Abstract
Glycosylation plays a major role in the structural diversification of plant natural products. It influences the properties of molecules by modifying the reactivity and solubility of the corresponding aglycones, so influencing cellular localization and bioactivity. Glycosylation of plant natural products is usually carried out by uridine diphosphate(UDP)-dependent glycosyltransferases (UGTs) belonging to the carbohydrate-active enzyme glycosyltransferase 1 (GT1) family. These enzymes transfer sugars from UDP-activated sugar moieties to small hydrophobic acceptor molecules. Plant GT1s generally show high specificity for their sugar donors and recognize a single UDP sugar as their substrate. In contrast, they are generally promiscuous with regard to acceptors, making them attractive biotechnological tools for small molecule glycodiversification. Although microbial hosts have traditionally been used for heterologous engineering of plant-derived glycosides, transient plant expression technology offers a potentially disruptive platform for rapid characterization of new plant glycosyltransferases and biosynthesis of complex glycosides.
Collectively, plants synthesize a diverse array of chemicals, most likely as a means of survival in diverse ecological niches. These molecules have been variously associated with abiotic stress resistance (Trossat et al. 1998; Nuccio et al. 1999), defense against herbivores and pathogens (Osbourn 1996; Vetter 2000; Howe and Jander 2008), establishment of symbiotic interactions (Oldroyd 2013), allelopathy (Weston and Mathesius 2013), and attraction of pollinators (Ogata et al. 2005; Theis and Raguso 2005; Yu and Utsumi 2009). Plant-specialized metabolites are derived from a repertoire of different types of scaffolds. These scaffolds commonly undergo further modification (e.g., oxidation, glycosylation, methylation, and acylation) to generate a wealth of chemical diversity, with around one million specialized metabolites being reported from plants so far (Afendi et al. 2012). Among these modifications, glycosylation has a profound impact on the physicochemical and bioactive properties of phytochemicals, affecting solubility, cellular localization, and bioactivity.
Here we review recent developments in understanding the enzymes involved in plant natural product glycosylation. We also consider the various heterologous expression hosts used for metabolic engineering of plant glycosides. Finally, we discuss the potential for harnessing enzymes from plants for glycodiversification of small molecules for medicinal, agronomic, and industrial applications.
GLYCOSYLATION OF PLANT-SPECIALIZED METABOLITES
The Impact of Glycosylation on Bioactivity
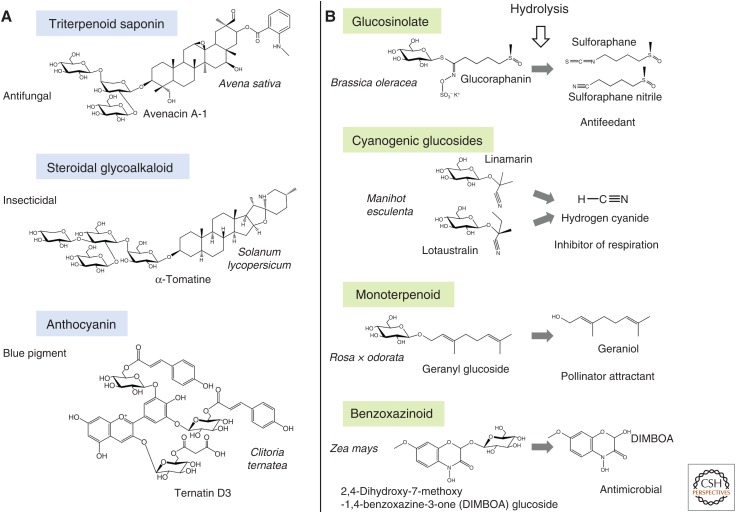
Many plant-specialized metabolites accumulate as glycosides. Glycosylation is often a means of storage of bioactive molecules (e.g., endogenous phytoanticipins or xenobiotics). While some plant glycosides are biologically active in their glycosylated form, in other cases partial or complete hydrolysis of sugars may activate or enhance bioactivity (Fig. 1). In scenarios where glycosylation is required for bioactivity, complex glycosylation patterns are often observed. For example, triterpenoid saponins and steroidal glycoalkaloids often contain oligosaccharide chains consisting of different types of sugar units (Vincken et al. 2007). Such compounds protect plants against attack by pathogenic microbes, herbivores, and competing plant species (Augustin et al. 2011; Sawai and Saito 2011). Two examples, avenacin A-1 from oat and α-tomatine from tomato, are shown in Figure 1A. Glycosylation is critical for the ability of triterpenoid saponins and steroidal glycoalkaloids to integrate into and permeabilize plasma membranes, and also for bioactivity (Bowyer et al. 1995). Glycosylation is also important for flavonoid pigment stability, enabling intra- and intermolecular association of anthocyanins with other flavonoids, metal ions (metalloanthocyanins), or aromatic organic acids under conducive pH conditions. The central role of flavonoid glycosidic moieties in formation of such hydrophobic stacking structures has recently been demonstrated by the engineering of blue chrysanthemums (Noda et al. 2017). Ternatin D3, one of the anthocyanins responsible for the blue coloration of Clitoria ternatea, is also shown in Figure 1A.
Figure 1.
Examples of glycosylated plant-specialized metabolites. Compounds that are active in their fully glycosylated forms (A) or upon hydrolysis (B) are shown.
Glucosinolates and cyanogenic glucosides (Fig. 1B) are examples of situations where glycosylated plant natural products are activated by removal of sugars to give deterrent or toxic breakdown products that provide protection against pest and pathogen attack. The enzymes that catalyze deglucosylation (thiospecific glucohydrolases myrosinases or specific β-glycosidases, respectively) are normally stored away from their substrates in separate subcellular compartments or cell types but are released on tissue damage (Hopkins et al. 2009; Gleadow and Møller 2014). Other examples of activation of plant natural products by hydrolysis of sugars include glycoside-bound volatiles (e.g., monoterpenes, sesquiterpenes, phenolics) with various roles in pollinator attraction, biotic/abiotic stress tolerance, and communication between plants (e.g., geranyl glucoside) and benzoxazinoid defense compounds (e.g., 2,4-dihydroxy-7-methoxy-1,4-benzoxazinone-3-one (DIMBOA) glucoside) (Fig. 1B; de Bruijn et al. 2018; Song et al. 2018). Those compounds that are activated by hydrolysis, in general, have relatively simple glycosidic moieties and are usually mono- or diglycosides, typically containing d-glucose.
Glycosyl Transferase Family 1 (GT1): An Engine for Glycodiversification
Enzymes that build or break down the considerable diversity of glycosylated structures found in living organisms (e.g., proteins, lipids, polysaccharides) are collectively referred to as carbohydrate-active enzymes (CAZymes) (Cantarel et al. 2009). CAZymes include the glycosyltransferases (GTs), a large enzyme superfamily that has been classified into 106 different enzyme families (www.cazy.org/glycosyltransferases.html). The enzymes that are primarily responsible for glycosylation of plant natural products belong to glycosyltransferase family 1 (GT1). Members of the GT1 enzyme family in plants use uridine diphosphate (UDP)-activated sugar donors to transfer sugar units onto small molecules and are therefore also referred to as UDP-dependent glycosyltransferases (UGTs).
GT1 enzymes make major contributions to the glycodiversification of plant-specialized metabolites (Vogt and Jones 2000; Bowles et al. 2006). They are involved in the production of important defense compounds such as terpenoid glycosides, glucosinolates, cyanogenic glycosides, and flavonoid glycosides (Sønderby et al. 2010; Sawai and Saito 2011; Gleadow and Møller 2014). The stability of pigments (e.g., anthocyanins), the taste of fruit (e.g., flavonoids, diterpenoids), and the retention of aromas in flowers or fruits (e.g., monoterpenes, phenyl alcohols) are also modulated via GT1-mediated glycosylation (Noda 2018; Song et al. 2018). GT1 enzymes are also involved in regulation of plant growth and development via modulation of phytohormone homeostasis (Piotrowska and Bajguz 2011). They further enable xenobiotic detoxification through glucoconjugation, a step that precedes transfer and subsequent storage of the modified xenobiotic in the vacuole (Brazier-Hicks et al. 2018).
GT1 PHYLOGENY
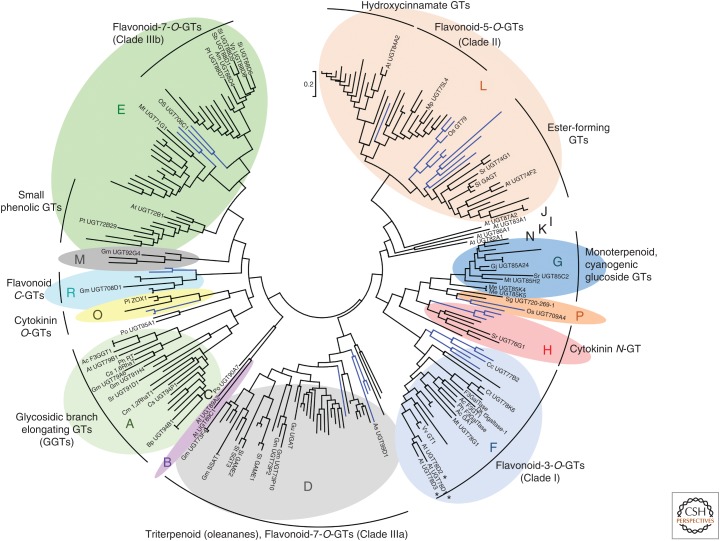
The family 1 GTs are one of the largest groups of natural product-decorating enzymes in plants. The expansion of this family in higher plants likely reflects chemical diversification during adaptation to life on land (Yonekura-Sakakibara and Hanada 2011; Caputi et al. 2012). Mining of the complete genome sequence of Arabidopsis thaliana has identified 107 predicted GT1 genes. These GT1 enzymes have been classified into 14 monophyletic groups (groups A to N) (Ross et al. 2001). Three new phylogenetic groups (O, P, and Q), which are not found in A. thaliana but are present in other plant species, have also subsequently been reported (Fig. 2; Caputi et al. 2012; Li et al. 2014).
Figure 2.
Phylogenetic tree of characterized plant glycosyltransferases 1 (GT1s). Reconstruction of GT1 phylogeny from a collection of 246 biochemically characterized GT1 protein sequences. The groups are delineated as defined by Ross et al. (2001) and Caputi et al. (2012). Discrete clusters of enzymes with similar activities are indicated. Monocot-specific branches are shown in blue. The tree was constructed with Mega 6.06 (Tamura et al. 2013) using the maximum likelihood method from a protein alignment obtained with MUSCLE 3.8 (Edgar 2004). *, Closely related Arabidopsis thaliana family F GT1s with different sugar specificities (see main text). The scale bar indicates 0.2 substitutions per site at the amino acid level.
Most plant GT1 enzymes use UDP glucose as their sugar donor. However, the ability to use alternative sugar donors appears to have evolved multiple times, and closely related enzymes may use different sugar donors. This is illustrated by three closely related A. thaliana group F flavonoid GTs, UGT78D1, UGT78D2, and UGT78D3 (indicated by asterisks in Fig. 2), each of which use a different UDP sugar donor (Jones et al. 2003; Tohge et al. 2005; Yonekura-Sakakibara et al. 2008). The discovery that a small number of key amino acid residues are critical for sugar donor specificity may explain the rapid evolution of sugar donor specificity observed throughout the phylogeny.
Although some light has been shed on determinants of sugar donor specificity, the determinants of acceptor recognition remain obscure and the link between phylogeny and acceptor utilization is not always straightforward. However, careful examination of the phylogenetic reconstruction of functionally characterized GT1s does reveal discrete clustering of GT1s with structurally related acceptors or functions (Fig. 2). Early phylogenetic studies of comprehensive collections of plant GT1 enzymes have focused mainly on flavonoid GTs. The tree topology of flavonoid GT1s broadly recapitulates their regiospecificity (Vogt and Jones 2000; Lim et al. 2004), suggesting that regiospecificity emerged prior to speciation. Different types of flavonoid GTs are located in four distinct (sub)clades that correlate with their respective regiospecificities (Fig. 2; Vogt and Jones 2000; Noguchi et al. 2009). Another group (group A) contains GT1s that extend flavonoid sugar chains, and also enzymes that add sugars to the sugar chains of a variety of other types of scaffolds, collectively referred to as glycosidic branch elongating GTs (GGTs) (Fig. 2). Additional discrete clusters of GT1s that recognize structurally similar acceptors have also been recently identified (Augustin et al. 2011; Yonekura-Sakakibara and Hanada 2011). An important monophyletic group composed exclusively of C-GTs has emerged in recent work (Nagatomo et al. 2014), labeled as group R in Figure 2. However, there are many exceptions to the premise that GT1 function can be predicted based on phylogeny alone. For example, numerous examples of enzymes that add sugars to the sugar chains of plant natural products have been found outside of group A (Mohamed et al. 2011; Sayama et al. 2012; Itkin et al. 2013). Similarly, whereas a number of triterpenoid GT1s are present in group D, there are numerous examples of triterpenoid GT1s that belong to other GT1 families (Achnine et al. 2005; Jung et al. 2014; Wei et al. 2015).
A growing number of monocot GT1s have now been characterized. Some of these fall into monocot-specific GT1 subfamilies within the larger monophyletic groups (D, E, and L) (Fig. 2; Caputi et al. 2012). Most of these GT1s seem to have similar acceptor specificity to their eudicot relatives within the same monophyletic group, although further work is needed to establish whether these monocot-specific subfamilies are functionally equivalent to their eudicot counterparts.
GT1s—PROMISCUOUS BIOCATALYSTS FOR SCAFFOLD GLYCOSYLATION
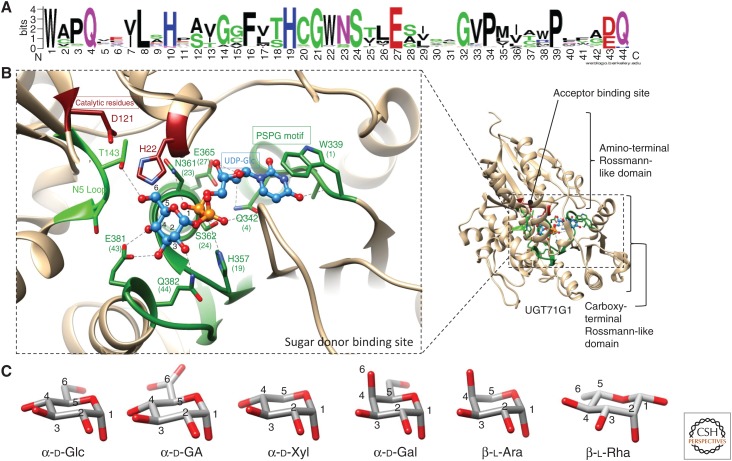
UGTs catalyze the transfer of a sugar from a UDP sugar donor to an acceptor (usually a lipophilic molecule). GT1s generally show high specificity for their sugar donors and a single activated sugar is efficiently recognized as the substrate (Osmani et al. 2009). Plant GT1 enzymes accommodate their sugar donors primarily via a highly conserved motif (Fig. 3A) called the plant secondary product glycosyltransferase (PSPG) motif (Hughes and Hughes 1994; Ross et al. 2001). In contrast to the observed specificity for particular sugar donors, in vitro studies using recombinant enzymes from A. thaliana have highlighted the promiscuity of many GT1s with regard to acceptor recognition (Vogt and Jones 2000; Lim et al. 2002; Caputi et al. 2008). For example, UGT85K4 and UGT85K5 from cassava (Manihot esculenta) (group G) are involved in cyanogenic glucoside biosynthesis. The ability of these enzymes to glycosylate precursors of the cyanogenic glycosides linamarin and lotaustralin in vitro is consistent with their role in planta. However, these enzymes also recognize a wide range of other acceptors in in vitro assays, including flavonoids, simple alcohols, and various hydroxynitriles (Kannangara et al. 2011). In contrast, other GT1s show high specificity toward one or a few structurally related compounds. For example, UGT85A24 from Gardenia jasminoides, which is part of an iridoid biosynthetic pathway, shows high specificity toward the iridoid 7-deoxyloganetin but does not exhibit activity toward the immediate precursor 7-deoxyloganetic acid (Nagatoshi et al. 2011). Most GT1s act on hydroxyl or carboxyl groups, but N-, S-, or C-glycosylation can also occur (Jones and Vogt 2001; Grubb et al. 2004; Hou et al. 2004; Brazier-Hicks et al. 2007b, 2009; Wang et al. 2011).
Figure 3.
Determinants of sugar specificity of plant glycosyltransferases 1 (GT1s). (A) Consensus plant secondary product glycosyltransferase (PSPG) motif generated by weblogo (weblogo.berkeley.edu) from an alignment of characterized GT1s. (B) The sugar donor-binding site for the crystal structure of the GT1 enzyme UGT71G (a flavonoid/triterpenoid O-glucosyltransferase Medicago truncatula) in complex with uridine diphosphate glucose (UDP-Glc) (PDB code 2ACW). The PSPG motif is shown in dark green, the N5 loop in light green, and catalytic residues in dark red. UDP-Glc is shown as a ball and stick model and colored in blue. Proposed hydrogen bonds are shown as dashed lines. (C) Compared structures of most common sugar donors of plant GT1s.
STRUCTURES OF PLANT GT1 ENZYMES
To date, a total of nine crystal structures of plant GT1s have been solved (Table 1; Shao et al. 2005; Offen et al. 2006; Brazier-Hicks et al. 2007a; Li et al. 2007; Modolo et al. 2009; Hiromoto et al. 2015; Wetterhorn et al. 2016; George Thompson et al. 2017; Hsu et al. 2018). Although those enzymes have relatively low amino acid sequence identity (typically 25%–45%), they share very similar 3D structures (Osmani et al. 2009; Wang 2009). This common structure is called the GT-B fold and is formed of two Rossmann-like domains, composed of central β-strands surrounded by several α-helices (Fig. 3B). The catalytic site is localized in a cleft between the two domains.
Table 1.
Plant GT1 enzymes for which crystal structures have been determined
| Gene name | Species | PDB entry | Activity | References |
|---|---|---|---|---|
| UGT71G1 | Medicago truncatula | 2ACW | Flavonoid/triterpenoid O-glucosyltransferase | Shao et al. 2005 |
| VvGT1 | Vitis vinifera | 2C1Z | Flavonoid 3-O-glucosyltransferase | Offen et al. 2006 |
| UGT72B1 | Arabidopsis thaliana | 2VCH | Chlorinated phenols N/O-glucosyltransferase | Brazier-Hicks et al. 2007a |
| UGT85H2 | M. truncatula | 2PQ6 | Flavonoid 3-O-glucosyltransferase | Li et al. 2007 |
| UGT78G1 | M. truncatula | 3HBF | Flavonoid 3-O-glucosyltransferase | Modolo et al. 2009 |
| UGT78K6 | Clitoria ternatea | 4REM | Flavonoid 3-O-glucosyltransferase | Hiromoto et al. 2015 |
| Os79 | Oryza sativa | 5TME | Deoxynivalenol O-glucosyltransferase | Wetterhorn et al. 2016 |
| UGT74F2 | A. thaliana | 5U6M | Salicylic acid O-glucosyltransferase | George Thompson et al. 2017 |
| UGT72B29 | Persicaria tinctoria | 5NLM | Indoxyl O-glucosyltransferase | Hsu et al. 2018 |
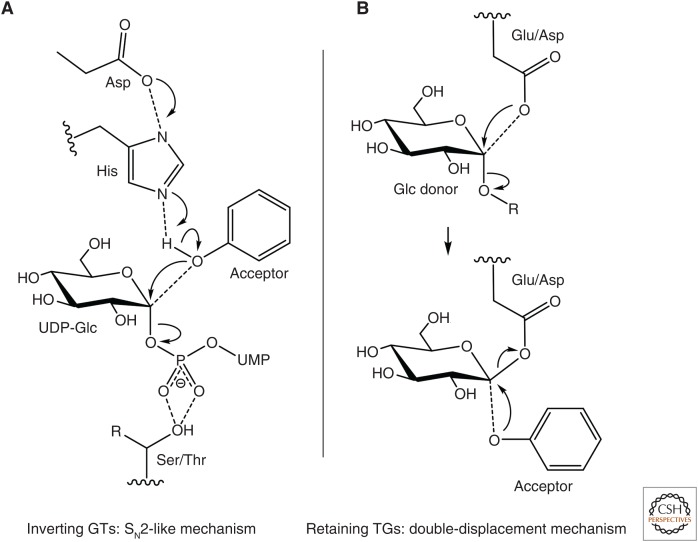
In plant GT1s, two highly conserved residues play a crucial part in the SN2-like mechanism in which the stereochemistry of the C1 anomeric carbon is inverted during the reaction, making them inverting GTs (Fig. 4A; Wang 2009). A histidine positioned around the 20th residue (Fig. 3A) acts as a general base to deprotonate the acceptor. Nucleophilic attack of the C1 carbon of the UDP-sugar is achieved by the deprotonated acceptor (Fig. 4A; Lairson et al. 2008; Wang 2009). A conserved aspartate residue in proximity to the histidine has been proposed to help to balance the charge from the histidine to form an acceptor-His-Asp catalytic triad (Fig. 4A). However, the existence of rare examples where those residues are not conserved suggests that alternative catalytic mechanisms might take place (Noguchi et al. 2007; Wang et al. 2011, 2013; Wilson et al. 2017). The crystal structure of the ester-forming salicylic acid glucosyltransferase UGT74F2 from A. thaliana suggests that other residues could be used to help deprotonate the acceptor for nucleophilic attack by a carboxylate oxygen (George Thompson et al. 2017). Similar mechanisms may also exist in ester-forming UGT families that lack the catalytic aspartate (Wilson et al. 2017). The recent crystallization of the deoxynivalenol-glucosyltransferase Os79 from rice, coupled with site-directed mutagenesis, suggests that Thr291 plays a critical role in either positioning the β-phosphate of UDP or protonating it (Wetterhorn et al. 2016). This residue is extremely well conserved in all UGTs as either a serine or threonine (Ser/Thr in Fig. 4A), with a few exceptions, including the Lamiales flavonoid 7-O-glucuronosyltransferase UGT88Ds (replaced by arginine), the A. thaliana arabinosyltransferase UGT78D3 (replaced by arginine), the majority of the C-glucosyltransferases from group Q (replaced by asparagine), and the cassava α-hydroxynitrile glucosyltransferases UGT85K4 and UGT85K5 (replaced by cysteine). These striking amino acid substitutions in GT1s with noncanonical substrates could suggest a pivotal role for this residue position in determining the substrate specificity and catalytic properties of GT1s.
Figure 4.
Proposed catalytic mechanisms of retaining glycosyltransferases 1 (GT1s) (A) and inverting glycosyl hydrolase family 1 (GH1) transglucosidases (TGs) (B). UMP, Uridine monophosphate.
ACCEPTOR RECOGNITION
The determinants of acceptor recognition in GT1s are poorly understood (Osmani et al. 2009; Wang 2009). GT1 acceptor promiscuity is reflected by the nature of the acceptor-binding site, which consists primarily of a large apolar pocket within the amino-terminal domain (Fig. 3B). Residues within the acceptor-binding pocket are located in regions that are poorly conserved in plant GT1s (Osmani et al. 2009). Little is known about the mechanisms underlying the correct orientation of the acceptor molecule and therefore the regiospecificity of glycosylation catalyzed by GT1s. Hydrophobic stacking of apolar acceptors by aromatic residues, plus the steric constraints relative to the shape and volume of the pocket, seem to be the primary determinants of acceptor recognition. The regiospecificity of particular GT1 isoforms appears to be defined by local subtle variations in the polarity and electronic surroundings of the binding pocket, allowing interactions of GT1s with their ligands (He et al. 2006; Li et al. 2007; Modolo et al. 2009). This is particularly exemplified by the crystal complexes of UGT78K6, an anthocyanidin 3-O-glucosyltransferase from C. ternatea, where polar residues surrounding the binding site allow for regiospecificity of the enzyme for diverse flavonoid acceptors (Hiromoto et al. 2015). The success of engineering strategies directed at altering acceptor specificity by modifying the overall volume and shape of the acceptor-binding pocket also point in this direction (Bai et al. 2016; Olsson et al. 2016; Wetterhorn et al. 2017).
A sequence-based computational tool that aims to predict GT1 function has recently been developed (Yang et al. 2018). By coupling machine learning with a comprehensive functional dataset (biochemical characterization of 54 recombinant A. thaliana GT1s over 17 sugar donors and 91 acceptors), algorithms were trained to predict GT1 function. This sequence-based prediction tool, GT-predict, may in future guide the curation and discovery of the burgeoning number of predicted GT1s emerging from large-scale plant genome-sequencing initiatives. This tool does not, however, consider the 3D orientation of the chemical groups relative to the acceptor scaffold, and so does not take into account regiochemical biases that could sterically favor/hinder ligand binding.
SUGAR DONOR SPECIFICITY
The sugar donor-binding pocket is located on the carboxy-terminal domain of the enzyme and is mainly composed of the PSPG motif. Highly conserved residues within the PSPG motif that interact with the UDP part of the sugar donor have been identified from crystal structures (Fig. 3). The first tryptophan (W1) in the PSPG motif forms a hydrophobic platform that stacks with the uracil ring of UDP (this residue is labeled W339 in Fig. 3B). The glutamine (Q) at the fourth position of the PSPG motif and the glutamic acid (E) at position 27 (Q342 and E365 in Fig. 3B) form hydrogen bonds with ribose hydroxyl groups. The histidine (H) at position 19 and the serine (S) at position 24 of the PSPG motif (H357 and S362 in Fig. 3B) interact with the oxygens of the two phosphates. Two further residues of the PSPG motif (D/E43 and Q44) are implicated in sugar recognition by interacting directly with the hydroxyl groups on position C2, C3, and C4 of the sugar (Shao et al. 2005; Wang 2009).
UDP-α-d-glucose (UDP-Glc) is the most common sugar donor recognized by GT1 enzymes. Over 80% of plant GT1 enzymes characterized so far are glucosyltransferases, and all the available crystal structures are for glucosyltransferases (Table 1). However, plant GT1 enzymes that use less common nucleotide-activated sugars have also been reported (Table 2). As mentioned earlier, sugar specificity seems to be determined by a few crucial positions, enabling rapid evolution from UDP-Glc to less common UDP-sugars. The pivotal role of the final residue of the PSPG motif for d-glucose/d-galactose selectivity has been highlighted (Table 2; Kubo et al. 2004). The replacement of the conserved Q44 by H44 determines not only galactosylation activity but also arabinosylation (Table 2; Fig. 3; Han et al. 2014; Louveau et al. 2018). Site-directed mutagenesis experiments have confirmed the importance of this residue and suggest a role in selection of the C4 stereochemistry (the C4 hydroxy of l-Ara and d-Gal being in an axial position, as opposed to the equatorial position of d-Glc C4 hydroxy; Fig. 3C). Q44 is also replaced by asparagine or histidine in some rhamnosyltransferases and xylosyltransferases but the role of these residues has not been investigated (Table 2). Characterization of xylosyltransferases has highlighted the importance of the N5 loop in sugar specificity (the fifth loop on the amino-terminal domain of GT1s, as defined by Osmani et al. 2009; Fig. 3B). Site-directed mutagenesis of xylosyltransferases and arabinosyltransferases has demonstrated the crucial role of certain residues in the N5 loop for selection between the hexoses d-glucose and d-galactose versus the corresponding structurally related pentoses, d-xylose and l-arabinose (Table 2; Fig. 3C). Crystal structures have also confirmed the proximity of the N5 loop with the d-glucose C6 hydroxy group, which is absent in pentoses (Fig. 3C) and its role in stabilizing GT1/UDP-Glc conformers in VvGT1 (T141) and Os79 (S142; Table 2). The central residue of the N5 loop has also been implicated in glucuronosylation activity shown by Lamiale UGT88Ds (Table 2; Noguchi et al. 2009). Nevertheless, the prevalent determinant in glucuronosylation activity appears to be the presence of a charged residue that is able to stabilize UDP-d-glucuronic acid (UDP-GA) binding by balancing the negative charge of GA C6 carboxylate. Mutation of the arginine residues R25 and R350 of UGT94B1 and UGT88D7 (flavonoid glucuronosyltransferases from Bellis perennis and Perilla frutescens), respectively, impaired GA selectivity (Table 2).
Table 2.
GT1 enzymes that add sugars other than d-glucose
| Gene name | Species | Accession No. | Sugar donors | Acceptor regiospecificity | Final PSPG residues | Structural evidence for sugar donor specificity | References | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| UF3GaT | Vigna mungo | Q9ZWS2 | UDP-α-d-Gal | Flavonoid-3-OH | G | D | H | G | L | None | Mato et al. 1998 |
| F3GalTase | Petunia x hybrida | Q9SBQ8 | UDP-α-d-Gal | Flavonoid-3-OH | G | D | H | Q | L | None | Miller et al. 1999 |
| AcGaT | Aralia cordata | BAD06514 | UDP-α-d-Gal | Flavonoid-3-OH | G | D | H | H | I | Mutation H374Q enhance affinity for Glc | Kubo et al. 2004 |
| GAME1 | Solanum lycopersicum | ADQ37964 | UDP-α-d-Gal | Steroidal alkaloid-3-OH | A | D | Q | F | Y | None | Itkin et al. 2011 |
| UGT73P2 | Glycine max | D4Q9Z4 | UDP-α-d-Gal | Triterpenoid glycoside | A | E | H | F | F | None | Shibuya et al. 2010 |
| F3galtase | Diospyros kaki | BAI40148 | UDP-α-d-Gal | Flavonoid 3-OH | G | D | H | Q | L | None | Ikegami et al. 2009 |
| F3GT1 | Actinidia chinensis | ADC34700 | UDP-α-d-Gal | Anthocyanidin 3-OH | G | D | H | P | M | None | Shibuya et al. 2010 |
| UGT78D3 | Arabidopsis thaliana | AED92375 | UDP-β-l-Ara | Flavonoid-5-OH | G | D | H | A | I | Mutation H380Q enhance activity with Xyl | Han et al. 2014 |
| UGT99D1 | Avena strigosa | AZQ26921 | UDP-β-l-Ara | Triterpenoid-3-OH | S | D | H | F | V | P154 in N5 loop could be involved in hydrophobic interaction with C5 of Ara; H404Q-P154S double mutant has enhanced activity with Glc | Louveau et al. 2018 |
| UGT73P10 | G. max | KRH74847 | UDP-β-l-Ara | Triterpenoid glucuronoside | A | E | H | F | F | None | Takagi et al. 2018 |
| GmSSAT1 | G. max | I1KZ41 | UDP-β-l-Ara | Triterpenoid-22-OH | G | D | H | F | L | None | Louveau et al. 2018 |
| ZOX1 | Phaseolus vulgaris | P56725 | UDP-α-d-Xyl | Cytokinin | S | D | Q | P | R | None | Martin et al. 1999 |
| GAME2 | S. lycopersicum | ADQ37966 | UDP-α-d-Xyl | Steroidal glycoalkaloid | S | D | N | F | Y | None | Itkin et al. 2013 |
| UGT73F4 | G. max | BAM29363 | UDP-α-d-Xyl | Triterpenoid glycoside | A | D | Q | F | Y | S138 in N5 loop may prevent interaction between T139 and C6 of Glc; mutation S138G enhances activity with Glc and Gal | Sayama et al. 2012 |
| F3GGT1 | Actinidia chinensis | AYJ72754 | UDP-α-d-Xyl | Anthocyanidin galactoside | G | D | Q | I | I | None | Montefiori et al. 2011 |
| GAGT | S. lycopersicum | CAI62049 | UDP-α-d-Xyl | Gentisic acid | S | D | Q | P | T | None | Tárraga et al. 2010 |
| UGT89A2 | A. thaliana | ABH04465 | UDP-α-d-Xyl | Dihydroxybenzoic acid | A | D | Q | F | V | I153 in N5 loop may prevent interaction between S152 and C6 of Glc; mutation I153S enhanced affinity for Glc | Chen and Li 2017 |
| UGT79B1 | A. thaliana | Q9LVW3 | UDP-α-d-Xyl | Anthocyanin | G | E | Q | I | L | I142 in N5 loop could be involved in hydrophobic interaction with C5 of Xyl; mutation I142T enhanced activity with Glc | Wang et al. 2018 |
| UGT94P1 | Camellia sinensis | BAO51835 | UDP-α-d-Xyl | Monoterpenoid glycoside | Y | E | Q | P | L | I141 in N5 loop could be involved in hydrophobic interaction with C5 of Xyl; mutation I141S enhanced activity with Glc | Ohgami et al. 2015 |
| UGT88D1 | Scutellaria baicalensis | B9A9D5 | UDP-α-d-GA | Flavonoid-7-OH | A | E | Q | R | L | R339 inside the PSPG motif and S111 in the N5 loop | Nagashima et al. 2000 |
| UGT88D5 | Scutellaria laeteviolacea | BAG31946 | UDP-α-d-GA | Flavonoid-7-OH | A | E | Q | K | L | R350 inside the PSPG motif and S127 in the N5 loop | Noguchi et al. 2009 |
| UGT88D7 | Perilla frutescens | BAG31948 | UDP-α-d-GA | Flavonoid-7-OH | A | E | Q | R | M | R350 inside the PSPG motif and S127 in the N5 loop may directly interact with COOH of GA; double mutant S127T R350W enhanced activity with Glc | Noguchi et al. 2009 |
| UGT88D6 | Sesamum indicum | BAG31947 | UDP-α-d-GA | Flavonoid-7-OH | A | E | Q | R | M | R350 inside the PSPG motif and S127 in the N5 loop | Noguchi et al. 2009 |
| UGT88D4 | Antirrhinum majus | BAG31945 | UDP-α-d-GA | Flavone-7-OH | A | E | Q | R | M | R355 inside the PSPG motif and S127 in the N5 loop | Noguchi et al. 2009 |
| UGT88D8 | Veronica persica | BAH47552 | UDP-α-d-GA | Apigenin-7-OH | A | E | Q | R | M | R352 inside the PSPG motif and S128 in the N5 loop | Ono et al. 2010 |
| GuUGAT | Glycyrrhiza uralensis | ANJ03631 | UDP-α-d-GA | Triterpenoid | G | E | Q | F | Y | None | Xu et al. 2016 |
| UGT94B1 | Bellis perennis | Q5NTH0 | UDP-α-d-GA | Anthocyanin | F | D | Q | P | Y | R25 could interact directly with COOH of GA, balancing the negative charge; R25S has increased activity with Glc | Sawada et al. 2005 |
| UGT77B2 | Crocosmia x crocosmiiflora | A0A2Z5CVA1 | UDP-α-l-Rha | Flavonol-3-OH | A | D | Q | K | T | None | Irmisch et al. 2018 |
| UGT78D1 | A. thaliana | AAF19756 | UDP-α-l-Rha | Flavonoid-5-OH | A | D | N | R | L | None | Jones et al. 2003 |
| UGT89C1 | A. thaliana | Q9LNE6 | UDP-α-l-Rha | Flavonol glucoside-7-OH | A | D | H | F | F | None | Yonekura-Sakakibara et al. 2007 |
| UGT79A6 | G. max | BAN91401 | UDP-α-l-Rha | Flavonol glucoside | G | D | Q | F | F | None | Rojas Rodas et al. 2014 |
| SGT3 | Solanum tuberosum | ABB84472 | UDP-α-l-Rha | Steroidal glycoalkaloid | S | D | N | F | Y | None | McCue et al. 2007 |
| RT | Petunia x hybrida | Q43716 | UDP-α-l-Rha | Anthocyanidin-3-OH | G | D | Q | I | L | None | Brugliera et al. 1994 |
| Cs1,6RhaT | Citrus sinensis | ABA18631 | UDP-α-l-Rha | Flavonoid glycoside | G | D | Q | F | L | None | Frydman et al. 2013 |
| UGT91H4 | G. max | D4Q9Z5 | UDP-α-l-Rha | Triterpenoid glycoside | L | D | Q | C | L | None | Shibuya et al. 2010 |
| Cm1,2RhaT | Citrus maxima | Q8GVE3 | UDP-α-l-Rha | Flavonoid glycoside | Y | E | Q | P | S | None | Frydman et al. 2004 |
PSPG, Plant secondary product glycosyltransferase.
Several plant UGT rhamnosyltransferases have been identified, but the lack of commercial sources of UDP-Rha has hindered structure/function analysis of these enzymes. The amino acid residues that determine l-Rha recognition have not as yet been identified, although some of the residues mentioned above could be involved (Table 2). The recent report of the crystallization of A. thaliana rhamnosyltransferase UGT89C1 may shed light on the determinants of rhamnose specificity (Zong et al. 2019).
In summary, the recent elucidation of several GT1 crystal structures along with site-directed mutagenesis have highlighted the central role of some residues in determining sugar specificity. As seen above, two residues are particularly important in this regard: the final residue of the PSPG motif and the central residue of the N5 loop. The convergent evolution of sugar specificity implies that alternative routes exist and this is shown by the lack of H44 in the tomato galactosyltransferase GAME1, which initiates the addition of the C3 sugar chain of the steroidal glycoalkaloid α-tomatine (Table 2; Itkin et al. 2011).
Enzymes capable of adding certain types of sugars that are commonly found in sugar chains of specialized metabolites (e.g., d-fucose or d-mannose) are still missing from the collection of functionally characterized GT1s from plants. However, the increasing body of knowledge about determinants of sugar specificity may enable such enzymes to be predicted based on the presence of unusual amino acid residues at specific positions that might suggest use of alternative sugar donors (nonglucose).
A ROLE FOR TRANSGLYCOSIDASES IN GLYCOSYLATION OF SPECIALIZED METABOLITES IN PLANTS
Assembly of glycosidic moieties attached to plant specialized metabolites remains poorly understood and only a limited number of enzymes that carry out such modifications have been characterized to date. Within the last 10 years, transglucosidases (TGs) have also been shown to have roles in glycosylation of small molecules. TGs have evolved from β-glucosidases belonging to the CAZY glycosyl hydrolase family 1 (GH1). GH1s are retaining glycosidases that hydrolyze glycosidic linkage via a double-displacement reaction involving a covalently bound glycosyl-enzyme intermediate (Fig. 4B; Lairson et al. 2008). GH1s usually release the sugar unit by transferring it onto a water molecule. In the case of TGs, water is replaced by a metabolite.
The TGs that have been characterized so far are scattered throughout the GH1 phylogeny and are more closely related to GHs than to other TGs. Modification of the transition state structures, mechanistic restriction of water access to the active site, and increased substrate specificity via binding site modifications have been proposed as factors that distinguish TGs from the prototype GH1 enzymes and promote transglycosylation activity (Bissaro et al. 2015). Plant TGs have been shown to glycosylate anthocyanins using acyl sugar substrates as sugar donors (Matsuba et al. 2010; Miyahara et al. 2012, 2014). A rice TG has also been identified that glucosylates flavonols, phytohormones, and phenylpropanoids (Luang et al. 2013).
PROSPECTING FOR NEW ENZYMES AND BIOSYNTHETIC PATHWAYS
As highlighted earlier, many glycosylated plant-specialized metabolites have important bioactive properties and a wide range of potential medicinal, agricultural, and industrial applications. The rapidly growing body of plant genome and transcriptome data (Xiao et al. 2013; Matasci et al. 2014; Kersey 2019) is opening up unprecedented opportunities to mine for genes encoding new sugar transferases to expand the glycosylation toolkit available for glycodiversification of small molecules using metabolic engineering approaches. The recent discovery that the genes for natural product pathways are sometimes organized in biosynthetic gene clusters in plant genomes is enabling the development of algorithms for prediction of new pathways encoded by suites of physically linked genes (e.g., plantiSMASH, PhytoClust), and are rapidly accelerating pathway discovery (Nützmann et al. 2016; Kautsar et al. 2017; Owen et al. 2017; Töpfer et al. 2017). The limiting factor now is not so much gene discovery as elucidation of the functions of new predicted enzymes and pathways.
Engineering Plant Glycoside Biosynthesis in Heterologous Hosts
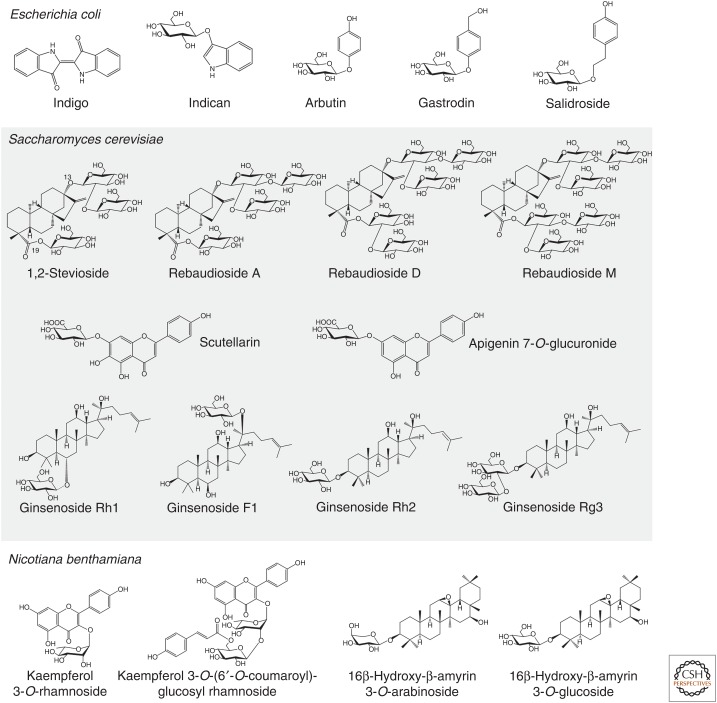
Plant glycosides have a wealth of medicinal, agricultural, and industrial applications. Many of the plant glycosides that are currently used commercially are sourced by extraction from the producing plant species, as these compounds are too challenging to be made by synthetic chemistry, at least at commercial scale. Given that many of the source species have not been domesticated, this presents concerns about the environmental sustainability of such processes. Cell suspension or hairy root cultures are used for production of some plant natural products, including the anticancer drug taxol (Howat et al. 2014). However, these systems suffer from instability of the production lines. Furthermore, purification of the target compound is often confounded by the fact that these cultured lines often produce a plethora of isomers or related structures, making purification extremely difficult. Metabolic engineering of plant natural product pathway genes into heterologous expression systems can enable not only the recapitulation of pathways for known molecules but also the systematic glycodiversification of natural product scaffolds to generate new-to-nature compounds. Microbial systems such as the Gram-negative bacterium Escherichia coli and Baker's yeast (Saccharomyces cerevisiae) are often used as the preferred hosts for metabolic engineering of plant natural product pathways. However transient expression in the wild relative of tobacco, Nicotiana benthamiana, is now opening up new opportunities for gram-scale production of plant natural products and analogs. Some examples of glycosylated plant compounds that have been generated by metabolic engineering in these various hosts are shown in Figure 4.
E. coli only makes a limited set of sugar donors (UDP-Glc, UDP-GA, UDP-4-deoxy-4-formamido-l-Ara, and TDP-sugars), which presents limitations for heterologous expression of pathways for small molecules with diverse sugars attached. An important contribution has been the engineering of E. coli for the production of additional sugar donors, allowing the introduction of new sugar moieties by GT1s expressed in E. coli. The Ahn laboratory has successfully produced flavonoid rhamnosides, xylosides, and arabinosides as well as flavonoids bearing other sugars, including deoxy-sugars, with yields in the region of a few hundred mg L−1 (Kim et al. 2012, 2013; Yoon et al. 2012; Han et al. 2014). Other examples of plant glycosides produced by metabolic engineering in E. coli include indican (the direct precursor of indigo) and various phenolic glucosides (arbutin, salidroside, gastrodin; Fig. 5).
Figure 5.
Examples of plant glycosides produced by metabolic engineering in heterologous hosts. Examples shown are from data in Hsu et al. (2018), Shen et al. (2017), Xue et al. (2016), Chung et al. (2017), Bai et al. (2016), Louveau et al. (2018), Liu et al. (2018), Irmisch et al. (2018), Olsson et al. (2016), and Zhuang et al. (2017).
S. cerevisiae has traditionally been the host of choice for heterologous production of terpenoids and has been used successfully for the semisynthetic production of the antimalarial sesquiterpenoid drug artemisinin (Paddon and Keasling 2014). Artemisinin is not glycosylated. However, industrial-scale production of several terpenoid glycosides has been successfully achieved in yeast, including triterpenoid glycosides known as ginsenosides, the active constituents of the herbal medicine ginseng (Mancuso and Santangelo 2017). In initial experiments the ginsenoside variants Rh1, F1, Rh2, and Rg3 (Fig. 4) accumulated at levels of >100 mg L−1 (Jung et al. 2014; Wang et al. 2015; Wei et al. 2015). Zhuang et al. (2017) were subsequently able to achieve yields of ∼300 mg L−1 ginsenoside Rh2 in a 5 L bioreactor by engineering and repurposing a promiscuous yeast ergosterol GT1ScUGT51. High-throughput colorimetric screening of a mutant library enabled the catalytic efficiency of ScUGT51 to be increased by ∼1800-fold. These developments now offer a credible and sustainable alternative to sourcing ginsenosides from the Panax species.
There has been considerable recent interest in biotechnological approaches to produce natural sweeteners. The search for low-calorie sweeteners to reduce sucrose consumption has been driven by awareness of the risks associated with excessive calorie intake, including obesity and diabetes. Efforts have been made to understand the biosynthetic pathways for important sweeteners from plants and to engineer their production in heterologous hosts (Seki et al. 2018). Engineering of the pathway for steviol glycosides from sweetleaf (Stevia rebaudiana) (Fig. 4) currently represents the most advanced example of commercial production of natural sweeteners in yeast. Rebaudiosides D and M (C19 di- and trisaccharides, respectively) are less bitter than 1,2-stevioside and rebaudioside A, the most abundant steviol glycosides found in S. rebaudiana leaves (C19 monosaccharides). A synthetic biology approach was taken to increase the ratio of rebaudiosides D/M to 1,2-stevioside/rebaudioside A in yeast. This was achieved by targeted optimization of one of the GT1s required for steviol glycoside biosynthesis to reshape the acceptor-binding site, and therefore modulating acceptor accommodation (Olsson et al. 2016). This work, as well as further pathway optimization (U.S. Patent No. 9,957,540 B2), allows improved production of rebaudioside D/M and reduced accumulation of undesirable bitter steviosides in yeast. These improved Stevia sweeteners (developed by the company Evolva) have been sold commercially under the brand name EverSweet since March 2018.
d-Glucose is readily available in yeast and most of the work reported here has involved the production of glucosides. However, efforts to engineer alternative UDP sugar pathways into yeast have also been undertaken. Liu et al. (2018) recently reported the production of two flavonoid glucuronosides, including 108 mg L−1 of scutellarin and 185 mg L−1 of apigenin 7-O-glucuronide in yeast (Fig. 4). Metabolic engineering was undertaken to increase the pull of malonyl-CoA (a phenylpropanoid precursor) as well as to provide the required sugar donor UDP-galactose via expression of the UDP-glucose dehydrogenase from the Chinese plant Erigeron breviscapus.
Plants have a number of advantages as hosts for heterologous expression of enzymes and pathways of plant origin. There is no need for sequence recoding, they support correct messenger RNA (mRNA) and protein processing, protein localization, and metabolic compartmentalization, and have many of the necessary metabolic precursors and coenzymes. Transient expression in N. benthamiana is proving to be a very rapid and powerful method for metabolic engineering of plant natural product pathways (Reed et al. 2017; Reed and Osbourn 2018; Stephenson et al. 2018). This method involves infiltrating Agrobacterium tumefaciens containing expression constructs for the genes of interest into N. benthamiana leaves. This system is also eminently scalable and is currently being used for commercial production of flu vaccines (Marsian and Lomonossoff 2016). The process takes only 5–6 days from agroinfiltration to extraction of plant tissue. Furthermore, combinations of genes can be expressed together simply by coinfiltrating multiple A. tumefaciens strains containing different expression constructs (Lau and Sattely 2015; Reed and Osbourn 2018). Transient plant expression has been used to generate gram-scale quantities of pure triterpenoid and is highly amenable to metabolic engineering of more complex glycosides.
Production of triterpenoid glucosides (Khakimov et al. 2015; Louveau et al. 2018), cyanogenic glycosides (Takos et al. 2011), and glucosinolates (Geu-Flores et al. 2009; Pfalz et al. 2011; Crocoll et al. 2016) have been reported in N. benthamiana. This platform is particularly attractive for production of complex glycosides. Recent publications have confirmed the amenability of N. benthamiana for heterologous production of metabolites conjugated to various sugar units. The production of triterpenoid arabinosides, xylosides, and galactosides as well as flavonoid rhamnosides has been reported (Fig. 5; Irmisch et al. 2018; Louveau et al. 2018).
CONCLUDING REMARKS
As we have seen in this review, glycosidic moieties impact substantially on the bioactivity and bioavailability of plant specialized metabolites. By harnessing an array of plant GT enzymes that are able to add single and multiple sugars, it should be possible in the future to carry out systematic glycodiversification of small molecule scaffolds. This will be facilitated by the ever-growing amount of available plant genome and transcriptome data, the development and refinement of algorithms that enable the discovery of new sugar transferase enzymes, and by synthetic biology (e.g., DNA synthesis, standardization of parts, modular cloning systems) (Owen et al. 2017). Better understanding of the structure/activity relationships of plant GTs will enable rational design of new biocatalysts. These advances, in combination with optimized microbial and plant heterologous expression platforms and the advanced tools offered by modern molecular biology (e.g., affordable synthetic genes, standardization of DNA assembly technologies, CRISPR) have the potential to drive the development of a new generation of biobased pharmaceuticals, food additives, personal care products, agrichemicals, and other industrially important compounds.
ACKNOWLEDGMENTS
We acknowledge the support of the Biotechnological and Biological Sciences–funded Institute Strategic Programme Grant “Molecules from Nature” (BB/P012523/1) and the John Innes Foundation.
Footnotes
Editor: Pamela C. Ronald
Additional Perspectives on Engineering Plants for Agriculture available at www.cshperspectives.org
REFERENCES
- Achnine L, Huhman DV, Farag MA, Sumner LW, Blount JW, Dixon RA. 2005. Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J 41: 875–887. 10.1111/j.1365-313X.2005.02344.x [DOI] [PubMed] [Google Scholar]
- Afendi FM, Okada T, Yamazaki M, Hirai-Morita A, Nakamura Y, Nakamura K, Ikeda S, Takahashi H, Altaf-Ul-Amin M, Darusman LK, et al. 2012. KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol 53: e1 10.1093/pcp/pcr165 [DOI] [PubMed] [Google Scholar]
- Augustin JM, Kuzina V, Andersen SB, Bak S. 2011. Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry 72: 435–457. 10.1016/j.phytochem.2011.01.015 [DOI] [PubMed] [Google Scholar]
- Bai Y, Yin H, Bi H, Zhuang Y, Liu T, Ma Y. 2016. De novo biosynthesis of gastrodin in Escherichia coli. Metab Eng 35: 138–147. 10.1016/j.ymben.2016.01.002 [DOI] [PubMed] [Google Scholar]
- Bissaro B, Monsan P, Fauré R, O'Donohue MJ. 2015. Glycosynthesis in a waterworld: New insight into the molecular basis of transglycosylation in retaining glycoside hydrolases. Biochem J 467: 17–35. 10.1042/BJ20141412 [DOI] [PubMed] [Google Scholar]
- Bowles D, Lim EK, Poppenberger B, Vaistij FE. 2006. Glycosyltransferases of lipophilic small molecules. Annu Rev Plant Biol 57: 567–597. 10.1146/annurev.arplant.57.032905.105429 [DOI] [PubMed] [Google Scholar]
- Bowyer P, Clarke BR, Lunness P, Daniels MJ, Osbourn AE. 1995. Host range of a plant pathogenic fungus determined by a saponin detoxifying enzyme. Science 267: 371–374. 10.1126/science.7824933 [DOI] [PubMed] [Google Scholar]
- Brazier-Hicks M, Offen WA, Gershater MC, Revett TJ, Lim EK, Bowles DJ, Davies GJ, Edwards R. 2007a. Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc Natl Acad Sci 104: 20238–20243. 10.1073/pnas.0706421104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazier-Hicks M, Edwards LA, Edwards R. 2007b. Selection of plants for roles in phytoremediation: The importance of glucosylation. Plant Biotechnol J 5: 627–635. [DOI] [PubMed] [Google Scholar]
- Brazier-Hicks M, Gershater M, Dixon D, Edwards R. 2018. Substrate specificity and safener inducibility of the plant UDP-glucose-dependent family 1 glycosyltransferase super-family. Plant Biotechnol J 16: 337–348. 10.1111/pbi.12775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugliera F, Holton TA, Stevenson TW, Farcy E, Lu CY, Cornish EC. 1994. Isolation and characterization of a cDNA clone corresponding to the Rt locus of Petunia hybrida. Plant J 5: 81–92. 10.1046/j.1365-313X.1994.5010081.x [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The carbohydrate-active enzymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res 37: D233–D238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi L, Lim EK, Bowles DJ. 2008. Discovery of new biocatalysts for the glycosylation of terpenoid scaffolds. Chemistry 14: 6656–6662. 10.1002/chem.200800548 [DOI] [PubMed] [Google Scholar]
- Caputi L, Malnoy M, Goremykin V, Nikiforova S, Martens S. 2012. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J 69: 1030–1042. 10.1111/j.1365-313X.2011.04853.x [DOI] [PubMed] [Google Scholar]
- Chen HY, Li X. 2017. Identification of a residue responsible for UDP-sugar donor selectivity of a dihydroxybenzoic acid glycosyltransferase from Arabidopsis natural accessions. Plant J 89: 195–203. 10.1111/tpj.13271 [DOI] [PubMed] [Google Scholar]
- Chung D, Kim SY, Ahn JH. 2017. Production of three phenylethanoids, tyrosol, hydroxytyrosol, and salidroside, using plant genes expressing in Escherichia coli. Sci Rep 7: 2578 10.1038/s41598-017-02042-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocoll C, Mirza N, Reichelt M, Gershenzon J, Halkier BA. 2016. Optimization of engineered production of the glucoraphanin precursor dihomomethionine in Nicotiana benthamiana. Front Bioeng Biotechnol 4: 14 10.3389/fbioe.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn WJC, Gruppen H, Vincken JP. 2018. Structure and biosynthesis of benzoxazinoids: Plant defence metabolites with potential as antimicrobial scaffolds. Phytochemistry 155: 233–243. 10.1016/j.phytochem.2018.07.005 [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman A, Weisshaus O, Bar-Peled M, Huhman DV, Sumner LW, Marin FR, Lewinsohn E, Fluhr R, Gressel J, Eyal Y. 2004. Citrus fruit bitter flavors: Isolation and functional characterization of the gene Cm1,2RhaT encoding a 1,2 rhamnosyltransferase, a key enzyme in the biosynthesis of the bitter flavonoids of citrus. Plant J 40: 88–100. 10.1111/j.1365-313X.2004.02193.x [DOI] [PubMed] [Google Scholar]
- Frydman A, Liberman R, Huhman DV, Carmeli-Weissberg M, Sapir-Mir M, Ophir R, Sumner LW, Eyal Y. 2013. The molecular and enzymatic basis of bitter/non-bitter flavor of citrus fruit: Evolution of branch-forming rhamnosyl-transferases under domestication. Plant J 73: 166–178. 10.1111/tpj.12030 [DOI] [PubMed] [Google Scholar]
- George Thompson AM, Iancu CV, Neet KE, Dean JV, Choe JY. 2017. Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana. Sci Rep 7: 46629 10.1038/srep46629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geu-Flores F, Nielsen MT, Nafisi M, Møldrup ME, Olsen CE, Motawia MS, Halkier BA. 2009. Glucosinolate engineering identifies a γ-glutamyl peptidase. Nat Chem Biol 5: 575–577. [DOI] [PubMed] [Google Scholar]
- Gleadow RM, Møller BL. 2014. Cyanogenic glycosides: Synthesis, physiology, and phenotypic plasticity. Annu Rev Plant Biol 65: 155–185. 10.1146/annurev-arplant-050213-040027 [DOI] [PubMed] [Google Scholar]
- Grubb CD, Zipp BJ, Ludwig-Müller J, Masuno MN, Molinski TF, Abel S. 2004. Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J 40: 893–908. [DOI] [PubMed] [Google Scholar]
- Han SH, Kim BG, Yoon JA, Chong Y, Ahn JH. 2014. Synthesis of flavonoid O-pentosides by Escherichia coli through engineering of nucleotide sugar pathways and glycosyltransferase. Appl Environ Microbiol 80: 2754–2762. 10.1128/AEM.03797-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XZ, Wang X, Dixon RA. 2006. Mutational analysis of the Medicago glycosyltransferase UGT71G1 reveals residues that control regioselectivity for (iso)flavonoid glycosylation. J Biol Chem 281: 34441–34447. 10.1074/jbc.M605767200 [DOI] [PubMed] [Google Scholar]
- Hiromoto T, Honjo E, Noda N, Tamada T, Kazuma K, Suzuki M, Blaber M, Kuroki R. 2015. Structural basis for acceptor-substrate recognition of UDP-glucose: Anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea. Protein Sci 24: 395–407. 10.1002/pro.2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins RJ, van Dam NM, van Loon JJ. 2009. Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu Rev Entomol 54: 57–83. 10.1146/annurev.ento.54.110807.090623 [DOI] [PubMed] [Google Scholar]
- Hou B, Lim EK, Higgins GS, Bowles DJ. 2004. N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J Biol Chem 279: 47822–47832. [DOI] [PubMed] [Google Scholar]
- Howat S, Park B, Oh IS, Jin YW, Lee EK, Loake GJ. 2014. Paclitaxel: Biosynthesis, production and future prospects. N Biotechnol 31: 242–245. 10.1016/j.nbt.2014.02.010 [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. 2008. Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66. 10.1146/annurev.arplant.59.032607.092825 [DOI] [PubMed] [Google Scholar]
- Hsu TM, Welner DH, Russ ZN, Cervantes B, Prathuri RL, Adams PD, Dueber JE. 2018. Employing a biochemical protecting group for a sustainable indigo dyeing strategy. Nat Chem Biol 14: 256–261. 10.1038/nchembio.2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J, Hughes MA. 1994. Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons. DNA Seq 5: 41–49. 10.3109/10425179409039703 [DOI] [PubMed] [Google Scholar]
- Ikegami A, Akagi T, Potter D, Yamada M, Sato A, Yonemori K, Kitajima A, Inoue K. 2009. Molecular identification of 1-Cys peroxiredoxin and anthocyanidin/flavonol 3-O-galactosyltransferase from proanthocyanidin-rich young fruits of persimmon (Diospyros kaki Thunb.). Planta 230: 841–855. 10.1007/s00425-009-0989-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmisch S, Jo S, Roach CR, Jancsik S, Man Saint Yuen M, Madilao LL, O'Neil-Johnson M, Williams R, Withers SG, Bohlmann J. 2018. Discovery of UDP-glycosyltransferases and BAHD-acyltransferases involved in the biosynthesis of the antidiabetic plant metabolite montbretin A. Plant Cell 30: 1864–1886. 10.1105/tpc.18.00406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M, Rogachev I, Alkan N, Rosenberg T, Malitsky S, Masini L, Meir S, Iijima Y, Aoki K, de Vos R, et al. 2011. GLYCOALKALOID METABOLISM1 is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. Plant Cell 23: 4507–4525. 10.1105/tpc.111.088732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M, Heinig U, Tzfadia O, Bhide AJ, Shinde B, Cardenas PD, Bocobza SE, Unger T, Malitsky S, Finkers R, et al. 2013. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341: 175–179. 10.1126/science.1240230 [DOI] [PubMed] [Google Scholar]
- Jones P, Vogt T. 2001. Glycosyltransferases in secondary plant metabolism: Tranquilizers and stimulant controllers. Planta 213: 164–174. [DOI] [PubMed] [Google Scholar]
- Jones P, Messner B, Nakajima J, Schaffner AR, Saito K. 2003. UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J Biol Chem 278: 43910–43918. 10.1074/jbc.M303523200 [DOI] [PubMed] [Google Scholar]
- Jung SC, Kim W, Park SC, Jeong J, Park MK, Lim S, Lee Y, Im WT, Lee JH, Choi G, et al. 2014. Two ginseng UDP-glycosyltransferases synthesize ginsenoside Rg3 and Rd. Plant Cell Physiol 55: 2177–2188. 10.1093/pcp/pcu147 [DOI] [PubMed] [Google Scholar]
- Kannangara R, Motawia MS, Hansen NK, Paquette SM, Olsen CE, Moller BL, Jorgensen K. 2011. Characterization and expression profile of two UDP-glucosyltransferases, UGT85K4 and UGT85K5, catalyzing the last step in cyanogenic glucoside biosynthesis in cassava. Plant J 68: 287–301. 10.1111/j.1365-313X.2011.04695.x [DOI] [PubMed] [Google Scholar]
- Kautsar SA, Suarez Duran HG, Blin K, Osbourn A, Medema MH. 2017. PlantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res 45: W55–W63. 10.1093/nar/gkx305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey PJ. 2019. Plant genome sequences: Past, present, future. Curr Opin Plant Biol 48: 1–8. 10.1016/j.pbi.2018.11.001 [DOI] [PubMed] [Google Scholar]
- Khakimov B, Kuzina V, Erthmann PO, Fukushima EO, Augustin JM, Olsen CE, Scholtalbers J, Volpin H, Andersen SB, Hauser TP, et al. 2015. Identification and genome organization of saponin pathway genes from a wild crucifer, and their use for transient production of saponins in Nicotiana benthamiana. Plant J 84: 478–490. 10.1111/tpj.13012 [DOI] [PubMed] [Google Scholar]
- Kim BG, Kim HJ, Ahn JH. 2012. Production of bioactive flavonol rhamnosides by expression of plant genes in Escherichia coli. J Agric Food Chem 60: 11143–11148. 10.1021/jf302123c [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim BG, Ahn JH. 2013. Regioselective synthesis of flavonoid bisglycosides using Escherichia coli harboring two glycosyltransferases. Appl Microbiol Biotechnol 97: 5275–5282. 10.1007/s00253-013-4844-7 [DOI] [PubMed] [Google Scholar]
- Kubo A, Arai Y, Nagashima S, Yoshikawa T. 2004. Alteration of sugar donor specificities of plant glycosyltransferases by a single point mutation. Arch Biochem Biophys 429: 198–203. 10.1016/j.abb.2004.06.021 [DOI] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG. 2008. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem 77: 521–555. 10.1146/annurev.biochem.76.061005.092322 [DOI] [PubMed] [Google Scholar]
- Lau W, Sattely ES. 2015. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science 349: 1224–1228. 10.1126/science.aac7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Modolo LV, Escamilla-Trevino LL, Achnine L, Dixon RA, Wang X. 2007. Crystal structure of Medicago truncatula UGT85H2–insights into the structural basis of a multifunctional (iso)flavonoid glycosyltransferase. J Mol Biol 370: 951–963. 10.1016/j.jmb.2007.05.036 [DOI] [PubMed] [Google Scholar]
- Li Y, Li P, Wang Y, Dong R, Yu H, Hou B. 2014. Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays). Planta 239: 1265–1279. 10.1007/s00425-014-2050-1 [DOI] [PubMed] [Google Scholar]
- Lim EK, Doucet CJ, Li Y, Elias L, Worrall D, Spencer SP, Ross J, Bowles DJ. 2002. The activity of Arabidopsis glycosyltransferases toward salicylic acid, 4-hydroxybenzoic acid, and other benzoates. J Biol Chem 277: 586–592. 10.1074/jbc.M109287200 [DOI] [PubMed] [Google Scholar]
- Lim EK, Ashford DA, Hou B, Jackson RG, Bowles DJ. 2004. Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol Bioeng 87: 623–631. 10.1002/bit.20154 [DOI] [PubMed] [Google Scholar]
- Liu X, Cheng J, Zhang G, Ding W, Duan L, Yang J, Kui L, Cheng X, Ruan J, Fan W, et al. 2018. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat Commun 9: 448 10.1038/s41467-018-02883-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louveau T, Orme A, Pfalzgraf H, Stephenson MJ, Melton R, Saalbach G, Hemmings AM, Leveau A, Rejzek M, Vickerstaff RJ, et al. 2018. Analysis of two new arabinosyltransferases belonging to the carbohydrate-active enzyme (CAZY) glycosyl transferase family1 provides insights into disease resistance and sugar donor specificity. Plant Cell 30: 3038–3057. 10.1105/tpc.18.00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luang S, Cho JI, Mahong B, Opassiri R, Akiyama T, Phasai K, Komvongsa J, Sasaki N, Hua YL, Matsuba Y, et al. 2013. Rice Os9BGlu31 is a transglucosidase with the capacity to equilibrate phenylpropanoid, flavonoid, and phytohormone glycoconjugates. J Biol Chem 288: 10111–10123. 10.1074/jbc.M112.423533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso C, Santangelo R. 2017. Panax ginseng and Panax quinquefolius: From pharmacology to toxicology. Food Chem Toxicol 107: 362–372. 10.1016/j.fct.2017.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsian J, Lomonossoff GP. 2016. Molecular pharming—VLPs made in plants. Curr Opin Biotechnol 37: 201–206. 10.1016/j.copbio.2015.12.007 [DOI] [PubMed] [Google Scholar]
- Martin RC, Mok MC, Mok DW. 1999. A gene encoding the cytokinin enzyme zeatin O-xylosyltransferase of Phaseolus vulgaris. Plant Physiol 120: 553–558. 10.1104/pp.120.2.553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matasci N, Hung LH, Yan Z, Carpenter EJ, Wickett NJ, Mirarab S, Nguyen N, Warnow T, Ayyampalayam S, Barker M, et al. 2014. Data access for the 1,000 Plants (1KP) project. Gigascience 3: 17 10.1186/2047-217X-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mato M, Ozeki Y, Itoh Y, Higeta D, Yoshitama K, Teramoto S, Aida R, Ishikura N, Shibata M. 1998. Isolation and characterization of a cDNA clone of UDP-galactose: Flavonoid 3-O-galactosyltransferase (UF3GaT) expressed in Vigna mungo seedlings. Plant Cell Physiol 39: 1145–1155. 10.1093/oxfordjournals.pcp.a029315 [DOI] [PubMed] [Google Scholar]
- Matsuba Y, Sasaki N, Tera M, Okamura M, Abe Y, Okamoto E, Nakamura H, Funabashi H, Takatsu M, Saito M, et al. 2010. A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the petals of carnation and delphinium. Plant Cell 22: 3374–3389. 10.1105/tpc.110.077487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCue KF, Allen PV, Shepherd LV, Blake A, Maccree MM, Rockhold DR, Novy RG, Stewart D, Davies HV, Belknap WR. 2007. Potato glycosterol rhamnosyltransferase, the terminal step in triose side-chain biosynthesis. Phytochemistry 68: 327–334. 10.1016/j.phytochem.2006.10.025 [DOI] [PubMed] [Google Scholar]
- Miller KD, Guyon V, Evans JN, Shuttleworth WA, Taylor LP. 1999. Purification, cloning, and heterologous expression of a catalytically efficient flavonol 3-O-galactosyltransferase expressed in the male gametophyte of Petunia hybrida. J Biol Chem 274: 34011–34019. 10.1074/jbc.274.48.34011 [DOI] [PubMed] [Google Scholar]
- Miyahara T, Takahashi M, Ozeki Y, Sasaki N. 2012. Isolation of an acyl-glucose-dependent anthocyanin 7-O-glucosyltransferase from the monocot Agapanthus africanus. J Plant Physiol 169: 1321–1326. 10.1016/j.jplph.2012.05.004 [DOI] [PubMed] [Google Scholar]
- Miyahara T, Tani T, Takahashi M, Nishizaki Y, Ozeki Y, Sasaki N. 2014. Isolation of anthocyanin 7-O-glucosyltransferase from Canterbury bells (Campanula medium). Plant Biotechnol 31: 555–559. 10.5511/plantbiotechnology.14.0908a [DOI] [Google Scholar]
- Modolo LV, Li L, Pan H, Blount JW, Dixon RA, Wang X. 2009. Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids. J Mol Biol 392: 1292–1302. 10.1016/j.jmb.2009.08.017 [DOI] [PubMed] [Google Scholar]
- Mohamed AA, Ceunen S, Geuns JM, Van den Ende W, De Ley M. 2011. UDP-dependent glycosyltransferases involved in the biosynthesis of steviol glycosides. J Plant Physiol 168: 1136–1141. 10.1016/j.jplph.2011.01.030 [DOI] [PubMed] [Google Scholar]
- Montefiori M, Espley RV, Stevenson D, Cooney J, Datson PM, Saiz A, Atkinson RG, Hellens RP, Allan AC. 2011. Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). Plant J 65: 106–118. 10.1111/j.1365-313X.2010.04409.x [DOI] [PubMed] [Google Scholar]
- Nagashima S, Hirotani M, Yoshikawa T. 2000. Purification and characterization of UDP-glucuronate: Baicalein 7-O-glucuronosyltransferase from Scutellaria baicalensis Georgi. cell suspension cultures. Phytochemistry 53: 533–538. 10.1016/S0031-9422(99)00593-2 [DOI] [PubMed] [Google Scholar]
- Nagatomo Y, Usui S, Ito T, Kato A, Shimosaka M, Taguchi G. 2014. Purification, molecular cloning and functional characterization of flavonoid C-glucosyltransferases from Fagopyrum esculentum M. (buckwheat) cotyledon. Plant J 80: 437–448. 10.1111/tpj.12645 [DOI] [PubMed] [Google Scholar]
- Nagatoshi M, Terasaka K, Nagatsu A, Mizukami H. 2011. Iridoid-specific glucosyltransferase from Gardenia jasminoides. J Biol Chem 286: 32866–32874. 10.1074/jbc.M111.242586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda N. 2018. Recent advances in the research and development of blue flowers. Breed Sci 68: 79–87. 10.1270/jsbbs.17132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda N, Yoshioka S, Kishimoto S, Nakayama M, Douzono M, Tanaka Y, Aida R. 2017. Generation of blue chrysanthemums by anthocyanin B-ring hydroxylation and glucosylation and its coloration mechanism. Sci Adv 3: e1602785 10.1126/sciadv.1602785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi A, Saito A, Homma Y, Nakao M, Sasaki N, Nishino T, Takahashi S, Nakayama T. 2007. A UDP-glucose:isoflavone 7-O-glucosyltransferase from the roots of soybean (Glycine max) seedlings. Purification, gene cloning, phylogenetics, and an implication for an alternative strategy of enzyme catalysis. J Biol Chem 282: 23581–23590. 10.1074/jbc.M702651200 [DOI] [PubMed] [Google Scholar]
- Noguchi A, Horikawa M, Fukui Y, Fukuchi-Mizutani M, Iuchi-Okada A, Ishiguro M, Kiso Y, Nakayama T, Ono E. 2009. Local differentiation of sugar donor specificity of flavonoid glycosyltransferase in Lamiales. Plant Cell 21: 1556–1572. 10.1105/tpc.108.063826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccio ML, Rhodes D, McNeil SD, Hanson AD. 1999. Metabolic engineering of plants for osmotic stress resistance. Curr Opin Plant Biol 2: 128–134. 10.1016/S1369-5266(99)80026-0 [DOI] [PubMed] [Google Scholar]
- Nützmann HW, Huang A, Osbourn A. 2016. Plant metabolic clusters—From genetics to genomics. New Phytol 211: 771–789. 10.1111/nph.13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ. 2006. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J 25: 1396–1405. 10.1038/sj.emboj.7600970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata J, Kanno Y, Itoh Y, Tsugawa H, Suzuki M. 2005. Plant biochemistry: Anthocyanin biosynthesis in roses. Nature 435: 757–758. 10.1038/nature435757a [DOI] [PubMed] [Google Scholar]
- Ohgami S, Ono E, Horikawa M, Murata J, Totsuka K, Toyonaga H, Ohba Y, Dohra H, Asai T, Matsui K, et al. 2015. Volatile glycosylation in tea plants: Sequential glycosylations for the biosynthesis of aroma β-primeverosides are catalyzed by two Camellia sinensis glycosyltransferases. Plant Physiol 168: 464–477. 10.1104/pp.15.00403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE. 2013. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263. 10.1038/nrmicro2990 [DOI] [PubMed] [Google Scholar]
- Olsson K, Carlsen S, Semmler A, Simon E, Mikkelsen MD, Moller BL. 2016. Microbial production of next-generation stevia sweeteners. Microb Cell Fact 15: 207 10.1186/s12934-016-0609-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono E, Ruike M, Iwashita T, Nomoto K, Fukui Y. 2010. Co-pigmentation and flavonoid glycosyltransferases in blue Veronica persica flowers. Phytochemistry 71: 726–735. 10.1016/j.phytochem.2010.02.008 [DOI] [PubMed] [Google Scholar]
- Osbourn A. 1996. Saponins and plant defense—A soap story. Trends Plant Sci 1: 4–9. 10.1016/S1360-1385(96)80016-1 [DOI] [Google Scholar]
- Osmani SA, Bak S, Møller BL. 2009. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 70: 325–347. 10.1016/j.phytochem.2008.12.009 [DOI] [PubMed] [Google Scholar]
- Owen C, Patron NJ, Huang A, Osbourn A. 2017. Harnessing plant metabolic diversity. Curr Opin Chem Biol 40: 24–30. 10.1016/j.cbpa.2017.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon CJ, Keasling JD. 2014. Semi-synthetic artemisinin: A model for the use of synthetic biology in pharmaceutical development. Nat Rev Microbiol 12: 355–367. 10.1038/nrmicro3240 [DOI] [PubMed] [Google Scholar]
- Pfalz M, Mikkelsen MD, Bednarek P, Olsen CE, Halkier BA, Kroymann J. 2011. Metabolic engineering in Nicotiana benthamiana reveals key enzyme functions in Arabidopsis indole glucosinolate modification. Plant Cell 23: 716–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowska A, Bajguz A. 2011. Conjugates of abscisic acid, brassinosteroids, ethylene, gibberellins, and jasmonates. Phytochemistry 72: 2097–2112. 10.1016/j.phytochem.2011.08.012 [DOI] [PubMed] [Google Scholar]
- Reed J, Osbourn A. 2018. Engineering terpenoid production through transient expression in Nicotiana benthamiana. Plant Cell Rep 37: 1431–1441. 10.1007/s00299-018-2296-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J, Stephenson MJ, Miettinen K, Brouwer B, Leveau A, Brett P, Goss RJM, Goossens A, O'Connell MA, Osbourn A. 2017. A translational synthetic biology platform for rapid access to gram-scale quantities of novel drug-like molecules. Metab Eng 42: 185–193. 10.1016/j.ymben.2017.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas Rodas F, Rodriguez TO, Murai Y, Iwashina T, Sugawara S, Suzuki M, Nakabayashi R, Yonekura-Sakakibara K, Saito K, Kitajima J, et al. 2014. Linkage mapping, molecular cloning and functional analysis of soybean gene Fg2 encoding flavonol 3-O-glucoside (1→6) rhamnosyltransferase. Plant Mol Biol 84: 287–300. 10.1007/s11103-013-0133-1 [DOI] [PubMed] [Google Scholar]
- Ross J, Li Y, Lim E, Bowles DJ. 2001. Higher plant glycosyltransferases. Genome Biol 2: reviews3004.3001–3004.3006. 10.1186/gb-2001-2-2-reviews3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada S, Suzuki H, Ichimaida F, Yamaguchi MA, Iwashita T, Fukui Y, Hemmi H, Nishino T, Nakayama T. 2005. UDP-glucuronic acid:anthocyanin glucuronosyltransferase from red daisy (Bellis perennis) flowers. Enzymology and phylogenetics of a novel glucuronosyltransferase involved in flower pigment biosynthesis. J Biol Chem 280: 899–906. 10.1074/jbc.M410537200 [DOI] [PubMed] [Google Scholar]
- Sawai S, Saito K. 2011. Triterpenoid biosynthesis and engineering in plants. Front Plant Sci 2: 25 10.3389/fpls.2011.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayama T, Ono E, Takagi K, Takada Y, Horikawa M, Nakamoto Y, Hirose A, Sasama H, Ohashi M, Hasegawa H, et al. 2012. The Sg-1 glycosyltransferase locus regulates structural diversity of triterpenoid saponins of soybean. Plant Cell 24: 2123–2138. 10.1105/tpc.111.095174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki H, Tamura K, Muranaka T. 2018. Plant-derived isoprenoid sweeteners: Recent progress in biosynthetic gene discovery and perspectives on microbial production. Biosci Biotechnol Biochem 82: 927–934. 10.1080/09168451.2017.1387514 [DOI] [PubMed] [Google Scholar]
- Shao H, He X, Achnine L, Blount JW, Dixon RA, Wang X. 2005. Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17: 3141–3154. 10.1105/tpc.105.035055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Wang J, Wang J, Chen Z, Yuan Q, Yan Y. 2017. High-level De novo biosynthesis of arbutin in engineered Escherichia coli. Metab Eng 42: 52–58. 10.1016/j.ymben.2017.06.001 [DOI] [PubMed] [Google Scholar]
- Shibuya M, Nishimura K, Yasuyama N, Ebizuka Y. 2010. Identification and characterization of glycosyltransferases involved in the biosynthesis of soyasaponin I in Glycine max. FEBS Lett 584: 2258–2264. 10.1016/j.febslet.2010.03.037 [DOI] [PubMed] [Google Scholar]
- Sønderby IE, Geu-Flores F, Halkier BA. 2010. Biosynthesis of glucosinolates—Gene discovery and beyond. Trends Plant Sci 15: 283–290. 10.1016/j.tplants.2010.02.005 [DOI] [PubMed] [Google Scholar]
- Song C, Härtl K, McGraphery K, Hoffmann T, Schwab W. 2018. Attractive but toxic: Emerging roles of glycosidically bound volatiles and glycosyltransferases involved in their formation. Mol Plant 11: 1225–1236. 10.1016/j.molp.2018.09.001 [DOI] [PubMed] [Google Scholar]
- Stephenson MJ, Reed J, Brouwer B, Osbourn A. 2018. Transient expression in Nicotiana benthamiana leaves for triterpene production at a preparative scale. J Vis Exp 138: 58169 10.3791/58169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi K, Yano R, Tochigi S, Fujisawa Y, Tsuchinaga H, Takahashi Y, Takada Y, Kaga A, Anai T, Tsukamoto C, et al. 2018. Genetic and functional characterization of Sg-4 glycosyltransferase involved in the formation of sugar chain structure at the C-3 position of soybean saponins. Phytochemistry 156: 96–105. 10.1016/j.phytochem.2018.09.002 [DOI] [PubMed] [Google Scholar]
- Takos AM, Knudsen C, Lai D, Kannangara R, Mikkelsen L, Motawia MS, Olsen CE, Sato S, Tabata S, Jørgensen K, et al. 2011. Genomic clustering of cyanogenic glucoside biosynthetic genes aids their identification in Lotus japonicus and suggests the repeated evolution of this chemical defence pathway. Plant J 68: 273–286. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tárraga S, Lisón P, López-Gresa MP, Torres C, Rodrigo I, Bellés JM, Conejero V. 2010. Molecular cloning and characterization of a novel tomato xylosyltransferase specific for gentisic acid. J Exp Bot 61: 4325–4338. 10.1093/jxb/erq234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis N, Raguso RA. 2005. The effect of pollination on floral fragrance in thistles. J Chem Ecol 31: 2581–2600. 10.1007/s10886-005-7615-9 [DOI] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al. 2005. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42: 218–235. 10.1111/j.1365-313X.2005.02371.x [DOI] [PubMed] [Google Scholar]
- Töpfer N, Fuchs LM, Aharoni A. 2017. The PhytoClust tool for metabolic gene clusters discovery in plant genomes. Nucleic Acids Res 45: 7049–7063. 10.1093/nar/gkx404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trossat C, Rathinasabapathi B, Weretilnyk EA, Shen TL, Huang ZH, Gage DA, Hanson AD. 1998. Salinity promotes accumulation of 3-dimethylsulfoniopropionate and its precursor S-methylmethionine in chloroplasts. Plant Physiol 116: 165–171. 10.1104/pp.116.1.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter J. 2000. Plant cyanogenic glycosides. Toxicon 38: 11–36. 10.1016/S0041-0101(99)00128-2 [DOI] [PubMed] [Google Scholar]
- Vincken JP, Heng L, de Groot A, Gruppen H. 2007. Saponins, classification and occurrence in the plant kingdom. Phytochemistry 68: 275–297. 10.1016/j.phytochem.2006.10.008 [DOI] [PubMed] [Google Scholar]
- Vogt T, Jones P. 2000. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci 5: 380–386. 10.1016/S1360-1385(00)01720-9 [DOI] [PubMed] [Google Scholar]
- Wang X. 2009. Structure, mechanism and engineering of plant natural product glycosyltransferases. FEBS Lett 583: 3303–3309. 10.1016/j.febslet.2009.09.042 [DOI] [PubMed] [Google Scholar]
- Wang J, Ma XM, Kojima M, Sakakibara H, Hou BK. 2011. N-glucosyltransferase UGT76C2 is involved in cytokinin homeostasis and cytokinin response in Arabidopsis thaliana. Plant Cell Physiol 52: 2200–2213. 10.1093/pcp/pcr152 [DOI] [PubMed] [Google Scholar]
- Wang J, Ma XM, Kojima M, Sakakibara H, Hou BK. 2013. Glucosyltransferase UGT76C1 finely modulates cytokinin responses via cytokinin N-glucosylation in Arabidopsis thaliana. Plant Physiol Biochem 65: 9–16. 10.1016/j.plaphy.2013.01.012 [DOI] [PubMed] [Google Scholar]
- Wang P, Wei Y, Fan Y, Liu Q, Wei W, Yang C, Zhang L, Zhao G, Yue J, Yan X, et al. 2015. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab Eng 29: 97–105. 10.1016/j.ymben.2015.03.003 [DOI] [PubMed] [Google Scholar]
- Wang H, Wang C, Fan W, Yang J, Appelhagen I, Wu Y, Zhang P. 2018. A novel glycosyltransferase catalyses the transfer of glucose to glucosylated anthocyanins in purple sweet potato. J Exp Bot 69: 5444–5459. 10.1093/jxb/ery305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Wang P, Wei Y, Liu Q, Yang C, Zhao G, Yue J, Yan X, Zhou Z. 2015. Characterization of Panax ginseng UDP-glycosyltransferases catalyzing protopanaxatriol and biosyntheses of bioactive ginsenosides F1 and Rh1 in metabolically engineered yeasts. Mol Plant 8: 1412–1424. 10.1016/j.molp.2015.05.010 [DOI] [PubMed] [Google Scholar]
- Weston LA, Mathesius U. 2013. Flavonoids: Their structure, biosynthesis and role in the rhizosphere, including allelopathy. J Chem Ecol 39: 283–297. 10.1007/s10886-013-0248-5 [DOI] [PubMed] [Google Scholar]
- Wetterhorn KM, Newmister SA, Caniza RK, Busman M, McCormick SP, Berthiller F, Adam G, Rayment I. 2016. Crystal structure of Os79 (Os04g0206600) from Oryza sativa: A UDP-glucosyltransferase involved in the detoxification of deoxynivalenol. Biochemistry 55: 6175–6186. 10.1021/acs.biochem.6b00709 [DOI] [PubMed] [Google Scholar]
- Wetterhorn KM, Gabardi K, Michlmayr H, Malachova A, Busman M, McCormick SP, Berthiller F, Adam G, Rayment I. 2017. Determinants and expansion of specificity in a trichothecene UDP-glucosyltransferase from Oryza sativa. Biochemistry 56: 6585–6596. 10.1021/acs.biochem.7b01007 [DOI] [PubMed] [Google Scholar]
- Wilson AE, Feng X, Ono NN, Holland D, Amir R, Tian L. 2017. Characterization of a UGT84 family glycosyltransferase provides new insights into substrate binding and reactivity of galloylglucose ester-forming UGTs. Biochemistry 56: 6389–6400. 10.1021/acs.biochem.7b00946 [DOI] [PubMed] [Google Scholar]
- Xiao M, Zhang Y, Chen X, Lee EJ, Barber CJ, Chakrabarty R, Desgagne-Penix I, Haslam TM, Kim YB, Liu E, et al. 2013. Transcriptome analysis based on next-generation sequencing of non-model plants producing specialized metabolites of biotechnological interest. J Biotechnol 166: 122–134. 10.1016/j.jbiotec.2013.04.004 [DOI] [PubMed] [Google Scholar]
- Xu GJ, Cai W, Gao W, Liu CS. 2016. A novel glucuronosyltransferase has an unprecedented ability to catalyse continuous two-step glucuronosylation of glycyrrhetinic acid to yield glycyrrhizin. New Phytol 212: 123–135. 10.1111/nph.14039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue F, Guo H, Hu Y, Liu R, Huang L, Lv H, Liu C, Yang M, Ma L. 2016. Expression of codon-optimized plant glycosyltransferase UGT72B14 in Escherichia coli enhances salidroside production. Biomed Res Int 2016: 9845927 10.1155/2016/9845927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Fehl C, Lees KV, Lim EK, Offen WA, Davies GJ, Bowles DJ, Davidson MG, Roberts SJ, Davis BG. 2018. Functional and informatics analysis enables glycosyltransferase activity prediction. Nat Chem Biol 14: 1109–1117. 10.1038/s41589-018-0154-9 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Hanada K. 2011. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J 66: 182–193. 10.1111/j.1365-313X.2011.04493.x [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Niida R, Saito K. 2007. Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J Biol Chem 282: 14932–14941. 10.1074/jbc.M611498200 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K. 2008. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176. 10.1105/tpc.108.058040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JA, Kim BG, Lee WJ, Lim Y, Chong Y, Ahn JH. 2012. Production of a novel quercetin glycoside through metabolic engineering of Escherichia coli. Appl Environ Microbiol 78: 4256–4262. 10.1128/AEM.00275-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Utsumi R. 2009. Diversity, regulation, and genetic manipulation of plant mono- and sesquiterpenoid biosynthesis. Cell Mol Life Sci 66: 3043–3052. 10.1007/s00018-009-0066-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Yang GY, Chen X, Liu Q, Zhang X, Deng Z, Feng Y. 2017. Biosynthesis of plant-derived ginsenoside Rh2 in yeast via repurposing a key promiscuous microbial enzyme. Metab Eng 42: 25–32. 10.1016/j.ymben.2017.04.009 [DOI] [PubMed] [Google Scholar]
- Zong G, Li J, Gao Y, Fei S, Liu X, Wang X, Shen Y. 2019. Overexpression, purification, biochemical and structural characterization of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana. Protein Expr Purif 156: 44–49. 10.1016/j.pep.2018.12.007 [DOI] [PubMed] [Google Scholar]