 2003; Weissman, 2000). The decision for erythroid and myeloid fates depends mainly on two transcription factors, GATA1 and SPI1 (also known as PU.1), that show a cross-inhibitory relationship resulting in physical interaction and direct competition between them for target genes (Nerlov et al., 2000; Rekhtman et al., 1999). However, there are many controversies about the factors responsible for terminal erythroid and myeloid differentiation and many unknown pathways probably being involved in its regulation (Cantor and Orkin, 2002; Hoppe et al., 2016). These unidentified pathways might have important clinical implications, because hematopoietic lineage bias is associated with increased incidence of diseases with prominent inflammatory components including atherosclerosis, autoimmunity, neurodegenerative disease, and carcinogenesis (Elias et al., 2017).
The inflammasomes are part of innate immune system and as intracellular receptors and sensors, they regulate the activation of inflammatory caspases, namely caspase-1 and caspase-11, which induce inflammation in response to infectious microbes and endogenous danger signals (Latz et al., 2013; Martinon et al., 2009). Typically inflammasome multiprotein complexes contain sensor proteins (nucleotide binding domain and leucine rich repeat gene family, NLRs), adaptor proteins (apoptosis-associated speck-like protein containing a CARD, ASC), and effector caspases in a zymogen form, all being able to interact among themselves by homotypic interactions (Broz and Monack, 2011; Sharma and Kanneganti, 2016). Recently, it has been shown that also guanylate binding protein (GBP) protein family forms part of these multiprotein complexes (Pilla et al., 2014; Santos et al., 2018; Tyrkalska et al., 2016; Wallet et al., 2017; Zwack et al., 2017). Oligomerization of pro-caspases and their autoproteolytic maturation lead to the processing and secretion of the pro-inflammatory cytokines interleukin-1β (IL-1β) and IL-18, and the induction of a form of programmed cell death called pyroptosis (Lamkanfi and Dixit, 2014). Lately, it has been reported that inflammasomes play crucial roles not only in infection and sterile inflammation but also in maintaining the basic cellular functions and controlling cellular homeostasis (Rathinam and Fitzgerald, 2016). Hence, additional uncovered regulatory functions for the inflammasomes have been shown in cell metabolism, proliferation, gene transcription and tumorigenesis (Rathinam and Fitzgerald, 2016; Sharma and Kanneganti, 2016). Although up to date little is known about the impact of the inflammasomes on hematopoiesis in general, it has been shown that the master erythroid transcription factor GATA1 can be cleaved in vitro by many caspases and in vivo by caspase-3 (De Maria et al., 1999).
Zebrafish has recently arisen as a powerful and useful model to study hematopoiesis (Berman et al., 2012; Ellett and Lieschke, 2010). Moreover, the genetic programs controlling hematopoiesis in the zebrafish are conserved with mammals, including humans, making them clinically relevant model systems (Jagannathan-Bogdan and Zon, 2013). Here we show the critical role played by the inflammasome in the regulation of erythroid and myeloid cell-fate decision, and terminal erythroid differentiation. Furthermore, the results also have important clinical implications, since pharmacological inhibition of the inflammasome rescued zebra-fish disease models of neutrophilic inflammation and anemia.
RESULTS
Inflammasome Inhibition Decreases the Number of Neutrophils and Macrophages in Zebrafish Larvae
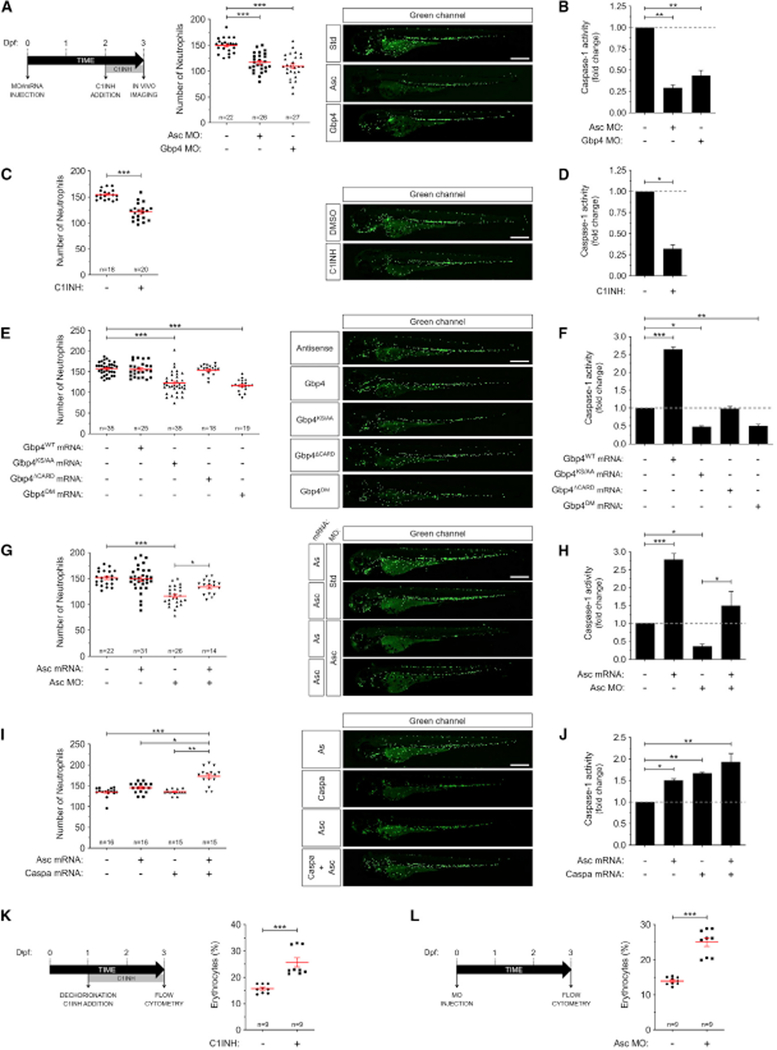
Using zebrafish transgenic lines with green fluorescent neutrophils Tg(mpx:eGFP)i114 or macrophages Tg(mpeg1:eGFP)gl22, we quantitated the total number of both cell populations in whole larvae at 72 hpf. Genetic inhibition of several inflammasome components, namely Gbp4 and Asc resulted in significant decreased numbers of both neutrophils (Figures 1A and 1B) and macrophages (Figures S1A and S1B). Similarly, pharmacological inhibition of caspase-1 with the irreversible inhibitor Ac-YVAD-CMK (Tyrkalska et al., 2016) also resulted in decreased numbers of myeloid cells (Figures 1C, 1D, S1C, S1D). These results were confirmed using an independent transgenic line Tg(lyz:dsRED)nz50 with labeled neutrophils (Figures S2A–S2D). Similarly, forced expression of the GTPase-deficient mutant of Gbp4 (KS/AA) as well as its double mutant (DM: KS/AA; ΔCARD), both of which behave as dominant negatives (DN) and inhibit inflammasome-dependent caspase-1 activation (Tyrkalska et al., 2016), resulted in decreased neutrophil number (Figures 1E and 1F). In addition, although activation of the inflammasome by forced expression of either Gbp4 or Asc failed to increase neutrophil (Figures 1E–1H) or macrophage (Figures S1E and S1F) numbers, it was able to rescue myeloid cell number and caspase-1 activity in Asc-deficient fish (Figures 1G and 1H). Notably, however, simultaneous expression of Asc and Caspa, the functional homolog of mammalian CASP1 (Kuri et al., 2017; Masumoto et al., 2003; Tyrkalska et al., 2016), significantly increased the number of neutrophils (Figures 1l and 1J) and macrophages (Figures S1E and S1F).
Figure 1. Inflammasome Inhibition Results in Decreased Neutrophil but Increased Erythrocyte Numbers in Zebrafish.
Tg(mpx:eGFP) (A-J) and Tg(lcr:eGFP) (L) zebrafish one-cell embryos were Injected with standard control (Std), Asc, or Gbp4 MOs (A, B, G, H, L), and/or with antisense (As), Gbp4WT, Gbp4KS/AA, Gbp4ΔCARD, Gbp4DM, Asc, or Caspa mRNAs (E-H). Alternatively, Tg(mpx:eGFP) (C, D, I, J) and Tg(lcr:eGFP) (K) embryos left uninjected were manually dechorionated at 24 or 48 hpf and treated by immersion with DMSO or the irreversible caspase-1 inhibitor Ac-YVAD-CMK (C1INH). Each dot represents the number of neutrophils (A, C, E, G, I) from a single larva or the percentage of erythrocytes from each pool of 50 larvae (K, L), while the mean ± SEM for each group is also shown. The sample size (n) is indicated for each treatment. Representative images of green channels of whole larvae for the different treatments are also shown. Scale bars, 500 μm. Caspase-1 activity in whole larvae was determined for each treatment at 72 hpf (one representative caspase-1 activity assay out of the three carried out is shown) (B, D, F, H, J). *p < 0.05; **p < 0.01; ***p < 0.001 according to ANOVA followed by Tukey multiple range test. See also Figures S1-S4.
The Inflammasome Regulates HSPC Differentiation but Is Dispensable for Their Emergence
The differentiation of hematopoietic stem and progenitor cells (HSPC) into various blood cell types is controlled by multiple extrinsic and intrinsic factors and the deregulation in hematopoiesis can result in a number of hematological abnormalities (Morrison et al., 1997; Yang et al., 2007). Chronic inflammatory disorders are usually associated to neutrophilia and anemia, the so-called anemia of chronic diseases (ACD). Therefore, we next examined whether the inflammasome also regulated erythropoiesis using a zebrafish transgenic line Tg(lcr:eGFP), which has specific erythroid GFP expression (Ganis et al., 2012). The results showed that inflammasome activity had the inverse effect on erythrocytes than on myeloid cells; that is, erythrocyte abundance increased following genetic and pharmacological inflammasome inhibition, as assayed by flow cytometry (Figures 1K and 1L and S3). However, the expression of cmyb and runx1, which begins by 36chpf and marks emerging definitive HSPC (Burns et al., 2005), was unaffected in guanylate binding protein-4 (Gbp4)- and Asc-deficient larvae at 48 hpf, as assayed by whole-mount in situ hybridization (WISH) (Figure S4). Similarly, the expression of rag1, which is expressed in differentiated thymic T cells, was apparently unaffected by 5 dpf in inflammasome-deficient larvae (Figure S4). Collectively, these results suggest a specific role of the inflammasome in the regulation of the balance between myelopoiesis and erythropoiesis.
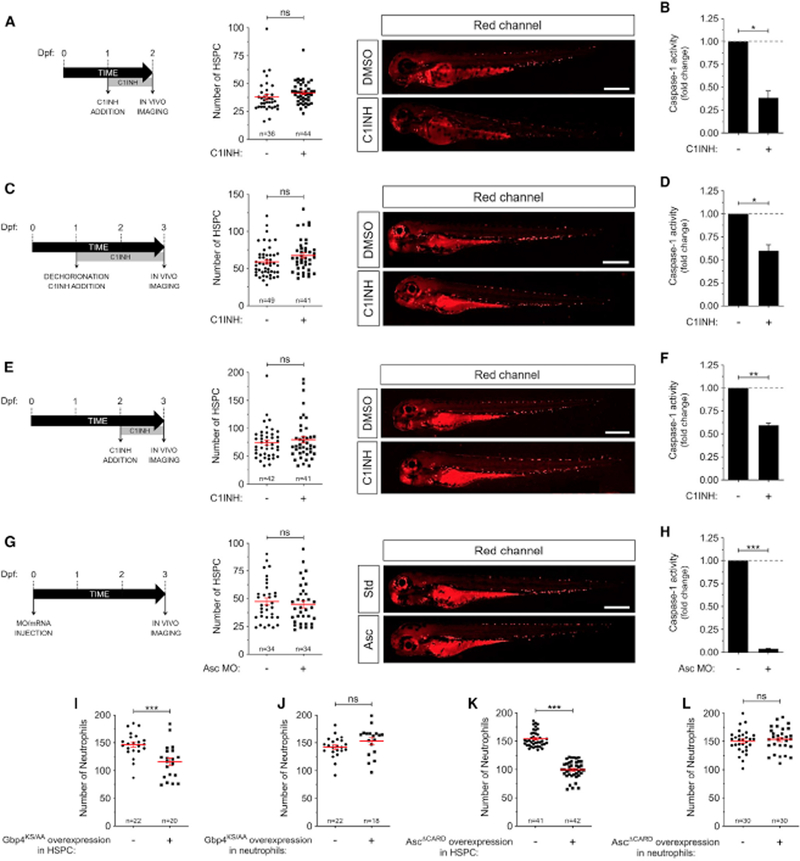
To further confirm the role of the inflammasome in HSPC differentiation, we quantitated the number of HSPC in the transgenic line Tg(runx1:GAL4; UAS:nfsB-mCherry), which has fluorescently labeled HSPC (Tamplin et al., 2015), upon genetic or pharmacological inhibition of the inflammasome at different developmental stages (24 and 48 hpf). Inhibition of caspase-1 resulted in no changes in HSPC number at any point of the treatment, the result being confirmed in Asc-deficient larvae (Figures 2A–2H). Furthermore, genetic inhibition of the inflammasome in neutrophils and HSPC by forced expression of DN forms of Asc (AscΔCARD) or Gbp4 (Gbp4KS/AA) (Tyrkalska et al., 2016) using the specific promoters mpx and runx1, respectively, showed that the number of neutrophils declined in HSPC, but not in neutrophil, inflammasome-deficient larvae (Figures 2I–2L). Collectively, these results confirm the dispensability of the inflammasome for HSPC emergence and renewal, but that is intrinsically required for HSPC differentiation.
Figure 2. The Inflammasome Is Intrinsically Required for HSPC Differentiation but Is Dispensable for Their Emergence in Zebrafish.
(A-H) Tg(runx1:GAL4; UAS:nfsb-mCherry) zebrafish embryos were manually dechorionated at 24 or 48 hpf and treated by immersion with DMSO or the irreversible caspase-1 inhibitor Ac-YVAD-CMK (C1INH) for 24 or48 h (A-F). Alternatively, Tg(runx1:GAL4; UAS:nfsb-mCherry) one-cell embryos were injected with standard control (Std) or Asc MOs (G-H). Each dot represents the number of HSPCs from a single larva, while the mean ± SEM for each group is also shown. The sample size (n) is indicated for each treatment. Representative images of red channels of whole larvae for the different treatments are also shown (A, C, E, G). Scale bars, 500 μm. Caspase-1 activity was determined for each treatment from 48 or 72 hpf larvae (one representative caspase-1 activity assay out of the three carried out is shown) (B, D, F, H).
(I-L) Tg(runx1:gal4; UAS:Gbp4KS/AA) (I), Tg(mpx:gal4; UAS:Gbp4KS/AA) (J), Tg(runx1 :gal4; UAS:AscΔCARD) (K), Tg(mpx:ga!4; UAS:AscΔCARO) (L) larvae were fixed at 72 hpf and stained with Sudan black for the detection of neutrophils. Each dot represents the number of neutrophils from a single larva, while the mean ± SEM for each group is also shown. The sample size (n) is indicated for each treatment. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001 according to Student t test. See also Figure S4.
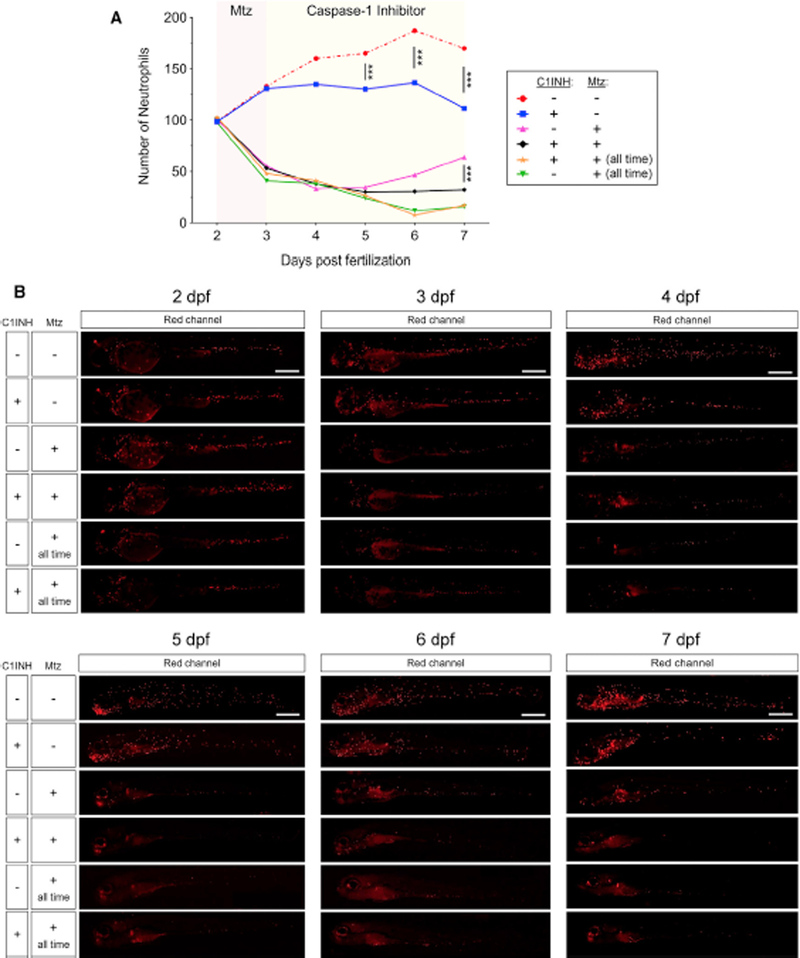
Zebrafish is an elegant model for cell ablation by use of the specific transgenic lines that expresses the bacterial nitroreductase, encoded by the nfsB gene, under the control of specific promoters (Davison et al., 2007). The nitroreductase enzyme converts the drug metronidazole (Mtz) to a cytotoxic product, which induces cell death in expressing cells to achieve tissue-specific ablation having no effect on other cell populations (Curado et al., 2007, 2008; Prajsnar et al., 2012). Using this approach, we ablated neutrophils in Tg(mpx:Gal4; UASnfsB-mCherry) zebrafish by applying Mtz for 24 h and then analyzed neutrophil recovery in the presence or absence of the cas-pase-1 inhibitor for 6 days (Figure 3). Mtz robustly reduced neutrophil numbers, which began to recover by 4 days postablation in control larvae (Figure 3). However, pharmacological inhibition of the inflammasome impaired neutrophil recovery upon ablation and strongly decreased neutrophil abundance in non-ablated larvae (Figure 3). As expected, continuous Mtz treatment resulted in drastic neutrophil decline but did not show any toxic effect on control larvae that did not express the nitroreductase (Figure 3). These results indicate that the inflammasome is indispensable for myeloid differentiation of HSPC.
Figure 3. Inflammasome Activity Is Indispensable for Myelopoiesis in Zebrafish.
Tg(mpx:GAL4; UAS:nsfb-mCherry) zebrafish larvae were manually dechorionated at 48 hpf and treated by immersion with metronidazole (Mtz) for 24 h and then with DMSO or the irreversible caspase-1 inhibitor Ac-YVAD-CMK (C1INH) for the next 4 days. Control groups were treated for 5 days with Mtz (all time).
(A) Each dot represents the number of neutrophils from a single larva, while the mean ± SEM for each group is also shown. Scale bars, 500 μm. ***p < 0.001 according to ANOVA followed by Tukey multiple range test.
(B) Representative images of red channels of whole larvae for the different treatments and time points are also shown.
Inflammasome Inhibition Impairs Demand-Driven Myelopoiesis
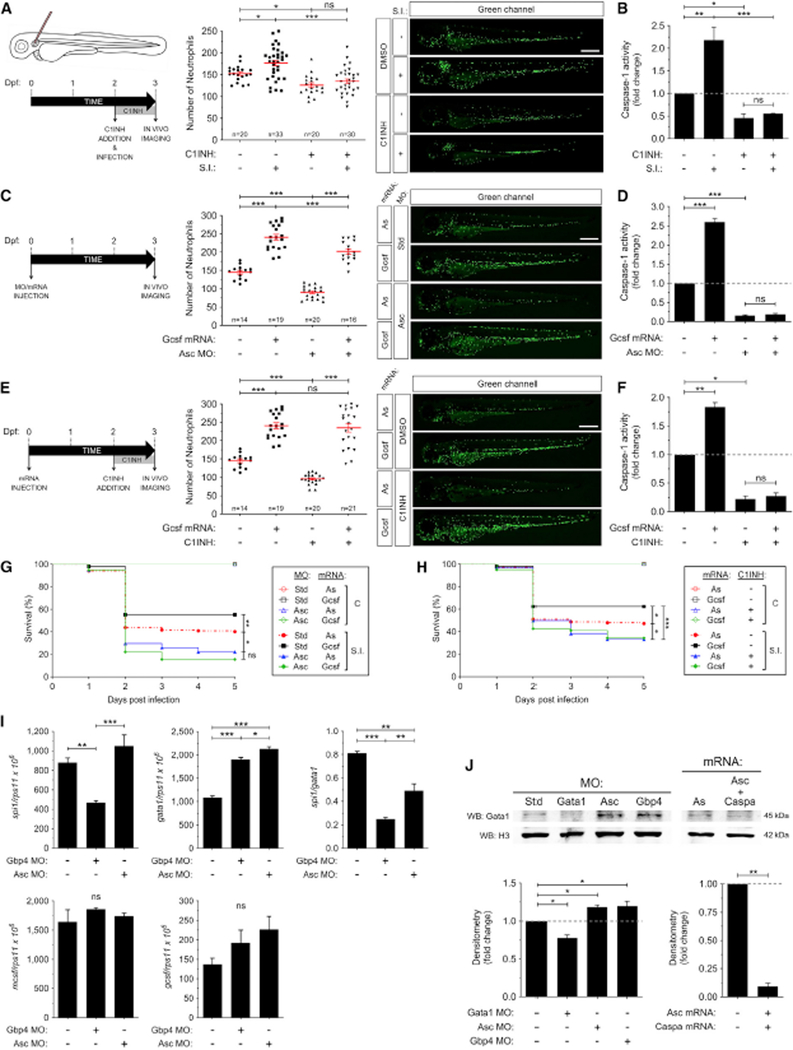
In response to infection, the hematopoietic tissue enhances production and mobilization of neutrophils, which have short lifespan and are needed in large numbers to fight infections. This process is called demand-driven or emergency hematopoiesis (Hall et al., 2012). To check whether only steady-state or also demand-driven hematopoiesis were regulated by the inflammasome, we infected 48 hpf larvae with Salmonella enterica serovar Typhimurium in the otic vesicle and counted total neutrophil numbers at 24 hpi in the presence or absence of the irreversible caspase-1 inhibitor Ac-YVAD-CMK. It was observed that pharmacological inhibition of the inflammasome was able to abrogate infection-driven myelopoiesis, which resulted in increased number of neutrophils in infected larvae (Figures 4A and 4B). Notably, forced expression of granulocyte colony-stimulating factor (Gcsf), which stimulates both steady-state and demand-driven granulopoiesis in zebrafish (Hall et al., 2012; Stachura et al., 2013), increased neutrophil number to similar amounts in wild-type and Asc-deficient larvae, as well as in larvae treated with the caspase-1 inhibitor, without affecting cas-pase-1 activity (Figures 4C–4F). Nevertheless, Gcsf was unable to rescue the higher susceptibility to S. Typhimurium infection of Asc-deficient and caspase-1 inhibitor-treated larvae (Figures 4G and 4H), confirming previous results in Gbp4-deficient larvae (Tyrkalska et al., 2016). All these results also suggest that the inflammasome regulates the myeloid and erythroid fate decision besides the function of mature myeloid cells.
Figure 4. Infection Is Unable to Bypass the Inflammasome Requirement for Neutrophil Production in Zebrafish.
(A-H) Tg(mpx:eGFP) zebrafish one-cell embryos were Injected with standard control (Std), Gbp4, or Asc MOs In combination with antisense (As), Gcsfa, Asc, Caspa mRNAs(C, D, G, H-J) or left uninjected, manually dechorionated at 48 hpf and treated by immersion with DMSO or the irreversible caspase-1 inhibitor Ac-YVAD-CMK (C1INH) (A, B, E, F). Larvae were then infected at 48 hpf with S. Typhimurium (S.I.) in the otic vesicle (A, B) or the yolk sac (G, H) and the number of neutrophils was counted in the whole body at 24 hpi (A, B) or 72 hpf (C-F) and the survival was determined during 5 days after the infection (G, H). Each dot represents the number of neutrophils from a single larva, while the mean ± SEM for each group is also shown. The sample size (n) is indicated for each treatment. Note than the 4 non-infected group showed no mortality and the 4 lines are overlapping. Representative images of green channels of whole larvae for the different treatments are shown (A-F). Scale bars, 500 μm. Caspase-1 activity was determined in whole larvae for each treatment at 72 hpf (one representative caspase-1 activity assay out of the three carried out is shown) (B, D, F).
(I-J) The mRNA amounts of spi1b, gata1a, mcsf, and gcsf in larval tails were measured by RT-qPCR at 24 hpf (I), while the protein amounts of Gata1a and histone H3 were determined using western blot in larval tails at 24 hpf (J). A densitometry analysis was performed to check the differences between treatments. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001 according to ANOVA followed byTukey multiple range test (A-F, I, J) or log rank test with Bonferroni correction (G, H).
The Inflammasome Shifts the Spi1/Gata1 Balance Favoring Myeloid Differentiation
The regulation of Spi1 and Gata1 has been shown to be critical for the differentiation of myeloid and erythroid cells, respectively, in all vertebrates. As inhibition of the inflammasome resulted in a hematopoietic lineage bias, that is, reduced myeloid and increased erythroid blood cells, we next analyzed spi1 and gata1 transcript amounts by RT-qPCR and whole-mount in situ hybridization (WISH). We observed decreased spi1/gata1 transcript ratio at 24 hpf in Gbp4- and Asc-deficient larvae, while the transcript amounts of the genes encoding Spi1-downstream pivotal macrophage and neutrophil growth factors, namely macrophage and granulocyte colony-stimulating factors (mcsf and gcsf genes), were unaffected (Figures 4I and S4). Importantly, Gata1 protein amounts were also fine-tuned by the inflammasome, because genetic inhibition of either Asc or Gbp4 was able to increase Gata1, while forced expression of Asc and Caspa, which resulted in increased number of neutrophils and macrophages (Figure 1I, 1J, S1E, S1F), robustly decreased Gata1 (Figure 4J). Therefore, the inflammasome regulates HSPC fate decision through fine-tuning Gata1 amounts.
The Regulation of HSPC Differentiation by the Inflammasome Is Evolutionarily Conserved
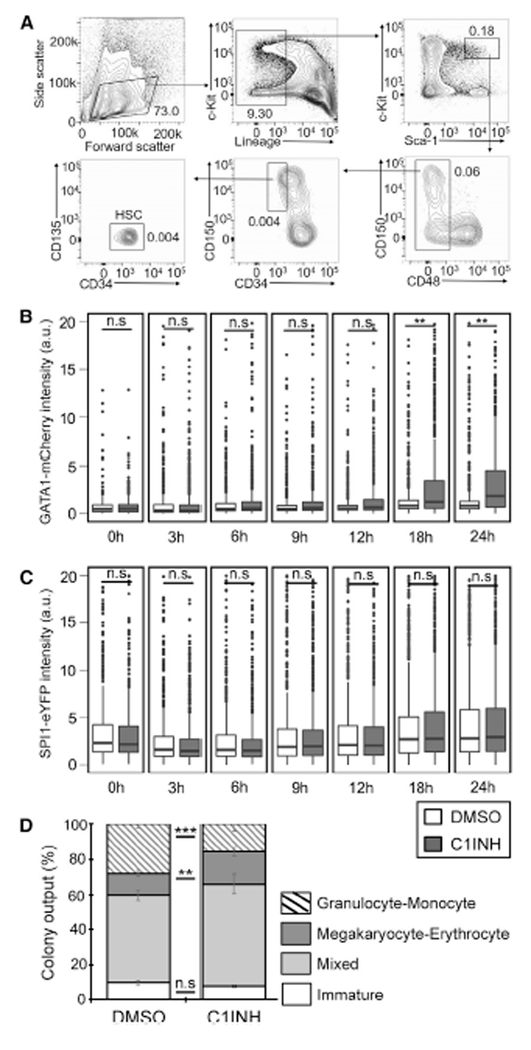
We next sought to determine whether the inflammasome also regulates mouse hematopoiesis. We quantified the impact of CASP1 inhibition on GATA1 and SPI1 protein amounts in single mouse hematopoietic stem cells (HSC) using time-lapse microscopy immediately following their isolation. Time-lapse movies were acquired for 24 h to quantify early dynamics in GATA1 amounts before the first cell division using a homozygous and extensively validated GATA1 and SPI1 reporter mouse line expressing a fusion of GATA1 and monomeric Cherry (mCherry) and SPI1 and enhanced yellow fluorescent protein (eYFP) from the endogenous Gata1 and Spi1 genomic loci, respectively (Hoppe et al., 2016). Inhibition of CASP1 upregulated GATA1-mCherry protein in differentiating HSC within 18 h, while SPI1-eYFP protein amounts were unaffected (Figures 5A–5C). In line with these results, CASP1 inhibition increased megakaryocyte-erythrocyte (MegE) colony output of mouse HSCs at expense of granulocyte-monocyte (GM) colonies (Figure 5D). These data demonstrate that at the time of normal lineage decision making in HSC, the manipulation of GATA1 protein amounts through the inflammasome can alter lineage choice, further confirming our in vivo studies in zebrafish.
Figure 5. Inflammasome Inhibition Increases GATA1 Protein Amounts and Megakaryocyte-Erythrocyte Colony Output in Mouse HSCs.
(A) Flow cytometry gating scheme used for isolation of mouse HSCs. HSCs are sorted as Lin−cKit+Sca1+CD48−CD34−CD135−CD150+. Numbers in the plots indicate % of lineage-depleted BM cells.
(B and C) Caspase1-inhibitor (C1INH) treatment increases GATA1 protein amounts (B) without affecting SPI1 protein amounts (C) in mouse HSCs. Data were acquired by time-lapse imaging of freshly-sorted HSCs (DMSO = 605, C1INH = 749 HSCs) from 12-week old SPI1-eYFP and GATA1-mCherry fluorescent protein fusion reporter mice in IMDM + BIT + SCF + Epo + Tpo + IL3 + IL6 supplemented with or without 100 mM of the irreversible caspase-1 inhibitor Ac-YVAD-CMK (2 biological replicates).
(D) Caspase1-inhibitor treatment increased MegE colony output at the expense of GM colonies. HSCs from SPI1-eYFP and GATA1-mCherry reporter mice were single-cell sorted into 384 well plates in IMDM +BIT+SCF +Epo + Tpo + IL-3 + IL-6 supplemented with or without 100 μM Ac-YVAD-CMK. At day 8, color conjugated CD41-APC and CD16/32-BV421 antibodies were added to the colonies and colonies were imaged and manually scored using morphology, SPI1-eYFP and CD16/32 signal to indicate GM colonies and GATA1-mCherry and CD41 signal to indicate MegE colonies. Data represent mean percentage of types of colonies formed from HSCs from 4 independent experiments (244 total colonies scored, error bars = SEM). ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001 according to two tailed Student’s t test (A, B) and Chi-square test (C).
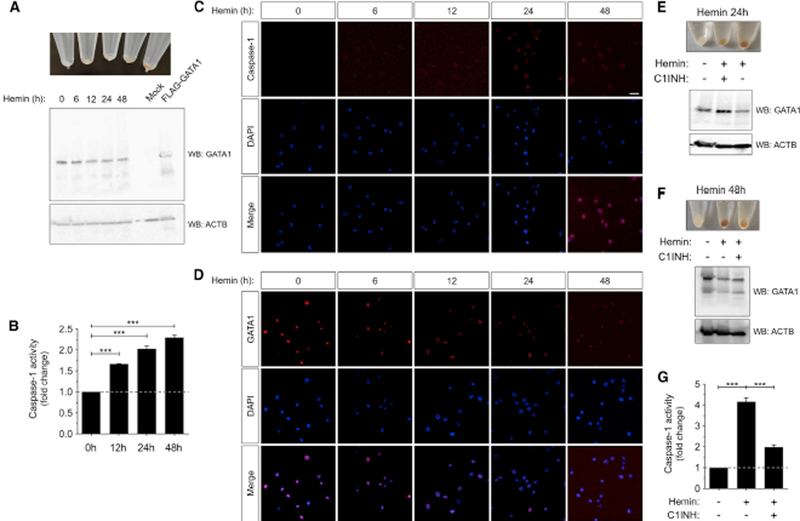
To further explore the relevance of the inflammasome in erythroid differentiation, we then used the human erythroleuke-mic K562 cell line, which can be differentiated to erythrocytes in the presence of hemin (Andersson et al., 1979; Koeffler and Golde, 1980). GATA1 amounts and activity were found to increase in the early stages of erythropoiesis, while they decreased in the late phase to allow terminal erythroid differentiation (Ferreira et al., 2005; Whyatt et al., 2000). As expected, we observed that hemin promoted gradual hemoglobin accumulation and decreased GATA1 protein amounts from 0 to 48 h (Figures 6A and 6D). Notably, the transcript amounts of NLRC4, NLRP3, and CASP1 gradually increased, while those of PYCARD peaked at 12 h and then declined to basal amounts (Figure S5). Furthermore, CASP1 activity (Figure 6B) and protein amounts (Figure 6C) progressively increased during erythroid differentiation, and CASP1 was uniformly distributed in both the cytosol and the nucleus (Figure 6C). In addition, pharmacological inhibition of CASP1 in K562 cells impaired hemin-induced erythroid differentiation, assessed as hemoglobin accumulation, and inhibited GATA1 decline at both 24 (Figure 6E) and 48 h (Figures 6F and 6G). As the Ac-YVAD-CMK inhibitor may also inhibit CASP4, we used the CASP4 and CASP5 inhibitor Ac-LEVD-CHO and found that it failed to affect the erythroid differentiation of K562 cells and their GATA1 amounts (Figure S6A).
Figure 6. Pharmacological Inhibition of Caspase-1 Impairs Erythroid Differentiation of K562 Cells.
K562 cells were Incubated with 50 μM hemin for the Indicated times In the presence or absence of the Irreversible caspase-1 Inhibitor Ac-YVAD-CMK (C1INH, 100 μM) and the cell pellets Imaged (A, E, F), lysed and resolved by SDS-PAGE and immunoblotted with anti-GATAI and anti-ACTB antibodies (A, E, F), processed for the quantification of caspase-1 activity using the fluorogenic substrate Z-YVAD-AFC (B, G) and for Immunofluorescence using anti-CASPI (C) and anti-GATAI (D) antibodies. Cell extracts from HEK293T transfected with GATA1-FLAG and empty FLAG were Included as mobility controls In (A). Nuclei were stained with DAPI. One representative caspase-1 activity (B, G), western blot (A, E, F) and hemoglobin accumulation (A, E, F) assay out of the three carried out Is shown, while one representative Immunofluorescence staining (C, D) assay out of the two carried out Is shown. Scale bars, 5 mm. ***p < 0.001 according to ANOVA followed by Tukey multiple range test. See also Figures S5–S7.
CASP1 may target several proteins to regulate HSPC differentiation. One possibility is that CASP1 directly cleaves GATA1, as it has been reported for CASP3, which negatively regulates erythropoiesis through GATA1 cleavage (De Maria et al., 1999). Therefore, we studied whether recombinant human CASP1 was able to cleave human GATA1 in vitro. The results showed that recombinant CASP1 cleaved GATA1 generating N- and C-terminal proteolytic fragments of about 30 and 15 kDa, respectively (Figures S7A–S7C). CASP1 cleavage of GATA1 at residues D276 and/or D300 may generate the obtained fragments, so we obtained single and double CASP1 mutants (D276E and D300E) and found that CASP1 was only able to cleave GATA1 at residue D300 (Figure S7D). Collectively, all these results uncover an evolutionarily conserved role of the inflammasome in the regulation of erythroid versus myeloid fate decision and terminal erythroid differentiation via cleavage of GATA1.
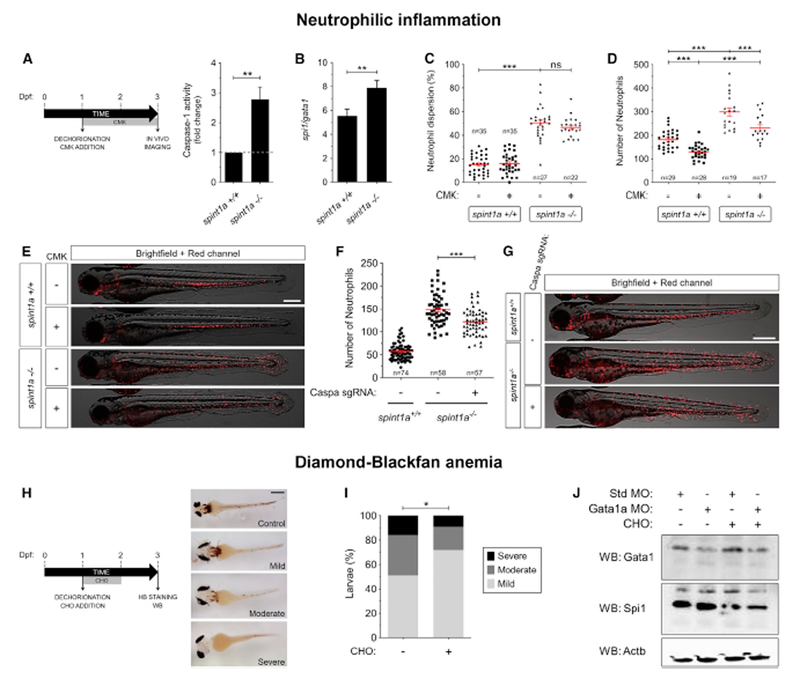
Pharmacological Inhibition of the Inflammasome Rescues Zebrafish Models of Neutrophilic Inflammation and Anemia
Hematopoietic lineage bias is associated with chronic inflammatory diseases, cancer, and aging (Elias et al., 2017; Marzano et al., 2018; Wu et al., 2014). Neutrophilic dermatoses are a group of diseases characterized by the accumulation of neutrophils in the skin (Marzano et al., 2018). We used a zebrafish Spint1a-deficient line as model of neutrophilic dermatosis, since it is characterized by strong neutrophil infiltration into the skin (Carney et al., 2007; Mathias et al., 2007). It was found that Spint1a-deficient larvae had increased caspase-1 activity (Figure 7A) and an altered spi1/gata1 ratio (Figure 7B). Notably, although pharmacological inhibition of caspase-1 failed to rescue neutrophil skin infiltration of Spint1a-deficient animals (Figures 7C and 7E), it was able to rescue their robust neutrophilia (Figures 7D and 7E). Similarly, genetic inactivation of caspa with CRISPR-Cas9 also rescued neutrophilia, but not neutrophil infiltration, of Spint1a-deficient animals (Figures 7F and 7G). However, CASP4 and CASP5 inhibition failed to rescue both neutrophilia and neutrophil infiltration in this animal model (Figure S6B).
Figure 7. Pharmacological Inhibition of Caspase-1 Rescues Zebrafish Models of Neutrophilic Inflammation and Anemia.
(A-G) Wild-type and spint1a mutant larvaewere manually dechorionated and treated from 1–3 dpf with the irreversible caspase-1 inhibitor Ac-YVAD-CMK(CMK, 100 μM) (A-E) or one-cell embryos injected with control (std) or caspa sgRNA and recombinant Cas9 (F, G). Caspase-1 activity (A), the spi1b/gata1a transcript expression ratio (B), neutrophil dispersion (C), and the number of neutrophils (D-G) were then determined. Each dot represents the number of neutrophils from a single larva, while the mean ± SEM for each group is also shown. The sample size (n) is indicated for each treatment. Representative overlay images of green and bright field channels of whole larvae for the different treatments are shown (E, G). Scale bar, 500 μm.
(H-J) Zebrafish one-cell embryos were injected with standard control (Std) or Gata1a MOs, manually dechorionated at 24 hpf and treated by immersion with DMSO or the reversible caspase-1 inhibitor Ac-YVAD-CHO (CHO) for 24–48 hpf. The inhibitor was then washed off and the larvae incubated until 72 hpf. Representative pictures of Gata1a-deficient larvae with mild, moderate and severe anemia (H), quantification of the phenotype of larval treated with DMSO or Ac- YVAD-CHO (I) and immunoblot of larval extracts with anti-Gata1a, anti-Spi1b and anti-Actb antibodies (J). One representative caspase-1 activity (A) and western blot (J) assay out of the three and two, respectively, carried out is shown. ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001 according toto Student t test (A, B), ANOVA followed by Tukey multiple range test (C, D, F) and Fisher’s exact test (I).
We next model Diamond-Blackfan anemia, a ribosomopathy caused by inefficient translation of GATA1 (Danilova and Gazda, 2015), in zebrafish larvae by reducing Gata1 amounts using a specific morpholino. We first titrated the morpholino and found that 1.7 ng/egg resulted in larvae with mild, moderate, and severe anemia (Figure 7H), while 0.85 ng/egg and 3.4 ng/egg had little or drastic effects, respectively (data not shown). We thus examined whether pharmacological inhibition of caspase-1 could rescue hemoglobin alterations of Gata1-deficient larvae. For these experiments, we treated larvae with the reversible inhibitor of caspase-1 Ac-YVAD-CHO for 24 to 48 hpf and analyzed hemoglobin at 72 hpf to allow terminal erythroid differentiation in the absence of caspase-1 inhibition. The results show that treatment of larvae for 24 h with this reversible inhibitor of caspase-1 partially rescued hemoglobin defects in Gata1-deficient larvae and Spi1/Gata1 protein ratio (Figures 7I and 7J). These results taken together demonstrate that pharmacological inhibition of caspase-1 rescues hematopoietic lineage bias in vivo.
DISCUSSION
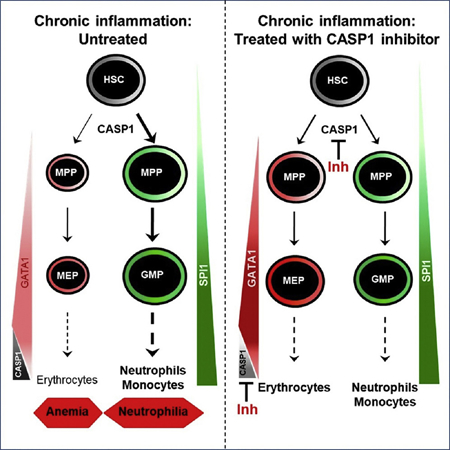
We report here an evolutionarily conserved signaling pathway that links the inflammasome with HSPC differentiation. Although previous reports have shown that proinflammatory signals are indispensable for HSPC emergence (Espin-Palazon et al., 2018), the roles of these signals, and in particular the inflammasome in HSPC formation, maintenance, and differentiation, are largely unknown. During periods of hematopoietic stress induced by chemotherapy or viral infection, activation of mouse NLRPIa prolongs cytopenia, bone-marrow hypoplasia, and immunosuppression (Masters et al., 2012). This effect is mediated by the CASPI-dependent, but ASC-independent, pyroptosis of hematopoietic progenitor cells (Masters et al., 2012). In addition, the NLRP3 inflammasome has been found to drive clonal expansion and pyroptotic cell death in myelodysplastic syndromes (Basiorka et al., 2016). Our results demonstrate that although the inflammasome is dispensable for HSPC emergence in zebrafish, it cell-intrinsically regulates HSPC differentiation in homeostasis conditions at two different levels: erythroid versus myeloid cell fate decision and terminal erythroid differentiation. Although CASP1 may target several proteins to regulate both processes, one plausible scenario is the cleavage of GATA1 at residue D300byCASP1, which would result in the quick degradation of GATA1, since we were unable to detect processed GATA1 in zebrafish larvae or K562 cells. The rapid induction of GATA1 protein amounts in mouse HSC upon pharmacological inhibition of CASP1 supports this hypothesis. Although SPI1 protein amounts were unaffected by CASP1 inhibition in HSC, SPI1 expression will be reduced at later stages of differentiation, but as a consequence of reduced GM differentiation, not as a reason for it (Strasser et al., 2018). Our model is compatible with our recent report showing that the expression of SPI1 is not relevant for the erythroid versus myeloid switch, since sometimes is already downregulated or off when GATA1 expression starts but sometimes is still expressed (Hoppe et al., 2016; Strasser et al., 2018). However, once GATA1 starts to be expressed, the HSPC always differentiate into MegE with GATA1 high and SPI1 low or off (Hoppe et al., 2016). Therefore, reduced GATA1 amounts upon inflammasome activation lead to reduced erythropoiesis and thus increased myelopoiesis. At the same time, inflammatory signaling through tumor necrosis factor α (TNFα) and IL1-β has recently been shown to directly upregulate SPI1 protein directly in HSCs in vitro and in vivo (Etzrodt et al., 2018; Pietras et al., 2016).
Similarly, terminal erythroid differentiation also requires GATA1 cleavage by CASP1. Thus, we observed that pharmacological inhibition of CASP1 leads to GATA1 accumulation and altered erythroid differentiation of K562 cells. This is not unexpected, since GATA1 inhibits terminal erythroid differentiation in vitro (Whyatt et al., 1997) and in vivo (Whyatt et al., 2000). Although it remains to be elucidated the signals responsible for the activation of the inflammasome in erythroid versus myeloid cell fate decision and terminal erythroid differentiation, as well as the inflammasome components involved, our genetic studies in zebrafish show that Gbp4 and Asc are both intrinsically required in vivo by HSPC to regulate their differentiation. A mild activation of CASP1 is anticipated to avoid the pyroptotic cell death of hematopoietic cells. This may be achieved by the assembly of small ASC specks and/or the low abundance of caspase-1 in hematopoietic progenitor cells and erythroid precursors, as occurs in neutrophils that exhibit sustained IL-1β release without pyroptosis compared to macrophages (Boucher et al., 2018; Chen et al., 2014).
Hematopoietic lineage bias is associated to increased incidence of diseases with prominent inflammatory components, including atherosclerosis, autoimmunity, neurodegenerative disease and carcinogenesis (Elias et al., 2017). In particular, neutrophilic dermatosis is characterized by the accumulation of neutrophils in the skin and cutaneous lesions (Marzano et al., 2018). We observed that the robust neutrophilia of a zebrafish model of skin inflammation is reversed by pharmacological and genetic inhibition of Caspa, despite skin lesions and neutrophil infiltration are largely unaffected. Therefore, inflammasome activation alters granulopoiesis through reducing Gata1 expression and, more importantly, its pharmacological inhibition restores Gata1 amounts and neutrophil counts. Furthermore, the critical role of the inflammasome in the regulation of Gata1 has also been pointed out by the ability of pharmacological inhibition of Caspa to restore erythroid hemoglobin and Gata1 amounts, and to decline Spi 1 amounts, in a zebrafish model of reduced Gata1, as occurs in Dia-mond-Blackfan anemia (Danilova and Gazda, 2015). Collectively, all these results point out to the ability of inflammasome inhibition as a therapeutic approach to treat human diseases with associated hematopoietic lineage bias, such as neutrophilic inflammation (Marzano et al., 2018; Ray and Kolls, 2017), ACD (Weiss, 2015), and chemotherapy-induced anemia (Testa et al., 2015). The availability of an orally active CASP1 inhibitor, VX-765, with high specificity, excellent pharmacokinetic properties, and efficacy in rheumatoid arthritis and skin inflammation mouse models (Wannamaker et al., 2007), further supports the clinical testing of CASP1 inhibitors in hematopoietic lineage bias disorders.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Victoriano Mulero (vmulero@um.es).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish (Danio rerio H.) were obtained from the Zebrafish International Resource Center and mated, staged, raised and processed as described (Westerfield, 2000). The lines roya9/a9; nacrew2/w2 (casper) (White et al., 2008), Tg(mpx:eGFP)i114 (Renshaw et al., 2006), Tg(mpeg1:eGFP)g/22, Tg(mpeg1:GAL4)gl25 (Ellett et al., 2011), Tg(lyz:dsRED)nz50 (Hall et al., 2007), Tg(mpx:Gal4.VP16)i222 (Davison et al., 2007), Tg(lcr:eGFP)cz3325(Ganis et al., 2012), Tg(runx1:GAL4)utn6 (Tamplin et al., 2015), T9(UAS:nfsB-mCherry)c264 (Davison et al., 2007) and Tg(spint1a)hi2217 (Carney et al., 2007; Mathias et al., 2007) have been previously described. The experiments performed comply with the Guidelines of the European Union Council (Directive 2010/63/EU) and the Spanish RD 53/2013. Experiments and procedures were performed as approved by the Bioethical Committees of the University of Murcia (approval numbers #75/2014, #216/2014 and 395/2017).
Mouse experiments were performed with 12–16 week old, male, SPI1-eYFP and GATA1-mCherry reporter mice (C57BL/6J background). Animal experiments were approved according to Institutional guidelines of ETH Zurich and Swiss Federal Law by veterinary office of Canton Basel-Stadt, Switzerland (approval number #2655).
METHOD DETAILS
DNA Construct and Generation of Transgenics
The uas:AscΔCARD-GFP construct was generated by MultiSite Gateway assemblies using LR Clonase II Plus (Life Technologies) according to standard protocols and using Tol2kit vectors described previously (Kwan et al., 2007). The expression constructs Gbp4, Gbp4KS→AA, Gbp4ΔCARD, Gbp4KS→AA and ΔCARD (double mutant, DM) and uas:gbp4KS→AA(Tyrkalska et al., 2016); Asc-Myc and Caspa (Masumoto et al., 2003); and Gcsfa (Liongue et al., 2009) were previously described.
The line Tg(UAS:gbp4KS→AA)ums3 was previously described (Tyrkalska et al., 2016). Tg(UAS:ascΔCARD-GFP)ums4 was generated by microinjecting 0.5–1cnl into the yolk sac of one-cell-stage embryos a solution containing 100 ng/μl uas:ascΔCARD-GFP and uas:gbp4KS→AA constructs, respectively, and 50 ng/μl Tol2 RNA in microinjection buffer (× 0.5 Tango buffer and 0.05% phenol red solution) using a microinjector (Narishige).
Morpholino, RNA and Protein Injection and Chemical Treatments of Zebrafish Larvae
Specific morpholinos (Gene Tools) were resuspended in nuclease-free water at 1 mM (Table S1). In vitro-transcribed RNA was obtained following the manufacturer’s instructions (mMESSAGE mMACHINE kit, Ambion). Morpholinos and RNA were mixed in microinjection buffer and microinjected into the yolk sac of one-cell-stage embryos using a microinjector (Narishige) (0.5–1 nL per embryo). The same amount of MOs and/or RNA was used in all experimental groups.
For genetic inactivation of caspa, injection mixes were prepared with 500 ng/μl EnGen® Cas9 NLS from Streptococcus pyogenes and 100 ng/μl tracrRNA with negative control or caspa (5′-GAACCAATTCCGAAGGATCC-′) crRNAs in 300 mM KCl buffer, incubated for 5 min at 37°C and used directly without further storage (Burger et al., 2016).
In some experiments, 1–2 dpf embryos were manually dechorionated and treated for 1 to 3 dpf at 28°C by bath immersion with the caspase-1 inhibitors Ac-YVAD-CMK (irreversible) or Ac-YVAD-CHO (reversible), and the reversible caspase-4 and caspase-5 inhibitor Ac-LEVD-CHO (100 μM, Peptanova) diluted in egg water supplemented with 1% DMSO or with Metronidazole (Mtz, 5 mM, Sigma-Aldrich).
Live Imaging, Sudan Black Staining of Neutrophils, Neutrophil Ablation and Erythrocyte Determination in Zebrafish Larvae
At 48 and 72 hpf, larvae were anesthetized in tricaine and mounted in 1% (wt/vol) low-melting-point agarose (Sigma-Aldrich) dissolved in egg water (de Oliveira et al., 2013). Images were captured with an epifluorescence Lumar V12 stereomicroscope equipped with green and red fluorescent filters while animals were kept in their agar matrixes at 28.5°C. All images were acquired with the integrated camera on the stereomicroscope and were used for subsequently counting the total number of neutrophils, macrophages or HSPC in whole larvae.
In order to decrease pigmentation and improve the signal from Sudan black staining, 24 hpf larvae were incubated in 200 mM 1-phenyl 2-thiourea (PTU) until 72 hpf, when they were anesthetized in buffered tricaine and fixed overnight at 4°C in 4% methanol-free formaldehyde. On the next day, all the larvae were rinsed with PBS thrice, incubated for 15 min with Sudan black and washed extensively in 70% EtOH in water. After that a progressive rehydration was performed: 50% EtOH in PBS and 0.1% Tween 20 (PBT) (Sigma-Aldrich), 25% EtOH in PBT and PBT alone. Finally, the larvae were visualized immediately using a Scope.A1 stereomicroscope equipped with a digital camera (AxioCam ICc 3, Zeiss) (Le Guyader et al., 2008).
For neutrophil ablation, larvae Tg(mpx:Gal4.VP16; UAS:nsfb-mCherry) were treated at 2 dpf with 5 mM Mtz and kept in dark. At 72 hpf the drug was removed and larvae were treated up to 7 dpf with 1% DMSO alone or containing Ac-YVAD-CMK (100 μM). The inhibitor was refreshed every 24 h and the larvae were imaged once a day up to 7 dpf and the number of neutrophils determined (Davison et al., 2007; Halpern et al., 2008).
Erythrocyte counts were determined by flow cytometry. At 3 dpf, pools of 50 Tg(lcr:eGFP) larvae were anesthetized in tricaine, minced with a razor blade and incubated at 28°C for 30 min with 0.077 mg/mL Liberase (Roche). Afterward, 10% FBS in PBS was added to inactivate liberase and the resulting cell suspension passed through a 40 μm cell strainer. Flow cytometric acquisitions were performed on a FACSCALIBUR (BD) and analysis was based on forward scatter and side scatter, duplicate exclusion, exclusion of dead cells by addition of SYTOX Blue to a final concentration of 1 μM, and GFP fluorescence. Before analyzing Tg(lcr:eGFP) zebrafish cell suspensions, the flow cytometry gates were set with suspensions from the same number of 3-dpf GFP-negative wild-type larvae of the same background. Analyses were performed using FlowJo software (Treestar).
Infection Assays of Zebrafish Larvae
For infection experiments, Salmonella enterica serovar Typhimurium strain 12023 (wild-type) provided by Prof. Holden was used. Overnight cultures in Luria-Bertani medium (LB) were diluted 1/5 in LB with 0.3 M NaCl, incubated at 37°C until 1.5 optical density at 600 nm was reached, and finally diluted in sterile PBS. Larvae of 2 dpf were anesthetized in embryo medium with 0.16 mg/mL tricaine and 10 bacteria were injected into the yolk sac or otic vesicle. Larvae were allowed to recover in egg water at 28–29°C, and monitored for clinical signs of disease or mortality over 5 days and neutrophil recruitment up to 24 hpi (Tyrkalska et al., 2016).
Whole-Mount RNA In Situ Hybridization (WISH) in Zebrafish Larvae
Transparent Casper embryos were used for WISH (Thisse et al., 1993). gatala, spilb, gcsfr, cmyb, runxl and ragl sense and antisense RNA probes were generated using the DIG RNA Labeling Kit (Roche Applied Science) from linearized plasmids. Embryos were imaged using a Scope.AI stereomicroscope equipped with a digital camera (AxioCam ICc 3, Zeiss).
K562 Cell Culture and Erythroid Differentiation Assays
K562 cells were maintained in RPMI supplemented with 10% FCS, 2 mM Glutamin, and 1% penicillin-streptomycin. Cells were maintained and split before confluence every 72h. For the differentiation, cells were treated with 50 mM hemin, prepared as previously described (Smith et al., 2000), in the presence of 0.1% DMSO alone or containing 100 μM Ac-YVAD-CMK or Ac-LEVD-CHO. Cells were collected at different time points (0, 6, 12, 24, 48 hours post-hemin addition), centrifuged, washed with PBS and stored at −80° C.
Caspase-1 Activity Assay
The caspase-1 activity was determined with the fluorometric substrate Z-YVAD-AFC as described previously (Angosto et al., 2012; Lopez-Castejon et al., 2008; Tyrkalskaet al., 2016). In brief, 30 pooled zebrafish larvae and 8×105 K562 cells were lysed in hypotonic cell lysis buffer [25 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), 5 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA), 5 mM dithiothreitol (DTT), 1:20 protease inhibitor cocktail, pH 7.5] on ice for 10 min. For each reaction, 80 μg protein were incubated for 90 min at 23°C with 50 μM Z-YVAD-AFC and 50 ml of reaction buffer [0.2% 3-[(3-cholami-dopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 0.2 M HEPES, 20% sucrose, 29 mM DTT, pH 7.5]. After the incubation, the fluorescence of the AFC released from the Z-YVAD-AFC substrate was measured with a FLUOstart spectofluorometer (BGM, LabTechnologies) at an excitation wavelength of 405 nm and an emission wavelength of 492 nm. One representative caspase-1 activity assay out of the three carried out is shown accompanying each cell count.
Laser Confocal Microscopy
Cells were seeded in Poly-L-Lys Cellware 12mm cover (Corning), 50,000 cells in 100 μl were allowed to attach to the cover during 10 min at room temperature, then medium and treatment were added. After hemin treatment cells were washed with PBS, fixed with 4% paraformaldehyde in PBS 10 min, incubated 20 min at room temperature with 20 mM glycine, permeabilized with 0.5% NP40 and blocked for 1 h with 2% BSA. Cells were then labeled with corresponding primary antibody, followed by Alexa 568-conjugated secondary antibody (Thermo Fisher Scientific). Samples were mounted using a mounting medium from Dako and examined with a Leica laser scanning confocal microscope AOBS and software (Leica Microsystems). The images were acquired in a 1,024 × 1,024 pixel format in sequential scan mode between frames to avoid cross-talk. The objective used was HCX PL APO CS × 63 and the pinhole value was 1, corresponding to 114.73 μm.
Immunoblotting
Lysis buffer for mammalian cell lysis contained 50 mMTris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% (w/v) NP-40and fresh protease inhibitor (1/20, P8340, Sigma-Aldrich), while for zebrafish larvae lysis contained 1% SDS. Protein quantification was done with BCA kit using BSA as a standard. Cell lysates (40 μg) in SDS sample buffer were subjected to electrophoresis on a polyacrylamide gel and transferred to PVDF membranes. The membranes were incubated for 1 h with TTBS containing 5% (w/v) skimmed dried milk powder or 2% (w/v) BSA. The membranes were immunoblotted in the same buffer 16 h at 4°C with the indicated primary antibodies. The blots were then washed with TTBS and incubated for 1 h at room temperature with secondary HRP-conjugated antibodies diluted 2,500-fold in 5% (w/v) skimmed milk in TTBS. After repeated washes, the signal was detected with the enhanced chemiluminescence reagent and ChemiDoc XRS Biorad. The primary antibodies used are: rabbit polyclonal to human GATA1 (1/200, #sc1234, Santa Cruz Biotechnology) for confocal assay, rabbit mAbto human GATA1 (1/200, #3535, Cell Signaling) for immunoblotting, rabbit polyclonal to CASP1 (1/200, #sc56036 Santa Cruz Biotechnology) for confocal assay, rabbit polyclonal to zebrafish Gata1a and Spi1b (1/2000, #GTX128333 and GTX128266, GeneTex), rabbit polyclonal to histone H3 (1/200, #ab1791, Abcam) and Monoclonal ANTI-FLAG® M2-Peroxidase (HRP) antibody produced in mouse. Densitometry analysis has been performed using Fiji ImageJ software (Schindelin et al., 2012).
Immunoprecipitation and Recombinant Caspase-1 Assay
Pull down assays were also performed as described previously (Tyrkalska et al., 2017), with small modifications. Cells were washed twice with PBS, solubilized in lysis buffer (50 mM Tris-HCl, pH 7.7, 150 mM NaCl, 1% NP-40 and 1:20 protease inhibitor cocktail) during 30 min in agitation and centrifuged (13,000 × g, 10 min). Cell lysate (1 mg) was incubated for 2 h at 4°C under gentle agitation with 40 ml of slurry of ANTIFLAG® M2. The immunoprecipitates were washed four times with lysis buffer containing 0.15 M NaCl, washed twice with PBS and incubated with 10 IU recombinant caspase-1 in reaction buffer (50 mM HEPES, pH 7.2, 50mM NaCl, 0.1% Chaps, 10 mM EDTA, 5% Glycerol, and 10 mM DTT) during 2 h at 37°C. The resin was boiled in SDS sample buffer 5 min at 95°C and the bound proteins were resolved on 4%−15% SDS-PAGE and transferred to PVDF membranes for 1h at 300 mA. Blots were probed with antibodies to FLAG and GATA1 (see above).
Analysis of Gene Expression
Total RNA was extracted from 106 K562 cells, whole embryos or larvae (60) or larval tails (100) with TRIzol reagent (Thermo Fisher Scientific) following the manufacturer’s instructions and treated with DNase I, amplification grade (1 U/mg RNA; Invitrogen). SuperScript III RNase H− Reverse Transcriptase (Invitrogen) was used to synthesize first-strand cDNA with oligo(dT)18 primer from 1 μg of total RNA at 50°C for 50 min. Real-time PCR was performed with an ABI PRISM 7500 instrument (Applied Biosystems) using Power SYBR Green Master Mix. Reaction mixtures were incubated for 10 min at 95°C, followed by 40 cycles of 15 s at 95°C, 1 min at 60°C, and finally 15 s at 95°C, 1 min 60°C and 15 s at 95°C. For each mRNA, gene expression was normalized to the ribosomal protein S11 (rps11) for zebrafish or β-actin (ACTB) for human cells content in each sample following the Pfaffl method (Pfaffl, 2001). The primers used are shown in (Table S2). In all cases, each PCR was performed with triplicate samples and repeated with at least two independent samples.
Isolation of Mouse HSCs
Male SPI1-eYFP and GATA1-mCherry mice were euthanized and isolation of HSCs was performed according to previously described protocols (Cabezas-Wallscheid et al., 2014; Hoppe et al., 2016; Kiel et al., 2005). Briefly, femurs, tibiae and vertebrae of adult mice were isolated and crushed in FACS buffer (2% FCS (PAA) + 1mM EDTA in PBS). Bone marrow suspension was subjected to ACK lysing buffer for 2 min followed by lineage depletion steps including incubation with biotinylated antibodies cocktail of CD3e, CD19, B220, CD11b, Gr-1 and Ter-119 for 7 min, streptavidin-conjugated beads (Roche) for 7 min and immune-magnetic (Stem Cell Technologies) separation for 7 min. Lineage depleted cells were stained with color-conjugated primary antibodies for 90 min. FACS sorting of HSCs was performed on FACS ARIA III (BD Biosciences) using the Lineage− Sca1+ cKit+ CD34− CD48− CD135− CD150+. All steps were performed at 4°C.
Mouse Single-Cell Liquid Culture Colony Assay
Single-cell sort of HSCs was performed in plastic-bottom 384 well plates (Greiner Bio-one) using FACS ARIA III under standard permissive culture media (IMDM (GIBCO) + 5% BIT (Stem Cell Technologies) + P/S + SCF + Epo + Tpo + IL3 + IL6) with or without 100 μM Ac-YVAD-CMK. Plates were incubated at 37°C and 5% CO2. At day 8, color-conjugated antibodies against lineage markers (CD41-APC + CD16/32-BV421) were added (1:5000 dilution) in wells, incubated for 3 hours at 37°C and 5% CO2 and imaging of hematopoietic colonies was performed on Nikon Eclipse Ti-E microscope. Colonies were scored manually. Granulocyte-monocyte colonies were indicated by morphology, CD16/32 and SPI-eYFP expression while megakaryocyte-erythrocyte colonies were indicated by morphology, CD41 and GATA1-mCherry expression as previously described (Hoppe et al., 2016).
Time-Lapse Imaging of Mouse HSCs
HSCs were sorted using FACS ARIA III and seeded in plastic-bottom 384 well plates (Greiner Bio-one) in multi-lineage supporting culture media as described (Hoppe et al., 2016) (IMDM + 5% BIT + P/S + SCF + Epo + Tpo + IL3 + IL6) with or without 100mM Ac-YVAD-CMK. Time-lapse imaging and quantification of SPI1-eYFP and GATA1-mCherry in HSCs was performed using previously established protocols (Etzrodt et al., 2018; Hilsenbeck et al., 2017; Hilsenbeck et al., 2016; Hoppe et al., 2016).
Quantification and Statistical Analysis
Data are shown as mean ± SEM and were analyzed by analysis of variance (ANOVA) and a Tukey or Bonferroni multiple range test to determine differences among groups. The differences between two samples were analyzed by the Student t test. Fisher’s exact and Chi-square tests were used for the analysis of contingency tables. A log rank test with the Bonferroni correction for multiple comparisons was used to calculate the statistical differences in the survival of the different experimental groups.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal to human GATA1 | Santa Cruz Biotechnology | Cat#sc1234, RRID:AB_2263157 |
| Rabbit mAb to human GATA1 | Cell Signaling | Cat#3535, RRID:AB_2108288 |
| Rabbit polyclonal to CASP1 | Santa Cruz Biotechnology | Cat#sc56036, RRID:AB_781816 |
| Rabbit polyclonal to zebrafish Gatal a | GeneTex | Cat#GTX128333 |
| Rabbit polyclonal to zebrafish Spi1 b | GeneTex | Cat#GTX128266 |
| Rabbit polyclonal to histone H3 | Abcam | Cat#ab1791, RRID:AB_302613 |
| Monoclonal ANTI-FLAG® M2-Peroxidase (HRP) antibody produced in mouse | Sigma-Aldrich | Cat#A8592, RRID:AB_439702 |
| Streptavidin-BV711 | BD Biosciences | Cat#563262 |
| anti-Sca1 conjugated with BV510 | Biolegend | Cat#108129, RRID:AB_2561593 |
| anti-cKIT conjudated with BV421 | Biolegend | Cat#105828, RRID:AB_11204256 |
| anti-CD135 conjugated with PerCPeFL710 | Thermo Fisher Scientific | Cat#46-1351-82, RRID:AB_10733393 |
| anti-CD34 conjugated wth eFL660 | Thermo Fisher Scientific | Cat#50-0341-82, RRID:AB_10596826 |
| anti-CD48 conjugated wth APCeFL780 | Thermo Fisher Scientific | Cat#47-0481-82, RRID:AB_2573962 |
| anti-CD150 conjugated with BV650 | Biolegend | Cat#115932, RRID:AB_2715765 |
| anti-B220-biotin | Thermo Fisher Scientific | Cat#13-0452-86, RRID:AB_466451 |
| anti-CD19-biotin | Thermo Fisher Scientific | Cat#13-0191-86, RRID:AB_466386 |
| antiCd3e-biotin | Thermo Fisher Scientific | Cat#13-0031-85, RRID:AB_466320 |
| anti-CD11b-biotin | Thermo Fisher Scientific | Cat#13-0112-85, RRID:AB_466360 |
| anti-Gr1-biotin | Thermo Fisher Scientific | Cat#13-5931-85, RRID:AB_466801 |
| anti-Ter119-biotin | Thermo Fisher Scientific | Cat#13-5921-85, RRID:AB_466798 |
| anti-CD41-APC | Thermo Fisher Scientific | Cat#17-0411-82, RRID:AB_1603237 |
| anti-CD16/32-BV421 | Biolegend | Cat#101332, RRID:AB_2650889 |
| Bacterial and Virus Strains | ||
| Salmonella enterica serovar Typhimurium, strain 12023 (wild type) | Prof. David Holden | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| EnGen® Cas9 NLS from Streptococcus pyogenes | New England Biolabs | Cat#M0646 |
| Alt-R® CRISPR-Cas9 tracrRNA | Integrated DNA Technologies | Cat#1072532 |
| Alt-R® CRISPR-Cas9 Negative Control crRNA | Integrated DNA Technologies | Cat#1072544 |
| Ac-YVAD-CMK | Peptanova | Cat#3180-v |
| Ac-YVAD-CHO | Peptanova | Cat#3165-v |
| Ac-LEVD-CHO | Peptanova | Cat#864-42-v |
| Metronidazole | Sigma Aldrich | Cat#M1547 |
| Sudan black | Sigma-Aldrich | Cat#380B-1KT |
| Z-YVAD-AFC | Merck | Cat#688225 |
| ANTIFLAG® M2 | Sigma-Aldrich | Cat#A2220 |
| Recombinant caspase-1 | GeneTex | Cat#GTX65025 |
| 4–15% SDS-PAGE | BioRad | Cat#456-1084 |
| DNase I, amplification grade | ThermoFisher Scientific | Cat#18068015 |
| SuperScript III RNase H− Reverse Transcriptase | ThermoFisher Scientific | Cat#18080085 |
| Power SYBR Green Master Mix | ThermoFisher Scientific | Cat#4368708 |
| Liberase TM research Grade | Sigma-Aldrich | Cat#05401119001 |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat#P8340 |
| Hemin | Sigma-Aldrich | Cat#16009-13-5 |
| ACK lysing buffer | Lonza | Cat#10-548E |
| RPMI-1640 | Sigma-Aldrich | Cat#R0883 |
| FCS | Cultek | Cat#91S181 D-050 |
| Penicillin/Streptomycin | Sigma-Aldrich | Cat#P4333 |
| Glutamine | Sigma-Aldrich | Cat#G7513 |
| IMDM | ThermoFisher Scientific | Cat# 21056023 |
| BIT | Stem Cell Technologies | Cat# 09500 |
| IL3 | Peprotech | Cat#213–13 |
| IL6 | Peprotech | Cat#216–16 |
| SCF | Peprotech | Cat#250–03 |
| EPO | Peprotech | Cat#100–65 |
| TPO | Peprotech | Cat#315–14 |
| Experimental Models: Cell Lines | ||
| K562 | ATCC | Cat#CCL243 |
| Experimental Models: Organisms/Strains | ||
| Zebrafish casper line (roya9/a9; nacrew2/w2) | Prof. LI Zon | |
| Tg(mpx:eGFP)i114 | Prof. SA Renshaw | |
| T9(mpe91:eGFP)gl22 | Prof. G Lieschke | |
| T9(mpeg1:GAL4)9125 | Prof. G Lieschke | |
| Tg(lyz:dsRED)nz50 | Prof. P Crosier | |
| T9(mpx:Gal4.VP16)i222 | Prof. SA Renshaw | |
| T9(lcr:eGFP)cz3325 | Prof. LI Zon | |
| T9(runx1:GAL4)utn6 | Prof. LI Zon | |
| T9(UAS:nfsB-mCherry)c264 | Prof. M. Halpern | |
| T9(spint1a)hi2217 | Prof. M. Hammerschmidt | |
| SPI1-eYFP/GATA1-mCherry1 reporter mice (C57BL/6J background) | Prof. Timm Schroeder | |
| Recombinant DNA | ||
| Tol2kit | Dr. K. Kwan | http://tol2kit.genetics.utah.edu/index.php/Main_Page |
Highlights.
The inflammasome regulates the erythroid-myeloid decision in HSC
Caspase-1 inhibition rapidly upregulates GATA1 protein in HSC
Caspase-1 regulates terminal erythroid differentiation
Caspase-1 inhibition rescues neutrophilic inflammation and anemia
ACKNOWLEDGMENTS
We strongly acknowledge I. Fuentes and P. Martinez for their excellent technical assistance with the zebrafish experiments. We also thank Profs.
S.A. Renshaw, P. Crosier, G. Lieschke, M. Hammerschmidt, and M. Halpern for the zebrafish lines, N. Inohara for the zebrafish Asc-Myc and Caspa constructs, C. Hall for the zebrafish Gcsfa construct, D. Holden for the ST strain, and P. Pelegrin and A. Baroja-Mazo for critical reading of the manuscript. This work was supported by the Spanish Ministry of Science, Innovation and Universities (grants BIO2014-52655-R and BIO2017-84702-R to V.M. and PI13/0234 to M.L.C., PhD fellowship to F.J.M.-M. and Juan de la Cierva postdoctoral contract to F.A.-P.), all co-funded with Fondos Europeos de Desarrollo Regional/European Regional Development Funds), Fundacion Seneca-Murcia (grant 19400/PI/14 to M.L.C.), the University of Murcia (postdoctoral contracts to A.B.-P.O. and D.G.-M., and PhD fellowship to F.J.M.-M.), SNF grant 179490 to T.S., and the European 7th Framework Initial Training Network Fish For Pharma (PhD fellowship to S.D.T., PITG-GA-2011-289209). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
DECLARATION OF INTEREST
L.I.Z. is a founder and stockholder of Fate Therapeutics, Inc., Scholar Rock, and Camp4 Therapeutics. A patent forthe use of caspase-1 inhibitorsto treat anemia has been registered by Universidad de Murcia, Boston Children’s Hospital and Instituto Murciano de Investigation Biosanitaria (#P201831288).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.immuni.2019.05.005.
REFERENCES
- Andersson LC, Nilsson K, and Gahmberg CG (1979). K562-a human erythroleukemic cell line. Int. J. Cancer 23, 143–147. [DOI] [PubMed] [Google Scholar]
- Angosto D, López-Castejón G, López-Muñoz A, Sepulcre MP, Arizcun M, Meseguer J, and Mulero V. (2012). Evolution of inflammasome functions in vertebrates: Inflammasome and caspase-1 trigger fish macrophage cell death but are dispensable for the processing of IL-1 p. Innate Immun. 18, 815–824. [DOI] [PubMed] [Google Scholar]
- Basiorka AA, McGraw KL, Eksioglu EA, Chen X, Johnson J, Zhang L, Zhang Q, Irvine BA, Cluzeau T, Sallman DA, et al. (2016). The NLRP3 inflammasome functions as a driver of the myelodysplastic syndrome phenotype. Blood 128, 2960–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J, Payne E, and Hall C. (2012). The zebrafish as a tool to study hematopoiesis, human blood diseases, and immune function. Adv. Hematol. 2012,425345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birbrair A, and Frenette PS (2016). Niche heterogeneity in the bone marrow. Ann. N Y Acad. Sci. 1370, 82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher D, Monteleone M, Coll RC, Chen KW, Ross CM, Teo JL, Gomez GA, Holley CL, Bierschenk D, Stacey KJ, et al. (2018). Caspase-1 self-cleavage is an intrinsic mechanism to terminate inflammasome activity. J. Exp. Med. 215, 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, and Monack DM (2011). Molecular mechanisms of inflammasome activation during microbial infections. Immunol. Rev. 243, 174–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger A, Lindsay H, Felker A, Hess C, Anders C, Chiavacci E, Zaugg J, Weber LM, Catena R, Jinek M, et al. (2016). Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development 143, 2025–2037. [DOI] [PubMed] [Google Scholar]
- Burns CE, Traver D, Mayhall E, Shepard JL, and Zon LI (2005). Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 19, 2331–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas-Wallscheid N, Klimmeck D, Hansson J, Lipka DB, Reyes A, Wang Q, Weichenhan D, Lier A, von Paleske L, Renders S, et al. (2014). Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 15, 507–522. [DOI] [PubMed] [Google Scholar]
- Cantor AB, and Orkin SH (2002). Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21, 3368–3376. [DOI] [PubMed] [Google Scholar]
- Carney TJ, von der Hardt S, Sonntag C, Amsterdam A, Topczewski J, Hopkins N, and Hammerschmidt M. (2007). Inactivation of serine protease Matriptase 1a by its inhibitor Hai1 is required for epithelial integrity of the zebrafish epidermis. Development 134, 3461–3471. [DOI] [PubMed] [Google Scholar]
- Chen KW, Groß CJ, Sotomayor FV, Stacey KJ, Tschopp J, Sweet MJ, and Schroder K. (2014). The neutrophil NLRC4 inflammasome selectively promotes IL-1β maturation without pyroptosis during acute Salmonella challenge. Cell Rep. 8, 570–582. [DOI] [PubMed] [Google Scholar]
- Cumano A, and Godin I. (2007). Ontogeny of the hematopoietic system. Annu. Rev. Immunol. 25, 745–785. [DOI] [PubMed] [Google Scholar]
- Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, and Stainier DY (2007). Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev. Dyn. 236, 1025–1035. [DOI] [PubMed] [Google Scholar]
- Curado S, Stainier DY, and Anderson RM (2008). Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat. Protoc. 3, 948–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N, and Gazda HT (2015). Ribosomopathies: how a common root can cause a tree of pathologies. Dis. Model. Mech. 8, 1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, and Parsons MJ (2007). Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev. Biol. 304, 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maria R, Zeuner A, Eramo A, Domenichelli C, Bonci D, Grignani F, Srinivasula SM, Alnemri ES, Testa U, and Peschle C. (1999). Negative regulation of erythropoiesis by caspase-mediated cleavage of GATA-1. Nature 401, 489–493. [DOI] [PubMed] [Google Scholar]
- de Oliveira S, Reyes-Aldasoro CC, Candel S, Renshaw SA, Mulero V, and Calado A. (2013). Cxcl8 (IL-8) mediates neutrophil recruitment and behaviorin the zebrafish inflammatory response. J. Immunol. 190,4349–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias HK, Bryder D, and Park CY (2017). Molecular mechanisms underlying lineage bias in aging hematopoiesis. Semin. Hematol. 54, 4–11. [DOI] [PubMed] [Google Scholar]
- Ellett F, and Lieschke GJ (2010). Zebrafish as a model for vertebrate hematopoiesis. Curr. Opin. Pharmacol. 10, 563–570. [DOI] [PubMed] [Google Scholar]
- Ellett F, Pase L, Hayman JW, Andrianopoulos A, and Lieschke GJ (2011). mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49–e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espin-Palazon R, Weijts B, Mulero V, and Traver D. (2018). Proinflammatory Signals as Fuel for the Fire of Hematopoietic Stem Cell Emergence. Trends Cell Biol. 28, 58–66. [DOI] [PubMed] [Google Scholar]
- Etzrodt M, Ahmed N, Hoppe PS, Loeffler D, Skylaki S, Hilsenbeck O, Kokkaliaris KD, Kaltenbach HM, Stelling J, Nerlov C, and Schroeder T. (2018). Inflammatory signals directly instruct PU.1 in HSCs via TNF. Blood 133, 816–819. [DOI] [PubMed] [Google Scholar]
- Ferreira R, Ohneda K, Yamamoto M, and Philipsen S. (2005). GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol. Cell. Biol. 25, 1215–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganis JJ, Hsia N, Trompouki E, de Jong JL, DiBiase A, Lambert JS, Jia Z, Sabo PJ, Weaver M, Sandstrom R, et al. (2012). Zebrafish globin switching occurs in two developmental stages and is controlled by the LCR. Dev. Biol. 366, 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore AV, Pillay LM, Venero Galanternik M, and Weinstein BM (2018). The zebrafish: A fintastic model for hematopoietic development and disease. Wiley Interdiscip. Rev. Dev. Biol. 7, e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C, Flores MV, Storm T, Crosier K, and Crosier P. (2007). The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 7, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CJ, Flores MV, Oehlers SH, Sanderson LE, Lam EY, Crosier KE, and Crosier PS (2012). Infection-responsive expansion of the hematopoietic stem and progenitor cell compartment in zebrafish is dependent upon inducible nitric oxide. Cell Stem Cell 70, 198–209. [DOI] [PubMed] [Google Scholar]
- Halpern ME, Rhee J, Goll MG, Akitake CM, Parsons M, and Leach SD (2008). Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish 5, 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilsenbeck O, Schwarzfischer M, Skylaki S, Schauberger B, Hoppe PS, Loeffler D, Kokkaliaris KD, Hastreiter S, Skylaki E, Filipczyk A, et al. (2016). Software tools for single-cell tracking and quantification of cellular and molecular properties. Nat. Biotechnol. 34, 703–706. [DOI] [PubMed] [Google Scholar]
- Hilsenbeck O, Schwarzfischer M, Loeffler D, Dimopoulos S, Hastreiter S, Marr C, Theis FJ, and Schroeder T. (2017). fastER: a user-friendly tool for ultrafast and robust cell segmentation in large-scale microscopy. Bioinformatics 33, 2020–2028. [DOI] [PubMed] [Google Scholar]
- Hoppe PS, Schwarzfischer M, Loeffler D, Kokkaliaris KD, Hilsenbeck O, Moritz N, Endele M, Filipczyk A, Gambardella A, Ahmed N, et al. (2016). Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature 535, 299–302. [DOI] [PubMed] [Google Scholar]
- Jagannathan-Bogdan M, and Zon LI (2013). Hematopoiesis. Development 140, 2463–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, and Morrison SJ (2005). SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121. [DOI] [PubMed] [Google Scholar]
- Koeffler HP, and Golde DW (1980). Human myeloid leukemia cell lines: a review. Blood 56, 344–350. [PubMed] [Google Scholar]
- Kondo M (2010). Lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. Immunol. Rev. 238, 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, Shizuru JA, and Weissman IL (2003). Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu. Rev. Immunol. 21, 759–806. [DOI] [PubMed] [Google Scholar]
- Kuri P, Schieber NL, Thumberger T, Wittbrodt J, Schwab Y, and Leptin M. (2017). Dynamics of in vivo ASC speck formation. J. Cell Biol. 216, 2891–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, and Chien CB (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, and Dixit VM (2014). Mechanisms and functions of inflammasomes. Cell 157, 1013–1022. [DOI] [PubMed] [Google Scholar]
- Latz E, Xiao TS, and Stutz A. (2013). Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 13, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guyader D, Redd MJ, Colucci-Guyon E, Murayama E, Kissa K, Briolat V, Mordelet E, Zapata A, Shinomiya H, and Herbomel P. (2008). Origins and unconventional behavior of neutrophils in developing zebrafish. Blood 111, 132–141. [DOI] [PubMed] [Google Scholar]
- Liongue C, Hall CJ, O’Connell BA, Crosier P, and Ward AC (2009). Zebrafish granulocyte colony-stimulating factor receptor signaling promotes myelopoiesis and myeloid cell migration. Blood 113, 2535–2546. [DOI] [PubMed] [Google Scholar]
- López-Castejón G, Sepulcre MP, Mulero I, Pelegrín P, Meseguer J, and Mulero V. (2008). Molecular and functional characterization of gilthead seabream Sparus aurata caspase-1: the first identification of an inflammatory caspase in fish. Mol. Immunol. 45, 49–57. [DOI] [PubMed] [Google Scholar]
- Martinon F, Mayor A, and Tschopp J. (2009). The inflammasomes: guardians of the body. Annu. Rev. Immunol. 27, 229–265. [DOI] [PubMed] [Google Scholar]
- Marzano AV, Borghi A, Wallach D, and Cugno M (2018). A Comprehensive Review of Neutrophilic Diseases. Clin. Rev. Allergy Immunol. 54, 114–130. [DOI] [PubMed] [Google Scholar]
- Masters SL, Gerlic M, Metcalf D, Preston S, Pellegrini M, O’Donnell JA, McArthur K, Baldwin TM, Chevrier S, Nowell CJ, et al. (2012).NLRP1 inflammasome activation induces pyroptosis of hematopoietic progenitor cells. Immunity 37, 1009–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto J, Zhou W, Chen FF, Su F, Kuwada JY, Hidaka E, Katsuyama T, Sagara J, Taniguchi S, Ngo-Hazelett P, et al. (2003). Caspy, a zebrafish caspase, activated by ASC oligomerization is required for pharyngeal arch development. J. Biol. Chem. 278, 4268–4276. [DOI] [PubMed] [Google Scholar]
- Mathias JR, Dodd ME, Walters KB, Rhodes J, Kanki JP, Look AT, and Huttenlocher A. (2007). Live imaging of chronic inflammation caused by mutation of zebrafish Hai1. J. Cell Sci. 120, 3372–3383. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Shah NM, and Anderson DJ (1997). Regulatory mechanisms in stem cell biology. Cell 88, 287–298. [DOI] [PubMed] [Google Scholar]
- Nerlov C, Querfurth E, Kulessa H, and Graf T. (2000). GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood 95, 2543–2551. [PubMed] [Google Scholar]
- Pfaffl MW (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras EM, Mirantes-Barbeito C, Fong S, Loeffler D, Kovtonyuk LV, Zhang S, Lakshminarasimhan R, Chin CP, Techner JM, Will B, et al. (2016). Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18, 607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilla DM, Hagar JA, Haldar AK, Mason AK, Degrandi D, Pfeffer K, Ernst RK, Yamamoto M, Miao EA, and Coers J. (2014). Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc. Natl. Acad. Sci. USA 111, 6046–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajsnar TK, Hamilton R, Garcia-Lara J, McVicker G, Williams A, Boots M, Foster SJ, and Renshaw SA (2012). A privileged intraphagocyte niche is responsible for disseminated infection of Staphylococcus aureus in a zebrafish model. Cell. Microbiol. 14, 1600–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, and Fitzgerald KA (2016). Inflammasome Complexes: Emerging Mechanisms and Effector Functions. Cell 765, 792–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, and Kolls JK (2017). Neutrophilic Inflammation in Asthma and Association with Disease Severity. Trends Immunol. 38, 942–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekhtman N, Radparvar F, Evans T, and Skoultchi AI (1999). Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 13, 1398–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, and Whyte MK (2006). Atransgenic zebrafish model of neutrophilic inflammation. Blood 108, 3976–3978. [DOI] [PubMed] [Google Scholar]
- Santos JC, Dick MS, Lagrange B, Degrandi D, Pfeffer K, Yamamoto M, Meunier E, Pelczar P, Henry T, and Broz P (2018). LPS targets host guanylate-binding proteins to the bacterial outer membrane for non-canonical inflammasome activation. EMBO J. 37, e98089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J,Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, and Kanneganti TD (2016). The cell biology of inflammasomes: Mechanisms of inflammasome activation and regulation. J. Cell Biol. 213, 617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RD, Malley JD, and Schechter AN (2000). Quantitative analysis of globin gene induction in single human erythroleukemic cells. Nucleic Acids Res. 28, 4998–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachura DL, Svoboda O, Campbell CA, Espín-Palazón R, Lau RP, Zon LI, Bartunek P, and Traver D. (2013). The zebrafish granulocyte colony-stimulating factors (Gcsfs): 2 paralogous cytokines and their roles in hematopoietic development and maintenance. Blood 122, 3918–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser MK, Hoppe PS, Loeffler D, Kokkaliaris KD, Schroeder T, Theis FJ, and Marr C. (2018). Lineage marker synchrony in hematopoietic genealogies refutes the PU.1/GATA1 toggle switch paradigm. Nat. Commun. 9, 2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamplin OJ, Durand EM, Carr LA, Childs SJ, Hagedorn EJ, Li P, Yzaguirre AD, Speck NA, and Zon LI (2015). Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 160, 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa U, Castelli G, and Elvira P. (2015). Experimental and investigational therapies for chemotherapy-induced anemia. Expert Opin. Investig. Drugs 24, 1433–1445. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, and Postlethwait JH (1993). Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 119, 1203–1215. [DOI] [PubMed] [Google Scholar]
- Tyrkalska SD, Candel S,Angosto D, Gómez-Abellan V, Martín-Sánchez F, García-Moreno D, Zapata-Perez R, Sanchez-Ferrer A, Sepulcre MP, Pelegrín P, and Mulero V. (2016). Neutrophils mediate Salmonella Typhimurium clearance through the GBP4 inflammasome-dependent production of prostaglandins. Nat. Commun. 7, 12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrkalska SD, Candel S, Pérez-Oliva AB, Valera A, Alcaraz-Pérez F, García-Moreno D, Cayuela ML, and Mulero V. (2017). Identification of an Evolutionarily Conserved Ankyrin Domain-Containing Protein, Caiap, Which Regulates Inflammasome-Dependent Resistance to Bacterial Infection. Front. Immunol. 8, 1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallet P, Benaoudia S, Mosnier A, Lagrange B, Martin A, Lindgren H, Golovliov I, Michal F, Basso P, Djebali S, et al. (2017). IFN-γ extends the immune functions of Guanylate Binding Proteins to inflammasome-independent antibacterial activities during Francisella novicida infection. PLoS Pathog. 13, e1006630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannamaker W, Davies R, Namchuk M, Pollard J, Ford P, Ku G, Decker C, Charifson P, Weber P, Germann UA, et al. (2007). (S)-1-((S)-2-{[1-(4-amino-3-chloro-phenyl)-methanoyl]-amino}−3,3-dimethyl-butanoy l)-pyrrolidine-2-carboxylic acid ((2R,3S)-2-ethoxy-5-oxo-tetrahydro-furan-3-yl)-amide (VX-765), an orally available selective interleukin (IL)-converting enzyme/caspase-1 inhibitor, exhibits potent anti-inflammatory activities by inhibiting the release of IL-1beta and IL-18. J. Pharmacol. Exp. Ther. 321, 509–516. [DOI] [PubMed] [Google Scholar]
- Weiss G. (2015). Anemia of Chronic Disorders: New Diagnostic Tools and New Treatment Strategies. Semin. Hematol. 52, 313–320. [DOI] [PubMed] [Google Scholar]
- Weissman IL (2000). Translating stem and progenitor cell biology to the clinic: barriers and opportunities. Science 287, 1442–1446. [DOI] [PubMed] [Google Scholar]
- Westerfield M. (2000). The Zebrafish Book A Guide for the Laboratory Use of Zebrafish Danio* (Brachydanio) rerio (Eugene, OR: University of Oregon Press; ). [Google Scholar]
- White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, Bourque C, Dovey M, Goessling W, Burns CE, and Zon LI (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyatt DJ, Karis A, Harkes IC, Verkerk A, Gillemans N, Elefanty AG, Vairo G, Ploemacher R, Grosveld F, and Philipsen S. (1997). The level of the tissue-specific factor GATA-1 affects the cell-cycle machinery. Genes Funct. 1, 11–24. [DOI] [PubMed] [Google Scholar]
- Whyatt D, Lindeboom F, Karis A, Ferreira R, Milot E, Hendriks R, de Bruijn M, Langeveld A, Gribnau J, Grosveld F, and Philipsen S. (2000). An intrinsic but cell-nonautonomous defect in GATA-1-overexpressing mouse erythroid cells. Nature 406, 519–524. [DOI] [PubMed] [Google Scholar]
- Wu WC, Sun HW, Chen HT, Liang J, Yu XJ, Wu C, Wang Z, and Zheng L. (2014). Circulating hematopoietic stem and progenitor cells are myeloid-biased in cancer patients. Proc. Natl. Acad. Sci. USA 111, 4221–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wang L, Kalfa TA, Cancelas JA, Shang X, Pushkaran S, Mo J, Williams DA, and Zheng Y. (2007). Cdc42 critically regulates the balance between myelopoiesis and erythropoiesis. Blood 110, 3853–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwack EE, Feeley EM, Burton AR, Hu B, Yamamoto M, Kanneganti TD, Bliska JB, Coers J, and Brodsky IE (2017). Guanylate Binding Proteins Regulate Inflammasome Activation in Response to Hyperinjected Yersinia Translocon Components. Infect. Immun. 85, e00778–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.