SUMMARY
The DUX4 transcription factor is briefly expressed in the early cleavage-stage embryo, where it induces an early wave of zygotic gene transcription, whereas its mis-expression in skeletal muscle causes the muscular dystrophy facioscapulohumeral dystrophy (FSHD). Here, we show that DUX4 induces the expression of the histone variants H3.X and H3.Y. We have used a myoblast cell line with doxycycline-inducible DUX4 to show that these histone variants are incorporated throughout the body of DUX4-induced genes. Following a brief pulse of DUX4, these histones contribute to greater perdurance and to enhanced re-activation of DUX4 target gene expression. These findings provide a model for H3.X/Y as a chromatin mechanism that facilitates the expression of DUX4 target genes subsequent to a brief pulse of DUX4 expression.
Graphical Abstract
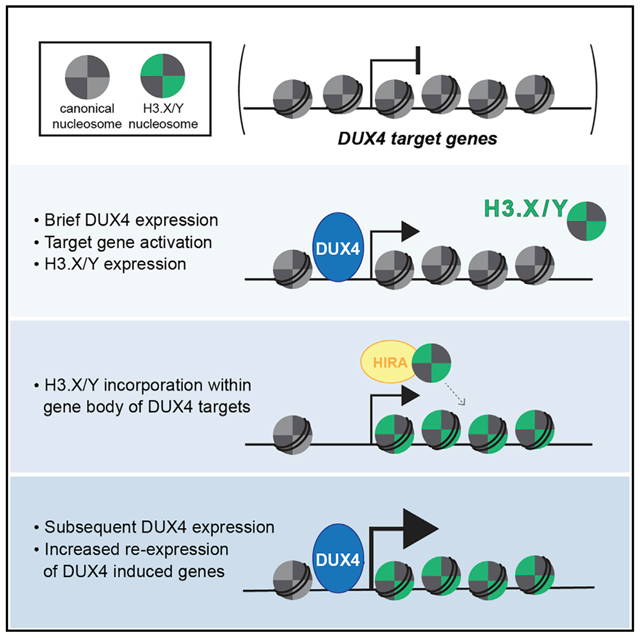
In Brief
Resnick et al. show that the DUX4-induced histone variants, H3.X and H3.Y, incorporate into the chromatin of DUX4-induced genes, making them more sensitive to subsequent expression. This suggests a mechanism for how the brief expression of DUX4 can establish a memory of its transcriptional network.
INTRODUCTION
Mis-expression of the double homeobox transcription factor DUX4 in skeletal muscle is the cause of facioscapulohumeral muscular dystrophy (FSHD) (Tawil et al., 2014). In cultured FSHD muscle cells, there is a de-repression of the DUX4 retrogene, resulting in a burst of DUX4 expression from a minority of myonuclei. In contrast to the toxicity of DUX4 expression in skeletal muscle cells, DUX4 is normally expressed in the early cleavage-stage embryo, where it regulates zygotic genome activation (De Iaco et al., 2017; Hendrickson et al., 2017; Whiddon et al., 2017). In both of these cases, the burst expression of DUX4 results in a perdurant developmental or pathological phenotype. This could be due to the initiation of a transcription factor cascade, an induced chromatin memory, or both.
Histone variants play critical roles in early development, such as the recently demonstrated requirement for H3.3 in paternal genome activation in mouse preimplantation embryos (Kong et al., 2018) as well as for retroelement silencing in embryonic stem cells (Elsässeret al., 2015). Although canonical H3 is incorporated into newly synthesized DNA, H3.3 and H3.3 variants are made throughout the cell cycle (Ahmad and Henikoff, 2002a) and use either the ATRX/DAXX complex to incorporate into repressed regions (Drané et al., 2010; Goldberg et al., 2010; Lewis et al., 2010) or the HIRA chaperone to incorporate into transcriptionally active DNA (Ahmad and Henikoff, 2002b; Tagami et al., 2004). Canonical H3 and H3.3 are extremely well conserved and differ by only 4 to 5 amino acids. More divergent histone variants, such as CENP-A and H3t, have more specialized roles in designating centromeres or facilitating the transition from histones to protamines during spermiogenesis, respectively (Howman et al., 2000; Tachiwana et al., 2010).
Histone variants H3.X and H3.Y were recently identified in the human genome as a multicopy gene family interspersed with the TAF11-like macrosatellite repeat (Wiedemann et al., 2010). Biochemical studies of H3.Y nucleosomes showed that they resulted in a more relaxed chromatin configuration than H3.3 nucleosomes, excluded linker histone H1, were incorporated at transcriptional start sites (TSSs) of actively transcribed genes, and that H3.Y used the HIRA chaperone (Kujirai et al., 2016; Zink et al., 2017). Collectively, these data suggest that H3.Y, and possibly H3.X as well, might be incorporated at active genes and help to maintain an open chromatin conformation.
Here, we show that DUX4 induces the expression of H3.X and H3.Y and that these histone variants are incorporated in highly transcribed DUX4 target genes. A short pulse of DUX4 that mimics its developmental expression pattern induced H3.X/Y expression and the majority of DUX4-regulated genes but was not cytotoxic, permitting the analysis of longer term consequences of DUX4 expression. DUX4-induced expression of H3.X/Y resulted in greater perdurance of DUX4 target gene expression and enhanced activation with a second pulse of DUX4. Together, these results indicate that incorporation of H3.X/Y at DUX4-induced genes contributes to prolonged expression and sensitizes these genes to subsequent induction.
RESULTS
DUX4 Induces the Expression of Histone Variants H3.X and H3.Y
To study the transcriptional network activated by DUX4, we have used a well-characterized cellular model system of human myoblasts with a doxycycline-inducible DUX4 transgene (MB135iDUX4 cells; Jagannathan et al., 2016). Induction of DUX4 in these cells has been shown to induce many DUX4-regulated genes belonging to the transcriptional program characteristic of the early cleavage-stage embryo (Hendrickson et al., 2017; Whiddon et al., 2017) and recapitulates the transcriptional consequences of endogenous DUX4 expression in FSHD cells (Jagannathan et al., 2016; Yao et al., 2014). As such, it is a validated model system for the identification of DUX4-regulated genes and the biological consequences of DUX4 expression.
Further analysis of our previous RNA sequencing datasets (Jagannathan et al., 2016) revealed high expression of unannotated transcripts in the region of the TAF11-like macrosatellite repeat array on chromosome 5 that were not detected in the absence of DUX4 induction. Some of these sequences correspond to histone variants H3.X and H3.Y (Wiedemann et al., 2010), as well as a previously unreported related sequence we designated H3.Z (Figure S1A). Chromatin immunoprecipitation sequencing (ChIP-seq) (Geng et al., 2012) showed DUX4 binding near the TSS of H3.X, H3.Y, and H3.Z loci (Figures 1A and S1D), suggesting they are direct targets of DUX4. Compared to H3.X or H3.Y, H3.Z has a frameshift mutation that disrupts the histone fold and produces a longer protein (Figures S1A and S1B). Although overexpression of H3.X or H3.Y in myoblasts resulted in nuclear staining, overexpression of H3.Z did not (Figure S1C), suggesting H3.Z does not generate a functional histone protein. Therefore, moving forward, we focused our efforts on the characterization of DUX4-induced expression of H3.X and H3.Y.
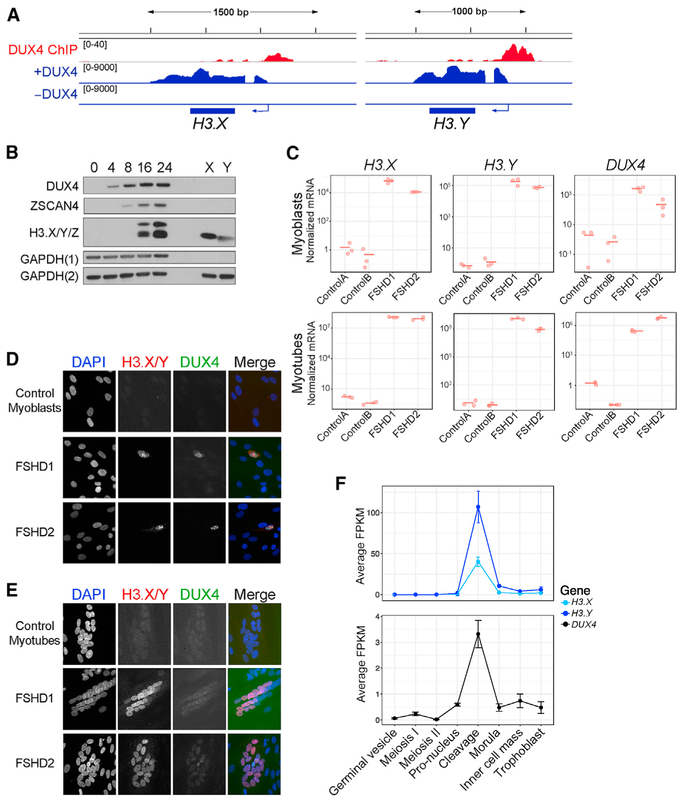
Figure 1. DUX4 Induces the Expression of Histone Variants H3.X and H3.Y.
(A) RNA-seq (blue) and ChIP-seq (red) tracks from Jagannathan et al. (2016) and Geng et al. (2012), respectively, showing that H3.X and H3.Y were induced with DUX4 induction and that DUX4 was bound upstream of H3.X and H3.Y.
(B) Western blot analysis of MB135iDUX4 time course after DUX4 induction. H3.X and H3.Y migrate at slightly different sizes as shown by in vitro translated protein (lanes X and Y). The larger band corresponds to the predicted size for H3.Z. GAPDH(1) and GAPDH(2) represent loading controls for DUX4 or ZSCAN4 and H3.X/Y/Z, respectively.
(C) qRT-PCR analysis of DUX4, H3.X, and H3.Y in control, FSHD1, and FSHD2 myoblasts and myotubes performed in biological triplicates. Values were normalized to the average of the control samples. ControlA = 54-1, ControlB = MB135, FSHD1 = 54-2, and FSHD2 = MB200.
(D and E) Immunofluorescence of H3.X/Y (red) and DUX4 (green) in FSHD1, FSHD2, and control myoblasts and myotubes.
(E) DUX4 and H3.X/Y co-stain rare FSHD myoblast cells and nuclei within myotubes, with no staining seen in control cells. DAPI channel in myoblasts and H3.X/Y and DUX4 channels in myotubes were brightened equally for all samples.
(F) Average fragments per kilobase of transcript per million mapped reads (FPKM) of reads from indicated stages of early human embryos (Hendrickson et al., 2017) that map to H3.X, H3.Y, and DUX4. Expression data displayed as mean with standard deviation of replicates.
See also Figure S1.
Western analysis with an antibody to an epitope shared by H3.X, H3.Y, and H3.Z (Wiedemann et al., 2010) detected both H3.X and H3.Y between 8 and 16 h after DUX4 induction in MB135iDUX4 cells, with levels increasing up to 24 h (Figure 1B). H3.X is slightly larger than H3.Y, as shown by in vitro translated protein, generating a closely spaced doublet. In addition to H3.X and H3.Y (see lanes labeled X and Y in Figure 1B), a band migrating at the size of H3.Z was also detected.
To determine whether endogenous DUX4 also regulated H3.X/Y, we used myoblast cell lines derived from individuals with FSHD1 and FSHD2, the two forms of the disease (Tawil et al., 2014), which show sporadic de-repression of DUX4 in ~0.1% of cells (Snider et al., 2010), with increasing frequency and amount of DUX4 expression upon differentiation to myotubes (Jones et al., 2012; Krom et al., 2012; Snider et al., 2010). qRT-PCR detected elevated levels of DUX4, H3.X, H3.Y, and H3.Z in both FSHD1 and FSHD2 myoblast cultures, but not in controls, with increased expression in the FSHD myotubes (Figures 1C and S1E). Immunofluorescence showed strong nuclear H3.X/Y staining in FSHD myoblasts and myotubes, which also co-localized with DUX4 staining, whereas no control myoblasts or myotubes stained positively for either H3.X/Y or DUX4 (Figures 1D and 1E).
During embryonic development, DUX4 is expressed in a short burst during the cleavage stage (Hendrickson et al., 2017). Re-analysis of human embryo RNA sequencing (RNA-seq) data (Hendrickson et al., 2017) revealed that expression of H3.X and H3.Y coincided with DUX4 expression (Figure 1F). Together, these data identify H3.X and H3.Y as genes regulated by DUX4 and show that they are co-expressed with endogenous DUX4 in biologically relevant contexts, i.e., FSHD muscle cells and the cleavage-stage human embryo. In addition, we have previously shown DUX4 expression in the testis (Snider et al., 2010), where H3.X/Y expression has also been reported (Wiedemann et al., 2010).
H3.X/Y Are Incorporated in Expressed Regions of the Genome
Previous studies demonstrated that exogenously expressed H3.Y was incorporated into nucleosomal DNA by the HIRA chaperone complex (Zink et al., 2017) at the TSSs of highly expressed genes (Kujirai et al., 2016; Zink et al., 2017). To determine the incorporation pattern of endogenous H3.X/Y in DUX4-expressing cells, we induced DUX4 in MB135iDUX4 myoblasts and used the antibody-targeted micrococcal nuclease (MNase) CUT&RUN assay (Skene et al., 2018; Skene and Henikoff, 2017) to map H3.X/Y incorporation genome-wide (schematic in Figure 2A). H3.X/Y localized in domains ranging from 500 bp to nearly 100 kb (Figure S2A), with ~75% of domains overlapping genic regions and the remaining 25% intergenic (Figure 2B), similar to what was seen in a previous study using ChIP-seq of tagged H3.Y expressed in HeLa cells (Zink et al., 2017).
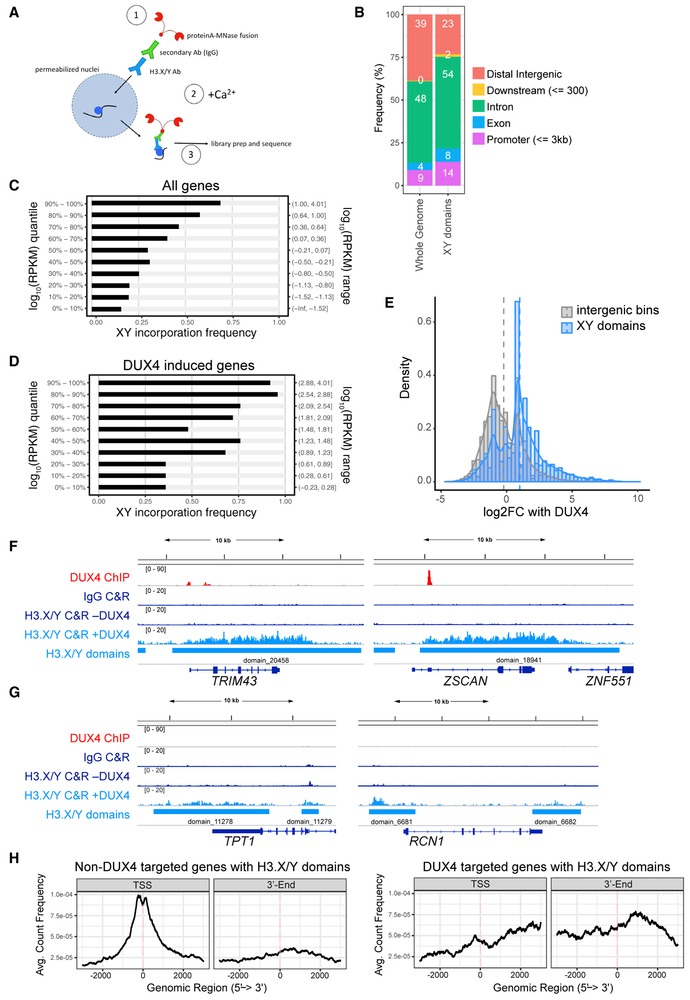
Figure 2. H3.X/Y Are Incorporated in Expressed Regions of the Genome.
(A) Schematic of CUT&RUN protocol. H3.X/Y nucleosome is represented by a blue circle. CUT&RUN has been used successfully with antibodies that do not perform well in standard chromatin immunoprecipitation (ChIP) assays (Liu et al., 2018), and we used it for this study because the antibody to H3.X/Y did not work for standard ChIP.
(B) Distribution of genomic annotations containing H3.X/Y domains.
(C) Bar graph depicting association of XY incorporation and all DUX4-induced genes (Jagannathan et al., 2016). Gene expression quantiles (log_10 RPKM quantile; y axis) are plotted against the percentage of genes in each quantile interval overlapping with H3.X/Y domains (x axis). Pearson correlation = 0.958.
(D) Same as (C) for robust DUX4 target genes (n = 251 with adjusted p < 0.05, corresponding to H_0: ∣lfc∣ < 4). Pearson correlation = 0.976.
(E) Histogram of intergenic region expression change with DUX4 induction. Comparing intergenic H3.X/Y domains with random intergenic bins of comparable size shows an association between H3.X/Y domains and DUX4-induced transcription. Vertical dashed lines mark mean log2 fold change with DUX4 for each group.
(F) DUX4 ChIP-seq (Geng et al., 2012), immunoglobulin G (IgG), or H3.X/Y CUT&RUN (C&R) and called H3.X/Y domains at DUX4 target genes TRIM43 and ZSCAN4.
(G) Same as (F) but for genes constitutively expressed (TPT and RCN1).
(H) Analysis of the distribution of H3.X/Y CUT&RUN reads within genes. Left panel shows highly expressed, non-DUX4 target genes with H3.X/Y domains (n = 679); right panel shows DUX4 target genes only (n = 191).
See also Figure S2.
H3.X/Y were preferentially incorporated at highly expressed genes (Figure 2C) and at DUX4-induced target genes (Figure 2D). Similarly, 80% of intergenic DUX4-induced long noncoding RNAs (lncRNAs, n = 380) showed H3.X/Y incorporation, and the presence of an intergenic H3.X/Y domain correlated with induction by DUX4 as compared to random intergenic bins of comparable length (Figure 2E). Specific examples of DUX4-induced genes and constitutively expressed genes are shown in Figures 2F and 2G, respectively. Averaging over larger sets of genes showed greater H3.X/Y enrichment at the TSSs of constitutively expressed genes (Figure 2H, left), consistent with previous reports (Kujirai et al., 2016; Zink et al., 2017), whereas H3.X/Y were enriched throughout the entire transcribed region of DUX4-induced genes (Figure 2H, right).
A Pulse of DUX4 Activates Target Gene Expression with Little Cell Toxicity
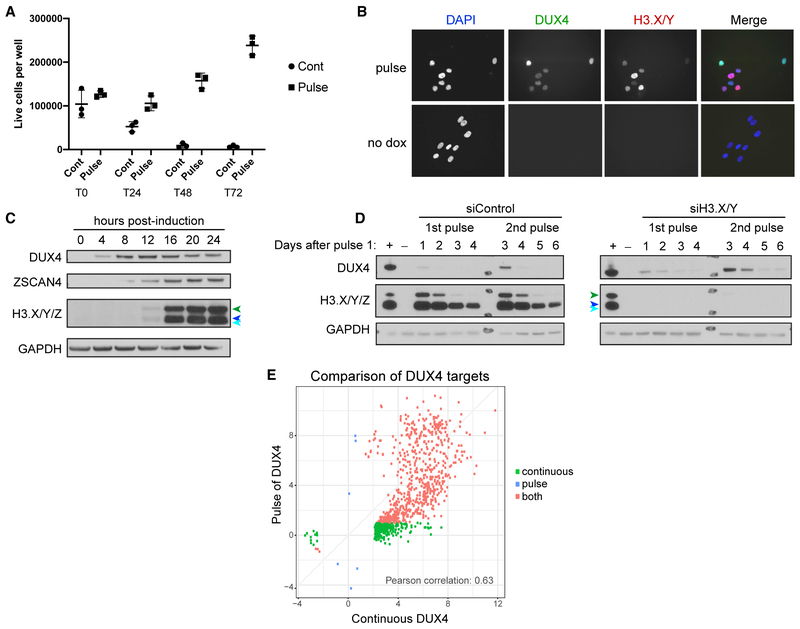
Mis-expression of DUX4 induces apoptotic cell death in nearly every cell type tested (Kowaljow et al., 2007; Rickard et al., 2015; Wallace et al., 2011), yet DUX4 is expressed in the germline and early embryo (Hendrickson et al., 2017; Snider et al., 2010), and the Dux mouse ortholog was shown to be important for early embryo development (De Iaco et al., 2017). Unlike in cell culture models, where toxicity occurs with continuous DUX4 expression, DUX4 is only briefly expressed in the early embryo. To test whether cells might survive transient expression of DUX4, we treated MB135iDUX4 myoblasts with a 4- to 6-h “pulse” of doxycycline to induce transient DUX4 expression. In contrast to the continuous expression of DUX4 that resulted in the death of nearly the entire cell population by 48–72 h, a 4-h pulse of doxycycline resulted in only a small reduction of the cell population at 24 h followed by expansion of that population through the 72-h time point (Figure 3A). Immunodetection showed that a short pulse of doxycycline induced DUX4 expression in nearly all nuclei (Figure 3B). Western analysis confirmed that DUX4 protein and DUX4 targets ZSCAN4 and H3.X/Y/Z were detectable at similar time points as observed for continuous DUX4 expression and persisted for at least 24 h (Figure 3C; compare to Figure 1B). Remarkably, the H3.X and H3.Y proteins remained detectable even 4 days after the initial pulse (Figure 3D). Based on RNA-seq analysis, the transcriptome 24 h after a pulse of DUX4 largely recapitulated the transcriptional changes characterized by the continuous expression of DUX4 seen in Jagannathan et al. (2016); R2 = 0.63; Figure 3E. Of the 941 genes that increased by a log2 fold-change > 2.0 (adjusted p < 0.05) in the continuous sample, 673 (72%) showed greater than a two-fold change (log2 fold-change > 1.0) 24 h after beginning a 4-h pulse of DUX4 (Table S1 and red dots in Figure 3E). These results suggest that the duration of DUX4 expression might be a major determinant of toxicity rather than the cell type and that a brief pulse of DUX4 results in robust activation of its transcriptional program and prolonged presence of H3.X/Y proteins.
Figure 3. A Pulse of DUX4 Activates Target Gene Expression with Little Cell Toxicity.
(A) Counts of live MB135iDUX4 cells (cells that exclude trypan blue) before and at daily time points after continuous exposure to doxycycline (cont) or a 4-h pulse of doxycycline (pulse). T0, T24, T48, and T72 indicate the hours following initial doxycycline addition.
(B) DUX4 and H3.X/Y immunofluorescence in MB135iDUX4 cells 8 h after the start of a 4-h pulse of doxycycline (pulse) shows induction in nearly all cells, with no staining in uninduced (no dox) cells.
(C) Western blot analysis of MB135iDUX4 cells up to 24 h after DUX4 pulse. Cells were induced from 0 to 4 h. H3.X/Y/Z are identified with colored arrowheads: H3.Z (green); H3.X (dark blue); and H3.Y (light blue).
(D) MB135iDUX4 cells with control or H3.X/Y knockdown with 1 or 2 pulses of DUX4 on days 0 and 2, respectively, and harvested 1–4 days after each pulse (days 1–4 for 1st pulse; days 3–6 for 2nd pulse). +, continuous dox overnight; –, uninduced day 0 cells. H3.X/Y/Z are identified as in (C).
(E) Comparison of DUX4-induced genes from RNA-seq datasets in MB135iDUX4 cells after continuous (from Jagannathan et al., 2016) or pulsed DUX4 expression (log2-fold change over no DUX4 induction with adjusted p < 0.05). Axes show degree of gene induction (log2-fold change over no DUX4) with adjusted p < 0.05 corresponding to H_0: ∣log2-fold change∣ < 2. Green indicates genes activated less than 2-fold (log2-fold change < 1.0) in the pulse condition, red indicates genes induced more than 2-fold in the pulse, and blue indicates genes induced by the pulse but less than 2-fold in the continuous.
H3.X/Y Incorporation Increases the Perdurance and Re-expression of DUX4 Target Genes
To determine whether H3.X/Y incorporation at DUX4 target genes could increase perdurance of gene expression and/or facilitate subsequent gene expression, we induced a pulse of DUX4 followed by a second pulse 2 days later, when H3.X/Y are present in chromatin (Figure 4A). We compared gene expression with or without H3.X/Y by using small interfering RNAs (-siRNAs) that targeted both H3.X and H3.Y (siXY) or control siRNA (siControl; Figures 4A-4D). RNA for the DUX4 targets ZSCAN4 and TRIM43, both of which overlap with H3.X/Y incorporation (see Figure 2F), were robustly induced 24 h after the first pulse of DUX4 expression with decreased levels by 48 h after the pulse. A second pulse of DUX4 2 days after the first pulse invoked a super-induction of ZSCAN4 and TRIM43 24 h later (day 3), with RNA levels roughly six-fold greater than after the single pulse. Despite similar, or slightly higher, levels of DUX4 expression (Figure 4B), H3.X/Y knockdown prevented this super-induction after the second pulse (Figure 4C). Following either the first pulse or second pulse of DUX4, ZSCAN4 and TRIM43 had greater perdurance of expression in siControl-treated cells compared to siXY-treated cells based on both the amount of RNA and the slope of the decline of mRNA (Figure 4D). This suggests that incorporation of H3.X/Y facilitated the increased inducibility of these genes and the persistence of their RNA expression. In contrast to these DUX4 target genes, constitutively expressed genes that incorporated H3.X/Y, e.g., TPT1 and XRCC5, were unaffected by H3.X/Y knockdown (Figures S2B and S2C). Overexpression of FLAG-tagged H3.X or H3.Y in the absence of DUX4 did not induce DUX4 target gene expression (Figures S3A-S3C), indicating that H3.X/Y were enhancing the response to DUX4 rather than acting alone to induce a transcriptional response. Similar to the results in myoblasts, H3.X/Y expression was necessary for enhanced induction of DUX4 target genes following a second pulse of DUX4 in differentiated muscle cells (Figures S4A and S4B), consistent with replication-independent nucleosome incorporation mediated by the HIRA chaperone. Furthermore, H3.X/Y knockdown in FSHD1 and FSHD2 myoblasts or myotubes decreased both DUX4 expression and DUX4 target gene expression (Figures S4C-S4E).
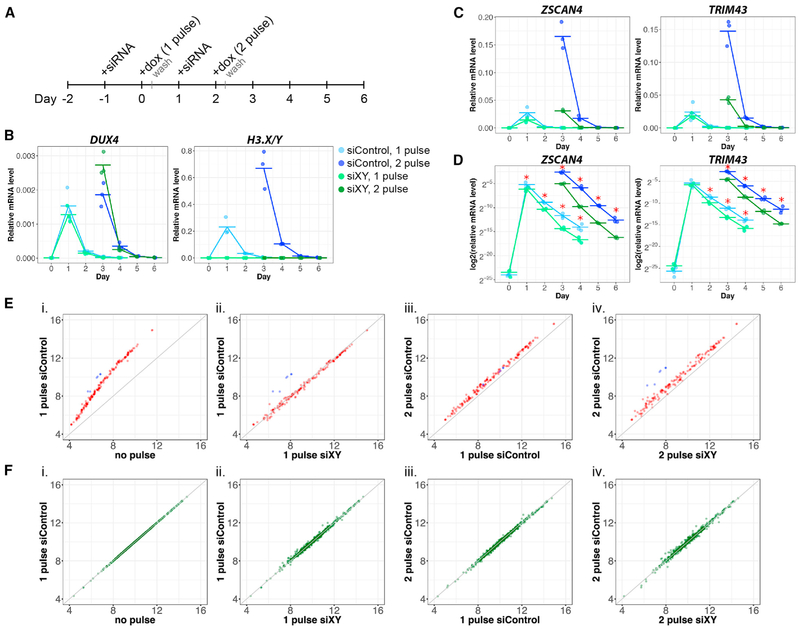
Figure 4. H3.X/Y Incorporation Increases the Perdurance and Re-expression of DUX4 Target Genes.
(A) Schematic of experimental design.
(B–D) qRT-PCR in MB135iDUX4 cells with 1 or 2 pulses of DUX4 and treatment with siH3.X/Y (green) or siControl (blue). Cells were harvested before induction and 1–4 days after each pulse, with 3 biological replicates for each sample shown, relative to RPL-27.
(D) Data from (C) plotted on a log scale illustrate differential perdurance of DUX4 target gene expression in siH3.X/Y relative to siControl samples (*p < 0.05; one-tailed Wilcoxon rank sum test). Based on a functional t test on the null distribution built by permutations (see STAR Methods), the difference of the slopes between siControl and siH3.XY is significant (p < 1e–12).
(E) Expression of DUX4 targets measured by RNA-seq in MB135iDUX4 cells, shown as average log2 normalized read counts of biological triplicates. Sequences in the H3.X/Y family targeted by siXY are shown in blue. Null model (no difference between conditions) is shown in gray. (1) Induction of DUX4 targets, (2) comparison of H3.X/Y and control knockdown with a single pulse, (3) super-induction of DUX4 targets with a second pulse in control knockdown samples, and (4) knockdown of H3.X/Y eliminating super-induction are shown.
(F) Expression of genes with H3.X/Y domains that are unaffected by DUX4, plotted as in (E).
See also Figures S2 and S4.
Expanding these experiments genome-wide, we performed RNA-seq 24 h after the 1-pulse and 2-pulse time points for the conditions shown in Figure 4A. Differential gene expression analysis revealed that H3.X/Y were necessary to sensitize nearly all DUX4-induced genes for subsequent super-induction. A single pulse of DUX4 showed induction of target genes (Figure 4Ei) that were not affected by H3.X/Y knockdown (Figure 4Eii), whereas super-induction of DUX4 target genes with a second pulse (Figure 4Eiii) was prevented by H3.X/Y knockdown (Figure 4Eiv). In contrast, constitutively expressed non-DUX4 targets with H3.X/Y incorporation were unaffected by pulses of DUX4 and knockdown treatments (Figure 4F). These results demonstrate that H3.X and H3.Y were incorporated into DUX4-induced genes and that this enhanced future expression of these genes.
DISCUSSION
Together, our data demonstrate that H3.X and H3.Y are induced by DUX4 in human muscle cells, are induced coincident with DUX4 expression in human embryos and in FSHD muscle cells, are incorporated at genes induced by DUX4, and that their incorporation both promotes the perdurance of DUX4 target gene expression and facilitates their subsequent induction. Previously, other groups have described important biochemical properties of H3.X and H3.Y, mostly through in vitro and mis-expression studies (Kujirai et al., 2016, 2017; Wiedemann et al., 2010; Zink et al., 2017). In particular, the in vitro biochemical studies predicting a more relaxed chromatin state with less efficient H1 binding (Kujirai et al., 2016) are consistent with our studies in a DUX4-inducible myoblast cell line showing enhanced transcription of genes incorporating H3.X/Y. In this way, our study builds on these important advances and characterizes biological consequences of endogenous H3.X/Y expression. Future studies will be needed to determine the role of H3.X/Y incorporation during embryogenesis and whether H3.X/Y contribute to FSHD pathophysiology.
In contrast to DUX4 target genes, constitutively expressed genes were mostly unaffected by incorporation of H3.X/Y. This was also associated with a different pattern of incorporation. Whereas H3.X/Y were incorporated throughout the gene body of DUX4 target genes, incorporation at constitutively expressed genes was largely flanking the TSSs, as was previously reported in studies that mis-expressed H3.Y in HeLa cells (Kujirai et al., 2016; Zink et al., 2017). It is possible that this different pattern of incorporation represents the difference between a constitutively expressed gene and an induced but previously silent gene. Because the majority of genes robustly regulated by DUX4 are not expressed in myoblasts, their nucleosomes would likely have canonical H3 histones that would be replaced with H3.X/Y and H3.3 when actively transcribed. In contrast, constitutively expressed genes would already have replaced canonical H3 histones with H3.3 and there might be less turnover in the gene body compared to newly induced genes, restricting H3.X/Y incorporation to nucleosomes flanking the TSS that undergo more rapid turnover. Although speculative, this difference might account for the specificity of perdurance and hyper-induction at DUX4-induced genes that incorporate H3.X/Y.
Another important finding of our study is that skeletal muscle cells survive the DUX4-induced transcriptional program following a transient burst of DUX4 expression. Using a well-characterized cell culture model (Jagannathan et al., 2016) with inducible DUX4, it was possible to mimic the kinetics of embryonic DUX4 expression with short pulses, leading to expression of targets, including H3.X/Y, without inducing cell death. This made it feasible to study the effects of DUX4 expression over several days. This finding has interesting implications for both stem cell biology and FSHD muscular dystrophy. In stem cell biology, brief expression of DUX4 similar to that in the early embryo occurs in a subset of embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs), where it also induces a transcriptional program similar to the cleavage-cell state (Hendrickson et al., 2017; Whiddon et al., 2017; De Iaco et al., 2017). Therefore, our findings suggest that the difference between the early embryo or ESCs/iPSCs that survive DUX4 expression and FSHD cells or other somatic cells that die when DUX4 is constitutively expressed might be the duration of expression rather than a protective factor unique to the early developmental program.
In this regard, skeletal muscle might be particularly susceptible to repeated bursts of DUX4 expression. As our study has demonstrated, incorporation of H3.X/Y in DUX4 target genes increased the perdurance of their expression and enhanced subsequent activation (Figure S4F). Because H3.X/Y use the HIRA chaperone (Kujirai et al., 2016; Zink et al., 2017), they are incorporated into actively transcribed regions independent of DNA replication. As skeletal muscle is post-mitotic and multinucleated, H3.X/Y incorporation could create a prolonged sensitivity for DUX4 target expression, both in the nucleus that initially expressed DUX4 and in adjacent nuclei that received DUX4 and H3.X/Y from their shared cytoplasm. In this model, stochastic bursts of DUX4 in different nuclei in a myotube might result in progressive accumulation of H3.X/Y in an expanding nuclear domain and progressive enhancement of expression of DUX4 target genes, resulting in toxicity like that seen with the constitutive expression of DUX4 (Figure S4G, right). In contrast, incorporation of H3.X/Y following the embryonic burst of DUX4 would be diluted by subsequent cell divisions (Figure S4G, left). Initially, this could result in greater perdurance of the DUX4-induced transcriptional program but ultimately not reach toxic levels. It is important to emphasize that these models depict possible biological implications of H3.X/Y function based on data from cell culture studies of DUX4 expression in an engineered cell line and FSHD muscle cells. It will require future studies to verify the details of each model. For example, the burst of DUX4 at the four-cell stage in human embryos is documented (Hendrickson et al., 2017), whereas subsequent bursts of DUX4 in the progeny of these cells has not been described and has yet to be carefully evaluated. Similarly, although H3.X/Y appear to have a role in DUX4 target gene expression in our cell culture models of FSHD, it remains to be shown whether H3.X/Y contribute to FSHD pathophysiology.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Stephen Tapscott (stapscot@fredhutch.org). In some cases, the Fred Hutchinson Cancer Research Center might require a standard Material Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The following cell lines were used in this study: MB135 (control, female, (Geng et al., 2012), 54-1 (control, male, (Krom et al., 2012), MB073 (FSHD1, male, Fields Center for FSHD and Neuromuscular Research), 54-2 (FSHD1, male, (Krom et al., 2012), MB200 (FSHD2, male, Fields Center for FSHD and Neuromuscular Research), and 2453 (FSHD2, male, Fields Center for FSHD and Neuromuscular Research). FSHD, control, and MB135iDUX4 myoblasts (described in Jagannathan et al., 2016) were grown in F10 growth medium (GIBCO Life Technologies) supplemented with 10% FBS (HyClone), 1% pen/strep (Life Technologies), 10pg/mL fgf (Life Technologies), 1 μM dexamethasone (Sigma), and 2 μg/mL puromycin or 10 μg/mL blasticidin as appropriate to maintain inducibility of the DUX4 transgene. FSHD and control cells were differentiated by growing to confluence and changing to differentiation medium: DMEM (GIBCO) with 1% horse serum (Life Technologies), 1% pen/strep, and 10 μM each transferrin and insulin for 72 h. Pulsed MB135iDUX4 cells were treated with 1 μg/mL doxycycline in growth medium for 6 h for RNA-seq experiments or 4 h for all other experiments, rinsed with PBS, and fresh growth medium added. Cells with continuous induction were treated with 1 μg/mL doxycycline in growth medium overnight or as specified. Differentiated MB135iDUX4 cells were treated with 2μg/mL doxycycline in differentiation medium for 8 h. All cell lines were cultured at 37°C in a humidified incubator supplied with 5% CO2.
METHOD DETAILS
siRNA treatment
siRNAs for H3.X/Y were designed using the Dharmacon siDESIGN Center. Two siRNAs that gave >90% knockdown were pooled for all experiments to minimize off-target effects with a non-targeting siRNA (Dharmacon D-001210-02-05) used as a control. For pulse experiments, cells were transfected with 50 pg siRNA in OPTIMEM with 7.5 μL Lipofectamine RNAiMax 16 h before each doxycycline treatment. For knockdown experiments in FSHD cells, a double transfection protocol was followed to ensure efficient depletion of pre-existing proteins. FSHD myoblasts were seeded in six-well plates and transfected the next day with 7.5 μL Lipofectamine RNAiMAX and 50pmol of either H3.X/Y-specific siRNA or a non-silencing control siRNA (Dharmacon D-001810-01) diluted in 500 μL OptiMEM Reduced Serum Medium (Thermo Fisher Scientific). Sixteen-hours post-transfection, media was replaced with either supplemented F10 growth media for myoblasts or serum-free differentiation media to promote myotube formation. Forty-eight h following the first transfection, cells were transfected with siRNAs a second time. Fresh media was added 16-hours post-transfection. Cells were harvested for RNA analysis or fixed for immunofluorescence 32 h later.
CUT&RUN-sequencing
MB135iDUX4 cells were treated with or without doxycycline for 18 h before harvesting. Protocol was followed as in Skene et al. (2018) with modifications to scale up for 5 million cells per sample. 100 μL beads were used per sample and wash/incubation volumes were increased to 300-500 μL. 0.05% digitonin was used in the wash buffer. 15 μL H3.X/Y primary antibody (clone 8H6-2111, Active Motif 61161) was used per H3.X/Y sample and incubated for 2 hr. 25 μL rabbit anti-rat secondary (ab6703) was added to each sample for 1 hr. After MNase digestion, fragments were liberated for 20 min and DNA was then purified using the phenol/chloroform method. Three biological replicates were performed for H3.X/Y samples, both with and without doxycycline, and one replicate for each IgG condition.
RNA isolation and sequencing
MB135iDUX4 cells were treated with siRNA knockdown as described above followed by DUX4 pulsing, either once or twice, in triplicate, and harvested 24 h after the start of a pulse. Untreated cells were also harvested from triplicate wells as negative controls. The NucleoSpin RNA kit (Macherey-Nagel) was used to extract RNA from whole cells, following the manufacturer’s instructions. RNA-seq libraries were prepared using the Illumina TruSeq RNA Sample Prep v2 Kit and a PerkinElmer Sciclone NGSx Workstation. All 15 libraries were pooled and sequenced on two flow lanes. The in-house R package and bioinformatics analysis were done with R-3.4.3/Bioconductor-3.5.
Western blotting
Protein was directly lysed from tissue culture plates using 2X Laemlli Buffer with 4% beta mercaptoethanol, sonicated, and boiled for 10 min. Samples were loaded on 4%–12% polyacrylamide gel (Novex) and run with MES buffer, then transferred to a PVDF membrane. Membranes were blocked in 5% milk in PBST for 1 h and incubated with primary antibody in 5% milk overnight at 4° (see KEY RESOURCES TABLE for details on antibodies). After washing, membranes were incubated with appropriate secondary antibodies for 1 h, washed, and detected with chemiluminescence on film.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| GAPDH (6C5) | GeneTex | GTX28245; RRID: AB_37067 |
| DUX4 | Geng et al., 2011 | E14-3 |
| ZSCAN4 | ThermoFisher | PA5-32106; RRID: AB_2549579 |
| H3.X/Y | Wiedemann et al., 2010 | clone 8H6-2111; Active Motif Cat# 61161, RRID:AB_2793533 |
| FLAG | Sigma | F3165 |
| Goat anti-rat | Jackson ImmunoResearch | 112-035-068 |
| Peroxidase AffiniPure Goat Anti-Rabbit (H+L) | Jackson ImmunoResearch | 111-035-144; RRID: AB_2307391 |
| Peroxidase AffiniPure Goat Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | 115-035-146; RRID: AB_2307392 |
| Rhodamine (TRITC)-AffiniPure Donkey Anti-Rat IgG | Jackson ImmunoResearch | 712-025-150 |
| Fluorescein (FITC)-AffiniPure Donkey Anti-Rabbit IgG | Jackson ImmunoResearch | 711-095-152 |
| Rhodamine (TRITC)-AffiniPure Donkey Anti-Mouse IgG | Jackson ImmunoResearch | 715-025-151 |
| Bacterial and Virus Strains | ||
| Stbl3 Competent E. coli | Generated in-lab | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DNase Amp grade | Invitrogen | 18068015 |
| RNaseOUT Recombinant Ribonuclease Inhibitor | Invitrogen | 10777019 |
| Oligo(dT) 12-18 Primer | Invitrogen | 18418012 |
| Superscript III | Invitrogen | 18080044 |
| Recombinant human basic fibroblast growth factor | Promega | G5071 |
| Dexamethasone | Sigma-Aldrich | D4902 |
| Puromycin dihydrochloride | Sigma-Aldrich | P833 |
| Blasticidin | GIBCO | R21001 |
| Penicillin/streptomycin | GIBCO | 15140122 |
| Doxycycline hyclate | Sigma-Aldrich | D9891 |
| Insulin | Sigma-Aldrich | I1882 |
| Transferrin | Sigma-Aldrich | T-0665 |
| Polybrene | Sigma-Aldrich | 107689 |
| Critical Commercial Assays | ||
| Lipofectamine RNAiMAX | Life Technologies | 13778150 |
| Lipofectamine 2000 | ThermoFisher | 11668019 |
| OptiMEM Reduced Serum Medium | ThermoFisher | 31985070 |
| iTaq SYBR Green Supermix | Bio-Rad | 1725124 |
| NucleoSpin RNA kit | Machery-Nagel | 740955 |
| Illumina TruSeq RNA Sample Prep v2 Kit | Illumina | RS-122-2001 |
| Deposited Data | ||
| DUX4 ChIP | Geng et al., 2012; NCBI GEO | GSE33838 |
| continuous DUX4 RNA-seq | Jagannathan et al., 2016; NCBI GEO | GSE85461 |
| early human embryo RNA-seq | Hendrickson et al., 2017; NCBI GEO | GSE72379 |
| RNA-seq data for this study | NCBI Gene Expression Omnibus | GSE119403 |
| CUT&RUN data for this study | NCBI Gene Expression Omnibus | GSE119403 |
| Mendeley Dataset for this study | https://data.mendeley.com/ | https://doi.org/10.17632/8mvjj5rw6r.1 |
| Experimental Models: Cell Lines | ||
| MB135 (female) | Geng et al., 2012 | N/A |
| 54-1 (male) | Krom et al., 2012 | N/A |
| MB073 (male, FSHD1) | Fields Center for FSHD and Neuromuscular Research | https://www.urmc.rochester.edu/neurology/fields-center.aspx |
| 54-2 (male, FSHD1) | Krom etal., 2012 | N/A |
| MB200 (male, FSHD2) | Fields Center for FSHD and Neuromuscular Research | https://www.urmc.rochester.edu/neurology/fields-center.aspx |
| 2453 (male, FSHD2) | Fields Center for FSHD and Neuromuscular Research | https://www.urmc.rochester.edu/neurology/fields-center.aspx |
| MB135iDUX4 (female) | Jagannathan et al., 2016 | N/A |
| MB135iFLAG-H3.3 (female) | This Study | N/A |
| MB135iFLAG-H3.X (female) | This Study | N/A |
| MB135iFLAG-H3.Y (female) | This Study | N/A |
| MB135iH3.Z (female) | This Study | N/A |
| MB135iGFP (female) | This Study | N/A |
| Oligonucleotides | ||
| Primers for qPCR and cloning (see Table S2) | This Study | N/A |
| siRNAs targeting H3.X/Y: UCAAGAAGCCUCACCGCUAUU, GCGGGAAAUCAGAAAGUACUU | This Study (Dharmacon Custom) | N/A |
| siGENOME non-Targeting #2 Control siRNA | Dharmacon | D-001210-02-20 |
| ON-TARGETplus Non-targeting #1 Control siRNA | Dharmacon | D-001810-01 |
| Recombinant DNA | ||
| pCW57.1 | Addgene | 41397; RRID: Addgene_41397 |
| pMD2.G | Addgene | 12259 |
| psPAX2 | Addgene | 12260 |
| pCW57.1-FLAG:H3.3 | This Study | N/A |
| pCW57.1-FLAG:H3.X | This Study | N/A |
| pCW57.1-FLAG:H3.Y | This Study | N/A |
| pCW57.1-H3.Z | This Study | N/A |
| pCW57.1-GFP | This Study | N/A |
| Software and Algorithms | ||
| Bowtie2-2.2.6 | Langmead et al., 2009 | RRID: SCR_005476; https://github.com/BenLangmead/bowtie/ |
| R package domainCalling | This Study | https://github.com/TapscottLab/domainCalling |
| ChIPseeker/Bioconductor-3.5 | Yu et al., 2015 | https://www.bioconductor.org/packages/release/bioc/html/ChIPseeker.html |
| csaw/Bioconductor-3.5 | Lun and Smyth, 2016 | https://bioconductor.org/packages/release/bioc/html/csaw.html |
| Genomic Alignments/Bioconductor-3.5 | Lawrence et al., 2013 | https://bioconductor.org/packages/release/bioc/html/GenomicAlignments.html |
| edgeR/Bioconductor-3.5 | Robinson et al., 2010 | RRID:SCR_012802; http://bioconductor.org/packages/release/bioc/html/edgeR.html/ |
| DESeq2/Bioconductor-3.5 | Love et al., 2014 | RRID:SCR_000154; http://bioconductor.org/packages/release/bioc/html/DESeq.html |
| Tophat-2.1.0 | Trapnell et al., 2009 | RRID:SCR_013035; http://ccb.jhu.edu/software/tophat/index.shtml |
| Ensembl v88 | Zerbino et al., 2018 | RRID:SCR_002344; http://www.ensembl.org//useast.ensembl.org/?redirectsrc=//www.ensembl.org%2F/ |
| CRAN: ggplot2 | Wickham, 2016 | RRID: SCR_014601; https://cran.r-project.org/web/packages/ggplot2/index.html |
Reverse transcription and qPCR
RNA was extracted as described for RNA-seq, treated with DNase I (ThermoFisher), and heat inactivated. 500ng-1 μg of RNA was reverse transcribed using SuperScript III First Strand cDNA Synthesis (ThermoFisher) according to manufacturer’s instructions, using oligo-dT priming. A no-enzyme sample was also run with a mix of all RNA samples as a control. qPCR was performed using 1x iTaq SYBR Green Master Mix (Bio-Rad) and primers at 1 μM each. Primers listed in Table S2.
Immunofluorescence
Briefly, cells were fixed in 1% formaldehyde for 15 min, permeabilized in PBST, and incubated overnight with primary antibodies. Plates were then washed with PBS, incubated 1 h with fluorescent secondary antibody, counter-stained with DAPI, and imaged using an immersion lens.
Cloning and polyclonal transgenic cell lines
A putative H3.Z sequence was identified from RNA-seq reads and used to design primers slightly outside this region. Amplicons from cDNA of DUX4-expressing cells were individually subcloned using the TOPO system, miniprepped using the NucleoSpin Plasmid kit (Macherey-Nagel) according to manufacturer’s instructions, and Sanger sequenced to generate the final H3.Z sequence. FLAG-tagged H3.X, FLAG-tagged H3.Y, H3.Z, and GFP were cloned into pCW57.1 (Addgene #41393). Lentivirus with inducible transgenes were generated by co-transfecting 293T cells with FLAG-tagged H3 variant constructs, pMD2.G (Addgene #12259) and psPAX2 (Addgene #12260) using lipofectamine 2000 (ThermoFisher). To generate polyclonal lines, MB135 cells were transduced with lentivirus. Stable cell lines were selected and maintained in 2 μg/mL puromycin.
QUANTIFICATION AND STATISTICAL ANALYSIS
Relative quantitation of gene expression
Quantitative PCR was carried out on a QuantStudio 7 Flex (Applied Biosystems). Relative expression levels of target genes were normalized to that of reference housekeeping gene RPL-27 using the relative standard curve method (Figures 4A-4C, S2B, S2C, and S4B) or by using the Comparative Ct Method (ΔΔCt; Figures 1C, S1E, S4D, and S4E). For Figure 1C, samples were normalized to the average of all control replicates, and any samples from control lines that had undetectable signal were set to 0 and not used for normalizing. Information about statistical details can be found in the figure legends. Throughout, graphs represent means with error bars representing standard deviation (SD) of biological triplicate measurements.
CUT&RUN domain calling
Our CUT&RUN data consist of 25 bps long, paired-end reads with average fragment length of 180 bps. We aligned reads to both human genome hg38 and spike-in genome dm6 using Bowtie2-2.2.6 with the following comment: bowtie2–local–very-sensitive-local–no-unal–no-mixed–no-discordant -q-phred33 -I 10 -X 700.
Since H3.X/Y were incorporated in many large regions within gene bodies, conventional peak calling algorithms such as MACS2 were not applicable. We thus developed an in-house R/Bioconductor package domainCalling (https://github.com/TapscottLab/domainCalling). The major functionalities of domainCalling include spike-in factor normalization and domain (broad peak) detection. To call the domains, the algorithm starts by counting reads overlapping with a sliding window throughout the genome for non-background (DOX+) and background (IgG or DOX−) samples. This window-based counting scheme is implemented by the csaw R/Bioconductor package (Lun and Smyth, 2016). After the counts are normalized by spike-in normalization, it filters out uninteresting windows if the average abundance of the non-background samples is (1) less than three-fold above background or (2) does not exceed the threshold, which is three reads per window. Finally, the retained windows are merged with neighbors within 2kbp distance. H3.X/Y domains were called for merged regions longer than 500 bps.
RNA-seq data analysis of pulsed samples
To preprocess the RNA-seq reads, we filtered out unqualified reads and aligned the reads to human genome hg38 using Tophat-2.1.0. We then profiled the gene expression using features collected from Ensembl v88 and the hit-counting function summarizeOverlaps() from Bioconductor’s GenomicAlignments package. To identify robust DUX4 target genes, we used DESeq2 comparing pulse1 siControl samples to negative controls. The alternative hypothesis is set to ∣β∣ > 4, where β denotes log2 fold change. 170 genes with adjusted p value < 0.05 were determined as robust DUX4 targets.
Statistical analysis of qPCR data
To test the significance of the perdurance effect of H3.X/Y (Figure 4D), we used functional data analysis (https://cran.r-project.org/web/packages/fda/index.html) treating the time course RT-qPCR expression data as a function or graph. The method started with registering the feature of the graph for each treatment, which was the day 1 data where the slope turned from positive to negative. Next it aligned the graphs of two groups of treatments at the value of the feature, and then, for each treatment, constructed a linear combination of functions interpolating the aligned time-course data starting from day 1 to day 4. Finally taken the null distribution built by 250 permutations, we applied the functional t test (fda::tperm.fd) to determine the difference between two groups of functions.
Software
The bioinformatics analysis was mostly performed on R-3.4.3/Bioconductor-3.5. The major infrastructure packages used include csaw, edgeR, GenomicAlignments, ChIPseeker and ggplot2.
DATA AND CODE AVAILABILITY
The MB135iDUX4 pulsed RNA-seq data and CUT&RUN data generated during this study are available at the Gene Expression Omnibus (GEO accession number GSE119403). Other datasets used in this study include DUX4 ChIP-seq (GEO accession number GSE33838), continuous DUX4 RNA-seq (GEO accession number GSE85461), and early embryo RNA-seq (GEO accession number GSE72379). The H3.Z sequence shown in Figure S1A has been submitted to GenBank and is awaiting an accession number. Code availability is detailed in the Key Resources Table with the following references: (Langmead et al., 2009; Lawrence et al., 2013; Love et al., 2014; Robinson et al., 2010; Shadle et al., 2017; Yu et al., 2015; Zerbino et al., 2018).
Supplementary Material
Highlights.
The cleavage-stage transcription factor DUX4 induces histone variants H3.X and H3.Y
H3.X/Y incorporate into genes transcriptionally induced by DUX4
H3.X/Y incorporation results in enhanced re-activation of DUX4-regulated genes
H3.X/Y are necessary to amplify the DUX4 network in FSHD muscle cells
ACKNOWLEDGMENTS
This work was supported by R01AR045203 (S.J.T.), P01NS069539 (S.J.T. and S.M.v.d.M.), Friends of FSH Research (S.J.T.), F31NS101773 (R.R.), T32HG000035 (R.R.), T32HD007183 (R.R.), and T32GM007266 (R.R.). Thank you to Christine Codomo for preparing CUT&RUN libraries and Alyssa Dawson, Elizabeth Jensen, and the rest of the Fred Hutch Genomics Shared Resource for sequencing.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.10.025.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ahmad K, and Henikoff S (2002a). Histone H3 variants specify modes of chromatin assembly. Proc. Natl. Acad. Sci. USA 99 (Suppl 4), 16477–16484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K, and Henikoff S (2002b). The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9, 1191–1200. [DOI] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, and Trono D (2017). DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet 49, 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drané P, Ouararhni K, Depaux A, Shuaib M, and Hamiche A (2010). The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24, 1253–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsässer SJ, Noh KM, Diaz N, Allis CD, and Banaszynski LA (2015). Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522, 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng LN, Tyler AE, and Tapscott SJ (2011). Immunodetection of Human Double Homeobox 4. HYBRIDOMA 30, 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, van der Maarel SM, Ruzzo WL, Gentleman RC, Tawil R, and Tapscott SJ (2012). DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell 22, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. (2010). Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet 49, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, and Choo KH (2000). Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA 97, 1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan S, Shadle SC, Resnick R, Snider L, Tawil RN, van der Maarel SM, Bradley RK, and Tapscott SJ (2016). Model systems of DUX4 expression recapitulate the transcriptional profile of FSHD cells. Hum. Mol. Genet 25, 4419–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TI, Chen JC, Rahimov F, Homma S, Arashiro P, Beermann ML, King OD, Miller JB, Kunkel LM, Emerson CP Jr., et al. (2012). Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum. Mol. Genet 21, 4419–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Banaszynski LA, Geng F, Zhang X, Zhang J, Zhang H, O’Neill CL, Yan P, Liu Z, Shido K, et al. (2018). Histone variant H3.3-mediated chromatin remodeling is essential for paternal genome activation in mouse preimplantation embryos. J. Biol. Chem 293, 3829–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowaljow V, Marcowycz A, Ansseau E, Conde CB, Sauvage S, Mattéotti C, Arias C, Corona ED, Nuñez NG, Leo O, et al. (2007). The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul. Disord 17, 611–623. [DOI] [PubMed] [Google Scholar]
- Krom YD, Dumonceaux J, Mamchaoui K, den Hamer B, Mariot V, Negroni E, Geng LN, Martin N, Tawil R, Tapscott SJ, et al. (2012). Generation of isogenic D4Z4 contracted and noncontracted immortal muscle cell clones from a mosaic patient: a cellular model for FSHD. Am. J. Pathol 181, 1387–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujirai T, Horikoshi N, Sato K, Maehara K, Machida S, Osakabe A, Kimura H, Ohkawa Y, and Kurumizaka H (2016). Structure and function of human histone H3.Y nucleosome. Nucleic Acids Res. 44, 6127–6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujirai T, Horikoshi N, Xie Y, Taguchi H, and Kurumizaka H (2017). Identification of the amino acid residues responsible for stable nucleosome formation by histone H3.Y. Nucleus 8, 239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, and Carey VJ (2013). Software for computing and annotating genomic ranges. PLoS Comput. Biol 9, e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, and Allis CD (2010). Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA 107, 14075–14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Hargreaves VV, Zhu Q, Kurland JV, Hong J, Kim W, Sher F, Macias-Trevino C, Rogers JM, Kurita R, et al. (2018). Direct promoter repression by BCL11A controls the fetal to adult hemoglobin switch. Cell 173, 430–42.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun AT, and Smyth GK (2016). csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 44, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard AM, Petek LM, and Miller DG (2015). Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum. Mol. Genet 24, 5901–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadle SC, Zhong JW, Campbell AE, Conerly ML, Jagannathan S, Wong CJ, Morello TD, van der Maarel SM, and Tapscott SJ (2017). DUX4-induced dsRNA and MYC mRNA stabilization activate apoptotic pathways in human cell models of facioscapulohumeral dystrophy. PLoS Genet. 13, e1006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, and Henikoff S (2017). An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff JG, and Henikoff S (2018). Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc 13, 1006–1019. [DOI] [PubMed] [Google Scholar]
- Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, and Miller DG (2010). Facioscapulohumeral dystrophy: incomplete suppression of a retro-transposed gene. PLoS Genet. 6, e1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Osakabe A, Kawaguchi K, Shiga T, Hayashi-Takanaka Y, Kimura H, and Kurumizaka H (2010). Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T. Proc. Natl. Acad. Sci. USA 107, 10454–10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, and Nakatani Y (2004). Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61. [DOI] [PubMed] [Google Scholar]
- Tawil R, van der Maarel SM, and Tapscott SJ (2014). Facioscapulohumeral dystrophy: the path to consensus on pathophysiology. Skelet. Muscle 4, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg S (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace LM, Garwick SE, Mei W, Belayew A, Coppee F, Ladner KJ, Guttridge D, Yang J, and Harper SQ (2011). DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann. Neurol 69, 540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiddon JL, Langford AT, Wong CJ, Zhong JW, and Tapscott SJ (2017). Conservation and innovation in the DUX4-family gene network. Nat. Genet 49, 935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis (New York: Springer-Verlag; ). [Google Scholar]
- Wiedemann SM, Mildner SN, Bönisch C, Israel L, Maiser A, Matheisl S, Straub T, Merkl R, Leonhardt H, Kremmer E, et al. (2010). Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J. Cell Biol 190, 777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Snider L, Balog J, Lemmers RJ, Van Der Maarel SM, Tawil R, and Tapscott SJ (2014). DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum. Mol. Genet 23, 5342–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, and He QY (2015). ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et al. (2018). Ensembl 2018. Nucleic Acids Res. 46 (D1), D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink LM, Delbarre E, Eberl HC, Keilhauer EC, Bönisch C, Pünzeler S, Bartkuhn M, Collas P, Mann M, and Hake SB (2017). H3.Y discriminates between HIRA and DAXX chaperone complexes and reveals unexpected insights into human DAXX-H3.3-H4 binding and deposition requirements. Nucleic Acids Res. 45, 5691–5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MB135iDUX4 pulsed RNA-seq data and CUT&RUN data generated during this study are available at the Gene Expression Omnibus (GEO accession number GSE119403). Other datasets used in this study include DUX4 ChIP-seq (GEO accession number GSE33838), continuous DUX4 RNA-seq (GEO accession number GSE85461), and early embryo RNA-seq (GEO accession number GSE72379). The H3.Z sequence shown in Figure S1A has been submitted to GenBank and is awaiting an accession number. Code availability is detailed in the Key Resources Table with the following references: (Langmead et al., 2009; Lawrence et al., 2013; Love et al., 2014; Robinson et al., 2010; Shadle et al., 2017; Yu et al., 2015; Zerbino et al., 2018).