Abstract
BRAF p.V600E mutations are detected in greater than 50% of pediatric Langerhans cell histiocytosis (LCH) lesions. However, the use of mutation-specific BRAF V600E immunohistochemistry (IHC) as a surrogate for molecular testing in pediatric LCH is unknown. We tested the mutation-specific BRAF V600E monoclonal antibody (clone VE1) in formalin-fixed, paraffin-embedded LCH samples from 26 pediatric patients (14 males and 12 females, ages 7 mo–17 y) using allele-specific real-time polymerase chain reaction (PCR) with a limit of detection of 0.5% as the comparative gold standard. BRAF VE1 staining was scored for both intensity (0–3+) and percentage of immunoreactive tumor cells (0%−100%). BRAF VE1 immunoreactivity was determined using both lenient (≥1+, ≥1%) and stringent (≥2+, ≥10%) scoring criteria. Using lenient-scoring criteria, we found that the sensitivity and specificity of IHC compared with allele-specific real-time PCR were 100.0% and 18.2%, respectively. The poor specificity of lenient IHC analysis was attributable to weak, 1+ staining in both BRAF-mutated and wild-type LCH. Using stringent-scoring criteria, we found that specificity improved to 100.0% at the expense of sensitivity that decreased to 80.0%. Stringent scoring generated 3 false-negative results, but in all cases, neoplastic tissue comprised less than 5% of the stained section and/or the specimen was decalcified. In conclusion, highly sensitive molecular assays remain the gold standard for BRAF mutation analysis in LCH paraffin-embedded lesions. To avoid false-positive results, unequivocal VE1 staining of 2+ intensity in greater than or equal to 10% neoplastic histiocytes is required. However, negative VE1 results require additional studies to exclude false-negatives, and stringent-scoring criteria may not be optimal for scant or decalcified specimens.
Keywords: BRAF, immunohistochemistry, LCH, pediatric, V600E, VE1
1 |. INTRODUCTION
Langerhans cell histiocytosis (LCH) is a proliferative disorder that arises from the myeloid dendritic cell precursors.1,2 Langerhans cell histiocytosis commonly presents in childhood and has a highly variable clinical presentation ranging from isolated skin or bone lesions that typically follow an indolent course, to potentially lethal-disseminated lesions in the bone marrow, the liver, or the spleen.3–5 Histologically, LCH lesions are characterized by an aberrant proliferation of neoplastic histiocytes with large reniform and grooved nuclei and a mixed inflammatory infiltrate with abundant eosinophils and occasional multinucleated giant cells. The neoplastic Langerhans cell histiocytes typically express CD1a and CD207 (Langerin), and this characteristic immunophenotype helps distinguish LCH from other histiocyte-rich lesions.2,6
The BRAF p.V600E mutation is detected in greater than 50% of pediatric LCH cases,7–9 and other V600 codon mutations such asp.V600K, p.V600D, or p.V600R are typically absent.10 Recurrent MAP2K1 alterations have also been reported in approximately one-third of BRAF wild-type LCH cases highlighting the importance of extracellular signal-regulated kinase (ERK) activation in this neoplastic disease.11–13 In children, BRAF p.V600E mutations have been associated with an increased risk of recurrence9,14 and high-risk LCH in 1 series.14 Thus, BRAF mutation analysis of the blood and bone marrow samples is an effective modality to monitor disease and can assist with oncologic therapeutic decision making.5,9
BRAF V600E–specific immunohistochemistry (IHC) using a monoclonal antibody specific to the mutated V600E epitope (clone VE1) has been applied to formalin-fixed, paraffin-embedded (FFPE) tissue sections from multiple BRAF p.V600E–positive tumor types.15 However, the requisite sensitivity and specificity of BRAF VE1 when compared with gold-standard DNA-diagnostic methods are often lacking, and the performance varies considerably among tumor types.16–21 The variability of BRAF VE1 sensitivity and specificity is partly attributable to tissue and tumor heterogeneity, nonstandardized scoring criteria, and the chosen comparative molecular method.22–26
BRAF mutations are typically detected clinically by DNA-based sequencing assays (eg, Sanger sequencing, pyrosequencing, or next generation sequencing) or allele-specific real-time polymerase chain reaction (AS-qPCR). However, the sensitivities of these assays can vary widely; Sanger sequencing has a limit of detection of approximately 15%, pyrosequencing of approximately 5%, and next generation sequencing of approximately 1% to 2% for detecting BRAF missense mutations27,28 while AS-qPCR and digital droplet PCR can achieve detection limits29 of less than 1%. There is a known association between neoplasia and inflammation,30,31 and LCH lesions harbor prominent inflammatory components via hypothesized cell autonomous and nonautonomous etiologies.8,9 Therefore, highly sensitive molecular assays such as AS-qPCR or digital droplet PCR are needed to detect BRAF mutations in LCH tissue sections, since BRAF-positive cells may comprise a small fraction of the inflammatory cellular lesion.
At our institution, all LCH FFPE patient samples are tested for BRAF p.V600E mutations using a sensitive AS-qPCR assay that can detect 1 mutant allele in 200 wild-type alleles (a limit of detection of 0.5%). On occasion, if molecular studies are inconclusive because of insufficient tissue, decalcification, or PCR inhibition, BRAF VE1 IHC is requested to determine the mutation status. However, there is limited literature regarding the use and performance of BRAF VE1 in pediatric LCH tissue lesions, and of the few studies in the literature that have investigated VE1 in LCH, none have detailed the performance of VE1 relative to its molecular comparison method.32–34 Moreover, the use of VE1 IHC as a surrogate for molecular testing is not well established. Therefore, we tested the commercially available VE1 monoclonal antibody on 26 pediatric FFPE LCH tissue samples and compared the IHC results to the corresponding highly sensitive clinical AS-qPCR molecular results to address its use in pediatric LCH.
2 |. METHODS
2.1 |. Patient samples
The study was conducted in accordance with protocols approved by the Institutional Review Board at Baylor College of Medicine and Texas Children’s Hospital (TCH). From the TCH pathology archives, we retrieved 28 FFPE tissue blocks from 15 male and 13 female LCH patients that had been clinically tested for BRAF p.V600E mutations in theTCH Molecular Oncology laboratory using AS-qPCR. One hematoxylin and eosin (H&E) and 2 unstained slides were cut for tissue adequacy and IHC analysis, respectively. After assessment of H&E section adequacy by a board-certified pathologist (MJH or KEF), 2 patient samples (1 female and 1 male) were excluded from analysis because of insufficient remaining neoplastic tissue.
In total, 26 patients were included in the study: 14 males and 12 females, ages 7 months to 17 years at time of diagnosis. Seven of 11 bone specimens were decalcified using a formic acid/formalin solution (Cal-Ex II Fixative/Decalcifier, ThermoFisher, Waltham, Massachusetts) for a maximum of 2 hours maximum prior to sectioning. Single system or multisystem disease was determined from retrospective review of provided clinical history on the laboratory test requisitions.
2.2 |. BRAF p.V600E AS-qPCR
Molecular BRAF p.V600E testing was performed using a clinically validated BRAF RGQ AS-qPCR assay (Qiagen, Valencia, California) that preferentially amplifies the single nucleotide c.1799T > A transversion in the BRAF (NM_004334.4) gene. Genomic DNA was extracted from FFPE tissue samples using the QIAamp DNA mini kit (Qiagen) per the manufacturer’s recommendations. Input DNA was quantified using the NanoDrop spectrophotometer (ThermoFisher), and thermal cycling was performed using the Roche LightCycler 480 instrument and associated software (Roche Diagnostics, Indianapolis, Indiana).
Mutant BRAF amplification, detection, and interpretation were performed using established laboratory protocols.35 Briefly, a control, nonpolymorphic region of BRAF exon 3 was amplified in separate wells to ensure DNA integrity and normalize the BRAF p.V600E results. Tissue sample ΔCt values were calculated as the difference between the Ct (crossing threshold) of the p.V600E mutation reaction and the exon 3 control reaction. Samples were determined to be BRAF p.V600E mutation positive if the ΔCt was less than 10, where a ΔCt of greater of equal to 10 was interpreted as negative for the p.V600E mutation. The lower limit of detection was established as 0.5% (eg, 1 BRAF homozygous mutant cell in approximately 200 BRAF wild-type cells) per cell-mixing studies.
2.3 |. BRAF V600E (VE1) immunohistochemistry
Immunohistochemistry was performed using the BOND-III automated IHC/ISH stainer (Leica Biosystems, Buffalo Grove, Illinois) and a commercially available BRAF V600E monoclonal antibody (clone VE1; Ventana, Tucson, Arizona) at a 1:45 dilution in Ventana antibody diluent, antigen retrieval with ER2 solution for 40 minutes, and diaminobenzidine as a chromogen. Three BRAF p.V600E–negative cases (2 papillary thyroid carcinomas, 1 melanoma) and 3 BRAF p.V600E–positive cases (2 papillary thyroid carcinomas, 1 sessile-serrated adenoma) were used for antibody optimization and included as IHC controls.
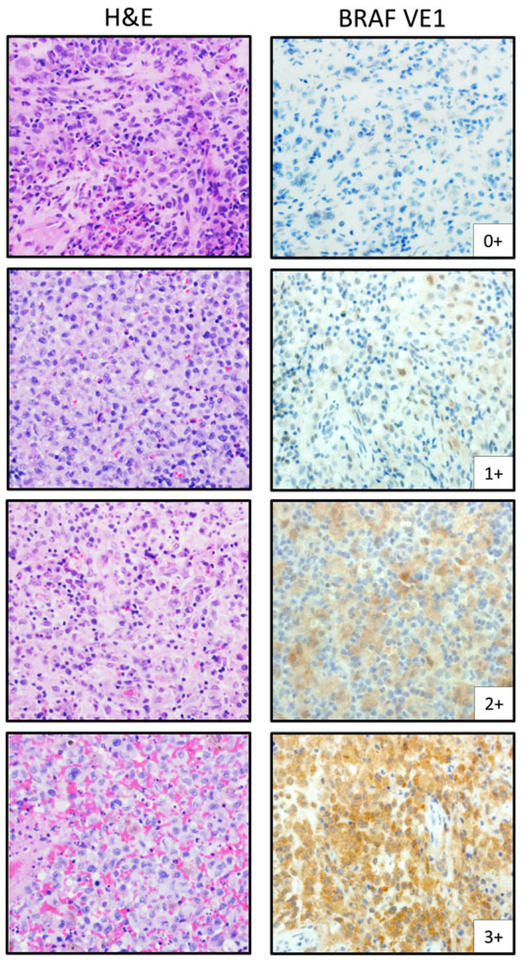
Slides were evaluated and scored for VE1-staining intensity (0–3+) and percentage of tumor cells positive for VE1 staining (0%−100% scored in 5% increments) to produce a composite score. BRAF VE1 immunoreactivity was determined by both lenient (≥1+, ≥1%) and stringent (≥2+, ≥10%) scoring criteria as described.24 VE1-staining intensity was determined as follows: 0+, absence of staining; 1+, faint staining appreciable only at 400× magnification; 2+, moderate staining apparent at 100× magnification with minimal variability in intensity; and 3+, strong staining apparent at 40× magnification (Figure 1). Three board-certified pathologists blinded to molecular status interpreted and scored VE1 IHC (LYB, KEF, and MJH), and consensus scoring was achieved for all cases by concomitant review.
FIGURE 1.
Semiquantitative IHC-scoring assessment for BRAF VE1 in pediatric LCH. A semiquantitative IHC-scoring scale (0–3+) was combined with percent staining of neoplastic cell cells (0%−100%) to deduce a composite score (see Methods for additional details). Lenient-scoring criteria considered any cytoplasmic staining of 1+ or greater intensity in any amount of neoplastic tissue (≥1%) as positive. Stringent-scoring criteria required greater than or equal to 2+ staining intensity in greater than or equal to 10% neoplastic cells to be considered positive. Light microscopic images acquired at 400× magnification to highlight 1+ VE1 staining. H&E, hematoxylin and eosin, IHC, immunohistochemistry
3 |. RESULTS
Twenty-six pediatric LCH samples (14 males and 12 females; 7 mo–17 y) with corresponding BRAF p.V600E AS-qPCR results (15 molecular-positive and 11 molecular-negative) were included in the study. The pertinent characteristics of each patient sample are summarized in Table 1. Samples were obtained from bone (n = 11), soft tissue (n = 6), lymph nodes (n = 5), skin (n = 2), the liver (n = 1), and the spleen (n = 1). Fourteen patients had multisystem LCH, and 12 patients had single system LCH. Clinical data were limited to the clinical information provided on pathology test requisitions that precluded additional clinical characterization.
TABLE 1.
Clinicopathologic characteristics of pediatric LCH study cohort (n = 26)
| IHC/MOL Concordance | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Age/Gender | Clinical Classification | LCH,% of section | Specimen | Decal | VE1 0–3+ | VE1 0%–100% | AS-qPCR | Lenient IHC(≥1+, ≥1%) | Stringent IHC(≥2+, ≥10%) |
| LCH-51 | 11/M | MS-LCH | 40 | Bone | Yes | 0+ | 0 | NEG | NEG | NEG |
| LCH-31 | 1/F | SS-LCH | 80 | Bone | Yes | 0+ | 0 | NEG* | NEG | NEG |
| LCH-55 | 10/M | SS-LCH | 30 | Bone | Yes | 1+ | 10 | NEG* | POS-FP | NEG |
| LCH-6 | 11/M | SS-LCH | 5 | Bone | Yes | 1+ | 50 | NEG | POS-FP | NEG |
| LCH-16 | 2/M | MS-LCH | 95 | Bone | No | 1+ | 70 | NEG | POS-FP | NEG |
| LCH-32 | 3/M | SS-LCH | 90 | Bone | No | 1+ | 80 | NEG | POS-FP | NEG |
| LCH-34 | 1/M | SS-LCH | 90 | Bone | No | 1+ | 30 | NEG | POS-FP | NEG |
| LCH-35 | 1/F | MS-LCH | 20 | Skin | No | 1+ | 60 | NEG | POS-FP | NEG |
| LCH-57 | 7 mo/F | MS-LCH | 70 | LN | No | 1+ | 70 | NEG | POS-FP | NEG |
| LCH-58 | 9 mo/F | SS-LCH | 50 | LN | No | 1+ | 90 | NEG | POS-FP | NEG |
| LCH-59 | 10/M | SS-LCH | 80 | Bone | No | 1+ | 70 | NEG | POS-FP | NEG |
| LCH-2 | 1/M | MS-LCH | <5 | Soft tissue | No | 1+ | 30 | POS | POS | NEG-FN |
| LCH-33 | 9 mo/M | MS-LCH | 30 | Bone | Yes | 1+ | 20 | POS | POS | NEG-FN |
| LCH-13 | 8/F | SS-LCH | <5 | Bone | Yes | 1+ | 50 | POS | POS | NEG-FN |
| LCH-53 | 4/M | SS-LCH | 25 | Bone | Yes | 2+ | 60 | POS | POS | POS |
| LCH-9 | 1/M | MS-LCH | 85 | LN | No | 2+ | 80 | POS | POS | POS |
| LCH-36 | 2/F | MS-LCH | 40 | Skin | No | 2+ | 50 | POS | POS | POS |
| LCH-47 | 2/F | MS-LCH | 70 | LN | No | 2+ | 90 | POS | POS | POS |
| LCH-3 | 9 mo/F | MS-LCH | 75 | Liver | No | 3+ | 40 | POS | POS | POS |
| LCH-5 | 2/F | MS-LCH | 70 | Soft tissue | No | 3+ | 80 | POS | POS | POS |
| LCH-7 | 11 mo/M | SS-LCH | 60 | Soft tissue | No | 3+ | 50 | POS | POS | POS |
| LCH-30 | 17/M | SS-LCH | 60 | Soft tissue | No | 3+ | 80 | POS | POS | POS |
| LCH-48 | 5/F | MS-LCH | 70 | LN | No | 3+ | 30 | POS | POS | POS |
| LCH-54 | 1/F | SS-LCH | 80 | Soft tissue | No | 3+ | 80 | POS | POS | POS |
| LCH-60 | 10/M | MS-LCH | 40 | Soft tissue | No | 3+ | 60 | POS | POS | POS |
| LCH-62 | 3/F | MS-LCH | 90 | Spleen | No | 3+ | 60 | POS | POS | POS |
The asterisks (*) denote decalcified samples that failed AS-qPCR in FFPE sections. Molecular results for these 2 specimens were obtained from nondecalcified sections from the same specimen.
Abbreviations: AS-qPCR, allele-specific real-time PCR; Decal, decalcification; F, female; FN, false-negative; FP, false-positive; IHC, immunohistochemistry; LCH, Langerhans cell histiocytosis; LN, lymph node; M, male; mo, month; MOL, molecular; MS-LCH, multisystem LCH; NEG, negative; POS, positive; SS-LCH, single system LCH.
Seven bone specimens were subjected to decalcification prior to sectioning. Two decalcified FFPE tissue samples failed AS-qPCR initially, so corresponding nondecalcified sections from the same specimen were tested to determine molecular BRAF positivity (Table 1, denoted by *). Hematoxylin and eosin slides and immunohistochemical stains for CD207 and CD1a were reviewed concurrently to confirm both the diagnosis of LCH and amount of lesional tissue available for scoring. The estimated amount of neoplastic LCH contained in each FFPE section ranged from less than 5% to 95% by H&E light microscopic examination.
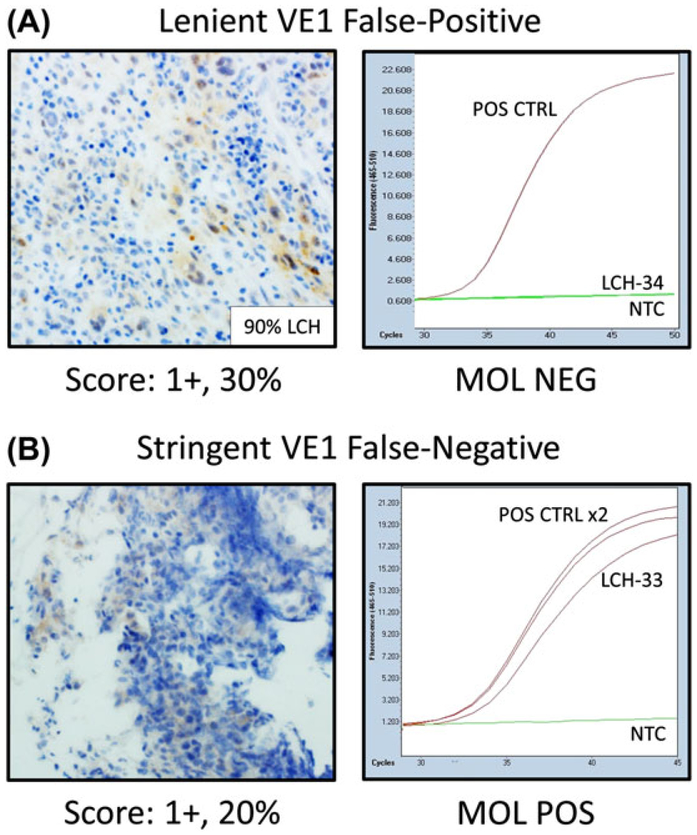
The original report,18 a recent white paper,36 and the manufacturer’s package insert recommend that cytoplasmic staining of 1+ or greater intensity in tumor cells with the VE1 monoclonal antibody be interpreted as positive although no consensus requirement for percent of immunoreactive tumor cells is given (referred to hereafter as lenient scoring criteria: ≥1+, ≥1%). Using these criteria, we found that VE1 staining was absent in 2 cases, and the remaining 24 cases exhibited at least weak (1+) staining in greater than or equal to 10% of neoplastic cells (Table 1). Thus, the sensitivity and specificity of IHC compared with AS-qPCR using lenient-scoring criteria were 100.0% and 18.2%, respectively (Table 2). The poor specificity of lenient IHC analysis was directly attributable to the weak cytoplasmic staining detected in lesional Langerhans cell histiocytes regardless of BRAF mutation status (Figure 2A).
TABLE 2.
Sensitivity and specificity of lenient and stringent IHC-scoring criteria
| Lenient IHC Scoring (≥1+, ≥1%) | ||
|---|---|---|
| MOL NEG | MOL POS | |
| IHC NEG | 2 | 0 |
| IHC POS | 9 | 15 |
| Sensitivity | 100.0% | |
| Specificity | 18.2% | |
| Stringent IHC Scoring (≥2+, ≥10%) | ||
| MOL NEG | MOL POS | |
| IHC NEG | 11 | 3 |
| IHC POS | 0 | 12 |
| Sensitivity | 80.0% | |
| Specificity | 100.0% | |
Abbreviations: IHC, immunohistochemistry; MOL, molecular; NEG, negative POS, positive.
FIGURE 2.
VE1-scoring criteria can influence comparison results. A, A lenient VE1 false-positive sample (LCH-34) showing weak, 1+ staining in 30% of neoplastic histiocytes but no amplification in the AS-qPCR molecular assay. B, A stringent VE1 false-negative sample (LCH-33) showing weak, 1+ staining in 20% of neoplastic histiocytes and amplification in the AS-qPCR molecular assay. Light microscopic images acquired at 400× magnification to highlight 1+ VE1 staining. CTRL, control; IHC, immunohistochemistry; LCH, Langerhans cell histiocytosis; MOL, molecular; NEG, negative; NTC, no template control; POS, positive; VE1, BRAF p.V600E immunohistochemistry
To control for the lack of specificity secondary to weak staining, we elected to reanalyze the data using previously established stringent criteria (scored as IHC positive if 2+ or 3+ VE1 staining was detected in ≥10% of neoplastic histiocytes).24 Using these scoring criteria, we found that 12 cases were scored as VE1 positive, and 14 cases were scored as VE1 negative. Stringent-scoring criteria improved specificity (100.0% vs 18.2%) at the expense of sensitivity (80.0% vs 100.0%, Table 2). Notably, 3 VE1 false-negative results were observed (Figure 2B), and all contained less than 5% neoplastic tissue and/or were decalcified (Tables 1 and 3).
TABLE 3.
VE1 IHC in decalcified LCH bone samples (n = 7)
| IHC/MOL Concordance | |||||
|---|---|---|---|---|---|
| Decalcified Bone Samples | LCH, % of section | Composite VE1 Score, % | AS-qPCR | Lenient IHC (≥1+, ≥1%) | Stringent IHC (≥2+, ≥10%) |
| LCH-51 | 40 | 0+, 0 | NEG | NEG | NEG |
| LCH-31 | 80 | 0+, 0 | NEG* | NEG | NEG |
| LCH-55 | 30 | 1+, 10 | NEG* | POS-FP | NEG |
| LCH-6 | 5 | 1+, 50 | NEG | POS-FP | NEG |
| LCH-33 | 30 | 1+, 20 | POS | POS | NEG-FN |
| LCH-13 | <5 | 1+, 50 | POS | POS | NEG-FN |
| LCH-53 | 25 | 2+, 60 | POS | POS | POS |
The asterisks (*) denote decalcified samples that failed AS-qPCR in FFPE sections. Molecular results for these 2 specimens were obtained from nondecalcified sections from the same specimen.
Abbreviations: AS-qPCR, allele-specific real-time PCR; FN, false-negative; FP, false-positive; IHC, immunohistochemistry; MOL, molecular; NEG, negative; POS, positive.
Langerhans cell histiocytosis involves the bone in approximately 90% of pediatric cases,37 and bone specimens may undergo decalcification prior to histological processing. Importantly, decalcification has been shown to affect DNA quality and interferes with nucleic acid–based assays,38 although some PCR applications can be optimized for decalcified specimens.39–41 We reasoned that if decalcified samples might produce inconclusive molecular results, VE1 may have a certain degree of use in such cases. Therefore, we also analyzed the performance of the VE1 antibody in the 7 Cal-Ex II-decalcified LCH samples included in our study (3 molecular-positive and 4 molecular-negative). The results are summarized in Table 3.
Both cases with complete absence of staining had been decalcified and demonstrated concordance with corresponding negative molecular studies (Table 3). One VE1-positive case (2+, 60%) was also molecular-positive and concordant. The remaining 4 decalcified cases demonstrated weak, 1+ staining in variable percentages of neoplastic cells with VE1 immunoreactivity (range, 10%−50%), and were scored VE1-negative using stringent criteria. However, 2 decalcified VE1-negative cases harbored BRAF mutations by molecular analysis (2 false-negatives). Lastly, the 2 decalcified specimens that required repeat molecular testing due to inconclusive initial results demonstrated very weak VE1 staining (absent and 1+, 10%, respectively), but were concordant with negative molecular results seen in nondecalcified sections from the same specimen.
4 |. DISCUSSION
In this study, we tested FFPE samples from 26 pediatric patients (≤18 y of age) with a tissue diagnosis of LCH for BRAF p.V600E mutations using a BRAF V600E-specific monoclonal antibody (clone VE1). We compared VE1 results to highly sensitive AS-qPCR results (lower limit of detection of 0.5%) using 2 separate IHC-scoring criteria and assessed the staining properties of this antibody in decalcified LCH FFPE samples or samples with scant neoplastic infiltrate. The study highlights some important considerations to avoid BRAF false-negative and false-positive results when using molecular or VE1 IHC analyses in pediatric LCH tissue sections.
4.1 |. BRAF p.V600E mutation detection by AS-qPCR
The reported prevalence of BRAF p.V600E mutations in pediatric LCH varies widely from 21%42 to 69%43 with an average prevalence of approximately 50%44. We observed BRAF mutations in 57.7% of samples using an AS-qPCR assay capable of detecting BRAF mutations with high sensitivity (0.5%, 1 mutated allele in 200 wild-type alleles). For comparison, published sensitivities for BRAF mutation detection using Sanger sequencing, pyrosequencing, and next generation sequencing are approximately 10% to 20%, 5%, and 1% to 2%, respectively.27,28 Interestingly, the frequency of 57% is identical to the original publication by Badalian-Very et al who used next generation sequencing methods on CD1a + cells purified from FFPE specimens.7
BRAF wild-type inflammatory cells are recruited to the LCH lesional milieu via several proposed cell autonomous and nonautonomous mechanisms.8,9 Therefore, unlike other BRAF-positive malignancies such as melanoma, colorectal carcinoma, or papillary thyroid carcinoma, mutated dendritic cells may comprise only a minor fraction of the composite neoplastic lesion and are typically not amenable to microdissection. Using Sanger sequencing, lower BRAF mutation rates (<30%) were detected in several adult and pediatric LCH studies raising the distinct possibility of false-negative results.34,45,46 Therefore, we recommend that LCH BRAF mutation testing be performed with highly sensitive molecular assays to account for the low-mutant allele fractions intrinsic to LCH and limit potential false-negative molecular results. Additional studies are needed to define the optimal method(s) and requisite lower limit of detection for LCH BRAF mutation testing.
4.2 |. Immunohistochemistry scoring algorithm: lenient vs stringent scoring
Although thorough technical optimization of the BRAF antibody using control tissue occurred prior to the study, it was clear that optimization of the IHC interpretation for LCH was also required; optimized scoring criteria and highly sensitive comparative molecular gold standards are required to reliably assess the sensitivity and specificity of VE1.24,25 In 2 previous studies of BRAF VE1 IHC in LCH, Roden et al considered any cytoplasmic staining as positive staining,33 and Mehes and colleagues used a semiquantitative (0–2+) intensity scale.32 In both studies, false-positive VE1 results were observed. Likewise in our study, weak, nonspecific cytoplasmic staining was observed in 24 of 26 cases and yielded 9 false-positive VE1 results (Table 2). This nonspecific staining and corresponding suboptimal specificity precluded any meaningful comparisons using lenient IHC-scoring criteria (1+, ≥1%).
Using a stringent scoring algorithm devised for melanoma samples that combined staining intensity (0–3+) and percentage staining of tumor cells (0%−100%), we found that unequivocal IHC staining (2+, ≥10%) was 100.0% specific for an underlying BRAF mutation. However, sensitivity suffered with these criteria, as false-negative results were seen in approximately 20% of cases. False-negative IHC results can be seen in tumors harboring non-p.V600E codon mutations (eg, p.V600K) that cross-react in molecular assays, but not with the mutant antibody epitope.47 However, non-p.V600E codon mutations are rarely detected in LCH; in-frame BRAF exon 12 deletions48 or mutations in other MAP kinase pathway genes such as MAP2K1, MAP3K1, and ARAF are far more common.12,13,49 Thus, our data and the data of others highlight that VE1 negative samples require additional testing to exclude false-negative results. Also, consensus BRAF VE1-scoring algorithms are needed to standardize IHC results across laboratories for LCH and other tumor types.
4.3 |. Low neoplastic cell content and decalcification impact BRAF immunohistochemistry results
Although molecular assays are highly sensitive, sufficient concentrations of extracted, intact nucleic acid from neoplastic nuclei are required for valid results. Langerhans cell histiocytosis biopsies with rare neoplastic cells and/or decalcified specimens may be acceptable for histopathologic and immunophenotypic diagnoses, but are often inadequate for molecular testing. Thus, we evaluated the ability to use VE1 as a complement to molecular testing in these specimen types.
In the 3 IHC false-negative cases by stringent-scoring criteria, 2 cases harbored very low-LCH content (<5%, Table 1). In our experience, stringent IHC-scoring criteria are best suited for FFPE sections with abundant tumor to make an aggregate assessment of staining intensity and percentage of positive tumor cells; scant neoplastic tissue may underestimate IHC intensity and/or percentage of neoplastic cells because of the inherent heterogeneity of the tumor and or FFPE sections.23,26 In such cases, a binary positive-scoring/negative-scoring assessment would be favored, but the high false-positive rate nullifies any potential benefit.
In addition to the detrimental effects on nucleic acid–based molecular assays,38 decalcification has been associated with decreased IHC-staining intensity of breast cancer markers.50 Interestingly, VE1-staining intensity was attenuated in almost all decalcified samples. Only 1 of three (33%) AS-qPCR–positive decalcified bone samples demonstrated greater than 1+ VE1 staining (Table 3). Both samples with complete absence of VE1 staining showed concurrent PCR negative results, including 1 sample that initially failed molecular analysis due to insufficient DNA integrity. The other decalcified sample that failed initial testing and was repeated with nondecalcified material (LCH-55) also demonstrated 1+ staining and yielded negative AS-PCR results. Additionally, 2 AS-qPCR–positive bone samples, LCH-33 and LCH-6 with 30% and less than 5% LCH, respectively, demonstrated false-negative VE1 results by stringent criteria (Table 3).
Collectively, these data suggest that scant and decalcified tissue specimens predispose to false-negative IHC results secondary to scoring inaccuracies and compromised VE1-staining intensity, respectively. For these reasons, VE1 offered no definitive advantage over molecular testing in decalcified LCH tissue sections in our study. We use a formic acid/formalin solution–based chelation (Cal-Ex II) solution for short duration to minimize nucleic acid hydrolysis and acknowledge that the type of decalcification reagent may have influenced the IHC and molecular results presented here. Therefore, a more extensive study is needed to definitely assess the effects of low-neoplastic tissue content and the effects of various decalcification solutions on the performance of VE1 in LCH using a semiquantitative scoring algorithm.
5 |. CONCLUSION
In this study, we assessed the performance of mutation-specific VE1 using optimized stringent-scoring criteria and compared results to a highly sensitive AS-qPCR molecular method in FFPE tissue samples from 26 pediatric LCH patients. Nonspecific, 1+ cytoplasmic staining in both BRAF-mutated and wild-type LCH initially hindered IHC analysis, but stringent-scoring criteria improved specificity to 100.0%. Stringent-scoring criteria yielded false-negative results in 20% of cases, and false-negative results were enriched in both decalcified samples and samples with scant neoplastic LCH content. We conclude that strong, 2+, staining in greater than or equal to 10% neoplastic histiocytes is indicative of an underlying BRAF p.V600E mutation, but all negative VE1 results require additional testing to exclude false-positives. Also, caution should be used when interpreting either decalcified specimens or specimens with scant neoplastic tissue content. Importantly, highly sensitive molecular assays capable of detecting low-level mutations remain the gold standard for BRAF mutation analysis in LCH lesion.
ACKNOWLEDGEMENTS
The authors would like to thank Angela Major for her excellent technical assistance. Portions of this work were presented at the Association of Molecular Pathology 2016 Annual Meeting in Austin, Texas, and the 32nd Annual Meeting of the Histiocytosis Society in Dublin, Ireland. No external funding was applied to this study.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Collin M, Bigley V, McClain KL, Allen CE. Cell(s) of origin of Langerhans cell histiocytosis. Hematol Oncol Clin North Am. 2015;29: 825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emile JF, Abla O, Fraitag S, et al. Revised classification of histiocytoses and neoplasms of the macrophage-dendritic cell lineages. Blood. 2016;127:2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gadner H, Minkov M, Grois N, et al. Therapy prolongation improves outcome in multisystem Langerhans cell histiocytosis. Blood. 2013;121:5006–5014. [DOI] [PubMed] [Google Scholar]
- 4.Hicks J, Flaitz CM. Langerhans cell histiocytosis: current insights in a molecular age with emphasis on clinical oral and maxillofacial pathology practice. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2005;100: S42–S66. [DOI] [PubMed] [Google Scholar]
- 5.Simko SJ, Garmezy B, Abhyankar H, et al. Differentiating skin-limited and multisystem Langerhans cell histiocytosis. J Pediatr. 2014;165:990–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau SK, Chu PG, Weiss LM. Immunohistochemical expression of Langerin in Langerhans cell histiocytosis and non-Langerhans cell histiocytic disorders. Am J Surg Pathol. 2008;32:615–619. [DOI] [PubMed] [Google Scholar]
- 7.Badalian-Very G, Vergilio JA, Degar BA, et al. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 2010;116: 1919–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badalian-Very G, Vergilio JA, Fleming M, Rollins BJ. Pathogenesis of Langerhans cell histiocytosis. Annu Rev Pathol. 2013;8:1–20. [DOI] [PubMed] [Google Scholar]
- 9.Berres ML, Lim KP, Peters T, et al. BRAF-V600E expression in precursor versus differentiated dendritic cells defines clinically distinct LCH risk groups. J Exp Med. 2014;211:669–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahm F, Capper D, Preusser M, et al. BRAFV600E mutant protein is expressed in cells of variable maturation in Langerhans cell histiocytosis. Blood. 2012;120:e28–e34. [DOI] [PubMed] [Google Scholar]
- 11.Brown NA, Furtado LV, Betz BL, et al. High prevalence of somatic MAP2K1 mutations in BRAF V600E–negative Langerhans cell histiocytosis. Blood. 2014;124:1655–1658. [DOI] [PubMed] [Google Scholar]
- 12.Chakraborty R, Hampton OA, Shen X, et al. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood. 2014;124:3007–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson DS, van Halteren A, Quispel WT, et al. MAP2K1 and MAP3K1 mutations in Langerhans cell histiocytosis. Genes Chromosomes Cancer. 2015;54:361–368. [DOI] [PubMed] [Google Scholar]
- 14.Heritier S, Emile JF, Barkaoui MA, et al. BRAF mutation correlates with high-risk Langerhans cell histiocytosis and increased resistance to first-line therapy. Journal Clin Oncol. 2016;34:3023–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritterhouse LL, Barletta JA. BRAF V600E mutation-specific antibody: a review. Semin Diagn Pathol. 2015;32:400–408. [DOI] [PubMed] [Google Scholar]
- 16.Adackapara CA, Sholl LM, Barletta JA, Hornick JL. Immunohistochemistry using the BRAF V600E mutation-specific monoclonal antibody VE1 is not a useful surrogate for genotyping in colorectal adenocarcinoma. Histopathology. 2013;63:187–193. [DOI] [PubMed] [Google Scholar]
- 17.Andrulis M, Penzel R, Weichert W, von Deimling A, Capper D. Application of a BRAF V600E mutation-specific antibody for the diagnosis of hairy cell leukemia. Am J Surg Pathol. 2012;36:1796–1800. [DOI] [PubMed] [Google Scholar]
- 18.Capper D, Preusser M, Habel A, et al. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011;122:11–19. [DOI] [PubMed] [Google Scholar]
- 19.Feller JK, Yang S, Mahalingam M. Immunohistochemistry with a mutation-specific monoclonal antibody as a screening tool for the BRAFV600E mutational status in primary cutaneous malignant melanoma. Mod Pathol. 2013;26:414–420. [DOI] [PubMed] [Google Scholar]
- 20.Fisher KE, Neill SG, Ehsani L, Caltharp SA, Siddiqui MT, Cohen C. Immunohistochemical investigation of BRAF p.V600E mutations in thyroid carcinoma using 2 separate BRAF antibodies. Appl Immunohistochem Mol Morphol. 2014;22:562–567. [DOI] [PubMed] [Google Scholar]
- 21.Ilie M, Long E, Hofman V, et al. Diagnostic value of immunohistochemistry for the detection of the BRAFV600E mutation in primary lung adenocarcinoma Caucasian patients. Ann Oncol. 2013;24:742–748. [DOI] [PubMed] [Google Scholar]
- 22.Anwar MA, Murad F, Dawson E, Abd Elmageed ZY, Tsumagari K, Kandil E. Immunohistochemistry as a reliable method for detection of BRAF-V600E mutation in melanoma: a systematic review and meta-analysis of current published literature. J Surg Res. 2016;203:407–415. [DOI] [PubMed] [Google Scholar]
- 23.Boursault L, Haddad V, Vergier B, et al. Tumor homogeneity between primary and metastatic sites for BRAF status in metastatic melanoma determined by immunohistochemical and molecular testing. PLoS One. 2013;8:e70826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisher KE, Cohen C, Siddiqui MT, Palma JF, Lipford EH 3rd, Longshore JW. Accurate detection of BRAF p.V600E mutations in challenging melanoma specimens requires stringent immunohistochemistry scoring criteria or sensitive molecular assays. Human Pathol. 2014;45: 2281–2293. [DOI] [PubMed] [Google Scholar]
- 25.Kuan SF, Navina S, Cressman KL, Pai RK. Immunohistochemical detection of BRAF V600E mutant protein using the VE1 antibody in colorectal carcinoma is highly concordant with molecular testing but requires rigorous antibody optimization. Human Pathol. 2014;45: 464–472. [DOI] [PubMed] [Google Scholar]
- 26.Yancovitz M, Litterman A, Yoon J, et al. Intra- and inter-tumor heterogeneity of BRAF(V600E)mutations in primary and metastatic melanoma. PLoS One. 2012;7:e29336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ihle MA, Fassunke J, Konig K, et al. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non-p.V600E BRAF mutations. BMC Cancer. 2014;14:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richter A, Grieu F, Carrello A, et al. A multisite blinded study for the detection of BRAF mutations in formalin-fixed, paraffin-embedded malignant melanoma. Sci Rep. 2013;3:1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reid AL, Freeman JB, Millward M, Ziman M, Gray ES. Detection of BRAF-V600E and V600 K in melanoma circulating tumour cells by droplet digital PCR. Clin Biochem. 2015;48:999–1002. [DOI] [PubMed] [Google Scholar]
- 30.Clevers H. At the crossroads of inflammation and cancer. Cell. 2004;118:671–674. [DOI] [PubMed] [Google Scholar]
- 31.Schottenfeld D, Beebe-Dimmer J. Chronic inflammation: a common and important factor in the pathogenesis of neoplasia. CA Cancer J Clin. 2006;56:69–83. [DOI] [PubMed] [Google Scholar]
- 32.Mehes G, Irsai G, Bedekovics J, et al. Activating BRAF V600E mutation in aggressive pediatric Langerhans cell histiocytosis: demonstration by allele-specific PCR/direct sequencing and immunohistochemistry. Am J Surg Pathol. 2014;38:1644–1648. [DOI] [PubMed] [Google Scholar]
- 33.Roden AC, Hu X, Kip S, et al. BRAF V600E expression in Langerhans cell histiocytosis: clinical and immunohistochemical study on 25 pulmonary and 54 extrapulmonary cases. Am J Surg Pathol. 2014;38: 548–551. [DOI] [PubMed] [Google Scholar]
- 34.Sasaki Y, Guo Y, Arakawa F, et al. Analysis of the BRAFV600E mutation in 19 cases of Langerhans cell histiocytosis in Japan. Hematol Oncol. 2016; Epub ahead of print]. doi: 10.1002/hon.2293 [DOI] [PubMed] [Google Scholar]
- 35.Ballester LY, Sarabia SF, Sayeed H, et al. Integrating molecular testing in the diagnosis and management of children with thyroid lesions. Pediatr Dev Pathol. 2016;19:94–100. [DOI] [PubMed] [Google Scholar]
- 36.Dvorak K. Evaluation of BRAF (V600E) mutation by immunohistochemical staining with anti-BRAF V600E (VE1) antibody: a comparison with Sanger sequencing. http://www.ventana.com/documents/N4850A_Evaluation_of_BRAF_WP.pdf. Accessed on September 8, 2016. [Google Scholar]
- 37.Lee JW, Shin HY, Kang HJ, et al. Clinical characteristics and treatment outcome of Langerhans cell histiocytosis: 22 years’ experience of 154 patients at a single center. Pediatric Hematol Oncol. 2014;31:293–302. [DOI] [PubMed] [Google Scholar]
- 38.Goswami RS, Luthra R, Singh RR, et al. Identification of factors affecting the success of next-generation sequencing testing in solid tumors. Am J Clin Pathol. 2016;145:222–237. [DOI] [PubMed] [Google Scholar]
- 39.Chen H, Luthra R, Goswami RS, Singh RR, Roy-Chowdhuri S. Analysis of pre-analytic factors affecting the success of clinical next-generation sequencing of solid organ malignancies. Cancers (Basel). 2015;7: 1699–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lassmann S, Gerlach UV, Technau-Ihling K, Werner M, Fisch P. Application of BIOMED-2 primers in fixed and decalcified bone marrow biopsies: analysis of immunoglobulin H receptor rearrangements in B-cell non-Hodgkin’s lymphomas. J Mol Diagn. 2005;7:582–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mangham DC, Williams A, McMullan DJ, et al. Ewing’s sarcoma of bone: the detection of specific transcripts in a large, consecutive series of formalin-fixed, decalcified, paraffin-embedded tissue samples using the reverse transcriptase-polymerase chain reaction. Histopathology. 2006;48:363–376. [DOI] [PubMed] [Google Scholar]
- 42.Alayed K, Medeiros LJ, Patel KP, et al. BRAF and MAP2K1 mutations in Langerhans cell histiocytosis: a study of 50 cases. Human Pathol. 2016;52:61–67. [DOI] [PubMed] [Google Scholar]
- 43.Satoh T, Smith A, Sarde A, et al. B-RAF mutant alleles associated with Langerhans cell histiocytosis, a granulomatous pediatric disease. PLoS One. 2012;7:e33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bubolz AM, Weissinger SE, Stenzinger A, et al. Potential clinical implications of BRAF mutations in histiocytic proliferations. Oncotarget. 2014;5(12):4060–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Go H, Jeon YK, Huh J, et al. Frequent detection of BRAF(V600E) mutations in histiocytic and dendritic cell neoplasms. Histopathology. 2014;65:261–272. [DOI] [PubMed] [Google Scholar]
- 46.Tong C, Jia X, Jia Y, He Y. Langerhans cell histiocytosis in Chinese adults: absence of BRAF mutations and increased FOXP3(+) regulatory T cells. Int J Clin Exp Pathol. 2014;7:3166–3173. [PMC free article] [PubMed] [Google Scholar]
- 47.Jabbar KJ, Luthra R, Patel KP, et al. Comparison of next-generation sequencing mutation profiling with BRAF and IDH1 mutation-specific immunohistochemistry. Am J Surg Pathol. 2015;39:454–461. [DOI] [PubMed] [Google Scholar]
- 48.Chakraborty R, Burke TM, Hampton OA, et al. Alternative genetic mechanisms of BRAF activation in Langerhans cell histiocytosis. Blood. 2016;128:2533–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson DS, Quispel W, Badalian-Very G, et al. Somatic activating ARAF mutations in Langerhans cell histiocytosis. Blood. 2014;123: 3152–3155. [DOI] [PubMed] [Google Scholar]
- 50.Gertych A, Mohan S, Maclary S, et al. Effects of tissue decalcification on the quantification of breast cancer biomarkers by digital image analysis. Diagn Pathol. 2014;9:213. [DOI] [PMC free article] [PubMed] [Google Scholar]