Abstract
One of the major functions of the protein homeostasis system is to maintain proteins in their soluble states, and indeed several human disorders are associated with the aberrant aggregation of proteins. An active involvement of the protein homeostasis system is necessary to avoid aggregation because proteins are expressed at levels close to their solubility limits, hence being poorly soluble. The mechanisms by which the protein homeostasis system acts to control protein aggregation are, however, still not known in much detail. To facilitate systematic investigations of these mechanisms, we describe here the CamSol method of predicting protein solubility, and illustrate its initial applications. We anticipate that with the advent of powerful proteomics and transcriptomic methods, in combination with the use of the CamSol method and related approaches to predict the solubility and other biophysical properties of proteins, it will become possible to increase our understanding of the principles of protein homeostasis related to the maintenance of the proteome in its soluble form.
PROTEIN SOLUBILITY
The solubility of a substance is defined by the value of the concentration at which soluble and insoluble phases are in equilibrium (Chaikin et al. 1995). The solubility therefore fundamentally depends on the physical and chemical properties of the substance itself, as well as on the properties of its environment, including in particular the composition of the solvent, the temperature, the pH, and the ionic strength. Operatively, the solubility of a substance can be measured by its saturation concentration, also known as critical concentration, which is the concentration of the soluble fraction of the substance in the presence of an insoluble fraction. One can also think about the solubility as the particular concentration for which adding more substance does not increase its concentration in solution, but triggers instead its precipitation.
As a wide variety of proteins are functional in their soluble forms, and are associated with disease in their insoluble forms, protein solubility is a major aspect of protein homeostasis (Vendruscolo et al. 2011). Defining the solubility of proteins, however, is difficult both theoretically and experimentally (Jain et al. 2017; Wolf Pérez et al. 2019). As given above, the definition of solubility is rigorous, but it only applies in a fully quantitative manner to substances that have well-defined soluble and insoluble phases. The vast majority of proteins, however, can populate a wide range of structurally heterogeneous states, including small and large oligomers, and it is thus challenging to distinguish between soluble and insoluble assemblies (Sormanni et al. 2015). In principle, oligomers capable of growing into larger aggregates can be considered as insoluble, but it is, in practice, difficult to measure the concentrations of growth-competent oligomers in a given sample. Despite this problem, it is still useful to consider protein solubility measurements in the study of protein homeostasis because the overall concentration of the oligomeric species is typically much smaller than that of monomers and large aggregates, so that the contribution of these oligomeric species to the value of the solubility is relatively small (Sormanni et al. 2015).
PROTEIN SOLUBILITY, PROTEIN HOMEOSTASIS, AND HUMAN DISEASE
The phenomenon of protein aggregation is associated with a wide range of human disorders, including Alzheimer's and Parkinson's diseases (Dobson 1999; Balch et al. 2008; Eisenberg and Jucker 2012; Knowles et al. 2014). These diseases can have different symptoms and affect different organs and tissues, but are all characterized by the dysfunctional aggregation of specific proteins. Although these diseases are associated with proteins with different sequences and native structures, most of these proteins seem to follow a generic behavior in misfolding and aggregation (Dobson 1999; Knowles et al. 2014). Such behavior involves the formation of many different types of aggregates, from small oligomers to large amyloid fibrils, yet all these categories of aggregates appear to share common structural features that do not depend on the particular proteins they originated from (Dobson 1999; Balch et al. 2008; Eisenberg and Jucker 2012; Knowles et al. 2014). Indeed, it is now well-established that almost every protein, and not just those that are disease-related, can aggregate in vitro under appropriate experimental conditions, and the resulting aggregates can be toxic for cells (Bucciantini et al. 2002; Selkoe 2003; Eisenberg and Jucker 2012; Chiti and Dobson 2017). Depending on the protein under scrutiny, these conditions may be more or less harsh in terms of pH, concentration, temperature, or presence of chemicals (e.g., trifluoroethanol, TFE), but essentially all proteins can aggregate (Dobson 1999; Knowles et al. 2014).
The fundamental nature of the aggregated state and its manifestations in living organisms has been increasingly recognized and has led to the observation of the phenomenon of widespread protein aggregation, whereby a large fraction of the proteome undergoes aggregation in vivo (David et al. 2010; Olzscha et al. 2011; Reis-Rodrigues et al. 2012; Ciryam et al. 2013; Walther et al. 2015). It has been realized that the origin of this phenomenon is that proteins are expressed at levels close to their solubility limits (Tartaglia et al. 2007; Baldwin et al. 2011), therefore being supersaturated (Ciryam et al. 2015). Taken together, all these observations indicate that the ability to aggregate and form fibrils can be regarded as an intrinsic property of polypeptide chains, even if the aggregation rates can vary dramatically among different proteins. To respond to the constant risk of aggregation, a sophisticated protein homeostasis system has evolved to maintain the proteome in a functional state (Balch et al. 2008; Hartl et al. 2011; Labbadia and Morimoto 2015).
Folded proteins can aggregate through at least two possible paths, depending on the protein under scrutiny and the experimental conditions (Chiti and Dobson 2017). In one path, protein molecules unfold at least partially before associating with each other. This process results in the formation of small oligomers, which can then grow in size forming more structured aggregates. Examples of proteins in this class are serpins, where aggregation is believed to initiate from a folding intermediate (Carrell and Lomas 1997). In an alternative path, proteins may aggregate directly from their native structures forming a first assembly that can remain functional. This is the case, for instance, for most antibody molecules, and other proteins that expose “sticky” regions for functional reasons, to bind other proteins or to form complexes (Pechmann et al. 2009). Once a critical number of molecules is present, so that the enthalpy associated with their ordered stacking overcomes the corresponding loss of entropy, the aggregates may evolve into protofibrils and ultimately into amyloid fibrils (Knowles et al. 2014). In this context we note that increasing amounts of evidence suggest that the small oligomers, rather than the large aggregates or amyloid fibrils, may be the most neurotoxic species (Lambert et al. 1998; Bucciantini et al. 2002; Haass and Selkoe 2007; Benilova et al. 2012; Jongbloed et al. 2015; Cline et al. 2018).
BIOPHYSICAL PRINCIPLES FOR THE PREDICTION OF PROTEIN AGGREGATION PROPENSITY
Although amyloid fibrils and amorphous aggregates are fundamental states of polypeptide chains (Knowles et al. 2014), the propensity to aggregate under physiological conditions varies substantially from protein to protein (Walther et al. 2015). To uncover the principles underlying this propensity, major advances have been made in understanding how the amino acid sequences determine the conformational properties of proteins. Three factors in particular have been shown to greatly affect the propensity to aggregate. The first one is the sequence composition, as the biophysical properties of the residues, in particular hydrophobicity, charge, and secondary structure preferences, have been shown to be the main determinant of the aggregation rates (Chiti et al. 2003; DuBay et al. 2004; Fernandez-Escamilla et al. 2004; Pawar et al. 2005; Tartaglia et al. 2008; Tartaglia and Vendruscolo 2008). The second one, which is particularly important for those proteins that tend to form native-like oligomers, is represented by the amino acids that are solvent-exposed on the surface of the native state (Tartaglia et al. 2008; Tartaglia and Vendruscolo 2008). Most, although not all, proteins tend to bury aggregation-promoting residues in the core of their three-dimensional structures. Yet, when such residues are exposed to the solvent for functional reasons, they can be the major driving force of aggregation (Pechmann et al. 2009). The third is the thermodynamic stability of the native state itself against unfolding. Less stable proteins, through large conformational fluctuations under native conditions, have more chances to expose hydrophobic residues that can promote aggregation (Tartaglia et al. 2008; Tartaglia and Vendruscolo 2008). These principles have been used for the development of a wide range of methods for predicting the aggregation propensity (Fernandez-Escamilla et al. 2004; Tartaglia and Vendruscolo 2008; Maurer-Stroh et al. 2010; Zambrano et al. 2015; Pallarès and Ventura 2016) and solubility (Magnan et al. 2009; Agostini et al. 2012; Smialowski et al. 2012) of proteins. Here we describe in particular the work that has led to the introduction of the CamSol method (Sormanni et al. 2015) and illustrate some of its applications.
THE CamSOL METHOD
Biophysical Principles of Protein Solubility and the Development of the CamSol Method
The pioneering work by Chiti and coworkers demonstrated that the changes in the aggregation rates of mutational variants of peptides can be accurately predicted using a linear combination of biophysical properties of their amino acid sequences (Chiti et al. 2003). Statistically significant correlations were observed between the changes in the aggregation rates resulting from single amino acid mutations and the corresponding changes in three biophysical properties of the polypeptide chain—hydrophobicity, charge, and the propensity to adopt α-helical and β-sheet secondary structures. The linear combination of biophysical properties was expressed as
| (1) |
where k is the aggregation rate of the wild-type peptide, k′ is the one carrying the single mutation, log denotes the natural logarithm and ΔIhydr, ΔIss, ΔIch are, respectively, the differences between the mutant and wild-type in hydrophobicity (Ihydr), secondary-structure propensity (Iss), and charge (Ich). This formula reproduced to a very good extent (r = 0.85) the changes in the aggregation rates measured experimentally for single amino acid substitutions for a series of peptides and unstructured proteins (Chiti et al. 2003).
This approach was then extended to predict the absolute aggregation rates of proteins, not just their changes upon amino acid substitutions (DuBay et al. 2004). These absolute aggregation rates strongly depend on factors that are extrinsic to the amino acid sequence, including pH, ionic strength, temperature, and, in particular, the concentration of the aggregating peptide or protein, and therefore these factors should be included in the calculations (DuBay et al. 2004).
Further developments of this approach resulted in the Zyggregator method, which extended the applicability and the accuracy of the predictions by incorporating new terms, such as the presence of hydrophobic/hydrophilic pattern of residues (Ipat) and of gatekeeper residues (Igk), which are charged residues of the same sign that flank hydrophobic regions (Pawar et al. 2005; Tartaglia and Vendruscolo 2008; Tartaglia et al. 2008). Importantly, this approach was also generalized to enable the identification of aggregation-prone regions within protein or peptides sequences, with a particular focus of predicting amyloid-promoting regions within disease-related proteins (Pawar et al. 2005; Tartaglia and Vendruscolo 2008; Tartaglia et al. 2008).
By building on these advances, in 2015 we introduced the CamSol method for the prediction of protein solubility (Sormanni et al. 2015). Although the solubility and the aggregation rate of a protein are related, and both dependent on the biophysical properties of amino acid sequences, they are not entirely equivalent. The solubility of a protein depends on the free energy difference between the native and the aggregated states, whereas its aggregation rate depends on the free energy barrier between these two states (Sormanni et al. 2015). To obtain accurate predictions, CamSol first calculates a solubility profile, which consists of a score for each residue in the sequence and is highly sensitive to the local context, and then it calculates an overall solubility score from the profile itself. The solubility profile can be calculated directly from the amino acid sequence (intrinsic profile) or from a structure of the protein under scrutiny (structurally corrected profile) (Sormanni et al. 2015).
The Intrinsic Solubility Profile
In the CamSol method, a linear combination of biophysical properties similar to Equation 1 is first employed to calculate a solubility score for each residue (Sormanni et al. 2015)
| (2) |
where , , , and are, respectively, the hydrophobicity, the charge at neutral pH, the α-helix and β-strand propensities of residue i, while the a coefficients are the parameters of the linear combination. Then, a smoothing average is carried out, whereby each score is replaced by a central moving average over a window of seven amino acids, thus effectively replacing the contribution of the biophysical properties of individual residues with that of seven-residue fragments centered around the residue under scrutiny
| (3) |
In this expression, accounts for the presence of specific patterns of alternating hydrophobic and hydrophilic residues, and takes into account the gatekeeping effect of individual charges
| (4) |
where Ci + j is the charge of the amino acid i + j and b is a parameter that defines the length scale over which the effects of the gatekeeper residues are relevant. The smoothing average and the additional terms ensure that the predicted effect of an amino acid substitution on the solubility profile will depend both on the difference between the biophysical properties of the old and the new amino acids, and on the local context in which the mutation is carried out.
The Structurally Corrected Solubility Profile
If a three-dimensional structure or structural model is available for the protein of interest, it is possible to calculate a structurally corrected profile (Sormanni et al. 2015). This profile is defined by projecting the intrinsic solubility profile onto the surface and smoothing over a surface patch of size A and dimension rA. The structurally corrected solubility propensity score of residue i can be written as
| (5) |
where the sum is extended over all the residues of the protein within a distance rA from residue i, excluding the residues that are contiguous along the sequence, as their proximity effect is already encompassed by the intrinsic solubility score. and are, respectively, the “exposure weight,” which depends on the solvent exposure of residue j, and the “smoothing weight,” defined as
| (6) |
where dij is the distance of residue j from residue i. This definition implies that neighboring residues contribute more to the local surface aggregation propensity than more distant ones. Furthermore, the smoothing weight does not bias toward a preselected surface patch size. rA is set to be 8 Å, as this value is consistent with the seven amino acids window implemented in the prediction of the intrinsic solubility profile (a distance of 8 Å in fact spans approximately three residues in a compact globular protein).
The exposure weight is defined as
| (7) |
where xj is the relative exposure of residue j, that is, the solvent-accessible surface area (SASA) of this residue in the given structure divided by the SASA of same residue in a Gly-Xxx-Gly peptide in an extended conformation. The Heaviside step function, ϑ, is employed so that residues with <5% solvent-exposed are not taken into account. Equation 7 thus describes a sigmoidal function where a and b are parameters tuned so that the weight grows slowly to a relative exposure x ≈ 20% and then grows linearly reaching 1 at x ≈ 50%; this is accomplished by setting a = −10 and b = 0.3. When a residue is 50% solvent-exposed, half of it faces inward in the structure, whereas the other half, facing the solvent, already provides the largest surface for potential aggregation partners. With this correction, residues not exposed to the surface, such as those buried in the hydrophobic core and essential for the folding of a protein, are assigned a score close to zero and, consequently, are not considered in the subsequent steps of the CamSol algorithm.
The quantity in Equation 5 is the intrinsic solubility of residue j computed using a modified version of Equation 3, which reads
| (8) |
where the average over the seven-residue window in Equation 3 has been replaced here by a weighted average (over the same window) with weights , which are the relative exposures of residue j linearly rescaled in the range [0.25, 1], so division by zero never occurs.
Similarly, embodies the same idea as in Equation 4, but the gatekeeping effect of charges of the same sign is now computed in the three-dimensional space
| (9) |
where Cj is the net charge of residue j at neutral pH, the smoothing weight is computed here using twice the patch radius rA and the exposure weight using the relative exposure of the charged atom in residue j.
Although the calculation of the structurally corrected solubility profile requires the knowledge of the structure of the protein, there is no need for particularly high resolution. The predictions are accurate as long as the solvent exposure of the amino acids and their relative Cα distances are correctly represented. This fact makes the CamSol procedure applicable to a large number of cases where only the sequence is known, as a structure good enough to enable the solubility prediction can be obtained by standard techniques, such as homology modeling.
The CamSol Solubility Score
A solubility score for the whole protein is then calculated from the intrinsic profile by accounting for the contribution of poorly soluble regions as well as that of highly soluble ones. From the intrinsic solubility profile we derive an overall solubility score for the whole protein (Sormanni et al. 2017)
| (10) |
where Si is the value of the intrinsic solubility profile for the amino acid i and N is the length of the input sequence. The upper and lower thresholds thup and thlow, as well as the coefficients ωup, ωlow, γ, and δ are fitted with a Monte Carlo procedure aimed at maximizing both the absolute value of the correlation coefficient of SP with measurements of aggregation rates from the literature, and its ability to discriminate between nonaggregating and aggregating peptides and proteins collected through a systematic literature search, which contained totally unrelated sequences rather than mutational variants of the same protein (Sormanni et al. 2017). Since the scores computed with Equation 10 are dimensionless numbers, they are rescaled so that the mean value and standard deviation calculated among more than 106 random sequences are 0 and 1, respectively (Sp = [Sp − μrandom]/σrandom). Random sequences used in this calculation were generated with the same amino acid frequency and length distribution of the human proteome. In an initial validation (Sormanni et al. 2015) we demonstrated that this solubility score was highly accurate (R = 0.98) in recapitulating the effect on the solubility of a single-domain antibody of a maximum of three simultaneous mutations. Subsequently, to carry out proteome-wide analyses we expanded the applicability of the solubility score using the aforementioned equation so that it could quantitatively rank the solubility of more distantly related proteins (Walther et al. 2015), while retaining the possibility of carrying out highly quantitative screenings of mutational libraries (Sormanni et al. 2017).
APPLICATIONS OF THE CamSol METHOD
Fast Solubility Screening of Libraries of Protein Variants
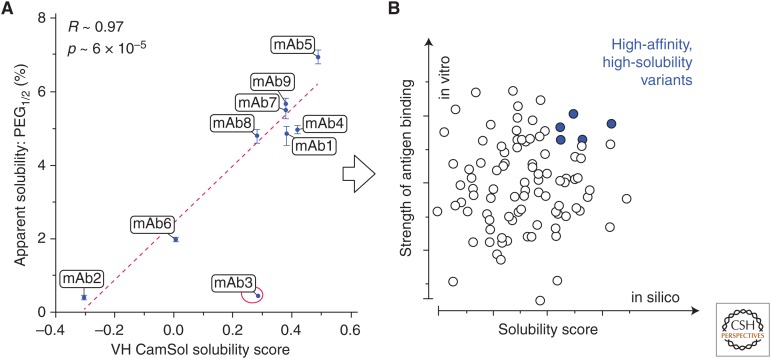
The ability of CamSol to rank the solubility of protein variants was tested using a phage-display-derived monoclonal antibody (mAb) library from MedImmune (Sormanni et al. 2017). The mAbs that we analyzed differ by up to 32 mutations in the Fv region, and a strong correlation was observed between calculated scores and corresponding solubility measurements (Fig. 1A). Similarly, a statistically significant correlation between CamSol predictions and solubility measurements was also reported for mutational variants of a troublesome mAb (Shan et al. 2018). Furthermore, in a recent study on a library of 17 mAbs, CamSol predictions were compared with a battery of commonly used developability assays and solubility measurements, and the correlations between CamSol and these experimental readouts were at par or better than those among the different assays with each other (Wolf Pérez et al. 2018). These results indicate that, since CamSol solubility predictions quickly run on laptops requiring only the amino acid sequences as input, this approach enables one to select from the beginning high-affinity (from library panning), high-solubility (from CamSol) antibodies, as shown in Figure 1B.
Figure 1.
(A) Scatterplot of the measured apparent solubility (expressed as the value of PEG1/2) of a directed-evolution-derived mAb library (Sormanni et al. 2017) as a function of the intrinsic CamSol solubility score calculated from the sequences of the heavy chain variable domain (VH) only, as the vast majority of mutations are found there. Regression lines, reported Pearson's coefficients of correlation (R), and corresponding p values (p) were calculated by excluding the outlier point circled in red (mAb3). (B) The computational prediction in A enabled the identification of the most soluble mAbs. Therefore, the screening of antibodies derived from an in vitro discovery experiment (e.g., phage display) may be performed using two parameters: (1) the measured binding strength (e.g., binding affinity or off-rate on the y-axis), and (2) the predicted solubility score calculated from the sequence (e.g., the CamSol-intrinsic solubility on the x-axis). The latter is readily computed from the amino acid sequence, thus enabling the selection of lead antibodies with high affinity and solubility from the very early stages of antibody discovery (adapted from Sormanni et al. 2017).
Identification of Aggregation-Promoting Hotspots
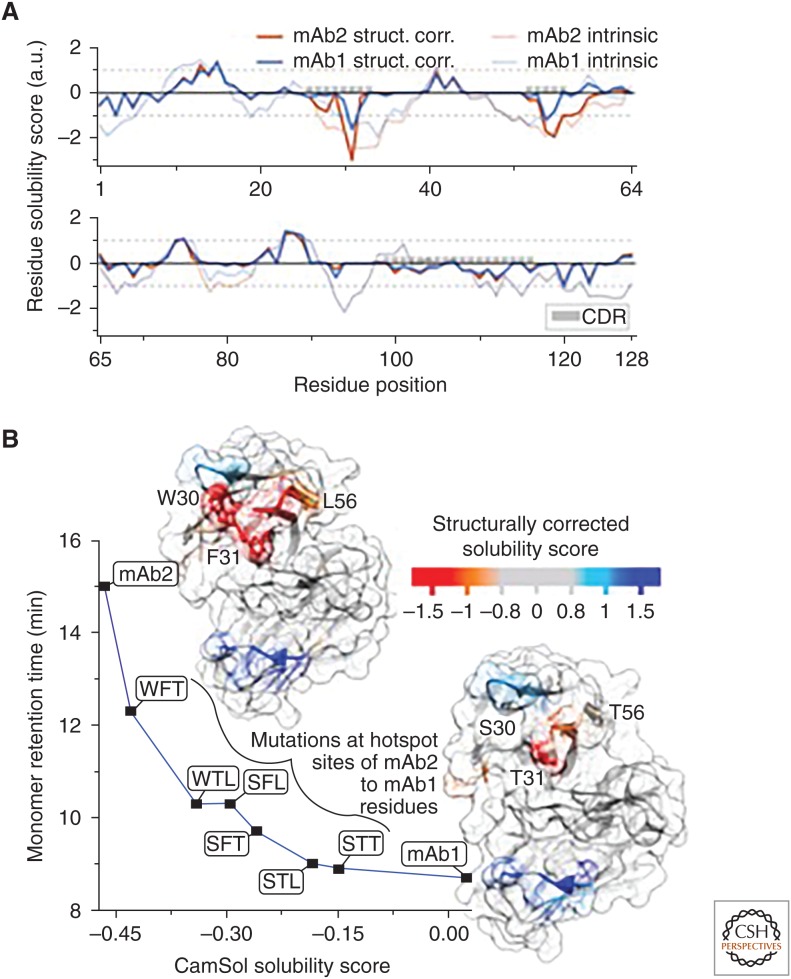
In addition to the results presented above, the structurally corrected CamSol calculation can be employed to identify aggregation-promoting hotspots on protein surfaces. In the example in Figure 2, the amino acids responsible for the increased self-association of mAb2 with respect to mAb1 were experimentally identified with a structural proteomics approach (Dobson et al. 2016) as W30, F31, L561, in perfect agreement with the CamSol predictions.
Figure 2.
(A) Structurally corrected (solid lines) and intrinsic (broken lines) CamSol solubility profiles for the VH domain of mAb1 (blue) and mAb2 (red), which are two of the monoclonal antibodies analyzed in Sormanni et al. (2017). The positions of the complemetarity-determining regions (CDRs) are highlighted with gray boxes. (B) The structurally corrected solubility profile is color coded on the surface of homology models of the VH/VL domains of mAb2 (top left) and mAb1 (lower right). The labeled residue positions on mAb2 (W30, F31, L57) are those that have been experimentally identified as aggregation hotspots (Dobson et al. 2016). Aggregation-promoting regions are in orange/red, whereas aggregation-protecting regions are in light blue/blue. The plot shows the measured high-performance size exclusion chromatography monomer retention time (Dobson et al. 2016) for various mAb variants as a function of their combined-chain solubility score calculated from the sequence alone. mAb2 has the residue types W, F, and L at the hotspot positions 30, 31, and 57, respectively, while mAb1 has S, T, and T. The six variants between mAb2 and mAb1 are named according to which mAb2 positions have been mutated to the corresponding mAb1 amino acid (e.g., WFT is mAb2 L57T). The line serves as a visual guide (adapted from Sormanni et al. 2017).
Rational Design of Protein Mutants with Enhanced Solubility
The CamSol method can be used to design antibodies or proteins with enhanced solubility starting from problematic molecules. In essence, the structurally corrected predictions reveal candidate sites for amino acid substitution (or in some cases insertions), and the intrinsic prediction can be used to quickly test all possible mutations at those sites, which will yield variants with improved solubility (Sormanni et al. 2015; Camilloni et al. 2016).
Stability and Solubility Trade-Offs and Link with Protein Aggregation
A large number of mutations on many unrelated proteins has been identified as leading to aggregation in vivo and to human diseases. Broadly speaking, these mutations can cause aggregation through two different pathways. In one case, the mutation destabilizes the native state, causing its partial or complete unfolding. As a consequence, poorly soluble regions usually buried in the hydrophobic core become exposed to the solvent thus triggering aggregation. In the other case, the mutation may occur on the surface of the protein or within a disordered region, thus impacting directly the solubility without substantially altering the native structure. The effect of mutations in the second class can readily be predicted with the CamSol method by using the amino acid sequence alone, as demonstrated for example by the solubility screening of the antibody library (Fig. 1), where all molecules were known to be folded and functional. However, the effect of mutations in the first class cannot be predicted by looking only at the solubility, as the aggregation is triggered by the fact that the solubility of the unfolded state is typically much lower than that of the native state, disregarding the impact on solubility of the specific mutation under scrutiny. However, when such disease-related mutations are already known, the CamSol method can be used to predict whether they belong the first or second class.
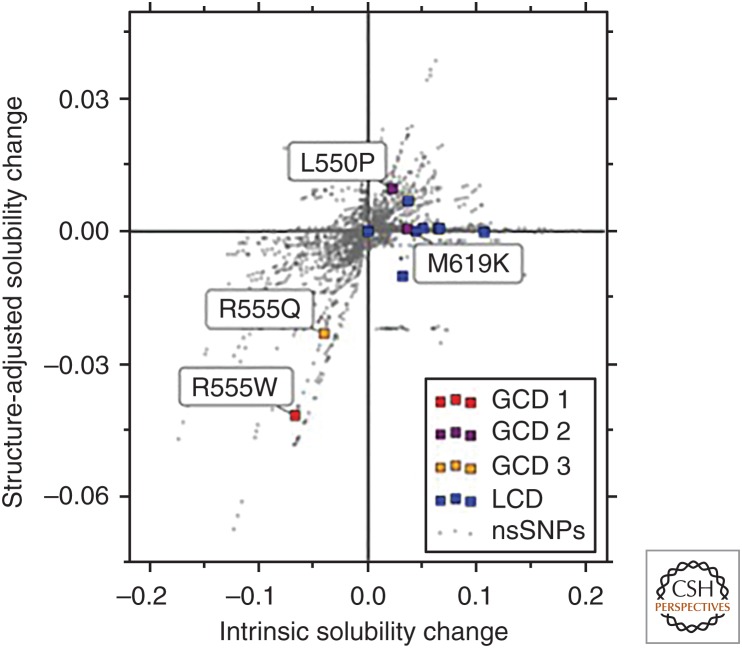
As an example, Figure 3 shows the predicted effect of all possible nonsynonymous single-nucleotide polymorphisms (nsSNPs) within the Fas1-4 domain of the corneal protein TGFBIp, which has some disease-related mutations leading to opaque extracellular deposits and corneal dystrophies (CDs) (Stenvang et al. 2018). The intrinsic score on the x-axis considers only the amino acid sequence, while the structure-adjusted score on the y-axis also includes information about whether residues are exposed or buried in the native state of monomeric TGFBIp. Out of all possible 771 point mutations, the mutation R555W associated with granular CD is the highest ranked known disease-related mutation in terms of both its predicted decrease in intrinsic and structure-adjusted solubility. Similarly, the mutation R555Q is predicted to decrease both solubility predictions more than any other known CD-associated mutation apart from R555W. Experimental data confirmed that both of these mutational variants are well folded and stable (Stenvang et al. 2018), thus indicating that aggregate formation and pathology in vivo are likely the result of reduced solubility of the folded state. Conversely, the other known disease-related mutations (colored points) fall on or near the x-axis suggesting that these mutations increase the aggregation propensity indirectly by decreasing the stability of the native fold, which is fully consistent with in vitro stability measurements of these mutants, as well as with the coexistence of amorphous and amyloid aggregates in this class of mutations (Stenvang et al. 2018).
Figure 3.
Predicted changes, using the CamSol method, in the intrinsic (x-axis) and structure-adjusted (y-axis) solubilities for all mutations from nonsynonymous single-nucleotide polymorphisms (nsSNPs) in Fas1-4 (Stenvang et al. 2018). Mutations associated with lattice corneal dystrophy (LCD) are shown in blue, and those associated with three subtypes of granular corneal dystrophy (GCD 1-3) are in red, purple, and orange for subtypes 1, 2, and 3, respectively. All LCD and GCD 2 mutations have little or no effect on the structure-adjusted solubility, consistent with the notion that they drive aggregation by indirectly increasing the aggregation propensity of Fas1-4 through destabilization of the native state. The GCD 1 mutation R555W is an extreme outlier in terms of decrease in solubility, according to both intrinsic and structure-adjusted predictions, and the GCD 3 mutation is predicted to decrease solubility more than any other corneal-dystrophy-associated mutation other than R555W, suggesting that GCD 1 and GCD 3 phenotypes are driven by loss of solubility of the folded Fas1-4 domain (adapted with permission from Stenvang et al. 2018).
USING CamSol TO INVESTIGATE THE LINKS BETWEEN PROTEIN SOLUBILITY AND PROTEIN HOMEOSTASIS
Proteome-Wide Predictions of Protein Solubility
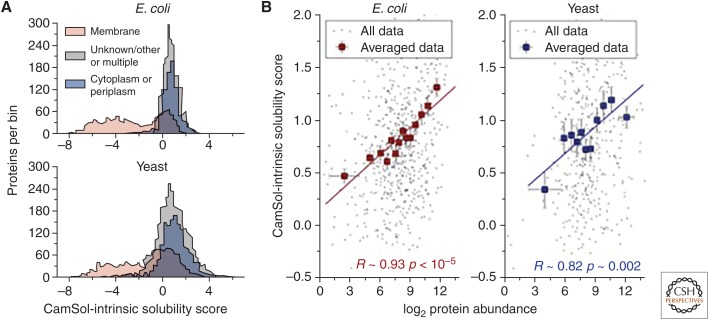
The results that we have described above show that the CamSol method is highly quantitative in predicting solubility changes upon mutations and in solubility screenings of protein libraries of similar sequences. In addition to these applications, CamSol can also be used to carry out proteome-wide studies so that useful insights can be obtained when considering average behaviors of groups of proteins. For example, running the CamSol-intrinsic prediction on complete proteomes readily reveals a bimodal distribution of solubility scores (Fig. 4) where membrane proteins form a group of low-solubility sequences while cytosolic and other proteins form a second group of higher solubility.
Figure 4.
(A) Distribution of CamSol solubility scores for the whole proteome of Escherichia coli (top) and yeast (lower). Proteins are separated according to their subcellular location (see legend) as annotated in the UniProt database (UniProt Consortium 2018). (B) Scatterplots between protein abundance as determined with mass-spectrometry measurements (x-axis) and CamSol solubility scores (y-axis). Only nonmembrane proteins for which abundance data were available (Leuenberger et al. 2017) are included in these plots. Individual data points are in black, whereas corresponding averaged points are in red for E. coli (left) and blue for yeast (right). Averaged points report the mean CamSol score and abundance level of groups of ∼50 proteins binned according to their abundance. Error bars are standard errors on the mean, and the reported Pearson's correlation coefficients (R) and associated p values (p) are calculated for the averaged data.
It was previously shown that protein solubility and cellular protein concentration correlate, indicating that protein solubility is tuned to their cellular concentration (Tartaglia et al. 2007). Indeed, the comparison of CamSol-intrinsic predictions with in vivo protein-abundance data measured by mass spectrometry reveals a very strong correlation (R = 0.93 and 0.82 for Escherichia coli and yeast, respectively) once proteins are binned according to their abundance levels. Conversely, when all data are considered individually, the correlation is much weaker, with Pearson coefficients of 0.3 and 0.2 for E. coli (663 nonmembrane proteins with measured abundance, p < 10−15) and yeast (529 points, p ∼ 3 × 10−5), respectively. Several factors may determine the lower correlation obtained when considering individual proteins. First, each data point in Figure 4B corresponds to one amino acid sequence for which abundance measurements were available, and the solubility prediction employed neglects the structural context (i.e., the fact that unrelated proteins expose their highly soluble and poorly soluble regions in different ways). In addition, many such sequences form complexes inside the cells, and the solubility of the formed complex is likely to be different from that of its monomeric subunits. It is also well known that protein abundance levels can vary substantially in response to external stimuli or environmental changes as well as during different phases of the cell cycle. In this context, because of the competing effects of random mutational drift, which on average decreases solubility, and of natural selection, which selects solubilizing mutations but only until each protein has become good enough to perform its function, a correlation should be expected only between the maximum concentration at which a protein can be expressed and its solubility. Finally, the correlation between total protein abundance and cellular concentration may not be perfect, as the relatively low abundance level may correspond to a very high protein concentration in some cellular compartments (Tartaglia and Vendruscolo 2009), and such proteins would need to be highly soluble to avoid aggregation.
It is therefore remarkable that on average such a strong correlation is observed between predicted solubility and measured protein abundance, which suggests that some of the potential sources of error discussed above cancel each other out when taking averages of groups of proteins. A similar correlation obtained with binned data (R = 0.77) was also reported when considering the number of aggregation-prone regions in place of the solubility (Tartaglia and Vendruscolo 2009; Ganesan et al. 2016).
CONCLUSIONS
We have summarized our current understanding of the biophysical principles of protein solubility and described how we used these principles to develop the CamSol method to enable quantitative predictions of this property. We anticipate that the development of approaches of this type, in combination with increasingly powerful proteomics and transcriptomic methods, will facilitate systematic investigations of the mechanisms by which the protein homeostasis system controls the aggregation of proteins at the proteome level.
Footnotes
Editors: Richard I. Morimoto, F. Ulrich Hartl, and Jeffery W. Kelly
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
- Agostini F, Vendruscolo M, Tartaglia GG. 2012. Sequence-based prediction of protein solubility. J Mol Biol 421: 237–241. 10.1016/j.jmb.2011.12.005 [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. 2008. Adapting proteostasis for disease intervention. Science 319: 916–919. 10.1126/science.1141448 [DOI] [PubMed] [Google Scholar]
- Baldwin AJ, Knowles TP, Tartaglia GG, Fitzpatrick AW, Devlin GL, Shammas SL, Waudby CA, Mossuto MF, Meehan S, Gras SL. 2011. Metastability of native proteins and the phenomenon of amyloid formation. J Am Chem Soc 133: 14160–14163. 10.1021/ja2017703 [DOI] [PubMed] [Google Scholar]
- Benilova I, Karran E, De Strooper B. 2012. The toxic Aβ oligomer and Alzheimer's disease: An emperor in need of clothes. Nat Neurosci 15: 349–357. 10.1038/nn.3028 [DOI] [PubMed] [Google Scholar]
- Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. 2002. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416: 507–511. 10.1038/416507a [DOI] [PubMed] [Google Scholar]
- Camilloni C, Sala BM, Sormanni P, Porcari R, Corazza A, De Rosa M, Zanini S, Barbiroli A, Esposito G, Bolognesi M, et al. 2016. Rational design of mutations that change the aggregation rate of a protein while maintaining its native structure and stability. Sci Rep 6: 25559 10.1038/srep25559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell RW, Lomas DA. 1997. Conformational disease. Lancet 350: 134–138. 10.1016/S0140-6736(97)02073-4 [DOI] [PubMed] [Google Scholar]
- Chaikin PM, Lubensky TC, Witten TA. 1995. Principles of condensed matter physics. Cambridge University Press, Cambridge. [Google Scholar]
- Chiti F, Dobson CM. 2017. Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu Rev Biochem 86: 27–68. 10.1146/annurev-biochem-061516-045115 [DOI] [PubMed] [Google Scholar]
- Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. 2003. Rationalization of the effects of mutations on peptide andprotein aggregation rates. Nature 424: 805–808. 10.1038/nature01891 [DOI] [PubMed] [Google Scholar]
- Ciryam P, Tartaglia GG, Morimoto RI, Dobson CM, Vendruscolo M. 2013. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep 5: 781–790. 10.1016/j.celrep.2013.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P, Kundra R, Morimoto RI, Dobson CM, Vendruscolo M. 2015. Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases. Trends Pharmacol Sci 36: 72–77. 10.1016/j.tips.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline EN, Bicca MA, Viola KL, Klein WL. 2018. The amyloid-β oligomer hypothesis: Beginning of the third decade. J Alzheimers Dis 64: S567–S610. 10.3233/JAD-179941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. 2010. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol 8: e1000450 10.1371/journal.pbio.1000450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM. 1999. Protein misfolding, evolution and disease. Trends Biochem Sci 24: 329–332. 10.1016/S0968-0004(99)01445-0 [DOI] [PubMed] [Google Scholar]
- Dobson CL, Devine PW, Phillips JJ, Higazi DR, Lloyd C, Popovic B, Arnold J, Buchanan A, Lewis A, Goodman J, et al. 2016. Engineering the surface properties of a human monoclonal antibody prevents self-association and rapid clearance in vivo. Sci Rep 6: 38644 10.1038/srep38644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBay KF, Pawar AP, Chiti F, Zurdo J, Dobson CM, Vendruscolo M. 2004. Prediction of the absolute aggregation rates of amyloidogenic polypeptide chains. J Mol Biol 341: 1317–1326. 10.1016/j.jmb.2004.06.043 [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Jucker M. 2012. The amyloid state of proteins in human diseases. Cell 148: 1188–1203. 10.1016/j.cell.2012.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. 2004. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol 22: 1302–1306. 10.1038/nbt1012 [DOI] [PubMed] [Google Scholar]
- Ganesan A, Siekierska A, Beerten J, Brams M, Van Durme J, De Baets G, Van der Kant R, Gallardo R, Ramakers M, Langenberg T. 2016. Structural hot spots for the solubility of globular proteins. Nat Commun 7: 10816 10.1038/ncomms10816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. 2007. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer's amyloid β-peptide. Nat Rev Mol Cell Biol 8: 101–112. 10.1038/nrm2101 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. 2011. Molecular chaperones in protein folding and proteostasis. Nature 475: 324–332. 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- Jain T, Sun T, Durand S, Hall A, Houston NR, Nett JH, Sharkey B, Bobrowicz B, Caffry I, Yu Y. 2017. Biophysical properties of the clinical-stage antibody landscape. Proc Natl Acad Sci 114: 944–949. 10.1073/pnas.1616408114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongbloed W, Bruggink KA, Kester MI, Visser P-J, Scheltens P, Blankenstein MA, Verbeek MM, Teunissen CE, Veerhuis R. 2015. Amyloid-β oligomers relate to cognitive decline in Alzheimer's disease. J Alzheimers Dis 45: 35–43. 10.3233/JAD-142136 [DOI] [PubMed] [Google Scholar]
- Knowles TP, Vendruscolo M, Dobson CM. 2014. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol 15: 384–396. 10.1038/nrm3810 [DOI] [PubMed] [Google Scholar]
- Labbadia J, Morimoto RI. 2015. The biology of proteostasis in aging and disease. Annu Rev BIochem 84: 435–464. 10.1146/annurev-biochem-060614-033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert MP, Barlow A, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan T, Rozovsky I, Trommer B, Viola KL. 1998. Diffusible, nonfibrillar ligands derived from Aβ1-42 are potent central nervous system neurotoxins. Proc Natl Acad Sci 95: 6448–6453. 10.1073/pnas.95.11.6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuenberger P, Ganscha S, Kahraman A, Cappelletti V, Boersema PJ, von Mering C, Claassen M, Picotti P. 2017. Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science 355: eaai7825 10.1126/science.aai7825 [DOI] [PubMed] [Google Scholar]
- Magnan CN, Randall A, Baldi P. 2009. SOLpro: Accurate sequence-based prediction of protein solubility. Bioinformatics 25: 2200–2207. 10.1093/bioinformatics/btp386 [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Debulpaep M, Kuemmerer N, De La Paz ML, Martins IC, Reumers J, Morris KL, Copland A, Serpell L, Serrano L. 2010. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat Methods 7: 237–242. 10.1038/nmeth.1432 [DOI] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. 2011. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144: 67–78. 10.1016/j.cell.2010.11.050 [DOI] [PubMed] [Google Scholar]
- Pallarès I, Ventura S. 2016. Understanding and predicting protein misfolding and aggregation: Insights from proteomics. Proteomics 16: 2570–2581. [DOI] [PubMed] [Google Scholar]
- Pawar AP, Dubay KF, Zurdo J, Chiti F, Vendruscolo M, Dobson CM. 2005. Prediction of “aggregation-prone” and “aggregation-susceptible” regions in proteins associated with neurodegenerative diseases. J Mol Biol 350: 379–392. 10.1016/j.jmb.2005.04.016 [DOI] [PubMed] [Google Scholar]
- Pechmann S, Levy ED, Tartaglia GG, Vendruscolo M. 2009. Physicochemical principles that regulate the competition between functional and dysfunctional association of proteins. Proc Natl Acad Sci 106: 10159–10164. 10.1073/pnas.0812414106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Rodrigues P, Czerwieniec G, Peters TW, Evani US, Alavez S, Gaman EA, Vantipalli M, Mooney SD, Gibson BW, Lithgow GJ. 2012. Proteomic analysis of age-dependent changes in protein solubility identifies genes that modulate lifespan. Aging Cell 11: 120–127. 10.1111/j.1474-9726.2011.00765.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. 2003. Folding proteins in fatal ways. Nature 426: 900–904. 10.1038/nature02264 [DOI] [PubMed] [Google Scholar]
- Shan L, Mody N, Sormanni P, Rosenthal KL, Damschroder MM, Esfandiary R. 2018. Developability assessment of engineered monoclonal antibody variants with a complex self-association behavior using complementary analytical and in silico tools. Mol Pharm 10.1021/acs.molpharmaceut.8b00867 [DOI] [PubMed] [Google Scholar]
- Smialowski P, Doose G, Torkler P, Kaufmann S, Frishman D. 2012. PROSO II–a new method for protein solubility prediction. FEBS J 279: 2192–2200. 10.1111/j.1742-4658.2012.08603.x [DOI] [PubMed] [Google Scholar]
- Sormanni P, Aprile FA, Vendruscolo M. 2015. The CamSol method of rational design of protein mutants with enhanced solubility. J Mol Biol 427: 478–490. 10.1016/j.jmb.2014.09.026 [DOI] [PubMed] [Google Scholar]
- Sormanni P, Amery L, Ekizoglou S, Vendruscolo M, Popovic B. 2017. Rapid and accurate in silico solubility screening of a monoclonal antibody library. Sci Rep 7: 8200 10.1038/s41598-017-07800-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvang M, Schafer NP, Malmos KG, Pérez AMW, Niembro O, Sormanni P, Basaiawmoit RV, Christiansen G, Andreasen M, Otzen DE. 2018. Corneal dystrophy mutations drive pathogenesis by targeting TGFBIp stability and solubility in a latent amyloid-forming domain. J Mol Biol 430: 1116–1140. 10.1016/j.jmb.2018.03.001 [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Vendruscolo M. 2008. The Zyggregator method for predicting protein aggregation propensities. Chem Soc Rev 37: 1395–1401. 10.1039/b706784b [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Vendruscolo M. 2009. Correlation between mRNA expression levels and protein aggregation propensities in subcellular localisations. Mol BioSyst 5: 1873–1876. [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Pechmann S, Dobson CM, Vendruscolo M. 2007. Life on the edge: A link between gene expression levels and aggregation rates of human proteins. Trends Biochem Sci 32: 204–206. 10.1016/j.tibs.2007.03.005 [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Pawar AP, Campioni M, Dobson CM, Chiti F, Vendruscolo M. 2008. Prediction of aggregation-prone regions in structured proteins. J Mol Biol 380: 425–436. 10.1016/j.jmb.2008.05.013 [DOI] [PubMed] [Google Scholar]
- UniProt Consortium. 2018. UniProt: the universal protein knowledgebase. Nucleic Acids Res 46: 2699 10.1093/nar/gky092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendruscolo M, Knowles TP, Dobson CM. 2011. Protein solubility and protein homeostasis: A generic view of protein misfolding disorders. Cold Spring Harbor Perspect Biol 3: a010454 10.1101/cshperspect.a010454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther DM, Kasturi P, Zheng M, Pinkert S, Vecchi G, Ciryam P, Morimoto RI, Dobson CM, Vendruscolo M, Mann M, et al. 2015. Widespread proteome remodeling and aggregation in aging C. elegans. Cell 161: 919–932. 10.1016/j.cell.2015.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf Pérez AM, Sormanni P, Andersen JS, Sakhnini LI, Rodriguez-Leon I, Bjelke JR, Gajhede AJ, De Maria L, Otzen DE, Vendruscolo M, Lorenzen N. 2019. In vitro and in silico assessment of the developability of a designed monoclonal antibody library. mAbs 10.1080/19420862.2018.1556082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano R, Jamroz M, Szczasiuk A, Pujols J, Kmiecik S, Ventura S. 2015. AGGRESCAN3D (A3D): Server for prediction of aggregation properties of protein structures. Nucleic Acids Res 43: W306–W313. 10.1093/nar/gkv359 [DOI] [PMC free article] [PubMed] [Google Scholar]