Abstract
Cryptic transcription is widespread and generates a heterogeneous group of RNA molecules of unknown function. To improve our understanding of cryptic transcription, we investigated their transcription start site (TSS) usage, chromatin organization, and posttranscriptional consequences in Saccharomyces cerevisiae. We show that TSSs of chromatin-sensitive internal cryptic transcripts retain comparable features of canonical TSSs in terms of DNA sequence, directionality, and chromatin accessibility. We define the 5′ and 3′ boundaries of cryptic transcripts and show that, contrary to RNA degradation–sensitive ones, they often overlap with the end of the gene, thereby using the canonical polyadenylation site, and associate to polyribosomes. We show that chromatin-sensitive cryptic transcripts can be recognized by ribosomes and may produce truncated polypeptides from downstream, in-frame start codons. Finally, we confirm the presence of the predicted polypeptides by reanalyzing N-terminal proteomic data sets. Our work suggests that a fraction of chromatin-sensitive internal cryptic promoters initiates the transcription of alternative truncated mRNA isoforms. The expression of these chromatin-sensitive isoforms is conserved from yeast to human, expanding the functional consequences of cryptic transcription and proteome complexity.
Genomes are pervasively transcribed, producing a wide diversity of coding and noncoding RNAs (for reviews, see Wei et al. 2011; Jensen et al. 2013; Pelechano and Steinmetz 2013; Kaikkonen and Adelman 2018), raising the question of the biological significance of such transcriptional activity (Jensen et al. 2013). Some of those transcripts are functionally relevant, such as the well-characterized long noncoding RNAs, antisense transcripts, or alternative isoforms (for reviews, see Jensen et al. 2013; Pelechano and Steinmetz 2013; Pelechano 2017; Kaikkonen and Adelman 2018). However, it remains unclear which fraction of these transcripts exerts a biological role (direct or regulatory). This question is particularly difficult to address when these transcriptional units arise within, or in close proximity to, protein coding genes in the same strand. Thus, their transcription signals are difficult to distinguish from the nearby or even overlapped protein coding genes. Among pervasively produced transcripts, so-called cryptic transcripts constitute a particularly heterogeneous group. Cryptic transcription is typically defined as the production of noncanonical transcripts of unknown function (Wei et al. 2011), whereas canonical transcripts can be interpreted as those encoding a full-length functional protein. The breadth of this definition shows that, despite their abundance and potential relevance for gene expression, our knowledge of this process remains limited.
Cryptic transcripts can be classified according to the mechanisms by how cells control their abundance: Cryptic transcripts levels may be modulated either by restricting transcription initiation or by selectively degrading them (for review, see Jensen et al. 2013). For simplicity, we will refer to the first class of processes as “chromatin sensitive” and to the second class as “RNA degradation sensitive.” A classical example of chromatin-sensitive mechanisms is the emergence of cryptic transcripts from within gene bodies when nucleosome positioning is altered by impairing the function of histone chaperons such as Spt6p (Kaplan et al. 2003; Doris et al. 2018). Spt6p depletion causes decreased expression of most genic promoters while increasing the expression of intragenic ones, thus suggesting a potential competition for initiation factors (Doris et al. 2018). Likewise, the disruption of histone deacetylation patterns also leads to the appearance of intragenic cryptic transcripts. Specifically, interfering with the activity of the Rpd3S deacetylase complex, which recognizes histone 3 Lys36 trimethylation (H3K36me3) deposited by the histone methyltransferase Set2 during RNA polymerase II elongation, leads to intragenic cryptic transcription (Carrozza et al. 2005; Lickwar et al. 2009; Churchman and Weissman 2011; Chabbert et al. 2015; Malabat et al. 2015; Kim et al. 2016). In contrast, the second class of cryptic transcripts (RNA degradation sensitive) are constitutively produced and degraded by the cell and thus become detectable only when RNA degradation is impaired (Jensen et al. 2013). For instance, cryptic unstable transcripts (CUTs) are identified in mutant cells with depletion of the nuclear RNA exosome (e.g., rrp6Δ) (Neil et al. 2009; Xu et al. 2009).
Because of their proximity to or even overlap with protein-coding genes, dissecting the function of cryptic transcription units is especially complicated. In some contexts, cryptic transcription has been associated with “opportunistic transcription,” whereby RNA polymerase II is recruited to any open chromatin region, generating spurious molecules. However, annotating cryptic transcripts as functional or spurious is not trivial. This has been exemplified in multiple instances in which either the RNA product itself or the transcriptional activity per se may have a clear functional impact. For example, the act of transcription itself can regulate the expression of neighboring genes through chromatin modulation (Martens et al. 2004; Hainer et al. 2011; Xu et al. 2011; Kim et al. 2012; van Werven et al. 2012; Chia et al. 2017; Brown et al. 2018). On the other hand, previous reports have shown that cryptic promoters can drive the expression of alternative isoforms with different posttranscriptional regulation or even encode alternative protein isoforms (Carlson et al. 1983; Cheung et al. 2008; Fournier et al. 2012; Arribere and Gilbert 2013; Pelechano et al. 2013; Gupta et al. 2014; Lycette et al. 2016).
To improve the classification of such events and further improve our understanding of cryptic transcription, we performed a comprehensive characterization of transcription start sites (TSSs) in Saccharomyces cerevisiae. We performed the analysis of both the biogenesis of cryptic transcripts and their posttranscriptional life with a focus on those derived from chromatin-sensitive mechanisms (i.e., set2Δ, rco1Δ, and eaf3Δ). As a comparison, we also examined the biogenesis of the RNA degradation–sensitive CUTs (rrp6Δ; RNA degradation sensitive). We identified their TSSs and investigated their sequence preference and chromatin organization. To assess the posttranscriptional life of cryptic transcripts and better define their boundaries, we examined the association between TSS and polyadenylation site usage by transcript isoform sequencing (TIF-seq) (Pelechano et al. 2013). To investigate their coding potential, we performed polyribosome fractionation followed by 5′ cap sequencing to investigate the association of cryptic transcripts with polyribosomes. We examined the ribosome protection pattern of cryptic transcripts measured by 5PSeq (Pelechano et al. 2015), focusing on the signature associated with internal methionine codons predicted to act as novel start codons. Finally, we validate our prediction using available N-terminal mass spectrometry (MS) data (Varland et al. 2018). Our work aims to investigate the functional relevance of chromatin-sensitive cryptic transcripts.
Results
Chromatin-sensitive and RNA degradation–sensitive cryptic transcripts show distinct TSS profiles
To understand how cryptic transcripts are generated, we performed a genome-wide mapping of their TSSs in S. cerevisiae. We conducted 5′ cap sequencing (Pelechano et al. 2016), which enables a precise identification of the 5′ end of transcripts in a wild-type strain (BY4741) and multiple mutants associated with cryptic transcription (Fig. 1A). To illustrate chromatin-sensitive cryptic transcription, we examined the TSS profile of cells lacking Set2, the histone methyltransferase responsible for the cotranscriptional deposition of H3K36me3 (Carrozza et al. 2005). We also investigated the TSS profile of strains deficient in Rco1 and Eaf3, components of the Rpd3S histone deacetylase complex acting downstream from Set2. Furthermore, we examined the emergence of cryptic TSSs in cells deficient for Set1, the histone methyltransferase responsible for H3K4 methylation and associated with cryptic transcription from promoter-proximal regions (Kim et al. 2012; van Werven et al. 2012). Finally, we conducted a comparative analysis of the TSS profiles of CUTs that emerge upon depletion of the nuclear RNA exosome subunit Rrp6 (rrp6Δ) (Neil et al. 2009; Xu et al. 2009) as an example of RNA degradation–sensitive cryptic transcription.
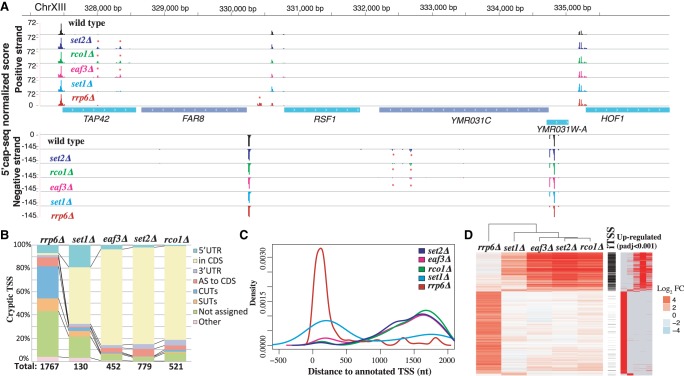
Figure 1.
Genome-wide identification of chromatin- and RNA degradation–sensitive TSSs. Detected chromatin-sensitive cryptic transcripts tend to overlap coding genes in the same orientation. (A) Representative 5′ cap sequencing track. Score (normalized counts) of collapsed replicates is shown (see Methods). Significantly differential expressed TSSs clusters are marked by * (P-adj <0.001). (B) Classification of differentially expressed TSSs in respect to annotated features. Annotation of stable unannotated transcripts (SUTs), CUTs, and UTR lengths are from Xu et al. (2009). (C) Distribution of differentially expressed TSSs in respect to annotated ORF-T TSSs. ORF-T refer to transcripts associated with canonical ORFs as described by strand-specific tiling arrays (Xu et al. 2009). (D) Relationship between TSSs identified in the analyzed strains. Each horizontal line represents an identified TSS cluster. On the left side, we display the relative fold change enrichment (FC) with respect to the wild-type strain in log2 (red, up-regulated, to blue, down-regulated). In black, we indicate which of those identified TSSs can be classified as iTSSs. Finally, significantly differentially expressed TSSs compared to wild type are shown at the right (in red). Only TSSs identified as differentially expressed with respect to the wild type in at least one condition are shown.
In total, 44,963 TSS clusters were identified across all data sets (Supplemental Table S1). We used information from our biological replicates and unique molecular identifiers (UMIs) to identify differentially expressed TSSs across strains (adjusted P-val < 0.001; see Methods) (Fig. 1B; Supplemental Fig. S1A). The disruption of the nuclear exosome (rrp6Δ) led to the highest number of up-regulated TSS clusters in comparison to the wild type (1767), whereas deletion of SET1 had a modest effect (130 TSS up-regulated clusters). The other mutant strains, set2Δ, rco1Δ, and eaf3Δ, presented an intermediate phenotype (i.e., with 779, 521, and 452 up-regulated TSS clusters, respectively). Up-regulated rrp6Δ-sensitive TSSs were detected in close proximity to the annotated TSSs of coding genes (often in opposite orientation to annotated TSSs) (Neil et al. 2009; Xu et al. 2009), whereas detectable set2Δ-, rco1Δ-, and eaf3Δ-sensitive TSSs occurred preferentially within the body of genes (Fig. 1C; Supplemental Fig. S1B; Carrozza et al. 2005; Lickwar et al. 2009). Strains with mutations affecting the same pathway (e.g., set2Δ, rco1Δ, and eaf3Δ) shared a high number of up-regulated cryptic TSSs, whereas cryptic TSSs resulting from disruption of the nuclear exosome (CUTs, rrp6Δ) were detected mainly outside of the coding regions (Fig. 1D; Supplemental Fig. S1C). We then characterized the intragenic up-regulated TSSs (iTSSs) that occur inside the coding region of genes and mostly originate from the Set2-Rco1-Eaf3 pathway (Fig. 1D). Our strand-specific detection approach enabled us to determine that most chromatin-sensitive cryptic iTSSs are detected in the same orientation as the corresponding ORF. This contrasts with what is observed for the RNA degradation–sensitive ones that arise more often antisense to the CDS than in the same orientation (red vs. yellow in Fig. 1B). Previous strand-specific RNA-seq analysis of the set2Δ strain has identified the presence of internal Set2-repressed antisense transcripts (SRATs) (Venkatesh et al. 2016). Our work confirms their finding (SRATs displayed in red in Fig. 1B; Supplemental Figs. S1A,E, S2) but further reveals that the vast majority of stable cryptic transcription overlaps the main transcript in the same orientation (yellow in Fig. 1B), a feature difficult to detect with conventional RNA-seq. To investigate the origin of the directionality of the chromatin-sensitive cryptic iTSSs, we reanalyzed NET-seq (Churchman and Weissman 2011), RNA-seq (Venkatesh et al. 2016), and alternative TSS data sets (Malabat et al. 2015) in addition to our data (Supplemental Figs. S1D,E, S2). This revealed that although nascent transcription arises bidirectionally from cryptic promoters, cryptic transcripts in the same orientation as the main ORF are more stable and thus accumulate to a higher level. In fact, chromatin-sensitive iTSSs can also be detected, albeit at a much lower level, in wild-type conditions (see below). The Winston laboratory has recently investigated the appearance of intragenic promoters upon Spt6p depletion (spt6-1004) (Doris et al. 2018). We compared up to what degree spt6-1004 up-regulated intragenic promoters overlap with the chromatin-sensitive cryptic iTSSs defined in this study (Supplemental Fig. S3). As can be observed, chromatin-sensitive cryptic iTSSs are only slightly increased in spt6-1004, whereas the vast majority of spt6-1004 up-regulated intragenic promoters are not up-regulated in a set2Δ strain (Supplemental Fig. S3A). Additionally, spt6-1004 has a clear effect decreasing the expression of canonical genic promoters, whereas set2Δ has a more punctuated effect in the body of the genes (Supplemental Fig. S3B). This suggest that, although related, these two pathways control different subsets of cryptic promoters that are only partially overlapping. To gain a better understanding of the regulation of the chromatin-sensitive iTSSs, we decided to focus our analysis on those iTSSs occurring in the same orientation as their overlapping coding gene.
Characterization of cryptic iTSS promoters
After identification of the putative promoter regions with cryptic iTSSs, we compared these with the canonical TSSs of protein-coding genes. iTSSs in all analyzed strains present a similar sequence composition to canonical TSSs, with a pyrimidine enrichment at the −1 and adenine at the 0 and −8 positions (Fig. 2A; Supplemental Fig. S4A; Zhang and Dietrich 2005; Pelechano et al. 2013). Please note that transcript position 0 as referred here (first nucleotide of the transcript) is traditionally referred also as +1, when using a scale without zero. Molecules derived from cryptic iTSSs can also be detected in wild-type cells, although at a lower level (Supplemental Fig. S1D). This suggests that cryptic iTSSs are used by at least a fraction of cells in normal growing conditions.
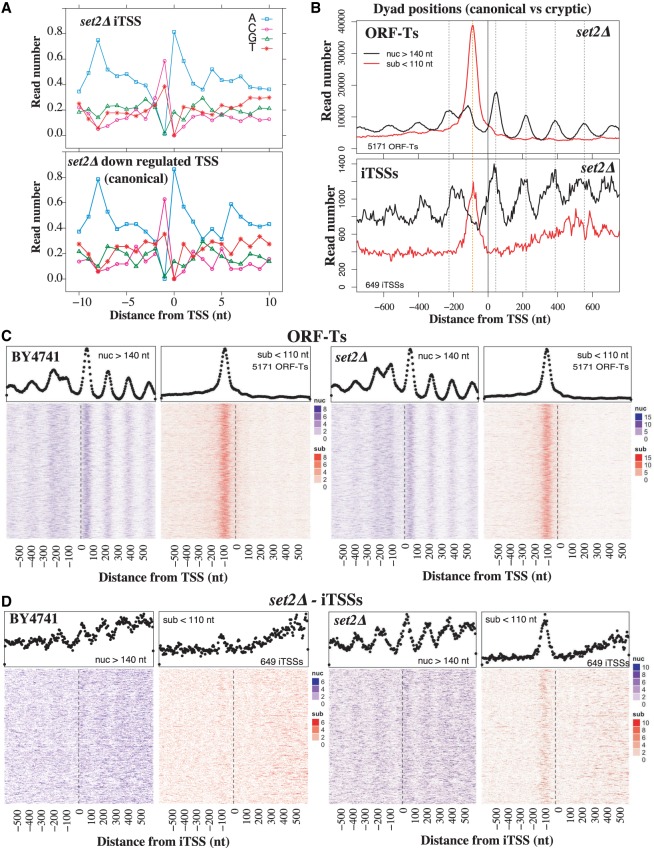
Figure 2.
The sequence and chromatin features of iTSSs resemble those of canonical TSSs. (A) Sequence preference of set2Δ iTSSs compared with canonical TSSs (set2Δ down-regulated that often overlap with canonical TSSs). (B) MNase protection pattern for canonical ORF-T TSSs. MNase fragments are distributed in nucleosome protection fragments (nuc) and subnucleosomal ones (sub) according to their length. Vertical dotted lines depict canonical dyad nucleosome axes (in black) and putative TF binding sites (in red). (C) Heatmaps depicting in detail the MNase protection pattern for canonical ORF-T TSSs in the wild-type strain and set2Δ. Each line of the heatmaps corresponds to an analyzed region for nucleosome fragments (in blue) and subnucleosomal fragments (in red) ordered by gene expression (Xu et al. 2009). The metagene with aggregation of all the heatmap information is shown above in black dots. (D) Heatmaps depicting in detail the MNase protection pattern for set2Δ iTSSs as in C. Chromatin data are reanalyzed from Chabbert et al. (2015). Heatmap sorted by iTSS expression level.
Given that chromatin-sensitive iTSSs resemble canonical gene-coding TSSs in their base composition and directionality, we assessed whether this also applies to their chromatin organization. We used information on nucleosomal and subnucleosomal fractions from our previous high-throughput ChIP-seq experiments (Fig. 2C; Supplemental Figs. S4B, S5A; Chabbert et al. 2015, 2018) to analyze the MNase protection pattern around cryptic iTSSs. Cryptic iTSSs present the same MNase protection architecture as canonical TSSs, with an organized nucleosome array downstream from the TSS and a subnucleosomal protection site overlapping the region at which transcription factors (TFs) would typically associate (Fig. 2D; Supplemental Fig. S5B; Henikoff et al. 2011). This is particularly evident for the Set2-Eaf3-Rco1–sensitive iTSSs, as they are further away from canonical TSSs (Fig. 1C) and thus easier to disentangle from the MNase pattern associated with canonical promoters (Fig. 2D). A similar, although more discrete pattern (i.e., nucleosome array and upstream subnucleosomal protection pattern) can also be observed around the same iTSSs in the wild-type strain (Fig. 2D; Supplemental Fig. S5B). The subnucleosomal fragments are only apparent when analyzing whole-cell extract, and are depleted after histone immunoprecipitation (Supplemental Fig. S6A). This suggests either that histones are not bound to those fragments or that they cannot be efficiently immunoprecipitated in our experimental conditions. The distance between the iTSSs and the first nucleosome downstream (analogous to the +1 nucleosome) is similar to the distance present in canonical TSSs and the dyad axis (Fig. 2B; Supplemental Fig. S5). However, the nucleosome-depleted regions (NDRs) commonly associated with promoters are a bit smaller in the case of the “cryptic promoters” of iTSSs. In our experimental conditions, we estimate that canonical NDRs are ∼275 nt, whereas iTSS NDRs are 215 nt and the distance between + 1/+2 nucleosome dyads is 165 nt (Supplemental Fig. S5). The presence of a periodic nucleosome organization in gene bodies around an internal “NDR” upstream of the cryptic iTSSs, suggest that iTSSs tend to occur or contribute to synchronizing, regular nucleosome arrays that are detectable even in mixed cell populations. This, together with the detection of a basal level of cryptic iTSS expression (Supplemental Fig. S1D), suggests that a small proportion of cells are expressing these cryptic transcripts even under normal conditions. This is in agreement with the fact that iTSS NDRs are longer than the average distance between nucleosome pairs even in a wild-type strain (Supplemental Fig. S5B). This suggests that factors or genome features may actively make these internucleosome regions distinct. Additionally, our observation that cryptic iTSSs may be bound by TF at low levels even in normal conditions is in agreement with recent evidence suggesting that TFs such as Gcn4 can also bind and activate internal promoters (Rawal et al. 2018).
To confirm that cryptic iTSSs present the canonical marks associated with promoter activity, we analyzed other chromatin features. We focus on chromatin-sensitive iTSSs that in general are distant from the canonical TSSs and thus not obscured by canonical promoter marks (Fig. 1C). We observed an increased signal of H3K4me3 at the first nucleosome (+1 nucleosome) downstream from the iTSSs in set2Δ that decreases downstream from the cryptic promoters (Supplemental Fig. S6B). As expected, this is only apparent in this mutant strain as cryptic transcripts are expressed at a sufficient level to be detectable.
Posttranscriptional life of iTSS-derived transcripts
Once we confirmed that iTSSs present a canonical promoter structure, we sought to determine the complete length of the transcripts derived from iTSSs in order to gain information on their posttranscriptional life. We applied our previously developed TIF-seq (Pelechano et al. 2013) approach that allows to jointly and unambiguously determine the start and end sites (TTSs) of each RNA molecule within a sample. We thus identified the start and end sites of all transcripts, including the chromatin-sensitive transcripts initiating from iTSSs. We further compared the TSSs and TTSs of iTSS-initiated transcripts to those of canonical transcripts. We identified that most transcripts originating from an iTSS in the set2Δ strain use the same polyadenylation sites as the canonical mRNAs. This was observed at both the individual and genome-wide levels (Fig. 3).
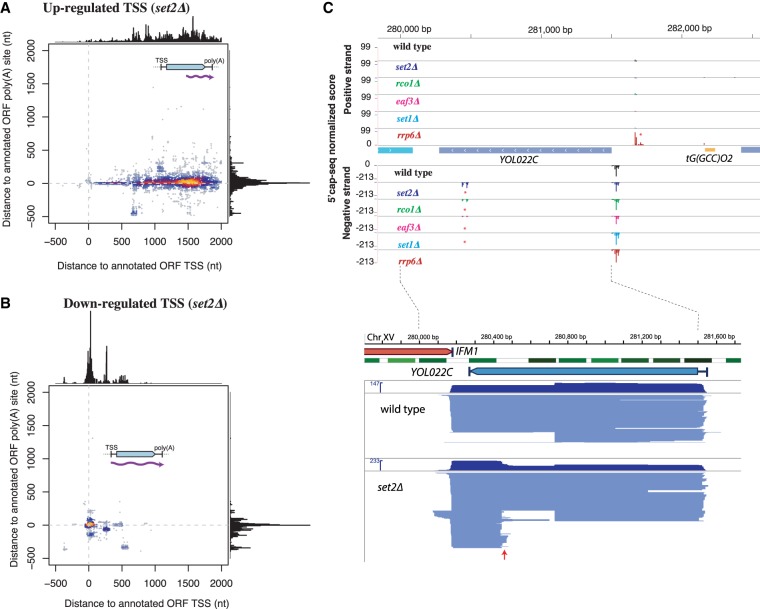
Figure 3.
Full-lengths of set2Δ iTSS-derived transcripts use canonical polyadenylation sites. (A) The TSS and TTS comparison between set2Δ iTSSs-initiated transcripts and annotated ORF-T boundaries (Xu et al. 2009). set2Δ iTSS-derived transcripts originate within the body of the gene (internal 5′) but use canonical 3′ polyadenylations sites. (B) Down-regulated TSSs in set2Δ use canonical 5′ and 3′ sites. (C) Example of TIFSeq coverage for the YOL022C gene as an example. The upper part shows TSS mapping (as in Fig. 1A). In the bottom part, we show full-length transcript in blue. Each line connecting between one identified TSS and poly(A) site represents one full-length transcript. The red arrow indicates the appearance of a set2Δ-sensitive iTSS. Nucleosomes are shown in green (Venters and Pugh 2009).
Specifically, most transcripts emerging from an iTSS in set2Δ originate within the gene body but use the canonical polyadenylation sites (Fig. 3A). This confirms and expands previous evidence from northern blot analysis (Kaplan et al. 2003; Carrozza et al. 2005). In contrast, TSSs down-regulated in set2Δ, which in the vast majority correspond to canonical mRNA TSSs, generate transcripts that also use the canonical polyadenylation sites (Fig. 3B; Xu et al. 2009). These suggest that stable chromatin-sensitive cryptic transcripts have the potential to encode N-terminal truncated proteins.
As most chromatin-sensitive cryptic iTSSs can produce 5′ truncated mRNAs, we further investigated if they are associated with ribosomes. We investigated their ribosome association as those molecules are present at low levels even in wild-type conditions, which could function as alternative mRNA isoforms. Additionally, previous work showed that a fraction of internal cryptic transcripts is degraded thought nonsense mediated decay (NMD), and thus putatively interacts with the translation machinery enough to be surveyed by NMD (Malabat et al. 2015). To measure association with ribosomes of the stable chromatin-sensitive cryptic transcripts, we combined isolation of polyribosomes by sucrose fractionation with 5′ cap sequencing (Supplemental Table S2). As expected, ORF-T TSSs are associated with polyribosome fractions, whereas noncoding RNAs such as stable unannotated transcripts (SUTs) or CUTs are much less associated (Fig. 4A; Supplemental Fig. S7A,B; Xu et al. 2009). Although the bulk of CUTs and SUTs is not preferentially associated with ribosomes, a fraction of them could encode peptides (see below). mRNA molecules originating from chromatin-sensitive cryptic iTSSs are also enriched in the heavy polyribosome fractions that are associated with active translation, and this association does not seem to depend on the length of the cryptic 5′ UTR (Supplemental Fig. S7C). This suggests that cryptic transcripts, especially those originating from chromatin-sensitive cryptic promoters, associate with ribosomes and have the potential to produce truncated proteins.
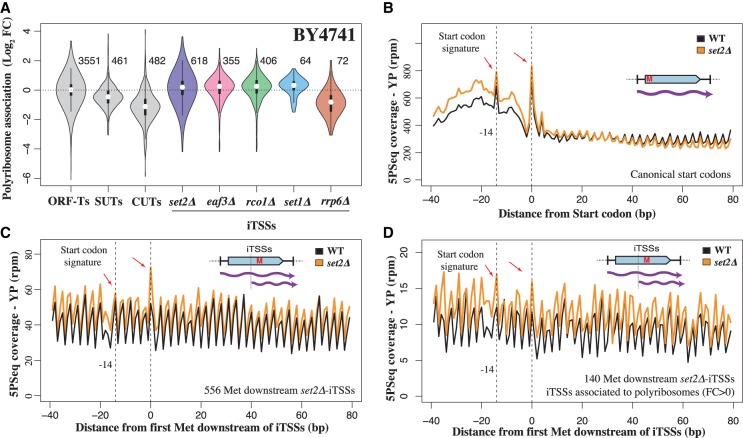
Figure 4.
A fraction of iTSS-derived transcripts associate to ribosomes, and the internal methionine can be recognized as a novel start codon. (A) Relative association with polyribosome fraction after sucrose fractionation versus total extract. Analyzed events (present at a sufficient level in the wild-type strain) are indicated to the right of each plot. (B) Example of 5PSeq start codon–associated signature after glucose depletion for coding genes. To decrease the effect of potential outliers, we assigned a value corresponding to the 95th percentile to values that were over this threshold at each distance from the start codon. (C) Start codon–associated signature after glucose depletion for predicted novel start codons in set2Δ iTSS-derived transcripts. Those positions are expected to behave as internal methionines in a wild-type strain. (D) As in C, but showing the subset of cryptic start codons in which mRNAs are more associated with polyribosomes (fold change >0 in A).
To assess whether ribosome-bound cryptic transcripts also are engaged in active translation, we assayed the ability of ribosomes to recognize such cryptic transcripts. To this aim, we used our previously developed 5PSeq approach, which measures ribosome dynamics by sequencing cotranslational mRNA degradation intermediates (Pelechano et al. 2015; 2016). We have previously shown that yeast cells in slow growth conditions such as growth in minimal media or stationary phase present a characteristic ribosome protection pattern at the translation start codon consistent with inhibition of translation initiation (Pelechano et al. 2015; Pelechano and Alepuz 2017). To distinguish the translation of the canonical full-length mRNAs from the shorter overlapping transcripts derived from iTSSs, we applied 5PSeq in glucose starvation to test if iTSS-initiated transcripts show a translation start codon pattern (Zid and O'Shea 2014). In fact, we identify a 5PSeq protection pattern at −14 nt and at the start codon (Fig. 4B; Supplemental Fig. S8). Initially, we tried to enhance the start codon signature using cycloheximide treatment, as it leads to a sharp increase of protection at −14 nt. However, as expected for an inhibitor of translation elongation, cycloheximide also leads to a massive increase of internal 5PSeq protection that obscures the signature of any internal cryptic translation start site (Supplemental Fig. S8E,F). To enhance the observed start codon signature, we exposed cells to a glucose-free media for 5 min. By limiting translation initiation, we increased the start codon signature and allowed the ribosomes engaged in translation to run-off the mRNA (Z Zhang and V Pelechano, in prep.), an effect that can be readily observed at the canonical start codons of annotated protein-coding genes (Fig. 4B; Supplemental Fig. S8A,B). We then analyzed the ribosome pattern associated with internal methionines and focused on those in-frame that could potentially be recognized as new start codons in transcripts derived from cryptic iTSSs but not in full-length mRNAs. We observed the start codon signature in the set2Δ strain but not in the wild-type strain (Fig. 4C,D). The detection of these start codon signatures is challenging as, even in the set2Δ strain, the ribosome protection pattern is a composite of the translation signatures of both canonical and iTSSs-derived transcripts. This result suggests not only that cryptic transcripts are associated with polyribosomes but that ribosomes can identify new start codons as canonical ones. Our 5PSeq analysis of the RNA degradation–sensitive transcripts (CUTs; up-regulated in rrp6Δ) revealed that those also could encode peptides (Supplemental Fig. S8C,D). The first predicted ORFs downstream from the CUT TSSs present a clear translation initiation signature and also a protection peak at 17 nt upstream of the stop codon (as expected from a terminating ribosome). This effect was especially clear in those CUTs not overlapping with canonical transcripts (i.e., non-iTSSs).
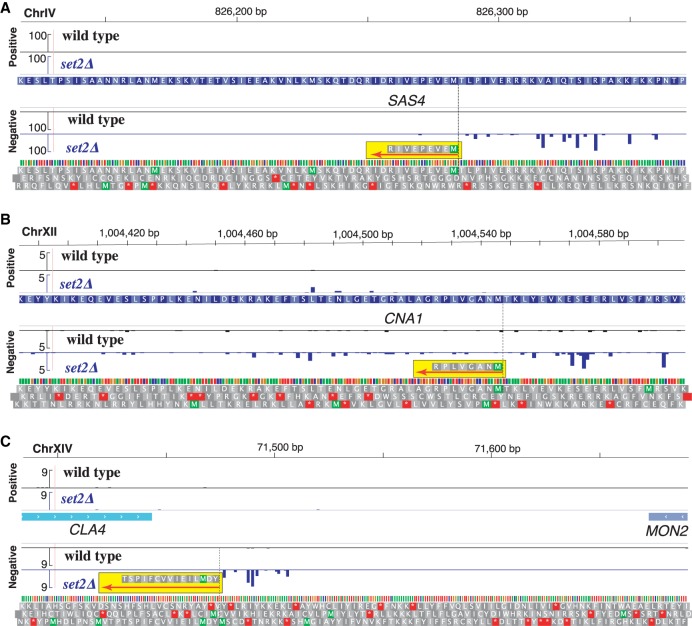
Finally, we analyzed whether our predicted truncated polypeptides matched acetylated N termini of proteins using a recently published proteomics data set as a reference (Varland et al. 2018). In the original study, the investigators identify 1056 canonical protein N-terminal sites in a wild-type strain using N-terminal COFRADIC, which is a technique that maps modified N termini of proteins on a global scale (Staes et al. 2011). As chromatin-sensitive iTSSs are expressed, even to a lower level, also in a wild type, we were able to detect after proteomic reanalysis seven iTSS-derived polypeptides (for details, see Methods) (Supplemental Table S3). Specifically, we confirmed the expression of truncated proteins for SAS4, ORC1, SWC4, CNA1, NST1, and SMC5 (Fig. 5A,B; Supplemental Fig. S9) and the expression of an iTSS-dependent peptide encoded in the 3′ UTR of MON2 (Fig. 5C). In addition, by comparing our 5′cap data set with the one obtained by Doris et al. (2018) for spt6-1004, we can identify truncated transcripts previously shown by western blot to produce also truncated proteins (Cheung et al. 2008).
Figure 5.
Chromatin-sensitive iTSSs encode peptides that can be detected by MS. Sequencing tracks display the 5′ cap sequence score (normalized counts) of collapsed replicates for wild type (in black) and Δset2 (in blue). Identified N-terminal peptides are highlighted in yellow, and their orientations are displayed using a red arrow. We display in gray the three potential translations of DNA in the same orientation of the detected peptide. (A) Truncation of SAS4 (MEVEPEVIR). (B) Truncation of CNA1 (MNAGVLPR). (C) Chromatin-sensitive transcript encoding a peptide in the 3′ UTR of MON2 (YDMLIEIVVCFIPST). N-terminal COFRADIC data from Varland et al. (2018).
Discussion
Here, we have shown that chromatin-sensitive cryptic promoters present multiple features similar to canonical gene-coding promoters. We focused on set2Δ-, rco1Δ-, and eaf3Δ-sensitive internal cryptic TSSs and showed that their DNA sequence, transcription directionality, and chromatin organization are similar to those of canonical promoters (Figs. 1, 2). This is in line with the characterization of cryptic promoters in the chaperone mutant spt6-1004 that was published during the review of this manuscript (Doris et al. 2018). Our MNase footprint analysis showed that those promoters present a canonical nucleosome array organization and suggested that canonical TFs bind upstream of the iTSSs in the body of genes and are associated with the appearance of intergenic NFR (Fig. 2). Our observations are in agreement with recent reports that show how Gcn4 binds frequently in coding regions and can activate transcription from internal promoters (Mittal et al. 2017; Rawal et al. 2018). This suggests that a significant fraction of the cryptic promoters are in fact alternative promoters, whose expression under standard conditions is restricted by the chromatin organization or the absence of a particular transcription factor. Previous studies have shown that a significant number of Set2-repressed cryptic promoters can be regulated by carbon sources (Kim et al. 2016). Altogether, this suggests that our classification of cryptic and canonical promoters may be influenced by the environmental conditions under which cells are profiled.
To assess to what degree these cryptic iTSSs could represent bona fide alternative transcript isoforms, we investigated their full boundaries (Fig 3). By using our previously developed TIF-seq approach, we identified that most of them use the canonical polyadenylation sites used by full-length isoforms. Previous work from the Jacquier laboratory has shown that, by studying the double mutant upf1Δ set2Δ, a proportion of internal cryptic transcripts are degraded by NMD (Malabat et al. 2015). Here we focused on the molecules that are present at a detectable level with the active NMD pathway and are thus more likely to have a posttranscriptional effect. We found that, even in a wild-type strain, chromatin-sensitive iTSSs are typically associated with polyribosome fractions. To further dissect if these short isoforms are not only bound to polyribosomes but actually translated, we applied an optimized version of our 5PSeq approach. We identified that the first methionine in the truncated transcripts presents a ribosome protection signature characteristic of translation start sites. In contrast, this signal is not detected in the wild-type strain, in which truncated isoforms are expressed at low levels. Finally, we reanalyzed a proteomics data set of N-terminally acetylated protein N termini expressed in wild-type cells (Varland et al. 2018), and we found newly truncated protein isoforms based on our isoform predictions. Our observation extends on previous observations from our group and others showing that variations in the transcripts’ 5′ boundaries potentially leading to truncated proteins are common in yeast. Our results are in line with seminal work from the Winston group showing that the histone chaperone mutant spt6-1004 can produce truncated proteins as analyzed by western blot (Cheung et al. 2008). These variations may be environmentally regulated or occur simultaneously in a apparently homogenous population of cells (Carlson et al. 1983; Fournier et al. 2012; Pelechano et al. 2013; Lycette et al. 2016; Varland et al. 2018). Independently of our results, in the future it will be necessary to further confirm the existence of all predicted truncated proteins by direct methods such as MS and characterize their functional relevance in the particular cell systems studied.
N-terminal proteomics approaches showed that downstream in-frame methionines often define alternative N termini in the budding yeast proteome (Fournier et al. 2012; Lycette et al. 2016; Varland et al. 2018). These alternative proteoforms can be detected even in standard laboratory conditions, suggesting that their expression coexists with the full-length proteoforms. However, most studies focused their analysis on the transcripts’ first 100 nt and thus did not investigate the downstream truncations that were commonly disregarded as cryptic transcripts. A similar phenomenon has been described in human cells, in which alternative N-terminal proteoforms can lead to different protein stability (Gawron et al. 2016; Na et al. 2018). Regardless of their origin, it is clear that truncated proteins can have significant phenotypical impacts such as changes in protein localization (Carlson et al. 1983) or may even act as dominant-negative factors opposing the function of the full-length protein (Ungewitter and Scrable 2010). Our results also reveal that a fraction of CUTs have also the potential of encoding peptides. As CUTs are naturally unstable, the potential production of peptides would be in principle also transient. In the future, further characterizing the abundance and functionality of alternative proteoforms derived from previously considered “cryptic” transcripts will be extremely valuable.
Although we focused our study on budding yeast, our conclusion that chromatin-sensitive cryptic iTSSs may act as alternative canonical TSSs have further implications. In mammals, alternative TSSs and TTSs, rather than alternative splicing, accounts for the majority of isoform differences across tissues (Reyes and Huber 2018). This highlights the importance of TSS selection in the definition of the transcriptome. It has been recently reported that the treatment of human cancer cell lines with DNA methyltransferase and histone deacetylase inhibitors (DNMTi and HDACi, respectively) results in the appearance of thousands of unannotated TSSs (TINATs) (Brocks et al. 2017). TINATs frequently splice into coding-protein exons and, in some cases, are associated with polyribosomes. Thus, disruption of the epigenome by the DNMTi and HDACi treatments leads to the expression of cryptic TSSs similar to the chromatin-sensitive iTSSs defined here, both in terms of biogenesis and potential posttranscriptional consequences. This suggests that the expression of cryptic TSSs is likely to be evolutionary conserved and a source of alternative (functional or aberrant) proteoforms that should be further investigated. The study of chromatin-sensitive cryptic promoter regulation will help to better distinguish spurious transcripts from those functionally relevant although only expressed in a subpopulation of cells or under specific environmental conditions.
Methods
Cell growth
All S. cerevisiae strains used in this study were derived from BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). BY4741, rrp6Δ (rrp6::kanMX4), set2Δ (set2::kanMX4), rco1Δ (rco1::kanMX4), and eaf3Δ (eaf3::kanMX4) were obtained from Euroscarf. set1Δ (set1::kanMX4) was generated using standard yeast chemical transformation as previously described (Chabbert et al. 2015). Cells were grown in YPD (1% yeast extract, 2% peptone, 2% glucose, and 40 mg/L adenine) and harvested at OD600 ∼ 1. For 5PSeq start codon identification, cells were shifted for 5 min to YP media without glucose (1% yeast extract, 2% peptone) before harvesting. For 5PSeq in presence of cycloheximide, 0.1 mg/mL final cycloheximide was added for 10 min before harvesting. Total RNA was phenol extracted using standard methods, and contaminant DNA was removed by DNase treatment (Turbo DNA-free kit, Ambion) (Pelechano et al. 2012).
5′ cap library preparation
Identification of 5′ capped mRNAs was performed as previously described (Pelechano et al. 2016). In brief, 10 µg total RNA was treated with calf intestinal alkaline phosphatase (NEB) to remove 5′P from fragmented and noncapped molecules. After purification, mRNA caps were removed using 3.75 units of Cap-Clip (Biozyme) exposing a 5′P in those molecules previously capped. Samples were ligated overnight at 16°C with a DNA/RNA oligo (rP5_RND: TTTCCCTACACGACGCTCTTCCGATrCrUrNrNrNrNrNrNrNrN) using T4 RNA ligase 1 (NEB). RNA integrity after ligation was assayed by agarose gel electrophoresis, and poly(A) RNA was purified using oligo dT magnetic beads. After this, ligated mRNA was fragmented for 5 min at 80°C in the presence of RNA fragmentation buffer (40 mM Tris-acetate at pH 8.1, 100 mM KOAc, 30 mM MgOAc). Ligated RNA was subjected to reverse transcription using random hexamers with SuperScript II (Life Technologies) with the following program: 10 min at 25°C, 50 min at 42°C, and heat inactivated for 15 min at 72°C. Second-strand cDNA synthesis was performed by a single PCR cycle (1 min at 98°C; 2 min at 50°C, and 15 min at 72°C) using a Phusion high-fidelity PCR master mix (NEB). A biotinylated oligo (BioNotI-P5-PET: [Btn]TATAGCGGCCGCAATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT) was added during the generation of the second cDNA strand. Double-stranded cDNA was purified using Ampure XP (Beckman Coulter) or HighPrep (MagBio) beads. After the samples were bound to streptavidin-coated magnetic beads (M-280 Dynabeads, Life Technologies) and subjected to standard Illumina end-repair, dA addition and adapter ligation were performed as previously described (Pelechano et al. 2016). Libraries were enriched by PCR and sequenced in an Illumina HiSeq 2000 instrument.
TIF-seq sequencing
TIF-seq libraries were performed as previously described (Pelechano et al. 2013) using 60 µg of DNA-free total RNA as input. In brief, 5′ noncapped molecules were dephosphorylated using 6 units of shrimp alkaline phosphatase (Fermentas). RNA was phenol purified, and the 5′P of capped molecules was exposed by treatment with 5 units of tobacco acid pyrophosphatase (Epicentre). RNA samples were ligated with the TIF-seq DNA/RNA 5oligo cap using T4 RNA ligase 1 (NEB). Full-length cDNA (FlcDNA) was generated with SuperScript III reverse transcriptase and amplified by PCR with HF Phusion master mix (Finnzymes).
FlcDNA was digested with NotI (NEB) to generate cohesive ends. Samples were subjected to intramolecular ligation using T4DNA ligase. TIF-seq chimeras were controlled mixing two aliquots of differentially barcoded FlcDNA during the ligation, as described in the original TIF-seq manuscript. Noncircularized molecules were degraded using exonuclease III and exonuclease I (NEB). Circularized FlcDNA was fragmented by sonication using a Covaris S220 (4 min, 20% duty cycle, intensity 5, 200 cycles/burst). Fragmented DNA was purified, and biotin-containing fragments were captured with streptavidin-conjugated Dynabeads M-280 (Invitrogen). Forked barcoded adapters were added using the standard Illumina DNA-seq library generation protocols. Libraries were enriched by 20 cycles of PCR Phusion polymerase (Finnzymes); 300-bp libraries were isolated using e-Gel 2% SizeSelect (Invitrogen) and sequenced in an Illumina HiSeq 2000 instrument (105 paired-end sequencing).
Polyribosome fractionation
One hundred milliliters of S. cerevisiae cells at OD600 ∼ 1 was treated with cycloheximide for 5 min (100 µg/mL, final concentration), harvested by centrifugation, and transferred to ice. Pellets were washed with ice-cold lysis buffer and resuspended in 700 µL lysis buffer. Lysis buffer contains 20 mM Tris-HCl (pH 8), 140 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 1% Triton X-100, 100 µg/mL cycloheximide, 500 µg/mL heparin, and complete EDTA-free protease inhibitor (one tablet per 10 mL, Sigma-Aldrich). For cell lysis, samples were transferred to precooled 1.5-mL screw-tubes with 300-µL glass beads and supplemented with 100 units of RNase inhibitor (RNasin plus, Promega). Cells were lysed using a FastPrep-24 shaker (6.0 m/s for 15 sec, MP biomedicals). Supernatant was recovered after 5-min centrifugation at 2300g and cleared with an additional centrifugation at 5900g. Extracts were supplemented with glycerol (5% final v/v) and stored at −70C, and 10%–50% sucrose gradients were prepared with a gradient master BIOCOMP (Nycomed Pharma). Sucrose solution contains 20 mM Tris-HCl (pH 8), 140 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 100 µg/mL cycloheximide, and sucrose (from 10% to 50%). Cleared cell extracts were ultracentifuged at 34,400 rpm for 2 h 40 min at 4°C using a C-1000 XP centrifuge with SW40 rotor (Beckman Coulter). Gradient UV absorption at 254 nm was measured, and selected fractions were selected for 5′ cap library preparation (5 µg purified RNA per sample). Polyribosome fraction (i.e., 2n+) was compared with the total extract before fractionation).
5PSeq
5PSeq libraries were prepared as previously described (Pelechano et al. 2015; 2016). 5PSeq protocol is the same as the one described for 5′ cap sequencing (see above) with variations only for the RNA ligation and rRNA depletion. Specifically, 6 µg of total RNA was directly ligated with a DNA/RNA oligo (rP5_RND). In that way, only molecules with a 5′P in the original sample are ligated. Ribosomal RNAs were depleted using ribo-zero magnetic gold kit (Illumina). Samples were sequenced in an Illumina NextSeq 500 instrument.
Bioinformatic analysis
For 5′ cap sequencing reads, random barcodes were first extracted and added to the reads name. The reads were aligned to yeast genome (S. cerevisiae genome (SGD R64- 1-1; sacCer3) with Novoalign (http://www.novocraft.com) using default setting. A customized script adapted from UMI-tools (Smith et al. 2017) was used for removing PCR duplicates (Supplemental Code S1). Specifically, we allowed 1-bp shifting at the beginning of 5′ ends. CAGEr was used for clustering the 5′ cap TSSs of BY4741 wild-type strain and the mutants (Haberle et al. 2015). TSS counts in different samples were normalized to match a common reference power-law distribution. Low-fidelity tags supported by less than two normalized counts in all samples were filtered out before clustering. In each sample, neighboring tags within 20 bp were spatially clustered into larger tag clusters. If the tag clusters were within 10 bp apart, they were aggregated together into nonoverlapping consensus clusters across all samples. The raw expression counts of the consensus clusters were further exported to the DESeq2 (Love et al. 2014) for differential expression analysis, comparing between mutants and wild-type strain. Polyribosome-derived 5′ cap sequencing reads were assigned to the consensus clusters by featureCounts (Liao et al. 2014), with read counting based on the 5′-most base. Differential expression analysis of polyribosome fractionation against total extract was performed using DESeq2.
Bar-ChIP sequencing data were processed as described previously (Chabbert et al. 2015).
TIF-seq sequencing data were processed as described previously (Pelechano et al. 2013). In general, all reads were first demultiplexed, and random barcodes were extracted. Pairs of transcript 5′ and 3′ end reads were mapped to yeast genome (S. cerevisiae genome; SGD R64- 1-1, sacCer3) with Novoalign (http://www.novocraft.com) using a default setting separately. Only transcripts with both ends mapped in that same chromosome at a length ranging from 40 to 5000 bp were used for further analysis.
5PSeq reads were mapped to the S. cerevisiae (genome R64-1-1) using STAR 2.5.3a (Dobin et al. 2013) with default parameters except AlignIntronMax (2500). PCR duplicates were removed as described for 5′ cap sequencing. Reads were aligned to either the start codon or the first in-frame methionine downstream from set2Δ-specific iTSS.
We analyzed the MS raw data from Varland et al. 2018 (PRIDE: PXD004326), including our additional predictions. MS/MS peak lists were searched essentially as described by Varland et al. (2018) using the Sequest database (Thermo Fisher Scientific). Spectral searches were performed using the UniProtKB S. cerevisiae database (version 2018_08) supplemented with the putative truncated proteins encoded by in-frame methionines downstream from iTSSs. To maximize our ability to detect iTSS-derived N-terminal peptides expressed also in the wild-type strain, we relaxed the stringency of the iTSS selection to P-adjusted <0.05. 13C2D3-acetylation of lysine side-chains, carbamidomethylation of cysteine, and methionine oxidation to methionine-sulfoxide were set as fixed modifications. 13C2D3-acetylation, acetylation of protein N termini, and pyroglutamate formation of N-terminal glutamine were set as a variable modification. Mass tolerances on precursor ions were set to 10 ppm and on fragment ions to 0.5 Da. The estimated false-discovery rate by searching decoy databases was <1%. Similar results were obtained using the Mascot search database (version 2.5, Matrix Science).
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE119114, GSE119160, GSE118758, GSE119134, and GSE128599.
Competing interest statement
C.D.C. is a full-time employee of Roche and a stakeholder in AstraZeneca.
Supplementary Material
Acknowledgments
We thank all members of Steinmetz, Pelechano, and Wei laboratories for discussion. We thank Petra Jakob, Manu Tekkedil, and Sandra Clauder-Münster for technical assistance. We thank Bruno Galy for his help with polyribosome fractionation. This study was technically supported by the European Molecular Biology Laboratory (EMBL) Genomics Core Facility, the Science for Life Laboratory (Sweden), and computational resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX). This study was financially supported by the National Key R&D Program of China (2017YFC0908405) and National Natural Science Foundation of China (grant no. 81870187) to W.W.; by the U.S. National Institutes of Health (NIH grant P01 HG000205), Deutsche Forschungsgemeinschaft (1422/4-1), and a European Research Council Advanced Investigator Grant to L.M.S.; and by the Swedish Research Council (VR 2016-01842), a Wallenberg Academy Fellowship (KAW 2016.0123), the Swedish Foundations’ Starting Grant (Ragnar Söderberg Foundation), and Karolinska Institutet (SciLifeLab Fellowship, SFO, and KI funds) to V.P. V.P. and W.W. acknowledge the support from a joint China–Sweden mobility grant from STINT (CH2018-7750) and the National Natural Science Foundation of China (grant no. 81911530167), respectively; B.P.H. and S.H.A. were supported by a fellowship from the EMBL Interdisciplinary Postdoctoral (EIPOD) program under Marie Sklodowska-Curie Actions COFUND (grant no. 291772). Y.Z. is funded by a fellowship from the China Scholarship Council. C.D.C. was supported by a PhD fellowship from the Boehringer Ingelheim Fonds.
Author contributions: Conceptualization was by V.P., W.W., and L.S.M. Acquisition of data and interpretation were by V.P., B.P.H., Y.Z., Y.P.S., C.D.C., and S.H.A. Computational analysis and interpretation were by W.W., V.P., J.W., I.P., and C.D.C. Writing—original draft preparation—was by V.P. and W.W. Writing—review and editing—was by V.P., W.W., B.P.H., J.W., Y.Z., Y.P.S., C.D.C., S.H.A., I.P., and L.S.M. Supervision was by V.P. and L.S.M. Funding acquisition was by L.S.M., V.P., and W.W.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.243378.118.
References
- Arribere JA, Gilbert WV. 2013. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res 23: 977–987. 10.1101/gr.150342.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocks D, Schmidt CR, Daskalakis M, Jang H-S, Shah NM, Li D, Li J, Zhang B, Hou Y, Laudato S, et al. 2017. DNMT and HDAC inhibitors induce cryptic transcription start sites encoded in long terminal repeats. Nat Genet 49: 1052–1060. 10.1038/ng.3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T, Howe FS, Murray SC, Wouters M, Lorenz P, Seward E, Rata S, Angel A, Mellor J. 2018. Antisense transcription-dependent chromatin signature modulates sense transcript dynamics. Mol Syst Biol 14: e8007 10.15252/msb.20178007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, Taussig R, Kustu S, Botstein D. 1983. The secreted form of invertase in Saccharomyces cerevisiae is synthesized from mRNA encoding a signal sequence. Mol Cell Biol 3: 439–447. 10.1128/MCB.3.3.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia W-J, Anderson S, Yates J, Washburn MP, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592. 10.1016/j.cell.2005.10.023 [DOI] [PubMed] [Google Scholar]
- Chabbert CD, Adjalley SH, Klaus B, Fritsch ES, Gupta I, Pelechano V, Steinmetz LM. 2015. A high-throughput ChIP-Seq for large-scale chromatin studies. Mol Syst Biol 11: 777 10.15252/msb.20145776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabbert CD, Adjalley SH, Steinmetz LM, Pelechano V. 2018. Multiplexed ChIP-Seq using direct nucleosome barcoding: a tool for high-throughput chromatin analysis. Methods Mol Biol 1689: 177–194. 10.1007/978-1-4939-7380-4_16 [DOI] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol 6: e277 10.1371/journal.pbio.0060277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia M, Tresenrider A, Chen J, Spedale G, Jorgensen V, Ünal E, van Werven FJ. 2017. Transcription of a 5′ extended mRNA isoform directs dynamic chromatin changes and interference of a downstream promoter. eLife 6: e27420 10.7554/eLife.27420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. 2011. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373 10.1038/nature09652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doris SM, Chuang J, Viktorovskaya O, Murawska M, Spatt D, Churchman LS, Winston F. 2018. Spt6 is required for the fidelity of promoter selection. Mol Cell 72: 687–699.e6. 10.1016/j.molcel.2018.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier CT, Cherny JJ, Truncali K, Robbins Pianka A, Lin MS, Krizanc D, Weir M. 2012. Amino termini of many yeast proteins map to downstream start codons. J Proteome Res 11: 5712–5719. 10.1021/pr300538f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawron D, Ndah E, Gevaert K, Van Damme P. 2016. Positional proteomics reveals differences in N-terminal proteoform stability. Mol Syst Biol 12: 858 10.15252/msb.20156662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta I, Clauder-Münster S, Klaus B, Järvelin AI, Aiyar RS, Benes V, Wilkening S, Huber W, Pelechano V, Steinmetz LM. 2014. Alternative polyadenylation diversifies post-transcriptional regulation by selective RNA–protein interactions. Mol Syst Biol 10: 719 10.1002/msb.135068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V, Forrest ARR, Hayashizaki Y, Carninci P, Lenhard B. 2015. CAGEr: precise TSS data retrieval and high-resolution promoterome mining for integrative analyses. Nucleic Acids Res 43: e51 10.1093/nar/gkv054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer SJ, Pruneski JA, Mitchell RD, Monteverde RM, Martens JA. 2011. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev 25: 29–40. 10.1101/gad.1975011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S. 2011. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci 108: 18318–18323. 10.1073/pnas.1110731108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TH, Jacquier A, Libri D. 2013. Dealing with pervasive transcription. Mol Cell 52: 473–484. 10.1016/j.molcel.2013.10.032 [DOI] [PubMed] [Google Scholar]
- Kaikkonen MU, Adelman K. 2018. Emerging roles of non-coding RNA transcription. Trends Biochem Sci 43: 654–667. 10.1016/j.tibs.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099. 10.1126/science.1087374 [DOI] [PubMed] [Google Scholar]
- Kim T, Xu Z, Clauder-Münster S, Steinmetz LM, Buratowski S. 2012. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell 150: 1158–1169. 10.1016/j.cell.2012.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee BB, Oh YM, Zhu C, Steinmetz LM, Lee Y, Kim WK, Lee SB, Buratowski S, Kim T. 2016. Modulation of mRNA and lncRNA expression dynamics by the Set2–Rpd3S pathway. Nat Commun 7: 13534 10.1038/ncomms13534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lickwar CR, Rao B, Shabalin AA, Nobel AB, Strahl BD, Lieb JD. 2009. The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS One 4: e4886 10.1371/journal.pone.0004886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycette BE, Glickman JW, Roth SJ, Cram AE, Kim TH, Krizanc D, Weir MP. 2016. N-terminal peptide detection with optimized peptide-spectrum matching and streamlined sequence libraries. J Proteome Res 15: 2891–2899. 10.1021/acs.jproteome.5b00996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malabat C, Feuerbach F, Ma L, Saveanu C, Jacquier A. 2015. Quality control of transcription start site selection by nonsense-mediated-mRNA decay. eLife 4: e06722 10.7554/eLife.06722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574. 10.1038/nature02538 [DOI] [PubMed] [Google Scholar]
- Mittal N, Guimaraes JC, Gross T, Schmidt A, Vina-Vilaseca A, Nedialkova DD, Aeschimann F, Leidel SA, Spang A, Zavolan M. 2017. The Gcn4 transcription factor reduces protein synthesis capacity and extends yeast lifespan. Nat Commun 8: 457 10.1038/s41467-017-00539-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na CH, Barbhuiya MA, Kim M-S, Verbruggen S, Eacker SM, Pletnikova O, Troncoso JC, Halushka MK, Menschaert G, Overall CM, et al. 2018. Discovery of noncanonical translation initiation sites through mass spectrometric analysis of protein N termini. Genome Res 28: 25–36. 10.1101/gr.226050.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457: 1038–1042. 10.1038/nature07747 [DOI] [PubMed] [Google Scholar]
- Pelechano V. 2017. From transcriptional complexity to cellular phenotypes: lessons from yeast. Yeast 34: 475–482. 10.1002/yea.3277 [DOI] [PubMed] [Google Scholar]
- Pelechano V, Alepuz P. 2017. eIF5A facilitates translation termination globally and promotes the elongation of many nonpolyproline-specific tripeptide sequences. Nucleic Acids Res 45: 7326–7338. 10.1093/nar/gkx479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Steinmetz LM. 2013. Gene regulation by antisense transcription. Nat Rev Genet 14: 880–893. 10.1038/nrg3594 [DOI] [PubMed] [Google Scholar]
- Pelechano V, Wilkening S, Järvelin AI, Tekkedil MM, Steinmetz LM. 2012. Genome-wide polyadenylation site mapping. Meth Enzymol 513: 271–296. 10.1016/B978-0-12-391938-0.00012-4 [DOI] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM. 2013. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 497: 127–131. 10.1038/nature12121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM. 2015. Widespread co-translational RNA decay reveals ribosome dynamics. Cell 161: 1400–1412. 10.1016/j.cell.2015.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM. 2016. Genome-wide quantification of 5′-phosphorylated mRNA degradation intermediates for analysis of ribosome dynamics. Nat Protoc 11: 359–376. 10.1038/nprot.2016.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawal Y, Chereji RV, Valabhoju V, Qiu H, Ocampo J, Clark DJ, Hinnebusch AG. 2018. Gcn4 binding in coding regions can activate internal and canonical 5′ promoters in yeast. Mol Cell 70: 297–311.e4. 10.1016/j.molcel.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Huber W. 2018. Alternative start and termination sites of transcription drive most transcript isoform differences across human tissues. Nucleic Acids Res 46: 582–592. 10.1093/nar/gkx1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T, Heger A, Sudbery I. 2017. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res 27: 491–499. 10.1101/gr.209601.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staes A, Impens F, Van Damme P, Ruttens B, Goethals M, Demol H, Timmerman E, Vandekerckhove J, Gevaert K. 2011. Selecting protein N-terminal peptides by combined fractional diagonal chromatography. Nat Protoc 6: 1130–1141. 10.1038/nprot.2011.355 [DOI] [PubMed] [Google Scholar]
- Ungewitter E, Scrable H. 2010. Δ40p53 controls the switch from pluripotency to differentiation by regulating IGF signaling in ESCs. Genes Dev 24: 2408–2419. 10.1101/gad.1987810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, Primig M, Amon A. 2012. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell 150: 1170–1181. 10.1016/j.cell.2012.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varland S, Aksnes H, Kryuchkov F, Impens F, Van Haver D, Jonckheere V, Ziegler M, Gevaert K, Van Damme P, Arnesen T. 2018. N-terminal acetylation levels are maintained during acetyl-CoA deficiency in Saccharomyces cerevisiae. Mol Cell Proteomics 17: 2309–2323. 10.1074/mcp.RA118.000982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S, Li H, Gogol MM, Workman JL. 2016. Selective suppression of antisense transcription by Set2-mediated H3K36 methylation. Nat Commun 7: 13610 10.1038/ncomms13610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF. 2009. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res 19: 360–371. 10.1101/gr.084970.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Pelechano V, Järvelin AI, Steinmetz LM. 2011. Functional consequences of bidirectional promoters. Trends Genet 27: 267–276. 10.1016/j.tig.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037. 10.1038/nature07728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Clauder-Münster S, Smolik M, Huber W, Steinmetz LM. 2011. Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol 7: 468 10.1038/msb.2011.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Dietrich FS. 2005. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res 33: 2838–2851. 10.1093/nar/gki583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, O'Shea EK. 2014. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature 514: 117–121. 10.1038/nature13578 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.