Abstract
The reprogramming of human somatic cells into induced pluripotent stem cells (iPSCs) a little over a decade ago raised exciting prospects to transform the study and potentially also the therapy of human diseases. iPSC models have now been created for a multitude of hematologic diseases, including malignancies. Here we discuss practical aspects of iPSC modeling of malignant diseases, review recent studies, and discuss the new opportunities that iPSC models offer, as well as their current limitations and prospects for future development.
The successful demonstration that human pluripotent stem cells (hPSCs) identical to human embryonic stem cells (hESCs), termed induced pluripotent stem cells (iPSCs), can be generated from somatic cells through the ectopic expression of a defined set of genes in 2007 was quickly succeeded by several proof-of-principle studies highlighting the potential of using iPSCs derived from patients to study and potentially cure inherited genetic diseases via gene and cell therapy and to test drugs (Hanna et al. 2007; Takahashi et al. 2007; Yu et al. 2007; Park et al. 2008; Lee et al. 2009). Modeling more complex diseases and high-throughput screening of small molecule libraries to identify lead compounds with iPSCs was demonstrated within the next few years (Brennand et al. 2011; Lee et al. 2012; Yang et al. 2013). In 2013, the development of the CRISPR/Cas9 system as a versatile and user-friendly genome editing tool took biomedical research by storm and paired with iPSC technology in a perfect marriage. More recent studies exploring the opportunities that iPSCs offer to study malignancies, with blood cancers featuring most prominently among them, are breaking new ground in cancer research (Chao et al. 2017; Kotini et al. 2017).
TECHNICAL ASPECTS OF iPSC MODELING OF BLOOD MALIGNANCIES
Reprogramming Malignant Cells
In contrast to the generation of iPSC models of inherited genetic diseases—for which the choice of starting cell type is solely based on availability and convenience and encompasses any cell type of the human body—in the case of malignant diseases the composition of the starting cell population is of utmost importance. The malignant cells that iPSC models seek to capture are contained within the bone marrow (BM) and peripheral blood (PB) of patients with leukemias. These samples typically contain an admixture of normal and malignant cells with varying degrees of clonal heterogeneity of the latter. These characteristics necessitate careful genetic characterization of the derivative iPSCs to establish their provenance in relation to the different clones present in the starting cell population.
Reprogramming effectively resets the epigenome and erases any leukemia-related epigenetic abnormalities. Thus, genetic tracking is the only guide to ascertain provenance of iPSC lines from malignant cells as opposed to residual normal cells in the sample and to assign them to specific clones and subclones. Thus, although routine reprogramming of nonmalignant cells entails random picking of a small number of iPSC colonies (4–6) and, after further characterization, establishment of three or more iPSC lines, reprogramming malignant cells requires more stringent procedures to be successful. Our group has devised a reprogramming strategy tailored to the specific considerations of leukemic samples—namely, their genetic complexity and clonal heterogeneity. First, we perform comprehensive genetic characterization of the starting sample, which includes karyotype, mutational analysis with comprehensive gene panels, fluorescence in situ hybridization (FISH) for common chromosomal translocations, and potentially comparative genomic hybridization (CGH) to characterize chromosomal deletions. Second, we develop patient-specific polymerase chain reaction (PCR) (classic or quantitative)-based assays for genotyping, which enables us to easily genotype iPSC colonies in real time, as they emerge, in relatively high throughput. This, in turn, and in combination with efficient reprogramming methods—namely, Sendai virus or lentiviral vectors—enables “deep reprogramming” (i.e., the generation and screening of large numbers [which can reach the hundreds] of iPSC colonies in a single reprogramming experiment). This allows us to derive iPSC lines representing as many clones as possible, as well as normal cells. The latter typically have a reprogramming advantage over malignant cells and can most often be captured in iPSCs even if they are very rare in the starting cell sample. For the same reasons, premalignant clones can often be captured even if their representation in the starting cell sample is small or undetectable by bulk genetic analyses. This is, however, not a universal rule, as we have encountered instances in which leukemia cells reprogram with very high efficiency, surpassing that of normal cells (Kotini et al. 2017). TP53 inactivation has been documented to enhance reprogramming efficiency, and, thus, this higher reprogramming propensity may be related to TP53 activation status (Banito et al. 2009; Hong et al. 2009; Kawamura et al. 2009; Li et al. 2009; Marión et al. 2009; Utikal et al. 2009). Although oftentimes more than one clone and additionally normal cells can be captured in iPSCs, the clonal representation captured by reprogramming is often skewed (Chao et al. 2017; Kotini et al. 2017). This strongly implies that reprogramming efficiency is affected by the genetics of leukemia, including mutations, chromosomal abnormalities, and the presence or absence of genetic instability, but the underlying rules are not currently well understood.
The reprogramming efficiency of leukemic cells can be controlled by several mechanisms. Epigenetic alterations, such as DNA methylation, may very well impact the reprogramming efficiency, as aberrant DNA methylation patterns have been well documented in acute myeloid leukemia (AML) and profound methylation changes need to occur during reprogramming to pluripotency, including demethylation of promoters of key pluripotency genes (Figueroa et al. 2010; Apostolou and Stadtfeld 2018). Moreover, the capacity of leukemic cells for ex vivo growth also impacts greatly their ability to reprogram. Cell division is critically required both for efficient transduction with reprogramming vectors to initiate reprogramming, as well as for the epigenome remodeling required to complete reprogramming to pluripotency. Thus, inability to enter a proliferative state severely hampers a cell's reprogramming ability (Hanna et al. 2009; Ruiz et al. 2011; Guo et al. 2014). For example, the ability for ex vivo proliferation of myelodysplastic syndrome (MDS) and AML cells varies considerably across samples, but is generally poor. In contrast, cells from patients with myeloproliferative neoplasms (MPNs) typically grow robustly in culture, at least for a limited time period, and can be reprogrammed into iPSCs with high efficiencies (Hu et al. 2011; Kumano et al. 2012; Gandre-Babbe et al. 2013; Hosoi et al. 2014; Ye et al. 2014; Mulero-Navarro et al. 2015; Miyauchi et al. 2018). The cellular pathways underlying the capacity of leukemic cells to grow ex vivo and their putative correlation with specific genetic groups remain unknown. It is possible that an inverse correlation exists with a proapoptotic phenotype (Potter and Letai 2016). Because of all the reprogramming barriers mentioned above, several investigators have reported very low success rates of reprogramming leukemia samples, excluding MPNs (Muñoz-López et al. 2016; Lee et al. 2017). It is likely that these low success rates can be significantly increased with tailored, more efficient, and genetically informed reprogramming strategies. In our hands, approximately half of MDS and AML samples that can be induced to enter cell division by cytokine stimulation yield at least one or a few malignant iPSC lines. As more leukemias of diverse genetic groups and specific mutations are reprogrammed, principles of how reprogramming efficiency is affected by different factors discussed above will likely begin to emerge and may even provide some insights into the pathogenesis and signaling dependences of different leukemias.
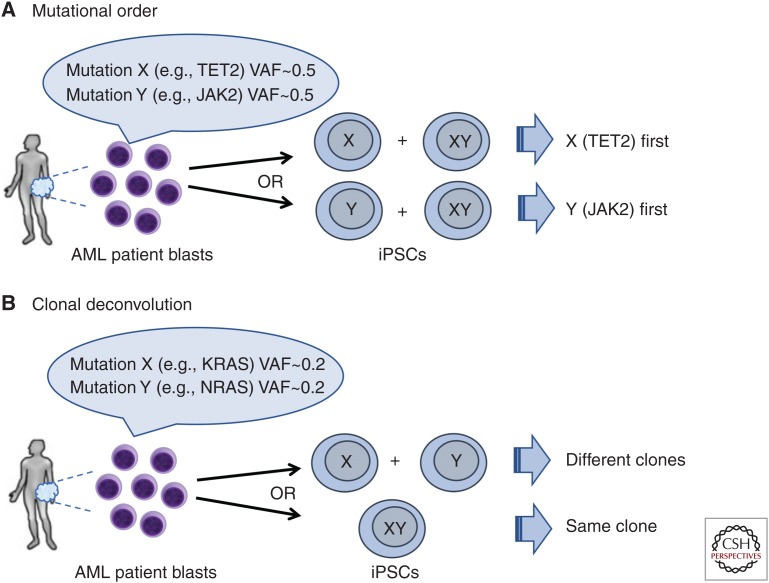
The process of reprogramming per se can also be informative in elucidating the clonal architecture and evolution of a leukemia sample, in a way akin to what has been previously done using colony forming assays in methylcellulose (Fig. 1; Jan et al. 2012; Ortmann et al. 2015). First, reprogramming can determine the order of mutation acquisition (Fig. 1A). The order by which driver mutations are acquired may affect clinical features and response to therapy (Kent and Green 2017; Levine et al. 2019). Genetic analyses of the bulk tumor cannot always determine the mutational order through inference based on variant allele fractions (VAFs). Such instances may include cases in which two mutations arose close together in time and their VAFs are very similar to each other or cases of advanced disease in which the earlier clones have been almost entirely replaced by the most evolved ones. In the latter case, rare cells of a parental clone may be captured in iPSCs if they have a reprogramming advantage, which they often do, over the fully leukemic clone. Second, reprogramming can help clonal deconvolution in cases with low mutational burden, in which bulk sequencing cannot discriminate whether two mutations are present in the same or in separate clones (Fig. 1B). Capturing these clones by reprogramming can unambiguously determine the clonal composition of the starting sample. For reprogramming to inform on the clonal parameters of the starting leukemia, however, it is critical that iPSC derivation is performed in conditions ensuring and preserving clonality. These include line establishment and passaging techniques (e.g., manual picking of a single colony under a microscope, single-cell subcloning in case of mixed clonality) and mutational analyses. If integrating vectors were used for reprogramming, integration site analysis can provide an additional means to ascertain clonality (Kotini et al. 2017).
Figure 1.
Mutational order reconstruction and clonal deconvolution by reprogramming. (A) Reprogramming into induced pluripotent stem cells (iPSCs) can decipher the order of mutation acquisition and thus illuminate the clonal history of the disease. In the example shown, two mutations (X and Y, e.g., TET2 and JAK2) have similar allele burden at presentation precluding bulk genetic analyses from determining the order by which they were acquired. iPSCs can capture the precursor single-mutant clone and thus determine which mutation occurred first. (B) Generation of iPSCs can deconvolute the clonal composition of a sample in the case of mutations with low variant allele fractions (VAFs) from the bulk analysis, which cannot determine whether they are present in the same or different clones. In this case, KRAS and NRAS mutations can frequently arise in divergent clonal evolution (example on top) or, alternatively, coexist in the same clone (example in the bottom).
Hematopoietic Specification of iPSCs
Virtually all phenotypic and molecular assays are performed and all readouts are obtained following the in vitro directed differentiation of iPSCs into hematopoietic cells. The efficiency and robustness of this process weighs heavily on the quality of all iPSC-based studies. Most evidence to date suggests that in vitro hPSC-derived hematopoiesis consists of cells corresponding to different developmental stages, without a clear temporal separation of their emergence (Choi et al. 2012; Pearson et al. 2015). Thus, hematopoietic lineages of all three waves that are successively generated during normal development of the hematopoietic system may be present in in vitro differentiation cultures. These include a first wave, referred to as “primitive,” giving rise only to erythroid, megakaryocyte, and macrophage lineages; a second wave that consists primarily of erythro-myeloid progenitors (EMPs) and some lymphoid progenitors; and a third wave that—unlike the first two, which are extraembryonic—arises in the embryo proper and gives rise to hematopoietic stem cells (HSCs) (Ivanovs et al. 2017; Lacaud and Kouskoff 2017). The terms “primitive” and “definitive” hematopoiesis were originally coined based on pronounced differences of the erythroid products of the first and last wave (size, enucleation, and globin gene expression). With better characterization of the EMPs, the term “definitive” was extended to include the second wave (Frame et al. 2013). However, inconsistency in the use of the term remains in the literature, as some investigators save the term “definitive” exclusively for adult-type HSC-generating hematopoiesis. Notably, this confusion extends to the description of the developmental stage of hPSC-derived hematopoietic products. Earlier protocols for the in vitro hematopoietic differentiation of hPSCs yielded mostly primitive hematopoiesis. More recently, however, the findings that definitive lineages could be generated through activin inhibition or WNT stimulation, acting at the level of early mesoderm patterning, has enabled contemporary protocols to derive primarily definitive- and not primitive-type cells (Kennedy et al. 2012; Sturgeon et al. 2014). However, it is still not clear if these newer protocols better capture the second or third wave of hematopoiesis or a mixture of the two. As mentioned earlier, HSCs are only generated in the third developmental wave, but it is now believed that all hematopoietic lineages can be generated by more restricted progenitors of the second wave. The fact that hPSC-derived hematopoietic cells can produce all mature lineages, but do not engraft in immunocompromised recipients (as further discussed below), has prompted the speculation that they may more closely represent EMP-type hematopoiesis.
An important limitation of hematopoietic differentiation protocols is the current inability to derive true HSCs with long-term multilineage engraftment capability, potentially related to the developmental immaturity of hPSC-derived hematopoiesis, discussed above (Vo and Daley 2015). This is a technical and not biological limitation, as multilineage engraftable hematopoiesis can be derived from human iPSCs via teratoma formation (Amabile et al. 2015; Suzuki et al. 2015). Whereas this method is impractical and not clinically translatable, it shows that HSCs can, in principle, be derived from hPSCs once the appropriate conditions are worked out, which may also include in vivo niche-derived signals. The unavailability of robust cell culture conditions to maintain and expand primary HSCs in vitro despite years of effort by several investigators is a related problem and attests to the challenge of this endeavor. Although bona-fide HSCs cannot be derived through hPSC-directed differentiation in vitro, mature cells of all hematopoietic lineages, and thus hematopoietic progenitor cells (HPCs) of all lineages, can be derived from them. However, the timing of their emergence, lineage potential, and developmental stage have not been well characterized and likely differ among differentiation protocols. Mapping the HPC populations in terms of uni- or multilineage potential and developmental origin with relation to timing of emergence and surface marker expression would greatly help boost the efficiency and reproducibility and harmonize findings across studies using diverse differentiation methods.
Genome Editing of iPSCs
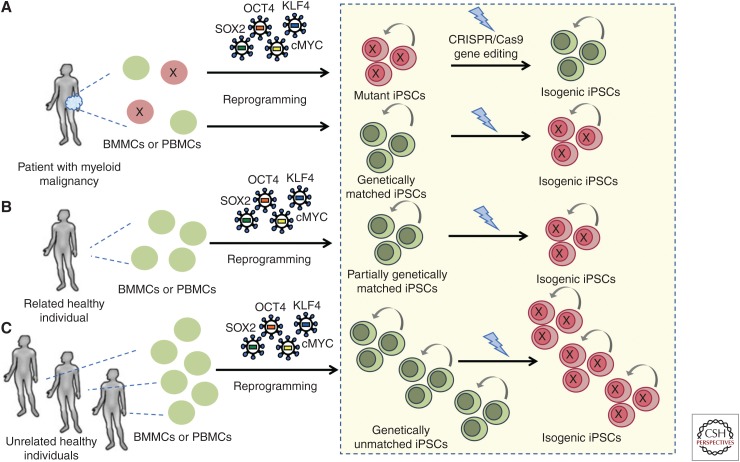
Whereas for some time hPSCs were thought to be rather refractory to genetic modifications, particularly those involving homologous recombination, advances in their culture—crucially the enhancement of survival and clonal growth of hPSCs by Rho kinase (Rock) inhibition—together with the advent of more efficient gene delivery and gene editing methods, like the CRISPR/Cas9 system, has now turned the genetic engineering of hPSCs into an almost routine practice (Watanabe et al. 2007; Kim and Kim 2014; Hockemeyer and Jaenisch 2016). The genetic engineering of iPSCs uses techniques and principles from the field of genetically engineered mouse models and can, similarly, be used to create reporter, knockout, and knock-in lines. Most crucially, gene editing of iPSCs offers the unique opportunity to derive isogenic lines (i.e., lines differing in only one gene that are otherwise genetically identical) either by introducing specific mutations found in hematologic malignancies in normal iPSCs, by correcting them in patient-derived iPSCs, or, ideally, by both strategies (Chang et al. 2018). Isogenic pairs of lines are more superior controls than unrelated or even patient-matched normal lines, as the latter will still have multiple genetic differences from the disease lines, both pathogenic and nonpathogenic (Fig. 2). The CRISPR/Cas9 system can be used both to inactivate genes to model common loss-of-function mutations in leukemias (e.g., TET2, RUNX1, EZH2, TP53, cohesin) through nonhomologous end joining repair of Cas9-mediated double strand DNA breaks and to introduce hotspot mutations in oncogenic driver genes (e.g., splicing factors, JAK2 V617F, DNMT3A R882H, NRAS G12D, NPM1c, FLT3-ITD) through homology-directed repair from a donor DNA template.
Figure 2.
Control induced pluripotent stem cell (iPSC) lines. Normal iPSCs derived from the same patient (from either hematopoietic or other types of somatic cells) can be used as genetically matched controls (A). If these are derived from hematopoietic cells, careful genetic analyses should clarify whether they represent completely normal cells or a precursor premalignant or clonal hematopoiesis (CH) clone, by ascertaining that they harbor none of the pathogenic mutations of the patient's malignant clone. These controls share the same genetic background with the disease iPSCs derived from the same patient, but are not technically isogenic, as they almost certainly differ from the malignant iPSCs in more than one gene. Alternatively, normal lines can be derived from healthy family members of the patient, if such material is available (B). Finally, existing lines derived from unrelated healthy donors can be used as normal controls (C). In the latter case, multiple lines should be used to control for variation because of differences in the genetic background. In all cases, gene editing can be used to develop isogenic matched lines by either correcting a given mutation in patient iPSCs or introducing it in normal iPSCs. Isogenic lines are far superior controls and should always be preferred over nonisogenic controls.
STUDIES USING iPSC MODELS OF HEMATOLOGIC MALIGNANCIES
MPNs, including chronic myeloid leukemia (CML), essential thrombocytopenia (ET), polycythemia vera (PV), and primary myelofibrosis (PMF), were the first malignant blood diseases that were modeled with patient-derived iPSCs (Ye et al. 2009, 2014; Carette et al. 2010; Hu et al. 2011; Kumano et al. 2012; Bedel et al. 2013; Gandre-Babbe et al. 2013; Saliba et al. 2013; Hosoi et al. 2014; Amabile et al. 2015; Mulero-Navarro et al. 2015; Suknuntha et al. 2015; Gomez Limia et al. 2017; Liu et al. 2017; Sloma et al. 2017; Miyauchi et al. 2018; Takei et al. 2018). In one study, CML-iPSCs could produce mature erythroid and myeloid cells and could not engraft leukemia in mice upon in vitro differentiation, suggesting that reprogramming and/or directed differentiation impaired their oncogenic potential. This phenotype might be related to the finding of decreased DNA methylation levels in the CML-iPSCs and the hematopoietic cells differentiated from them, compared to the primary CML cells (Amabile et al. 2015). In contrast, the dependency of CML cells on BCR-ABL could be recapitulated in patient-derived iPSCs (Carette et al. 2010; Kumano et al. 2012; Bedel et al. 2013; Suknuntha et al. 2015; Miyauchi et al. 2018). CML-iPSCs were not responsive to imatinib at the pluripotent state, but sensitivity was reestablished in hematopoietic cells in vitro differentiated from them. The finding that BCR-ABL, while expressed, does not confer dependency at the pluripotent state, highlights the requirement of cooperation between an oncogenic genetic lesion and the appropriate cellular context for malignant features to manifest. MPN patient-derived iPSCs with heterozygous and homozygous JAK2 V617F, MPL, and CALR mutations have been generated, reflecting the ease of reprogramming of MPN cells, but these studies have not gone beyond reproducing known cellular phenotypes from ex vivo cultured MPN cells, mainly their disease-defining ability for cytokine-independent colony formation in methylcellulose (Saliba et al. 2013; Ye et al. 2014; Gomez Limia et al. 2017; Liu et al. 2017; Takei et al. 2018). The possibility of modeling responses of JAK2 V617F MPN to JAK inhibitors was demonstrated in one study (Ye et al. 2014).
Juvenile myelomonocytic leukemia (JMML), a pediatric MDS/MPN overlap syndrome caused by various signaling mutations, has also been modeled in iPSCs (Gandre-Babbe et al. 2013; Mulero-Navarro et al. 2015; Gagne et al. 2018; Tasian et al. 2019). Specifically, patient-derived iPSCs with PTPN11 and CBL mutations were generated and shown to recapitulate disease phenotypes, such as hypersensitivity to growth factors and increased proliferation. JMML-iPSC-derived myeloid cells were also shown to model differential signaling pathway activation and sensitivity to kinase inhibitors: PTPN11-mutant JMML-iPSC-derived hematopoietic cells exhibited constitutive activation of RAS/MAPK signaling and were sensitive to MEK inhibition, whereas CBL-mutant JMML-iPSC-derived hematopoietic cells showed JAK/STAT signaling activation and sensitivity to JAK inhibitors (Tasian et al. 2019).
In contrast to MPN cells, MDS and AML cells are relatively refractory to reprogramming. Our group derived the first MDS-iPSCs from two patients with chromosome 7q deletion (del7q) (Kotini et al. 2015). We found that all del7q lines had a markedly diminished potential for generation of CD34+/CD45+ HPCs upon in vitro differentiation and almost absent ability to generate all types of hematopoietic colonies in methylcellulose assays. By studying the hematopoietic potential of a series of iPSC lines with corrected chr7q dosage, as well as lines with engineered deletions spanning various chr7q regions, we were able to pinpoint a critical region whose hemizygosity was sufficient to confer this loss of differentiation potential. We then selected candidate haploinsufficient genes by means of reduced expression in the hemizygous compared to normal lines and further prioritized hits of a pooled rescue screen of 75 candidate cDNAs. Four genes, EZH2, LUC7L2, HIPK2, and ATP6V0E2, were validated to partially rescue the del7q phenotype. Interestingly, the first three genes have also been found to harbor monoallelic loss-of-function mutations in MDS, in further support of a function as haploinsufficient tumor suppressor genes in MDS. This work highlights how iPSCs combined with sophisticated genetic engineering strategies can be used to functionally map critically lost regions within recurrent chromosomal deletions, which cannot easily be modeled in the mouse or other organisms because of lack of conservation of synteny. A subsequent study focused on modeling a hotspot mutation in a splicing factor gene, SRSF2 P95L, in MDS-patient-derived iPSCs together with del7q and showed proof of principle of the usefulness of iPSC modeling to connect specific driver genetic lesions with cellular phenotypes and drug responses, such as sensitivity of SRSF2-mutant iPSC-derived hematopoietic cells to splicing inhibitors (Chang et al. 2018). Studies like this can inform precision medicine approaches, whereas the knowledge of which cellular phenotypes are conferred by a specific mutation (and which are not) can guide assay development for drug testing and drug discovery.
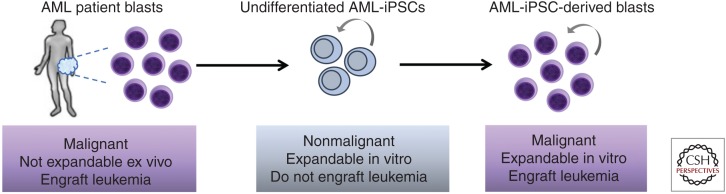
More recently, our group reported a larger collection of iPSC lines derived from patients with low-risk MDS, high-risk MDS, and secondary AML (sAML), one of which also harbored a germline GATA2 mutation, conferring predisposition to MDS/AML (Kotini et al. 2017). Guided by comprehensive mutational analyses of the starting patient cells and the derived iPSC lines, we assembled a panel of lines capturing the different stages of disease, from familial predisposition to low-risk MDS, high-risk MDS, and sAML. By characterizing their hematopoiesis with a battery of assays, we constructed a framework of cellular phenotypes characterizing the distinct stages of myeloid malignancy from preleukemia to AML through an MDS stage. Using this system and phenotypic map we showed that we can model transitions between stages, including disease progression and reversal, by introducing and correcting, respectively, progression-associated mutations by CRISPR/Cas9. We also uncovered potential disease stage-specific effects of 5-azacytidine, a drug used as frontline therapy for MDS. Furthermore, transcriptome analyses of iPSC-derived CD34+ hematopoietic progenitors of all stages revealed gene expression signatures associated with disease progression and enrichment for gene sets derived from primary AML patient cells. Surprisingly, given the inability of iPSC-derived hematopoiesis to engraft in immunodeficient mice, we found that AML-iPSC-derived hematopoietic cells robustly engrafted into NSG mice via intravenous injection, giving rise to a serially transplantable lethal myeloid leukemia. Similar findings were reported at the same time by the group of Ravi Majeti, who derived iPSCs from patients with MLL-translocated AML (Chao et al. 2017). This study also reported serial engraftment of AML-iPSC-derived hematopoietic cells administered intravenously, as well as orthotopically into NSG mice harboring human ossicles that mimic a humanized hematopoietic niche (Reinisch et al. 2016). This study additionally demonstrated, through detailed transcriptome and DNA methylation analyses, that AML-associated epigenetic changes were erased upon reprogramming, as AML-iPSCs were very similar to normal control iPSCs and clearly distinct from primary AML cells. Consistent with this, AML-iPSCs were able to differentiate into tissues of all germ layers without overt signs of malignancy in vitro and in teratomas. However, in stark contrast, their hematopoietic progeny were similar to primary AML blasts in their gene expression and DNA methylation patterns, consistent with reacquisition of phenotypic leukemic features in vitro and in vivo. No evidence of “epigenetic memory” of leukemia was found in the undifferentiated AML-iPSCs, suggesting that the reestablishment of leukemia was exclusively driven by the AML genetic lesion (MLL rearrangement) in conjunction with the acquisition of the correct cellular identity. The aforementioned studies thus uniquely highlight the sufficiency of genetic lesions for leukemia establishment on one hand and the importance of the appropriate cellular context for leukemic properties to manifest on the other (Fig. 3). In both studies, iPSCs could be derived both from major AML clones and subclones with KRAS mutations. These iPSCs shared all other leukemia-associated genetic lesions except for the KRAS mutation, highlighting the opportunities that iPSC modeling presents to isolate clones and study their properties and differential drug susceptibilities.
Figure 3.
Induced pluripotent stem cells (iPSCs) derived from acute myeloid leukemia (AML) patients. AML-iPSCs do not exhibit malignant features at the undifferentiated pluripotent state, but only once they are differentiated along the hematopoietic lineage. Unlike primary AML blasts that have limited growth potential in vitro, iPSC-derived blasts can be extensively, potentially indefinitely, expanded in culture.
In contrast to myeloid malignancies, no studies of iPSC modeling of lymphoid leukemias or lymphomas have been reported to date. Although it is possible that lymphoid blasts are even harder to reprogram than myeloid blasts, the scarcity of such studies likely also reflects the challenges in the derivation of lymphoid lineages through directed in vitro differentiation of hPSCs (Muñoz-López et al. 2016; Montel-Hagen and Crooks 2019a). Of note, a recent study used gene targeting of normal human iPSCs to model childhood acute B-lymphoblastic leukemia harboring the ETV6-RUNX1 translocation (Böiers et al. 2018). The authors first characterized B lymphoid development in first-trimester human embryos and provided evidence of an IL-7R+ progenitor cell population. They next showed that hPSCs recapitulate this developmental transition and that ETV6-RUNX1 expression specifically affects it, causing expansion of the IL-7R+ progenitor compartment, block of B lineage commitment, and aberrant myeloid gene expression signatures in pro-B cells. These findings support the idea that ETV6-RUNX1 targets a specific susceptible progenitor cell type that is uniquely present in embryonic and fetal life for preleukemic initiation, which can explain why ETV6-RUNX1 translocations are rarely found in adult acute lymphoblastic leukemia (ALL).
Familial forms of predisposition to MDS/AML caused by inherited mutations have also been modeled in iPSCs. Our group described subtle phenotypic changes consistent with a preleukemic state in iPSC-derived hematopoietic cells from a patient with a germline GATA2 mutation and modeled progression to low-risk or high-risk MDS by engineering additional genetic lesions that frequently occur in AML arising on the grounds of inherited GATA2 deficiency—namely, mutational inactivation of the second GATA2 allele and del7q (Kotini et al. 2017). Familial platelet disorder (FPD), caused by germline mutations of RUNX1 and characterized by thrombocytopenia, platelet dysfunction, and predisposition to MDS/AML, has been modeled with iPSCs by several groups (Connelly et al. 2014; Sakurai et al. 2014; Antony-Debre et al. 2015; Iizuka et al. 2015). A study using iPSCs from different FPD pedigrees showed that RUNX1 haploinsufficiency causes only megakaryocytic defects, whereas dominant negative mutations confer additional phenotypic changes, supporting the idea that only the latter confer risk of leukemia progression (Antony-Debre et al. 2015). iPSC models of several inherited BM failure syndromes, which are often accompanied by predisposition to leukemia development, have been created, including Fanconi anemia, Shwachman–Diamond syndrome, Diamond–Blackfan anemia, dyskeratosis congenita, and severe congenital neutropenia. These studies mostly focused on modeling gene correction strategies and providing proof of principle of rescue of disease-relevant phenotypes by gene therapy rather than on the propensity to malignancy and have been reviewed elsewhere (Georgomanoli and Papapetrou 2019).
NEW OPPORTUNITIES AFFORDED BY iPSCs FOR THE STUDY OF LEUKEMIA BIOLOGY AND PRECLINICAL RESEARCH
Arguably the most appealing property of iPSC modeling for hematologic malignancies is the possibility to create precise genetic models. The capturing of intact human leukemic genomes enables modeling of disease-driving mutations and other genetic lesions in their native genomic context, in the appropriate cellular setting, following differentiation to the desired cell type, and in isogenic conditions. This can be particularly valuable in instances of mutations in genes whose functions are not well conserved among species or for the modeling of large-scale structural abnormalities, such as translocations, large chromosomal deletions, trisomies, or complex karyotypes that cannot be easily engineered or modeled in the mouse because of synteny issues (Kotini et al. 2015). A wealth of information on recurrent gene mutations in MDS and AML has become available in recent years, and currently no good models exist for several of them. Conventional immortalized cell lines (ICLs) with mutations such as IDH1 and IDH2 are lacking. Similarly, there are no hematopoietic cell lines with splicing factor mutations. Although ectopic expression of mutant genes or in situ introduction of mutations in the endogenous locus through CRISPR is often used, these approaches still suffer from uncontrolled levels of expression and stoichiometry imbalances, as most ICLs are aneuploid and contain variable numbers of copies of most endogenous genes. Altered stoichiometry and levels of expression of splicing factors can artificially affect their RNA-binding properties. Furthermore, alternative splicing and intronic sequences are not well conserved between mouse and human. Thus, the modeling of splicing factor mutations and other heterozygous hotspot mutations in iPSCs, through reprogramming or gene editing, can provide a physiological genomic context to study their downstream effects (Chang et al. 2018). Finally, the reprogramming of entire leukemic genomes offers the opportunity to capture the patient's genetic background including any inherited variation and other unknown mutations or polymorphisms that may modify the phenotype or disease risk (Kilpivaara et al. 2009).
Reprogramming technology also presents exciting opportunities to study the relative contribution of genetic and epigenetic factors in leukemogenesis. The studies of AML-iPSCs, discussed above, clearly established the requirement for an interplay between the leukemia genome and an appropriate cellular milieu (Chao et al. 2017; Kotini et al. 2017). They also showed that a leukemia genome is sufficient to establish leukemia in the correct cell type, as reprogramming erases all preexisting leukemic epigenetic alterations (Chao et al. 2017). Furthermore, these studies showed that AML-iPSC-derived blasts can model at least to some extent the transcriptome and epigenome of primary cells, as signatures shared with primary AML were found. However, more detailed comparisons between equivalent populations of iPSC-derived and primary AML blasts are required to understand how faithfully the former recapitulate the chromatin and gene expression landscape of the latter. Also, although current evidence suggests that epigenetic changes that are genetically determined by mutations in epigenetic modifiers (e.g., TET2, DNMT3A, ASXL1, EZH2, and others) should be reestablished in iPSCs upon differentiation, formal demonstration is still missing. The finding that, in contrast to AML-iPSCs, CML-iPSCs do not exhibit overt leukemic features upon differentiation is puzzling and may reflect a requirement for a specific cell type (e.g., HSCs) that was not produced in the differentiation cultures (Amabile et al. 2015). This may provide an example of how reprogramming could be used to determine the cell of origin of specific leukemias and to investigate the cell type requirements of cancers more broadly. iPSC models of familial cancer forms with different tissue preponderance may prove particularly useful for this (Lee et al. 2015).
iPSCs offer unique opportunities to model disease progression and clonal evolution of malignancy. Reprogramming can be performed in clonal conditions and often favors normal or premalignant over malignant cells (Kotini et al. 2017). Thus, unlike ICLs that can only be derived from malignant cells, iPSCs can be derived from normal and preleukemic cells harboring initiating mutations without the full set of late mutations. Additionally, the ease of stepwise addition of targeted mutations by CRISPR and the recent wealth of information on patterns of mutational cooperation open the possibility of modeling clonal evolution through synthetic biology approaches (Papaemmanuil et al. 2016).
One of the most highly advertised uses of iPSC technology from the outset was its use in drug testing and screening. Responses to DOT1L inhibitors in MLL-rearranged AML, to rigosertib and MEK inhibitors in the context of AML with KRAS mutations, and to splicing modulators in SRSF2-mutant MDS have been modeled in iPSC-derived cells, with cytotoxicity as the main readout (Chao et al. 2017; Kotini et al. 2017; Chang et al. 2018). Efforts to model drug resistance to imatinib in CML and to cytarabine-based chemotherapy in AML have also been reported (Chao et al. 2017; Miyauchi et al. 2018). Because they provide a theoretically unlimited source of cells, iPSC models can be used to develop platforms for high-throughput small-molecule screens. Additionally, they offer the possibility to perform phenotype-driven screens in cases in which there are no known targets, empowered by the availability of isogenic normal controls (Chang et al. 2018). Current work on drug screening in iPSC hematopoietic derivatives has not yet moved past proof-of-principle studies and is limited by scalability, as will be further discussed below.
COMPARISON TO OTHER PATIENT-DERIVED MODELS (IMMORTALIZED CELL LINES, PATIENT-DERIVED XENOGRAFTS)
Once established, iPSC lines can be expanded indefinitely at the pluripotent state, cryopreserved, and shared with other investigators. Thus, iPSC modeling enables experimentation with patient-derived material that, in contrast to primary cells, is unlimited, allowing for robust and well-controlled experiments that were hitherto only possible with ICLs. However, iPSCs are fundamentally different than ICLs. iPSCs are maintained in a self-renewing state by a pluripotency gene network that is self-sustaining and that, although some of its components may be shared with cancer, is clearly distinct from oncogenic signaling that maintains ICLs, which are dependent on strong viral or cellular oncogenes. iPSCs maintain normal diploid karyotypes and are genetically more stable than ICLs, which are typically aneuploid. As mentioned above, unlike ICLs that can only be derived from fully transformed cells and typically aggressive cancers, iPSCs can be derived from normal and premalignant cells. Although acquisition of genetic lesions over time is a problem with all cultured cells, measures for its mitigation are much more readily applied in iPSC research. The stem cell community is more aware of the risks of genetic diversification of cultured lines and their monitoring is a routine practice, whereas passage number is almost never tracked or reported in studies using ICLs. Aneuploid status and copy-number variation, present even among individual cells within the same ICL, can pose problems with modeling leukemia-associated mutations. Furthermore, copy-number aberrations may confound the results of CRISPR screens, as multiple Cas9-mediated DNA breaks can impair cell proliferation in a gene-independent manner, a phenomenon referred to as the CRISPR copy-number effect. While the argument can be made that iPSCs could replace ICLs in many applications, the latter are still popular because of low cost and ease of use. Generation of iPSC-derived expandable progenitor cell lines, further discussed later, may overcome most of the practical obstacles to the broader use of iPSCs.
iPSCs can be derived from leukemia cells passaged through xenotransplantation and, conversely, hematopoietic cells derived from AML-iPSCs can generate xenografts. Such “secondary” patient-derived xenografts (PDXs) can offer opportunities not afforded by primary PDXs—namely, the generation of clonal PDX models devoid of genetic drifts and amenable to extensive genetic manipulations prior to transplantation. For other applications (e.g., use as patient-specific models or avatars) in which capturing the genetic and epigenetic heterogeneity of the primary AML is desirable, primary PDXs may be preferable. Finally, how epigenetically similar iPSC xenografts are to primary PDXs and to primary AML cells is yet unknown. iPSC xenografts should thus be more valuable for genotype-to-phenotype studies, for which similarity to primary tumor is less relevant than the faithful representation of genotypes in the correct genomic environment.
MISCONCEPTIONS AND TRUE LIMITATIONS OF iPSC MODELS
As experience with iPSC modeling is building, some misconceptions are still plaguing the field, whereas important current limitations of the approach provide opportunities for future improvements.
Genetic Stability of iPSC Lines
Early studies reported a high degree of chromosome aberrations, copy-number variation, and point mutations in iPSCs and stirred serious concerns that reprogrammed cells accumulate multiple abnormalities at the chromosomal, subchromosomal, and single-base level (Mayshar et al. 2010; Gore et al. 2011; Hussein et al. 2011; Laurent et al. 2011). Subsequent studies revealed that the majority of these genetic variants are preexistent in the starting somatic cells (Abyzov et al. 2012; Cheng et al. 2012; Young et al. 2012). Numerous other studies have now established that reprogramming in itself is not mutagenic and that iPSCs are not inherently genetically unstable, as they acquire mutations during expansion in culture or during differentiation at a rate similar to that of normal adult somatic cells and consistent with spontaneous mutation acquisition rates during cell division (Cheng et al. 2012; Liang and Zhang 2013; Peterson and Loring 2014; Tapia and Schöler 2016). However, several real issues remain and need to be considered in iPSC experiments. First, reprogramming can cause “founder effects,” akin to those in ICLs and other patient-derived models, by selecting for rare cells of the starting population. This can skew the representation of the primary tumor and may even select for “ultrafit” clones—for example, cells with epigenetic inactivation of TP53. Second, mutations will always accumulate with increased passage. Importantly, the selection forces driving genetic evolution in culture, as well as in PDX models, seem to be different than those acting in patients, leading to increased genomic divergence between the models and the primary leukemia over time (Hussein et al. 2011; Ben-David et al. 2017, 2018). In particular, aneuploidies and copy-number alterations tend to disappear in iPSCs and other models over time. In our study, we observed a growth disadvantage and strong selection pressure for correction of chr7q dosage in del7q MDS-iPSC lines over increasing passage, whereas del7q cells presumably have a selection advantage in the in vivo patient setting (Kotini et al. 2015). Third, while normal iPSCs are not more unstable than any other cell type, cancer driving mutations may involve genes regulating genome integrity and may impact genomic stability. Importantly, as further discussed below, with awareness and adherence to good laboratory practices most of these issues can be resolved.
Developmental Stage of hPSC-Derived Hematopoiesis and Its Impact on Disease Modeling
iPSC differentiation to desired cell types mimics development, and this has severe implications for disease modeling. First, developmental phenotypes may confound disease-relevant phenotypes. For example, some recurrent MDS and AML mutations, such as GATA2, RUNX1, and others, affect genes with important roles in the development of the hematopoietic system and may block the differentiation of iPSCs at early stages. For example, RUNX1 mutations may impair or abolish the generation of HPCs from hemogenic endothelium (HE). This problem is analogous to the problem of embryonic lethality in knockout mouse models and, similarly, it could be overcome by engineering conditional alleles. Second, as discussed above, current protocols yield developmentally immature and mixed cells. Despite this, it is encouraging that several studies have shown that disease-relevant phenotypes—such as low clonogenicity, reduced proliferation, and increased apoptosis in MDS; cytokine independency in MPN; and differentiation block and increased self-renewal in AML—can be recapitulated in iPSC models (Ye et al. 2014; Kotini et al. 2015, 2017; Chao et al. 2017; Chang et al. 2018). Developmental immaturity may pose more of a problem for modeling disease features critically dependent on aging. Artificial induction of aging in iPSCs through expression of progerin could be used to reveal age-related disease manifestations (Miller et al. 2013). Most crucially, better protocols that can specify adult-type hematopoiesis exclusively or at least temporally separated from earlier waves and/or markers for purification of the former are needed. To this end, protocols that early on induce the right type of mesoderm and HE with potential for adult-type hematopoiesis will need to be devised (Kennedy et al. 2012; Sturgeon et al. 2014; Ditadi et al. 2015; Guibentif et al. 2017).
Line-to-Line Variation
Reports of large variability in differentiation propensity and phenotypes among diverse normal iPSC lines have spurred concerns over the robustness of iPSC modeling in general. Given that humans are genetically very diverse, it is hardly a surprise that most of the variation in phenotypes, differentiation propensity, gene expression, DNA methylation, and epigenetic marks has now been attributed to differences in genetic background (Kyttälä et al. 2016; DeBoever et al. 2017; Kilpinen et al. 2017; Pashos et al. 2017). Variation among genetically matched iPSC lines can also be present because of epigenetic differences established and fixed upon reprogramming (Liang and Zhang 2013). These can be further exacerbated if differentiation protocols and assays for readouts allow for a high degree of noise. Because such noise may be mistaken as disease-specific phenotypes, it is imperative that the variation of the differentiation procedure or phenotypic assay used does not exceed the effect size of the reported disease-relevant phenotype. The former can be measured by estimating the variance in the phenotypic measures among independent differentiations of the same iPSC line and among different lines of the same genotype and comparing it to the phenotypic difference between normal and disease lines (Kotini et al. 2015).
AREAS FOR IMPROVEMENT
Good Practices
Important in all areas of research, good practices are particularly critical in hPSC research and, consequently, stem cell researchers are typically alert to factors that can compromise their results and ways to mitigate the risks. Genetic diversification caused by selection imposed by culture conditions or genetic drifts can be detected by frequent karyotyping and periodic authentication of iPSC lines by DNA fingerprinting and mutational status of known mutations and prevented by routine tracking and reporting of passage number, cryopreservation, and storage of large stocks of early passage lines. Also, maintaining cultured lines at a low passage number, performing experiments of one entire study with cells at similar passage number, and avoiding unnecessary passaging and introduction of bottlenecks, such as multiple freeze–thaw cycles, extreme cell number reductions during passaging, and unnecessary subcloning, are advisable. Bottlenecks cannot be altogether avoided—for example, those imposed during initial line establishment or during genetic engineering (antibiotic selection, single-cell cloning)—and are a feature of all cancer models, including ICLs, PDXs, and organoid cultures (Ben-David et al. 2019). iPSCs offer the advantage of indefinite expansion, allowing for any inadvertent genetic changes to be traced back in time and “corrected” by retrieval of earlier passage stocks. Additional good practices include maintaining standardized cell culture conditions to decrease selection pressure, regular testing for mycoplasma contamination status, and alertness to any abrupt phenotypic changes in division time, morphological features, or any phenotype. Any such changes should be readily investigated as they often indicate underlying genetic changes. For example, our discovery of spontaneously corrected del7q MDS-iPSC clones was prompted by the observation of phenotypic rescue of hematopoiesis in the corrected clones (Kotini et al. 2015). Finally, the opportunity to have tailored control lines presents an important asset of iPSC modeling that should be harnessed (Fig. 2). Using multiple genetically identical and independently propagated lines, which can be relatively easily generated in one round of reprogramming or gene editing, can solve most issues of reproducibility arising from genetic divergence.
Future Advances
Three significant current barriers to iPSC modeling of blood cancers present opportunities for future advances.
Generation of Expandable HPCs (eHPCs)
Currently, iPSC models remain inaccessible to most of the hematology community because of a lack of hPSC culture and differentiation expertise, which are both quite laborious and costly processes. One way by which this could be sidestepped is the generation of iPSC-derived eHPCs (i.e., HPCs that can be maintained in a self-renewing state), thus bypassing the hPSC culture and differentiation steps. These eHPCs could be passaged as simple suspension cultures in media and conditions akin to those used for conventional ICLs, expanded extensively or even indefinitely, cryopreserved, and terminally differentiated on demand, while preserving all or most of their mutation-specific characteristics.
Doulatov et al. discovered five factors (HOXA9, ERG, RORA, SOX4, and MYB) that could maintain eHPCs. Discontinuation of factor expression through doxycycline withdrawal enabled differentiation along the myeloid and erythroid lineage (Doulatov et al. 2013). Furthermore, these progenitors could give rise to short-term myeloerythroid engraftment in immunocompromised mice. More recently, our group showed that a combination of 13 factors can maintain CD34+/CD45+ eHPCs long-term in vitro (Chang et al. 2018). Apart from opening up iPSC modeling to the broader community, eHPC approaches can also, very importantly, overcome limits of scale of hematopoietic differentiation and dramatically increase the throughput of assays made possible. The latter can include biochemical assays requiring large cell numbers, proteomics, and high-throughput small-molecule or genetic (e.g., CRISPR-based) synthetic lethality screens to identify therapeutic targets. For example, five-factor eHPCs derived from iPSC models of DBA enabled an unbiased chemical screen, which would not have been possible otherwise, and led to the discovery of a new small molecule stimulating erythropoiesis through induction of autophagy (Doulatov et al. 2017).
Generation of Transplantable HSCs
Although hematopoietic cells from AML-iPSCs from two different AML genotypes (MLL-rearranged and del7q) were shown to engraft in immunodeficient mice, it is currently unclear if this is a generalizable property of all AML-iPSCs. Furthermore, hematopoietic cells from CML, MDS, and normal iPSCs clearly do not engraft (Amabile et al. 2015; Chao et al. 2017; Kotini et al. 2017). This limitation, which might be at least in part a result of the developmental immaturity of hPSC-derived hematopoiesis, discussed earlier, restricts experimentation in the in vivo setting. The limited success in establishing differentiation conditions to generate engraftable hematopoiesis by many investigators over two decades has been reviewed elsewhere (Vo and Daley 2015; Rowe et al. 2016; Wahlster and Daley 2016). More recently, the Daley laboratory accomplished long-term multilineage engraftment of iPSC-derived cells, albeit at relatively modest efficiency, by forced expression of seven transcription factors (ERG, HOXA5, HOXA9, HOXA10, LCOR, RUNX1, and SPI1) (Sugimura et al. 2017).
Incorporation of Microenvironment Components and Three-Dimensional Cultures
iPSCs mainly read cellular phenotypes and do not capture nongenetic and non-cell-autonomous features of disease. Thus, future enhancements to the capabilities of the system could include cocultures with other cell types of the BM niche, such as stromal and endothelial cells. The latter could be sourced from primary cultures, autologous or allogeneic, or differentiated from iPSCs. Such cultures could be valuable to, for example, model clonal advantage conferred by specific CH- and MDS- associated mutations, which is currently not captured in vitro. For example, cells with isolated del7q or splicing factor mutations, in the absence of mutations driving overt AML, exhibit a growth disadvantage in vitro, although they clonally expand in vivo (Kotini et al. 2015, 2017; Chang et al. 2018). This discrepancy is likely a result of the in vitro conditions, as primary MDS cells also grow poorly ex vivo and ICLs with splicing factor mutations have slower division rates than wild-type cells. Culture conditions better mimicking an in vivo environment with presumably different selection pressures could enable better modeling of clonal dynamics and other disease phenotypes that are dependent on interaction with other cell types and extracellular components. 3D cultures could further enhance the capabilities of iPSC models, for example, by promoting developmental maturation and more faithful establishment of epigenetic disease components. Although the importance of 3D organoid-type cultures may be lesser for liquid tumors than solid tumors, they may enable modeling of lymphoid malignancies in the future—for example, by tissue engineering approaches recapitulating germinal center-like structures and thymic organoids to allow positive selection and maturation of T cells (Parent et al. 2013; Sun et al. 2013; Montel-Hagen et al. 2019b; Rowe and Daley 2019).
CONCLUDING REMARKS
Recent studies indicate that leukemia research can benefit tremendously from iPSC technology. Cultural aspects of hematology, like a long tradition of banking and making good use of patient material and, consequently, the size and breadth of tissue banks already in place, render iPSC modeling all the more appealing for this discipline. iPSC modeling is still a young field, and although experience is building, misconceptions are also lingering. Like all models, iPSCs have limitations, but also unique strengths. Being human models, the findings made with them can be anticipated to better translate to the clinic. As patient-derived models, iPSCs can engage in a back and forth with primary cells (i.e., hypotheses generated from clinical observations can be tested in iPSCs and findings in iPSCs can be validated in primary cells from the same patient). Because iPSCs can faithfully model human genomes, large biobanks representative of all major genetic groups and specific genotypes of blood cancers can be envisioned. These can uniquely support genotype-to-phenotype studies, target identification and validation, drug testing, screening and repurposing, and toxicity testing. Moving on from 2D single-lineage cultures to 3D and in vivo systems, capturing tissue and organ level phenotypes, in the future, can bring on the best of both worlds. In a balancing act amid doomsday scenarios and extreme hype, today iPSC technology is moving steadily toward fulfilling its promise to revolutionize the study of human physiology and disease pathogenesis.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH) Grants Nos. R01HL121570, R01HL137219, R01HL132071, and R01CA225231, by grants from the Edward P. Evans Foundation, the New York State Stem Cell Board, the Henry and Marilyn Taub Foundation, the Alex's Lemonade Stand Foundation, and the RUNX1 research program; by a Pershing Square Sohn Prize from the Pershing Square Sohn Cancer Research Alliance; and by a Leukemia and Lymphoma Society Scholar award.
Footnotes
Editors: Michael G. Kharas, Ross L. Levine, and Ari M. Melnick
Additional Perspectives on Leukemia and Lymphoma: Molecular and Therapeutic Insights available at www.perspectivesinmedicine.org
REFERENCES
- Abyzov A, Mariani J, Palejev D, Zhang Y, Haney MS, Tomasini L, Ferrandino AF, Rosenberg Belmaker LA, Szekely A, Wilson M, et al. 2012. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature 492: 438–442. 10.1038/nature11629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amabile G, Di Ruscio A, Müller F, Welner RS, Yang H, Ebralidze AK, Zhang H, Levantini E, Qi L, Martinelli G, et al. 2015. Dissecting the role of aberrant DNA methylation in human leukaemia. Nat Commun 6: 7091 10.1038/ncomms8091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony-Debre I, Manchev VT, Balayn N, Bluteau D, Tomowiak C, Legrand C, Langlois T, Bawa O, Tosca L, Tachdjian G, et al. 2015. Level of RUNX1 activity is critical for leukemic predisposition but not for thrombocytopenia. Blood 125: 930–940. 10.1182/blood-2014-06-585513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou E, Stadtfeld M. 2018. Cellular trajectories and molecular mechanisms of iPSC reprogramming. Curr Opin Genet Dev 52: 77–85. 10.1016/j.gde.2018.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al. 2009. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev 23: 2134–2139. 10.1101/gad.1811609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedel A, Pasquet JM, Lippert E, Taillepierre M, Lagarde V, Dabernat S, Dubus P, Charaf L, Beliveau F, de Verneuil H, et al. 2013. Variable behavior of iPSCs derived from CML patients for response to TKI and hematopoietic differentiation. PLoS ONE 8: e71596 10.1371/journal.pone.0071596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Ha G, Tseng YY, Greenwald NF, Oh C, Shih J, McFarland JM, Wong B, Boehm JS, Beroukhim R, et al. 2017. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet 49: 1567–1575. 10.1038/ng.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Siranosian B, Ha G, Tang H, Oren Y, Hinohara K, Strathdee CA, Dempster J, Lyons NJ, Burns R, et al. 2018. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 560: 325–330. 10.1038/s41586-018-0409-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Beroukhim R, Golub TR. 2019. Genomic evolution of cancer models: Perils and opportunities. Nat Rev Cancer 19: 97–109. 10.1038/s41568-018-0095-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böiers C, Richardson SE, Laycock E, Zriwil A, Turati VA, Brown J, Wray JP, Wang D, James C, Herrero J, et al. 2018. A human IPS model implicates embryonic B-myeloid fate restriction as developmental susceptibility to B acute lymphoblastic leukemia-associated ETV6-RUNX1. Dev Cell 44: 362–377.e7. 10.1016/j.devcel.2017.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, et al. 2011. Modelling schizophrenia using human induced pluripotent stem cells. Nature 473: 221–225. 10.1038/nature09915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Pruszak J, Varadarajan M, Blomen VA, Gokhale S, Camargo FD, Wernig M, Jaenisch R, Brummelkamp TR. 2010. Generation of iPSCs from cultured human malignant cells. Blood 115: 4039–4042. 10.1182/blood-2009-07-231845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CJ, Kotini AG, Olszewska M, Georgomanoli M, Teruya-Feldstein J, Sperber H, Sanchez R, DeVita R, Martins TJ, Abdel-Wahab O, et al. 2018. Dissecting the contributions of cooperating gene mutations to cancer phenotypes and drug responses with patient-derived iPSCs. Stem Cell Reports 10: 1610–1624. 10.1016/j.stemcr.2018.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MP, Gentles AJ, Chatterjee S, Lan F, Reinisch A, Corces MR, Xavy S, Shen J, Haag D, Chanda S, et al. 2017. Human AML-iPSCs reacquire leukemic properties after differentiation and model clonal variation of disease. Cell Stem Cell 20: 329–344.e7. 10.1016/j.stem.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Hansen NF, Zhao L, Du Y, Zou C, Donovan FX, Chou BK, Zhou G, Li S, Dowey SN, et al. 2012. Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell 10: 337–344. 10.1016/j.stem.2012.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KD, Vodyanik MA, Togarrati PP, Suknuntha K, Kumar A, Samarjeet F, Probasco MD, Tian S, Stewart R, Thomson JA, et al. 2012. Identification of the hemogenic endothelial progenitor and its direct precursor in human pluripotent stem cell differentiation cultures. Cell Rep 2: 553–567. 10.1016/j.celrep.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JP, Kwon EM, Gao Y, Trivedi NS, Elkahloun AG, Horwitz MS, Cheng L, Liu PP. 2014. Targeted correction of RUNX1 mutation in FPD patient-specific induced pluripotent stem cells rescues megakaryopoietic defects. Blood 124: 1926–1930. 10.1182/blood-2014-01-550525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoever C, Li H, Jakubosky D, Benaglio P, Reyna J, Olson KM, Huang H, Biggs W, Sandoval E, D'Antonio M, et al. 2017. Large-scale profiling reveals the influence of genetic variation on gene expression in human induced pluripotent stem cells. Cell Stem Cell 20: 533–546e7. 10.1016/j.stem.2017.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditadi A, Sturgeon CM, Tober J, Awong G, Kennedy M, Yzaguirre AD, Azzola L, Ng ES, Stanley EG, French DL, et al. 2015. Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat Cell Biol 17: 580–591. 10.1038/ncb3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S, Vo LT, Chou SS, Kim PG, Arora N, Li H, Hadland BK, Bernstein ID, Collins JJ, Zon LI, et al. 2013. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell 13: 459–470. 10.1016/j.stem.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S, Vo LT, Macari ER, Wahlster L, Kinney MA, Taylor AM, Barragan J, Gupta M, McGrath K, Lee HY, et al. 2017. Drug discovery for Diamond-Blackfan anemia using reprogrammed hematopoietic progenitors. Sci Transl Med 9: aah5645 10.1126/scitranslmed.aah5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, Schifano E, Booth J, van Putten W, Skrabanek L, et al. 2010. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 17: 13–27. 10.1016/j.ccr.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame JM, McGrath KE, Palis J. 2013. Erythro-myeloid progenitors: “Definitive” hematopoiesis in the conceptus prior to the emergence of hematopoietic stem cells. Blood Cells Mol Dis 51: 220–225. 10.1016/j.bcmd.2013.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne AL, Maguire JA, Gandre-Babbe S, Chou ST, Tasian SK, Loh ML, Weiss MJ, Gadue P, French DL. 2018. Generation of a human Juvenile myelomonocytic leukemia iPSC line, CHOPi001-A, with a mutation in CBL. Stem Cell Res 31: 157–160. 10.1016/j.scr.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S, Paluru P, Aribeana C, Chou ST, Bresolin S, Lu L, Sullivan SK, Tasian SK, Weng J, Favre H, et al. 2013. Patient-derived induced pluripotent stem cells recapitulate hematopoietic abnormalities of juvenile myelomonocytic leukemia. Blood 121: 4925–4929. 10.1182/blood-2013-01-478412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgomanoli M, Papapetrou EP. 2019. Modeling blood diseases with human induced pluripotent stem cells. Dis Model Mech 12: dmm039321 10.1242/dmm.039321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Limia CE, Devalle S, Reis M, Sochacki J, Carneiro M, Madeiro da Costa R, D'Andrea M, Padilha T, Zalcberg IR, Solza C, et al. 2017. Generation and characterization of a human induced pluripotent stem (iPS) cell line derived from an acute myeloid leukemia patient evolving from primary myelofibrosis carrying the CALR 52 bp deletion and the ASXL1 p.R693X mutation. Stem Cell Res 24: 16–20. 10.1016/j.scr.2017.08.006 [DOI] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, et al. 2011. Somatic coding mutations in human induced pluripotent stem cells. Nature 471: 63–67. 10.1038/nature09805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guibentif C, Rönn RE, Böiers C, Lang S, Saxena S, Soneji S, Enver T, Karlsson G, Woods NB. 2017. Single-cell analysis identifies distinct stages of human endothelial-to-hematopoietic transition. Cell Rep 19: 10–19. 10.1016/j.celrep.2017.03.023 [DOI] [PubMed] [Google Scholar]
- Guo S, Zi X, Schulz VP, Cheng J, Zhong M, Koochaki SH, Megyola CM, Pan X, Heydari K, Weissman SM, et al. 2014. Nonstochastic reprogramming from a privileged somatic cell state. Cell 156: 649–662. 10.1016/j.cell.2014.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, et al. 2007. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 318: 1920–1923. 10.1126/science.1152092 [DOI] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. 2009. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462: 595–601. 10.1038/nature08592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Jaenisch R. 2016. Induced pluripotent stem cells meet genome editing. Cell Stem Cell 18: 573–586. 10.1016/j.stem.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. 2009. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460: 1132–1135. 10.1038/nature08235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoi M, Kumano K, Taoka K, Arai S, Kataoka K, Ueda K, Kamikubo Y, Takayama N, Otsu M, Eto K, et al. 2014. Generation of induced pluripotent stem cells derived from primary and secondary myelofibrosis patient samples. Exp Hematol 42: 816–825. 10.1016/j.exphem.2014.03.010 [DOI] [PubMed] [Google Scholar]
- Hu K, Yu J, Suknuntha K, Tian S, Montgomery K, Choi KD, Stewart R, Thomson JA, Slukvin II. 2011. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood 117: e109–119. 10.1182/blood-2010-07-298331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Närvä E, Ng S, Sourour M, Hämäläinen R, Olsson C, et al. 2011. Copy number variation and selection during reprogramming to pluripotency. Nature 471: 58–62. 10.1038/nature09871 [DOI] [PubMed] [Google Scholar]
- Iizuka H, Kagoya Y, Kataoka K, Yoshimi A, Miyauchi M, Taoka K, Kumano K, Yamamoto T, Hotta A, Arai S, et al. 2015. Targeted gene correction of RUNX1 in induced pluripotent stem cells derived from familial platelet disorder with propensity to myeloid malignancy restores normal megakaryopoiesis. Exp Hematol 43: 849–857. 10.1016/j.exphem.2015.05.004 [DOI] [PubMed] [Google Scholar]
- Ivanovs A, Rybtsov S, Ng ES, Stanley EG, Elefanty AG, Medvinsky A. 2017. Human haematopoietic stem cell development: From the embryo to the dish. Development 144: 2323–2337. 10.1242/dev.134866 [DOI] [PubMed] [Google Scholar]
- Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR, Majeti R. 2012. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med 4: 149ra118 10.1126/scitranslmed.3004315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisua Belmonte JC. 2009. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460: 1140–1144. 10.1038/nature08311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M, Awong G, Sturgeon CM, Ditadi A, LaMotte-Mohs R, Zúñiga-Pflücker JC, Keller G. 2012. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Rep 2: 1722–1735. 10.1016/j.celrep.2012.11.003 [DOI] [PubMed] [Google Scholar]
- Kent DG, Green AR. 2017. Order matters: The order of somatic mutations influences cancer evolution. Cold Spring Harb Perspect Med 7: a027060 10.1101/cshperspect.a027060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, Bala S, Bensaddek D, Casale FP, Culley OJ, et al. 2017. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546: 370–375. 10.1038/nature22403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpivaara O, Mukherjee S, Schram AM, Wadleigh M, Mullally A, Ebert BL, Bass A, Marubayashi S, Heguy A, Garcia-Manero G, et al. 2009. A germline JAK2 SNP is associated with predisposition to the development of JAK2V617F-positive myeloproliferative neoplasms. Nat Genet 41: 455–459. 10.1038/ng.342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim JS. 2014. A guide to genome engineering with programmable nucleases. Nat Rev Genet 15: 321–334. 10.1038/nrg3686 [DOI] [PubMed] [Google Scholar]
- Kotini AG, Chang CJ, Boussaad I, Delrow JJ, Dolezal EK, Nagulapally AB, Perna F, Fishbein GA, Klimek VM, Hawkins RD, et al. 2015. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat Biotechnol 33: 646–655. 10.1038/nbt.3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotini AG, Chang CJ, Chow A, Yuan H, Ho TC, Wang T, Vora S, Solovyov A, Husser C, Olszewska M, et al. 2017. Stage-specific human induced pluripotent stem cells map the progression of myeloid transformation to transplantable leukemia. Cell Stem Cell 20: 315–328.e7. 10.1016/j.stem.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano K, Arai S, Hosoi M, Taoka K, Takayama N, Otsu M, Nagae G, Ueda K, Nakazaki K, Kamikubo Y, et al. 2012. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood 119: 6234–6242. 10.1182/blood-2011-07-367441 [DOI] [PubMed] [Google Scholar]
- Kyttälä A, Moraghebi R, Valensisi C, Kettunen J, Andrus C, Pasumarthy KK, Nakanishi M, Nishimura K, Ohtaka M, Weltner J, et al. 2016. Genetic variability overrides the impact of parental cell type and determines iPSC differentiation potential. Stem Cell Reports 6: 200–212. 10.1016/j.stemcr.2015.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacaud G, Kouskoff V. 2017. Hemangioblast, hemogenic endothelium, and primitive versus definitive hematopoiesis. Exp Hematol 49: 19–24. 10.1016/j.exphem.2016.12.009 [DOI] [PubMed] [Google Scholar]
- Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, et al. 2011. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell 8: 106–118. 10.1016/j.stem.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, Ganat YM, Menon J, Shimizu F, Viale A, et al. 2009. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature 461: 402–406. 10.1038/nature08320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Ramirez CN, Kim H, Zeltner N, Liu B, Radu C, Bhinder B, Kim YJ, Choi IY, Mukherjee-Clavin B, et al. 2012. Large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds that rescue IKBKAP expression. Nat Biotechnol 30: 1244–1248. 10.1038/nbt.2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DF, Su J, Kim HS, Chang B, Papatsenko D, Zhao R, Yuan Y, Gingold J, Xia W, Darr H, et al. 2015. Modeling familial cancer with induced pluripotent stem cells. Cell 161: 240–254. 10.1016/j.cell.2015.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Salci KR, Reid JC, Orlando L, Tanasijevic B, Shapovalova Z, Bhatia M. 2017. Brief report: Human acute myeloid leukemia reprogramming to pluripotency is a rare event and selects for patient hematopoietic cells devoid of leukemic mutations. Stem Cells 35: 2095–2102. 10.1002/stem.2655 [DOI] [PubMed] [Google Scholar]
- Levine AJ, Jenkins NA, Copeland NG. 2019. The roles of initiating truncal mutations in human cancers: The order of mutations and tumor cell type matters. Cancer Cell 35: 10–15. 10.1016/j.ccell.2018.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Cañamero M, Blasco MA, Serrano M. 2009. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460: 1136–1139. 10.1038/nature08290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Zhang Y. 2013. Genetic and epigenetic variations in iPSCs: Potential causes and implications for application. Cell Stem Cell 13: 149–159. 10.1016/j.stem.2013.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Ye Z, Gao Y, He C, Williams DW, Moliterno A, Spivak J, Huang H, Cheng L. 2017. Generation of human iPSCs from an essential thrombocythemia patient carrying a V501L mutation in the MPL gene. Stem Cell Res 18: 57–59. 10.1016/j.scr.2016.12.012 [DOI] [PubMed] [Google Scholar]
- Marión RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. 2009. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460: 1149–1153. 10.1038/nature08287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. 2010. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell 7: 521–531. 10.1016/j.stem.2010.07.017 [DOI] [PubMed] [Google Scholar]
- Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, Mandal PK, Vera E, Shim JW, Kriks S, et al. 2013. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell 13: 691–705. 10.1016/j.stem.2013.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi M, Koya J, Arai S, Yamazaki S, Honda A, Kataoka K, Yoshimi A, Taoka K, Kumano K, Kurokawa M. 2018. ADAM8 is an antigen of tyrosine kinase inhibitor-resistant chronic myeloid leukemia cells identified by patient-derived induced pluripotent stem cells. Stem Cell Reports 10: 1115–1130. 10.1016/j.stemcr.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montel-Hagen A, Crooks GM. 2019a. From pluripotent stem cells to T cells. Exp Hematol 71: 24–31. 10.1016/j.exphem.2018.12.001 [DOI] [PubMed] [Google Scholar]
- Montel-Hagen A, Seet CS, Li S, Chick B, Zhu Y, Chang P, Tsai S, Sun V, Lopez S, Chen HC, et al. 2019b. Organoid-induced differentiation of conventional T cells from human pluripotent stem cells. Cell Stem Cell 24: 376–389.e8. 10.1016/j.stem.2018.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero-Navarro S, Sevilla A, Roman AC, Lee DF, D'Souza SL, Pardo S, Riess I, Su J, Cohen N, Schaniel C, et al. 2015. Myeloid dysregulation in a human induced pluripotent stem cell model of PTPN11-associated juvenile myelomonocytic leukemia. Cell Rep 13: 504–515. 10.1016/j.celrep.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-López A, Romero-Moya D, Prieto C, Ramos-Mejía V, Agraz-Doblas A, Varela I, Buschbeck M, Palau A, Carvajal-Vergara X, Giorgetti A, et al. 2016. Development refractoriness of MLL-rearranged human B cell acute leukemias to reprogramming into pluripotency. Stem Cell Reports 7: 602–618. 10.1016/j.stemcr.2016.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann CA, Kent DG, Nangalia J, Silber Y, Wedge DC, Grinfeld J, Baxter EJ, Massie CE, Papaemmanuil E, Menon S, et al. 2015. Effect of mutation order on myeloproliferative neoplasms. N Engl J Med 372: 601–612. 10.1056/NEJMoa1412098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, et al. 2016. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 374: 2209–2221. 10.1056/NEJMoa1516192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent AV, Russ HA, Khan IS, LaFlam TN, Metzger TC, Anderson MS, Hebrok M. 2013. Generation of functional thymic epithelium from human embryonic stem cells that supports host T cell development. Cell Stem Cell 13: 219–229. 10.1016/j.stem.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. 2008. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451: 141–146. 10.1038/nature06534 [DOI] [PubMed] [Google Scholar]
- Pashos EE, Park Y, Wang X, Raghavan A, Yang W, Abbey D, Peters DT, Arbelaez J, Hernandez M, Kuperwasser N, et al. 2017. Large, diverse population cohorts of hiPSCs and derived hepatocyte-like cells reveal functional genetic variation at blood lipid-associated loci. Cell Stem Cell 20: 558–570.e10. 10.1016/j.stem.2017.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson S, Cuvertino S, Fleury M, Lacaud G, Kouskoff V. 2015. In vivo repopulating activity emerges at the onset of hematopoietic specification during embryonic stem cell differentiation. Stem Cell Reports 4: 431–444. 10.1016/j.stemcr.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Loring JF. 2014. Genomic instability in pluripotent stem cells: implications for clinical applications. J Biol Chem 289: 4578–4584. 10.1074/jbc.R113.516419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter DS, Letai A. 2016. To prime, or not to prime: That is the question. Cold Spring Harb Symp Quant Biol 81: 131–140. 10.1101/sqb.2016.81.030841 [DOI] [PubMed] [Google Scholar]
- Reinisch A, Thomas D, Corces MR, Zhang X, Gratzinger D, Hong WJ, Schallmoser K, Strunk D, Majeti R. 2016. A humanized bone marrow ossicle xenotransplantation model enables improved engraftment of healthy and leukemic human hematopoietic cells. Nat Med 22: 812–821. 10.1038/nm.4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe RG, Daley GQ. 2019. Induced pluripotent stem cells in disease modelling and drug discovery. Nat Rev Genet 20: 377–388. 10.1038/s41576-019-0100-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe RG, Mandelbaum J, Zon LI, Daley GQ. 2016. Engineering hematopoietic stem cells: Lessons from development. Cell Stem Cell 18: 707–720. 10.1016/j.stem.2016.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S, Panopoulos AD, Herrerías A, Bissig KD, Lutz M, Berggren WT, Verma IM, Izpisua Belmonte JC. 2011. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr Biol 21: 45–52. 10.1016/j.cub.2010.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai M, Kunimoto H, Watanabe N, Fukuchi Y, Yuasa S, Yamazaki S, Nishimura T, Sadahira K, Fukuda K, Okano H, et al. 2014. Impaired hematopoietic differentiation of RUNX1-mutated induced pluripotent stem cells derived from FPD/AML patients. Leukemia 28: 2344–2354. 10.1038/leu.2014.136 [DOI] [PubMed] [Google Scholar]
- Saliba J, Hamidi S, Lenglet G, Langlois T, Yin J, Cabagnols X, Secardin L, Legrand C, Galy A, Opolon P, et al. 2013. Heterozygous and homozygous JAK2V617F states modeled by induced pluripotent stem cells from myeloproliferative neoplasm patients. PLoS ONE 8: e74257 10.1371/journal.pone.0074257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloma I, Mitjavila-Garcia MT, Feraud O, Griscelli F, Oudrhiri N, El Marsafy S, Gobbo E, Divers D, Proust A, Smadja DM, et al. 2017. Whole-genome analysis reveals unexpected dynamics of mutant subclone development in a patient with JAK2-V617F-positive chronic myeloid leukemia. Exp Hematol 53: 48–58. 10.1016/j.exphem.2017.05.007 [DOI] [PubMed] [Google Scholar]
- Sturgeon CM, Ditadi A, Awong G, Kennedy M, Keller G. 2014. Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat Biotechnol 32: 554–561. 10.1038/nbt.2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura R, Jha DK, Han A, Soria-Valles C, da Rocha EL, Lu YF, Goettel JA, Serrao E, Rowe RG, Malleshaiah M, et al. 2017. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545: 432–438. 10.1038/nature22370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suknuntha K, Ishii Y, Tao L, Hu K, McIntosh BE, Yang D, Swanson S, Stewart R, Wang JYJ, Thomson J, et al. 2015. Discovery of survival factor for primitive chronic myeloid leukemia cells using induced pluripotent stem cells. Stem Cell Res 15: 678–693. 10.1016/j.scr.2015.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Xu J, Lu H, Liu W, Miao Z, Sui X, Liu H, Su L, Du W, He Q, et al. 2013. Directed differentiation of human embryonic stem cells into thymic epithelial progenitor-like cells reconstitutes the thymic microenvironment in vivo. Cell Stem Cell 13: 230–236. 10.1016/j.stem.2013.06.014 [DOI] [PubMed] [Google Scholar]
- Suzuki NM, Niwa A, Yabe M, Hira A, Okada C, Amano N, Watanabe A, Watanabe K, Heike T, Takata M, et al. 2015. Pluripotent cell models of Fanconi anemia identify the early pathological defect in human hemoangiogenic progenitors. Stem Cells Transl Med 4: 333–338. 10.5966/sctm.2013-0172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Takei H, Edahiro Y, Mano S, Masubuchi N, Mizukami Y, Imai M, Morishita S, Misawa K, Ochiai T, Tsuneda S, et al. 2018. Skewed megakaryopoiesis in human induced pluripotent stem cell-derived haematopoietic progenitor cells harbouring calreticulin mutations. Br J Haematol 181: 791–802. 10.1111/bjh.15266 [DOI] [PubMed] [Google Scholar]
- Tapia N, Schöler HR. 2016. Molecular obstacles to clinical translation of iPSCs. Cell Stem Cell 19: 298–309. 10.1016/j.stem.2016.06.017 [DOI] [PubMed] [Google Scholar]
- Tasian SK, Casas JA, Posocco D, Gandre-Babbe S, Gagne AL, Liang G, Loh ML, Weiss MJ, French DL, Chou ST. 2019. Mutation-specific signaling profiles and kinase inhibitor sensitivities of juvenile myelomonocytic leukemia revealed by induced pluripotent stem cells. Leukemia 33: 181–190. 10.1038/s41375-018-0169-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. 2009. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460: 1145–1148. 10.1038/nature08285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo LT, Daley GQ. 2015. De novo generation of HSCs from somatic and pluripotent stem cell sources. Blood 125: 2641–2648. 10.1182/blood-2014-10-570234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlster L, Daley GQ. 2016. Progress towards generation of human haematopoietic stem cells. Nat Cell Biol 18: 1111–1117. 10.1038/ncb3419 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Nishikawa S, Muguruma K, et al. 2007. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol 25: 681–686. 10.1038/nbt1310 [DOI] [PubMed] [Google Scholar]
- Yang YM, Gupta SK, Kim KJ, Powers BE, Cerqueira A, Wainger BJ, Ngo HD, Rosowski KA, Schein PA, Ackeifi CA, et al. 2013. A small molecule screen in stem-cell-derived motor neurons identifies a kinase inhibitor as a candidate therapeutic for ALS. Cell Stem Cell 12: 713–726. 10.1016/j.stem.2013.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Zhan H, Mali P, Dowey S, Williams DM, Jang YY, Dang CV, Spivak JL, Moliterno AR, Cheng L. 2009. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood 114: 5473–5480. 10.1182/blood-2009-04-217406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Liu CF, Lanikova L, Dowey SN, He C, Huang X, Brodsky RA, Spivak JL, Prchal JT, Cheng L. 2014. Differential sensitivity to JAK inhibitory drugs by isogenic human erythroblasts and hematopoietic progenitors generated from patient-specific induced pluripotent stem cells. Stem Cells 32: 269–278. 10.1002/stem.1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MA, Larson DE, Sun CW, George DR, Ding L, Miller CA, Lin L, Pawlik KM, Chen K, Fan X, et al. 2012. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell 10: 570–582. 10.1016/j.stem.2012.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920. 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]