SUMMARY
Hematopoiesis provides an accessible system for studying the principles underlying cell-fate decisions in stem cells. Proposed models of hematopoiesis suggest that quantitative changes in lineage-specific transcription factors (LS-TFs) underlie cell-fate decisions. However, evidence for such models is lacking as TF levels are typically measured via RNA expression rather than by analyzing temporal changes in protein abundance. Here, we used single-cell mass cytometry and absolute quantification by mass spectrometry to capture the temporal dynamics of TF protein expression in individual cells during human erythropoiesis. We found that LS-TFs from alternate lineages are co-expressed, as proteins, in individual early progenitor cells and quantitative changes of LS-TFs occur gradually rather than abruptly to direct cell-fate decisions. Importantly, upregulation of a megakaryocytic TF in early progenitors is sufficient to deviate cells from an erythroid to a megakaryocyte trajectory, showing that quantitative changes in protein abundance of LS-TFs in progenitors can determine alternate cell fates.
In Brief
Brand and colleagues used CyTOF with temporal barcoding and quantitative mass spectrometry to reconstitute a trajectory of human erythropoiesis. By measuring endogenous TF protein levels in single cells over time, they show that TFs from competing lineages are co-expressed in bipotential progenitors, and changes in their abundance underlie cell fate decisions.
Graphical Abstract

INTRODUCTION
Hematopoiesis provides an ideal model to understand the principles underlying cell fate choices in stem cells (Bresnick et al., 2018; Doulatov et al., 2012; Orkin and Zon, 2008). Recent studies using this system are changing our interpretation of the mechanism underlying cell fate decisions from a “stepwise” model, in which cells are thought to differentiate by jumping from one stable state to the next, to a “continuous” model, in which lineage commitment occurs gradually along divergent trajectories (Laurenti and Göttgens, 2018). However, lineage fate decisions have only been analyzed at the level of RNAs encoding lineage-specific transcription factors (LS-TFs) in “snapshots” of populations or individual cells without temporal measurements (Olsson et al., 2016; Tusi et al., 2018; Zheng et al., 2018). It is currently not known whether the proteins representing LS-TFs of alternate lineages are co-expressed in single hematopoietic stem and progenitor cells (HSPCs) or whether the levels of such proteins change over time as cells differentiate. It therefore remains to be determined whether quantitative changes in the abundance of LS-TF proteins expressed throughout the time course of differentiation play a role in establishing and/or maintaining lineage trajectories.
Based on RNA analyses, lineage choice has long been proposed to occur in bipotential progenitors through quantitative changes in the relative levels of LS-TFs (Graf and Enver, 2009; Orkin, 2000). Although several pairs of LS-TFs have been proposed to mediate cell fate decisions (e.g., GATA1 vs PU.1 at the erythroid vs myeloid branch point; Huang et al., 2007; KLF1 vs FLI1 at the erythroid vs megakaryocyte branch point; Bouilloux et al., 2008; Siripin et al., 2015), a more recent study, using fluorescently tagged TFs, concluded that LS-TFs associated with alternative cell fates are not co-expressed in hematopoietic progenitors (Hoppe et al., 2016). However, endogenous LS-TFs have not been measured at the protein level in single cells, and thus, the question remains whether LS-TFs from alternate lineages are co-expressed in hematopoietic progenitors.
Here, we studied changes in the expression of key LS-TFs as HSPCs differentiate along the pathway to erythroid cells using mass cytometry time of flight (CyTOF) (Spitzer and Nolan, 2016), which allowed us to simultaneously measure 27 proteins (16 LS-TFs and 11 cell surface markers) in single cells. Furthermore, temporal barcoding (Bodenmiller et al., 2012; Zunder et al., 2015) also enabled us to perform multiplex analysis of these proteins at 13 sequential time points during erythropoiesis. This provided us with an unprecedented opportunity to effectively capture the temporal and quantitative dynamics of TFs at the protein level as multipotent hematopoietic cells undergo lineage specification and differentiate into erythroid cells.
RESULTS
Time Course Analysis of Human Erythropoiesis by Mass Cytometry
Although erythropoiesis has been studied using single-cell RNA sequencing (RNA-seq) in mice (Tusi et al., 2018), models derived from this study have not incorporated temporal protein abundance measurements, and thus, the dynamics of erythroid lineage progression remains unclear. To address this, we performed a time course experiment whereby cord-blood-derived human CD34+ HSPCs were differentiated toward the erythroid lineage as previously described (Palii et al., 2011). This system fully recapitulates the various stages of erythropoiesis (Figure 1A). Cells were collected every 2 days between the formation of early HSPCs and terminally differentiated erythroid cells (22 days in total). At each time point, cells were barcoded with palladium isotopes (Bodenmiller et al., 2012) and pooled into a single tube prior to staining with a cocktail of 27 antibodies selected to cover a broad range of hematopoietic (Majeti et al., 2007; Notta et al., 2011) and erythropoietic (Hu et al., 2013) markers (Table S1). Such temporal barcoding allowed us to compare the levels of key proteins between previously defined stages of erythropoiesis (Figure S1A). A graph-based, unsupervised clustering algorithm PhenoGraph (Chen et al., 2016; Levine et al., 2015) was applied to the pooled time points, identifying 18 subpopulations (Figures 1B and S1B) that represent all previously characterized stages of erythroid differentiation (Table S2). Subsequent barcode deconvolution of single cells revealed the temporal appearance and disappearance of these populations, effectively capturing the dynamics of progression along the erythroid lineage (Figures 1C–1E). Interestingly, our data also reveal considerable heterogeneity within the previously defined colony-forming unit-erythroid (CFU-e) population. Indeed, in addition to early CFU-e1 (cluster 13) and late CFU-e2 (cluster 10) that differ by CD38 expression, we identified CFU-e3 (cluster 11), a population expressing the same combination of proteins as CFU-e2 except at a lower level, and a CFU-e-like population (cluster 7) that is highly similar to CFU-e3 with the exception of having lost CD36 and CD71 markers (Figures 1D and S1B). The exact role of these diverse CFU-e-like populations remains to be established.
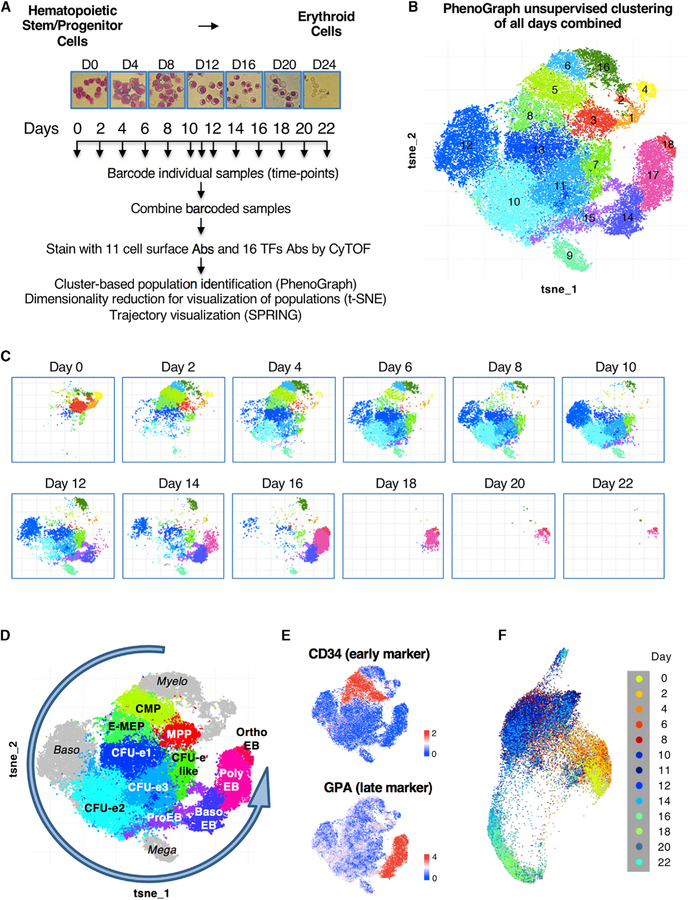
Figure 1. Time Course Analysis of Human Erythropoiesis by Mass Cytometry.
(A) Schematic of sample collection, temporal barcoding, and mass cytometry analyses. CD34+ HSPCs were isolated from cord blood and differentiated ex vivo along the erythroid lineage. Cells stained with May-Grünwald-Giemsa are shown (magnification 40×).
(B) Cell subset identification by unsupervised clustering with the PhenoGraph algorithm. Each dot represents a cell (48,076 cells total). t-stochastic neighbor embedding (t-SNE) plot of colored PhenoGraph clusters is shown.
(C) Temporal deconvolution of PhenoGraph clusters.
(D) Reconstitution of the human erythroid trajectory based on temporal appearance of PhenoGraph clusters (Figure S1B). MPP → CMP → E-MEP → CFU-e1 → CFU-e2 → ProEB → Baso_EB → Poly_EB → Ortho_EB. Non-erythroid clusters are shown in gray.
(E) Expression level plots highlighting the hematopoietic early marker CD34 and the erythroid marker GPA.
(F) Single-cell population trends over time visualized by the algorithm SPRING applied to mass cytometry data. Each dot represents a cell (20,314 cells total) and is color coded to indicate the day of identification.
Although clustering analyses define discrete cell populations, we noticed that the levels of some proteins vary between cells within a designated population, suggesting that there may not be a clear-cut separation between populations and that cells may merge from one cell population to another. To address this, we used the K-nearest neighbor algorithm SPRING (Weinreb et al., 2018a). SPRING visualization of the pooled CyTOF time series suggests a continuum of differentiation with no clear separation of cell populations from early hematopoietic progenitors to late erythroid cells (Figure 1F). Although cells appear to pass gradually along the erythroid trajectory, transient cell populations also accumulate at specific stages (Figure S1C). Furthermore, temporal coloring of the SPRING plot (Figure 1F) revealed the dynamics of differentiation, which correlates remarkably well with the previously defined erythroid trajectory. Thus, through unbiased clustering, trajectory visualization, and temporal analysis of high-dimensional protein data, we were able to define a hierarchical trajectory that includes the major known as well as some novel stages of erythropoiesis from early hematopoietic progenitors to late erythroid cells.
In addition to the main erythroid trajectory, our data reveal a minor “myeloid” trajectory characterized by expression of early myeloid markers (PU.1 and CD45RA—clusters 6 and 16), a minor “megakaryocytic” trajectory characterized by elevated levels of CD49f and CD41 (cluster 9), and a population of basophils (CD44high and GATA2high—cluster 12) recently proposed to emerge from a common erythroid-megakaryocyte-basophil progenitor (Tusi et al., 2018; Figures 1B, 1D, S1B, and S1C). The kinetics of these minor trajectories (i.e., they appear after day 2) shows that, even in an erythroid-promoting environment, hematopoietic progenitors retain the potential to initiate other hematopoietic lineages.
Lineage-Specific Transcription Factors Are Co-expressed in Individual Bipotential Progenitors
Our temporal, multi-dimensional CyTOF dataset includes measurements for 16 endogenous TFs and therefore provides an unprecedented opportunity to examine co-expression of putative LS-TFs at the protein level in single cells along the hematopoietic and erythroid trajectory. Erythroid-biased megakaryocyte-erythroid progenitor (E-MEP) cells (identified by unbiased clustering analysis; Figure 1D) were previously shown to possess differentiation potential toward both erythroid and megakaryocyte lineages in single-cell assays (Psaila et al., 2016). Furthermore, KLF1 and FLI1 were predicted to be key regulators of the erythroid vs megakaryocytic decision point using single cell RNA sequencing (scRNA-seq) data (Tusi et al., 2018). To determine whether KLF1 and FLI1 are co-expressed at the protein level in individual progenitors, we gated E-MEP cells using a previously established strategy (Psaila et al., 2016; Figure S2A). As observed by unbiased clustering (Figure 1C), cells at the E-MEP stage are most abundant between days 2 and 4 (Figure 2A, top). Most importantly, CyTOF data show that the majority of E-MEPs co-express KLF1 and FLI1 proteins in individual cells (Figure 2A, bottom) demonstrating co-expression of TF proteins from competing hematopoietic lineages at the single-cell level in bipotential progenitors. We similarly found single-cell co-expression of the GATA1 and PU.1 pair of LS-TFs at the common myeloid progenitor (CMP) differentiation stage (Figure 2D).
Figure 2. Co-expression of Lineage-Specific Transcription Factors in Individual Bipotential Progenitors.
(A) Biaxial dot plots of mass cytometry data showing co-expression of KLF1 (erythroid) and FLI1 (megakaryocytic) proteins in E-MEPs.
(B) 2D scatterplots showing gradual changes in KLF1 and FLI1 protein levels in individual cells across time and populations.
(C) Ridge plots showing relative expression of KLF1 and FLI1 proteins in temporally ordered cell populations along the erythroid trajectory. (B) and (C) use PhenoGraph cell populations.
(D) Biaxial dot plots of mass cytometry data showing co-expression of GATA1 and PU.1 proteins in CMPs.
See also Figure S2.
Protein Levels of LS-TFs Change Gradually (Rather Than in a Switch-like Manner) along the Erythroid Trajectory
We next examined quantitative changes in LS-TFs as cells progress along the erythroid trajectory. In contrast to the stepwise model predicting that cell fate decisions are made via switches in LS-TFs occurring in bipotential progenitors, we did not observe an abrupt switch in KLF1 or FLI1 proteins immediately after the E-MEP stage. Instead, we found that the levels of both TFs change gradually (and oppositely) as cells progress along the erythroid trajectory, with both factors being co-expressed until the pro-erythroblast (proEB) stage (Figures 2B, 2C, S2B, and S2C).
Although CyTOF allows one to measure quantitative changes in KLF1 and FLI1 proteins during erythroid differentiation, it does not provide information on the relative levels between the two proteins. To address this, we used a targeted mass spectrometry approach (stable isotope dilution [SID]-selected reaction monitoring [SRM]) to quantify the absolute concentration of the proteins (Picotti and Aebersold, 2012). Nuclear extracts were prepared at regular intervals during the course of erythropoiesis, and SID-SRM assays were performed to quantify FLI1 and KLF1 proteins at each time point (Figure 3). These data show that KLF1 and FLI1 proteins are present at equimolar levels (i.e., ~3,000 molecules of each TF per nucleus) around day 2, which corresponds to the time point with the highest proportion of cells at the E-MEP stage. SID-SRM quantification at subsequent time points indicates a gradual increase vs decrease in KLF1 vs FLI1 protein levels, respectively, consistent with the CyTOF data. Thus, single-cell CyTOF and SID-SRM-based mass spectrometry independently confirm a gradual change in KLF1 and FLI1 protein levels as hematopoietic cells progress along the erythroid trajectory.
Figure 3. KLF1 and FLI1 Protein Levels Change Gradually during Erythropoiesis.
(A) Absolute quantification of KLF1 and FLI1 proteins across time by SID-SRM mass spectrometry.
(B and C) Representative results showing quantification of the indicated KLF1 (B) and FLI1 (C) peptides. The main picture shows co-eluting peptides—the spiked-in “heavy” standard peptides in blue and the endogenous “light” peptides in red. Inset picture shows specific transitions measured for each light peptide. These transitions are summed to produce the red peak in the main graph.
Overexpression of FLI1 in Bipotential Progenitors Deviates the Erythroid Trajectory toward a Megakaryocytic Fate
Our finding that KLF1 and FLI1 proteins are co-expressed in single cells at the E-MEP stage together with the observation that their levels gradually change in opposite directions as cells pass along the erythroid trajectory suggests that quantitative changes in KLF1 and FLI1 proteins underlie the erythroid versus megakaryocytic cell fate choice. However, the question remains whether such changes direct cell fate decisions or whether they merely reinforce lineage identity. To test this, we overexpressed the putative megakaryocyte-promoting LS-TF (FLI1) in E-MEPs and asked whether this deviated progenitors from the erythroid trajectory. We reasoned that, if LS-TFs are only involved in reinforcing lineage decisions, overexpression of a LS-TF from an alternate lineage might interfere with differentiation, but not promote an alternate lineage. On the other hand, if quantitative changes in the levels of LS-TFs play a role in establishing cell fate, overexpression of alternate TFs in progenitors should direct differentiation toward an alternate lineage trajectory. Briefly, cells at the E-MEP stage (day 2) were infected with lentiviruses expressing a FLAG-tagged version of the non-erythroid TF FLI1 (FLAG-FLI1) (Figure 4A). Again, we performed CyTOF with temporal barcoding whereby introduction of a FLAG antibody allowed us to track the fate of thousands of cells over time upon ectopic FLI1 expression (along with thousands of unperturbed cells), enabling us to directly assess the consequences of quantitative changes in the level of a TF protein on cell fate decision in single cells. In addition to the expected erythroid trajectory, SPRING analysis revealed a second trajectory with a similar kinetic (Figure 4B) characterized by gradual upregulation of megakaryocytic markers (e.g., CD41, CD49f, and CXCR4) instead of erythroid markers (e.g., CD71 and GPA; Figures 4E, S3D, and S4A) showing that cells along this trajectory have deviated from an erythroid fate toward a megakaryocytic fate. Strikingly, this alternative trajectory is almost exclusively composed of cells that express the ectopic FLAG-FLI1 protein (Figures 4C, 4D, and S3C), strongly arguing that FLI1 not only interferes with erythroid differentiation but also promotes a megakaryocytic fate.
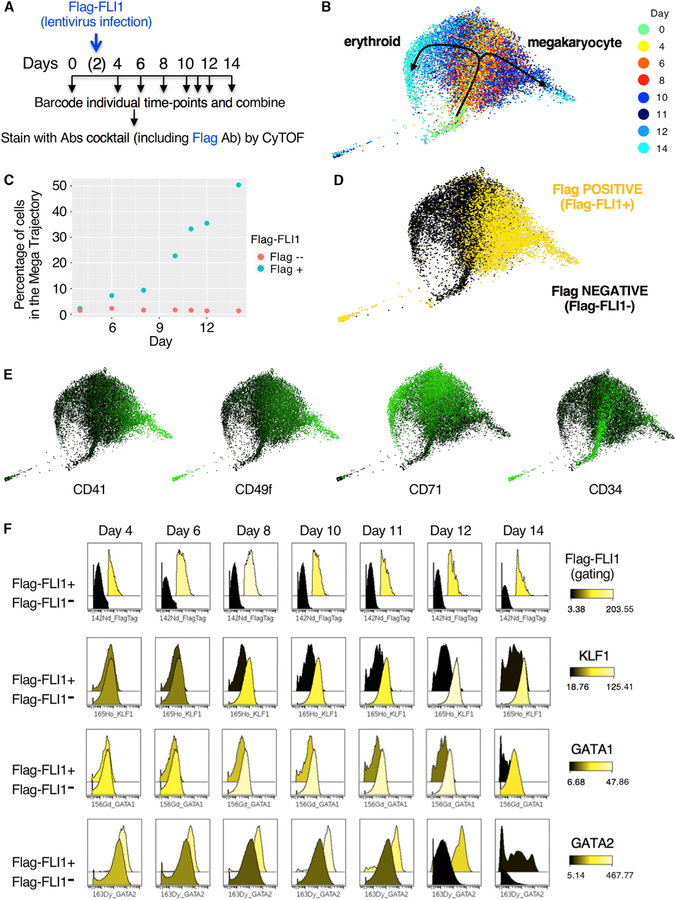
Figure 4. Overexpression of FLI1 in Bipotential Progenitors Is Sufficient to Deviate the Erythroid Trajectory toward a Megakaryocytic Fate.
(A) Schematic of sample collection, temporal barcoding, and mass cytometry analyses upon expression of a FLAG-tagged FLI1 protein in early progenitors.
(B) Single-cell population trends over time visualized by SPRING applied to CyTOF data. Each dot represents a cell (9,000 cells total), and its color indicates the measured day.
(C) Graph indicating the percentage of FLAG-negative and FLAG-positive cells that follows a megakaryocytic path (defined as CD41+) over time.
(D) SPRING plot from (B) colored for FLAG expression (yellow for positive; black for negative).
(E) SPRING plots from (B) colored for the indicated markers. Coloring scale from black (no expression) to green (high expression) is shown.
(F) Cytobank histogram overlays of CyTOF data showing temporal variations in the relative level of each indicated marker in cells expressing FLAG-FLI1 (back histogram) versus FLAG-FLI1-negative cells (front histogram). Peaks are shaded using a color scale based on the raw values of medians for each x axis channel.
See also Figures S3 and S4 and Table S3.
In addition, gating for FLAG-positive and FLAG-negative cells allowed us to directly compare dynamic changes in the levels of TFs between the two cell populations over time (Figures 4F, S3, and S4). In cells that do not express FLAG-FLI1, we observe the expected time-dependent increase in KLF1 and GATA1 proteins that occurs during erythroid differentiation (Figure 4F, front rows). In contrast, FLAG-FLI1-expressing cells fail to upregulate these TFs (Figure 4F, back rows) or other erythroid markers (i.e., IKZF1, TAL1, CD71, and GPA; Figure S3D, back rows), suggesting decreased erythroid differentiation. Furthermore, we found that GATA2 protein levels increase with time in cells expressing FLAG-FLI1, more so than in their FLAG-FLI1-negative counterparts (Figure 4F). This is consistent with GATA2’s role in favoring megakaryocytic over erythroid differentiation (Ikonomi et al., 2000). In addition, we found that FLAG-FLI1-expressing cells upregulate megakaryocytic markers, such as MafG, CD41, CD49f, and CXCR4 (Figure S4A), showing an active role of FLAG-FLI1 in promoting megakaryocyte differentiation. Thus, even in a strongly erythroid-promoting environment, ectopic expression of FLI1 in bipotential progenitors can deviate cells from their preferred erythroid trajectory toward an alternate megakaryocytic fate. Collectively, these data strongly argue for an active role of a LS-TF (FLI1) protein in the initiation of a cell fate decision by promoting megakaryocytic differentiation at the expense of an erythroid fate.
DISCUSSION
In the present study, we used single-cell proteomics to define the temporal hierarchy of human erythropoiesis. Through the development of a unique approach that integrates time-dependent, single-cell protein measurements, we were able to effectively capture the dynamic progression and hierarchical relationships of hematopoietic cells as they transition between multiple stages along the erythroid trajectory. Importantly, we provide these data as an interactive web tool to facilitate visualization and further analyses.
The classical stepwise model of hematopoiesis whereby homogeneous populations of progenitors are faced with a succession of binary choices between mutually exclusive lineages has recently been replaced by a continuous model of differentiation whereby lineage-biased hematopoietic stem and progenitor cells gradually differentiate along a continuum of lineage-specific trajectories without clearly defined cell populations (Laurenti and Göttgens, 2018). However, these trajectories rely on transcriptomic rather than proteomic measurements, and they are based on algorithms that use snapshot scRNA-seq measurements to predict lineage trajectories by organizing cells along a pseudo-time axis according to their relative phenotypic similarity (Kester and van Oudenaarden, 2018). Although valuable and insightful, such static data have a number of known limitations (Weinreb et al., 2018b), and the lack of temporal information makes it more difficult to assess true ascendant and descendant relationships. For instance, there is some uncertainty on the positioning of megakaryocytes within the hematopoietic hierarchy, as some studies have suggested that they originate directly from hematopoietic stem cells (HSCs) (Notta et al., 2016) and other studies have proposed that they originate from MEPs (Tusi et al., 2018), and yet other studies have not been able to conclude (Zheng et al., 2018). Here, our analyses incorporate time-dependent single-cell proteomic measurements, allowing us to define a trajectory that is based not only on protein similarity but also on the time-dependent appearance of different cell types. As such, we were able to observe that, in the system analyzed here, megakaryocytes originate from MEPs, not HSCs. Furthermore, although our results are consistent with a continuous rather than a stepwise model of erythropoiesis, our data reveal a timely ordered appearance and disappearance of transient cell populations or stages that accumulate at various positions along the erythroid trajectory with cells undergoing gradual transitions between these stages. Notably, these stages include previously defined cell subpopulations that, despite their transient nature, can be isolated from cord blood using combinations of cell surface markers (An et al., 2014; Chen et al., 2014; Psaila et al., 2016), providing in vivo validation of our temporal hierarchy of human erythropoiesis.
Quantitative changes in the relative amounts of LS-TFs have historically been considered as a key mechanism for lineage determination. Indeed, in the stepwise hematopoiesis model, cell fate choice was thought to occur through quantitative on or off switches between LS-TFs that are co-expressed in bipotential progenitors (Graf and Enver, 2009; Orkin, 2000). However, a study using knocked in fluorescently labeled pairs of LS-TFs Gata1 and Pu1 in mice reported an inability to detect co-expression of the two fluorescent markers in a large number of hematopoietic progenitors (Hoppe et al., 2016), raising doubt whether LS-TFs are in fact co-expressed in single cells at the protein level. We note, however, that expression of TFs from different lineages is expected to be very low in early progenitors, possibly below the limit of detection for fluorescent markers. Furthermore, fluorescent tags may alter protein stability and disruption of genomic loci during tag insertion may tamper with cis-regulatory elements, thereby affecting mRNA expression and/or stability. Our temporal, single-cell measurements of 16 endogenous unmodified TFs at the protein level during human erythropoiesis reveal that at least 2 previously proposed pairs of LS-TFs are co-expressed as proteins in individual hematopoietic progenitors, namely PU1 and GATA1 at the CMP stage, and KLF1 and FLI1 at the E-MEP stage. However, in contrast to what has been proposed in the stepwise model, there is no abrupt switch in protein levels between these LS-TFs following the progenitor stages. Instead, quantitative changes in TFs occur gradually along the hematopoietic and erythroid trajectory. Despite being gradual, quantitative changes in lineage-determining TFs actively contribute to cell fate decisions, as single-cell temporal tracking of progenitors overexpressing FLI1 shows a clear deviation from the erythroid trajectory to take on a megakaryocyte cell fate. Thus, our data strongly support the concept that the choice to differentiate along a specific lineage trajectory is intimately linked to changes in the abundance of LS-TFs, even though this process occurs gradually and takes longer (i.e., several cell stages) than previously anticipated in the stepwise model.
We have established a resource for analyzing human erythropoiesis at the level of single cells. Using this, we now describe a dynamic model of human erythropoiesis that is based on temporal single-cell proteomics measurements and demonstrate that quantitative changes in the abundance of LS-TFs protein levels are key for cell fate decisions in the context of continuous lineage trajectories. It seems likely that these principles established in the hematopoietic system will apply to similar cell fate decisions that occur in many other tissues throughout development and differentiation.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Marjorie Brand (mbrand@ohri.ca).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Hematopoietic stem/progenitor cells isolation
Umbilical cord blood from full-term deliveries was obtained from Canadian Blood Services “Cord Blood for Research program” (CBR-2014–001). CD34+ hematopoietic stem/progenitor cells were isolated as previously described (Palii et al., 2011) using a 2-step protocol. For the first step, fresh cord blood was incubated with 5 μl/ml RosetteSep Human Cord Blood CD34 Pre-Enrichment Cocktail (STEMCELL Technologies cat#15631) for 10 min at RT, then diluted with an equal volume of the recommended medium (PBS supplemented with 2% Fetal Bovine Serum and 1 mM EDTA) and layered on the top of the Ficoll-Paque™PLUS (GE Healthcare, cat# 17-1440-03). After centrifugation for 30 min at 400 × g at RT, with the brake off, CD34-pre-enriched mononuclear cells were recovered from the Ficoll-Paque™ PLUS: plasma interface, washed two times with the above recommended medium and either frozen or subjected to the EasySep Human CD34 Positive Selection (STEMCELL Technologies cat#18096). For the second step, CD34-pre-enriched mononuclear cells were resuspended at 2 × 108 cells/ ml in the above recommended medium and incubated with 100 μl/ml EasySep Positive Selection Cocktail for 15 min at RT, followed by a second incubation with 50 μl/ml Mix EasySep Magnetic Nanoparticles for 10 min at RT. Cell suspension was diluted to a total volume of 5 mL (for < 108 cells) or 10 mL (for 1 × 108 − 8.5 × 108 cells) in the same recommended medium and placed into “The Big Easy” Silver EasySep Magnet (STEMCELL Technologies cat#18001) for 5 min. After 4 washes of 5 min each in the same recommended medium, CD34-positive cells were recovered from the magnet. Purified cells were analyzed by FACS for CD34 expression using the PE Mouse Anti-Human CD34 antibody (BD Biosciences, cat# 555822) and either cryopreserved in 10% DMSO or cultured directly as described below. All procedures were approved by the Ottawa Health Science Network Research Ethics Board (2007804–01H).
Ex vivo human erythropoiesis and cell harvest
CD34+ cells were differentiated along the erythroid lineage using a previously described 4-step protocol (Palii et al., 2011). The first step (Day0 to Day11) consists of cultivating CD34+ cells in serum-free IMDM medium supplemented with 1% penicillin/streptomycin, 4×10−3 M L-glutamine, 40 ug/ml inositol, 10 ug/ml folic acid, 1.6×10−4 M monothioglycerol, 90 ng/ml ferrous nitrate, 900 ng/ml ferrous sulfate, 20% albumin-insulin-transferrin (BIT), also containing the following cytokines: 10−6 M hydrocortisone (HC), 100 ng/ml stem cell factor (SCF), 5 ng/ml interleukin 3 (IL-3) and 3 IU/ml erythropoietin (EPO) for 8 days followed by 3 days in supplemented IMDM medium containing only SCF and EPO. For the second step (Day12 to Day14), cells were co-cultured on a layer of stromal MS-5 cells in the supplemented IMDM medium containing only EPO. For the third step (Day15 to Day18), cells were co-cultured on a layer of MS-5 cells in the supplemented IMDM medium with no cytokines. For the fourth step (Day19 to Day24), cells were co-cultured on a layer of MS-5 cells in the supplemented IMDM medium in the presence of 10% fetal bovine serum. Every second day, cells were counted and monitored for viability (trypan blue exclusion), cell surface expression of the following markers: CD34 (CD34-PE, BD Biosciences, cat# 555822), CD36 (CD36-PE, BD Biosciences, cat# 555455), CD71 (CD71-FITC, Beckman Coulter, cat# IM0483U), GPA (CD235a-PE, BD Biosciences, cat# 555570) and LDS751 (Molecular Probes, cat# L7585) by FACS and hemoglobin production (benzidine staining). Cells were harvested at regular intervals (as indicated) during the course of differentiation and cryopreserved in their respective culture media supplemented with 10% DMSO.
Lentivirus preparation and infection
Lentiviral particles expressing Flag-tagged FLI1 (Flag-FLI1) were prepared as previously described (Palii et al., 2011). Specifically, 293T cells were transfected with the pMD2.G envelope vector (Addgene #12259), the psPAX2 packaging vector (Addgene #12260) and the pTRIPdU3-MND-FlagFLI1-IRES-GFP lentiviral expression vector using calcium phosphate precipitation. To generate the lentiviral vector, a C-terminally Flag tagged human FLI1 (NM_002017) cDNA was obtained from GenScript (plasmid name: pcDNA3.1-Flag-FLI1) and subcloned into the pTRIPdU3-MND-IRES-GFP lentiviral expression vector (Renou et al., 2017). Lentiviral particles were harvested, concentrated by ultracentrifugation and used to infect cells at the Day2 time-point with a MOI of 20. Lentiviral infection was repeated 24h later in the same conditions. Cells were washed and differentiation was left to proceed in the same conditions.
To test for potential effects of lentiviral infection on erythroid and/or megakaryocyte differentiation in the absence of Flag-FLI1 expression, the above experiment was repeated using either the pTRIPdU3-MND-FlagFLI1-IRES-GFP lentiviral vector (to induce the expression of GFP and Flag-FLI1 proteins) or the pTRIPdU3-MND-IRES-GFP lentiviral vector (to induce the expression of GFP only) as a control. Cells expressing the megakaryocytic marker CD41 or the erythroid marker GPA were measured by FACS within the infected cell population (gated for GFP expression) at regular time-points for up to 14 days after infection. This experiment revealed that most GFP+ cells infected with the Flag-FLI1 vector express CD41 after 14 days. In contrast, GFP+ cells infected with the non-Flag-FLI1 expressing vector do not express CD41 (data not shown) indicating that lentiviral infection itself (in the absence of Flag-FLI1 expression) does not trigger megakaryocytic differentiation. Furthermore, cells infected with the non-Flag-FLI1 expressing vector express GPA, consistent with proper erythroid differentiation (data not shown). Collectively, these results indicate that lentiviral infection per se does not perturb erythroid or megakaryocyte differentiation.
METHOD DETAILS
May-Grünwald Giemsa staining
Cells at the indicated time-points were harvested, washed with PBS, cytospun and fixed in ethanol for 2 min prior to staining with May-Grünwald (SIGMA cat# 63590) for 5 min and Giemsa stain modified solution (SIGMA cat#48900) for 10 min.
Antibody labeling with metal conjugates
Antibodies were purchased pre-conjugated when commercially available. Otherwise, purified antibodies were conjugated to specific lanthanide metals using Maxpar X8 Antibody Labeling Kit according to manufacturer instruction (Fluidigm cat#PRD002 Version 7) as described below. Partial reduction of 100 μg of each antibody was performed by incubation in 4mM TCEP for 30 min at 37°C. Partially reduced antibodies were then conjugated with lanthanide-loaded polymer for 1h at 37°C. Conjugated antibodies were quantified by measuring the absorbance at 280 nm prior to storage at 4°C in Candor Biosciences Antibody Stabilizer (cat#130050) supplemented with 0.05% sodium azide. Antibody cocktails used for mass cytometry are listed in Tables S1 and S3.
Temporal barcoding and mass cytometry
Cryopreserved cells at the different time-points (3 × 106 cells per time-point) were thawed and incubated in their respective original culture media (including cytokines) for 1h at 37°C. Barcoding and staining were then performed according to Fluidigm’s instructions as described below. Briefly, cells were transferred into polystyrene tubes (10 mL capacity), washed with Maxpar PBS Buffer (Fluidigm cat#201058) and stained with 0.5 μM final cisplatin (Fluidigm cat#201064) in 200 μl Maxpar PBS for 5 min at room temperature (RT) to exclude dead cells. The reaction was quenched by adding Maxpar Cell Staining Buffer (CSB; Fluidigm cat#201068). After 2 washes with CSB, cells were fixed for 30 min at RT in 1 mL Maxpar Nuclear Antigen Staining Buffer 1X (Fluidigm cat#201063). Cells were then permeabilized by washing twice with 2 mL Barcode Perm Buffer 1X (Fluidigm cat#201057) and resuspended in 400 μl Barcode Perm Buffer 1X. 60 μl of unique Palladium Barcodes (Fluidigm; Cell-ID 20-Plex Pd Barcoding Kit cat#201060) were then added to cells at each time point prior to incubation for 30 min at RT. Cells were then washed twice with 2 mL Nuclear Antigen Staining Perm (1X) Buffer (NP Buffer; Fluidigm cat#201063) and resuspended in 100 μl NP Buffer. Cells from all time-points were then combined into a single tube, counted, resuspended at 2–6 × 106 cells/ml in NP Buffer and stained for 45 min at RT with an equal volume of antibody cocktail (see Tables S1 and S3). Cells were then washed once with NP Buffer, and further stained with 156Gd-A-PE secondary antibody for 30 min at RT. After staining, cells were washed twice in CSB and incubated overnight at 4°C in 31.25 nM final Cell-ID Intercalator-Ir (Fluidigm cat#201192A) in Maxpar Fix and Perm Buffer (Fluidigm cat#201067). Samples were washed twice in milliQ H2O filtered into 30 μm strained cap tubes and diluted in milliQ H2O containing 10% EQ Four Element Calibration Beads (Fluidigm cat#201078) to a concentration of 0.5 × 106 cells/ml. Samples were acquired on Helios Mass Cytometer (Fluidigm). Helios was tuned and performance was checked before each run according to Fluidigm recommendation.
Nuclear extraction and targeted mass spectrometry
Cryopreserved cells at the different time-points were thawed and washed using IMDM supplemented medium. Cells were then washed in ice-cold PBS buffer, resuspended in ice-cold Swelling Buffer (10 mM HEPES K+ pH7.9; 1.5 mM MgCl2; 10 mM KCl; 0.1% (v/v) NP40; protease inhibitor cocktail) and incubated on ice for 30 min. During incubation, cells were vortexed every 5 min to allow cell lysis. Nuclei were then pelleted by centrifugation for 5 min at 1,500 rpm (4°C) and resuspended in 1 vol. of 37°C pre-heated Extraction Buffer 1 (50 mM HEPES K+ pH7.9; 1 mM MgCl2; 150 mM NaCl; 0.5% Na deoxycholate; 50 ng/μl Benzonase Millipore, cat# 70746; protease inhibitor cocktail) prior to incubation at 37°C on a Thermomixer (14,000 rpm) for 15 min. Proteins were extracted first by 6 passages through a 27 ½ gauge needle prior to addition of 1 vol. of 37°C pre-heated Extraction Buffer 2 (50 mM HEPES K+ pH7.9; 150 mM NaCl; 9.5% Na deoxycholate; 1 mM EDTA; protease inhibitor cocktail). The mixture was heated at 70°C for 5 min and proteins were extracted further by 6 passages through a 27 ½ gauge needle prior to incubation at 40°C on a Thermomixer (14,000 rpm) for 15 min. Nuclear extracts were recovered by centrifugation at 13,000 rpm for 15 min and snap frozen. Prior to mass spectrometry analysis, extracted nuclear proteins were digested with Lys-C and Trypsin. Isotopically heavy peptide standards (Pierce/Thermo Scientific) were spiked into samples after digestion, then peptides were purified together by mixed cation exchange (MCX; Waters) chromatography. Selected reaction monitoring (SRM) targeted mass spectrometry was performed on an Agilent 6490 triple quadrupole mass spectrometer, equipped with a chip cube interface. Peptides were separated online in a reversed-phase microfluidics HPLC chip (160nL trap; Agilent) by application of a 3%–25% acetonitrile gradient over 60 min. Optimized transitions (precursor-fragment ion pairs) were monitored with a 5 min retention time window, and cycle times were set to yield a minimum dwell time of 12 ms.
QUANTIFICATION AND STATISTICAL ANALYSIS
Mass cytometry data analyses
Files were processed following Fluidigm recommendation, including randomization and normalization using EQ Beads signal. Then files were concatenated, debarcoded and randomized according to Fluidigm’s instructions using the CyTOF® Software. Gating to identify and export live cells (see Figure S2A for the gating strategy) was done in Cytobank (Kotecha et al., 2010). Histograms (Figures 4F, S1A, S3D, and S4) and biaxial dot plots (Figures 2A and 2D) of mass cytometry data were generated in Cytobank. Clustering analysis was done with PhenoGraph (Levine et al., 2015) as part of the R Bioconductor package Cytofkit (Chen et al., 2016) using the markers listed in Table S1 with up to 5,000 cells sampled per time point and the number of nearest neighbors equal to 30. The transformation method used was cytofAsinh, and the visualization method was t-SNE. The 2D scatterplots and ridge plots (Figures 2B and 2C) were generated using ggplot2 (RStudio) with PhenoGraph-defined cell populations. The K-nearest neighbor algorithm SPRING (Weinreb et al., 2018a) was used for trajectory analyses with k = 5 and up to 2,000 cells sampled per time-point. Trajectories were built using the markers listed in Table S1 without temporal data. Temporal coloring (Figure 1F) or population coloring (Figure S1C) were subsequently added to the graph. Similarly, for the Flag-FLI1 overexpression analysis, trajectories were first generated using the markers listed in Table S3 without temporal data or Flag-expression data. Temporal information (Figure 4B), Flag expression (Figure 4D) and markers expression (Figure 4E) were subsequently added to the plot.
Annotation of cell populations identified by PhenoGraph
Cell clusters corresponded to well-defined cell populations, and were manually annotated based on their expression of previously characterized differentiation markers (Table S2 and Figure S1B). At Day 0, the main population consists of hematopoietic multipotent progenitors (MPP, defined as CD34+ CD38low, CD90− CD45RA− CD49f−; cluster 3) that are progressively replaced by common myeloid progenitors (CMP, defined as CD34+ CD38+ CD45RA− CD123 (IL3-Ra)low; cluster 5) and erythroid-biased megakaryocyte-erythroid progenitors (E-MEP, defined as CD34+ CD38+ CD45RA− CD123 (IL3-Ra)− CD44mod CD71+ CD41− (Psaila et al., 2016); cluster 8) at Day 2. From Day 4 to Day 10, we observe the progressive disappearance of these early CD34-positive hematopoietic progenitors (Figure 1E top panel) that are replaced by a succession of CD34-negative erythroid progenitor populations previously defined as colony-forming unit erythroid (CFU-e) that we named CFU-e1 (CD34− CD38+ CD45RA− CD123 (IL3-Ra)− CD36+ CD71+ CD235a (GPA)−; cluster 13) and CFU-e2 (CD34− CD38− CD45RA− CD123 (IL3-Ra)− CD36+ CD71+ CD235a (GPA)−; cluster 10) followed by pro-erythroblasts (ProEB, defined as CD34− CD38− CD45RA− CD123 (IL3-Ra)− CD36+ CD71+ CD235a (GPA)+/−; cluster 15). At Day 14, we observe the emergence of the first cell population expressing high levels of the terminal erythroid marker GPA (also called CD235ab) (Figure 1E bottom panel), namely basophilic erythroblasts (Baso_EB, defined as CD34− CD38− CD45RA− CD123 (IL3-Ra)− CD36++ CD71++ CD235a (GPA)+; cluster 14), followed by polychromatic erythroblasts (Poly_EB, defined as CD34-CD38− CD45RA− CD123 (IL3-Ra)− CD36+ CD71+ CD235a (GPA)+; cluster 17) and finally orthochromatic erythroblasts (Ortho_EB, defined as CD34− CD38− CD45RA− CD123 (IL3-Ra)− CD36− CD71low CD235a (GPA)+; cluster 18), which correspond to the last stage of erythroid differentiation prior to enucleation. (Figures 1B–1E and S1).
Mass spectrometry data analyses
Raw data was processed using Skyline (MacLean et al., 2010) and peaks were manually verified. Absolute quantification was performed with stable isotope dilution of AQUA peptides and the generation of standard curves based on the method described by Aebersold and colleagues (Ludwig et al., 2012; Schubert et al., 2013). For this, the peak areas of the two most intense, interference free transitions were summed together (quantifier transitions). Additional transitions (1–2) were used to confirm correct peak identification (qualifier transitions). Quantifications were generated from biological duplicates.
DATA AND SOFTWARE AVAILABILITY
Mass cytometry data is deposited in the FlowRepository database under accession numbers FR-FCM-ZYPS and FR-FCM-ZYPT. http://flowrepository.org/public_experiment_representations
ADDITIONAL RESOURCES
An interactive web tool to visualize protein changes along the hematopoietic/erythroid trajectory is provided at the following link: http://trena.systemsbiology.net/login
user: cytof
password: cytof
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE conjugated Mouse Anti-CD34 (clone 581) | BD Biosciences | Cat#555822; RRID:AB_396151 |
| PE conjugated Mouse Anti-Human CD36 (clone CB38) | BD Biosciences | Cat#555455; RRID:AB_395848 |
| FITC conjugated Mouse Anti-Human CD71 (clone YDJ1.2.2) | Beckman Coulter | Cat#IM0483U; RRID:AB_2756301 |
| PE conjugated Mouse Anti-Human CD235a (GPA) (clone GA-R2) | BD Biosciences | Cat#555570; RRID:AB_395949 |
| Antibody cocktail 1 (for CyTOF experiment) | This paper | Table S1 |
| Antibody cocktail 2 (for CyTOF experiment) | This paper | Table S3 |
| Biological Samples | ||
| Umbilical cord blood | Canadian Blood Services (CBR-2014–001) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| IMDM medium | SIGMA | Cat#I3390 |
| Penicillin/streptomycin | ThermoFisher | Cat#15140122 |
| L-glutamine | GIBCO | Cat#25030 |
| Inositol | SIGMA | Cat#I5125 |
| Folic acid | SIGMA | Cat#F7876 |
| Monothioglycerol | SIGMA | Cat#M6145 |
| Ferrous nitrate | SIGMA | Cat#8508 |
| Ferrous sulfate | SIGMA | Cat#F8633 |
| Albumin-insulin-transferrin (BIT) | STEMCELL Technologies | Cat#9500 |
| Hydrocortisone (HC) | SIGMA | Cat#H2270 |
| Stem cell factor (SCF) | PeproTech | Cat#300–07 |
| Interleukin 3 (IL-3) | PeproTech | Cat#200–03 |
| Erythropoietin (EPO) | PeproTech | Cat#100–64 |
| LDS751 | Molecular Probes | Cat#L7585 |
| Benzidine | SIGMA | Cat#B3503 |
| Benzonase | Millipore | Cat#70746 |
| May-Grunwald | SIGMA | Cat#63590 |
| Giemsa | SIGMA | Cat#48900 |
| Antibody Stabilizer | Candor Biosciences | Cat#130050 |
| Maxpar PBS Buffer | Fluidigm | Cat#201058 |
| Cisplatin | Fluidigm | Cat#201064 |
| Maxpar Cell Staining Buffer (CSB) | Fluidigm | Cat#201068 |
| Maxpar Nuclear Antigen Staining Buffer | Fluidigm | Cat#201063 |
| Barcode Perm Buffer | Fluidigm | Cat#201057 |
| Cell-ID 20-Plex Pd Barcoding Kit (Palladium Barcodes) | Fluidigm | Cat#201060 |
| Nuclear Antigen Staining Perm Buffer (NP Buffer) | Fluidigm | Cat#201063 |
| Cell-ID Intercalator-Ir | Fluidigm | Cat#201192A |
| Maxpar Fix and Perm Buffer | Fluidigm | Cat#201067 |
| EQ Four Element Calibration Beads | Fluidigm | Cat#201078 |
| Critical Commercial Assays | ||
| RosetteSep Human Cord Blood CD34 Pre-Enrichment Cocktail | STEMCELL Technologies | Cat#15631 |
| EasySep Human CD34 Positive Selection Kit | STEMCELL Technologies | Cat#18096 |
| Maxpar X8 Antibody Labeling Kit | Fluidigm | Cat#PRD002 Version 7 |
| Deposited Data | ||
| CyTOF data (Human Erythropoiesis - Cord Blood) | This paper | FlowRepository: FR-FCM-ZYPS |
| CyTOF data (Human Erythropoiesis - Cord Blood - FLI1 overexpression) | This paper | FlowRepository: FR-FCM-ZYPT |
| Experimental Models: Cell Lines | ||
| MS-5 | DSMZ | Cat#ACC 441 |
| 293T | ATCC | Cat#CRL-3216 |
| Recombinant DNA | ||
| pMD2.G envelope vector | Didier Trono | Cat#12259; RRID:Addgene_12259 |
| psPAX2 packaging vector | Didier Trono | Cat#12260; RRID:Addgene_12260 |
| pTRIPdU3-MND-FlagFLI1-IRES-GFP lentiviral expression vector | This paper | N/A |
| pcDNA3.1-Flag-FLI1 vector (containing the C-terminally Flag tagged human FLI1 (NM_002017) cDNA) | GenScript | N/A |
| pTRIPdU3-MND-IRES-GFP lentiviral expression vector | Renou et al., 2017 | N/A |
| Software and Algorithms | ||
| CyTOF Software | Fluidigm | https://www.fluidigm.com/software |
| Cytobank | Kotecha et al., 2010 | https://mrc.cytobank.org |
| Cytofkit | Chen et al., 2016 | https://bioconductor.org/packages/3.4/bioc/html/cytofkit.html |
| SPRING | Weinreb et al., 2018a | https://kleintools.hms.harvard.edu/tools/spring.html |
| Skyline | MacLean et al., 2010 | https://skyline.ms/project/home/software/Skyline/begin.view |
| Other | ||
| Web tool for single-cell visualization of proteins during human erythropoiesis | This paper | http://trena.systemsbiology.net/login |
Highlights.
Human erythropoiesis reconstituted from temporal protein measurements in single cells
TFs from competing hematopoietic lineages are co-expressed in bipotential progenitors
Protein levels of TFs change gradually along the erythroid trajectory
Quantitative changes of lineage-specific TFs in early progenitors mediate cell fate decisions
ACKNOWLEDGMENTS
We thank J. Bieker of Icahn School of Medicine at Mount Sinai (New York), F. Pflumio of UMR967, INSERM/CEA (Paris), and D. Trono of EPFL (Lausanne) for providing reagents; C. Fisher of WIMM for experimental help; A. Chu and K. Sivaraman of OHRI for help with bioinformatic analyses; and F.J. Dilworth of OHRI for review of the manuscript. This work was supported by the NIH grant 1R01DK098449-01A1 (to M.B. and J.A.R.) and the Canada-UK Foundation (to M.B.). The WIMM Mass Cytometry Facility is supported by the MRC HIU core-funded project (MC_UU_00008) and the Oxford Single Cell Biology Consortium.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found with this article online at https://doi.org/10.1016/j.stem.2019.02.006.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- An X, Schulz VP, Li J, Wu K, Liu J, Xue F, Hu J, Mohandas N, and Gallagher PG (2014). Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood 123, 3466–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, Simonds EF, Bendall SC, Sachs K, Krutzik PO, and Nolan GP (2012). Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol 30, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouilloux F, Juban G, Cohet N, Buet D, Guyot B, Vainchenker W, Louache F, and Morlé F (2008). EKLF restricts megakaryocytic differentiation at the benefit of erythrocytic differentiation. Blood 112, 576–584. [DOI] [PubMed] [Google Scholar]
- Bresnick EH, Hewitt KJ, Mehta C, Keles S, Paulson RF, and Johnson KD (2018). Mechanisms of erythrocyte development and regeneration: implications for regenerative medicine and beyond. Development 145, dev151423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Kostadima M, Martens JHA, Canu G, Garcia SP, Turro E, Downes K, Macaulay IC, Bielczyk-Maczynska E, Coe S, et al. (2014). Transcriptional diversity during lineage commitment of human blood progenitors. Science 345, 1251033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lau MC, Wong MT, Newell EW, Poidinger M, and Chen J (2016). Cytofkit: a bioconductor package for an integrated mass cytometry data analysis pipeline. PLoS Comput. Biol 12, e1005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S, Notta F, Laurenti E, and Dick JE (2012). Hematopoiesis: a human perspective. Cell Stem Cell 10, 120–136. [DOI] [PubMed] [Google Scholar]
- Graf T, and Enver T (2009). Forcing cells to change lineages. Nature 462, 587–594. [DOI] [PubMed] [Google Scholar]
- Hoppe PS, Schwarzfischer M, Loeffler D, Kokkaliaris KD, Hilsenbeck O, Moritz N, Endele M, Filipczyk A, Gambardella A, Ahmed N, et al. (2016). Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature 535, 299–302. [DOI] [PubMed] [Google Scholar]
- Hu J, Liu J, Xue F, Halverson G, Reid M, Guo A, Chen L, Raza A, Galili N, Jaffray J, et al. (2013). Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood 121, 3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Guo YP, May G, and Enver T (2007). Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev. Biol 305, 695–713. [DOI] [PubMed] [Google Scholar]
- Ikonomi P, Rivera CE, Riordan M, Washington G, Schechter AN, and Noguchi CT (2000). Overexpression of GATA-2 inhibits erythroid and promotes megakaryocyte differentiation. Exp. Hematol 28, 1423–1431. [DOI] [PubMed] [Google Scholar]
- Kester L, and van Oudenaarden A (2018). Single-cell transcriptomics meets lineage tracing. Cell Stem Cell 23, 166–179. [DOI] [PubMed] [Google Scholar]
- Kotecha N, Krutzik PO, and Irish JM (2010). Web-based analysis and publication of flow cytometry experiments. Curr. Protoc. Cytom Chapter 10, Unit1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E, and Göttgens B (2018). From haematopoietic stem cells to complex differentiation landscapes. Nature 553, 418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el-A.D., Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. (2015). Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig C, Claassen M, Schmidt A, and Aebersold R (2012). Estimation of absolute protein quantities of unlabeled samples by selected reaction monitoring mass spectrometry. Mol. Cell Proteomics 11, M111.013987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, and MacCoss MJ (2010). Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti R, Park CY, and Weissman IL (2007). Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell 1, 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, and Dick JE (2011). Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science 333, 218–221. [DOI] [PubMed] [Google Scholar]
- Notta F, Zandi S, Takayama N, Dobson S, Gan OI, Wilson G, Kaufmann KB, McLeod J, Laurenti E, Dunant CF, et al. (2016). Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 351, aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, and Grimes HL (2016). Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537, 698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH (2000). Diversification of haematopoietic stem cells to specific lineages. Nat. Rev. Genet 1, 57–64. [DOI] [PubMed] [Google Scholar]
- Orkin SH, and Zon LI (2008). Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palii CG, Pasha R, and Brand M (2011). Lentiviral-mediated knockdown during ex vivo erythropoiesis of human hematopoietic stem cells. J. Vis. Exp 2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, and Aebersold R (2012). Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods 9, 555–566. [DOI] [PubMed] [Google Scholar]
- Psaila B, Barkas N, Iskander D, Roy A, Anderson S, Ashley N, Caputo VS, Lichtenberg J, Loaiza S, Bodine DM, et al. (2016). Single-cell profiling of human megakaryocyte-erythroid progenitors identifies distinct megakaryocyte and erythroid differentiation pathways. Genome Biol. 17, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renou L, Boelle PY, Deswarte C, Spicuglia S, Benyoucef A, Calvo J, Uzan B, Belhocine M, Cieslak A, Landman-Parker J, et al. (2017). Homeobox protein TLX3 activates miR-125b expression to promote T-cell acute lymphoblastic leukemia. Blood Adv. 1, 733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert OT, Mouritsen J, Ludwig C, Röst HL, Rosenberger G, Arthur PK, Claassen M, Campbell DS, Sun Z, Farrah T, et al. (2013). The Mtb proteome library: a resource of assays to quantify the complete proteome of Mycobacterium tuberculosis. Cell Host Microbe 13, 602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siripin D, Kheolamai P, U-Pratya Y, Supokawej A, Wattanapanitch M, Klincumhom N, Laowtammathron C, and Issaragrisil S (2015). Transdifferentiation of erythroblasts to megakaryocytes using FLI1 and ERG transcription factors. Thromb. Haemost 114, 593–602. [DOI] [PubMed] [Google Scholar]
- Spitzer MH, and Nolan GP (2016). Mass cytometry: single cells, many features. Cell 165, 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusi BK, Wolock SL, Weinreb C, Hwang Y, Hidalgo D, Zilionis R, Waisman A, Huh JR, Klein AM, and Socolovsky M (2018). Population snapshots predict early haematopoietic and erythroid hierarchies. Nature 555, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb C, Wolock S, and Klein AM (2018a). SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics 34, 1246–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb C, Wolock S, Tusi BK, Socolovsky M, and Klein AM (2018b). Fundamental limits on dynamic inference from single-cell snapshots. Proc. Natl. Acad. Sci. USA 115, E2467–E2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Papalexi E, Butler A, Stephenson W, and Satija R (2018). Molecular transitions in early progenitors during human cord blood hematopoiesis. Mol. Syst. Biol 14, e8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunder ER, Finck R, Behbehani GK, Amir el-A.D., Krishnaswamy S, Gonzalez VD, Lorang CG, Bjornson Z, Spitzer MH, Bodenmiller B, et al. (2015). Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc 10, 316–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass cytometry data is deposited in the FlowRepository database under accession numbers FR-FCM-ZYPS and FR-FCM-ZYPT. http://flowrepository.org/public_experiment_representations