Abstract
Some of the most widely used drugs, like aspirin and penicillin, are covalent drugs. Covalent binding can improve potency, selectivity and duration of the effects, but the intrinsic reactivity represents a potential liability and may result in idiosyncratic toxicity. For decades, the cons were believed to outweigh the pros, and covalent targeting was deprioritized in drug discovery. Recently, several covalent inhibitors have been approved for cancer treatment, thus rebooting the field. In this review, we briefly reflect on the history of selective covalent targeting, and provide a comprehensive overview of emerging developments from a chemical biology stand-point. Our discussion will reflect on efforts to validate irreversible covalent ligands, expand the scope of targets, and discover new ligands and warheads. We conclude with a brief commentary of remaining limitations and emerging opportunities in selective covalent targeting.
eTOC blurb
In this review, Zhang et al. provide a chemical biology perspective on the field of selective covalent targeting. The authors highlight approaches to robust validation and standards for irreversible covalent ligands, and comment on recent studies that expand the scope of targets, ligands and warheads.
Deep down, under all those Western blots and microscopy images, many chemical biologists are lovers and practitioners of chemistry, a scientific discipline that is centrally interested in reactivity. Thus, many in the field have been exploiting chemical reactivity between a small molecule and a biomolecule to create tools for biological research and agents for disease treatment. This second area of interest has, in part, been inspired by examples of approved drugs that although not developed as covalent have since been shown to exert their therapeutic effects by covalently binding their targets. Most notable examples of these are aspirin and penicillin, which target cyclooxygenases and bacterial DD-transpeptidase, respectively (Singh et al., 2011). More recently, a range of rationally designed covalent inhibitors has received FDA approval, causing a resurgence of interest in this field (Byrd et al., 2016) (Kisselev et al., 2012) (Kwong et al., 2011) (Rotella, 2013) (Li et al., 2008) (Yver, 2016).
Interestingly, the idea that selective covalent inhibitors could be valuable is not a new one. As a review from the 1960’s illustrates, reactions between nucleophilic side chains of proteinogenic amino acids and electrophilic warheads of small molecule inhibitors have already been considered decades ago (Baker, 1964). The advantages of irreversible inhibition that this review noted remain relevant today and include: (a) improved effectiveness of irreversible vs. reversible compounds; and (b) the potential for higher specificity over reversible compounds given that irreversible ligands form a covalent bond with a relatively unique nucleophile on the target. On the other hand, the noted challenges we still consider relevant were: (a) achieving target selectivity given the use of reactive warheads; (b) ensuring that reactivity of the irreversible inhibitors does not interfere with tissue distribution and/or intracellular delivery; and (c) community skepticism surrounding the idea of selective covalent targeting.
The recent drug approvals may have minimized some of the community skepticism; however, further efforts are needed to address issues surrounding limited number of available warheads with suitable reactivity and selectivity, as well as stability and compatibility with in vivo use. Here, we will discuss the importance of validating selective irreversible ligands, and comment on the standards that need to be satisfied before using these compounds as chemical probes. We will then comment on emerging opportunities in selective irreversible covalent targeting and conclude by reflecting on some of the limitations and current challenges. An important aspect of this topic that will not be covered here is the target selection process and how to optimize it in order to achieve maximum potency and selectivity by taking into account not only the nature of the available reactive sites but target’s half-life as well. We feel that this issue deserves to be covered separately and hope to see it written about in the near future.
We would also like to note that many excellent reviews on different aspects of this topic have recently been published (Jackson et al., 2017) (Bandyopadhyay and Gao, 2016) (De Cesco et al., 2017) (Lagoutte et al., 2017) (Mukherjee and Grimster, 2018) (Shannon and Weerapana, 2015) (Pettinger et al., 2017) (Lonsdale and Ward, 2018) (Chaikuad et al., 2018) (Hallenbeck et al., 2017) (Zhao and Bourne, 2018) (Cuesta and Taunton, 2019). Our main goal here is to provide a chemical biology perspective on this topic, as a complementary viewpoint to primarily drug development and medicinal chemistry discussions present in the current literature.
Validating irreversible covalent tool compounds
Over the last decade, chemical biology community has developed a set of guidelines for chemical probes, also known as tool compounds (Arrowsmith et al., 2015) (here, we will use term tool compounds to avoid confusion with agents referred to as “covalent probes “ that are used for activity-based protein profiling (ABPP)). Although these guidelines have been defined for noncovalent ligands, they do provide a basic framework that can be applied to covalent tool compounds. In this section, we will comment on how to expand the existing guidelines, and discuss strategies to characterize and validate selective covalent ligands.
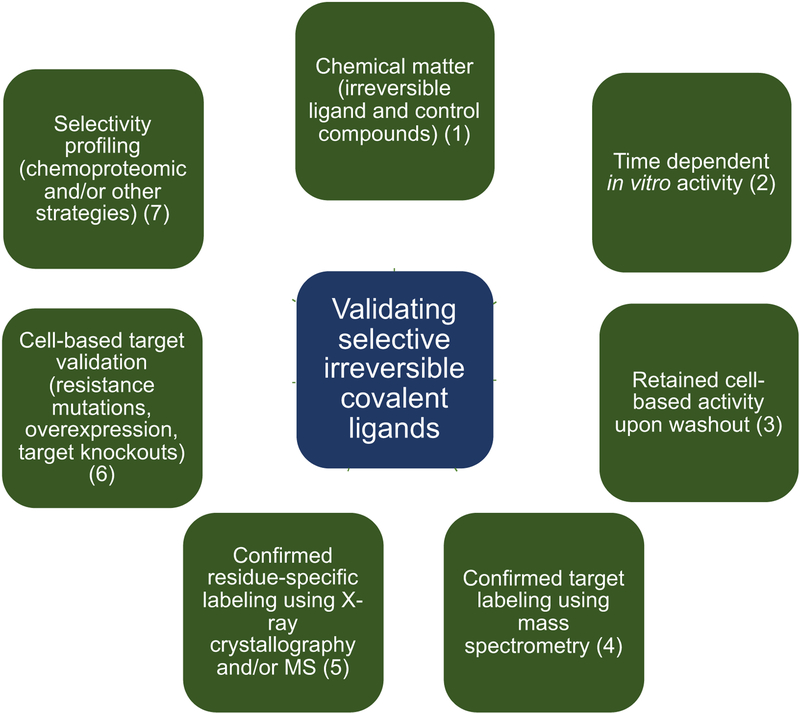
Key questions that validation process for a chemical probe has to address are: (1) how potent is the compound in biochemical and cellular assays?; (2) what is the mode of binding? (3) are suitable negative control compounds available and is the compound chemically stable under given conditions?; (4) is the probe engaging the correct target in cells and are observed biological effects due to the on-target engagement?; (5) how selective is the compound?; and (6) (for probes to be used in living organisms) what are the PK/PD properties? (Müller et al., 2018). In addition, three central questions for validating irreversible covalent ligands are: (a) is the ligand engaging covalently and irreversibly with the target?; (b) are any biological effects due to non-covalent interactions with the target and/or off-targets; and (c) are any biological effects of an irreversible covalent ligand due to off-target reactivity? The validation process we developed to help answer these questions is shown in Figure 1, and we will use an example from our laboratory to illustrate the type of experiments that we commonly employ.
Figure 1. Selective covalent ligand validation workflow.
Structured process for irreversible covalent inhibitors validation is based on using in vitro and cell-based strategies to assess compound potency, covalent mode of binding, target selectivity, and link observed phenotype to direct covalent engagement between the compound and the target. Although no single experiment is sufficient to validate a molecule as a selective covalent tool compound, collective evidence accumulated through this validation process can be used to judge a quality of a tool compound.
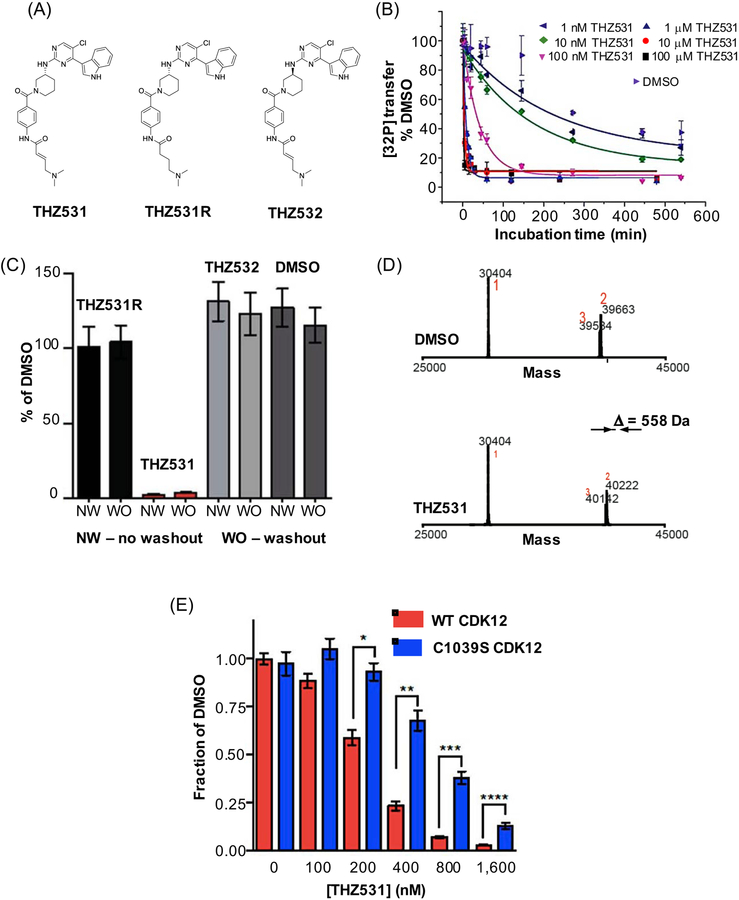
The majority of efforts to develop selective irreversible covalent ligands have been designed to take advantage of the intrinsic nucleophilic nature of proteinogenic amino acid side chains, most notably the thiol group (–SH) of cysteines (Cys). Along these lines, our illustrative example THZ531 was developed as an irreversible covalent inhibitor for cyclin-dependent kinases 12 and 13 (CDK12/13) (Figure 2A) (Zhang et al., 2016). In addition to THZ531, we also synthesized THZ531R, a compound where α,β-unsaturated carbonyl was reduced thus eliminating the Cys-reactive Michael acceptor, and THZ532, an inactive enantiomer (Figure 2A). We used both THZ531R and THZ532 as negative controls throughout our validation process, thus fulfilling recommended step 1 in our covalent inhibitor validation process (Figure 1).
Figure 2. Validating THZ531, a CDK12/13 irreversible covalent inhibitor.
(A) chemical structures of THZ531, a CDK12/13 inhibitor, and two negative control compounds, an enantiomer THZ532, and a reversible, noncovalent binder THZ531R; (B) preincubation time-dependent in vitro activity is a signature behavior of irreversible covalent inhibitors. In this experiment in vitro kinase activity assay of CDK12-cyclin K was measured using different concentrations of THZ531 (1 nM to 100 μM) and varying preincubation times and expressed as relative [32P] transfer; (C) irreversible covalent inhibitors retain activity in cell-based assays upon washout as shown for THZ531 and two negative control compounds, THZ531R and THZ532. Jurkat cells were treated with the indicated compounds for 6 hrs, inhibitor was washed out and cells were allowed to grow for the remainder of the 72 hr (washout, WO). This growth was compared to the growth of cells treated with inhibitors for the full 72 hrs (no washout, NW); (D) intact mass spectrometry (MS) offers evidence for covalent adduct formation based on mass difference between DMSO treated and THZ531 treated CDK12. 558 Da difference observed here corresponds to Cys-directed THZ531 adduct; and (E) 72-hour antiproliferation assay using WT and Cys1039Ser cells and different concentrations of THZ531. Under all concentrations tested, mutant cell lines display resistance to THZ531, highlighting that Cys1039 is important for mediating THZ531 effects. Panels (B), (C), (D) and (F) have been reproduced and/or modified from Zhang et al., 2016 with premission, Copyright 2016, Springer Nature.
In general, covalent inhibitors display concentration-dependent and incubation time-dependent activity in in vitro enzymatic assays (Strelow, 2017). Therefore, one of the steps in our validation process is measuring loss of activity as a function of preincubation times. In this experiment, we varied the length of incubation time with THZ531, and quantified kinase activity using a radiometric assay that measures the ability of recombinant CDK12 to phosphorylate a Pol II CTD-peptide substrate in the presence of its cofactor, cyclin K, normalized to the relative [32P] transfer under DMSO control (Figure 1, recommended step 2; Figure 2B). Although incubation time-dependence of activity can be due to factors other than covalent target binding, we use these results as indicators of covalent inhibition. Additionally, it is recommended that potency of irreversible inhibitors is expressed as kinact/KI, where kinact is the maximal rate of inactivation and KI is the reversible binding constant (Strelow, 2017). Contributions of affinity and reactivity to the overall potency need to be considered separately, and an in vitro method for characterizing these two components has been described (Schwartz et al., 2014). However, it is worth pointing out that in vitro characterization may not translate to in vivo conditions.
Washout experiments, where cells are first exposed to the inhibitor, then washed out and allowed to grow in inhibitor-free media, are also an important step in the validation process. Here, the growth rates with washout are compared to “no washout” conditions. The sustained effect of covalent inhibitors subjected to the washout experiments is attributed to the irreversible nature of their target engagement. For THZ531 validation we used Jurkat T-cell acute lymphoblastic leukemia cells and demonstrated that THZ531 maintained the effects 72 hours post-washout, whereas negative controls, including the reversible compound THZ531R, had no effect (Figure 1, recommended step 3; Figure 2C). Moreover, we also used a biotinylated analog of THZ531 (bioTHZ531) to treat Jurkat cell lysates and identify cellular targets that affinity purify with bioTHZ531 after extensive washouts to remove nonspecific and noncovalent targets. These pull down experiments provided evidence that CDK12-cyclin K and CDK13-cyclin K are the main covalent targets. A potential caveat when using tagged analogs, such as bioTHZ531, is that the tag itself may introduce non-specific interactions, and these effects need to be taken into account. Overall, experiments like measuring incubation time dependence of activity, loss of activity upon removal of the reactive warhead, and cellular washout experiments provide multiple lines of supporting evidence of irreversible mechanism of action.
More direct methods for confirming and visualizing covalent binding in vitro are mass spectrometry (MS) and X-ray crystallography. We employed both of these strategies to validate that THZ531 covalently binds CDK12 and CDK13 in vitro. MS experiments showed the formation of the covalent adduct (+558 Da corresponding to the addition of THZ531; Figure 1, recommended step 4; Figure 2D), and, upon proteolysis, identified a peptide fragment containing the exact site (Cys1039 on CDK12) of modification. A 2.7 Å crystal structure of CDK12-cyclin K bound to THZ531 confirmed these findings (Figure 1, recommended step 5).
To further establish activity and selectivity in vivo, an essential step in validating covalent ligands are cell-based experiments that use resistance mutations. For example, cysteines are commonly mutated to a serine or an alanine, and the presumed target protein harboring the point mutation is introduced to cells either exogenously or using CRISPR/Cas9 knock-in technology. Our cellular work using Cys1039Ser CDK12 mutant created using CRISPR/Cas9 demonstrated that this mutation was sufficient to make CDK12 refractory to covalent affinity pull down and led to partial restoration of cellular proliferation and reduced apoptosis (Figure 1, recommended step 6; Figure 2F). Overall, cell-based experiments using resistance mutations provide essential evidence that a given phenotype induced by a covalent inhibitor is on-target and on-mechanism, something that neither biochemical experiments described above nor chemoproteomics experiments be describe below can address.
There are several methods that can be used to map selectivity of covalent ligands. For example, THZ531 was profiled using kinase panel assays Ambit™ for in vitro characterization and KiNativ™ for cell-based profiling (Du et al., 2009). Both sets of data indicated that CDK12 and 13 are the primary, but not the only, targets of THZ531. The major limitation of profiling strategy like this is that it includes only kinases with a varied number of targets in the profiling panel. A range of chemoproteomic approaches has been developed to allow interrogations of a broader target space. For example, ABPP in combination with MS-based proteomics, like SILAC (for Stable Isotope Labeling with Amino Acids in Cell Culture) or tandem mass tagging (TMT) click-chemistry pull down experiments, can be used to map selectivity (Drewes and Knapp, 2018). We recently contributed to the development of a chemoproteomic method called CITe-Id (for Covalent Inhibitor Target-site Identification) (Browne et al., 2019), which is able to capture, identify and quantify dose-dependent covalently bound Cys sites in cell lysates. Using CITe-Id we were able to demonstrate that a covalent kinase inhibitor THZ1, originally developed to target CDK7, binds covalently to additional proteins, including non-kinase targets. Although useful, chemoproteomic strategies have number of limitations, including incomplete coverage of the proteome, and can lead to both false positives, by identifying proteins as targets when they are not, and false negatives, by not detecting the binding event. Therefore, as mentioned above, any comprehensive validation process must include cell-based target confirmation experiments, such as the use of knockdowns, target overexpression, and evolution of resistant mutants.
The range of proteins for which covalent ligands targeting Cys have been described now extends into the area of “undruggable” targets such as KRASG12C, a common oncogenic mutant of the small GTPase KRAS (Patricelli et al., 2016) (Janes et al., 2018). These two reports describe development and validation of covalent agents ARS-853 and ARS-1620, respectively, both inspired by prior work that demonstrated that covalent inhibitors targeting Cys12 in KRASG12C were feasible (Ostrem et al., 2013). Whereas ARS-853 was shown to be a cell based KRASG12C inhibitor, ARS-1620 is active in vivo. Both compounds exhibit a strict requirement for Cys12 for their activity, as wild-type (WT) and mutant cell lines where residue 12 is not Cys (such as KRASG12S, KRASG12V or KRASG12D) were insensitive. Additionally, both compounds inhibit GDP-bound form of KRASG12C and display a narrow selectivity window, with ARS-853 binding RTN4 and FAM213A in addition to KRASG12C, and ARS-1620 hitting FAM213A and additionally AHR. Selectivity profiles were measured by cysteine reactivity profiling, a chemoproteomic competition-based method, which profiled 2,740 surface exposed Cys belonging to 1,584 annotated proteins for ARS-853, and 8,501 Cys residues belonging to 3,012 annotated proteins for ARS-1620. Lastly, ARS-1620 analysis included the use of an inactive control, an atropisomer. Together, these studies demonstrate the value of covalent targeting as a way to discriminate between WT and disease-associated mutant proteins.
Targeting serines (Ser) and threonines (Thr) has also been successfully exploited for tool compound and drug development. Although hydroxy group (–OH) of Ser and Thr is relatively inert under physiological conditions, activated Ser and Thr are found in active sites of many enzymes. In fact, ABPP was originally developed for profiling serine hydrolases, a large family of diverse enzymes that use catalytic (activated) Ser to hydrolyze an amide, an ester or a thioester bond (Bachovchin and Cravatt, 2012). Additionally, a number of approved covalent drugs target Ser and Thr residues, including: aspirin (cyclooxygenase inhibitor), penicillin (bacterial DD-transpeptidase inhibitor), telaprevir and boceprevir (HCV protease inhibitors), avibactam (β-lactamase inhibitor), carfilzomib, bortezomib and ixazomib (proteasome inhibitors), rivastigmine (acetylcholinesterase inhibitor), and saxagliptin (dipeptidyl peptidase-4 (DPP-4) inhibitor). Selective covalent targeting of activated Ser and Thr will not be further discussed here as this topic was covered in more detail in recent reviews (Mukherjee and Grimster, 2018) (Shannon and Weerapana, 2015).
In the following sections we will discuss emerging strategies for targeting lysines, tyrosines, histidines and methionines, and comment on opportunities and limitations. This topic, targeting sites beyond cysteines, has been a subject of several recent reviews (Mukherjee and Grimster, 2018) (Pettinger et al., 2017) (Jones, 2018). We refer those interested in covalent lysine targeting that includes discussion of aldehyde-containing warheads and reversible covalent inhibition to a very recent review by Cuesta and Taunton, as those topics will not be discussed here (Cuesta and Taunton, 2019).
Selective covalent targeting of lysines
Lysine (Lys) side chains are sites of numerous post-translational modifications (PTMs), such as methylation, ubiquitination, SUMOylation and a range of acylations, with acetylation being the most well-established (Jones, 2018). However, targeting Lys ε-amino group with irreversible electrophilic ligands has been challenging due to its high pKa (~10) resulting in full protonation under physiological pH (7.4). In addition, most lysine-targeting agents display low compatibility with in vivo applications, therefore limiting their use as pharmacological tools. Recent chemoproteomic profiling of lysines reactivity suggests that human proteome may contain a number of lysine residues that could be targeted using covalent strategy (Hacker et al., 2017). Importantly, many of the lysine sites documented in that study were found in proteins for which no small molecule ligands are currently available, therefore suggesting a potential opportunity. Below we will describe several more recently described Lys-directed covalent ligands, focusing on literature examples that include validation processes that are similar to the one we propose in Figure 1.
In an example from the kinase field, Anscombe et al. used vinyl sulfone as a warhead and coupled it to a reversible CDK2 inhibitor with a purine scaffold, to achieve covalent inhibition of CDK2, a cyclin dependent kinase that belongs to a large family of closely related kinases (Anscombe et al., 2015). The covalent inhibitor, NU6300 (Figure 3A), was found to bind Lys89, one of the two lysine residues located in a solvent exposed region at the vicinity to the ATP binding pocket, as confirmed by mutagenesis combined with mass spectrometry (MS)-based analysis, and high-resolution crystal structure. In addition to preparing NU6300, the authors also synthesized NU6310, where the vinyl group of NU6300 was replaced by an ethyl group thus resulting in non-covalent ATP-competitive inhibitor NU6310. To distinguish between covalent and non-covalent inhibition, the authors incubated CDK2/cyclin A (cyclin A is a cofactor required for CDK2 activity) overnight with either NU6300 or NU6310, then dialyzed to remove unbound inhibitor, and checked for activity using a peptide substrate derived from a known cellular substrate of CDK2, retinoblastoma protein RB. Whereas NU6310 was fully washed out by this treatment, NU6300 remained bound and therefore inhibited CDK2 activity post dialysis. Furthermore, in a complementary in vitro experiment NU6300 activity varied with the preincubation time, as expected from a covalent inhibitor. In cells, treatment with NU6300 inhibited phosphorylation of RB protein and maintained the inhibition after a washout experiment whereas another non-covalent inhibitor NU6302 showed diminished activity upon washout, strongly supporting the irreversible covalent mode of inhibition. In terms of selectivity, NU6300 was accessed across a limited kinase panel (131 kinases out of about 518 human kinases (Manning et al., 2002)) under conditions that don’t distinguish noncovalent from covalent inhibition, and subsequently tested under preincubation conditions. The limited selectivity profiling suggested that Aurora A, STK3/MST2, and MAP4K3 may represent potential off-targets. However, since no broader selectivity profiling was conducted, additional off-targets may exist. Lastly, cell-based validation did not include experiments to demonstrate target engagement and on-target mechanism, which are caveats that follow up work will need to address.
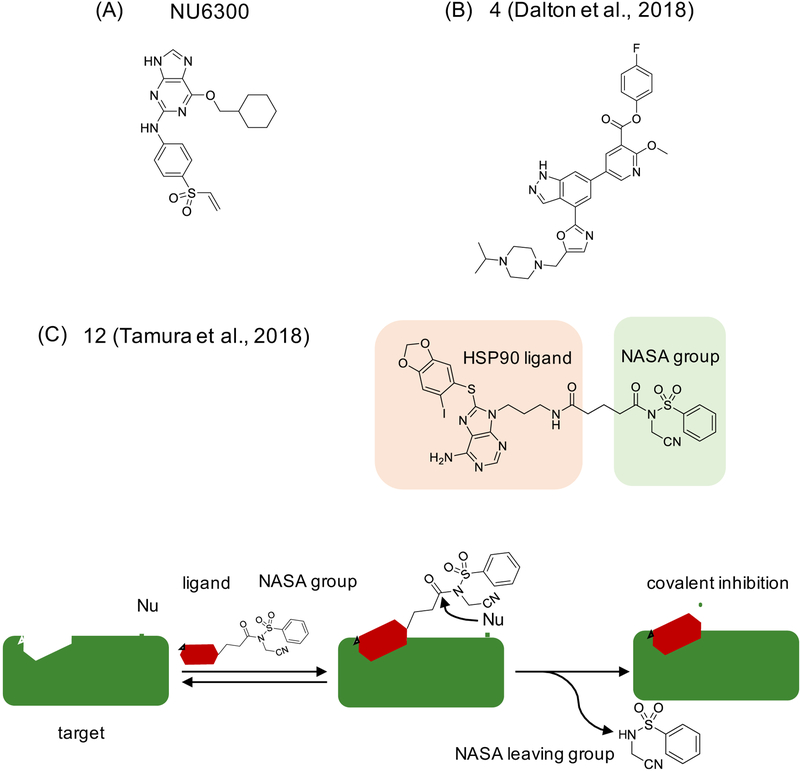
Figure 3. Examples of covalent ligands developed to target lysines.
(A) NU6300 targets CDK2 at Lys89 (Anscombe et al., 2015), (B) compound 4 targets highly conserved Lys779 PI3Kd (Dalton et al., 2018), and (C) compound 12 targets Lys58 on Hsp90 using ligand-directed (LD) protein labeling and N-acyl-N-alkyl sulfonamide (NASA) warhead. The LD-NASA process, as used for selective covalent inhibition, is depicted below. The initial reversible binding step is mediated by ligand-target recognition, while the second, irreversible step is mediated by the nucleophilic attack by a side chain residue resulting in release of NASA warhead and the ligand covalent attachment (Tamura et al., 2018).
A more recent example in this area describes development of a selective covalent inhibitor of lipid kinase phosphoinositide 3-kinase delta (PI3Kδ) that targets the highly conserved lysine (Lys779 in PI3Kδ) (Dalton et al., 2018). Dalton et al. used a potent reversible PI3K inhibitor as a starting point and employed structure-guided design to introduce an activated phenolic ester as the electrophile that targets Lys779. Through rounds of optimization, 4-fluorobenzoic ester emerged as the best performing warhead and yielded compound 4 (Figure 3B) that readily labeled Lys779 of PI3Kδ, based on kinetic, MS and structural analysis. Interestingly, although compound 4 was able to covalently bind PI3Kα and PI3Kβ in ATP-free condition, only PI3Kδ remained covalently inhibited in the presence of 1 mM ATP. Selectivity profiling using a kinase panel (total 10 lipid kinases and 140 protein kinases) and chemoproteomics experiments using TMT labeling in human cells showed limited off-target binding. In chemoproteomics experiments the authors used competition experiment mode, where cells were first exposed to the inhibitor which was then competed out with an azide-carrying probe that enabled clicking to biotin and affinity enrichment, followed by SDS-elution to identify only covalently bound targets, in this case PI3Kα, PI3Kβ, and PI3K protein Vps34. Cell-based washout experiments with compound 4 and noncovalent control compound were also included in validation. Although characterization did not include data to formally link compound 4/PI3Kδ binding and the observed phenotype, the results are of interest as they suggest that selective Lys-directed targeting of a highly conserved residue is feasible.
Using a version of a strategy called ligand-directed (LD) protein labeling, Tamura et al. have recently reported N-acyl-N-alkyl sulfonamide (NASA) as a rapid labeling warhead that can be incorporated into a covalent inhibitor design (Tamura et al., 2018). In brief, LD is a protein labeling strategy that uses a target recognition handle to bind to the protein of interest, and improve labeling on otherwise less reactive sites through proximity effect (Tamura and Hamachi, 2019). In the original setup, the reaction that takes place results in labeling of a specific amino acid side chain and dissociation of the target recognition handle. Tamura et al. describe a different application of LD whereby a reversible HSP90 inhibitor PU-H71 (Caldas-Lopes et al., 2009) was combined with NASA, resulting in compound 12 (Figure 3C) that readily labeled Lys58 on HSP90 based on MS/MS data. The TMT-labeling quantitative MS analysis identified Hsp90α, Hsp90β and Grp94 (also knowns as HSP90B1) as targets, all of which contain the conserved lysine residue, as well as three unrelated proteins (tubulin-α, solute carrier family member SLC25A6, and a mitochondrial protein HADHA). The cell-based washout experiments, done using PU-H71 as a control, provided additional evidence of covalent target engagement and prolonged effects of compound 12, including lasting effects on destabilization of Hsp90 client proteins (21h post-washout). These results suggest that LD-NASA strategy could lead to further development of Lys-directed selective covalent ligands. However, this proof-of-principle study did not address whether NASA leaving group (see Figure 3C) has additional downstream effects, which would be worthwhile examining in follow up work.
Sulfonyl fluorides (SFs) have been described as privileged warheads for targeting a range of amino acid side chains, including lysines, as we will illustrate here, and tyrosines, which will be discussed in the next section (Narayanan and Jones, 2015). The label “privileged” may be in part due to SF chemical and thermodynamic stability under aqueous conditions. Another beneficial aspect of SFs is the non-toxic and unreactive nature of the fluoride leaving group. Although SF containing compounds have been in routine use as biochemical reagents (pan-serine protease inhibitors PMSF and AEBSF, for example), attempts to use SF as a warhead for selective covalent ligand development have emerged more recently. The most notable example of Lys-directed SF ligands developed with selectivity in mind is a series reported to target Lys15 on transthyretin (TTR) (Grimster et al., 2013). TTR is a thyroid hormone (T4) transporter that forms a stable homotetramer with two T4 binding sites located at the dimer-dimer interface. Dissociation of the tetramer leads to misfolding and aggregation, and developing covalent compounds that act as kinetic stabilizers of the tetramer by targeting Lys15 has been proposed as a pharmacological strategy (Choi et al., 2010). Grimster et al. employed aromatic SF functional group as an electrophile proposed to be activated for Lys15 attack by a neighboring water molecule or a protein hydrogen bond donor. The authors used structure-based design to develop a 1,3,4-oxadiazole series with aromatic SF functional group, and presented in vitro and structural evidence of sulfonamide formation, as well as in vitro evidence that this binding prevents aggregation and amyloid formation by stabilizing the tetramer. The authors also show that reaction of SF compounds is about three orders of magnitude faster than their previous Lys15-targeting series (Choi et al., 2010). However, their results using Lys15Ala TTR mutant did show that compounds in their series could label additional sites, and the authors also observed that some of the members of this compound series were sensitive to hydrolysis. Moreover, although the authors did observe labeling of TTR in plasma at the levels that were shown to prevent aggregation in vitro, the study at this point did not go further to demonstrate broader reactivity and characterize biological effects. Still, for the reasons stated above, SFs remain attractive as warheads and we will describe their use for tyrosine targeting in the next section.
Covalent targeting of tyrosines
In 2013, SF probes were introduced as tools for chemoproteomic profiling, and their use led to an insight that glutathione transferases (GSTs) contain a reactive tyrosine residue (Gu et al., 2013). Since then there have been additional reports supporting the notion that GSTs have targetable tyrosines, such as Tyr108 located in the G-site of GSTP-1 that was addressed using an SF warhead (Shishido et al., 2017), as well as dichlorotriazine warhead (Crawford and Weerapana, 2016). These results are notable given that very few small molecules have been designed to target GSTP-1’s G-site mainly due to a barrier of competing with high concentration GSH in cells, a challenge that covalent ligands may be able to overcome through irreversible binding. However, given a somewhat limited extent of validation provided in these two studies, we would consider the two lead molecules (compound 4 from Shishido et al., 2017 and Las17 developed by Crawford and Weerapana, 2016) as initial proof-of-concept molecules.
In an earlier study, SF warhead was systematically introduced to ortho-, meta- and para-positions of the benzylic ring of diaminoquinazolines (DAQs) (Hett et al., 2015), compounds that reversibly inhibited mRNA decapping scavenger protein DcpS, a potential target for spinal muscular atrophy (SMA). Introducing an SF warhead led to a hundred-fold enhanced activity in in vitro assays relative to the parent compound. Interestingly, meta- and ortho- isomers were found to label Tyr113, whereas para- reacted with Tyr143 based on MS and X-ray crystallography data (Figure 4A). The authors proposed that proximity of residues that facilitates deprotonation of Tyr –OH is needed to direct reactivity of the SF warhead towards these tyrosines over lysines. Although Hett et al. showed DcpS engagement in peripheral blood mononucleated cells (PBMCs), the extent of characterization included in this work was limited and the authors noted the need for future work to establish selectivity and improve ability to predict tyrosine reactivity.
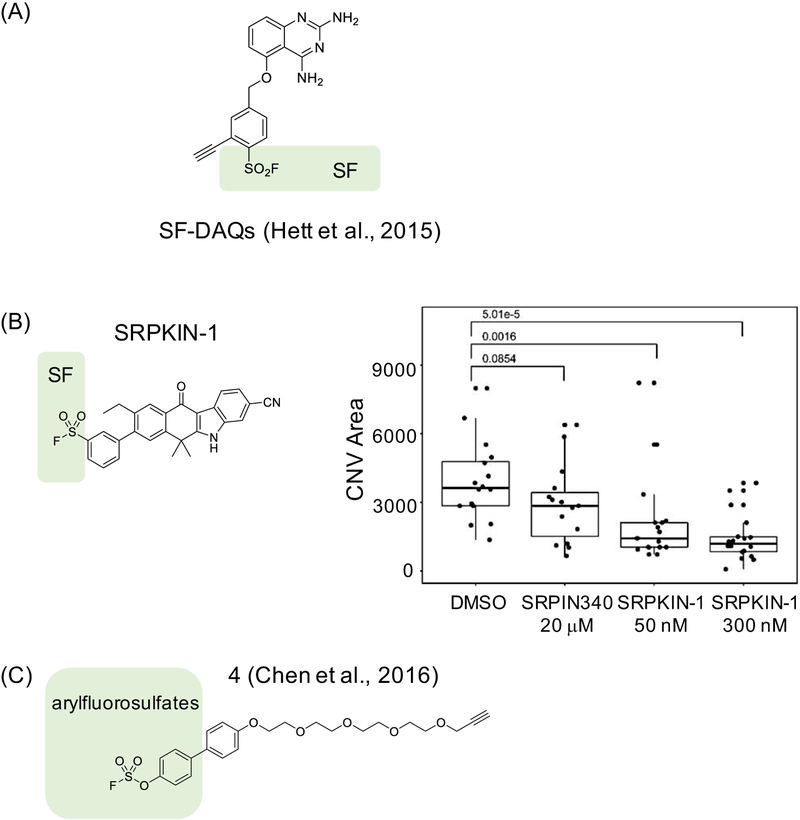
Figure 4. Examples of covalent ligands developed to target tyrosines.
(A) A series of sulfonyl fluoride (SF)-containing diaminoquinazolines (DAQs) compounds that irreversibly inhibit mRNA decapping scavenger protein DcpS by targeting Tyr113 (meta- and ortho-isomers) or Tyr143 (para-isomers) (Hett et al., 2015), (B) SRPKIN-1 is a Tyr-directed inhibitor with an SF warhead that forms a covalent bond with Tyr227 on SRPK1/2. Biological effects induced by SRPKIN-1 and a reversible control compound SRPIN-340 in a mouse model of wet age-related macular degeneration (wet AMD) display clear differences, supporting that covalent binding is critical for bioactivity. The experiment shown was conducted using laser-induced choroidal neovascularization (CNV) at concentrations of compounds as indicated, and the extent of the effect was quantified as CNV area – here inhibition of SRPK with SRPKIN-1 leads to changes in VEGF splicing resulting in production of anti-angiogenic VEGF-A165b isoform and decrease in new blood vessel formation as measured by the size of CNV area; and (C) arylfluorosulfate-containing probe 4 targeting Tyr134 on CRABP2 (Chen et al., 2016).
Recently, we have rationally designed a tyrosine-reactive covalent inhibitor for SRPK1/2, kinases that regulate splicing by phosphorylating the family of serine/arginine (SR)-rich splicing factors (Hatcher et al., 2018). We noted that SRPK1 is one of the off-targets of alectinib, an ALK inhibitor, and used a structure of the SRPK-alectinib complex as a basis for covalent inhibitor design. We identified Tyr227 as a potential targetable site due to its proximity to the 4-morpholinopiperidine group on alectinib, and we installed an SF group to generate SRPKIN-1 (Figure 4B). We confirmed that SRPKIN-1 covalently binds to Tyr227 using capillary electrophoresis-MS (CE-MS) to detect adduct formation, and tryptic digestion followed by CEMS to identify the site of modification. We also documented a sustained inhibition upon washout, as expected for a compound with an irreversible mode of action. Selectivity of SPRKIN-1 was examined using KiNativ™ before and after washout in HeLa cells, with SRPK1/2 as the only two targets inhibited above 90% upon washout. One of the known alternatively spliced genes regulated by SRPK is gene coding for VEGF, where high expression of SRPK diminishes level of 165b, an antiangiogenises protein, and knock-down of SRPK restores 165b levels. The biological effects induced by SRPKIN-1 are consistent with selective covalent inhibition of SRPK1; we observed restoration of VEGF 165b splicing isoforms with a nanomolar dose in cell-based assays, as well as suppression of angiogenesis in a mouse model. We also documented difference between effects of SRPKIN-1 and a reversible negative control compound, including in a mouse model of wet age-related macular degeneration (wet AMD). The irreversible SRPK1 inhibitor was shown to block angiogenesis in the AMD model, while negative control compound had no effect (Figure 4B). Potential limitations of this work are the absence of information about any non-kinase off-targets, as well as a possibility that phenotypic effects are due to binding to a residue other than Tyr227 as no mutant data was included.
Recently, arylfluorosulfates were proposed as alternative warheads for covalent modifications of some nucleophiles, including tyrosines, acting via an acid-base catalyzed sulfur (VI) fluoride exchange (SuFEx) reaction (Chen et al., 2016). This work led to development of probe 4, which covalently modified cellular retinoic acid binding protein 2 (CRABP2) on Tyr134 (Figure 4C). Probe 4 was comprehensively validated, both in vitro and in cells. Biochemical, mutagenesis and structural analysis suggest that, as with SF warhead, target Tyr –OH needs to be activated by the local environment for reaction to occur. In the case of CRABP2, apparent pKa of Tyr134 –OH to ~7.6 due to the proximity of two arginines. Overall, this study suggested that arylfluorosulfate warheads could be attractive for further development, although care needs to be taken given their ability to react with nucleophiles beyond tyrosines (Martín-Gago and Olsen, 2019).
Covalent targeting of histidines and methionines
Histidine is not a common target for covalent modifications; however, histidine is basic under physiological conditions and can act as a nucleophile. Probably the best known historic example of serendipitous discovery of His-binding agent is natural product fumagillin that was shown to inhibit methionine aminopeptidase-2 (MetAP-2) by binding to His231 (Griffith et al., 1998) (Liu et al., 1998). Fumagillin possesses a spiroepoxide as a reactive warhead, which led to exploring this functional group for His-targeting in number of fumagillin derivatives, as well as targeting of lysines (Evans et al., 2007).
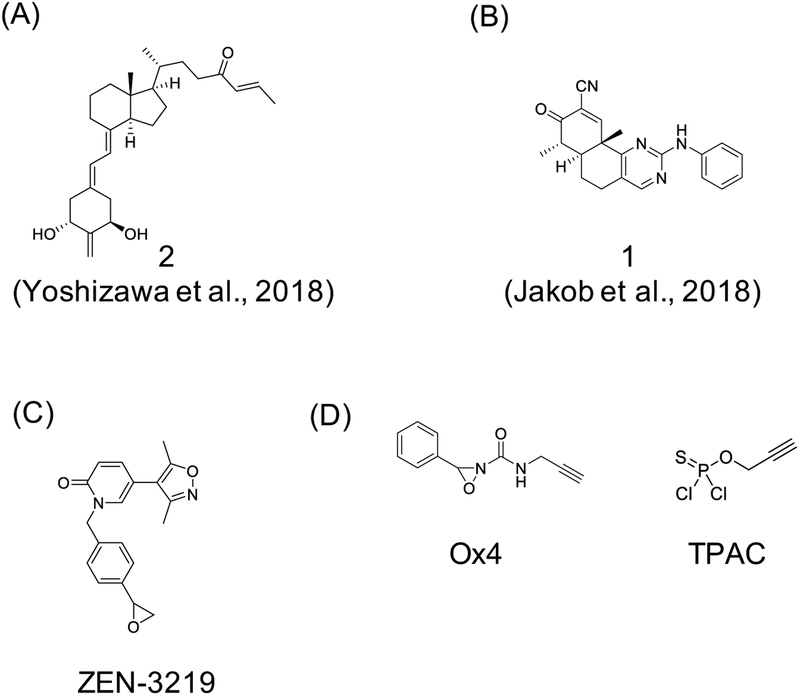
In more recent efforts, Yoshizawa et al. designed His-targeting analogs of 1α,25-dihydroxyvitamin D3 (1,25D3) (Yoshizawa et al., 2018). The 25-hydroxyl group of 1,25D3 forms hydrogen bonds with His301 and His393 of the vitamin D receptor (VDR), and Yoshizawa et al. introduced a β-enone at position C26 corresponding to the oxygen of the 25-hydroxy group of 1,25D3 and prepared four compounds with different warheads (example of full agonist 2 is shown in Figure 5A). Covalent modification was confirmed by ESI-MS analysis, and irreversible nature of adducts was confirmed by incubating VDR ligand binding domain (VDR-LBD) with the β-enone containing compounds for 24 hours followed by a wash and incubation with 1,25D3 for another 24 hours. Subsequent ESI-MS analysis revealed only covalently modified peaks for the VDR-LBD, and crystal structures supported a covalent mode of binding. Taken together, these results represent in vitro proof-of-concept for targeting histidines using an electrophilic enone group. More recently, α,β-unsaturated enone compound 1 (Figure 5B) was reported as a reversible covalent inhibitor that targets His315 of Isocitrate Dehydrogenase 1 (IDH1) (Jakob et al., 2018).
Figure 5. Examples of covalent ligands developed to target histidines and methionines.
(A) full agonist 2 uses electrophilic enone group to target histidine on vitamin D receptor (VDR) (Yoshizawa et al., 2018), (B) α,β-unsaturated enone compound 1 targets His315 of Isocitrate Dehydrogenase 1 (IDH1) (Jakob et al., 2018), (C) ZEN-3219 employs an epoxide warhead to target Met149 located in the acetyl-Lys binding site within bromodomain 1 (BD1) of BRD4 (Kharenko et al., 2018), and (D) oxaziridine-based probe Ox4 developed for reactive methionine profiling using ReACT (redox-activated chemical tagging) (Lin et al., 2017), and thio-phosphoro alkyne dichloridate (TPAC) probe for activity profiling of histidines (Jia et al., 2019).
Similarly, strategies for selective covalent targeting of methionines are in the early development and proof-of-concept stages. Recently, Kharenko et al. described a covalent inhibitor of BRD4 that targets Met149 located in the acetyl-Lys binding site within bromodomain 1 (BD1) (Kharenko et al., 2018). The authors employed structure-guided design based on a noncovalent ligand to generate a compound series with an epoxide warhead (see ZEN-3219 as a representative structure from this series; Figure 5C). Covalent modification was confirmed via MALDI-TOF experiments that identified an increase in +325 Da indicating formation of a covalent sulfonium linkage, which was also observed in high resolution co-crystal structures. The validation process included use of negative control compound and washout experiments, as well as binding competition assays done in the presence of free methionine and cysteine. Although targets outside the narrow range of proteins tested (other members of BRD family as well as a handful of additional non-BET family bromodomain proteins) may exist, this report represents a rare example of a methionine targeting compound. It is also relevant to note that some of the experimental approaches used for validation of covalent ligands we outlined above may not be suitable due to potential lower stability and/or reversibility of sulfonium ion formed in this reaction.
Given the redox sensitivity, methionines may be targetable via redox-based mechanisms. Lin et al. took advantage of this property to develop a Met-directed strategy they call ReACT (for redox-activated chemical tagging) (Lin et al., 2017). ReACT approach is based on optimizing oxaziridine (Ox) compounds to prefer sulfur imidation reaction, resulting in conversion of methionine into a sulfimide derivative (Figure 5D). The authors provided examples for using this strategy for bioconjugation purposes, and as chemoproteomic tools to map hyper-reactive methionines. Using ReACT in combination with tandem orthogonal proteolysis-activity-based-protein profiling (TOP-ABPP; (Speers and Cravatt, 2005)), Lin et al. identified more than 100 oxidation-sensitive methionines in HeLa cell lysates treated with low dose of one of their probes, Ox4 (Figure 5D). These results suggest that human proteome contains redox-sensitive methionines that could potentially be selectively targeted with covalent agents. Interestingly, the same group has recently tackled histidine labeling by developing thiophosphorodichloridate reagents that mimic naturally occurring histidine phosphorylation (Jia et al., 2019). Optimization of phosphorus-based electrophiles yielded thio-phosphoro alkyne dichloridate (TPAC; Figure 5D) as the best performing reagent that led to selective histidine labeling in HeLa cell lysates. Encouraged by these results, the authors are now working towards expanding TPAC and related tools into chemoselective probes, thus opening a new window of opportunity to map and target reactive histidines.
Emerging strategies for selective covalent ligand discovery
Beyond serendipity, and structure-based optimization of noncovalent ligands, there is only a handful of strategies for discovery of novel selective covalent ligands. Most of them use some form of screening, and we will discuss fragment-based ligand discovery (FBLD), application of nucleic acid encoded libraries, and covalent docking in more detail below. We will also highlight several examples where discovery of selective covalent ligands was coupled with pursuing molecules that modulate protein stability or assembly. We will also include a brief description of recently described warheads, and conclude by providing an example of how nucleophilic warheads are being introduced into the covalent ligand tool box.
Screening strategies for discovery of selective covalent ligands
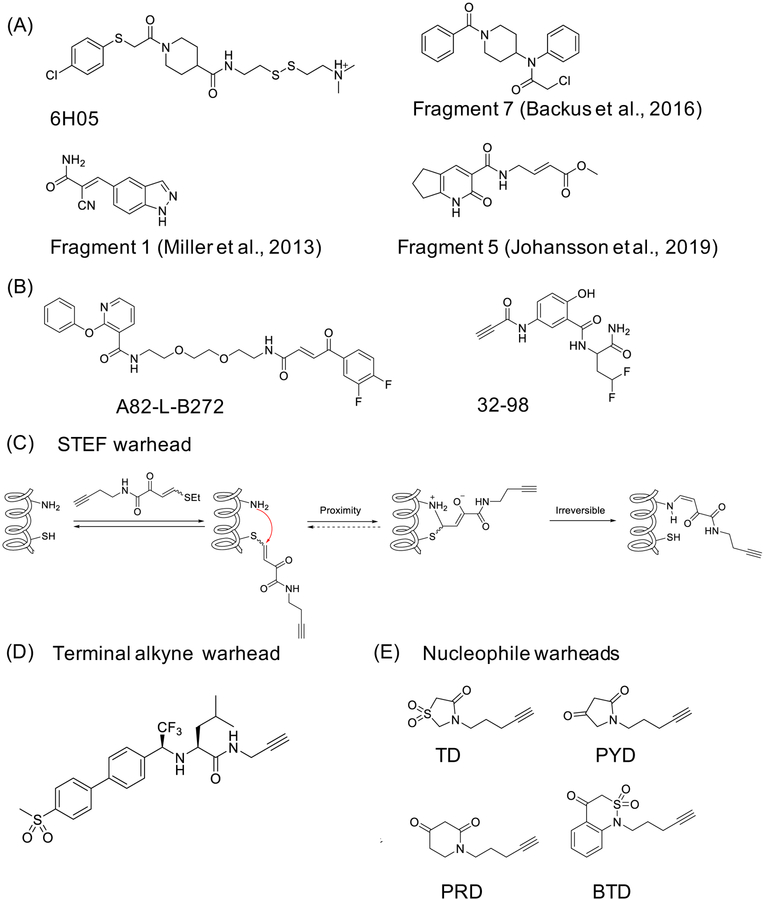
Compounds that include reactive functional groups are usually filtered out of the libraries used for high-throughput screening campaigns, as the reactive groups are considered to be hallmarks of pan-assay interference (PAINS) compounds (Baell and Nissink, 2018). Therefore, screening for covalent ligands usually requires specialized library design. Covalent FBLD uses low molecular weight (MW) compounds (typically below 250 Da). Due to their small size, the fragments may interact with cryptic or hidden pockets that would otherwise be inaccessible and therefore open new targeting opportunities. An early example of using FBLD applied to covalent ligands achieved discovery of an allosteric site on KRASG12C (Ostrem et al., 2013). Ostrem et al. used disulfide tethering to screen a library of 480 disulfide-containing fragments. The readout for this screen was in vitro intact MS and the efforts resulted in identification of a fragment 6H05 (Figure 6A). Structural analysis showed that the fragment interacted with a previously “invisible” pocket, and subsequent optimization, including introduction of irreversible acrylamide and vinyl sulfonamide warheads, led to the first series of mutant selective, covalent KRASG12C inhibitors. About the same time, Miller et al. described their efforts that employed fragments equipped with a reversible warhead, cyanoacrylamide, to discover lead inhibitors for MSK/RSK family kinases (fragment 1, Figure 6A) (Miller et al., 2013). On a larger scale, a library of 100 fragments containing aminomethyl methyl acrylate warhead was screened against cysteine protease papain as a proof of concept (Kathman et al., 2014). This study showed that FBLD is useful for discovery of covalent ligands that target highly reactive catalytic cysteine sites, such as the one found in papain. However, the authors noted that there might be limitations to this strategy when employed to finding fragments that target less reactive cysteines.
Figure 6. Selective covalent ligand discovery through library screening and warhead development.
(A) Fragment-based ligand discovery (FBLD) efforts led to discovery of the following fragments: 6H05 for KRASG12C (Ostrem et al., 2013), fragment 1 for MSK/RSK (Miller et al., 2013), fragment 7 for CASP8 and CASP10 (Backus et al., 2016), and fragment 5 for HOIP (Johansson et al., 2019); (B) example compounds developed using DNA-encoded chemical libraries that target JNK1 (Zimmermann et al., 2017), and ERBB2 (Zambaldo et al., 2016); (C) b-heteroatom substituted acrylamides warheads STEFs, and their mechanism (Hansen et al., 2019); (B) terminal alkyne as a warhead, here included into a potent cathepsin K (CatK) inhibitor, odanacatib (Mons et al., 2019); and (C) nucleophilic warheads that target sulfenic acid (Cys-SOH): benzo[c][1,2]thiazine (BTD), pyrrolidine-2,4-dione (PYD), piperidine-2,4-dione (PRD), and thiazolidine-4-one 1,1-dioxide (TD) (Gupta et al., 2017).
Another limitation of FBLD that applies to both noncovalent and covalent fragments, is that the strategy works only in vitro, with purified target proteins. To address this issue, Backus et al. conducted a competitive isoTOP-ABPP based screen of a library containing approximately 60 electrophilic fragments in two human cell lines to profile in-cell reactivity (Backus et al., 2016). Interestingly, although the screening was conducted using high concentrations of fragments (500 μM), most of them displayed restricted and non-overlapping reactivity, suggesting that this type of analysis could lead to discovery of both new chemical probes and novel ligandable sites. As an illustration of this idea, Backus et al. followed up on one of their fragments that blocked caspase 8 (CASP8) and caspase 10 (CASP10) activity by binding to inactive zymogen forms (7, Figure 6A). The authors elaborated fragment 7 into CASP8 selective ligand and used both compounds to examine the roles of CASP8 and CASP10 in human T cells.
Very recently, Johansson et al. employed a library of 104 amide fragments containing α,β-unsaturated methyl ester electrophiles and screened it against RBR (ring-between-ring fingers) domain of an E3 ubiquitin ligase, HOIP, which led to identification of fragment 5 (Figure 6A) (Johansson et al., 2019). Fragment 5 was shown to block formation of E3-Ub thioester intermediate in vitro, and structural analysis revealed that the fragment binds covalently to Cys885 on HOIP. Further studies with a more potent analog, in combination with inactive compounds in cell-based assays, including Cys885Ala mutant cells, support covalent binding-dependent on-target activity. ABPP and quantitative MS studies identified 11 other proteins with no other E3 ligases as off-targets for the unoptimized fragment.
Another limitation of the FBLD examples we highlighted thus far is the limited size of fragment libraries employed. To address this issue, Resnick et al. describe a strategy to expand size of covalent fragment libraries (Resnick et al., 2019) The authors acquired 993 commercially available low molecular weight compounds (92% of compounds below 300 Da), featuring either acrylamide or chloroacetamide warheads, and limited to those compounds that adhere to ‘rule-of-three’, an empirical way to prioritize quality of fragments (Jhoti et al., 2013). An in vitro LC-MS screen against a panel of ten proteins, including those without an available chemical probe, as well as BSA as a negative control, resulted in hits for 7 of the targets. Follow up studies demonstrated that more reactive fragments are not necessarily more promiscuous.
Nucleic acid-encoded libraries represent an alternative library design (Neri and Lerner, 2018). Although not originally developed for covalent ligand discovery, this strategy did yield covalent inhibitors in the past, albeit serendipitously (Chan et al., 2017) (Cuozzo et al., 2017). Recently, DNA-encoded library, constructed using building blocks that are reactive towards different amino acids, was used for covalent inhibitor discovery (Zimmermann et al., 2017). The resulting library contained almost 150 thousand compounds and was screened against JNK1, a kinase target with known covalent inhibitors. The screen yielded two synergistic building blocks that were then connected via a chemical linker into A82-L-B272 (Figure 6B). Covalent binding of A82-L-B272 to JNK1 was confirmed in vitro, but no further validation was conducted. Another example combined peptide nucleic acid (PNA) encoding with electrophilic compounds, and hybridization of PNA molecules onto a DNA array (Zambaldo et al., 2016). This allowed the target to be screened against the array, and a denaturing step was introduced to distinguish between noncovalent and covalent inhibitors. This strategy was applied to a library of 10,000 covalent binders that were screened against two kinases, ERBB2 and MEK2, and resulted in a potential lead compound (32–98) (Figure 6B). However, the lead compound was not validated beyond the KinomeScan, which showed that, in addition to ERBB2, the compound inhibited MEK4, JAK3 and DAPK3.
Another approach that can be used to screen large numbers of compounds is structure-based docking. Optimizing docking screens for discovery of covalent ligands needs to overcome several obstacles, namely the construction of robust and diverse libraries that include electrophilic compounds, and the development of docking protocols that take into account both binding energetics and bond formation. In an early example, London et al. constructed a library of 650,000 electrophilic compounds that featured a range of warheads (aldehydes, alkyl halides, boronic acids, carbamates, α-cyanoacrylamides, epoxides, α-ketoamides, and α,β-unsaturated carbonyls) (London et al., 2014). The docking was performed using DOCKovalent. The method led to discovery of boronic acid inhibitors of AmpC b-lactamase that bind covalently to the active site serine, and several cysteine-directed cyanoacrylamide inhibitors of RSK2, MSK1, and JAK3. Since the early report, DOCKovalent was used to discover covalent inhibitors for KRASG12C (Nnadi et al., 2018), and MKK7, an upstream activator of JNK kinases (Shraga et al., 2019). Although ligand validation process was incomplete in the case of KRASG12C work, both studies support the value of using docking as a part of covalent ligand discovery workflow. However, as noted by London et al., further development of docking methods designed to model covalent bond formation, and allow the target to be dynamic is needed.
Discovery of novel warheads
The nature of a warhead impacts general reactivity of the covalent ligand, as well as its selectivity, potency, tissue distribution, cellular localization, and metabolic stability, and development of new warheads remains a priority. Many warheads, such as sulfonyl fluorides (SFs) and spiroepoxides, are reactive towards multiple side chains, and selectivity mapping studies need to address not only the range of targets but the range of nucleophilic sites on proteins that a given warhead reacts with, as discussed in a recent perspective (Gehringer and Laufer, 2019). Here, we would like to comment on two warheads reported in the short time since the publication of that perspective, semi‐oxamide vinylogous thioesters (Hansen et al., 2019) and alkynes (Mons et al., 2019). Hansen et al. use b-heteroatom substituted acrylamides as a starting point for developing a class of electrophiles that reacts via an addition-elimination reaction named STEFs (Figure 6C). The results presented suggest that STEFs form a reversible bond with a cysteine, then subsequently irreversibly react with a proximal amine. Preliminary tests conducted in cell lysates identified 114 protein targets, and follow up work identified 45 binding sites, all of which were lysines, including sites on KHSRP, RPS2 and APPL1-BAR-PH domain. Additionally, the authors show that STEFs can be incorporated into complex scaffolds, for example by replacing the warhead in an EGFR inhibitor afatinib. However, it remains unclear whether STEFs will be useful for selective covalent ligand development. On the other hand, Mons et al. used a terminal alkyne group as the warhead. The terminal alkyne behaves as a latent electrophile, therefore potentially minimizing off-target reactivity. This work demonstrates that installing a terminal alkyne onto a potent cathepsin K (CatK) inhibitor, odanacatib, results in a covalent inhibitor that targets the catalytic cysteine in this cysteine protease (Figure 6D). The study introduces terminal alkynes as a potential new warhead for covalent ligand design, as well as urges caution when using this functional group as a bioorthogonal handle.
The final example of different warheads we would like to highlight is the use of carbon nucleophiles as warheads. One type of electrophilic sites on proteins that has been documented are formed by reactive oxygen species (ROS)-mediated oxidation of the cysteine’s thiol group to sulfenic acid (Cys-SOH). This reversible modification has been linked to regulation of protein function, including in kinases, such as EGFR, and protein tyrosine phosphatases, such as PTP1B, SHP2 and YopH. Recently, Gupta et al. screened a library of about 100 cyclic carbon nucleophile containing compounds with a range of scaffolds in RKO colon adenocarcinoma cells using a chemoproteomics workflow for target identification and reactivity mapping (Gupta et al., 2017). The authors demonstrated a range of reactivities, with benzo[c][1,2]thiazine (BTD) warhead displaying the highest reaction rate in preliminary reactivity assays, and highest level of reactivity in RKO cell lysates, and warheads like pyrrolidine-2,4-dione (PYD), piperidine-2,4-dione (PRD), and thiazolidine-4-one 1,1-dioxide (TD) displaying narrower target space (Figure 6E). Overall, Cys-SOH directed nucleophilic probes labeled 1283 sites on 761 proteins, including proteins previously reported to be S-sulfenylated, like protein tyrosine phosphatases. It is premature to conclude that selective targeting of protein electrophiles is feasible, as the extent (in terms of scope and critically stoichiometry), nature and biological roles (if any) of these sites is unexplored. However, a recent chemoproteomic study does suggest that a range of electrophilic sites on proteins may exist in human cells. Therefore, we consider that the evidence for these sites has begun to emerge and we expect to see more work done in this area in the future.
Beyond inhibition: covalent ligands that modulate target assembly or stability
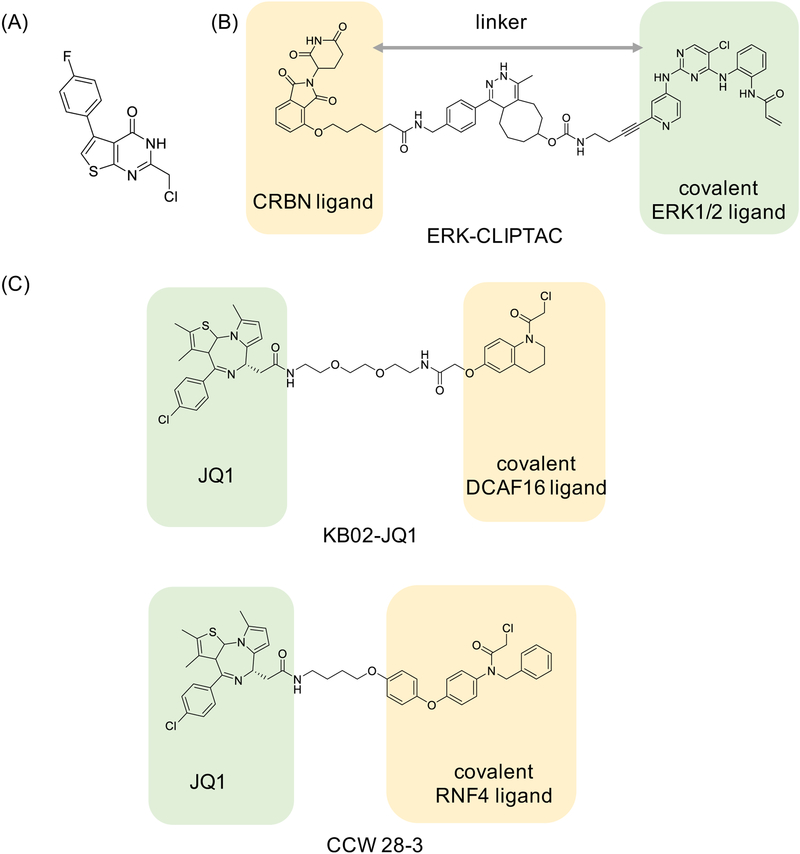
Small molecules, noncovalent and covalent alike, can modulate biological functions in ways beyond inhibiting (or activating activity), including via changing protein-protein interactions, inducing dimerization, or leading to selective degradation. We will briefly comment on several recent disclosures of covalent ligands that exert effects through these additional mechanisms. For example, very recent work on tryparedoxin (Tpx) from Trypanosoma brucei, a causative agent of African sleeping sickness, has reported a selective covalent ligand that binds a cysteine residue on Tpx (Figure 7A), and leads to Tpx dimer formation, with small molecule forming the important part of the interface (Wagner et al., 2019). Dimer formation sequesters Tpx from its role in peroxide detoxification, regulating reactive oxygen species (ROS) and other functions. The work is an example of chemically induced dimerization achieved via a covalent ligand.
Figure 7. Examples of selective covalent ligands that modulate target assembly and/or stability.
(A) A small molecule dimerizer of tryparedoxin (Tpx) from Trypanosoma brucei (Wagner et al., 2019); (B) ERK-CLIPTAC, potentially the first degrader molecule that includes a covalent inhibitor into its design (Lebraud et al., 2016); and (C) KB02-JQ1 degrader (PROTAC) molecule that recruits DCAF16 E3 ligase via covalent KB02 warhead (Zhang et al., 2019), and CCW 28–3 that uses CCW 16 as covalent RNF4 E3 ligase recruiter (Ward et al., 2019).
Another kind of chemical induced dimerization that triggers well-defined downstream effects is targeted protein degradation (Cromm and Crews, 2017). The most commonly used small molecule degraders are bispecific compounds, called PROTACs, which include an E3 ubiquitin ligase recruiting warhead, and a target binding warhead. The proximity of the ligase and the target results in ubiquitination and proteasomal degradation of the target. One of the stated advantages of the degraders is that they can be used at substochiometric (catalytic) concentrations as the degrader molecule is “recycled” in the process. This suggests that PROTACs with a covalent handle on the target recruiting side may not be beneficial, while a covalent handle on the E3 ligase recruiting side may offer improvements. For example, a study that examined PROTAC-mediated degradation of Bruton’s Tyrosine Kinase (BTK) concluded that covalent binding to the target protein has a negative impact on PROTAC’s ability to degrade the target (Tinworth et al., 2019). On the other hand, a study that described CLIPTACs, PROTACs that form intracellularly via click chemistry, included an example of ERK1/2 CLIPTAC based on the covalent inhibitor (Figure 7B). The resulting CLIPTAC led to ERK1/2 degradation within 4 hours of treatment, and the effect was sustained to up to 24 hours (Lebraud et al., 2016). Additionally, an existing strategy for targeted degradation of tagged proteins called HaloPROTAC is based on the use of chloroalkene functional group in the HaloPROTAC molecule. This group forms a covalent bond with the tag, an engineered bacterial dehalogenase fused to the protein of interest. An E3 ubiquitin ligase recruiting arm then brings the ligase to the close proximity of the tagged protein, resulting in ubiquitination and degradation of the entire construct (Buckley et al., 2015). Therefore, at least in principle, covalent PROTACs are possible, including those that use electrophilic warheads for E3 ligase recruitment (Figure 7C) (Zhang et al., 2019) (Ward et al., 2019). However, these examples represent early proof-of-principle and additional studies must be undertaken to better understand the dynamic changes in the system caused by covalent PROTACs, consequences of prolonged PROTAC persistence, and the potentially increased off-target space when compared to noncovalent ones. Moreover, as with other covalent ligands, extent of the effects will greatly depend on the covalent target’s half-life (E3 ligase or protein of interest), and although not formally shown yet, it is expected that targets with long half-lives will exhibit most pronounced effects.
Concluding remarks
In conclusion, to be useful as research tools or drug leads, covalent ligands have to be rigorously validated. The covalent inhibitor validation process we use includes number of complementary strategies aimed at establishing evidence for covalent binding and on-target mechanism of action. Additionally, as with other tool compounds, the decision to use a specific covalent tool compound must be made based on the specific biological question that the tool is meant to help answer. Here, special care needs to be taken to match the target and the tool compound with the specific biological context and understanding the optimal conditions of use, such as concentration range, cell line selection and similar (Blagg and Workman, 2017). Having said that, we recognize that some targets and systems may be challenging to work with or refractive to certain types of experiments (for example, crystallography), thus limiting the extent of validation that can be performed. We also appreciate the cost of extensive validation, as well as the fact that equipment and resources we have at our disposal may not be routinely available elsewhere. In those instances, cell-based validation experiments may offer cost-effective way to provide essential insights into on-target and on-mechanism activity. Moreover, missing validation steps should not necessarily prevent disclosures of novel covalent ligands, provided that limitations and caveats are clearly stated and discussed. Therefore, reports of new targeting modalities especially if they aim to expand the scope of targetable sites, although potentially more preliminary, may still serve to fuel further discovery.
Overall, as the conversation surrounding selective covalent bindings moves away from liabilities and towards possibilities, we expect to see new innovations and collaborations emerge. Armed with chemical ingenuity, robust characterization and validation, and improved methodologies, we expect covalent ligands to remain relevant both as tools for basic research and leads for drug development.
Acknowledgements
This review covers a rapidly evolving field and our overview is limited to the literature as published on or before February 2019. We therefore apologize to anyone who has published important results since then. We are grateful to Dr. Fleur M. Ferguson for providing helpful feedback. Additionally, we would like to thank the editors of Cell Chemical Biology for giving us this opportunity, and our copy editor who polished the revised work. We would also like to thank our three reviewers who invested a lot of efforts into evaluating the initial and revised manuscript. Their comments have significantly improved our review. This work was supported by the Linde Program in Chemical Biology and NIH grants R01 CA136851-07 (N.G.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest
Nathanael S. Gray is a founder, science advisory board member (SAB) and equity holder in Gatekeeper, Syros, Petra, C4, B2S and Soltego. The Gray lab receives or has received research funding from Novartis, Takeda, Astellas, Taiho, Janssen, Kinogen, Voronoi, Her2llc, Deerfield and Sanofi.
Milka Kostic is a consulting editor at Life Science Editors.
Lead Contact: Further information and requests should be directed to and will be fulfilled by the Lead Contact, Nathanael Gray (nathanael_gray@dfci.harvard.edu)).
References
- Anscombe E, Meschini E, Mora-Vidal R, Martin MP, Staunton D, Geitmann M, Danielson UH, Stanley WA, Wang LZ, Reuillon T, Golding BT, Cano C, Newell DR, Noble MEM, Wedge SR, Endicott JA, Griffin RJ, 2015. Identification and Characterization of an Irreversible Inhibitor of CDK2. Chem. Biol 22, 1159–1164. 10.1016/j.chembiol.2015.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith CH, Audia JE, Austin C, Baell J, Bennett J, Blagg J, Bountra C, Brennan PE, Brown PJ, Bunnage ME, Buser-Doepner C, Campbell RM, Carter AJ, Cohen P, Copeland RA, Cravatt B, Dahlin JL, Dhanak D, Edwards AM, Frederiksen M, Frye SV, Gray N, Grimshaw CE, Hepworth D, Howe T, Huber KVM, Jin J, Knapp S, Kotz JD, Kruger RG, Lowe D, Mader MM, Marsden B, Mueller-Fahrnow A, Müller S, O’Hagan RC, Overington JP, Owen DR, Rosenberg SH, Roth Bryan, Roth Brian, Ross R, Schapira M, Schreiber SL, Shoichet B, Sundström M, Superti-Furga G, Taunton J, Toledo-Sherman L, Walpole C, Walters MA, Willson TM, Workman P, Young RN, Zuercher WJ, 2015. The promise and peril of chemical probes. Nat. Chem. Biol 11, 536–541. 10.1038/nchembio.1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin DA, Cravatt BF, 2012. The pharmacological landscape and therapeutic potential of serine hydrolases. Nat. Rev. Drug Discov. 11, 52–68. 10.1038/nrd3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backus KM, Correia BE, Lum KM, Forli S, Horning BD, González-Páez GE, Chatterjee S, Lanning BR, Teijaro JR, Olson AJ, Wolan DW, Cravatt BF, 2016. Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574. 10.1038/nature18002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell JB, Nissink JWM, 2018. Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017—Utility and Limitations. ACS Chem. Biol 13, 36–44. 10.1021/acschembio.7b00903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BR, 1964. Factors in the design of active-site directed irreversible inhibitors. J. Pharm. Sci 53, 347–364. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Gao J, 2016. Targeting biomolecules with reversible covalent chemistry. Curr. Opin. Chem. Biol 34, 110–116. 10.1016/j.cbpa.2016.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagg J, Workman P, 2017. Choose and Use Your Chemical Probe Wisely to Explore Cancer Biology. Cancer Cell 32, 9–25. 10.1016/j.ccell.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne CM, Jiang B, Ficarro SB, Doctor ZM, Johnson JL, Card JD, Sivakumaren SC, Alexander WM, Yaron TM, Murphy CJ, Kwiatkowski NP, Zhang T, Cantley LC, Gray NS, Marto JA, 2019. A Chemoproteomic Strategy for Direct and Proteome-Wide Covalent Inhibitor Target-Site Identification. J. Am. Chem. Soc 141, 191–203. 10.1021/jacs.8b07911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley DL, Raina K, Darricarrere N, Hines J, Gustafson JL, Smith IE, Miah AH, Harling JD, Crews CM, 2015. HaloPROTACS: Use of Small Molecule PROTACs to Induce Degradation of HaloTag Fusion Proteins. ACS Chem. Biol 10, 1831–1837. 10.1021/acschembio.5b00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas-Lopes E, Cerchietti L, Ahn JH, Clement CC, Robles AI, Rodina A, Moulick K, Taldone T, Gozman A, Guo Y, Wu N, de Stanchina E, White J, Gross SS, Ma Y, Varticovski L, Melnick A, Chiosis G, 2009. Hsp90 inhibitor PU-H71, a multimodal inhibitor of malignancy, induces complete responses in triple-negative breast cancer models. Proc. Natl. Acad. Sci 106, 8368–8373. 10.1073/pnas.0903392106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaikuad A, Koch P, Laufer SA, Knapp S, 2018. The Cysteinome of Protein Kinases as a Target in Drug Development. Angew. Chem. Int. Ed Engl. 57, 4372–4385. 10.1002/anie.201707875 [DOI] [PubMed] [Google Scholar]
- Chan AI, McGregor LM, Jain T, Liu DR, 2017. Discovery of a Covalent Kinase Inhibitor from a DNA-Encoded Small-Molecule Library × Protein Library Selection. J. Am. Chem. Soc 139, 10192–10195. 10.1021/jacs.7b04880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Dong J, Plate L, Mortenson DE, Brighty GJ, Li S, Liu Y, Galmozzi A, Lee PS, Hulce JJ, Cravatt BF, Saez E, Powers ET, Wilson IA, Sharpless KB, Kelly JW, 2016. Arylfluorosulfates Inactivate Intracellular Lipid Binding Protein(s) through Chemoselective SuFEx Reaction with a Binding Site Tyr Residue. J. Am. Chem. Soc 138, 7353–7364. 10.1021/jacs.6b02960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Connelly S, Reixach N, Wilson IA, Kelly JW, 2010. Chemoselective small molecules that covalently modify one lysine in a non-enzyme protein in plasma. Nat. Chem. Biol 6, 133–139. 10.1038/nchembio.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromm PM, Crews CM, 2017. Targeted Protein Degradation: from Chemical Biology to Drug Discovery. Cell Chem. Biol 24, 1181–1190. 10.1016/j.chembiol.2017.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta A, Taunton J, 2019. Lysine-Targeted Inhibitors and Chemoproteomic Probes. Annu. Rev. Biochem 88, 365–381. 10.1146/annurev-biochem-061516-044805 [DOI] [PubMed] [Google Scholar]
- Cuozzo JW, Centrella PA, Gikunju D, Habeshian S, Hupp CD, Keefe AD, Sigel EA, Soutter HH, Thomson HA, Zhang Y, Clark MA, 2017. Discovery of a Potent BTK Inhibitor with a Novel Binding Mode by Using Parallel Selections with a DNA-Encoded Chemical Library. ChemBioChem 18, 864–871. 10.1002/cbic.201600573 [DOI] [PubMed] [Google Scholar]
- Dalton SE, Dittus L, Thomas DA, Convery MA, Nunes J, Bush JT, Evans JP, Werner T, Bantscheff M, Murphy JA, Campos S, 2018. Selectively Targeting the Kinome-Conserved Lysine of PI3Kδ as a General Approach to Covalent Kinase Inhibition. J. Am. Chem. Soc 140, 932–939. 10.1021/jacs.7b08979 [DOI] [PubMed] [Google Scholar]
- De Cesco S, Kurian J, Dufresne C, Mittermaier AK, Moitessier N, 2017. Covalent inhibitors design and discovery. Eur. J. Med. Chem 138, 96–114. 10.1016/j.ejmech.2017.06.019 [DOI] [PubMed] [Google Scholar]
- Drewes G, Knapp S, 2018. Chemoproteomics and Chemical Probes for Target Discovery. Trends Biotechnol. 36, 1275–1286. 10.1016/j.tibtech.2018.06.008 [DOI] [PubMed] [Google Scholar]
- Du J, Bernasconi P, Clauser KR, Mani DR, Finn SP, Beroukhim R, Burns M, Julian B, Peng XP, Hieronymus H, Maglathlin RL, Lewis TA, Liau LM, Nghiemphu P, Mellinghoff IK, Louis DN, Loda M, Carr SA, Kung AL, Golub TR, 2009. Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nat. Biotechnol 27, 77–83. 10.1038/nbt.1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Morris GM, Wu J, Olson AJ, Sorensen EJ, Cravatt BF, 2007. Mechanistic and structural requirements for active site labeling of phosphoglycerate mutase by spiroepoxides. Mol. Biosyst 3, 495–506. 10.1039/b705113a [DOI] [PubMed] [Google Scholar]
- Gehringer M, Laufer SA, 2019. Emerging and Re-Emerging Warheads for Targeted Covalent Inhibitors: Applications in Medicinal Chemistry and Chemical Biology. J. Med. Chem 10.1021/acs.jmedchem.8b01153 [DOI] [PubMed] [Google Scholar]
- Griffith EC, Su Z, Niwayama S, Ramsay CA, Chang YH, Liu JO, 1998. Molecular recognition of angiogenesis inhibitors fumagillin and ovalicin by methionine aminopeptidase 2. Proc. Natl. Acad. Sci. U. S. A 95, 15183–15188. 10.1073/pnas.95.26.15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimster NP, Connelly S, Baranczak A, Dong J, Krasnova LB, Sharpless KB, Powers ET, Wilson IA, Kelly JW, 2013. Aromatic sulfonyl fluorides covalently kinetically stabilize transthyretin to prevent amyloidogenesis while affording a fluorescent conjugate. J. Am. Chem. Soc 135, 5656–5668. 10.1021/ja311729d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Shannon DA, Colby T, Wang Z, Shabab M, Kumari S, Villamor JG, McLaughlin CJ, Weerapana E, Kaiser M, Cravatt BF, van der Hoorn RAL, 2013. Chemical Proteomics with Sulfonyl Fluoride Probes Reveals Selective Labeling of Functional Tyrosines in Glutathione Transferases. Chem. Biol 20, 541–548. 10.1016/j.chembiol.2013.01.016 [DOI] [PubMed] [Google Scholar]
- Gupta V, Yang J, Liebler DC, Carroll KS, 2017. Diverse Redoxome Reactivity Profiles of Carbon Nucleophiles. J. Am. Chem. Soc 139, 5588–5595. 10.1021/jacs.7b01791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker SM, Backus KM, Lazear MR, Forli S, Correia BE, Cravatt BF, 2017. Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem 9, 1181–1190. 10.1038/nchem.2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallenbeck KK, Turner DM, Renslo AR, Arkin MR, 2017. Targeting Non-Catalytic Cysteine Residues Through Structure-Guided Drug Discovery. Curr. Top. Med. Chem 17, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen BK, Loveridge CJ, Thyssen S, Wørmer GJ, Nielsen AD, Palmfeldt J, Johannsen M, Poulsen TB, 2019. STEFs: Activated Vinylogous Protein-Reactive Electrophiles. Angew. Chem. Int. Ed Engl. 58, 3533–3537. 10.1002/anie.201814073 [DOI] [PubMed] [Google Scholar]
- Hatcher JM, Wu G, Zeng C, Zhu J, Meng F, Patel S, Wang W, Ficarro SB, Leggett AL, Powell CE, Marto JA, Zhang K, Ki Ngo JC, Fu X-D, Zhang T, Gray NS, 2018. SRPKIN-1: A Covalent SRPK1/2 Inhibitor that Potently Converts VEGF from Pro-angiogenic to Anti-angiogenic Isoform. Cell Chem. Biol 25, 460–470.e6. 10.1016/j.chembiol.2018.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hett EC, Xu H, Geoghegan KF, Gopalsamy A, Kyne RE, Menard CA, Narayanan A, Parikh MD, Liu S, Roberts L, Robinson RP, Tones MA, Jones LH, 2015. Rational targeting of active-site tyrosine residues using sulfonyl fluoride probes. ACS Chem. Biol 10, 1094–1098. 10.1021/cb5009475 [DOI] [PubMed] [Google Scholar]
- Jackson PA, Widen JC, Harki DA, Brummond KM, 2017. Covalent Modifiers: A Chemical Perspective on the Reactivity of α,β-Unsaturated Carbonyls with Thiols via Hetero-Michael Addition Reactions. J. Med. Chem 60, 839–885. 10.1021/acs.jmedchem.6b00788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob CG, Upadhyay AK, Donner PL, Nicholl E, Addo SN, Qiu W, Ling C, Gopalakrishnan SM, Torrent M, Cepa SP, Shanley J, Shoemaker AR, Sun CC, Vasudevan A, Woller KR, Shotwell JB, Shaw B, Bian Z, Hutti JE, 2018. Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315. J. Med. Chem 61, 6647–6657. 10.1021/acs.jmedchem.8b00305 [DOI] [PubMed] [Google Scholar]
- Janes MR, Zhang J, Li L-S, Hansen R, Peters U, Guo X, Chen Y, Babbar A, Firdaus SJ, Darjania L, Feng J, Chen JH, Li Shuangwei, Li Shisheng, Long YO, Thach C, Liu Yuan, Zarieh, A, Ely T, Kucharski JM, Kessler LV, Wu T, Yu K, Wang Y, Yao Y, Deng X, Zarrinkar PP, Brehmer D, Dhanak D, Lorenzi MV, Hu-Lowe D, Patricelli MP, Ren P, Liu Yi, 2018. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell 172, 578–589.e17. 10.1016/j.cell.2018.01.006 [DOI] [PubMed] [Google Scholar]
- Jhoti H, Williams G, Rees DC, Murray CW, 2013. The “rule of three” for fragment-based drug discovery: where are we now? Nat. Rev. Drug Discov. 12, 644–645. 10.1038/nrd3926-c1 [DOI] [PubMed] [Google Scholar]
- Jia S, He D, Chang CJ, 2019. Bioinspired Thiophosphorodichloridate Reagents for Chemoselective Histidine Bioconjugation. J. Am. Chem. Soc 141, 7294–7301. 10.1021/jacs.8b11912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson H, Isabella Tsai Y-C, Fantom K, Chung C-W, Kümper S, Martino L, Thomas DA, Eberl HC, Muelbaier M, House D, Rittinger K, 2019. Fragment-Based Covalent Ligand Screening Enables Rapid Discovery of Inhibitors for the RBR E3 Ubiquitin Ligase HOIP. J. Am. Chem. Soc 141, 2703–2712. 10.1021/jacs.8b13193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LH, 2018. Reactive Chemical Probes: Beyond the Kinase Cysteinome. Angew. Chem. Int. Ed 57, 9220–9223. 10.1002/anie.201802693 [DOI] [PubMed] [Google Scholar]
- Kathman SG, Xu Z, Statsyuk AV, 2014. A fragment-based method to discover irreversible covalent inhibitors of cysteine proteases. J. Med. Chem 57, 4969–4974. 10.1021/jm500345q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharenko OA, Patel RG, Brown SD, Calosing C, White A, Lakshminarasimhan D, Suto RK, Duffy BC, Kitchen DB, McLure KG, Hansen HC, van der Horst EH, Young PR, 2018. Design and Characterization of Novel Covalent Bromodomain and Extra-Terminal Domain (BET) Inhibitors Targeting a Methionine. J. Med. Chem 61, 8202–8211. 10.1021/acs.jmedchem.8b00666 [DOI] [PubMed] [Google Scholar]
- Kisselev AF, van der Linden WA, Overkleeft HS, 2012. Proteasome inhibitors: an expanding army attacking a unique target. Chem. Biol 19, 99–115. 10.1016/j.chembiol.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong AD, Kauffman RS, Hurter P, Mueller P, 2011. Discovery and development of telaprevir: an NS3–4A protease inhibitor for treating genotype 1 chronic hepatitis C virus. Nat. Biotechnol 29, 993–1003. 10.1038/nbt.2020 [DOI] [PubMed] [Google Scholar]
- Lagoutte R, Patouret R, Winssinger N, 2017. Covalent inhibitors: an opportunity for rational target selectivity. Curr. Opin. Chem. Biol 39, 54–63. 10.1016/j.cbpa.2017.05.008 [DOI] [PubMed] [Google Scholar]
- Lebraud H, Wright DJ, Johnson CN, Heightman TD, 2016. Protein Degradation by In-Cell Self-Assembly of Proteolysis Targeting Chimeras. ACS Cent. Sci 2, 927–934. 10.1021/acscentsci.6b00280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Ambrogio L, Shimamura T, Kubo S, Takahashi M, Chirieac LR, Padera RF, Shapiro GI, Baum A, Himmelsbach F, Rettig WJ, Meyerson M, Solca F, Greulich H, Wong K-K, 2008. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene 27, 4702–4711. 10.1038/onc.2008.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Yang X, Jia S, Weeks AM, Hornsby M, Lee PS, Nichiporuk RV, Iavarone AT, Wells JA, Toste FD, Chang CJ, 2017. Redox-based reagents for chemoselective methionine bioconjugation. Science 355, 597–602. 10.1126/science.aal3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Widom J, Kemp CW, Crews CM, Clardy J, 1998. Structure of human methionine aminopeptidase-2 complexed with fumagillin. Science 282, 1324–1327. 10.1126/science.282.5392.1324 [DOI] [PubMed] [Google Scholar]
- London N, Miller RM, Krishnan S, Uchida K, Irwin JJ, Eidam O, Gibold L, Cimermančič P, Bonnet R, Shoichet BK, Taunton J, 2014. Covalent docking of large libraries for the discovery of chemical probes. Nat. Chem. Biol 10, 1066–1072. 10.1038/nchembio.1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale R, Ward RA, 2018. Structure-based design of targeted covalent inhibitors. Chem. Soc. Rev 47, 3816–3830. 10.1039/c7cs00220c [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S, 2002. The protein kinase complement of the human genome. Science 298, 1912–1934. 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- Martín-Gago P, Olsen CA, 2019. Arylfluorosulfate-Based Electrophiles for Covalent Protein Labeling: A New Addition to the Arsenal. Angew. Chem. Int. Ed Engl. 58, 957–966. 10.1002/anie.201806037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RM, Paavilainen VO, Krishnan S, Serafimova IM, Taunton J, 2013. Electrophilic fragment-based design of reversible covalent kinase inhibitors. J. Am. Chem. Soc 135, 5298–5301. 10.1021/ja401221b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mons E, Jansen IDC, Loboda J, van Doodewaerd BR, Hermans J, Verdoes M, van Boeckel CAA, van Veelen PA, Turk B, Turk D, Ovaa H, 2019. The Alkyne Moiety as a Latent Electrophile in Irreversible Covalent Small Molecule Inhibitors of Cathepsin K. J. Am. Chem. Soc 141, 3507–3514. 10.1021/jacs.8b11027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee H, Grimster NP, 2018. Beyond cysteine: recent developments in the area of targeted covalent inhibition. Curr. Opin. Chem. Biol 44, 30–38. 10.1016/j.cbpa.2018.05.011 [DOI] [PubMed] [Google Scholar]
- Müller S, Ackloo S, Arrowsmith CH, Bauser M, Baryza JL, Blagg J, Böttcher J, Bountra C, Brown PJ, Bunnage ME, Carter AJ, Damerell D, Dötsch V, Drewry DH, Edwards AM, Edwards J, Elkins JM, Fischer C, Frye SV, Gollner A, Grimshaw CE, IJzerman A, Hanke T, Hartung IV, Hitchcock S, Howe T, Hughes TV, Laufer S, Li VM, Liras S, Marsden BD, Matsui H, Mathias J, O’Hagan RC, Owen DR, Pande V, Rauh D, Rosenberg SH, Roth BL, Schneider NS, Scholten C, Singh Saikatendu K, Simeonov A, Takizawa M, Tse C, Thompson PR, Treiber DK, Viana AY, Wells CI, Willson TM, Zuercher WJ, Knapp S, Mueller-Fahrnow A, 2018. Donated chemical probes for open science. eLife 7 10.7554/eLife.34311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A, Jones LH, 2015. Sulfonyl fluorides as privileged warheads in chemical biology. Chem. Sci 6, 2650–2659. 10.1039/c5sc00408j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri D, Lerner RA, 2018. DNA-Encoded Chemical Libraries: A Selection System Based on Endowing Organic Compounds with Amplifiable Information. Annu. Rev. Biochem 87, 479–502. 10.1146/annurev-biochem-062917-012550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nnadi CI, Jenkins ML, Gentile DR, Bateman LA, Zaidman D, Balius TE, Nomura DK, Burke JE, Shokat KM, London N, 2018. Novel K-Ras G12C Switch-II Covalent Binders Destabilize Ras and Accelerate Nucleotide Exchange. J. Chem. Inf. Model 58, 464–471. 10.1021/acs.jcim.7b00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM, 2013. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551. 10.1038/nature12796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli MP, Janes MR, Li L-S, Hansen R, Peters U, Kessler LV, Chen Y, Kucharski JM, Feng J, Ely T, Chen JH, Firdaus SJ, Babbar A, Ren P, Liu Y, 2016. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discov. 6, 316–329. 10.1158/2159-8290.CD-15-1105 [DOI] [PubMed] [Google Scholar]
- Pettinger J, Jones K, Cheeseman MD, 2017. Lysine-Targeting Covalent Inhibitors. Angew. Chem. Int. Ed Engl. 56, 15200–15209. 10.1002/anie.201707630 [DOI] [PubMed] [Google Scholar]
- Resnick E, Bradley A, Gan Jinrui, Douangamath A, Krojer T, Sethi R, Geurink PP, Aimon A, Amitai G, Bellini D, Bennett J, Fairhead M, Fedorov O, Gabizon R, Gan Jin, Guo J, Plotnikov A, Reznik N, Ruda GF, Díaz-Sáez L, Straub VM, Szommer T, Velupillai S, Zaidman D, Zhang Y, Coker AR, Dowson CG, Barr HM, Wang C, Huber KVM, Brennan PE, Ovaa H, von Delft F, London N, 2019. Rapid Covalent-Probe Discovery by Electrophile-Fragment Screening. J. Am. Chem. Soc 141, 8951–8968. 10.1021/jacs.9b02822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotella DP, 2013. The discovery and development of boceprevir. Expert Opin. Drug Discov. 8, 1439–1447. 10.1517/17460441.2013.843525 [DOI] [PubMed] [Google Scholar]
- Schwartz PA, Kuzmic P, Solowiej J, Bergqvist S, Bolanos B, Almaden C, Nagata A, Ryan K, Feng J, Dalvie D, Kath JC, Xu M, Wani R, Murray BW, 2014. Covalent EGFR inhibitor analysis reveals importance of reversible interactions to potency and mechanisms of drug resistance. Proc. Natl. Acad. Sci. U. S. A 111, 173–178. 10.1073/pnas.1313733111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon DA, Weerapana E, 2015. Covalent protein modification: the current landscape of residue-specific electrophiles. Curr. Opin. Chem. Biol 24, 18–26. 10.1016/j.cbpa.2014.10.021 [DOI] [PubMed] [Google Scholar]
- Shraga A, Olshvang E, Davidzohn N, Khoshkenar P, Germain N, Shurrush K, Carvalho S, Avram L, Albeck S, Unger T, Lefker B, Subramanyam C, Hudkins RL, Mitchell A, Shulman Z, Kinoshita T, London N, 2019. Covalent Docking Identifies a Potent and Selective MKK7 Inhibitor. Cell Chem. Biol 26, 98–108.e5. 10.1016/j.chembiol.2018.10.011 [DOI] [PubMed] [Google Scholar]
- Singh J, Petter RC, Baillie TA, Whitty A, 2011. The resurgence of covalent drugs. Nat. Rev. Drug Discov. 10, 307–317. 10.1038/nrd3410 [DOI] [PubMed] [Google Scholar]
- Speers AE, Cravatt BF, 2005. A tandem orthogonal proteolysis strategy for high-content chemical proteomics. J. Am. Chem. Soc 127, 10018–10019. 10.1021/ja0532842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelow JM, 2017. A Perspective on the Kinetics of Covalent and Irreversible Inhibition. SLAS Discov. Adv. Life Sci. RD 22, 3–20. 10.1177/1087057116671509 [DOI] [PubMed] [Google Scholar]
- Tamura T, Hamachi I, 2019. Chemistry for Covalent Modification of Endogenous/Native Proteins: From Test Tubes to Complex Biological Systems. J. Am. Chem. Soc 141, 2782–2799. 10.1021/jacs.8b11747 [DOI] [PubMed] [Google Scholar]
- Tamura T, Ueda T, Goto T, Tsukidate T, Shapira Y, Nishikawa Y, Fujisawa A, Hamachi I, 2018. Rapid labelling and covalent inhibition of intracellular native proteins using ligand-directed N-acyl-N-alkyl sulfonamide. Nat. Commun 9, 1870 10.1038/s41467-018-04343-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinworth CP, Lithgow H, Dittus L, Bassi ZI, Hughes SE, Muelbaier M, Dai H, Smith IED, Kerr WJ, Burley GA, Bantscheff M, Harling JD, 2019. PROTAC-Mediated Degradation of Bruton’s Tyrosine Kinase Is Inhibited by Covalent Binding. ACS Chem. Biol 14, 342–347. 10.1021/acschembio.8b01094 [DOI] [PubMed] [Google Scholar]
- Wagner A, Le TA, Brennich M, Klein P, Bader N, Diehl E, Paszek D, Weickhmann AK, Dirdjaja N, Krauth-Siegel RL, Engels B, Opatz T, Schindelin H, Hellmich UA, 2019. Inhibitor-Induced Dimerization of an Essential Oxidoreductase from African Trypanosomes. Angew. Chem. Int. Ed 58, 3640–3644. 10.1002/anie.201810470 [DOI] [PubMed] [Google Scholar]
- Ward CC, Kleinman JI, Brittain SM, Lee PS, Chung CYS, Kim K, Petri Y, Thomas JR, Tallarico JA, McKenna JM, Schirle M, Nomura DK, 2019. Covalent Ligand Screening Uncovers a RNF4 E3 Ligase Recruiter for Targeted Protein Degradation Applications. ACS Chem. Biol 10.1021/acschembio.8b01083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CC, Kleinman JI, Brittain SM, Lee PS, Chung CYS, Kim K, Petri Y, Thomas JR, Tallarico JA, McKenna JM, Schirle M, Nomura DK, 2018. Covalent Ligand Screening Uncovers a RNF4 E3 Ligase Recruiter for Targeted Protein Degradation Applications. bioRxiv. 10.1101/439125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa M, Itoh T, Hori T, Kato A, Anami Y, Yoshimoto N, Yamamoto K, 2018. Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands. J. Med. Chem 61, 6339–6349. 10.1021/acs.jmedchem.8b00774 [DOI] [PubMed] [Google Scholar]
- Yver A, 2016. Osimertinib (AZD9291)—a science-driven, collaborative approach to rapid drug design and development. Ann. Oncol 27, 1165–1170. 10.1093/annonc/mdw129 [DOI] [PubMed] [Google Scholar]
- Zambaldo C, Daguer J-P, Saarbach J, Barluenga S, Winssinger N, 2016. Screening for covalent inhibitors using DNA-display of small molecule libraries functionalized with cysteine reactive moieties. MedChemComm 7, 1340–1351. 10.1039/C6MD00242K [DOI] [Google Scholar]
- Zhang T, Kwiatkowski N, Olson CM, Dixon-Clarke SE, Abraham BJ, Greifenberg AK, Ficarro SB, Elkins JM, Liang Y, Hannett NM, Manz T, Hao M, Bartkowiak B, Greenleaf AL, Marto JA, Geyer M, Bullock AN, Young RA, Gray NS, 2016. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat. Chem. Biol 12, 876–884. 10.1038/nchembio.2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Crowley VM, Wucherpfennig TG, Dix MM, Cravatt BF, 2019. Electrophilic PROTACs that degrade nuclear proteins by engaging DCAF16. Nat. Chem. Biol 15, 737–746. 10.1038/s41589-019-0279-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Crowley VM, Wucherpfennig TG, Dix MM, Cravatt BF, 2018. Electrophilic PROTACs that degrade nuclear proteins by engaging DCAF16. bioRxiv. 10.1101/443804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Zhang X, Li H, 2018. Beyond histone acetylation-writing and erasing histone acylations. Curr. Opin. Struct. Biol 53, 169–177. 10.1016/j.sbi.2018.10.001 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Bourne PE, 2018. Progress with covalent small-molecule kinase inhibitors. Drug Discov. Today 23, 727–735. 10.1016/j.drudis.2018.01.035 [DOI] [PubMed] [Google Scholar]
- Zimmermann G, Rieder U, Bajic D, Vanetti S, Chaikuad A, Knapp S, Scheuermann J, Mattarella M, Neri D, 2017. A Specific and Covalent JNK-1 Ligand Selected from an Encoded Self-Assembling Chemical Library. Chem. Weinh. Bergstr. Ger 23, 8152–8155. 10.1002/chem.201701644 [DOI] [PMC free article] [PubMed] [Google Scholar]