Abstract
Innate immunity and adaptive immunity consist of highly specialized immune lineages that depend on transcription factors for both function and development. In this review, we dissect the similarities between two innate lineages, innate lymphoid cells (ILCs) and dendritic cells (DCs), and an adaptive immune lineage, T cells. ILCs, DCs, and T cells make up four functional immune modules and interact in concert to produce a specified immune response. These three immune lineages also share transcriptional networks governing the development of each lineage, and we discuss the similarities between ILCs and DCs in this review.
Keywords: transcription factors, hematopoiesis, dendritic cells, innate lymphoid cells, T cells
INTRODUCTION
A potent immune response requires cross talk and collaboration between the innate and adaptive immune systems, both of which contain highly specialized immune lineages. Innate immune responses involve many different types of innate cells, such as neutrophils, monocytes, and (specifically discussed in this review) innate lymphoid cells (ILCs) and dendritic cells (DCs). Innate cells recognize pathogens through their germline-encoded receptors and first initiate a proinflammatory response that aims to contain and rapidly clear the infection. Importantly, the innate cells direct the specific type of adaptive immune response that is most effective at clearing the particular type of infection by secreting cytokines and chemokines to alert the adaptive immune response. The adaptive immune response can be divided into three types of immunity, which are specific to the type of pathogen that evokes the response. Type I immunity is in response to intracellular microbes, such as bacteria and viruses, while type II immunity protects against helminths and environmental substances. Type III immunity is involved in protection against extracellular bacteria and fungi. Each of these immune responses corresponds to a specific type of T cell, which expresses antigen-specific receptors, and to specific innate cells, ILCs and DCs, that together effectively respond to a pathogen.
ILCs are often considered to be innate T cells, as both cell types share functional and developmental similarities. Like T cells, ILCs can be divided into several distinct subsets that correspond to the three types of immunity that cells elicit. However, unlike T cells, which have antigen-specific receptors, undergo clonal selection, and expand when stimulated, ILCs do not have antigen-specific receptors or the ability to undergo clonal selection. ILCs instead rapidly respond to the pathogen and secrete cytokines to control the infection. The developmental similarities between T cells and ILCs have been discussed in detail in other reviews (Eberl et al. 2015, Robinette & Colonna 2016) and are discussed only briefly in this review.
DCs are characterized as professional antigen presenting cells and are responsible for priming T cells for potent T cell activation. DCs encode a number of receptors that allow for antigen uptake, antigen processing, and antigen presentation to MHC molecules on T cells. Initially thought of as one family, DCs are now recognized for their heterogeneity in both location and function, with distinct subsets specialized for specific response. Two sets of classical/conventional DCs (cDCs), cDC1 and cDC2, as well as plasmacytoid DCs (pDCs) and epidermal Langerhans cells have been identified (Cella et al. 1999, Schuler et al. 1985, Siegal et al. 1999, Steinman & Witmer 1978).
This review focuses on the similarities between T cells and their innate counterparts, ILCs and DCs, in promoting a specialized immune response. We present a model in which these three immune cell types are distinguished by the major immune effector module they promote, and we then discuss recent progress in understanding the development of ILC and DC subsets. Both cell lineages share an unrecognized transcriptional network similarity that deepens our understanding of innate cell lineage communication with adaptive immunity.
COMMON EFFECTOR MODULES BETWEEN T CELLS, ILCS, AND DCS
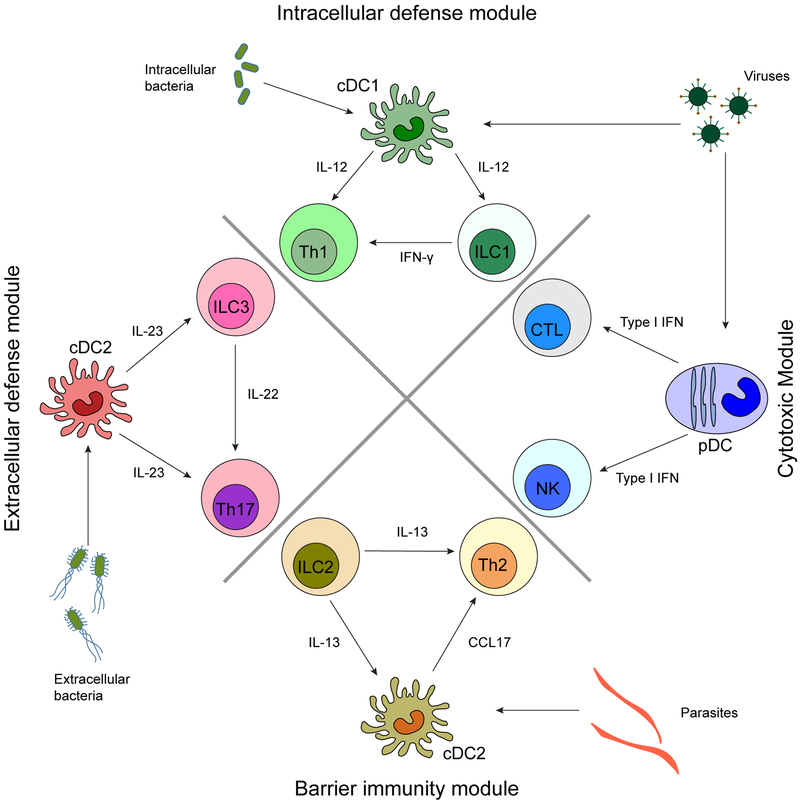
As discussed above, there are three broad types of immune responses, types I, II, and III, where each type of immunity corresponds to specific ILC and DC subsets to elicit innate immunity, which in turn leads to a coordinated effort to prime a T cell subset for the proper adaptive immune response. Type I immunity protects against intracellular pathogens and can be divided into two types of responses: (a) a cytotoxic response led by NK cells, pDCs, and CD8+ T cells and (b) an intracellular defense module held by ILC1s, cDC1s, and TH1 cells. Type II immunity protects against helminths and environmental substances and is elicited by ILC2, a subset of cDC2, and TH2 cells. Finally, type III immunity is involved in protection against extracellular bacteria and fungi and involves ILC3s, a subset of cDC2, and TH17 cells. Figure 1 shows a schematic of the types of immunity of each immune module and the cell types involved. Another subset of T cells, termed regulatory T cells defined by the transcription factor Foxp3, are not discussed in this review, as inhibitory innate counterparts are not well known.
Figure 1.
Four core immune modules shared between ILCs, DCs, and T cells. An immune response specific to a particular pathogen involves cross talk and collaboration between two innate cell lineages, ILCs and DCs, and an adaptive cell lineage, T cells, by secretion of cytokines and chemokines. Each cell type in this module is governed by specific transcription factors that influence the effector functions. Abbreviations: cDC, classical/conventional dendritic cell; CTL, cytotoxic T lymphocyte; DC, dendritic cell; IFN, interferon; ILC, innate lymphoid cell; pDC, plasmacytoid dendritic cell.
Type I Immunity Involves Cytotoxic and Intracellular Defense Modules
NK cells, ILC1s, pDCs, and cDC1s interact with CD8+ T cells and TH1 cells to defend against viruses and intracellular pathogens.
NK cells, pDCs, and CD8+ T cells compose the cytotoxic module to protect against viruses.
NK cells and pDCs are among the first cells to initiate and mount a response to viral infection. NK cells express activating or inhibitory receptors that can recognize ligands on infected cells and can directly lyse the infected cells through granules containing either pore-forming enzymes termed perforins or serine proteases termed granzymes. NK cells also express receptors for the proinflammatory cytokines IL-12, IL-15, IL-18, and type I interferons (IFNs), all of which are produced by other lineages during viral infection and are important for NK cell activation. Importantly, NK cells produce large amounts of IFN-γ, which activates macrophages for phagocytosis and prevents viral spread.
pDCs sense viruses through surface receptors such as Toll-like receptors 7 and 9 (TLRs 7 and 9) and cytosolic sensors such as RIG-I and MDA5. The main effector function of pDCs is to secrete large amounts of type I interferon to rapidly clear infected cells. Several studies have selectively ablated pDCs in vivo to demonstrate their function in regulating the antiviral immune response. Conditionally deleting the transcription factor Tcf4 (E2–2) using an Itgax-cre that allows for deletion in CD11c+ immune cells results in an impaired innate response to mouse hepatitis virus and an impaired adaptive response to lymphocytic choriomeningitis virus (LCMV) (Cervantes-Barragan et al. 2012). Specifically, pDCs are vital in sustaining a cytotoxic T lymphocyte (CTL) response against chronic viral infection (Cervantes-Barragan et al. 2012). Another study in which pDC-specific expression of diphtheria toxin receptor was achieved using a human BDCA2 promoter indicated that lacking pDCs at the early stage of response against murine cytomegalovirus (MCMV) infection leads to lowered type I IFN production and attenuated NK cell activation (Takagi et al. 2011). pDCs may recruit and activate NK cells through their cytokine production (Guillerey et al. 2012).
CD8+ T cells are essential for antiviral immunity, as they can both directly lyse cells through perforin and granzyme and provide memory to the immune system for protection against reinfection. In corroboration with earlier studies, Brewitz et al. (2017) showed that pDCs promote CD8+ T cell help in response to viral infection and moreover that pDCs are recruited to CD8+ T cell priming sites through the chemokines CCL3 and CCL4. Type I IFN produced by pDCs bolstered cDC1 maturation and cross-presentation efficacy (Brewitz et al. 2017).
Development of both NK cells and CD8+ T cells depends on the transcription factors T-bet (encoded by Tbx21) and Eomesodermin (Eomes, encoded by Eomes) (Gordon et al. 2012, Intlekofer et al. 2005). Both transcription factors belong to the T-box transcription factor family (Papaioannou 2014). Like other members in the family, these transcription factors share a highly similar T-box domain, suggesting that they bind to the same DNA motifs (Zhang et al. 2018). However, they possess divergent N and C termini, suggesting that they interact with distinct partners for activity (Pearce et al. 2003). In NK and CD8+ T cell function, T-bet and Eomes play different roles but can only partially compensate for each other (Kaech & Cui 2012, Pearce et al. 2003). In T cell development, T-bet appears to act before Eomes (Cruz-Guilloty et al. 2009). While T-bet modulates the expression of IFN-γ and IL-12Rβ2, Eomes nonredundantly drives perforin and granzyme B production (Pearce et al. 2003, Townsend et al. 2004). Additionally, studies have suggested that T-bet and Eomes mutually regulate each other to maintain a balance between NK cells and ILC1 populations (Daussy et al. 2014, Pikovskaya et al. 2016). The regulation of both transcription factors remains largely unclear, and studies have indicated that exogenous factors, such as TGF-β, may modulate Eomes and T-bet expression (Cortez et al. 2016).
ILC1s, cDC1s, and TH1 cells belong to the intracellular defense module.
ILC1s and cDC1s provide inflammatory signals to activate TH1 cells and promote an immune response against intracellular pathogens, such as Toxoplasma gondii. ILC1s are found in nearly all tissues and secrete large amounts of IFN-γ (Sojka et al. 2014). While both NK cells and ILC1s produce IFN-γ, ILC1s produce IFN-γ more quickly than do NK cells in response to several viruses, such as MCMV, SeV, and PR8 influenza virus, in part due to cross talk with cDC1 (Artis & Spits 2015, Serafini et al. 2015, Weizman et al. 2017). cDC1s are the main and nonredundant producers of IL-12, a cytokine responsible for activating ILC1s, for driving IFN-γ production, and for instigating a TH1 response (Bliss et al. 1999, 2000; Gazzinelli et al. 1994; Hou et al. 2011; Liu et al. 2006; Mashayekhi et al. 2011; Pepper et al. 2008; Reis e Sousa et al. 1997).
cDC1s are required for protection against intracellular pathogens and for rejection of tumors. Studies demonstrating the importance of this cDC subset have been done with Batf3−/− mice, which specifically lack cDC1 (Hildner et al. 2008). Studies in which these mice were infected with T. gondii showed that cDC1 produce IL-12 and furthermore that IL-12 can restore cDC1 in infected Batf3−/− mice (Mashayekhi et al. 2011, Tussiwand et al. 2012). Mice lacking all Batf family members (Batf−/− Batf2−/− Batf3−/−) could not restore the cDC1 population after administration of IL-12, suggesting that Batf family members may be able to compensate for the lack of Batf3 in cases of infection (Mashayekhi et al. 2011, Tussiwand et al. 2012). cDC1s also promote production of IFN-γ by activating invariant NK T (iNKT) cells, as iNKT cells can produce IFN-γ after interacting with cDC1 pulsed with α-galactosylceramide (α-GalCer), a glycolipid antigen presented on CD1d molecules that are highly expressed on activated cDC1s (Arora et al. 2014; Fujii et al. 2002, 2004; Kawano et al. 1997). cDC1s are also uniquely capable of cross-presentation, which is crucial for antitumor immunity (Diamond et al. 2011, Fuertes et al. 2011, Hildner et al. 2008). Recent work has indicated a necessary role for the protein Wdfy4 in cross-presentation, but the precise mechanism by which this protein or other proteins play in the antigen presentation process remains unclear (Theisen et al. 2018).
T-bet is also important for the development of ILC1 and TH1 cells (Daussy et al. 2014, Robinette et al. 2015). These cells are characterized by their shared function in producing IFN-γ and TNF-α and by their reliance on IL-12, IL-15, and IL-18 for activation. IL-12 signaling through STAT4 activation results in TH1 differentiation in vivo but is dispensable for ILC1 polarization (Hsieh et al. 1993, O’Sullivan et al. 2016). In T cell development, T-bet directly represses TH2 specification, but it is unclear whether an analogy can be drawn in the ILC population where T-bet represses ILC2 polarization (Hwang et al. 2005, Zhu et al. 2012). TH1s and ILC1s primarily participate in the immune response against viral infection and intracellular pathogens but are also associated with certain inflammatory bowel diseases and type I diabetes (Eberl et al. 2015). T-bet drives IFN-γ production by directly binding to the regulatory elements of the IFNG gene and inducing the expression of Runx3, another transcription factor that drives IFN-γ production. Recent studies indicate that Eomes is induced upon TH1 cell activation and that it is expressed at steady state in certain ILC1 subsets (Bernink et al. 2015, Knox et al. 2014, Lupar et al. 2015). These data suggest that Eomes may play a role in intracellular defense module under certain circumstances, such as infection.
Type II Immunity Protects Against Helminths and Environmental Substances
ILC2s and a subset of cDC2 defined by the transcription factor Klf4, along with TH2 cells, are important in maintaining barrier immunity.
ILC2s, Klf4-dependent cDC2s, and TH2 cells contribute to maintaining barrier immunity.
The third immune module is characterized by immunity to helminth infection and maintenance of barrier function. ILC2s and Klf4-dependent cDC2 are important innate cells for this type of immunity, and TH2 cells provide the adaptive help. This type of immunity is modulated by the cytokines IL-5, IL-9, and IL-13. ILC2s are the major innate sources of IL-5 and IL-13 (von Moltke & Locksley 2014). These cytokines bolster the production of IL-4, which is required for TH2 differentiation. This group of cells is further characterized by their expression of the transcription factor GATA3. GATA3 is essential for the development for both lineages and drives the production of IL-5 and IL-13 by binding directly to the promoter region of these genes, and conditional deletion of GATA3 results in the reduction of IL-5 and IL-13 production (Hoyler et al. 2012; Mjosberg et al. 2012a; Yagi et al. 2011, 2014; Zheng & Flavell 1997; Zhu et al. 2004, 2006). Upon induction, GATA3 needs to overcome a repressive threshold maintained by FOG-1 expression to stabilize the polarization of both lineages via autoactivation (Ouyang et al. 2000, Zhou et al. 2001). Such polarization can also be achieved via cytokine production. In particular, ILC2s in mouse small intestine produce IL-13 to actively drive differentiation of and IL-25 production by Tuft cells. IL-25 in turn further drives IL-13 production by ILC2s and maintains lineage stability (von Moltke et al. 2016).
While the cDC1 population appears to be homogeneous, the cDC2 population seems to be heterogeneous. Conditional deletion of Kruppel-like factor 4 (Klf4) results in the loss of CD24+ CD172+ cDC2s in the lung and lymph node, as well as a decrease in a progenitor population in the bone marrow (Tussiwand et al. 2015). Moreover, Klf4-deficient mice showed impaired protection against Schistosoma mansoni infection but not herpes simplex virus, T. gondii, or Citrobacter rodentium infections, indicating a specific defect in type II but not CTL, type I, or type III responses (Tussiwand et al. 2015). Klf4 is a transcription activator or repressor and modulates the development of multiple lineages in epithelial tissues such as skin, lung, and intestine (Alder et al. 2008, Dang et al. 2000, Feinberg et al. 2007, Ghaleb et al. 2005, Katz et al. 2002, Kurotaki et al. 2013, McConnell & Yang 2010, Segre et al. 1999, Yamanaka 2008, Yoshida & Hayashi 2014, Zheng et al. 2009). However, the specific function and target of KLF4 in cDC2 remain unclear. Several studies argue that cDC2s may modulate TH2 responses to house dust mite (HDM) antigen (Hammad et al. 2010, Williams et al. 2013). Upon HDM challenge, cDC2s are rapidly recruited to lung airways and migrate to the lymph node to induce type II immunity (Mesnil et al. 2012). Also, IL-13 produced by ILC2s induce CCL17 production by lung and dermal cDC2s to attract memory TH2 cells in response to allergen (Halim et al. 2016).
Type III Immunity Protects Against Extracellular Bacteria and Fungi
ILC3s and a subset of cDC2s dependent on Notch2 are required for immunity against extracellular pathogens and fungi.
ILC3s, Notch2-dependent cDC2s, and TH17 cells protect against extracellular pathogens and fungi.
ILC3s and Notch2-dependent cDC2s compose the innate lineages for the fourth immune module. These cells are associated with defense against extracellular pathogens and fungi and can contribute to tissue homeostasis. Deregulation of these cells often results in autoimmune diseases such as multiple sclerosis, psoriasis, and Crohn’s disease. STAT3 is required for proper response to IL-23 in ILC3s (Guo et al. 2014, Rankin et al. 2016), whereas it is wholly required for IL-23 signaling and subsequent in vivo TH17 polarization.
Conditional deletion of Notch2 in CD11c+ cells revealed that cDC2 nonredundantly produce IL-23 in response to the extracellular bacteria C. rodentium, a mouse model for enteropathogenic Escherichia coli (Satpathy et al. 2013). Notch2 is a member of Notch family transcription factors that has four members in mammals, Notch 1–4. Members of this family of transcription factors function through ligand-mediated activation. Upon binding of ligands such as Delta-like family proteins, sequential proteolytic cleavages release the Notch intracellular domain (NICD). NICD then enters the nucleus and drives the expression of target genes in cooperation with several cofactors, including RBPJ and Mam. Differential CX3CR1 and ESAM expression reveals two subsets within the cDC2 population, and Notch2 deficiency results in the specific loss of the CX3CR1lo ESAMhi subset in the spleen (Lewis et al. 2011, Mesnil et al. 2012). Mice with conditional deletion of Notch2 in cDCs using Itgax-cre have shown a decrease in TH17 numbers in the MLN (Lewis et al. 2011), and loss of Irf4 results in impaired TH17 differentiation induced by small intestine CD103+CD11b+ cDC2s (Persson et al. 2013, Schlitzer et al. 2013). The same phenomenon occurs with the specific deletion of small intestine lamina propria CD103+CD11b+ cDC2s using a human Langerin-DTA transgenic mouse model that also leads to a decrease in TH17 cell numbers (Welty et al. 2013). Although ESAMhi cDC2s are required for resistance to C. rodentium, mice lacking expression of Irf4 or CCR7 and thereby having DCs with impaired migration capacity do not exhibit significant susceptibility to C. rodentium, suggesting that IL-23 production by lamina propria resident cDC2s is sufficient for the effective control of this pathogen (Bajana et al. 2012, Satpathy et al. 2013). As mentioned above, IL-23 is required for ILC3 secretion of IL-22, and therefore cDC2s modulate intestine type III immunity by targeting both TH17 and ILC3s.
The master regulator for this module is transcription factor RORγt, and a defect in this factor results in the complete absence of both lineages in vivo (Ivanov et al. 2016, Sawa et al. 2010). They also share a reliance on the transcription factor AHR, a receptor that binds various ligands with high risk of exposure in daily life, including dietary metabolites and pollutants (Cella & Colonna 2015). AHR is also essential for IL-22 production by this module (Basu et al. 2012, Kiss et al. 2011, Qiu et al. 2012).
ILC, DC, AND T CELL PLASTICITY IN IMMUNE MODELS
Although all three cell types reviewed here can be divided into subsets, each contributing to a specific type of immunity, recent work has shown that considerable plasticity can exist between the subsets. This imparts an important role for the local microenvironment in shaping an immune response. Plasticity has also been observed in both mouse and human.
The ability of some T cell subsets to convert to other subsets has been extensively studied (DuPage & Bluestone 2016, Murphy & Stockinger 2010). IL-2 and IL-4, the instructive signals for TH1 and TH2 polarization, respectively, drive lineage conversion. Recently polarized TH1 cells can start to produce IL-4, cease IFN-γ production, and convert to TH2-like cells upon IL-4 treatment in vitro or helminth infection in vivo (Panzer et al. 2012, Szabo et al. 1995). TH2 cells are relatively more stable than TH1s, partially because of GATA3 autoactivation and the mutual exclusion between GATA3 and T-bet (Ouyang et al. 2000). However, under circumstances in which IL-12Rβ expression is restored in TH2 polarized cells by type I IFNs, IL-12 treatment can result in conversion to the TH1 phenotype (Hegazy et al. 2010). Plasticity has also been demonstrated in the TH17 lineage, which may be the most plastic member of the T helper family (Stockinger & Omenetti 2017). Another study demonstrated that IL-12 treatment induces TH17 to downregulate RORγt and IL-17 expression and start to express T-bet and IFN-γ and retains a TH1-like gene expression profile (Lee et al. 2009a,b). In humans, a population of cells expressing both RORγt and T-bet that could produce both type I and type III cytokines was identified in patients with Crohn disease (Annunziato et al. 2007).
ILC plasticity largely mirrors T cell plasticity, with most ILC plasticity in ILC3s (Cherrier et al. 2018, Murphy & Stockinger 2010). In vitro treatment of ILC3 with IL-2 or IL-5 can transform ILC3s into IFN-γ-producing ILC1-like cells (Bernink et al. 2015, Cella et al. 2010). In mice, an increase in T-bet expression and Notch signaling together with a decrease in RORγt expression converts CCR6− NKp46+ ILC3s into NK1.1+ ILC1s that can produce IFN-γ (Vonarbourg et al. 2010). This conversion was later shown to be T-bet dependent (Klose et al. 2013, Rankin et al. 2013). ILC2s also show some degree of plasticity. ILC2s can produce IL-17 and be converted into ILC3s by injection of IL-25 or exposure to Notch ligand (Huang et al. 2015, Zhang et al. 2017). These converted ILC2s also induce RORγt expression (Zhang et al. 2017). Multiple groups have also shown that ILC2s secrete IFN-γ in response to IL-12 and IL-1B and convert to ILC1s (Lim et al. 2016, Ohne et al. 2016, Silver et al. 2016).
Many of the studies of DC plasticity have focused on functional plasticity without investigating changes in transcriptional profile (Pulendran et al. 2008). In this sense, plasticity refers to altered ability to stimulate T cell response. For example, DCs cultured with IFN-γ can induce TH1 responses (Ochsenbein et al. 2001), while thymic stromal lymphopoietin (TSLP) can strongly activate and modulate DCs to stimulate TH2 responses in an OX40-L-dependent manner (Ito et al. 2005). However, these studies failed to determine whether the differential ability to induce a T cell response is simply because the same subset of DCs is responding to different extracellular signaling or whether there is actually subset conversion driven by differential expression of lineage-defining transcription factors such as Irf8, Batf3, and Klf4 induced by environmental cues.
TRANSCRIPTIONAL BASIS OF EARLY ILC AND DC DEVELOPMENT
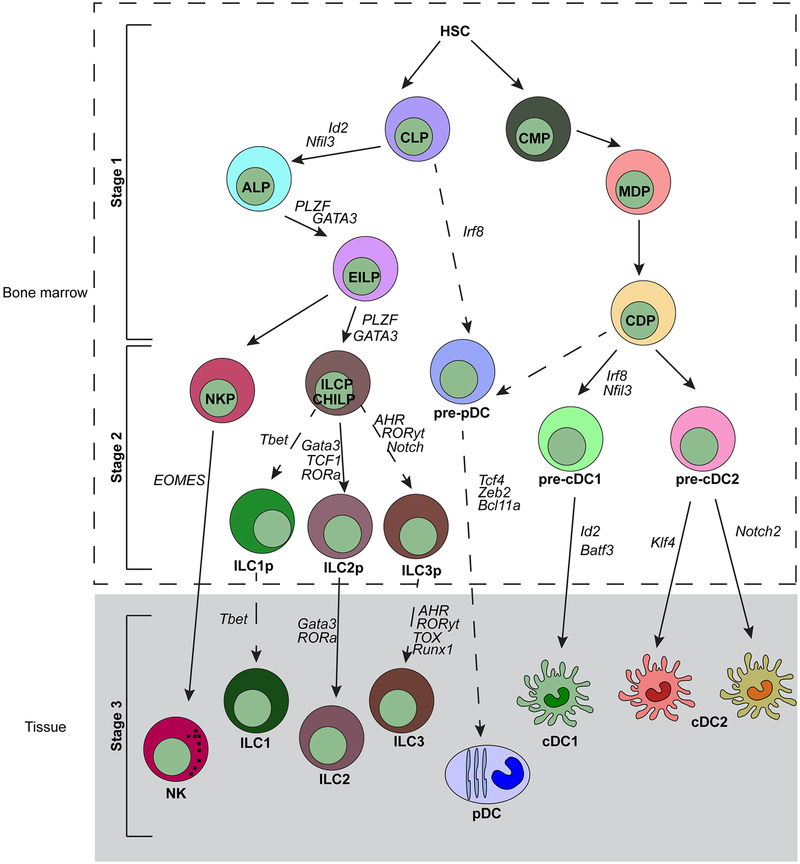
We now focus on the transcriptional networks governing ILC and DC development. Models of ILC and DC development can be divided into three distinct stages, as discussed for ILCs in a recent review by Serafini et al. (2015). Briefly, stage 1 is the specification of common precursors from a multipotent progenitor that has not excluded other cell fates. Stage 2 is the commitment of the precursors to their mature counterparts. Both stages 1 and 2 normally occur in the bone marrow. Stage 3 involves the maintenance and regulation of the mature cell subsets in tissues. Figure 2 shows both ILC development and DC development.
Figure 2.
ILC and DC development can be divided into three stages. Stage 1 refers to specification of common precursors from multipotent progenitors that have not yet excluded other cell lineage fates. Stage 2 is the commitment of those common precursors to the mature cell. Stage 3 is the maintenance of those cells in tissues. Many transcription factors influence either specification or commitment, and the precise roles for those factors are still unknown. Abbreviations: ALP, all-lymphoid progenitor; cDC, classical/conventional dendritic cell; CDP, common dendritic progenitor; CHILP, common helper-like ILC progenitor; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; EILP, early innate lymphoid progenitor; HSC, hematopoietic stem cell; ILC, innate lymphoid cell; ILCP, ILC progenitor; MDP, macrophage/DC progenitor; NKP, NK progenitor; pDC, plasmacytoid dendritic cell.
ILC Development
All subsets of ILCs are found in nearly all organs and tissues in the body, but ILC progenitors develop in the fetal liver and bone marrow. In the fetal liver, ILC progenitors that are phenotypically similar to LTis arrive on day E12.5–13.5 and subsequently express lymphotoxins to support lymphoid structure development. ILC progenitors in the bone marrow, which are a subset of the common lymphoid progenitor (CLP) that do not express the surface marker Ly6D, arise from the all-lymphoid progenitor (ALP) and from the IL-7Ra+ lymphoid-primed multipotent progenitor (LMPP) (Cherrier & Eberl 2012, Ghaedi et al. 2016, Inlay et al. 2009, Ishizuka et al. 2016, Klose et al. 2014, Moro et al. 2010, Possot et al. 2011, Yang et al. 2011). Another faction of the CLP, designated by positive expression of the integrin α4β7, is thought to include the first uncommitted ILC progenitor, but further studies are needed to identify this progenitor (Seillet et al. 2016). Briefly, stage 1 is the specification of common ILC precursors from the CLP, which include the αLP, the CHILP (the common progenitor to the helper-like ILC lineages), and the ILCP (the common precursor to ILCs). These progenitors have largely excluded B and T cell potentials, making them distinct from the CLP, but may not represent fully committed ILC precursors. Within the αLP progenitor, only the CXCR6+ subset excludes T cell potential, but it does give rise to conventional NK (cNK) and non-NK ILC1, ILC2, and ILC3 cell types. However, the CXCR6− subset can develop into T cells as well (Yu et al. 2014). This αLP may be the most uncommitted ILC progenitor found thus far, but further studies are required to dissect this population. The next progenitor, the CHILP, was discovered in 2014. The CHILP gives rise only to non-NK ILC1s, ILC2s, and ILC3s. ILC2, ILC3, and non-NK ILC1 cells (Klose et al. 2014). [←**AU: Are these non-NK also?**yes,] The last progenitor—the ILCP—was also discovered in 2014 and expresses high levels of the transcription factor PLZF (Constantinides et al. 2014). This progenitor is committed to all three ILC lineages but excludes LTi and NK cell potentials (Constantinides et al. 2014). Early innate lymphoid progenitors (EILPs) were identified in 2015 using a reporter mouse specific for the transcription factor TCF-1, which is expressed by all ILC progenitors (Yang et al. 2015). Recently, EILPs were established as intermediate progenitors between ALPs and ILCPs because certain transcription factors, such as PLZF and GATA-3, were expressed at intermediate levels between ALPs and ILCPs (Harly et al. 2018). Importantly, EILPs are a functionally distinct cell type relative to ALPs and ILCPs because they are specified but not committed to the ILC lineage (Harly et al. 2018). Progenitors identified in stage 1 represent cells specified to the ILC lineage but, because they can give rise to other cell lineages under in vivo or in vitro conditions, do not represent committed progenitors.
Commitment to specific ILC subsets occurs in stage 2 of ILC development. In this stage, identification of progenitors specific to NK cells, ILC1, ILC2, and ILC3 [←**AU: are these non-NK?**] is aided by knowledge of T helper subsets, as ILCs and T helper cells are similar in both development and function. Recent studies have attempted to find a committed progenitor to the ILC1 lineage that is distinct from a proposed progenitor of cNK cells. Both lineages express T-bet and Eomes. T-bet is important for the development of ILC1 and cNK cells, as T-bet−/− mice do not have liver or intestinal ILC1 (Daussy et al. 2014, Gordon et al. 2012, Klose et al. 2014, Townsend et al. 2004). ILCs express T-bet, but only cNK cells express Eomes (Pikovskaya et al. 2016). Additionally, while a committed progenitor of the non-NK cell ILC1 is yet to be defined, fate mapping studies of PLZF suggest that PLZF governs the divergence between ILC1 and NK cells (Constantinides et al. 2015).
A precursor committed to the ILC2 lineage, termed ILC2p, has been identified by high expression of GATA3 (Hoyler et al. 2012). While GATA3 is also expressed in the CHILP, continuous expression of GATA3 is required for ILC2 maturation and function, as described in GATA3 reporter mice (Hoyler et al. 2012). GATA3−/− mice lack ILC1, ILC2, and a subset of ILC3, so while GATA3 might not be a commitment factor for ILC2, it is critical for the maintenance of this ILC subset (Yagi et al. 2014). Other transcription factors necessary for ILC2 development are TCF-1 (encoded by Tcf7) and RORα. Tcf7−/− mice lack ILC2 in the lung and lack immature ILC2 in the bone marrow (Mielke et al. 2013, Yang et al. 2013). These mice also have reduced numbers of RORγt+ ILCs, suggesting that TCF-1 is required for the full development of more than one ILC subset. However, TCF-1 is absolutely required for ILC2 development in a cell-intrinsic manner (Mielke et al. 2013, Yang et al. 2013). Likewise, RORα is a transcription factor required for ILC2 development in a cell-intrinsic manner (Halim et al. 2012, Wong et al. 2012). The latter two transcription factors may depend on Notch signaling, similar to their dependence in the T cell lineage, but that has not been explored completely.
ILC3 development depends on the transcription factor RORγt, which is analogous to the requirement of this transcription factor in TH17 development. RORγt is necessary for LTi cells and for NKp46+ ILC3s (Cherrier & Eberl 2012, Eberl et al. 2004, Vonarbourg et al. 2010). The transcription factor TOX is also necessary for full development of LTi cells and for complete differentiation into NK cells (Aliahmad et al. 2010). AHR deficiency also affects the same populations as RORγt and TOX deficiency (Halim et al. 2012; Kiss et al. 2011; Lee et al. 2012a,b; Qiu et al. 2012).
Stage 3 of ILC development is about ILC maintenance and regulation in peripheral tissues. We do not go into much detail in this review, but studies have suggested that cytokines and some transcription factors are required for control of ILC populations and can contribute to ILC plasticity, as described above.
DC Development
In this review, we draw a comparison between ILC and DC development and believe that, like ILC development, DC development can also be divided into three distinct stages. Furthermore, both ILC development and DC development rely on some of the same transcription factors, and we explore where those similarities occur and what role the transcription factor plays in either lineage.
Stage 1 begins with the specification of DC precursors from progenitor cells that are multipotent for myeloid potential [termed the common myeloid progenitor (CMP)] to cells that are multipotent for macrophage and DC potentials [termed the macrophage/DC progenitor (MDP)] to progenitors that retain only DC potential [termed the common dendritic cell progenitor (CDP)]. DCs comprise three subsets—pDCs, cDC1, and cDC2—and the CDP gives rise to all three subsets. Stage 2 is defined by the commitment of the specified progenitor to specific DC subsets. Recent work has defined progenitors that are committed to either the cDC1 or the cDC2 fate, and newer work has aimed to elucidate a progenitor that is committed to pDC fate. Stage 3 is the maintenance of DCs in peripheral tissues.
Stage 1 is the specification of multipotent progenitors that have lymphoid, granulocyte, and myeloid potentials to progenitors that retain only DC potential. Early progenitors that can give rise to DCs in vivo are the CMP and the LMPP (Naik et al. 2013, Onai et al. 2007, Schlenner et al. 2010, Traver et al. 2000). The next progenitor thought to arise from the CMP and still retain DC potential is the granulocyte/macrophage progenitor (GMP), but recent studies have shown that the GMP cannot develop into DCs (Yanez et al. 2017). The MDP arises from the CMP and produces only macrophages and DCs both in vitro and in vivo (Auffray et al. 2009a,b; Fogg et al. 2006; Yanez et al. 2017). The exact transcriptional mechanisms that cause the divergence and the exclusion of these progenitors from neutrophil fate are not known. However, the transcription factors PU.1 and IRF8, along with members of the CEBP family, are thought to influence the development of these cell lineages. The MDP is thought to give rise to the CDP, but how the MDP gives rise to the CDP and how macrophage potential is lost are unanswered questions. The LMPP can give rise to other lymphoid progenitors, such as the CLP, which has the potential to give rise to pDCs (Rodrigues et al. 2018, Sathe et al. 2014, Yang et al. 2005). pDCs have been thought to arise from lymphoid cells, as they can be traced with IL7R, and recent work has shown that the majority of pDCs come from the CLP rather than the CDP (Rodrigues et al. 2018).
Stage 2 of DC development is the commitment of the CDP to clonogenic progenitors that give rise to cDC1s, cDC2, or pDCs. Clonogenic progenitors for cDC1s and cDC2, namely pre-cDC1s and pre-cDC2s, were identified in 2015 (Grajales-Reyes et al. 2015, Schlitzer et al. 2015), and a progenitor of pDCs was elucidated in 2018 (Rodrigues et al. 2018). Many transcription factors have been identified as important factors for cDC1, cDC2, and pDC development, and recent work has identified how transcription factors interact in the cDC1 lineage.
cDC1 development depends on expression of the transcription factors IRF8, Batf3, Nfil3 (nuclear factor interleukin 3 regulated, also known as E4bp4), and Id2 (inhibitor of differentiation 2) and on suppression of the transcription factor Zeb2 (Grajales-Reyes et al. 2015, Hildner et al. 2008, Schiavoni et al. 2002, Scott et al. 2016, Sichien et al. 2016, Tamura et al. 2005, Wu et al. 2016). IRF8 is a lineage-defining factor for cDC1, and Irf8−/− lack CDPs, pre-cDC1, and cDC1. Progenitors deficient in IRF8 diverted toward the granulocyte lineage and produced more neutrophils, indicating a role for IRF8 in regulating myeloid/granulocyte potential (Schönheit et al. 2013). Batf3, a transcription factor belonging to the Batf family, has a leucine zipper domain that heterodimerizes with JUN and IRF factors (Vinson et al. 2002). Studies aiming to understand the relationship between IRF8 and Batf3 began in 2015, when the pre-cDC1 was identified as a lineage−CD117intCD135+MHC-IIlow-intCD11c+SiglecH− cell that was either CD24+ or Zbtb46gfp+ (Grajales-Reyes et al. 2015). Zbtb46, a transcription factor belonging to the Broad Complex, Tramtrack, Bric-a-Brac, and Zinc Finger family, is selectively expressed in cDCs and their progenitors but is not required for their development (Satpathy et al. 2012a,b). The pre-cDC1 is present in Batf3−/− mice, but not in Irf8−/− mice, indicating that specification of the pre-cDC1 could occur in the absence of Batf3. Batf3−/− pre-cDC1s fail to maintain IRF8 expression, causing it to divert into the cDC2 lineage (Grajales-Reyes et al. 2015). High expression of IRF8 is necessary for the cDC1 lineage, and Batf3 is required to maintain IRF8 autoactivation following specification to the cDC1 fate (Grajales-Reyes et al. 2015). ChIP-seq analysis identified a +32-kb Irf8 enhancer that contains several AP1-IRF composite elements and that binds IRF8 and BATF3 in cDC1s in vivo. Recently, CRISPR-mediated deletion of the +32-kb Irf8 enhancer in mice (Irf8 +32–/–) suggests that Batf3 supports IRF8 autoactivation using this enhancer (V. Durai, P. Bagadia, J.M. Granja, A.T. Satpathy, D. Kulkarni, et al., submitted). Like Batf3−/− mice, Irf8 +32−/− mice lack mature cDC1 but maintain pre-cDC1 development in vivo. Development of this progenitor instead depends upon a +41-kb Irf8 enhancer, which binds E proteins and is active in mature pDCs and cDC1 progenitors, but not in mature cDC1s. Deletion of this enhancer eliminated IRF8 expression in pDCs and completely eliminated development of the specified pre-cDC1. This enhancer activity requires E proteins to induce sufficient levels of IRF8 during specification of the pre-cDC1, but it is still unclear why mature cDC1s require BATF3 and the +32-kb Irf8 enhancer to maintain IRF8 expression.
Recent work from our lab has organized the transcription factors Nfil3, Id2, and Zeb2 into a transcriptional network that promotes cDC1 fate (P. Bagadia, X. Huang, T. Liu, V. Durai, G. Grajales-Reyes, et al., submitted). Nfil3, a basic leucine zipper (bZIP) transcription repressor (Cowell et al. 1992, Zhang et al. 1995), is expressed in and is required for cDC1, but not for cDC2 or pDC, development (Kashiwada et al. 2011, Seillet et al. 2013). Id2 is a known inhibitor of E proteins and is expressed in cDC1 and cDC2, but not in pDCs, and is required only for cDC1 development (Hacker et al. 2003, Kusunoki et al. 2003). Current models propose that Id2 excludes the pDC fate in DC progenitors by blocking the activity of E proteins, particularly E2–2 (Tcf4), required for pDCs (Ghosh et al. 2010, Grajkowska et al. 2017, Watowich & Liu 2010). The transcription repressor Zeb2 is expressed in pDCs and cDC2s, but not cDC1s. It suppresses cDC1 development and is required for pDC development, perhaps through inhibition of Id2 transcription (Scott et al. 2016, Wu et al. 2016).
We found that the CDP originates in a Zeb2hi and Id2lo state in which IRF8 expression is maintained by the +41-kb Irf8 enhancer. Single-cell RNA sequencing of the CDP identified a fraction of the CDP that is already specified to the cDC1 fate, in a stage earlier than the pre-cDC1. This fraction expresses transcription factors required for cDC1 development, such as Id2, Batf3, and Zbtb46, and excludes transcription factors required for pDC development, such as Zeb2 and Tcf4. This fraction’s development arises when Nfil3 induces a transition into a Zeb2lo and Id2hi state. A circuit of mutual Zeb2-Id2 repression stabilizes states before and after this transition. Id2 expression in the specified pre-cDC1 inhibits E proteins, blocking activity of the +41-kb Irf8 enhancer and thereby imposing a new requirement for Batf3 for maintaining IRF8 expression via the +32-kb Irf8 enhancer (P. Bagadia, X. Huang, T. Liu, V. Durai, G. Grajales-Reyes, et al., submitted).
The transcriptional mechanisms governing cDC2 and pDC development are less known, but progenitors for each lineage have been identified. The pre-cDC2 was identified in 2015 as a lineage−CD117lowCD135+CD115+MHC-II−CD11c+Zbtb46gfp+ cell in the bone marrow (BM) (Grajales-Reyes et al. 2015). Although the pre-cDC2 expresses IRF8, mature cDC2 expresses only IRF4. However, Irf4−/− mice do not lack cDC2, although the cDC2 that develops exhibits defective migration (Bajana et al. 2012). As discussed above, two transcription factors, Klf4 and Notch2, have selectively ablated specific cDC2 populations, but how heterogeneity in the cDC2 lineage occurs and how the pre-cDC2 specifies to each cDC2 population remain unclear. Additionally, how the pre-cDC2 loses IRF8 to become IRF4 dependent is unknown.
pDC development depends on the transcription factors Tcf4, Zeb2, and Bcl11a (Cisse et al. 2008; Ghosh et al. 2010; Wu et al. 2013, 2016). pDCs also express high levels of IRF8 but are present in Irf8−/− mice with altered phenotype and functionality (Sichien et al. 2016). The basis for lineage divergence between pDCs and cDCs from the CDP is not known, but analysis of the +41-kb Irf8 enhancer suggests that a shared progenitor between pDCs and cDC1s may exist. Alternatively, pDCs may have arisen completely separately from the CLP, as suggested in a recent work that identified a pre-pDC (Rodrigues et al. 2018). This study characterized a pre-pDC as a lineage−CD16/32−B220−Ly6C−CD117int/loCD135+CD115−CD127+SiglecH+Ly6D+ cell (Rodrigues et al. 2018). Pre-pDCs express high levels of IRF8 and, once matured, express Tcf4.
Stage 3 of DC development concerns maintenance and regulation in peripheral tissues. Studies have suggested that cytokines and some transcription factors are required for control of DC populations and may regulate DC plasticity in tissues, as discussed above.
SIMILARITIES BETWEEN ILC AND DC DEVELOPMENT
ILC development and DC development depend on some of the same transcription factor families and often the same transcription factors. Here, we discuss the similarities between the development of these lineages by studying E proteins/Id proteins, Nfil3, and Zeb2.
E protein and Id protein family member interactions are often portrayed as transcriptional switches in the development or function of specific immune subsets. Both types of proteins belong to the basic-helix-loop-helix (bHLH) family of transcription factors and exert both transcription activation and transcription repression roles in the immune system. bHLH family members contain two protein domains that are highly conserved but functionally different. The N terminus of the proteins contains the basic region, which allows for binding to DNA at a specific sequence, known as an E box. The C terminus of the proteins contains the HLH domain, which allows for hetero- or homodimeric binding to other protein subunits. bHLH proteins also contain two activation domains, ADI and ADII, which map to regions that are distinct from the zipper (Aronheim et al. 1993). These activation domains were identified in the N-terminal half of E2A and are conserved in the E protein subfamily, and they can function independently of bHLH and employ different roles. ADI is active in many cell types (Aronheim et al. 1993) and can recruit the SAGA chromatin-remodeling complex (Massari et al. 1999). ADII is thought to direct transcription activation, as site-directed mutagenesis at this region decreased transactivation potential (Aronheim et al. 1993).
E proteins are a member of the class I bHLH family and canonically bind to the DNA sequence CAnnTG. There are three known E proteins in mouse: E2A, HEB, and E2–2. The E2a gene encodes for two proteins, E12 and E47, by alternative splicing. E47 can homodimerize, while E12 can bind to other members of the bHLH family. HEB can also be alternatively spliced to produce HEBAlt and HEBCan.
Id proteins are a member of the class V bHLH family and lack the basic DNA binding domain present in other bHLH family members. There are four known Id proteins in mouse: Id1, Id2, Id3, and Id4. Their primary function is to inhibit the activity of E proteins by sequestering E proteins and acting as dominant-negative inhibitors of E protein function. The HLH domain of Id proteins can heterodimerize with the bHLH domain of E proteins, thus causing nonfunctional heterodimers.
Id and E proteins are required in the initial stages of ILC development. Id2 is required in the ILC lineage to extinguish T cell potential and is required for all ILC subsets. The discovery of the CHILP as a lineage−IL-7R+CD135−α4β7+CD25−Id2high progenitor cell elucidated the fate-determining role of Id and E proteins in the ILC/T cell lineage split. Id2 is required for total ILC development (Verykokakis et al. 2014, Yokota et al. 1999) but is necessary only for proper CD4/CD8 T cell function. Id2 is also required specifically for the differentiation of ILC22s and type 2 ILCs and for the induction of α4β7 (Hwang & McKenzie 2013). Overexpression of Id2, in contrast, prevents the development of T and B cells, as well as of pDCs, and promotes NK cell and ILC differentiation (Mjosberg et al. 2012b). Sequestration of E47, a protein subunit of E2a, by Id2 promotes mature NK and LTi cell development (Mjosberg et al. 2012b), but loss of both E2a and Id2 in doubly deficient mice can restore mature NK cells in the bone marrow and LTi development (Boos et al. 2007). These particular results suggested that Id2 does have a function in mature NK cell development but has other requirements in the bone marrow and thymus.
In DCs, Id2 is absolutely required for cDC1 development and is, as discussed above, required for early cDC1 specification. Work done in our lab has elucidated a newfound role for E proteins in the cDC1 lineage. E proteins are required for the activity of the +41-kb Irf8 enhancer, but Id2 expression in the cDC1-specified fraction of the CDP blocks activity at this enhancer and allows for a cDC1 fate through the activation of the +32-kb Irf8 enhancer. In summary, for both ILC and DC lineages, E proteins and Id proteins act as switches to specify one subtype over the other.
Nfil3 is a bZIP transcriptional regulator and regulates many diverse biological processes (Keniry et al. 2013). The N-terminal portion of the bZIP domain contains a basic motif, which directly binds to DNA. The C-terminal portion of the bZIP domain contains the leucine zipper region, which is responsible for homodimerization. The Nfil3 protein also contains a unique transcription repression domain that is transferable since its fusion with the GAL4 DNA binding domain leads to transcription repression in reporter assays.
In the hematopoietic system, Nfil3 is essential for NK cell development. Nfil3 was the first transcription factor shown to be selectively and critically required for NK cell development, as Nfil3−/− mice lack those populations but do not lack B cells, T cells, or NKT cells (Gascoyne et al. 2009, Kamizono et al. 2009). The defect in NK cells is intrinsic in nature, which leads to failure to eliminate major histocompatibility complex class I–deficient target cells and to produce IFN-γ. However, further studies suggest that the NK cell population is far more complex and may have several different origins. Firth et al. (2013) showed that the MCMV/recombinant virus expressing the viral m157 glycoprotein could induce a Ly49H+ NK cell population in Nfil3−/− mice. The virus-induced Ly49H+ NK cells are fully functional with respect to IFN-γ production and cytotoxicity and could produce long-lived memory NK cells comparable to NK cells from wild-type mice. Even at steady state, the development of several tissue-resident NK cells, particularly in mucosal sites, such as salivary gland NK cells (Cortez et al. 2014, Erick et al. 2016), kidney tissue-resident NK cells (Victorino et al. 2015), and uterine NK cells (Redhead et al. 2016), is Nfil3 independent. Nfil3 is required for the formation of Eomes-expressing NK cells, whereas Eomes− NK cells develop independently of Nfil3 (Crotta et al. 2014, Seillet et al. 2014b).
Nfil3 acts early in NK cell specification, as ablation of Nfil3 in immature NK cells in bone marrow or mature peripheral NK cells through the use of Ub- or Nkp46-cre lines in combination with Nfil3 floxed mice does not influence NK lineage maintenance or homeostasis (Firth et al. 2013). A later study examined the different stages of NK cell progenitors in Nfil3−/− mice and showed that Nfil3 is required at the NK lineage commitment point when NK progenitors develop from CLPs (Male et al. 2014). Both studies conclude that Nfil3 acts early in NK cell specification.
How Nfil3 regulates NK cell development is an open question. Male et al. (2014) showed that Eomes, T-bet, and Id2 can rescue NK production from Nfil3−/− progenitors because Nfil3 binds directly to the regulatory regions of both Eomes and Id2, promoting their transcription. Nandakumar et al. (2013) further demonstrated that the histone H2A deubiquitinase MYSM1 interacts with Nfil3 and recruits Nfil3 to the Id2 locus. They observed that MYSM1 is involved in maintaining active chromatin at the Id2 locus to promote NK cell development (Nandakumar et al. 2013). In addition, Brady and colleagues showed that Notch1 is another novel Nfil3 target gene (Kostrzewski et al. 2018). While abrogation of Notch signaling impedes NK cell production, Notch peptide ligands could rescue NK cell development from Nfil3−/− progenitors (Kostrzewski et al. 2018).
In addition to having a role in NK cell development, Nfil3 is also essential for the development of nearly all ILC subtypes. All ILC subsets exhibit high Nfil3 expression, and thus Nfil3 deficiency leads to compromised development of all ILC subsets in a cell-intrinsic manner (Geiger et al. 2014; Seillet et al. 2014a,b). The only known ILC subtype that does not require Nfil3 for its development is uterine ILC3, which participates in maintaining tissue homeostasis and barrier immunity during pregnancy (Doisne et al. 2015).
Nfil3 directs the differentiation of a committed ILC progenitor and acts transiently to enforce ILC lineage commitment. Nfil3 is required for the development of the earliest ILC lineage progenitors: the Id2+ CHILP and the α4β7hiPLZF+ ILC progenitor (Xu et al. 2015, Yu et al. 2014). Geiger et al. (2014) generated Nfil3fl/fl × Nkp46iCre mice and showed that they have normal numbers of ILC3, indicating that Nfil3 is not required for lineage maintenance. Id2 has been reported to be a target of Nfil3 for NK cell development. Indeed, Nfil3 also directly binds to the Id2 locus, promotes Id2 expression in the CHILP, and orchestrates CHILP emergence from CLPs. Ectopic Id2 expression in Nfil3−/− progenitors also rescues all ILC lineage development. Recently, Belz and colleagues generated Nfil3fl/flId2-CreERT2+/T mice to spatiotemporally delete Nfil3 in Id2-expressing cells (Seillet et al. 2016). Their results show that all ILC lineages develop normally from those mice (Seillet et al. 2016). Thus, Nfil3 is a key factor for ILC lineage commitment, but its expression is only transiently required before Id2 expression.
For DC lineages, Nfil3 was shown to be specifically required for cDC1 development at steady state. Nfil3−/− mice display impaired cross-presentation to CD8+ T cells against cell-associated antigens (Kashiwada et al. 2011). However, a later study suggested that cDC1 can be induced in an Nfil3-independent manner in short-term bone marrow reconstitution (Seillet et al. 2013). Mechanically, Nfil3−/− mice have normal numbers of pre-cDC progenitors, which have significantly reduced Batf3 levels (Kashiwada et al. 2011). Our recent work with Nfil3 in cDC1 commitment shows that Nfil3 is required for early specification, perhaps initiation, of the cDC1 lineage (P. Bagadia, X. Huang, T. Liu, V. Durai, G. Grajales-Reyes, et al., submitted). As such, evidence for the role of Nfil3 in both ILC and DC lineages suggests that Nfil3 acts early in lineage specification.
Zeb2 is a zinc-finger transcription repressor that was first shown to be a regulator of epithelial-mesenchymal transition via interaction with Smad family proteins (Comijn et al. 2001, Remacle et al. 1999, van Grunsven et al. 2003, Verschueren et al. 1999). Zeb2 has two clusters of zinc fingers for DNA binding, one at each terminus. Both clusters bind to the CACCT sequence and are necessary for repression (Comijn et al. 2001, Remacle et al. 1999). Zeb2 can exert its repressive function by directly interacting with C-terminal binding proteins (CtBPs), a known corepressor family, via its CtBP interaction domain. Most notably, Zeb2 is a known partner of Smad family proteins and can bind with Smads 1, 2, 3, 5, and 8, known as R-Smads (van Grunsven et al. 2003). Germline deletion of Zeb2 leads to embryonic lethality in mice (Higashi et al. 2002, Van de Putte et al. 2003). Zeb2 performs a wide range of functions in multiple systems, ranging from dysregulation in several cancers (Vandewalle et al. 2009) to modulating myelination of oligodendrocytes (Weng et al. 2012). In 2011, the role of Zeb2 in the hematopoietic system was first demonstrated, as Zeb2 deletion using Tie-2-cre or Vav-cre resulted in defects in hematopoietic stem cell differentiation and homing to bone marrow (Goossens et al. 2011).
Zeb2 is required for NK cell terminal maturation, as shown through a NK cell–specific deletion of Zeb2 (Ncr1icre) (van Helden et al. 2015). NK cell–specific Zeb2 deletion resulted in reduced survival for mature NK cells, in defects in their exit from BM, and in increased susceptibility to B16F10 melanomas (van Helden et al. 2015). T-bet was shown to be necessary and sufficient to induce Zeb2 expression in NK cells, and Zeb2-deficient mature NK cells phenocopied their T-bet-deficient counterparts.
Several studies have associated Zeb2 with T cell terminal differentiation and memory formation. In response to LCMV infection, Zeb2 is upregulated by KLRG1hi effector CD8+ T cells, and loss of Zeb2 expression in these cells results in the loss of antigen-specific CD8+ effector cells and in the impairment of generation of effector memory cells, while the formation of central memory T cells was accelerated (Dominguez et al. 2015, Omilusik et al. 2015). Later studies further demonstrate that coordinated expression of Zeb2 and its family member, Zeb1, is critical for CD8+ T cell fate decision. Zeb2 promotes terminal T cell differentiation, whereas Zeb1 is critical for memory T cell survival and function, with TGF-β signaling selectively inducing Zeb1 and repressing Zeb2 (Guan et al. 2018).
Within the DC compartment, Zeb2 is required for pDC development (Scott et al. 2016, Wu et al. 2016). One study argued that Zeb2 is also required for cDC2 development (Scott et al. 2016), while another indicated that Zeb2 is dispensable for cDC2 development and is instead required to actively repress generation of cDC1 progenitors (Wu et al. 2016). Our recent work shows that Zeb2 forms a mutually repressive loop with Id2 and is repressed by Nfil3 in the CDP to allow for overall expression of Id2 for cDC1 specification (P. Bagadia, X. Huang, T. Liu, V. Durai, G. Grajales-Reyes, et al., submitted). The mechanisms by which Zeb2 influences ILC and DC lineage fates are less clear than the mechanisms by which Id2 and Nfil3 might influence fate, but work is currently being done in both fields to understand where and when Zeb2 acts.
CONCLUSION
Major questions remain regarding how ILC and DC subsets exert distinct effector functions and how transcription factors necessary for the development of each subset function at the molecular and genetic levels. ILC and DC subsets share transcription factors, and a transcriptional network similar to the one that exists in DCs may exist in ILCs.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Alder JK, Georgantas RW III, Hildreth RL, Kaplan IM, Morisot S, et al. 2008. Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J. Immunol 180(8):5645–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliahmad P, de la Torre B, Kaye J. 2010. Shared dependence on the DNA-binding factor TOX for the development of lymphoid tissue–inducer cell and NK cell lineages. Nat. Immunol 11(10):945–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, et al. 2007. Phenotypic and functional features of human Th17 cells. J. Exp. Med 204(8):1849–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronheim A, Shiran R, Rosen A, Walker MD. 1993. The E2A gene product contains two separable and functionally distinct transcription activation domains. PNAS 90(17):8063–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P, Baena A, Yu KO, Saini NK, Kharkwal SS, et al. 2014. A single subset of dendritic cells controls the cytokine bias of natural killer T cell responses to diverse glycolipid antigens. Immunity 40(1):105–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D, Spits H. 2015. The biology of innate lymphoid cells. Nature 517(7534):293–301 [DOI] [PubMed] [Google Scholar]
- Auffray C, Fogg DK, Narni-Mancinelli E, Senechal B, Trouillet C, et al. 2009a. CX3CR1+ CD115+ CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation. J. Exp. Med 206(3):595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C, Sieweke MH, Geissmann F. 2009b. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu. Rev. Immunol 27:669–92 [DOI] [PubMed] [Google Scholar]
- Bajana S, Roach K, Turner S, Paul J, Kovats S. 2012. IRF4 promotes cutaneous dendritic cell migration to lymph nodes during homeostasis and inflammation. J. Immunol 189(7):3368–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R, O’Quinn DB, Silberger DJ, Schoeb TR, Fouser L, et al. 2012. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity 37(6):1061–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernink JH, Krabbendam L, Germar K, de Jong E, Gronke K, et al. 2015. Interleukin-12 and −23 control plasticity of CD127+ group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity 43(1):146–60 [DOI] [PubMed] [Google Scholar]
- Bliss SK, Butcher BA, Denkers EY. 2000. Rapid recruitment of neutrophils containing prestored IL-12 during microbial infection. J. Immunol 165(8):4515–21 [DOI] [PubMed] [Google Scholar]
- Bliss SK, Zhang Y, Denkers EY. 1999. Murine neutrophil stimulation by Toxoplasma gondii antigen drives high level production of IFN-gamma-independent IL-12. J. Immunol 163(4):2081–88 [PubMed] [Google Scholar]
- Boos MD, Yokota Y, Eberl G, Kee BL. 2007. Mature natural killer cell and lymphoid tissue–inducing cell development requires Id2-mediated suppression of E protein activity. J. Exp. Med 204(5):1119–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewitz A, Eickhoff S, Dahling S, Quast T, Bedoui S, et al. 2017. CD8+ T cells orchestrate pDC-XCR1+ dendritic cell spatial and functional cooperativity to optimize priming. Immunity 46(2):205–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M, Colonna M. 2015. Aryl hydrocarbon receptor: linking environment to immunity. Semin. Immunol 27(5):310–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M, Jarrossay D, Facchetti F, Alebardi O, Nakajima H, et al. 1999. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat. Med 5(8):919–23 [DOI] [PubMed] [Google Scholar]
- Cella M, Otero K, Colonna M. 2010. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1beta reveals intrinsic functional plasticity. PNAS 107(24):10961–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L, Lewis KL, Firner S, Thiel V, Hugues S, et al. 2012. Plasmacytoid dendritic cells control T-cell response to chronic viral infection. PNAS 109(8):3012–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrier DE, Serafini N, Di Santo JP. 2018. Innate lymphoid cell development: a T cell perspective. Immunity 48(6):1091–103 [DOI] [PubMed] [Google Scholar]
- Cherrier M, Eberl G. 2012. The development of LTi cells. Curr. Opin. Immunol 24(2):178–83 [DOI] [PubMed] [Google Scholar]
- Cisse B, Caton ML, Lehner M, Maeda T, Scheu S, et al. 2008. Transcription factor E2–2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 135(1):37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, et al. 2001. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell 7(6):1267–78 [DOI] [PubMed] [Google Scholar]
- Constantinides MG, Gudjonson H, McDonald BD, Ishizuka IE, Verhoef PA, et al. 2015. PLZF expression maps the early stages of ILC1 lineage development. PNAS 112(16):5123–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. 2014. A committed precursor to innate lymphoid cells. Nature 508:397–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez VS, Cervantes-Barragan L, Robinette ML, Bando JK, Wang Y, et al. 2016. Transforming growth factor-beta signaling guides the differentiation of innate lymphoid cells in salivary glands. Immunity 44(5):1127–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez VS, Fuchs A, Cella M, Gilfillan S, Colonna M. 2014. Salivary gland NK cells develop independently of Nfil3 in steady-state. J. Immunol 192(10):4487–91 [DOI] [PubMed] [Google Scholar]
- Cowell IG, Skinner A, Hurst HC. 1992. Transcriptional repression by a novel member of the bZIP family of transcription factors. Mol. Cell. Biol 12(7):3070–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotta S, Gkioka A, Male V, Duarte JH, Davidson S, et al. 2014. The transcription factor E4BP4 is not required for extramedullary pathways of NK cell development. J. Immunol 192(6):2677–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, et al. 2009. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med 206(1):51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang DT, Pevsner J, Yang VW. 2000. The biology of the mammalian Kruppel-like family of transcription factors. Int. J. Biochem. Cell. Biol 32(11–12):1103–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, et al. 2014. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med 211(3):563–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, et al. 2011. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J. Exp. Med 208(10):1989–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doisne JM, Balmas E, Boulenouar S, Gaynor LM, Kieckbusch J, et al. 2015. Composition, development, and function of uterine innate lymphoid cells. J. Immunol 195(8):3937–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez CX, Amezquita RA, Guan T, Marshall HD, Joshi NS, et al. 2015. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J. Exp. Med 212(12):2041–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M, Bluestone JA. 2016. Harnessing the plasticity of CD4+ T cells to treat immune-mediated disease. Nat. Rev. Immunol 16(3):149–63 [DOI] [PubMed] [Google Scholar]
- Eberl G, Colonna M, Di Santo JP, McKenzie AN. 2015. Innate lymphoid cells: a new paradigm in immunology. Science 348(6237):aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR. 2004. An essential function for the nuclear receptor RORγt in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol 5(1):64–73 [DOI] [PubMed] [Google Scholar]
- Erick TK, Anderson CK, Reilly EC, Wands JR, Brossay L. 2016. NFIL3 expression distinguishes tissue-resident NK cells and conventional NK–like cells in the mouse submandibular glands. J. Immunol 197(6):2485–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, et al. 2007. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J 26(18):4138–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth MA, Madera S, Beaulieu AM, Gasteiger G, Castillo EF, et al. 2013. Nfil3-independent lineage maintenance and antiviral response of natural killer cells. J. Exp. Med 210(13):2981–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg DK, Sibon C, Miled C, Jung S, Aucouturier P, et al. 2006. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science 311(5757):83–87 [DOI] [PubMed] [Google Scholar]
- Fuertes MB, Kacha AK, Kline J, Woo SR, Kranz DM, et al. 2011. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8α+ dendritic cells. J. Exp. Med 208(10):2005–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Liu K, Smith C, Bonito AJ, Steinman RM. 2004. The linkage of innate to adaptive immunity via maturing dendritic cells in vivo requires CD40 ligation in addition to antigen presentation and CD80/86 costimulation. J. Exp. Med 199(12):1607–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Shimizu K, Kronenberg M, Steinman RM. 2002. Prolonged IFN-gamma-producing NKT response induced with alpha-galactosylceramide-loaded DCs. Nat. Immunol 3(9):867–74 [DOI] [PubMed] [Google Scholar]
- Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, et al. 2009. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol 10(10):1118–24 [DOI] [PubMed] [Google Scholar]
- Gazzinelli RT, Wysocka M, Hayashi S, Denkers EY, Hieny S, et al. 1994. Parasite-induced IL-12 stimulates early IFN-gamma synthesis and resistance during acute infection with Toxoplasma gondii. J. Immunol 153(6):2533–43 [PubMed] [Google Scholar]
- Geiger TL, Abt MC, Gasteiger G, Firth MA, O’Connor MH, et al. 2014. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J. Exp. Med 211(9):1723–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaedi M, Steer CA, Martinez-Gonzalez I, Halim TYF, Abraham N, Takei F. 2016. Common-lymphoid-progenitor-independent pathways of innate and T lymphocyte development. Cell Rep 15(3):471–80 [DOI] [PubMed] [Google Scholar]
- Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. 2005. Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res 15(2):92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh HS, Cisse B, Bunin A, Lewis KL, Reizis B. 2010. Continuous expression of the transcription factor E2–2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity 33(6):905–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens S, Janzen V, Bartunkova S, Yokomizo T, Drogat B, et al. 2011. The EMT regulator Zeb2/Sip1 is essential for murine embryonic hematopoietic stem/progenitor cell differentiation and mobilization. Blood 117(21):5620–30 [DOI] [PubMed] [Google Scholar]
- Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, et al. 2012. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity 36(1):55–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajales-Reyes GE, Iwata A, Albring J, Wu X, Tussiwand R, et al. 2015. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α+ conventional DC clonogenic progenitor. Nat. Immunol 16(7):708–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajkowska LT, Ceribelli M, Lau CM, Warren ME, Tiniakou I, et al. 2017. Isoform-specific expression and feedback regulation of E protein TCF4 control dendritic cell lineage specification. Immunity 46(1):65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T, Dominguez CX, Amezquita RA, Laidlaw BJ, Cheng J, et al. 2018. ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8+ T cell fates. J. Exp. Med 215(4):1153–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillerey C, Mouries J, Polo G, Doyen N, Law HK, et al. 2012. Pivotal role of plasmacytoid dendritic cells in inflammation and NK-cell responses after TLR9 triggering in mice. Blood 120(1):90–99 [DOI] [PubMed] [Google Scholar]
- Guo X, Qiu J, Tu T, Yang X, Deng L, et al. 2014. Induction of innate lymphoid cell–derived interleukin-22 by the transcription factor STAT3 mediates protection against intestinal infection. Immunity 40(1):25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker C, Kirsch RD, Ju XS, Hieronymus T, Gust TC, et al. 2003. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat. Immunol 4(4):380–86 [DOI] [PubMed] [Google Scholar]
- Halim TY, Hwang YY, Scanlon ST, Zaghouani H, Garbi N, et al. 2016. Group 2 innate lymphoid cells license dendritic cells to potentiate memory TH2 cell responses. Nat. Immunol 17(1):57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim TY, MacLaren A, Romanish MT, Gold MJ, McNagny KM, Takei F. 2012. Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity 37(3):463–74 [DOI] [PubMed] [Google Scholar]
- Hammad H, Plantinga M, Deswarte K, Pouliot P, Willart MA, et al. 2010. Inflammatory dendritic cells—not basophils—are necessary and sufficient for induction of Th2 immunity to inhaled house dust mite allergen. J. Exp. Med 207(10):2097–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harly C, Cam M, Kaye J, Bhandoola A. 2018. Development and differentiation of early innate lymphoid progenitors. J. Exp. Med 215(1):249–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegazy AN, Peine M, Helmstetter C, Panse I, Frohlich A, et al. 2010. Interferons direct Th2 cell reprogramming to generate a stable GATA-3+T-bet+ cell subset with combined Th2 and Th1 cell functions. Immunity 32(1):116–28 [DOI] [PubMed] [Google Scholar]
- Higashi Y, Maruhashi M, Nelles L, Van de PT, Verschueren K, et al. 2002. Generation of the floxed allele of the SIP1 (Smad-interacting protein 1) gene for Cre-mediated conditional knockout in the mouse. Genesis 32(2):82–84 [DOI] [PubMed] [Google Scholar]
- Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, et al. 2008. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science 322(5904):1097–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou B, Benson A, Kuzmich L, DeFranco AL, Yarovinsky F. 2011. Critical coordination of innate immune defense against Toxoplasma gondii by dendritic cells responding via their Toll-like receptors. PNAS 108(1):278–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, et al. 2012. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity 37(4):634–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O’Garra A, Murphy KM. 1993. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science 260(5107):547–49 [DOI] [PubMed] [Google Scholar]
- Huang Y, Guo L, Qiu J, Chen X, Hu-Li J, et al. 2015. IL-25-responsive, lineage-negative KLRG1hi cells are multipotential ‘inflammatory’ type 2 innate lymphoid cells. Nat. Immunol 16(2):161–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. 2005. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science 307(5708):430–33 [DOI] [PubMed] [Google Scholar]
- Hwang YY, McKenzie AN. 2013. Innate lymphoid cells in immunity and disease. Adv. Exp. Med. Biol 785:9–26 [DOI] [PubMed] [Google Scholar]
- Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, et al. 2009. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev 23(20):2376–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, et al. 2005. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol 6(12):1236–44 [DOI] [PubMed] [Google Scholar]
- Ishizuka IE, Constantinides MG, Gudjonson H, Bendelac A. 2016. The innate lymphoid cell precursor. Annu. Rev. Immunol 34:299–316 [DOI] [PubMed] [Google Scholar]
- Ito T, Wang YH, Duramad O, Hori T, Delespesse GJ, et al. 2005. TSLP-activated dendritic cells induce an inflammatory T helper type 2 cell response through OX40 ligand. J. Exp. Med 202(9):1213–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S, Scallan JP, Kim KW, Werth K, Johnson MW, et al. 2016. CCR7 and IRF4-dependent dendritic cells regulate lymphatic collecting vessel permeability. J Clin. Investig 126(4):1581–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Cui W. 2012. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol 12(11):749–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, et al. 2009. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med 206(13):2977–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada M, Pham NL, Pewe LL, Harty JT, Rothman PB. 2011. NFIL3/E4BP4 is a key transcription factor for CD8α+ dendritic cell development. Blood 117:6193–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, et al. 2002. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 129(11):2619–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, et al. 1997. CD1d-restricted and TCR-mediated activation of vα14 NKT cells by glycosylceramides. Science 278(5343):1626–29 [DOI] [PubMed] [Google Scholar]
- Keniry M, Pires MM, Mense S, Lefebvre C, Gan B, et al. 2013. Survival factor NFIL3 restricts FOXO-induced gene expression in cancer. Genes Dev 27(8):916–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, et al. 2011. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334(6062):1561–65 [DOI] [PubMed] [Google Scholar]
- Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, et al. 2014. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell 157(2):340–56 [DOI] [PubMed] [Google Scholar]
- Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, et al. 2013. A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature 494(7436):261–65 [DOI] [PubMed] [Google Scholar]
- Knox JJ, Cosma GL, Betts MR, McLane LM. 2014. Characterization of T-bet and Eomes in peripheral human immune cells. Front. Immunol 5:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrzewski T, Borg AJ, Meng Y, Filipovic I, Male V, et al. 2018. Multiple levels of control determine how E4bp4/Nfil3 regulates NK cell development. J. Immunol 200(4):1370–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotaki D, Osato N, Nishiyama A, Yamamoto M, Ban T, et al. 2013. Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood 121(10):1839–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusunoki T, Sugai M, Katakai T, Omatsu Y, Iyoda T, et al. 2003. TH2 dominance and defective development of a CD8+ dendritic cell subset in Id2-deficient mice. J. Allergy Clin. Immunol 111(1):136–42 [DOI] [PubMed] [Google Scholar]
- Lee JS, Cella M, Colonna M. 2012a. AHR and the transcriptional regulation of type-17/22 ILC. Front. Immunol 3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, et al. 2012b. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol 13(2):144–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Mukasa R, Hatton RD, Weaver CT. 2009a. Developmental plasticity of Th17 and Treg cells. Curr. Opin. Immunol 21(3):274–80 [DOI] [PubMed] [Google Scholar]
- Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, et al. 2009b. Late developmental plasticity in the T helper 17 lineage. Immunity 30(1):92–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KL, Caton ML, Bogunovic M, Greter M, Grajkowska LT, et al. 2011. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity 35(5):780–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AI, Menegatti S, Bustamante J, Le Bourhis L, Allez M, et al. 2016. IL-12 drives functional plasticity of human group 2 innate lymphoid cells. J. Exp. Med 213(4):569–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CH, Fan YT, Dias A, Esper L, Corn RA, et al. 2006. Dendritic cells are essential for in vivo IL-12 production and development of resistance against Toxoplasma gondii infection in mice. J. Immunol 177(1):31–5 [DOI] [PubMed] [Google Scholar]
- Lupar E, Brack M, Garnier L, Laffont S, Rauch KS, et al. 2015. Eomesodermin Expression in CD4+ T cells restricts peripheral Foxp3 induction. J. Immunol 195(10):4742–52 [DOI] [PubMed] [Google Scholar]
- Male V, Nisoli I, Kostrzewski T, Allan DS, Carlyle JR, et al. 2014. The transcription factor E4bp4/Nfil3 controls commitment to the NK lineage and directly regulates Eomes and Id2 expression. J. Exp. Med 211(4):635–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashayekhi M, Sandau MM, Dunay IR, Frickel EM, Khan A, et al. 2011. CD8α+ dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity 35(2):249–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Grant PA, Pray-Grant MG, Berger SL, Workman JL, Murre C. 1999. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol. Cell 4(1):63–73 [DOI] [PubMed] [Google Scholar]
- McConnell BB, Yang VW. 2010. Mammalian Kruppel-like factors in health and diseases. Physiol. Rev 90(4):1337–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnil C, Sabatel CM, Marichal T, Toussaint M, Cataldo D, et al. 2012. Resident CD11b+Ly6C− lung dendritic cells are responsible for allergic airway sensitization to house dust mite in mice. PLOS ONE 7(12):e53242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke LA, Groom JR, Rankin LC, Seillet C, Masson F, et al. 2013. TCF-1 controls ILC2 and NKp46+RORγt+ innate lymphocyte differentiation and protection in intestinal inflammation. J. Immunol 191(8):4383–91 [DOI] [PubMed] [Google Scholar]
- Mjosberg J, Bernink J, Golebski K, Karrich JJ, Peters CP, et al. 2012a. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity 37(4):649–59 [DOI] [PubMed] [Google Scholar]
- Mjosberg J, Bernink J, Peters C, Spits H. 2012b. Transcriptional control of innate lymphoid cells. Eur. J. Immunol 42(8):1916–23 [DOI] [PubMed] [Google Scholar]
- Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, et al. 2010. Innate production of TH2 cytokines by adipose tissue–associated c-Kit+Sca-1+ lymphoid cells. Nature 463(7280):540–44 [DOI] [PubMed] [Google Scholar]
- Murphy KM, Stockinger B. 2010. Effector T cell plasticity: flexibility in the face of changing circumstances. Nat. Immunol 11(8):674–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik SH, Perie L, Swart E, Gerlach C, van Rooij N, et al. 2013. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature 496(7444):229–32 [DOI] [PubMed] [Google Scholar]
- Nandakumar V, Chou Y, Zang L, Huang XF, Chen SY. 2013. Epigenetic control of natural killer cell maturation by histone H2A deubiquitinase, MYSM1. PNAS 110(41):E3927–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenbein AF, Sierro S, Odermatt B, Pericin M, Karrer U, et al. 2001. Roles of tumour localization, second signals and cross priming in cytotoxic T-cell induction. Nature 411(6841):1058–64 [DOI] [PubMed] [Google Scholar]
- Ohne Y, Silver JS, Thompson-Snipes L, Collet MA, Blanck JP, et al. 2016. IL-1 is a critical regulator of group 2 innate lymphoid cell function and plasticity. Nat. Immunol 17(6):646–55 [DOI] [PubMed] [Google Scholar]
- Omilusik KD, Best JA, Yu B, Goossens S, Weidemann A, et al. 2015. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med 212(12):2027–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai N, Obata-Onai A, Schmid MA, Ohteki T, Jarrossay D, Manz MG. 2007. Identification of clonogenic common Flt3+ M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nature 8(11):1207–16 [DOI] [PubMed] [Google Scholar]
- O’Sullivan TE, Rapp M, Fan X, Weizman OE, Bhardwaj P, et al. 2016. Adipose-resident group 1 innate lymphoid cells promote obesity-associated insulin resistance. Immunity 45(2):428–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W, Lohning M, Gao Z, Assenmacher M, Ranganath S, et al. 2000. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity 12(1):27–37 [DOI] [PubMed] [Google Scholar]
- Panzer M, Sitte S, Wirth S, Drexler I, Sparwasser T, Voehringer D. 2012. Rapid in vivo conversion of effector T cells into Th2 cells during helminth infection. J. Immunol 188(2):615–23 [DOI] [PubMed] [Google Scholar]
- Papaioannou VE. 2014. The T-box gene family: emerging roles in development, stem cells and cancer. Development 141(20):3819–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, et al. 2003. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science 302(5647):1041–43 [DOI] [PubMed] [Google Scholar]
- Pepper M, Dzierszinski F, Wilson E, Tait E, Fang Q, et al. 2008. Plasmacytoid dendritic cells are activated by Toxoplasma gondii to present antigen and produce cytokines. J. Immunol 180(9):6229–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson EK, Uronen-Hansson H, Semmrich M, Rivollier A, Hagerbrand K, et al. 2013. IRF4 transcription-factor-dependent CD103+CD11b+ dendritic cells drive mucosal T helper 17 cell differentiation. Immunity 38(5):958–69 [DOI] [PubMed] [Google Scholar]
- Pikovskaya O, Chaix J, Rothman NJ, Collins A, Chen YH, et al. 2016. Eomesodermin is sufficient to direct type 1 innate lymphocyte development into the conventional NK lineage. J. Immunol 196(4):1449–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possot C, Schmutz S, Chea S, Boucontet L, Louise A, et al. 2011. Notch signaling is necessary for adult, but not fetal, development of RORγt+ innate lymphoid cells. Nat. Immunol 12(10):949–58 [DOI] [PubMed] [Google Scholar]
- Pulendran B, Tang H, Denning TL. 2008. Division of labor, plasticity, and crosstalk between dendritic cell subsets. Curr. Opin. Immunol 20(1):61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, et al. 2012. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 36(1):92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin LC, Girard-Madoux MJ, Seillet C, Mielke LA, Kerdiles Y, et al. 2016. Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat. Immunol 17(2):179–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin LC, Groom JR, Chopin M, Herold MJ, Walker JA, et al. 2013. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat. Immunol 14(4):389–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redhead ML, Portilho NA, Felker AM, Mohammad S, Mara DL, Croy BA. 2016. The transcription factor NFIL3 is essential for normal placental and embryonic development but not for uterine natural killer (UNK) cell differentiation in mice. Biol. Reprod 94(5):101. [DOI] [PubMed] [Google Scholar]
- Reis e Sousa C, Hieny S, Scharton-Kersten T, Jankovic D, Charest H, et al. 1997. In vivo microbial stimulation induces rapid CD40 ligand-independent production of interleukin 12 by dendritic cells and their redistribution to T cell areas. J. Exp. Med 186(11):1819–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle JE, Kraft H, Lerchner W, Wuytens G, Collart C, et al. 1999. New mode of DNA binding of multi-zinc finger transcription factors: DeltaEF1 family members bind with two hands to two target sites. EMBO J 18(18):5073–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette ML, Colonna M. 2016. Immune modules shared by innate lymphoid cells and T cells. J. Allergy Clin. Immunol 138(5):1243–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, et al. (Immunological Genome Consortium). 2015. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol 16(3):306–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues PF, Alberti-Servera L, Eremin A, Grajales-Reyes GE, Ivanek R, Tussiwand R. 2018. Distinct progenitor lineages contribute to the heterogeneity of plasmacytoid dendritic cells. Nat. Immunol 19(7):711–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe P, Metcalf D, Vremec D, Naik SH, Langdon WY, et al. 2014. Lymphoid tissue and plasmacytoid dendritic cells and macrophages do not share a common macrophage–dendritic cell–restricted progenitor. Immunity 41(1):104–15 [DOI] [PubMed] [Google Scholar]
- Satpathy AT, Briseno CG, Lee JS, Ng D, Manieri NA, et al. 2013. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol 14(9):937–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy AT, Kc W, Albring JC, Edelson BT, Kretzer NM, et al. 2012a. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med 209(6):1135–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy AT, Wu X, Albring JC, Murphy KM. 2012b. Re(de)fining the dendritic cell lineage. Nat. Immunol 13(12):1145–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S, Cherrier M, Lochner M, Satoh-Takayama N, Fehling HJ, et al. 2010. Lineage relationship analysis of RORγt+ innate lymphoid cells. Science 330(6004):665–69 [DOI] [PubMed] [Google Scholar]
- Schiavoni G, Mattei F, Sestili P, Borghi P, Venditti M, et al. 2002. ICSBP is essential for the development of mouse type I interferon–producing cells and for the generation and activation of CD8α+ dendritic cells. J. Exp. Med 196(11):1415–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenner SM, Madan V, Busch K, Tietz A, Laufle C, et al. 2010. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity 32(3):426–36 [DOI] [PubMed] [Google Scholar]
- Schlitzer A, McGovern N, Teo P, Zelante T, Atarashi K, et al. 2013. IRF4 transcription factor–dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity 38(5):970–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A, Sivakamasundari V, Chen J, Sumatoh HR, Schreuder J, et al. 2015. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol 16(7):718–28 [DOI] [PubMed] [Google Scholar]
- Schönheit J, Kuhl C, Gebhardt ML, Klett FF, Riemke P, et al. 2013. PU.1 level-directed chromatin structure remodeling at the Irf8 gene drives dendritic cell commitment. Cell Rep 3(5):1617–28 [DOI] [PubMed] [Google Scholar]
- Schuler G, Romani N, Steinman RM. 1985. A comparison of murine epidermal Langerhans cells with spleen dendritic cells. J. Investig. Dermatol 85(Suppl. 1):99–106 [DOI] [PubMed] [Google Scholar]
- Scott CL, Soen B, Martens L, Skrypek N, Saelens W, et al. 2016. The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J. Exp. Med 213(6):897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. 1999. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet 22(4):356–60 [DOI] [PubMed] [Google Scholar]
- Seillet C, Huntington ND, Gangatirkar P, Axelsson E, Minnich M, et al. 2014a. Differential requirement for Nfil3 during NK cell development. J. Immunol 192(6):2667–76 [DOI] [PubMed] [Google Scholar]
- Seillet C, Jackson JT, Markey KA, Brady HJ, Hill GR, et al. 2013. CD8α+ DCs can be induced in the absence of transcription factors Id2, Nfil3, and Batf3. Blood 121(9):1574–83 [DOI] [PubMed] [Google Scholar]
- Seillet C, Mielke LA, Amann-Zalcenstein DB, Su S, Gao J, et al. 2016. Deciphering the innate lymphoid cell transcriptional program. Cell Rep 17(2):436–47 [DOI] [PubMed] [Google Scholar]
- Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, et al. 2014b. Nfil3 is required for the development of all innate lymphoid cell subsets. J. Exp. Med 211(9):1733–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini N, Vosshenrich CA, Di Santo JP. 2015. Transcriptional regulation of innate lymphoid cell fate. Nat. Rev. Immunol 15(7):415–28 [DOI] [PubMed] [Google Scholar]
- Sichien D, Scott CL, Martens L, Vanderkerken M, Van Gassen S, et al. 2016. IRF8 transcription factor controls survival and function of terminally differentiated conventional and plasmacytoid dendritic cells, respectively. Immunity 45(3):626–40 [DOI] [PubMed] [Google Scholar]
- Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, et al. 1999. The nature of the principal type 1 interferon–producing cells in human blood. Science 284(5421):1835–37 [DOI] [PubMed] [Google Scholar]
- Silver JS, Kearley J, Copenhaver AM, Sanden C, Mori M, et al. 2016. Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat. Immunol 17(6):626–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, et al. 2014. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife 3:e01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM, Witmer MD. 1978. Lymphoid dendritic cells are potent stimulators of the primary mixed leukocyte reaction in mice. PNAS 75(10):5132–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger B, Omenetti S. 2017. The dichotomous nature of T helper 17 cells. Nat. Rev. Immunol 17(9):535–44 [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Jacobson NG, Dighe AS, Gubler U, Murphy KM. 1995. Developmental commitment to the Th2 lineage by extinction of IL-12 signaling. Immunity 2(6):665–75 [DOI] [PubMed] [Google Scholar]
- Takagi H, Fukaya T, Eizumi K, Sato Y, Sato K, et al. 2011. Plasmacytoid dendritic cells are crucial for the initiation of inflammation and T cell immunity in vivo. Immunity 35(6):958–71 [DOI] [PubMed] [Google Scholar]
- Tamura T, Tailor P, Yamaoka K, Kong HJ, Tsujimura H, et al. 2005. IFN regulatory factor-4 and −8 govern dendritic cell subset development and their functional diversity. J. Immunol 174(5):2573–81 [DOI] [PubMed] [Google Scholar]
- Theisen DJ, Davidson JT, Briseno CG, Gargaro M, Lauron EJ, et al. 2018. WDFY4 is required for cross-presentation in response to viral and tumor antigens. Science 362(6415):694–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, et al. 2004. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity 20(4):477–94 [DOI] [PubMed] [Google Scholar]
- Traver D, Akashi K, Manz M, Merad M, Miyamoto T, et al. 2000. Development of CD8α-positive dendritic cells from a common myeloid progenitor. Science 290(5499):2152–54 [DOI] [PubMed] [Google Scholar]
- Tussiwand R, Everts B, Grajales-Reyes GE, Kretzer NM, Iwata A, et al. 2015. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity 42(5):916–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, Wumesh KC, et al. 2012. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 490(7421):502–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Putte T, Maruhashi M, Francis A, Nelles L, Kondoh H, et al. 2003. Mice lacking Zfhx1b, the gene that codes for Smad-interacting protein-1, reveal a role for multiple neural crest cell defects in the etiology of Hirschsprung disease–mental retardation syndrome. Am. J. Hum. Genet 72(2):465–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Grunsven LA, Michiels C, Van de Putte T, Nelles L, Wuytens G, et al. 2003. Interaction between Smad-interacting protein-1 and the corepressor C-terminal binding protein is dispensable for transcriptional repression of E-cadherin. J. Biol. Chem 278(28):26135–45 [DOI] [PubMed] [Google Scholar]
- van Helden MJ, Goossens S, Daussy C, Mathieu AL, Faure F, et al. 2015. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J. Exp. Med 212(12):2015–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandewalle C, Van Roy F, Berx G. 2009. The role of the ZEB family of transcription factors in development and disease. Cell. Mol. Life Sci 66(5):773–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, et al. 1999. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. J. Biol. Chem 274(29):20489–98 [DOI] [PubMed] [Google Scholar]
- Verykokakis M, Zook EC, Kee BL. 2014. ID’ing innate and innate-like lymphoid cells. Immunol. Rev 261(1):177–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victorino F, Sojka DK, Brodsky KS, McNamee EN, Masterson JC, et al. 2015. Tissue-resident NK cells mediate ischemic kidney injury and are not depleted by anti-asialo-GM1 antibody. J. Immunol 195(10):4973–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. 2002. Classification of human B-ZIP proteins based on dimerization properties. Mol. Cell. Biol 22(18):6321–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J, Ji M, Liang HE, Locksley RM. 2016. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529(7585):221–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J, Locksley RM. 2014. I-L-C-2 it: type 2 immunity and group 2 innate lymphoid cells in homeostasis. Curr. Opin. Immunol 31:58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, et al. 2010. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor–expressing RORγt+ innate lymphocytes. Immunity 33(5):736–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watowich SS, Liu YJ. 2010. Mechanisms regulating dendritic cell specification and development. Immunol. Rev 238(1):76–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weizman OE, Adams NM, Schuster IS, Krishna C, Pritykin Y, et al. 2017. ILC1 confer early host protection at initial sites of viral infection. Cell 171(4):795–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welty NE, Staley C, Ghilardi N, Sadowsky MJ, Igyarto BZ, Kaplan DH. 2013. Intestinal lamina propria dendritic cells maintain T cell homeostasis but do not affect commensalism. J. Exp. Med 210(10):2011–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Q, Chen Y, Wang H, Xu X, Yang B, et al. 2012. Dual-mode modulation of Smad signaling by Smad-interacting protein Sip1 is required for myelination in the central nervous system. Neuron 73(4):713–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JW, Tjota MY, Clay BS, Vander LB, Bandukwala HS, et al. 2013. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat. Commun 4:2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SH, Walker JA, Jolin HE, Drynan LF, Hams E, et al. 2012. Transcription factor RORα is critical for nuocyte development. Nat. Immunol 13(3):229–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Briseno CG, Grajales-Reyes GE, Haldar M, Iwata A, et al. 2016. Transcription factor Zeb2 regulates commitment to plasmacytoid dendritic cell and monocyte fate. PNAS 113(51):14775–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Satpathy AT, Kc W, Liu P, Murphy TL, Murphy KM. 2013. Bcl11a controls Flt3 expression in early hematopoietic progenitors and is required for pDC development in vivo. PLOS ONE 8(5):e64800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Domingues RG, Fonseca-Pereira D, Ferreira M, Ribeiro H, et al. 2015. NFIL3 orchestrates the emergence of common helper innate lymphoid cell precursors. Cell Rep 10(12):2043–54 [DOI] [PubMed] [Google Scholar]
- Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, et al. 2014. The transcription factor GATA3 is critical for the development of all IL-7Rα-expressing innate lymphoid cells. Immunity 40(3):378–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Zhu J, Paul WE. 2011. An updated view on transcription factor GATA3-mediated regulation of Th1 and Th2 cell differentiation. Int. Immunol 23(7):415–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S 2008. Induction of pluripotent stem cells from mouse fibroblasts by four transcription factors. Cell Prolif 41(Suppl. 1):51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez A, Coetzee SG, Olsson A, Muench DE, Berman BP, et al. 2017. Granulocyte-monocyte progenitors and monocyte–dendritic cell progenitors independently produce functionally distinct monocytes. Immunity 47(5):890–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Bryder D, Adolfsson J, Nygren J, Mansson R, et al. 2005. Identification of Lin−Sca1+ki+CD34+Flt3− short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood 105(7):2717–23 [DOI] [PubMed] [Google Scholar]
- Yang M, Li D, Chang Z, Yang Z, Tian Z, Dong Z. 2015. PDK1 orchestrates early NK cell development through induction of E4BP4 expression and maintenance of IL-15 responsiveness. J. Exp. Med 212(2):253–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Monticelli LA, Saenz SA, Chi AW, Sonnenberg GF, et al. 2013. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity 38(4):694–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Saenz SA, Zlotoff DA, Artis D, Bhandoola A. 2011. Natural helper cells derive from lymphoid progenitors. J. Immunol 187(11):5505–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, et al. 1999. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature 397(6721):702–6 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Hayashi M. 2014. Role of Kruppel-like factor 4 and its binding proteins in vascular disease. J. Atheroscler. Thromb 21(5):402–13 [DOI] [PubMed] [Google Scholar]
- Yu X, Wang Y, Deng M, Li Y, Ruhn KA, et al. 2014. The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. eLife 3:e04406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Marotel M, Fauteux-Daniel S, Mathieu AL, Viel S, et al. 2018. T-bet and Eomes govern differentiation and function of mouse and human NK cells and ILC1. Eur. J. Immunol 48(5):738–50 [DOI] [PubMed] [Google Scholar]
- Zhang K, Xu X, Pasha MA, Siebel CW, Costello A, et al. 2017. Notch signaling promotes the plasticity of group-2 innate lymphoid cells. J. Immunol 198(5):1798–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhang J, Kornuc M, Kwan K, Frank R, Nimer SD. 1995. Molecular cloning and characterization of NF-IL3A, a transcriptional activator of the human interleukin-3 promoter. Mol. Cell. Biol 15(11):6055–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Pritchard DM, Yang X, Bennett E, Liu G, et al. 2009. KLF4 gene expression is inhibited by the Notch signaling pathway that controls goblet cell differentiation in mouse gastrointestinal tract. Am. J. Physiol. Gastrointest. Liver Physiol 296(3):G490–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Flavell RA. 1997. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89(4):587–96 [DOI] [PubMed] [Google Scholar]
- Zhou M, Ouyang W, Gong Q, Katz SG, White JM, et al. 2001. Friend of GATA-1 represses GATA-3-dependent activity in CD4+ T cells. J. Exp. Med 194(10):1461–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Jankovic D, Oler AJ, Wei G, Sharma S, et al. 2012. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity 37(4):660–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, et al. 2004. Conditional deletion of Gata3 shows its essential function in TH1-TH2 responses. Nat. Immunol 5(11):1157–65 [DOI] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Cote-Sierra J, Guo L, Paul WE. 2006. GATA-3 promotes Th2 responses through three different mechanisms: induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell–specific factors. Cell Res 16(1):3–10 [DOI] [PubMed] [Google Scholar]