Summary
An individual’s genetic makeup plays a large role in determining susceptibility to Alzheimer’s disease (AD), but has largely been ignored in preclinical studies. To test the hypothesis that incorporating genetic diversity into mouse models of AD would improve translational potential, we combined a well-established mouse model of AD with a genetically diverse reference panel to generate mice that harbor identical high-risk human mutations but differ across the remainder of their genome. We first show that genetic variation profoundly modifies the impact of human AD mutations on both cognitive and pathological phenotypes. We then validate this complex AD model by demonstrating high degrees of genetic, transcriptomic, and phenotypic overlap with human AD. Overall, work here both introduces a novel AD mouse population as an innovative and reproducible resource for the study of mechanisms underlying AD and provides evidence that preclinical models incorporating genetic diversity may better translate to human disease.
eTOC Blurb
Neuner et al. describe the generation and validation of the AD-BXDs, the first reproducible set of AD mouse models incorporating genetic diversity. These models recapitulate aspects of human AD and provide a new resource to identify genes and pathways associated with risk and resilience to AD.
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disorder characterized by both dementia and the accumulation of neuropathological amyloid plaques and tau tangles (Selkoe, 1991). Mutations that drive overproduction of beta-amyloid (Aβ) have been shown to cause early onset familial AD (FAD), leading to a model in which production and accumulation of Aβ is thought to be an initiating event in a sequence leading to memory loss, neurodegeneration, gliosis, and synaptic dysfunction (Hardy and Higgins, 1992). However, strategies to directly target amyloid for clearance have failed to translate into successful treatments and the number of deaths attributable to AD and costs associated with the disease continue to rise.
Although age is the greatest risk factor for developing AD, it is increasingly clear that genetics and family history play a large role. The heritability of AD is estimated to be in the range of 50–80%, indicating an individual’s susceptibility or resilience to disease is, at least in part, determined by heritable DNA variants (Gatz et al., 1997). Even among patients with FAD mutations, the age at first symptom onset is widely variable, with some patients exhibiting symptoms decades later than predicted based on mutation status. This suggests additional genetic factors exist that may provide protection from disease (Ryman et al., 2014). While number of genetic risk factors have been identified (Lambert et al., 2013), besides APOE and TREM2, effect sizes of identified variants are generally small. As such, a large proportion of variation in disease risk and severity remains undefined and unexploited (Ridge et al., 2013). In addition, the study design utilized in most genome wide association tests (e.g. case-control) is primed to identify variants associated with risk of AD, rather than variants that modify an individual’s disease trajectory and/or delay the onset of disease and provide protection from AD. This is due primarily to the fact that asymptomatic individuals rarely enter the clinic for treatment, and even if they are included in a study, are likely to be enrolled as cognitively normal controls.
Identifying genetic variants and pathways involved in protection from AD will provide valuable targets for new therapeutics to prevent or delay the onset of symptoms. Individuals with high-risk genotypes but who fail to present with clinical symptoms of AD represent an ideal population in which to study resilience. However, causal mutations in APP and PSEN1/2 (i.e. high-risk genotypes) are rare in humans, greatly limiting statistical power and opportunity for analysis. In addition, access to brain tissue at early disease time points, before overt symptom onset, is limited for obvious reasons in human studies, precluding the identification of causal versus collateral molecular mechanisms. Thus, mouse models harboring causal AD mutations are important tools that present many advantages including defined high-risk genotypes, early access to brain tissue, and precise environmental control.
That said, there are caveats to traditional AD mouse models, including the fact that most mouse models of AD are maintained on only a single or a few genetic backgrounds (Onos et al., 2016). This includes backgrounds of mixed origins, which may confound interpretation of results. Previous studies in mice have demonstrated that genetic background has a strong effect on Aβ levels, with several mapping studies in F2 populations identifying large genomic regions involved in modulating amyloid burden (Ryman et al., 2008; Sebastiani et al., 2006). However, these studies and individual strain-by-strain comparisons have not evaluated the effect of genetic background on cognitive performance (Jackson et al., 2015; Sipe et al., 1993). Given the impact of genetics on cognitive decline in human populations (Gatz et al., 1997), we hypothesized genetic background was also playing an important role in modifying cognitive decline in animal models of disease, and that inclusion of genetic diversity would improve translational validity of AD mouse models.
In order to directly test this hypothesis, and to aid in the identification of specific genes involved in modifying resilience to AD, we developed the first AD transgenic mouse reference panel. This panel, which we term the AD-BXDs, combines two well-established resources: 1) the 5XFAD transgenic line on an otherwise fully inbred C57BL/6J (B6) background that recapitulates various aspects of the human disease, including amyloid-β42 accumulation, cognitive deficits, and neuron loss (Oakley et al., 2006), and 2) the BXD genetic reference panel, the largest and best-characterized series of recombinant inbred strains derived from the two common inbred strains B6 and DBA/2J (D2) (Peirce et al., 2004; Taylor et al., 1999). The BXD panel segregates for more than 4.8 million sequence variants, including many in genes known to confer risk for AD (Wang et al., 2016). The resulting panel of F1 hybrids represent a novel and fully isogenic resource to monitor phenotypic outcomes in individuals harboring identical high-risk FAD mutations in human APP and PSEN1 genes, raised in controlled environments, but whose allelic contributions differ across the remainder of the genome.
The general aim of the work described here is to build and test the validity of the AD-BXDs as a resource that will enable the research community to systematically identify sets of genetic variants and pathways involved in determining individual susceptibility or resilience to AD. As both parental lines of the AD-BXD panel are fully inbred, the resulting panel also provides a reproducible resource to efficiently evaluate gene-by-environment-by-treatment effects to test, triage, and translate therapeutics more quickly and accurately. To validate this design as a model of AD, here we show that our AD-BXD panel faithfully recapitulates key aspects of the human disease, including phenotypic variation in disease onset and severity, sensitivity to genetic variation in genes known to confer risk for human late-onset Alzheimer’s disease (LOAD), and a high level of concordance with transcriptional aspects of human disease. Thus, we present the AD-BXD panel as a new mouse model of human AD with high translational potential for both understanding the complex etiology of FAD and sporadic LOAD and discovering new genetic and molecular pathways associated with AD risk and resilience.
Results
Genetic background modifies expressivity of FAD mutations
In order to evaluate the influence of genetic background on the impact of causal FAD mutations on behavioral and molecular phenotypes, we generated a panel of 28 genetically diverse F1 mouse strains with and without FAD mutations. Female B6 mice heterozygous for the autosomal dominant 5XFAD transgene (Oakley et al., 2006) were crossed to males from the BXD genetic reference panel (Peirce et al., 2004) to generate F1 progeny carrying the 5XFAD transgene (ADBXDs) or non-transgenic littermates (Ntg-BXDs; Figure 1A). Working memory and body weight were monitored bi-monthly and more in-depth phenotyping that included tests of motor function and anxiety was performed at both 6 and 14 months of age (Figure 1B). A subset of mice was subsequently tested for long-term spatial learning and memory function using a contextual fear conditioning (CFC) paradigm (Fanselow, 2000; Neuner et al., 2016). This subset was immediately harvested following CFC testing and tissue was collected for biobanking and later use, including RNA-sequencing and enzyme-linked immunosorbent assays (ELISAs) as described below. This time point (immediately following testing) was chosen in order to capture molecular changes corresponding to differences in learning-related intrinsic neuronal excitability reported previously (Kaczorowski and Disterhoft, 2009; Kaczorowski et al., 2011).
Figure 1: Genetic background modifies AD symptoms in a novel transgenic reference panel.
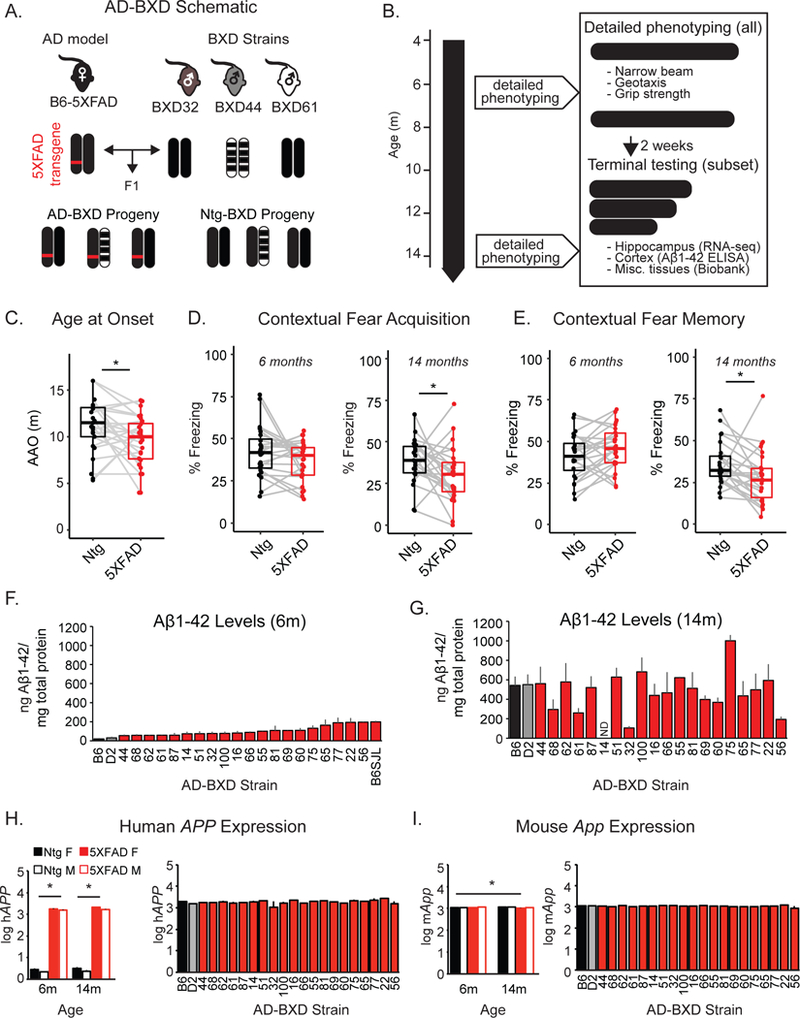
(A) Female B6 mice heterozygous for the dominant 5XFAD transgene were bred to males from 27 BXD strains to generate genetically diverse but isogenic F1 offspring. (B) Body weight and working memory on the y-maze were measured bi-monthly, and at 6 and 14 months more detailed phenotyping was performed. (C) As expected, onset of working memory deficits was significantly earlier in AD-BXDs compared to Ntg-BXDs [AD-BXDs: n = 223 (123 females/100 males) across 28 strains vs Ntg-BXDs, n = 168 mice (107 female/61 male) across 25 strains, one-tailed t(1,51) = 2.1, p = 0.02]. (D) AD-BXD mice exhibited contextual fear acquisition (CFA) comparable to Ntg-BXD mice at 6m [left, one-tailed t(1, 48) = 1.4, p = 0.08] but are impaired by 14m [right, one-tailed t(1, 49) = 2.0, p = 0.03] months. Within AD-BXD mice, background strain significantly modified the impact of the transgene on CFA [effect of strain F(26, 354) = 3.3, p < 0.001]. (E) AD-BXD mice exhibit recall comparable to Ntg-BXDs during the contextual fear memory (CFM) task at 6 months [left, one-tailed t(1,48) = 1.4, p = 0.08] but are impaired by 14 months [right, one-tailed t(1, 49) = 1.9, p = 0.03]. Within AD-BXD mice, background strain significantly modified the impact of the transgene on CFM [effect of strain F(26, 354) = 3.5, p < 0.001]. For D and E, 146 6m AD-BXD (102 females/44 males) and 209 14m AD-BXD (111 females/98 males) across 26 strains were used, along with 114 6m Ntg-BXD (83 females/31 males) across 24 strains and 167 14m Ntg-BXD mice (106 females/61 males) across 27 strains. (F) and (G) Aβ42, as measured by ELISA, increased drastically from 6 to 14m [effect of age F(1,153) = 128.0, p < 0.001] but varied significantly across genetic backgrounds [effect of strain F(22,153) = 2.0, p = 0.01]. n = 154 mice (89 female/65 male) across 23 strains. ND = no data. Strain B6SJL represents the original background strain described in Oakley et al. 2006 for comparison. (H, left) Transgene expression was assessed in subset of AD and Ntg-BXD lines [n = 293 (177 females/116 males across 28 strains)]. RNA sequencing reads from the hippocampus were aligned to the human mutant sequence of APP, quantified by number of transcripts per million reads (TPM), and log transformed. AD-BXD mice exhibited significantly greater hAPP expression [t(1, 291) = 92.3, p < 0.001]. Across the AD-BXDs, there was no significant effects of age, sex, or (H, right) background strain. Only strains with Aβ42 data are shown here for comparison to F and G. (I, left) Same analysis was done for reads aligned to the mouse endogenous App. Across the panel, 5XFAD mice exhibited slight but significant reduction in App [t(1,291) = 2.6, p = 0.01]. However, within AD-BXD mice there was no effect of age, sex, or (I, right) background strain. For plots C-E, each point represents a strain average. All t-tests in C-E were one-tailed tests based on prior data assessing effects of the 5XFAD transgene on cognitive function (Kaczorowski et al., 2011; Oakley et al., 2006; Ohno, 2009) ; *p < 0.05. See also Figure S1, S2, S3, and S4.
As expected (Kaczorowski et al., 2011; Oakley et al., 2006; Ohno, 2009), the 5XFAD transgene accelerated the age at onset (AAO) of working memory deficits in AD-BXD mice relative to Ntg-BXD mice (Figure 1C) and exacerbated contextual fear acquisition (CFA) and contextual fear memory (CFM) deficits, particularly by 14 months of age (Figure 1D-E). However, the impact of causal FAD mutations on cognitive performance varied widely depending on the specific background strain evaluated. Notably, this variation in cognitive function parallels the variation observed in human patients harboring FAD mutations (Ryman et al., 2014) and was not correlated with strain-specific variation in activity, pain sensitivity, sensorimotor abilities, or anxiety (Figure S1). These results suggest the observed variation in cognitive function is regulated, in part, by genetic variants that segregate across the AD-BXD panel. In support, heritability (h2RIx̅ ) estimates comparing between-strain variance (due to genetic diversity) to total sample variance (due to both genetic and environmental factors), given the average number of biological replicates per strain (Belknap, 1998), demonstrate there is a significant genetic component underlying observed variation (Table 1).
Table 1:
Heritability estimates for phenotypic traits in AD- and Ntg-BXDs. Heritability (h2RI̅) was determined by calculating the ratio of between-strain variance (i.e. genetic variance) to total sample variance (within-strain variance due to technical/environmental factors plus between-strain variance), given the average number of biological replicates per strain according to established methods (Belknap, 1998).
| Non-transgenic (Ntg)-BXDs | ||||
|---|---|---|---|---|
| Trait | Between strain variance |
Av. within strain variance |
Av. n/strain |
Heritability (h2RIx̅) |
| Age at onset | 7.7 | 15.7 | 7.5 | 0.8 |
| 6m CFA | 181.2 | 433.1 | 5.1 | 0.7 |
| 6m CFM | 196.6 | 340.5 | 5.1 | 0.7 |
| 14m CFA | 146.6 | 520.7 | 7.2 | 0.7 |
| 14m CFM | 168.0 | 321.9 | 7.2 | 0.8 |
|
6m
Sensorimotor composite |
0.3 | 2.1 | 8.7 | 0.6 |
|
14m
Sensorimotor composite |
1.7 | 6.8 | 7.4 | 0.6 |
|
6m EPM % Time in Open
Arms |
7.2 | 62.7 | 8.7 | 0.5 |
|
14m EPM % Time in Open
Arms |
29.2 | 149.6 | 7.4 | 0.6 |
| AD-BXDs | ||||
| Trait |
Between strain variance |
Av. within strain variance |
Av. n/strain |
Heritability (h2RIx̅) |
| Age at onset | 5.7 | 15.7 | 8.8 | 0.8 |
| 6m CFA | 142.9 | 293.7 | 5.6 | 0.7 |
| 6m CFM | 163.3 | 376.8 | 5.6 | 0.7 |
| 14m CFA | 172.0 | 360.4 | 9.0 | 0.8 |
| 14m CFM | 141.5 | 299.8 | 9.0 | 0.8 |
|
6m
Sensorimotor composite |
1.2 | 3.8 | 10.8 | 0.7 |
|
14m
Sensorimotor composite |
2.4 | 9.3 | 9.2 | 0.7 |
|
6m EPM % Time in Open
Arms |
38.8 | 322.7 | 10.7 | 0.6 |
|
14m EPM % Time in Open
Arms |
266.6 | 625.6 | 8.8 | 0.8 |
| 6m Amyloid (ELISA) | 2570.6 | 2050.5 | 3.3 | 0.8 |
| 14m Amyloid (ELISA) | 36141.9 | 64897.9 | 3.9 | 0.7 |
Abbreviations: Av: average, m: months, CFA: contextual fear acquisition, CFM: contextual fear memory, EPM: Elevated Plus Maze, WM: Decline Slope, ELISA: enzyme-linked immunosorbent assay
Human FAD mutations in APP and PSEN1 included in the 5XFAD transgene increase production of the toxic 42 amino-acid length amyloid beta species (Aβ1–42), thought to be an initiating factor in a cascade of symptoms eventually leading to neuron loss and dementia (Hardy and Higgins, 1992). To assess the impact of genetic background on the levels of Aβ1–42 across the panel, brain extracts from 23 AD-BXD strains were assayed in duplicate on human Aβ1–42-specific sandwich ELISAs (Oakley et al., 2006). Variation in human Aβ1–42 levels was heritable (Table 1), and overall levels increased with age [effect of age F(1,153) = 128.0, p < 0.001] (Figure 1F-G). A significant main effect of strain was observed [F(22,153) = 2.0, p = 0.01], indicating that genetic background significantly modified human Aβ1–42 levels across the panel. In order to test whether elevated amyloid levels corresponded to an increase in plaque density, we performed immunohistochemistry (IHC) analysis on a subset of fixed hemibrains and observed robust plaque deposition in both the hippocampus and cortex of AD-BXD strains, each of which significantly correlated with amyloid levels as measured by ELISA (Figure S2A-C). As expected, human Aβ1–42 was not reliably detected in 8 Ntg-BXD brains by ELISA, or in 3 Ntg-BXD brains by IHC (Figure S2D), suggesting that at least by 6 months of age, Ntg-BXDs do not develop deposition of human Aβ42 compared to their 5XFAD isogenic counterparts. Similar to what is observed in human populations, no significant correlation was observed between amyloid levels and cognitive function (Figure S3), suggesting partially independent mechanisms work to regulate the extent of cognitive decline and amyloid accumulation.
Differences in cognitive function and Aβ1–42 pathology were not explained by an effect of age, sex, or background strain on the transcription of the 5XFAD transgene itself, as measured by alignment of RNA-sequencing reads from the hippocampus to the mutated human APP (Figure 1H, Figure S4A) or PSEN1 (Figure S4B) sequences that make up the 5XFAD transgene (Oakley et al., 2006). The lack of a sex difference on either transgene expression or amyloid levels is in contrast to a previous report using a single genetic background demonstrating the 5XFAD transgene is differentially expressed based on sex (Sadleir et al., 2015), suggesting that sex-specific effects may vary across genetic backgrounds. In addition, across the AD-BXD panel, there was no effect of genetic background, age, or sex on expression of endogenous App (Figure 1I, Figure S4C) or Psen1 (Figure S4D). Overall, these results suggest that naturally occurring variants segregating across the AD-BXD panel, rather than artificial differences due to transgene expression, play a significant role in determining susceptibility and/or resilience to changes in cognitive function and amyloid deposition caused by high-risk FAD mutations.
Cognitive function in the AD-BXDs is sensitive to known AD risk variants
To test the hypothesis that the inclusion of genetic diversity would better model human AD, we first evaluated whether the AD-BXD panel is sensitive to variation in genes known to confer risk for LOAD. Because the apolipoprotein E gene (APOE) is the best characterized risk gene for LOAD in human patients and is relatively well conserved in the mouse (Liao et al., 2015), we queried variants in mouse Apoe. One single nucleotide polymorphism (SNP) in Apoe segregates across the BXD panel (Figure 2A), occurring near the receptor-binding region (Mahley et al., 2009). Based on sequence alignment, this SNP causes a switch from glutamate to aspartate at mouse position 163 (Zerbino et al., 2018). While the exact functional consequences of this SNP are unknown, and likely depend on the context of surrounding amino acids, we predicted the D allele of Apoe would represent a susceptibility allele across the AD-BXDs based on sequence homology.
Figure 2: AD-BXD panel is sensitive to variation in known AD risk gene Apoe.

(A) The D allele harbors only a single E > D missense SNP at mouse 163 (red). (B) Across AD-BXD mice, there was a significant effect of Apoe allele [F(1, 354) = 4.7, p = 0.03], age [F(1,354) = 12.3, p = 0.001], and sex [F(1,354) = 17.9, p < 0.001] on contextual fear acquisition (CFA). There was a trend toward Apoe having a more significant effect on CFA in females than in males [strain*sex interaction, F(1, 354) = 3.2, p = 0.08]. (C) Across AD-BXD mice, there was a significant effect of Apoe allele [F(1, 354) = 20.9, p < 0.001], age [F(1,354) = 86.2, p < 0.001], sex [F(1, 354) = 4.9, p = 0.03], and an age by sex interaction [F(1, 354) = 7.6, p = 0.006] on contextual fear memory (CFM), demonstrating that while strains carrying the D allele at Apoe do indeed perform more poorly on this task, all female AD mice are more susceptible to AD-related cognitive decline as measured by CFM. See also Figure S5.
To test this hypothesis, we first identified genotyping markers flanking Apoe across the AD- BXDs and then determined the allelic composition of Apoe in each strain. A significant effect of Apoe allele was observed on CFA [F(1,354) = 4.7, p = 0.03], indicating that strains carrying one copy of the D allele of Apoe performed worse on this task (Figure 2B). We also observed a significant effect of age [F(1,354) = 12.3, p = 0.001] and sex [F(1, 354) = 17.9, p < 0.001] on CFA, as well as a trend toward an interaction between sex and Apoe genotype [F(1, 354) = 3.2, p = 0.08]. Together, these results indicate that while most mice exhibited age-related decline in acquisition, female mice generally performed worse on the task and were also particularly susceptible to the effects of the D allele of Apoe. The Apoe effect was even more pronounced when we considered CFM; a significant main effect of Apoe allele was again detected [F(1,355) = 20.9, p < 0.001], along with significant effects of sex [F(1,355) = 4.9, p = 0.03], age [F(1,355) = 86.2, p < 0.001], and a sex by age interaction [F(1,355) = 7.6, p = 0.006] (Figure 2C). These results indicate first that mice harboring a single copy of the D allele of Apoe exhibited poorer CFM, and second that female mice are more susceptible to AD-related cognitive decline with age. No effect of Apoe genotype was observed on working memory traits. Across Ntg-BXDs, Apoe genotype exhibited either a less robust effect, or no effect, on cognitive performance on CFA and CFM tasks, respectively (Figure S5).
Overall, the above data demonstrates that variation at the Apoe locus in mice, particularly those harboring the 5XFAD transgene, is associated with cognitive outcomes. In humans, additional genes have been identified that play small, although significant, roles in regulating susceptibility to AD (Lambert et al., 2013). Recent studies suggest that information about genetic variation at these additional loci, in the form of a genetic risk score (GRS), can better predict an individual’s risk of developing AD (Chouraki et al., 2016). In order to evaluate whether naturally occurring variants in genes associated with LOAD risk in humans are associated with cognitive outcomes in the AD-BXD panel, we computed a GRS for each of our strains similar to the method described by Chouraki and colleagues in 2016 (Chouraki et al., 2016). First, we stratified strains into impaired and unimpaired groups based on 6 month-old CFM (Figure 3A). We then identified the genotype of each strain at 21 LOAD risk genes (across 19 genetic markers, Table 2) and classified the risk allele of each gene as that allele which appeared more frequently in the impaired group. Odds ratios were calculated and transformed based on risk allele dosage to obtain a final GRS for each strain, which was normally distributed across the panel (Shapiro-Wilk test for normality p = 0.7, Figure 3B).
Figure 3: Genetic risk score calculated from genotype at known AD risk genes predicts cognitive decline.
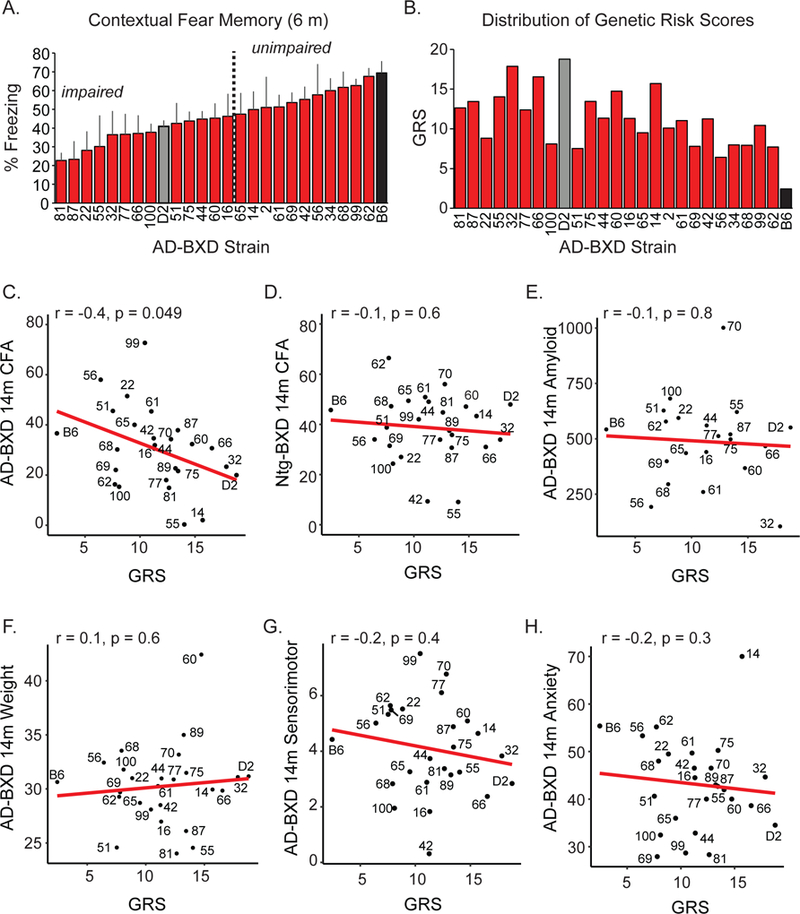
(A) Strains were stratified into impaired (below population average) and unimpaired (above population average) based on 6m CFM performance. (B) Genetic risk scores (GRS) were calculated for each strain based on allelic composition of 21 genes known to confer risk for AD. The risk allele of each gene was defined as that which appeared more frequently in the impaired population pictured in (A). (C) GRS significantly predicts how a given AD-BXD strain will perform on CFA at 14m. (D) No relationship between GRS and CFA in 14m Ntg-BXD mice was observed. (E) No relationship between GRS and non-cognitive traits across AD-BXDs including 14m amyloid load, (F) 14m weight, (G) 14m sensorimotor performance, (H) or 14m anxiety as measured by percent open entries on the elevated plus maze was observed. See also Figure S6.
Table 2:
Genes known to confer risk of Alzheimer’s disease in humans vary across the AD-BXD panel and confer various degrees of risk in our mouse population; this information was used to create a genetic risk score for each strain.
| Gene | 6m CFM AD | |||||||
|---|---|---|---|---|---|---|---|---|
| Mouse Chr. |
SNP Density (SNP/Kb) |
High impact changes |
Risk allele |
Odds ratio |
95 % CI | Z stat | Pval | |
| Inpp5d | 1 | 1.24 | NMD SNP + indel |
B | 1.30 | 0.28 – 6.3 | 0.36 | 0.72 |
| Cr1l | 1 | 0.06 | - | D | 2.00 | 0.41 – 9.8 | 0.85 | 0.39 |
| Celf1 | 2 | 0.70 | - | D | 1.50 | 0.30 – 7.4 | 0.50 | 0.62 |
| Cass4 | 2 | 0.08 | - | D | 1.50 | 0.30 – 7.4 | 0.50 | 0.62 |
| Zcwpw1 | 5 | 0.11 | - | D | 1.63 | 0.34 – 8.0 | 0.61 | 0.54 |
| Epha1 | 6 | 0.00 (indel) | - | D | 1.60 | 0.33 – 7.8 | 0.58 | 0.56 |
| Cd33 | 7 | 3.17 | MS, Stop gained |
D | 1.67 | 0.30 – 9.2 | 0.59 | 0.56 |
| Picalm | 7 | 1.75 | - | D | 3.60 | 0.71 – 18.3 | 1.55 | 0.12 |
| Sorl1 | 9 | 12.49 | MS, SRV, SAV |
D | 2.50 | 0.50 – 12.6 | 1.11 | 0.27 |
| Abca7 | 10 | 0.05 | - | D | 1.17 | 0.24 – 5.6 | 0.19 | 0.85 |
| Slc24a4 | 12 | 4.07 | SRV | D | 3.60 | 0.71 – 18.3 | 1.55 | 0.12 |
| Rin3 | 12 | 4.95 | MS | Located within same region as Slc24a4 | ||||
| Mef2c | 13 | 0.15 | - | B | 0.86 | 0.18 – 4.1 | 0.19 | 0.85 |
| Nme8 | 13 | 3.68 | SRV | D | 1.40 | 0.30 – 6.6 | 0.42 | 0.67 |
| Clu | 14 | 0.00 (indel) | NMD | D | 5.50 | 0.84 – 36.2 | 1.77 | 0.08 |
| Ptk2b | 14 | 1.85 | MS, SRV | Located within same region as Clu | ||||
| Fermt2 | 14 | 2.40 | - | D | 1.83 | 0.32 – 10.6 | 0.68 | 0.50 |
| Cd2ap | 17 | 4.85 | MS, SRV | - | 1.00 | 0.21 – 4.7 | 0.00 | 1.00 |
| H2-Eb1 | 17 | 21.41 | MS, SRV, stop gained |
D | 1.05 | 0.22 – 5.0 | 0.06 | 0.95 |
| Trem2 | 17 | 0.13 | - | B | 1.20 | 0.25 – 5.8 | 0.23 | 0.82 |
| Bin1 | 18 | 0.21 | MS | D | 1.33 | 0.28 – 6.3 | 0.36 | 0.72 |
Abbreviations: SNP: single nucleotide polymorphism, indel: insertion/deletion, Chr: chromosome, SRV: splice region variant, SAV: splice acceptor variant, MS: missense
Once each GRS was calculated, we then asked how well a strain’s score predicted cognitive outcomes as measured on an uncorrelated task in a separate cohort of AD-BXD mice (i.e. 14m CFA). Although no individual risk gene significantly differentiated impaired vs unimpaired strains at 6m, when taken together, the GRS was significantly associated with cognitive outcomes in AD-BXD mice (Figure 3C). Notably, the GRS was not associated with cognition in Ntg-BXDs, suggesting genes used to create the GRS exhibit more specificity toward mediating AD-related decline (Figure 3D). We repeated this entire process with 1,000 sets of 19 randomly selected genetic markers and determined the correlation of the GRS and 14m AD-BXD CFA was among the top 5% of all observed permutations, suggesting the additive association of LOAD risk genes with 5XFAD-related cognitive decline is much greater than a set of genes randomly distributed across the genome. In addition, a GRS derived from genotypes at the same risk alleles, but using the distribution of ‘impaired’ and ‘unimpaired’ Ntg-BXD strains, rather than AD-BXD strains, to define odds ratios for each individual LOAD risk gene showed no relationship with late-disease cognitive outcomes in either 14m Ntg-BXDs or AD-BXDs (Figure S6), further demonstrating these genes uniquely interact with the 5XFAD transgene. Finally, the original GRS (Figure 3B) showed no association to noncognitive traits such as amyloid levels, weight, sensorimotor abilities, or anxiety (Figure 3E-H). Overall, these results demonstrate 1) the AD-BXD panel is sensitive to variation in known LOAD risk variants, and 2) the CFA task is particularly sensitive to this variation, and has the potential to be used as a translationally relevant cognitive assay in preclinical AD studies.
AD-BXD transcriptome shows concordance with late-onset AD signature
We next decided to investigate whether or not the AD-BXD panel shared similarities with human AD at the transcriptional level. We first performed RNA-sequencing on hippocampal tissue from a subset of AD-BXDs and Ntg-BXDs and evaluated the expression of genes known to be misregulated in AD. As expected from studies of post-mortem human tissue, the 5XFAD transgene significantly altered the expression of a number of these genes, particularly Bin1, Clu, Cd33 (Karch et al., 2012), Trem2 (Piccio et al., 2016), and C1qa (Hong et al., 2016) (Figure 4A). Similar to what we observed for behavioral and pathological phenotypes, risk gene expression varied across the ADBXD panel. This suggests genetic background may influence AD susceptibility by altering underlying transcriptional networks, so to gain a mechanistic understanding of functional categories altered in AD-BXDs relative to Ntg-BXDs, we performed differential expression analysis using DESeq2 (Love et al., 2014) followed by gene set enrichment analysis (GSEA) (Subramanian et al., 2005) (Table S1). As expected, the gene ontology (GO) functional categories most significantly enriched among genes observed to be downregulated in AD largely related to neuronal activity, structure, and function (Figure 4B, left) while the GO functional categories most significantly enriched among genes observed to be upregulated in AD related largely to immune response (Figure 4B, right). Together, these data highlight the maintenance of neuron activity, particularly the activity of select ion channels and receptors, as pathways that may be augmented to promote resilience, while immune pathways as those that may need to be suppressed to promote resilience.
Figure 4: Genetic background modifies AD-associated transcriptome.

(A) Genes known to be associated with AD are differentially expressed in our panel, n = 132 mice (65 females/67 males across 15 strains). Each point represents a single genotype/strain/age/sex averaged sample, **p < 0.05 two-tailed t-test. (B) List of top gene ontology (GO) functional categories enriched among genes (left) downregulated and (right) upregulated in all AD-BXDs relative to all Ntg-BXDs. See also Table S1.
To further evaluate whether observed changes in our AD-BXD model paralleled those observed in human patients, we next performed a series of cross-species comparative analyses using aged brain tissue (14m AD-BXD mice) to best parallel the tissue available from human patients. First, we evaluated the expression of a set of 60 core genes previously defined as a human AD consensus signature, primarily enriched for downregulated mitochondrial and neuronal genes [Table S2, (Hargis and Blalock, 2017)]. We observed higher concordance between our mouse panel and this human AD signature (Figure 5A) than that reported for other AD models on a single genetic background (Hargis and Blalock, 2017). This effect replicated in 3 independent human datasets tested (Figure S7). Second, we noted that the significant upregulation of immune-related pathways in our AD-BXD mice (Figure 4B) paralleled the significant association of immune-related genes with human AD, both at the transcriptional and genetic level (International Genomics of Alzheimer’s Disease, 2015; Zhang et al., 2013). To test whether the identity of genes driving this association were similar across mice and humans, we used GeneWeaver (Baker et al., 2016) to calculate overlap of genes upregulated in aged 14m AD-BXD mice (Table S3) and two gene lists associated with human AD. First, we utilized a list of genes belonging to the transcriptional co-expression module most highly associated with human AD identified by Zhang and colleagues (Zhang et al., 2013), and second, a list of 151 highly connected AD-related genes identified by Jones et. al. (International Genomics of Alzheimer’s Disease, 2015). Each of these lists were significantly enriched for genes with immune-related annotations. In both cases, the overlap between mouse and human signatures was significant (Figure 5B).
Figure 5: Aged AD-BXD transcriptome shows high concordance with late-onset human AD signature.
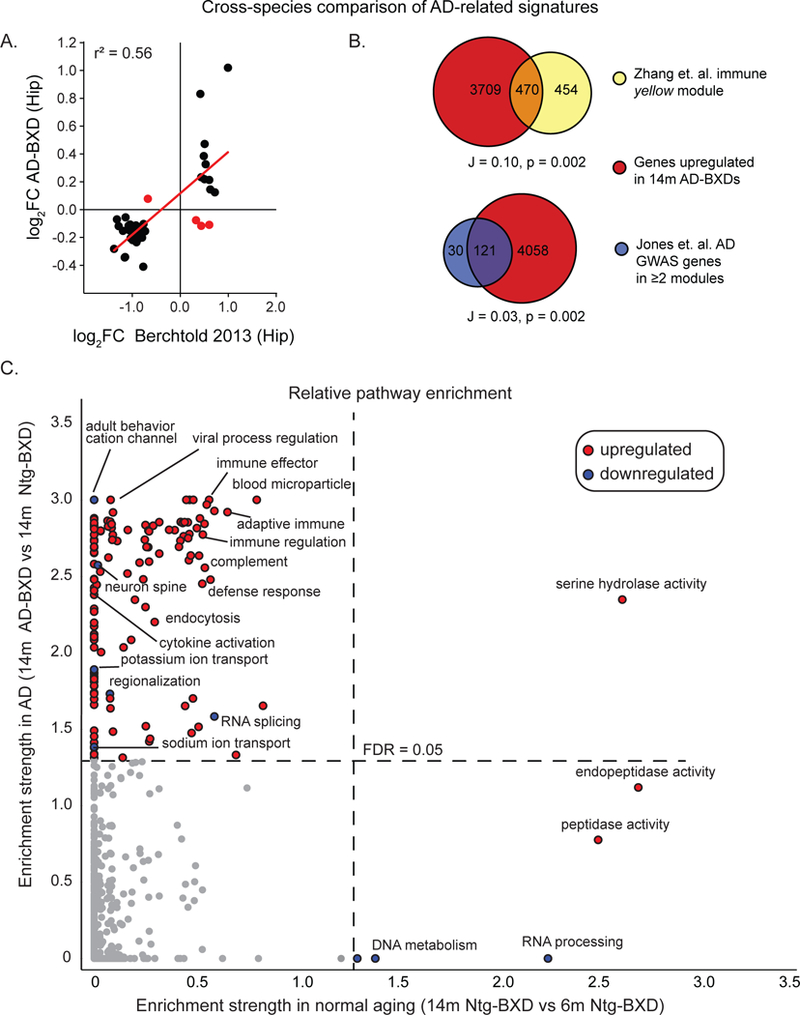
(A) 39 out of 60 (65%) of AD signature genes identified by and Blalock (Hargis and Blalock, 2017) show concordant significant changes in expression across mouse and human transcriptomes. The log2 fold change (FC) of significantly differentially expressed genes between 14m AD- and Ntg-BXDs is plotted on the y-axis, while the log2FC of gene expression between human AD patients and controls from study by a study by Berchtold and colleagues (Berchtold et al., 2013) is plotted on the x-axis. Each point represents a single gene; discordant genes with log2 fold changes with opposite direction have been highlighted in red. (B) Genes upregulated in 14m ADBXDs relative to Ntg-BXDs were compared to genes associated with human AD by (top) Zhang and colleagues (Zhang et al., 2013) and (bottom) Jones and colleagues (International Genomics of Alzheimer’s Disease, 2015). A significant overlap was identified in both cases. (C) Graph of enrichment strength of gene ontology (GO) categories across (y-axis) mouse AD or (x-axis) normal aging. Gene set enrichment analysis was performed on genes identified to be differentially expressed relative to 5XFAD carrier status or normal aging in Ntg-BXDs (Table S3). For GO terms that were identified in both scenarios, the FDR q-values were transformed to obtain a measure of enrichment strength and scores were plotted against each other to identify unique and/or common differentially regulated GO terms. As such, each axis can be thought of significance; the upper left quadrant highlights pathways that are uniquely significantly altered in AD-BXDs relative to Ntg-BXDs, while the bottom right quadrant highlights pathways that are uniquely significantly altered in normal aging (14m Ntg-BXDs vs 6m Ntg-BXDs). Data points are colored based on directionality of enrichment score calculated by GSEA: red = genes belonging to this category were significantly upregulated in given scenario, blue = genes belonging to this category were significantly downregulated in given scenario. Dotted lines represent enrichment scores for FDR q-value = 0.05. See also Figure S6 and Tables S2-S4.
Finally, we tested whether the AD-specific enrichment of immune-related pathways observed in human AD, but not normal aging (Raj et al., 2017), was preserved across our AD and Ntg-BXDs. To do this, we identified GO terms enriched among those genes significantly differentially expressed between 14m AD and Ntg-BXDs (Table S3, 5XFAD-related genes) and those enriched among genes significantly differentially expressed between 6m and 14m Ntg-BXDs (Table S3, normal aging-related genes). To enable comparison across datasets, we identified those GO terms with enough genes to be identified in each set and compared enrichment strength across AD and normal aging in our mouse panel (Figure 5C and Table S4). Enrichment of immune-related terms was exclusively observed among our list of 5XFAD-related genes, and not normal aging-related genes. A similar trend was observed in neuron and ion-channel related terms, suggesting downregulation of neuron structure, function, and/or activity to also be a unique feature of AD relative to normal aging in the mouse. Changes unique to normal aging include DNA metabolism, RNA processing, and peptidase activity (Figure 5C, bottom right). Overall, the incorporation of genetic diversity into a mouse model of AD resulted in a transcriptomic profile that more closely matched human AD than previous AD models with limited genetic background variation (Hargis and Blalock, 2017).
Discussion
AD-BXD panel represents a new translational model of human AD
It has long been recognized that Alzheimer’s disease is a complex and polygenic disease, likely influenced by multiple variants, some with relatively small effect sizes (Lambert et al., 2013). Here, we introduce the first genetically diverse population of AD mice - the AD-BXDs - as a more translational model of human AD by demonstrating a high level of concordance between the AD-BXDs and both familial and sporadic forms of human AD at the molecular and behavioral level. In particular, the observed variation in AAO mirrors the variation in human patients reported by Ryman and colleagues (Ryman et al., 2014), suggesting our panel captures a portion of the phenotypic heterogeneity observed in human FAD patients. In addition, our female AD-BXD mice appear to be more susceptible to Apoe risk and AD-related cognitive decline (Figure 2B-C) despite comparable levels of amyloid deposition (Figure 1F-G), 5XFAD transgene expression (Figure 1H), and endogenous App levels (Figure 1I), similar to epidemiological trends observed in human patients (Altmann et al., 2014; Mielke et al., 2014; Zokaei et al., 2017). At the genetic level, we demonstrate that the extent of AD-related cognitive decline is influenced by a given strain’s specific allele distribution across a set of 21 loci associated with sporadic LOAD. Transcriptionally, the changes occurring in the AD-BXDs relative to Ntg-BXDs, particularly at aged time points, show a high level of overlap with transcriptional changes occurring in human AD patients relative to age-matched controls, both in terms of up-regulated inflammatory pathways and down-regulated neuronal signatures, suggesting some common molecular mechanisms exist between mouse and human. Overall, results here demonstrate the critical role genetic background plays in determining susceptibility to disease and present the AD-BXD panel as a useful tool that will enable the identification of modifier genes more likely to translate to human patients.
Genetically diverse isogenic mice as a resource for experimental precision medicine
In recent years, there has been growing skepticism regarding the utility of AD mouse models, in part because research using these models have failed to translate into successful treatments. There are a variety of reasons that may explain this failure, many of which have been discussed previously (Onos et al., 2016). However, a common theme of many traditional models is a lack of genetic diversity. Regardless of the specific strain used, the use of a single genetic background precludes the ability to understand the impact of individual genetic diversity on disease trajectory. Here we introduce the AD-BXD panel as a new preclinical resource that will allow future studies to investigate the influence of genetic complexity on the behavioral, molecular, and pathological phenotypes in AD, as well as response to interventions. By layering multiple scales of data collected from the AD-BXDs (phenotype, transcriptomics, proteomics, etc.), we may ultimately be able to identify subtypes of AD and begin to develop personalized therapeutic interventions. Given the advantages represented by this experimental design, and the increasing awareness that genetic background impacts a variety of complex traits (Sittig et al., 2016), the approach used here should be of broad interest across scientific disciplines. We hypothesize the incorporation of genetic diversity into preclinical studies of various complex diseases will greatly enhance the overall translational potential of mouse models. In addition, our experimental design is likely to be broadly applicable to mouse models of human disease that incorporate a dominantly inherited high-risk genotype in the form of a transgene or other genetic perturbation. In these cases, the ultimate identification of genetic factors that modify disease onset and/or severity will provide insight into pathways critical for regulating disease pathogenesis.
The role of modifier genes in normal aging and resilience to AD
The creation of the genetically diverse Ntg and AD-BXD panel enables the use of genetic mapping to identify modifier alleles that influence the onset and severity of cognitive decline in both normal aging and AD. The inclusion of both Ntg and AD lines is particularly powerful, as the extent to which genetic mechanisms that underlie normal cognitive aging and AD overlap is still unclear. Comparison of mapping results across genotypes will identify alleles that either act as general modifiers and contribute to a phenotype regardless of disease status, or specific AD modifiers that exhibit an epistatic relationship with the 5XFAD transgene. To this end, our results indicate that the B6 background strain may contribute modifiers that increase resilience to high-risk 5XFAD mutations, creating an ideal opportunity to model resilience for the first time. In particular, the B6 background appears to attenuate the impact of 5XFAD mutations on cognitive traits, despite moderate-to-high levels of Aβ42 (Figure 1G). In addition, across 21 genes known to confer risk for LOAD, the B allele represents the protective allele in 17 cases (Table 2). Future studies will utilize these resources to perform genetic mapping to identify genomic regions involved in the regulation of cognitive decline, and precise genes present in the B6 background that contribute to resilience. Due to large haplotype blocks segregating among the BXD strains, it is likely additional Ntg- and AD-BXD strains will need to be incorporated to narrow in on causal variants, genes, and pathways regulating quantitative traits across the panel. As the functional interrogation of all candidate genes will require large investments of time and resources, it is likely the development of new disease-modifying strategies will be a community effort.
Improving rigor and reproducibility in preclinical studies
In addition to providing an ideal preclinical resource to identify modifiers of susceptibility and resilience to AD, the AD-BXD panel presents a multitude of additional advantages. Notably, as each parental line (C57BL/6J and BXDs) is fully inbred, the F1 mice described here can be recreated across time and laboratories, maximizing the utility of our characterization of individual lines as either cognitively resilient or susceptible and enhancing rigor and reproducibility of the approach. The use of genetically identical, or isogenic, F1 mice also allows for calculation of heritability across a number of diverse traits, ranging from body weight and sensorimotor abilities to anxiety and cognitive function. Traits with high heritability are, by definition, largely influenced by genetics rather than environmental or technical factors and thus are more likely to replicate across time and laboratories. Identification of traits that are robust to environmental influences will enhance reproducibility of research and confidence that results will translate from the bench to clinic more rapidly. Traditional studies that utilize only a single inbred strain to study disease are useful for understanding basic mechanisms associated with symptom onset, but are equivalent to study disease in a single human. In contrast, results from studies that utilize diverse genetic backgrounds are a better model of complex disease across individuals, and are therefore, more likely to generalize across patient populations.
Consideration of tau pathology
One of the limitations of our study is the lack of consideration of tau pathology. Since the 5XFAD transgene is not generally thought to induce significant tau pathology (Oakley et al., 2006), we did not evaluate tau pathology across the AD-BXDs. As hyperphosphorylated tau is one of the main pathological events observed in AD, this is a general shortcoming of most currently available mouse models of AD. Although beyond the scope of this study, naturally occurring variants in Mapt [see Sanger Mouse Genomes project (Keane et al., 2011)] segregate across the BXD panel and may influence the production of hyperphosphorylated tau (and ultimately, neurofibrillary tangles). As such, it is possible that some genetic backgrounds do exhibit tau pathology. These strains (if identified) would provide ideal models for preclinical screens in future studies and contribute to our understanding of the genetic variants modifying production and/or clearance of hyperphosphorylated tau, as tau and amyloid neuropathologies do not often occur together in currently used mouse models (Kitazawa et al., 2012). As tau pathology has been reported to be more strongly associated with cognitive function in AD than other pathologies (Brier et al., 2016), the AD-BXD panel may provide new opportunities to study the relationship between tau, amyloid, and cognitive function in the context of genetic diversity.
Conclusions and future directions
The ultimate goal of mouse studies relating to AD is the eventual translation of identified candidates into viable human therapeutics or biomarkers of disease. Our results suggest the AD-BXD panel is a valuable resource to do just that, as we demonstrate high levels of overlap between the ADBXDs and human AD at the genetic, transcriptional, and phenotypic levels. In addition, we validate contextual fear conditioning, particularly the acquisition phase, as a translationally valid task that is likely to share some of the same underlying mechanisms as current cognitive tests used in the human clinic. Together, the combination of new tools (i.e. AD-BXDs) and valid tasks (contextual fear conditioning), used at the appropriate time points, may enable, for the first time, the identification of genetic modifiers of AD susceptibility that can be targeted as new therapeutic opportunities. Future studies will use the resources described here for genetic mapping and integration of both gene and protein expression profiling to accomplish this goal.
Star Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Catherine C. Kaczorowski (catherine.kaczorowski@jax.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Female congenic C57BL/6J mice hemizygous for the dominant 5XFAD transgene (Oakley et al., 2006), which consists of 5 human mutations known to cause familial AD [three in amyloid precursor protein (APP; Swedish: K670N, M671L, Florida: I716V, and London: V717I) and two in presenilin 1 (PSEN1; M146L and L286V)], were obtained from The Jackson Laboratory (JAX MMRRC Stock No: 34848-JAX). These mice were bred with 28 males from a set of genetically diverse recombinant inbred strains from the well-established BXD genetic reference panel (Peirce et al., 2004). By selecting the same maternal background strain (i.e. 5XFAD-C57BL/6J) across the panel for cross with male BXD strains, we were able to introduce variants in the nuclear DNA, hold the mitochondrial genome constant, and control for strain-specific differences in maternal behavior on offspring behavior. The F1 progeny resulting from this B6–5XFAD by BXD cross are isogenic recombinant inbred backcross mice, each harboring one maternally derived B allele and either a B or D paternally derived allele at any given genomic locus. As expected from a Mendelian pattern of inheritance, ~50% of these F1 mice carry the 5XFAD transgene (termed AD-BXDs) and ~50% are non-transgenic (Ntg) littermate controls referred to Ntg- BXDs. Male and female offspring were group housed (2–5 per cage) and maintained on a 12 hr light/dark cycle with ad libitum access to food and water. All mice were genotyped for the 5XFAD transgene through a combination of in-house genotyping according to The Jackson Laboratory protocols for strain #34848-JAX and outside services (Transnetyx, TN, USA, and The Jackson Laboratory Transgenic Genotyping Services). Working memory and body weights were monitored longitudinally, and more detailed phenotyping occurred at 6 and 14m. These time points were selected to obtain an adult phenotype (6m) and a middle-aged to aged time point (14m) that captured variation in disease symptoms before the mice exhibited severe health-related problems that confounded behavioral testing. All mouse experiments occurred at University of Tennessee Health Science Center and were carried out in accordance with the standards of the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC), as well as the recommendations of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Tennessee Health Science Center.
METHOD DETAILS
Y-maze
For all behavioral testing, mice were habituated to transport and to the testing room for three days prior to testing. The y-maze test of spontaneous alternation was performed as described previously (Oakley et al., 2006). The y-maze used for testing was made of clear acrylic with arms that were 2” wide x 12” long x 2” high. The maze was placed on a table in a dimly lit room and spatial cues were displayed on walls around the table. Mice were placed in a randomized start arm and video tracking software was used to monitor arm entries (ANY-maze, Stoelting Co., IL, USA). An arm entry was called when the mouse’s entire body, including the two back feet, entered the arm. The sequence and total number of arms entered was recorded, and the percentage of successful alternations was calculated as follows: number of alternations/maximum possible alternations (total number of arms entered – 2) x 100. For each animal that was measured longitudinally (i.e. not harvested at the early 6m time point), the age at which each animal became ‘impaired’, or performed below chance levels (50%), was recorded and used as the animals “age at onset” [AD-BXDs: n = 226 (126 females/100 males) across 28 strains vs Ntg-BXDs, n = 171 mice (108 females/63 males) across 25 strains]. Strain averages for age at onset were then calculated.
Sensorimotor battery
At 6m [AD-BXDs n = 284 (185 females/90 males) across 28 strains, Ntg-BXDs n = 220 (158 females/62 males) across 27 strains] and 14m [AD-BXDs n = 222 (104 females/106 males) across 26 strains, Ntg-BXDs n = 172 (109 females/63 males) across 25 strains], mice were subjected to a sensorimotor battery consisting of three tasks. First, mice were placed in the center of a 3-foot long narrow (0.5”) beam elevated 20.75” off a table surface and the time taken for the mouse to cross the narrow beam onto a safe platform on either side was measured. Second, mice were placed face-down on a wire mesh grid (holes were 1cm x 1cm) that was placed at a 45° angle. The time taken for a mouse to right itself (negative geotaxis) was recorded. A 3 minute maximum time limit was imposed for both the narrow beam and incline screen tests. If a mouse fell from the narrow beam, the maximum score of 180s was given. Third, grip strength was measured using a standard grip strength meter (Colbourn Instruments). Each of these three tasks were repeated in triplicate and the average score across three trials was used. For each mouse, a z-score based on the 6m population average was calculated for each task and the three z-scores were summed to derive a sensorimotor composite score, which was used here to relate sensorimotor performance to cognitive abilities.
Elevated plus maze
At 6m [AD-BXDs n = 280 (191 females/89 males) across 28 strains, Ntg-BXDs n = 220 (158 females/62 males) across 27 strains] and 14m [AD-BXDs n = 221 (116 females/105 males) across 26 strains, Ntg-BXDs n = 173 (110 females/63 males) across 25 strains], anxiety was evaluated using an elevated plus maze task. Mice were placed in the center of the maze and allowed to explore for 6 minutes. Video tracking software (ANY-maze, Stoelting Co.) was used to track the mouse and calculate the time spent in open versus closed arms of the maze as well as the number of arm entries into either open or closed arms, the total number of arm entries, and the total distance travelled in the maze.
Contextual fear conditioning
Following 3 days of habituation to transport and to the testing room, mice were trained on a standard contextual fear conditioning (CFC) paradigm as previously described (Neuner et al., 2015). Training consisted of a 180s baseline period followed by four mild foot shocks (1s, 0.9mA), separated by 115 ± 20s. A 40s interval following each foot shock was defined as the post-shock interval, and the percentage of time spent freezing during each of these intervals was measured using FreezeFrame software (Coulbourn Instruments, PA, USA). The percentage of time spent freezing during the final post-shock interval (PS4) was used as an index of contextual fear acquisition (CFA). Twenty-four hours later, hippocampus-dependent contextual fear memory (CFM) was tested by returning the mouse to the testing chamber for 10 min. The percentage of time spent freezing during the testing trial was measured using FreezeFrame software and used as an index of CFM. For CFC, 146 6m ADBXD (102 females/44 males) and 209 14m AD-BXD (111 females/98 males) across 26 strains were used, along with 114 6m Ntg-BXD (83 females/31 males) across 24 strains and 167 14m Ntg-BXD mice (106 females/61 males) across 27 strains. Pain sensitivity was evaluated in a subset of mice by recording the length of activity burst following each shock. An average post-shock reactivity score was calculated by averaging the length of each activity burst following the four training shocks.
Enzyme-linked immunosorbent assay (ELISA)
Brains were removed immediately following CFC at appropriate time points (6m or 14m) and hemisected. One half of the brain was immediately dissected, snap frozen, and stored at −80°C until use. Beta-amyloid 1–42 (Aβ42) levels were quantified from sections of temporal cortex [6m n = 72 mice (46 female/46 male) across 22 AD-BXD strains, 14m n = 82 mice (43 female/33 male) across 21 AD-BXD strains] as previously described (Oakley et al., 2006). Briefly, tissue was homogenized in 1X PBS + 1% Triton-X 100 using the TissueLyser II system (Qiagen) and sonicated 2×10s on low power. Protein concentration was determined using a NanoDrop 2000 UV-Vis Spectrophotometer (ThermoScientific, USA). Brain homogenates (10 mg/ml) were extracted in a final concentration of 5M GuHCl overnight at 4°C. Samples were then diluted appropriately and run in duplicate on Aβ42specific sandwich colorimetric ELISAs according to the manufacturer’s protocol (Cat# 298–92401, Wako Chemicals, Richmond, VA). Optical densities at 450 nm were read on a Biotek plate reader (BioTek, USA) and Aβ42 concentration was determined by comparison with Aβ42 standard curves. Only readings in the linear range of the standard curve were included in analysis. Duplicates were averaged to determine concentration of Aβ42 in each sample. Finally, Aβ42 concentrations were normalized to total protein concentration and are reported as nanograms of Aβ42 per milligrams of total protein.
Immunohistochemistry and plaque quantification
At harvest, the half brain not used for fresh dissection was placed in 4% paraformaldehyde and kept at 4C until further use. In order to minimize technical variation in immunohistochemistry, brains were sent to Neuroscience Associates (Knoxville, TN), where 40 hemibrains were embedded, processed, and stained simultaneously. Briefly, the brains were freeze-sectioned coronally at 40 µm intervals (not including cerebellum) and staining for Aβ1–42 was performed on every 24th section spaced at 960 µm, yielding approximately 9 sections per hemibrain. For analysis, images of each section containing hippocampus were collected on a Nikon Eclipse 90i microscope using NISElements Advanced Research program. Images were taken using a 2x objective with computer automated focusing. Approximately 4 images were captured for each hemibrain and stitched together using NIS-Elements Advanced Research program. ImageJ particle analysis was used to automate detection of plaques (Hurtado et al., 2010). Regions of interest (hippocampus and cortex) in each image were manually outlined and pixel size of each region calculated and used to determine the percentage of each area covered by amyloid plaques, controlling for regional size differences. Neuroscience Associates also performed scanning of each slide at 20x using a Huron scanner and these images are used for illustrative purposes in Figure S2.
Heritability estimates
Heritability estimates for each phenotype (Table 1) were calculated according to established methods (Belknap, 1998). Briefly, we compared between-strain variance (due to genetic diversity, VG) to total sample variance (due to both genetic and environmental factors, VE) given the average number of biological replicates per strain (n) according to the following formula: h2RIx̅ = VG/(VG + VE/n). The average number of mice per strain was used to represent n and is reported in Table 1. VE was calculated by summing between-strain variance and within-strain variance, as within-strain variance should capture all variation not due to genetic diversity. As heritability was calculated using both males and females, within-strain variance will also capture variation due to sex. However, as we calculated heritability independently for each trait of interest across AD- and Ntg-BXDs, our heritability estimates do not capture variation due to age or genotype.
RNA sequencing
Snap frozen hippocampi from AD-BXD strains and Ntg-BXD littermate controls at 6m [AD-BXDs n = 33 (15 females/18 males) across 13 strains, Ntg-BXDs n = 31 (17 females/14 males) across 14 strains] and 14m [AD-BXDs n = 36 (16 female/20 male) across 14 strains, Ntg-BXDs n = 33 (17 female/16 male) across 15 strains] were used for RNA sequencing. RNA was isolated on a Qiacube using the RNeasy mini kit (Qiagen) and treated with DNase to remove contaminating DNA. RNA quality was confirmed using a BioAnalyzer (Agilent Technologies). All samples had RNA Integrity Numbers (RIN values) > 8.0. Sequencing libraries were prepared from 1 µg RNA with the Truseq Stranded mRNA Sample Preparation Kit (Illumina Inc) following the manufacturer’s protocol. Final PCR-enriched fragments were validated on a 2200 Tapestation Instrument using the D1000 ScreenTape (Agilent Technologies) and quantified by qPCR using a Universal Library Quantification Kit (Kapa Biosystems) on the QuantStudio 6 Flex (ThermoFisher Scientific). Final library pools were sequenced by 75bp paired-end sequencing on a HiSeq2500 (Illumina Inc). Because both C57BL/6J and DBA/2J alleles segregate within our panel, the GBRS/EMASE pipeline developed by the Churchill group at The Jackson Laboratory was used in order to align reads to a diploid transcriptome (https://emase.readthedocs.io/en/latest/). An expectation maximization algorithm was used in order to align reads to the correct allele, allowing for the quantification of both total reads assigned to a gene and the number of reads assigned to either the B or D allele. For final by-strain analysis, samples belonging to the same strain/sex/age/genotype group were averaged. Differential expression analysis was conducted using the DESeq2 package (Love et al., 2014). For evaluation of transgene expression and its effect on endogenous App and Psen1 expression, RNA-sequencing reads from a larger subset of AD- and Ntg-BXDs [n = 293 (177 females/116 males across 28 strains)] were sequenced according to identical methods and were additionally aligned to the mutated human APP and PSEN1 sequences. Expression was quantified using transcripts per million and then log transformed to compare expression across groups. Transgene and endogenous App and Psen1 expression is available as Table S5.
Comparison of AD-BXD and human transcriptomes
In order to evaluate how well the AD-BXD transcript profile matches that of human AD, we utilized a dataset recently published by Hargis and Blalock (Hargis and Blalock, 2017) comparing existing mouse models of AD to human AD. They identified a consensus AD signature consisting of 60 genes derived from the top 10% commonly upregulated and downregulated genes across three human AD datasets (Table S2). In order to see how the transcriptome from our AD-BXD panel compared to normal expression patterns, differential expression analysis comparing hippocampal gene expression from 14m AD-BXD lines to non-carrier littermate controls was performed using DESeq2 (Love et al., 2014). The log2 fold change (log2FC) for each of the 60 AD consensus genes that were significantly differentially expressed (nominal p-value < 0.05) across AD and Ntg-BXDs was identified and used for comparison across human and mouse datasets obtained from Hargis et. al. (Hargis and Blalock, 2017). To evaluate similarities between immune-enriched genes upregulated in AD-BXDs and gene lists identified as associated with AD from Zhang et. al. (Zhang et al., 2013) and Jones et. al. (International Genomics of Alzheimer’s Disease, 2015), all three gene lists were uploaded into GeneWeaver (Baker et al., 2016) and Jaccardian similarity indexes were calculated and evaluated for significance.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed as previously described (Neuner et al., 2016; Subramanian et al., 2005). Briefly, significantly differentially expressed genes from the comparison of interest (Table S1: All AD-BXD vs All Ntg-BXD, Table S3: 14m AD-BXD vs 14m Ntg-BXD and 6m Ntg-BXD vs 14m Ntg-BXD) were ranked according to log2 fold change. Gene ontology (GO) gene sets were obtained from the Broad Institute’s Molecular Signatures Database (MSigDB) (Liberzon et al., 2015) and ranked gene lists were tested for enrichment using GSEA’s GSEAPreranked feature, version 3.0. To compare functional annotation enrichment among differentially expressed genes across AD and normal aging (Figure 5), GO terms identified in each comparison (late-stage AD: 14m AD-BXD vs 14m Ntg-BXD, Table S3 and normal aging: 6m NtgBXD vs 14m Ntg-BXD, Table S3) were extracted from GSEA results. A score for enrichment strength was calculated by transforming the FDR q-values generated by GSEA using the following formula: -log10(FDRq + 0.001), similar to that described in (Raj et al., 2017). The values calculated in each scenario (AD and normal aging) were then plotted against each other in Figure 5 to identify those pathways with stronger enrichment in AD than normal aging, and vice versa.
Calculation of a genetic risk score
To evaluate whether the AD-BXD panel was sensitive to variation in known AD risk loci, we derived a genetic risk score for each strain, similar to that described by Chouraki and colleagues in 2016 (Chouraki et al., 2016). Strains were first stratified into impaired (below the AD-BXD population average) and unimpaired (above the AD-BXD population average) based on CFM performance at 6m. We then identified the genotype of each strain at 21 genes known to contribute risk for AD and identified the risk allele of each gene (i.e. the allele that appeared more frequently in the impaired group, Table 2). Note as some genes appeared in the same linkage block, only 19 genotypes were used in the calculation of the GRS. The odds ratio for each gene was calculated and log transformed to determine an individual risk score per gene. These individual risk scores were used to derive an overall genetic risk score for each strain that reflected how many copies of each risk allele were present. Overall genetic risk scores were transformed based on previous methods (Chouraki et al., 2016) using the following formula: total risk score * (# of markers tested/sum of individual gene risk scores). The GRS was then correlated to cognitive traits as reported. To avoid influence from our original definition of ‘impaired’ versus ‘unimpaired’ using 6m CFM, we correlated GRS to uncorrelated, independent cognitive tasks. As contextual fear conditioning is a cross-sectional task, and we wanted to investigate the extent to which these genes regulated cognitive decline, we focused on cognitive tasks from a separate cohort of aged AD-BXDs, particularly 14m CFA. Finally, to empirically estimate the null distribution for the correlation of our genetic risk score and cognitive traits of interest, we randomly sampled 1000 sets of 19 markers across the genome and repeated the derivation of GRS. We created 1000 GRS from randomly sampled data, and correlated each random GRS to strain-matched cognitive performance, which illustrated the correlation between 14m CFA and our derived GRS was stronger than the correlation observed for 95% of randomly sampled genes. As an additional control, we repeated the process but based on allelic distribution of risk alleles across Ntg-BXDs. We defined Ntg-BXD strains as ‘impaired’ vs ‘unimpaired’ based on 6m CFM performance, identified the risk allele for each of the 21 genes listed in Table 2, calculated the odds ratio for each, and derived a Ntg-based GRS. This GRS showed no relationship with cognitive outcomes in either Ntg- or AD-BXDs, or any non-cognitive traits tested. Table 2, gene lengths were obtained using start and end positions listed in Ensembl version 92 and SNP counts were obtained from Sanger, release REL-1505 (Keane et al., 2011).
QUANTIFICATION AND STATISTICAL ANALYSIS
All experiments and data analysis were conducted with experimenters blind to strain background and genotype (5XFAD vs Ntg) where appropriate. Statistical analysis was performed using SPSS software Version 23 (IBM), R, and Excel. Distribution was evaluated for normality using ShipiroWilkes test. Additional analyses included independent unpaired t-tests, univariate ANOVAs, Pearson correlation, and Jaccard index to test similarity. Correction for multiple comparisons was also used where appropriate (i.e differential expression analysis). Data values reported in both the main text and figure legends are given as mean ± standard error of the mean unless otherwise stated. Outliers were identified based on a pre-defined criteria of average values ± 3 SD outside the mean.
DATA AND SOFTWARE AVAILABILITY
Genotypes from the BXD strains are publically available on GeneNetwork.org. RNA-sequencing from the hippocampus of a subset of AD-BXD strains is available on Gene Expression Omnibus (GEO) under accession number GSE101144. Data including raw phenotype information will also be deposited in the AMP-AD Knowledge Portal (doi: 10.7303/syn17016211). EMASE software used for alignment of RNA sequencing reads to a diploid transcriptome is available online at: (https://emase.readthedocs.io/en/latest/).
Supplementary Material
Highlights.
Genetically diverse AD mouse models parallel complexity of human AD
Comparative analyses identify the C56BL/6J background strain as resilient
Reproducible AD mouse lines and cognitive assays valuable for preclinical studies
Resource design serves as blueprint for improved models of polygenic diseases
Acknowledgments:
This study is part of the National Institute on Aging Resilience-AD program and is supported through the NIA grant award R01AG057914 to CCK. This work was also supported by the BrightFocus Foundation (A2016397S to CCK), the National Institute on Aging (R01AG054180 to CCK, F31AG050357 to SMN), and the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK102918 to KMSO). Additional support was provided by the Evnin Family and the University of Tennessee Health Science Center Neuroscience Institute the Translational Genomics Research Institute. The authors thank Dr. Lynda Wilmott and Thomas Shapaker for collection of behavioral data and Kwangbom Choi, Matthew de Both, Ryan Richholt, and Ashley Siniard for assistance with RNA-sequencing. The authors would also like to thank Dr. Rob Williams for thoughtful input on the project design, and Drs. Vivek Philip and Ji-Gang Zhang for assistance with data analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Altmann A, Tian L, Henderson VW, Greicius MD, and Alzheimer’s Disease Neuroimaging Initiative, I. (2014). Sex modifies the APOE-related risk of developing Alzheimer disease. Annals of neurology 75, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker E, Bubier JA, Reynolds T, Langston MA, and Chesler EJ (2016). GeneWeaver: data driven alignment of cross-species genomics in biology and disease. Nucleic acids research 44, D555559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belknap JK (1998). Effect of within-strain sample size on QTL detection and mapping using recombinant inbred mouse strains. Behavior genetics 28, 29–38. [DOI] [PubMed] [Google Scholar]
- Berchtold NC, Coleman PD, Cribbs DH, Rogers J, Gillen DL, and Cotman CW (2013). Synaptic genes are extensively downregulated across multiple brain regions in normal human aging and Alzheimer’s disease. Neurobiology of aging 34, 1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blalock EM, Buechel HM, Popovic J, Geddes JW, and Landfield PW (2011). Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient Alzheimer’s disease. Journal of chemical neuroanatomy 42, 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, and Landfield PW (2004). Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proceedings of the National Academy of Sciences of the United States of America 101, 2173–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brier MR, Gordon B, Friedrichsen K, McCarthy J, Stern A, Christensen J, Owen C, Aldea P, Su Y, Hassenstab J, et al. (2016). Tau and Abeta imaging, CSF measures, and cognition in Alzheimer’s disease. Science translational medicine 8, 338ra366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouraki V, Reitz C, Maury F, Bis JC, Bellenguez C, Yu L, Jakobsdottir J, Mukherjee S, Adams HH, Choi SH, et al. (2016). Evaluation of a Genetic Risk Score to Improve Risk Prediction for Alzheimer’s Disease. Journal of Alzheimer’s disease : JAD 53, 921–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanselow MS (2000). Contextual fear, gestalt memories, and the hippocampus. Behavioural brain research 110, 73–81. [DOI] [PubMed] [Google Scholar]
- Gatz M, Pedersen NL, Berg S, Johansson B, Johansson K, Mortimer JA, Posner SF, Viitanen M, Winblad B, and Ahlbom A (1997). Heritability for Alzheimer’s disease: the study of dementia in Swedish twins. The journals of gerontology Series A, Biological sciences and medical sciences 52, M117–125. [DOI] [PubMed] [Google Scholar]
- Hardy JA, and Higgins GA (1992). Alzheimer’s disease: the amyloid cascade hypothesis. Science 256, 184–185. [DOI] [PubMed] [Google Scholar]
- Hargis KE, and Blalock EM (2017). Transcriptional signatures of brain aging and Alzheimer’s disease: What are our rodent models telling us? Behavioural brain research 322, 311–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokama M, Oka S, Leon J, Ninomiya T, Honda H, Sasaki K, Iwaki T, Ohara T, Sasaki T, LaFerla FM, et al. (2014). Altered expression of diabetes-related genes in Alzheimer’s disease brains: the Hisayama study. Cerebral cortex 24, 2476–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, Merry KM, Shi Q, Rosenthal A, Barres BA, et al. (2016). Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado DE, Molina-Porcel L, Iba M, Aboagye AK, Paul SM, Trojanowski JQ, and Lee VM (2010). A{beta} accelerates the spatiotemporal progression of tau pathology and augments tau amyloidosis in an Alzheimer mouse model. The American journal of pathology 177, 1977–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Genomics of Alzheimer’s Disease, C. (2015). Convergent genetic and expression data implicate immunity in Alzheimer’s disease. Alzheimer’s & dementia : the journal of the Alzheimer’s Association 11, 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson HM, Onos KD, Pepper KW, Graham LC, Akeson EC, Byers C, Reinholdt LG, Frankel WN, and Howell GR (2015). DBA/2J genetic background exacerbates spontaneous lethal seizures but lessens amyloid deposition in a mouse model of Alzheimer’s disease. PLoS One 10, e0125897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczorowski CC, and Disterhoft JF (2009). Memory deficits are associated with impaired ability to modulate neuronal excitability in middle-aged mice. Learning & memory 16, 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczorowski CC, Sametsky E, Shah S, Vassar R, and Disterhoft JF (2011). Mechanisms underlying basal and learning-related intrinsic excitability in a mouse model of Alzheimer’s disease. Neurobiology of aging 32, 1452–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, and Goate AM (2012). Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PloS one 7, e50976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. (2011). Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa M, Medeiros R, and Laferla FM (2012). Transgenic mouse models of Alzheimer disease: developing a better model as a tool for therapeutic interventions. Current pharmaceutical design 18, 1131–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, et al. (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nature genetics 45, 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao F, Zhang TJ, Jiang H, Lefton KB, Robinson GO, Vassar R, Sullivan PM, and Holtzman DM (2015). Murine versus human apolipoprotein E4: differential facilitation of and colocalization in cerebral amyloid angiopathy and amyloid plaques in APP transgenic mouse models. Acta neuropathologica communications 3, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, and Tamayo P (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell systems 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW, Weisgraber KH, and Huang Y (2009). Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. Journal of lipid research 50 Suppl, S183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke MM, Vemuri P, and Rocca WA (2014). Clinical epidemiology of Alzheimer’s disease: assessing sex and gender differences. Clin Epidemiol 6, 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuner SM, Garfinkel BP, Wilmott LA, Ignatowska-Jankowska BM, Citri A, Orly J, Lu L, Overall RW, Mulligan MK, Kempermann G, et al. (2016). Systems genetics identifies Hp1bp3 as a novel modulator of cognitive aging. Neurobiology of aging 46, 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuner SM, Wilmott LA, Hope KA, Hoffmann B, Chong JA, Abramowitz J, Birnbaumer L, O’Connell KM, Tryba AK, Greene AS, et al. (2015). TRPC3 channels critically regulate hippocampal excitability and contextual fear memory. Behavioural brain research 281, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley H, Cole SL, Logan S, Maus E, Shao P, Craft J, Guillozet-Bongaarts A, Ohno M, Disterhoft J, Van Eldik L, et al. (2006). Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. The Journal of neuroscience : the official journal of the Society for Neuroscience 26, 10129–10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M (2009). Failures to reconsolidate memory in a mouse model of Alzheimer’s disease. Neurobiology of learning and memory 92, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onos KD, Sukoff Rizzo SJ, Howell GR, and Sasner M (2016). Toward more predictive genetic mouse models of Alzheimer’s disease. Brain research bulletin 122, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, and Williams RW (2004). A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC genetics 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccio L, Deming Y, Del-Aguila JL, Ghezzi L, Holtzman DM, Fagan AM, Fenoglio C, Galimberti D, Borroni B, and Cruchaga C (2016). Cerebrospinal fluid soluble TREM2 is higher in Alzheimer disease and associated with mutation status. Acta neuropathologica 131, 925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj T, Chibnik LB, McCabe C, Wong A, Replogle JM, Yu L, Gao S, Unverzagt FW, Stranger B, Murrell J, et al. (2017). Genetic architecture of age-related cognitive decline in African Americans. Neurology Genetics 3, e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge PG, Mukherjee S, Crane PK, Kauwe JS, and Alzheimer’s Disease Genetics C (2013). Alzheimer’s disease: analyzing the missing heritability. PloS one 8, e79771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryman D, Gao Y, and Lamb BT (2008). Genetic loci modulating amyloid-beta levels in a mouse model of Alzheimer’s disease. Neurobiology of aging 29, 1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryman DC, Acosta-Baena N, Aisen PS, Bird T, Danek A, Fox NC, Goate A, Frommelt P, Ghetti B, Langbaum JB, et al. (2014). Symptom onset in autosomal dominant Alzheimer disease: a systematic review and meta-analysis. Neurology 83, 253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadleir KR, Eimer WA, Cole SL, and Vassar R (2015). Abeta reduction in BACE1 heterozygous null 5XFAD mice is associated with transgenic APP level. Molecular neurodegeneration 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani G, Krzywkowski P, Dudal S, Yu M, Paquette J, Malo D, Gervais F, and Tremblay P (2006). Mapping genetic modulators of amyloid plaque deposition in TgCRND8 transgenic mice. Human molecular genetics 15, 2313–2323. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ (1991). The molecular pathology of Alzheimer’s disease. Neuron 6, 487–498. [DOI] [PubMed] [Google Scholar]
- Sipe JD, Carreras I, Gonnerman WA, Cathcart ES, de Beer MC, and de Beer FC (1993). Characterization of the inbred CE/J mouse strain as amyloid resistant. The American journal of pathology 143, 1480–1485. [PMC free article] [PubMed] [Google Scholar]
- Sittig LJ, Carbonetto P, Engel KA, Krauss KS, Barrios-Camacho CM, and Palmer AA (2016). Genetic Background Limits Generalizability of Genotype-Phenotype Relationships. Neuron 91, 1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BA, Wnek C, Kotlus BS, Roemer N, MacTaggart T, and Phillips SJ (1999). Genotyping new BXD recombinant inbred mouse strains and comparison of BXD and consensus maps. Mamm Genome 10, 335–348. [DOI] [PubMed] [Google Scholar]
- Wang X, Pandey AK, Mulligan MK, Williams EG, Mozhui K, Li Z, Jovaisaite V, Quarles LD, Xiao Z, Huang J, et al. (2016). Joint mouse-human phenome-wide association to test gene function and disease risk. Nat Commun 7, 10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Giron CG, et al. (2018). Ensembl 2018. Nucleic acids research 46, D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, et al. (2013). Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zokaei N, Giehl K, Sillence A, Neville MJ, Karpe F, Nobre AC, and Husain M (2017). Sex and APOE: A memory advantage in male APOE epsilon4 carriers in midlife. Cortex; a journal devoted to the study of the nervous system and behavior 88, 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotypes from the BXD strains are publically available on GeneNetwork.org. RNA-sequencing from the hippocampus of a subset of AD-BXD strains is available on Gene Expression Omnibus (GEO) under accession number GSE101144. Data including raw phenotype information will also be deposited in the AMP-AD Knowledge Portal (doi: 10.7303/syn17016211). EMASE software used for alignment of RNA sequencing reads to a diploid transcriptome is available online at: (https://emase.readthedocs.io/en/latest/).


