SUMMARY
Telomerase is a DNA polymerase that extends the 3′ ends of chromosomes by processively synthesizing multiple telomeric repeats. It is a unique ribonucleoprotein (RNP) containing a specialized telomerase reverse transcriptase (TERT) and telomerase RNA (TER) with its own template and other elements required with TERT for activity (catalytic core), as well as species-specific TER-binding proteins important for biogenesis and assembly (core RNP); other proteins bind telomerase transiently or constitutively to allow association of telomerase and other proteins with telomere ends for regulation of DNA synthesis. Here we describe how nuclear magnetic resonance (NMR) spectroscopy and X-ray crystallography of TER and protein domains helped define the structure and function of the core RNP, laying the groundwork for interpreting negative-stain and cryo electron microscopy (cryo-EM) density maps of Tetrahymena thermophila and human telomerase holoenzymes. As the resolution has improved from ∼30 Å to ∼5 Å, these studies have provided increasingly detailed information on telomerase architecture and mechanism.
1. INTRODUCTION
The linear chromosomes of higher organisms present a special problem for DNA replication. DNA replication normally starts with an RNA primer that pairs to the opposite strand and provides the free 3′-OH required for DNA polymerases to initiate synthesis. The RNA primers are subsequently removed and replaced by DNA. At the 3′ ends of a replicating chromosome, the removal of the RNA primer would result in loss of ∼20 nucleotides (nt) for each round of DNA replication. Eventually, this loss of DNA leads to cellular senescence (Armanios and Blackburn 2012). This “end replication problem” led to the search for a specialized enzyme that might maintain the DNA at the ends of chromosomes and the discovery of telomerase (Greider and Blackburn 1989). The DNA at the ends of eukaryotic chromosomes has a repetitive sequence, the telomeric repeat, that is generally a 6–8-nt G-rich sequence (e.g., TTGGGG in ciliates, TTAGGG in vertebrates) on the 3′-end strand (G-strand) and ends with a single-stranded overhang of variable length (Blackburn and Collins 2011). Telomerase extends the 3′ end by synthesizing multiple copies of the telomere repeat using its specialized telomerase reverse transcriptase (TERT) and its telomerase RNA (TER), which contains the template that directs repeat synthesis (Blackburn et al. 2006; Blackburn and Collins 2011). The TER template is complementary to 1.5–1.8 telomere repeats and comprises an alignment region followed by the templating region (Greider and Blackburn 1987; Podlevsky and Chen 2016). TERT contains conserved reverse transcriptase (RT, palm and fingers) and carboxy-terminal element (CTE, thumb) domains found in other reverse transcriptases, as well as an RNA-binding domain (RBD), that together form a TERT ring structure (Gillis et al. 2008; Chan et al. 2017; Wang and Feigon 2017). It also contains a telomerase amino-terminal domain (TEN) that is connected to the RBD by a long linker. In contrast to the relatively conserved TERT, TER is highly divergent in length (varying from ∼150 nt in ciliates to >2000 nt in some yeasts) and structural motifs (Podlevsky et al. 2008). This is explained by TER being a rapidly evolving noncoding (nc)RNA (Nelson and Shippen 2015; Podlevsky and Chen 2016). TER is an RNA polymerase III transcript in ciliates but is transcribed by RNA polymerase II in vertebrates, yeasts, and plants (Greider and Blackburn 1989; Egan and Collins 2012). This divergence has allowed multiple solutions for biogenesis and processing of TER, assembly of TER with TERT, cellular localization, and recruitment to telomeres (Fig. 1). However, TERs have two regions that are conserved to interact with TERT—the template/pseudoknot domain (t/PK), which forms a circle including the template and a pseudoknot, and the stem-terminus element (STE), which includes a hairpin (Figs. 1 and 2A,B) (Theimer and Feigon 2006; Zhang et al. 2011; Podlevsky and Chen 2016).
Figure 1.
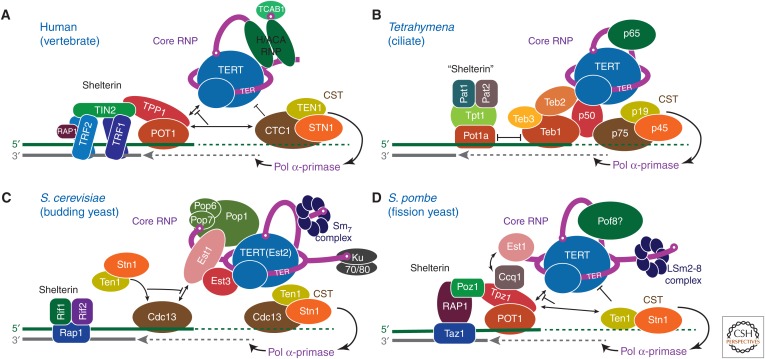
Telomerase ribonucleoprotein (RNP) complexes and their interactions at telomeres. Telomerase RNPs and telomere-associated proteins and interactions are illustrated schematically for (A) human, (B) Tetrahymena thermophila, (C) budding yeast (Saccharomyces cerevisiae), and (D) fission yeast (Schizosaccharomyces pombe). Protein identities are indicated and colors chosen to highlight structural or functional conservation between species. Three principal complexes are illustrated: The telomerase core RNP (TER–TERT catalytic core and TER-binding proteins), telomerase accessory proteins (CST complexes, p50/TPP1/Est3, TEB/POT1), and shelterin/shelterin-like complexes. Black lines with arrows represent recruitment and/or enhancement of activity, whereas blunted black lines denote inhibition or termination of activity.
Figure 2.
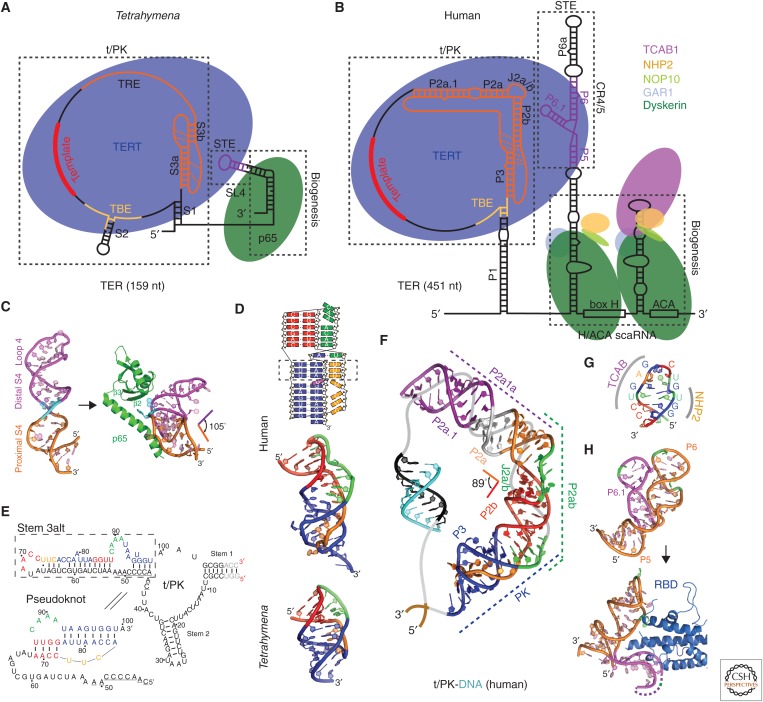
Tetrahymena and human telomerase core RNP and structures of TER domains. Schematics of telomerase core RNP of (A) Tetrahymena and (B) human telomerase. Proteins are shown at their approximate relative size and interactions, and the shape of TER is based on structural studies described in the text. (C) Nuclear magnetic resonance (NMR) structure of the free Tetrahymena stem-terminus element (STE; stem loop 4) (Protein Data Bank [PDB]: 2FEY) and model of the Tetrahymena p65 atypical RNA recognition motif (xRRM)–stem loop 4 complex based on the crystal structure of p65 xRRM–stem 4 (PDB: 4ERD) and NMR structure of loop 4 (PDB: 2M21). (D) Solution structures of minimal pseudoknots (PKs) from human (PDB: 2K95) and Tetrahymena (PDB: 5KMZ). Secondary structure schematic of the human P2b–P3 minimal PK is shown on the top. Dashed rectangle highlights the three U-A-U triples. (E) Comparison of secondary structures of free Tetrahymena TER and the folded PK, determined by NMR (Cash and Feigon 2017). The template residues are underlined. (F) Molecular model of human template/pseudoknot domain (t/PK). The solution NMR structure of the minimal PK (P2b/P3), P2b–J2a/b–P2a (P2ab), and residual dipolar coupling (RDC)–MC-Sym model of P2a.1–J2a.1–P2a (P2a1a) were computationally combined to model the full-length P2/P3 pseudoknot (Zhang et al. 2010). The single-stranded region (gray) containing the template (black) is shown bound to telomeric DNA (cyan). (G) Solution NMR structure of the 3′ apical stem loop (conserved region 7 [CR7]) containing the TCAB1 and NHP2 binding interfaces, determined by NMR and mutagenesis (Theimer et al. 2007). (H) NMR structure of free medaka (Oryzias latipes) STE (CR4/5) (PDB: 2MHI) compared with crystal structure of the CR4/5–RNA-binding domain (RBD) complex (PDB: 4O26).
An understanding of telomerase structure and mechanism has become increasingly important as it has been recognized as a key determinant of human health, longevity, and tumorigenesis (Marrone et al. 2005; Fujii et al. 2009; Artandi and DePinho 2010; Armanios and Blackburn 2012; Bernardes de Jesus and Blasco 2013; Shay 2016). Even slight imbalances in telomerase levels can lead to debilitating diseases in humans. Telomerase insufficiencies are associated with premature aging and stem cell renewal disorders including dyskeratosis congenita, aplastic anemia, and pulmonary fibrosis (Townsley et al. 2014; Sarek et al. 2015; Shay 2016). In contrast, telomerase is up-regulated in >80% of human cancers, rendering tumor cells immortal and driving runaway cell proliferation (Blasco 2003; Stewart and Weinberg 2006; Artandi and DePinho 2010).
Despite its enormous medical interest, telomerase has been and remains a challenging target for structural studies, in part because of the low cellular abundance of telomerase in most cells (e.g., approximately 240 in human cells) (Xi and Cech 2014), the propensity for TERT to aggregate, and difficulties in assembling telomerase from its components in vitro. It has also been challenging to experimentally define the telomerase components and their stoichiometry from different organisms. The catalytic core of TERT and TER is sufficient in vitro for processive telomere repeat synthesis, but in vivo activity requires other species-specific proteins that bind alone or as complexes to TER, forming the core ribonucleoprotein (RNP) (Schmidt and Cech 2015; Wu et al. 2017b). Additional proteins associate constitutively or transiently during telomeric DNA synthesis for G-strand handling, recruitment of proteins for C-strand synthesis, and termination (Fig. 1) (Nandakumar and Cech 2013; Schmidt and Cech 2015; Chan et al. 2017; Wu et al. 2017b). Regions of TER outside the t/PK and STE vary greatly among organisms, as TER has evolved different solutions for telomerase biogenesis, assembly, and localization. In ciliates, a La-related protein group 7 (LARP7) (Bousquet-Antonelli and Deragon 2009) protein (p65 in Tetrahymena thermophila) (Witkin and Collins 2004) functions in 3′-end protection and assembly of TER with TERT. Human TER has a specialized H/ACA scaRNA domain (Mitchell et al. 1999a) that binds the H/ACA scaRNP proteins (Egan and Collins 2010). The much larger fission and budding yeast TERs bind different sets of proteins (Wu et al. 2017b), including various Sm and LSm proteins and, for budding yeast the Pop1–Pop6–Pop7 subcomplex from mitochondrial ribonuclease (RNase) P (Fig. 1C) (Lin et al. 2015; Lemieux et al. 2016), which will not be discussed further here. With the exception of a Saccharomyces cerevisiae Ku70/80–TER hairpin crystal structure (Chen et al. 2018), most have been studied structurally only in contexts other than telomerase, and no electron microscopy (EM) structures for yeast telomerase have been reported to date. For function at telomeres, human telomerase core RNP binds to telomere-associated proteins that enhance processivity, TPP1–POT1 (which are also components of telomere shelterin complex) (de Lange 2005; Palm and de Lange 2008), or inhibit telomerase and recruit DNA polymerase α-primase for synthesis of the C-strand dCCCTAAn, CTC1–STN1–TEN1 (CST) (Fig. 1A) (Chen et al. 2012; Chen and Lingner 2013). As revealed by structural studies described below, homologs of these proteins associate constitutively with Tetrahymena telomerase (Fig. 1B) (Jiang et al. 2015; Chan et al. 2017).
The first studies of telomerase structure focused on defining TER domains, almost exclusively by nuclear magnetic resonance (NMR) spectroscopy, and TERT domains, by X-ray crystallography (Chan et al. 2017). As other protein components of telomerase were discovered and TER–TERT domain complexes defined, structural studies were extended to them, and recently many new protein domain structures have been reported (Chan et al. 2017; Chen et al. 2018). These studies have been paralleled by structural studies of telomere-interacting proteins (Price et al. 2010; Wan et al. 2015; Hoffman and Skordalakes 2016), which are beyond the scope of this review; some of these are also associated with telomerase at least during telomeric DNA synthesis. In 2013, the first negative-stain EM structures of telomerase, from Tetrahymena and human, at ∼25–30 Å resolution, were reported (Jiang et al. 2013; Sauerwald et al. 2013). EM overcomes in part the problem of low cellular abundance of telomerase because it requires orders of magnitude less sample than NMR or crystallography. A cryo-electron microscopy (cryo-EM) structure of Tetrahymena telomerase at a resolution of 9 Å followed two years later in 2015 (Jiang et al. 2015). Cryo-EM has undergone a resolution revolution during the past few years, leading in favorable cases to atomic resolution structures of complexes such as the ribosome and spliceosome (Fica and Nagai 2017; Frank 2017). In 2018, both the first cryo-EM structure of human telomerase holoenzyme with DNA substrate, at a resolution of 8–10 Å (Nguyen et al. 2018), and a 4.8 Å resolution cryo-EM structure of Tetrahymena telomerase with telomeric DNA (Jiang et al. 2018), were published. In the following, we discuss how structural studies using NMR, crystallography, and EM of Tetrahymena and human telomerase have been applied to and evolved our understanding of telomerase RNP structure and function.
2. SOLUTION NMR STUDIES OF TER STRUCTURE AND DYNAMICS
In parallel with the identification of a consensus structure for TER elements required for catalysis (Chen and Greider 2004), comprising the t/PK and STE domains that independently bind TERT (Fig. 2A,B), the first structures of domains of TER were reported (Fig. 2C–H) (Comolli et al. 2002; Leeper et al. 2003; Theimer et al. 2003; Leeper and Varani 2005; Theimer et al. 2005; Chen et al. 2006; Richards et al. 2006a; Richards et al. 2006b; Kim et al. 2008). NMR has proven particularly useful for structural studies of TER domains, although it was clear from the start that care needed to be taken to ensure that these structures, removed from the context of their protein partners and the full-length TER, were biologically relevant. In a typical study, the biological importance of structural features of the TER domain being studied were investigated by assaying the effect of nucleotide substitutions on structure, telomerase activity in the context of TERT and TER assembled in vitro and/or holoenzyme assembled in vivo, telomere length in vivo, and assembly with TERT using electrophoretic mobility shift assays.
Early studies identified a minimal pseudoknot (PK) in human TER, with most of the conserved nucleotides in the full-length PK, that contains the stem loop interactions (P2b/P3 in vertebrates) (Theimer et al. 2005). Ciliates have the smallest PK (∼30 nt), whereas vertebrates and yeasts have PKs with an extended stem 1 (P2 in human) and loop 2 that are outside the minimal PK (Fig. 2A,B,D) (Theimer et al. 2005; Kim et al. 2008; Cash et al. 2013; Wang et al. 2016; Cash and Feigon 2017). In human TER, the full-length PK includes the P2b/P3 minimal PK, a central stem region with a 5–6-nt asymmetric internal loop (P2a–J2a/b–P2b), and an irregular helical extension (P2a.1–J2a.1) (Fig. 2B,F). The solution NMR structure of the human minimal PK established that its fold is stabilized by tertiary interactions between the loops and stems and also highlighted the structural importance of U-A-U base triples within the PK (Fig. 2D) (Theimer et al. 2005; Kim et al. 2008; Zhang et al. 2010). A correlation between PK stability and telomerase activity was shown by mutagenesis and thermodynamic analysis (Theimer et al. 2005). Structural and functional studies on ciliate, yeast, and other vertebrate (e.g., medaka fish [Oryzias latipes]) TER PKs have confirmed the importance of base triples (triple helix) for PK stability and telomerase function across various species (Shefer et al. 2007; Qiao and Cech 2008; Cash et al. 2013; Wang et al. 2016; Cash and Feigon 2017). The recent cryo-EM structures of Tetrahymena and human telomerase show that the solution NMR structures of the PKs fit well to the EM density maps of the holoenzymes (Jiang et al. 2015, 2018; Nguyen et al. 2018), confirming that the structures of the isolated domains obtained by NMR are biologically relevant. NMR structures of the other helical domains of Tetrahymena TER, stem loop 2 (Richards et al. 2006a), and stem loop 4 (Chen et al. 2006; Richards et al. 2006b) were also determined. The p65 carboxy-terminal domain binds and bends stem 4 at a conserved GA bulge (Fig. 2C) (Singh et al. 2012), as discussed in Section 3.
The structures and dynamics of the remaining subdomains of the TER full-length PK in vertebrates (human and medaka fish) have also been extensively investigated by solution NMR (Kim et al. 2010; Zhang et al. 2010; Kim et al. 2014; Wang et al. 2016). The human P2ab domain consists of P2a, P2b, and a 5-nt J2a/b pyrimidine-rich asymmetric bulge (Fig. 2B), conserved in location but not sequence. Its NMR structure showed that the J2a/b bulge loop has a characteristic S-shape and creates an ∼90° bend with a surprisingly low twist (∼10°) between the flanking helices (Zhang et al. 2010). The J2a/b bulge loop is intrinsically flexible but the interhelical motions across the loop are remarkably restricted. Nucleotide substitutions in J2a/b that affect the bend angle, direction, and interhelical dynamics are correlated with telomerase activity. The other PK subdomain, P2a.1, which was proposed to be specific to mammals, is connected to P2 by J2a.1. This region, which contains a flexible internal loop that forms an irregular helix, was modeled by combining the RNA modeling program MC-Sym (Parisien and Major 2008) with residual dipolar couplings (RDCs). A model of the complete human PK was then calculated by computationally combining the NMR structures encompassing the three subdomains (Fig. 2F). This model structure showed that the J2a/b bulge defines the overall arc-shaped topology of the full-length PK. The NMR analysis of medaka PK showed it has a similar topology, despite differences in sequence and size, including a cryptic P2a.1 (Wang et al. 2016).
Other vertebrate TER catalytic core domains that have been studied by NMR are the human and medaka conserved region 4/5 (CR4/5) (Kim et al. 2014), which rearranges when in complex with medaka TERT RBD (Fig. 2H) (Huang et al. 2014), and the P6.1 region of human CR4/5, which contains two pseudouracils (Kim et al. 2010). In the H/ACA domain, a structure–function study of the CR7 stem loop identified conserved elements for TCAB1 and NHP2 binding (Fig. 2G) (Theimer et al. 2007).
Recent NMR studies of Tetrahymena TER showed that the Tetrahymena PK does not form in the free TER. Instead, part of the PK sequence pairs with and sequesters the template (stem 3alt) (Fig. 2E) (Cash and Feigon 2017). This alternative structure may function in vivo to shield template and PK residues from unproductive interactions or cleavage by endonucleases while still leaving the high-affinity TER RBD-binding sites available for assembly with TERT. Sequence analysis indicates that human t/PK may also form an alternative structure, and this is supported by single-molecule fluorescence resonance energy transfer (FRET) studies (Hengesbach et al. 2012).
3. USING NMR TO STUDY PROTEIN–RNA INTERACTIONS AND OPTIMIZE CONSTRUCTS FOR CRYSTALLOGRAPHY: p65–TER COMPLEX
NMR has also been used to study telomerase protein domains and interactions with TER and has proven useful in optimizing constructs for crystallography. Good examples are the studies of the Tetrahymena telomerase biogenesis and assembly protein p65 and its interaction with TER (Singh et al. 2012). The LARP7 protein p65 associates with the 3′ end of TER and is essential for assembly in vivo (Prathapam et al. 2005; O’Connor and Collins 2006; Singh et al. 2013). In addition to a La module (La motif and RNA recognition motif [RRM]) (Bousquet-Antonelli and Deragon 2009) that binds the TER 3′-UUUU tail, it has a carboxy-terminal domain that binds to and bends stem 4 (Stone et al. 2007; Berman et al. 2010; Akiyama et al. 2012; Singh et al. 2012). Many different protein constructs of the carboxy-terminal domain were made and NMR was used to evaluate whether they were stably folded, identify flexible regions, and define domain boundaries (Singh et al. 2012). The initial assay for this is a simple 1H–15N heteronuclear single-quantum correlation (HSQC) experiment, which provides one cross peak for each peptide NH. Once good spectra are obtained, backbone assignments can be determined from standard triple-resonance experiments. In the case of p65, a flexible carboxy-terminal tail and a large unstructured internal loop were deleted and truncated, respectively, ultimately yielding a construct that was suitable for NMR structure determination. Binding and assembly assays using isothermal titration calorimetry (ITC) and electrophoretic mobility shift showed that the carboxy-terminal tail, but not the internal loop, is important for TER binding and assembly of p65–TER with TERT. The solution structure identified the domain as a new type of atypical RRM, named xRRM, and the partially deleted internal loop at a highly flexible (but dispensable) region connecting the β2 and β3 strands (Fig. 2C) (Singh et al. 2013). NMR was also used to assay RNA constructs for structure and binding. This information guided the design of protein constructs to obtain a crystal structure of the p65 xRRM with TER. The carboxy-terminal tail, which is essential for RNA binding, was retained and various constructs with partial or full deletion of the β2–β3 loop were screened, ultimately yielding a crystal structure with TER stem 4, which revealed the structural basis of the conformational change in stem loop 4 induced by p65 that facilitates assembly of p65–TER with TERT (Fig. 2C).
Another example of the complementarity of NMR and crystallography are the studies of TERT TEN (Fig. 3E) (Jacobs et al. 2005; O’Connor et al. 2005; Jacobs et al. 2006). Tetrahymena TEN was identified as a soluble domain of TERT by high-throughput screening and characterized as an independently folded domain by NMR (Jacobs et al. 2005). It was subsequently crystallized (Jacobs et al. 2006). Recently the structure of another TEN domain from the thermotolerant yeast Hansenula polymorpha (hpTEN) was determined by both crystallography and NMR (Polshakov et al. 2016; Petrova et al. 2018). The protein core in the NMR and crystal structures has the same overall organization, with a backbone atom root-mean-square deviation (RMSD) of 0.8 Å, but there are flexible regions that differ between the two structures, including helices α5 and α6 present in only the crystal or NMR structure, respectively.
Figure 3.
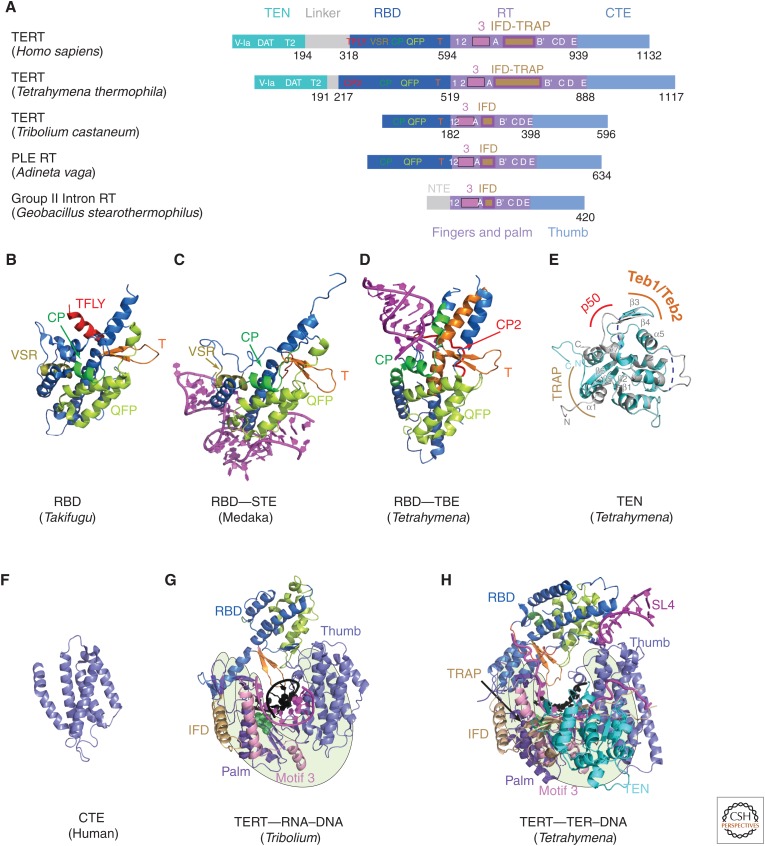
TERT domains and structures. (A) Schematics comparing domains of reverse transcriptases (RTs) from human telomerase, Tetrahymena telomerase, Tribolium castaneum telomerase, Penelope-like element (PLE; Adineta vaga), and Group II intron (Geobacillus stearothermophilus). Domains and conserved motifs are aligned using the RT domains. Note that Tribolium “TERT” is more similar to the A. vaga PLE RT than to the true TERTs from human and Tetrahymena. Crystal structures (B–G) of (B) Takifugu rubripes RBD (PDB: 4LMO), (C) medaka RBD–STE complex (PDB: 4O26), (D) Tetrahymena RBD–template boundary element (TBE) complex (PDB: 5C9H), and (E) free Tetrahymena telomerase amino-terminal domain (TEN) (PDB: 2B2A, cyan) superimposed with cryo-EM structure of TEN in Tetrahymena telomerase holoenzyme (PDB: 6D6V) (gray). Regions interacting with p50, TEB, and TRAP are indicated. Blue dashed lines represent the missing loops in the crystal structure. (F) Human carboxy-terminal element (CTE) (PDB: 5UGW) and (G) Tribolium TERT with an RNA–DNA hairpin mimicking a template–DNA duplex (PDB: 3KYL). (H) Cryo-EM structure of Tetrahymena TERT–TER with template–DNA duplex (PDB: 6D6V). TRAP is mostly covered by TEN, so is difficult to see in this view. The TRAP and TEN domains are unique to TERT, whereas motif 3 and IFD are found in closely related RTs from group II introns and PLEs. For G and H, the polymerase “hand” view is shown.
4. X-RAY CRYSTALLOGRAPHY OF TERT DOMAINS AND COMPLEXES WITH TER
TERT needs to be assembled with TER to be functional in synthesizing telomeric DNA repeats. In vitro, telomerase activity can be recovered from TERT and TER assembled in rabbit reticulocyte lysate (Min and Collins 2010); however, the vast majority of the protein is aggregated and not functional. Not surprisingly then, TERT has been notoriously difficult to purify and crystallize.
Almost all TERTs contain four major domains, two of which are unique to TERT (i.e., the TEN domain and RBD) and two domains found in other reverse transcriptases (i.e., RT [palm and fingers] and CTE [thumb]) (Fig. 3A) (Kelleher et al. 2002; Autexier and Lue 2006; Mason et al. 2011). A major breakthrough was the crystal structure of the putative TERT from the flour beetle Tribolium castaneum reported in 2008 (and with an RNA–DNA hairpin mimicking a template–DNA duplex in 2010), which revealed that the RBD–RT–CTE domains form a ring structure (Fig. 3G) (Gillis et al. 2008; Mitchell et al. 2010). Tribolium TERT is atypical in that it lacks a TEN domain (Fig. 3A) and it has no known TER. The genome sequence of Tribolium revealed that its telomeres are composed of a mix of retrotransposons and simple repeats that are mostly, but not exclusively, TCAGG, suggesting that telomerase, as a telomere maintenance mechanism, was lost in Tribolium (Tribolium Genome Sequencing Consortium et al. 2008); thus, Tribolium TERT is unlikely to be a true TERT. Nevertheless, the Tribolium TERT ring structure has proven useful for modeling into negative-stain and cryo-EM maps of Tetrahymena and human telomerase (Jiang et al. 2013, 2015; Nguyen et al. 2018). The Tribolium TERT structure has also provided structural and biochemical insights into the mechanism of nucleotide addition, revealing the location of many features found in other reverse transcriptases such as the primer grip, thumb loop, and thumb helix, as well as two previously unknown motifs, the insertion in fingers domain (IFD) (Lingner et al. 1997; Lue et al. 2003) and motif 3 (Xie et al. 2010) in the RT domain (Gillis et al. 2008; Mitchell et al. 2010). However, the two IFD helices and motif 3, originally thought to be TERT–specific, were subsequently also found in group II intron (Qu et al. 2016; Zhao and Pyle 2016; Stamos et al. 2017) and some Penelope-like element (PLE) RTs (Fig. 3A,G) (Gladyshev and Arkhipova 2007). Notably, structures of the TERT ring alone do not reveal how template boundaries are determined or how translocation occurs (e.g., what separates the template–DNA duplex and what keeps the telomeric DNA from dissociating during translocation). Comparison with the structure of the Tetrahymena TERT–TER catalytic core from the 4.8 Å cryo-EM map (discussed later) highlights the structural differences of a true TERT containing the telomerase unique TRAP and TEN domains, a larger RBD, and an integral TER (Fig. 3A,G,H).
The TERT RBD has distinct binding sites for the TER template boundary element (TBE) and STE domains (Lai et al. 2001), and these have been structurally characterized by EM (Jiang et al. 2015, 2018) and in greater atomic detail by crystallography (Fig. 3B–D) (Harkisheimer et al. 2013; Huang et al. 2014; Jansson et al. 2015). RNA-binding motifs were suggested from crystal structures of Tetrahymena and vertebrate fish (Takifugu rubripes and O. latipes) RBD, as well as sequence analysis and mutagenesis (Harkisheimer et al. 2013; Huang et al. 2014; Jansson et al. 2015). A 3.0 Å crystal structure of Tetrahymena RBD bound by the TBE shows that the single-stranded nucleotides on either side of stem 2 bind two sides of the RBD, making a network of polar contacts to three conserved motifs in the RBD (i.e., CP2 [TFLY in vertebrates], CP, and T) (Fig. 3D) (Jansson et al. 2015). These interactions anchor the TBE onto TERT, thereby determining the 5′-template boundary by physically preventing nucleotides beyond the last template nucleotide from moving into the active site.
The TER STE (distal stem loop 4 in Tetrahymena and P6.1 in CR4/5 in vertebrates) (Fig. 2A,B) binds to both the RBD and CTE (Bley et al. 2011; Huang et al. 2014; Jiang et al. 2015, 2018), which interface to close the TERT ring in the absence of TER in Tribolium TERT. A 3.0 Å crystal structure of medaka RBD bound to the CR4/5 domain provided details of the RBD part of the interaction (Fig. 3C) (Huang et al. 2014). The conserved motif QFP and the amino-terminal region of helix α2 interact extensively at a unique three-way junction in CR4/5. However, density for the P6.1 loop is missing and it appears that the missing loop could interact with the CTE (Nguyen et al. 2018).
The 2.2 Å crystal structure of the Tetrahymena TERT TEN domain revealed a mixed α/β-fold with a highly basic patch proposed to be a DNA-binding groove (Fig. 3E) (Jacobs et al. 2006; Zaug et al. 2008; Wu and Collins 2014). TEN has been proposed to function in binding TER (Tetrahymena loop 4 and the TRE) (O’Connor et al. 2005), as a telomeric DNA anchor site (Jacobs et al. 2006; Zaug et al. 2008; Wyatt et al. 2009; Jurczyluk et al. 2011; Eckert and Collins 2012), and more recently in stabilization of short template–DNA duplexes (Robart and Collins 2011; Wu and Collins 2014; Akiyama et al. 2015; Shastry et al. 2018). TERT TEN domain is connected to the RBD by a long at least partially unstructured linker, but its location relative to other TERT domains and TER was established only by the cryo-EM studies, which revealed unexpected interactions and functions (see Sec. 7) (Jiang et al. 2015, 2018).
Besides the RBD, the only other domain of vertebrate TERT for which a crystal structure has been reported is of a partial human CTE, missing the amino-terminal region containing the thumb loop, at 2.3 Å resolution (Fig. 3F) (Hoffman et al. 2017). The all-helical bundle structure is highly similar to the Tribolium CTE (thumb domain) and revealed the locations of some human mutations associated with the telomerase insufficiency disease dyskeratosis congenita.
5. NEGATIVE-STAIN EM OF TELOMERASE
Reports of negative-stain EM structures of human and Tetrahymena telomerase in 2013 at 25–30 Å resolution marked a major breakthrough in structural studies of telomerase (Jiang et al. 2013; Sauerwald et al. 2013), in part because they showed that sufficient quantities of functional enzymes could be purified at least for EM studies and suggested the possibility of higher resolution structures to follow (Akiyama and Stone 2013; Miracco et al. 2014). The negative-stain EM structure of Tetrahymena telomerase provided the architecture of a telomerase with locations of TERT, TER domains, p65, and other known accessory proteins (Fig. 4A) (Jiang et al. 2013). Although negative-stain EM structures have largely been supplanted by cryo-EM structures obtained with direct electron detectors, it is useful to discuss the method and what was learned from it, as it made possible the higher resolution cryo-EM structures subsequently reported. In addition, negative-stain EM is often useful to screen samples for homogeneity and concentration, especially because it may be difficult to find good cryogenic conditions for RNPs (e.g., sometimes RNPs do not survive the freezing process intact). Negative-stain EM is also useful for obtaining an initial unbiased model of a complex (i.e., no assumed starting model is used) (Jiang et al. 2013). For negative-stain EM, aqueous samples are applied to an EM grid and then stained with a heavy metal solution such as uranyl formate.
Figure 4.
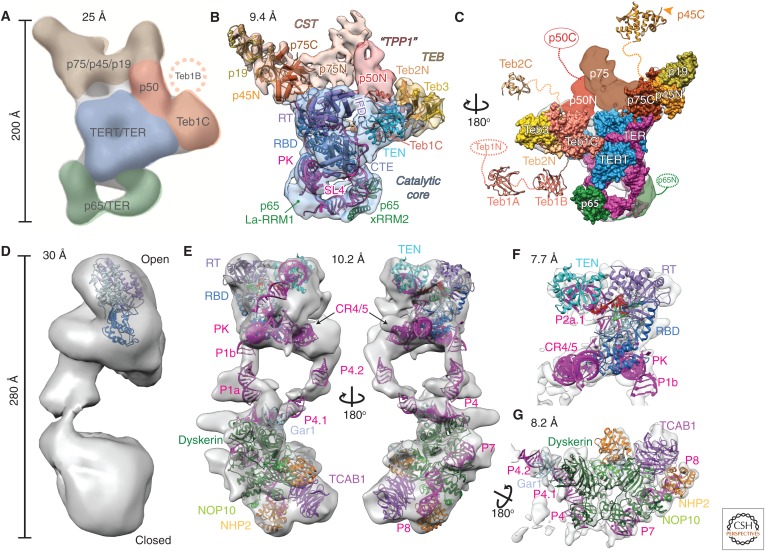
Comparison of negative-stain and cryo-EM density maps and models of Tetrahymena and human telomerase. (A–C) EM density maps and models of Tetrahymena telomerase. (A) A 25 Å negative-stain EM map with locations and estimated boundaries of subunits colored, based on affinity tagging of subunits, comparison of particles lacking all or part of a subunit, and modeling of the catalytic core (Jiang et al. 2013) (EMDB: 5804). (B) A 9.4 Å resolution cryo-EM map (core RNP, blue; CST, tan; TEB, straw; and p50, red) and pseudoatomic models of the core RNP and TEB and CST trimerization domains of three OB folds (Jiang et al. 2015). (C) An 180° rotated view of B with modeled domains shown as space-fill on the cryo-EM map. Model of the catalytic core and TEB is based on the 8.9 Å resolution map. Additional domains of Teb1, Teb2, p45, p65, and p50 are not visible in the cryo-EM map because of positional dynamics and are shown as crystal structures (Teb1A [PDB: 3U4V], Teb1B [PDB: 3U4Z], p45C [PDB: 5DFN]), homology models (Teb2C based on PDB: 1DPU), or ovals. (D–G) EM density maps and models of human telomerase. (D) A 30 Å negative-stain EM map originally proposed to be a dimer (EMDB: 2310) (Sauerwald et al. 2013). Tribolium TERT was automatically fit into the map using UCSF Chimera (Pettersen et al. 2004). (E) Two views of the 10.2 Å resolution cryo-EM map of human telomerase holoenzyme (EMDB: 7521) with a modeled catalytic core (top) and H/ACA scaRNP (bottom) (Nguyen et al. 2018). (F) A 7.7 Å resolution cryo-EM map (EMDB: 7518) from focused refinement and model of the catalytic core. (G) A 8.2 Å resolution map (EMDB: 7519) from focused refinement and model of the H/ACA scaRNP. TER is magenta in all models and TERT is blue. Proteins and TER domains are labeled.
Analysis of 3D reconstructions from negative-stain EM data on human and Tetrahymena telomerase provided the first views of the overall shape of telomerase (Fig. 4A,D). For Tetrahymena telomerase, detailed information on the locations of the seven known proteins and one RNA was obtained by collecting negative-stain EM data on telomerase holoenzymes with affinity tags on individual subunits (Fig. 4A). Strains with individual proteins tagged at the endogenous locus (in most cases) with tandem 3×FLAG (F) and two protein A (ZZ) tags separated by a TEV protease cleavage site allowed purification and biochemical characterization of an endogenously expressed enzyme (Min and Collins 2009). A method was developed to bind the antigen-binding fragment (Fab) of an anti-FLAG antibody during the purification, and one to three bound Fabs could then be visualized in negative-stain EM 2D class averages and 3D reconstructions, thereby revealing the location of the carboxyl (or amino, for ZZF tags) terminus of the protein where the Fab(s) bound. A different affinity tag, MS2 coat protein, identified the location of TER stem loop 2, where the loop was replaced by the MS2 coat protein-binding site. Using this method, locations of all the subunits except p45 were determined, whose tag was not visible in the class averages; this process was enhanced by comparison of 3D reconstructions of particles lacking all or part of a subunit. With the general subunit locations identified, known crystal and NMR structures of telomerase subunits or homology models (Tribolium TERT RT and CTE, Tetrahymena RBD and TEN, p65–stem loop 4, and Teb1C) could be fit into the negative-stain EM map. This analysis was notably successful; locations of all tagged subunits (except p45) were subsequently shown by cryo-EM to be correct.
Major discoveries in the negative-stain EM study were the identification of p50, previously thought to be substoichiometric, as a central hub binding TERT, Teb1C, and the p75–p45–p19 complex (Fig. 4A), and that p50 was essential to the processivity enhancement conferred by Teb1 (Hong et al. 2013; Jiang et al. 2013). Teb1 (Min and Collins 2009) is paralogous to the large subunit of replication protein A (RPA), a single-stranded DNA-binding trimeric complex that is involved in all aspects of DNA replication and repair (Prakash and Borgstahl 2012; Sugitani and Chazin 2015). The EM map also provided the location of p65 relative to TERT, defining the overall architecture of the core RNP. An interesting footnote to the affinity labeling of p45 is the subsequent discovery that the Fab was in fact bound but not visible in the EM 2D class averages attributable to domain dynamics (i.e., it was bound to a carboxy-terminal domain of p45 separated by a flexible linker to its amino-terminal domain) (Fig. 4C). In images of single particles, three Fabs could be seen in various locations relative to the rest of the particle (Jiang et al. 2015).
Human telomerase used for the negative-stain EM structure (Sauerwald et al. 2013) was purified from HEK293T cells transiently transfected with human TER and TERT (Cristofari et al. 2007). Analysis of mass spectrometry data identified TERT, dyskerin, and NOP10 as components of the enzyme. Based on molecular weight estimated from native polyacrylamide gel electrophoresis (PAGE) and activity assays on telomerases with two different affinity tags on TERT, the investigators concluded that human telomerase is an obligate dimer with two TERT and two TER molecules plus dyskerin and NOP10. The 3D reconstruction from negative-stain EM and tomography revealed a bilobal structure with an “open” and “closed” lobe at the two ends bridged by density ∼25 Å in diameter (Fig. 4D). The location of TERT was identified from the position of nanogold-labeled bound telomeric DNA, which showed up as dots in none (13.5%), one (50%), or both (36.5%) of the lobes in the single particles. TERT was modeled into the computationally isolated open monomer, and it was concluded that there was one TERT in each lobe with the connecting density proposed to be a TER helix. However, studies from the Collins laboratory where each of the H/ACA scaRNP proteins had been individually tagged provided strong evidence that human telomerase was a monomer with one TERT and TER and two sets of H/ACA proteins bound to the scaRNA domain, which would be about the same molecular weight as the proposed dimer (Egan and Collins 2010). This was found to be unambiguously the case for the recently reported cryo-EM structure of human telomerase (Nguyen et al. 2018), discussed in detail below. Comparison of the negative-stain EM maps with the cryo-EM maps of human telomerase (Fig. 4) indicates that they are very similar when the difference in resolution is accounted for, although for the negative-stain EM sample it may be a mixture of active and inactive TERT assemblies (Nguyen et al. 2018).
6. SUBNANOMETER RESOLUTION CRYO-EM OF TELOMERASE
After the negative-stain EM structures of Tetrahymena and human telomerase reported in 2013, it took another two and five years, respectively, for cryo-EM structures, at 8–10 Å resolution, to be reported (Jiang et al. 2015; Nguyen et al. 2018).
6.1. Tetrahymena Telomerase
Taking advantage of the improvements in data collection for cryo-EM and in particular the emergence of direct detector cameras, cryo-EM structures of Tetrahymena telomerase holoenzyme were solved to 9.4 Å resolution for the entire particle (Fig. 4B,C) and to 8.9 Å resolution excluding the p75–p45–p19 complex, which negative-stain EM 3D reconstructions had shown to be flexibly positioned around p50 (Jiang et al. 2015). Processing of cryo-EM data using a focused refinement is useful for increasing the resolution of the part of the particle within a soft mask that excludes other regions, and was used for both Tetrahymena and human telomerase. It is also important to note that the reported EM resolutions use the “gold standard” approach with a Fourier shell correlation FSC = 0.143 cutoff (Rosenthal and Henderson 2003; Scheres and Chen 2012), but the resolution in different regions of the EM map varies depending on flexibility and other factors.
The identification of protein subunits and a pseudoatomic model of Tetrahymena telomerase catalytic core and most of the accessory proteins in the cryo-EM map was a tour de force that combined NMR spectroscopy, X-ray crystallography, and mass spectrometry with the cryo-EM data. The localization of subunits by affinity tagging and the unbiased model obtained from the negative-stain EM structure (Jiang et al. 2013) were crucial for obtaining and interpreting the 9 Å cryo-EM map. At this resolution, protein helices and β-barrels and RNA helices can be discerned in the density and crystal structures and the homology models fit with certainty. For the Tetrahymena telomerase core RNP, the known structures and models for TER were helical domains (including a newly determined NMR structure of the PK) (Richards et al. 2006a; Cash and Feigon 2017), the p65 xRRM–stem 4 complex (Singh et al. 2012) and loop 4 (Fig. 2) (Richards et al. 2006b), and for TERT the RT and CTE domains from the Tribolium TERT crystal structure (Mitchell et al. 2010) and crystal structures of Tetrahymena RBD (Rouda and Skordalakes 2007) and TEN domain (Fig. 3) (Jacobs et al. 2006). Rigid-body fitting of these TER, TERT, and p65 domains provided a pseudoatomic model of the RNP catalytic core, which showed that the t/PK encircles the TERT RBD–RT–CTE ring. The PK is positioned over the CTE and the TEN domain is stacked over the CTE on the other side of the TERT ring with the 3′ template–adjacent single-stranded TER running between the two (Fig. 4B). Stem 1 and stem loop 4 form a U-shape with only loop 4 contacting TERT, apparently inserting at the RBD–CTE interface. Loop 4 and the PK, both of which had been previously proposed to contribute directly to catalysis (O’Connor et al. 2005; Qiao and Cech 2008), are far from the active site. Based on their positions in the cryo-EM structure, the PK was proposed instead to act like a watchband ratchet clasp, with the PK fully folding only after the t/PK assembled onto TERT (Jiang et al. 2015), and loop 4 was proposed to stabilize closure of the TERT ring.
For the rest of the particle, only Teb1C had a previously known structure and the other proteins were thought to be specific to Tetrahymena (or ciliates). We note that although Teb1 has four domains, Teb1A, B, and N are connected by flexible linkers and are invisible in the cryo-EM map (Fig. 4C). Teb1C is a large OB fold with a Zn-ribbon motif that confers a characteristic C-shape (Zeng et al. 2011). Fitting of Teb1C into the cryo-EM map revealed a surprise—although its carboxyl terminus had been correctly located by anti-FLAG Fab tagging (Jiang et al. 2013), it fit only part of the assigned density in a “knob” and the remaining density could not be accounted for by any known Tetrahymena telomerase protein. Because Teb1 is an RPA1 paralog, the investigators surmised that the remaining density might arise from previously undiscovered RPA2 and RPA3 paralogs. A crystal structure of the RPA heterotrimer of three OB-folds (Fan and Pavletich 2012) fit the density, supporting this hypothesis, and the presence of these two previously unknown subunits (named Teb2 and Teb3) was confirmed by liquid chromatography–mass spectrometry (LC–MS/MS). A subsequent study showed that whereas Teb1 is unique to Tetrahymena telomerase, Teb2 and Teb3 are shared subunits with Tetrahymena RPA (Upton et al. 2017). Another surprise was the identification of the p75–p45–p19 complex as Tetrahymena homologs of human CST. CST is another RPA-related complex, whose STN1 and TEN1 subunits are similar to RPA2 and RPA3 proteins, respectively, whereas CTC1 (Cdc13 in yeasts) is more divergent from RPA1 (Chan et al. 2017). Crystal structures of p19 and of p45 carboxy-terminal domain revealed an OB-fold and winged helix (WH)–WH, respectively, that are structurally homologous to TEN1 and STN1 carboxy-terminal domain. An exhaustive 6D search algorithm for automated fitting revealed that the RPA trimeric core also fits into the EM density assigned to p75–p45–p19 in addition to the TEB density. The identification of p75–p45–p19 as CST was independently verified by structural and functional studies (Wan et al. 2015). Finally, p50 was identified as a β-barrel, and its location near TEN, Teb1, and p75 suggested that it is a homolog of TPP1, although it could not be modeled. In humans, TEN interacts directly with a region of TPP1 called the TEL patch, and this interaction is required for recruitment to telomeres (Zaug et al. 2010; Nandakumar et al. 2012; Sexton et al. 2012; Zhong et al. 2012; Schmidt et al. 2014). Thus, the integrative structural biology studies of Tetrahymena telomerase revealed that it is constitutively assembled with homologs of CST and TPP1–POT1 that in humans and yeasts only temporally associate with telomerase at telomeres. This makes Tetrahymena an ideal model organism for investigating how telomerase interacts with telomere-associated proteins (Feigon et al. 2016; Chan et al. 2017).
6.2. Human Telomerase
The cryo-EM structure of human telomerase with substrate DNA (T12TTAGGG) at sub-nanometer resolution represents another landmark study, which defined the subunit composition and overall architecture of human telomerase and also provided the first model for a eukaryotic H/ACA RNP (Fig. 4E–G) (Nguyen et al. 2018). To overcome the problem of low cellular levels of telomerase, telomerase was reconstituted in vivo from human cells transiently transfected with tagged ZZ–TEV–Twin Strep–TERT and TER, similar to what was performed for the negative-stain EM structure, and purified. A key step was identifying the eluted telomerase fractions that were homogeneous by negative-stain EM, the so-called active telomerase. Another important step was identifying the locations of the catalytic core and the scaRNP protein TCAB1/WDR79 (tryptophan–aspartic acid repeat protein 79) in negative-stain 3D reconstructions by comparison with samples prepared with TER lacking the H/ACA domain and a TCAB1 mutant that does not bind the H/ACA RNP, respectively. The TER interface between the H/ACA RNP and the catalytic core of human telomerase is highly dynamic, limiting the overall resolution of a complete cryo-EM map to 10.2 Å (Fig. 4E). Focused refinement provided 3D reconstructions of the catalytic core at 7.7 Å (Fig. 4F) and the H/ACA scaRNP at 8.2 Å (Fig. 4G) resolution. The catalytic core was modeled by rigid-body fitting of crystal structures of RT and CTE from Tribolium TERT (Mitchell et al. 2010), RBD with TER CR4/5 (P5–P6.1–P6a) from medaka (Huang et al. 2014), and TEN from Tetrahymena (Fig. 3) (Jacobs et al. 2006), and of NMR structures from the human t/PK model described above (Fig. 2) (Zhang et al. 2010, 2011). A model of the human telomerase catalytic core, built by homology modeling human TERT and fitting the human t/PK NMR model of TERT based on the TER path in the cryo-EM structure of Tetrahymena telomerase (Wang et al. 2016; Wang and Feigon 2017), fits the human telomerase cryo-EM map remarkably well. Although the human t/PK has a much larger helical region, it encircles TERT in a similar way as does Tetrahymena t/PK, with the minimal PK on the CTE and the single-stranded template-adjacent TER between the TERT ring and TEN domain (Fig. 4E,F). The human STE element, CR4/5 domain, interacts with the RBD. In the cryo-EM maps of Tetrahymena telomerase, the STE element loop 4 inserts between the RBD and CTE (Jiang et al. 2015, 2018). Although the P6.1 stem loop of the CR4/5 domain is not modeled in the cryo-EM map of human telomerase, it is proposed to insert in a similar manner (Nguyen et al. 2018).
The H/ACA RNP, which is the human TER biogenesis domain, was first identified by Collins (Mitchell et al. 1999a), leading to the proposal that dyskerin mutations in dyskeratosis congenita are due to telomerase insufficiency (Mitchell et al. 1999b). Numerous other disease mutations associated with telomerase insufficiency have since been identified in most human telomerase components (Fogarty et al. 2003; Angrisani et al. 2014; Holohan et al. 2014; Wegman-Ostrosky and Savage 2017). Despite many structures of H/ACA proteins and various complexes (Hamma and Ferre-D’Amare 2004; Manival et al. 2006; Liang et al. 2007; Duan et al. 2009; Koo et al. 2011; Li et al. 2011a; Li et al. 2011b), as well as a complete single-hairpin archaeal H/ACA snoRNP used in the modeling (Li and Ye 2006), no structures of a complete eukaryotic H/ACA RNP have been previously reported. The structure of the human telomerase H/ACA RNP domain with its two sets of H/ACA proteins and TCAB1 (found in H/ACA scaRNPs) thus provides the first view of how the two hairpins are oriented with respect to each other and the intersubunit interactions (Fig. 4G). The 5′ and 3′ hairpins are approximately perpendicular to one another, and the two dyskerin molecules have an interacting surface that maps to several disease mutations, providing a structural basis for understanding their disease phenotype. Although dyskerin, NOP10, and NHP2 all contact the 3′ hairpin RNA, only dyskerin directly binds the 5′ hairpin, which lacks an NHP2-binding loop and is extended to form the STE. The human telomerase H/ACA RNP structure may also explain how Alu H/ACA RNPs with especially short or long 5′ hairpins assemble two sets of H/ACA proteins (Ketele et al. 2016).
7. APPROACHING ATOMIC RESOLUTION: TELOMERASE WITH TELOMERIC DNA
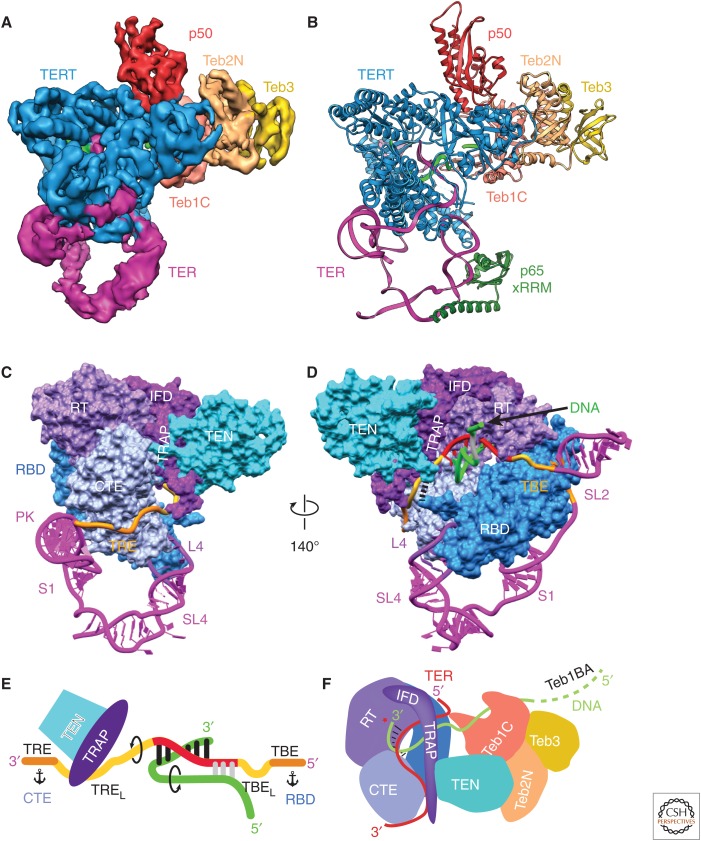
The cryo-EM structures of human and Tetrahymena telomerase discussed above show that the particle is remarkably dynamic; this has so far limited the resolution that can be obtained from cryo-EM maps. It took two and a half years after the ∼9 Å structure of Tetrahymena telomerase was reported to achieve a resolution of 4.8 Å with telomeric DNA and 6.4 Å without DNA (Fig. 5A) (Jiang et al. 2018). In the 4.8 Å cryo-EM structure the telomeric DNA (GTTGGG)3G is bound midway through a cycle of telomere repeat synthesis, and the presence of DNA appears to stabilize the catalytic core somewhat. A soft mask was used to improve the resolution, and in the resulting density maps the dynamic p75–p45–p19 (Tetrahymena CST) complex was masked out. The cryo-EM maps were calculated starting with more than three million particles of which <2% were in the final data set for 3D reconstruction. At 4.8 Å resolution, it is just possible to do de novo modeling of secondary structure elements in the regions of the map where there is no starting crystal or NMR structure. Fortunately, related structures (albeit distantly for some) exist for most of the protein subunits or domains of Tetrahymena telomerase as described above, including the catalytic subunit TERT. Taking advantage of these evolutionary relationships in EM density-guided computational modeling using the Rosetta package (Song et al. 2013) allowed an ensemble of molecular models to be generated. Visual inspection of the highest scoring solutions, together with rebuilding select regions and subsequent real-space refinement of protein and modeled TER using PHENIX (Adams et al. 2010), ultimately allowed the determination of a complete nearly atomic model of the DNA-bound TERT-TER catalytic core, p50N, and Teb1C-Teb2N-Teb3 (Fig. 5B). This model provided the first complete structure of a true TERT, previously unknown details of the architecture of the catalytic core, structures and interactions of p50-TEB, the path of TER on TERT (Fig. 5C,D), and the path of telomeric DNA from the six base-pair template DNA duplex to its exit on Teb1C (Fig. 5E,F).
Figure 5.
Cryo-EM of Tetrahymena telomerase at 4.8 Å resolution provides insights into the mechanism. (A) A 4.8 Å resolution cryo-EM density map of Tetrahymena telomerase with telomeric DNA. (B) Atomic model of the TERT–TER catalytic core, p65 xRRM, p50, and TEB. The model of p65 xRRM is based on the crystal structure (PDB: 4ERD). (C-D) Two views of the structure of the telomerase catalytic core with TERT as spacefill and TER as ribbon. TERT and TER domains are labeled. TEN and TRAP are connected to the RBD and RT domains, respectively, from opposite sides of the TERT ring, physically interlocking the t/PK and TERT. In D, black dashes between the TEN and RBD domains denote the linker between them. The nascent telomeric DNA is shown exiting the template. (E) Cartoon of template recognition element (TRE)–template–TBE interactions with TERT and telomeric DNA. Arrows indicate directions of rotation of TER and DNA during telomere repeat synthesis, and anchors correspond to TER anchor sites that define the template boundaries. TRE linker (TREL) and the TBE loop (TBEL) are the single-stranded regions between the template and TRE and TBE, respectively. (F) Cartoon illustrating the paths of TRE–template–TBE on TERT and of telomeric DNA from the TERT active site to TEB; the TRAP–TEN interaction; the TRAP–TRE interaction. The protein p50 is omitted from this schematic for clarity. (Figure panels are modified from Jiang et al. 2018.)
In addition to providing much more detail than the lower-resolution cryo-EM model of DNA-free telomerase, two previously unknown features of the catalytic core were identified. First, TERT and TER are intricately interlocked, with TER and TERT physically enclosing one another, explaining in part the difficulty of assembling telomerase in vitro (Fig. 5B–D). Second, a long linker between the two IFD helices, which among RTs is uniquely found in TERTs, was revealed to form a structured mostly β-strand interface with the TERT TEN domain (Fig. 5C–F). This region, named TRAP, explains how TEN, which is connected to the RBD by a long linker, can be stably positioned. The single-stranded 3′ template–adjacent TER (named TRE linker, TREL) runs between the CTE and TRAP, and the TRE binds a highly basic surface on CTE (Fig. 5C). The structure shows that roles proposed for TEN in regulating repeat addition processivity (Robart and Collins 2011; Wu and Collins 2014; Akiyama et al. 2015; Shastry et al. 2018) are mostly indirect through TRAP (Fig. 5E,F). The DNA–template duplex occupies a large cavity in the TERT ring. As the growing duplex exits toward TRAP, the looped out TREL appears to be captured by TRAP, whereas the single-stranded telomeric DNA makes a turn in the opposite direction toward Teb1C (Fig. 5E,F). Intriguingly, the single-stranded telomeric DNA seems to contact the 5′ end of the template, potentially forming another short helix. The opposing paths of the DNA and TRE suggest a mechanism to limit the template–DNA duplex to 6–7 base pairs (Wang et al. 1998; Forstemann and Lingner 2005; Wu et al. 2017a), because of destabilization of the helix end and accumulation of torsional stress (Fig. 5E). On the 5′ side of the template the TBE is anchored to the RBD (Jansson et al. 2015), and the TBE loop (TBEL) could go from looped out to maximally stretched during synthesis of a telomere repeat. Thus, TRE–TREL and TBEL–TBE are proposed to define the 3′ and 5′ template boundaries during each telomere repeat synthesis cycle by means of TRAP capture and release of TREL and TBEL stretch and compaction, respectively, adjacent to anchor sites (TRE on the CTE and TBE on RBD) on each side. Other elements of TER apparently function to stabilize the closed structure of the TERT ring (loop 4) and in assembly of the t/PK on TERT (PK). The PK has a limited interaction surface with a basic patch on the CTE. This overall architecture appears to be conserved in human telomerase (Wang et al. 2016; Nguyen et al. 2018).
In addition to forming a complex with TRAP that physically encloses the t/PK on the TERT ring, the TEN domain binds p50 in the same way as predicted for human TEN–TPP1 recruitment of telomerase to telomeres. The modeled structure of p50 reveals that it contains an OB-fold oriented to interact with TEN in the same way as predicted for the TPP1 TEL-patch. The p50 interacting surface of TEN changes conformation significantly compared with the crystal structure of the TEN domain (Fig. 3E). The protein p50 also binds Teb1, similar to TPP1–POT1, although details of the interaction appear to be different (Chen et al. 2017; Wang and Feigon 2017). The TEB complex is further anchored to TERT through interactions with TEN (Fig. 3E), an interaction that likely does not exist for POT1. In summary, the 4.8 Å cryo-EM structure of Tetrahymena telomerase with telomeric DNA provided key new insights into telomerase mechanism and DNA handling.
8. CONCLUDING REMARKS: TELOMERASE IS A UNIQUE RNP
Unlike catalytic RNPs such as the ribosome and spliceosome in which the proteins are the scaffold and the RNA does the catalysis (Cech and Steitz 2014), it is now clear that the TERT protein does the actual catalysis. But in this unique RNP, the RT has evolved to require domains specific to TERT and elements of TER in addition to the template that work together for processive synthesis of telomere repeats (Jiang et al. 2018). Another unusual feature of telomerase is the species-specific divergence of TER biogenesis domains, which have co-opted protein complexes from other RNPs for specialized function in telomerase. The first substrate bound cryo-EM structures of Tetrahymena and human telomerase have provided a treasure trove of details of RNP architecture, mechanism, and interaction at telomeres. In the not too distant future, we expect atomic resolution structures of telomerase in which all side chains can be modeled to provide atomic details, elucidating mechanisms and enabling drug targeting and understanding of disease mutations.
ACKNOWLEDGMENTS
Telomerase studies in the Feigon laboratory are funded by National Institutes of Health (NIH) GM048123 and National Science Foundation (NSF) MCB1517625 grants. We acknowledge Department of Energy (DOE) grant DE-FC0202ER63421 for support of the UCLA-DOE NMR, X-ray, and crystallization cores.
Footnotes
Editors: Thomas R. Cech, Joan A. Steitz, and John F. Atkins
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama BM, Stone MD. 2013. Structural biology: A solution to the telomerase puzzle. Nature 496: 177–178. [DOI] [PubMed] [Google Scholar]
- Akiyama BM, Loper J, Najarro K, Stone MD. 2012. The C-terminal domain of Tetrahymena thermophila telomerase holoenzyme protein p65 induces multiple structural changes in telomerase RNA. RNA 18: 653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama BM, Parks JW, Stone MD. 2015. The telomerase essential N-terminal domain promotes DNA synthesis by stabilizing short RNA-DNA hybrids. Nucleic Acids Res 43: 5537–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angrisani A, Vicidomini R, Turano M, Furia M. 2014. Human dyskerin: Beyond telomeres. Biol Chem 395: 593–610. [DOI] [PubMed] [Google Scholar]
- Armanios M, Blackburn EH. 2012. The telomere syndromes. Nat Rev Genet 13: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi SE, DePinho RA. 2010. Telomeres and telomerase in cancer. Carcinogenesis 31: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autexier C, Lue NF. 2006. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem 75: 493–517. [DOI] [PubMed] [Google Scholar]
- Berman AJ, Gooding AR, Cech TR. 2010. Tetrahymena telomerase protein p65 induces conformational changes throughout telomerase RNA (TER) and rescues telomerase reverse transcriptase and TER assembly mutants. Mol Cell Biol 30: 4965–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardes de Jesus B, Blasco MA. 2013. Telomerase at the intersection of cancer and aging. Trends Genet 29: 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Collins K. 2011. Telomerase: An RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol 3: a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Greider CW, Szostak JW. 2006. Telomeres and telomerase: The path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med 12: 1133–1138. [DOI] [PubMed] [Google Scholar]
- Blasco MA. 2003. Telomeres and cancer: A tale with many endings. Curr Opin Genet Dev 13: 70–76. [DOI] [PubMed] [Google Scholar]
- Bley CJ, Qi X, Rand DP, Borges CR, Nelson RW, Chen JJ. 2011. RNA-protein binding interface in the telomerase ribonucleoprotein. Proc Natl Acad Sci 108: 20333–20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Deragon JM. 2009. A comprehensive analysis of the La-motif protein superfamily. RNA 15: 750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash DD, Feigon J. 2017. Structure and folding of the Tetrahymena telomerase RNA pseudoknot. Nucleic Acids Res 45: 482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash DD, Cohen-Zontag O, Kim NK, Shefer K, Brown Y, Ulyanov NB, Tzfati Y, Feigon J. 2013. Pyrimidine motif triple helix in the Kluyveromyces lactis telomerase RNA pseudoknot is essential for function in vivo. Proc Natl Acad Sci 110: 10970–10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. 2014. The noncoding RNA revolution—Trashing old rules to forge new ones. Cell 157: 77–94. [DOI] [PubMed] [Google Scholar]
- Chan H, Wang Y, Feigon J. 2017. Progress in human and Tetrahymena telomerase structure determination. Annu Rev Biophys 46: 199–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Greider CW. 2004. An emerging consensus for telomerase RNA structure. Proc Natl Acad Sci 101: 14683–14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Lingner J. 2013. CST for the grand finale of telomere replication. Nucleus 4: 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Fender J, Legassie JD, Jarstfer MB, Bryan TM, Varani G. 2006. Structure of stem-loop IV of Tetrahymena telomerase RNA. EMBO J 25: 3156–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Redon S, Lingner J. 2012. The human CST complex is a terminator of telomerase activity. Nature 488: 540–544. [DOI] [PubMed] [Google Scholar]
- Chen C, Gu P, Wu J, Chen X, Niu S, Sun H, Wu L, Li N, Peng J, Shi S, et al. 2017. Structural insights into POT1-TPP1 interaction and POT1 C-terminal mutations in human cancer. Nat Commun 8: 14929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xue J, Churikov D, Hass EP, Shi S, Lemon LD, Luciano P, Bertuch AA, Zappulla DC, Geli V, et al. 2018. Structural insights into yeast telomerase recruitment to telomeres. Cell 172: 331–343.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comolli LR, Smirnov I, Xu L, Blackburn EH, James TL. 2002. A molecular switch underlies a human telomerase disease. Proc Natl Acad Sci 99: 16998–17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Reichenbach P, Regamey PO, Banfi D, Chambon M, Turcatti G, Lingner J. 2007. Low- to high-throughput analysis of telomerase modulators with Telospot. Nat Methods 4: 851–853. [DOI] [PubMed] [Google Scholar]
- de Lange T. 2005. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110. [DOI] [PubMed] [Google Scholar]
- Duan J, Li L, Lu J, Wang W, Ye K. 2009. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol Cell 34: 427–439. [DOI] [PubMed] [Google Scholar]
- Eckert B, Collins K. 2012. Roles of telomerase reverse transcriptase N-terminal domain in assembly and activity of Tetrahymena telomerase holoenzyme. J Biol Chem 287: 12805–12814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ED, Collins K. 2010. Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol Cell Biol 30: 2775–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ED, Collins K. 2012. Biogenesis of telomerase ribonucleoproteins. RNA 18: 1747–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Pavletich NP. 2012. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev 26: 2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigon J, Chan H, Jiang J. 2016. Integrative structural biology of Tetrahymena telomerase—Insights into catalytic mechanism and interaction at telomeres. FEBS J 283: 2044–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Nagai K. 2017. Cryo-electron microscopy snapshots of the spliceosome: Structural insights into a dynamic ribonucleoprotein machine. Nat Struct Mol Biol 24: 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty PF, Yamaguchi H, Wiestner A, Baerlocher GM, Sloand E, Zeng WS, Read EJ, Lansdorp PM, Young NS. 2003. Late presentation of dyskeratosis congenita as apparently acquired aplastic anaemia due to mutations in telomerase RNA. Lancet 362: 1628–1630. [DOI] [PubMed] [Google Scholar]
- Forstemann K, Lingner J. 2005. Telomerase limits the extent of base pairing between template RNA and telomeric DNA. EMBO Rep 6: 361–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. 2017. Advances in the field of single-particle cryo-electron microscopy over the last decade. Nat Protoc 12: 209–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Shao L, Colmegna I, Goronzy JJ, Weyand CM. 2009. Telomerase insufficiency in rheumatoid arthritis. Proc Natl Acad Sci 106: 4360–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis AJ, Schuller AP, Skordalakes E. 2008. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 455: 633–637. [DOI] [PubMed] [Google Scholar]
- Gladyshev EA, Arkhipova IR. 2007. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci 104: 9352–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. 1987. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 51: 887–898. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337. [DOI] [PubMed] [Google Scholar]
- Hamma T, Ferre-D’Amare AR. 2004. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 Å resolution. Structure 12: 893–903. [DOI] [PubMed] [Google Scholar]
- Harkisheimer M, Mason M, Shuvaeva E, Skordalakes E. 2013. A motif in the vertebrate telomerase N-terminal linker of TERT contributes to RNA binding and telomerase activity and processivity. Structure 21: 1870–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengesbach M, Kim NK, Feigon J, Stone MD. 2012. Single-molecule FRET reveals the folding dynamics of the human telomerase RNA pseudoknot domain. Angew Chem Int Ed Engl 51: 5876–5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman H, Skordalakes E. 2016. Crystallographic studies of telomerase. Methods Enzymol 573: 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman H, Rice C, Skordalakes E. 2017. Structural analysis reveals the deleterious effects of telomerase mutations in bone marrow failure syndromes. J Biol Chem 292: 4593–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan B, Wright WE, Shay JW. 2014. Cell biology of disease: Telomeropathies: An emerging spectrum disorder. J Cell Biol 205: 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K, Upton H, Miracco EJ, Jiang J, Zhou ZH, Feigon J, Collins K. 2013. Tetrahymena telomerase holoenzyme assembly, activation, and inhibition by domains of the p50 central hub. Mol Cell Biol 33: 3962–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brown AF, Wu J, Xue J, Bley CJ, Rand DP, Wu L, Zhang R, Chen JJ, Lei M. 2014. Structural basis for protein-RNA recognition in telomerase. Nat Struct Mol Biol 21: 507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Podell ER, Wuttke DS, Cech TR. 2005. Soluble domains of telomerase reverse transcriptase identified by high-throughput screening. Protein Sci 14: 2051–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Podell ER, Cech TR. 2006. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol 13: 218–225. [DOI] [PubMed] [Google Scholar]
- Jansson LI, Akiyama BM, Ooms A, Lu C, Rubin SM, Stone MD. 2015. Structural basis of template-boundary definition in Tetrahymena telomerase. Nat Struct Mol Biol 22: 883–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Miracco EJ, Hong K, Eckert B, Chan H, Cash DD, Min B, Zhou ZH, Collins K, Feigon J. 2013. The architecture of Tetrahymena telomerase holoenzyme. Nature 496: 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan H, Cash DD, Miracco EJ, Ogorzalek Loo RR, Upton HE, Cascio D, O’Brien Johnson R, Collins K, Loo JA, et al. 2015. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science 350: aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Wang Y, Sušac L, Chan H, Basu R, Zhou ZH, Feigon J. 2018. Structure of telomerase with telomeric DNA. Cell 173: 1179–1190.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczyluk J, Nouwens AS, Holien JK, Adams TE, Lovrecz GO, Parker MW, Cohen SB, Bryan TM. 2011. Direct involvement of the TEN domain at the active site of human telomerase. Nucleic Acids Res 39: 1774–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher C, Teixeira MT, Forstemann K, Lingner J. 2002. Telomerase: Biochemical considerations for enzyme and substrate. Trends Biochem Sci 27: 572–579. [DOI] [PubMed] [Google Scholar]
- Ketele A, Kiss T, Jady BE. 2016. Human intron-encoded AluACA RNAs and telomerase RNA share a common element promoting RNA accumulation. RNA biology 13: 1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NK, Zhang Q, Zhou J, Theimer CA, Peterson RD, Feigon J. 2008. Solution structure and dynamics of the wild-type pseudoknot of human telomerase RNA. J Mol Biol 384: 1249–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NK, Theimer CA, Mitchell JR, Collins K, Feigon J. 2010. Effect of pseudouridylation on the structure and activity of the catalytically essential P6.1 hairpin in human telomerase RNA. Nucleic Acids Res 38: 6746–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NK, Zhang Q, Feigon J. 2014. Structure and sequence elements of the CR4/5 domain of medaka telomerase RNA important for telomerase function. Nucleic Acids Res 42: 3395–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BK, Park CJ, Fernandez CF, Chim N, Ding Y, Chanfreau G, Feigon J. 2011. Structure of H/ACA RNP protein Nhp2p reveals cis/trans isomerization of a conserved proline at the RNA and Nop10 binding interface. J Mol Biol 411: 927–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Mitchell JR, Collins K. 2001. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol 21: 990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeper TC, Varani G. 2005. The structure of an enzyme-activating fragment of human telomerase RNA. RNA 11: 394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeper T, Leulliot N, Varani G. 2003. The solution structure of an essential stem-loop of human telomerase RNA. Nucleic Acids Res 31: 2614–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux B, Laterreur N, Perederina A, Noel JF, Dubois ML, Krasilnikov AS, Wellinger RJ. 2016. Active yeast telomerase shares subunits with ribonucleoproteins RNase P and RNase MRP. Cell 165: 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ye K. 2006. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 443: 302–307. [DOI] [PubMed] [Google Scholar]
- Li S, Duan J, Li D, Ma S, Ye K. 2011a. Structure of the Shq1-Cbf5-Nop10-Gar1 complex and implications for H/ACA RNP biogenesis and dyskeratosis congenita. EMBO J 30: 5010–5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Duan J, Li D, Yang B, Dong M, Ye K. 2011b. Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase. Genes Dev 25: 2409–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Xue S, Terns RM, Terns MP, Li H. 2007. Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex. Nat Struct Mol Biol 14: 1189–1195. [DOI] [PubMed] [Google Scholar]
- Lin KW, McDonald KR, Guise AJ, Chan A, Cristea IM, Zakian VA. 2015. Proteomics of yeast telomerase identified Cdc48-Npl4-Ufd1 and Ufd4 as regulators of Est1 and telomere length. Nat Commun 6: 8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276: 561–567. [DOI] [PubMed] [Google Scholar]
- Lue NF, Lin YC, Mian IS. 2003. A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol Cell Biol 23: 8440–8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manival X, Charron C, Fourmann JB, Godard F, Charpentier B, Branlant C. 2006. Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5-aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity. Nucleic Acids Res 34: 826–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone A, Walne A, Dokal I. 2005. Dyskeratosis congenita: Telomerase, telomeres and anticipation. Curr Opin Genet Dev 15: 249–257. [DOI] [PubMed] [Google Scholar]
- Mason M, Schuller A, Skordalakes E. 2011. Telomerase structure function. Curr Opin Struct Biol 21: 92–100. [DOI] [PubMed] [Google Scholar]
- Min B, Collins K. 2009. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell 36: 609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B, Collins K. 2010. Multiple mechanisms for elongation processivity within the reconstituted Tetrahymena telomerase holoenzyme. J Biol Chem 285: 16434–16443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miracco EJ, Jiang J, Cash DD, Feigon J. 2014. Progress in structural studies of telomerase. Curr Opin Struct Biol 24: 115–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K. 1999a. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol 19: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K. 1999b. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402: 551–555. [DOI] [PubMed] [Google Scholar]
- Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E. 2010. Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol 17: 513–518. [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Cech TR. 2013. Finding the end: Recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol 14: 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. 2012. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492: 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AD, Shippen DE. 2015. Evolution of TERT-interacting lncRNAs: Expanding the regulatory landscape of telomerase. Front Genet 6: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen THD, Tam J, Wu RA, Greber BJ, Toso D, Nogales E, Collins K. 2018. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature 557: 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor CM, Collins K. 2006. A novel RNA binding domain in Tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol Cell Biol 26: 2029–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor CM, Lai CK, Collins K. 2005. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J Biol Chem 280: 17533–17539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. 2008. How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334. [DOI] [PubMed] [Google Scholar]
- Parisien M, Major F. 2008. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 452: 51–55. [DOI] [PubMed] [Google Scholar]
- Petrova OA, Mantsyzov AB, Rodina EV, Efimov SV, Hackenberg C, Hakanpaa J, Klochkov VV, Lebedev AA, Chugunova AA, Malyavko AN, et al. 2018. Structure and function of the N-terminal domain of the yeast telomerase reverse transcriptase. Nucleic Acids Res 46: 1525–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Podlevsky JD, Chen JJ. 2016. Evolutionary perspectives of telomerase RNA structure and function. RNA Biol 13: 720–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen JJ. 2008. The telomerase database. Nucleic Acids Res 36: D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polshakov VI, Petrova OA, Parfenova YY, Efimov SV, Klochkov VV, Zvereva MI, Dontsova OA. 2016. NMR assignments of the N-terminal domain of Ogataea polymorpha telomerase reverse transcriptase. Biomol NMR Assign 10: 183–187. [DOI] [PubMed] [Google Scholar]
- Prakash A, Borgstahl GE. 2012. The structure and function of replication protein A in DNA replication. Subcell Biochem 62: 171–196. [DOI] [PubMed] [Google Scholar]
- Prathapam R, Witkin KL, O’Connor CM, Collins K. 2005. A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat Struct Mol Biol 12: 252–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE. 2010. Evolution of CST function in telomere maintenance. Cell Cycle 9: 3157–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F, Cech TR. 2008. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol 15: 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu G, Kaushal PS, Wang J, Shigematsu H, Piazza CL, Agrawal RK, Belfort M, Wang HW. 2016. Structure of a group II intron in complex with its reverse transcriptase. Nat Struct Mol Biol 23: 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards RJ, Theimer CA, Finger LD, Feigon J. 2006a. Structure of the Tetrahymena thermophila telomerase RNA helix II template boundary element. Nucleic Acids Res 34: 816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards RJ, Wu H, Trantirek L, O’Connor CM, Collins K, Feigon J. 2006b. Structural study of elements of Tetrahymena telomerase RNA stem-loop IV domain important for function. RNA 12: 1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart AR, Collins K. 2011. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol Cell 42: 308–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. 2003. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333: 721–745. [DOI] [PubMed] [Google Scholar]
- Rouda S, Skordalakes E. 2007. Structure of the RNA-binding domain of telomerase: Implications for RNA recognition and binding. Structure 15: 1403–1412. [DOI] [PubMed] [Google Scholar]
- Sarek G, Marzec P, Margalef P, Boulton SJ. 2015. Molecular basis of telomere dysfunction in human genetic diseases. Nat Struct Mol Biol 22: 867–874. [DOI] [PubMed] [Google Scholar]
- Sauerwald A, Sandin S, Cristofari G, Scheres SH, Lingner J, Rhodes D. 2013. Structure of active dimeric human telomerase. Nat Struct Mol Biol 20: 454–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Chen S. 2012. Prevention of overfitting in cryo-EM structure determination. Nat Methods 9: 853–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Cech TR. 2015. Human telomerase: Biogenesis, trafficking, recruitment, and activation. Genes Dev 29: 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Dalby AB, Cech TR. 2014. Identification of human TERT elements necessary for telomerase recruitment to telomeres. eLife 3 10.7554/eLife.03563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton AN, Youmans DT, Collins K. 2012. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J Biol Chem 287: 34455–34464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastry S, Steinberg-Neifach O, Lue N, Stone MD. 2018. Direct observation of nucleic acid binding dynamics by the telomerase essential N-terminal domain. Nucleic Acids Res 46: 3088–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW. 2016. Role of telomeres and telomerase in aging and cancer. Cancer Discov 6: 584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer K, Brown Y, Gorkovoy V, Nussbaum T, Ulyanov NB, Tzfati Y. 2007. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol Cell Biol 27: 2130–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Wang Z, Koo BK, Patel A, Cascio D, Collins K, Feigon J. 2012. Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol Cell 47: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Choi CP, Feigon J. 2013. xRRM: A new class of RRM found in the telomerase La family protein p65. RNA Biol 10: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, Baker D. 2013. High-resolution comparative modeling with RosettaCM. Structure 21: 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamos JL, Lentzsch AM, Lambowitz AM. 2017. Structure of a thermostable group II intron reverse transcriptase with template-primer and its functional and evolutionary implications. Mol Cell 68: 926–939.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Weinberg RA. 2006. Telomeres: Cancer to human aging. Annu Rev Cell Dev Biol 22: 531–557. [DOI] [PubMed] [Google Scholar]
- Stone MD, Mihalusova M, O’Connor CM, Prathapam R, Collins K, Zhuang X. 2007. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature 446: 458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugitani N, Chazin WJ. 2015. Characteristics and concepts of dynamic hub proteins in DNA processing machinery from studies of RPA. Prog Biophys Mol Biol 117: 206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer CA, Feigon J. 2006. Structure and function of telomerase RNA. Curr Opin Struct Biol 16: 307–318. [DOI] [PubMed] [Google Scholar]
- Theimer CA, Finger LD, Trantirek L, Feigon J. 2003. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc Natl Acad Sci 100: 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer CA, Blois CA, Feigon J. 2005. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell 17: 671–682. [DOI] [PubMed] [Google Scholar]
- Theimer CA, Jady BE, Chim N, Richard P, Breece KE, Kiss T, Feigon J. 2007. Structural and functional characterization of human telomerase RNA processing and Cajal body localization signals. Mol Cell 27: 869–881. [DOI] [PubMed] [Google Scholar]
- Townsley DM, Dumitriu B, Young NS. 2014. Bone marrow failure and the telomeropathies. Blood 124: 2775–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium, Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, Gibbs R, Beeman RW, Brown SJ, Bucher G, et al. 2008. The genome of the model beetle and pest Tribolium castaneum. Nature 452: 949–955. [DOI] [PubMed] [Google Scholar]
- Upton HE, Chan H, Feigon J, Collins K. 2017. Shared subunits of Tetrahymena telomerase holoenzyme and replication protein A have different functions in different cellular complexes. J Biol Chem 292: 217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan B, Tang T, Upton H, Shuai J, Zhou Y, Li S, Chen J, Brunzelle JS, Zeng Z, Collins K, et al. 2015. The Tetrahymena telomerase p75-p45-p19 subcomplex is a unique CST complex. Nat Struct Mol Biol 22: 1023–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Feigon J. 2017. Structural biology of telomerase and its interaction at telomeres. Curr Opin Struct Biol 47: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Gilley D, Blackburn EH. 1998. A novel specificity for the primer-template pairing requirement in Tetrahymena telomerase. EMBO J 17: 1152–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yesselman JD, Zhang Q, Kang M, Feigon J. 2016. Structural conservation in the template/pseudoknot domain of vertebrate telomerase RNA from teleost fish to human. Proc Natl Acad Sci 113: E5125–E5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegman-Ostrosky T, Savage SA. 2017. The genomics of inherited bone marrow failure: From mechanism to the clinic. Br J Haematol 177: 526–542. [DOI] [PubMed] [Google Scholar]
- Witkin KL, Collins K. 2004. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev 18: 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RA, Collins K. 2014. Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. EMBO J 33: 921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RA, Tam J, Collins K. 2017a. DNA-binding determinants and cellular thresholds for human telomerase repeat addition processivity. EMBO J 36: 1908–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RA, Upton HE, Vogan JM, Collins K. 2017b. Telomerase mechanism of telomere synthesis. Annu Rev Biochem 86: 439–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt HD, Tsang AR, Lobb DA, Beattie TL. 2009. Human telomerase reverse transcriptase (hTERT) Q169 is essential for telomerase function in vitro and in vivo. PLoS One 4: e7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi L, Cech TR. 2014. Inventory of telomerase components in human cells reveals multiple subpopulations of hTR and hTERT. Nucleic Acids Res 42: 8565–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Podlevsky JD, Qi X, Bley CJ, Chen JJ. 2010. A novel motif in telomerase reverse transcriptase regulates telomere repeat addition rate and processivity. Nucleic Acids Res 38: 1982–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Cech TR. 2008. Mutation in TERT separates processivity from anchor-site function. Nat Struct Mol Biol 15: 870–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Nandakumar J, Cech TR. 2010. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev 24: 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Min B, Huang J, Hong K, Yang Y, Collins K, Lei M. 2011. Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proc Natl Acad Sci 108: 20357–20361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Kim NK, Peterson RD, Wang Z, Feigon J. 2010. Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. Proc Natl Acad Sci 107: 18761–18768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Kim NK, Feigon J. 2011. Architecture of human telomerase RNA. Proc Natl Acad Sci 108: 20325–20332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Pyle AM. 2016. Crystal structures of a group II intron maturase reveal a missing link in spliceosome evolution. Nat Struct Mol Biol 23: 558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE. 2012. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150: 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]