Abstract
Aging is a pleiotropic process affecting many aspects of mammalian physiology. Mammals are composed of distinct cell type identities and tissue environments, but the influence of these cell identities and environments on the trajectory of aging in individual cells remains unclear. Here, we performed single-cell RNA-seq on >50,000 individual cells across three tissues in young and old mice to allow for direct comparison of aging phenotypes across cell types. We found transcriptional features of aging common across many cell types, as well as features of aging unique to each type. Leveraging matrix factorization and optimal transport methods, we found that both cell identities and tissue environments exert influence on the trajectory and magnitude of aging, with cell identity influence predominating. These results suggest that aging manifests with unique directionality and magnitude across the diverse cell identities in mammals.
Aging is a gradual process of functional and homeostatic decline in living systems. This decline results in increased mortality risk and disease prevalence, eventually resulting in death. Aging appears to be a conserved feature of eukaryotic biology, affecting organisms as phylogenetically diverse as the single-celled Saccharomyces cerevisiae, the eutelic nematode Caenorhabditis elegans, mice, and humans (Kenyon 2010; Longo et al. 2012; Tissenbaum 2014). Despite the near universal nature of the aging process, the underlying causes of aging are poorly understood. Aging phenotypes have been observed and hypotheses have been proposed for more than a hundred years (Weismann et al. 1891; Kirkwood 1977; Gladyshev 2013; López-Otín et al. 2013), but we do not yet know the cellular and molecular players that cause aging or how they differ between biological contexts. Both the fundamental nature of aging and its negative effects provide motivation to enumerate these players and establish causal relationships among aging phenotypes.
Mammalian aging phenotypes manifest at the organismal, tissue, cellular, and molecular levels (Zhang et al. 2015). Extensive research has produced catalogs of aging phenotypes at the physiological level, providing functional and behavioral hallmarks of age-related decline. Likewise, molecular profiling of nucleic acids, proteins, and metabolites has provided a phenotypic description of aging in individual tissues. Transcriptomic analyses of aging tissues have revealed some common changes, such as increased inflammatory pathways in several tissues (Lee et al. 2002; Rodwell et al. 2004; O'Brown et al. 2015; Su et al. 2015; Benayoun et al. 2019). Proteomic analyses of aging tissues have also found changes in immunological and stress response pathways in multiple organs (Ori et al. 2015; Angelidis et al. 2019). Metabolomics have likewise revealed changes in fatty acids across multiple tissues (Houtkooper et al. 2011).
Additional lines of inquiry have aimed to address a classical question in aging biology: Do different tissues age in the same way? Transcriptomic analysis at the bulk tissue level revealed tissue-specific features of aging in several studies (Schumacher et al. 2008; Jonker et al. 2013; Ori et al. 2015; Benayoun et al. 2019). Proteomic analysis of brain and liver in young and old mice similarly suggests that most age-related changes are tissue-specific (Ori et al. 2015). Mass spectroscopy studies have reported heterogeneous proliferative histories within and between murine tissues, suggesting one means by which aging may manifest differently across tissues and cells (Drigo et al. 2019). Studies of DNA methylation patterns with aging also suggest that different tissues age in unique ways (Horvath 2013). However, the cellular origins of aging phenotypes within a tissue remain largely unknown (Rodwell et al. 2004; O'Brown et al. 2015; Benayoun et al. 2019).
Mammals contain a multitude of distinct cell types, each of which may be composed of multiple cell states. This combination of cell type and state together form a cell identity. Each of these cell identities shows specialized functions. In the mouse alone, recent cell atlas efforts have revealed more than 100 cell types (Han et al. 2018; The Tabula Muris Consortium 2018). These surveys have cataloged diverse murine cell identities, but the plasticity of these identities and their contributions to tissue- and organism-level pathology remain unknown.
At both the molecular and functional level, a host of aging phenotypes and associated mechanisms have been revealed in individual cell types (Shaw et al. 2010; Chakkalakal et al. 2012; Keyes et al. 2013; Liu et al. 2013; Flach et al. 2014; Blau et al. 2015; Brack and Muñoz-Cánoves 2016; Keyes and Fuchs 2018). Although some of these studies present unique features of aging within individual cell identities, it is difficult to compare them systematically because of differences in experimental conditions and assay methodology. Using traditional molecular biology assays, it is difficult to measure high-dimensional molecular phenotypes across multiple cell identities, making large-scale comparisons of aging phenotypes across cell identities intractable. The recent development of single-cell RNA-sequencing (scRNA-seq) has ameliorated this limitation, allowing for measurement of transcriptional features across all prevalent cell identities in a tissue in a single experiment.
Although the technology has only recently matured, scRNA-seq experiments in individual tissues have already revealed novel aspects of the aging process. In a pioneering scRNA-seq study of hematopoietic progenitors, the axis of aging was shown to be opposite the axis of differentiation (Kowalczyk et al. 2015). Multiple investigations have reported that cell–cell heterogeneity (Enge et al. 2017; Angelidis et al. 2019) and gene expression variance (Martinez-Jimenez et al. 2017) increase with age. However, the specific influence of cell identity and tissue environment on the trajectory and magnitude of aging has yet to be resolved.
Here, we used scRNA-seq to generate a set of molecular profiles in which we can compare aging phenotypes across cell identities. By profiling 50,000+ cells from three tissues in young and old mice, we identified common features of aging that span cell identities, as well as features unique to each identity. We found that changes in protein localization gene sets and increased inflammatory gene expression occur across cell identities, whereas other changes are more specific to individual cell identities. For example, phospholipid metabolism gene sets were elevated in old lung type II pneumocytes, and muscle-related gene sets were decreased in old lung stromal cells. Using matrix factorization and optimal transport methods, we computed trajectories of aging for each cell identity and assessed the influence of identity and environment on these trajectories.
Results
Single-cell RNA-sequencing identifies a diversity of cell types and states in young and old mouse tissue
We collected transcriptional profiles of young and old cells of many identities by isolating single cells from the kidney, lung, and spleen of n = 4 young (7 mo) and n = 3 old (22–23 mo) C57Bl/6J mice. We note that our “young” animals are at the later portion of mature adulthood, as compared to the early portion of mature adulthood (3–5 mo) used in some studies. All three tissues were collected from the same animals (Methods; Supplemental Note 1, tissue selection).
Isolations were performed at the same time of day to limit circadian variation, which affects the expression of nearly half of all murine genes (Zhang et al. 2014). After single-cell isolation, cells were immediately encapsulated and barcoded for library preparation using the 10x Genomics microfluidics system, followed by sequencing (Fig. 1A). We recovered 55,293 individual cell transcriptomes (Methods; Supplemental Table S1).
Figure 1.
scRNA-seq reveals that nonimmune cell-type proportions are preserved with age. (A) Schematic representation of the experimental design. We isolated kidney, lung, and spleen tissue simultaneously from each young (7–8 mo) and old (22–23 mo) mouse. After generating single-cell suspensions, we prepared cells for scRNA-seq using the 10x Chromium system. (B) UMAP embeddings of each tissue investigated in our data set. Colors represent cell type annotations. Cell types were derived using a deep neural network classifier trained on the Tabula Muris data set, followed by manual validation using marker genes. (C) Matching UMAP embeddings depicting the age of each cell. (D) For each animal, we computed the proportion of cells in each cell identity. The mean proportion of each cell identity for each age across animals is presented as a bar graph. The underlying proportions observed in individual animals are overlaid as black dots. We observed an increased frequency of immune cell types in kidney and lung.
We determined cell type and state identity by leveraging annotations provided in the Tabula Muris (The Tabula Muris Consortium 2018) following the “cell ontology” structure (Bakken et al. 2017). Some age-related changes may be unique to individual states within a cell type (i.e., CD4 vs. CD8 T cells). To ensure that we can explicitly detect these cell state changes, we manually annotated cell states within each cell type in the Tabula Muris (Methods; Supplemental Fig. S1). We use the term “cell identity” to refer to the combination of cell type and state labels, such that CD4 T cells and CD8 T cells are different cell identities (Fig. 1B; for abbreviation legend, see Supplemental Table S2).
We trained deep neural networks to classify cell types based on these annotations, then used these networks to predict cell types in our data (Methods; Fig. 1B). We validated classifications by inspecting marker gene expression post hoc (Supplemental Figs. S2, S3B) and computing correlations between cell identities in our data and the Tabula Muris (Supplemental Fig. S4). We found that all distinct cell clusters expressed marker genes corresponding to an annotated cell type in the Tabula Muris, such that we did not recover any new or previously unseen cell types here (Supplemental Fig. S3B). From these cell identity annotations, we identified 19 unique cell types and 28 unique cell states across the three tissues.
Comparing our cell types to the Tabula Muris, we recovered all but one of the cell types identified (kidney loop of Henle epithelial cells) (Supplemental Figs. S3A, S5). The cell-type proportions that we recovered differ from the Tabula Muris (e.g., we recovered comparatively more immune cells in the kidney and lung), but are not outside expected ranges based on previous comparisons between scRNA-seq data sets (Park et al. 2018; The Tabula Muris Consortium 2018). We also used an orthogonal cell type identification method (Kiselev et al. 2018) and found predictions to be largely consistent with our neural network–derived, manually validated annotations (Supplemental Fig. S6). In both UMAP and principal component analysis (PCA) projections, we found that cells segregate by cell type and, to a lesser degree, age, rather than by experimental batch (Fig. 1C; Supplemental Figs. S7, S8).
Immune cells are more prevalent in old kidneys and lungs, while nonimmune cell-type proportions are preserved
One prospective way in which aging may influence tissue function is by altering the proportion of each cellular identity within the tissue. To investigate this possibility, we quantified the proportion of each cell type within each tissue across ages.
Lymphocytes were significantly more abundant in the kidneys and lungs of old animals (t-test on additive log ratio [ALR] transformed proportions, Q < 0.05) (Fig. 1D; Supplemental Note 2). This may reflect increasing immune infiltration of the nonlymphoid tissues with age, as suggested in previous studies of kidney, lung, and other nonlymphoid tissues (Rodwell et al. 2004; Aoshiba and Nagai 2007; Toapanta and Ross 2009; Lumeng et al. 2011; O'Brown et al. 2015). For the spleen and only the nonimmune cells in the kidney and lung, differences in cell-type proportions between young and old animals were not statistically significant (|log2(old/young)| < 1, Q > 0.05, t-test of ALR-transformed proportions) (Fig. 1D; Supplemental Fig. S9B). However, we cannot rule out that recovery of specific cell types was confounded by an interaction of aging with our isolation procedures. These results therefore present a hypothesis of increased immune infiltration with age but require confirmation by in situ cell type quantification methods (Supplemental Note 2).
Shifting cell state proportions within a cell type may be an alternative mechanism by which aging phenotypes manifest. Examples of this phenomenon are present in the literature, such as a decrease in naive CD8 T cells relative to other T cell states (Decman et al. 2012; Goronzy et al. 2015) and the shift from highly regenerative to less regenerative stem cell states in the blood and muscle (Chakkalakal et al. 2012; Bernitz et al. 2016). To address whether cell state proportions change with age, we quantified the proportion of cells in each distinct state for all cell types. We found a decreased proportion of spleen CD8 T cells relative to other spleen T cells (Quinn et al. 2016, 2018) and changes in the cell state proportions of kidney collecting duct epithelial cells (Supplemental Fig. S9; Supplemental Note 2). These results suggest that changes in cell state proportions within a cell type can occur with age. If different cell states have distinct functional roles, these shifts in cell state proportions may contribute to age-related phenotypes.
Cycling cells are similarly rare in young and old animals
Previous reports highlight cell cycle activity changes with age in multiple cell populations (Chakkalakal et al. 2012; Kowalczyk et al. 2015; Nalapareddy et al. 2017). To investigate changes in cell cycle frequency in our data, we evaluated cell cycle activity by scoring the expression of S phase–associated genes and G2M-associated genes (Methods; Tirosh et al. 2016). We observed only small changes in either of these cell cycle module scores with age across cell identities, with only a handful of statistically significant differences (Supplemental Fig. S10A; Supplemental Note 3). Thus, cell cycle rates appear not to be dramatically changed with age in our data.
Previous studies also reported accumulation of noncycling senescent cells in aging tissues (Childs et al. 2017). We find few cells expressing proposed senescence marker Cdkn2a, with no significant differences in expression between young and old cells (Supplemental Note 4; Supplemental Fig. S11A).
Changes in cell–cell variation with age depend on cell identity
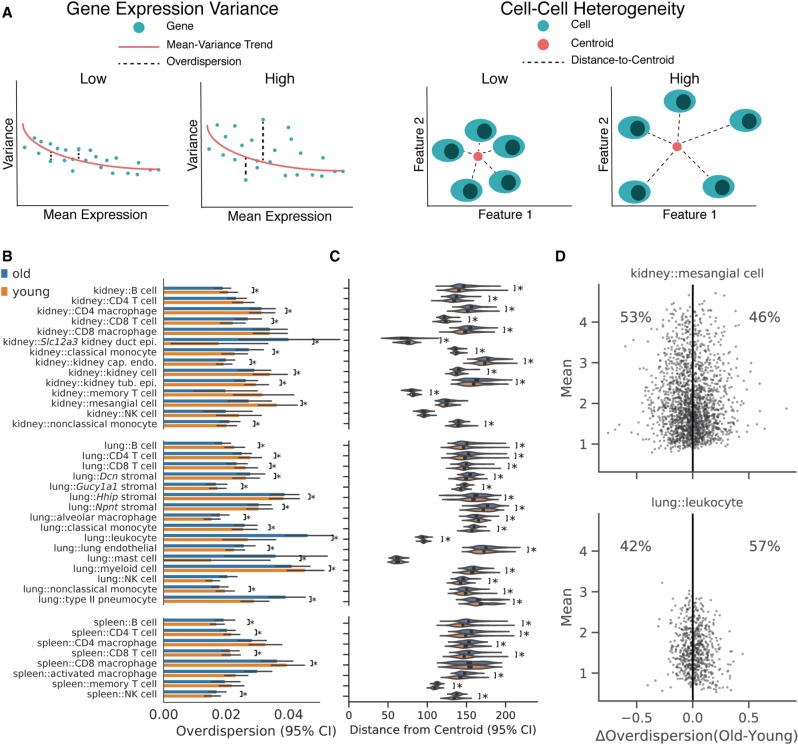
Single-cell analysis measures not only the mean expression of each gene, but also the variation within a cell population. Previous studies have reported that both gene expression variance and cell–cell heterogeneity increase with age using RNA-seq (Enge et al. 2017; Martinez-Jimenez et al. 2017; Angelidis et al. 2019) and quantitative PCR methods (Bahar et al. 2006). Gene expression variance quantifies the mean dispersion across genes in the transcriptome, such that each gene contributes equally. In contrast, cell–cell heterogeneity measures the average distance in transcriptional space between cells in a population (Fig. 2A; Supplemental Note 5). Both transcriptional variation and cell–cell heterogeneity have important implications for cell physiology and function, as explored in seminal studies of transcriptional noise in cell fate selection (Blake et al. 2006; Süel et al. 2007) and bet hedging (Cohen 1966; Slatkin 1974; Kussell et al. 2005; Beaumont et al. 2009; Altschuler and Wu 2010). To determine if age-related changes in variation and heterogeneity depend on cell identity and tissue environment, we evaluated both properties across the many combinations that we observed.
Figure 2.
Changes in cell–cell heterogeneity with age depend on cell identity. (A) Diagram illustrating the difference between gene expression variance (left) and cell–cell heterogeneity metrics (right). Gene expression variance measures the variance of the average gene in a manner that controls for mean expression levels. Cell–cell heterogeneity measures the average difference between cells across genes. Examples of data providing high and low values for each metric are schematized. (B) Overdispersion values for each cell identity conditioned on age. We computed overdispersion as the residual dispersion of each gene after accounting for mean expression level with a rolling median dispersion spline (difference from the median method). Each point in the underlying data represents a single gene. Many cell identities do not show a substantial shift in the overdispersion distribution. Some identities show increased mean overdispersion with age, whereas others show a decrease: (*) Wilcoxon rank-sum test, Q < 0.05. (C) Cell–cell heterogeneity measurements based on Euclidean distances to the population centroid for each cell identity and age. We computed Euclidean distances to the centroid for each cell using all measured genes. Each point in the underlying data represents a single cell. Most cell identities showed increased cell–cell heterogeneity in old cells: (*) Wilcoxon rank-sum test, q < 0.05. However, some identities show decreased heterogeneity with age (kidney::CD8 T cell; kidney::classical monocyte). (D) Difference in overdispersion between old and young cells (ΔOverdispersion) as a function of mean gene expression value in VIM-positive kidney capillary endothelial cells (upper) and lung leukocytes (lower). Each point represents the change in overdispersion and mean expression value for a single gene. Shading reflects the density of points. We found that genes within a cell identity show heterogeneous changes in overdispersion with age.
We evaluated transcriptional variation using the difference from the median (DM) method (Kolodziejczyk et al. 2015) to estimate “overdispersion” (Methods; Supplemental Note 5). Across cell identity/environment combinations, we found that transcriptional variation changes subtly (median Cohen's d = 0.031) (Fig. 2B). Individual cell identities can be identified that show either increased (lung leukocytes) or decreased (kidney mesangial cells) variance with age (Fig. 2D).
We quantified cell heterogeneity in each cell identity/environment combination using the distance to the centroid method (Enge et al. 2017; Angelidis et al. 2019). Cell–cell heterogeneity appears to increase for many cell identities (Wilcoxon rank-sum test, Q < 0.05, median Cohen's d = 0.473), including B cells across all three tissues, and lung stromal cells. We also observed decreased heterogeneity with age in some cell identities, such as lung type II pneumocytes and kidney CD8 T cells (Fig. 2C). Quantifying cell–cell heterogeneity using alternative gene sets and distance metrics yielded similar results (Methods; Supplemental Note 5; Supplemental Fig. S12).
Taken together, these results indicate that changes in gene expression variance and cell–cell heterogeneity with age depend on cell identity. These results do not contradict previous observations of increased variation with age in some cell identities and contexts (Enge et al. 2017; Martinez-Jimenez et al. 2017), but rather suggest a nuanced view of changes in noise as a hallmark of aging at the single-cell level (Supplemental Note 5; Todhunter et al. 2018).
Differential expression reveals aging phenotypes common across cell identities
Previous studies have revealed differentially expressed genes within whole tissues or individual cell types in aging (Rodwell et al. 2004; Jonker et al. 2013; Cosgrove et al. 2014; O'Brown et al. 2015; Su et al. 2015; White et al. 2015; Keyes et al. 2016; Benayoun et al. 2019). However, it remains unclear to what degree age-related transcriptional changes are shared or unique across cell identities. To address this outstanding question, we performed differential expression analysis within each cell identity between young and old mice.
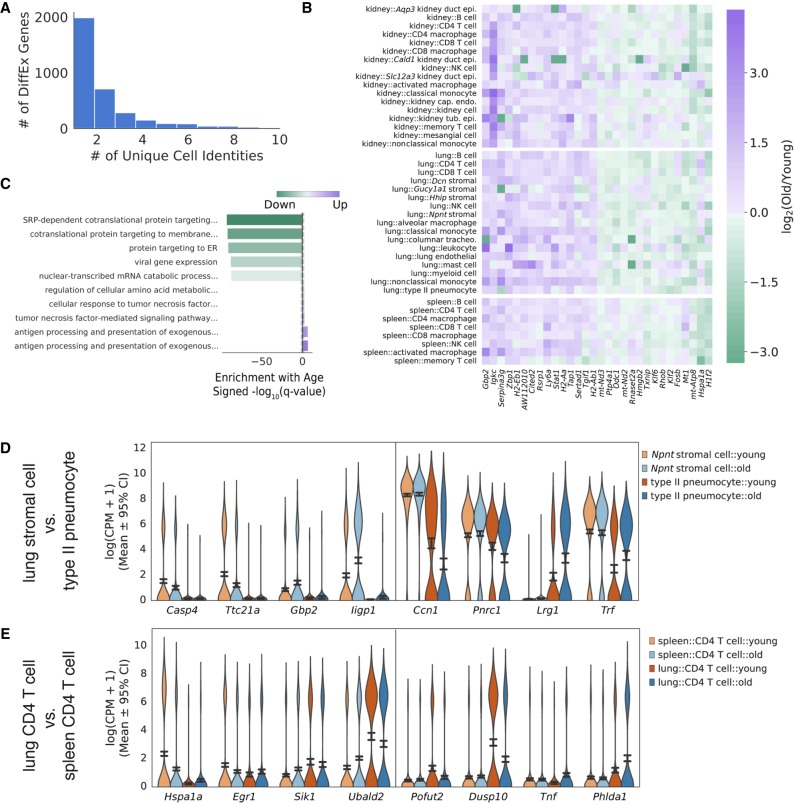
We identified differentially expressed genes for each cell identity/tissue environment combination using the rank sums method (Methods). For each differentially expressed gene, we counted how many unique cell identities differentially express that gene. We considered the same cell type in different tissues to have the same cell identity. For example, a gene significantly up-regulated in lung natural killer cells, kidney natural killer cells, and spleen CD8 T cells is considered to be differentially expressed in two cell identities rather than three. The majority of differentially expressed genes with age are specific to one or a few cell identities (Fig. 3A).
Figure 3.
Differential expression analysis identifies common age-related changes across cell identities and tissue environments. (A) Distribution of the number of genes differentially expressed in at least k cell identities. We counted identical cell identities in different tissues (e.g., lung B cells, spleen B cells) as only one unique cell identity. We selected genes differentially expressed in k > 5 unique cell identities as common differentially expressed genes. (B) Heatmap of top 15 genes significantly changed in each direction across more than five cell identities. Fold changes between old and young cells are presented for each cell type in each tissue. Although no gene is universally changed across cell identities, each gene is changed across multiple tissues and developmental lineages. (C) Top five enriched Gene Ontology terms for genes that are down-regulated (negative values) and up-regulated (positive values) with aging across more than five cell identities. Dotted gray lines near 0 represent the α = 0.05 significance threshold. Antigen processing and metabolic pathways appear to be up-regulated, whereas protein translation and translocation pathways appear to be down-regulated with aging. (D) Violin plot of genes that are uniquely changed between two cell states in the lung (Wilcoxon rank-sum test, Q < 0.05): Npnt stromal cells and type II pneumocytes. Each gene presented is significantly up-regulated or down-regulated in one cell identity and does not change in the same direction (log2 [old/young] <0.1) in the other cell identity. Confidence intervals were computed by bootstrapping. For each cell state, we show the top two specific down-regulated genes and up-regulated genes. (E) Violin plot of genes that are uniquely changed between CD4 T cells isolated from the spleen and the lung. Genes were selected as in D.
However, 261 genes were differentially expressed across k > 5 cell identities and consistently changed across multiple cell identities (Fig. 3B). We chose five cell identities as a cutoff for common differentially expressed genes to reduce the number of genes identified owing to common differential expression in highly similar cell identities (i.e., monocytes and macrophages). Thus, some aspects of transcriptional aging are common to many cell identities. Using Gene Ontology (GO) enrichment analysis for biological processes (Kuleshov et al. 2016), we identified SRP-dependent protein localization and protein translocation to the ER as commonly down-regulated across cell identities (Fig. 3C, upper). This observation is consistent with the observed interaction of protein translocation systems with aging (Steffen and Dillin 2016; Hendrickson et al. 2018). Antigen processing and inflammatory pathways were significantly up-regulated with age, a result that has also been observed across tissues in previous reports (Rodwell et al. 2004; O'Brown et al. 2015; Ori et al. 2015; Benayoun et al. 2019). Our gene set enrichment results were not sensitive to the specific number of cell identities (k) that we required to consider a gene commonly differentially expressed (Supplemental Fig. S13). Hierarchical clustering of this common set of differentially expressed genes revealed gene clusters enriched for specific inflammatory processes (type I interferon signaling, cytokine secretion), suggesting that more than one immunological pathway changes with age (Supplemental Fig. S14).
Previous investigations using bulk transcriptional assays reported increased inflammation-associated gene expression with age in multiple nonlymphatic tissues but could not identify the source cell type(s). Both an increase in the number of immune cells in these tissues, or an increase in the expression of inflammation-associated genes in nonlymphoid cells may explain this observation. Bulk transcriptional assays cannot independently assess these possibilities, but our single-cell data allowed us to assess both changes in cell-type proportions and cell-type–specific gene expression. In addition to the increased number of immune cells in kidney and lung, we found that inflammatory gene expression increased with age in both immune and nonimmune cell types (Supplemental Note 6; Supplemental Fig. S15A,B). This result suggests an interrelated transition of the tissue's cell populations toward an inflamed state with age.
Many genes changed expression uniquely in individual cell identities, even within the same tissue. For instance, Npnt lung stromal cells showed up-regulated and down-regulated genes and gene sets that are unchanged in lung type II pneumocytes and vice versa (Fig. 3D; Supplemental Fig. S15C). We also observed genes that are differentially expressed in a tissue-specific manner. For example, CD4 T cells in the lung down-regulated and up-regulated genes and gene sets that are unchanged in CD4 T cells of the spleen and vice versa (Fig. 3E; Supplemental Fig. S15D). Collectively, these results indicate that cell identity and tissue environment both influence differential gene expression. Although most differentially expressed genes are specific to individual cell identities, a subset of changes appears to be common across many identities.
Aging manifests novel B cell states in the spleen
Beyond shifting cell state proportions, aging could promote the formation of novel cell states unseen in young animals. Previous studies have reported the emergence of novel cell states with age in the hematopoietic system (Kirschner et al. 2017), and senescence phenotypes may be considered novel cell states from this perspective (Sousa-Victor et al. 2014; Sharpless and Sherr 2015; Baker et al. 2016). Here, we observed another example of a cell state that arises with age in spleen B cells.
Spleen B cells in our experiments showed three distinct clusters that do not map well to a canonical subtyping scheme (Fig. 4A). We identified these clusters using Louvain community detection and maximized the variance ratio criterion (VRC) (Calinski and Harabasz 1974) to find an optimal partition (Supplemental Fig. S16). One of these clusters was dominated by cells from old animals (P < 0.05, χ2 contingency table) (Fig. 4A,C). We performed differential expression analysis on each of these clusters to identify marker genes and noted greater Apoe in the larger cluster and greater B2m in the other.
Figure 4.
Aging manifests novel neoplastic B cell states in the spleen. (A) UMAP embeddings of the spleen B cell compartment, colored by age (left). Louvain clustering identified three subpopulations within the B cell compartment of the spleen, named by their marker genes (right). (B) Marker genes for each cluster and a set of lymphoma-specific genes presented as violin plots. All markers were significantly enriched in the Apoe-high and B2m-high cluster. (C) Proportions of cells in each cluster as a function of age. The Apoe-high and B2m-high states are occupied predominantly by aged cells (t-test on ALR-transformed proportions, Q < 0.05). (D) Enrichment of MSigDB Hallmark gene sets based on differentially expressed genes in the Apoe-high cluster relative to the normal B cell cluster. Myc targets, mTOR signaling, and DNA repair pathways were up-regulated, whereas TRP53 signaling was down-regulated, suggesting neoplasia. Gray dotted lines represent the α = 0.05 significance threshold. (E) UMAP embedding of the Apoe cell cluster alone. A second Louvain clustering iteration reveals two clusters within this Apoe-high group (right). Visualizing the animal of origin for each cell, it is apparent that Cluster 1 is dominated by a single old animal (left). (F) Marker genes for each of the subclusters within the Apoe-high state presented as violin plots. Cluster 0 is enriched for lymphoma-associated gene Zeb2, and Cluster 1 is enriched for Plac8. These results similarly suggest neoplasia.
Previous reports point out that the C57Bl/6 mice used in this study die with an unusually high incidence of lymphoma (Blackwell et al. 1995; Pettan-Brewer and Treuting 2011; Brayton et al. 2012). To ask if these clusters dominated by old cells potentially represent lymphomas, we examined the expression of several genes associated with B cell lymphomas in a previous study (Lenburg et al. 2007). We identified several lymphoma-associated genes significantly up-regulated in each of these clusters (Q < 0.05, Wilcoxon rank-sum test) (Fig. 4B, rows 4–9). Performing gene set enrichment analysis using the MSigDB Hallmark gene sets (Liberzon et al. 2015), we observed that Myc target genes and DNA repair pathways were up-regulated, whereas TRP53 pathway genes were down-regulated (Fig. 4D). This pattern of differential gene expression suggests that the Apoe-high population is preneoplastic.
At a finer level of detail, the Apoe-high cluster appears to have two distinct lobes. One of these two lobes seems to contain cells almost entirely derived from a single old animal (1 of 3) (Fig. 4E), whereas the other lobe contains cells found in all of the old animals observed. To determine what differentiates these lobes, we examined marker genes and identified differences in the expression of Zeb2, the B cell marker Cd72, Plac8, and Cd24a as discriminating features (Q < 0.05, Wilcoxon rank-sum test) (Fig. 4F). Each of these genes has previously been associated with neoplasia (Kinsey et al. 2014; Li et al. 2017; Zhao et al. 2019). We believe these cell states may therefore represent a lymphoma or prelymphoma, although exact diagnosis is difficult because of the histopathological definition of neoplastic malignancies.
These emergent cell states support the notion that some aging phenotypes manifest not by shifting the distribution or location of youthful states, but by creating new states altogether. Neoplastic cells are an extreme case of such a phenomenon. The observation of a cell state that is generally ubiquitous in old mice and a related state that is present in only one animal also highlights the commonalities and stochastic animal–animal heterogeneity present in aging studies.
Cell identity determines the trajectory of aging
Our differential expression and heterogeneity analyses suggest that cell identities age differently and the same cell identity ages differently across tissues. How much do cell identity and tissue environment influence the trajectory of aging? To answer this question quantitatively, we computed an aging trajectory for each cell identity in each tissue based on their transcriptional profiles.
We performed this analysis using an embedding space derived by non-negative matrix factorization (NMF). NMF embeddings of transcriptional data have been shown to recover relationships between genes, such that each component of the embedding may be interpreted as a gene expression program (Brunet et al. 2004; Shao and Höfer 2017; Kotliar et al. 2019). Likewise, cell values for each component quantify activity of those programs. NMF embedding allowed us to assess aging trajectories at a level of abstraction above individual genes.
To compute aging trajectories, we first embedded all cells observed across tissues in a 20-dimensional NMF space (Methods; Fig. 5B; Supplemental Fig. S17). To assign semantic meaning to the embedding dimensions, we identified genes associated with each dimension by thresholding on the dimension loadings and analyzed gene set enrichment (Supplemental Fig. S18). We computed the aging trajectory for each cell identity/tissue combination as the distance between the centroid of the young cells and the centroid of the old cells in this embedding (Methods). For clarity, we computed separate aging trajectories for each cell identity/tissue combination, such that lung B cells and spleen B cells were treated independently. This procedure yields a 20-dimensional vector representing the trajectory of age-related change observed in each cell identity.
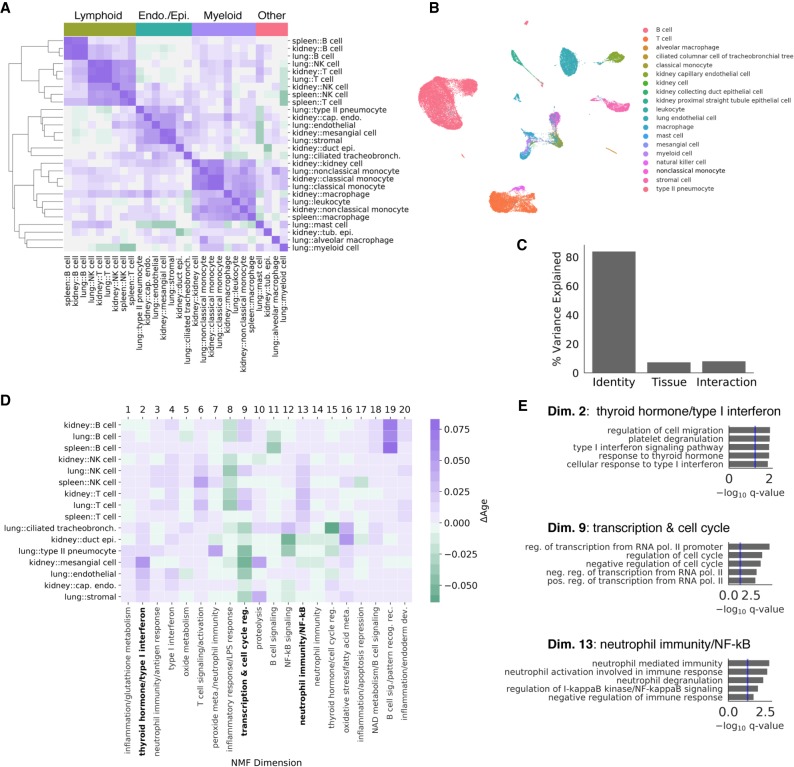
Figure 5.
Cell identity and tissue environment influence aging trajectories. We computed aging trajectories as the distance between young and old cell centroids in a non-negative matrix factorization (NMF) low-rank embedding of each cell. We embedded all cells in a common non-negative latent space, where each latent dimension is associated with the activity of particular genes. (A) Cosine similarities between the aging trajectories of each cell state in each tissue are compared in a heatmap. Hierarchical clustering of aging trajectories revealed that similar cell types have similar trajectories. In contrast, different cell types from the same tissue did not cluster together. We labeled clusters based on their constituent cell types above. (B) A UMAP visualization derived from the 20-dimensional NMF embedding with cell types overlaid as colors. The NMF embedding retains biological variation between cell types. (C) Variance in the aging vectors of immune cell types found in all three tissues explained by cell type, tissue environment, and their interaction (ANOVA). (D) Heatmap visualization of the aging vectors for lymphocyte and endothelial/epithelial cell types. Endothelial/epithelial cell types and lymphoid cell types were identified by clustering aging vectors, presented in A. Semantic descriptions of each embedding dimension derived from gene enrichment analysis are presented as column labels. Some expression programs show common changes with age across both groups of cell types, whereas others appear to be different between groups. We highlight three expression programs that show cell-identity-specific changes (bold). (E) Gene Ontology enrichment analysis results for selected dimensions of the NMF embedding.
We compared these trajectories using the cosine similarity. The cosine similarity is 1 if trajectories are in the same direction, 0 if they are orthogonal, and −1 if they are in opposite directions. Clustering by these cosine similarities, we found that qualitatively similar cell identities have similar aging trajectories, whereas dissimilar cell identities have orthogonal or dissimilar trajectories (Fig. 5A).
Examining the clustering partition, cells segregated into roughly four clusters: endothelial and epithelial cells (blue), myeloid cells (purple), lymphocytes (green), and a remaining cluster with both myeloid and epithelial cell types (red). Cell identities from the kidney and lung intermixed within the endothelial/epithelial cell cluster. Likewise, within the lymphocyte cluster B cells and T cells cluster more tightly by cell identity across tissues than by tissue of origin. This suggests that cell identity has a larger influence on aging trajectories than tissue environment.
To quantify the influence of both cell identity and tissue environment, we focused on the aging trajectories of immune cell types observed in multiple tissues (B cells, T cells, natural killer cells, macrophages, monocytes). For these cell types, we constructed linear models and performed an analysis of variance (ANOVA) to determine the proportion of variation explained by cell identity, tissue environment, and their interaction (Robinson et al. 2015). Consistent with qualitative observations, cell identity explained a markedly larger fraction of variance than tissue environment (Fig. 5C). These results were robust to bootstrap resampling across cells (Supplemental Fig. S19) or across animals (Methods; Supplemental Fig. S20) and were statistically significant when evaluated with permutation tests (Methods; Supplemental Fig. S19). Results were also robust using a PCA embedding rather than NMF (Supplemental Fig. S21; Supplemental Note 7).
Endothelial cells and lymphocytes show distinct aging trajectories
We next asked how endothelial/epithelial and lymphocyte aging trajectories differed. Using the clustering assignments from Figure 5A, we compared aging trajectories for each cell identity in the endothelial and lymphocyte clusters. The two clusters showed consistent differences in the magnitude of change within multiple NMF dimensions (Fig. 5D). Based on Gene Ontology enrichment within these dimensions, endothelial/epithelial cell types showed an increase in thyroid hormone and type I interferon signaling (Dimension 2) (Fig. 5E) and a decrease in transcriptional machinery and cell cycle regulation relative to immune cell types (Dimension 9). In contrast, lymphocytes showed a larger increase in NF-kB signaling and related immune response pathways (Dimension 13). B cell activation associated pathways were increased with age in a B cell–specific manner (Dimension 19). Both clusters increased NAD metabolism and B cell signaling pathways (Dimension 18), as well as T cell signaling and activation (Dimension 6). These results are consistent with literature observing changes in type I interferon activity (Li et al. 2015) and thyroid hormone signaling with aging (Biondi and Cooper 2008; Gesing et al. 2012; Visser et al. 2016), as well as our differential expression analysis.
Optimal transport analysis indicates that cell identity determines the magnitude of aging
Do some cell identities or tissue environments change more with age? To answer this question, we estimated the magnitude of aging using optimal transport distances in an NMF embedding, as described above. Here, we used an embedding with 500 latent dimensions to capture more variation within the data (Supplemental Fig. S17B). Optimal transport is a technique for measuring distances between equally sized samples that has been applied to a wide variety of data analysis tasks, including scRNA-seq (Schiebinger et al. 2019).
The discrete optimal transport distance that we applied here measures the minimum amount of change needed to make one group of cells match another. This metric captures differences in the covariance structure and modality of a cell population, in addition to differences in the population means (Supplemental Fig. S22A,B). We compared the distance between two cell populations by sampling n = 300 cells from each of them a number of times (300) and averaging the distance across the samples (Methods). This bootstrap sampling scheme allowed us to meet the equal sample size requirement for optimal transport distances, even when we observed different numbers of young and old cells.
We compared the distance between young and old cells from each cell identity and tissue environment in the NMF embedding. For each cell identity, we made three distinct comparisons. We made a heterochronic comparison between young and old cells to estimate the magnitude of aging. This process is schematized in spleen B cells to provide intuition (Fig. 6A). We also made two isochronic comparisons, comparing young cells to young cells and old cells to old cells (Fig. 6B). These isochronic comparisons serve as a null distribution, estimating the distance we would expect to see between random samples of cells in the absence of age-related change. We normalized the heterochronic (young–old) comparison values for each cell identity by dividing by the mean of the larger isochronic distance. We expect this isochronic distance to scale with the heterogeneity of the cell population (Supplemental Fig. S22C), such that this normalization accounts for differences heterogeneity across cell types.
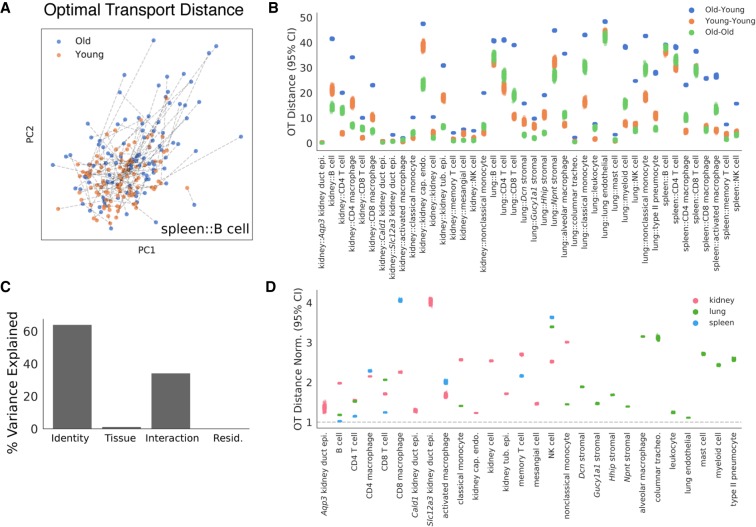
Figure 6.
Optimal transport (OT) estimates the magnitude of aging across cell identities. To estimate a magnitude of age-related change in each cell population, we computed an OT distance between random samples of young and old cells from each cell identity in a rank 500 NMF embedding. (A) Young and old spleen B cells are presented in a PCA projection of the NMF embedding. Dashed lines overlaid indicate the globally optimal partners for each young and old cell, collectively representing the OT solution. The sum of distances along each dashed line is the OT distance. OT distances were computed independently for each cell identity across 300 random samples. (B) OT distances for each cell type in each tissue are presented. For each cell identity, we computed distances for heterochronic samples of cells (old-young) and isochronic samples of cells (young-young, old-old). The latter isochronic comparisons serve as negative controls. Distributions across n = 300 random samples each are presented for each of these comparisons. (C) Variance in normalized OT distances explained by cell identity and tissue environment (ANOVA). We fit a linear model with normalized OT distance as the response and cell identity, tissue environment, and their interaction as the inputs. We found that cell identity explains the majority of variation in aging magnitudes, whereas tissue explains little. The residual represents variation across the random samples in which we measured the OT distance. (D) Heterochronic OT distances for each cell identity. Each dot represents the normalized OT distance for a single sample. Values are normalized to the largest mean value of the isochronic negative controls. The gray dotted line marks a normalized distance of 1, which indicates that a heterochronic comparison shows similar distances to an isochronic comparison in that cell identity.
We interpreted these normalized optimal transport distances as an estimate of the magnitude of age-related change. Aging magnitudes varied greatly across the cell identities that we observed (Fig. 6D). Some cell identities showed little more distance between young and old cells than between two isochronic groups of cells (normalized distance of ≈ 1), whereas others showed three- to fourfold larger differences in heterochronic comparisons. Lung and spleen natural killer (NK) cells and spleen CD8 macrophages showed some of the greatest optimal transport distances, whereas lung and spleen B cells and lung endothelial cells showed some of the smallest.
To quantify the relative contributions of cell identity and tissue environment to aging magnitude, we used the same linear modeling and ANOVA approach as above. These models revealed that cell identity explains the majority of variation in aging magnitudes, but tissue environment explains little (Fig. 6C). The residual in this model represents the small amount of variation caused by the random sampling procedure that we used to compute OT distances. These results were robust across separate NMF optimization runs (Supplemental Fig. S23A) and optimal transport parameters (Supplemental Fig. S23B,C).
We found that different cell states within the same cell type can have notable differences in aging magnitude. For instance, spleen CD8 macrophages showed a larger aging magnitude than spleen CD4 macrophages. Similarly, lung Dcn stromal cells showed a larger aging magnitude than other stromal cell counterparts. Across all three tissues, CD8 T cells showed a larger aging magnitude than CD4 T cells. These observations collectively indicate that cellular identity can have a notable impact on the magnitude of age-related change, such that even different cell states within the same cell type show differences in aging magnitude. In contrast, tissue environments appear to influence aging magnitude less, suggesting that cell identity composition drives most of the difference in aging magnitude between tissues.
Discussion
Aging occurs across varied mammalian species, each composed of diverse cell types and states. Although aging phenotypes have been cataloged at the organismal and tissue level, their constituent cell identities have been less explored. To understand the causes of aging, we aim to construct a causal network of molecular players and their relationships. If these players and relationships are dependent on cell identity, as evidence suggests for tissues (Schumacher et al. 2008; Jonker et al. 2013; Ori et al. 2015; Benayoun et al. 2019), an accurate representation of the causal network requires resolution of the organismal building blocks—individual cells. Here, we used single-cell RNA-seq to investigate aging across a diverse set of murine cell identities in three tissues.
We found that cell identities differentially express unique genes with aging, consistent with previous reports of cell-identity-specific aging phenotypes (Angelidis et al. 2019). Similar cell types (e.g., kidney capillary endothelial cells and lung endothelial cells) showed broadly similar aging trajectories across tissues, and distinct cell types from the same tissue (e.g., B cells and type II pneumocytes in the lung) had dissimilar trajectories. This suggests that cell identities and aging trajectories are coupled, and distinct cell types may be undergoing independent aging processes. Consistent with this notion, cell identity explained the majority of variation in these aging trajectories. Tissue environment explained a lesser amount of variation, but its influence is also present in our data in small sets of unique differentially expressed genes in the same cell identity across tissues. Collectively, our results indicate that the molecular manifestations of aging differ between cell identities and tissue environments. A causal network for aging phenotypes must therefore be conditioned on both cell identity and environment to accurately reflect biology.
Although most changes are unique to tissues or cell types, we identified a shared core of differentially expressed genes with age. This core is characterized by decreased expression of genes involved in SRP-dependent protein translation and protein targeting to the ER and increased expression of genes involved in inflammation, consistent with previous observations in S. cerevisiae (Hendrickson et al. 2018) and multiple studies of aging tissues (Amador-Noguez et al. 2004; Rodwell et al. 2004; O'Brown et al. 2015; Benayoun et al. 2019). Observation of these changes across many cell identities and tissue environments suggests that they are consistent molecular players in the causal network of aging. However, functional studies modulating these pathways in multiple cell identities are necessary to establish causal links to other aging phenotypes.
We present an optimal transport metric to estimate the magnitude of age-related change between two cell populations. Although differential expression and aging trajectories reveal specific gene and pathway changes, the optimal transport distance provides an integrated read-out, allowing us to estimate the magnitude of age-related transcriptional change across cell identities. Using this approach, cell identities showed multifold differences in aging magnitude, and cell identity again explained the majority of variation. Our results are conceptually consistent with previous reports of large transcriptional changes with age in some cell identities and more subtle changes in others (Kowalczyk et al. 2015; Keyes et al. 2016; Davie et al. 2018; Keyes and Fuchs 2018). The magnitude of gene expression changes has been reported to correlate with functional impact (Wawer et al. 2014; Subramanian et al. 2017), suggesting that cell identity aging magnitudes may highlight identities with larger age-related functional changes.
Our results indicate that cell identity influences multiple aspects of aging, highlighting the importance of aging studies at the single-cell level. However, it remains difficult to identify which age-related changes are causal and link molecular changes at the level of individual cell types to physiological aging phenotypes, like reduced glomerular filtration rate or decreased pulmonary regeneration. Future single-cell studies may focus on collecting additional time points and phenotypes throughout the aging process, allowing for time series–based causal inference methods (Granger 1969; Bar-Joseph et al. 2012; Finkle et al. 2018; Qiu et al. 2018; Lu et al. 2019) to reveal the relationships between the molecular players of aging. Functional challenges, such as the differentiation of stem cells during regeneration or the stimulation of immune cells during infection, would also help dissect how transcriptional aging magnitudes and differential gene expression influence tissue function. Single-cell measurements collected during functional challenges may also reveal the dynamics of perturbation and subsequent return to homeostasis necessary to evaluate “resilience” in a given cell type (Kirkland et al. 2016; Hadley et al. 2017).
Methods
Animals
Young (29–34-wk-old) and old (88–93-wk-old) male C57Bl/6J mice were used for all experiments. Young mice were acquired from the Jackson Laboratory at 26 wk of age and allowed to acclimate for at least 3 wk before sacrifice. Old mice were likewise acquired for the Jackson Laboratory at 51 wk of age and aged at Calico Life Sciences to 88–93 wk. Our young mice were at the older extent of the “mature adult” life stage, as compared to some other studies using animals at the early side of the “mature adult” life stage (roughly 12–16 wk of age). By using animals at the end of the mature adult phase, we hoped to minimize contrasts between our young and old animals owing to the late phases of ontogeny, rather than aging. Mice were housed communally with a standard 12 h dark cycle and fed ad libitum. Mice were killed by administration of carbon dioxide at a controlled flow rate and at the same time each day to minimize circadian variation, shortly after the beginning of the light cycle. Kidneys, lungs, and spleen were collected from each experimental animal and weighed. Downstream cell isolation for each tissue proceeded immediately.
Cell isolation
We removed each tissue and washed each in HBSS, then dissected the tissue into small pieces using a razor blade. Briefly, kidney and lung were digested using Liberase DL and TM (Roche), respectively. Spleen was mechanically dissociated. Cells were filtered using 40-µm cell strainers and diluted to 106 cells/mL before single-cell library preparation (for details, see Supplemental Methods).
Single-cell RNA-sequencing experiments
We prepared libraries (individual lanes on the 10x Chromium) with the 10x Single Cell 3′ v2 kit using 6000 cells per lane on the 10x Chromium microfluidics device (10x Genomics). We sequenced libraries at a target depth of 50 million reads/sample on an Illumina HiSeq 4000 (Illumina). For each sample, we performed two technical replicates by preparing two separate libraries from the same cell suspension across two channels of the Chromium microfluidic device.
Read alignment and gene expression quantification
We aligned reads to the mm10 reference genome obtained from Ensembl. We used gene annotations from the GENCODE vM20 release with slight modification (Supplemental Methods). We used 10x Genomics Cell Ranger 3.0.2 to perform alignment, UMI demultiplexing, and cell barcode identification, generating a cells × genes count matrix (Supplemental Methods).
Quality control
We removed libraries that contained a low sequencing depth (less than 10 million reads) or very few cells detected (less than 500 cells) from subsequent analyses as likely experimental errors. Based on this metric, only one technical replicate failed QC, so all tissues from all animals in the experimental design proceeded to our subsequent QC steps. We leveraged the SCANPY toolkit (Wolf et al. 2018) in subsequent analyses. We quantified the total number of UMIs, total number of genes detected, fraction of UMIs mapping to the mitochondrial genome, and fraction of reads mapping to the Rn45s repeat annotation for each cell. We filtered cells with fewer than 1000 UMIs or fewer than 500 unique genes detected, as described for the Tabula Muris (The Tabula Muris Consortium 2018). Additionally, we filtered out cells with a high fraction of reads mapping to the mitochondrial genome (>10%) or the Rn45s repeat (>5%) as likely dead cells (Ilicic et al. 2016).
We identified one sample with large batch effects (young animal 1, lung) and removed it from downstream analysis, leaving three young and three old animals for lung aging comparisons. We identified batch effects based on segregation of the sample into unique Louvain clusters and visible segregation of the sample in the latent space. Lung-specific cell types (alveolar macrophage, endothelial, stromal, type II pneumocyte) either showed large batch effects or were unidentified in this sample.
Dimensionality reduction and embedding
We normalized the cells × genes matrix by the total number of UMIs in each cell and scaled by 106 to yield counts per million (CPM). We natural log transformed this matrix after the addition of a pseudocount of 1 to avoid undefined values. We removed genes that were not detected across replicates in a tissue and genes that were expressed in less than 10 cells in a tissue before dimensionality reduction.
To enable unsupervised clustering and cell type identification, we performed dimensionality reduction with principal component analysis (PCA) on the combined set of samples for each tissue after selection of highly variable genes (Supplemental Methods). Once embedded in this PCA space, we constructed a nearest neighbor graph identifying the k = 15 nearest neighbors for each cell. We derived UMAP embeddings presented for visualization from this nearest neighbor graph using a minimum distance of 0.5 and a spread of 1.0 (Becht et al. 2019).
Clustering and cell type identification
We applied Louvain community detection (Blondel et al. 2008; Satija et al. 2015) to the nearest neighbor graph constructed in PCA space to define a cluster partition. To infer cell types, we trained a neural network classifier to predict cell ontology classes given single-cell RNA-seq mRNA abundance profiles (Supplemental Methods). As a training set, we used the Tabula Muris compendium, which provides expert cell type annotations in the mouse (The Tabula Muris Consortium 2018). In addition to these annotations, we manually added cell state annotations to the Tabula Muris data to provide a level of granularity below cell ontology classes (Supplemental Fig. S1). Example cell states include categorizing T cells into CD4 T cell and CD8 T cell subgroups, as well as the addition of subgroup labels to heterogeneous cell types such as lung stromal cells in the Tabula Muris. After classification, we performed a manual validation of cell type annotation consistent with previous studies (Supplemental Methods; Park et al. 2018; The Tabula Muris Consortium 2018; Angelidis et al. 2019). We also classified cell types using the scmap-cell approach as an orthogonal cell type identification method (Kiselev et al. 2018) and found results largely agreed, but failed to recover correct annotations for several cell types (Supplemental Fig. S6; Supplemental Methods).
Differential cell-type proportion analysis
To determine whether cell-type proportions differed between old and young animals, we performed an additive log ratio (ALR) transform on the observed cell type frequencies and assessed significant changes for each cell type using a t-test. We replaced 0 counts with 1 before the closure operation to avoid undefined values (Aitchison 1982; Martín-Fernández et al. 2003). Within a given cell type, we performed a χ2 test of the age × cell state contingency table to determine if the proportions of cell states change with age.
Differential variability analysis
We measured differences in transcriptional variation between young and old animals in two distinct ways. The first method evaluates changes in the variability of each gene between young and old animals and attempts to identify a shift in the distribution of genewise variation. We assessed gene-specific variability by measuring the “overdispersion” of each gene, as computed using the difference from the median (DM) method (Supplemental Methods; Kowalczyk et al. 2015). We performed DM analysis for each cell state in each tissue in our data set separately.
The second method we used evaluates cell–cell heterogeneity based on the Euclidean distance between cells in expression space, as introduced previously (Enge et al. 2017). For each cell state in each tissue, we computed the centroid of the cell state in gene expression space. We computed the Euclidean distance from each cell to this centroid as a metric of cell–cell heterogeneity within each cell state. We also considered alternative measures of cell–cell heterogeneity using heuristically selected subsets of genes as in previous reports (Enge et al. 2017; Angelidis et al. 2019) and found that results were highly correlated with our primary method (Supplemental Fig. S12; Supplemental Methods).
Differential expression analysis
We computed differentially expressed genes between two groups of cells A and B using the Wilcoxon rank-sum test with the Benjamini-Hochberg procedure for FDR control (Benjamini and Hochberg 1995). We performed differential expression between young and old cells within each cell state in each tissue independently. In addition to comparing mean expression values with the rank sums test, we also computed the proportion of cells expressing each gene in each group. Cells with ≥1 UMIs for a given gene are considered to be expressing the gene, whereas cells with 0 UMIs are considered to not be expressing the gene.
Identification of common and unique differentially expressed genes
To identify common differentially expressed genes with age across cell identities, we computed for each gene the number of cell identities in which that gene is significantly differentially expressed. Note that we counted each cell identity as a single entry in this score, regardless of how many tissues it appears in. For example, if a gene is differentially expressed in lung CD4 T cells and kidney CD4 T cells, we counted this as only a single identity for the purposes of identifying common differentially expressed genes. We then selected genes that are differentially expressed in the same direction in k > 5 cell identities and consider this gene set to be “commonly differentially expressed” with age.
To identify genes that are uniquely differentially expressed between two cell identities A and B, we computed genes which are significantly differentially expressed in A and are (1) not-significantly differentially expressed in B, and (2) have a log2 fold change <0.1 in B.
Gene Ontology enrichment analysis
We used Enrichr (Kuleshov et al. 2016) to perform gene set enrichment analysis against the Gene Ontology Biological Process 2018 version gene set collection. We also used the MSigDB Hallmark gene sets (Liberzon et al. 2015), for which we computed enrichment scores using Fisher's exact test. In both cases, we corrected for multiple hypothesis testing using the Benjamini-Hochberg procedure.
Cell cycle scoring
We estimated cell cycle activity by scoring the expression of a set of S phase–associated and G2/M phase–associated genes, as shown previously (Tirosh et al. 2016) and as implemented in Seurat (Supplemental Methods; Satija et al. 2015) . For our analysis of cell cycle variation with scLVM (Buettner et al. 2015), we fit scLVM models with k = 1 latent variable to explain variation in genes contained in the cell cycle GO term (GO:0007049), as described in the scLVM documentation (Supplemental Methods).
Analysis of variance in transcriptional space
To determine the proportion of variance in transcriptional space (genewise UMI counts, NMF embedding dimensions) explained by experimental factors in our data, we used the linear modeling approach of Robinson et al. (2015) (Supplemental Methods). For each of our linear models (Figs. 5, 6), we treated cell identity (e.g., CD4 T cell) and tissue environment (e.g., lung) as separate factors. We also considered the interaction term of cell identity and tissue environment in these models. We can therefore assess the variance explained by cell identity, tissue environment, and their interaction. The residual variation in linear models fit with aging magnitude as a response reflects variation introduced by our random sampling procedure used to compute the OT distance. Likewise, the residual variation in models fit to aging trajectories derived from bootstrapped samples reflects variation introduced by the random sampling of the bootstrap procedure.
Non-negative matrix factorization embedding
We performed NMF using a standard multiplicative update optimization and random initialization (Lee and Seung 2001), as implemented in the NIMFA package (Zitnik and Zupan 2012). We performed NMF optimization using all cells observed across all three tissues after ln(CPM + 1) normalization. The NMF embedding was fit to a set of highly variable genes, identified as described above. We chose a rank k = 20 for the embedding used for aging trajectories by selecting the “knee” in a plot of rank versus explained variance. We used an NMF embedding of rank k = 500 for optimal transport distance calculation to capture more variation in the data, also chosen based on a later “knee” in the rank versus explained variance curve. We assigned semantic meaning to NMF dimensions using Gene Ontology enrichment analysis on a set of genes with high loadings for a given latent variable (Supplemental Methods).
Aging trajectory calculation
We computed aging trajectories for each cell type/tissue environment combination individually. For example, we computed separate aging trajectories for lung B cells and spleen B cells. For each cell identity, we computed the centroid c of the young cells and old cells in the NMF embedding and computed the vector from the young cells to the old cells . We compared aging trajectories across tissue::cell type groups using the cosine similarity. To assess the statistical significance of these results, we performed permutation testing and bootstrap resampling (Supplemental Methods).
Optimal transport estimation of aging magnitude
We used a discrete optimal transport (OT) distance to estimate the magnitude of difference between two cell populations A and B. Because of the conservation of mass assumption in the optimal transport formulation, we took random samples of the same size n from each population for comparison. We computed the OT distance as minimum cost solution to the linear sum assignment problem, solved using the Munkres algorithm (Munkres 1957). For each comparison of populations A and B, we performed 300 random samples of size n = 300 cells and computed the average to reduce the variability inherent in this stochastic sampling approach. In the data set examined here, multiple populations have <300 cells for sampling. In this circumstance, we set n = 0.8 min(||A||, ||B||) and used the same repeated sampling approach as above.
To estimate the “magnitude of aging,” we computed OT distances for three distinct comparisons. We made a heterochronic comparison of young cells to old cells as a measure of the difference between these populations (old-young comparison). As negative controls, we computed both isochronic comparisons, measuring distances between random samples from pool of young cells (young-young comparison), and measuring distances between samples from the pool of old cells (old-old comparison). The isochronic comparisons serve as an estimate of the distance we would expect between random samples simply attributable to heterogeneity within the population (Supplemental Fig. S22C).
We normalized the heterochronic old-young comparison by dividing these measurements by the mean of the largest isochronic distance (i.e., if young-young distance is larger than old-old, we divide by the young-young mean, and vice versa). This normalization scheme is conservative, using the upper bound estimate of differences caused by cell–cell heterogeneity as our baseline for noting an effect caused by aging. Following normalization, we therefore interpreted old-young OT distances > 1 as indicative of differences caused by aging and interpret a larger value of this normalized distance as reflecting a larger magnitude of age-related change. To ensure these distance estimates are robust to different optimizations of the NMF embedding, we performed NMF optimization using 10 distinct random initializations and compute OT distances in each of these embedding spaces.
Software tools
We used GNU parallel (Tange 2011), the SciPy computing environment (Oliphant 2007), and the Seaborn plotting package for several analyses (https://seaborn.pydata.org/).
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE132901. Additionally, we provide access to relevant data and analysis code on our website (https://mca.research.calicolabs.com/) and as Supplemental Code.
Competing interest statement
The authors are current and former employees of Calico Life Sciences. Funding for experiments was provided by Calico Life Sciences, LLC.
Supplementary Material
Acknowledgments
We thank the Calico Physiology group, including Chunlian Zhang, Adam Kennedy, and Ganesh Kolumam for animal experiment support. We thank the Calico Genomics group led by Margaret Roy for assistance with library preparation and sequencing. We thank Allon Klein and Daniel Gottschling for helpful discussions and support. Organ icons in Figure 1 were designed by Eucalyp from Flaticon.
Author contributions: J.C.K. performed analysis and wrote the paper. L.P. conceived the study and performed experiments. N.D.R. performed analysis. D.G.H. performed analysis and provided experimental support. D.R.K. supervised research and wrote the paper. A.Z.R. conceived the study, supervised research, and performed experiments.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.253880.119.
Freely available online through the Genome Research Open Access option.
References
- Aitchison J. 1982. The statistical analysis of compositional data. J R Stat Soc Ser B (Methodological) 44: 139–160. 10.1111/j.2517-6161.1982.tb01195.x [DOI] [Google Scholar]
- Altschuler SJ, Wu LF. 2010. Cellular heterogeneity: Do differences make a difference? Cell 141: 559–563. 10.1016/j.cell.2010.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador-Noguez D, Yagi K, Venable S, Darlington G. 2004. Gene expression profile of long-lived Ames dwarf mice and Little mice. Aging Cell 3: 423–441. 10.1111/j.1474-9728.2004.00125.x [DOI] [PubMed] [Google Scholar]
- Angelidis I, Simon LM, Fernandez IE, Strunz M, Mayr CH, Greiffo FR, Tsitsiridis G, Ansari M, Graf E, Strom TM, et al. 2019. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat Commun 10: 963 10.1038/s41467-019-08831-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoshiba K, Nagai A. 2007. Chronic lung inflammation in aging mice. FEBS Lett 581: 3512–3516. 10.1016/j.febslet.2007.06.075 [DOI] [PubMed] [Google Scholar]
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dollé ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, et al. 2006. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature 441: 1011–1014. 10.1038/nature04844 [DOI] [PubMed] [Google Scholar]
- Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, Saltness RA, Jeganathan KB, Verzosa GC, Pezeshki A, et al. 2016. Naturally occurring p16Ink4a-positive cells shorten healthy lifespan. Nature 530: 184–189. 10.1038/nature16932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken T, Cowell L, Aevermann BD, Novotny M, Hodge R, Miller JA, Lee A, Chang I, McCorrison J, Pulendran B, et al. 2017. Cell type discovery and representation in the era of high-content single cell phenotyping. BMC Bioinformatics 18: 559 10.1186/s12859-017-1977-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Joseph Z, Gitter A, Simon I. 2012. Studying and modelling dynamic biological processes using time-series gene expression data. Nat Rev Genet 13: 552–564. 10.1038/nrg3244 [DOI] [PubMed] [Google Scholar]
- Beaumont HJ, Gallie J, Kost C, Ferguson GC, Rainey PB. 2009. Experimental evolution of bet hedging. Nature 462: 90–93. 10.1038/nature08504 [DOI] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, Newell EW. 2019. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 37: 38–44. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Benayoun BA, Pollina EA, Singh PP, Mahmoudi S, Harel I, Casey KM, Dulken BW, Kundaje A, Brunet A. 2019. Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res 29: 697–709. 10.1101/gr.240093.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bernitz JM, Kim HS, MacArthur B, Sieburg H, Moore K. 2016. Hematopoietic stem cells count and remember self-renewal divisions. Cell 167: 1296–1309.e10. 10.1016/j.cell.2016.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi B, Cooper DS. 2008. The clinical significance of subclinical thyroid dysfunction. Endocr Rev 29: 76–131. 10.1210/er.2006-0043 [DOI] [PubMed] [Google Scholar]
- Blackwell BN, Bucci TJ, Hart RW, Turturro A. 1995. Longevity, body weight, and neoplasia in ad libitum-fed and diet-restricted C57BL6 mice fed NIH-31 open formula diet. Toxicol Pathol 23: 570–582. 10.1177/019262339502300503 [DOI] [PubMed] [Google Scholar]
- Blake WJ, Balázsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. 2006. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell 24: 853–865. 10.1016/j.molcel.2006.11.003 [DOI] [PubMed] [Google Scholar]
- Blau HM, Cosgrove BD, Ho AT. 2015. The central role of muscle stem cells in regenerative failure with aging. Nat Med 21: 854–862. 10.1038/nm.3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. 2008. Fast unfolding of communities in large networks. J Stat Mech Theory Exp 2008: P10008 10.1088/1742-5468/2008/10/P10008 [DOI] [Google Scholar]
- Brack AS, Muñoz-Cánoves P. 2016. The ins and outs of muscle stem cell aging. Skelet Muscle 6: 1 10.1186/s13395-016-0072-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayton CF, Treuting PM, Ward JM. 2012. Pathobiology of aging mice and GEM: background strains and experimental design. Vet Pathol 49: 85–105. 10.1177/0300985811430696 [DOI] [PubMed] [Google Scholar]
- Brunet JP, Tamayo P, Golub TR, Mesirov JP. 2004. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci 101: 4164–4169. 10.1073/pnas.0308531101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, Theis FJ, Teichmann SA, Marioni JC, Stegle O. 2015. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol 33: 155–160. 10.1038/nbt.3102 [DOI] [PubMed] [Google Scholar]
- Calinski T, Harabasz J. 1974. A dendrite method for cluster analysis. Commun Stat 3: 1–27. 10.1080/03610927408827101 [DOI] [Google Scholar]
- Chakkalakal JV, Jones KM, Basson MA, Brack AS. 2012. The aged niche disrupts muscle stem cell quiescence. Nature 490: 355–360. 10.1038/nature11438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs BG, Gluscevic M, Baker DJ, Laberge RM, Marquess D, Dananberg J, van Deursen JM. 2017. Senescent cells: an emerging target for diseases of ageing. Nat Rev Drug Discov 16: 718–735. 10.1038/nrd.2017.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D. 1966. Optimizing reproduction in a randomly varying environment. J Theor Biol 12: 119–129. 10.1016/0022-5193(66)90188-3 [DOI] [PubMed] [Google Scholar]
- Cosgrove BD, Gilbert PM, Porpiglia E, Mourkioti F, Lee SP, Corbel SY, Llewellyn ME, Delp SL, Blau HM. 2014. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat Med 20: 255–264. 10.1038/nm.3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie K, Janssens J, Koldere D, De Waegeneer M, Pech U, Kreft Ł, Aibar S, Makhzami S, Christiaens V, González-Blas CB, et al. 2018. A single-cell transcriptome atlas of the aging Drosophila brain. Cell 174: 982–998.e20. 10.1016/j.cell.2018.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decman V, Laidlaw BJ, Doering TA, Leng J, Ertl HC, Goldstein DR, Wherry EJ. 2012. Defective CD8 T cell responses in aged mice are due to quantitative and qualitative changes in virus-specific precursors. J Immunol 188: 1933–1941. 10.4049/jimmunol.1101098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drigo R, Lev-Ram V, Tyagi S, Ramachandra R, Deerinck T, Bushong E, Phan S, Orphan V, Lechene C, Ellisman MH, et al. 2019. Age mosaicism across multiple scales in adult tissues. Cell Metab 30: 343–351.e3. 10.1016/j.cmet.2019.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enge M, Arda HE, Mignardi M, Beausang J, Bottino R, Kim SK, Quake SR. 2017. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell 171: 321–330.e14. 10.1016/j.cell.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkle JD, Wu JJ, Bagheri N. 2018. Windowed Granger causal inference strategy improves discovery of gene regulatory networks. Proc Natl Acad Sci 115: 2252–2257. 10.1073/pnas.1710936115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach J, Bakker ST, Mohrin M, Conroy PC, Pietras EM, Reynaud D, Alvarez S, Diolaiti ME, Ugarte F, Forsberg EC, et al. 2014. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 512: 198–202. 10.1038/nature13619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesing A, Lewiński A, Karbownik-Lewińska M. 2012. The thyroid gland and the process of aging; what is new? Thyroid Res 5: 16 10.1186/1756-6614-5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladyshev VN. 2013. The origin of aging: Imperfectness-driven non-random damage defines the aging process and control of lifespan. Trends Genet 29: 506–512. 10.1016/j.tig.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goronzy JJ, Fang F, Cavanagh MM, Qi Q, Weyand CM. 2015. Naive T cell maintenance and function in human aging. J Immunol 194: 4073–4080. 10.4049/jimmunol.1500046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger CWJ. 1969. Investigating causal relations by econometric models and cross-spectral methods. Econometrica 37: 424 10.2307/1912791 [DOI] [Google Scholar]
- Hadley EC, Kuchel GA, Newman AB, Allore HG, Bartley JM, Bergeman CS, Blinov ML, Colon-Emeric CS, Dabhar FS, Dugan LL, et al. 2017. Report: NIA workshop on measures of physiologic resiliencies in human aging. J Gerontol A Biol Sci Med Sci 72: 980–990. 10.1093/gerona/glx015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, et al. 2018. Mapping the mouse cell atlas by microwell-seq. Cell 173: 1307 10.1016/j.cell.2018.05.012 [DOI] [PubMed] [Google Scholar]
- Hendrickson DG, Soifer I, Wranik BJ, Kim G, Robles M, Gibney PA, McIsaac RS. 2018. A new experimental platform facilitates assessment of the transcriptional and chromatin landscapes of aging yeast. eLife 7: e39911 10.7554/eLife.39911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. 2013. DNA methylation age of human tissues and cell types. Genome Biol 14: R115 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Argmann C, Houten SM, Cantó C, Jeninga EH, Andreux PA, Thomas C, Doenlen R, Schoonjans K, Auwerx J. 2011. The metabolic footprint of aging in mice. Sci Rep 1: 9–11. 10.1038/srep00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilicic T, Kim JK, Kolodziejczyk AA, Bagger FO, McCarthy DJ, Marioni JC, Teichmann SA. 2016. Classification of low quality cells from single-cell RNA-seq data. Genome Biol 17: 29 10.1186/s13059-016-0888-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonker MJ, Melis JP, Kuiper RV, van der Hoeven TV, Wackers PFK, Robinson J, van der Horst GT, Dollé ME, Vijg J, Breit TM, et al. 2013. Life spanning murine gene expression profiles in relation to chronological and pathological aging in multiple organs. Aging Cell 12: 901–909. 10.1111/acel.12118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. 2010. The genetics of ageing. Nature 464: 504–512. 10.1038/nature08980 [DOI] [PubMed] [Google Scholar]
- Keyes BE, Fuchs E. 2018. Stem cells: aging and transcriptional fingerprints. J Cell Biol 217: 79–92. 10.1083/jcb.201708099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes BE, Segal JP, Heller E, Lien WH, Chang CY, Guo X, Oristian DS, Zheng D, Fuchs E. 2013. Nfatc1 orchestrates aging in hair follicle stem cells. Proc Natl Acad Sci 110: E4950–E4959. 10.1073/pnas.1320301110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes BE, Liu S, Asare A, Naik S, Levorse J, Polak L, Lu CP, Nikolova M, Pasolli HA, Fuchs E. 2016. Impaired epidermal to dendritic T cell signaling slows wound repair in aged skin. Cell 167: 1323–1338.e14. 10.1016/j.cell.2016.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey C, Balakrishnan V, O'Dell MR, Huang JL, Newman L, Whitney-Miller CL, Hezel AF, Land H. 2014. Plac8 links oncogenic mutations to regulation of autophagy and is critical to pancreatic cancer progression. Cell Rep 7: 1143–1155. 10.1016/j.celrep.2014.03.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland JL, Stout MB, Sierra F. 2016. Resilience in aging mice. J Gerontol A Biol Sci Med Sci 71: 1407–1414. 10.1093/gerona/glw086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood TBL. 1977. Evolution of ageing. Nature 270: 301–304. 10.1038/270301a0 [DOI] [PubMed] [Google Scholar]
- Kirschner K, Chandra T, Kiselev V, Cruz DFS, Macaulay IC, Park HJ, Li J, Kent DG, Kumar R, Pask DC, et al. 2017. Proliferation drives aging-related functional decline in a subpopulation of the hematopoietic stem cell compartment. Cell Rep 19: 1503–1511. 10.1016/j.celrep.2017.04.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev VY, Yiu A, Hemberg M. 2018. scmap: projection of single-cell RNA-seq data across data sets. Nat Methods 15: 359–362. 10.1038/nmeth.4644 [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk AA, Kim JK, Tsang JC, Ilicic T, Henriksson J, Natarajan KN, Tuck AC, Gao X, Bühler M, Liu P, et al. 2015. Single cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell 17: 471–485. 10.1016/j.stem.2015.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotliar D, Veres A, Nagy MA, Tabrizi S, Hodis E, Melton DA, Sabeti PC. 2019. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. eLife 8: e43803 10.7554/eLife.43803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, Schneider RK, Wagers AJ, Ebert BL, Regev A. 2015. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res 25: 1860–1872. 10.1101/gr.192237.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussell E, Kishony R, Balaban NQ, Leibler S. 2005. Bacterial persistence: a model of survival in changing environments. Genetics 169: 1807–1814. 10.1534/genetics.104.035352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DD, Seung HS. 2001. Algorithms for non-negative matrix factorization. In Advances in neural information processing systems 13 (NIPS 2000) (ed. Leen TK, et al. ), pp. 535–541, Denver MIT Press, Cambridge, MA. [Google Scholar]
- Lee CK, Allison DB, Brand J, Weindruch R, Prolla TA. 2002. Transcriptional profiles associated with aging and middle age-onset caloric restriction in mouse hearts. Proc Natl Acad Sci 99: 14988–14993. 10.1073/pnas.232308999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenburg ME, Sinha A, Faller DV, Denis GV. 2007. Tumor-specific and proliferation-specific gene expression typifies murine transgenic B cell lymphomagenesis. J Biol Chem 282: 4803–4811. 10.1074/jbc.M605870200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ju J, Weyand CM, Goronzy JJ. 2015. Age-associated failure to adjust type I IFN receptor signaling thresholds after T cell activation. J Immunol 195: 865–874. 10.4049/jimmunol.1402389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Mar BG, Zhang H, Puram RV, Vazquez F, Weir BA, Hahn WC, Ebert B, Pellman D. 2017. The EMT regulator ZEB2 is a novel dependency of human and murine acute myeloid leukemia. Blood 129: 497–508. 10.1182/blood-2016-05-714493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. 2015. The molecular signatures database hallmark gene set collection. Cell Syst 1: 417–425. 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cheung TH, Charville GW, Hurgo BM, Leavitt T, Shih J, Brunet A, Rando TA. 2013. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep 4: 189–204. 10.1016/j.celrep.2013.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Shadel GS, Kaeberlein M, Kennedy B. 2012. Replicative and chronological aging in saccharomyces cerevisiae. Cell Metab 16: 18–31. 10.1016/j.cmet.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. 2013. The hallmarks of aging. Cell 153: 1194–1217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Dumitrascu B, McDowell IC, Jo B, Barrera A, Hong LK, Leichter SM, Reddy TE, Engelhardt BE. 2019. Causal network inference from gene transcriptional time series response to glucocorticoids. bioRxiv 10.1101/587170 [DOI] [PMC free article] [PubMed]
- Lumeng CN, Liu J, Geletka L, Delaney C, Delproposto J, Desai A, Oatmen K, Martinez-Santibanez G, Julius A, Garg S, et al. 2011. Aging is associated with an increase in T cells and inflammatory macrophages in visceral adipose tissue. J Immunol 187: 6208–6216. 10.4049/jimmunol.1102188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Fernández JA, Barceló-Vidal C, Pawlowsky-Glahn V. 2003. Dealing with zeros and missing values in compositional data sets using nonparametric imputation. Math Geol 35: 253–278. 10.1023/A:1023866030544 [DOI] [Google Scholar]
- Martinez-Jimenez CP, Eling N, Chen HC, Vallejos CA, Kolodziejczyk AA, Connor F, Stojic L, Rayner TF, Stubbington MJT, Teichmann SA, et al. 2017. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science 355: 1433–1436. 10.1126/science.aah4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkres J. 1957. Algorithms for the assignment and transportation problems. J Soc Ind Appl Math 5: 32–38. 10.1137/0105003 [DOI] [Google Scholar]
- Nalapareddy K, Nattamai KJ, Kumar RS, Karns R, Wikenheiser-Brokamp KA, Sampson LL, Mahe MM, Sundaram N, Yacyshyn MB, Yacyshyn B, et al. 2017. Canonical Wnt signaling ameliorates aging of intestinal stem cells. Cell Rep 18: 2608–2621. 10.1016/j.celrep.2017.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brown ZK, Van Nostrand EL, Higgins JP, Kim SK. 2015. The inflammatory transcription factors NFκB, STAT1 and STAT3 drive age-associated transcriptional changes in the human kidney. PLoS Genet 11: e1005734 10.1371/journal.pgen.1005734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant TE. 2007. Python for scientific computing. Comput Sci Eng 9: 10–20. 10.1109/MCSE.2007.58 [DOI] [Google Scholar]
- Ori A, Toyama BH, Harris MS, Bock T, Iskar M, Bork P, Ingolia NT, Hetzer MW, Beck M. 2015. Integrated transcriptome and proteome analyses reveal organ-specific proteome deterioration in old rats. Cell Syst 1: 224–237. 10.1016/j.cels.2015.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, Li M, Barasch J, Suszták K. 2018. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360: 758–763. 10.1126/science.aar2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettan-Brewer C, Treuting PM. 2011. Practical pathology of aging mice. Pathobiol Aging Age Relat Dis 1: 7202 10.3402/pba.v1i0.7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Rahimzamani A, Wang L, Mao Q, Durham T, Figueroa JLM, Saunders L, Trapnell C, Kannan S. 2018. Towards inferring causal gene regulatory networks from single cell expression measurements. bioRxiv 10.1101/426981 [DOI] [PMC free article] [PubMed]
- Quinn KM, Zaloumis SG, Cukalac T, Kan WT, Sng XY, Mirams M, Watson KA, McCaw JM, Doherty PC, Thomas PG, et al. 2016. Heightened self-reactivity associated with selective survival, but not expansion, of naïve virus-specific CD8+ T cells in aged mice. Proc Natl Acad Sci 113: 1333–1338. 10.1073/pnas.1525167113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn KM, Fox A, Harland KL, Russ BE, Li J, Nguyen THO, Loh L, Olshanksy M, Naeem H, Tsyganov K, et al. 2018. Age-related decline in primary CD8+ T cell responses is associated with the development of senescence in virtual memory CD8+ T cells. Cell Rep 23: 3512–3524. 10.1016/j.celrep.2018.05.057 [DOI] [PubMed] [Google Scholar]
- Robinson DG, Wang JY, Storey JD. 2015. A nested parallel experiment demonstrates differences in intensity-dependence between RNA-seq and microarrays. Nucleic Acids Res 43: e131 10.1093/nar/gkv636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodwell GE, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang L, Xiao W, Mindrinos M, Crane E, Segal E, et al. 2004. A transcriptional profile of aging in the human kidney. PLoS Biol 2: e427 10.1371/journal.pbio.0020427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A. 2015. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33: 495–502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebinger G, Shu J, Tabaka M, Cleary B, Subramanian V, Solomon A, Gould J, Liu S, Lin S, Berube P, et al. 2019. Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell 176: 928–943.e22. 10.1016/j.cell.2019.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, van der Pluijm I, Moorhouse MJ, Kosteas T, Robinson AR, Suh Y, Breit TM, van Steeg H, Niedernhofer LJ, van IJcken W, et al. 2008. Delayed and accelerated aging share common longevity assurance mechanisms. PLoS Genet 4: e1000161 10.1371/journal.pgen.1000161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Höfer T. 2017. Robust classification of single-cell transcriptome data by nonnegative matrix factorization. Bioinformatics 33: 235–242. 10.1093/bioinformatics/btw607 [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Sherr CJ. 2015. Forging a signature of in vivo senescence. Nat Rev Cancer 15: 397–408. 10.1038/nrc3960 [DOI] [PubMed] [Google Scholar]
- Shaw AC, Joshi S, Greenwood H, Panda A, Lord JM. 2010. Aging of the innate immune system. Curr Opin Immunol 22: 507–513. 10.1016/j.coi.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. 1974. Hedging one's evolutionary bets. Nature 250: 704–705. 10.1038/250704b0 [DOI] [Google Scholar]
- Sousa-Victor P, Gutarra S, Garcia-Prat L. 2014. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506: 316–321. 10.1038/nature13013 [DOI] [PubMed] [Google Scholar]
- Steffen KK, Dillin A. 2016. A ribosomal perspective on proteostasis and aging. Cell Metab 23: 1004–1012. 10.1016/j.cmet.2016.05.013 [DOI] [PubMed] [Google Scholar]
- Su J, Ekman C, Oskolkov N, Lahti L, Ström K, Brazma A, Groop L, Rung J, Hansson O. 2015. A novel atlas of gene expression in human skeletal muscle reveals molecular changes associated with aging. Skelet Muscle 5: 35 10.1186/s13395-015-0059-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, et al. 2017. A Next Generation Connectivity Map: L1000 platform and the first 1,000,000 profiles. Cell 171: 1437–1452.e17. 10.1016/j.cell.2017.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. 2007. Tunability and noise dependence in differentiation dynamics. Science 315: 1716–1719. 10.1126/science.1137455 [DOI] [PubMed] [Google Scholar]
- The Tabula Muris Consortium. 2018. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562: 367–372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange O. 2011. GNU Parallel: the command-line power tool. The USENIX Magazine 36: 42–47. [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. 2016. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352: 189–196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA. 2014. Using C. elegans for aging research. Invertebr Reprod Dev 59: 59–63. 10.1080/07924259.2014.940470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toapanta FR, Ross TM. 2009. Impaired immune responses in the lungs of aged mice following influenza infection. Respir Res 10: 112 10.1186/1465-9921-10-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todhunter ME, Sayaman RW, Miyano M, LaBarge MA. 2018. Tissue aging: the integration of collective and variant responses of cells to entropic forces over time. Curr Opin Cell Biol 54: 121–129. 10.1016/j.ceb.2018.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser WE, Bombardieri CR, Zevenbergen C, Barnhoorn S, Ottaviani A, van der Pluijm I, Brandt R, Kaptein E, van Heerebeek R, van Toor H, et al. 2016. Tissue-specific suppression of thyroid hormone signaling in various mouse models of aging. PLoS One 11: e0149941 10.1371/journal.pone.0149941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawer MJ, Li K, Gustafsdottir SM, Ljosa V, Bodycombe NE, Marton MA, Sokolnicki KL, Bray MA, Kemp MM, Winchester E, et al. 2014. Toward performance-diverse small-molecule libraries for cell-based phenotypic screening using multiplexed high-dimensional profiling. Proc Natl Acad Sci 111: 10911–10916. 10.1073/pnas.1410933111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weismann A, Poulton EB, Schönland S, Shipley AE. 1891. Essays upon heredity and kindred biological problems. Clarendon Press, Oxford, UK. [Google Scholar]
- White RR, Milholland B, MacRae SL, Lin M, Zheng D, Vijg J. 2015. Comprehensive transcriptional landscape of aging mouse liver. BMC Genomics 16: 899. 10.1186/s12864-015-2061-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, Theis FJ. 2018. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19: 15 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. 2014. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci 111: 16219–16224. 10.1073/pnas.1408886111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Chen HZ, Liu DP. 2015. The four layers of aging. Cell Syst 1: 180–186. 10.1016/j.cels.2015.09.002 [DOI] [PubMed] [Google Scholar]
- Zhao S, Molina A, Yu A, Hanson J, Cheung H, Li X, Natkunam Y. 2019. High frequency of CD74 expression in lymphomas: implications for targeted therapy using a novel anti-CD74-drug conjugate. J Pathol Clin Res 5: 12–24. 10.1002/cjp2.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitnik M, Zupan B. 2012. NIMFA: a Python library for nonnegative matrix factorization. J Mach Learn Res 13: 849–853. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.