SUMMARY
An abnormal number of chromosomes, or aneuploidy, accounts for most spontaneous abortions, causes developmental defects, and is associated with aging and cancer. The molecular mechanisms by which aneuploidy disrupts cellular function remain largely unknown. Here, we show that aneuploidy disrupts the morphology of the nucleus. Mutations that increase the levels of long-chain bases suppress nuclear abnormalities of aneuploid yeast independent of karyotype identity. Quantitative lipidomics indicates that long-chain bases are integral components of the nuclear membrane in yeast. Cells isolated from patients with Down syndrome also show that abnormal nuclear morphologies and increases in long-chain bases not only suppress these abnormalities but also improve their fitness. We obtained similar results with cells isolated from patients with Patau or Edward syndrome, indicating that increases in long-chain bases improve the fitness of aneuploid cells in yeast and humans. Targeting lipid biosynthesis pathways represents an important strategy to suppress nuclear abnormalities in aneuploidy-associated diseases.
In Brief
The cellular defects associated with aneuploidy are not well defined. Hwang et al. show that aneuploid yeast and human cells have abnormal nuclear morphology. Targeting ceramide synthesis suppresses nuclear abnormalities and improves the proliferation of aneuploid cells, including cells isolated from patients with Down syndrome.
Graphical Abstract
INTRODUCTION
The incidence of aneuploidy in human germ cells increases with age, leading to a higher risk of spontaneous abortions, stillbirths, and infants born with chromosomal abnormalities, including trisomies for chromosomes 13, 18, or 21, which cause Patau, Edward, or Down syndrome, respectively (Edwards et al., 1960; Lejeune et al., 1959; Nagaoka et al., 2012; Patau et al., 1960). Among these, only patients with Down syndrome live to adulthood but show cognitive disabilities and several pathological conditions associated with premature aging (Antonarakis, 2017). About 1 out of every 700 babies are born with Down syndrome each year, making this syndrome the most common genetic disease among humans (https://www.cdc.gov). While it is thought that pathologies associated with Down syndrome are driven by the expression and activity of genes present on chromosome 21, it has proven difficult to show that an extra copy of a specific gene is solely responsible for a given phenotype in patients with Down syndrome (Antonarakis, 2017). An alternative, yet not mutually exclusive, hypothesis is that cellular defects associated with trisomy 21 may be caused by the disruption of cellular homeostasis due to the presence of the extra chromosome, that is, the aneuploid status of the cell. However, cellular defects in human trisomies driven by the presence of the extra chromosome independent of the genes encoded within it remain unknown. Thus, strategies to ameliorate clinical symptoms in patients with Down syndrome associated with aneuploidy do not exist.
To study the physiological consequences of aneuploidy at the cellular level, we generated a series of isogenic yeast strains, each harboring an extra copy of a different chromosome (called disomes) (Torres et al., 2007). Previous studies revealed several aneuploidy-associated phenotypes in the disomes independent of the identity of the extra chromosome (Dephoure et al., 2014; Sheltzer et al., 2011; Torres et al., 2007, 2010). These include lowered viability, altered metabolism, genomic instability, and loss of protein homeostasis. Importantly, these phenotypes are also present in aneuploid human cell lines and trisomic mouse embryonic fibroblasts (MEFs), indicating that the cellular responses to aneuploidy are conserved in yeast and humans (Donnelly et al., 2014; Passerini et al., 2016; Santaguida et al., 2015; Stingele et al., 2013; Williams et al., 2008). Loss of protein homeostasis is mainly driven by the mRNA expression of the genes present on the extra chromosomes, which in turn leads to increased protein synthesis, folding, and turnover. In support of this hypothesis, aneuploid cells are sensitive to drugs that inhibit protein degradation pathways. However, increasing protein degradation by the loss of the deubiquitinating enzyme UBP6 improves the fitness of aneuploid yeast cells independent of the identity of the extra chromosome (Dephoure et al., 2014). Thus, targeting protein degradation pathways is a strategy to specifically affect the fitness of aneuploid cells.
Aneuploidy is thought to affect cellular metabolism due to the synthesis of biomolecules and energy demands associated with increased protein synthesis. Aneuploid yeast cells show increased glucose utilization and strictly rely on the biosynthesis of the amino acid serine, a key molecule that is used for the synthesis of nucleotides, proteins, and lipids (Hwang et al., 2017; Torres et al., 2007). Although the metabolic requirements of human trisomies are not well characterized, a conserved metabolic pathway that is affected by aneuploidy in both yeast and mammalian cells is the biosynthesis of sphingolipids (Hwang et al., 2017; Tang et al., 2017). Aneuploid yeast, trisomic MEFs, and aneuploid colon cancer cells show dysregulation of sphingolipid synthesis and are sensitive to drugs that lead to the accumulation of ceramides. Importantly, the physiological consequences or cellular processes affected by altered sphingolipid levels in aneuploid yeast cells, MEFs, or cancer cells are not understood.
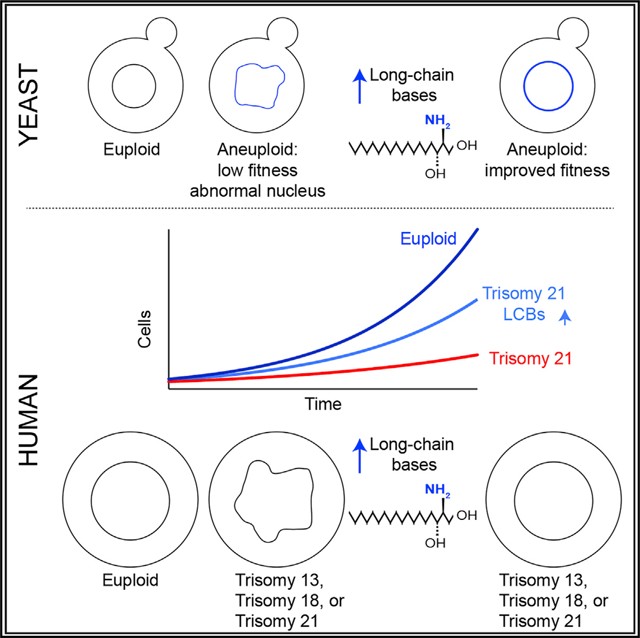
The mechanisms that cause lower viability or genomic instability in aneuploid cells are also not well understood. Furthermore, how the presence of an extra chromosome affects organelle function in yeast or human cells has not been studied. Here we found that aneuploidy disrupts nuclear integrity in yeast and primary fibroblasts from patients with Patau, Edward, or Down syndrome. Remarkably, impairing ceramide synthesis, which leads to the accumulation of long-chain bases, improves fitness and suppresses nuclear abnormalities in both aneuploid yeast and human cells. Our results indicate that long-chain bases are integral structural components of the nuclear membrane and their levels are important in maintaining nuclear integrity. Because abnormal nuclear morphology is a hallmark of aging, the discovery that the presence of an extra chromosome disrupts nuclear morphology raises the hypothesis that aneuploidy drives pathologies associated with premature aging in patients with Down syndrome.
RESULTS
Aneuploidy Disrupts Nuclear Morphology in Yeast
The effects of aneuploidy on the morphology and integrity of the nucleus have not been studied. We previously showed that the volume of the cell increases in the disomes and that these increases correlate with the size of the extra chromosome (Torres et al., 2007). These results raise the possibility that the volume of the nucleus is also affected by the presence of an extra chromosome. To investigate this, we measured nuclear volumes in the disomes by tagging the inner nuclear membrane protein Heh1 with GFP at its endogenous locus. Using live-cell microscopy, we analyzed the surface area of nuclei by measuring the perimeter of the nucleus of wild-type cells and the disomes. We found that the nuclei of the disomes display significant increases in surface area compared with wild-type cells (Figures 1A and 1B). These results indicate that the volume of the nucleus is affected by the presence of an extra chromosome.
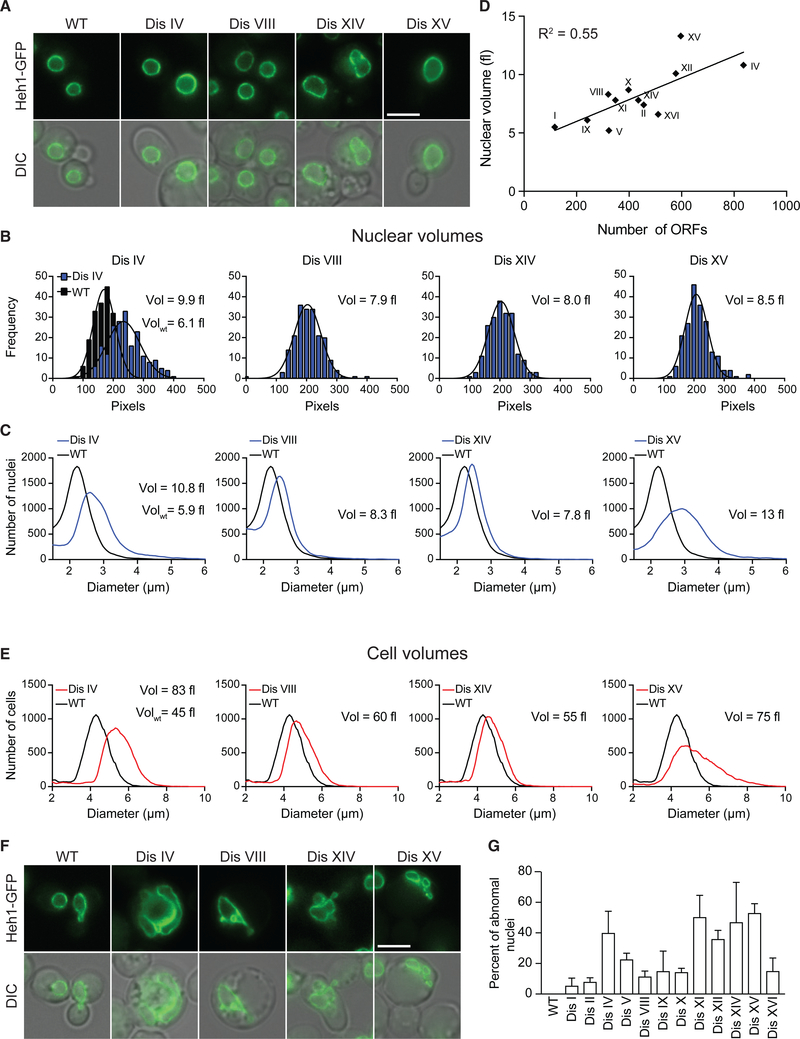
Figure 1. Aneuploidy Disrupts the Nuclear Morphology of Yeast.
(A) Representative live-cell images of wild-type cells and 4 disomes expressing Heh1 tagged with GFP (Heh1-GFP) at its endogenous locus showing round nuclei. Differential interference contrast (DIC) microscopy is shown below. Scale bar, 5 μm.
(B) Histograms of the surface area of nuclei from aneuploid yeast strains compared with wild-type cells (n = 200). Nuclear volumes calculated from surface area measurements of images from live-cell microscopy are shown (see STAR Methods).
(C) Representative Coulter counter profiles of the diameters of purified nuclei from wild-type cells and 4 disomes (n = 30,000).
(D) Linear correlation of nuclear volumes measured using a Coulter counter and the number of open reading frames in the duplicated chromosomes (R2 = 0.55).
(E) Representative Coulter counter histograms of the diameters of wild-type cells and 4 disomes (n = 30,000).
(F) Representative live-cell images of wild-type cells and 4 disomes showing abnormal nuclear morphology. DIC microscopy is shown below. Scale bar, 5 μm.
(G) Percentage of cells showing abnormal nuclear morphologies in aneuploid yeasts, shown as mean ± SD (n = 200, 3 biological replicates).
To validate these results, we isolated nuclei from yeast cells and measured their volume in solution. Briefly, yeast cells were lysed enzymatically, and nuclei were isolated in a sucrose gradient and reconstituted in PBS (see STAR Methods). Of note, we avoided the use of detergents, because they would undoubtedly interfere with the lipid composition and structure of the nuclear membrane. Using a cell counter analyzer (Coulter counter), we confirmed that nuclear volumes of yeast cells increase by the presence of an extra chromosome (Figures 1C, 1D, and S1A). Interestingly, increases in nuclear volumes of the disomes show a linear correlation with the number of genes encoded in the extra chromosome (Figure 1D; R2 = 0.55). Furthermore, nuclear volumes correlate with increases in cell volume, indicating that the nuclear-to-cytoplasmic ratio of aneuploid cells remained constant (Figures 1E and S1B).
Strikingly, during the analysis of nuclear volumes, we found that the nuclear morphology of a substantial portion of the population in all aneuploid yeast strains was severely compromised (from 5% in disome I to 53% in disome XV; Figures 1F, 1G, and S1C). Whereas wild-type cells show round, smooth, and continuous nuclear morphologies, the shape of the nucleus of the disomes is highly irregular and discontinuous, indicating that the presence of an extra chromosome disrupts the integrity of the nuclear membrane. These results revealed that aneuploidy causes defects in nuclear morphology independent of the identity of the extra chromosome. Furthermore, these data suggest that abnormal nuclear morphology may contribute to several cellular phenotypes associated with aneuploidy, including lower viability, genomic instability, and shortened lifespan (Sheltzer et al., 2011; Sunshine et al., 2016). Abnormal morphology of the nucleus constitutes an aneuploidy-associated phenotype that has not been previously described.
Interestingly, yeast cells that harbor yeast artificial chromosomes (YACs) containing heterochromatic human DNA show minor increases in cell or nuclear volumes and show typical nuclear morphologies (Figure S1A). These results indicate that the presence of extra DNA is not sufficient to disrupt the nucleus in yeast cells.
Mutations that Elevate the Levels of LCBs Suppress Nuclear Abnormalities of Aneuploid Yeasts
We previously identified mutations that improve the fitness of aneuploid yeast cells irrespective of the identity of the extra chromosome (Torres et al., 2010). Among these were several mutations in genes that regulate the biosynthesis of long-chain bases (LCBs: DHS [dihydrosphingosine] and PHS [phytosphingosine], also called sphingoid bases) and ceramide, key molecules essential for viability and proliferation. One of the mutations that improved the fitness of all aneuploid yeast strains analyzed was the loss of function of ceramide synthase LAG1 (Hwang et al., 2017). Ceramide synthase converts LCBs into ceramides; therefore, lower enzymatic activity leads to the accumulation of LCBs (Figure 2A). Using quantitative mass spectrometry, we first confirmed that loss of LAG1 minimally affects the levels of ceramides while the levels of PHS increased by 7.1-fold in yeast cells (3.9 pmol/106 cells in WT to 28.2 pmol/106 cells in lag1Δ; Figure 2B; Table S1). Ceramide levels remain mostly unchanged due to the activity of LAC1, the other ceramide synthase paralog in yeast. Consistently, deletion of both LAG1 and LAC1 causes lethality in yeast.
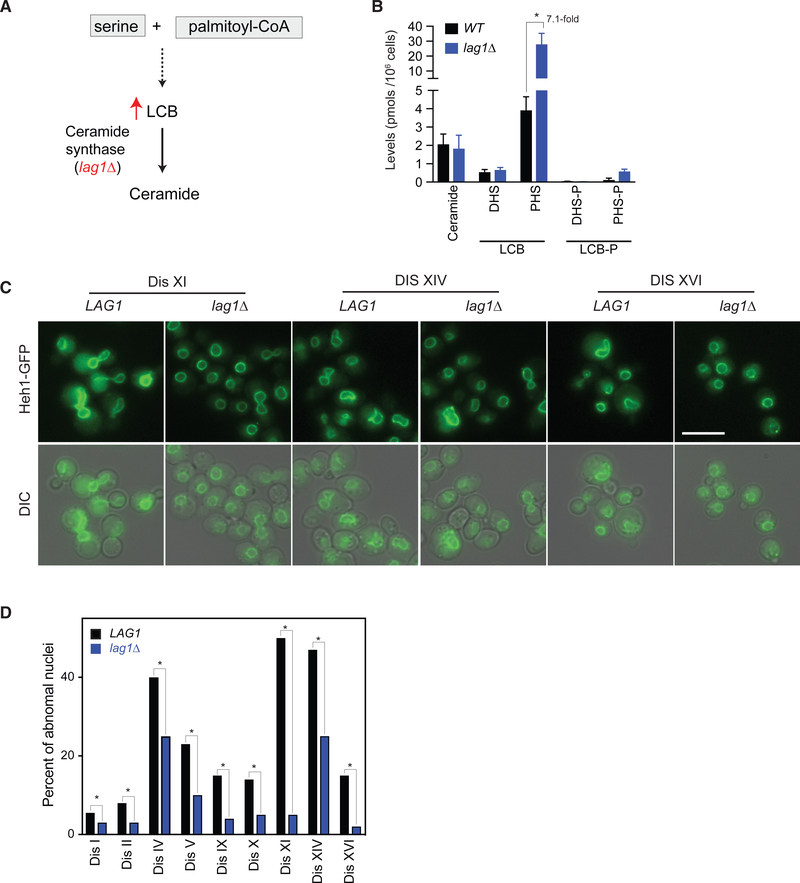
Figure 2. Increasing the Levels of Long-Chain Bases Suppresses the Nuclear Abnormalities of Aneuploid Yeast.
(A) Simplified schematic of the biochemical pathway of the synthesis of long-chain bases (LCBs) and ceramide.
(B) Quantitative mass spectrometry ofthe levels of LCBs and ceramides in wild-type cells and cells harboring lag1Δ (error bars represent SD, n = 3 samples). DHS, dihydrosphingosine; PHS, phytosphingosine; DHS-P, phosphorylated DHS; PHS-P; phosphorylated PHS.
(C) Representative live-cell images of disomes XI, XIV, and XVI expressing Heh1-GFP and harboring lag1Δ. Scale bar, 10 μm.
(D) Comparison of nuclear abnormalities of disomes versus disomes harboring lag1Δ (n = 200 cells, *p < 0.01, unpaired two-tailed Student’s t test).
Interestingly, transcriptional analyses revealed that higher LCB levels in aneuploid cells result in the upregulation of a cluster of genes associated with components of the nucleus (Hwang et al., 2017). We therefore investigated whether the nuclear abnormalities we observed in aneuploid yeasts are affected by the loss of LAG1. We found that irrespective of the identity of the extra chromosome, nuclear defects of aneuploid yeast strains are significantly suppressed by lag1Δ (Figures 2C and 2D). These results indicate that increases in LCB levels improve the fitness of aneuploid cells due to the suppression of nuclear abnormalities and raise the possibility that changes in LCB levels upon the loss of ceramide synthase accumulate in the nuclear membrane.
To validate these results, we analyzed nuclear morphologies of disomes harboring the deletion of sphingosine kinase LCB4 (lcb4Δ), which also significantly improves the fitness of aneuploid yeasts independent of karyotype (Hwang et al., 2017). This mutation raises LCB levels by impairing their phosphorylation, which is required for their degradation by the lipid lyase DPL1 (Figure S2A). Consistently, we found that loss of LCB4 also suppresses nuclear defects in the disomes (Figure S2B). Because lcb4Δ reduces phosphorylated LCBs and does not affect ceramide levels, our results indicate that increases in LCBs are responsible for improving the fitness of aneuploid yeasts.
A third mutation in the sphingolipid pathway that improves the fitness of the disomes independent of karyotype is the loss of sphingosine-1-phosphate phosphatase, or LCB3. Loss of LCB3 leads to increased levels of LCBs and their phosphorylated forms while minimally affecting ceramide levels. Consistently, we found that loss of LCB3 also improves the nuclear morphology of the disomes (Figure S2C). Although the loss of the deubiquitinating enzyme UBP6 improves the fitness of aneuploid strains by enhancing protein degradation, this mutation does not suppress the nuclear abnormalities of aneuploid cells (Figure S2D). These results indicate that suppression of nuclear defects is specific to changes in the levels of LCBs and that the mechanisms by which loss of UBP6 improve the fitness of aneuploid cells are distinct. Indeed, changes in LCB levels show additive effects with the loss of UBP6 to improve the fitness of aneuploid cells (Hwang et al., 2017). Interestingly, we found that while increasing LCB levels suppresses nuclear defects in the disomes, these lipids do not affect the cell or nuclear volumes of the disomes (Figures S2E and S2F). Our findings support the hypothesis that increases in LCB levels play a critical role in improving the integrity of the nuclear membrane in aneuploid cells. Furthermore, they provide a mechanism by which different mutations in the sphingolipid biosynthetic pathway, all of which elevate LCB levels, improve the fitness of aneuploid cells independent of the identity of the extra chromosome.
De Novo Synthesis of LCBs Is Required to Maintain Nuclear Integrity in Yeast
Our results raise the possibility that the biosynthesis of LCBs plays an essential role in maintaining nuclear morphology in yeast cells. To investigate this, we analyzed the effects of targeting the de novo synthesis of sphingolipids in wild-type cells. Serine palmitoyltransferase (SPT), the first and rate-limiting enzyme in the de novo synthesis of LCBs, consists of 2 essential subunits, LCB1 and LCB2, and a third non-essential small regulatory subunit, TSC3 (Figure S2A). This complex localizes to the nuclear envelope (NE) and the endoplasmic reticulum (ER) (Figure 3A). We found that inhibition of SPT by myriocin severely disrupts the nuclear morphology of yeast cells and that these effects do not occur upon translation inhibition by rapamycin (Figures 3B and 3C). Consistently, loss of TSC3, which impairs SPT activity and lowers LCB levels, also disrupted nuclear morphology and integrity (Figures 3D–3G and S3A–S3D). These defects were significantly attenuated by the addition of exogenous LCBs, indicating that impaired sphingolipid synthesis is responsible for the nuclear abnormalities of tsc3Δ (Figures 3F and 3G). Using electron microscopy, we found that the well-defined double lipid bilayer that constitutes the nuclear membrane in wild-type cells is significantly affected in cells harboring tsc3Δ (Figures 3H and S3E). These results show that SPT activity is necessary to maintain the integrity of the nuclear membrane in yeast.
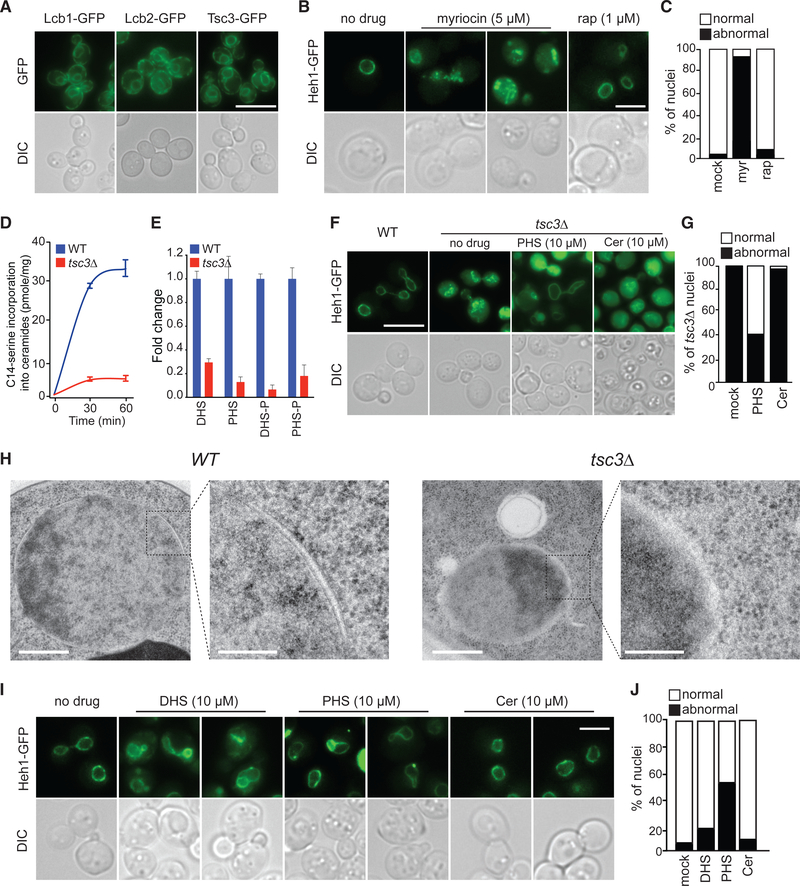
Figure 3. De Novo Synthesis of Long-Chain Bases Is Essential for Nuclear Integrity in Yeast.
(A) Representative images of cells expressing Lcb1, Lcb2, or Tsc3 tagged with GFP. Scale bar, 10 μm.
(B) Representative images of wild-type cells expressing Heh1-GFP treated with myriocin or rapamycin for 3 h. Scale bar, 10 μm.
(C) Percentage of abnormal nuclei of wild-type cells treated with myriocin or rapamycin (n = 200).
(D) Kinetics of the serine palmitoyltransferase (SPT) enzymatic activity in wild-type cells and tsc3Δ (see STAR Methods). Error bars represent SD (n = 3).
(E) Fold change of the levels in LCBs upon tsc3Δ compared with wild-type cells, shown as mean ± SD (n = 3 biological replicates).
(F) Representative live-cell images of tsc3Δ cells expressing Heh1-GFP alone or treated with PHS or ceramide for 3 h. Scale bar, 10 μm.
(G) Percentage of nuclear abnormalities of tsc3Δ upon PHS or ceramide treatment (n = 200 cells).
(H) Representative electron micrograph images of the nuclear envelope in wild-type yeast and cell harboring tsc3Δ. Scale bar, 500 nm; zoom scale bar, 100 nm.
(I) Representative images of wild-type cells expressing Heh1-GFP treated with DHS, PHS, or ceramide for 3 h. Scale bar, 5 μm.
(J) Percentage of abnormal nuclei in wild-type cells expressing Heh1-GFP treated with DHS, PHS, or ceramide (n = 200 cells).
Next, we tested the effects of the addition of excess LCBs to the growth medium on the morphology of the nucleus. Wildtype cells cultured in medium containing high concentrations of LCBs display deformed nuclei with significant membrane expansions (Figures 3I and 3J). However, excess ceramide did not affect the morphology of yeast nuclei or suppress nuclear abnormalities of tsc3Δ (Figures 3F, 3G, 3I, and 3J). These results indicate that the cellular levels of LCBs, not ceramides, play an essential role in maintaining the morphology of the nucleus in yeast. Thus, robust regulatory mechanisms must exist to maintain homeostatic levels of LCBs in the cell, because either decreasing or increasing these levels can significantly affect the morphology of the nucleus.
LCBs, Not Ceramides, Are Enriched in the Nuclear Membrane
The results showing that the morphology of the nucleus is sensitive to changes in LCB levels suggest that these lipids may play a fundamental role in the integrity of the nuclear membrane. To determine whether LCBs are integral components of the nuclear membrane, we measured their levels in the nucleus using quantitative mass spectrometry. To this end, we purified nuclei without detergents in a sucrose gradient. Using a particle analyzer and microscopy, we confirmed that the nuclear preparations were homogeneous and did not include whole or broken cells or cell debris (Figures 1C and 4A). Using the reticulon Rtn1 as a marker for the ER, we estimated that about 20% of the ER membrane co-purified with the nuclei (Figure 4B). By comparing the amounts of LCBs in nuclei versus whole cells, we found that up to 50% of cellular LCBs localize to the nucleus (Figure 4C, and 2.3 of 4.6 pmol/106 cells; Table S1).
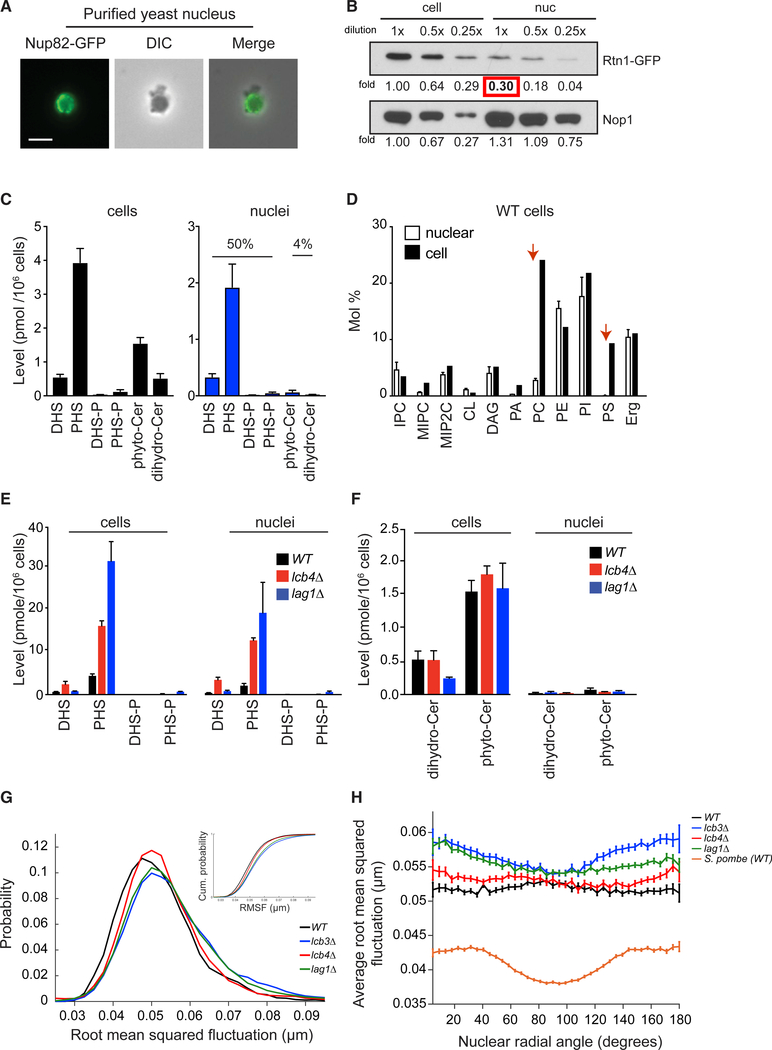
Figure 4. Long-Chain Bases, Not Ceramides, Are Enriched in the Yeast Nucleus.
(A) Representative image of a purified nucleus from yeast cells. Scale bar, 2 μm.
(B) Western blots of the reticulon protein Rtn1 tagged with GFP. Nop1 is a nucleolar protein used for loading control.
(C) Levels of LCBs (DHS, PHS, DHS-P, and PHS-P) and ceramides (phytoceramide and dihydroceramide) in whole yeast cells (left, black bars) and in nuclei (right, blue bars) are shown as mean ± SD (n = 3).
(D) Global lipidomics of whole cells and purified nuclei in yeast. IPC, inositol-P-ceramide; MIPC, mannosyl-inositol-phosphorylceramide; M(IP)2C, mannosyl-diinositol-phosphorylceramide; CL, cardiolipin; DAG, diacylglycerol; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine; Erg, ergosterol. Means ± SD are shown (n = 3).
(E) Levels of LCBs in lcb4Δ and lag1Δ cells and purified nuclei are shown as mean ± SD (n = 3).
(F) Levels of ceramides in lcb4Δ or lag1Δ cells and purified nuclei are shown as mean ± SD (n = 3).
(G) Probability distribution of root-mean-square (average) nuclear envelope fluctuation sizes for wild-type, lcb3Δ, lcb4Δ, and lag1Δ cells (cumulative probability distribution in subset), n = ~75 for each cell type.
(H) Average root-mean-square nuclear envelope fluctuation size as a function of nuclear radial angle for the same population of cells (S. pombe nuclear envelope fluctuations are shown for comparison).
In comparison, only 4% of total ceramides are found in the nucleus (Figure 4C, and 0.09 of 2.1 pmol/106 cells). These results indicate that LCB levels are ~25-fold higher than ceramide levels in the nucleus. Considering a nuclear volume of 6 fL, we estimate the concentration of nuclear LCBs to be around 400 μM, indicating that LCBs are abundant in the nuclear membrane. Notably, global lipidomics did not reveal enrichment for any other major class of lipids in the yeast nucleus (Figure 4D; Table S2). Instead, it demonstrated that the nuclear levels of phosphatidylserine (PS) and phosphatidylcholine (PC) are significantly lower than expected when compared with their levels in whole cells. PS and PC constitute only 0.1% and 2.7% of the nuclear lipids, respectively, compared with 10% or 24% of the cellular lipids, respectively (Figure 4D). These results indicate that LCBs are enriched in the nuclear membrane and that ceramides, the products of ceramide synthase, an enzyme that localizes to the NE and ER, are rapidly transported out of the nucleus. Furthermore, our results support the hypothesis that the enrichment of LCBs in the yeast nuclear membrane has an essential structural role in maintaining membrane integrity.
Mutations that Improve the Fitness of Aneuploid Cells Increase the Levels of LCBs in the Nucleus
In addition to suppressing the nuclear defects of aneuploid cells, loss of ceramide synthase extends the lifespan of yeast (D’mello etal., 1994). Our results raise the possibility that lag1Δ could promote longevity by maintaining nuclear integrity due to elevated levels of LCBs in the nuclear membrane. To investigate whether lag1Δ affects the lipid composition of the nucleus, we performed quantitative lipidomics of whole cells and purified nuclei harboring lag1Δ. We found that most increases in the levels of LCBs elicited by the loss of LAG1 accumulate in the nucleus (Figures 4E and S4A; Table S1). Nuclear DHS and PHS levels increase 2.1- and 9.2-fold, respectively, in lag1Δ compared with wild-type cells. In addition, we found that up to 65% of total LCBs are in the nucleus in lag1Δ cells (19 of 29 pmol/106 cells). As with total ceramides, the nuclear levels of ceramides are minimally affected by the loss of LAG1 (Figures 4F and S4A).
Similar results were obtained when nuclear lipids were quantified from cells harboring lcb4Δ (Figures 4E, 4F, and S4A). Nuclear DHS and PHS levels increase 9.4- and 6.1-fold, respectively, in lcb4Δ compared with wild-type cells, and up to 88% of total LCBs are in the nucleus (15 of 17 pmol/106 cells). Total or nuclear levels of ceramides are not significantly affected by lcb4Δ (Figure 4F). Furthermore, global lipidome analysis showed that neither the loss of LAG1 nor that of LCB4 affects the nuclear levels of any other major class of lipids (Figure S4B; Table S2). We conclude that these mutations increase the levels of LCBs in the nucleus to ameliorate nuclear defects associated with aneuploidy. In addition, these results support the hypothesis that raising the levels of LCBs in the nucleus plays an important role in extending the lifespan of cells harboring lag1Δ.
Global quantitative lipidomics showed that the levels of both LCBs and ceramides are elevated in aneuploid yeasts compared with wild-type cells (Hwang etal., 2017). Interestingly, increases in LCB levels in aneuploid yeasts range from 1.2-fold in disome I to 2.5-fold in disome IV, raising the possibility that LCB levels are altered in the nucleus of the disomes. Indeed, quantitative lipidomics of purified nuclei of 3 representative disomes revealed that the increases in LCBs accumulate in the nucleus (Figure S4C). Aneuploid cells rely on the increases’ synthesis of LCBs for viability; however, these increases are not sufficient to completely prevent their nuclear abnormalities. Loss of LAG1 or LCB4 improves the fitness of aneuploid cells by further increasing nuclear levels of LCBs without the toxic accumulation of ceramides.
Higher Levels of LCBs Increase Nuclear Envelope Fluctuations In Vivo
Because increases in LCBs do not affect the volume of the nucleus, we decided to assess whether LCBs alter physical properties of the nuclear membrane in vivo. To this end, we used an assay to measure NE fluctuations using high-resolution fluorescence microscopy of live cells (Schreiner et al., 2015; Williams et al., 2019). In short, to visualize the NE, we tagged Nup82 with GFP at its endogenous locus in wild-type cells and cells harboring lag1Δ. To quantitatively determine nuclear fluctuations, we reconstructed a 3D contour of the NE, and using spherical coordinates centered on the nucleus at each time point, each NE fluctuation was defined as the difference between the radial coordinate of the contour and its time-average radial value. The frequency and magnitude of the fluctuations were determined by computing the root-mean-square fluctuation (RMSF). Comparison of the distribution of RMSF from a population of nuclei over time clearly shows that nuclei from lag1Δ cells have larger fluctuations compared with nuclei from wild-type cells (Figure 4G). Analysis of the angle at which these fluctuations take place (using a polar coordinate system based on bright-field imaging and an axis defined by bisecting the cell volume centered on the bud location) shows that loss of LAG1 increases NE fluctuations throughout the nuclear membrane (Figure 4H). We obtained similar results when we analyzed NE fluctuations of cells harboring the lcb4 or lcb3 deletions, although lcb4Δ showed smaller changes. These results indicate that all three mutants increase NE fluctuations in yeast and support the hypothesis that higher levels of LCBs in the nuclear membrane lead to increased membrane dynamics. Increased NE fluctuations may reflect higher membrane compliance that may help tolerate the presence of an extra chromosome in the nucleus of aneuploid cells.
De Novo Synthesis of LCBs Is Required to Maintain Nuclear Integrity in Human Cells
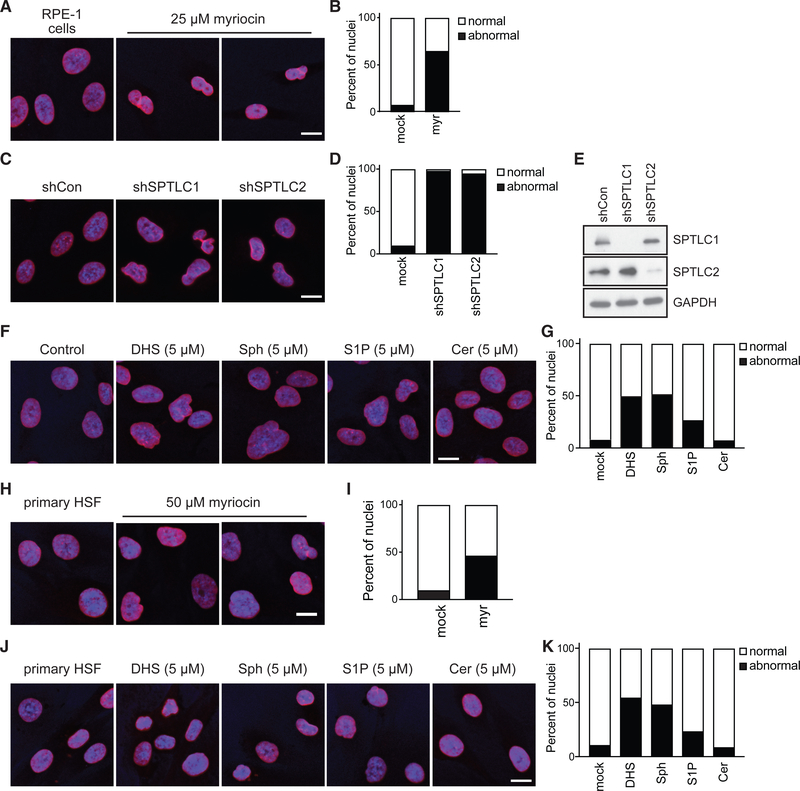
The biochemical pathway of the de novo synthesis of LCBs and ceramide is conserved from yeast to humans. Human SPT consists of 2 subunits, SPTLC1 or SPTLC2/3, and a small subunit, SPTSSA/B, to generate DHS (Figure S5A). Six human genes encode for ceramide synthases, CerS1–CerS6. Expression of each gene is restricted to different tissues, and their activities are thought to confer specificity for lipid molecules of varying length. Sphingosine, the other major LCBs in human cells, is generated by the catabolism of ceramide by ceramidases. To study the effects of changing LCB levels on the morphology of the nucleus in human cells, we initially turned to hTERT-immortalized, non-transformed, human retinal pigment epithelial (RPE-1) cells. These cells are mostly diploid and display homogeneous, smooth, and mostly circular or elliptical nuclear morphologies (Figure 5A). Using immunofluorescence of lamin B1, we found that the nuclear morphology of RPE-1 cells is severely compromised upon SPT inhibition by myriocin (Figures 5A and 5B). Nuclei show abnormal morphologies and significant decreases in volume upon myriocin treatment.
Figure 5. Long-Chain Bases Are Essential for Maintaining Nuclear Integrity in Human Cells.
(A) Representative images of RPE-1 nuclei mock treated and treated with myriocin for 24 h. Immunofluorescence for lamin B1 is red, and the nucleus is blue stained with Hoechst 33342.
(B) Percentage of nuclear abnormalities of RPE-1 cells treated with myriocin (n = 100 cells).
(C) Representative images of RPE-1 nuclei upon knockdown of SPTLC1 or SPTLC2 after 72 h.
(D) Percentage of nuclear abnormalities of RPE-1 cells upon knockdown of SPTLC1 or SPTLC2 (n = 100 cells).
(E) Western blot analysis of SPTLC1 and SPTLC2 protein levels in RPE-1 cells.
(F) Representative images of RPE-1 nuclei treated with DHS, sphingosine (Sph), sphingosine-1-phosphate (S1P), or ceramide (Cer) for 24 h.
(G) Percentage of RPE-1 cells showing abnormal nuclear morphology when treated with DHS, sphingosine (Sph), sphingosine-1-phosphate (S1P), or ceramide (n = 100 cells).
(H) Representative images of primary human skin fibroblast (HSF) nuclei mock treated and treated with myriocin for 24 h.
(I) Percentage of nuclear abnormalities of primary HSF treated with myriocin (n = 100 cells).
(J) Representative images of primary HSF nuclei treated with DHS, sphingosine (Sph), sphingosine-1-phosphate (S1P), or ceramide for 24 h.
(K) Percentage of primary HSF showing abnormal nuclear morphology when treated with DHS, sphingosine (Sph), sphingosine-1-phosphate (S1P), or ceramide (Cer) (n = 100 cells).
All images are representative of three biological replicates. Scale bars (A, C, F, H, and J), 10 μm.
Next, we took a genetic approach and found that knockdown of either of the main SPT subunits (SPTLC1 or SPTLC2) dramatically affects nuclear morphology and decreases nuclear volumes in RPE-1 cells (Figures 5C–5E). As observed in yeast, we found that addition of exogenous LCBs to the growth medium disrupts the morphology of the nucleus in RPE-1 cells, often causing significant increases in volume without affecting karyotype, while ceramides have little effect (Figures 5F, 5G, and S5B). Interestingly, sphingosine-1-phosphate (S1P) also affected the nuclear morphology of RPE-1 cells, but to a lesser degree (Figures 5F and 5G). These results indicate that either lowering or increasing the levels of LCBs affects the morphology of human epithelial cells.
To validate these results, we performed similar experiments with primary human skin fibroblasts isolated from apparently healthy donors (Figures 5H–5K). Consistently, we found that inhibition of SPT by myriocin or addition of excess LCBs to the growth medium disrupts nuclear morphology of primary human fibroblasts. Lastly, we obtained similar results with hTERT-immortalized diploid human foreskin fibroblasts (Figures S5C and S5D). Altogether, these results establish an essential role for LCBs in maintaining the integrity of the nucleus in human epithelial cells and fibroblasts.
Aneuploidy Disrupts Nuclear Morphology of Fibroblasts Isolated from Patients with Down Syndrome
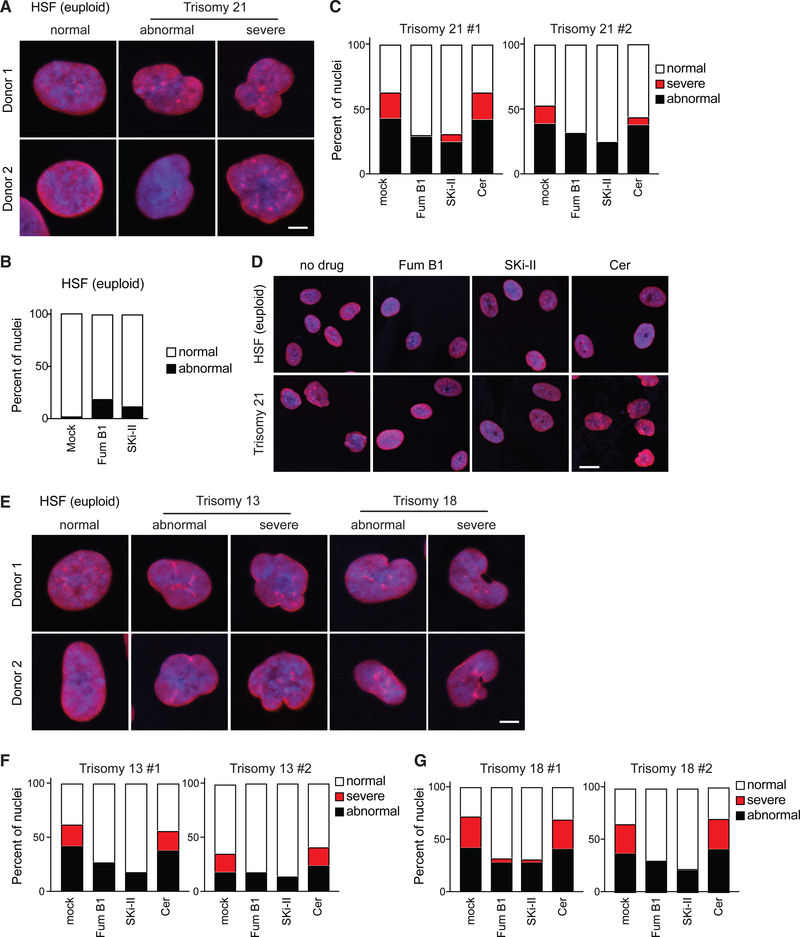
The cellular defects driven by aneuploidy in cells from patients with Down syndrome are not known. Because most aneuploidy-associated phenotypes discovered in yeast cells have subsequently been shown to be present in human aneuploid cells, we hypothesized that aneuploidy might disrupt nuclear morphology in human cells trisomic for chromosome 21. To investigate the effects of aneuploidy on the morphology of the nucleus in human cells, we initially obtained early-passage primary trisomy 21 fibroblasts from two independent donors with Down syndrome (Coriell catalog numbers GM04616 and GM04592) and fibroblasts from two euploid apparently healthy individuals, hereafter called controls (GM00969 and GM08447). To assess nuclear morphology, we cultured all cell lines using standard tissue culture conditions and performed immunofluorescence using an antibody against lamin B1. Remarkably, we found that compared with controls, the nuclei of both trisomy 21 cell lines displayed abnormal nuclear morphologies (Figure 6A). While 98% of the nuclei from control fibroblasts show a smooth circular or elliptical nuclear shapes, up to 60% of nuclei from fibroblasts of patients with Down syndrome show irregular and distorted shapes (Figures 6A–6C).
Figure 6. Long-Chain Bases Suppress Nuclear Abnormalities Associated with Aneuploidy in Human Cells.
(A) Representative images of primary human fibroblasts from 2 euploid donors and 2 patients with Down syndrome. Immunofluorescence for lamin B1 is red, and the nucleus is blue stained with Hoechst 33342. Scale bar, 2.5 μm.
(B) Percentage of fibroblasts from euploid donors showing abnormal nuclear morphology when treated with fumonisin B1 or SKi-II (n = 100).
(C) Percentage of fibroblasts from 2 patients with Down syndrome showing abnormal nuclear morphology treated with fumonisin B1, SKi-II, or ceramide (Cer) (n = 100).
(D) Representative images of trisomy 21 nuclei treated with fumonisin B1, SKi-II, or ceramide. Scale bar, 10 μm.
(E) Representative images of primary human fibroblasts from 2 euploid donors and 2 patients with Patau syndrome(trisomy 13) or Edward syndrome (trisomy 18). Immunofluorescence for lamin B1 is red, and the nucleus is blue stained with Hoechst 33342. Scale bar, 2.5 μm.
(F) Percentage of fibroblasts from 2 patients with Patau syndrome showing abnormal nuclear morphology treated with fumonisin B1, SKi-II, orceramide (n = 100).
(G) Percentage of fibroblasts from 2 patients with Edward syndrome showing abnormal nuclear morphology treated with fumonisin B1, Ski-II, or ceramide (n = 100).
Interestingly, as seen in aneuploid yeast cells, we observe significant phenotypic heterogeneity in nuclear morphologies of trisomy 21 cells, with some cells showing normal morphologies (40%–50%), others showing a mildly affected nucleus (abnormal, 40%–45%), and a third population showing dramatic changes in nuclear morphology (severe, 15%–20%) (Figures 6A and 6C). We hypothesize that this heterogeneity is associated with the phenotypic variability and non-genetic individuality previously described as hallmarks of aneuploid cells (Beach et al., 2017; Pavelka et al., 2010). Indeed, analyses of nuclear morphologies of fibroblasts from five other patients with Down syndrome show abnormal nuclear morphologies to varying degrees independent of the age or sex of the donor (Figure S6A). These results indicate that the presence of an extra copy of chromosome 21 causes nuclear abnormalities in human fibroblasts and supports the hypothesis that aneuploidy disrupts nuclear morphology in yeast and humans.
An abnormal nuclear morphology is a hallmark of premature aging (Scaffidi and Misteli, 2006). Because several clinical symptoms in patients with Down syndrome are associated with premature aging, we hypothesize that the nuclear morphology of trisomy 21 fibroblasts may resemble nuclei of cells from patients with progeroid syndromes. Therefore, we obtained primary fibroblasts from 2 patients with Hutchinson-Gilford progeria syndrome (HGPS) through the Progeria Research Foundation Cell and Tissue Bank (catalog numbers HGADFN167 and HGADFN367). Analysis of nuclear morphologies revealed that nuclei of trisomy 21 cells display abnormalities strikingly similar to those observed in nuclei from fibroblasts of HGPS patients (Figures S6B and S6C). Therefore, it is plausible that an abnormal nuclear morphology is associated with premature aging phenotypes of patients with Down syndrome.
Inhibition of Ceramide Synthase Suppresses Nuclear Abnormalities of Fibroblasts from Patients with Down Syndrome
Our findings showing that lag1Δ suppresses the nuclear abnormalities of aneuploid yeast raise the possibility that increasing the levels of LCBs may improve the nuclear defects of fibroblasts from patients with Down syndrome. To investigate this, we turned to fumonisin B1, a toxin that specifically inhibits ceramide synthases. Using quantitative mass spectrometry, we first confirmed that micromolar concentrations of fumonisin B1 cause a significant increase in the levels of LCBs in human cells (Figure S6D; Table S3). Interestingly, DHS, not sphingosine, is elevated by fumonisin B1, indicating that this drug inhibits ceramide synthase activity associated with the de novo synthesis pathway of LCBs.
Whereas fumonisin B1 minimally affected the nuclei of control fibroblasts, it significantly suppressed the nuclear abnormalities of fibroblasts from patients with Down syndrome (Figures 6B–6D). To confirm that elevated levels of LCBs suppress the nuclear abnormalities of trisomy 21 cells, we inhibited LCB degradation by pharmacological inhibition of sphingosine kinases. Consistently, we found that although control cells were minimally affected, the nuclear abnormalities of trisomy 21 cells are significantly suppressed upon treatment with the sphingosine kinase inhibitor SKi-II (Figures 6B–6D). Notably, we found that ceramide does not alter nuclear morphology and does not improve the nuclear shape of trisomy 21 fibroblasts, indicating that the effects are specific to changes in the levels of LCBs (Figures 6C and 6D). Next, we examined whether increasing LCB levels suppressed the nuclear abnormalities of primary fibroblasts from patients with HGPS, which show abnormal nuclear morphologies due to point mutations in lamin A. We found that neither fumonisin B1 or SKi-II had an effect on the nucleus of fibroblasts from patients with HGPS (Figures S6B and S6C). Altogether, these results indicate that aneuploidy disrupts the morphology of the nucleus due to insufficient levels of LCBs. Consequently, increasing cellular levels of LCBs is beneficial to cells from patients with Down syndrome, but not to those from patients with other laminopathies.
Primary Fibroblasts Isolated from Patients with Edward or Patau Syndrome Show Abnormal Nuclear Morphologies
To determine whether abnormal nuclear morphologies of fibroblasts from patients with Down syndrome are specific to trisomy 21 or aneuploidy affects the morphology of human cells independent of the identity of the extra chromosome, we analyzed the nuclear morphologies of other human trisomies. To this end, we obtained primary fibroblast cell lines from two independent donors with Patau syndrome (trisomy 13, GM00526 and GM02948) or Edward syndrome (trisomy 18, GM00734 and GM03538). We found that the nuclei of these cells show abnormal morphologies similar to those of cells from patients with Down syndrome or HGPS. In both trisomies, up to 60% of the cells show abnormal nuclear morphologies (Figures 6E–6G). These results support the hypothesis that the morphology of the nucleus is affected by aneuploidy, independent of the identity of the extra chromosome, in yeast and human. Furthermore, we found that either fumonisin B1 or SKi-II suppressed the nuclear defects of trisomies 13 and 18, whereas ceramides had little effect. These results provide strong evidence that increasing LCB levels suppresses nuclear abnormalities of aneuploid human cells independent of the abnormal karyotype.
To investigate whether the acquisition of an extra copy of a given chromosome affects the nucleus in human cells, we analyzed the nuclear morphology of a series of human trisomic cell lines constructed by micronuclei-mediated chromosome transfer into RPE-1 cells (Stingele et al., 2012). As observed in primary fibroblasts, we found that up to 40% of the RPE-1 cells harboring extra copies of chromosome 7 (RPE-1 7/3), chromosome 8 (RPE-1 8/3), or chromosome 21 (RPE-1 21/3) also display abnormal nuclei compared with their control counterparts (Figures S6E and S6F). These results show that trisomy in human cells affects the morphology of the nucleus in primary fibroblasts and immortalized epithelial cell lines.
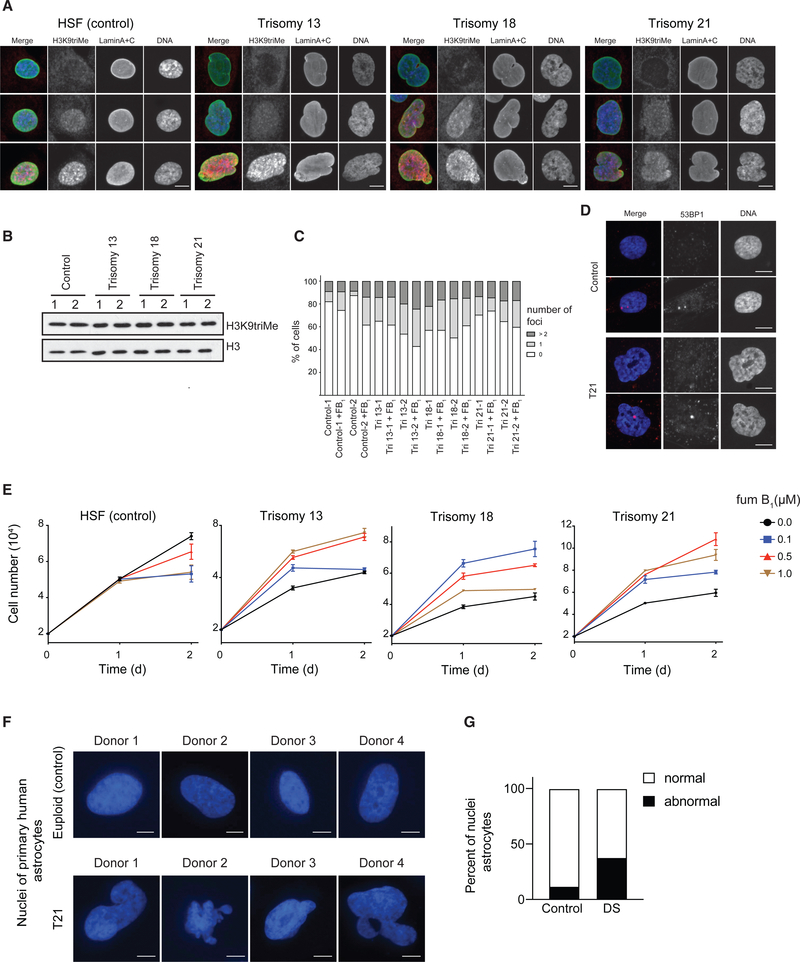
Increases in LCB Levels Improve the Fitness of Human Aneuploid Cells
To gain insight into the physiological consequences of harboring a nucleus with abnormal morphology, we initially assessed whether the association between NE and chromatin was affected in aneuploid cells. Using immunofluorescence, we found that the levels of the tri-methylation of lysine 9 of histone H3 (H3K9triMe), an established marker of lamina-associated domains, were heterogeneous in control primary human fibroblasts. The levels of this histone mark varied from no signal to intermediate to high levels in the nuclei of control fibroblasts (Figure 7A). Notably, we found that this heterogeneity was also present in trisomy 21 fibroblasts, because nuclei with abnormal morphologies show varying levels of H3K9triMe. These results indicate that the levels of this histone mark do not correlate with altered nuclear morphology (Figure 7A). We obtained similar results when we assessed the levels of H3K9triMe of fibroblasts trisomic for chromosomes 13 or 18 (Figure 7A). Consistently, global analysis of H3K9triMe by western blots showed no significant changes between control and trisomic fibroblasts (Figure 7B).
Figure 7. Suppressing Aneuploidy-Associated Phenotypes Improves the Proliferation of Human Trisomies.
(A) Representative images of primary human fibroblasts from euploid donors and patients with Patau syndrome (trisomy 13), Edward syndrome (trisomy 18), or Down syndrome (trisomy 21). Immunofluorescence is shown for lamin A/C, H3K9triMe, and Hoechst 33342. In the merge images, lamin A/C is green, H3K9triMe is red, and DNA is blue. Scale bar, 5 μm.
(B) Western blot analysis of H3K9triMe in human fibroblasts.
(C) Quantification of the number of 53BP1 foci in human fibroblasts mock treated or in the presence of fumonisin B1 (n = 100).
(D) Representative images of primary human fibroblasts from euploid donors and patients with Down syndrome. Immunofluorescence is shown for 53BP1 (red) and Hoechst 33342 (blue). There is no correlation between abnormal nuclear morphology and number of 53BP1 foci. Scale bar, 5 μm.
(E) Growth curves of primary human fibroblasts from 2 euploid donors and 2 patients with Patau (trisomy13), Edward (trisomy18), or Down syndrome(trisomy 21) with increasing concentrations of fumonisin B1.
(F) Representative images of primary human astrocytes from 4 euploid donors and 4 donors with Down syndrome. Immunofluorescence for the nucleus is blue stained with Hoechst 33342. Scale bar, 5 μm.
(G) Percentage of astrocytes from euploid and trisomy 21 donors showing abnormal nuclear morphology (n > 200).
Aneuploid cells show increased signs of replication stress assessed by the number of 53BP1 foci (Santaguida et al., 2017). Indeed, analysis of the number of 53BP1 foci revealed that human trisomies 21, 18, and 13 show increased replication stress when compared with controls (Figure 7C). However, the number of foci did not correlate with nuclear abnormalities in the aneuploid cells. Furthermore, the number of 53BP1 foci did not change when cells were grown in the presence of fumonisin B1 (Figure 7D). These data confirm that trisomy 21 fibroblasts show increased signs of replication stress; however, there seems to be no direct functional connection between abnormal nuclear morphology and 53BP1 foci.
To assess the physiological relevance of suppressing nuclear abnormalities in cells from patients with Down, Edward, or Patau syndrome, we measured the proliferation rates of cells treated with fumonisin B1. In the absence of this drug, human trisomic cells proliferate more slowly than control counterparts, supporting the hypothesis that aneuploidy hampers cell proliferation in yeast and human cells. Remarkably, we found that although fumonisin B1 minimally affected the proliferation of control cells, it significantly improved the proliferation rates of all three human trisomies independent of karyotype (Figure 7E). These results indicate that abnormal nuclear morphologies are associated with impaired proliferation, and targeting ceramide synthesis represents an important strategy to suppress aneuploidy-driven phenotypes in human cells.
Astrocytes Isolated from Down Syndrome Patients Show Abnormal Nuclear Morphologies
In addition to fibroblasts, is the nuclear morphology of other cell types affected by trisomy 21? To address this, we analyzed the nuclear morphologies of primary astrocytes derived from the fetal brain with Down syndrome (Busciglio et al., 2002; Helguera et al., 2013). DNA staining revealed that the nuclear morphologies of four independent primary astrocytes established from brains with trisomy 21 are significantly affected when compared with controls (Figures 7F and 7G). These results indicate that the nuclear integrity of several cell types and tissues might be affected by trisomy 21. Altogether, our results provide an important paradigm for Down syndrome research to study aneuploidy-driven cellular defects and discover novel approaches to improve their fitness.
DISCUSSION
Abnormal nuclear morphology is a hallmark of premature aging, neurodegenerative diseases, and cancer (Frost et al., 2016; Liu et al., 2012; Scaffidi and Misteli, 2006; Zink et al., 2004). The shape of the nucleus also changes dramatically during senescence and healthy aging (Campisi, 2013; Lopez-Otin et al., 2013; Scaffidi and Misteli, 2006). However, the factors that lead to an abnormal nuclear shape remain mostly unknown. During aging, cells not only accumulate thousands of mutations but also show an increased frequency of chromosomal abnormalities. The incidence of aneuploidy is strongly associated with increased age, and studies in yeast showed that aneuploidy shortens the lifespan. Our results lead to the hypothesis that aneuploidy may promote aging by disrupting nuclear integrity.
Important insights into the aging process have come from studies of human diseases associated with premature aging. The identification of mutations in the lamin A/C gene LMNA as a cause of HGPS was a significant breakthrough that led to the development of effective therapies (Gordon et al., 2016). Silent mutations in LMNA affect splicing, leading to the expression of a dominant-negative truncated version of lamin named progerin. Progerin accumulates at the inner nuclear membrane and affects the morphology and function of the NE. Interestingly, although the genetic bases of HGPS and progeroid syndromes are entirely unrelated to trisomy 21 in Down syndrome, several clinical symptoms are shared between these two conditions. These include growth retardation, skeletal and craniofacial anomalies, heart defects, abnormal skin, low muscle tone, gastrointestinal issues, and lower immunity. Our studies show that early-passage human fibroblasts from patients with Down syndrome show abnormal nuclei reminiscent to those from patients with HGPS. Based on these results, we propose that aneuploidy plays a causative role in the deterioration of cellular function during healthy aging and that this process is accelerated in patients with Down syndrome by the presence of the extra chromosome.
The risk of neurodegenerative diseases increases with age in humans. Notably, patients with Down syndrome develop neurodegenerative diseases similar to Alzheimer’s disease, and most develop dementia by the age of 60 years. Although there is a strong association between increased incidence of aneuploidy and neurodegenerative diseases, the molecular mechanisms underlying this connection are not known. In addition to the accumulation of protein plaques, a hallmark of the histopathology of neurodegeneration is the disruption of nuclear morphology. Affected neurons show abnormal nuclear shapes and altered chromatin staining. Furthermore, pathologists use abnormal nuclear characteristics to diagnose and stage several other human diseases, including cancer. The events causing disruption of nuclear integrity in human disease are not well understood. Our studies provide an optimal platform to determine the molecular mechanisms by which aneuploidy disrupts the nucleus and to discover novel approaches that ameliorate the aneuploidy-associated phenotypes in several human diseases.
Our results indicate that a concomitant increase in the biosynthesis of lipids has a critical role in helping cells tolerate the presence of an extra chromosome in the nucleus. We show that increasing the levels of LCBs improves fitness and suppresses nuclear abnormalities in human trisomies, indicating that these lipids play an essential role in the integrity of the nuclear membrane. In yeast, we were able to show that LCBs, not ceramides, are enriched in the nuclear membrane. Previous studies showed that both LCB and ceramide levels increase in aneuploid yeast, trisomic MEFs, and aneuploid cancer cells (Hwang et al., 2017; Tang et al., 2017). Lipidomics analysis confirmed that these lipids also increased in primary fibroblasts isolated from patients with Down, Patau, or Edward syndrome compared with controls (Figure S7D). In response to aneuploidy, cells increase the levels of LCBs, which presumably help maintain nuclear membrane integrity. However, these increases are not sufficient to fully tolerate the presence of an extra autosome and are accompanied by increases in the levels of ceramides. Ceramide levels are tightly coupled to the LCB synthesis, and at least in the context of aneuploidy, a mechanism that prevents the accumulation of ceramide, a lipid molecule associated with cell-cycle arrest and apoptosis, does not seem to be engaged. A genetic screen pointed to gene mutations that do precisely this: increase LCBs without further increases in ceramides. Remarkably, increasing LCBs by either genetic or chemical approaches improves fitness and suppresses nuclear abnormalities of aneuploid cells in yeast and humans. Our findings also indicate that in addition to functioning as signaling molecules, LCBs play an essential structural role in the nucleus due to their unique biophysical properties. LCBs are single-chain amphipathic molecules with propensities for tight packing and high curvature. Thus, the link between LCB levels and nuclear integrity points toward potential direct or indirect effects on the physiology of the cell. Moreover, such implications open new avenues for studying the long-known association between lifespan and sphingolipids.
Interestingly, immunofluorescence and western blot analysis indicate that the cellular levels of lamin proteins are not affected in human trisomic cells (Figures 6A, 6E, and S7A). Furthermore, actin cables show typical staining in cells harboring nuclei with abnormal morphology (Figure S7B). Interestingly, here we show that extra DNA alone is not sufficient to disrupt nuclear morphology. Yeast cells harboring YACs with human DNA and fibroblasts from patients with triple X syndrome show normal nuclear morphologies (Figure S7C). The amount of the DNA increases in triple X syndrome (156.0 Mb) is more significant than that of trisomies 21, 18, and 13 (46.7, 80.4, and 114.3 Mb, respectively), yet patients with triple X syndrome show minor clinical symptoms, with up to 90% of cases not diagnosed. The extra chromosome X is silenced by Xist, supporting the hypothesis that the presence of extra DNA is not as deleterious to the physiology of the cell as gaining an extra autosome. Thus, we hypothesize that the active autosome and increases in the cellular machinery involved in chromatin regulation and increased gene expression may disrupt the morphology of the nucleus, perhaps by creating biophysical stress in the nuclear membrane. In support of this hypothesis, we show in yeast that mutations that suppress nuclear abnormalities increase the fluctuations of the NE, which may represent increased membrane compliance, helping the cell cope with the presence of the extra chromosome.
Lastly, our work provides an important framework for Down syndrome research. In addition to increased expression of the genes located on chromosome 21, cells from patients with Down syndrome are affected by aneuploidy, which disrupts cellular homeostasis at several levels. We show in this paper that targeting ceramide synthesis improves the fitness of aneuploid cells, including primary fibroblasts isolated from patients with Down syndrome. Interestingly, fumonisin B1 is a food contaminant commonly present in our diet, and at low levels, it seems to be well tolerated by humans. The potential to use fumonisin B1 or other strategies to suppress aneuploidy-associated phenotypes in patients with Down syndrome represents an exciting area for future research.
STAR⋆METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Reagents were obtained from commercial sources as indicated. This study did not generate new unique reagents. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Eduardo Torres (eduardo. torres@umassmed.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast strains and growth conditions
All stains are derivatives of W303 (E187) and are listed below. All aneuploid strains used in this study were subjected to comparative genomic hybridization (CGH) to ensure that the additional chromosome was present in its entirety. All deletions were first introduced in wild-type cells by transformation and backcrossed into the disomes. One liter of synthetic medium (SM) consists of 1.7 g of yeast nitrogen base without amino acids or ammonium sulfate, 4.8 g of ammonium sulfate, and 2 g of amino acid mix, which consist of equal amounts of Ala, Arg, Asn, Asp, Cys, Gln, Glu, Gly, Iso, Leu, Lys, Met, Phe, Pro, Ser, Thr, Trp, Tyr, Val, adenine and uracil. Duplicated chromosomes are marked with HIS3 and KanMX6 cassette and the presence of the extra chromosome is ensured by selection in medium lacking His and the addition of kanamycin (200 μg/ml).
Culture of human cell lines
RPE1 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO) supplemented with 10% fetal bovine serum (FBS; sigma) and 2 mM L-glutamine. Primary human skin fibroblasts (HSF) from young healthy individuals (GM08447, GM056659, GM00969, and GM02036) and Trisomy fibroblasts (Trisomy 21: GM04616, GM04592, AG05397, AG06922, GM02767, AG08941, and AG08942; Trisomy 13: GM00526 and GM02948; Trisomy 18: GM00734 and GM03538) were purchased from Coriell Cell Repositories and used in passage between P6 to P15. They were grown in Minimum Essential Media (MEM; GIBCO) supplemented with 15% FBS and 2 mM L-glutamine. The HGPS fibroblasts from patients (HGADFN167 and HGADFN367) and the control primary fibroblasts from healthy gender-matched parents (HGFDFN168 and HGMDFN368) were obtained from The Progeria Research Foundation (PRF) Cell and Tissue Bank. These cells were grown in DMEM supplemented with 15% FBS and 2 mM L-glutamine and used in passage between P6 to P10. hTert-HFF cell was kindly provided by Jennifer A. Benanti (Benanti and Galloway, 2004). The aneuploid RPE1 cells generation and karyotypes are described in Dürrbaum et al. (2018).
Primary astrocytes
Primary human astrocytes cultures were established from four trisomy 21 (14–21 weeks of gestational age) and four age-matched control (diploid) fetal cortical specimens. Sex information was not collected from the specimens. The protocols for tissue processing complied with all federal and institutional guidelines. The cultures were maintained in DMEM supplemented with 10% FBS as described in Helguera et al. (2013).
METHOD DETAILS
Live-cell microscopy of yeast cells
Cells grown at 25°C in synthetic media were collected during early exponential phase (OD600 between 0.6 and 1.0) by centrifugation. After washing cells with 1 X phosphate-buffered saline (PBS), they were put on slide glass, covered with glass and images were immediately taken using Nikon Eclipse E400 microscope.
Quantification of nuclear volume from microscopy
Images were analyzed using imageJ software (https://imagej.nih.gov). Using the freehand tool the surface area (SA) of 200 non-dividing nuclei was measured in pixels. The mean value was calculated by fitting the histogram to a normal distribution using the GraphPad Prism 7 software. The volume was calculated assuming a perfect circle. Volume = 4/3*pi*r3 where r = sqrt (SA/pi). Pixels were converted to μm using a scale bar of 10 μm (factor 10 = 60 pixels).
Volume quantification of purified nuclei using Coulter Counter
Yeast cells grown at 25°C in synthetic media were collected during early exponential phase (OD between 0.6 and 1.0). 108 cells were harvested and washed twice with water. Cells were lysed in 1 mL of lysis buffer (1.1 M sorbitol, 20 mM KCl, 0.5 mM MgCl2, pH 7.4) with 50 mg/ml of zymolase and 10 mM DTT for 1 hour at 30°C. Spheroplasts were spun down at 2K rpm for 5 minutes, resuspended in hypotonic solution (8% PVP, 10 mM Tris-HCl pH 6.4,0.5 mM MgCl2), and incubated for 10 minutes at room temperature. Nuclei were spun down at 2K rpm for 5 minutes and resuspended in PBS right before measuring their volume in a Coulter Counter (n = 30,000).
Quantification of abnormal nuclear morphology in yeast
To quantify the number of abnormal nuclei, 200 non-dividing cells expressing Heh1-GFP were analyzed by live-microscopy. Normal nuclei which are always observed in wild-type cells show mostly circular, smooth and continuous GFP signal. Any nucleus that showed deformed shape, broken nuclei, aggregated GFP signal, invaginations, membrane expansions or discontinuous GFP signal was considered abnormal. Due to the heterogeneity and severity of the nuclear abnormalities of the aneuploid cells, we could not use any software that would automatically quantify different geometrical parameters (e.g., circularity, sphericity index) of the shape of the abnormal nuclei.
Electron microscopy of wild-type cells and cells harboring tsc3Δ was outsourced to the The Harvard Medical School EM Facility (https://electron-microscopy.hms.harvard.edu/).
In vitro serine palmitoyltransferase assay
SPT activity was measured using methods described by Harmon et al. (2013) and Rütti et al. (2009). Briefly, 3 × 106 of exponentially growing yeast cells were used for each assay. Cells were broken in 500 μL of cold 50 mM HEPES/NaOH (pH 8.1) and 1 mM EDTA using French Press G-M. After removing cell debris by centrifugation at 10,000 ×g for 5 minutes at 4°C, the supernatant was spun at 100,000 × g in a TLA 120.2 (Beckman) rotor for 30 minutes at 4°C to obtain microsomes. Microsomes were incubated in 200 μl of reaction buffer containing 50 mM HEPES/NaOH (pH 8.1), 50 μM pyridoxy phosphate, 100 μM palmitoyl-CoA, 5 mM L-serine and 0.5 mM L-[U-C14] serine (50 μCi/ml) at 37°C. C14-labeled lipids were extracted by addition of 0.5 mL of methanol/KOH: CHCl3 (4:1) with vortexing. After sequential additions of 0.5 mL of CHCl3, 0.5 mL of 0.2 mN NH4OH and 100 μl of 2 N NH4OH, mix them and centrifuge at 12,000 × g for 1 min at room temperature. The lower phase (CHCl3) was washed twice with 0.9 mL of 0.2 mN NH4OH and transferred in new tube. The CHCl3 was dried under N2 gas flow and resuspended in 5 mL of scintillation cocktail. The radioactivity from the isolated lipids was determined by scintillation counter.
Yeast nuclear preparation for mass spectrometry
Diploid of WT, lcb4Δ, lag1Δ with Nup82-GFP cells and WT with Rtn1-GFP were used for preparation of yeast nuclei. The methods are described in Schreiner etal. (2015). Briefly, exponentially growing yeast cells were taken and treated with 100 mM of Tris-HCl (pH 9.4) and 10 mM DTT for 10 minutes at 25°C. After washing cells with enzyme buffer (1.1 M sorbitol, 20 mM of KCl, 0.5 mM of MgCl2, pH 7.4 with NaOH), cells were incubated with 200 μg/mL of zymolase (MP biomedicals) and 100 μL of glusulase (PerkinElmer) in enzyme buffer for 1 hour at 30°C. Spheroplasts were applied on Ficoll-sorbitol cushion (7.5% of Ficoll and 1.1 M sorbitol) and centrifuged at 10,000 × g for 15 minutes at 4°C. After harvesting spheroplasts, cells were broken using Dounce homogenizer (Wheaton, type B) in 8% Polyvinylpyrrolidone, 10 mM of Tris-HCl (pH 6.4), 0.5 mM of MgCl2 and proteinase inhibitor cocktail. Lysates were loaded on three density step sucrose gradients (1.8, 2.3 and 2.5 M) and centrifuged at 15,000 × g for 1.5 hours at 4°C. Nuclei were harvested between 1.8 and 2.3 M sucrose gradient and then the number of nuclei were counted using a Coulter Counter (Beckman).
Quantitative lipidomics of LCBs and ceramides
Mass spectrometry quantifications of long-chain bases and ceramides were outsourced to the Lipidomics Core Facility of the Medical University of South Carolina. The methods are described in Bielawski et al. (2009). 108 yeast cells grown in SC media at room temperature were harvested by centrifugation and frozen in liquid nitrogen before shipment in dry ice. Raw data of the total lipid levels were divided by the number of cells measured using a Coulter Counter.
Global lipodomics of yeast nuclei
Quantification of the global lipid composition of purified nuclei were outsourced to Lipotype (https://www.lipotype.com). They use quantitative shotgun lipidomic analysis previously described in Ejsing et al. (2009) and Kloseet al. (2012). Briefly, samples were mixed with 30 μl of internal lipid standard mixture, providing a total spike of 24 pmol DAG 17:0–17:0, 22 pmol PA 17:0–14:1, 41 pmol PE 17:0–14:1,41 pmol PS 17:0–14:1,42 pmol PC 17:0–14:1,40 pmol PI 17:0–14:1, 14 pmol CL 15:0–15:0–15:0–16:1,22 pmol ceramide 18:0;3/18:0;0, 37 pmol IPC 18:0;2/26:0;0, 36 pmol MIPC 18:0;2/26:0;0, 31 pmol M(IP)2C 18:0;2/26:0;0, and 57 pmol cholesterol-D7. Samples were subsequently subjected to two-step lipid extraction (1 mL of solvent in each step) executed at 4°C. The lower organic phases were collected and evaporated in a vacuum evaporator at 4°C. Lipid extracts were dissolved in 100 μL chloroform/methanol (1:2; vol/vol) and analyzed by mass spectrometry using a LTQ Orbitrap XL (Thermo Fisher Scientific) equipped with a robotic nanoflow ion source TriVersa NanoMate (Advion Biosciences). PA, PS, PE, PC, CL, PI, IPC, MIPC and M(IP)2C, DAG and lysolipid species were monitored by negative ion mode FT MS analysis, whereas TAG and ceramide species were monitored by positive ion mode FT MS analysis.
Nuclear envelope dynamics
S. cerevisiae of WT, lcb4Δ, lag1Δ with Nup82-GFP and S. pombe with Cut11-GFP were used for measuring nuclear envelope fluctuations. Methods to quantify nuclear envelope fluctuations are described in detailed in Schreiner et al. (2015) and Williams et al. (2019).
Nuclear staining of primary astrocytes
For nuclear staining, the cultures were fixed with 4% PFA and incubated with Hoechst 33342 (Molecular Probes). Fluorescence images were generated with Zeiss Axiovert 200 inverted microscope (objx63) and AxioVision Rel.4.8. software.
Immunofluorescence microscopy
Cells were fixed with 4% paraformaldehyde in 1X PBS for 10 minutes and permeabilized with 0.1% of Triton X-100 in 1X PBS for 8 minutes at room temperature. After blocking with 1 % BSA and horse serum in 1X PBS for 1 hour at room temperature, cells were incubated with 0.2 μg/mL of primary antibodies in 1X PBS for 2 hours at room temperature. Primary antibodies and nuclei were visualized by donkey anti-mouse IgG H&L Alexa Fluor® 488 (Abcam), donkey anti-rabbit IgG H&L Alexa Fluor®568 (Abcam) and Hoechst 33342 (Invitrogen). Phalloidin-iFluor 488 Reagent (Abcam) was used for visualizing actin polymerization. Then fluorescence signals were detected by LSM700 (Zeiss). A representative z-slice from the image stack was chosen for figures.
Western blot assay
For yeast samples, same number of spheroplasts and nuclei was lysed with 1 X sample loading buffer (50 mM Tris-HCl pH 6.8, 2% SDS, 100 mM β-mercaptoethanol, 0.2% bromophenol blue) and heated on 95°C for 10 minutes. For human samples, cells were lysed in lysis buffer (25 mM Tris-HCl pH 7.5,150 mM NaCl, 1% Triton X-100, 0.1% SDS, and 0.5% deoxycholate with protease inhibitors). Lysate was separated on SDS polyacrylamide gels, and then proteins were transferred onto a PVDF membrane and analyzed with antibodies against GFP (Invitrogen), Nop1(Abcam), Lamin A/C (Abcam), Lamin B1 (Abcam), SPTLC1 (Abcam), SPTLC2 (Abcam), Histone 3 (Abcam), H3K9triMe (Abcam), 53BP1 (Cell signal) and GAPDH (Millipore). Immunoreactive signals were detected by the Super Signal reagent (Pierce).
QUANTIFICATION AND STATISTICAL ANALYSIS
Averages and standard deviations were calculated using Excel; error bars represent standard deviation of the average of 3 independent experiments. Statistical significance was defined by p values < 0.05 from an unpaired t test. Images were analyzed using imageJ software (https://imagej.nih.gov). Using the freehand tool, the surface area (SA) of 200 non-dividing nuclei was measured in pixels. The mean value was calculated by fitting the histogram to a normal distribution using the GraphPad Prism 7 software. Further information for different experiments can be found in the figure legends.
DATA AND CODE AVAILABILITY
This paper did not generate any unique datasets or core. The lipidomics measurements are presented in Tables S1, S2, and S3.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-GFP antibody | ThermoScientific | MA5–15256; RRID:AB_10979281 |
| Anti-NOP1 (Fibrillarin) antibody | Abcam | ab4575; RRID:AB_304531 |
| Anti-SPTLC1 antibody | Abcam | ab176706 |
| Anti-SPTLC2 antibody | Abcam | ab23696; RRID:AB_447617 |
| Anti-Lamin A/C antibody | Abcam | ab190380; RRID:AB_2747781 |
| Anti-Lamin B1 antibody | Abcam | ab16048; RRID:AB_10107828 |
| Anti-H3K9triMe antibody | Abcam | ab8898; RRID:AB_306848 |
| Anti-53BP1 antibody | Cell signal | #4937; RRID:AB_10694558 |
| Anti-GAPDH | Millipore | AB2302; RRID:AB_10615768 |
| Donkey Anti-Mouse IgG H&L (Alexa Fluor® 488) | Abcam | ab150105; RRID:AB_2732856 |
| Donkey Anti-Rabbit IgG H&L (Alexa Fluor® 568) | Abcam | ab175470; RRID:AB_2783823 |
| Goat Anti Rabbit antibody | BioRad | 170–6515; RRID:AB_11125142 |
| Goat Anti Mouse antibody | BioRad | 172–1011; RRID:AB_11125936 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Myriocin | Sigma | M1177 |
| Sphinganine (DHS) | Avanti Polar Lipids | 860498P |
| Phytosphingosine (PHS) | Avanti Polar Lipids | 860499 |
| C16-ceramide | Avanti Polar Lipids | 860516P |
| C18-ceramide | Avanti Polar Lipids | 860518P |
| Sphingosine (SPH) | Cayman | 10007907 |
| Sphingosine-1-phosphate (S1P) | Cayman | 62570 |
| Rapamycin | Sigma | R8781 |
| Fumonisin B1 (FB1) | Cayman | 62580 |
| Sphingosine Kinase inhibitor (SKi)-II | Cayman | 10009222 |
| Latrunculin B1 | Cayman | 10010631 |
| Fetal Bovine Serum (FBS) | Sigma | F2442 |
| Penicillin/Streptomycin | GIBCO | 15140–122 |
| Trypsin-EDT solution | GIBCO | 25200–056 |
| Glutamine | GIBCO | A2916801 |
| Immobilon-P PVDF membrane | Millipore | IPVH00010 |
| Protease inhibitor cocktail tablets | Roche | 11836170001 |
| L-[U-C14] serine | PerkinElmer | NEC286E050UC |
| EcoLite(+) Liquid Scintillation Cocktail | MP Biomedicals | 188247501 |
| Zymolase | USBiological | Z1004 |
| Glusulase | PerkinElmer | Z1000 |
| Phalloidin-iFluor 488 Reagent | Abcam | ab176753 |
| Hoechst 33342 | Invitrogen | H3570 |
| Experimental Models: Cell Lines | ||
| Euploid human skin fibroblast (Control)_1 | Coriell Institute | GM00969; RRID:CVCL_7311 |
| Euploid human skin fibroblast (Control)_2 | Coriell Institute | GM08447; RRID:CVCL_7487 |
| Euploid human skin fibroblast (Control)_3 | Coriell Institute | GM05659; RRID:CVCL_7434 |
| Euploid human skin fibroblast (Control)_4 | Coriell Institute | GM02036; RRID:CVCL_7348 |
| Human Trisomy 13 fibroblast_1 | Coriell Institute | GM00526; RRID:CVCL_X075 |
| Human Trisomy 13 fibroblast_2 | Coriell Institute | GM02948; RRID:CVCL_X275 |
| Human Trisomy 18 fibroblast_1 | Coriell Institute | GM00734; RRID:CVCL_X236 |
| Human Trisomy 18 fibroblast_2 | Coriell Institute | GM03538; RRID:CVCL_X290 |
| Human Trisomy 21 fibroblast_1 | Coriell Institute | GM04616; RRID:CVCL_V475 |
| Human Trisomy 21 fibroblast_2 | Coriell Institute | GM04592; RRID:CVCL_V473 |
| Human Trisomy 21 fibroblast_3 | Coriell Institute | AG05397; RRID:CVCL_L780 |
| Human Trisomy 21 fibroblast_4 | Coriell Institute | AG06922; RRID:CVCL_X793 |
| Human Trisomy 21 fibroblast_5 | Coriell Institute | GM02767; RRID:CVCL_V469 |
| Human Trisomy 21 fibroblast_6 | Coriell Institute | AG08941; RRID:CVCL_X871 |
| Human Trisomy 21 fibroblast_7 | Coriell Institute | AG08942; RRID:CVCL_X872 |
| Human Triple X fibroblast_1 | Coriell Institute | GM00254; RRID:CVCL_7276 |
| Human Triple X fibroblast_2 | Coriell Institute | GM04626; RRID:CVCL_X310 |
| hTert-HFF | Benanti and Galloway, 2004 | N/A |
| RPE1 | Stingele et al., 2012 | N/A |
| RPE1–7/3 | Stingele et al., 2012 | N/A |
| RPE1–8/3 | Stingele et al., 2012 | N/A |
| RPE1–21/3 | Stingele et al., 2012 | N/A |
| HGPS fibroblasts from patient_1 | The Progeria Research Foundation (PRF) Cell and Tissue Bank | HGADFN167; RRID:CVCL_1Y92 |
| HGPS fibroblasts from patient_2 | The Progeria Research Foundation (PRF) Cell and Tissue Bank | HGADFN367; RRID:CVCL_1Y99 |
| Euploid human astrocyte (Control)_1 | Helguera et al., 2013 | N/A |
| Euploid human astrocyte (Control)_2 | Helguera et al., 2013 | N/A |
| Euploid human astrocyte (Control)_3 | Helguera et al., 2013 | N/A |
| Euploid human astrocyte (Control)_4 | Helguera et al., 2013 | N/A |
| Human Trisomy 21 astrocyte_1 | Helguera et al., 2013 | N/A |
| Human Trisomy 21 astrocyte_2 | Helguera et al., 2013 | N/A |
| Human Trisomy 21 astrocyte_3 | Helguera et al., 2013 | N/A |
| Human Trisomy 21 astrocyte_4 | Helguera et al., 2013 | N/A |
| Experimental Models: Organisms/Strains | ||
| S. cerevisiae: Strain background: W303: see Table S1 | This paper | This paper |
| S. pombe: cut11-GFP::ura4+ h+ ura4-D18 leu1–32 | Schreiner et al., 2015 | MKSP10 |
| Recombinant DNA | ||
| pLKO.1:TRCN0000035010 targeting SPTLC1 | Open Biosystems | TRCN0000035010 |
| pLKO.1:TRCN0000034973 targeting SPTLC2 | Open Biosystems | TRCN0000034973 |
| pLKO.1-scramble | Addgene | #1864; RRID:Addgene_1864 |
| pCMV-dR8.2 | Addgene | #12263 |
| pMD2.G | Addgene | #12259; RRID:Addgene_12259 |
| Software and Algorithms | ||
| Graphpad Prism 8.0 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/index.html |
Highlights.
Aneuploidy disrupts nuclear morphology in yeast and human cells
Long-chain bases (LCBs) are integral structural components of the nuclear membrane
Increasing the levels of LCBs suppresses nuclear abnormalities in aneuploid cells
Inhibition of ceramide synthesis improves the fitness of trisomy 21 cells
ACKNOWLEDGMENTS
We are grateful to Jennifer Benanti for reagents. We are grateful to Maria Ericsson for technical assistance in the analysis of the electron micrograph images. This research was supported by NIH grant 1R01GM118481–01A1 to E.M.T. and NIA grant AG056850 to J.B.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.10.059.
REFERENCES
- Antonarakis SE (2017). Down syndrome and the complexity of genome dosage imbalance. Nat. Rev. Genet 18, 147–163. [DOI] [PubMed] [Google Scholar]
- Beach RR, Ricci-Tam C, Brennan CM, Moomau CA, Hsu PH, Hua B, Silberman RE, Springer M, and Amon A (2017). Aneuploidy Causes Non-genetic Individuality. Cell 189, 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benanti JA, and Galloway DA (2004). Normal human fibroblasts are resistant to RAS-induced senescence. Mol. Cell. Biol 24, 2842–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski J, Pierce JS, Snider J, Rembiesa B, Szulc ZM, and Bielawska A (2009). Comprehensive quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods Mol. Biol 579, 443–67. [DOI] [PubMed] [Google Scholar]
- Busciglio J, Pelsman A, Wong C, Pigino G, Yuan M, Mori H, and Yankner BA (2002). Altered metabolism of the amyloid beta precursor protein is associated with mitochondrial dysfunction in Down’s syndrome. Neuron 33, 677–688. [DOI] [PubMed] [Google Scholar]
- Campisi J (2013). Aging, cellularsenescence, and cancer. Annu. Rev. Physiol 75, 685–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’mello NP, Childress AM, Franklin DS, Kale SP, Pinswasdi C, and Jazwinski SM (1994). Cloning and characterization of LAG1, a longevity-assurance gene in yeast. J. Biol. Chem 269, 15451–15459. [PubMed] [Google Scholar]
- Dephoure N, Hwang S, O’Sullivan C, Dodgson SE, Gygi SP, Amon A, and Torres EM (2014). Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. eLife 3, e03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly N, Passerini V, Dürrbaum M, Stingele S, and Storchová Z (2014). HSF1 deficiency and impaired HSP90-dependent protein folding are hallmarks of aneuploid human cells. EMBO J. 33, 2374–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürrbaum M, Kruse C, Nieken KJ, Habermann B, and Storchová Z (2018). The deregulated microRNAome contributes to the cellular response to aneuploidy. BMC Genomics 19, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JH, Harnden DG, Cameron AH, Crosse VM, and Wolff OH (1960). A newtrisomic syndrome. Lancet 1, 787–790. [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, and Shevchenko A (2009). Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA 106, 2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Bardai FH, and Feany MB (2016). Lamin Dysfunction Mediates Neurodegeneration in Tauopathies. Curr. Biol 26, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon LB, Kleinman ME, Massaro J, D’Agostino RB Sr., Shappell H, Gerhard-Herman M, Smoot LB, Gordon CM, Cleveland RH, Nazarian A, et al. (2016). Clinical Trial ofthe Protein Farnesylation Inhibitors Lonafarnib, Pravastatin, and Zoledronic Acid in Children With Hutchinson-Gilford Progeria Syndrome. Circulation 134, 114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon JM, Bacikova D, Gable K, Gupta SD, Han G, Sengupta N, Somashekarappa N, and Dunn TM (2013). Topological and functional characterization ofthe ssSPTs, small activating subunits of serine palmitoyltransferase. J. Biol. Chem 288, 10144–10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helguera P, Seiglie J, Rodriguez J, Hanna M, Helguera G, and Busciglio J (2013). Adaptive downregulation of mitochondrial function in down syndrome. Cell Metab. 17, 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Gustafsson HT, O’Sullivan C, Bisceglia G, Huang X, Klose C, Schevchenko A, Dickson RC, Cavaliere P, Dephoure N, and Torres EM (2017). Serine-Dependent Sphingolipid Synthesis Is a Metabolic Liability of Aneuploid Cells. Cell Rep. 21, 3807–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C, Surma MA, Gerl MJ, Meyenhofer F, Shevchenko A, and Simons K (2012). Flexibility of a eukaryotic lipidome—insights from yeast lip- idomics. PLoS ONE 7, e35063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune J, Gautier M, and Turpin R (1959). [Study of somatic chromosomes from 9 mongoloid children]. C. R. Hebd. Seances Acad. Sci 248, 1721–1722. [PubMed] [Google Scholar]
- Liu GH, Qu J, Suzuki K, Nivet E, Li M, Montserrat N, Yi F, Xu X, Ruiz S, Zhang W, et al. (2012). Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature 491, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, and Kroemer G (2013). The hallmarks ofaging. Cell 153, 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ, and Hunt PA (2012). Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet 13, 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passerini V, Ozeri-Galai E, de Pagter MS, Donnelly N, Schmalbrock S, Kloosterman WP, Kerem B, and Storchová Z (2016). The presence of extra chromosomes leads to genomic instability. Nat. Commun 7, 10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patau K, Smith DW, Therman E, Inhorn SL, and Wagner HP (1960). Multiple congenital anomaly caused by an extra autosome. Lancet 1,790–793. [DOI] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, and Li R (2010). Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468, 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rütti MF, Richard S, Penno A, von Eckardstein A, and Hornemann T (2009). An improved method to determine serine palmitoyltransferase activity. J. Lipid Res. 50,1237–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Vasile E, White E, and Amon A (2015). Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev. 29, 2010–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Richardson A, Iyer DR, M’Saad O, Zasadil L, Knouse KA, Wong YL, Rhind N, Desai A, and Amon A (2017). Chromosome Mis-segregation Generates Cell-Cycle-Arrested Cells with Complex Karyotypes that Are Eliminated by the Immune System. Dev. Cell 41, 638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, and Misteli T (2006). Lamin A-dependent nuclear defects in human aging. Science 312, 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner SM, Koo PK, Zhao Y, Mochrie SG, and King MC (2015). The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat. Commun 6, 7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, and Amon A (2011). Aneuploidy drives genomic instability in yeast. Science 333, 1026–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, and Storchova Z (2012). Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol. Syst. Biol 8, 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, and Storchova Z (2013). Activation of autophagy in cells with abnormal karyotype. Autophagy 9, 246–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunshine AB, Ong GT, Nickerson DP, Carr D, Murakami CJ, Wasko BM, Shemorry A, Merz AJ, Kaeberlein M, and Dunham MJ (2016). Aneuploidy shortens replicative lifespan in Saccharomyces cerevisiae. Aging Cell 15,317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Yuwen H, Wang K, Bruno PM, Bullock K, Deik A, Santaguida S, Trakala M, Pfau SJ, Zhong N, et al. (2017). Aneuploid Cell Survival Relies upon Sphingolipid Homeostasis. Cancer Res. 77, 5272–5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, and Amon A (2007). Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317, 916–924. [DOI] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A,Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, and Amon A (2010). Identification of aneuploidy-tolerating mutations. Cell 143, 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, and Amon A (2008). Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322, 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JF, Mochrie SGJ, and King MC (2019). A versatile image analysis platform for three-dimensional nuclear reconstruction. Methods 157, 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Fischer AH, and Nickerson JA (2004). Nuclear structure in cancer cells. Nat. Rev. Cancer 4, 677–687. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper did not generate any unique datasets or core. The lipidomics measurements are presented in Tables S1, S2, and S3.