Abstract
Personalized cancer medicine offers the promise of more effective treatments that are tailored to an individual’s own dynamic cancer phenotype. Meanwhile, tissue-engineering approaches to modeling tumors may complement these advances by providing a powerful new approach to understanding the adaptation dynamics occurring during treatment. However, in both of these areas new tools will be required to gain a full picture of the genetic and epigenetic regulators of phenotype dynamics occurring in the small populations of cells that drive resistance. In this study, we perform epigenomic analysis of brain tumor cells that are collected from micro-engineered three-dimensional tumor models, overcoming the challenges associated with the small numbers of cells contained within these micro-tissue niches, in this case collecting ~1,000 cells per sample. Specifically, we use a high-resolution epigenomic analysis method known as microfluidic-oscillatory-washing-based chromatin immunoprecipitation with sequencing (MOWChIP-seq) to analyze histone methylation patterns (H3K4me3). We identified gene loci that are associated with the H3K4me3 modification, which is generally a mark of active transcription. We compared methylation patterns in standard 2D cultures and 3D cultures based on type I collagen hydrogels, under both normoxic and hypoxic conditions. We found that culture dimensionality drastically impacted the H3k4me3 profile and resulted in differential modifications in response to hypoxic stress. Differentially H3K4me3-marked regions under the culture conditions used in this study have important implications for gene expression differences that have been previously observed. In total, our work illustrates a direct connection between cell culture or tissue niche condition and genome-wide alterations in histone modifications, providing the first steps towards analyzing the spatiotemporal variations in epigenetic regulation of cancer cell phenotypes. This study, to our knowledge, also represents the first time broad-spectrum epigenomic analysis has been applied to small cell samples collected from engineered micro-tissues.
Keywords: MOWChIP-seq, micro-tissue model, phenotypic heterogeneity, microenvironment, epigenetics
Graphical Abstract
Introduction
Despite tens of billions of dollars of investment in the United States alone into the development of new therapies, cancer is still a leading cause of death worldwide.1–3 One area where improvement is needed is a greater appreciation and understanding of the highly variable and dynamic abnormalities that may be present in any one type of cancer or even within a single tumor.4–5 Heterogeneity can occur in bulk tumor tissue, circulating tumor cells, disseminated/metastasized tumor cells, and cancer stem cells. Phenotypic variations usually happen at the single cell level and include alterations in protein expression, treatment response, and growth rate.6–7 The current standard of therapy is to treat the average profile of selected biopsied tumor cells, however, cancer is an extremely complex disease and there is an emerging realization that not taking a complete account of the molecular, phenotypic, and genetic subgroups present within a patient’s cancer will lead to the selection of therapies that are unlikely to yield more than incremental benefits.4–5, 8–9 Additionally, most targeted therapeutic strategies focus on genetic markers and mutations, but emerging research has shown that epigenetic abnormalities also play a role in regulating cancer progression.10 The development and implementation of high-resolution analysis of the genetic and epigenetic profile of a patient’s specific cancer is needed in order to improve the efficacy of current therapies.
Epigenetic alterations, which regulate the activation or silencing of cancer-associated genes, have been shown to be an important mechanism driving the development of phenotypic variability within a tumor. Histone modifications (e.g. acetylation, methylation, or phosphorylation of histone proteins) in particular have the potential to play a major role in regulating cell phenotype by altering transcription, DNA replication, DNA repair, and genomic stability within the cell.11–13 Next-generation sequencing (NGS) is a valuable tool that can be employed to analyze the profile of these histone modifications among cancer subgroups. Although NGS can be used for the targeted sequencing of well characterized cancer associated genes, whole-exome or whole-genome sequencing can be utilized to discover disease-causing gene variants in genetically heterogeneous or rare diseases.14 Epigenome profiling in particular could provide a better understanding of the pathogenesis of tumors and enable the observation of variable responses to anti-cancer drugs.15 Links have been established between epigenetic alterations and the prognosis of prostate cancer, the development of drug resistance in glioblastoma, and mechanisms of invasion and metastasis in breast cancer.16–19 Epigenetics also contribute, in part, to cellular responses to altering extracellular matrix (ECM) mechanics and could be essential in obtaining a complete understanding of the relationship between cancer cells and their microenvironment.20
The aim of this study was to characterize the global H3K4me3 profiles of U-251 MG Glioblastoma cells (U251) cultured under various microenvironmental stresses, which included 2D, 3D, normoxic, and hypoxic culture conditions. U251 cells were selected as a model of a highly aggressive cancer that has particularly few treatment options. However, research aimed at GBM heterogeneity has largely been focused on the role of clonal evolution and cancer stem cells and little is known about how microenvironmental stresses could be regulating the development of tumor subpopulations.21 The reliance on 2D culture screening platforms has been largely implicated in the mere 9.6% success rate of a cancer therapy to pass through all three phases of a clinical trial and has led to the development of more advanced preliminary screening platforms that can better model the complexity of the in vivo tumor and predict therapeutic responses.3, 22 Culture dimensionality has a significant impact on cell phenotype, as evidenced by altered cell phenotypes, genetic mechanisms, and responses to therapeutics in monolayer versus 3D culture systems that have been widely observed.23–25 The introduction of an ECM mimic provides a vastly different and more physiologically relevant environment than traditional 2D cultures for cell-cell and cell-ECM interactions as well. Along with culture dimensionality, hypoxia has also been shown to drive phenotypic changes, particularly in the context of cancer, where tumor cells under hypoxic conditions have been shown to have increased angiogenic potential and therapeutic resistance.26 However the role of epigenetics in these observed phenotypic changes remains unknown, and this epigenomic profiling was therefore the target of our study. The histone modification profile we measured was obtained using our recently invented highly sensitive microfluidic-oscillatory-washing based chromatin immunoprecipitation (MOWChIP) platform that reduces nonspecific adsorption to obtain high yields of highly enriched DNA, enabling the collection and analysis of approximately 180 picograms of ChIP DNA from 1,000 cells, where traditional ChIP protocols were only able to obtain tens of picograms of DNA from 10,000 cells.27 MOWChIP was followed by NGS using Illumina sequencing. The successful use of the MOWChIP platform in this study serves as a proof-of-concept to utilize this highly sensitive platform in the future to better understand current barriers to generating effective therapies, such as characterizing intratumoral heterogeneity, which is composed of small subpopulations of cell phenotypes, and analyzing the phenotypic dynamics of the sparse but crucial populations of cancer stem cells. We will analyze the implications of the disparities we observed between culture conditions in the global H3K4me3 profile of U251 cells. We will then discuss the significance of this study in the advancement of methods to improve personalized cancer care efficiency.
Materials and Methods
Cell culture
U251-MG human glioblastoma cells (Sigma-Aldrich, St. Louis, MO) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS) (Life Technologies, Grand Island, NY), 1% penicillin/streptomycin. (Lonza), and 0.1% non-essential amino acids (VWR, Radnor, PA). The cells were maintained in a 37˚C humidified incubator with 5% CO2.
Preparation of collagen stock
The collagen stock was prepared as published previously.28 Briefly, type I collagen stock was prepared from Sprague Dawley rat tails (BioreclamationIVT, Baltimore, MD). Tendons were resected from rat tails and added to 200 ml of 0.1% sterile acetic acid per gram of tendon. After at least 48 hours at 4˚C and occasional agitation (twice a day), the collagen solution (still containing bits of tissue) was centrifuged for 45 minutes at 30,000 × g at 4˚C in an Avanti J-25 centrifuge (Beckman Coulter, Brea, CA). The clear supernatant collagen solution was collected and the pellet was discarded. The collagen solution was then lyophilized and stored at −20˚C. The lyophilized collagen was re-suspended in 0.1% acetic acid at a stock concentration of 10 mg/ml. To re-dissolve the lyophilized material, the suspension was vortexed and stored at 4˚C for 48 hours to ensure that the collagen was completely dissolved (i.e. uniform solution with no visible collagen masses).
Formation of cellularized collagen hydrogels
Cellularized collagen scaffolds were formed in (poly)-dimethylsiloxane (PDMS) wells (Sylgard® 184 Silicone Elastomer Kit, Dow Corning, Midland, MI) of 10 mm diameter and 1 mm thickness. The PDMS wells were surface treated with 1% poly (ethylenimine) (PEI) for 10 minutes and 0.1% glutaraldehyde for 20 minutes (Fisher Scientific, Pittsburg, PA) to crosslink the collagen to the PDMS. Excess glutaraldehyde was rinsed off of the wells with DI water. Hydrogels with a final concentration of 5 mg/ml collagen were created from the 10 mg/ml stock solution. U251s were seeded within the collagen hydrogels at a density of 1 million cells/ml. The hydrogels were prepared by transferring the desired volume of collagen stock (50% of the final hydrogel volume) into a centrifuge tube kept on ice. Ice cold 10X DMEM (10% of final solution) and 1N NaOH (2% of final solution) were added to and mixed thoroughly with the collagen stock. In a separate centrifuge tube, U251s were added to 1X DMEM (volume needed to complete the remaining 50% of the final hydrogel volume). The U251 cell suspension was then added to the uncrosslinked collagen solution and slowly mixed on ice until cells were homogenously dispersed in the collagen solution. 10X DMEM and 1X DMEM were purchased from Sigma-Aldrich (St. Louis, MO) and NaOH was purchased from Fisher Scientific (Pittsburg, PA). Using a syringe, the neutralized collagen solution was transferred to the PDMS wells and allowed to polymerize at 37˚C at 5% CO2 for 30 minutes. After collagen was crosslinked, sufficient media was added to the wells to completely submerge the collagen scaffolds.
Cell sample collection
The day after U251s were seeded in the collagen hydrogels, media was refreshed and well plates or flasks were placed in an incubator at normoxic (20% O2) or hypoxic (1% O2) conditions. Samples were then incubated for 72 hours under these controlled conditions. Cells in flasks were trypsinized, rinsed once in ice cold PBS, then re-suspended in ice cold PBS at 1,000 cell/μl. Cells seeded in collagen hydrogels were obtained by digesting the collagen in a solution of 0.5% collagenase (Thermo Fisher, Waltham, MA) and 1% FBS in Hanks Buffered Salt Solution (HBSS) (Lonza). Collagen scaffolds were submerged in collagenase solution and incubated at 37˚C and 5% CO2 for 2 hours. Digested collagen solution containing released cells was collected, rinsed once in ice cold PBS, and re-suspended in ice cold PBS at 1,000 cell/μl. 1 μl of phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, St. Louis, MO) and 1 μl of protease inhibitor cocktail (1X concentration) (Sigma-Aldrich, St. Louis, MO) were added to a 100 μl aliquot of cell suspension for each culture condition.
MOWChIP-Seq
The custom microfluidic chip for cellular analysis was fabricated and prepared as previously described.27 The microfluidic chip was mounted on an inverted microscope (IX 71, Olympus). A data acquisition card (NI SCB-68, National Instruments) and a LabVIEW (LabVIEW 2012, National Instruments) program were employed to control the switching of the solenoid valve. 10 μl cell suspension containing 1,000 U251 cells was mixed with 10 μl of 2X lysis buffer (4% Triton X, 100 mM Tris, pH 7.5, 100 mM NaCl and 30 mM MgCl2) and incubated at room temperature for 10 min. 1 μl of 0.1 M CaCl2 and 2.5 μl of 10 U/μl MNase (88216,ThermoFisher) were rapidly mixed with the sample and incubated at room temperature for 10 min. 2.22 μl of 0.5 M EDTA (pH 8) was added and incubated on ice for 10 minutes. The sample was centrifuged at 16,100×g at 4°C for 5 minutes. Supernatant was transferred to a new microcentrifuge tube and stored on ice. Superparamagnetic Dynabeads Protein A (2.8 μm, 30 mg/ml, 1001D, Invitrogen) were used. 150 μg beads were mixed with 0.75 μg anti-H3K4me3 antibody (ab8505, Abcam) in 150 μl IP buffer (20 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% (w/v) sodium deoxycholate, 0.1% SDS, 1% (v/v) Triton X-100) at 4 °C on a rotator mixer at 24 r.p.m for 2 hours. Chromatin immunoprecipitation and oscillatory washing was conducted as previously described.27 Beads were suspended in 50 μl of freshly made elution buffer (1 mg/ml proteinase K, 10 mM Tris-HCl, pH 7.4, 50 mM NaCl, 10 mM EDTA, pH 8.0, and 0.03% SDS) and incubated on a thermocycler at 60 °C for 1 hour. DNA was purified using SPRIselect beads (Beckman Coulter) following the manufacturer’s instructions. Quantitative PCR (qPCR) was performed for selecting high quality ChIP samples. Samples with relative fold enrichment of positive loci VEGF over 15 were used for sequencing library construction. All ChIP-seq libraries were constructed using Accel-NGS 2S plus DNA library kit (Swift Bioscience) following the manufacturer’s instructions with minor modifications. During library preparation, ChIP DNA was end repaired and sequentially ligated with Illumina adapters. PCR amplification was used to increase the yield of indexed libraries. EvaGreen dye (1x, Biotium) was added to avoid PCR over-amplification. The fragment size of the library was determined by a TapeStation and quantified by KAPA qPCR library quantification kit. A volume of 1 μl of each diluted library were pooled together at a final concentration of 10 nM for sequencing by Illumina HiSeq 4000 with single-end 50 nucleotide read.
ChIP-seq data analysis
Sequencing reads were processed by AQUAS pipeline. Raw reads were mapped to hg19 genome. Duplicates and low quality mapped reads were filtered out. Filtered reads were used for narrow peak calling with MACS2 (q < 0.05). Cross-correlation scores and IDR (irreproducible discovery rate) were calculated. Signal tracks were generated and visualized on IGV (Integrative Genomics Viewer 3.0). Genome-wide differentially marked regions between sample groups were identified by DiffBind.29 The analysis was executed using DESeq2 and a p-value of 0.05 was set as a threshold. RPKM (reads per kilobase million) fold was used as a scoring method. The Venn diagram was plotted by InteractivVenn.30 Functional enrichment GO (Gene Ontology) terms were obtained by PANTHER overrepresentation tests.31 Pathway analysis was conducted using ConsensusPathDB. ChIP peak annotation was obtained by ChIPseeker with default settings. 32–33
Results
Culture dimensionality and oxygen status impact H3K4me3 modifications
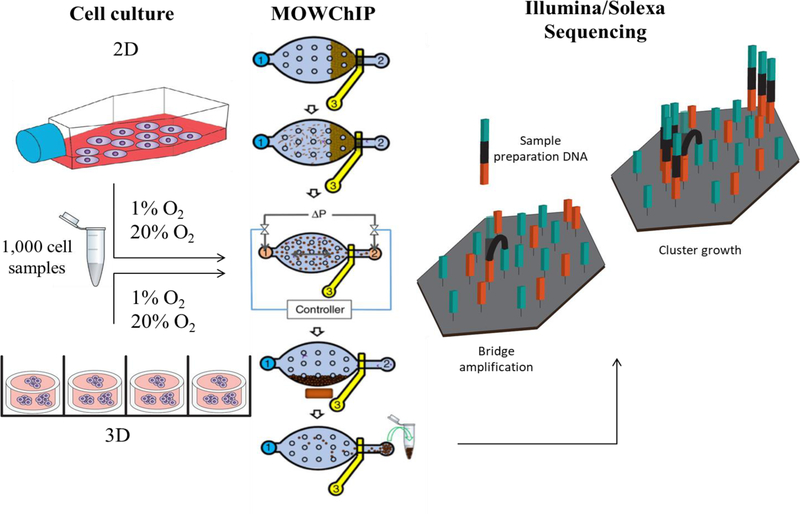
Hydrogels are the most widely used 3D tissue mimics due to their ease of fabrication, and capacity to provide a dynamic physiological microenvironment for modeling cell processes. Type I collagen is a popular ECM mimic because it is derived from native ECM and provides a bioactive microenvironment for cells.34 In order to examine the impact of culture dimensionality on the modifications of H3K4me3 across the genome of U251 GBM cells, we cultured the cells in a 2D monolayer on conventional tissue culture polystyrene in a cell flask or seeded them within type I collagen hydrogels for 72 hours. To analyze the impact of hypoxia on U251 H3K4me3 modifications, the cells were also cultured under normoxic (20% O2) or hypoxic (1% O2) conditions. Furthermore, our 3D hydrogels had a thickness of 1 mm, within the diffusion limits of oxygen, and U251s were seeded at a density of 1 million cells/ml to ensure the cells maintained within the 3D cultures were exposed to uniform oxygen concentrations.35 MOWChIP-seq was then performed using 1,000 cell sample sets to obtain a genome-wide profile of H3K4me3 for each culture condition (Figure 1). Each 3D scaffold contains ~37,000 cells, far below the cell numbers needed for traditional ChIP protocols. The use of an MOWChIP platform enabled us to use samples as small as 1,000 cells in this study, and NGS was performed using Illumina Solexa technology.27
Figure 1.
Schematic of sample culture and processing. U251s were cultured under 2D or 3D conditions in normoxic (20% O2) or hypoxic (1% O2) oxygen conditions, 1,000 cell sample sizes were collected and processed using a MOWChIP platform, figure adapted with permission from Nature27, followed by Illumina/Solexa sequencing technology
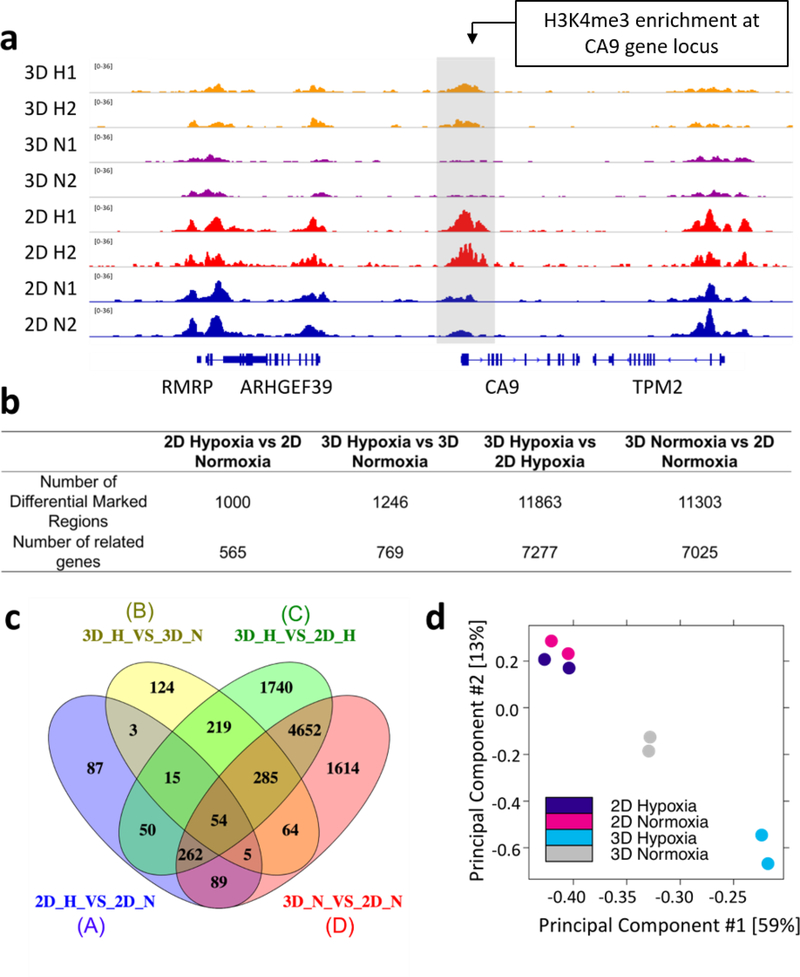
We observed extensive changes in H3K4me3 profiles when culture conditions such as dimensionality or oxygen level changed, as shown by the differential enrichment at the hypoxia-induced CA9 gene locus under hypoxic versus normoxic conditions (Fig. 2a). In order to determine the impact of hypoxia and culture dimensionality on H3K4me3 profiles, differential enrichment analysis of MOWChIP-seq peak data was performed. In general, genome-wide H3K4me3 varied much more substantially due to the alteration of culture dimensionality (3D vs. 2D) than alterations in oxygen status. 11,303 differentially marked regions were discovered under normoxic conditions and 11,863 under hypoxic conditions between 3D and 2D culture sets, compared to the 1,000 and 1,246 differentially marked regions identified under varying oxygen levels in 2D and 3D culture, respectively (Figure 2b). The majority of these differentially marked regions (11,371/11,863 or 96% for 2D versus 3D culture under hypoxia and 11,003/11,303 or 97% for 2D versus 3D culture under normoxia) experienced a decrease in H3K4me3 enrichment when the culture dimensionality changed from 2D to 3D (supplementary info). The genes associated with these differentially marked regions were also analyzed (Figure 2b and c). The majority of genes impacted by oxygen status (94%, 1,180 out of 1,257 genes as determined by dividing the number of genes in the non-overlapping regions of oval A and B by the total number of genes contained within the A and B ovals in Figure 2c) were unique to either 2D or 3D culture conditions. In contrast, only 42% (3,796 out of 9,049 as determined by dividing the number of genes in the non-overlapping regions of oval C and D by the total number of genes contained within the C and D ovals in Figure 2c) of the genes that were impacted by culture dimensionality were unique to either normoxic or hypoxic conditions. Principal component analysis of sequencing data from 2D and 3D samples cultured under normoxic and hypoxic conditions shows a clustering of samples within each treatment group indicating the reliability of the replicates to produce independent and consistent H3K4me3 profiles (Figure 2d).
Figure 2.
Peak analysis of H3K4me3 across the genome. (a) Normalized H3K4me3 MOWChIP-seq signals at CA9 gene locus. (b) Differential enrichment analysis of MOWChIP-seq peak data by DiffBind, P-value = 0.05 used as threshold, differentially marked regions were identified using DESeq2 (c) The Venn diagram displays the genes where differential H3K4me3 enrichment occurred between changes in culture dimensionality and oxygen status. Each oval represents the number of genes where differential H3K4me3 enrichment occurred between the culture conditions labeled A-D, (d) Principal Component Analysis shows group clustering based on differentially marked regions
Culture dimensionality impacts the H3K4me3 patterns on WNT, IL-1, and MAPK pathways and protein synthesis pathways
Functional enrichment analysis was performed to determine the enrichment of GO (Gene Ontology) terms, which represent biological pathways in which a significant number of genes within those pathways had differences in H3K4me3 enrichment between culture conditions. (Figure 3) Following the trends seen previously, the majority of the biological pathways that had changes in H3K4me3 enrichment were a result of altering culture dimensionality as opposed to oxygen status (121 vs 46 GO terms). It is also interesting to note that only 5 of the 41 pathways that had differences in enrichment under hypoxic vs normoxic conditions overlapped between 2D and 3D culture sets. There were 39 GO terms that had alterations in H3K4me3 enrichment when culture dimensionality was changed that overlapped between hypoxic and normoxic conditions. These processes are likely to be the ones solely impacted by culture dimensionality. Among these pathways include alterations in the regulation of Wnt signaling, IL-1-mediated signaling, and the MAPK cascade which have been highlighted in Figure 3. We also found that certain pathways had decreased H3K4me3 marking under 3D culture conditions compared to 2D culture under both hypoxia and normoxia. Those pathways included G-protein coupled receptor signaling pathways, mRNA splicing via spliceosome, rRNA processing and positive regulation of transcription by RNA polymerase II; the majority of which regulate protein synthesis (supplementary info).
Figure 3.
GO term analysis of H3K4me3 differential binding on biological pathways. Functional enrichment analysis obtained by PANTER Overrepresentation Tests on the sets of differentially marked regions using a GO database released February 2, 2018. LOG10(P)= −3.5 was used as a cutoff value. Heatmap shows the biological pathways that had differential H3K4me3 binding between the culture conditions indicated above each column.
Culture dimensionality impacts the distribution of H3K4me3 within gene structures
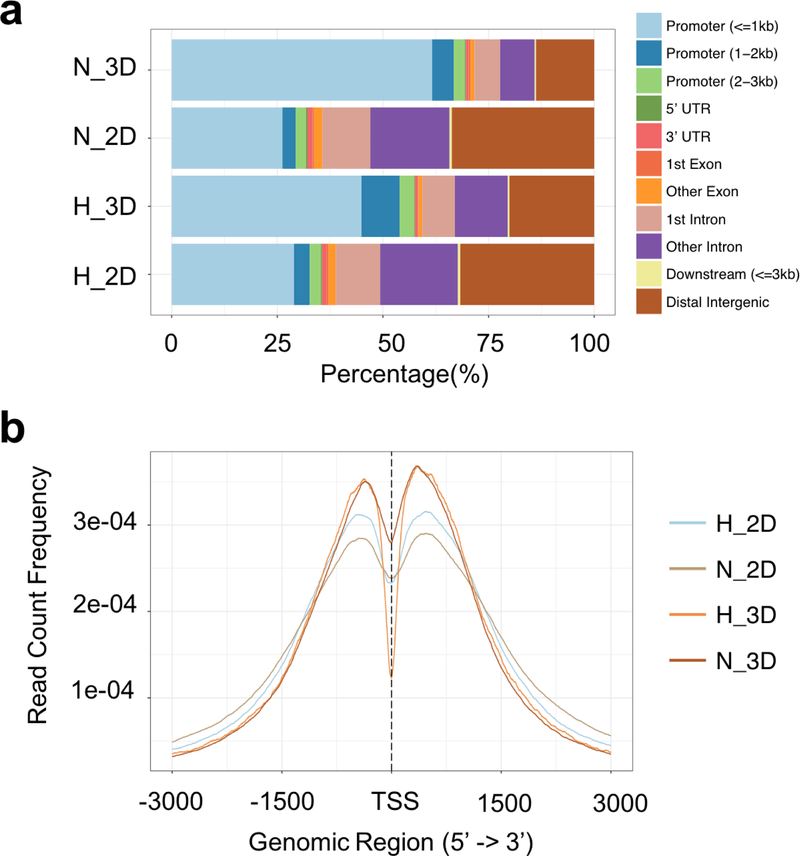
To examine the broader characteristics of the H3K4me3 profile, the location of the histone modifications within gene structures was analyzed. The 3D cultured cells under both normoxic and hypoxic conditions showed a greater concentration of H3K4me3 in gene promoter regions, whereas in 2D cultured cells there was a more uniform distribution of H3K4me3 between the promoter and intergenic regions (Figure 4a). Differences between 2D and 3D cultured cells were also observed in analyzing the H3K4me3 enrichment around the transcription start sites (TSS). There was a greater enrichment of H3K4me3 near TSS in 3D cultured cells under both normoxic and hypoxic conditions (Figure 4b).
Figure 4.
Analysis of H3K4me3 distribution along gene structure. (a) genomic annotation among MOWChIP-seq data. The 3D cultured cells showed a greater concentration of H3K4me3 in gene promoter regions, whereas in 2D cultured cells there was a more uniform distribution of H3K4me3 between the promoter and intergenic regions, (b) average profiles of ChIP peaks among different culture conditions, replicates were analyzed and pooled using AQUAS pipeline. There was a greater enrichment of H3K4me3 near TSS in 3D cultured cells under both normoxic and hypoxic conditions
Discussion
To the best of our knowledge, this is the first study that combines an in vitro 3D micro-tissue culture model with ChIP-seq to analyze the global histone modifications of U251 glioblastoma cells under varying culture conditions. A major barrier to overcome has been the small number of cells typically contained within these platforms. 3D micro-tissue platforms provide the power to analyze dynamic responses to complex spatiotemporal gradients, there may only be tens to hundreds of cells within localized niches. For this study we have used MOWChIP-seq to perform epigenomic analysis on 1,000 cell samples, characteristic of the number of cells contained within our micro-tissues, although cell number restrictions were artificial in this study, it serves as a first step towards highly localized epigenomic analysis in spatiotemporally varying tissues. Such capabilities could be invaluable in analyzing the rapid resistance dynamics that may be related to the relatively fast timescales of epigenetic responses to therapies or microenvironmental changes.18, 20
For this study, we elected to direct our analysis towards epigenetic alterations in the U251 glioblastoma multiforme cell line. As mentioned previously, GBM is a highly heterogeneous cancer with few effective treatment options. GBM is characterized by tumor recurrence of aggressive cell phenotypes and it has been shown to develop epigenetic mechanisms of therapeutic resistance.18, 36 Obtaining a better understanding of how the microenvironment may be facilitating the generation and support of the highly migratory and aggressive tumor cell phenotype that recurs after treatment, as well as how cues from the microenvironment may be shaping the development of chemoresistance, could elucidate potential new treatment targets. It has been shown that pediatric gliomas are biologically distinct from adult gliomas.37 The observation that certain pediatric gliomas harbor very few recurrent genetic events and instead are often regulated by epigenetic events such as CpG hypermethylation and post-translational histone modifications further underscores the importance of understanding the epigenetic events occurring within a tumor and throughout its progression in order to develop effective treatment strategies.38 Not only could studies such as this elucidate the mechanisms behind GBM cellular heterogeneity and the development of therapeutic resistance overall, but could also help to better differentiate these processes in pediatric versus adult glioblastomas.
We used traditional 2D culture methods along with our 3D micro-tissue models, based on type I collagen as a widely used physiological tissue mimic, to assess the impact of 3D culture on the H3K4me3 profile. The impact of hypoxia on the global H3K4me3 histone profile was also assessed by culturing the U251s under normoxic (20% O2) and hypoxic (1% O2) conditions. Many studies have elucidated the significant impact of the microenvironment on cell behavior, particularly in the context of cancer, where alterations in the surrounding ECM and the presence of hypoxia can affect tumor cell phenotype and drive tumor progression.24, 39–43 Our culture system enabled us to expose U251 GBM cells to these microenvironmental stresses and analyze how histone modifications are impacted to help answer questions that have arisen surrounding the role of epigenetics in tumor initiation and progression.
Using MOWChIP-seq peak data analysis, we observed that both dimensionality and oxygen status impacted H3K4me3 enrichment across the respective genomes. The variation in H3K4me3 between cells cultured in 2D versus 3D was approximately ten-fold greater than the differential enrichment that occurred between normoxic and hypoxic conditions in the same culture platform. This may indicate that certain microenvironmental factors such as the culture dimension, substrate, or ECM composition that cells are exposed to may have a greater impact on cell phenotype than other factors, such as oxygen concentration. Interestingly, this is consistent with previous work which analyzed global gene expression changes resulting from alterations in culture dimensionality.44 It was also observed that, overall, the intensity of H3K4me3 peaks decreased under 3D culture conditions compared to 2D. The decrease in global H3K4me3 levels under 3D culture conditions could indicate a decrease in overall gene expression. This global reduction in H3K4me3 levels in 3D culture could be a result of differences in cell shape or demethylase activity or both between 2D and 3D culture conditions, which would agree with the increase in demethylase expression, such as KDM5B, that has been observed in glioma tissues.45 Previous work has also shown that cell shape can regulate global histone modifications. In the study by Le Beyec et al. cell rounding was a sufficient stimulus to induce histone deacetylation and chromatin condensation in the absence of biochemical signals transduced from the ECM.46
Following what was observed regarding the number of differentially marked regions, a greater number of signaling pathways were also found to be impacted by culture dimensionality than oxygen concentration. GO term analysis identified the pathways that had the greatest changes in H3K4me3 when culture conditions were manipulated. These pathways include the Wnt signaling pathway, which is involved in cell proliferation and migration, and the MAPK cascade which regulates cell cycle entry and proliferation. Both signaling pathways play a major role in many cancers and alterations in histone modifications on these pathways may have a significant impact on cell behavior and cancer progression. Furthermore, we found that the IL-1 mediated signaling pathway had variations in H3K4me3 enrichment when culture dimensionality was changed, which agrees with previous reports that inflammatory signaling is altered between 2D and 3D culture conditions.44 The alterations in inflammatory signaling in the 2D and 3D cultures could have implications in the cellular response to hypoxia and regulation of angiogenesis.47 H3K4me3 levels increased in 2D cultures, regardless of oxygen status, on the mRNA splicing via spliceosome, rRNA processing, and positive regulation of transcription by RNA polymerase II biological pathways. Each of these pathways involve protein synthesis and are regulators of gene expression, pathways that are characteristically upregulated in traditional 2D in vitro cultures compared to cells in vivo.48 Interestingly, this over-proliferative phenotype of 2D cells is thought to be a major cause of the failure of these traditional cultures to accurately predict patient responses to cancer therapeutics.49 The lack of overlapping terms between 2D and 3D culture sets when oxygen status was altered illustrates varying epigenetic responses to hypoxia depending on culture condition, potentially signifying that culture dimensionality alters what histone modifications occur in response to other environmental stresses. Previous work that has analyzed the downstream gene expression profiles of cells cultured under similar conditions found that the majority of hypoxia-associated genes were unique to either 2D or 3D cultures.44 Although the oxygen diffusion rates within the 3D scaffold could slightly alter the timescale over which the cells would be exposed to changes in oxygen concentration versus 2D cultures, uniform oxygen conditions in the 3D cultures should be present within an hour thereby minimizing related experimental variations.35 Through our differential enrichment and GO term analysis, we have shown that the epigenetic response to microenvironmental stresses is highly dependent on the culture dimensionality and the H3K4me3 differential enrichment patterns on oxygenation-independent biological pathways agree with important phenotypic differences (e.g. protein synthesis and gene expression) that have been observed between 2D and 3D cultures. However, further analysis of epigenetics in vitro and in vivo is needed in order to validate current in vitro culture models, which was not ultimately the goal of this present study.
We next analyzed the distribution of H3K4me3 peaks along the gene structure. Previous work has noted that lower levels of histone modifications correlate to a poorer prognosis in certain cancers.17 This poorer prognosis could be due to a re-distribution of histone modifications to a limited number of genes that confer a more aggressive phenotype to the tumor or the decrease in global histone modifications could indicate a switch from a euchromatic to a heterochromatic state, that protects the cells against genotoxic stress by limiting DNA exposure.16–17 The significance of the histone modification location has also been shown by Lee et al. and Qin et al., where the region at which the modification occurs on the gene structure (e.g. coding versus regulatory region) impacts whether gene activation or repression is induced.50–51 Therefore, not only are the specific genes where histone modifications are occurring important, but so is the location of the histone modifications along that gene. Our analysis shows that culturing cells within 3D platforms results in a greater concentration of H3K4me3 on the gene promoter regions compared with cells cultured in 2D. The enrichment profile we observed in 3D could potentially impact both gene transcription and chromatin configuration as H3K4me3 concentrated at promoter regions has been associated with gene activation, but a reduction in overall H3K4me3 and concentration at specific sites could also alter the conformation of the chromatin, negatively regulating gene transcription but protecting the DNA from toxic stresses. We also observed a greater concentration of H3K4me3 around TSS in 3D versus 2D cultured cells. Previous work has shown that an increase in H3K4me3 distribution frequency near TSS was positively correlated with gene expression.52 However, future downstream gene expression analysis is needed in order to better understand the implications of the enrichment profiles we have observed in this study.
Overall our study has shown that histone modifications are strongly regulated by culture conditions and that the ability to engineer and tune culture microenvironments will greatly contribute to furthering our understanding of how the tumor microenvironment could drive cell phenotypic alterations. Progression towards an invasive and ultimately metastatic state is characterized by a host of changes in the tumor microenvironment, and alterations in both tissue oxygenation and ECM composition and density are key among these.39, 53 We observed that histone modification profiles are strongly regulated by tuning these specific culture conditions (e.g. normoxic versus hypoxic and 2D versus 3D). Yet there also still remains the question of whether the epigenomic profiles we have characterized are truly more representative of the in vivo phenotypic state. Along with the epigenomic differences between 2D and 3D cultures presented in this proof-of-concept work, transcriptomic variations have been found when culture dimensionality and 3D scaffold composition were altered.54 These discoveries emphasize the need to investigate specific cell-ECM interactions in future work, where perhaps the use of a more brain-specific matrix material, like hyaluronan, would have resulted in epigenomic profiles that more closely represent what is seen in vivo than those we characterized with the use of collagen hydrogels.
The capabilities of the high-resolution MOWChIP platform, which has been used to analyze as few as 100 cells, will also be a great asset in analyzing the epigenetic heterogeneity present within cell samples from the same culture.27 We have shown it is possible to analyze small numbers of cells obtained from in vitro model tissue samples, which is a promising start to the analysis of the small cancer cell subpopulations that exist in vivo, such as cancer stem cells or circulating cancer cells, which are a major contributor to disease progression and therapeutic resistance. However, the ability to generate and maintain cellular heterogeneity within our 3D culture models will be important to validate in future studies. The integration of engineered in vitro tumor mimics with high resolution epigenetic analysis that was utilized in this study will help to further the understanding of how microenvironmental epigenetic regulation of cell phenotype works in concert with genetic variations to generate cellular heterogeneity within a tumor, and the characterization of these processes will enable more effective personalized therapeutic strategies to be developed.
Conclusion
The current study helps to elucidate how microenvironmental signals can impact histone modification profiles across a human genome. The use of an engineered 3D micro-tissue model in concert with traditional 2D culture methods and varying oxygen concentrations enabled us to analyze the impact of culture dimensionality and oxygen status on H3K4me3 enrichment. Our findings illustrated that microenvironmental stresses directly impact histone modification profiles. Culture dimensionality is an essential regulator of the histone modifications that occur in response to microenvironmental signals and the global methylation profile. The H3K4me3 biological pathway binding profile also suggests that the phenotype of cells cultured in the 3D platforms more closely resembles what is seen in vivo compared with the 2D counterparts. This was also the first study that has analyzed small sample sizes of cells culture in 3D micro-tissue models using a high-resolution ChIP-seq platform. The methods employed in this work could be a key tool to understanding how the tumor microenvironment regulates tumor cell phenotype, particularly characterizing the formation of heterogeneous tumor cell subpopulations. Utilizing sensitive cell characterization assays such as this could help to improve the efficacy of personalized cancer therapy strategies.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health [grant numbers R21EB019123, R01CA213423, R01EB017235, and R33CA214176], the Institute for Critical Technology and Applied Science (ICTAS) Center for Engineered Health of Virginia Tech, and NSF CAREER Award [grant number CBET-1652112].
Footnotes
Supporting Information
The raw data required to reproduce these findings are available to download from Gene Expression Omnibus. ChIP-seq data are deposited under accession number GSE113824 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113824). Supplementary information includes raw data of cell counts collected from 3D hydrogel platforms, images of cells in the experimental culture conditions, further pathway enrichment analysis, differential binding analysis of H3K4me3 across genome, a summary of H3K4me3 MOWChIP-seq data on 1,000 U251 cells, and a list of gene IDs from GO database for overlapped genes between culture conditions.
References
- 1.Håkanson M; Cukierman E; Charnley M, Miniaturized pre-clinical cancer models as research and diagnostic tools. Advanced drug delivery reviews 2014, 69, 52–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DiMasi JA; Hansen RW; Grabowski HG, The price of innovation: new estimates of drug development costs. Journal of health economics 2003, 22 (2), 151–185. [DOI] [PubMed] [Google Scholar]
- 3.Cox MC; Reese LM; Bickford LR; Verbridge SS, Toward the broad adoption of 3D tumor models in the cancer drug pipeline. ACS Biomaterials Science & Engineering 2015, 1 (10), 877–894. [DOI] [PubMed] [Google Scholar]
- 4.Rodon J; Soria J; Berger R; Batist G; Tsimberidou A; Bresson C; Lee J; Rubin E; Onn A; Schilsky R, Challenges in initiating and conducting personalized cancer therapy trials: perspectives from WINTHER, a Worldwide Innovative Network (WIN) Consortium trial. Annals of Oncology 2015, 26 (8), 1791–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerlinger M; Rowan AJ; Horswell S; Larkin J; Endesfelder D; Gronroos E; Martinez P; Matthews N; Stewart A; Tarpey P, Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. New England journal of medicine 2012, 366 (10), 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian M; Wang DC; Chen H; Cheng Y. In Detection of single cell heterogeneity in cancer, Seminars in cell & developmental biology, Elsevier: 2017; pp 143–149. [DOI] [PubMed] [Google Scholar]
- 7.Navin NE; Hicks J, Tracing the tumor lineage. Molecular oncology 2010, 4 (3), 267–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciriello G; Miller ML; Aksoy BA; Senbabaoglu Y; Schultz N; Sander C, Emerging landscape of oncogenic signatures across human cancers. Nature genetics 2013, 45 (10), 1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munoz J; Swanton C; Kurzrock R, Molecular profiling and the reclassification of cancer: divide and conquer. Am Soc Clin Oncol Educ Book 2013, 2013, 127–34. [DOI] [PubMed] [Google Scholar]
- 10.Jones PA; Baylin SB, The epigenomics of cancer. Cell 2007, 128 (4), 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner BM, Reading signals on the nucleosome with a new nomenclature for modified histones. Nature structural & molecular biology 2005, 12 (2), 110. [DOI] [PubMed] [Google Scholar]
- 12.Shahbazian MD; Grunstein M, Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem 2007, 76, 75–100. [DOI] [PubMed] [Google Scholar]
- 13.Kim YZ, Altered histone modifications in gliomas. Brain tumor research and treatment 2014, 2 (1), 7–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khotskaya YB; Mills GB; Mills Shaw KR, Next-generation sequencing and result interpretation in clinical oncology: challenges of personalized cancer therapy. Annual review of medicine 2017, 68, 113–125. [DOI] [PubMed] [Google Scholar]
- 15.Farlik M; Sheffield NC; Nuzzo A; Datlinger P; Schönegger A; Klughammer J; Bock C, Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell reports 2015, 10 (8), 1386–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurdistani S, Histone modifications as markers of cancer prognosis: a cellular view. British journal of cancer 2007, 97 (1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seligson DB; Horvath S; Shi T; Yu H; Tze S; Grunstein M; Kurdistani SK, Global histone modification patterns predict risk of prostate cancer recurrence. Nature 2005, 435 (7046), 1262. [DOI] [PubMed] [Google Scholar]
- 18.Kitange GJ; Mladek AC; Carlson BL; Schroeder MA; Pokorny JL; Cen L; Decker PA; Wu W; Lomberk G; Gupta SK, Inhibition of histone deacetylation potentiates the evolution of acquired temozolomide resistance linked to MGMT upregulation in glioblastoma xenografts. Clinical Cancer Research 2012, clincanres. 0560.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westcott JM; Prechtl AM; Maine EA; Dang TT; Esparza MA; Sun H; Zhou Y; Xie Y; Pearson GW, An epigenetically distinct breast cancer cell subpopulation promotes collective invasion. The Journal of clinical investigation 2015, 125 (5), 1927–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y; Tang CB; Kilian KA, Matrix Mechanics Influence Fibroblast–Myofibroblast Transition by Directing the Localization of Histone Deacetylase 4. Cellular and Molecular Bioengineering 2017, 10 (5), 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonavia R; Cavenee WK; Furnari FB, Heterogeneity maintenance in glioblastoma: a social network. Cancer research 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mullard A, Parsing clinical success rates. Nature Publishing Group: 2016. [DOI] [PubMed] [Google Scholar]
- 23.Sutherland RM; McCredie JA; Inch WR, Growth of multicell spheroids in tissue culture as a model of nodular carcinomas. Journal of the National Cancer Institute 1971, 46 (1), 113–120. [PubMed] [Google Scholar]
- 24.Theodorescu D; Sheehan C; Kerbel R, TGF-β gene expression depends on tissue architecture. In Vitro Cellular & Developmental Biology-Animal 1993, 29 (2), 105–108. [DOI] [PubMed] [Google Scholar]
- 25.Croix BS; FLøRENES VA; Rak JW; Flanagan M; Bhattacharya N; Slingerland JM; Kerbel RS, Impact of the cyclin–dependent kinase inhibitor p27Kip1 on resistance of tumor cells to anticancer agents. Nature medicine 1996, 2 (11), 1204–1210. [DOI] [PubMed] [Google Scholar]
- 26.Harris AL, Hypoxia—a key regulatory factor in tumour growth. Nature Reviews Cancer 2002, 2 (1), 38. [DOI] [PubMed] [Google Scholar]
- 27.Cao Z; Chen C; He B; Tan K; Lu C, A microfluidic device for epigenomic profiling using 100 cells. Nature methods 2015, 12 (10), 959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cross VL; Zheng Y; Choi NW; Verbridge SS; Sutermaster BA; Bonassar LJ; Fischbach C; Stroock AD, Dense type I collagen matrices that support cellular remodeling and microfabrication for studies of tumor angiogenesis and vasculogenesis in vitro. Biomaterials 2010, 31 (33), 8596–8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ross-Innes CS; Stark R; Teschendorff AE; Holmes KA; Ali HR; Dunning MJ; Brown GD; Gojis O; Ellis IO; Green AR, Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 2012, 481 (7381), 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heberle H; Meirelles GV; da Silva FR; Telles GP; Minghim R, InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC bioinformatics 2015, 16 (1), 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas PD; Campbell MJ; Kejariwal A; Mi H; Karlak B; Daverman R; Diemer K; Muruganujan A; Narechania A, PANTHER: a library of protein families and subfamilies indexed by function. Genome research 2003, 13 (9), 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamburov A; Wierling C; Lehrach H; Herwig R, ConsensusPathDB—a database for integrating human functional interaction networks. Nucleic acids research 2008, 37 (suppl_1), D623–D628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu G; Wang L-G; He Q-Y, ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31 (14), 2382–2383. [DOI] [PubMed] [Google Scholar]
- 34.Page H; Flood P; Reynaud EG, Three-dimensional tissue cultures: current trends and beyond. Cell and tissue research 2013, 352 (1), 123–131. [DOI] [PubMed] [Google Scholar]
- 35.Ehsan SM; George SC, Nonsteady state oxygen transport in engineered tissue: implications for design. Tissue Engineering Part A 2013, 19 (11–12), 1433–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sottoriva A; Spiteri I; Piccirillo SG; Touloumis A; Collins VP; Marioni JC; Curtis C; Watts C; Tavaré S, Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proceedings of the National Academy of Sciences 2013, 110 (10), 4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones C; Baker SJ, Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nature reviews Cancer 2014, 14 (10), 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mack S; Witt H; Piro R; Gu L; Zuyderduyn S; Stütz A; Wang X; Gallo M; Garzia L; Zayne K, Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 2014, 506 (7489), 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu P; Weaver VM; Werb Z, The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol 2012, 196 (4), 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weaver VM; Lelièvre S; Lakins JN; Chrenek MA; Jones JC; Giancotti F; Werb Z; Bissell MJ, β4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer cell 2002, 2 (3), 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen-Ngoc K-V; Cheung KJ; Brenot A; Shamir ER; Gray RS; Hines WC; Yaswen P; Werb Z; Ewald AJ, ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proceedings of the National Academy of Sciences 2012, 201212834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Axelson H; Fredlund E; Ovenberger M; Landberg G; Påhlman S. In Hypoxia-induced dedifferentiation of tumor cells–a mechanism behind heterogeneity and aggressiveness of solid tumors, Seminars in cell & developmental biology, Elsevier: 2005; pp 554–563. [DOI] [PubMed] [Google Scholar]
- 43.Joanne LY; Rak JW; Carmeliet P; Nagy A; Kerbel RS; Coomber BL, Heterogeneous vascular dependence of tumor cell populations. The American journal of pathology 2001, 158 (4), 1325–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DelNero P; Lane M; Verbridge SS; Kwee B; Kermani P; Hempstead B; Stroock A; Fischbach C, 3D culture broadly regulates tumor cell hypoxia response and angiogenesis via pro-inflammatory pathways. Biomaterials 2015, 55, 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dai B; Hu Z; Huang H; Zhu G; Xiao Z; Wan W; Zhang P; Jia W; Zhang L, Overexpressed KDM5B is associated with the progression of glioma and promotes glioma cell growth via downregulating p21. Biochemical and biophysical research communications 2014, 454 (1), 221–227. [DOI] [PubMed] [Google Scholar]
- 46.Le Beyec J; Xu R; Lee S-Y; Nelson CM; Rizki A; Alcaraz J; Bissell MJ, Cell shape regulates global histone acetylation in human mammary epithelial cells. Experimental cell research 2007, 313 (14), 3066–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voronov E; Carmi Y; Apte RN, The role IL-1 in tumor-mediated angiogenesis. Frontiers in physiology 2014, 5, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Birgersdotter A; Sandberg R; Ernberg I. In Gene expression perturbation in vitro—a growing case for three-dimensional (3D) culture systems, Seminars in cancer biology, Elsevier: 2005; pp 405–412. [DOI] [PubMed] [Google Scholar]
- 49.Edmondson R; Broglie JJ; Adcock AF; Yang L, Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay and drug development technologies 2014, 12 (4), 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S; Lee J; Chae S; Moon Y; Lee H-Y; Park B; Yang EG; Hwang D; Park H, Multi-dimensional histone methylations for coordinated regulation of gene expression under hypoxia. Nucleic acids research 2017, 45 (20), 11643–11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qin L; Wu X; Block ML; Liu Y; Breese GR; Hong JS; Knapp DJ; Crews FT, Systemic LPS causes chronic neuroinflammation and progressive neurodegeneration. Glia 2007, 55 (5), 453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barski A; Cuddapah S; Cui K; Roh T-Y; Schones DE; Wang Z; Wei G; Chepelev I; Zhao K, High-resolution profiling of histone methylations in the human genome. Cell 2007, 129 (4), 823–837. [DOI] [PubMed] [Google Scholar]
- 53.Pupa SM; Ménard S; Forti S; Tagliabue E, New insights into the role of extracellular matrix during tumor onset and progression. Journal of cellular physiology 2002, 192 (3), 259–267. [DOI] [PubMed] [Google Scholar]
- 54.Tekin H; Simmons S; Cummings B; Gao L; Adiconis X; Hession CC; Ghoshal A; Dionne D; Choudhury SR; Yesilyurt V, Effects of 3D culturing conditions on the transcriptomic profile of stem-cell-derived neurons. Nature Biomedical Engineering 2018, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.