Abstract
The faster a disease can be diagnosed, the sooner effective treatment can be initiated, motivating a drive to replace standard laboratory techniques with point-of-care technologies that return answers in minutes rather than hours. Thus motivated, we describe the development of an E-DNA scaffold sensor for the rapid and convenient measurement of antibodies diagnostic of syphilis. To achieve this (and in contrast to previous sensors of this class, which relied on single, linear epitopes for detection), we utilized a near full-length antigen as the sensor’s recognition element, allowing us to simultaneously display multiple epitopes. The resultant sensor is able to detect antibodies against Treponema pallidum pallidum, the causative agent of syphilis, at clinically relevant concentrations in samples in less than 10 min. Preliminary results obtained using sero-positive and sero-negative human samples suggest the clinical sensitivity and specificity of the approach compare well to current gold-standard tests, while being simple and rapid enough to deploy at the point of care.
Introduction
The more rapidly a disease is diagnosed, the sooner treatment can be initiated, improving both compliance and outcomes, rendering it desirable to achieve diagnosis within the timeframe of a single patient/clinician interaction1. This is particularly true for infectious diseases, as single-visit diagnosis allows for the treatment of patients who would otherwise be lost to follow up, allowing healthcare providers to intervene immediately to change behaviors and limit transmission2–5. Accomplishing this, however, necessitates diagnostic tools that are simple enough to use at the point of care and are capable of returning answers within minutes rather than hours, attributes that the current, largely laboratory-centered approaches to molecular diagnostics fail to achieve6.
The potential value of improved point-of-care diagnostics is illustrated by the “gold-standard” approach for diagnosing syphilis, a disease for which the incidence rate has more than doubled in the United States during the past decade7. The current standard of care for syphilis diagnosis involves two serological tests performed in sequence. The first, the “nontreponemal” test, detects the presence of cardiolipids associated with cell damage and is used to determine whether a patient has an active infection8–10. The second, “treponemal” test, detects antibodies specifically indicative of exposure to pathogenic bacteria from the Treponema genus such as Treponema pallidum pallidum, the causative agent of syphilis11. This two-pronged approach12 is employed because the specificity of the two individual tests is poor; other underlying illnesses or pregnancy can cause false positives in the non-treponemal test and antibodies remaining from past exposure (rather than ongoing infection) can cause false positives in the treponemal test13, 14. However, while the two-pronged approach improves clinical specificity, it slows down diagnosis. Specifically, while the non-treponemal test can be performed rapidly at the point of care using assays such as the rapid plasma reagin test, the treponemal test relies on traditional serological assays (e.g., hemagglutination, Western blotting, or enzyme linked immunosorbent assays), thus necessitating specialized lab facilities15, 16. Given this, a rapid treponemal test capable of being deployed at the point of care could help to limit the spread of syphilis by eliminating the gap between a patient’s initial visit and a positive diagnosis, allowing clinicians to intervene immediately.
Motivated by the general need for improved, point-of-care serological tests, we have recently developed a general platform for rapid (< 10 min), convenient measurement of the concentration of specific antibodies in unprocessed serum [Figure 1] and have adapted it here to the treponemal test. Electrochemical DNA (E-DNA) biosensors have already proven to be a versatile platform for detection of analytes in complex media, such as undiluted serum17–19. Here we use a modification of the E-DNA scaffold platform20, which is comprised of a nucleic acid duplex (“scaffold”) bound to the surface of an electrode via a flexible linker. On the distal end of this scaffold, a redox reporter is conjugated to one end of the DNA backbone, while a recognition element, such as an antigenic protein, is bound to the other. Binding of an antibody to the recognition element reduces the efficiency with which the redox reporter approaches the electrode surface, resulting in a change in electron transfer rate that can rapidly and conveniently be measured using standard electrochemical approaches.
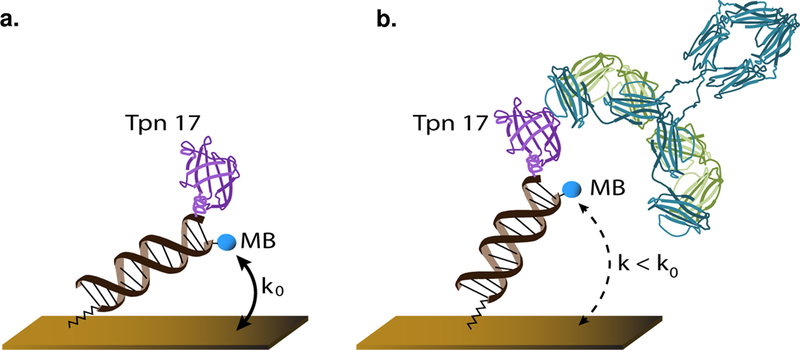
Figure 1.
(a) E-DNA scaffold sensors are comprised of a nucleic acid “scaffold” bound to the surface of a gold electrode via a flexible linker (Cash et al., 2009). The distal end of the scaffold is modified with a redox reporter (here methylene blue; MB), and a nitrilotriacetic acid (NTA) that, in the presence of copper, tightly binds a hexa-his tag on the antigen (here TpN17). (b) Binding of the target antibody to the recognition element reduces the efficiency with which the redox reporter approaches the surface of the electrode, reducing the electron transfer rate and, in turn, the current observed when the sensor is interrogated using square wave voltammetry.
Experimental
Gene design, overexpression, and protein purification:
We retrieved the nucleotide sequence encoding residues 33–156 of Treponema pallidum TpN17 (residue numbering from UniProt entry P29722) from the European Nucleotide Archive (accession ID M74825). We introduced a hexahistidine tag and two serine residues on the amino terminus for purification purposes, substituted Cys58 with Ser (leaving a unique cysteine at position 42), and codon optimized for overexpression in Escherichia coli, yielding the gene sequence:
ATGCACCACCACCACCACCACAGCAGCGGCAAGGCGAAAGCGGAGAAGGTGGAATGCGCGCTGAAAGGTGGCATTTTCCGTGGTACCCTGCCGGCGGCGGACAGCCCGGGTATTGATACCACCGTGACCTTTAACGCGGACGGCACCGCGCAGAAGGTTGAGCTGGCGCTGGAAAAGAAAAGCGCGCCGAGCCCGCTGACCTACCGTGGTACCTGGATGGTTCGTGAGGACGGCATCGTGGAACTGAGCCTGGTTAGCAGCGAGCAAAGCAAGGCGCCGCACGAGAAAGAACTGTACGAACTGATTGATAGCAACAGCGTGCGTTATATGGGTGCGCCGGGTGCGGGCAAGCCGAGCAAAGAGATGGCGCCGTTCTATGTTCTGAAGAAAACCAAGAAATAA.
Codon optimization, synthesis, subcloning into a pET-3a vector using 5′ Nde1 and 3′ BamH1 restriction sites, and sequencing of the final construct was performed by a commercial vendor (GenScript, USA).
We transformed the expression construct into E. coli BL21(DE3) cells (New England Biolabs, USA) using standard heat shock. Gene overexpression was for 5 h in lysogeny broth with 100 μg/mL of carbenicillin at 37°C, 220 rpm, induced by addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside at OD600 > 0.5. Cells were harvested by centrifugation and resuspended in 20 mM sodium phosphate, 40 mM imidazole, 500 mM NaCl, pH 7.4. Protein purification was at 4° C. Cells were lysed by ultrasonication in presence of DNase (Sigma-Aldrich, USA) and RNase (Roche, Switzerland). Cell debris was removed by centrifugation at 11,000 rpm for 1 h. TpN17 was purified from the 0.2-μm-filtered supernatant on a HisTrap HP column (GE Life Sciences, USA), eluting the protein via a linear imidazole gradient up to 500 mM. Pure TpN17 fractions were identified on SDS-PAGE gels by SafeStain staining (Thermo Fisher, USA) and dialyzed into 1x phosphate buffered saline (PBS), pH 7.4 (Sigma-Aldrich, USA), and protein concentration was determined by UV/visible spectroscopy.
Sensor fabrication:
Gold disc electrodes (2 mm diameter) were first mechanically polished in both a 1 μm diamond and a 0.05 μm aluminum oxide slurry, followed by electrochemical cleaning by successive cycling in both 0.5 M NaOH and 0.5 M H2SO4. An anchor DNA strand which had been thiol and methylene blue (MB) modified (HS(CH2)6-CAG TCA GTC AGT CAG TCA GTC AGT-MB)) was reduced in a 10 mM TCEP solution for 1 h before being diluted to a working concentration of 16 nM in 1xPBS. As this anchor strand we employed the same sequence our group has used previously for other scaffold-type sensors20–24. Electrodes were incubated in the DNA anchor solution for 1 h and then rinsed briefly with deionized water. We next coated any remaining exposed gold on the electrode with a protective alkane-thiol monolayer by immersing them in a 10 mM solution of 6-mercapto-1-hexanol overnight at 4° C.
Successful deposition of both the monolayer and anchor strand was confirmed by placing the electrodes in a 1x PBS solution and measuring the methylene blue reduction peak with square wave voltammetry using a 25 mV, 60 Hz, signal. A nitrilotriacetic acid (NTA)-modified complimentary DNA strand was then diluted to 100 nM and the electrodes incubated in this solution for 30 min. Binding of the complementary DNA was verified by measuring the reduction in magnitude of the MB peak. Following this, TpN17 was bound to the assembled scaffold using a His-NTA complex. The electrodes were incubated in a 100 μM CuSO4 solution in 1x PBS for 15 min. After this, a 15 μL drop of 10 μM His-tagged TpN17 was placed on the tip of the electrode and incubated for 45 min. The resulting sensors were rinsed, and the attachment of the protein verified by again scanning using square wave voltammetry.
Electrochemical measurements:
Comparative measurements of the anchor strands were performed in 1x PBS buffer. We prepared three electrodes for each of our constructs (DNA/DNA, PEG-DNA/DNA, DNA/PNA). Prior to depositing the anchor strand, we determined the surface area of each electrode by immersing the electrodes into 0.05 M H2SO4 and measuring the area the gold oxide reduction peak. After depositing the anchor strands and forming the alkane-thiol monolayer, we used square wave voltammetry (60 Hz, 25 mV signal) to measure the methylene blue reduction peak of each construct, using a linear baseline subtraction to account for any current difference between the more positive and more negative sides of the potential window. We then measured the signal reduction after adding in the complementary NTA-labeled oligonucleotide for each electrode as well as the signal reduction due to the addition of the TpN17 protein. Finally, we added 100 nM of mouse monoclonal anti-TpN 17 antibodies (Clone B1707M, Catalog # MBS319589, MyBioSource, USA) in order to verify that the signal change due to saturating antibody concentrations remained constant amongst all of the sensor constructs.
As with the anchor strand measurements, we performed the antibody titrations in 1x PBS. We began by measuring the baseline methylene blue peak current for three electrodes. Monoclonal antibodies were then added every subsequent 20 min and the reduction in peak current measured. In order to verify that the signal change was not due to degradation of the sensor over time, three additional electrodes were prepared and measurements in 1x PBS (without the addition of antibodies) were performed contemporaneously with the titration measurements.
For measurements of clinical samples, human serum samples for both healthy and syphilis positive patients were obtained from a commercial source (Bioreclamation IVT, USA). The infection status of each of the positive patients was confirmed via a rapid plasma regain test by the vendor. Serum measurements were performed by first diluting samples into their appropriate buffers and then placing sensors in a dilute serum solution and immediately beginning to scan using square wave voltammetry at a 25 mV amplitude and 30 Hz. ELISAs (Zeus Scientific, USA) were purchased and measurements performed according to manufacturer’s instructions.
Results
The first step in fabricating antibody-detecting scaffold sensors is the identification of an appropriate antibody-recognizing epitope or antigen. All previous examples of sensors in this class employed relatively short (< 18 amino acid) linear epitopes or low molecular weight haptens as these recognition elements20–23. Due to the difference in size between the recognition peptide and corresponding antibody these have the advantage of producing large signal gain (relative change in signal upon the addition of saturating target)24. They nevertheless suffer from two potentially significant limitations. First, not all antigens contain linear epitopes, reducing the generality of the approach. Second, not all patients seroconvert (i.e., generate detectable antibodies) against any given epitope. In contrast, the use of full-length or near-full-length antigens as recognition elements supports the simultaneous presentation of multiple linear and conformational epitopes, which should improve clinical sensitivity by expanding the range of diagnostically relevant antibodies that can be detected and increasing the likelihood that the patient will have seroconverted for one of the epitopes present.
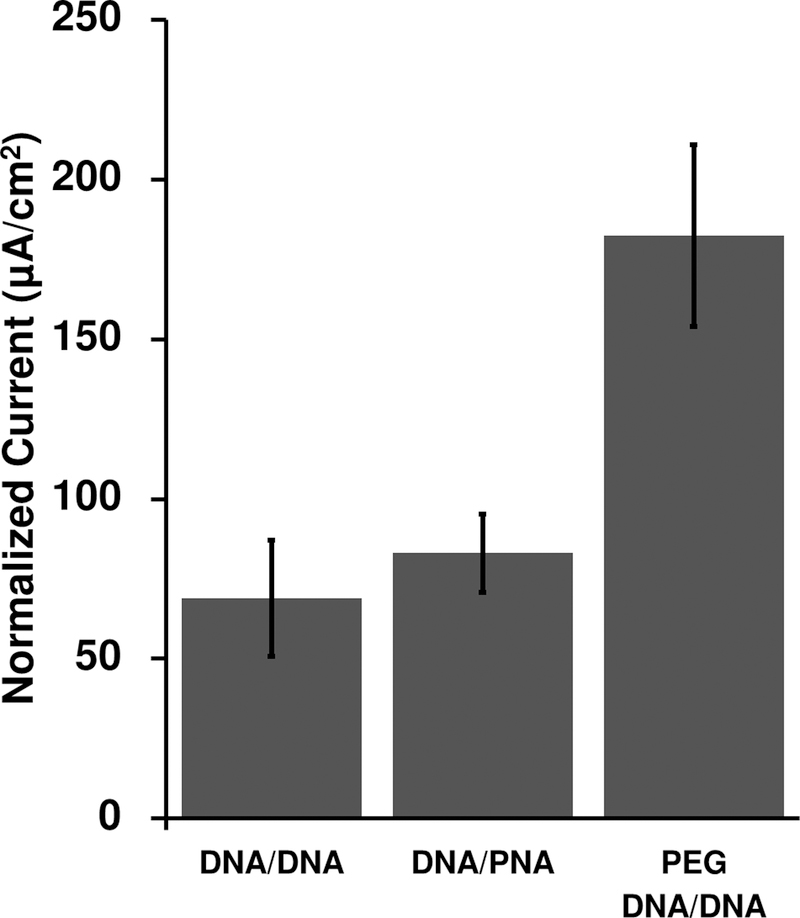
As their antibody-recognizing elements, most commercial treponemal assays employ a combination of up to four full-length T. pallidum membrane proteins ranging in size from 15 to 47 kDa (ref25). ELISAs employing TpN17 as their sole antigen, however, have been shown to achieve good clinical sensitivity and specificity for the diagnosis of syphilis26. Because this antigen is rather large, however, we expected the resulting sensor to produce only relatively small gain and peak current, reducing the signal-to-noise ratio24. In response we pursued two, complementary methods of improving gain and signaling current. First, we engineered our recombinant TpN17 protein to include only the part of TpN17 corresponding to a previously reported crystal structure27, reducing the protein’s molecular weight to 15 kDa. Second, we explored several methods of improving the scaffold’s electron transfer rate, which should lead to improved peak currents24. Specifically, we investigated three constructs differing in scaffold flexibility: (1) the relatively rigid, fully-double-stranded DNA scaffold we have used previously; (2) a peptide-nucleic-acid (PNA)/DNA hybrid scaffold, which we assume is more flexible based off of the conformational flexibility of PNA28; and (3) a double stranded DNA scaffold attached to the SAM via a flexible, 18-unit polyethylene glycol (PEG) linker. At each step of the fabrication process, we interrogated the sensors with square wave voltammetry, measured the peak current, and compared the magnitude of the current between constructs as normalized by the surface area of the sensing electrode [Figure 2]. While the PNA/DNA scaffold provided a slightly higher peak current than the double stranded DNA scaffold, the PEG linker scaffold easily outperformed both, producing more than twice the peak current of the double-stranded DNA scaffold. Based on these results and on the ease of its fabrication we employed the PEG linker scaffold in our subsequent experiments.
Figure 2.
Here we compare the signaling of three sensor architectures varying in scaffold flexibility: a fully-double-stranded DNA scaffold (DNA/DNA), a DNA/peptide-nucleic-acid (PNA) scaffold, and a double stranded scaffold connected to the surface via a flexible polyethylene glycol (PEG) linker. The PEG DNA/DNA scaffold provides approximately twice the current (for a given sized sensor) as our previously employed, DNA/DNA scaffold, and thus we have employed it here. For each construct, we measured the peak current of nine individually fabricated electrodes and normalized each by the surface area of the individual electrode.
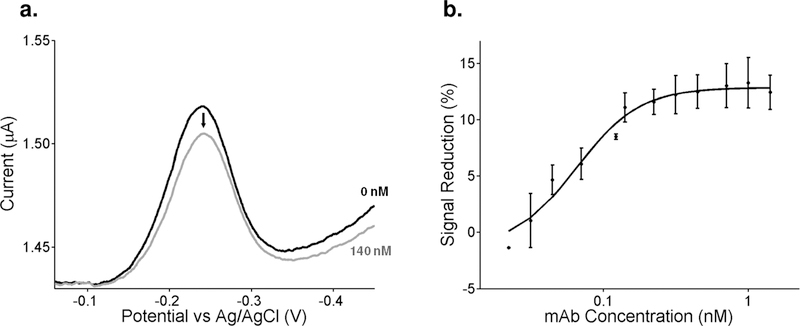
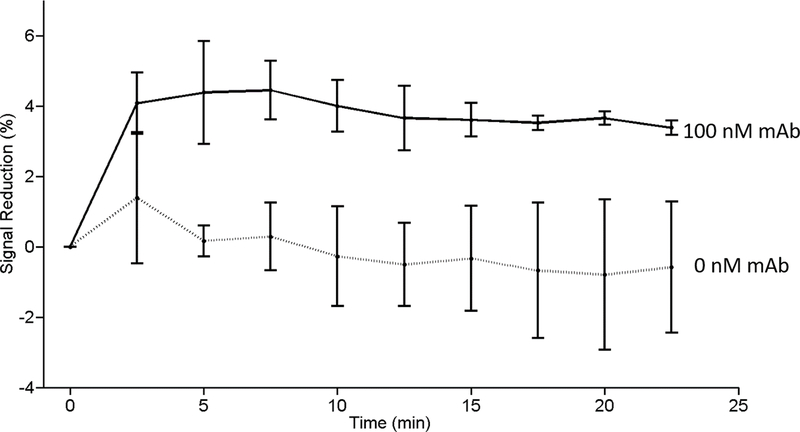
When tested using monoclonal anti-TpN17 antibodies we see a monotonic relationship between signaling current and antibody concentration. To see this, we first established a baseline current in buffer solutions (i.e., without antibodies present), followed by titration with monoclonal antibodies and recording the signal change after a 20 min equilibration at each concentration [Figure 3a]. Fitting the titration curve to a Langmuir isotherm yielded a dissociation constant of 3 nM with the signal change saturating at a gain of ~13% after addition of effectively saturating (20 nM) antibody [Figure 3b]. Having established that we could detect monoclonal antibodies in buffer, we next challenged the sensor against anti-TpN17 monoclonal antibodies spiked into healthy (sero-negative) human serum. One of the chief challenges working in such a complex media is combatting non-specific adsorption of material to the surface of the sensor, which can cause anomalous signal changes resulting in baseline drift. To minimize such effects, we diluted the serum 20-fold with 1 M NaCl. After the addition of 100 nM monoclonal antibody the sensor effectively equilibrated within 5 min and the signal remained stable thereafter [Figure 4]. The signal gain we observe under these conditions, however, is lower than that seen in buffer. As expected, in the absence of an antibody challenge the sensors exhibited no significant signal change.
Figure 3.
We accomplish antibody detection through electrochemical interrogation of the sensor. (a) To determine the concentration of antibody present in our sample, we measure the magnitude of the peak methylene blue current. Shown here are representative electrochemical response curves (i.e., square wave voltammagrams) collected in the absence of target and in the presence of 140 nM of the monoclonal antibody. After linear baseline subtraction (to account for any difference in current not due to the reduction of methylene blue), we used the heights of these peaks to generate the associated binding curve. (b) Upon the addition of increasing concentrations of monoclonal antibody, we observe a binding curve that is well fit by the expected Langmuir isotherm (R2 = 0.98). The error bars represent the standard deviation of three independently fabricated electrodes.
Figure 4.
The sensor rapidly detects monoclonal antibody added to 1:20 diluted syphilis-negative human serum. Shown is the signal change seen in the absence and presence of a monoclonal anti-TnP17 antibody. The error bars represent the standard deviation of three independently fabricated electrodes.
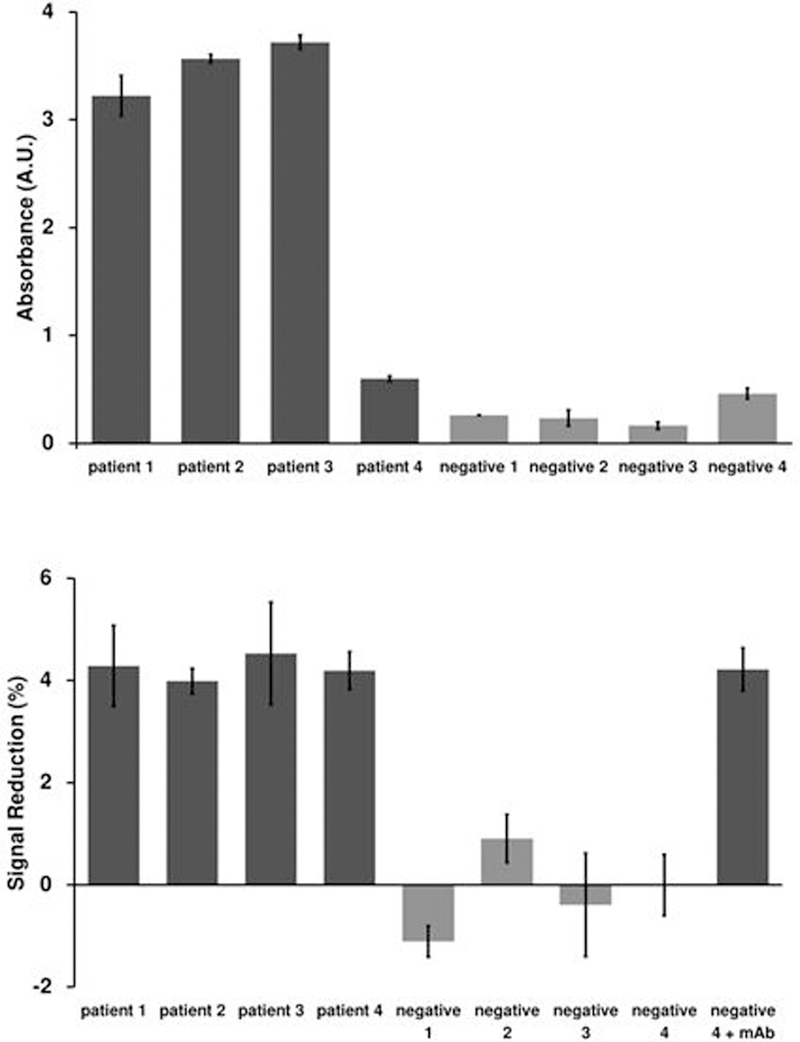
As a demonstration of the clinical sensitivity and specificity of our platform we next challenged the platform with differentiating between syphilis-positive and syphilis-negative human serum samples. To do this we first confirmed the status of four commercially sourced, putatively syphilis-positive human samples and four equivalent syphilis-negative human samples using a commercially available ELISA [Figure 5a]. One of the syphilis-positive patients, identified as patient 4, only responded weakly to the ELISA. Nevertheless, the sample absorbance was still above the cutoff threshold determined using the calibration samples included in the kit. This was likely a result of the patient having a low antibody titer, although without further information from the ELISA manufacturer regarding the components of their kit this must remain speculation. We then characterized each of the eight samples using our sensors. Specifically, for each of the patient samples we prepared electrodes and sample dilutions as described above. We then placed the electrodes into the sample and performed an immediate baseline current measurement followed by a second measurement after 10 min. In every case the signal change between these two measurements differentiated the syphilis-positive samples from the negative samples with good statistical significance [Figure 5b], including patient 4, which was only weakly reactive by ELISA.
Figure 5.
The E-DNA platform can detect endogenous anti-syphilis antibodies in syphilis-positive human serum with clinical sensitivity comparable to that of a commercial ELISA. (a) We performed an ELISA on human serum samples according to the manufacturer’s instructions in order to verify the infection status of our samples and controls. While patient four responded relatively weakly for a positive sample and negative four responded strongly for a negative sample, both fell within the appropriate cutoff values as defined by the standard samples provided in the ELISA kit. Error bars represent the standard deviation of three measurements. (b) We then measured the same four patients and four negative controls utilizing our scaffold sensor and were able to detect antibodies in all four samples, including a low-titer sample (patient 4). A positive control comprised of 100 nM monoclonal antibody spiked into a negative sample (negative 4) presents, as expected, as positive. The values presented represent the signal change after 10 min exposure, while the error bars represent the standard deviation of five independently manufactured sensors.
Discussion
Here we have shown that an E-DNA scaffold sensor employing a near-full-length antigen as its recognition element can detect diagnostically relevant antibodies at clinically relevant concentrations in human serum samples. Despite the relatively large size of the antigenic protein it employs, this new treptonemal test easily differentiates syphilis positive human serum samples from those of healthy control patients in as little as ten minutes via a simple assay that requires no reagents other than the dilution buffer. The clinical sensitivity of the test compares favorably to that a commercially available ELISA, one of the current gold standards for serological diagnosis of syphilis, while taking a fraction of the time (minutes as opposed to hours) and being far less complex (far fewer steps) to perform.
Given that not all patients will produce antibodies against any single epitope and that many antibodies recognize conformational rather than linear epitopes, the use of a folded antigen as our recognition element expands the potential range of diseases that can be diagnosed using sensors in this class and opens the door for a more general platform which can be used to adapt established clinical assays into point-of-care tests. This increase in generality, however, comes at the cost of signal gain. Working in buffer solutions we have previously established that recognition elements up to ~25 kDa in size can produce appreciable signal change upon binding. While our results in buffer are in line with this observation, our results from serum samples lead us to believe that biofouling and the associated loss in signal change might reduce the maximum possible size of the recognition element for scaffold sensors operating in complex media closer to 15 kDa. Even with this restriction in mind, this proof of principle opens the door to a wide range of potentially useful full-length and near full-length antigen E-DNA sensors based off of this platform, and the precise molecular-weight-limit is likely to depend on the specific geometry of the antibody-antigen complex.
Acknowledgements
This work was supported by the NIH through grant R01AI107936. The authors would like to thank Dr. Claudio Parolo for insights and advice.
Footnotes
Conflicts of interest
The authors have no conflicts to declare.
References
- 1.Gift T, Pate M, Hook E, and Kassler W, Sexually Transmitted Diseases, 1999, 26, 232–240 [DOI] [PubMed] [Google Scholar]
- 2.Huang W, Gaydos CA, Barnes MR, Jett-Goheen M, and Blake DR, Sexually Transmitted Infections, 2013, 89, 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swain GR, McDonald RA, Pfister JR, Gradus MS, Sedmak GV, and Singh A, Clinical Medicine & Research, 2004, 2, 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huppert J, Hesse E, and Gaydos CA, Point of Care, 2010, 9, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaydos C, and Hardick J, Expert Review of Anti-Infective Therapy, 2014, 12, 657–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortez KJ, and Greenwald MA Current Infectious Disease Reports, 2014, 16, 423. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention (2017). Sexually Transmitted Disease Surveillance 2016. Atlanta: U.S: Department of Health and Human Services. [Google Scholar]
- 8.Morshed MG, and Singh AE, Clinical and Vaccine Immunology, 2015, 22, 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soreng K, Levy R, and Fakile Y, Clinical Microbiology Newsletter, 2014, 36, 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falcone VH, Stout GW, and M.B. M Jr., Public Health Reports, 1964, 79, 491. [PMC free article] [PubMed] [Google Scholar]
- 11.Ratnam S, Can J Infect Dis Med Microbiol, 2005, 16, 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention, Morb Mortal Wkly Rep, 2010, 59, 1–116 [Google Scholar]
- 13.Larsen SA, Steiner BM, and Rudolph AH, Clin Microbiol Rev, 1995, 8, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seña AC, White BL, and Sparling PF, Clinical Infectious Diseases, 2010, 51, 700–708. [DOI] [PubMed] [Google Scholar]
- 15.Park BG, Yoon JG, Rim JH, Lee A, and Kim HS, Journal of Clinical Microbiology, 2016, 54, 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dicker L, Mosure D, Steece R Stone K, Sexually Transmitted Diseases, 2007, 34,41–46 [DOI] [PubMed] [Google Scholar]
- 17.Schoukroun-Barnes LR, Macazo FC, Gutierrez B, Lottermoser J, Liu J, and White RJ, Annual review of analytical chemistry (Palo Alto, Calif.), 2016, 9(1), 163–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao C, Zhang F, Goldys EM, Gao F, and Liu G, TrAC Trends in Analytical Chemistry, 2018, 102, 379–396 [Google Scholar]
- 19.Mettakoonpitak J, Boehle K, Nantaphol S, Teengam P, Adkins JA, Srisa-Art M, and Henry CS, Electroanalysis, 2016, 28, 1420 [Google Scholar]
- 20.Cash KJ, Ricci F, and Plaxco KW, Journal of the American Chemical Society, 2009, 131, 6955–6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cash KJ, Ricci F, and Plaxco KW, Chemical Communications, 2009, 0, 6222–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White RJ, Kallewaard HM, Hsieh W, Patterson AS, Kasehagen JB, Cash KJ, Uzawa T, Soh HT, and Plaxco KW, Analytical Chemistry, 2012, 84, 1098–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonham AJ, Paden NG, Ricci F, and Plaxco KW, Analyst, 2013, 138, 5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang D, Parolo C, Sun S, Ogden NE, Dahlquist FW, and Plaxco KW, ACS Sensors, 2018, 3, 1271–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malm K, Andersson S, Fredlund H, Norrgren H, Biague A, Månsson F, Ballard R, and Unemo M, Journal of the European Academy of Dermatology and Venereology, 2015, 29, 2369–2376. [DOI] [PubMed] [Google Scholar]
- 26.Marangoni A, Sambri V, Accardo S, Cavrini F, D’Antuono A, Moroni A, Storni E, and Cevenini R, Clinical and Vaccine Immunology, 2005, 12, 1231–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brautigam CA, Deka RK, Liu WZ, and Norgard MV, Protein Science, 2015, 24, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eriksson M, and Nielsen PE, Nature Structural Biology, 1996, 3, 410. [DOI] [PubMed] [Google Scholar]