Abstract
Transcription factor binding to target sites in vivo is a dynamic process that involves cycles of association and dissociation, with individual proteins differing in their binding dynamics. The dynamics at individual sites on a genomic scale have been investigated in yeast cells, but comparable experiments have not been done in multicellular eukaryotes. Here, we describe a tamoxifen-inducible, time-course ChIP-seq approach to measure transcription factor binding dynamics at target sites throughout the human genome. As observed in yeast cells, the TATA-binding protein (TBP) typically displays rapid turnover at RNA polymerase (Pol) II-transcribed promoters, slow turnover at Pol III promoters, and very slow turnover at the Pol I promoter. Turnover rates vary widely among Pol II promoters in a manner that does not correlate with the level of TBP occupancy. Human Pol II promoters with slow TBP dissociation preferentially contain a TATA consensus motif, support high transcriptional activity of downstream genes, and are linked with specific activators and chromatin remodelers. These properties of human promoters with slow TBP turnover differ from those of yeast promoters with slow turnover. These observations suggest that TBP binding dynamics differentially affect promoter function and gene expression, possibly at the level of transcriptional reinitiation/bursting.
Binding of transcription factors to specific genomic DNA sequences is required for accurate and regulated transcription by RNA polymerases. This ensures biologically appropriate levels of RNA transcripts for a wide variety of environmental and developmental conditions. Transcription factor binding in vivo is analyzed conventionally by chromatin immunoprecipitation (ChIP), which measures occupancy at target sites on a cell- and time-averaged basis (Struhl 2007). However, ChIP represents a static measurement that does not consider the dynamics of binding, namely the dissociation and reassociation of proteins with their target sites.
Fluorescence recovery after photobleaching (FRAP) experiments indicate that many transcription factors show highly dynamic binding with very rapid dissociation rates, while other proteins (e.g., histones) have much slower turnover (McNally et al. 2000; Houtsmuller 2005; Mueller et al. 2010). However, FRAP experiments typically measure the average dynamic properties of a given protein on all target sites, and it is not possible to distinguish unbound versus DNA-bound proteins in the bleached area. Using live-cell imaging, binding dynamics at specific DNA sites can be visualized on artificially tandem arrays of binding sites (McNally et al. 2000) or at genomic regions consisting of naturally occurring repeats (Karpova et al. 2008). More recent imaging technologies enable detection of binding on a specific endogenous gene at the single molecule level (Donovan et al. 2019). In yeast, binding dynamics on endogenous single-copy genes can be measured on an individual basis by using a quench flow apparatus to perform a formaldehyde time course on a subsecond scale (Poorey et al. 2013; Zaidi et al. 2017). However, none of these methods address whether binding dynamics are uniform or variable over the entire range of target sites.
Genome-scale, site-specific analysis of transcription factor binding dynamics has been performed in yeast using a competition-ChIP approach (Dion et al. 2007; van Werven et al. 2009; Lickwar et al. 2012). Expression of an epitope-tagged transcription factor is induced by the addition of galactose, and whole-genome ChIP measurements are made at various times after induction. The kinetics of binding by the induced protein (distinguished by its epitope tag) at each target site provide information on protein turnover at that site. Analyses of yeast TATA-binding protein (TBP), Rap1, and histone H3 by competition ChIP reveal that binding dynamics are variable at their target sites in a manner that is poorly correlated with occupancy levels determined by conventional ChIP (Dion et al. 2007; van Werven et al. 2009; Lickwar et al. 2012). Comparable experiments have not been performed in any multicellular organism.
Here, we describe a tamoxifen-inducible, time-course ChIP-seq analysis that permits the measurement of transcription factor binding dynamics at target sites on a genome-wide scale in human cells. Tamoxifen is an agonist of estrogen receptors, and it induces nuclear translocation of hybrid proteins containing the ligand-binding domain of the estrogen receptor. We apply this method to analyze the dynamics of the TATA-binding protein. In vivo, TBP is required for transcription from promoters mediated by all three nuclear RNA polymerases (Pol) (Cormack and Struhl 1992). These three classes of promoters are responsible for the synthesis of rRNA (Pol I), mRNA and other RNAs (Pol II), and tRNA and other RNAs (Pol III). In mammalian cells, TBP does not bind promoters on its own but rather as part of multiprotein complexes that are specific for the three promoter classes (SL1 for Pol I, TFIID for Pol II, and TFIIIB for Pol III) (Sharp 1992; Hernandez 1993; Struhl 1994). Our results indicate similarities and differences in promoter-specific TBP binding dynamics in human and yeast cells.
Results
Measuring dynamics of transcription factor binding to target sites in human cells
The genome-scale assay involves the rapid induction of an epitope-tagged protein and performing ChIP-seq analysis at various times after induction (Fig. 1A). The binding kinetics of the epitope-tagged protein provide a measurement of the turnover of the untagged, endogenous protein previously bound to the same sites. Although this method does not measure the absolute dissociation rate of the endogenous protein, it is suitable for measuring relative turnover rates at different loci. Specifically, the target sites are simultaneously assayed with the same mixture of epitope-tagged and untagged endogenous protein at each time point (Fig. 1A).
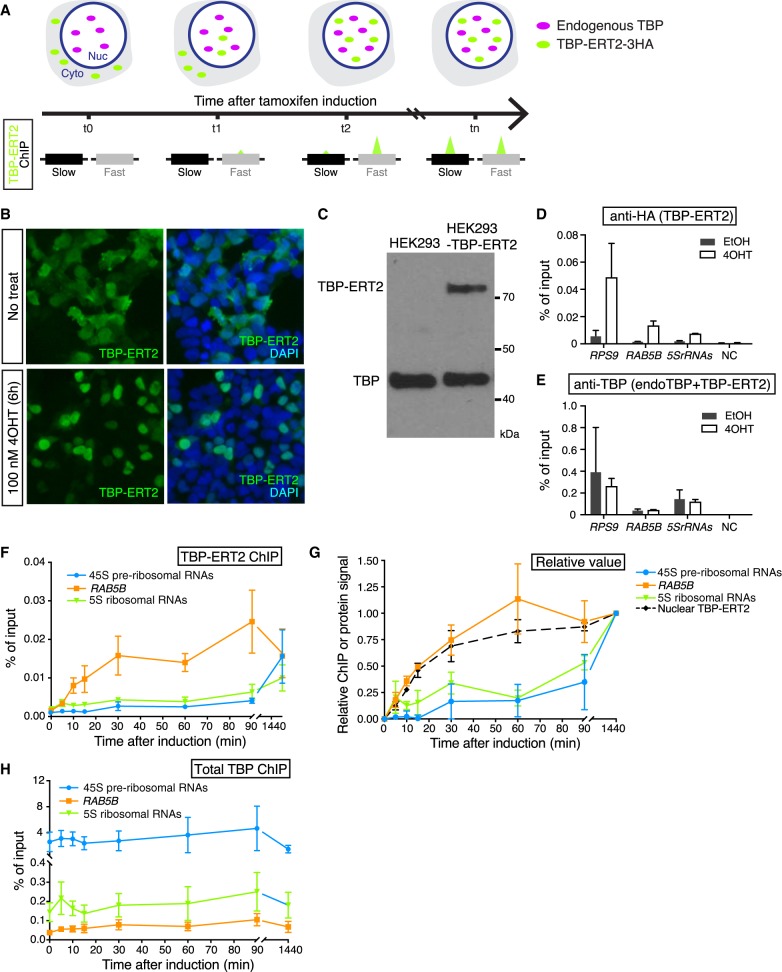
Figure 1.
Nuclear translocation of TBP-ERT2. (A) Schematic illustration of tamoxifen-inducible, time-course ChIP analysis. (B) Immunofluorescence image of TBP-ERT2 (green) and DAPI (blue) in HEK293 cells transiently transfected with TBP-ERT2-expressing vector that are treated or nontreated with 100 nM tamoxifen (4OHT) for 6 hr. (C) Western blot of whole-cell extracts from HEK293- and TBP-ERT2-expressing cells with antibody against endogenous TBP. (D,E) Binding at the indicated loci (x-axis) of TBP-ERT2 (D) and total TBP (endogenous TBP and TBP-ERT2) (E) in cells stably expressing TBP-ERT2. Error bars indicate SD (n = 3). (F) Binding of TBP-ERT2 at the indicated loci at the indicated times after tamoxifen addition. (G) Abundance of nuclear TBP-ERT2 (black dashed line) and binding relative to the value at 1440 min. Error bars indicate SD (n = 3). (H) Binding of total TBP at the indicated loci at the indicated times after tamoxifen addition.
For measuring TBP binding dynamics in HEK293 cells, we expressed a 3HA-tagged TBP derivative that contains ERT2, the ligand binding domain of the estrogen receptor that permits rapid nuclear translocation of the fused protein and binding to its target sites upon tamoxifen addition. As expected, tamoxifen induces translocation of TBP-ERT2 in the cytoplasm to the nucleus (Fig. 1B). We established a cell line that expresses TBP-ERT2 at a level that is 20% of endogenous TBP (Fig. 1C) and confirmed a significant increase of TBP-ERT2 association with target sites upon tamoxifen treatment (Fig. 1D). In contrast, target site association of total TBP (sum of endogenous TBP and TBP-ERT2) is similar before and after induction (Fig. 1E). The increase of the nuclear TBP-ERT2 amount starts within 5 min and is near steady-state within 30–60 min (Supplemental Fig. S1A,B), a time-frame comparable to that of GAL1 induction in yeast.
Different TBP turnover rates at Pol I, Pol II, and Pol III promoters
As an initial experiment, we analyzed promoters representing the three different RNA polymerases: 45S preribosomal rDNA (Pol I promoter); RAB5B (Pol II promoter); 5S ribosomal RNAs (Pol III promoter). TBP-ERT2 association at the RAB5B promoter quickly increases in a manner kinetically indistinguishable from the increase in nuclear TBP-ERT2 concentration (Fig. 1F,G), indicating a turnover rate of <5 min. In contrast, TBP-ERT2 association at the 45S pre-rRNAs and 5S rRNAs promoters increases much more slowly, indicating a much slower turnover rate at Pol I and Pol III promoters (Fig. 1F,G). The slow turnover of TBP-ERT2 at the 45S pre-rRNAs promoter is not due to slow access to the nucleolus (the site of ribosomal genes), because TBP-ERT2 localizes to the nucleolus at early time points when TBP-ERT2 is not bound at the 45S pre-rRNAs promoter (Supplemental Fig. S1C). In contrast to TBP-ERT2, the ChIP signal for total TBP at all these promoters remains constant throughout the time course (Fig. 1H). The fast turnover at Pol II promoters and slow turnover at Pol I and Pol III promoters in human cells is similar to what is observed in yeast (van Werven et al. 2009; Zaidi et al. 2017).
Genome-wide analysis of TBP binding dynamics
We performed time-course ChIP-seq analysis of TBP-ERT2 to examine the TBP binding dynamics at target sites on a genome-wide scale. To account for differences in immunoprecipitation efficiency among the samples, we added a constant amount of sonicated yeast chromatin prepared from a strain expressing 3HA-TBP as a spike-in control for each sample (Bonhoure et al. 2014; Orlando et al. 2014). We obtained 30–60 million reads/sample (Supplemental Table S1) and analyzed the 13,148 TBP-ERT2 peaks that met our criteria. TBP-ERT2-3HA ChIP signals are thus normalized with yeast 3HA-TBP signals at each time point. Approximately 85% of TBP-ERT2 peaks are located in promoter regions (−1 kb to +100 bp from transcription start sites [TSSs]) of annotated genes transcribed by Pol I, Pol II, and Pol III (Supplemental Fig. S2A); we presume the remaining 15% of target sites are mis- or unannotated promoters. As expected, levels of TBP-ERT2 association at all time points (except 0) are highly correlated, and they gradually increase over time (Fig. 2A; Supplemental Fig. S2B,C). Most peaks show greater than fourfold increased association between the final induced state compared to the noninduced state (Supplemental Fig. S2D).
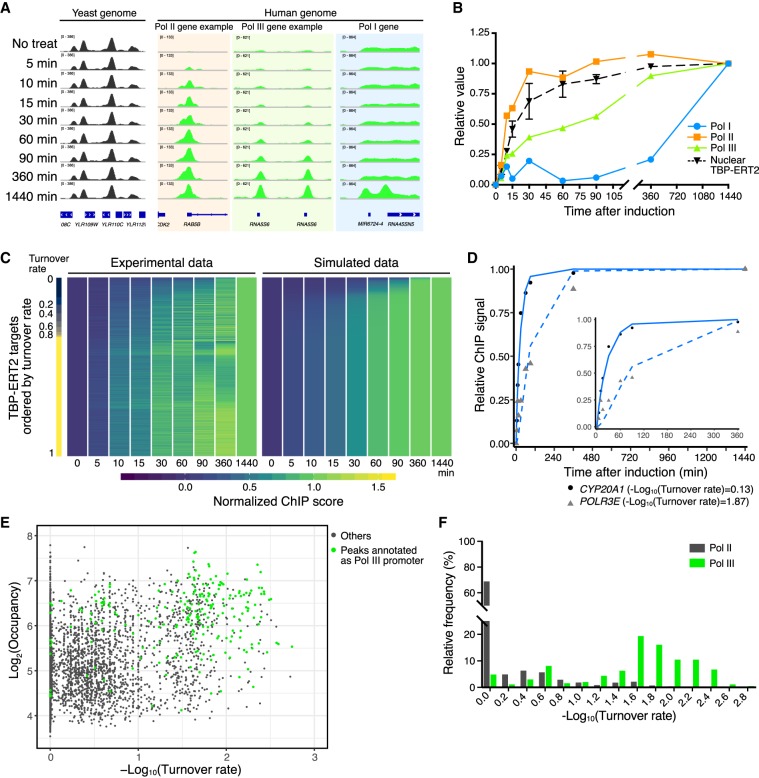
Figure 2.
Time-course ChIP-seq analysis. (A) Examples of human loci bound by TBP-ERT2 and yeast loci bound by 3HA-TBP (spike-in). (B) TBP-ERT2 binding to the composite average of Pol I, Pol II, and Pol III promoters relative to the 1440-min sample. (C) TBP-ERT2 binding relative to the 1440-min sample (left) and simulated relative TBP-ERT2 binding by using turnover rate (right) of all detected peaks. Peaks are ordered by turnover rate value (Lambda). (D) Examples of TBP-ERT2 binding and simulated data at the CYP20A1 (solid line) and POLR3E (dashed line) loci. (E) Scatter plot of log2 TBP-ERT2 occupancy versus −log10TBP-ERT2 binding turnover rate. (F) Histogram of the relative frequency of −log10TBP-ERT2 binding turnover rate.
To compare the occupancy of TBP-ERT2 at each locus over time, we first subtracted ChIP signals at 0 min from those at all the other time points and then calculated relative ChIP signals with respect to the endpoint (1440 min). On an overall basis and in accord with results on individual promoters, TBP-ERT2 occupancy at the majority of Pol II promoters is kinetically indistinguishable from nuclear translocation, whereas occupancy at Pol III (half-maximal occupancy at 90 min) and Pol I (half-maximal occupancy >6 h) is much slower (Fig. 2B).
TBP turnover rates among Pol II or Pol III promoters are not related to occupancy
Using relative nuclear TBP-ERT2 levels (Supplemental Fig. S3A) and time-course ChIP-seq data, we employed a mathematical model (described in Methods) to calculate binding turnover rates (λ) for each TBP-ERT2 target site. The different kinetics of TBP-ERT2 occupancy can be simulated by using different binding turnover rates (Supplemental Fig. S3B). Slower turnover rates (larger value of −log10[turnover rate]) indicates that a longer time is required for the ChIP signal to reach to the half-maximal steady-state level (Supplemental Fig. S3C). We extracted 6476 sites for which the goodness of fit (squared norm of the residual [Resnorm]) is <0.4 and used this subset for subsequent analysis (Supplemental Fig. S4). Overall, the experimental data are captured well by the simulated data (Fig. 2C,D). Although the ribosomal genes are highly repetitive, with individual copies being heterogeneous in transcriptional activity and chromatin state (Conconi et al. 1989; Cloix et al. 2002), the time-course data at these genes fits the mathematical model, arguing that the results are limited to active genes and against the idea of subpopulations of loci with different TBP binding dynamics.
Importantly, TBP-ERT2 binding turnover rates among Pol II promoters or among Pol III promoters can vary considerably, and these different turnover rates correlate poorly (r = 0.32) with the level of TBP-ERT2 occupancy (Fig. 2E,F). Thus, the TBP turnover rate at human promoters is an independent parameter that cannot be determined from the occupancy at one time point (Fig. 2E; Supplemental Fig. S3D).
Slow TBP turnover rates at Pol II promoters are linked to a consensus TATA sequence, certain transcription factors, and accessible chromatin
To address why some Pol II promoters have slower TBP binding turnover rates than others, we categorized TBP-ERT2-bound Pol II promoters into three classes based on their turnover rates (Fig. 3A) and searched for enriched DNA sequence motifs that differentiated the classes. Promoters with slow TBP turnover rates are strongly enriched for the TBP consensus binding motif, namely the TATA element (53% of the top motif, 28% of the center motif, and 37% of the bottom motif) (Fig. 3B). To independently verify this observation, we scanned each peak region for the sequence with the highest match score to the position weight matrix of the TBP consensus motif. The cumulative curve and the box plot of match scores indicate that the slow turnover sites have higher similarity to the TATA motif, the middle turnover sites have intermediate similarity, and the fast turnover sites have lowest similarity (Fig. 3C,D). Half of the slow turnover sites have at least one strong TBP consensus motif in the same direction of the downstream gene (forward motifs, Fig. 3E). Forward motifs in the middle or slow turnover classes are densely clustered around ±100 bp around the transcription start site of downstream genes, but forward motifs in the fast class or reverse motifs in all classes are more dispersed (Fig. 3F). In contrast to the TATA motif, other core promoter elements, including the initiator, are not enriched among any of the various turnover classes (Supplemental Fig. S5).
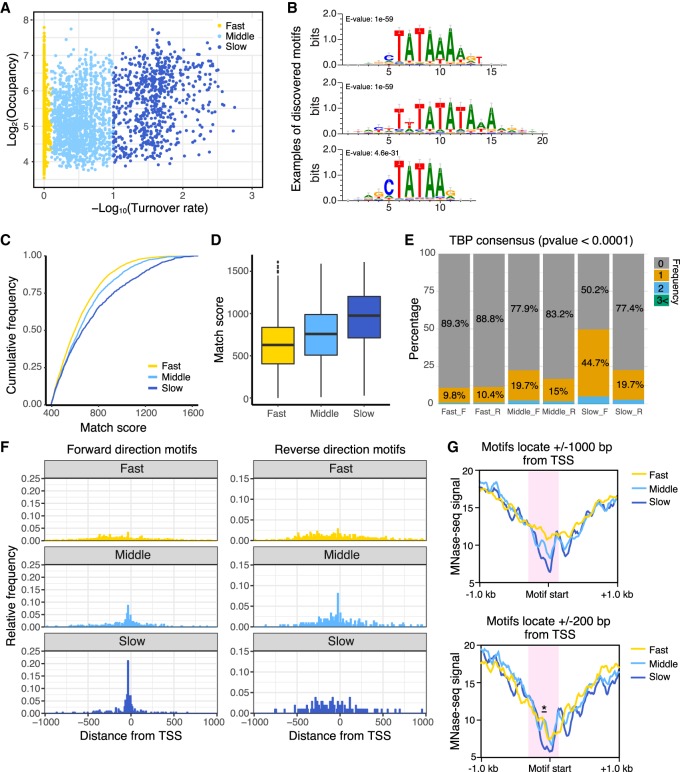
Figure 3.
Pol II promoters with slow TBP turnover preferentially contain the consensus TATA sequence and fewer nucleosomes. (A) Scatter plot of TBP-ERT2 peaks annotated as Pol II promoters in log2TBP-ERT2 occupancy versus −log10TBP-ERT2 binding turnover rate. Colors indicate the classes categorized based on the binding turnover rates. (B) Top three DNA motifs enriched in the slow turnover class. (C) Cumulative frequency of match score to the position weight matrix of TATA consensus motif. (D) Box plot of TATA consensus match scores to a forward-oriented consensus sequence for the indicated classes of promoters. (E) Percentage of promoters that have strong (P < 0.0001) hit sequences to TATA consensus motif, with colors indicating the number of TATA sequences in each promoter in the forward (F) or reverse (R) direction with respect to the downstream genes. (F) Position of TATA sequences in forward or reverse direction within 1 kb upstream of or downstream from the TSS. (G) MNase-seq (Sathira et al. 2010) signal around hit sequences located in −1 kb to +1 kb from the TSS (top) and −200 bp to +200 bp from the TSS (bottom). Red shaded areas represent regions having marked differences of MNase-seq signals between classes. Asterisk in the bottom panel represents the region showing significant differences (Student's t-test, P < 0.001).
To identify DNA-binding transcription factors that are linked to promoters with slow TBP turnover, we examined promoter regions (±500-bp region upstream of the TSS) for presence of all known TF binding motifs (JASPAR database). Specifically, we searched for motifs that are enriched in the slow class promoters as compared to those in fast class promoters. Motifs of homeobox transcription factors (CDX1, BARHL1, and LMX1B) were identified based on the significance (Fisher's exact test) (Supplemental Fig. S6). Most of these motifs have a high AT content (Supplemental Fig. S6A) and locate at the same position as the TBP consensus (Supplemental Fig. S6B). The majority of slow-class promoters with these consensus motifs also have the TBP consensus (Supplemental Fig. S6C).
Nucleosome occupancy (MNase-seq data) around the TBP consensus motifs, which are located ±1000 or ±200 bp from the transcription start site, is lowest in the slow turnover class and highest in the fast turnover class (Fig. 3G, red shaded area). As TBP can bind in a transcriptionally productive fashion to both TATA-containing and TATA-less promoters, these results indicate that TATA-containing promoters with low nucleosome occupancy are enriched in the slow turnover class. In addition, slow-class Pol II promoters that lack a strong TATA consensus motif tend to have a higher AT content in the region just upstream of the TSS compared to other classes (Supplemental Fig. S7).
High transcriptional activity is linked to both high TBP occupancy and slow TBP turnover
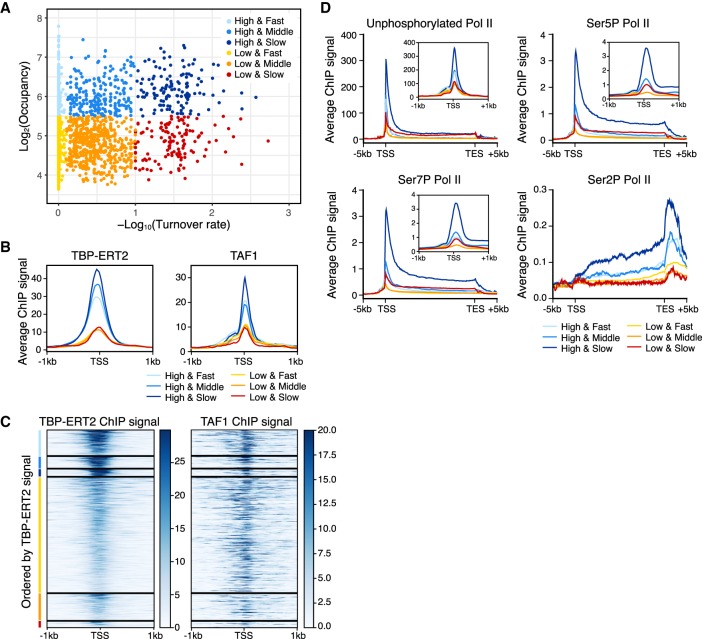
To investigate the relationships between TBP binding turnover rate, TBP occupancy, and transcriptional activity, we further subdivided TBP peaks at Pol II promoters into six groups based on TBP binding turnover rate (fast, middle, and slow) and occupancy (high and low; for simplicity, analysis was restricted to 3326 promoters having a sole annotation) (Fig. 4A–C). Although the correlation between occupancy and turnover rate is weak, average TBP occupancy is slightly higher in the slow class (Fig. 4B). Occupancy of TAF1 is strongly correlated to TBP occupancy among the three classes (Fig. 4B,C), presumably because it an obligate component of the TFIID complex and hence behaves similarly to TBP. However, promoters with high TBP occupancy and slow TBP turnover have much higher Pol II occupancy than observed in the other five classes (Fig. 4D). Increased Pol II occupancy occurs regardless of the CTD phosphorylation state, suggesting that each stage of transcription is enhanced in this group. Pol II occupancy at the other five classes is fairly similar, although it is slightly higher as a function of TBP occupancy and turnover rate.
Figure 4.
Strong transcription activity of genes with promoters with slow TBP turnover. Colors indicate the groups categorized based on the binding turnover rates and occupancy (n = 3326; High and Fast n = 441, High and Middle n = 202, High and Slow n = 126, Low and Fast n = 1985, Low and Middle n = 462, Low and Slow n = 110). (A) Scatter plot of TBP-ERT2 peaks annotated as Pol II promoters in log2TBP-ERT2 occupancy versus −log10TBP-ERT2 binding turnover rate. (B) Means of TBP-ERT2 and TAF1 binding between −1 kb to +1 kb from the TSS in each group. (C) Heat map of TBP-ERT2 and TAF1 binding at each target site within the indicated groups. (D) Means of unphosphorylated or phosphorylated Pol II binding (Fong et al. 2017) between the TSS and transcription end sites (TES) plus 5 kb upstream or downstream in each group. Plots in the insets are means of Pol II ChIP-seq signal along −1 kb to +1 kb from TSS in each group.
The high transcriptional activity of promoters with high TBP occupancy and slow TBP turnover is confirmed by PRO-seq experiments (Woo et al. 2018) that measure the amount of nascent RNA transcripts (Supplemental Fig. S8A–C). All six classes have a similar Pol II pausing index that represents the ratio of paused Pol II in the promoter-proximal region relative to the elongating Pol II throughout the coding region (Supplemental Fig. S8D). In addition, the high TBP occupancy and slow TBP turnover group has a low antisense:sense ratio compared to the other classes, indicating a strong preference for transcription in the sense direction in this group (Supplemental Fig. S8E,F). Thus, transcriptional activity is not simply dependent on TBP occupancy but rather is also strongly associated with TBP binding dynamics.
Although slow TBP turnover is linked to high transcriptional activity, some promoters with fast TBP turnover are highly transcribed. These promoter regions are enriched for certain transcription factor binding motifs as compared to highly active promoters with slow TBP turnover (Supplemental Fig. S9). We speculate that cognate transcription factors binding to these motifs recruit other factors that compensate for the fast turnover of TBP, thereby resulting in higher levels of transcription.
TBP binding dynamics are differentially associated with recruitment of chromatin-modifying factors
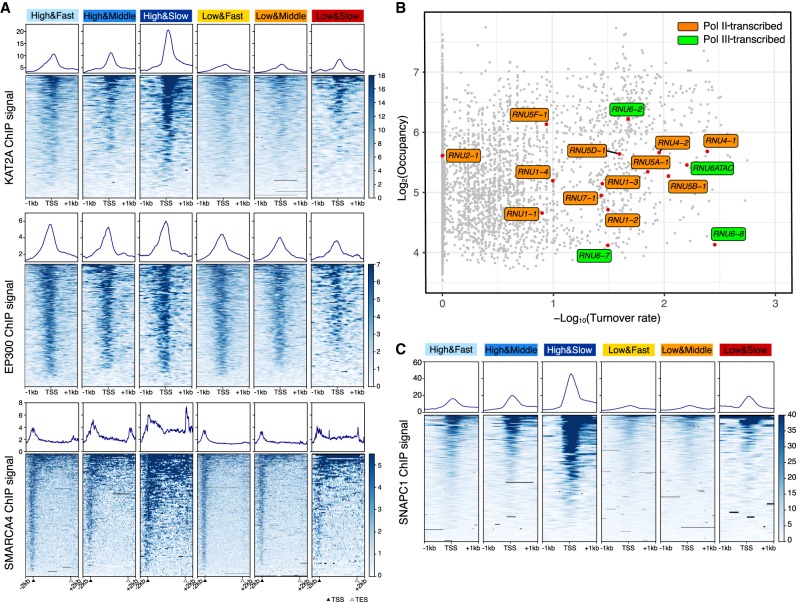
Using the same six classes of promoters, we analyzed the relationship of TBP occupancy and dynamics with respect to the recruitment of chromatin regulatory factors. KAT2A (also known as GCN5) histone acetylase (catalytic subunit of SAGA and other complexes) and SMARCA4 (also known as BRG1) (catalytic subunit of SWI/SNF and related nucleosome-remodeling complexes) are specifically enriched at promoters with slow TBP binding turnover (Fig. 5A). This result is observed at both high and low TBP occupancy, although recruitment of these chromatin-modifying complexes is lower at promoters with low TBP occupancy. In contrast, recruitment of EP300, a histone acetylase, does not vary as a function of TBP dynamics (Fig. 5A).
Figure 5.
KAT2A, SMARCA4, and SNAPC1 are enriched in genes with slow TBP binding dynamics. (A) Means and heat maps of KAT2A (top), EP300 (middle), and SMARCA4 (bottom) binding on each target site. (B) Distribution of snRNA genes (red) in the scatter plot of log2TBP-ERT2 occupancy versus log10TBP-ERT2 binding turnover rate. Orange and green, respectively, indicate Pol II- and Pol III-transcribed snRNA. (C) Means and heat maps of SNAPC1 binding (Baillat et al. 2012) on each target site (snRNA genes have been excluded).
snRNA gene promoters recognized by the SNAPC have slow TBP turnover
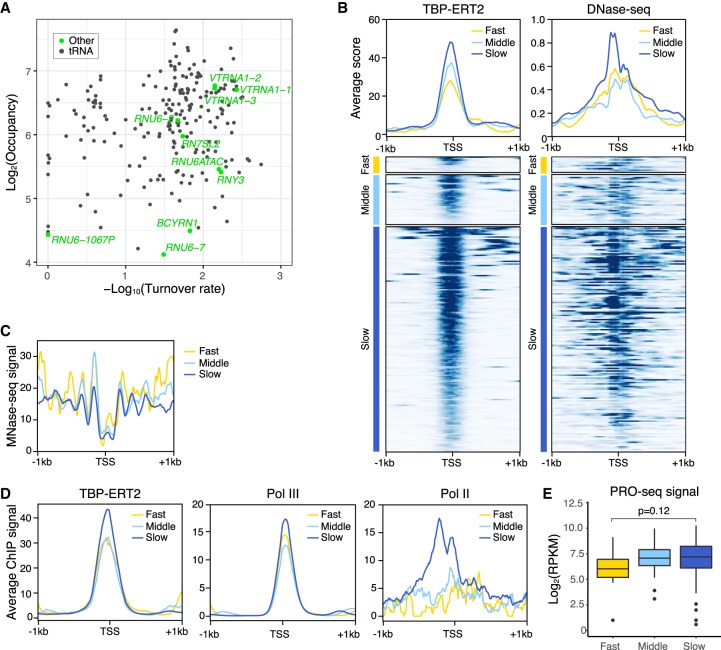
snRNA genes are transcribed by either Pol II or Pol III. All snRNA gene promoters contain a proximal sequence element (PSE) recognized by the snRNA-activating protein complex (SNAPC) that consists of five subunits. Consistent with typical Pol III-transcribed genes, slow TBP binding turnover is observed at the Pol III-transcribed snRNA genes RNU6 and RNU6ATAC. In addition, Pol II-transcribed snRNA genes (except for RNU2-1) also show slow TBP binding turnover (Fig. 5B). These results suggested that slow TBP binding turnover at snRNA genes may be related to SNAPC recruitment. To test this hypothesis, we examined the six classes of promoters for binding of SNAPC1, a component of SNAPC that binds to highly active Pol II genes in addition to snRNA genes (Baillat et al. 2012). SNAPC1 is strongly enriched at promoters with slow TBP turnover (Fig. 5C), strongly suggesting that SNAPC contributes to slow TBP dynamics at both Pol II and Pol III promoters.
Differential TBP turnover at Pol III promoters
Although TBP binding turnover on Pol III promoters is generally slower than on Pol II promoters, several Pol III promoters show fast turnover at speeds comparable to that of typical Pol II promoters. Almost all Pol III promoters that show fast TBP binding turnover are tRNA gene promoters, except for one case annotated as a RNU6 pseudogene (Fig. 6A). To address the basis of differential TBP turnover at Pol III promoters, we subdivided all Pol III promoters into three classes based on turnover rate and analyzed TBP occupancy and chromatin structure (MNase-seq and DNase-seq). Pol III promoters with slow TBP turnover have more open chromatin structure (higher DNase-seq signal and lower MNase-seq signal) (Fig. 6B,C) as compared to Pol III promoters with faster TBP turnover. In this regard, Pol III promoters behave similarly to Pol II promoters.
Figure 6.
Pol III promoters with slow TBP binding dynamics have less nucleosomes and higher Pol III recruitment. (A) Scatter plot of TBP-ERT2 peaks annotated as Pol III genes in log2TBP-ERT2 occupancy versus log10TBP-ERT2 binding turnover rate. (B) Means and heat maps of TBP-ERT2 binding and DNase-seq signal on each target site near Pol III genes. (C) Means of MNase-seq (Sathira et al. 2010) signal along −1 kb to +1 kb from the TSS in each class. (D) Means of TB P-ERT2, Pol III (POLR3A), and Pol II binding (Choquet et al. 2019) along −1 kb to +1 kb from the TSSs of tRNA genes in each class. (E) Box plot of PRO-seq signal (Woo et al. 2018) at tRNA genes in each class.
We investigated the association between the transcriptional activity and TBP occupancy or turnover rate with a subset of Pol III promoters (n = 130; for simplicity, we restricted these to tRNA genes having a sole annotation <1 kb from other genes). In contrast to the case of Pol II promoters, Pol III occupancy and RNA levels are quite similar between the tRNA gene promoters with slow or fast TBP turnover (Fig. 6D,E), and the consensus TATA sequence occurs equivalently on Pol III promoters of both turnover classes (Supplemental Fig. S10). The subtle increased levels of Pol III on the slow class promoters are likely due to slight increases in the level of TBP occupancy (Fig. 6D). Although Pol II binds close to active, but not inactive, Pol III promoters in human cells (Moqtaderi et al. 2010; Oler and Cairns 2010; Raha et al. 2010), Pol II enrichment is unexpectedly higher in the slow class promoters than the other classes (Fig. 6D).
Discussion
An ERT2-based, time-course ChIP method to measure dynamics of transcription factor binding at target sites on a genome-wide scale in human cells
Although transcription factor binding to chromatin is highly dynamic on an overall basis, binding dynamics at target loci in metazoan cells have not been investigated. Here, we describe an ERT2-based, inducible time-course ChIP-seq method to address the dynamics of transcription factor binding at target loci on a genome-wide scale. This method should be applicable to all transcription factors whose function is retained upon fusion to the ERT2 domain that mediates nuclear import upon treatment with tamoxifen. The turnover rate measured by this approach is determined by the dissociation rate of the endogenous transcription factor bound to promoters prior to tamoxifen addition and the association rate of the tamoxifen-induced ERT2 derivative. While our approach cannot directly distinguish between association and dissociation rates, we strongly suspect that the dissociation rate is the primary determinant. In all cases tested, slow turnover is associated with high chromatin accessibility, the opposite of what one would expect if association rates were important.
Relative TBP turnover rates at Pol I, Pol II, and Pol III promoters are evolutionarily conserved
Our results indicate that TBP turnover in human cells is very rapid at Pol II promoters, considerably slower at Pol III promoters, and extremely slow at Pol I promoters. This observation is very similar to what occurs in yeast cells (van Werven et al. 2009), indicating that relative TBP turnover rates at promoters transcribed by the three different transcriptional machineries are conserved from yeast to human. Although the Pol II and Pol III machineries and the relevant TBP complexes are highly conserved across eukaryotes, the yeast and human Pol I-specific TBP complexes are very different. Aside from TBP, the proteins in the human SL1 complex (Comai et al. 1992) have only weak sequence similarities with those in the yeast Core factor (Lin et al. 1996), even though both complexes recognize the core promoter element in the pre-rRNA gene promoter. The exceptionally low turnover rates of such different TBP complexes at the rDNA promoter strongly suggest that low TBP turnover is fundamentally important at Pol I promoters. As rRNA genes exist in long tandem copies, only some of which are active at any one time, this suggests that the active and inactive states of rRNA gene promoters are maintained for considerable time.
Although the induction kinetics of nuclear TBP are similar in yeast and human (saturation by 30–60 min), induced human TBP requires a longer time than yeast TBP to saturate TBP occupancy levels on Pol I and Pol III promoters (Pol I: >3 h in yeast and >24 h in human; Pol III: 60–90 min in yeast and >6 h in human) (van Werven et al. 2009). We suspect that this difference reflects the much longer time for cell division in human cells. Although the molecular basis for this difference is unclear, it might be due to clearance of DNA-bound TBP during DNA replication.
Determinants of fast versus slow TBP turnover rates among Pol II or Pol III promoters
Although Pol II promoters with slow TBP turnover tend to have higher TBP occupancy, TBP turnover rate at Pol II promoters is poorly correlated (r = 0.32) with TBP occupancy in human cells. At most Pol II promoters, the kinetics of TBP-ERT2 binding are indistinguishable from those of TBP-ERT2 translocation into the nucleus upon tamoxifen induction, indicating a very fast turnover rate. However, some Pol II promoters have a much slower turnover rate as well as distinct properties. Pol II promoters with slow TBP turnover (1) frequently contain consensus TATA motifs or AT-rich regions at the typical TATA location 25–30 bp upstream of the start site, (2) are associated with high levels of transcription of downstream genes, (3) have low nucleosome occupancy, and (4) show stronger recruitment of chromatin-modifying activities such as KAT2A histone acetylase and the SWI/SNF nucleosome remodeling complex. snRNA gene promoters, which specifically rely on SNAPC for transcription, also exhibit slow TBP turnover. These results demonstrate that TBP binding dynamics, not simply TBP occupancy, are important for transcriptional activity and chromatin structure around promoters.
These observations on TBP binding dynamics on Pol II promoters in human cells differ from what occurs in yeast cells. Yeast promoters with slow TBP turnover were reported to have higher mRNA levels (van Werven et al. 2009), but higher levels of Pol II occupancy on these genes is not evident in the same paper. Furthermore, a reanalysis of the same data using a different model and gene set suggested the opposite conclusion, that yeast promoters with fast TBP turnover have modestly higher levels of nascent transcription (Zaidi et al. 2017). In addition, TATA-containing yeast promoters are not enriched in the slow TBP turnover class and have modestly faster binding turnover of TBP compared to TATA-less promoters (van Werven et al. 2009; Zaidi et al. 2017).
Both Pol II and Pol III promoters with relatively slow TBP turnover rates are associated with more open chromatin structure. However, unlike the case for Pol II promoters, there is little if any relationship between turnover rates and transcriptional activity on Pol III promoters. In accord with the fact that Pol II binds closely to active, but not inactive, tRNA gene promoters in human cells (Moqtaderi et al. 2010; Oler and Cairns 2010; Raha et al. 2010), tRNA gene promoters with slow TBP turnover are associated with higher levels of Pol II binding. As tRNA and most other Pol III genes lack TATA sequences in their promoters, recruitment of Pol II and associated chromatin-modifying activities might provide an alternative mechanism to ensure stable TBP binding.
Implications for transcriptional mechanisms
TFIID binds to promoters via sequence-specific interactions between TBP and the TATA sequence as well as interactions of TAFs with initiator and downstream promoter elements. The simplest explanation for the properties of promoters with slow TBP turnover is that TBP interaction with a consensus TATA element stabilizes the TFIID complex on the promoter. As such, the weaker TBP-TATA interaction at so-called TATA-less promoters would lead to faster dissociation rates. In this regard, the transcription factors preferentially linked to Pol II promoters with slow TBP turnover might directly recruit TFIID to promoters and hence stabilize the TFIID-TATA interaction. Although Mediator is the direct target of most activators, there is a subset of activators in yeast (Kuras et al. 2000; Li et al. 2000) and human (Chen et al. 2013) cells that directly recruit TFIID.
A strong TBP-TATA interaction is important for high levels of transcription mediated by most activator proteins (Iyer and Struhl 1995; Lee and Struhl 1995; Struhl 1996), presumably because it is associated with increased transcriptional reinitiation (multiple rounds of transcription following preinitiation complex formation) (Yean and Gralla 1997, 1999; Yudkovsky et al. 2000; Wong et al. 2014) and transcriptional bursting (Zenklusen et al. 2008; Sanchez and Golding 2013). Gene looping between promoter and terminator regions is likely to facilitate Pol II recycling after transcription termination (O'Sullivan et al. 2004; Tan-Wong et al. 2012), and this may contribute to increased transcriptional reinitiation at promoters with slow TBP turnover. In this regard, we observed increased SWI/SNF occupancy at the 3′ regions of genes whose promoters show slow TBP turnover, similar to SWI/SNF occupancy at 3′ regions of genes observed in Arabidopsis (Archacki et al. 2017). The function of transcriptional activator proteins is linked to reduced nucleosome occupancy via recruitment of chromatin-modifying activities, and this might account for why promoters with slow TBP turnover show decreased nucleosome occupancy.
The differences between yeast and human Pol II promoters with respect to TBP dynamics are likely to reflect differences between transcriptional mechanisms in these two organisms. First, at human (and most other eukaryotic) promoters, Pol II pauses just downstream from the initiation site, but this pause does not occur in yeast cells because it lacks NELF, a critical factor for the pause (Adelman and Lis 2012; Zhou et al. 2012; Kwak and Lis 2013). This Pol II pause helps keep the chromatin open in the vicinity near the pause (Gilchrist et al. 2010). Second, yeast TBP exists in two distinct active forms for Pol II transcription, TFIID and a form lacking TAFs (probably free TBP) (Kuras et al. 2000; Li et al. 2000), whereas human TBP is predominantly (and probably exclusively) in the form of TFIID. One possible explanation is that yeast TBP has a faster turnover at yeast Pol II promoters than TFIID, thereby accounting for why TATA-containing promoters (which are favored by the free TBP form) have faster TBP turnover than TATA-lacking promoters. In human cells, where TFIID is presumably the exclusive form for TBP, slow turnover is simply due to increased binding affinity via the TBP-TATA interaction.
Our results showing that higher transcriptional activity is linked to slow TBP turnover beyond simple TBP occupancy are consistent with results with yeast Rap1 (Lickwar et al. 2012). We suggest that relationship reflects the fact that assembly of active transcription complexes takes some time. Proteins with fast turnover might initiate the transcription process but will be unable to complete it if they dissociate prior to full assembly of an active complex. Thus, while factor occupancy is clearly linked to transcriptional activity, the amount of time that the factor remains associated with the chromatin template has an independent effect that could be manifest on the preinitiation complex per se or transcriptional reinitiation that results in multiple transcripts for each assembled complex.
Methods
Plasmids, cell culture, and cell fractionation
pCMV-TBP-ERT2-3HA was constructed and stably introduced into HEK293 (ATCC) cells by G418 selection as described in Supplemental Methods. TBP-ERT2 nuclear translocation was induced by 4OHT (Sigma-Aldrich, H7904) addition to culture medium at the final concentration of 100 nM. Nuclear and nucleolar fractionation was performed by standard procedures detailed in the Supplemental Methods.
ChIP-seq analysis during the time course
For each chromatin sample from cells at various times of induction with tamoxifen, we added yeast chromatin prepared from a 3HA-TBP-expressing strain at the constant ratio (0.039 µg total protein in the lysate, that corresponds to 3% of total human protein amount in the lysates) as a spike-in control. These mixed lysates were immunoprecipitated with 0.3 µg of anti-HA antibody (Santa Cruz Biotechnology, #sc-7392) or 3 µg of anti-TBP antibody (Abcam, #ab51841) as described in the Supplemental Methods. Primers used for qPCR analysis are listed in Supplemental Table S2. Library preparation, DNA sequencing, and mapping of TBP-ERT2 binding sites were performed as described in Supplemental Methods.
Modeling of protein translocation to the nucleus
We assume that the increase of nuclear fraction of TBP-ERT2 corresponds to the decrease of cytoplasmic fraction of TBP-ERT2 at a given time point under the condition with an excess amount of 4OHT compared to the number of TBP-ERT2 molecules and that the amount of total TBP-ERT2 does not change over time. This implies the following constraint:
| (1) |
The dynamics of TBP can be described as follows:
| (2) |
Thus
| (3) |
We consider the situation where all the TBP-ERT2 are initially in the cytoplasm and eventually end up in the nucleus. Indeed
| (4) |
| (5) |
Using Equation 5, Equation 3 can be rewritten as
| (6) |
We assume that the measured nuclear TBP-ERT2 signal (mTBPnuc[t]) is sum of the true signal (TBPnuc[t]) and background (BG) derived from experimental errors, like imperfect separation of cytoplasmic and nucleus. Thus,
| (7) |
From Equation 4
| (8) |
Indeed, Equation 6 can be transformed as
| (9) |
We measured the nuclear amount of TBP-ERT2 at all time points (0, 5, 10, 15, 30, 60, 90, 360, 1440 min) using western blotting analysis and determined k in Equation 9 that yields the best fit to the measured data using the lsqcurvefit function of MATLAB (Supplemental Fig. S3A).
Turnover model
We used a simple model to describe the interaction between TBP and DNA with the assumption that (1) TBP detected by western blotting is mostly free in the soluble fraction, (2) the turnover rate of endogenous TBP and TBP-ERT2 are the same, and (3) when the TBP molecule dissociates from DNA, the other TBP molecule immediately comes to occupy the binding site. Thus, the dynamics of DNA-bound TBP-ERT2 can be described with the TBP turnover rate λ as follows:
| (10) |
We also assume that the measured TBP-ERT2 ChIP signal (mTBPbound[t]) is the sum of the true signal (TBPbound[t]) and background (BG). Thus
| (11) |
From Equation 4, theoretically
| (12) |
and thus
| (13) |
We define C(t) and W(t) as relative concentrations to the endpoint of DNA-bound TBP (measured by ChIP-seq) and TBP protein (signal detected by western blotting), respectively.
| (14) |
| (15) |
Indeed, Equation 10 can be written with relative concentrations
| (16) |
λ (turnover rate) for each locus was determined using ODE45 and the lsqcurvefit function of MATLAB to yield the best fit to the measured data. We used multiple initiation points to avoid local minima. We excluded the peaks that show low goodness of fit (residual sum of squares ≤ 0.4), and used the rest of the 6476 peaks for the later analysis. In this model, the speed of the protein translocation to the nucleus is the major limitation for measuring TBP binding kinetics. As shown in the simulated result (Supplemental Fig. S3B), in the cases that the TBP binding turnover rate exceeds 1, it is out of the range of the precise detection in our experimental system. Thus, we set a maximum turnover rate as 1 to lump together all the TBP target sites for which turnover rates exceed the detection limit as the “very fast” category.
Motif analysis
Based on their turnover rate, 6258 peaks which are not annotated as Pol III promoters or genes were categorized into three classes: Fast (0 ≤ log10[λ] < 0.1, n = 4480), Middle (0.1 ≤ log10[λ] < 1, n = 1299), Slow (1 ≤ log10[λ], n = 479). Two hundred seventeen peaks which are annotated as Pol III promoters or genes were also categorized into three classes based on the same criteria: Fast (n = 11), Middle (n = 35), Slow (n = 171). FASTA files of the DNA sequences of peaks in the Fast class and Slow class were used as input for RSAT peak-motifs (http://rsat.sb-roscoff.fr/peak-motifs_form.cgi) to search the Slow class enriched motifs. The match scores, P-values, and locations of TBP consensus motifs, INR, BREu, and BREd in the peaks or in the defined regions upstream of the start site were searched by PWMscan of PWMtools (https://ccg.epfl.ch/pwmtools/pwmscan.php) with scanning options (Score cut-off: 0). TBP PWM was taken from JASPAR (http://jaspar.genereg.net/, MA0108.2) and the other PWMs were taken from ElemeNT (http://lifefaculty.biu.ac.il/gershon-tamar/index.php/element-description). For the search of known TF binding motifs enriched in the Slow class, ±500-bp regions from the TSSs of genes classified as the Fast and Slow class were used as input for CentriMo (http://meme-suite.org/doc/centrimo.html).
Analysis of publicly available data
Publicly available data sets (Sathira et al. 2010; Baillat et al. 2012; The ENCODE Project Consortium 2012; Fong et al. 2017; Davis et al. 2018; Woo et al. 2018; Choquet et al. 2019) used in this study are listed in Supplemental Table S3. Heat maps and average plots were generated using deepTools. For the analysis of the pausing index, we defined proximal regions as −500 bp to +500 bp from the gene start site. Gene body regions were defined as +500 bp to the gene end, minus proximal regions of the internal gene start site of the isoforms.
Data access
All raw and processed sequencing data generated in this paper have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE133729.
Supplementary Material
Acknowledgments
This work was conceived and begun in the laboratory of Jason Lieb at the University of Chicago. We thank A. Stutzman for help with the plasmid construction, and S. Pott, K. Ikegami, A. Ruthenburg, and members of the Struhl laboratory for helpful comments and discussions. This work was supported by a fellowship from the University of Chicago Fellows Program to Y.H. and grants to K.S. from the National Institutes of Health (GM 30186 and CA 107486).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.254466.119.
References
- Adelman K, Lis JT. 2012. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13: 720–731. 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archacki R, Yatusevich R, Buszewicz D, Krzyczmonik K, Patryn J, Iwanicka-Nowicka R, Biecek P, Wilczynski B, Koblowska M, Jerzmanowski A, et al. 2017. Arabidopsis SWI/SNF chromatin remodeling complex binds both promoters and terminators to regulate gene expression. Nucleic Acids Res 45: 3116–3129. 10.1093/nar/gkw1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Gardini A, Cesaroni M, Shiekhattar R. 2012. Requirement for SNAPC1 in transcriptional responsiveness to diverse extracellular signals. Mol Cell Biol 32: 4642–4650. 10.1128/MCB.00906-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhoure N, Bounova G, Bernasconi D, Praz V, Lammers F, Canella D, Willis IM, Herr W, Hernandez N, Delorenzi M, et al. 2014. Quantifying ChIP-seq data: a spiking method providing an internal reference for sample-to-sample normalization. Genome Res 24: 1157–1168. 10.1101/gr.168260.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WY, Zhang J, Geng H, Du Z, Nakadai T, Roeder RG. 2013. A TAF4 coactivator function for E proteins that involves enhanced TFIID binding. Genes Dev 27: 1596–1609. 10.1101/gad.216192.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet K, Forget D, Meloche E, Dicaire MJ, Bernard G, Vanderver A, Schiffmann R, Fabian MR, Teichmann M, Coulombe B, et al. 2019. Leukodystrophy-associated POLR3A mutations down-regulate the RNA polymerase III transcript and important regulatory RNA BC200. J Biol Chem 294: 7445–7459. 10.1074/jbc.RA118.006271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloix C, Tutois S, Yukawa Y, Mathieu O, Cuvillier C, Espagnol MC, Picard G, Tourmente S. 2002. Analysis of the 5S RNA pool in Arabidopsis thaliana: RNAs are heterogeneous and only two of the genomic 5S loci produce mature 5S RNA. Genome Res 12: 132–144. 10.1101/gr.181301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Tanese N, Tjian R. 1992. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell 68: 965–976. 10.1016/0092-8674(92)90039-F [DOI] [PubMed] [Google Scholar]
- Conconi A, Widmer RM, Koller T, Sogo JM. 1989. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57: 753–761. 10.1016/0092-8674(89)90790-3 [DOI] [PubMed] [Google Scholar]
- Cormack BP, Struhl K. 1992. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell 69: 685–696. 10.1016/0092-8674(92)90232-2 [DOI] [PubMed] [Google Scholar]
- Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, et al. 2018. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 46: D794–D801. 10.1093/nar/gkx1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315: 1405–1408. 10.1126/science.1134053 [DOI] [PubMed] [Google Scholar]
- Donovan BT, Huynh A, Ball DA, Patel HP, Poirier MG, Larson DR, Ferguson ML, Lenstra TL. 2019. Live-cell imaging reveals the interplay between transcription factors, nucleosomes, and bursting. EMBO J 38: e100809 10.15252/embj.2018100809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Saldi T, Sheridan RM, Cortazar MA, Bentley DL. 2017. RNA Pol II dynamics modulate co-transcriptional chromatin modification, CTD phosphorylation, and transcriptional direction. Mol Cell 66: 546–557.e3. 10.1016/j.molcel.2017.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. 2010. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143: 540–551. 10.1016/j.cell.2010.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N. 1993. TBP, a universal eukaryotic transcription factor? Genes Dev 7: 1291–1308. 10.1101/gad.7.7b.1291 [DOI] [PubMed] [Google Scholar]
- Houtsmuller AB. 2005. Fluorescence recovery after photobleaching: application to nuclear proteins. Adv Biochem Eng Biotechnol 95: 177–199. [DOI] [PubMed] [Google Scholar]
- Iyer V, Struhl K. 1995. Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol Cell Biol 15: 7059–7066. 10.1128/MCB.15.12.7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova TS, Kim MJ, Spriet C, Nalley K, Stasevich TJ, Kherrouche Z, Heliot L, McNally JG. 2008. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science 319: 466–469. 10.1126/science.1150559 [DOI] [PubMed] [Google Scholar]
- Kuras L, Kosa P, Mencia M, Struhl K. 2000. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288: 1244–1248. 10.1126/science.288.5469.1244 [DOI] [PubMed] [Google Scholar]
- Kwak H, Lis JT. 2013. Control of transcriptional elongation. Annu Rev Genet 47: 483–508. 10.1146/annurev-genet-110711-155440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Struhl K. 1995. Mutations on the DNA-binding surface of TATA-binding protein can specifically impair the response to acidic activators in vivo. Mol Cell Biol 15: 5461–5469. 10.1128/MCB.15.10.5461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-Y, Bhaumik SR, Green MR. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288: 1242–1244. 10.1126/science.288.5469.1242 [DOI] [PubMed] [Google Scholar]
- Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD. 2012. Genome-wide protein–DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 484: 251–255. 10.1038/nature10985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Moorefield B, Payne J, Aprikian P, Mitomo K, Reeder RH. 1996. A novel 66-kilodalton protein complexes with Rrn6, Rrn7, and TATA-binding protein to promote polymerase I transcription initiation in Saccharomyces cerevisiae. Mol Cell Biol 16: 6436–6443. 10.1128/MCB.16.11.6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally JG, Muller WG, Walker D, Wolford R, Hager GL. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287: 1262–1265. 10.1126/science.287.5456.1262 [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K. 2010. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol 17: 635–640. 10.1038/nsmb.1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller F, Mazza D, Stasevich TJ, McNally JG. 2010. FRAP and kinetic modeling in the analysis of nuclear protein dynamics: What do we really know? Curr Opin Cell Biol 22: 403–411. 10.1016/j.ceb.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oler AJ, Cairns BR. 2010. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol 17: 620–628. 10.1038/nsmb.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Chen MW, Brown VE, Solanki S, Choi YJ, Olson ER, Fritz CC, Bradner JE, Guenther MG. 2014. Quantitative ChIP-seq normalization reveals global modulation of the epigenome. Cell Rep 9: 1163–1170. 10.1016/j.celrep.2014.10.018 [DOI] [PubMed] [Google Scholar]
- O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. 2004. Gene loops juxtapose promoters and terminators in yeast. Nat Genet 36: 1014–1018. 10.1038/ng1411 [DOI] [PubMed] [Google Scholar]
- Poorey K, Viswanathan R, Carver MN, Karpova TS, Cirimotich SM, McNally JG, Bekiranov S, Auble DT. 2013. Measuring chromatin interaction dynamics on the second time scale at single-copy genes. Science 342: 369–372. 10.1126/science.1242369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M. 2010. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci 107: 3639–3644. 10.1073/pnas.0911315106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A, Golding I. 2013. Genetic determinants and cellular constraints in noisy gene expression. Science 342: 1188–1193. 10.1126/science.1242975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathira N, Yamashita R, Tanimoto K, Kanai A, Arauchi T, Kanematsu S, Nakai K, Suzuki Y, Sugano S. 2010. Characterization of transcription start sites of putative non-coding RNAs by multifaceted use of massively paralleled sequencer. DNA Res 17: 169–183. 10.1093/dnares/dsq007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA. 1992. TATA-binding protein is a classless factor. Cell 68: 819–821. 10.1016/0092-8674(92)90023-6 [DOI] [PubMed] [Google Scholar]
- Struhl K. 1994. Duality of TBP, the universal transcription factor. Science 263: 1103–1104. 10.1126/science.8108728 [DOI] [PubMed] [Google Scholar]
- Struhl K. 1996. Chromatin structure and RNA polymerase II connection: implications for transcription. Cell 84: 179–182. 10.1016/S0092-8674(00)80970-8 [DOI] [PubMed] [Google Scholar]
- Struhl K. 2007. Interpreting chromatin immunoprecipitation experiments. In Evaluating techniques in biochemical research (ed. Zuk D), pp. 29–33. Cell Press, Cambridge, MA. [Google Scholar]
- Tan-Wong SM, Zaugg JB, Camblong J, Xu Z, Zhang DW, Mischo HE, Ansari AZ, Luscombe NM, Steinmetz LM, Proudfoot NJ. 2012. Gene loops enhance transcriptional directionality. Science 338: 671–675. 10.1126/science.1224350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, van Teeffelen HA, Holstege FC, Timmers HT. 2009. Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat Struct Mol Biol 16: 1043–1048. 10.1038/nsmb.1674 [DOI] [PubMed] [Google Scholar]
- Wong K-H, Jin Y, Struhl K. 2014. TFIIH phosphorylation of the Pol II CTD stimulates Mediator dissociation from the preinitiation complex and promoter escape. Mol Cell 54: 601–612. 10.1016/j.molcel.2014.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YM, Kwak Y, Namkoong S, Kristjánsdóttir K, Lee SH, Lee JH, Kwak H. 2018. TED-seq identifies the dynamics of poly(A) length during ER stress. Cell Rep 24: 3630–3641.e7. 10.1016/j.celrep.2018.08.084 [DOI] [PubMed] [Google Scholar]
- Yean D, Gralla J. 1997. Transcription reinitiation rate: a special role for the TATA box. Mol Cell Biol 17: 3809–3816. 10.1128/MCB.17.7.3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yean D, Gralla JD. 1999. Transcription reinitiation rate: a potential role for TATA box stabilization of the TFIID:TFIIA:DNA complex. Nucleic Acids Res 27: 831–838. 10.1093/nar/27.3.831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkovsky N, Ranish JA, Hahn S. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408: 225–229. 10.1038/35041603 [DOI] [PubMed] [Google Scholar]
- Zaidi HA, Auble DT, Bekiranov S. 2017. RNA synthesis is associated with multiple TBP-chromatin binding events. Sci Rep 7: 39631 10.1038/srep39631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH. 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15: 1263–1271. 10.1038/nsmb.1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH. 2012. RNA polymerase II elongation control. Annu Rev Biochem 81: 119–143. 10.1146/annurev-biochem-052610-095910 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.