Abstract
Despite recent advances in the treatment of multiple myeloma, patients with this disease still inevitably relapse and become refractory to existing therapies. Mutations in K-RAS, N-RAS and B-RAF are common in multiple myeloma, affecting 50% of patients at diagnosis and >70% at relapse. However, targeting mutated RAS/RAF via MEK inhibition is merely cytostatic in myeloma and largely ineffective in the clinic. We examined mechanisms mediating this resistance and identified histone deacetylase inhibitors as potent synergistic partners. Combining the MEK inhibitor AZD6244 (selumetinib) with the pan-histone deacetylase inhibitor LBH589 (panobinostat) induced synergistic apoptosis in RAS/RAF mutated multiple myeloma cell lines. Interestingly, this synergy was dependent on the pro-apoptotic protein BIM. We determined that while single-agent MEK inhibition increased BIM levels, the protein remained sequestered by antiapoptotic BCL-2 family members. LBH589 dissociated BIM from MCL-1 and BCL-XL, which allowed it to bind BAX/BAK and thereby initiate apoptosis. The AZD6244/LBH589 combination was specifically active in cell lines with more BIM:MCL-1 complexes at baseline; resistant cell lines had more BIM:BCL-2 complexes. Those resistant cell lines were synergistically killed by combining the BH3 mimetic ABT-199 (venetoclax) with LBH589. Using more specific histone deacetylase inhibitors, i.e. MS275 (entinostat) and FK228 (romidepsin), and genetic methods, we determined that concomitant inhibition of histone deacetylases 1 and 2 was sufficient to synergize with either MEK or BCL-2 inhibition. Furthermore, these drug combinations effectively killed plasma cells from myeloma patients ex vivo. Given the preponderance of RAS/RAF mutations, and the fact that ABT-199 has demonstrated clinical efficacy in relapsed/refractory multiple myeloma, these drug combinations hold prom ise as biomarker-driven therapies.
Introduction
Multiple myeloma (MM) is a cancer of differentiated plasma cells.1 It evolves from a premalignant condition called monoclonal gammopathy of undetermined significance, which affects 5.3% of adults over the age of 70.2 More than 30,000 people are projected to be diagnosed with MM in the USA in 2018.3 Despite improvements in survival, MM remains incurable.1,4 In addition, it is a clinically heterogeneous disease, with several major cytogenetic abnormalities that affect prognosis.5,6 Nevertheless, most patients receive uniform up-front treatment.1,6 Clearly, there is an unmet need for therapies that target particular drivers of the disease.
The RAS/RAF/MEK/ERK pathway is abnormally activated in MM through several mechanisms including oncogenic mutations and cytokines in the bone marrow microenvironment.7,8 Activating mutations in K-RAS, N-RAS and B-RAF have been reported in 50% of MM patients at diagnosis.7,9 Such mutations are present in <10% of patients with monoclonal gammopathy of undetermined significance, suggesting a role in disease progression.7,10 Furthermore, >70% of patients have RAS/RAF mutations present at relapse.11 It follows that directly targeting RAS/RAF/MEK/ERK in MM could be a promising strategy. However, MEK inhibition is merely cytostatic in MM in vitro.12 Furthermore, a clinical trial evaluating MEK inhibitor monotherapy in MM showed limited efficacy.13 Thus, it appears that for MEK inhibitors to be relevant in MM, they must be combined with other agents.
Recently, histone deacetylase (HDAC) inhibitors have shown significant activity in numerous tumor types, both in vitro and in the clinic.14 In fact, the pan-HDAC inhibitor LBH589 (panobinostat) was recently approved for treating relapsed/refractory MM patients in combination with bortezomib.15 As chemotherapeutic agents, HDAC inhibitors have been shown to inhibit cell survival and proliferation and enhance immune-mediated cytotoxicity.14,15
We hypothesized that LBH589 could induce enhanced apoptosis when combined with MEK inhibition in MM. Our hypothesis stemmed from two considerations: (i) MEK inhibitors induce apoptosis in several other RAS/RAF mutated cancers,16,17 suggesting MM-specific resistance factors, and (ii) HDAC inhibitors kill MM cells through several known mechanisms, including modulation of the pro- and antiapoptotic BCL-2 family members, which often mediate chemoresistance.15,18-20
In the present study, we show that MEK inhibition with AZD6244 (selumetinib), when combined with LBH589, synergistically drives intrinsic apoptotic cell death in MCL-1 “primed” RAS/RAF mutated MM cell lines. Mechanistically, MEK inhibition increases BIM levels; LBH589 acts as a de facto MCL-1 and BCL-XL inhibitor, dissociating BIM:MCL-1 and BIM:BCL-XL complexes. In contrast, we demonstrate that LBH589 synergizes with the BH3 mimetic ABT-199 (venetoclax) in BCL-2 “primed” cell lines, which are resistant to the AZD6244/LBH589 combination. Finally, we show that concomitant inhibition of HDAC1 and HDAC2 is sufficient to synergize with either MEK or BCL-2 inhibition in the same distinct MM cell lines.
Given that refractoriness to whole classes of drugs (e.g. proteasome inhibitors) is the final common endpoint for nearly all patients with MM,21 the agents in this study are felicitous because they work via alternative mechanisms of action, are already approved or in clinical development, and offer the tantalizing prospect of targeted therapy guided by RAS/RAF mutational status and MCL-1/BCL-2 functional dependence.
Methods
Ethics
This study was approved by the Mayo Clinic Institutional Review Board. Patients’ cells were collected after informed consent, in adherence to the Declaration of Helsinki.
Multiple myeloma cell lines and patients’ cells
DOX40, H929, KMS11, KMS18, KMS28BM, MM1S, MM1R, OPM1, OPM2, RPMI8226 and U266 were obtained (see Online Supplementary Methods).19,22 Briefly, all cell lines were cultured in RPMI 1640 medium (Mediatech Inc., Manassas, VA, USA) containing 10% fetal bovine serum (Mediatech, Inc.). Freshly obtained bone marrow aspirates from MM patients were collected after informed consent, then CD138+ or bone marrow stromal cells were sorted and cultured as previously described.19,22
Reagents
ABT-199 was generously provided by Abbvie (Chicago, IL, USA). AZD6244, MEK162, SCH772984, SAHA, LBH589, MS275 and FK228 were purchased from Selleckchem (Houston, TX, USA). Tubacin was purchased from Sigma-Aldrich (St. Louis, MO, USA). Stock solutions were made in dimethylsulfoxide, aliquoted and stored at −20°C.
MTT, proliferation, and apoptosis assays
Cellular viability was measured using 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrasodium bromide (MTT) (Chemicon International Inc., Temecula, CA, USA) colorimetric assays at the indicated time points. Proliferation arrest assays were completed using 3H-thymidine uptake as previously described.23 Apoptosis of patients’ cells was assayed using annexin/propidium iodide (PI) as previously described.22,23 Briefly, cells were washed twice with annexin binding buffer (ABB: 10 mM HEPES pH 7.4, 140 mM NaCl, 2.5 mM CaCl2) and then 100 μL cells (107/mL) were stained for 15 min at room temperature with 3 μL of annexin V-FITC (Caltag, Burlingame, CA, USA), then washed with ABB and re-suspended in 500 μL of ABB with 5 μL of 1 mg/mL PI (Sigma-Aldrich, St. Louis, MO, USA). The samples were then run on a Canto flow cytometer (BD Biosciences, San Jose, CA). All experiments with MM cell lines were performed in triplicate.
Western blotting
MM cell lines were lysed with NP40 buffer, 1 mM phenylmethyl-sulfonyl-fluoride (PMSF), protease inhibitor cocktail (PIC), and 1 mM HALT Phosphatase Inhibitor (Thermo Fisher Scientific, Rockford, IL, USA). Protein concentrations were measured using the BCA assay (Thermo Fisher). First, 20-25 μg were loaded on 4-20% Tris-Glycine gels and transferred onto polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA , USA). Antibodies for acetylated histone 3, BAK, BAX, BCL-2, BCL-XL, BIM, caspase 8, caspase 9, ERK, GAPDH, HDAC1, HDAC2, HDAC6, MCL-1, PARP, p-BCL-2 (S70), pERK, p-MCL-1 (S64), and p-MCL-1 (T163) were purchased from Cell Signaling Technology (Danvers, MA, USA) and used for probing as previously described.19,22 All western blot experiments were performed in triplicate with a representative blot shown.
Immunoprecipitation
Proteins (100-150 μg) were incubated in a total volume of 500 μL of NP40 buffer, 1 mM PMSF, 1 mM PIC, 1 mM HALT Phosphatase Inhibitor (Thermo Fisher Scientific), and a 1:100 dilution of the following primary antibodies for 4 h: BAX 6A7 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), BAX (Millipore Sigma, Burlington, MA, USA), and BCL-2, BCL-XL, BIM and MCL-1 (Cell Signaling Technology). Samples were then incubated with ChIP-Grade Protein G Magnetic Beads (Cell Signaling Technology) for 12 h, washed five times, boiled in 2x Laemmli Sample Buffer dye (Bio-Rad) at 100 °C for 5 min, then loaded on 4-20% Tris-Glycine gels (Bio-Rad) and probed as described above. All experiments were performed in triplicate with a representative blot shown.
Short interfering RNA transfection
Short interfering (si)RNA for BIM, BAX, BAK, HDAC1, HDAC2 and HDAC6 were purchased from Thermo Fisher Scientific. ERK1 and ERK2 siRNA were purchased from Santa Cruz Biotechnology. siRNA were electroporated into MM cell lines using the Lonza nucleofector kit V (Lonza, Basel, Switzerland). The manufacturer’s G-15 program was used for KMS18 and OPM2; O-23 was used for MM1S and KMS28. All experiments were performed in triplicate.
Isobologram analysis
The effects of combination treatments in MM cells were analyzed using the CalcuSyn™ software program (Biosoft, Ferguson, MO, USA), which is based on the Chou-Talalay method, as previously described.19,24
Results
Single-agent MEK inhibition does not induce cell death in multiple myeloma cell lines
Prior studies have documented the lack of induction of cytotoxicity by single-agent MEK inhibitors in MM.12 We aimed to confirm these findings using a panel of MM cell lines that are wild-type or mutated for RAS/RAF. We treated cell lines with increasing doses of the MEK inhibitor AZD6244 (selumetinib) and observed a lack of significant cytotoxicity, with IC50 not reached at doses up to 1500 nM (Online Supplementary Figure 1A). AZD6244 was slightly more capable at inducing proliferation arrest in the same panel of cell lines, but still largely ineffective (Online Supplementary Figure 1B). These results were also confirmed by performing annexin/PI staining after treating two RAS mutant MM cell lines with 5000 nM of AZD6244, which is far above the concentrations at which the kinase activity of MEK is inhibited (Online Supplementary Figure 1C). Thus, it became clear that despite commonly occurring oncogenic mutations in RAS and RAF, MEK inhibitors have limited scope as single-agents in MM.
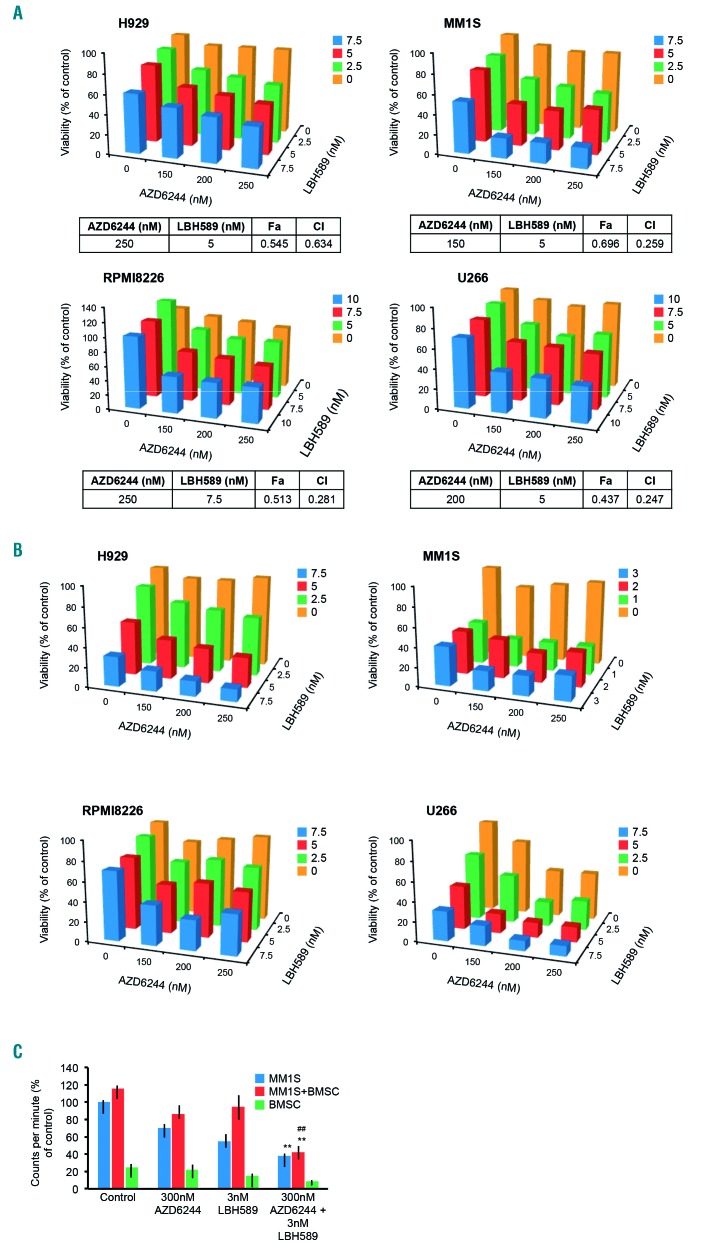
Figure 1.
MEK inhibition in combination with histone deacetylase inhibition synergistically kills multiple myeloma cell lines in vitro. (A) AZD6244 and the pan-histone deacetylase (HDAC) inhibitor LBH589 induced synergistic cytotoxicity, assayed using MTT at 72 h in the RAS-mutated human multiple myeloma (MM) cell lines H929, MM1S, and RPMI8226, and the RAF-mutated human MM cell line U266. Viability is shown as percent of control on the Y-axis. Combination index (CI) values <1.0, indicating synergy, are shown for each cell line. (B) AZD6244 and LBH589 induced proliferation arrest, assayed with 3H-thymidine incorporation, by 48 h in H929, MM1S, RPMI8226, and U266 cells, shown as percent of control on the Y-axis. (C) Bone marrow stromal cells (BMSC) derived from an MM patient were co-cultured with MM1S, and proliferation was assayed using 3H-thymidine incorporation after treatment for 48 h with AZD6244, LBH5589, or the combination. Error bars represent the standard error of the mean of triplicate experiments. Differences between groups were calculated with the Student t test. **P<0.001, ##P<0.01. All experiments were performed in triplicate.
MEK + HDAC inhibition induces synergistic cell death in multiple myeloma cell lines
We examined the ability of the recently approved pan-HDAC inhibitor LBH589 (panobinostat) to enhance cell death induced by AZD6244 in MM cell lines. Low doses of the AZD6244/LBH589 drug combination induced potent synergistic cytotoxicity (Figure 1A). Interestingly, the synergy was observed in cell lines with mutations in K-RAS and N-RAS (i.e. H929, MM1R, MM1S, RPMI8226) and B-RAF (i.e. U266), but not in cell lines that are wild-type for RAS and RAF (i.e. KMS11, KMS18, OPM2). We also observed significantly more potent proliferation arrest when the drugs were used in combination (Figure 1B).
Components of the bone marrow microenvironment such as bone marrow stromal cells play an indispensable role in MM disease progression and resistance to therapies.25 We therefore investigated whether AZD6244/LBH589 was able to overcome the protective effects of bone marrow stromal cells. To do this, we co-cultured MM1S cells with patient-derived bone marrow stromal cells and measured the proliferation rate after treatment with either single-agent AZD6244 or LBH589, or the drug combination. We observed that the AZD6244/LBH589 combination was able to inhibit the proliferation of MM1S even when co-cultured with bone marrow stromal cells (Figure 1C). We also noted synergistic cell death when we used another MEK inhibitor, MEK162 (binimetinib), or the ERK inhibitor SCH772984 in combination with LBH589 (Online Supplementary Figure 1D). Furthermore, AZD6244 synergistically killed MM cells when combined with the pan-HDAC inhibitor SAHA (data not shown).
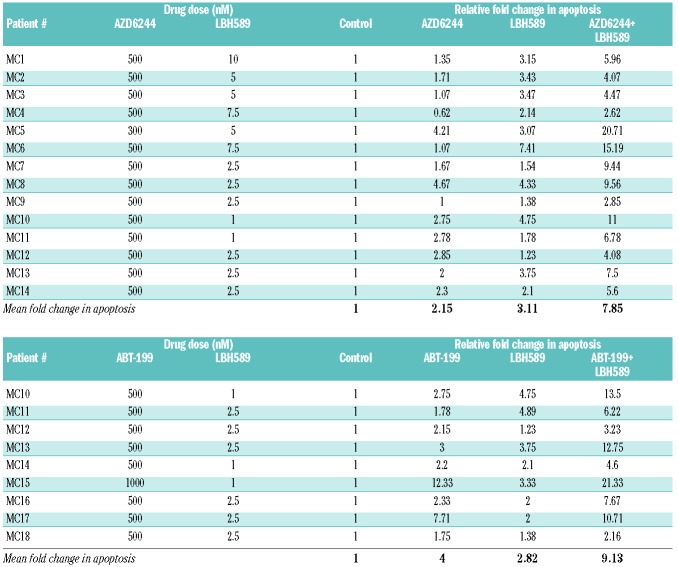
To understand whether the cytotoxicity caused by the AZD6244/LBH589 combination occurred through the apoptotic pathway, we performed annexin/PI staining. The drug combination clearly induced apoptotic cell death by 72 h (Figure 2A). We also examined whether the combination induced the cleavage of caspases and PARP, both of which are markers of apoptosis. The AZD6244/LBH589 combination induced potent cleavage of caspase 9 and PARP, but not caspase 8, suggesting that the cell death occurred through the mitochondrial intrinsic apoptotic pathway (Figure 2B). Next, we examined whether the drugs, at the doses used above, were able to inhibit their target proteins. As expected, AZD6244 inhibited pERK and LBH589 caused an increase in acetylated histone H3 levels (Figure 2C). Finally, to confirm that MEK/ERK pathway inhibition contributed to the synergy with LBH589, and to examine whether both isoforms of ERK need to be inhibited for the synergy to occur, we nucleofected isoform-specific ERK siRNA into MM1S and treated the cells with LBH589. We observed that ERK1 or ERK2 knockdown individually enhanced the cell death induced by LBH589 (Figure 2D, E). However, simultaneous knockdown of both isoforms led to even more pronounced cell death when used in combination with LBH589, supporting an important survival role for both ERK isoforms in this context. Finally, we treated plasma cells obtained from MM patients with the drug combination. The characteristics of these patients are detailed in Online Supplementary Table S1. It is worth noting that several patients had high-risk features including TP53 deletion, t(4;14) and refractoriness to multiple lines of therapy. We observed augmented apoptosis with the AZD6244/LBH589 drug combination compared to the effects of either drug alone (Table 1).
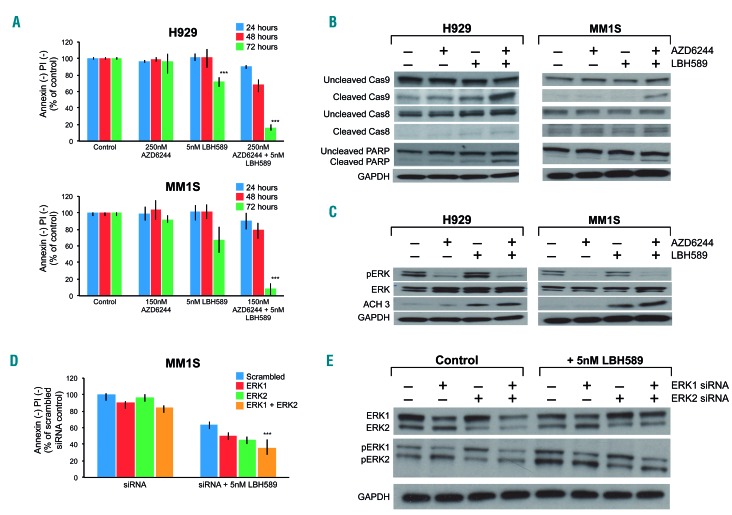
Figure 2.
MEK + histone deacetylase inhibition drives synergistic apoptosis in multiple myeloma cell lines and inhibits target proteins. (A) Flow cytometric cell viability of the RAS mutant human multiple myeloma (MM) cell lines H929 and MM1S measured as the proportion of annexin−/propidium iodide (PI)− cells, after 24, 48 and 72 h of treatment with AZD6244 and LBH589 at the indicated doses. Viability is shown as percent of control on the Y-axis. (B,C) H929 and MM1S were treated with AZD6244/LBH589 for 24 h, then whole-cell lysates were separated using sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to western blotting for the indicated proteins. (D) MM1S was electroporated with scrambled siRNA, ERK1, ERK2 siRNA or the combination, then left untreated or treated with 5 nM LBH589. At 72 h, cell viability was assessed using flow cytometry by analyzing the proportion of annexin−/PI− cells, shown as percent of control on the Y-axis. (E) Whole-cell lysates from these cells were separated using SDS-PAGE and subjected to western blotting to confirm ERK1 and ERK2 silencing. Error bars represent the standard error of mean of triplicate experiments. Differences between groups were calculated with the Student t test. **P<0.001. All experiments were performed in triplicate.
Table 1.
Changes in apoptosis of multiple myeloma patients’ plasma cells exposed to various drug combinations. Plasma cells sorted from patients with multiple myeloma were exposed to AZD6244, LBH589, and the combination, or ABT-199, LBH589, and the combination for 72 h. The proportion of cells undergoing apoptosis was measured using flow cytometry, and the relative fold change in apoptosis is indicated. The upper part of the table summarizes the results from the 14 patients treated with the AZD6244/LBH489 combination, while the lower part summarizes the results for the nine patients treated with the ABT-199/LBH589 combination.
MEK + HDAC inhibitor-induced synergistic apoptosis is mediated by BIM
Given that the MEK/ERK pathway is known to phosphorylate the pro-apoptotic BH3-only protein BIM at serine 69 (S69) to mark it for proteasomal degradation,26 we were not surprised to find that AZD6244 treatment increased BIM protein levels (Figure 3A). Although prior studies have shown that HDAC inhibition increases BIM expression in MM,18,19 we did not observe increased levels of BIM with the several-fold lower doses of LBH589 that were used in this study (Figure 3A). Even so, given that MEK inhibition increased BIM and that the drug combination induced potent activation of intrinsic apoptotic markers, we hypothesized that BIM might play an important role in the observed synergy. To investigate this, we nucleo fected MM1S cells with BIM siRNA and observed that BIM knockdown completely protected from the synergistic cell death induced by the MEK + HDAC inhibitor combination (Figure 3B).
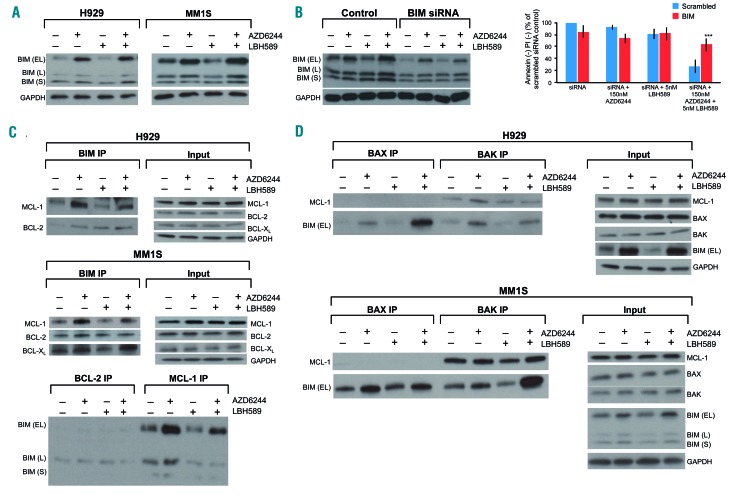
Figure 3.
MEK + histone deacetylase inhibitor induced synergistic apoptosis is mediated by BIM. (A) The RAS mutant human multiple myeloma (MM) cell lines H929 and MM1S were treated with AZD6244/LBH589 for 24 h, then whole-cell lysates were blotted for the indicated proteins. (B) MM1S was electroporated with scrambled siRNA or BIM siRNA and then left untreated or treated with 5 nM LBH589. At 72 h, cell viability was assessed using flow cytometry by analyzing the proportion of annexin−/propidium iodide (PI)− cells, shown as percent of control on the Y-axis. Furthermore, the whole-cell lysates were separated using sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to western blotting for the indicated proteins to confirm silencing. Error bars represent the standard error of mean of triplicate experiments. Differences between groups were calculated with the Student t test. **P<0.001. (C) (Upper) H929 and MM1S were treated with AZD6244 (250 nM and 150 nM, respectively) and LBH589 (5 nM) for 24 h. BIM immunoprecipitates were separated using SDS-PAGE and subjected to western blotting to examine BCL-2, MCL-1 and BCL-XL binding patterns. Whole cell lysates (input) were also separated and probed for the indicated proteins. (Lower) Immunoprecipitates from MM1S for BCL-2 and MCL-1 were also separated and probed to examine BIM binding. (D) H929 and MM1S were treated with AZD6244 (250 nM and 150 nM, respectively) and LBH589 (5 nM). BAX and BAK immunoprecipitation was performed and western blotting was used to examine levels of BIM and MCL-1 bound to BAX and BAK. Whole cell lysates (input) were also separated and probed for the indicated proteins. All experiments were performed in triplicate.
HDAC inhibition dissociates BIM from MCL-1 and BCL-XL
From the above results, it became clear that BIM plays an essential role in the synergistic apoptosis induced by the drug combination. Since BIM was upregulated by AZD6244, but not by LBH589, we reasoned that the increased levels of BIM induced by AZD6244 were unable to activate apoptosis due to sequestration by the antiapoptotic BCL-2 family members (i.e. BCL-2, BCL-XL and MCL-1). Thus, we hypothesized that the mechanism by which LBH589 synergizes with AZD6244 is by modulating the interactions of BIM with the antiapoptotic BCL-2 family proteins. In pursuit of demonstration of this, we immunoprecipitated BIM and examined its binding pattern with BCL-2, BCL-XL and MCL-1 before and after drug treatment. AZD6244 increased the relative amount of BIM bound to all three antiapoptotic proteins, which may partly explain why even though the drug markedly increases BIM levels, it has limited cytotoxic effects in MM as a single agent (Figure 3C). Interestingly, LBH589 dissociated BIM from MCL-1 and BCL-XL, but not BCL-2 (Figure 3C). This effect was particularly evident when the antiapoptotic proteins were “primed” with more BIM by AZD6244. We also noted this dissociation in the reciprocal experiment when pulling down with BCL-2 and MCL-1, and probing for BIM (Figure 3C). Furthermore, we observed increased BIM bound to both BAX and BAK after AZD6244 and LBH589 treatment individually, which was markedly increased after the combination treatment (Figure 3D). We also noted that LBH589 dissociated BAK from MCL-1 in a similar manner to BIM, which theoretically would facilitate increased BIM:BAK complexes in the presence of LBH589 to further activate the intrinsic apoptotic cascade (Figure 3D).
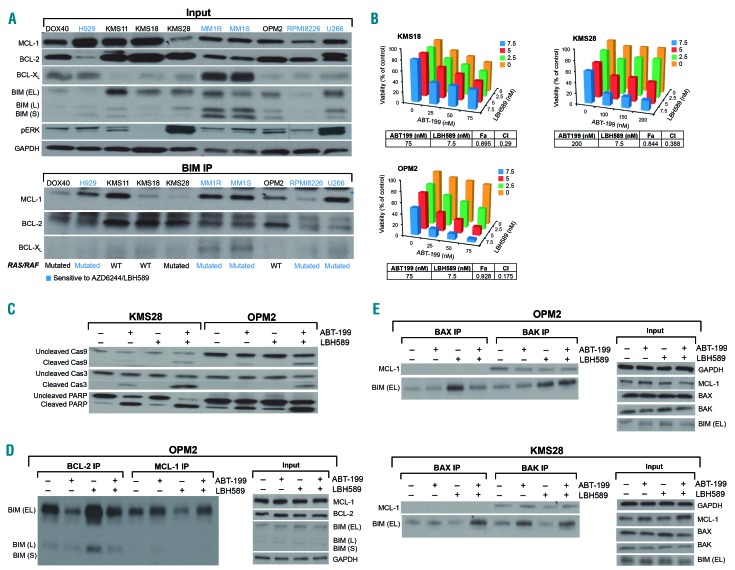
MCL-1/BCL-2 expression and BIM binding profile correlate with sensitivity to MEK + HDAC inhibition
In light of LBH589 dissociating BIM from MCL-1 and BCL-XL, but not from BCL-2, we wondered if the cell lines that were resistant to the MEK+HDAC inhibitor combination expressed higher baseline levels of BCL-2. Western blots confirmed that this was partly true, but many of the AZD6244/LBH589-sensitive cell lines, including MM1R, MM1S, RPMI8226 and U266, had high expression of both BCL-2 and MCL-1 (Figure 4A). To interrogate the possibility that expression is a poor indication of functional dependence on either antiapoptotic protein, we immunoprecipitated BIM and examined baseline levels of BIM:BCL-2, BIM:MCL-1 and BIM:BCL-XL complexes in cell lines that were either sensitive or resistant to the MEK + HDAC inhibitor combination. Notably, BIM was mostly bound to MCL-1 in all of the cell lines which were sensitive to the MEK + HDAC inhibitor combination, including the aforementioned MM1R, MM1S, RPMI8226 and U266, suggesting that MCL-1 that is “BIM-primed” is critical for the observed synergy to occur (Figure 4A). On the other hand, cell lines with BIM mostly bound to BCL-2 were resistant to the MEK + HDAC inhibitor combination (Figure 4A). It was noteworthy that BCL-XL was weakly expressed in most of the cell lines and there were few BIM:BCL-XL complexes at baseline (Figure 4A).
Figure 4.
Predominance of BIM:MCL1 complexes correlates with sensitivity to MEK + histone deacetylase (HDAC) inhibition; MEK + HDAC inhibitor-resistant cell lines that had predominantly BIM:BCL-2 complexes were synergistically killed by BCL-2 + HDAC inhibition. (A) Whole-cell lysates from a panel of human multiple myeloma (MM) cell lines were immunoprecipitated with BIM. Subsequently, western blotting was performed to examine baseline levels of BIM:BCL-2, BIM:BCL-XL and BIM:MCL-1 complexes. Whole cell lysates (input) were also separated with sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) and probed for the indicated proteins. Cell lines sensitive to the AZD6244/LBH589 combination are indicated in blue. The mutational status of the cell lines, i.e. mutated or wild-type (WT) for RAS/RAF, is shown. (B) The BH3 mimetic ABT-199 and the pan-HDAC inhibitor LBH589 induced synergistic cytotoxicity (assessed using MTT) by 72 h in the human MM cell lines KMS18, OPM2 (RAS/RAF wild-type) and KMS28 (RAS mutant). Viability is shown as percent of control on the Y-axis. Combination index (CI) values <1.0, indicating synergy, are shown for each cell line. (C) KMS28 and OPM2 were treated with ABT-199/LBH589 for 24 h, then whole-cell lysates were separated using SDS-PAGE and probed for the indicated proteins. (D) OPM2 was treated with 50 nM ABT-199 and 5 nM LBH589 for 24 h, then immunoprecipitates for BCL-2 and MCL-1, and whole cell lysates (input) were separated using SDS-PAGE and probed for the indicated proteins. (E) OPM2 and KMS28 were treated with ABT-199 (50 nM and 250 nM, respectively) and LBH589 (5 nM) for 12 h. BAX and BAK immunoprecipitates and whole-cell lysates (input) were separated using SDS-PAGE and subjected to western blotting for the indicated proteins. All experiments were performed in triplicate.
BCL-2 + HDAC inhibition induces synergistic apoptosis in MEK + HDAC inhibitor-resistant, BCL-2-primed cell lines
Since cell lines with more BIM:BCL-2 complexes relative to BIM:MCL-1 complexes were resistant to the MEK + HDAC combination, we hypothesized that these BCL-2-primed cell lines would be sensitive to BCL-2 inhibition with the BH3 mimetic ABT-199 (venetoclax) when used in combination with LBH589. Remarkably, BCL-2 + HDAC inhibition caused synergistic proliferation arrest and cytotoxicity in nearly all the cell lines that were resistant to the MEK + HDAC combination, including DOX40, KMS18, KMS28 and OPM2 (Figure 4B and data not shown). Pronounced induction of apoptosis was confirmed by annexin/PI staining (Online Supplementary Figure S2).
This coincided with cleavage of caspase 9/3 and PARP (Figure 4C). Immunoprecipitation experiments demonstrated that ABT-199 dissociated BIM from BCL-2, and LBH589 dissociated BIM from MCL-1 (Figure 4D). In addition, we found that ABT-199 and LBH589 both increased BIM bound to both BAX and BAK, an effect that was even more pronounced with the drug combination (Figure 4E). Moreover, BAX/BAK knockdown with siRNA protected from the ABT-199/LBH589-induced cytotoxicity, confirming that the observed synergistic cell death occurred via the mitochondrial intrinsic apoptotic pathway (Figure 4F). Finally, we noted enhanced apoptosis after treating plasma cells from MM patients with the ABT-199/LBH589 combination (Table 1).
Notably, the ABT-199/LBH589 combination did not synergistically induce cell death in the RAS/RAF mutant, AZD6244/LBH589-sensitive cell lines (i.e. H929, MM1R, MM1S, RPMI and U266). This dovetails with the finding that while LBH589 dissociated BIM:MCL-1 complexes in all the cell lines we tested, it did not shift BIM onto BCL-2 in RAS/RAF mutant cell lines (e.g. MM1S) (Figure 3C). However, LBH589 did shift BIM onto BCL-2 in the RAS/RAF wild-type cell lines (e.g. OPM2) (Figure 4D), which jibes with its potent synergistic effect when combined with ABT-199. In summary, it appears that AZD6244/LBH589 and ABT-199/LBH589 target two distinct subgroups of MM cell lines with different BCL-2 family binding proclivities.
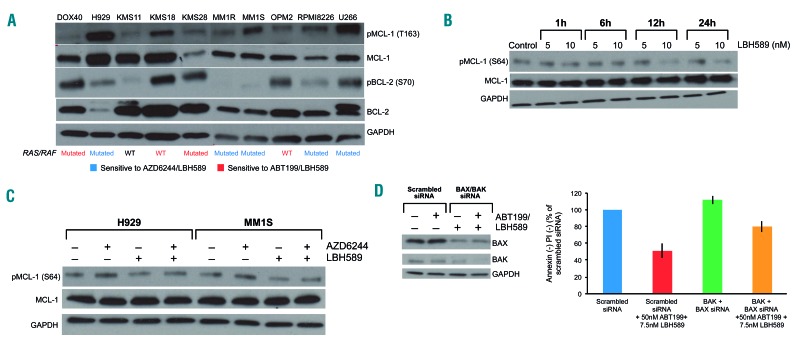
Baseline MCL-1/BCL-2 phosphorylation status correlates with sensitivity to MEK + HDAC or BCL-2 + HDAC inhibition
To determine why certain cell lines would preferentially have BIM bound to one antiapoptotic protein over another, i.e. BCL-2 versus MCL-1, we examined several phosphorylation sites known to affect the binding capacity and stability of the BCL-2 family to see if there was any correlation. Interestingly, cell lines with higher p-BCL-2 at serine 70 (S70) tended to have more BIM bound to BCL-2 (Figure 5A). p-BCL-2 (S70) is known to increase the antiapoptotic capacity of BCL-2, i.e. its ability to bind BAK and BH3-only proteins.27
Figure 5.
Phosphorylation status of MCL-1/BCL-2 correlates with sensitivity to MEK + histone deacetylase (HDAC) or BCL-2 + HDAC inhibition, and HDAC inhibition affects the phosphorylation of MCL-1. (A) Whole-cell lysates from a panel of human multiple myeloma (MM) cell lines were separated with sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to western blotting for the indicated proteins. Cell lines sensitive to the AZD6244/LBH589 combination are indicated in blue. Cell lines sensitive to the ABT-199/LBH589 combination are indicated in red. KMS11 was resistant to both drug combinations, and is indicated in black. Mutational status of the cell lines, either mutated or wild-type (WT) for RAS/RAF, is shown. (B) H929 was treated for 1, 6, 12, and 24 h with 5 and 10 nM LBH589. Whole cell lysates were separated with SDS-PAGE and subjected to western blotting for the indicated proteins. (C) H929 and MM1S were treated with AZD6244 (250 nM and 150 nM, respectively) and LBH589 (5 nM) for 24 h. Whole cell lysates from these cells were separated with SDS-PAGE and western blotting was performed for the indicated proteins. (D) KMS28 was electroporated with scrambled siRNA or BAX and BAK siRNA, then treated with 250 nM ABT-199 and 5nM LBH589. At 72 h, cell viability was assessed using flow cytometry by analyzing the proportion of annexin−/propidium iodide (PI)− cells, shown as percent of control on the Y-axis. In addition, whole-cell lysates were separated using SDS-PAGE and probed for the indicated proteins to confirm silencing. Error bars represent the standard error of mean of triplicate experiments. All experiments were performed in triplicate.
On the other hand, cell lines with BIM mostly bound to MCL-1 tended to have relatively low expression of p-BCL-2 (S70), as well as relatively high expression of p-MCL-1 at threonine 163 (T163) (Figure 5A). p-MCL-1 (T163) is a well-documented post-translational modification that stabilizes MCL-1, protecting it from proteasomal degradation.28 However, we did not observe a correlation between p-MCL-1 at serine 64 (S64) and BIM binding preference (data not shown). p-MCL-1 (S64) increases the binding capacity of MCL-1, but not its stability.29
To our surprise, p-BCL-2 (S70) and p-MCL-1 (T163) were nearly perfect in predicting sensitivity to either MEK + HDAC or BCL-2 + HDAC inhibition (Figure 5A). This was particularly striking in the case of DOX40, a doxorubicin-resistant cell line derived from RPMI8226. DOX40 expressed more p-BCL-2 (S70) than p-MCL-1 (T163), and was sensitive to the ABT-199/LBH589 combination, whereas its parental cell line RPMI8226 did not express p-BCL-2 (S70), but did express p-MCL-1 (T163), and was sensitive to the AZD6244/LBH589 combination (Figure 5A). Interestingly, the only cell line we tested that was resistant to both drug combinations was KMS11, and this line has low expression of both p-BCL-2 (S70) and p-MCL-1 (T163) (Figure 5A).
RAS/RAF mutational status predicts sensitivity to MEK + HDAC or BCL-2 + HDAC inhibition
Since MEK inhibitors target the pathway downstream of RAS/RAF, we were curious to determine whether sensitivity to the AZD6244/LBH589 combination correlates with RAS/RAF mutational status. This is a felicitous prospect because the MEK/ERK pathway in part controls p-MCL-1 (T163).28 Indeed, the RAS-mutated cell lines H929, MM1R, MM1S, RPMI8226, and the RAF-mutated cell line U266 were all sensitive to MEK + HDAC inhibition and had high baseline expression of p-MCL-1 (T163) (Figures 1A and 5A). On the other hand, the RAS/RAF wild-type cell lines KMS18 and OPM2 were resistant to MEK + HDAC inhibition, but sensitive to BCL-2 + HDAC inhibition. These cell lines also had high expression of p-BCL-2 (S70) (Figures 4B and 5A). However, the correlation between sensitivity to either drug combination and RAS/RAF mutational status was not perfect: DOX40 and KMS28 both have mutated RAS, but were sensitive to BCL-2 + HDAC inhibition rather than to MEK + HDAC inhibition. Finally, KMS11, which was resistant to both combinations, has wild-type RAS/RAF. Thus, we conclude that p-BCL-2 (S70), p-MCL-1 (T163) and RAS/RAF mutational status could all be useful biomarkers to predict for sensitivity to either the MEK + HDAC or BCL-2 + HDAC inhibitor combinations.
HDAC inhibition alters the phosphorylation of MCL-1
Given that HDAC inhibition dissociated BIM from MCL-1, we investigated whether LBH589 altered the expression of any BH3-only proteins that could theoretically bind to MCL-1 to displace BIM. However, at the doses at which we observed synergy, LBH589 did not appreciably alter the expression of any of the BH3-only proteins, including NOXA, PUMA, BAD, BID, BIK, BMF or HRK (data not shown). Since we found that baseline post-translational modifications on several of the antiapoptotic BCL-2 family members correlated with sensitivity to either drug combination, we studied whether LBH589 altered any post-translational modifications on MCL-1 or BCL-XL that could explain the BIM dissociation.
Interestingly, LBH589 down regulated p-MCL-1 (S64) in a time- and dose-dependent manner (Figure 5B). In addition, AZD6244 increased p-MCL-1 (S64), which may explain in part why MCL-1 becomes “primed” with BIM after AZD6244 treatment (Figure 5C). When the drugs were combined, LBH589 still decreased p-MCL-1 (S64) when compared to AZD6244 alone (Figure 5C). Taken together, it seems that LBH589 might facilitate apoptosis by decreasing the phosphorylation of MCL-1 at the S64 residue, making it less “sticky” to BIM, especially after it becomes primed with BIM in the context of MEK inhibitor treatment.29
p-MCL-1 (S64) is known to be driven by JNK,29 and CDK1/2.29,30 However, we were unable to replicate synergy when we used JNK or CDK inhibitors in combination with AZD6244 or ABT-199 (data not shown). Relatively less is known about the role of post-translational modifications on BCL-XL, but similar to BCL-2 and MCL-1, phosphorylation has been documented to modulate the antiapoptotic role of BCL-XL.20 However, LBH589 did not appreciably alter p-BCL-XL (S62) levels (data not shown).
HDAC6 inhibition does not enhance apoptosis induced by MEK inhibition
Next, we wanted to identify which HDAC(s) must be inhibited for the observed synergy to occur with MEK or BCL-2 inhibitors. First, we examined the role of HDAC6 given the relevance of inhibiting the aggresome pathway in MM and early clinical results obtained using HDAC6 inhibition in the relapsed/refractory setting.15,31 Treating cells with AZD6244 in combination with the HDAC6-specific inhibitor tubacin did not lead to synergistic cell death (Online Supplementary Figure S3A). These results were confirmed by knocking down HDAC6 using siRNA (Online Supplementary Figure S3B). Considering all the findings, we concluded that HDAC6 inhibition does not seem to be important for the synergistic cell death induced by the AZD6244/LBH589 combination.
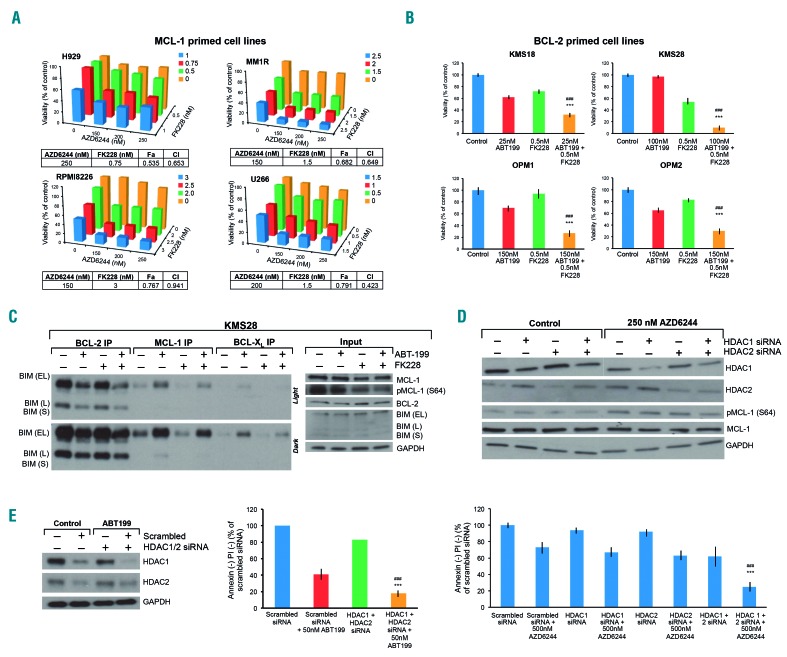
Simultaneous inhibition of HDAC1 and HDAC2 is sufficient to enhance cell death induced by MEK or BCL-2 inhibition
We next examined whether inhibiting class I HDAC augmented cell death in combination with MEK or BCL- 2 inhibitors. For this, we first used MS275 (entinostat), which inhibits HDAC1, 2 and 3. We observed potent synergy when MS275 was used with AZD6244 or ABT-199 in MM cell lines (Online Supplementary Figure S4A, B) and patients’ cells (Online Supplementary Table S2) similar to the synergy observed with LBH589. This synergy occurred even at very low doses of MS275 (i.e. 150 nM), which would be unlikely to inhibit HDAC3 (the IC50 for HDAC3 is 1.7 μM).32 To determine whether inhibiting HDAC1 and 2 would be sufficient to synergize with AZD6244 or ABT-199, we used the HDAC1 and 2 inhibitor FK228 (romidepsin) in combination with AZD6244 or ABT-199 and observed marked synergy in MM cell lines (Figure 6A, B) and patients’ cells (Online Supplementary Table S2). Using western blotting and immunoprecipitation, we also replicated the mechanistic finding that FK228 downregulates p-MCL-1 (S64) and causes BIM dissociation from MCL-1 and BCL-XL in KMS28 (Figure 6C) and H929 (Online Supplementary Figure S4C). This strongly suggested that HDAC1 and 2 are involved in the mechanism. To confirm these results, we performed knockdown studies with HDAC1- and HDAC2-specific siRNA. Knocking down HDAC1 and HDAC2 individually in combination with AZD6244 or ABT-199 caused minor increases in apoptosis. However, simultaneous knockdown of both HDAC1 and HDAC2, when combined with AZD6244 or ABT-199, caused significantly more apoptosis (Figure 6D, E). Inhibiting HDAC3 in addition to HDAC1 and HDAC2 did not augment the synergy (data not shown). Dovetailing with our results with pharmacological HDAC inhibitors, we observed p-MCL-1 (S64) downregulation when HDAC1 and HDAC2 were silenced in tandem (Figure 6D). We also noted that knocking down either HDAC1 or HDAC2 individually caused a reciprocal upregulation of HDAC2 and HDAC1, respectively. We speculate that this could be the reason why inhibition of HDAC1 or HDAC2 individually was not sufficient to synergize with either MEK or BCL-2 inhibition (Figure 6D, E). Taken together, these results showed that simultaneous inhibition of HDAC1 and HDAC2 is sufficient to markedly enhance the apoptosis induced by AZD6244 or ABT-199 in MM.
Figure 6.
Concomitant inhibition of histone deacetylases 1 and 2 replicates synergy with MEK or BCL-2 inhibition in multiple myeloma cell lines. (A) The histone deacetylase (HDAC)-1 and -2 inhibitor FK228 (romidepsin) was combined with AZD6244 in increasing doses in the MCL-1 primed, RAS/RAF mutant human multiple myeloma (MM) cell lines H929, MM1R, RPMI8226 and U266. Cellular viability was assessed using MTT at 72 h. Viability is shown as percent of control on the Y-axis. Combination index (CI) values <1.0, indicating synergy, are shown for each cell line. (B) FK228 was combined with ABT-199 in increasing doses in the BCL-2-primed human MM cell lines KMS18, OPM1, OPM2 (RAS/RAF wild-type) and KMS28 (RAS mutant). Cellular viability was assessed using flow cytometry by analyzing the proportion of annexin−/propidium iodide (PI)− cells, shown as percent of control on the Y-axis. (C) KMS28 was treated with 100 nM ABT-199 and 0.5 nM FK228 for 24 h, then immunoprecipitates for BCL-2, MCL-1 and BCL-XL, or whole cell lysates (input) were separated using sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) and probed for the indicated proteins. Light and dark film exposures are shown for the BCL-2, MCL-1 and BCL-XL immunoprecipitates so the BIM:MCL-1 and BIM:BCL-XL dissociations are apparent. (D) MM1S was electroporated with scrambled siRNA, HDAC1 siRNA, HDAC2 siRNA or HDAC1 and 2 siRNA, then left untreated or treated with 250 nM AZD6244. At 72 h, cell viability was assessed using flow cytometry by analyzing the proportion of annexin−/PI− cells, shown as percent of control on the Y-axis. In addition, whole-cell lysates were separated using SDS-PAGE and probed for the indicated proteins to confirm silencing. (E) KMS18 was electroporated with scrambled siRNA or HDAC1 and HDAC2 siRNA, then left untreated or treated with 50 nM ABT-199. At 72 h, cell viability was assessed using flow cytometry by analyzing the proportion of annexin−/PI− cells, shown as percent of control on the Y-axis. Also whole-cell lysates were separated using SDS-PAGE and probed for the indicated proteins to confirm silencing. Error bars represent the standard error of mean of triplicate experiments. Differences between groups were calculated with the Student t test. **P<0.001, ##P<0.01. All experiments were performed in triplicate.
Discussion
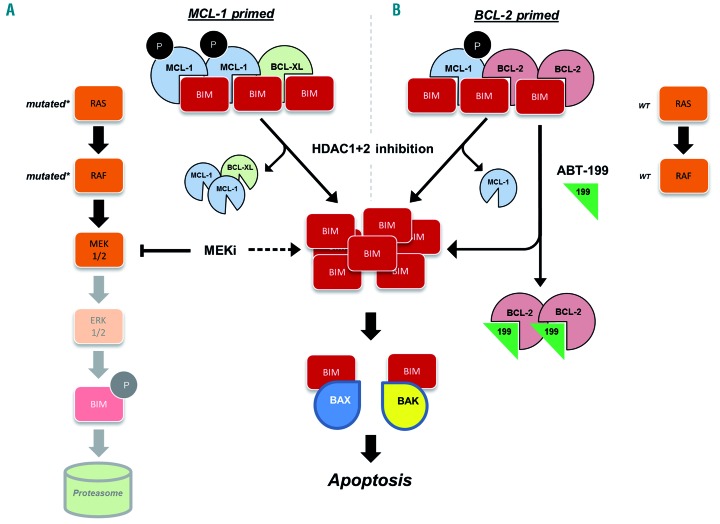
The sequestration of pro-apoptotic BCL-2 family proteins, such as BIM, by their antiapoptotic counterparts, i.e. BCL-2 and MCL-1, is a pervasive survival strategy in cancer.20 Hence, treatments that alter the pro/antiapoptotic BCL-2 family member ratio or modulate their binding dynamics hold considerable promise, particularly in hematologic malignancies. In the present study, we identified two drug combinations, i.e. MEK + HDAC and BCL-2 + HDAC inhibition, which target two distinct subgroups of MM: MCL-1 or BCL-2 primed, respectively (summarized in Figure 7). In support of these being discrete phenotypes, none of the MM cell lines that we tested was sensitive to both drug combinations. Furthermore, sensitivity aligned mostly based on RAS/RAF mutational status.
Figure 7.
Proposed mechanism of MEK or BCL-2 inhibition in combination with histone deacetylase inhibition in multiple myeloma. (A) MCL-1-primed lines, which all had mutated RAS/RAF, were sensitive to the MEK + histone deacetylase (HDAC) inhibitor combination. MEK inhibition increased BIM levels, and HDAC1+2 inhibition dissociated BIM:MCL-1 and BIM:BCL-XL complexes, the former perhaps by means of downregulation of p-MCL-1 (S64). (B) BCL-2-primed cell lines tended to be wild-type (WT) for RAS/RAF, and were sensitive to the BCL-2+HDAC inhibitor combination. Likewise, HDAC1+2 inhibition dissociated BIM:MCL-1 complexes. Both drug combinations in effect increased free BIM levels, which we found were able to engage BAX and BAK, ultimately leading to synergistic apoptotic cell death.
Mutations in the RAS/RAF pathway are present in nearly half of all malignant tumor types.33 In lieu of a direct way to inhibit RAS, MEK inhibitors have shown significant clinical benefit in several RAS/RAF-mutated cancers.34 Herein, we identified that RAS/RAF-mutated MM cells are particularly sensitive to MEK inhibition when this is used in combination with pan-HDAC or class I HDAC inhibitors. Our results demonstrate that while single-agent MEK inhibition increased BIM, this protein remained sequestered by antiapoptotic BCL-2 family members. Concomitant HDAC inhibition, specifically of class I HDAC, dissociated BIM:MCL-1 and BIM:BCL-XL complexes, but not BIM:BCL-2 complexes, thereby freeing BIM to activate apoptotic cell death (Figure 7A).
Given the prevalence of RAS/RAF mutations in MM, we anticipate that this combination could be highly clinically effective. While we did not investigate specific differences between N-RAS and K-RAS mutations here, a prior study showed different response durations after standard therapy, i.e. bortezomib, between these two subgroups; future research should examine the significance of these isoforms in the context of MEK + HDAC inhibition.35 In addition, given the marked intratumoral heterogeneity of MM, the impact of the variant allele frequency of RAS/RAF mutations must be evaluated in future studies.
Our mechanistic studies identified that MEK + HDAC inhibitor-sensitive MM cell lines expressed high levels of p-MCL-1 (T163), a post-translational modification that enhances the stability of MCL-1 and is driven in part by ERK.36 MCL-1 is unique among the antiapoptotic BCL-2 family because of its short half-life, but is often upregulated or stabilized in numerous cancers, including MM.37,38 Although p-MCL-1 (T163) does not affect the binding capacity of MCL-1, stability of the antiapoptotic BCL-2 family proteins is part and parcel of their function.39
Interestingly, we identified for the first time that the pan-HDAC inhibitor LBH589 and class I HDAC inhibitors MS275 and FK228 act as MCL-1 and BCL-XL inhibitors at relatively low doses. Furthermore, we suggest that the downregulation of p-MCL-1 at the S64 residue may be responsible for the former observation.29 Unlike other studies using several-fold higher doses of HDAC inhibitors in their experiments,18,40 HDAC inhibition did not increase levels of BIM, NOXA, or any other BH3-only proteins in our experiments. Thus, HDAC inhibitor-driven upregulation of BH3-only proteins does not appear to mediate synergy with MEK or BCL-2 inhibition, at least in MM. However, we were unable to elucidate specifically how HDAC inhibition downregulated p-MCL-1 (S64), or functionally prove that this was responsible for the observed BIM dissociation. In addition, further studies are required to determine how LBH589 dissociates BIM from BCL-XL.
MM cell lines that were resistant to MEK+HDAC inhibition had markedly more BIM:BCL-2 complexes at baseline. Combining ABT-199 (venetoclax) with HDAC inhibitors synergistically killed these cell lines (Figure 7B). ABT-199 is a highly potent BCL-2 inhibitor which, in just a few years, has altered the treatment landscape of chronic lymphocytic leukemia.41 It has also produced promising clinical responses in many other hematologic malignancies and several non-Hodgkin lymphomas.42 More relevant to our study, single-agent ABT-199 recently produced encouraging clinical responses in patients with relapsed/refractory MM, particularly those with t(11;14) who had high BCL2 expression.43 It is conceivable that HDAC inhibition could either expand the pool of patients who would be sensitive to BCL-2 inhibition, or deepen the responses of partially sensitive patients.
However, it should be noted that concomitant HDAC inhibition did not sensitize all MM cell lines to BCL-2 inhibition, as evidenced by the in vitro resistance of RAS/RAF mutant, MCL-1-primed MM cell lines to BCL-2 + HDAC inhibition. Even so, it is well documented that increased sequestration of BH3-only proteins by MCL-1 and BCL-XL is a major resistance mechanism of BH3 mimetics.44 Perhaps then, HDAC inhibition could re-sensitize patients, who relapse after ABT-199 treatment, to BCL-2 inhibition. It is also worth noting that several specific MCL-1 inhibitors are actively being developed for use in MM.45 Speculatively, cells that acquire resistance to MCL-1 inhibition could be investigated to determine whether they become more BCL-2-dependent, and thereby sensitive to the ABT-199/LBH589 combination.
Notably, we demonstrated markedly increased apoptosis in plasma cells sorted from MM patients with a wide variety of clinical characteristics after treatment with the MEK + HDAC and BCL-2 + HDAC inhibitor combinations ex vivo. However, there were too few patients’ samples to discern if the type or number of prior lines of therapy, or particular cytogenetic abnormalities predicted for sensitivity to either combination. Even so, it is conceivable that readily discernible subgroups of patients, beyond the subgroups formed on the basis of RAS/RAF mutational status, may be identified and that these subgroups could be more likely to respond to either combination therapy, akin to patients with t(11;14) treated with ABT-199.43
Using both pharmacological and genetic methods, we determined that simultaneous inhibition of HDAC1 and HDAC2 was sufficient to induce apoptosis when combined with either MEK or BCL-2 inhibition. This is of interest because the clinical utility of the pan-HDAC inhibitor LBH589 in MM has been limited by toxicities, mostly related to diarrhea, fatigue, lymphopenia and thrombocytopenia,15 which could perhaps be avoided with more specific HDAC inhibition. MS275 (entinostat), an HDAC1, 2, and 3 inhibitor, is actively being investigated in clinical trials for numerous tumor types.46 Moreover, FK228 (romidepsin), which mostly inhibits HDAC1 and HDAC2, has been approved by the Food and Drug Administration for the treatment of several T-cell lymphomas.
Specific HDAC1+2 inhibition has previously been shown to drive apoptosis in lymphoid cell lines.47 In MM, a prior study showed that the apoptosis induced by HDAC inhibitors is mainly mediated by HDAC class I inhibition.48 Interestingly, elevated HDAC1 expression has been correlated with poor prognosis in MM.49 In our study, we observed that both HDAC1 and HDAC2 must be inhibited for the synergistic effect with AZD6244 or ABT-199. Given the genetic and functional similarity between HDAC1 and 2, we think that it will be important to target both of these proteins simultaneously to attain meaningful clinical responses. It must be noted that while our approach using HDAC inhibition holds considerable promise in MM and other cancers, the pharmacological specificities of HDAC inhibitors have been inconsistently described to date, and often do not include specificity for particular HDAC1/2-containing multiprotein complexes such as CoREST.14 Speculatively, perhaps inhibition of these complexes is important for the mechanisms described in the present study, rather than inhibition of the isolated enzymatic activity of HDAC1 and 2. If so, compounds with greater specificity for CoREST or other HDAC1/2-containing complexes could achieve greater efficacy with potentially less toxicity.50
Acknowledgments
We thank Dr. Scott Kaufmann, M.D., Ph.D. for helpful discussions and commentary during the writing of this manuscript. We also thank the patients and their families for the gracious donation of their bone marrow aspirates to our laboratory. This work was funded by the Predolin Foundation, Mayo Clinic Hematology Small Grants Program, International Myeloma Foundation, CTSA grant number TL1TR002380 from the National Center for Advancing Translational Sciences (NCATS), Mayo Clinic Multiple Myeloma SPORE, Wendy Will Case Cancer Fund, Mayo Clinic Development Funds from the Myeloma, Amyloidosis and Dysproteinemia Disease Oriented Group (MADDOG), and the Mayo Clinic Office of Translation to Practice. Additionally, we would like to thank the American Association for Cancer Research for a Scholar-in-Training Award, and the American Society of Hematology for an HONORS award.
Footnotes
Parts of this manuscript were presented in abstract form at the American Association for Cancer Research (AACR) Annual Meeting on April 17, 2018 in Chicago, IL, USA.
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/10/2061
References
- 1.Rajkumar SV. Treatment of multiple myeloma. Nat Rev Clin Oncol. 2011;8(8):479-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kyle RA, Therneau TM, Rajkumar SV, et al. Prevalence of monoclonal gammopathy of undetermined significance. N Engl J Med. 2006;354(13):1362-1369. [DOI] [PubMed] [Google Scholar]
- 3.Noone AM, Howlader N, Krapcho M, et al. SEER Cancer Statistics Review, 1975-2015, National Cancer Institute; Bethesda, MD, https://seer.cancer.gov/csr/1975_2015/, based on November 2017 SEER data submission, posted to the SEER web site, April 2018. [Google Scholar]
- 4.Kumar SK, Dispenzieri A, Lacy MQ, et al. Continued improvement in survival in multiple myeloma: changes in early mortality and outcomes in older patients. Leukemia. 2014;28(5):1122-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonneveld P, Avet-Loiseau H, Lonial S, et al. Treatment of multiple myeloma with high-risk cytogenetics: a consensus of the International Myeloma Working Group. Blood. 2016;127(24):2955-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar SK, Rajkumar SV. The multiple myelomas — current concepts in cytogenetic classification and therapy. Nat Rev Clin Oncol. 2018;15(7):409-421. [DOI] [PubMed] [Google Scholar]
- 7.Chng WJ, Gonzalez-Paz N, Price-Troska T, et al. Clinical and biological significance of RAS mutations in multiple myeloma. Leukemia. 2008;22(12):2280-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lentzsch S, Chatterjee M, Gries M, et al. PI3-K/AKT/FKHR and MAPK signaling cascades are redundantly stimulated by a variety of cytokines and contribute independently to proliferation and survival of multiple myeloma cells. Leukemia. 2004;18(11): 1883-1890. [DOI] [PubMed] [Google Scholar]
- 9.Walker BA, Boyle EM, Wardell CP, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015;33(33):3911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasmussen T, Kuehl M, Lodahl M, Johnsen HE, Dahl IM. Possible roles for activating RAS mutations in the MGUS to MM transition and in the intramedullary to extramedullary transition in some plasma cell tumors. Blood. 2005;105(1):317-323. [DOI] [PubMed] [Google Scholar]
- 11.Kortüm KM, Mai EK, Hanafiah NH, et al. Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes. Blood. 2016;128(9):1226-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramakrishnan V, D’Souza A. Signaling pathways and emerging therapies in multiple myeloma. Curr Hematol Malig Rep. 2016;11(2):156-164. [DOI] [PubMed] [Google Scholar]
- 13.Holkova B, Zingone A, Kmieciak M, et al. A phase 2 trial of AZD6244 (selumetinib, ARRY-142886), an oral MEK1/2 inhibitor, in relapsed/refractory multiple myeloma. Clin Cancer Res. 2016;22(5):1067-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124(1):30-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laubach JP, Moreau P, San-Miguel JF, Richardson PG. Panobinostat for the treatment of multiple myeloma. Clin Cancer Res. 2015;21(21):4767-4773. [DOI] [PubMed] [Google Scholar]
- 16.Meng J, Fang B, Liao Y, Chresta CM, Smith PD, Roth JA. Apoptosis induction by MEK inhibition in human lung cancer cells is mediated by Bim. PLoS One. 2010;5(9):e13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang YF, Jiang CC, Kiejda KA, Gillespie S, Zhang XD, Hersey P. Apoptosis induction in human melanoma cells by inhibition of MEK is caspase-independent and mediated by the Bcl-2 family members PUMA, Bim, and Mcl-1. Clin Cancer Res. 2007;13(16): 4934-4942. [DOI] [PubMed] [Google Scholar]
- 18.Chen S, Dai Y, Pei XY, Grant S. Bim upregulation by histone deacetylase inhibitors mediates interactions with the Bcl-2 antagonist ABT-737: evidence for distinct roles for Bcl-2, Bcl-xL, and Mcl-1. Mol Cell Bio. 2009;29(23):6149-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramakrishnan V, Kimlinger T, Timm M, Haug J, Rajkumar SV, Kumar S. Multiple mechanisms contribute to the synergistic anti-myeloma activity of the pan-histone deacetylase inhibitor LBH589 and the rapalog RAD001. Leuk Res. 2014;38(11):1358-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Correia C, Lee S-H, Meng XW, et al. Emerging understanding of Bcl-2 biology: Implications for neoplastic progression and treatment. Biochim Biophys Acta. 2015; 1853(7):1658-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar SK, Dimopoulos MA, Kastritis E, et al. Natural history of relapsed myeloma, refractory to immunomodulatory drugs and proteasome inhibitors: a multicenter IMWG study. Leukemia. 2017;31(11): 2443-2448. [DOI] [PubMed] [Google Scholar]
- 22.Ramakrishnan V, Ansell S, Haug J, et al. MRK003, a g-secretase inhibitor exhibits promising in vitro pre-clinical activity in multiple myeloma and non-Hodgkin’s lymphoma. Leukemia. 2012;26(2):340-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S, Raje N, Hideshima T, et al. Antimyeloma activity of two novel N-substituted and tetraflourinated thalidomide analogs. Leukemia. 2005;19(7):1253-1261. [DOI] [PubMed] [Google Scholar]
- 24.Chou T-C, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27-55. [DOI] [PubMed] [Google Scholar]
- 25.Ghobrial IM, Detappe A, Anderson KC, Steensma DP. The bone-marrow niche in MDS and MGUS: implications for AML and MM. Nat Rev Clin Oncol. 2018;15(4):219-233. [DOI] [PubMed] [Google Scholar]
- 26.Cragg MS, Jansen ES, Cook M, et al. Treatment of B-RAF mutant human tumor cells with a MEK inhibitor requires Bim and is enhanced by a BH3 mimetic. J Clin Invest. 2008;118(11):3651-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai H, Ding H, Meng XW, Lee SH, Schneider PA, Kaufmann SH. Contribution of Bcl-2 phosphorylation to Bak binding and drug resistance. Cancer Res. 2013;73(23): 6998-7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Domina AM, Vrana JA, Gregory MA, Hann SR, Craig RW. MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene. 2004;23(31):5301-5315. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi S, Lee S-H, Meng XW, et al. Serine 64 phosphorylation enhances the antiapoptotic function of Mcl-1. J Biol Chem. 2007;282(25):18407-18417. [DOI] [PubMed] [Google Scholar]
- 30.Choudhary GS, Tat TT, Misra S, et al. Cyclin E/Cdk2-dependent phosphorylation of Mcl-1 determines its stability and cellular sensitivity to BH3 mimetics. Oncotarget. 2015;6(19):16912-16925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yee AJ, Bensinger WI, Supko JG, et al. Ricolinostat plus lenalidomide, and dexame -thasone in relapsed or refractory multiple myeloma: a multicentre phase 1b trial. Lancet Oncol. 2016;17(11):1569-1578. [DOI] [PubMed] [Google Scholar]
- 32.Tatamiya T, Saito A, Sugawara T, Nakanishi O. Isozyme-selective activity of the HDAC inhibitor MS-275. Cancer Res. 2004;64(7):567. [Google Scholar]
- 33.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018;173(2):321-37.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Adjei AA. The clinical development of MEK inhibitors. Nat Rev Clin Oncol. 2014;11(7):385-400. [DOI] [PubMed] [Google Scholar]
- 35.Mulligan G, Lichter DI, Di Bacco A, et al. Mutation of NRAS but not KRAS significantly reduces myeloma sensitivity to single-agent bortezomib therapy. Blood. 2004;123(5):632-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domina AM, Vrana JA, Gregory MA, Hann SR, Craig RW. MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene. 2004;23(31):5301-5315. [DOI] [PubMed] [Google Scholar]
- 37.Wuilleme-Toumi S, Robillard N, Gomez P, et al. Mcl-1 is overexpressed in multiple myeloma and associated with relapse and shorter survival. Leukemia. 2005;19(7):1248-1252. [DOI] [PubMed] [Google Scholar]
- 38.Gores GJ, Kaufmann SH. Selectively targeting Mcl-1 for the treatment of acute myel- ogenous leukemia and solid tumors. Genes Dev. 2012;26(4):305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rooswinkel RW, van de Kooij B, de Vries E, et al. Antiapoptotic potency of Bcl-2 proteins primarily relies on their stability, not binding selectivity. Blood. 2014;123(18): 2806-2815. [DOI] [PubMed] [Google Scholar]
- 40.Torres-Adorno AM, Lee J, Kogawa T, et al. Histone deacetylase inhibitor enhances the efficacy of MEK inhibitor through NOXA-mediated MCL1 degradation in triple-negative and inflammatory breast cancer. Clin Cancer Res. 2017;23(16):4780-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts AW, Davids MS, Pagel JM, et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med. 2016;374(4):311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davids MS, Roberts AW, Seymour JF, et al. Phase I first-in-human study of venetoclax in patients with relapsed or refractory non-Hodgkin lymphoma. J Clin Oncol. 2017;35(8):826-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S, Kaufman JL, Gasparetto C, et al. Efficacy of venetoclax as targeted therapy for relapsed/refractory t(11;14) multiple myeloma. Blood. 2017;130(22):2401-2409. [DOI] [PubMed] [Google Scholar]
- 44.Bose P, Gandhi V, Konopleva M. Pathways and mechanisms of venetoclax resistance. Leuk Lymphoma. 2017;58(9):1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caenepeel S, Brown SP, Belmontes B, et al. AMG 176, a selective MCL1 inhibitor, is effective in hematologic cancer models alone and in combination with established therapies. Cancer Discov. 2018;8(12):1582-1597. [DOI] [PubMed] [Google Scholar]
- 46.Surolia I, Bates SE. Entinostat finds a path: a new study elucidates effects of the histone deacetylase inhibitor on the immune system. Cancer. 2018;124(24):4597-4600. [DOI] [PubMed] [Google Scholar]
- 47.Stubbs MC, Kim W, Bariteau M, et al. Selective inhibition of HDAC1 and HDAC2 as a potential therapeutic option for B-ALL. Clin Cancer Res. 2015;21(10):2348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mithraprabhu S, Khong T, Jones SS, Spencer A. Histone deacetylase (HDAC) inhibitors as single agents induce multiple myeloma cell death principally through the inhibition of class I HDAC. Br J Haematol. 2013;162(4):559-562. [DOI] [PubMed] [Google Scholar]
- 49.Mithraprabhu S, Kalff A, Chow A, Khong T, Spencer A. Dysregulated class I histone deacetylases are indicators of poor prognosis in multiple myeloma. Epigenetics. 2014;9(11):1511-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalin JH, Wu M, Gomez AV, et al. Targeting the CoREST complex with dual histone deacetylase and demethylase inhibitors. Nat Commun. 2018;9(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]