Abstract
The small GTP-binding protein Rad (RRAD, Ras associated with diabetes) is the founding member of the RGK (Rad, Rem, Rem2, and Gem/Kir) family that regulates cardiac voltage-gated Ca2+ channel function. However, its cellular and physiological functions outside of the heart remain to be elucidated. Here we report that Rad GTPase function is required for normal bone homeostasis in mice, as Rad deletion results in significantly lower bone mass and higher bone marrow adipose tissue (BMAT) levels. Dynamic histomorphometry in vivo and primary calvarial osteoblast assays in vitro demonstrate that bone formation and osteoblast mineralization rates are depressed, while in vitro osteoclast differentiation is increased, in the absence of Rad. Microarray analysis revealed that canonical osteogenic gene expression (Runx2, osterix, etc.) is not altered in Rad−/− calvarial osteoblasts; instead robust up-regulation of matrix Gla protein (MGP, +11-fold), an inhibitor of extracellular matrix mineralization and a protein secreted during adipocyte differentiation, was observed. Strikingly, Rad deficiency also resulted in significantly higher marrow adipose tissue levels in vivo and promoted spontaneous in vitro adipogenesis of primary calvarial osteoblasts. Adipogenic differentiation of wildtype calvarial osteoblasts resulted in the loss of endogenous Rad protein, further supporting a role for Rad in the control of BMAT levels. These findings reveal a novel in vivo function for Rad and establish a role for Rad signaling in the complex physiological control of skeletal homeostasis and bone marrow adiposity.
Keywords: Ras GTPase, osteoblasts, osteogenesis, adipogenesis, bone marrow adipose tissue, matrix Gla protein
1. Introduction
Rad (RRAD, Ras associated with diabetes) is a monomeric G-protein that was originally identified as a gene up-regulated in the skeletal muscle of a subset of patients with type 2 diabetes [1] and is the founding member of the RGK (Rem, Rad, Rem2, and Gem/Kir) subfamily of Ras-related small GTPases [1–5]. The common structure for all RGK proteins consists of a conserved Ras-related core domain, a series of nonconservative amino acid substitutions within regions involved in guanine nucleotide binding and hydrolysis, a non-CAAX-containing C-terminal extension, and large N-terminal extensions relative to other Ras family proteins [6, 7]. Although RGK proteins display modest intrinsic GTPase activity, recent studies have questioned whether RGK proteins are predominantly controlled via the canonical GTP/GDP regulatory cycle [7, 8], and no guanine exchange factor (GEF) or GTPase activating protein (GAP) regulatory proteins have been identified to date. However, it is clear that both transcriptional and posttranscriptional control mechanisms, particularly phosphorylation, regulate RGK protein activity in response to a diversity of environmental stimuli [9–13]. Rad was initially identified as a gene overexpressed in the skeletal muscle of type II diabetic individuals [1], although analysis of Rad expression in Pima Indians and the Zucker diabetic rat model did not find such a correlation [14]. Rad expression is also up-regulated in regenerating limb muscle following amputation in the newt [15], in the myogenic progenitor cell population during skeletal muscle regeneration [16], denervated mouse muscle [17], and in vascular smooth muscle cells following balloon injury [18]. We have recently found that Rad protein levels are lowered in human patients with end-stage nonischemic heart failure [19]. In addition to these changes in Rad expression in muscle, Rad levels are also altered in other tissues that require further investigation. For instance, Rad expression is induced in the suprachiasmatic nucleus following light stimulation [20, 21], in human peripheral blood mononuclear cells after acute heat shock [22], in cirrhotic livers relative to normal livers [23], and in human placenta following hypoxia [24]. In each of these cases, the mechanism for up-regulation of Rad expression and its functional implications require further investigation.
Rad function has primarily been studied in the heart, where Rad has been shown to inhibit L-type calcium channel activity [25, 26] and attenuate β-adrenergic signaling [19, 26–28]; however, Rad is also expressed in non-excitable tissues [14], suggesting physiological roles for this G-protein beyond calcium channel regulation. Indeed, recent work suggests that Rad deficiency can lead to increased cardiac fibrosis [29], which arises from excess deposition of extracellular matrix (ECM) in the heart [30]. Rad was found to inhibit connective tissue growth factor expression in cardiomyocytes through its association with CCAAT-enhancer binding protein-δ (C/EBP-δ) to regulate ECM production [29]. Together with a literature indicating that RGK proteins, through interactions with both calmodulin and 14–3-3 proteins, undergo regulated nuclear transport [31–33], these data suggest that RGK proteins, including Rad, may play an underappreciated role in the regulation of gene expression.
RGK proteins have also been identified as novel regulators of cell differentiation [34–36]. Rem2, an RGK family member is highly expressed in embryonic stem cells and plays a key role in ectoderm differentiation and neuronal development [34, 35]. The observation that Rem2 may regulate cell differentiation was recently extended to include Rad [36]. Satija and colleagues recently reported that lithium treatment of human mesenchymal stem cells (MSCs) to enhance osteogenic differentiation elicited a robust increase in Rad expression [36]. Notably, siRNA-mediated Rad silencing reversed the osteogenic priming effect of lithium [36]. Hence, Rad may play a role in the regulation of osteogenesis that requires further investigation.
Mesenchymal stem cells (MSCs) present in the bone marrow are the precursors for osteoblasts, chondrocytes, and adipocytes [37, 38]. Interestingly, many conditions that can induce bone loss, such as estrogen insufficiency, anorexia, disuse, and hind limb unloading, are accompanied by increased bone marrow adipose tissue (BMAT) [39–43]. In patients with osteoporosis, bone marrow adiposity is significantly increased, and bone formation rates are inversely related to BMAT levels [44, 45]. One mechanism that has been proposed to explain the often inverse relationship between bone density and bone marrow adiposity is a shift in mesenchymal progenitors toward more adipogenic differentiation at the expense of osteoblast formation, but much remains to be determined [46]. However, the transcriptional programs that drive MSCs to adopt these two cell fates are well characterized, with C/EBP-α and peroxisome proliferator-activated receptor γ2 (PPARγ2) initiating expression of genes associated with mature adipocytes [47], and Runt-related transcription factor 2 (Runx2) and the downstream osteoblast-specific transcription factor osterix/Sp7 required for osteogenic differentiation [48–50]. Unexpectedly, and in contrast to white and brown adipocytes, bone marrow adipocytes were recently found to express osterix/Sp7 [51], suggesting that MSCs directed toward an osteogenic fate may be re-allocated toward an adipogenic one. While several Ras-related GTPases and mitogen activated protein kinases (MAPKs) have been demonstrated to contribute to proper osteoblast development [52–54], there is need for a deeper mechanistic understanding of the regulatory signaling pathways involved in osteogenesis versus adipogenesis, especially the signal transduction cascades controlling BMAT.
Given the importance of the balance between osteogenesis and adipogenesis in human disease, we seek in the present study to characterize the effects of Rad deletion on bone homeostasis and bone marrow adiposity in vivo and on osteoblast function in vitro using global Rad-knockout (Rad−/−) mice. We test the hypothesis that genetic deletion of Rad results in low bone mass through a decrease in bone formation by osteoblasts, and we postulate that Rad might be one of the elusive upstream regulators of the switch between osteogenesis and adipogenesis.
2. Materials and Methods
2.1. Animals
Mice lacking Rad expression (Rad−/−) were obtained from the Kahn laboratory at the Joslin Diabetes Center and have been described previously [55]. Rad−/− mice were backcrossed with C57/BL6 mice for 5 generations. All experimental procedures and methods used were approved by the Institutional Animal Care and Use Committee of the University of Kentucky and conformed to the National Institutes of Health Guide for Care and Use of Laboratory Animals.
The femora used in this study were obtained from male and female mice at four months of age. Mouse spleen cells were isolated from two-month-old male mice for osteoclast differentiation assays. Primary calvarial osteoblasts were isolated from pooled litters of three-day-old mice. For fluorochrome labeling, mice were injected intraperitoneally with calcein (pH 7.4, 30 mg/kg, Sigma) at 7 and 2 days prior to sacrifice.
2.2. Microcomputed tomography
Femora were fixed in 10% neutral buffered formalin and transferred to 70% ethanol prior to scanning. μCT scanning was performed using a Scanco Model 40 (Scanco Medical AG, Basserdorf, Switzerland) at 55 kV and 145 μA, 0.3-second integration time with a 10 μm isotropic voxel size in plane and a 10 μm thickness. The overall femur length was used to guide the cortical and trabecular analyses. Specifically, the trabecular ROI ranged from just proximal of the distal growth plate to 30% of the bone length as measured from the distal end. The trabecular output variables included total volume (TV), bone volume (BV), bone volume fraction (BV/TV), connectivity density (Conn.D), structural model index (SMI), trabecular number (Tb.N), trabecular thickness (Tb.Th), trabecular spacing (Tb.Sp), apparent density (Ap.Dens), material density (Mat.Dens), specific bone surface (BS/BV), and degree of anisotropy (DA). Trabecular analyses utilized a sigma value of 0.8, support of 1, and threshold at 270. Cortical data at the midshaft of the femur were also analyzed, including cortical bone area (Ct.Ar), total cross-sectional area (Tt.Ar), medullary area (Ma.Ar), cortical thickness (Ct.Th), and cortical area fraction (Ct.Ar/Tt.Ar). Cortical analyses utilized a sigma value of 1.5, support of 2, and threshold at 350. CT analysis of the distal femur was limited to female animals, whereas CT analysis at the diaphysis to obtain cortical geometry was performed on femora from both male and female mice.
2.3. Mechanical testing
Four-point bend testing was performed as previously described [56] to measure whole bone mechanical properties [57]. Briefly, the anterior surface of each femur was placed on two supports, and the femur was loaded at a rate of 2 mm/min until failure. Force-displacement curves allowed determination of structural properties including ultimate force, stiffness, displacement, and energy absorption for each specimen, and apparent material properties were derived using standard beam-bending equations for four-point bending. Diaphysis μCT was used to normalize the mechanical properties. The 0.2% offset criterion was used to define yield points, and a custom MATLAB (Version 11) program was used for all mechanical analyses [56, 58].
2.4. Histology
Femora were fixed in 10% neutral buffered formalin and embedded in methyl methacrylate. Thick sections (~80 μm) were cut at mid-diaphysis using a diamond wire saw. Thin sections (~4–10 μm) were cut at the distal femur using a tungsten-carbide knife.
For dynamic histomorphometry, the total surface and the surfaces with single and double labeling were measured along with the inter-label width on the periosteal and endocortical surfaces of cortical bone and on trabecular surfaces of the distal femur. A value of 0.1 μm per day was used for mineral apposition rate (MAR) when only one label was present to permit bone formation rate (BFR/BS) to be calculated.
For static histomorphometry, thin sections from the distal femora were deplasticized in acetone and subjected to staining for tartrate-resistant acid phosphatase (TRAP) to quantify osteoclast surface (Oc.S/BS) as previously described [59] or Von Kossa stain for mineral with Macneal’s tetrachrome counterstain [60]. All the terminology and units used follow the recommendations of the Histomorphometry Nomenclature Committee of the American Society for Bone and Mineral Research [61].
2.5. In vitro osteoclastogenesis assay
Mouse spleen cells were prepared as previously described [62]. Spleen cells were cultured in untreated Petri dishes for 5 days in α-MEM supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 10 ng/mL macrophage colony stimulating factor (M-CSF). Cells were then split into 24-well plates (25,000 cells/well) in α-MEM supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 5 ng/mL M-CSF, and 50 ng/mL receptor activator of nuclear factor kappa-B ligand (RANKL), and cultured for 7 days prior to osteoclast differentiation analysis. Tartrate-resistant acid phosphatase (TRAP) staining was performed (Takara, Cat No. MK300), and the number of TRAP-positive multinucleated cells (MNCs, at least 3 nuclei) per well were counted.
2.6. Calvarial osteoblast isolation and culture
Primary neonatal mouse calvarial osteoblast cultures were established as previously described [63]. The primary osteoblasts were plated in 10 cm dishes and maintained in α-MEM supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. After 72 hours, the cells were split into 6 well dishes at a density of 15,000 cells/cm2. Upon confluence, cells were either maintained in growth media or switched to osteogenic media (α–MEM + 10% FBS, 5 mM β-glycerophosphate, and 100 μg/mL ascorbic acid) or adipogenic media (α–MEM + 15% FBS, 5 μg/mL insulin, 50 μM indomethacin, 0.5 μM IBMX, and 1 μM dexamethasone). The media was changed every third day until endpoint assays were performed.
2.7. Alkaline phosphatase staining
Alkaline phosphatase activity of calvarial osteoblasts was measured using the premixed BCIP/NBT solution (Sigma) according to the manufacturer’s instructions on day 7 of incubation in osteogenic media or adipogenic media.
2.8. Alizarin Red S staining
Calvarial osteoblast calcium deposition was measured using Alizarin Red S (ARS, Sigma) staining on day 28 of incubation in osteogenic media. Cell monolayers were fixed with 10% neutral buffered formalin before incubation with 0.02% ARS (pH 4.2) for 45 minutes in the dark at room temperature. Cell monolayers were then washed several times with distilled water and imaged. For quantification, ARS stain was solubilized using 10% acetic acid, neutralized with 10% ammonium hydroxide, and centrifuged. The optical density of the supernatant at 405 nm was measured using a plate reader.
2.9. Oil Red O staining
The presence of adipocytes in calvarial osteoblast cultures was determined using Oil Red O (ORO, Sigma) staining at day 14 in standard growth media. A stock solution of 0.3% ORO in isopropanol was used to generate a working ORO solution by diluting 3 parts stock solution with 2 parts distilled water just before each experiment. Cell monolayers were fixed in 10% neutral buffered formalin and then incubated in 60% isopropanol for 5 minutes before staining with the working solution of ORO for 15 minutes. Cell monolayers were then washed with distilled water, incubated with Harris Hematoxylin solution for one minute to counterstain nuclei, then washed and maintained in PBS for imaging.
2.10. Western blotting
Calvarial osteoblasts were harvested in ice cold lysis buffer (20 mM Tris-HCl, pH 7.5; 250 mM NaCl; 10 mM MgCl2; 1% Triton X-100; and 1x protease inhibitor cocktail, Calbiochem) and subjected to SDS-PAGE and immunoblotting analysis using Goat anti-Rad (Everest BioTech) or Rabbit anti-Gapdh (Cell Signaling) primary antibody and peroxidase-conjugated mouse anti-goat IgG (Jackson ImmunoResearch) or donkey anti-rabbit (GE Healthcare) secondary antibody. Signals were developed using Hyglo chemiluminescent reagent (Denville Scientific) and detected using a ChemiDoc MP (Bio-Rad).
2.11. Microarray
Total RNA was isolated from confluent WT and Rad−/− calvarial osteoblast cultures using the standard Trizol and chloroform method, and RNA quality was assessed using RNA 6000 Nano-LabChip (Agilent). The University of Kentucky microarray core facility performed labeling of the RNA and hybridization to the chip. Total RNA (100 ng per sample) was labeled and hybridized onto the Affymetrix Clariom D mouse array. The arrays were hybridized for 16 hours at 45°C and 60 rpm. The arrays were washed and stained on the Affymetrix Fluidics 450 station and scanned on the Affymetrix GeneChip7G scanner to quantify the signal intensity of hybridized probes. Data were analyzed using the Affymetrix Command Console software.
2.12. Reverse transcriptase-polymerase chain reaction
For RT-PCR, cDNA was prepared from 1 μg of total RNA using the RT2 First Strand Kit (Qiagen). The following primers were utilized for RT-PCR: mouse matrix Gla protein (MGP), 5’-GGAGAAATGCCAACACCTTT-3’ (forward) and 5’-CGAAACTCCACAACCAAATG-3’ (reverse) and 18S, 5’-TAGAGGGACAAGTGG CGTTC-3’ (forward) and 5’-CGCTGAGCCAGTCAGTGT-3’ (reverse). PCR products were amplified using DreamTaq Green (Thermo Scientific) and resolved on 1% agarose gels. PCR products were imaged with Gel Logic 112 (Fisher Biotech), and band intensities were quantified using ImageJ.
2.13. EchoMRI
The body composition of live, unanesthetized mice was measured using an EchoMRI-100 whole body composition analyzer (Echo Medical System, Houston, TX).
2.14. Statistics
Statistical analysis was performed using Student’s t test, with p<0.05 considered significant, and all data are reported as mean +/− SEM.
3. Results
3.1. Lower trabecular and cortical bone density in Rad−/− femora
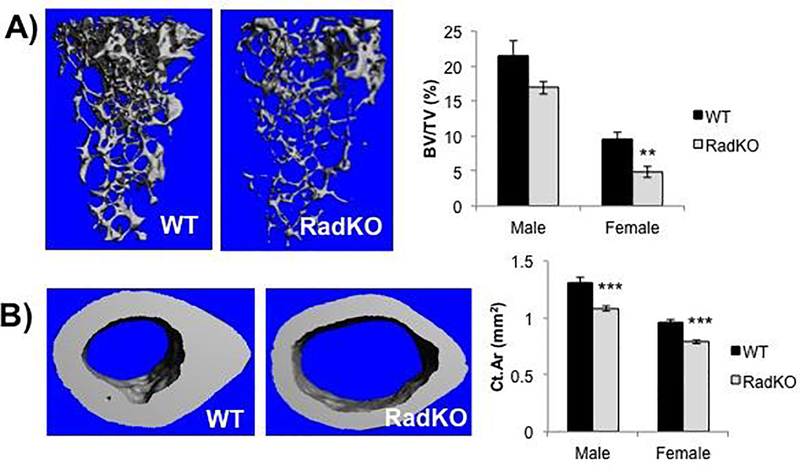
To explore the impact of Rad deficiency on bone density, WT and Rad−/− femora from both male and female mice were analyzed by microcomputed tomography (μCT). Rad−/− femora from female mice exhibited a significantly lower trabecular bone volume fraction and trabecular number, with a parallel increase in trabecular spacing relative to WT controls (Figure 1A and Table 1). A similar trend was observed in male Rad−/− femora (Figure 1A). Trabecular thickness was not significantly different from WT. Rad−/− femora also exhibited a significantly lower cortical bone area and thickness than WT, whereas the medullary area was significantly higher when compared to WT controls (Figure 1B and Table 1). Taken together, these data suggest that Rad GTPase contributes to the maintenance of normal bone density.
Figure 1. Lower trabecular and cortical bone volume in the femora of Rad−/− mice.
A) Representative images from μCT analysis of trabecular bone at the distal femora of WT and RadKO mice with accompanying quantification of the trabecular bone volume fraction (BV/TV). B) Representative images from μCT analysis of cortical bone at the midshaft of WT and RadKO mouse femora with corresponding quantification of the cortical bone area fraction (Ct.Ar/Tt.Ar). N=5–15 mice per group, 4 months of age. ** p<0.01, *** p<0.001 compared to WT by Student’s t test.
Table 1.
Trabecular and cortical geometry of 4-month-old female mouse femora
| Distal Femur | Wildtype (N=5) | Rad−/− (N=5) |
| TV (mm3) | 3.26 +/− 0.18 | 4.65 +/− 0.06*** |
| BV (mm3) | 0.316 +/− 0.043 | 0.224 +/− 0.038 |
| BV/TV (%) | 9.60 +/− 0.88 | 4.81 +/− 0.80** |
| Conn.D | 109.32 +/− 8.76 | 27.06 +/− 6.16*** |
| SMI | 2.43 +/− 0.09 | 3.20 +/− 0.08*** |
| Tb.Th (mm) | 0.045 +/− 0.002 | 0.047 +/− 0.002 |
| Tb.N (1/mm) | 3.49 +/− 0.12 | 2.72 +/− 0.11** |
| Tb.Sp (mm) | 0.287 +/− 0.008 | 0.368 +/− 0.015** |
| Ap.Dens | 278.56 +/− 9.72 | 204.50 +/− 11.25** |
| BS/BV | 61.31 +/− 2.21 | 63.38 +/− 3.30 |
| DA | 1.34 +/− 0.04 | 1.35 +/− 0.03 |
| Femoral Midshaft | Wildtype (N=5) | Rad−/− (N=5) |
| Ct.Ar (mm2) | 0.964 +/− 0.018 | 0.789 +/− 0.016*** |
| Tt.Ar (mm2) | 1.452 +/− 0.013 | 1.806 +/− 0.031*** |
| Ma.Ar (mm2) | 0.489 +/− 0.010 | 1.018 +/− 0.022*** |
| Ct.Th (mm) | 0.257 +/− 0.007 | 0.183 +/− 0.003*** |
| Ct.Ar/Tt.Ar (%) | 66.34 +/− 0.79 | 43.67 +/− 0.60*** |
p<0.01
p<0.001 compared to WT using Student’s t test
3.2. Rad−/− femora have altered mechanical properties
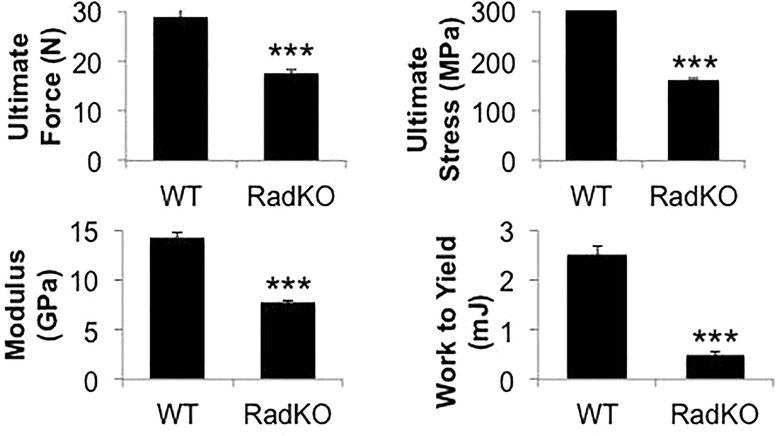
Rad−/− femora displayed a significant mechanical phenotype, including a significantly lower cortical bone ultimate force, stiffness, work to yield, ultimate stress, and elastic modulus compared to femora from WT controls (Figure 2). Total displacement, toughness, and total strain were all significantly higher in the absence of Rad (Table 2). These data indicate that Rad loss results in a unique mechanical phenotype characterized by weaker and more elastic bones, which is consistent with the lower bone density evident from μCT analysis.
Figure 2. Rad deletion results in altered mechanical properties.
Quantification of mechanical properties from four-point bending analysis of mouse femora. N=13–15 mice per genotype, 4 months of age. *** p<0.001 compared to WT by Student’s t test.
Table 2.
Whole bone structural and estimated material mechanical properties from femoral four-point bending
| Femur | Wildtype (N=13) | Rad −/−(N=15) |
| Yield Force (N) | 24.50 +/− 1.20 | 7.99 +/− 0.41*** |
| Ultimate Force (N) | 28.86 +/− 1.57 | 17.54 +/− 0.72*** |
| Displacement to Yield (mm) | 186.93 +/− 4.16 | 103.98 +/− 5.76*** |
| Postyield Displacement (mm) | 165.85 +/− 18.39 | 810.28 +/− 87.62*** |
| Total Displacement (mm) | 352.77 +/− 18.45 | 914.26 +/− 88.08*** |
| Stiffness (N/mm) | 195.73 +/− 7.99 | 127.17 +/− 5.38*** |
| Work to Yield (m J) | 2.52 +/− 0.16 | 0.51 +/− 0.05*** |
| Postyield Work (mJ) | 4.46 +/− 0.49 | 10.17 +/− 0.75*** |
| Total Work (mJ) | 6.98 +/− 0.52 | 10.68 +/− 0.75*** |
| Yield Stress (MPa) | 258.67 +/− 13.54 | 74.75 +/− 4.87*** |
| Ultimate Stress (MPa) | 301.56 +/− 12.34 | 160.64 +/− 4.52*** |
| Strain to Yield (mε) | 20380.8 +/− 935.8 | 11527.4 +/− 533.2*** |
| Total Strain (mε) | 38517.2 +/− 2681.7 | 101673.0 +/− 9722.0*** |
| Elastic Modulus (GPa) | 14.17 +/− 0.63 | 7.73 +/− 0.21*** |
| Resilience (MPa) | 2.94 +/− 0.26 | 0.53 +/− 0.05*** |
| Toughness (MPa) | 8.29 +/− 0.91 | 10.76 +/− 0.74* |
p<0.05
p<0.001 compared to WT using Student’s t test
3.3. Rad deletion enhances osteoclast differentiation in vitro
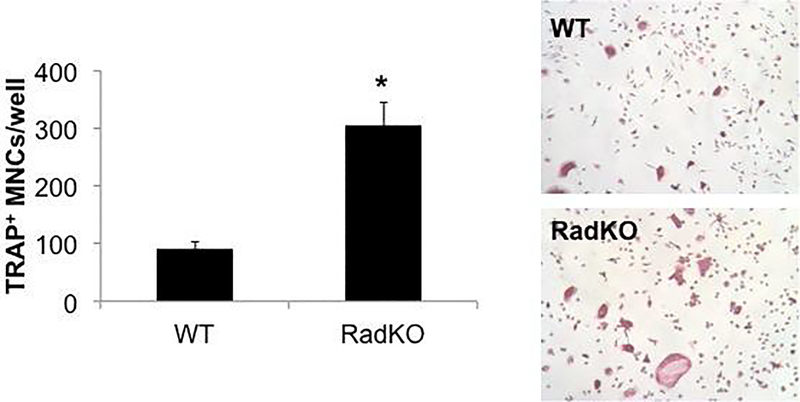
A decrease in bone density and strength could occur via an increase in bone resorption by osteoclasts, a decrease in bone formation by osteoblasts, or a combination of the two. To determine the impact of Rad deletion on osteoclast differentiation, mononuclear cells were isolated from the spleens of WT and Rad−/− mice and treated with M-CSF and RANKL to stimulate their differentiation toward multinucleated osteoclasts [62]. Staining for tartrate-resistant acid phosphatase (TRAP) and counting TRAP+ multinucleated cells (MNCs) indicated that loss of Rad significantly enhanced osteoclast differentiation in vitro (Figure 3).
Figure 3. In vitro osteoclast differentiation is enhanced in the absence of Rad.
Representative images and quantification of tartrate-resistant acid phosphatase (TRAP) stained osteoclasts derived from spleen cells. The number of TRAP-positive multinucleated cells (MNCs, at least 3 nuclei) was counted in each well of a 24-well plate. N=3 animals per genotype, male, 2 months of age, *p<0.05 compared to WT by Student’s t test.
3.4. Lower osteoclast surface in Rad−/− femora
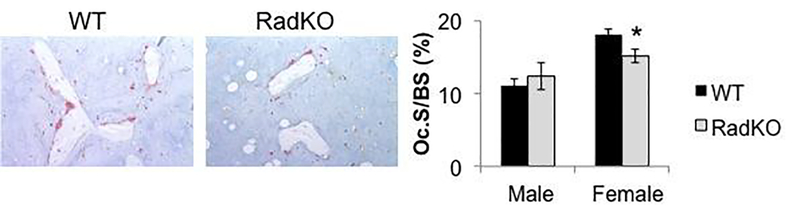
We next performed TRAP staining of WT and Rad−/− femora to evaluate osteoclast numbers in vivo. Osteoclast surface was unchanged in male Rad−/− animals, and a small but significant decrease in osteoclast surface was observed in the distal femora of female Rad−/− mice compared to WT (Figure 4). These data suggest that, despite the increase in in vitro osteoclastogenesis that we have observed, the low bone density observed in Rad−/− mice may not arise solely from increased osteoclast numbers.
Figure 4. Modest decrease in osteoclast surface in the absence of Rad.
Tartrate-resistant acid phosphatase (TRAP) staining of WT and RadKO distal femur thin sections and corresponding quantification of the percentage of the bone surface occupied by osteoclasts (Oc.S/BS). N=5–8 mice per group, 4 months of age. * p<0.05 compared to WT by Student’s t test.
3.5. Lower bone formation rate in Rad−/− femora
The reduction in osteoclast surface in Rad−/− femora suggested that the low bone mass phenotype might involve more than reduced osteoclast numbers and motivated us to ask whether Rad deletion might also alter osteoblast activity. To test this notion, we used dynamic histomorphometry to determine the rate of bone formation in WT and Rad−/− femora in vivo. In trabecular bone at the distal femur, we observed a significantly lower mineral apposition rate, but a higher percent mineralizing surface in Rad−/− femora compared to WT controls (Figure 5, Table 3). The latter observation may arise in part due to the significant decrease in total trabecular bone surface at the distal femora of Rad−/− mice compared to WT (Figure 5A). Normalization of MAR and MS/BS results in a downward trend in bone formation rate (BFR/BS) in trabecular bone of Rad−/− animals compared to WT (Figure 5B, Table 3).
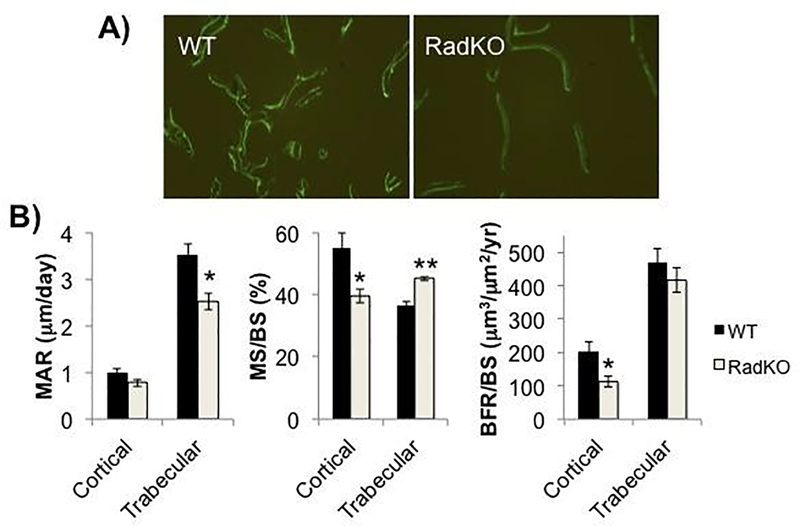
Figure 5. Lower bone formation rate in Rad−/− femora.
A) Representative images of calcein double labeling in distal femur trabecular bone of WT and RadKO mice (10X). B) Mineral apposition rate (MAR), mineralizing surface (MS/BS), and bone formation rate (BFR/BS) in the cortical bone (periosteal surface) and trabecular bone of WT and RadKO mice. N=5 mice per genotype, female, 4 months of age * p<0.05, ** p<0.01 compared to WT by Student’s t test.
Table 3.
Histomorphometry
| Distal Femur | Wildtype (N=5) | Rad−/− (N=5) |
| Trabecular MAR (μm/day) | 3.508 +/− 0.256 | 2.520 +/− 0.183 * |
| Trabecular MS/BS (%) | 36.59 +/− 1.42 | 45.17 +/− 0.64 ** |
| Trabecular BFR/BS (μm3/μm2/year) | 470.00 +/− 41.68 | 416.95 +/− 35.96 |
| Femur Diaphysis | Wildtype (N=5) | Rad−/− (N=5) |
| Ct.Ar (mm) | 0.923 +/− 0.024 | 0.737 +/− 0.014 *** |
| Periosteal MAR (μm/day) | 1.008 +/− 0.074 | 0.780 +/− 0.080 |
| Periosteal MS/BS (%) | 54.95 +/− 4.81 | 39.48 +/− 2.29 * |
| Periosteal BFR/BS (μm3/μm2/year) | 204.75 +/− 28.70 | 114.34 +/− 17.10 * |
| Endocortical MAR (μm/day) | 1.075 +/− 0.086 | 0.928 +/− 0.058 |
| Endocortical MS/BS (%) | 75.95 +/− 4.01 | 69.04 +/− 4.67 |
| Endocortical BFR/BS (μm3/μm2/year) | 299.13 +/− 32.78 | 237.54 +/− 30.37 |
p<0.05
p<0.01
p<0.001 compared to WT using Student’s t test
We also measured these parameters in cortical bone. Consistent with the μCT data, histology indicated a significantly lower cortical bone area at the mid-diaphysis of Rad−/− femora compared to WT. The mineralizing surface (MS/BS) and bone formation rate (BFR/BS) at the periosteal surface of Rad−/− femur diaphyses were significantly lower than in WT, and the periosteal mineral apposition rate (MAR) also trended downward in Rad−/− femora (Figure 5B, Table 3). These same measures at the endocortical surface of the femur diaphysis trended downward in Rad−/− animals but did not reach significance (Table 3). Overall, these data suggest that a decrease in osteoblast function may give rise to the lower bone mass observed in Rad−/− mice.
3.6. Rad−/− calvarial osteoblast function is blunted in vitro
To characterize the contribution of Rad GTPase signaling to osteoblast differentiation and function in vitro, the phenotype of osteoblasts derived from neonatal WT and Rad−/− calvaria was examined. Immunoblot analysis confirmed Rad expression in this cell population (Figure 6A). Consistent with the in vivo decrease in bone formation, osteoblast differentiation was impaired in Rad−/− calvarial cells as shown by a reduction in alkaline phosphatase activity, an enzymatic marker of osteoblast maturation (Figure 6B), and a significant decrease in mineralization as indicated by Alizarin Red S staining (Figure 6C) following osteogenic induction. Together these data indicate that osteoblast development and/or function is diminished in the absence of Rad.
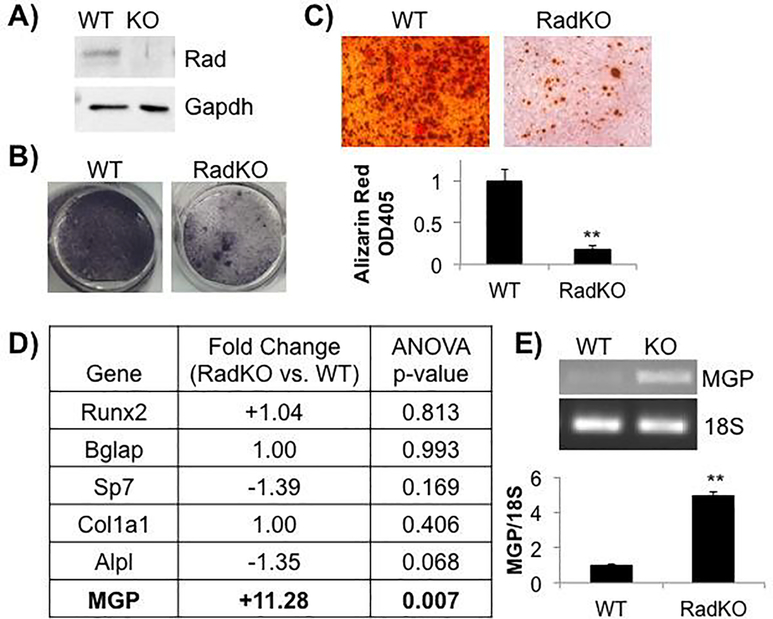
Figure 6. Less mineralization in Rad−/− osteoblasts in vitro.
A) Representative immunoblot of WT and RadKO calvarial osteoblast lysates confirms Rad expression in these cells. N=3 isolations per genotype. B) Representative images of WT and RadKO primary calvarial osteoblasts stained for alkaline phosphatase activity after 7 days in osteogenic media. N=3 isolations per genotype. C) Representative images of Alizarin Red S staining of WT and RadKO primary osteoblast monolayers after 28 days in osteogenic media. Staining was quantified by solubilization of the stain in acetic acid, neutralization, and optical density measurement at 405 nm. N=3–4 isolations per genotype. ** p<0.01 compared to WT by Student’s t test. D) Osteoblast marker gene expression from microarray analysis of WT and RadKO primary osteoblasts. N=2 isolations per genotype. E) RT-PCR analysis of MGP expression confirms microarray result. N=3 isolations per genotype.
3.7. Higher expression of matrix Gla protein in Rad−/− calvarial osteoblasts
To examine the molecular mechanisms underlying the decrease in osteoblast function upon Rad loss, microarray analysis was performed to compare the gene expression profile of naïve calvarial osteoblasts from WT and Rad−/− mice. Surprisingly, Rad deficiency had no effect on the expression of the canonical osteoblast marker genes Runt-related transcription factor 2 (Runx2), osteocalcin (Bglap), or type I collagen (Col1a1) (Figure 6D). Expression of the osteogenic transcription factor osterix (Sp7) and of alkaline phosphatase (Alpl) trended downwards but did not reach significance (Figure 6D). Instead, our profiling data indicated that matrix Gla protein (Mgp), a 15-kDa secreted protein that was initially isolated and identified from demineralized bovine bone and has since been found to inhibit bone mineralization [64–66], is markedly increased in Rad−/− calvarial osteoblasts compared to WT (+11.28-fold) (Figure 6D). This increase in matrix Gla protein (MGP) expression in Rad−/− osteoblasts was confirmed by RT-PCR (Figure 6E).
3.8. Rad−/− calvarial osteoblasts show a striking adipogenic phenotype
During the course of culturing primary calvarial cells, we unexpectedly observed a dramatic increase in the number of cells that appeared to have lipid droplets in the Rad−/− osteoblast preparations after ten days in culture when compared to WT osteoblasts. This observation, coupled with published work indicating that MGP not only inhibits mineralization but that its secretion increases ~30-fold during the in vitro differentiation of human pre-adipocytes [67], suggested that lower osteogenesis following Rad deletion might be linked to increased adipogenic differentiation. To test this possibility, WT and Rad−/− calvarial osteoblast monolayers were stained with Oil Red O (ORO) to confirm that these structures were lipid droplets. Consistent with a potential role for Rad in inhibiting adipogenesis, the number of ORO-positive cells was significantly higher in Rad−/− calvarial cultures compared to WT (Figure 7A).
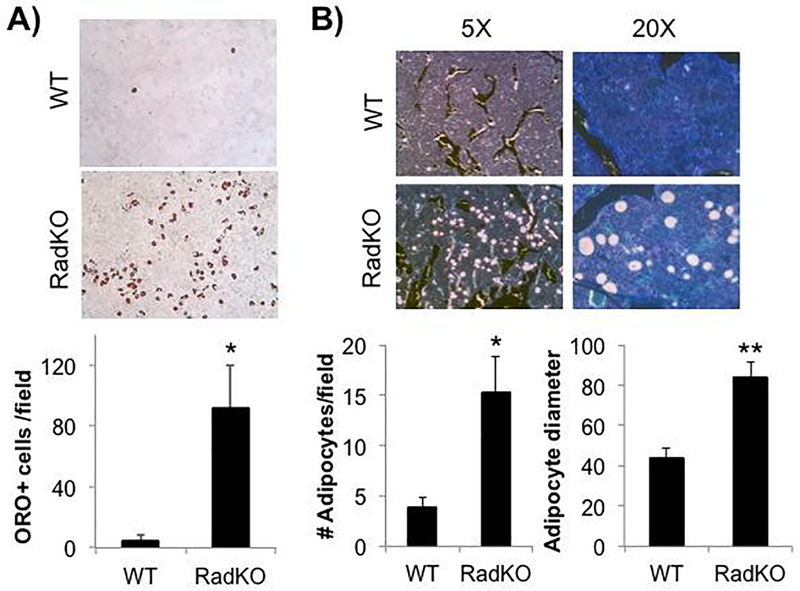
Figure 7. Loss of Rad confers an adipogenic phenotype.
A) Representative images of WT and RadKO primary calvarial osteoblasts stained with Oil Red O after 14 days in mineralizing conditions. The number of ORO-positive cells was counted for 3 random fields per isolation. N=3–4 isolations per genotype. B) Von Kossa/MacNeal’s staining of thin sections from WT and RadKO distal femora. Number of adipocytes per 20X field and the average adipocyte diameter in pixels were quantified. N=5 mice per genotype, female, 4 months of age. * p<0.05, ** p<0.01 by Student’s t test.
3.9. Increased BMAT at the distal femora of Rad−/− mice
The significant increase in adipogenesis observed in primary Rad−/− calvarial osteoblast cultures under normal growth conditions suggested that Rad deficiency might alter the in vivo balance of osteoblasts and adipocytes in the bone marrow compartment. Von Kossa/MacNeal’s tetrachrome staining of WT and Rad−/− distal femora was performed to evaluate the overall cell distribution and revealed a significant increase in BMAT at the Rad−/− distal femur compared to WT (Figure 7B). Rad deletion resulted in significantly higher adipocyte numbers as well as significantly larger adipocyte size compared to WT (Figure 7B). Notably, reexamination of TRAP-stained distal femora (Figure 3) was consistent, with unstained round structures resembling adipocytes frequently observed in Rad−/− femora.
3.10. Total body fat percentage is unchanged in Rad−/− mice
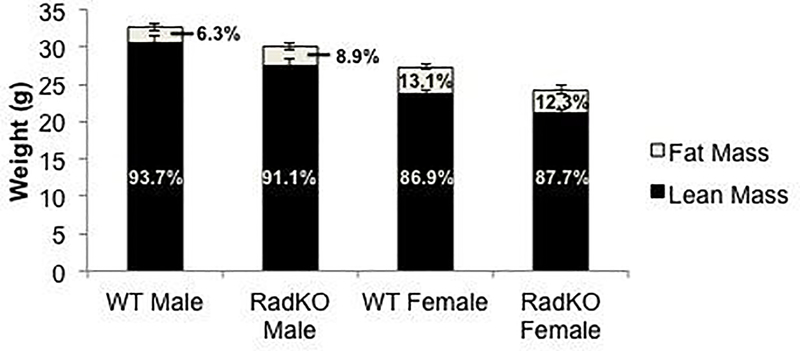
To determine whether Rad loss results in a global alteration in adipogenesis, we weighed and performed EchoMRI body composition analysis on 4-month-old WT and Rad−/− mice. These analyses showed that Rad−/− mice weigh less than WT littermates, but we observed no significant change in body fat percentage upon Rad deletion in either male or female mice (Figure 8); thus, the increase in adipogenesis in Rad−/− mice appears to be specific to BMAT.
Figure 8. No change in lean or fat mass percentage in Rad−/− mice.
EchoMRI analysis of body composition of WT and RadKO mice indicates no change in lean or fat mass percentage. N=8 WT male, 9 RadKO male, 21 WT female, 11 RadKO female mice, 4 months of age.
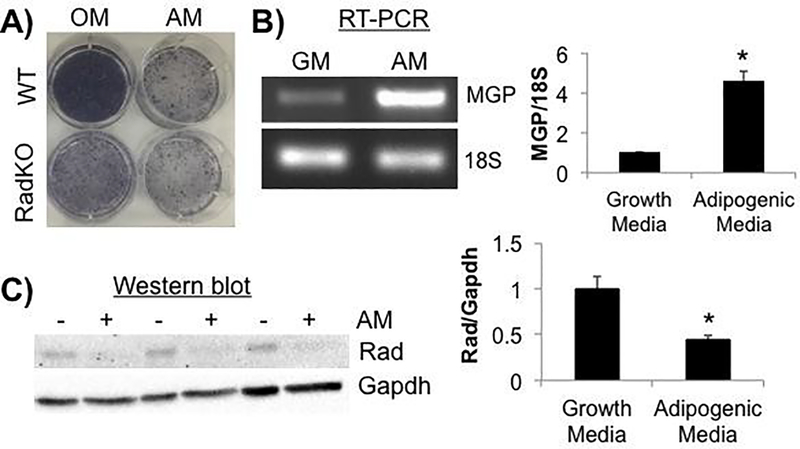
3.11. Reduction in Rad levels following adipogenic induction of calvarial osteoblasts
Rad gene expression increases following lithium stimulation of mesenchymal stem cells, which enhances osteogenic differentiation, and Rad silencing has been shown to attenuate osteogenic priming [36]. These data prompted us to examine whether Rad expression is altered in primary calvarial cells upon adipogenic differentiation. Treatment with adipogenic media for one week resulted in the accumulation of ORO+ lipid droplets in WT calvarial cultures (data not shown), as well as a reduction in alkaline phosphatase staining resembling that seen in Rad−/− cultures without adipogenic stimulation (Figure 9A). Adipogenic treatment also resulted in a significant rise in MGP gene expression (Figure 9B) and a significant reduction in endogenous Rad protein levels (Figure 9C) in WT calvarial osteoblasts. Taken together, these data suggest that dynamic control of Rad may play a role in directing differentiation towards the osteogenic versus adipogenic lineages.
Figure 9. Adipogenic induction results in lower endogenous Rad levels.
A) Representative images of alkaline phosphatase activity in WT and RadKO primary osteoblast monolayers following 7 days in osteogenic (OM) or adipogenic (AM) media. N=3 isolations per genotype. B) RT-PCR analysis of MGP expression levels normalized to 18S RNA. RNA was isolated from WT primary osteoblasts after 3 days in growth (GM) or adipogenic (AM) media. N=3 isolations. C) Western blotting of WT osteoblast lysates after 7 days in growth (−) media or adipogenic (+) media. N=3 isolations. * p<0.05 by Student’s t test.
4. Discussion
Bone is a dynamic tissue that undergoes continuous remodeling throughout life in response to changing demands on the skeleton and in order to maintain mineral homeostasis. Dysregulation of the bone remodeling process is one characteristic of age-related osteoporosis. In addition to low bone mass, osteoporosis is often characterized by an increase in bone marrow adiposity [68]. Osteoblasts and adipocytes share a common mesenchymal stem cell precursor, but the mechanisms by which these precursors are marked for an adipogenic versus an osteogenic cell fate have not been fully elucidated. In this study, we performed the first analysis of the bone physiology of Rad−/− mice and demonstrated that genetic loss of Rad GTPase results in low bone mass accompanied by a dramatic expansion in bone marrow adipose tissue (BMAT) that is similar to the presentation of age-related osteoporosis in humans. We also showed that Rad is significantly down regulated during the adipogenic differentiation of primary calvarial cells, suggesting that Rad signaling may play a previously unrecognized role in directing osteogenic versus adipogenic differentiation.
Following the report of a requirement for Rad function in lithium-mediated osteogenic priming of MSCs [36], we hypothesized that global Rad−/− mice would have lower bone density than WT controls, which we confirmed by μCT analysis. Further evaluation of these bones to examine their mechanical properties revealed that while Rad−/− femora have significantly lower strength and stiffness, which would typically render them more susceptible to fracture, they also have significantly longer displacement than femora from WT controls. Hence, Rad−/− femora are simultaneously weaker and more elastic, bending under smaller loads than WT but not overtly fracturing. Both collagen and water provide plasticity to bone [69], and the contribution of these variables to the Rad−/− mechanical phenotype could be explored in the future.
Bone dynamics are controlled by the coordinated actions of osteoclasts and osteoblasts [70], and we sought to define the cell type(s) responsible for the lower bone density in Rad−/− mice. Despite the observation of an increase in osteoclast differentiation in Rad−/− cells in vitro (Figure 5.4), osteoclast surface is not higher in Rad−/− distal femora in vivo at four months of age (Figure 5.5). These data tend to suggest that increased bone resorption is not the only mechanism responsible for the low bone mass seen in Rad−/− animals. Future studies to define levels of bone resorption markers and analysis of the bone density and osteoclast surface area of WT and Rad−/− femora from younger and older mice are needed to better inform the phenotype that we have observed. Additional work is also needed to understand the role of Rad in osteoclast differentiation.
Analysis of osteoblast function in vivo and in vitro suggested that bone formation is impaired in Rad−/− mice. The bone formation rate was significantly lower at the periosteal surface of cortical bone and trended downwards at the endocortical surface as well as in trabecular bone, indicating that Rad loss might result in decreased osteoblast differentiation and/or function. In vitro calvarial osteoblast assays corroborated this notion, as our studies indicated lower alkaline phosphatase activity and decreased mineralization in Rad−/− calvarial cultures following osteogenic induction compared to WT. Further studies, including differentiation studies using MSCs, are currently underway to determine whether the loss of Rad impacts osteoblast differentiation, function, or both.
Surprisingly, we did not see a change in the expression of canonical osteoblast marker genes in Rad−/− calvarial cells, as would be expected if osteoblast development was hindered in the absence of Rad. In part this might arise from our study design, using freshly isolated calvarial cells prior to osteogenic induction. It will be important to determine whether Rad deletion hinders this gene expression program following osteogenic induction. Our microarray analysis did, however, reveal a robust increase in matrix Gla protein (MGP) expression in Rad−/− calvarial cells compared to WT. MGP up-regulation may provide a novel mechanism for the decrease in osteogenesis as well as the increase in adipogenesis observed upon Rad deletion. MGP has been shown to prevent mineralization in the osteoblast-like cell line MC3T3-E1 [65, 66]. In keeping with its role as an inhibitor of mineralization, transgenic mice overexpressing MGP in osteoblasts have low bone density [71], and MGP-knockout mice exhibit profound calcification of the aorta and other arteries, as well as inappropriate calcification of cartilaginous structures like the growth plate and the tracheal rings [72, 73]. Interestingly, not only does MGP inhibit mineralization, but its secretion is robustly increased during adipocyte differentiation, second only to the body fat regulatory hormone leptin [67], and we observed induction of MGP gene expression following adipogenic differentiation of WT cells. Thus, the elevation in MGP gene expression in Rad−/− osteoblasts is likely important to the overall phenotype of increased adipogenesis at the expense of osteogenesis.
Our finding of a change in the extracellular matrix protein MGP expression in Rad−/− calvarial osteoblasts, along with the report that Rad loss drives cardiac fibrosis via changes in extracellular matrix [29], suggests that one role for Rad beyond calcium channel control is regulation of the ECM. However, it is interesting that Rad loss appears to drive distinct gene profiles in different tissues. In the heart, Rad has been found to inhibit C/EBP-δ activity to promote altered gene expression [29]. One possibility that we are currently exploring is that Rad loss in mesenchymal progenitors may alter C/EBP function, or the function of another transcription factor, to favor adipogenesis. Indeed, C/EBP transcriptional activity plays an important role in the early stages of adipocyte differentiation [74]. Studies are underway to determine whether Rad function is required for osteogenic differentiation of MSCs during adult bone homeostasis via inhibition of C/EBP family proteins, with the lack of Rad function promoting adipogenesis through enhanced C/EBP-dependent transcription.
The observation that Rad deletion increases bone marrow adiposity but not total body fat is intriguing. BMAT represents a significant fraction of total body adipose in humans, yet its origin and physiological functions remain to be fully characterized [75]. Marrow adipose has gained recent interest as a distinct fat depot that appears to have roles regulating bone homeostasis, hematopoiesis, and metabolism [76]. It is well known that adipokines and free fatty acids released by adipocytes can modulate bone remodeling and hematopoiesis [77]. BMAT quantity is associated with bone density loss in aging, menopause, and other metabolic conditions [39, 68, 78]. Indeed, BMAT has the potential to contribute to both local and systemic metabolic processes [77]. Notably, a growing cancer literature has linked Rad to regulation of tumor metabolism [79-82]. The increase in BMAT, but not of peripheral fat, in Rad−/− animals suggests that Rad function may serve as a novel regulator of BMAT development and regulation. This possibility is supported by a study showing that Rad protein levels in human skeletal muscle are correlated with measures of obesity and resting metabolic rate [83].
Unraveling the pathways that regulate the bifurcation between osteogenic and adipogenic differentiation is critical to understanding the disease progression of osteoporosis and identifying new therapies, as a shift in this balance favoring adipogenesis at the expense of osteogenesis may contribute to the increase in BMAT that accompanies low bone density in osteoporotic patients. Our observation that adipogenesis is correlated with a reduction in endogenous Rad protein levels, coupled with the report that Rad expression is up-regulated during osteogenic priming of MSCs [36], suggests that Rad levels may be dynamically regulated during differentiation as a means of regulating cell fate, with higher expression promoting osteogenesis and lower expression promoting adipogenesis. Given that Rad expression is changed in other settings, including heart failure [19, 55], diabetes [1], and cancer [79–82, 84–88], identifying the molecular mechanisms whereby Rad levels are regulated is an important future goal. Unlike most Ras-related small GTPases, Rad activity has not been shown to be controlled by classical GTP/GDP cycling [7]; therefore, alterations of total Rad levels may represent a novel means of regulation in a number of physiological settings. Cellular regulation of Rad function remains poorly characterized, and future studies will be aimed at determining whether signaling pathways known to direct bone marrow mesenchymal cell fate, such as parathyroid hormone [89], bone morphogenetic protein [90], and Wnt signaling [91, 92], impact Rad expression and/or function. We are in the process of generating a conditional Rad-knockout mouse, which will permit detailed mechanistic analysis of the impact of selective Rad loss on osteoblast and adipocyte lineages.
5. Conclusions
Rad GTPase plays an important, previously uncharacterized role in the regulation of bone homeostasis. Deletion of Rad in mice results in low bone density and gives rise to femora that are simultaneously weaker and more elastic. Analysis of this unique osteopenic phenotype demonstrated that Rad deficiency results in reduced rates of bone formation in vivo, lower in vitro osteoblast function, and higher rates of in vitro osteoclastogenesis. Additional studies are needed to characterize the molecular mechanism that underlies these changes. Moreover, higher BMAT levels are observed within Rad−/− femora without obvious expansion of peripheral adipose tissue, and adipocytes spontaneously arise in primary cultures from Rad−/− calvaria. These data suggest that Rad may alter osteogenic versus adipogenic lineage commitment, potentially via regulation of matrix Gla protein expression. Importantly, we show that endogenous Rad levels are decreased following adipogenic treatment of calvarial cells, complementing the previously reported increase in Rad expression during osteogenesis [36]. Taken together, these observations implicate Rad GTPase as a novel regulatory protein whose levels can be dynamically modulated to control the balance of osteogenesis and adipogenesis, and as such, studies into the mechanism of Rad action and regulation may present potential targets for osteoporosis research.
Highlights.
Rad GTPase deficiency in mice results in osteopenia, altering both osteoblast and osteoclast differentiation in vitro.
Rad loss promotes in vivo bone marrow adipose tissue accumulation and stimulates in vitro adipogenesis of primary calvarial osteoblasts.
Rad loss increases the expression of matrix Gla protein, an inhibitor of mineralization, without altering canonical osteoblast gene expression.
Adipogenesis results in reduced endogenous Rad, suggesting a potential linkage between Rad levels, bone density, and bone marrow adiposity.
Acknowledgments
The Microcomputed Tomography core facility at Rush University provided the μCT analysis for this study. The Bone Histology core facility at Indiana University School of Medicine processed and stained the bone sections for histology. Dr. Carole Moncman provided assistance with imaging analysis. Dr. Subramanya Pandruvada provided helpful discussion and technical advice.
Funding
This work was supported by National Institutes of Health grant HL072936 and HL131782 (DAA and JS) and by American Heart Association grant 16GRNT27790094 (JS and DAA). CNW was supported by NIH T32-HL072743, by National Science Foundation DGE-1247392, and by a PEO Scholar Award. The EchoMRI-100 is supported by National Institutes of Health grant 8 P20 GM103527.
Abbreviations
- ARS
Alizarin Red S
- BFR/BS
bone formation rate/bone surface
- BMAT
bone marrow adipose tissue
- C/EBP
CCAAT-enhancer binding protein
- MGP
matrix Gla protein
- MAR
mineral apposition rate
- MS/BS
mineralizing surface/bone surface
- MSC
mesenchymal stem cell
- Oc.S/BS
osteoclast surface/bone surface
- ORO
Oil Red O
- Rad
Ras associated with diabetes
- RGK
Rem, Rad, Rem2, Gem/Kir
- TRAP
tartrate resistant acid phosphatase
Footnotes
Conflicts of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reynet C, and Kahn CR, Rad: a member of the Ras family overexpressed in muscle of type II diabetic humans. Science, 1993. 262(5138): p. 1441–4. [DOI] [PubMed] [Google Scholar]
- 2.Finlin BS, and Andres DA, Rem is a new member of the Rad- and Gem/Kir Ras-related GTP-binding protein family repressed by lipopolysaccharide stimulation. J Biol Chem, 1997. 272: p. 21982–21988. [DOI] [PubMed] [Google Scholar]
- 3.Finlin BS, et al. , Rem2, a new member of the Rem/Rad/Gem/Kir family of Ras-related GTPases. Biochem J, 2000. 347: p. 223–231. [PMC free article] [PubMed] [Google Scholar]
- 4.Maguire J, et al. , Gem: an induced, immediate early protein belonging to the Ras family. Science, 1994. 265(5169): p. 241–244. [DOI] [PubMed] [Google Scholar]
- 5.Dorin D, et al. , Kir, a novel Ras-family G-protein, induces invasive pseudohyphal growth in Saccharomyces cerevisiae. Oncogene, 1995. 11(11): p. 2267–2271. [PubMed] [Google Scholar]
- 6.Bilan PJ, et al. , The ras-related protein rad associates with the cytoskeleton in a non-lipid-dependent manner. Exp Cell Res, 1998. 242(2): p. 391–400. [DOI] [PubMed] [Google Scholar]
- 7.Correll RN, et al. , The RGK family of GTP-binding proteins: regulators of voltage-dependent calcium channels and cytoskeleton remodeling. Cell Signal, 2008. 20(2): p. 292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang DD, and Colecraft HM, Rad and Rem are non-canonical G-proteins with respect to the regulatory role of guanine nucleotide binding in Ca(V)1.2 channel regulation. J Physiol, 2015. 593(23): p. 5075–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jhun BS, et al. , Adrenergic signaling controls RGK-dependent trafficking of cardiac voltage-gated L-type Ca2+ channels through PKD1. Circ Res, 2012. 110(1): p. 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moyers JS, et al. , Effects of phosphorylation on function of the Rad GTPase. Biochem J, 1998. 333(Pt 3): p. 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finlin BS, and Andres DA, Phosphorylation-dependent association of the Ras-related GTP-binding protein Rem with 14–3-3 proteins. Arch Biochem Biophys, 1999. 368(2): p. 401–412. [DOI] [PubMed] [Google Scholar]
- 12.Ward Y, et al. , Phosphorylation of critical serine residues in Gem separates cytoskeletal reorganization from down-regulation of calcium channel activity. Mol Cell Biol, 2004. 24(2): p. 651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghiretti AE, et al. , CaMKII-dependent phosphorylation of the GTPase Rem2 is required to restrict dendritic complexity. J Neurosci, 2013. 33(15): p. 6504–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paulik MA, et al. , Identification of Rad’s effector-binding domain, intracellular localization, and analysis of expression in Pima Indians. J Cell Biochem, 1997. 65(4): p. 527–41. [PubMed] [Google Scholar]
- 15.Shimizu-Nishikawa K, et al. , Identification and characterization of newt rad (ras associated with diabetes), a gene specifically expressed in regenerating limb muscle. Dev Dyn, 2001. 220(1): p. 74–86. [DOI] [PubMed] [Google Scholar]
- 16.Hawke TJ, et al. , Rad is temporally regulated within myogenic progenitor cells during skeletal muscle regeneration. Am J Physiol Cell Physiol, 2006. 290(2): p. C379–87. [DOI] [PubMed] [Google Scholar]
- 17.Magnusson C, et al. , Denervation-induced alterations in gene expression in mouse skeletal muscle. Eur J Neurosci, 2005. 21(2): p. 577–80. [DOI] [PubMed] [Google Scholar]
- 18.Fu M, et al. , Rad GTPase attenuates vascular lesion formation by inhibition of vascular smooth muscle cell migration. Circulation, 2005. 111(8): p. 1071–1077. [DOI] [PubMed] [Google Scholar]
- 19.Levitan BM, et al. , Rad-deletion phenocopies tonic sympathetic stimulation of the heart. J Cardiovasc Transl Res, 2016. 9(5–6): p. 432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porterfield VM, and Mintz EM, Temporal patterns of light-induced immediate-early gene expression in the suprachiasmatic nucleus. Neurosci Lett, 2009. 463(1): p. 70–3. [DOI] [PubMed] [Google Scholar]
- 21.Rovsing L, and Moller M, Photic stimulation of the suprachiasmatic nucleus via the non-visual optic system. A gene expression study in teh blind Crx−/− mouse. Cell Tissue Res, 2014. 358(1): p. 239–48. [DOI] [PubMed] [Google Scholar]
- 22.Sonna LA, et al. , Effect of acute heat shock on gene expression by human peripheral blood mononuclear cells. J Appl Physiol (1985), 2002. 92(5): p. 2208–2220. [DOI] [PubMed] [Google Scholar]
- 23.Kannangai R, et al. , Hepatic angiomyolipoma and hepatic stellate cells share a similar gene expression profile. Hum Pathol, 2005. 36(4): p. 341–7. [DOI] [PubMed] [Google Scholar]
- 24.Wyatt SM, et al. , The correlation between sampling site and gene expression in the term human placenta. Placenta, 2005. 26(5): p. 372–379. [DOI] [PubMed] [Google Scholar]
- 25.Finlin BS, et al. , Regulation of voltage-gated calcium channel activity by the Rem and Rad GTPases. Proc Natl Acad Sci, 2003. 100(24): p. 14469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manning JR, et al. , Rad GTPase deletion increases L-type calcium channel current leading to increased cardiac contraction. J Am Heart Assoc, 2013. 2(6): p. 000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manning JR, et al. , Loss of Rad-GTPase produces a novel adaptive cardiac phenotype resistant to systolic decline with aging. Am J Physiol Heart Circ Physiol, 2015. 309(8): p. H1336–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang G, et al. , Rad as a novel regulator of excitation-contraction coupling and beta-adrenergic signaling in heart. Circ Res, 2010. 106(2): p. 317–27. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, et al. , Rad GTPase inhibits cardiac fibrosis through connective tissue growth factor. Cardiovasc Res, 2011. 91(1): p. 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martos R, et al. , Diastolic heart failure: evidence of increased myocardial collagen turnover linked to diastolic dysfunction. Circulation, 2007. 115(7): p. 888–895. [DOI] [PubMed] [Google Scholar]
- 31.Mahalakshmi RN, et al. , Nuclear localization of endogenous RGK proteins and modulation of cell shape remodeling by regulated nuclear transport. Traffic, 2007. 8(9): p. 1164–78. [DOI] [PubMed] [Google Scholar]
- 32.Mahalakshmi RN, et al. , Nuclear transport of Kir/Gem requires specific signals and importin alpha5 and is regulated by calmodulin and predicted serine phosphorylations. Traffic, 2007. 8(9): p. 1150–1163. [DOI] [PubMed] [Google Scholar]
- 33.Beguin P, et al. , Nuclear sequestration of beta-subunits by Rad and Rem is controlled by 14–3-3 and calmodulin and reveals a novel mechanism for Ca2+ channel regulation. J Mol Biol, 2006. 355(1): p. 34–46. [DOI] [PubMed] [Google Scholar]
- 34.Edel MJ, et al. , Rem2 GTPase maintains survival of human embryonic stem cells as well as enhancing reprogramming by regulating p53 and cyclin D1. Genes Dev, 2010. 24(6): p. 561–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edel MJ, et al. , Rem2 GTPase controls proliferation and apoptosis of neurons during embryo development. Cell Cycle, 2010. 9(17): p. 3414–3422. [DOI] [PubMed] [Google Scholar]
- 36.Satija NK, et al. , High throughput transcriptome profiling of lithium stimulated human mesenchymal stem cells reveals priming towards osteoblastic lineage. PLoS One, 2013. 8(1): p. e55769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caplan AI, et al. , Mesenchymal stem cells: Building blocks for molecular medicine in the 21st century. Trends Mol Med, 2001. 7(6): p. 259–264. [DOI] [PubMed] [Google Scholar]
- 38.Jiang Y, et al. , Pluripotency of mesenchymal stem cells derived from adult marrow. Nature, 2002. 418(6893): p. 41–49. [DOI] [PubMed] [Google Scholar]
- 39.Bredella MA, et al. , Increased bone marrow fat in anorexia nervosa. J Clin Endocrinol Metab, 2009. 94(6): p. 2129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ecklund K, et al. , Bone marrow changes in adolescent girls with anorexia nervosa. J Bone Miner Res, 2010. 25(2): p. 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Misra M, and Klibanski A, Anorexia nervosa and osteoporosis. Rev Endocr Metab Disor, 2006. 7(1–2): p. 91–9. [DOI] [PubMed] [Google Scholar]
- 42.Vestergaard P, et al. , Fractures in patients with anorexia nervosa, bulimia nervosa, and other eating disorders-a nationwide register study. Int J Eat Disord, 2002. 32(3): p. 301–8. [DOI] [PubMed] [Google Scholar]
- 43.LeBlanc AD, et al. , Skeletal responses to space flight and the bed rest analog: a review. J Musculoskelet Neuronal Interact, 2007. 7(1): p. 33–47. [PubMed] [Google Scholar]
- 44.Verma S, et al. , Adipocytic proportion of bone marrow is inversely related to bone formation in osteoporosis. J Clin Pathol, 2002. 55(9): p. 693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeung DK, et al. , Osteoporosis is associated with increased marrow fat content and decreased marrow fat unsaturation: a proton MR spectroscopy study. J Magn Reson Imaging, 2005. 22(2): p. 279–85. [DOI] [PubMed] [Google Scholar]
- 46.Veldhuis-Vlug AG, and Rosen CJ, Mechanisms of marrow adiposity and its implications for skeletal health. Metabolism, 2017. 67: p. 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lefterova MI, et al. , PPARg and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev, 2008. 22: p. 2941–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komori T, et al. , Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell, 1997. 89: p. 755–764. [DOI] [PubMed] [Google Scholar]
- 49.Komori T, A fundamental transcription factor for bone and cartilage. Biochem Biophys Res Commun, 2000. 276: p. 813–816. [DOI] [PubMed] [Google Scholar]
- 50.Nakashima K, et al. , The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell, 2002. 108(1): p. 17–29. [DOI] [PubMed] [Google Scholar]
- 51.Song L, et al. , Loss of wnt/beta-catenin signaling causes cell fate shift of preosteoblasts from osteoblasts to adipocytes. J Bone Miner Res, 2012. 27(11): p. 2344–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Y, et al. , Rap1A regulates osteoblastic differentiation via the ERK and p38 mediated signaling. PLoS One, 2015. 10(11): p. e0143777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ge C, et al. , Critical role of the extracellular signal-regulated kinase-MAPK pathway in osteoblast differentiation and skeletal development. J Cell Biol, 2007. 176(5): p. 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez-Carballo E, et al. , p38 MAPK signaling in osteoblast differentiation. Front Cell Dev Biol, 2016. 4: p. 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang L, et al. , Rad GTPase deficiency leads to cardiac hypertrophy. Circulation, 2007. 116(25): p. 2976–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aref MW, et al. , Zoledronate treatment has different effects in mouse strains with contrasting baseline bone mechanical phenotypes. Osteoporos Int, 2016. 27(12): p. 3637–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jepsen KJ, et al. , Establishing biomechanical mechanisms in mouse models: practical guidelines for systematically evaluating phenotypic changes in the diaphyses of long bones. J Bone Miner Res, 2015. 30: p. 951–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berman AG, et al. , Raloxifene reduces skeletal fractures in an animal model of osteogenesis imperfecta. Matrix Biol, 2015. 52–54: p. 19–28. [DOI] [PubMed] [Google Scholar]
- 59.Erlebacher A, and Derynck R, Increased expression of TGF-b2 in osteoblasts results in an osteoporosis-like phenotype. J Cell Biol, 1996. 132: p. 195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schenck RK, et al. , Preparation of calcified tissues for light microscopy, in Methods of calcified tissue preparation, Dickson GR, Editor. 1984, Elsevier: Amsterdam: p. 1–56. [Google Scholar]
- 61.Dempster DW, et al. , Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res, 2013. 28(1): p. 2–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinn JM, et al. , A combination of osteoclast differentiation factor and macrophage-colony stimulating factor is sufficient for both human and mouse osteoclast formation in vitro. Endocrinology, 1998. 139(10): p. 4424–7. [DOI] [PubMed] [Google Scholar]
- 63.Jonason JH, and O’Keefe RJ, Isolation and culture of neonatal mouse calvarial osteoblasts. Methods Mol Biol, 2014. 1130: p. 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Price PA, et al. , Matrix Gla protein, a new gamma-carboxyglutamic acid-containing protein which is associated with the organic matrix of bone. Biochem Biophys Res Commun, 1983. 117(3): p. 765–71. [DOI] [PubMed] [Google Scholar]
- 65.Gopalakrishnan R, et al. , Matrix gamma-carboxyglutamic acid protein is a key regulator of PTH-mediated inhibition of mineralization in MC3T3-E1 osteoblast-like cells. Endocrinology, 2001. 142: p. 4379–4388. [DOI] [PubMed] [Google Scholar]
- 66.Gopalakrishnan R, et al. , Role of matrix Gla protein in parathyroid hormone inhibition of osteoblast mineralization. Cells Tissues Organs, 2005. 181: p. 166–175. [DOI] [PubMed] [Google Scholar]
- 67.Mutch DM, et al. , Using gene expression to predict the secretome of differentiating human preadipocytes. Int J Obes (Lond), 2009. 33(3): p. 354–63. [DOI] [PubMed] [Google Scholar]
- 68.Devlin MJ, and Rosen CJ, The bone-fat interface: basic and clinical implications of marrow adiposity. Lancet Diabetes Endocrinol, 2015. 3(2): p. 141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Granke M, et al. , The role of water compartments in the material properties of cortical bone. Calcif Tissue Int, 2015. 97(3): p. 292–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raggatt LJ, and Partridge NC, Cellular and molecular mechanisms of bone remodeling. J Biol Chem, 2010. 285(33): p. 25103–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Murshed M, et al. , Extracellular matrix mineralization is regulated locally; different roles of two gla-containing proteins. J Cell Biol, 2004. 165(5): p. 625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo G, et al. , Spontaneous calcification of arteries and cartilage in mice lacking matrix Gla protein. Nature, 1997. 386: p. 78–81. [DOI] [PubMed] [Google Scholar]
- 73.El-Maadawy S, et al. , Cartilage formation and calcification in arteries of mice lacking matrix Gla protein. Connect Tissue Res, 2003. 44 Suppl 1: p. 272–278. [PubMed] [Google Scholar]
- 74.Darlington GJ, et al. , The role of C/EBP genes in adipocyte differentiation. J Biol Chem, 1998. 273(46): p. 30057–30060. [DOI] [PubMed] [Google Scholar]
- 75.Sulston RJ, and Cawthorn WP, Bone marrow adipose tissue as an endocrine organ: close to the bone? Horm Mol Biol Clin Investig, 2016. 28(1): p. 21–38. [DOI] [PubMed] [Google Scholar]
- 76.Suchacki KJ, et al. , Bone marrow adipose tissue: formation, function and regulation. Curr Opin Pharmacol, 2016. 28: p. 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hardouin P, et al. , Bone marrow adipose tissue: to be or not to be a typical adipose tissue. Front Endocrinol (Lausanne), 2016. 7: p. 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rendina-Ruedy E, and Rosen CJ, Bone-fat interaction. Endocrinol Metab Clin N Am, 2017. 46: p. 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan Y, et al. , Ras-related associated with diabetes gene acts as a suppressor and inhibits Warburg effect in hepatocellular carcinoma. Onco Targets Ther, 2016. 9: p. 3925–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu J, et al. , RRAD inhibits the Warburg effect through negative regulation of the NF-kB signaling. Oncotarget, 2015. 6(17): p. 14982–14992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang C, et al. , Tumor suppressor p53 negatively regulates glycolysis stimulated by hypoxia through its target RRAD. Oncotarget, 2014. 5(14): p. 5535–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shang R, et al. , RRAD inhibits aerobic glycolysis, invasion, and migration and is associated with poor prognosis in hepatocellular carcinoma. Tumour Biol, 2016. 37(4): p. 5097–5105. [DOI] [PubMed] [Google Scholar]
- 83.Garvey WT, et al. , Muscle Rad expression and human metabolism: potential role of the novel Ras-related GTPase in energy expenditure and body composition. Diabetes, 1997. 46(3): p. 444–450. [DOI] [PubMed] [Google Scholar]
- 84.Wei CC, et al. , The pseudogene DUXAP10 promotes an aggressive phenotype through binding with LSD1 and repressing LATS2 and RRAD in non small cell lung cancer. Oncotarget, 2017. 8(3): p. 5233–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tseng YH, et al. , Regulation of growth and tumorigenicity of breast cancer cells by the low molecular weight GTPase Rad and nm23. Cancer Res, 2001. 61(5): p. 2071–2079. [PubMed] [Google Scholar]
- 86.Mo Y, et al. , Promoter hypermethylation of Ras-related GTPase gene RRAD inactivates a tumor suppressor function in nasopharyngeal carcinoma. Cancer Lett, 2012. 323(2): p. 147–54. [DOI] [PubMed] [Google Scholar]
- 87.Hsiao BY, et al. , Rad is a p53 direct transcriptional target that inhibits cell migration and is frequently silenced in lung carcinoma cells. J Mol Med (Berl), 2011. 89(5): p. 481–92. [DOI] [PubMed] [Google Scholar]
- 88.Jin Z, et al. , Aberrant methylation of the Ras-related associated with diabetes gene in human primary esophageal cancer. Anticancer Res, 2013. 33(11): p. 5199–203. [PubMed] [Google Scholar]
- 89.Fan Y, et al. , Parathyroid hormone directs bone marrow mesenchymal cell fate. Cell Metab, 2017. 25(3): p. 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Celil AB, et al. , BMP-2 and insulin-like growth factor-I mediate osterix (Osx) expression in human mesenchymal stem cells via the MAPK and protein kinase D signaling pathways. J Cell Biochem, 2005. 280(36): p. 31353–31359. [DOI] [PubMed] [Google Scholar]
- 91.Day TF, et al. , Wnt/b-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell, 2005. 8(5): p. 739–750. [DOI] [PubMed] [Google Scholar]
- 92.Augello A, and De Bari C, The regulation of differentiation in mesenchymal stem cells. Hum Gene Ther, 2010. 21(10): p. 1226–1238. [DOI] [PubMed] [Google Scholar]